Abstract
Direct delivery of proteins into plants represents a promising alternative to conventional gene delivery for probing and modulating cellular functions without the risk of random integration of transgenes into the host genome. This remains challenging, however, because of the lack of a protein delivery tool applicable to diverse plant species and the limited information about the entry mechanisms of exogenous proteins in plant cells. Here, we present the synthetic multidomain peptide (named dTat-Sar-EED4) for cytosolic protein delivery in various plant species via simple peptide-protein coincubation. dTat-Sar-EED4 enabled the cytosolic delivery of an active enzyme with up to ∼20-fold greater efficiency than previously described cell-penetrating peptides in several model plant systems. Our analyses using pharmacological inhibitors and transmission electron microscopy revealed that dTat-Sar-EED4 triggered a unique endocytic mechanism for cargo protein internalization. This endocytic mechanism shares several features with macropinocytosis, including the dependency of actin polymerization, sensitivity to phosphatidylinositol-3 kinase activity, and formation of membrane protrusions and large intracellular vesicles (>200 nm in diameter), even though macropinocytosis has not been identified to date in plants. Our study thus presents a robust molecular tool that can induce a unique cellular uptake mechanism for the efficient transport of bioactive proteins into plants.
Keywords: multidomain peptide, protein delivery, macropinocytosis, chemical biology, plant biotechnology
Introduction
Photosynthetic autotrophs such as plants represent promising platforms for the sustainable production of valuable chemicals,1 biomaterials,2 and biofuels using renewable resources including sunlight and CO2.3 Genetic engineering of plants, a process mainly achieved by delivering exogenous genes into host cells, can facilitate the biosynthesis of these materials and climatic adaptation. However, current gene delivery technologies face substantial challenges associated with the potential random integration of transgenes into the host genome,4 which may negatively affect plant traits and offset the benefits of genetic engineering. By contrast, direct delivery of proteins into plant cells offers a straightforward approach to modulating many biological processes and the associated plant traits without the risk of random transgene integration.5 However, protein delivery in plants remains challenging because of the dual barrier of the cell wall and plasma membrane. Delivering protein cargoes across this dual barrier typically requires particle bombardment, where heavy metal particles are coated with a protein cargo and physically shot into target plant cells.6 However, this method suffers from poor delivery efficiency, low throughput, the need for specialized equipment, and potential contamination by heavy metals.
Various molecular tools such as synthetic polymers, peptides, and other chemicals have been developed for the transport of protein cargoes into animal cells, a process often referred to as “protein transduction”.7 These molecular tools mainly utilize endocytic pathways for protein transduction across the plasma membrane,8 providing an opportunity for high-throughput, scalable protein delivery without the use of specialized equipment or heavy metals. The endocytic pathways responsible for macromolecule entry into animal cells are classified as clathrin-mediated endocytosis (CME) or clathrin-independent endocytosis (CIE).9 CME appears to have a size limit and surface charge preference for cargo uptake, possibly because of the properties of clathrin-coated vesicles,10,11 which mediate the intracellular transport of cargo during CME. Conversely, CIE (such as macropinocytosis) represents an indiscriminate uptake mechanism for various extracellular materials via a large endocytic vesicle termed the macropinosome (>200 nm in diameter).12 Macropinocytosis may be advantageous for the cellular uptake of various proteins with different shapes, sizes, and surface charges. The ability of a molecular tool to induce macropinocytosis plays a key role in efficient protein transduction.13,14
Compared with the advances made in animal systems, the implementation of molecular tool-mediated protein transduction in plants has lagged behind, and a versatile molecular tool enabling plant species-independent protein transduction has yet to emerge. Few studies have explored whether molecular tools are able to deliver a functional protein cargo in its active form into the desired compartments (e.g., the cytosol) of various plant cells. Additionally, very little is known about the pathways by which a molecular tool and its cargo enter plant cells. Specifically, macropinocytosis and other CIE pathways are poorly characterized compared with the well-studied CME,15 and their contributions to protein transduction in plants remain largely unexplored.
We previously described a synthetic peptide consisting of a d-arginine-rich domain (dTat: rrrqrrkkr),16 a hexameric sarcosine (Sar) linker and a hydrophobic endosomolytic domain (EED4: GWWG).17,18 This peptide, referred to as dTat-Sar-EED4 (rrrqrrkkr-(Sar)6-GWWG, Figure 1A), enhanced the cellular uptake and cytosolic translocation of a DNA–polycation peptide complex in plant protoplasts (isolated cells lacking cell walls). The complex could not enter protoplasts through CME since its diameter (∼150 nm) exceeded that of plant clathrin-coated vesicles (∼60 nm).19 This size mismatch led us to hypothesize that dTat-Sar-EED4 drives a non-CME mechanism allowing the efficient entry of exogenous cargoes into plant cells. Here, we explored the use of dTat-Sar-EED4 and its analogue (dTat-Sar-EED5: rrrqrrkkr-(Sar)6-GFWFG, Figure 1A) as protein delivery tools and investigated their cargo transduction mechanism in plants. We determined that dTat-Sar-EED peptides could transduce a functional enzyme in its active form into cells of different plant species, with a transduction efficiency up to ∼20-fold greater than that of previously reported protein transduction domains (i.e., cell-penetrating peptides, CPPs) in model plants. Confocal laser-scanning microscopy (CLSM) analyses using several pharmacological inhibitors and transmission electron microscopy (TEM) observations revealed that dTat-Sar-EED4 utilizes a macropinocytosis-like CIE pathway for cargo transduction in plants. Altogether, this study provides a promising molecular tool that can induce a newly discovered cell entry mechanism for introducing bioactive proteins into intact plants.
Figure 1.
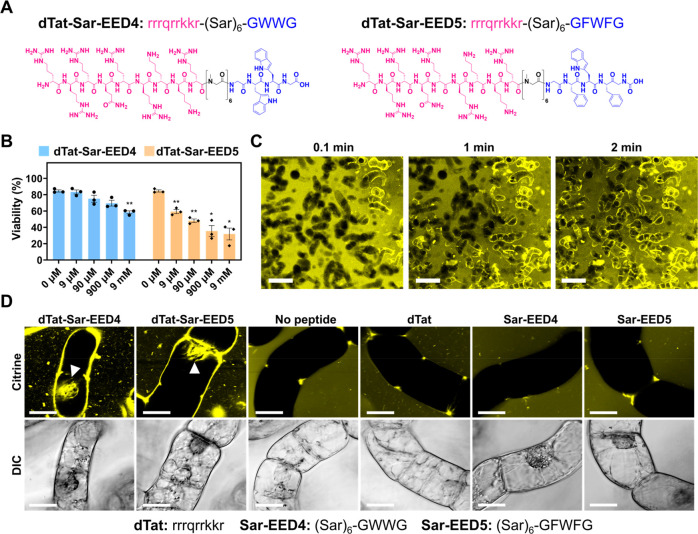
dTat-Sar-EED peptides mediate Citrine transduction into BY-2 cells. (A) Chemical structures of dTat-Sar-EED4 and dTat-Sar-EED5. (B) Viability of BY-2 cells determined by Evans blue assay after treatment for 1 h with various concentrations of either dTat-Sar-EED4 or dTat-Sar-EED5. Data from three biological replicates are represented as the mean ± standard error values. Statistical significance compared to the control (0 μM): *P < 0.05, **P < 0.01 based on Dunnett’s T3 test (n = 3). (C) Time-lapse confocal images of Citrine internalization into BY-2 cells treated with dTat-Sar-EED4 (90 μM). Scale bars, 2 mm. (D) Confocal images showing Citrine internalization into BY-2 cells treated for 1 h with Citrine alone (100 μg/mL, No peptide) or in combination with 90 μM dTat-Sar-EED4, dTat-Sar-EED5, dTat, Sar-EED4 or Sar-EED5. Differential interference contrast (DIC) images are shown below, with the amino acid sequence of each peptide below the images. White arrowheads indicate Citrine located in the nucleus. Scale bars, 20 μm.
Results
dTat-Sar-EED Peptides Mediate Protein Transduction into Plant Cells
We first assessed the cytotoxicity of the dTat-Sar-EED peptides to tobacco (Nicotiana tabacum) Bright Yellow-2 (BY-2) cells, which serve as a model plant system. We examined the viability of BY-2 cells by Evans blue staining after 1 h of treatment with either dTat-Sar-EED4 or dTat-Sar-EED5 at various concentrations. Both peptides caused more cell death with increasing concentrations (from 9 μM to 9 mM), and dTat-Sar-EED4 seemed to be less toxic than dTat-Sar-EED5 (Figure 1B): prolonged treatment (90 μM for up to 48 h) did not result in significant cytotoxicity (Figure S1). We then evaluated the peptide-mediated transduction of a protein cargo (the 27-kDa yellow fluorescent protein Citrine) into BY-2 cells through CLSM. A time-lapse observation revealed the rapid internalization of Citrine into BY-2 cells within a few minutes after the addition of dTat-Sar-EED4 (90 μM) (Figure 1C, Movies S1, S2), although no Citrine internalization was observed at lower peptide concentrations (9 nM–9 μM, SI Figure S2). We detected Citrine fluorescence in the nuclei of some cells after 1 h of incubation with either dTat-Sar-EED4 (90 μM) or dTat-Sar-EED5 (90 μM) (Figure 1D), but not in the absence of peptide. These observations indicated that both peptides are capable of cytosolic translocation of Citrine into cultured plant cells. We then investigated whether individual peptide domains (dTat, Sar-EED4, or Sar-EED5) possessed this ability. BY-2 cells exposed to 90 μM of each peptide domain for 1 h did not show Citrine uptake (Figure 1D), highlighting the importance of the fusion of the cationic dTat and hydrophobic EED domains in translocating protein cargo.
dTat-Sar-EED Peptides Mediate Protein Transduction into Diverse Targets
We next aimed to apply the dTat-Sar-EED peptides to model land plants, including cotyledons of Arabidopsis thaliana (a model dicot plant), calli of Oryza sativa (a model monocot crop plant) and sporelings of Marchantia polymorpha (a model bryophyte). We evaluated the cytotoxicity of the peptides to these plants via Evans blue assays. The dTat-Sar-EED peptides (particularly dTat-Sar-EED5) were toxic to A. thaliana cotyledons and O. sativa calli at concentrations of 90 μM or more, although they did not cause severe cell death in M. polymorpha sporelings, even at 9 mM (Figure 2A). We therefore employed 90 μM dTat-Sar-EED4 to achieve Citrine transduction into cells of all three model plants with only moderate cytotoxicity. Citrine internalization was observed for all model plants in the presence of dTat-Sar-EED4, but not in the absence of the peptide (Figure 2B). Citrine appeared to be transferred to the nucleus in A. thaliana cotyledon cells (Figure 2B), as also observed in BY-2 cells (Figure 1D), whereas such behavior was not detected in O. sativa calli (Figure 2B), suggesting that the nuclear translocation of Citrine may occur via another mechanism we have not yet identified.
Figure 2.
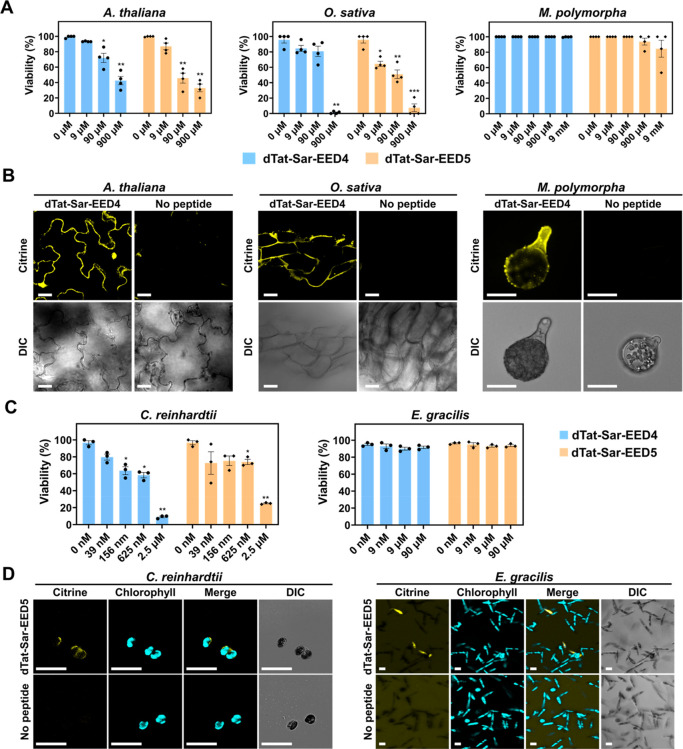
dTat-Sar-EED peptides mediate Citrine transduction into various types of cells. (A) Viability of A. thaliana cotyledons, O. sativa calli and M. polymorpha sporelings determined by Evans blue staining after treatment for 1 h with various concentrations of either dTat-Sar-EED4 or dTat-Sar-EED5. Data from four biological replicates are represented as the mean ± standard error values. Statistical significance compared with the control (0 μM): *P < 0.05, **P < 0.01, ***P < 0.001 based on Dunnett’s T3 test (n = 4). (B) Confocal images showing Citrine internalization into A. thaliana cotyledons, O. sativa calli, and M. polymorpha sporelings after 1 h incubation with Citrine (200 μg/mL) in the absence or presence of dTat-Sar-EED4 (90 μM). DIC images are shown below. Scale bars, 20 μm. (C) Viability of C. reinhardtii and E. gracilis determined by a colony formation assay and Evans blue staining after treatment for 20 min and 3 h, respectively, with various concentrations of either dTat-Sar-EED4 or dTat-Sar-EED5. Data from three biological replicates are represented as the mean ± standard deviation values. Statistical significance compared to the control (0 μM): *P < 0.05, **P < 0.01 based on Dunnett’s T3 test (n = 3). (D) Confocal images showing Citrine internalization into C. reinhardtii and E. gracilis treated with Citrine alone (100 μg/mL, No peptide) or in combination with dTat-Sar-EED5 (C. reinhardtii: 156 nM, 20 min; E. gracilis; 9 μM, 3 h). Scale bars, 20 μm.
The successful protein transduction in several land plant species using our method led us to explore the applicability of dTat-Sar-EED peptides to other photosynthetic autotrophs, such as algae, which are important hosts for sustainable manufacturing.20 We chose Chlamydomonas reinhardtii (a green alga with a cell wall) and Euglena gracilis (a secondary endosymbiotic alga possessing a pellicle, or protein-rich outer layer, instead of a cell wall) as model systems. Treatment with either peptide at 2.5 μM induced severe cell death in C. reinhardtii (Figure 2C). Unlike BY-2 cells and the investigated land plant species, dTat-Sar-EED4 appeared to be slightly more toxic to C. reinhardtii cells than dTat-Sar-EED5 at concentrations of 156 nM or more (Figure 2C). Neither peptide exhibited cytotoxicity toward E. gracilis at 9 nM–90 μM (Figure 2C). We treated C. reinhardtii and E. gracilis with 156 nM and 9 μM dTat-Sar-EED5, respectively, for Citrine transduction. At 3 h after treatment with the peptide, Citrine was internalized into the cells of both microalgae, but no Citrine internalization was observed without the peptide (Figure 2D), demonstrating the wide applicability of our peptide to algae in addition to land plants.
Peptide-Mediated Transduction of GUS Maintains Its Bioactivity
To investigate whether the dTat-Sar-EED peptides could transduce a functional protein in its active form into plants, we utilized the combination of β-glucuronidase from Escherichia coli (GUS, a ∼290-kDa tetrameric enzyme) and its fluorogenic substrate, dye-conjugated β-d-glucuronide (dye-βGlcU). An active form of GUS was able to catalyze the hydrolysis of nonfluorescent dye-βGlcU into a green fluorescent dye (Figure 3A). We selected BY-2 cells, A. thaliana cotyledons, and O. sativa calli as model plant systems. We treated the samples with GUS and dTat-Sar-EED4 (90 μM) for 30 min, washed out extracellular proteins and peptides, and treated them with dye-βGlcU for 15 min, which allowed cell entry of dye-βGlcU due to its capability for direct cell-membrane penetration (Figure 3A). Excess substrates were washed out before CLSM observation.
Figure 3.
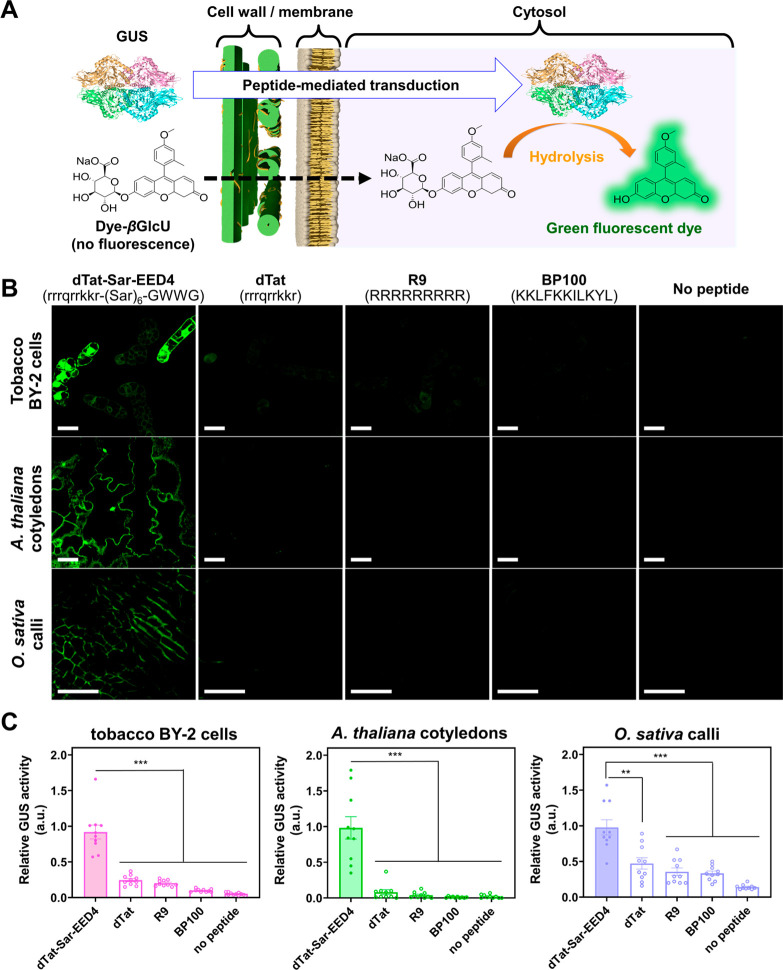
dTat-Sar-EED4 enables more efficient transduction of a bioactive protein than do previously reported CPPs in plants. (A) Schematic representation of the GUS activity assay. If GUS maintains its enzymatic activity following peptide-mediated cytosolic translocation, it catalyzes the hydrolysis of the cell-permeable dye-βGlcU, yielding a green fluorescent dye. Accordingly, the activity of internalized GUS can be monitored by detecting green fluorescence of the dye generated in cells. (B) Confocal images of BY-2 cells, A. thaliana cotyledons, and O. sativa calli treated with GUS alone (No peptide) or in combination with either 90 μM of dTat-Sar-EED4, dTat, R9 or BP100 for 30 min, washed with medium and then incubated with dye-βGlcU (10 μM) for 15 min. Excess dye-βGlcU was washed out from the samples before CLSM observation. Scale bars, 40 μm. DIC images corresponding to the fluorescent images are shown in Figure S5. (C) Transduction efficiency of each system based on relative GUS activity in BY-2 cells, A. thaliana cotyledons, and O. sativa calli. Data from 10 biological replicates are represented as the mean ± standard error values. Statistical significance compared to dTat-Sar-EED4: **P < 0.01, ***P < 0.001 based on Dunnett’s T3 test (n = 10).
Green fluorescence was clearly visible in the cytosol of BY-2 cells, A. thaliana cotyledons, and O. sativa calli treated with dTat-Sar-EED4, GUS, and dye-βGlcU, whereas no fluorescence was detected in the controls, in which the peptide, enzyme, or fluorogenic substrate was omitted (Figures 3B, S3). These observations indicate that GUS was cytosolically transduced to plant cells in its active form by dTat-Sar-EED4. Similar results were obtained using dTat-Sar-EED5 (Figure S4). Under the same experimental conditions, we compared the transduction efficiency of dTat-Sar-EED4 with that of several previously described CPPs, including dTat (rrrqrrkkr),16 R9 (RRRRRRRRR),21 and BP100 (KKLFKKILKYL)22 in BY-2 cells, A. thaliana cotyledons, and O. sativa calli. The efficiency of dTat-Sar-EED4 was up to ∼20-fold higher than that of the other CPPs in these model plant systems (Figures 3B,C and S5). These results indicate that dTat-Sar-EED4 enables the efficient transduction of biologically active proteins into different plant species.
dTat-Sar-EED4 Uses a Novel Transduction Mechanism
As CME is the only well-characterized endocytic pathway in plants, we aimed to investigate its involvement in dTat-Sar-EED4 internalization into A. thaliana root epidermal cells, which are often used to study plant endocytosis. We employed FM4-64, a styryl dye used to trace endocytic pathways, and Endosidin 9-17 (ES9-17), an inhibitor of clathrin heavy chain that specifically blocks CME in plants.23 The roots were treated with dimethyl sulfoxide (DMSO, 0.6%, v/v) or ES9-17 (30 μM) and then stained with FM4-64 (10 μM) in the presence or absence of dTat-Sar-EED4 (30 μM) for CLSM observations. We detected punctate FM4-64 fluorescence, which represented endosomal compartments, in the intracellular space of DMSO-treated samples regardless of the presence of dTat-Sar-EED4 (Figure 4A). Similarly, intracellular FM4-64 fluorescence was also observed in water-treated samples, indicating that FM4-64 was internalized via endocytosis, but not DMSO-induced membrane perturbation (Figure S6). In the absence of dTat-Sar-EED4, FM4-64 uptake was clearly reduced by ES9-17 treatment (Figure 4A, B) due to the effective inhibition of CME by ES9-17.23 However, dTat-Sar-EED4 counteracted the suppression of FM4-64 uptake by ES9-17 (Figure 4A,B). These results suggest that the peptide induces a non-CME mechanism for FM4-64 internalization. The enhancement of FM4-64 uptake by dTat-Sar-EED4 was slight in DMSO-treated samples but more pronounced in ES9-17-treated samples (Figure 4B). This observation suggests that the inhibition of CME by ES9-17 upregulates CIE pathways in root cells, as reported in mammalian cells.24
Figure 4.
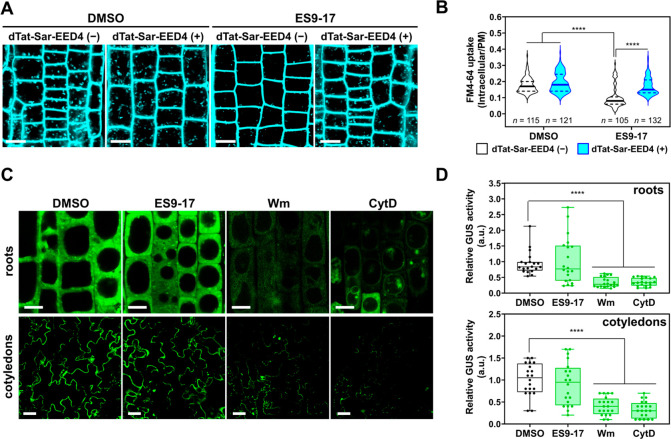
dTat-Sar-EED4 utilizes a clathrin-independent pathway for cargo transduction. (A) Confocal images of A. thaliana root cells treated for 30 min with DMSO or ES9-17 (30 μM) and then stained for 30 min with FM4-64 (10 μM) alone or in combination with dTat-Sar-EED4 (30 μM). Scale bars, 10 μm. (B) Violin plot representation of FM4-64 uptake determined by the ratio of intracellular/plasma membrane signal intensity of cells shown in (A). Solid lines and dashed lines in the plot represent the median and quartile values, respectively. Statistical significance: ****P < 0.000 based on Tukey’s multiple comparison test. More than 100 cells from eight biologically independent samples were analyzed for each system. (C) Confocal images of GUS-transduced A. thaliana roots and cotyledons using dTat-Sar-EED4 (roots, 30 μM, 15 min; cotyledons, 90 μM, 30 min) in the absence (DMSO) or presence of inhibitors such as ES9-17 (roots, 30 μM; cotyledons, 100 μM), Wm (40 μM) or CytD (40 μM). Scale bars: roots, 10 μm; cotyledons, 40 μm. (D) Boxplot representation of relative activity of GUS transduced by dTat-Sar-EED4 under different conditions in C: boxes represent the interquartile range, lines within the boxes represent the median values, and upper and lower whiskers represent the highest and lowest values, respectively. Statistical significance compared to the control (DMSO): *P < 0.05, ****P < 0.0001 based on Dunnett’s T3 test (n = 20 biological replicates).
We explored the dTat-Sar-EED4-mediated cargo transduction mechanisms by performing quantitative evaluation of the GUS transduction efficiency in epidermal cells of A. thaliana roots and cotyledons treated with several pharmacological inhibitors. The GUS transduction efficiency in roots and cotyledons was not significantly inhibited by ES9-17 treatment (Figure 4C,D). These results are similar to those obtained for FM4-64 internalization (Figure 4A,B). Conversely, we observed a significant suppression of GUS transduction in roots and cotyledons treated with wortmannin (Wm, an inhibitor of phosphatidylinositol-3 kinase [PI3K]) and cytochalasin D (CytD, an inhibitor of action polymerization). These inhibitors did not alter the enzymatic activity of GUS in vitro (Figure S7), confirming that they affected the internalization rather than the catalytic ability of GUS. The molecular mechanisms by which Wm and CytD inhibit their targets (PI3K and actin filaments) in plants are similar to those reported in animal systems.25,26 Our data suggest that the mechanism of dTat-Sar-EED4-mediated GUS transduction is clathrin-independent but sensitive to PI3K activity and actin polymerization.
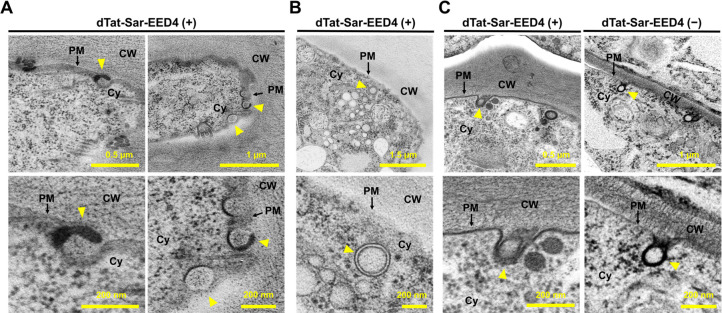
The cargo transduction mechanism driven by dTat-Sar-EED4 appears to share several features with macropinocytosis, such as its independence from clathrin and sensitivity to Wm and CytD,12 but the existence of macropinocytosis as such has not been established in plants. Additional features of macropinocytosis include the formation of plasma membrane protrusions and intracellular vesicles >200 nm in diameter.12 To investigate whether these characteristics can be found in plants, we treated A. thaliana roots and BY-2 cells with dTat-Sar-EED4 and subsequently fixed the samples by high-pressure freezing/frozen substitution for TEM. We detected the formation of plasma membrane protrusions in root cells treated with dTat-Sar-EED4 but not in untreated control cells (Figure 5A), along with unstructured objects in the vicinity of the membrane protrusions (Figure S8). Because these unstructured objects were stained with osmium tetroxide, we speculated that they might be aggregates of unsaturated fatty acids that dissociated from plasma membranes, possibly by interacting with the peptide. We also detected an intracellular vesicle with a diameter >200 nm in some treated cells (Figure 5B), although only a few such vesicles were observed, likely due to the endosome-disrupting ability of the EED4 domain in the peptide.27 Membrane invaginations, likely associated with CME, were observed in both treated and control cells (Figure 5C). Similar trends were also observed in BY-2 cells (Figure S9). These observations suggest that dTat-Sar-EED4 induces a macropinocytosis-like mechanism in plants.
Figure 5.
dTat-Sar-EED4 induces the formation of plasma membrane protrusions and a large intracellular vesicle in plant cells. (A,B) Formation of plasma membrane protrusions (A) and a large intracellular vesicle (B) observed in TEM images of high-pressure frozen A. thaliana root epidermal cells after treatment with 30 μM dTat-Sar-EED4 for 15 min. Yellow arrowheads in (A) indicate plasma membrane protrusions, while those in (B) represent intracellular vesicles (>200 nm in diameter). (C) Formation of membrane invaginations detected in TEM images of high-pressure frozen root epidermal cells after treatment with 30 μM dTat-Sar-EED4 or water for 15 min. Yellow arrowheads indicate the membrane invagination. CW, cell wall; PM, plasma membrane; Cy, cytoplasm.
Discussion
The delivery of proteins instead of DNA can overcome many drawbacks of current genetic engineering techniques for plants. However, a versatile tool for protein delivery in plants has been lacking, which poses a fundamental limit to plant bioengineering for diverse applications. Here, we described the multidomain peptides dTat-Sar-EED4 and dTat-Sar-EED5, which we successfully used to introduce an exogenous fluorescent protein (Citrine) into a wide range of target cells, including tobacco BY-2 cells (Figure 1C,D), A. thaliana cotyledons, O. sativa calli, M. polymorpha sporelings (Figure 2B), and C. reinhardtii and E. gracilis cells (Figure 2D). When we cytosolically delivered the enzyme GUS into cells using dTat-Sar-EED peptides, it remained functional and could catalyze the hydrolysis of its substrate in various plant species (Figure 3B,C). We also demonstrated that dTat-Sar-EED4 can induce a distinct clathrin-independent mechanism, which shares several characteristics with macropinocytosis, for cargo transduction in plants (Figures 4–5).
Although the delivery of bioactive proteins into plants currently relies on particle bombardment or polyethylene glycol-mediated protoplast transfection, their widespread use is limited by the need for specialized equipment, the difficulty of regenerating entire plants from protoplasts, and the large variation in delivery efficiency among different plant species.6,28 By contrast, our peptide-mediated approach does not require the use of specialized equipment or protoplasts and can be applied to a wide range of plant species as well as algae (Figure 2). The protein cargoes delivered by dTat-Sar-EED4 were mainly located in the cytosol but not seen in the vacuole (Figures 1D, 2B, and 3B), a plant organelle equivalent to the lysosome in mammalian cells. This is indicative of efficient endosomal escape of the cargoes into the cytosol most likely due to the endosome-disrupting domain (EED4) in the peptide, which we previously reported.18,27 dTat-Sar-EED4 enabled the cytosolic transduction of a bioactive protein cargo with up to ∼20-fold higher efficiency than previously described CPPs in several model plants (Figure 3B,C). These advantages will allow more convenient and higher-throughput delivery of bioactive proteins to diverse targets. An important application of bioactive protein delivery is transgene-free plant genome editing, which can be achieved by directly introducing engineered nucleases such as zinc-finger nucleases, transcription activator like effector nucleases (TALENs), and clustered regularly interspaced short palindromic repeats (CRISPR)-CRISPR associated protein 9 (Cas9) systems into plant cells.29−31 Recent efforts have focused on delivering pre-assembled CRISPR-Cas9 ribonucleoprotein (RNP) complexes into plant cells via particle bombardment or protoplast transfection.32 However, these approaches often yield poor genome editing efficiency, except in a few model plants,33 limiting the utility of transgene-free genome editing in plant breeding. Our peptide may be useful for addressing the issue of low editing efficiency via improvements in the delivery of RNP complexes to cells of various plant species.
A key finding of our study is that dTat-Sar-EED4 mainly utilizes a distinct CIE pathway for protein cargo transduction into A. thaliana root cells. This finding was supported by CLSM analysis using a plant CME-specific inhibitor (ES9-17) (Figure 4) and TEM observation (Figure 5). The entry mechanisms of exogenous macromolecules in plants, particularly those mediated by CIE, remain largely unexplored. By contrast, CIE pathways such as macropinocytosis play a central role in the cellular uptake of exogenous macromolecules in animal cells.34 Macropinocytosis is induced by arginine-rich peptides for cargo transduction in mammalian systems,35,36 but it has yet to be reported in plants. Our findings show that the CIE mechanism responsible for dTat-Sar-EED4-mediated protein transduction in plants shares three features with this pathway. First, we observed the formation of membrane protrusions, a characteristic of macropinocytosis, in TEM images of dTat-Sar-EED4-treated A. thaliana roots (Figure 5A). Second, the vesicles that we detected in treated roots appeared to be larger (with diameters greater than ∼200 nm, Figure 5B) than plant clathrin-coated vesicles (average diameter ∼60 nm; maximum diameter ∼110 nm)19 but similar in size to macropinosomes (>200 nm). Third, GUS transduction using dTat-Sar-EED4 was significantly blocked by inhibiting PI3K activity and actin polymerization through treatment with Wm and CytD, respectively (Figure 4C,D), which is in line with the finding that macropinocytosis is a PI3K-dependent and actin-driven process. Although further evidence is needed to confirm that macropinocytosis exists in plants, our results suggest that dTat-Sar-EED4 can drive a macropinocytosis-like mechanism to transduce a protein cargo into plant cells. We envision that dTat-Sar-EED4 could be used to study plant CIE as well as to deliver bioactive cargoes into cells. Pioneering studies have demonstrated the CIE-mediated internalization of nanobeads,37 a glucose analogue,38 and endogenous membrane-associated proteins into plants,39,40 but we still lack a clear understanding of why, when, and how CIE pathways operate in plants. Outstanding questions remain, including whether CIE pathways present in animal systems, such as macropinocytosis, are conserved in plants, as well as the physiological roles of plant CIE. The use of dTat-Sar-EED4 as a CIE inducer may offer opportunities to explore these important questions in fundamental plant science.
Conclusions
In summary, we described multidomain peptide-mediated protein transduction into plants through a macropinocytosis-like mechanism. The synthetic peptides presented here allow the cytosolic transduction of biologically active proteins in different plant species, which could help overcome the current limitations of plant bioengineering. Our study also revealed the significant contribution of a macropinocytosis-like endocytic pathway to peptide-mediated protein transduction. The macropinocytosis-like mechanism in plants shares several features with the well-established macropinocytosis in mammalian cells. However, the former mechanism does not appear to be driven by arginine-rich CPPs, such as dTat and R9 (Figure 3), even though these peptides have been reported to induce macropinocytosis in mammalian cells.16,35 This might be related to the different cell surface conditions and lipid membrane compositions between plant and mammalian cells. Heparan sulfate proteoglycans ubiquitously found on the surface of mammalian cells act as a receptor for the induction of macropinocytosis,41 but they are not present on the plant cell surface. Cholesterol in mammalian plasma membranes is known to be required for macropinocytosis,42 whereas it is not found in plant plasma membranes. Meanwhile, macropinocytosis in mammalian cells involves several proteins including PI3K, Na+/H+ exchangers, small GTPases, etc. Some of them also exist in plants, but their relation to the macropinocytosis-like mechanism found in this study remains unclear. Further studies on the molecular basis of this newly discovered pathway may provide design principles of more sophisticated protein delivery tools and advance our understanding of macromolecule entry mechanisms in plants.
Experimental Section
Peptides, Proteins, and Pharmacological Inhibitors
dTat-Sar-EED4 (rrrqrrkkr-(Sar)6-GWWG; Mw 2253), dTat-Sar-EED5 (rrrqrrkkr-(Sar)6-GFWFG; Mw 2361), dTat (rrrqrrkkr; Mw 1340), Sar-EED4 ((Sar)6-GWWG; Mw 931), Sar-EED5 ((Sar)6-GFWFG; Mw 1039), R9 (RRRRRRRRR; Mw 1424), and BP100 (KKLFKKILKYL; Mw 1422) were obtained from the Research Resources Center of RIKEN Brain Science Institute. The purity of each peptide was >95%, as determined by reverse-phase high-performance liquid chromatography. Citrine (27 kDa) was synthesized and purified as described previously.43 GUS was purchased from Nacalai Tesque. ES9-17 (Carbosynth), Wm (Fujifilm), and CytD (Fujifilm) were dissolved in DMSO (TCI) and stored at −30 °C.
Plant Materials and Growth Conditions
Tobacco (Nicotiana tabacum) BY-2 cell suspension cultures were purchased from the RIKEN BioResource Center. The BY-2 cells were maintained in modified Linsmaier and Skoog (mLS) medium in the dark at 26 °C, with shaking at 130 rpm, and subcultured at 1-week intervals as described previously.44 Exponentially growing cells (3–4 days after subculture) were diluted to an OD600 of 0.5 with mLS medium and used for analysis. Arabidopsis thaliana (Col-0) seedlings and Oryza sativa (Nipponbare) calli were generated under the same conditions used previously.18,45 For the experiments, cotyledons or roots were collected from seedlings (7–9 days after sowing) and calli were cultivated at 30 °C on a Petri plate with callus induction medium (N6D) for 1 week under constant light. Sporelings of the liverwort Marchantia polymorpha were obtained by crossing between the male Tak-1 and female Tak-2 strains.46 Sporelings from sporangia were incubated in 150 μL Milli-Q water for 4 days and then used for the experiments.
Strains and Culture of Algae
Chlamydomonas reinhardtii wild-type (cc125+) cells were cultivated in Tris Acetate Phosphate (TAP) liquid medium at 23 °C under constant light. C. reinhardtii cells in log phase (2 days after subculture) were used for the experiments. Euglena gracilis cells were maintained in CM medium (pH 3.5) at 26 °C, with shaking at 100 rpm, and subcultured at 1-week intervals according to a previously reported protocol.47 Cells were cultured to an OD730 of 0.23 and used for the experiments.
Cytotoxicity Test for Various Types of Plant and Algal Cells
BY-2 cell suspensions were treated with various concentrations (9 μM–9 mM) of dTat-Sar-EED4 or dTat-Sar-EED5. After incubation at 26 °C for 1 h, cell viability was determined via an Evans blue assay as described previously.48 The peptide-treated cells were incubated in aqueous solution containing Evans blue (150 μg/mL, Sigma-Aldrich) at 25 °C for 30 min and washed with water. The cells were then incubated with water/methanol mixture (50/50, v/v) containing sodium dodecyl sulfate (SDS, 1%, w/v) at 25 °C for 2 h to extract the Evans blue and subjected to spectrophotometric quantification at 600 nm. The viability of BY-2 cells boiled at 98 °C for 30 min was used as a control (100% dead cells).
A. thaliana cotyledons (obtained from three seedlings) and O. sativa calli (∼50 mg) were infiltrated with aqueous solution containing various concentrations of dTat-Sar-EED4 or dTat-Sar-EED5 by the vacuum/compression method as described previously.45 After 1 h of incubation, viability was evaluated using an Evans blue assay. The viability of samples boiled at 98 °C for 30 min was used as a control (100% dead cells).
Sporelings of the liverwort M. polymorpha were incubated at 22 °C for 1 h in the presence of various concentrations (9–900 μM) of either dTat-Sar-EED4 or dTat-Sar-EED5 and then treated with Evans blue (150 μg/mL). We counted the number of dead cells, which were stained with Evans blue, as well as live cells, which were not stained. Viability was determined based on the live/dead cell ratio, and the viability of sporelings boiled at 95 °C for 10 min was used as a control (100% dead cells).
C. reinhardtii cells were treated with various concentrations (39 nM–2.5 μM) of dTat-Sar-EED4 or dTat-Sar-EED5 for 20 min in TAP medium containing sucrose (40 mM). The cells were then washed and spread onto TAP agar plates. After incubation, the number of colonies that formed on the agar plates was counted. To calculate viability, the number of colonies with peptide treatment was divided by that without peptide treatment.
E. gracilis cells were suspended in solution containing various concentrations (9 nM–90 μM) of either dTat-Sar-EED4 or dTat-Sar-EED5 and incubated at 26 °C for 3 h. After centrifugation, cells were collected and resuspended in solution containing Evans blue (150 μg/mL). After 20 min of incubation, the cells were washed with water three times prior to microscopic observation to count the number of live and dead cells. The live/dead cell ratio was considered to represent viability, and the viability of untreated cells was used as a control.
Synthetic Peptide-Mediated Citrine Transduction into Various Types of Plant and Algal Cells
Citrine internalization into tobacco BY-2 cells was performed in a glass-bottom dish. The cell suspension (160 μL) was mixed with solution (40 μL) containing dTat-Sar-EED4 or dTat-Sar-EED5 (90 μM) in combination with Citrine (100 μg/mL) and incubated at 26 °C for 1 h. As a control, other cells were incubated with Citrine (100 μg/mL) alone or in combination with 90 μM of dTat, Sar-EED4 or Sar-EED5 at 26 °C for 1 h. Cellular internalization of Citrine was directly visualized from the dish under various magnifications at excitation/emission (Em/Ex) wavelengths of 488/515–580 nm using a confocal microscope (LSM 700/880, Carl Zeiss) and Zen 2011 operating software.
A. thaliana cotyledons and O. sativa calli were infiltrated with aqueous solution (100 μL) containing dTat-Sar-EED4 (90 μM) and Citrine (200 μg/mL) by the vacuum/compression method as described previously.45 Other cotyledons and calli were infiltrated with solution containing Citrine (200 μg/mL) alone as a control. After incubation (cotyledon: 22 °C, 1 h; callus: 30 °C, 1 h), the infiltrated samples were washed twice with half-strength Murashige and Skoog (1/2 MS) medium (100 μL) containing 1% sucrose (Fujifilm) and subjected to CLSM observation with an LSM 880. Citrine fluorescence was detected at Ex/Em wavelengths of 488/515–560 nm.
M. polymorpha sporelings were suspended in solution containing dTat-Sar-EED4 (90 μM) and Citrine (200 μg/mL) and incubated at 22 °C for 1 h. Other sporelings were incubated with only Citrine (200 μg/mL) at 22 °C for 1 h, as a control. To visualize Citrine fluorescence in sporelings, CLSM observation was performed using an SP8X system (Leica Microsystems) with a time-gated method (0.5–12.0 ns) according to a previous report.49 The detection of Citrine signal was performed at Ex/Em wavelengths of 510/546–566 nm.
C. reinhardtii cells were suspended in TAP medium containing dTat-Sar-EED5 (156 nM), Citrine (100 μg/mL) and sucrose (40 mM). The cells were incubated for 20 min, washed, and transferred onto a microscope slide. Other cells were incubated with Citrine (100 μg/mL) alone for 20 min and used as a control. CLSM observation was performed with an LSM 880 at Ex/Em wavelengths of 488/520–550 nm (for Citrine) and of 488/660–720 nm (for chlorophyll).
The internalization of Citrine into E. gracilis cells was performed in a 96-well microplate. Cell suspension (80 μL) was added to each well. The cells were then treated with Citrine (100 μg/mL) in the presence or absence of dTat-Sar-EED5 (9 μM). Culture medium was added to each well to a final volume of 100 μL and the plates incubated at 26 °C, with shaking at 100 rpm, for 3 h. CLSM observation was performed with an LSM 700. Fluorescent signals from Citrine and chlorophyll were detected at Ex/Em wavelengths of 488/520–550 nm and 488/660–720 nm, respectively.
Synthetic Peptide-Mediated GUS Transduction into Model Plant Cells
BY-2 cells were treated with GUS (100 μg/mL) in the presence or absence of either 90 μM of dTat-Sar-EED4 or dTat-Sar-EED5 at 26 °C for 1 h in mLS medium. Other cells were treated with GUS (100 μg/mL) in combination with 90 μM of dTat, R9 or BP100 at 26 °C for 1 h in mLS medium. After treatment, the cells were washed with mLS medium and treated with dye-βGlcU (Goryo Chemical) at 26 °C for 30 min in the dark. Excess dye-βGlcU was washed out with mLS medium prior to CLSM observation with an LSM 700. The delivered GUS activity was monitored by detecting the green fluorescent signal of the dye produced by GUS-catalyzed hydrolysis in cells with Ex/Em wavelengths of 488/510–550 nm.
A. thaliana cotyledons and O. sativa calli were infiltrated with aqueous solutions (200 μL) containing GUS (500 μg/mL) alone or in combination with each peptide either (90 μM of dTat-Sar-EED4, dTat-Sar-EED5, dTat, R9, or BP100) via the vacuum/compression method.45 After 1 h of incubation, the cotyledons and calli were washed with 1/2 MS medium containing 1% sucrose and then additionally treated with dye-βGlcU at 26 °C for 30 min. After being washed with 1/2 MS medium containing 1% sucrose, the samples were subjected to CLSM observation using an LSM 700 with Ex/Em wavelengths of 488/510–550 nm to monitor the delivered GUS activity.
The delivered GUS activity was evaluated by quantifying the green fluorescence intensity of the dye in cells using Fiji/ImageJ software. The means of relative green fluorescence intensity were determined from at least eight different regions of interest, which were manually selected using the freehand selection tool of Fiji/ImageJ in each confocal image, from 10 biologically independent samples. The background signal was subtracted from these fluorescence intensities to obtain the GUS activity levels in cells. The delivered GUS activity for each system was normalized to that of the dTat-Sar-EED4-treated samples.
FM4-64 Uptake into A. thaliana Roots
FM4-64 (Thermo Fisher Scientific) was dissolved in DMSO. A. thaliana roots were pretreated with DMSO (0.6% v/v, control) or ES9-17 (30 μM) at 25 °C for 30 min in 1/2 MS medium, and then FM4-64 (10 μM) was added alone or in combination with dTat-Sar-EED4 (90 μM). Following incubation at 25 °C for 30 min in the dark, CLSM observation was performed to detect FM4-64 uptake into cells using an LSM 700 with Ex/Em wavelengths of 488/620–680 nm.
To quantify the intracellular/plasma membrane (PM) fluorescence intensity ratio, the mean fluorescence intensities in the entire cytoplasm and apical plasma membrane regions, which were manually segmented using the polygonal selection tool of Fiji/ImageJ, were assigned to intracellular and PM fluorescence intensities, respectively. The intracellular/PM fluorescence intensity ratio was calculated using Fiji/ImageJ for more than 100 cells from 8 independent roots for each system.
Synthetic Peptide-Mediated GUS Transduction into A. thaliana Plants Treated with Pharmacological Inhibitors
A. thaliana roots and cotyledons were pretreated with ES9-17 (root, 30 μM; cotyledon, 100 μM), Wm (40 μM), CytD (40 μM) or DMSO (0.5% v/v, control) at 25 °C for 30 min in 1/2 MS medium (200 μL). The samples were treated with dTat-Sar-EED4 (90 μM) and GUS (500 μg/mL) in the presence of each inhibitor (ES9-17, 30 μM for root and 100 μM for cotyledon; Wm, 40 μM; CytD, 40 μM) or DMSO (0.5% v/v) at 25 °C for 30 min in 1/2 MS medium containing 1% sucrose (200 μL). The samples were washed with 1/2 MS medium containing 1% sucrose to remove extracellular peptides and enzymes and incubated with dye-βGlcU at 26 °C for 30 min in the dark. After being washed with 1/2 MS medium containing 1% sucrose to remove excess substrates, the samples were subjected to CLSM observation with an LSM 700 with Ex/Em wavelengths of 488/510–550 nm to evaluate the activity of the delivered GUS. The GUS activity delivered in each system was quantified as described above and normalized to that of the controls (DMSO-treated samples).
TEM of A. thaliana roots and BY-2 cells
A. thaliana roots and BY-2 cells were treated with dTat-Sar-EED4 (30 μM or 90 μM) at 25 °C for 15 or 10 min, respectively. As controls, other roots and cells were incubated at 25 °C in water (15 min) and MS medium (10 min), respectively. The root and cell samples were frozen in a high-pressure freezer (Leica EM ICE), and TEM observation were performed as previously described.50,51
Statistical Analysis and Generation of Graphs
Statistical tests and graphs were generated with GraphPad Prism 9.
Acknowledgments
This work was supported by Grants-in-Aid from the Japan Science and Technology Agency Exploratory Research for Advanced Technology (JST-ERATO; grant no. JPMJER1602 to K.N.) and from the Japan Society for the Promotion of Science for Scientific Research (JSPS-KAKENHI; grant no. JP19K15411 to T.M.). N. tabacum BY-2 cell line (rpc00001) was provided by the RIKEN BRC which is participating in the National BioResource Project of the MEXT/AMED, Japan. We thank the Support Unit for Bio-Material Analysis, RIKEN Center for Brain Science Research Resources Division, for performing peptide synthesis. We also thank Drs. Takashi Osanai, Yoshiki Nishimura, and Takayuki Kohchi for providing the E. gracilis, C. reinhardtii and M. polymorpha strains, respectively. We acknowledge Ms. Mio Hikawa and Ms. Hitomi Takahashi for technical support in the M. polymorpha experiment.
Supporting Information Available
The Supporting Information is available free of charge at https://pubs.acs.org/doi/10.1021/jacsau.1c00504.
Viability of BY-2 cells after prolonged treatment with dTat-Sar-EED4 for up to 48 h, confocal images showing negligible Citrine uptake in BY-2 cells treated with low concentrations of dTat-Sar-EED4 or dTat-Sar-EED5, confocal images of BY-2 cells, A. thaliana cotyledons and O. sativa calli after GUS transduction with dTat-Sar-EED4 under different conditions, confocal images of BY-2 cells, A. thaliana cotyledons and O. sativa calli after GUS transduction with dTat-Sar-EED5 under different conditions, confocal images of BY-2 cells, A. thaliana cotyledons and O. sativa calli after GUS transduction with or without various types of peptides, confocal images of A. thaliana root cells treated for 30 min with water or DMSO (0.6% v/v) and then stained for 30 min with FM4-64 (10 μM), GUS enzymatic activity in the presence of pharmacological inhibitors, transmission electron microscopic (TEM) images showing the formation of plasma membrane protrusions with unstructured objects in dTat-Sar-EED4-treated A. thaliana roots, TEM images of BY-2 cells treated without and with dTat-Sar-EED4 (PDF)
Movie S1. Citrine uptake by BY-2 cells in the presence of 90 μM dTat-Sar-EED4 (AVI)
Movie S2. Citrine uptake by BY-2 cells in the presence of 90 μM dTat as a control (AVI)
Author Contributions
T.M. and K.N. conceived the study and designed the experiments. T.M. and J.C. performed the experiments with BY-2 cells. T.M. performed the experiments with A. thaliana and O. sativa. J.C. and M.H-T. performed the experiments with E. gracilis. M.O. and Y.K. performed the experiments with C. reinhardtii and M. polymorpha, respectively. Y.G. and K.T. performed the TEM observations. Y.M. and T.K. prepared Citrine by cell-free synthesis. T.M. analyzed all the experimental data. T.M. and K.N. wrote the manuscript. All authors commented on the results and the manuscript.
The authors declare no competing financial interest.
Supplementary Material
References
- Barone R. P.; Knittel D. K.; Ooka J. K.; Porter L. N.; Smith N. T.; Owens D. K. The production of plant natural products beneficial to humanity by metabolic engineering. Curr. Plant Biol. 2020, 24, 100121. 10.1016/j.cpb.2019.100121. [DOI] [Google Scholar]
- Mohammadinejad R.; Shavandi A.; Raie D. S.; Sangeetha J.; Soleimani M.; Shokrian Hajibehzad S.; Thangadurai D.; Hospet R.; Popoola J. O.; Arzani A.; Gómez-Lim M. A.; Iravani S.; Varma R. S. Plant molecular farming: production of metallic nanoparticles and therapeutic proteins using green factories. Green Chem. 2019, 21, 1845–1865. 10.1039/C9GC00335E. [DOI] [Google Scholar]
- Rodionova M. V.; Poudyal R. S.; Tiwari I.; Voloshin R. A.; Zharmukhamedov S. K.; Nam H. G.; Zayadan B. K.; Bruce B. D.; Hou H. J. M.; Allakhverdiev S. I. Biofuel production: Challenges and opportunities. Int. J. Hydrogen Energy 2017, 42, 8450–8461. 10.1016/j.ijhydene.2016.11.125. [DOI] [Google Scholar]
- Cunningham F. J.; Goh N. S.; Demirer G. S.; Matos J. L.; Landry M. P. Nanoparticle-Mediated Delivery towards Advancing Plant Genetic Engineering. Trends Biotechnol. 2018, 36, 882–897. 10.1016/j.tibtech.2018.03.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Martin-Ortigosa S.; Peterson D. J.; Valenstein J. S.; Lin V. S. Y.; Trewyn B. G.; Lyznik L. A.; Wang K. Mesoporous Silica Nanoparticle-Mediated Intracellular Cre Protein Delivery for Maize Genome Editing via loxP Site Excision. Plant Physiol. 2014, 164, 537–547. 10.1104/pp.113.233650. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Que Q.; Chilton M.-D. M.; Elumalai S.; Zhong H.; Dong S.; Shi L.. Repurposing Macromolecule Delivery Tools for Plant Genetic Modification in the Era of Precision Genome Engineering. In Transgenic Plants: Methods and Protocols; Kumar S., Barone P., Smith M., Eds.; Springer New York: New York, 2019; pp 3–18. [DOI] [PubMed] [Google Scholar]
- Mitragotri S.; Burke P. A.; Langer R. Overcoming the challenges in administering biopharmaceuticals: formulation and delivery strategies. Nat. Rev. Drug Discovery 2014, 13, 655–672. 10.1038/nrd4363. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stewart M. P.; Sharei A.; Ding X.; Sahay G.; Langer R.; Jensen K. F. In vitro and ex vivo strategies for intracellular delivery. Nature 2016, 538, 183–192. 10.1038/nature19764. [DOI] [PubMed] [Google Scholar]
- Mosquera J.; García I.; Liz-Marzán L. M. Cellular Uptake of Nanoparticles versus Small Molecules: A Matter of Size. Acc. Chem. Res. 2018, 51, 2305–2313. 10.1021/acs.accounts.8b00292. [DOI] [PubMed] [Google Scholar]
- Harush-Frenkel O.; Debotton N.; Benita S.; Altschuler Y. Targeting of nanoparticles to the clathrin-mediated endocytic pathway. Biochem. Biophys. Res. Commun. 2007, 353, 26–32. 10.1016/j.bbrc.2006.11.135. [DOI] [PubMed] [Google Scholar]
- Tu Z.; Achazi K.; Schulz A.; Mülhaupt R.; Thierbach S.; Rühl E.; Adeli M.; Haag R. Combination of Surface Charge and Size Controls the Cellular Uptake of Functionalized Graphene Sheets. Adv. Funct. Mater. 2017, 27, 1701837. 10.1002/adfm.201701837. [DOI] [Google Scholar]
- Kerr M. C.; Teasdale R. D. Defining Macropinocytosis. Traffic 2009, 10, 364–371. 10.1111/j.1600-0854.2009.00878.x. [DOI] [PubMed] [Google Scholar]
- Arafiles J. V. V.; Hirose H.; Hirai Y.; Kuriyama M.; Sakyiamah M. M.; Nomura W.; Sonomura K.; Imanishi M.; Otaka A.; Tamamura H.; Futaki S. Discovery of a Macropinocytosis-Inducing Peptide Potentiated by Medium-Mediated Intramolecular Disulfide Formation. Angew. Chem., Int. Ed. 2021, 60, 11928–11936. 10.1002/anie.202016754. [DOI] [PubMed] [Google Scholar]
- Zhou S.; Huang Y.; Chen Y.; Liu S.; Xu M.; Jiang T.; Song Q.; Jiang G.; Gu X.; Gao X.; Chen J. Engineering ApoE3-incorporated biomimetic nanoparticle for efficient vaccine delivery to dendritic cells via macropinocytosis to enhance cancer immunotherapy. Biomaterials 2020, 235, 119795. 10.1016/j.biomaterials.2020.119795. [DOI] [PubMed] [Google Scholar]
- Paez Valencia J.; Goodman K.; Otegui M. S. Endocytosis and Endosomal Trafficking in Plants. Annu. Rev. Plant Biol. 2016, 67, 309–335. 10.1146/annurev-arplant-043015-112242. [DOI] [PubMed] [Google Scholar]
- Snyder E. L.; Meade B. R.; Saenz C. C.; Dowdy S. F. Treatment of terminal peritoneal carcinomatosis by a transducible p53-activating peptide. PLoS Biol. 2004, 2, e36 10.1371/journal.pbio.0020036. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lönn P.; Kacsinta A. D.; Cui X.-S.; Hamil A. S.; Kaulich M.; Gogoi K.; Dowdy S. F. Enhancing endosomal escape for intracellular delivery of macromolecular biologic therapeutics. Sci. Rep. 2016, 6, 32301. 10.1038/srep32301. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Miyamoto T.; Tsuchiya K.; Numata K. Dual Peptide-Based Gene Delivery System for the Efficient Transfection of Plant Callus Cells. Biomacromolecules 2020, 21, 2735–2744. 10.1021/acs.biomac.0c00481. [DOI] [PubMed] [Google Scholar]
- Narasimhan M.; Johnson A.; Prizak R.; Kaufmann W. A.; Tan S.; Casillas-Pérez B.; Friml J. Evolutionarily unique mechanistic framework of clathrin-mediated endocytosis in plants. eLife 2020, 9, e52067. 10.7554/eLife.52067. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chia S. R.; Ong H. C.; Chew K. W.; Show P. L.; Phang S.-M.; Ling T. C.; Nagarajan D.; Lee D.-J.; Chang J.-S. Sustainable approaches for algae utilisation in bioenergy production. AMA Arch. Int. Med. 2018, 129, 838–852. 10.1016/j.renene.2017.04.001. [DOI] [Google Scholar]
- Bilichak A.; Luu J.; Eudes F. Intracellular delivery of fluorescent protein into viable wheat microspores using cationic peptides. Front. Plant Sci. 2015, 6, 666. 10.3389/fpls.2015.00666. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Eggenberger K.; Mink C.; Wadhwani P.; Ulrich A. S.; Nick P. Using the peptide Bp100 as a cell-penetrating tool for the chemical engineering of actin filaments within living plant cells. ChemBioChem 2011, 12, 132–137. 10.1002/cbic.201000402. [DOI] [PubMed] [Google Scholar]
- Dejonghe W.; Sharma I.; Denoo B.; De Munck S.; Lu Q.; Mishev K.; Bulut H.; Mylle E.; De Rycke R.; Vasileva M.; Savatin D. V.; Nerinckx W.; Staes A.; Drozdzecki A.; Audenaert D.; Yperman K.; Madder A.; Friml J.; Van Damme D.; Gevaert K.; Haucke V.; Savvides S. N.; Winne J.; Russinova E. Disruption of endocytosis through chemical inhibition of clathrin heavy chain function. Nat. Chem. Biol. 2019, 15, 641–649. 10.1038/s41589-019-0262-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Meier O.; Boucke K.; Hammer S. V.; Keller S.; Stidwill R. P.; Hemmi S.; Greber U. F. Adenovirus triggers macropinocytosis and endosomal leakage together with its clathrin-mediated uptake. J. Cell Biol. 2002, 158, 1119–1131. 10.1083/jcb.200112067. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Takáč T.; Pechan T.; Šamajová O.; Ovečka M.; Richter H.; Eck C.; Niehaus K.; Šamaj J. Wortmannin Treatment Induces Changes in Arabidopsis Root Proteome and Post-Golgi Compartments. J. Proteome Res. 2012, 11, 3127–3142. 10.1021/pr201111n. [DOI] [PubMed] [Google Scholar]
- Ketelaar T.; de Ruijter N. C. A.; Emons A. M. C. Unstable F-Actin Specifies the Area and Microtubule Direction of Cell Expansion in Arabidopsis Root Hairs. Plant Cell 2003, 15, 285–292. 10.1105/tpc.007039. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Miyamoto T.; Tsuchiya K.; Numata K. Endosome-escaping micelle complexes dually equipped with cell-penetrating and endosome-disrupting peptides for efficient DNA delivery into intact plants. Nanoscale 2021, 13, 5679–5692. 10.1039/D0NR08183C. [DOI] [PubMed] [Google Scholar]
- Wang J. W.; Cunningham F. J.; Goh N. S.; Boozarpour N. N.; Pham M.; Landry M. P. Nanoparticles for protein delivery in planta. Curr. Opin. Plant Biol. 2021, 60, 102052. 10.1016/j.pbi.2021.102052. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Demirer G. S.; Silva T. N.; Jackson C. T.; Thomas J. B. W.; Ehrhardt D.; Rhee S. Y.; Mortimer J. C.; Landry M. P. Nanotechnology to advance CRISPR-Cas genetic engineering of plants. Nat. Nanotechnol. 2021, 16, 243–250. 10.1038/s41565-021-00854-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Odahara M.; Watanabe K.; Kawasaki R.; Tsuchiya K.; Tateishi A.; Motoda Y.; Kigawa T.; Kodama Y.; Numata K. Nanoscale Polyion Complex Vesicles for Delivery of Cargo Proteins and Cas9 Ribonucleoprotein Complexes to Plant Cells. ACS Appl. Nano Mater. 2021, 4, 5630–5635. 10.1021/acsanm.1c00695. [DOI] [Google Scholar]
- Bilichak A.; Sastry-Dent L.; Sriram S.; Simpson M.; Samuel P.; Webb S.; Jiang F.; Eudes F. Genome editing in wheat microspores and haploid embryos mediated by delivery of ZFN proteins and cell-penetrating peptide complexes. Plant Biotechnol. J. 2020, 18, 1307–1316. 10.1111/pbi.13296. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yin K.; Gao C.; Qiu J.-L. Progress and prospects in plant genome editing. Nat. Plants 2017, 3, 17107. 10.1038/nplants.2017.107. [DOI] [PubMed] [Google Scholar]
- Svitashev S.; Schwartz C.; Lenderts B.; Young J. K.; Mark Cigan A. Genome editing in maize directed by CRISPR-Cas9 ribonucleoprotein complexes. Nat. Commun. 2016, 7, 13274. 10.1038/ncomms13274. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hansen C. G.; Nichols B. J. Molecular mechanisms of clathrin-independent endocytosis. J. Cell Sci. 2009, 122, 1713–1721. 10.1242/jcs.033951. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nakase I.; Niwa M.; Takeuchi T.; Sonomura K.; Kawabata N.; Koike Y.; Takehashi M.; Tanaka S.; Ueda K.; Simpson J. C.; Jones A. T.; Sugiura Y.; Futaki S. Cellular Uptake of Arginine-Rich Peptides: Roles for Macropinocytosis and Actin Rearrangement. Mol. Ther. 2004, 10, 1011–1022. 10.1016/j.ymthe.2004.08.010. [DOI] [PubMed] [Google Scholar]
- Wadia J. S.; Stan R. V.; Dowdy S. F. Transducible TAT-HA fusogenic peptide enhances escape of TAT-fusion proteins after lipid raft macropinocytosis. Nat. Med. 2004, 10, 310–315. 10.1038/nm996. [DOI] [PubMed] [Google Scholar]
- Bandmann V.; Müller J. D.; Köhler T.; Homann U. Uptake of fluorescent nano beads into BY2-cells involves clathrin-dependent and clathrin-independent endocytosis. FEBS Lett. 2012, 586, 3626–3632. 10.1016/j.febslet.2012.08.008. [DOI] [PubMed] [Google Scholar]
- Bandmann V.; Homann U. Clathrin-independent endocytosis contributes to uptake of glucose into BY-2 protoplasts. Plant J. 2012, 70, 578–584. 10.1111/j.1365-313X.2011.04892.x. [DOI] [PubMed] [Google Scholar]
- Li R.; Liu P.; Wan Y.; Chen T.; Wang Q.; Mettbach U.; Baluška F.; Šamaj J.; Fang X.; Lucas W. J.; Lin J. A Membrane Microdomain-Associated Protein, Arabidopsis Flot1, Is Involved in a Clathrin-Independent Endocytic Pathway and Is Required for Seedling Development. Plant Cell 2012, 24, 2105–2122. 10.1105/tpc.112.095695. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Baral A.; Irani N. G.; Fujimoto M.; Nakano A.; Mayor S.; Mathew M. K. Salt-Induced Remodeling of Spatially Restricted Clathrin-Independent Endocytic Pathways in Arabidopsis Root. Plant Cell 2015, 27, 1297–1315. 10.1105/tpc.15.00154. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nakase I.; Tadokoro A.; Kawabata N.; Takeuchi T.; Katoh H.; Hiramoto K.; Negishi M.; Nomizu M.; Sugiura Y.; Futaki S. Interaction of Arginine-Rich Peptides with Membrane-Associated Proteoglycans Is Crucial for Induction of Actin Organization and Macropinocytosis. Biochemistry 2007, 46, 492–501. 10.1021/bi0612824. [DOI] [PubMed] [Google Scholar]
- Grimmer S.; van Deurs B.; Sandvig K. Membrane ruffling and macropinocytosis in A431 cells require cholesterol. J. Cell Sci. 2002, 115, 2953–2962. 10.1242/jcs.115.14.2953. [DOI] [PubMed] [Google Scholar]
- Ng K. K.; Motoda Y.; Watanabe S.; Sofiman Othman A.; Kigawa T.; Kodama Y.; Numata K. Intracellular delivery of proteins via fusion peptides in intact plants. PLoS One 2016, 11, e0154081. 10.1371/journal.pone.0154081. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nagata T.; Nemoto Y.; Hasezawa S.. Tobacco BY-2 cell line as the “HeLa” cell in the cell biology of higher plants. In Int. Rev. Cytol., Jeon K. W., Friedlander M., Eds.; Academic Press: Cambridge, 1992; Vol. 132, pp 1–30. [Google Scholar]
- Midorikawa K.; Kodama Y.; Numata K. Vacuum/compression infiltration-mediated permeation pathway of a peptide-pDNA complex as a non-viral carrier for gene delivery in planta. Sci. Rep. 2019, 9, 271. 10.1038/s41598-018-36466-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ogasawara Y.; Ishizaki K.; Kohchi T.; Kodama Y. Cold-induced organelle relocation in the liverwort Marchantia polymorpha L. Plant, Cell Environ. 2013, 36, 1520–1528. 10.1111/pce.12085. [DOI] [PubMed] [Google Scholar]
- Cramer M.; Myers J. Growth and photosynthetic characteristics of Euglena gracilis. Arch. Microbiol. 1952, 17, 384–402. 10.1007/BF00410835. [DOI] [Google Scholar]
- Iriti M.; Sironi M.; Gomarasca S.; Casazza A. P.; Soave C.; Faoro F. Cell death-mediated antiviral effect of chitosan in tobacco. Plant Physiol. Biochem. 2006, 44, 893–900. 10.1016/j.plaphy.2006.10.009. [DOI] [PubMed] [Google Scholar]
- Kodama Y. Time gating of chloroplast autofluorescence allows clearer fluorescence imaging in planta. PLoS One 2016, 11, e0152484. 10.1371/journal.pone.0152484. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Toyooka K.; Goto Y.; Asatsuma S.; Koizumi M.; Mitsui T.; Matsuoka K. A Mobile Secretory Vesicle Cluster Involved in Mass Transport from the Golgi to the Plant Cell Exterior. Plant Cell 2009, 21, 1212–1229. 10.1105/tpc.108.058933. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Toyooka K.; Sato M.; Kutsuna N.; Higaki T.; Sawaki F.; Wakazaki M.; Goto Y.; Hasezawa S.; Nagata N.; Matsuoka K. Wide-Range High-Resolution Transmission Electron Microscopy Reveals Morphological and Distributional Changes of Endomembrane Compartments during Log to Stationary Transition of Growth Phase in Tobacco BY-2 Cells. Plant Cell Physiol. 2014, 55, 1544–1555. 10.1093/pcp/pcu084. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.