Abstract
Background
Idiopathic pulmonary fibrosis (IPF) is the most common type of interstitial pneumonia. Lung cancer, mainly non-small cell lung cancer (NSCLC), is a complication of idiopathic pulmonary fibrosis. IPF is also an independent risk factor of lung cancer. Some studies have shown that the complement system can promote the progression of interstitial pulmonary fibrosis. In addition, C1q has also demonstrated to exert a tumor-promoting effect in many tumors. However, the role of C1q in idiopathic pulmonary fibrosis and lung cancer still remain unclear.
Methods
We selected common differentially expressed genes in IPF and non-small cell lung cancer using datasets from GEO, and investigated common hub gene. The hub genes were validated in IPF by establishing mouse model of IPF and using another four datasets from the GEO. Multiple databases were analyzed including those of Kaplan–Meier Plotter, Tumor Immune Estimation Resource (TIMER2.0) and the Human Protein Atlas (HPA) for NSCLC.
Results
In this study, 37 common DEGs were identified in IPF and NSCLC including 32 up-regulated genes and 5 down-regulated genes, and C1q was identified as common hub gene. The methylation status of C1q decreased and the expression levels of C1q increased in both lung cancer and idiopathic pulmonary fibrosis. The prognosis of non-small cell lung cancer and IPF patients with high levels of C1q is poor.
Conclusions
These results show that C1q participates in pulmonary fibrosis and non-small cell lung cancer, and may be a potential diagnostic / prognostic biomarker or a therapeutic target.
Supplementary Information
The online version contains supplementary material available at 10.1186/s12885-021-08912-3.
Keywords: Complement, C1q, Idiopathic pulmonary fibrosis, Non-small-cell lung cancer, Prognosis, DNA methylation
Background
Idiopathic pulmonary fibrosis (IPF) is the most common type of interstitial pneumonia [1]. It is a fatal, invariably progressive disease. Lung transplantation is the most effective treatment [2]. The incidence of IPF appears to be increasing, so early recognition and intervention with supportive pharmacologic agents are needed to prevent its progression. Lung cancer (LC) is one of the most common complications in IPF patients, and mortality among patients with IPF and LC is highest [3]. Likewise, IPF is an independent risk factor for LC [4]. Nintedanib, used as an anti-fibrotic drug for IPF, is also approved for non-small cell lung cancer (NSCLC) [5], which further suggests the close relationship between IPF and NSCLC. Thus, the pathogenic overlap of IPF and LC may help clinicians to better understand the molecular mechanisms involved in both diseases, and may contribute to provide therapeutic strategies.
C1q protein was first identified in 1961 and was described as a “11s thermolabile serum protein” that participated in immune hemolysis [6]. It is the first subcomponent of the C1 complex, which activates the classical pathway of the complement [7]. C1q is composed of three polypeptide chains (C1qa, C1qb and C1qc) and regulates immune and non-immunes responses. The complement system appears to accelerate the pathogenesis of IPF [8, 9], while studies about the relationship between IPF and C1q are limited. C1q can act as a promoting factor of cancer in the tumor microenvironment [10]. However, the role of C1q in IPF and LC has not been fully explored.
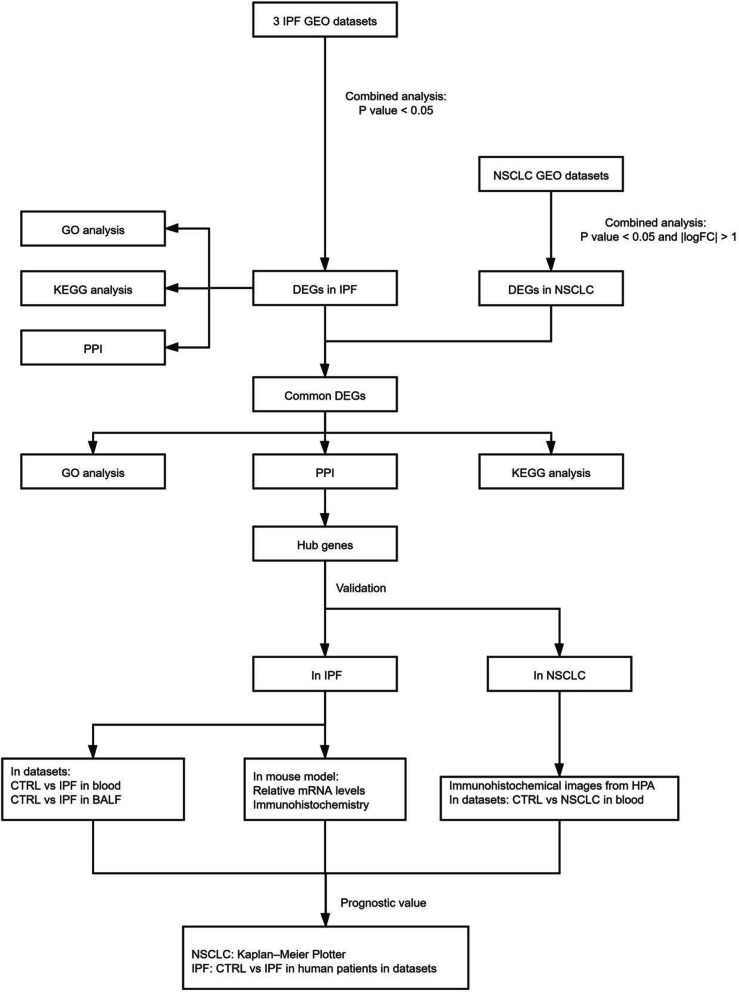
In our study, three mRNA microarray datasets were downloaded from the Gene Expression Omnibus (GEO) and 37 differentially expressed genes (DEGs) were obtained comparing lung tissue with pulmonary fibrosis and normal lung tissues. Next, we explored common DEGs in both IPF and NSCLC. Gene ontology (GO) terms and KEGG pathway enrichment analysis were performed using DAVID. Protein–protein interaction (PPI) network analysis was performed and hub genes were screened to aid better understand the relationships between DEGs. C1q was finally identified as a co-hub gene. We verified the role of C1q in IPF using another four datasets from the GEO and establish mouse model of IPF. Besides, we analyzed multiple databases including those of Kaplan–Meier Plotter, Tumor Immune Estimation Resource (TIMER2.0) and the Human Protein Atlas (HPA) to reveal the clinical significance and function of C1q in NSCLC especially in lung adenocarcinoma. Finally, SurvivalMeth platforms and GSE63704 was used to detecte methylation levels of C1q. The flow diagram of this study is shown in Fig. 1.
Fig. 1.
The flow diagram of this study
Methods
Mice
All experimental procedures involving animals were performed in accordance with the guidelines of the National Institutes of Health for the care and use of laboratory animals (NIH Publication, 8th Edition, 2011) and approved by the Animal Care and Use Committees of Shanghai Tenth People’s Hospital (Shanghai, China; permit number: SHDSYY-2019-2149). 12–16-week-old female mice were purchased from Charles River Laboratories. The murine pulmonary fibrosis model was induced with intratracheal injection of 1.5 U/kg BLM for 4 weeks, and control mice were given normal saline in same volume. After the study, all the mice were first anesthetized by allowing them to inhale 2.0% isoflurane and subjected to cervical dislocation. The study received approval by the Animal Care and Use Committees of Shanghai Tenth People’s Hospital for animal welfare.
Real-time RT-PCR
Total RNA from lung tissue was extracted by Trizol reagent (Life Technologies), and then reserve transcribed to generate cDNA by the PrimeScript reverse transcription reagent kit (TaKaRa Bio, Shiga, Japan). Quantitative real-time PCR was carried out using SYBR Green (Roche). The gene Glyceraldehyde-3-phosphate dehydrogenase (GAPDH) was used as an internal control. Primers were listed in Supplemental Table 1.
Histology and Immunohistology
Lung tissue samples were fixed by 4% of paraformaldehyde, dehydrated, and paraffin embedded for sectioning into 5-μm-thick sections and then used for Masson staining and immunohistochemical staining. The results of Masson staining were used to evaluate the cross-sectional area and the collagen volume respectively by light microscopy. Immunostainings for C1QA (A13659; ABclonal; 1:100), C1QB (A5339; ABclonal; 1:100) and C1QC (A9227; ABclonal; 1:100) was performed on paraffin embedded sections and incubated with goat anti-rabbit secondary antibodies to examine positive cells in lung tissue. All histopathological sections were measured with Image-Pro Plus 6.0 image analysis system.
Microarray data
Data were retrieved from the GEO database, including the GSE485 series on the GPL81 platform (Affymetrix Murine Genome U74A Version 2 Array), the GSE97546 series on the GPL6887 platform (Illumina MouseWG-6 v2.0 expression beadchip), the GSE37635 [11] series on the GPL6885 platform (Illumina MouseRef-8 v2.0 expression beadchip), and the GSE31013 [12] series on the GPL1261 platform (Affymetrix Mouse Genome 430 2.0 Array). We chose samples from established animal models that required more than 1 week for fibrosis to be established to ensure the IPF model was successful (Table 1). Datasets including GSE98468, GSE102751 and GSE124685 [13, 14] were used to confirme the role of C1q in IPF. GSE63704 was used to detecte methylation levels of C1q in IPF. Datasets including GSE135304 and GSE76033 were used to confirme the role of C1q in NSCLC (Table 2).
Table 1.
Characteristics of datasets for analysis
| Dataset ID | Tissue | Strains of mice | Gender | Number of samples | GPL | References |
|---|---|---|---|---|---|---|
| GSE37635 | Lung tissue | C57BL/6 | Female |
Control n = 7 1-week-IPF n = 7 2-week-IPF n = 6 3-week-IPF n = 6 4-week-IPF n = 6 5-week-IPF n = 6 |
GPL6885 | Stoop R et al. 2013 |
| GSE97546 | Lung tissue | BALB/C | Female |
Control n = 3 IPF n = 3 |
GPL6887 | LeBleu V et al. 2020 |
| GSE485 | Lung tissue | C57BL/6 | Male |
Control n = 2 2-week-IPF n = 2 |
GPL81 | Moller D et al. 2003 |
| GSE31013 | Lung tissue |
B6C3F1 (C57BL/6 J × C3H F1) |
Male |
Control n = 6 Lung tumor n = 6 |
GPL1261 | Sills RC et al. 2012 |
Table 2.
Characteristics of the datasets for validation
| Dataset ID | Tissue | Organism | NO. of samples | GPL | References |
|---|---|---|---|---|---|
| GSE98468 | Balf | Mus musculus |
Control n = 3 IPF n = 3 |
GPL19057 | Xie N et al. 2017 |
| GSE102751 | Blood | Homo sapiens |
Control n = 3 IPF n = 3 |
GPL11154 | Habiel DM et al. 2019 |
| GSE124685 | Lung tissue | Homo sapiens |
Control n = 35 IPF n = 49 |
GPL17303 | Kaminiski N et al. 2019 |
| GSE63704 | Lung tissue | Homo sapiens |
Control n = 37 IPF n = 43 |
GPL13534 | Wielscher M et al. 2015 |
| GSE135304 | Blood | Homo sapiens |
Control n = 88 NSCLC n = 404 |
GPL10558 | Showe L et al. 2020 |
| GSE76033 | Blood | Mus musculus |
Control n = 3 Lung cancer n = 3 |
GPL13112 | Poczobutt JM et al. 2016 |
Data processing of differentially expressed genes
GEO2R is an online tool that allows the comparison of groups in the GEO series to identify DEGs [15]. We identified DEGs comparing healthy mice and mice with lung fibrosis using GEO2R https://www.ncbi.nlm.nih.gov/geo/geo2r/ ). Genes without corresponding gene symbols were removed. Only one probe was kept for genes with different probes, and p < 0.05 was considered 143 for statistical significance. Co-differentially expressed p < 0.05 was considered for statistical significance. Co-differentially expressed genes were identified and Venn maps were drawn via the Venn online tool (http://bioinformatics.psb.ugent.be/webtools/Venn/ ).
Functional enrichment analysis of differentially expressed genes
DAVID ( https://david.abcc.ncifcrf.gov/ ) is an online bio-informatics tool composed of a comprehensive biological knowledge base and analytic tools. It can extract biological meaning of genes or proteins systematically [16]. We used DAVID to perform a functional enrichment analysis, including biological processes (BP), molecular functions (MF), and cellular components (CC), and KEGG pathway analysis of the overlapping DEGs( p < 0.05). The bubble chart was plotted by http://www.bioinformatics.com.cn , an online platform for data analysis and visualization.
Construction of the protein interaction network and modules selection
The String online database (https://string-db.org) was used to construct a protein–protein interaction (PPI) network for DEGs and a combined-score > 0.4 was considered a significant difference. The PPI network was visualized with Cytoscape (www.cytoscape.org). Cytoscape has been used for integrated models of biomolecular interaction networks [17]. The essential proteins were predicted using the Molecular Complex Detection (MCODE) within the PPI network as follows: MCODE score > 5, degree cutoff = 2, node density cutoff = 0.1, node score cutoff = 0.2, k-score = 2, and maximum depth = 100.
Survival analysis and validation of the hub genes
Kaplan–Meier Plotter (http://kmplot.com/analysis/index.php?p=service) was used to assess the prognostic value of hub genes in non-small-cell LC [18], which includes 1925 patients. We scanned the immunohistochemical images of lung adenocarcinoma and squamous cell carcinoma from The HPA (https://www.proteinatlas.org/), has open access allows free access to data for exploration of the human proteome [19]. TIMER2.0 [20] (http://timer.cistrome.org/) was used to analyze the correlation of C1q with the fibrosis indexes. SurvivalMeth (http://bio-bigdata.hrbmu.edu.cn/survivalmeth/) was used to analyze the effect of DNA methylation of C1q on LUAD prognosis [21].
Statistical analysis
Microarray data was analysed using Statistical Package for Social Sciences (SPSS, Version 20.0). Heatmap, box plot and violin plot were drawn by Graphpad prism 8. Continuous variables were presented as mean ± SEM. Comparisons between groups were made using unpaired two-tailed t test or one-way ANOVA when appropriate and p < 0.05 was considered to be significant.
Results
Identification and analysis of differentially expressed genes in idiopathic pulmonary fibrosis
Three microarray datasets were chosen to identify DEGs in pulmonary fibrosis from the GEO. The GSE485 dataset contained 2 control samples and 2 pulmonary fibrosis samples. The GSE97546 dataset contained 3 lung samples of healthy mice and 3 lung samples of mice with lung fibrosis. The GSE37635 included 6 timepoints: the control (n = 7), 1 week (n = 7), 2 weeks (n = 6), 3 weeks (n = 6), 4 weeks (n = 6), and 5 weeks (n = 6) after bleomycin-treatment to induce lung fibrosis. We first extracted gene expressions changes at each time points in the GSE37635 dataset (Fig. S1), then the results were intersected with the other two datasets.
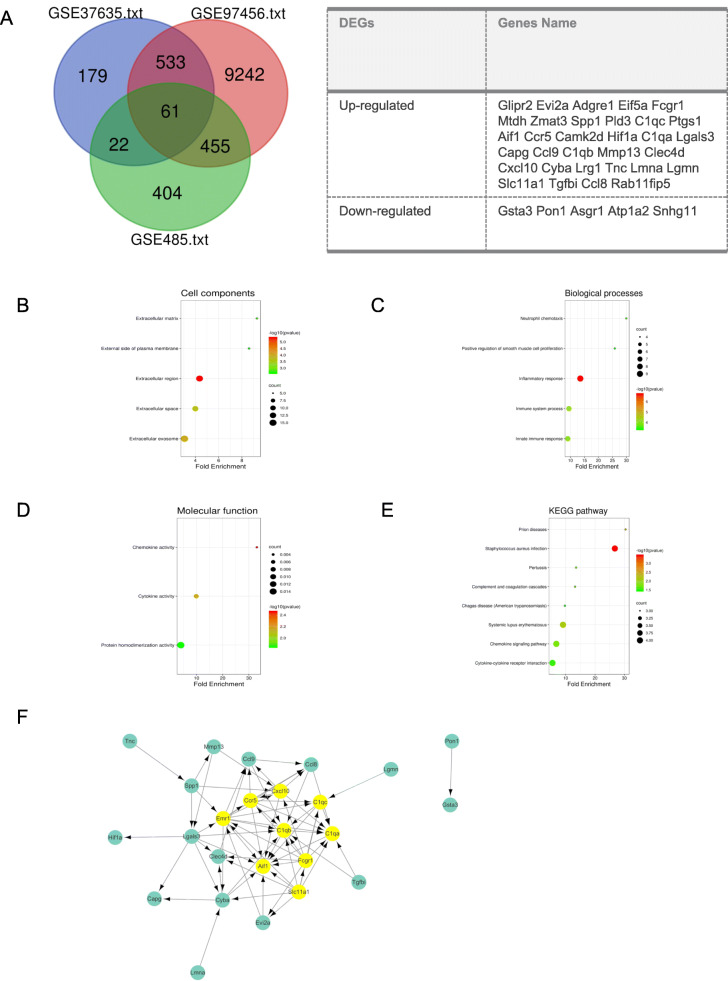
Sixty one genes overlapped across the three datasets as shown in the Venn map. Genes with opposite trends were then excluded. Finally, we identified the 37 overlapping DEGs in the 3 datasets, consisting of 32 genes that were up-regulated and 5 genes that were down-regulated (Fig. 2A).
Fig. 2.
Analysis of DEGs in pulmonary fibrosis. (A) Venn diagram of DEGs in mRNA expression profiling datasets GSE485, GSE37635, and GSE97546. (B-D) GO and KEGG enrichment analysis of the DEGs. (F) PPI network of DEGs. (G) A significant module selected from PPI network, all of them were upregulated genes
Differentially expressed gene ontology and KEGG pathway analysis in pulmonary fibrosis
DAVID was used to perform GO term and KEGG pathway enrichment analysis for further insight into the function of the identified DEGs. We selected the top 5 terms of biological processes and cell components according to p value. We found cell components of DEGs mainly enriched in extracellular regions, extracellular exosome and extracellular space (Fig. 2B). The biological processes were mainly related to inflammation and immune pathways (Fig. 2C). Further, molecular function mainly involved chemokine activity, cytokine activity and protein homodimerization activity (Fig. 2D). Moreover, eight KEGG pathways were overrepresented in DEGs, including staphylococcus aureus infection, prion diseases, systemic lupus erythematosus, chemokine signaling pathway, pertussis, complement and coagulation cascades, cytokine-cytokine receptor interaction and chagas disease (Fig. 2E).
Protein-protein interaction network construction and hub genes selection
The PPI network, which was consisted of 25 nodes and 74 edges, was constructed using STRING and was visualized by Cytoscape software to explore the association between the DEGs. The MCODE plugin was used to identify hub genes. C1qa, Fcgr1, C1qb, C1qc, Ccr5, Slc11a1, Aif1, Emr1 and Cxcl10 were identified as the most closely connected module which were highlighted in yellow, including 9 nodes and 32 edges (Fig. 2F), and the genes in this region were upregulated in pulmonary fibrosis.
Screening and analysis of genes related to hub co-expressed genes in lung cancer
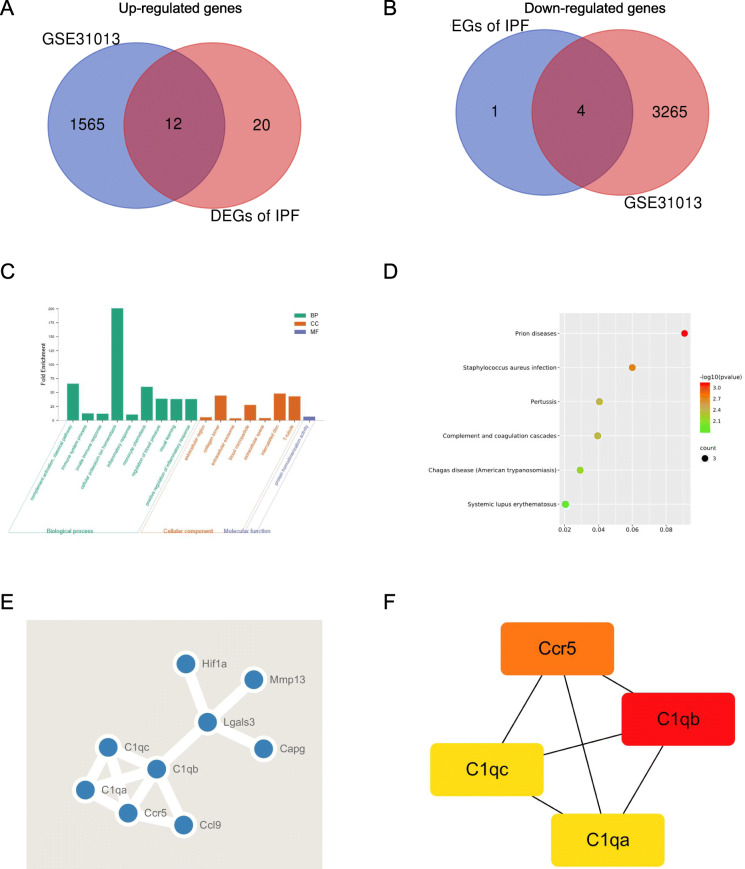
Considering IPF increased the risk of LC development especially NSCLC [22], we selected the GSE31013 dataset containing 6 samples of lung tumors and 6 samples of age-matched normal lung tissue. After the DEGs between lung tumors and control samples were identified via GEO2R online tools with |logFC| > 1 and p value < 0.05, it was surprising to find that nearly half of the DEGs in pulmonary fibrosis also exhibited had significant difference in spontaneous lung tumors formations (Fig. 3A-B, Table S2).
Fig. 3.
Analysis of commin DEGs in pulmonary fibrosis and non-small cell lung cancer. (A) Venn diagram of common DEGs in DEGs of IPF and GSE31013. (B) GO enrichment analysis of the common DEGs. (C) KEGG pathways of the common DEGs. (D) PPI network of common DEGs. (E) The most significant module related to common DEGs
The DEGs gene ontology and KEGG pathway of these genes were re-analyzed via DAVID to better understand their functions. The results showed that genes of the cellular component of this module were also mainly enriched in the extracellular region and the molecular function mainly enriched in protein homodimerization activity. The biological process analysis results suggested complement activation and cellular potassium ion homeostasis together with immune and inflammatory pathways might contribute to the occurrence and development of pulmonary fibrosis and LC (Fig. 3C-D). The top hub (C1qa, C1qb, C1qc and Ccr5) genes were also identified by the MCODE plugin, including 4 nodes and 6 edges (Fig. 3E-F).
Validation of C1q in IPF and lung cancer
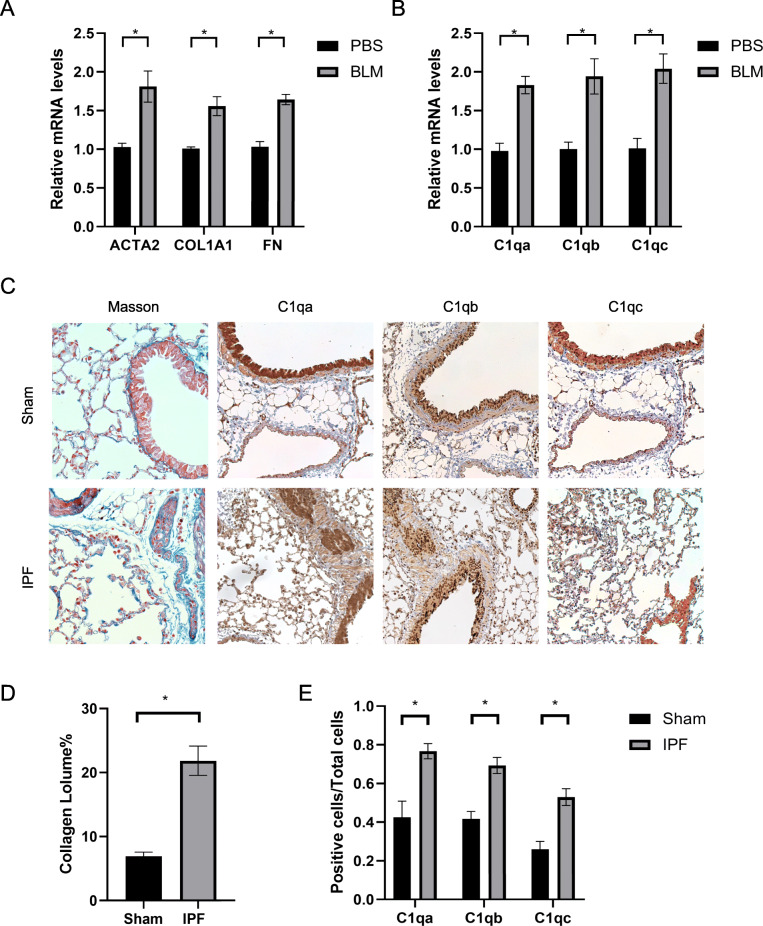
As C1q ranked high among differentially expressed genes (Table S3), we focused on the role of C1q in IPF and LC. First, pulmonary fibrosis was induced by intratracheal instillation of bleomycin, while the control group was given normal saline in the same condition. The levels of fibrosis were measured by QPCR (Fig. 4A) and Masson staining (Fig. 4C-D) in IPF group increased, indicating the accomplishment of modeling. We further detected the levels of C1q by QPCR and immunohistochemistry, and found that the expression of C1q increased in IPF (Fig. 4B-C, E), consistent with previous analysis. Besides, we found the concentrations of C1q in the blood and balf of IPF increased as well (Fig. S2, Fig. S3).
Fig. 4.
The expression of C1q (C1qa, C1qb, C1qc) in IPF. (A) Relative mRNA level analysis of fibrosis markers. (B) Relative mRNA level analysis of C1q. (C) Masson staining and immunostaining of C1q in IPF. *p < 0.05 versus Control group (n = 4 for each experimental group). All data are presented by mean ± SEM
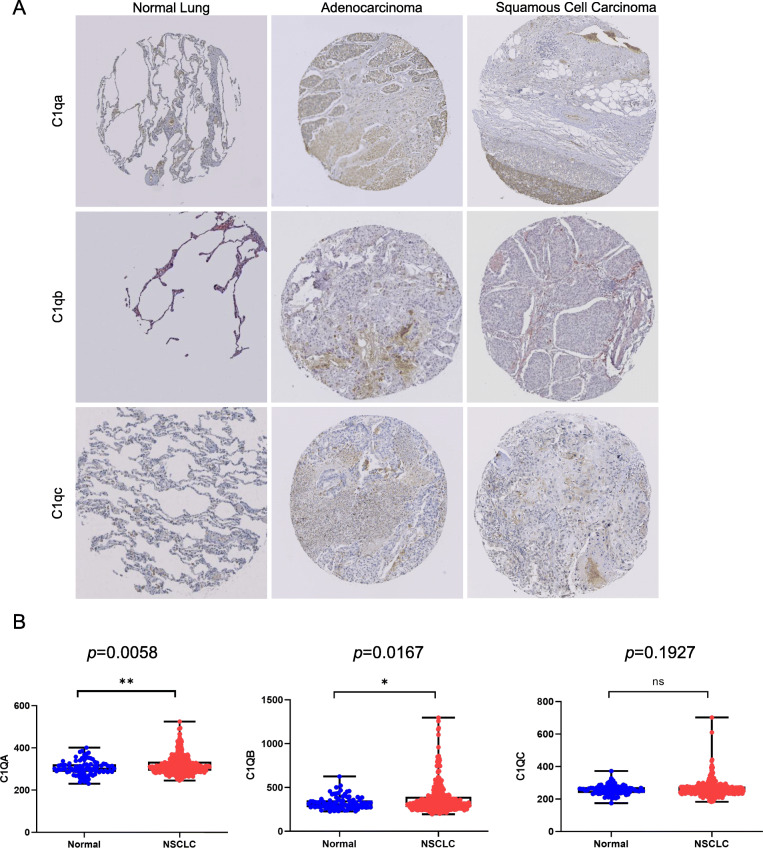
The immunohistochemical stainings from the HPA showed C1q expression of lung adenocarcinoma was higher than that of normal lung, while squamous cell carcinoma showed no significance up-regulated trend (Fig. 5A, Fig. S4). Furthermore, data in GSE135304 revealed that the levels of C1q also increased in blood of patients with malignant (MN) pulmonary (Fig. 5B). To confirm the result in blood, we tested the expression of C1q in blood monocytes by GSE76033 as C1q mainly expresses in macrophages (Fig. S5, Fig. S6).
Fig. 5.
The expression of C1q (C1qa, C1qb, C1qc) in NSCLC including LUAD and LUSC. (A) Immunohistochemistry data of C1q (C1qa, C1qb and C1qc) in lung tissue of patients of control, LUAD and LUSC from The Human Protein Atlas. (B) The expression of C1q in the blood of patients with malignant (MN) pulmonary and patients with no nodules
As methylation induced aberrant epigenetic regulation is an important common pathogenic mechanism of IPF and lung cancer [23], we explored DNA methylation levels of C1q in NSCLC (LUAD and LUSC) and IPF separately using the SurvivalMeth database and GSE63704. The results were shown in the box plot. We found most probes of hub genes decreased significantly in LUAD (n = 438) compared to normal samples (n = 32) (p < 0.05) (Fig. S7A-D). In LUSC, decreased probe of C1q also showed significant difference (Fig. S8A-D). Moreover, 4 probes of C1qa, 5 probes of C1qb, and 6 probes of C1qc were significantly decreased in IPF group (Fig. S9A-D). Common significant probes were listed in Table 3 and Table 4.
Table 3.
Summary of significant methylation probes of hub genes
| LUAD | LUSC | IPF | ||||
|---|---|---|---|---|---|---|
| Gene | Up-regulated | Down-regulated | Up-regulated | Down-regulated | Up-regulated | Down-regulated |
| C1qa | 0 | 5 | 0 | 6 | 0 | 9 |
| C1qb | 1 | 9 | 0 | 10 | 0 | 10 |
| C1qc | 1 | 8 | 2 | 8 | 0 | 9 |
| Ccr5 | 1 | 4 | 1 | 4 | 0 | 4 |
Table 4.
Characteristics of significant methylation probes of hub genes
| Gene | Probe ID | UCSC RefGene Group | SNP |
|---|---|---|---|
| C1qa | cg11505417 | 3’UTR | rs1044378 |
| cg00108454 | 5’UTR | – | |
| cg17465569 | 5’UTR | – | |
| cg08710757 | TSS1500 | rs45557836 | |
| cg10916651 | TSS200 | – | |
| C1qb | cg18763854 | 3’UTR | – |
| cg03941108 | 5’UTR | – | |
| cg22477971 | 5’UTR | – | |
| cg18366830 | Body | rs11549682 | |
| cg07012832 | TSS1500 | – | |
| cg10088685 | TSS1500 | – | |
| cg24931346 | TSS200 | – | |
| cg00215182 | TSS200 | – | |
| C1qc | cg17104151 | 5’UTR | – |
| cg00136477 | 5’UTR | – | |
| cg04161137 | Body | – | |
| cg11152959 | Body | rs45487196 | |
| cg04161137 | Body | – | |
| cg25600750 | Body | – | |
| cg11393848 | TSS1500 | rs12404537 | |
| cg12775742 | TSS200 | – | |
| Ccr5 | cg07616471 | 5’UTR | rs2856764 |
| cg00803692 | TSS200 | – | |
| cg04131610 | TSS200 | – |
The prognostic value of C1q in IPF and NSCLC
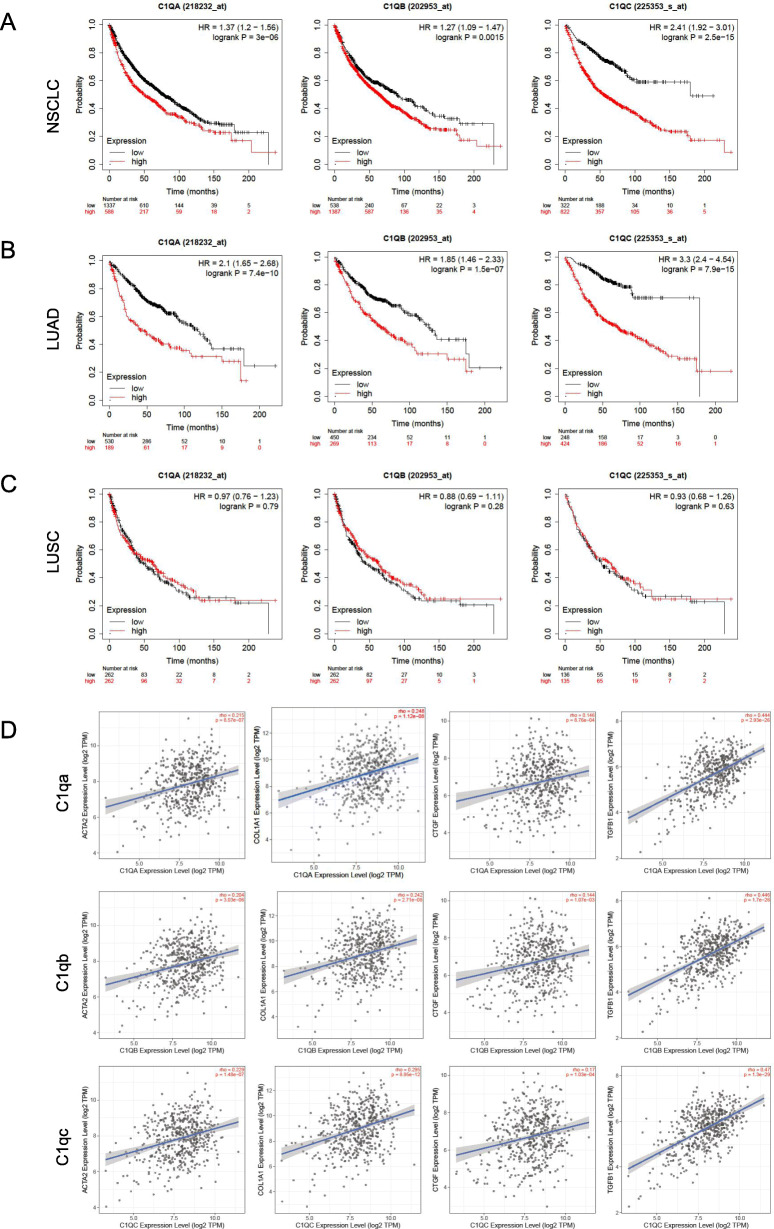
Expression of C1qa, C1qb and C1qc genes were examined and their clinical prognostic significance was investigated within the Kaplan–Meier Plotter database including 1925 cases of NSCLC. We noted that NSCLC patients with elevated C1q (C1qa, C1qb, C1qc) levels had lower OS (p < 0.05) (Fig. 6A). Patients with higher concentrations of Ccr5 showed higher survival probability while it was positively correlated with collagen fiber related indexes, which was inconsistent with previous results, so we didn’t focus on the role of Ccr5(Fig. S10, Fig. S11). Furthermore, we investigated the Kaplan–Meier curve in 672 patients with lung adenocarcinoma (LUAD) and 271 patients with lung squamous cell carcinoma (LUSC). The results demonstrated that high levels of C1q expression revealed worse prognosis of adenocarcinoma (p < 0.05) (Fig. 6B) compared with squamous cell carcinoma (p > 0.05) (Fig. 6C).
Fig. 6.
Overall survival analyses of C1q in NSCLC. (A) NSCLC patients with elevated C1q (C1qa, C1qb, C1qc) levels had lower OS (p < 0.05). (B) LUAD patients with elevated C1q levels had lower OS (p < 0.05). (C) The levels of C1q had no correlation with prognosis of LUSC. (D) C1q including C1qa, C1qb and C1qc were positive correlative with α-SMA, COL1A1, CTGF and TGF-β1 in lung adenocarcinoma at TIMER2.0 database
We detected a relationship between C1q and TGF-β1 as well as other mesenchymal markers (ACTA2, COL1A1, and CTGF) in lung adenocarcinoma using the online tool TIMER2.0. These factors involved in pulmonary fibrosis were positively related to the level of C1q expression (Fig. 6D).
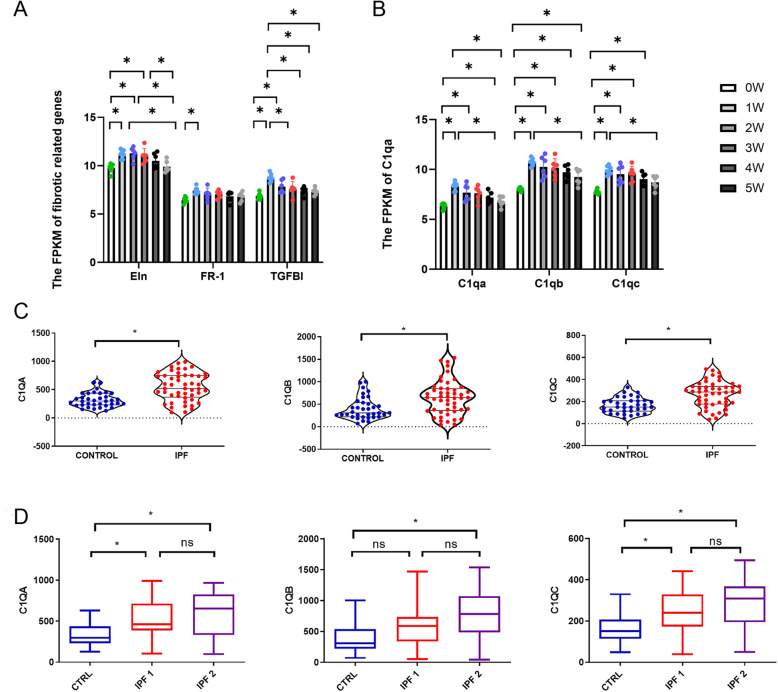
GSE37635 mentioned before was used to verify the role of C1q in IPF. It was found that elastin and collagen fiber related markers including elastin (Eln), Fgfr1 (FR1) and TGFBI peaked at first week and decreased with time, in line with the expression of C1q (Fig. 7A, B). We further investigated it in IPF human patients by GSE124685 which had 84 samples including control samples (n = 35) and IPF samples (n = 49) from 10 control patients and 6 IPF patients (Table S4). The results were consistent with previous studies. The FPKM of C1q was also significantly increased in IPF patients (Fig. 7C). And then we divided samples IPF into two groups according to alveolar surface density (ASD) which was negatively correlated with degree of fibrosis. We identified samples with ASD > 6/μm as early or progressive fibrosis and grouped them to IPF1. Other samples (ASD < 6/μm) with end-stage fibrosis were grouped to IPF2. Although the difference between the IPF1 and IPF2 was not significant, the average expression levels of C1q in IPF2 group had an uptrend compared to IPF1 group (Fig. 7D, Table S5). These results suggested that C1q might also have prognostic value for IPF progression.
Fig. 7.
Overall survival analyses of C1q in IPF. (A) The variations of the expression of collagen fiber related markers including elastin (Eln), Fgfr1 (FR1) and TGFBI with time in IPF mice. (B) The variations of the expression of C1q with time in IPF mice. (C) The violin plot in expression of C1q between control and IPF. (D) The box plot in expression of C1q among control, IPF1 and IPF2
Discussion
IPF is the most common of the idiopathic interstitial pulmonary disease. It is a fatal disease with a median survival of 4 years after diagnosis [24]. However, the underlying mechanisms and the treatment of IPF still remain unknown. Furthermore, growing epidemiologic evidence supports the association between IPF and LC. IPF can increase the risk of the development of LC by 7 to 20% [22]. Lung tumors in patients with IPF first occur adjacent to fibrotic areas. Compared with non-IPF-associated lung tumors, they have different histologic distribution and immunohistochemical features [23]. NSCLC is the predominant type in LC-IPF patients, of which adenocarcinoma (ADC) is the most common subtype [25]. Studies show that there are many mechanistic similarities between IPF and LC especially in NSCLC diseases, of which methylation is an important common mechanism [23, 26]. Myofibroblasts of IPF and cancer cells of LC are characterized by altered intercellular communication, enhanced migration and mobility, and increased invasion through the extracellular matrix. The fibroblasts in LC also exhibited mesenchymal-like features and have heterogeneous phenotypes [27].
In our study, a series of bioinformatics analyses were performed on three mRNA microarray datasets, and 37 common DEGs were identified including 32 up-regulated genes and 5 down-regulated genes. The cell component of DEGs was mainly enriched in the extracellular region and biological process were enriched in the inflammatory and immune responses, consistent with the previous study of IPF [28, 29]. It was found that among the DEGs, nine potential hub genes (C1qa, Fcgr1, C1qb, C1qc, Ccr5, Slc11a1, Emr1, Aif1 and Cxcl10) were obtained using the MCODE plug-in of Cytoscape.
To explore the deep correlation between pulmonary fibrosis and NSCLC, we screened co-expressed genes in the DEGs between pulmonary fibrosis and LC, and surprisingly found 16 out of the 37 initial DEGs were also significantly more expressed in LC. Four hub genes for both IPF and NSCLC were finally identified (C1qa, C1qb, C1qc, Ccr5). C1qa, C1qb, and C1qc are three polypeptide chains the composed C1q which is the first subcomponent of the classical pathway of the complement system [30]. Ccr5 is a chemokine receptor. It can mediate receptor activation by binding several endogenous chemokines [31].
We explored the methylation levels of hub genes, and found most of probes in IPF and NSCLC decreased which could give a good interpretation to the up regulated expression of C1q. In order to evaluate the diagnostic and prognostic value of the hub genes, we also determined the correlations between key genes and clinical data. The results demonstrated that the OS rate was significantly lower among NSCLC patients with high levels of C1q, while patients with high concentrations of Ccr5 showed higher survival probability. We further investigated the role C1q played in the survival rate of lung adenocarcinoma and squamous cell carcinoma respectively. There was a significant difference in lung adenocarcinoma but no significant difference in lung squamous cell carcinoma, consistent with the immunohistochemical images analyzed from the PHA. Kaplan–Meier curves and immunohistochemical and stained pictures also indicated that C1q was closely related to the prognosis of NSCLC particularly lung adenocarcinoma. TGF-β is the most important mediator of the pathogenesis and carcinogenesis of IPF [32]. C1q showed a positive correlation with LUAD as well as other fibrosis markers like a-SMA, CTGF and collagen-1. These findings suggested that representative molecules contributing to IPF might also be regulated by C1q in lung adenocarcinoma. We further investigated the expression of C1q in IPF human patients, and found C1q might also have prognostic value for IPF progression. Besides, we comfirmed the role of C1q in the blood and balf of IPF.
C1q is synthesized primarily by macrophages and dendritic cells [33]. The complement component C1q plays an important role in host immune responses. Previous studies mainly focused on its role in systemic lupus erythematosus. Deficiency of C1q leads to high susceptibility to systemic lupus erythematosus-like symptoms, and complications with skin and renal diseases [34]. However, the role and mechanism of C1q in IPF and LC still remain unclear.
Correlation studies on the contribution to IPF are lacking. Sakkas et al. found that high levels of anti-C1q autoantibodies could predict the occurrence of pulmonary fibrosis patients with SSc [35]. Besides, C1q was positive in 73% of IPF cases by investigating bronchoalveolar lavage fluids, higher than in hypersensitivity pneumonitis (40%) or in sarcoidosis (31%) [36]. These result from another standpoint indicate that C1q participates in the occurrence and development of pulmonary fibrosis.
Studies accumulated over the past few years suggest that C1q can promote tumor formation by facilitating the proliferation and migration of cancer cells along with angiogenesis and metastasis [37, 38]. Bulla et al. reported that C1q promoted the growth of malignant pleural mesothelioma. C1q could induce adhesion and proliferation of mesothelioma cells and bind to hyaluronic acid in the microenvironment of MPM [39]. Tedesco et al. discovered that C1q-deficient (C1qa−/−) mice exhibited inhibited melanoma tumor growth which led to a better prognosis. C1q is mainly expressed in mesenchymal elements and promotes tumor growth cooperating with fibronectin [10]. Besides, C1q was also found to stimulate the progression of hepatocellular tumor. It enhanced migratory and invasive phenotypes of liver cancer cells [40]. Furthermore, C1q had a negative prognostic effect on kidney carcinomas [41].
C1q has pro-fibrotic and immunosuppressive properties, which are two contributing factors to both of IPF and NSCLC. C1q binding in human lung fibroblasts was reported to be heterogeneous [42], and able to interact with fibronectin to mediate adhesion of fibroblasts to immune complexes, resulting in stimulation in collagen synthesis [43, 44]. Low circulating levels of C1q prevented proliferation of fibroblast cells via down regulation of Wnt/β-catenin signaling [45, 46]. In addition, C1q plays a part in immune dysregulation. C1q inhibited proliferation of the Th1 and Th17 T cell subset mediated by human macrophage and dendritic cells [47]. It also suppressed the activation, proliferation, and cytotoxic functions of CD8+ T cells in vitro [48]. Meanwhile, tumor cells could directly assemble with C1q, enabling the formation of complement activation and further promoting tumor growth [49]. Other complement components like C4d and C5a showed similar phenomena in LC [50, 51]. Considering complement components have been demonstrated to promote tumor growth, the inhibition of complement activation and binding have been proposed as effective treatment for cancer [52, 53].
Our study revealed that the concentrations of C1q were increased and took part in many physiological and pathological processes in IPF and lung cancer. The results of bioinformatics analysis highlighted that high concentrations of C1q have important clinical prognostic value in NSCLC, especially in adenocarcinoma. The levels of C1q in severe pulmonary fibrosis also have an uptrend. It’s the first time that we find methylation status of C1q decreases and the expression of C1q increases in IPF and NSCLC, which is positively correlated with pulmonary fibrosis. This may also be a connection between the two diseases. In conclusion, our findings can provide the basis of new targets for diagnosis, therapy, and prognosis of IPF and LC.
Supplementary Information
Additional file 1: Supplementary Fig. 1. Venn diagram of GSE37635. Common DEGs at 6 timepoints profiling GSE37635. Supplementary Fig. 2. The heatmap of GSE102751. It indicates the significant differences between the blood of control and IPF patients. The high expression and low expression are represented in red and green, respectively. Supplementary Fig. 3. The heatmap of GSE98468. It indicates the significant differences between the balf of control and IPF mice. The high expression and low expression are represented in red and green, respectively. Supplementary Fig. 4. Immunohistochemistry data of lung tissue. Immunohistochemistry data of C1q (C1qa, C1qb and C1qc) in normal lung, LUAD and LUSC from The Human Protein Atlas. Supplementary Fig. 5. The expression of C1q. C1q (C1qa, C1qb and C1qc) mainly express in macrophages in all cell types and lung tissue from The Human Protein Atlas. Supplementary Fig. 6. The expression of C1q in blood monocytes. The FPKM of C1q in blood monocytes significantly increased in 3-week tumor mice by GSE76033. Supplementary Fig. 7. DNA methylation levels of probes of hub genes in LUAD. (A) 5 methylation probes of C1qa showed significant difference in normal group and LUAD group; (B) 10 methylation probes of C1qb showed significant difference in normal group and LUAD group; (C) 9 methylation probes of C1qc showed significant difference in normal group and LUAD group;(D) 5 methylation probes of Ccr5 showed significant difference in normal group and LUAD group. Supplementary Fig. 8. Methylation levels of hub genes in LUSC. (A) 6 methylation probes of C1qa showed significant difference in normal group and LUSC group; (B) 10 methylation probes of C1qb showed significant difference in normal group and LUAD group; (C) 10 methylation probes of C1qc showed significant difference in normal group and LUSC group;(D) 4 methylation probes of Ccr5 showed significant difference in normal group and LUSC group. Supplementary Fig. 9. Methylation levels of hub genes in IPF. (A) 9 methylation probes of C1qa showed significant difference in normal group and IPF group; (B) 10 methylation probes of C1qb showed significant difference in normal group and IPF group; (C) 9 methylation probes of C1qc showed significant difference in normal group and IPF group;(D) 4 methylation probes of Ccr5 showed significant difference in normal group and IPF group. Supplementary Fig. 10. Overall survival analysis of Ccr5 in NSCLC patients. NSCLC patients with elevated Ccr5 levels had higher OS (p < 0.05). Supplementary Fig. 11. Relationship between Ccr5 and fibrosis in NSCLC. The expression of Ccr5 was positive correlative with α-SMA, COL1A, CTGF and TGF-β1 in NSCLC including LUAD and LUSC at TIMER2.0 database. Supplemental Table 1. Characteristics of the genes included in datasets. Supplemental Table 2. The logFC of DEGs. Up-regulated genes were labeled in red, down-regulated genes were labeled in black. Supplemental Table 3 Characteristics of the genes included in datasets. Supplemental Table 4 Characteristics of human subjects. Supplemental Table 5 The means and standard deviations of FPKM of C1q.
Acknowledgements
Not applicable.
Abbreviations
- IPF
Idiopathic pulmonary fibrosis
- LC
Lung cancer
- NSCLC
Non-small cell lung cancer
- GEO
Gene expression omnibus
- DEGs
Differentially expressed genes
- GO
Gene ontology
- KEGG
Kyoto encyclopedia of genes and genomes
- DAVID
Database for annotation, visualization and integrated discovery
- PPI
Protein–protein interaction
- TIMER2.0
Tumor immune estimation resource2.0
- HPA
The human protein atlas
- BP
Biological processes
- MF
Molecular functions
- CC
Cellular components
- MCODE
The molecular complex detection
- QPCR
Quantitative real-time PCR
- LUAD
Lung adenocarcinoma
- LUSC
Lung squamous cell carcinoma
- OS
Overall survival
- FPKM
Fragments per Kilobasekof exon model per million mapped fragments
- ASD
Alveolar surface density
- ADC
Adenocarcinoma
- MPM
Malignant pleural mesothelioma
Authors’ contributions
WX K and B L carried out all the bioinformatics evaluations. WX K and YF S wrote the manuscript. YF Z, JH Z and Q Y revised the manuscript. YW X and WH P conceived the study, participated in its design and coordination. All authors read and approved the final manuscript.
Funding
This work was supported by grants of The National Key Research and Development Progrm of China (No. 2016YFC1301202).
Availability of data and materials
All the datasets are available in a public, open access repository.
Databases Links
GSE37635 https://www.ncbi.nlm.nih.gov/geo/query/acc.cgi?acc=GSE37635
GSE97546 https://www.ncbi.nlm.nih.gov/geo/query/acc.cgi?acc=GSE97546
GSE485 https://www.ncbi.nlm.nih.gov/geo/query/acc.cgi?acc=GSE485
GSE31013 https://www.ncbi.nlm.nih.gov/geo/query/acc.cgi?acc=GSE31013
GSE98468 https://www.ncbi.nlm.nih.gov/geo/query/acc.cgi?acc=GSE98468
GSE102751 https://www.ncbi.nlm.nih.gov/geo/query/acc.cgi?acc=GSE102751
GSE124685 https://www.ncbi.nlm.nih.gov/geo/query/acc.cgi?acc=GSE124685
GSE63704 https://www.ncbi.nlm.nih.gov/geo/query/acc.cgi?acc=GSE63704
Kaplan–Meier Plotter http://kmplot.com/analysis/index.php?p=service
The HPA https://www.proteinatlas.org/
TIMER2.0 http://timer.cistrome.org/
SurvivalMeth http://bio-bigdata.hrbmu.edu.cn/survivalmeth/
Declarations
Ethics approval and consent to participate
The study received approval by the Animal Care and Use Committees of Shanghai Tenth People’s Hospital for animal welfare (SHDSYY-2019-2149). The study was carried out in compliance with the ARRIVE guidelines. A complete ARRIVE Guidelines checklist is included in Additional file.
Consent for publication
Not applicable.
Competing interests
There is no conflict of interest.
Footnotes
Publisher’s Note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Wenxin Kou and Bo Li contributed equally to this work.
Contributor Information
Yawei Xu, Email: xuyawei@tongji.edu.cn.
Wenhui Peng, Email: pwenhui@tongji.edu.cn.
References
- 1.Richeldi L, Collard HR, Jones MG. Idiopathic pulmonary fibrosis. Lancet (London, England) 2017;389(10082):1941–1952. doi: 10.1016/S0140-6736(17)30866-8. [DOI] [PubMed] [Google Scholar]
- 2.Lederer DJ, Martinez FJ. Idiopathic pulmonary fibrosis. N Engl J Med. 2018;378(19):1811–1823. doi: 10.1056/NEJMra1705751. [DOI] [PubMed] [Google Scholar]
- 3.Raghu G, Amatto VC, Behr J, Stowasser S. Comorbidities in idiopathic pulmonary fibrosis patients: a systematic literature review. Eur Respir J. 2015;46(4):1113–1130. doi: 10.1183/13993003.02316-2014. [DOI] [PubMed] [Google Scholar]
- 4.Saito A, Horie M, Micke P, Nagase T. The Role of TGF-β Signaling in Lung Cancer Associated with Idiopathic Pulmonary Fibrosis. Int J Mol Sci. 2018;19(11):3611. doi: 10.3390/ijms19113611. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Reck M, Kaiser R, Mellemgaard A, Douillard J-Y, Orlov S, Krzakowski M, von Pawel J, Gottfried M, Bondarenko I, Liao M, Gann CN, Barrueco J, Gaschler-Markefski B, Novello S, LUME-Lung 1 Study Group Docetaxel plus nintedanib versus docetaxel plus placebo in patients with previously treated non-small-cell lung cancer (LUME-lung 1): a phase 3, double-blind, randomised controlled trial. Lancet Oncol. 2014;15(2):143–155. doi: 10.1016/S1470-2045(13)70586-2. [DOI] [PubMed] [Google Scholar]
- 6.Muller-Eberhard HJ, Kunkel HG. Isolation of a thermolabile serum protein which precipitates gamma-globulin aggregates and participates in immune hemolysis. Proc Soc Exp Biol Med. 1961;106(2):291–295. doi: 10.3181/00379727-106-26313. [DOI] [PubMed] [Google Scholar]
- 7.Kishore U, Reid KB. C1q: structure, function, and receptors. Immunopharmacology. 2000;49(1–2):159–170. doi: 10.1016/S0162-3109(00)80301-X. [DOI] [PubMed] [Google Scholar]
- 8.Okamoto T, Mathai SK, Hennessy CE, Hancock LA, Walts AD, Stefanski AL, et al. The relationship between complement C3 expression and the MUC5B genotype in pulmonary fibrosis. Am J Physiol Lung Cell Mol Physiol. 2018;315(1):L1–L10. doi: 10.1152/ajplung.00395.2017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Gu H, Fisher AJ, Mickler EA, Duerson F, Cummings OW, Peters-Golden M, et al. Contribution of the anaphylatoxin receptors, C3aR and C5aR, to the pathogenesis of pulmonary fibrosis. FASEB J. 2016;30(6):2336–2350. doi: 10.1096/fj.201500044. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Bulla R, Tripodo C, Rami D, Ling GS, Agostinis C, Guarnotta C, Zorzet S, Durigutto P, Botto M, Tedesco F. C1q acts in the tumour microenvironment as a cancer-promoting factor independently of complement activation. Nat Commun. 2016;7(1):10346. doi: 10.1038/ncomms10346. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Blaauboer ME, Emson CL, Verschuren L, van Erk M, Turner SM, Everts V, Hanemaaijer R, Stoop R. Novel combination of collagen dynamics analysis and transcriptional profiling reveals fibrosis-relevant genes and pathways. Matrix Biol. 2013;32(7–8):424–431. doi: 10.1016/j.matbio.2013.04.005. [DOI] [PubMed] [Google Scholar]
- 12.Pandiri AR, Sills RC, Ziglioli V, Ton T-VT, Hong H-HL, Lahousse SA, et al. Differential transcriptomic analysis of spontaneous lung tumors in B6C3F1 mice: comparison to human non-small cell lung cancer. Toxicol Pathol. 2012;40(8):1141–1159. doi: 10.1177/0192623312447543. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Xie N, Cui H, Ge J, Banerjee S, Guo S, Dubey S, Abraham E, Liu RM, Liu G. Metabolic characterization and RNA profiling reveal glycolytic dependence of profibrotic phenotype of alveolar macrophages in lung fibrosis. Am J Physiol Lung Cell Mol Physiol. 2017;313(5):L834–LL44. doi: 10.1152/ajplung.00235.2017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.McDonough JE, Ahangari F, Li Q, Jain S, Verleden SE, Herazo-Maya J, et al. Transcriptional regulatory model of fibrosis progression in the human lung. JCI Insight. 2019;4(22). [DOI] [PMC free article] [PubMed]
- 15.Davis S, Meltzer PS. GEOquery: a bridge between the gene expression omnibus (GEO) and BioConductor. Bioinformatics. 2007;23(14):1846–1847. doi: 10.1093/bioinformatics/btm254. [DOI] [PubMed] [Google Scholar]
- 16.Huang DW, Sherman BT, Lempicki RA. Systematic and integrative analysis of large gene lists using DAVID bioinformatics resources. Nat Protoc. 2009;4(1):44–57. doi: 10.1038/nprot.2008.211. [DOI] [PubMed] [Google Scholar]
- 17.Shannon P, Markiel A, Ozier O, Baliga NS, Wang JT, Ramage D, Amin N, Schwikowski B, Ideker T. Cytoscape: a software environment for integrated models of biomolecular interaction networks. Genome Res. 2003;13(11):2498–2504. doi: 10.1101/gr.1239303. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Győrffy B, Surowiak P, Budczies J, Lánczky A. Online survival analysis software to assess the prognostic value of biomarkers using transcriptomic data in non-small-cell lung cancer. PLoS ONE. 2013;8(12):e82241. doi: 10.1371/journal.pone.0082241. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Uhlén M, Fagerberg L, Hallström BM, Lindskog C, Oksvold P, Mardinoglu A, et al. Proteomics. Tissue-based map of the human proteome. Science (New York, NY) 2015;347(6220):1260419. doi: 10.1126/science.1260419. [DOI] [PubMed] [Google Scholar]
- 20.Li T, Fu J, Zeng Z, Cohen D, Li J, Chen Q, Li B, Liu XS. TIMER2.0 for analysis of tumor-infiltrating immune cells. Nucleic Acids Res. 2020;48(W1):W509–WW14. doi: 10.1093/nar/gkaa407. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Zhang C, Zhao N, Zhang X, Xiao J, Li J, Lv D, et al. SurvivalMeth: a web server to investigate the effect of DNA methylation-related functional elements on prognosis. Brief Bioinform. 2020;22(3). 10.1093/bib/bbaa162. [DOI] [PubMed]
- 22.Ballester B, Milara J, Cortijo J. Idiopathic Pulmonary Fibrosis and Lung Cancer: Mechanisms and Molecular Targets. Int J Mol Sci. 2019;20(3). [DOI] [PMC free article] [PubMed]
- 23.Tzouvelekis A, Gomatou G, Bouros E, Trigidou R, Tzilas V, Bouros D. Common pathogenic mechanisms between idiopathic pulmonary fibrosis and lung Cancer. Chest. 2019;156(2):383–391. doi: 10.1016/j.chest.2019.04.114. [DOI] [PubMed] [Google Scholar]
- 24.Raghu G, Collard HR, Egan JJ, Martinez FJ, Behr J, Brown KK, Colby TV, Cordier JF, Flaherty KR, Lasky JA, Lynch DA, Ryu JH, Swigris JJ, Wells AU, Ancochea J, Bouros D, Carvalho C, Costabel U, Ebina M, Hansell DM, Johkoh T, Kim DS, King te Jr, Kondoh Y, Myers J, Müller NL, Nicholson AG, Richeldi L, Selman M, Dudden RF, Griss BS, Protzko SL, Schünemann HJ, ATS/ERS/JRS/ALAT Committee on Idiopathic Pulmonary Fibrosis An official ATS/ERS/JRS/ALAT statement: idiopathic pulmonary fibrosis: evidence-based guidelines for diagnosis and management. Am J Respir Crit Care Med. 2011;183(6):788–824. doi: 10.1164/rccm.2009-040GL. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Dela Cruz CS, Tanoue LT, Matthay RA. Lung cancer: epidemiology, etiology, and prevention. Clin Chest Med. 2011;32(4):605–644. doi: 10.1016/j.ccm.2011.09.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Hata A, Nakajima T, Matsusaka K, Fukuyo M, Morimoto J, Yamamoto T, Sakairi Y, Rahmutulla B, Ota S, Wada H, Suzuki H, Matsubara H, Yoshino I, Kaneda A. A low DNA methylation epigenotype in lung squamous cell carcinoma and its association with idiopathic pulmonary fibrosis and poorer prognosis. Int J Cancer. 2020;146(2):388–399. doi: 10.1002/ijc.32532. [DOI] [PubMed] [Google Scholar]
- 27.Horowitz JC, Osterholzer JJ, Marazioti A, Stathopoulos GT. "scar-cinoma": viewing the fibrotic lung mesenchymal cell in the context of cancer biology. Eur Respir J. 2016;47(6):1842–1854. doi: 10.1183/13993003.01201-2015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Kolahian S, Fernandez IE, Eickelberg O, Hartl D. Immune mechanisms in pulmonary fibrosis. Am J Respir Cell Mol Biol. 2016;55(3):309–322. doi: 10.1165/rcmb.2016-0121TR. [DOI] [PubMed] [Google Scholar]
- 29.Mercer PF, Chambers RC. Innate immune signaling and stem cell renewal in idiopathic pulmonary fibrosis. Nat Med. 2016;22(11):1210–1212. doi: 10.1038/nm.4230. [DOI] [PubMed] [Google Scholar]
- 30.Mangogna A, Agostinis C, Bonazza D, Belmonte B, Bulla R. Is the Complement Protein C1q a Pro- or Anti-tumorigenic Factor? Bioinformatics Analysis Involving Human Carcinomas. Front Immunol. 2019;10:865. doi: 10.3389/fimmu.2019.00865. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Blanpain C, Migeotte I, Lee B, Vakili J, Doranz BJ, Govaerts C, et al. CCR5 binds multiple CC-chemokines: MCP-3 acts as a natural antagonist. Blood. 1999;94(6):1899–1905. doi: 10.1182/blood.V94.6.1899. [DOI] [PubMed] [Google Scholar]
- 32.Ley B, Brown KK, Collard HR. Molecular biomarkers in idiopathic pulmonary fibrosis. Am J Physiol Lung Cell Mol Physiol. 2014;307(9):L681–LL91. doi: 10.1152/ajplung.00014.2014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Castellano G, Trouw LA, Fiore N, Daha MR, Schena FP, van Kooten C. Infiltrating dendritic cells contribute to local synthesis of C1q in murine and human lupus nephritis. Mol Immunol. 2010;47(11–12):2129–2137. doi: 10.1016/j.molimm.2010.02.006. [DOI] [PubMed] [Google Scholar]
- 34.Petry F, Botto M, Holtappels R, Walport MJ, Loos M. Reconstitution of the complement function in C1q-deficient (C1qa−/−) mice with wild-type bone marrow cells. Immunopharmacology. 2001;49(7):4033. doi: 10.4049/jimmunol.167.7.4033. [DOI] [PubMed] [Google Scholar]
- 35.Liaskos C, Rentouli S, Simopoulou T, Gkoutzourelas A, Norman GL, Brotis A, Alexiou I, Katsiari C, Bogdanos DP, Sakkas LI. Anti-C1q autoantibodies are frequently detected in patients with systemic sclerosis associated with pulmonary fibrosis. Br J Dermatol. 2019;181(1):138–146. doi: 10.1111/bjd.17886. [DOI] [PubMed] [Google Scholar]
- 36.Dall'Aglio PP, Pesci A, Bertorelli G, Brianti E, Scarpa S. Study of immune complexes in bronchoalveolar lavage fluids. Respiration. 1988;54(Suppl 1):36–41. doi: 10.1159/000195495. [DOI] [PubMed] [Google Scholar]
- 37.Markiewski MM, Lambris JD. Unwelcome complement. Cancer Res. 2009;69(16):6367–6370. doi: 10.1158/0008-5472.CAN-09-1918. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Loveland BE, Cebon J. Cancer exploiting complement: a clue or an exception? Nat Immunol. 2008;9(11):1205–1206. doi: 10.1038/ni1108-1205. [DOI] [PubMed] [Google Scholar]
- 39.Agostinis C, Vidergar R, Belmonte B, Mangogna A, Amadio L, Geri P, Borelli V, Zanconati F, Tedesco F, Confalonieri M, Tripodo C, Kishore U, Bulla R. Complement protein C1q binds to hyaluronic acid in the malignant pleural mesothelioma microenvironment and promotes tumor growth. Front Immunol. 2017;8:1559. doi: 10.3389/fimmu.2017.01559. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Lee J-H, Poudel B, Ki H-H, Nepali S, Lee Y-M, Shin J-S, Kim DK. Complement C1q stimulates the progression of hepatocellular tumor through the activation of discoidin domain receptor 1. Sci Rep. 2018;8(1):4908. doi: 10.1038/s41598-018-23240-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Mangogna A, Agostinis C, Bonazza D, Belmonte B, Zacchi P, Zito G, Romano A, Zanconati F, Ricci G, Kishore U, Bulla R. Is the complement protein C1q a pro- or anti-tumorigenic factor? Bioinforma Anal Involving Human Carcinomas Front Immunol. 2019;10:865. doi: 10.3389/fimmu.2019.00865. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Narayanan AS, Lurton J, Raghu G. Distribution of receptors of collagen and globular domains of C1q in human lung fibroblasts. Am J Respir Cell Mol Biol. 1997;17(1):84–90. doi: 10.1165/ajrcmb.17.1.2732. [DOI] [PubMed] [Google Scholar]
- 43.Rennard SI, Chen YF, Robbins RA, Gadek JE, Crystal RG. Fibronectin mediates cell attachment to C1q: a mechanism for the localization of fibrosis in inflammatory disease. Clin Exp Immunol. 1983;54(1):239–247. [PMC free article] [PubMed] [Google Scholar]
- 44.Fleming KA, McGee JO. The relationship between collagen and C1q biosynthesis in cultured human fibroblasts. Biochem J. 1982;206(2):287–293. doi: 10.1042/bj2060287. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Horii N, Uchida M, Hasegawa N, Fujie S, Oyanagi E, Yano H, Hashimoto T, Iemitsu M. Resistance training prevents muscle fibrosis and atrophy via down-regulation of C1q-induced Wnt signaling in senescent mice. FASEB J. 2018;32(7):3547–3559. doi: 10.1096/fj.201700772RRR. [DOI] [PubMed] [Google Scholar]
- 46.Mangolini M, Götte F, Moore A, Ammon T, Oelsner M, Lutzny-Geier G, Klein-Hitpass L, Williamson JC, Lehner PJ, Dürig J, Möllmann M, Rásó-Barnett L, Hughes K, Santoro A, Méndez-Ferrer S, Oostendorp RAJ, Zimber-Strobl U, Peschel C, Hodson DJ, Schmidt-Supprian M, Ringshausen I. Notch2 controls non-autonomous Wnt-signalling in chronic lymphocytic leukaemia. Nat Commun. 2018;9(1):3839. doi: 10.1038/s41467-018-06069-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Clarke EV, Weist BM, Walsh CM, Tenner AJ. Complement protein C1q bound to apoptotic cells suppresses human macrophage and dendritic cell-mediated Th17 and Th1 T cell subset proliferation. J Leukoc Biol. 2015;97(1):147–160. doi: 10.1189/jlb.3A0614-278R. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Ling GS, Crawford G, Buang N, Bartok I, Tian K, Thielens NM, et al. C1q restrains autoimmunity and viral infection by regulating CD8 T cell metabolism. Science (New York, NY) 2018;360(6388):558–563. doi: 10.1126/science.aao4555. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Roumenina LT, Daugan MV, Noé R, Petitprez F, Vano YA, Sanchez-Salas R, Becht E, Meilleroux J, Clec'h BL, Giraldo NA, Merle NS, Sun CM, Verkarre V, Validire P, Selves J, Lacroix L, Delfour O, Vandenberghe I, Thuilliez C, Keddani S, Sakhi IB, Barret E, Ferré P, Corvaïa N, Passioukov A, Chetaille E, Botto M, de Reynies A, Oudard SM, Mejean A, Cathelineau X, Sautès-Fridman C, Fridman WH. Tumor cells hijack macrophage-produced complement C1q to promote tumor growth. Cancer Immunol Res. 2019;7(7):1091–1105. doi: 10.1158/2326-6066.CIR-18-0891. [DOI] [PubMed] [Google Scholar]
- 50.Corrales L, Ajona D, Rafail S, Lasarte JJ, Riezu-Boj JI, Lambris JD, Rouzaut A, Pajares MJ, Montuenga LM, Pio R. Anaphylatoxin C5a creates a favorable microenvironment for lung cancer progression. J Immunol. 2012;189(9):4674–4683. doi: 10.4049/jimmunol.1201654. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Ajona D, Pajares MJ, Corrales L, Perez-Gracia JL, Agorreta J, Lozano MD, Torre W, Massion PP, de-Torres JP, Jantus-Lewintre E, Camps C, Zulueta JJ, Montuenga LM, Pio R. Investigation of complement activation product c4d as a diagnostic and prognostic biomarker for lung cancer. J Natl Cancer Inst. 2013;105(18):1385–1393. doi: 10.1093/jnci/djt205. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Markiewski MM, DeAngelis RA, Benencia F, Ricklin-Lichtsteiner SK, Koutoulaki A, Gerard C, et al. Modulation of the antitumor immune response by complement. Nat Immunol. 2008;9(11):1225–1235. doi: 10.1038/ni.1655. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Pio R, Ajona D, Lambris JD. Complement inhibition: a promising concept for cancer treatment. Semin Immunol. 2013;25(1):54. doi: 10.1016/j.smim.2013.04.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Additional file 1: Supplementary Fig. 1. Venn diagram of GSE37635. Common DEGs at 6 timepoints profiling GSE37635. Supplementary Fig. 2. The heatmap of GSE102751. It indicates the significant differences between the blood of control and IPF patients. The high expression and low expression are represented in red and green, respectively. Supplementary Fig. 3. The heatmap of GSE98468. It indicates the significant differences between the balf of control and IPF mice. The high expression and low expression are represented in red and green, respectively. Supplementary Fig. 4. Immunohistochemistry data of lung tissue. Immunohistochemistry data of C1q (C1qa, C1qb and C1qc) in normal lung, LUAD and LUSC from The Human Protein Atlas. Supplementary Fig. 5. The expression of C1q. C1q (C1qa, C1qb and C1qc) mainly express in macrophages in all cell types and lung tissue from The Human Protein Atlas. Supplementary Fig. 6. The expression of C1q in blood monocytes. The FPKM of C1q in blood monocytes significantly increased in 3-week tumor mice by GSE76033. Supplementary Fig. 7. DNA methylation levels of probes of hub genes in LUAD. (A) 5 methylation probes of C1qa showed significant difference in normal group and LUAD group; (B) 10 methylation probes of C1qb showed significant difference in normal group and LUAD group; (C) 9 methylation probes of C1qc showed significant difference in normal group and LUAD group;(D) 5 methylation probes of Ccr5 showed significant difference in normal group and LUAD group. Supplementary Fig. 8. Methylation levels of hub genes in LUSC. (A) 6 methylation probes of C1qa showed significant difference in normal group and LUSC group; (B) 10 methylation probes of C1qb showed significant difference in normal group and LUAD group; (C) 10 methylation probes of C1qc showed significant difference in normal group and LUSC group;(D) 4 methylation probes of Ccr5 showed significant difference in normal group and LUSC group. Supplementary Fig. 9. Methylation levels of hub genes in IPF. (A) 9 methylation probes of C1qa showed significant difference in normal group and IPF group; (B) 10 methylation probes of C1qb showed significant difference in normal group and IPF group; (C) 9 methylation probes of C1qc showed significant difference in normal group and IPF group;(D) 4 methylation probes of Ccr5 showed significant difference in normal group and IPF group. Supplementary Fig. 10. Overall survival analysis of Ccr5 in NSCLC patients. NSCLC patients with elevated Ccr5 levels had higher OS (p < 0.05). Supplementary Fig. 11. Relationship between Ccr5 and fibrosis in NSCLC. The expression of Ccr5 was positive correlative with α-SMA, COL1A, CTGF and TGF-β1 in NSCLC including LUAD and LUSC at TIMER2.0 database. Supplemental Table 1. Characteristics of the genes included in datasets. Supplemental Table 2. The logFC of DEGs. Up-regulated genes were labeled in red, down-regulated genes were labeled in black. Supplemental Table 3 Characteristics of the genes included in datasets. Supplemental Table 4 Characteristics of human subjects. Supplemental Table 5 The means and standard deviations of FPKM of C1q.
Data Availability Statement
All the datasets are available in a public, open access repository.
Databases Links
GSE37635 https://www.ncbi.nlm.nih.gov/geo/query/acc.cgi?acc=GSE37635
GSE97546 https://www.ncbi.nlm.nih.gov/geo/query/acc.cgi?acc=GSE97546
GSE485 https://www.ncbi.nlm.nih.gov/geo/query/acc.cgi?acc=GSE485
GSE31013 https://www.ncbi.nlm.nih.gov/geo/query/acc.cgi?acc=GSE31013
GSE98468 https://www.ncbi.nlm.nih.gov/geo/query/acc.cgi?acc=GSE98468
GSE102751 https://www.ncbi.nlm.nih.gov/geo/query/acc.cgi?acc=GSE102751
GSE124685 https://www.ncbi.nlm.nih.gov/geo/query/acc.cgi?acc=GSE124685
GSE63704 https://www.ncbi.nlm.nih.gov/geo/query/acc.cgi?acc=GSE63704
Kaplan–Meier Plotter http://kmplot.com/analysis/index.php?p=service
The HPA https://www.proteinatlas.org/
TIMER2.0 http://timer.cistrome.org/
SurvivalMeth http://bio-bigdata.hrbmu.edu.cn/survivalmeth/