Conspectus
Infectious diseases by pathogenic microorganisms are one of the leading causes of mortality worldwide. Healthcare and socio-economic development have been seriously affected for different civilizations because of bacterial and viral infections. According to the Centers for Disease Control and Prevention (CDC), pandemic in 1918 by the Influenza A virus of the H1N1 subtype was responsible for 50 to 100 million deaths worldwide. Similarly, the Asian flu pandemic in 1957, Hong Kong flu in 1968, and H1N1pdm09 flu pandemic in 2009 were responsible for more than 1 million deaths across the globe each time. As per the World Health Organization (WHO), the current pandemic by coronavirus disease 2019 (COVID-19) due to the severe acute respiratory syndrome coronavirus 2 (SARS-CoV-2) virus is responsible for more than 4.8M death worldwide till now. Since the gold standard polymerase chain reaction (PCR) test is more time-consuming, the health care system cannot test all symptomatic and asymptomatic covid patients every day, which is extremely important to tackle the outbreak. One of the significant challenges during the current pandemic is developing mass testing tools, which is critical to control the virus spread in the community. Therefore, it is highly desirable to develop advanced material-based approaches that can provide a rapid and accurate diagnosis of COVID-19, which will have the capability to save millions of human lives.
Aiming for the targeted diagnosis of deadly virus, researchers have developed nanomaterials with various sizes, shapes, and dimensions. These nanomaterials have been used to identify biomolecules via unique optical, electrical, magnetic, structural, and functional properties, which are lacking in other materials. Despite significant progress, nanomaterial-based diagnosis of biomolecules is still facing several obstacles due to low targeting efficiency and nonspecific interactions. To overcome these problems, the bio-conjugated nanoparticle has been designed via surface coating with polyethylene glycol (PEG) and then conjugated with antibodies, DNA, RNA, or peptide aptamers. Therefore, the current account summarized an overview of the recent advances in the design of bio-conjugated nanomaterial-based approached as effective diagnosis of SARS-CoV-2 virus, SARS-CoV-2 viral RNA, antigen, or antibody, with a particular focus on our work and other’s work related to this subject. First, we present how to tailor the surface functionalities of nanomaterials to achieve bio-conjugated material for targeted diagnosis of the virus. Then we review the very recent advances in the design of antibody/aptamer/peptide conjugated nanostructure, which represent a powerful platform for naked-eye colorimetric detection via plasmonic nanoparticles. We then discuss nanomaterial-based surface-enhanced Raman scattering (SERS) spectroscopy, which has the capability for very low-level fingerprint identification of virus, antigen, and antibody via graphene, plasmonic nanoparticle, and hetero-structure material. After that, we summarized about fluorescence and nanoparticle surface energy transfer (NSET) based on specific identification of SARS-CoV-2 infections via CNT, quantum dots (QDs), and plasmonic nanoparticles. Finally, we highlight the merit and significant challenges of nanostructure-based tools in infectious diseases diagnosis. For the researchers who want to engage in the new development of bio-conjugated material for our survival from the current and future pandemics, we hope that this account will be helpful for generating ideas that are scientifically stimulating and practically challenging.
Graphical Abstract
1. INTRODUCTION
Over the centuries infectious diseases caused by deadly viruses, bacteria, and different organisms have been among the leading causes of mortality in this world1–3. Around 103 years ago, the 1918 influenza pandemic, infected 500 million people worldwide and killed more than a half-million people in the United States and up to 50 million people worldwide1–3. The acquired immunodeficiency syndrome (AIDS) epidemic started 40 years before, has killed more than 35 million people till today1–3. As of March 2020, the world is currently dealing with a global outbreak of coronavirus disease 2019 (CIVID-19) caused by severe acute respiratory syndrome coronavirus 2 (SARS-CoV-2) virus4–8. The current pandemic has taken more than 4.2 M human lives worldwide till now6–13. It has also created vast volatility, uncertainty, and complexity in healthcare, education, transportation, and the financial industry in our world5–16. The current pandemic has highlighted the massive need for rapid and accurate diagnosis to control the spread of the virus by quarantining4–20. The current diagnosis techniques used in clinics for COVID-19 infections are reverse-transcription polymerase chain reaction (RT-PCR), reverse-transcription loop-mediated isothermal amplification (RT-LAMP), clustered, regularly interspaced short palindromic repeats (CRISPR), enzyme-linked immunosorbent assay (ELISA), lateral flow assay (LFA), etc. (Figure 1)10–40. The gold standard real-time polymerase chain reaction (RT-PCR), which targets different SARS-CoV-2 genomic regions such as nucleocapsid (N), spike (S) protein, or envelope (E) genes is the ultimate diagnostic method for the detection of SARS-CoV-2 (Figure 1)4–20. Since PCR procedures need 1–3 days to confirm the clinical data, clinicians cannot perform millions of tests per day, which is extremely important to control the outbreak5–25. Therefore, there are strong demand for the development of rapid diagnostic tests, which can have high sensitivity and specificity for measuring virus RNA, antigen (S or N protein) or antibodies (IgA, IgM, IgG) of symptomatic or asymptomatic patients10–30. Clinical testing facilities need novel point of care (POC) devices that can provide results very rapidly, maybe within 5 minutes.
Figure 1:
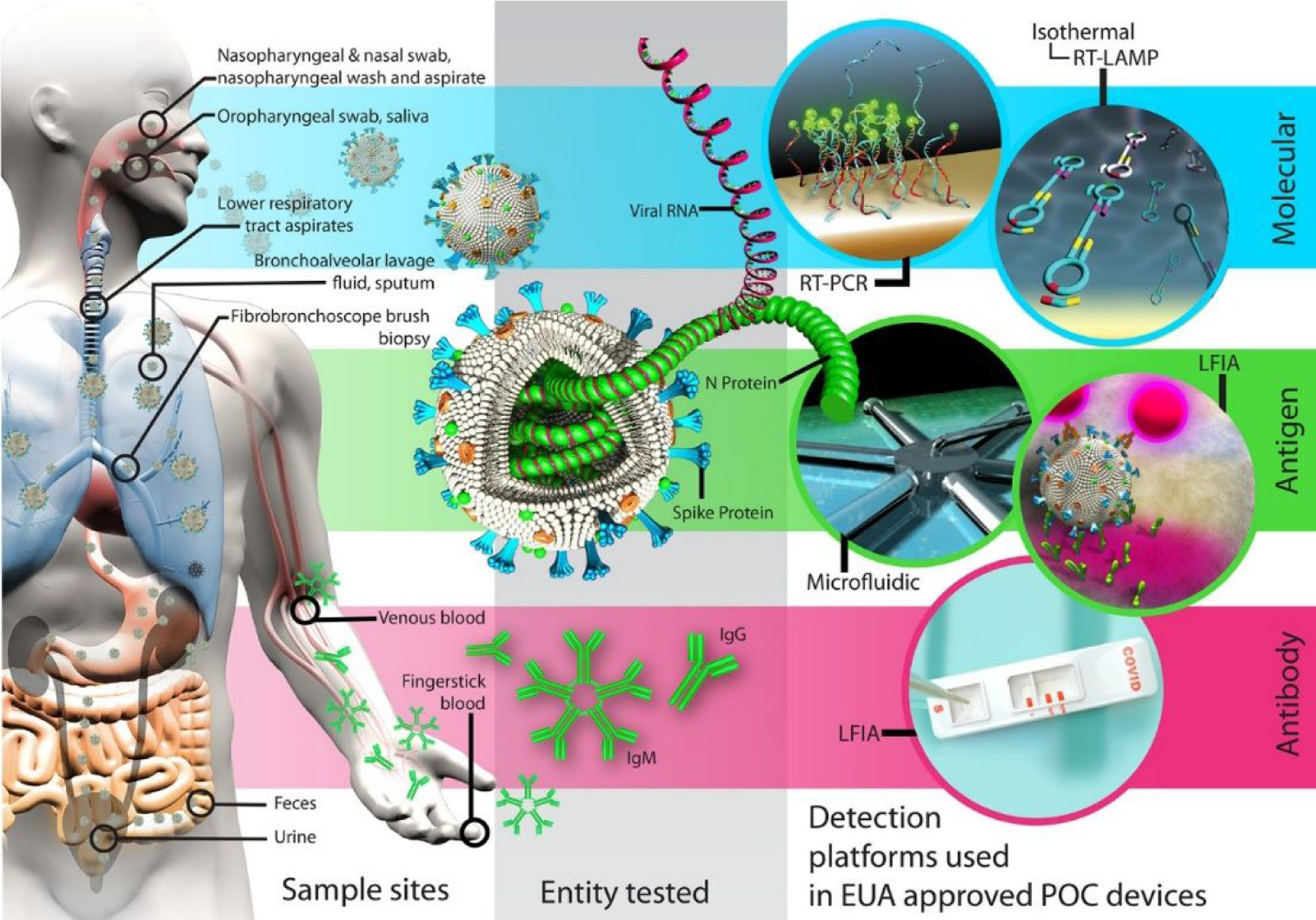
The current diagnostics methods for SARS-CoV-2 used in clinics. Reproduced with permission from Ref (8). Copyright 2021 American Chemical Society.
To overcome the problem, researchers have developed several nanomaterial-based approaches20–43. Since nanomaterials exhibit unique structural and functional properties, considerable efforts have been made in the last year on the rational design of nanoprobes to improve SARS-CoV-2 diagnosis (Figure 2)10–30. For reported diagnosis tools, various unique optical and electrical properties of nanomaterials have been used for signal transduction20–40. Since mass testing is fundamental to tackle the COVID-19 pandemic, several efforts are currently underway to develop easy-to-use colorimetric diagnostic tests that can be used in point-of-care settings or even for home daily use kits15–24. Since plasmonic gold nanoparticles have several orders of magnitude higher extinction coefficients than common dyes, it has been used heavily for sensitive colorimetric diagnosis purpose15–24. Similarly, high sensitivity and single molecular level detection capability diagnosis tools such as surface-enhanced Raman spectroscopy (SERS), nanomaterial-based surface energy transfer (NSET) have been developed using nanomaterials23–35. Since Raman scattering efficiency can be enhanced 108-1014 orders of magnitudes using plasmonic nanomaterials, researchers have used those materials for developing SERS active diagnosis tools for the identification of infectious virus40–53.
Figure 2:
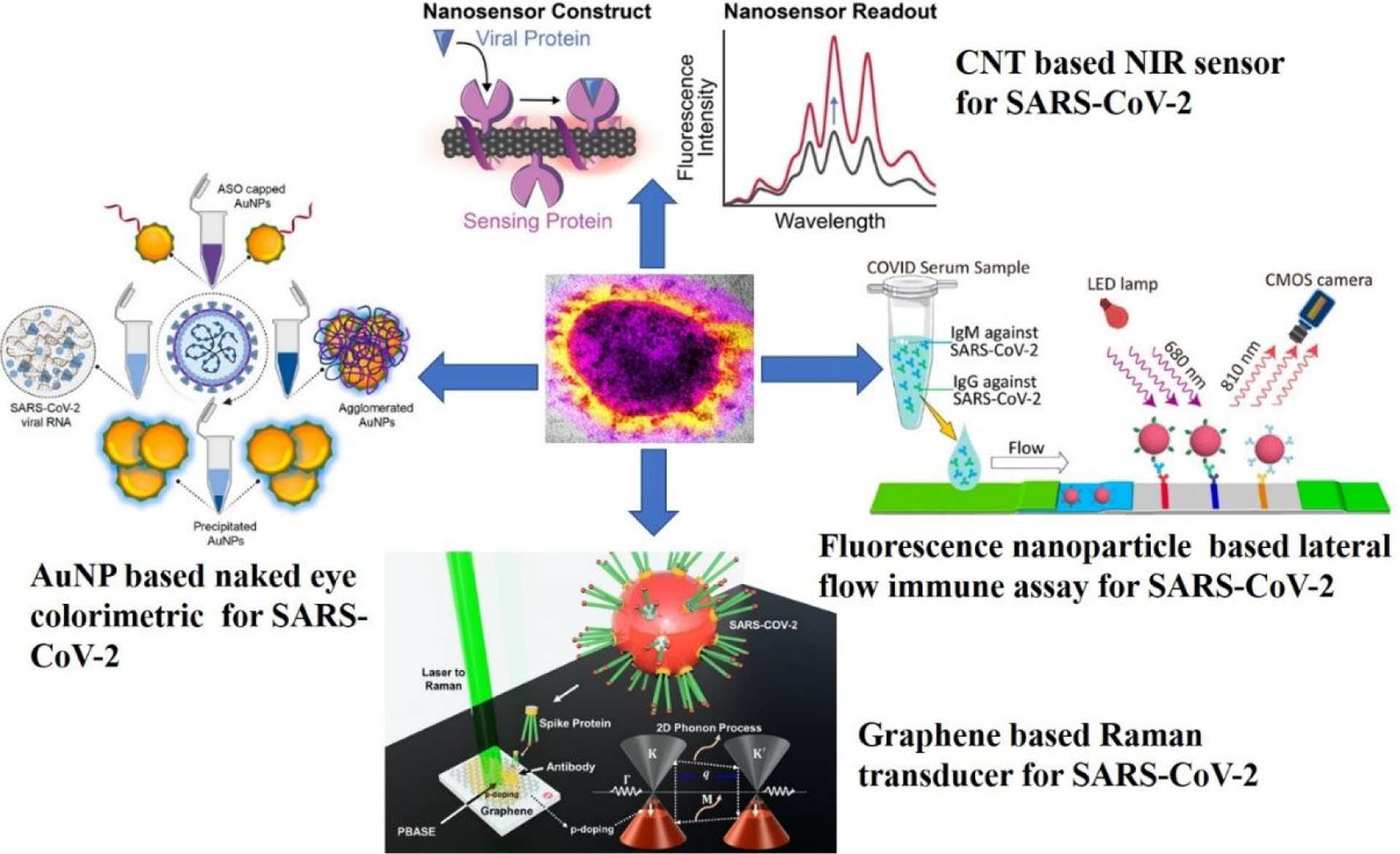
Schematic illustration of bio-conjugated nanoscale material-based tools which have been designed to diagnose SARS-CoV-2 RNA, antigen, and antibody. Reproduced with permission from Ref (16). Copyright 2020 American Chemical Society. Reproduced with permission from Ref (38). Copyright 2021 American Chemical Society. Reproduced with permission from Ref (19). Copyright 2021 American Chemical Society. Reproduced with permission from Ref (37). Copyright 2021 American Chemical Society.
Although researchers have made significant advances in this area, nanomaterial-based bio-diagnosis is still facing several obstacles. Since nanomaterials often bind nonspecifically with biomolecules, it significantly limits specificity and detection sensitivity44–50. In addition, since most of the nanoparticles possess positively or negatively charged surfaces to confer water solubility, they tend to exhibit strong nonspecific binding with protein and other biomolecules in biological fluids20–30,44–50. One way to overcome these problems is to design bio-conjugated nanomaterial by manipulating biomolecular recognition events (Figure 3A–3D)20–30,44–50. For this purpose, we and others used a novel strategy, which is initially coating the nanomaterial with polyethylene glycol (PEG) to reduce charge and nonspecific binding (Figure 3A–3B, 3D)20–3–,44–50. After that, the PEG-coated nanomaterials are conjugated with targeting agents such as SARS-CoV-2 specific antibody or DNA/RNA/peptides aptamer (Figure 3). Using the above strategy, we have created a bio-conjugated nanomaterial-based approach with minimum false alarms and it exhibits high sensitivity and specificity44–50.
Figure 3:
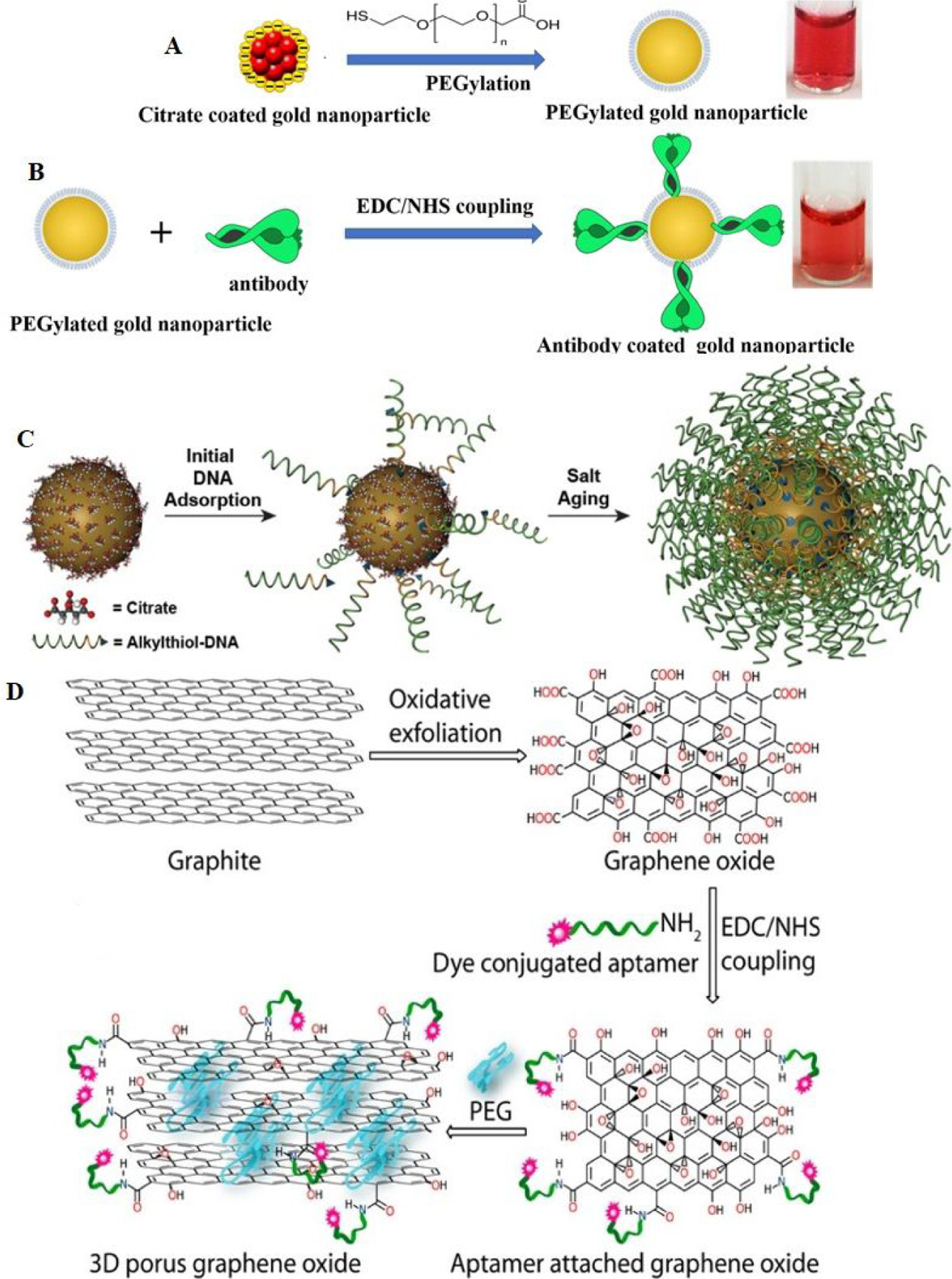
A) Design of PEG-coated gold nanoparticle. B) Design of antibody-conjugated PEG -coated gold nanoparticles. Reproduced with permission from Ref (23). Copyright 2021, Royal Society of Chemistry. C) Design of DNA-conjugated gold nanoparticles. Reproduced with permission from Ref (47). Copyright 2012 American Chemical Society. D) Design of peptide-conjugated graphene oxide. Reproduced with permission from Ref (44). Copyright 2015 American Chemical Society.
The fast-evolving research in this area has led to the development of bio-conjugated zero-dimensional (0D) spherical gold nanoparticle, quantum dots, one-dimensional (1D) carbon nanotubes (CNTs), nanorods, and two-dimensional (2D) graphene, and others for bio-diagnosis purposes 20–43. All reported nanomaterial-based SARS-CoV-2 diagnosis is based on the nucleic acid- or protein-based detection methodology20–43.
Along these lines, we have reported bio-conjugated nanomaterial-based naked eye colorimetric, highly sensitive SERS and NSET based specific and rapid diagnosis of SARS-CoV-2 virus, viral antigen, RNA, and other viruses23–24,45–46,52. In this account, we present an overview of the recent advances on the use of bio-conjugated nanoparticles as an effective diagnosis tool for SARS-CoV-2, which aims to help scientists to understand the use of bio-conjugated material for rapid diagnosis and facilitating its early realization of practical clinical applications. To provide the rationale behind these developments, we discuss general mechanisms for different types of bio-conjugated nanomaterial-based approaches for the diagnosis of SARS-CoV-2. We also highlight future opportunities and challenges of nanomaterial-based strategies for possible clinical uses.
2. Bio-conjugated plasmonic nanoparticle for targeted naked eye diagnosis
As per WHO, it is vital to expand the COVID-19 testing capacity up to 100-fold over existing RT-PCR tools1–10. For this purpose, easy to use, equipment-free, rapid, and low-cost colorimetric COVID-19 diagnostic tests can be an excellent choice for point-of-care applications if good sensitivity and specificity can be obtained. Naked eye colorimetric diagnostic tools utilize visual color change to detect targeted analytes16,21–25.
Dye-based colorimetric diagnosis is very well documented in biology from 1884, when Hans Christian Gram developed a naked eye sensor to provide distinct color for gram-positive and gram-negative bacteria20–30, 47–54. Since the absorption coefficient for organic dyes is relatively low, the dye-based colorimetric sensor has low sensitivity, and it exhibits poor selectivity20–30,47–54. To overcome the above problem, researchers have developed a bio-conjugated nanomaterial-based colorimetric diagnosis assay, using which selectivity and sensitivity can be enhanced by several orders of magnitude (Figures 4A–4D)23–27,47–54. Since for gold nanoparticles, the absorption and scattering occur in the visible region, and it became the golden choice for colorimetric sensor23–27,47–54. Due to the very high extinction coefficients (~109 for 20 nm gold nanoparticle), plasmonic gold nanoparticles absorb light millions of times stronger than the organic dye molecules As a result, the sensitivity of colorimetric diagnosis tools using gold nanoparticles will be excellent23–27,47–54. Benefiting from unique localized surface plasmon resonance (LSPR) optical properties, and biocompatible properties, gold nanoparticles are the commonly used plasmonic nanoparticles for the colorimetric diagnosis tools23–27,47–54.
Figure 4:
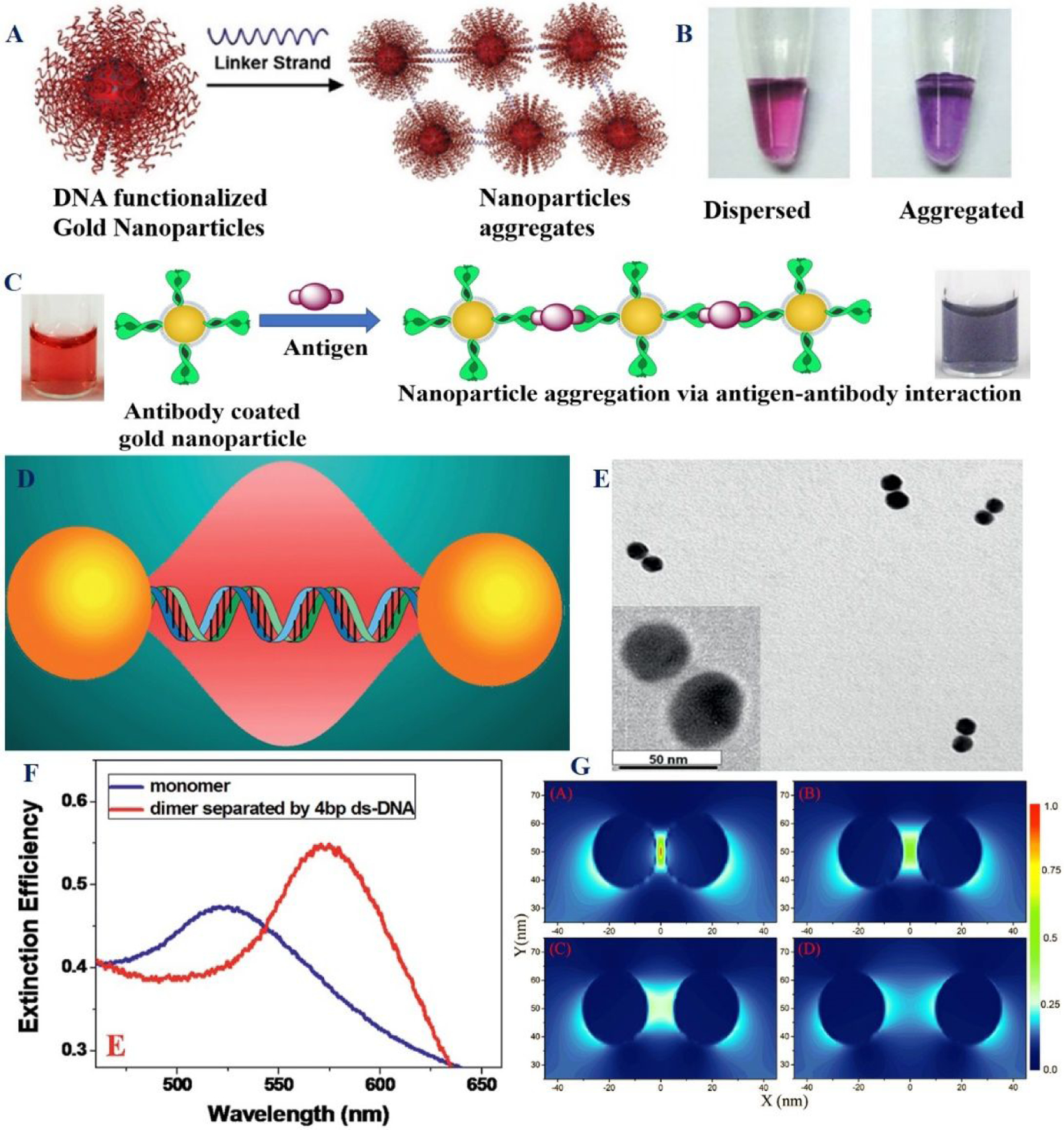
A) The scheme shows the aggregation and dispersion of spherical nucleic acid conjugated gold nanoparticles. B) Photograph shows visible red-to-purple color transition due to the aggregation. Reproduced with permission from Ref (47). Copyright 2012 American Chemical Society. C) The scheme shows the aggregation of antibody-conjugated gold nanoparticles in the presence of antigen. Reproduced with permission from Ref (23). Copyright 2021, Royal Society of Chemistry. D) The scheme shows the design of gold nanoparticle dimer where monomers are separated by -ds DNA. E) TEM image of gold nanodimer when monomers are separated by 8 nm through ds-DNA. F) Absorption spectra of monomer and dimer. G) Finite-difference time-domain (FDTD) simulation data show how electric field enhancement of dimer GNP varies with different spacing. Reproduced with permission from Ref (51). Copyright 2015 Royal Society of Chemistry.
Since its pioneered work by Mirkin and coworkers in 1997 47, researchers have developed a colorimetric assay using the bio-conjugated gold nanoparticles (Figures 4A–4C)23–24,45–57. Along these lines, we have reported bio-conjugated nanomaterial-based naked eye colorimetric diagnosis of COVID-19 antigen, SARS-CoV-2 virus, rotavirus, dengue virus, and different superbugs such as Carbapenem-resistant Enterobacteriaceae (CRE) E. coli, Salmonella DT104, and methicillin-resistant Staphylococcus aureus (MRSA)23–24,45–46,52–53.
2.1. Fundamentals of plasmonic nanoparticle based colorimetric assays.
Colorimetric assay’s working principle is based on LSPR properties of gold nanoparticles, which enables free electrons to oscillate collectively with the direction of the electric field of the incident light12,16–24,47–57. Due to the presence of LSPR, the boundary of plasmonic gold and silver nanoparticles hold a highly confined and enhanced electromagnetic field, which allows a strong absorption peak in the visible regions12,16–24,47–50. As a result, plasmonic nanoparticle absorption/extinction or color can be varied by forming aggregates via bio-conjugated nanoparticle interactions with analytes12,16–24,47–57. LSPR coupling among the nanoparticles can be manipulated by changing the distance between gold nanoparticles or by varying the degree of aggregation of nanoparticles (Figures 4D–4G)21–24, 47–57. To understand better about distance dependence of LSPR coupling mechanism, our group reported experimental and theoretical investigation by separating nanoparticles by double-strand DNA (Figures 4E–4G)51. Theoretically, finite difference time domain (FDTD) simulation investigation indicates that the plasmon coupling is highly dependent on the distance between two nanoparticles (Figure 4G)51.
2.2. Targeted naked eye colorimetric diagnosis of SARS-CoV-2 RNA, antigen and virus.
Inspired by the color variations after aggregations, researchers have designed the fast-developing field of naked eye colorimetric nanosensors to diagnose SARS-CoV-2 RNA, antigen, and the virus itself (Figures 5A–5G)16,21–25. In an exciting strategy, thiol-modified antisense oligonucleotides functionalized gold nanoparticle has been used for colorimetric detection of the N gene (nucleocapsid phosphoprotein) of SARS-CoV-2 (Figures 5A–5B)16. Bio-conjugated gold nanoparticles agglomerate selectively in the presence of RNA sequence of SARS-CoV-2 (Figure 5A)16, which can be diagnosed within 10 minutes.
Figure 5:
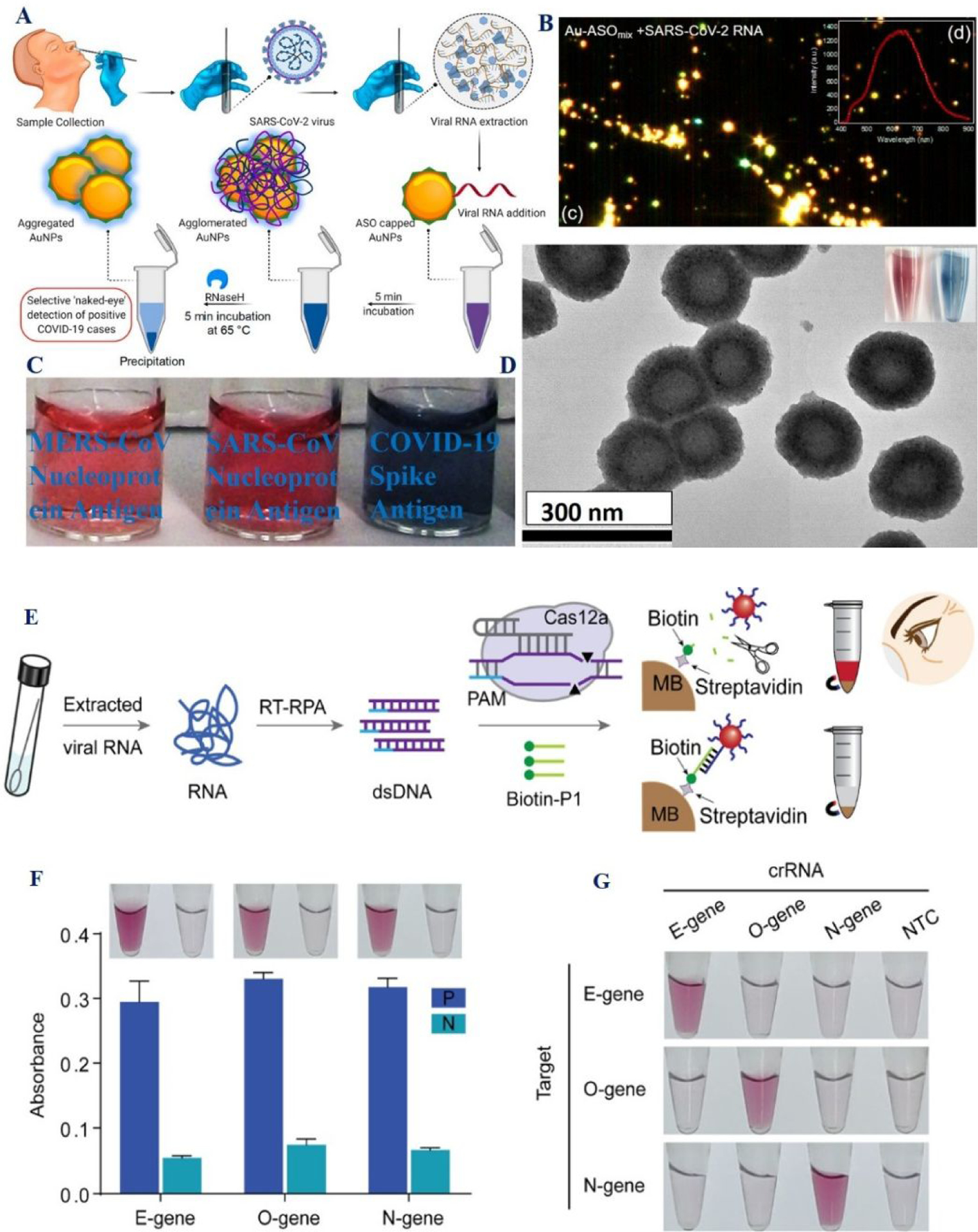
A) The scheme shows naked-eye sensing of SARS-CoV-2 RNA using gold nanoparticles capped with antisense oligonucleotides specific for N-gene. B) Enhanced dark-field microscope hyperspectral imaging of antisense oligonucleotides capped GNPs in the presence of SARS-CoV-2 RNA. Reproduced with permission from Ref (16). Copyright 2020 American Chemical Society. C) The photograph demonstrates the selectivity of bio-conjugated GNP based colorimetric assay for COVID-19 antigen. Reproduced with permission from Ref (23). D) TEM image of pseudoSARS-CoV-2 virus. Copyright 2021, Royal Society of Chemistry. E) Schematic shows CRISPR/Cas12a system assay with gold nanoparticle-DNA probes for detecting SARS-CoV-2 from clinical samples. F) Gold nanoparticle-CRISPR/Cas12a system-based detection of SARS-CoV-2 virus where P represents the virus RNA samples and N represents the blank control. G) Gold nanoparticle-CRISPR/Cas12a system-based detection of SARS-CoV-2 RNA, where NTC represents no-template control. Reproduced with permission from Ref (22). Copyright 2021 American Chemical Society.
The selectivity of the assay has been demonstrated using MERS-CoV viral RNA16. The reported detection limit was 0.18 ng/μL for targeted RNA (Table 1)16. Motivated by the simplicity of the assay, we have developed an anti-spike antibody attached GNP-based colorimetric assay for rapid COVID-19 antigen diagnosis23. In the presence of SARS-CoV-2 spike recombinant antigen, due to the antigen-antibody interaction, a colorimetric change from purple to bluish color is observed with naked eyes (Figures 5D). The SARS-CoV-2 antigen can be diagnosed within 10 minutes, and the detection limit for the colorimetric assay was obtained to be 1 nanogram (ng)/mL (Table 1)23. Furthermore, the selectivity for naked eye assay has been demonstrated using severe acute respiratory syndrome coronaviruses (SARS-CoV) nucleoprotein antigen and Middle East respiratory syndrome coronavirus (MERS-CoV) nucleoprotein antigen separately (Figure 5D)23. The sensitivity of the colorimetric assay for the identification of coronavirus was obtained as 1000 virus particles/mL23.
Table 1:
Bio-conjugated nanomaterials for the diagnosis of SARS-CoV-2 antigen or virus
| Target molecules | Detection platform | Bio-conjugated Nanostructure used | Detection time | Detection Sensitivity | References |
|---|---|---|---|---|---|
| SARS-CoV-2 N gene | Colorimetric | RNA/DNA attached gold nanoparticle | 10 minutes | .18 ng/μL | 16 |
| SARS-CoV-2 spike protein | Colorimetric | Antibody-conjugated gold nanoparticles | 5 minutes | 1 ng/mL | 23 |
| SARS-CoV-2 spike protein | Colorimetric | MNAzyme - conjugated gold nanoparticles | 5 minutes | 90% for clinical samples | 39 |
| SARS-CoV-2 S, N & P gene | Colorimetric | Antibody-conjugated gold nanoparticles | 20 minutes | 95% for clinical samples | 21 |
| SARS-CoV-2 IgM/IgG antibodies | Colorimetric based Lateral Flow | Nucleoprotein attached gold nanoparticles | 15 minutes | 93% for clinical samples | 27 |
| SARS-CoV-2 RNA | Colorimetric CRISPR/Cas | RNA/DNA attached gold nanoparticle | 15 minutes | 95.2% for clinical samples | 22 |
| SARS-CoV-2 spike protein | SERS | Antibody-conjugated gold nanoparticles | 10 minutes | 4 pg/mL | 23 |
| SARS-CoV-2 spike protein | SERS | Antibody attached graphene | 15 minutes | 3.75 fg/mL | 38 |
| SARS-CoV-2 IgM/IgG antibodies | SERS based LFA | Antigen attached SiO2@Ag nanoparticle | 20 minutes | 100% for clinical samples | 35 |
| SARS-CoV-2 spike protein | SERS | Antibody-conjugated gold nanoparticles | 20 minutes | 87.7% clinical sample | 42 |
| SARS-CoV-2 spike protein | NSET | Aptamer-conjugated gold nanoparticles | 10 minutes | 130 fg/mL | 24 |
| SARS-CoV-2 spike protein | Fluorescence | ACE2 attached SWCNT | 90 minutes | 12.6 nM | 37 |
| SARS-CoV-2 IgG and IgM | Fluorescence based LFA | Antigen/SiO2@Au@QD nanobeads | 15 minutes | 100% for clinical samples | 28 |
| SARS-CoV-2 spike protein | NSET | Antibody coated quantum dots | 20 minutes | 200 fg/mL | 30 |
| SARS-CoV-2 spike protein | Field effect transistor | Antibody coated MXene-graphene | 50 seconds | 1 fg/mL | 36 |
| SARS-CoV-2 RNA | RT-PCR | 6 hours | 97% for clinical samples | 8 | |
| SARS-CoV-2 RNA | CRISPR-Cas | 40 minutes | 95% for clinical samples | 11 | |
| SARS-CoV-2 antigen | LFA | 15 minutes | 95% for clinical samples | 8 |
Although CRISPR system is known from 1987, just five years before CRISPR-based diagnoses became popular for human pathogen diagnosis11,15,18–22. Recently scientists have developed CRISPR/ Cas12a and CRISPR/Cas13a, systems for the designing of rapid and sensitive detection of SARS-CoV-211,15,18–22. To, improve the sensitivity of CRISPR detection, the SPR properties of plasmonic gold nanoparticles have been used to pair with the CRISPR-Cas12a system (Figure 5E)22. Reported data indicates that CRISPR/Cas12a-assisted detection using gold nanoparticle as colorimetric probe has capability to distinguish the N gene and O gene of SARS-CoV-2 from two other closely related coronaviruses (Figures 5F–5G)22. Experimental data shows that that gold nanoparticle CRISPR-Cas12a system can be used to detect as low as 50 RNA copies22. From the reported data, we can conclude that the gold nanoparticle- CRISPR-Cas12a system has potential for SARS-CoV-2 screening in clinics where state-of-the-art facilities are lacking.
2.3. Naked eye diagnosis of SARS-CoV-2 from clinical sample
Using similar strategies, antibody-conjugated gold nanoparticle-based colorimetric assay has been used for SARS-CoV-2 identification in clinical samples from 45 positive to SARS-CoV-2 patients and 49 negative patients21. For this purpose, gold nanoparticles were attached with three surface proteins of SARS-CoV-2 (spike, envelope, and membrane) (Figures 6A–6B)21. The reported sensitivity and specificity of the colorimetric assay were higher than 95% (Table 1)21. For clinical samples, the GNP-based colorimetric essay can be compared with a real-time PCR. The viral load’s corresponding threshold cycle (Ct) = 36.5 can be detected using gold nanoparticle-based colorimetric assay21.
Figure 6:
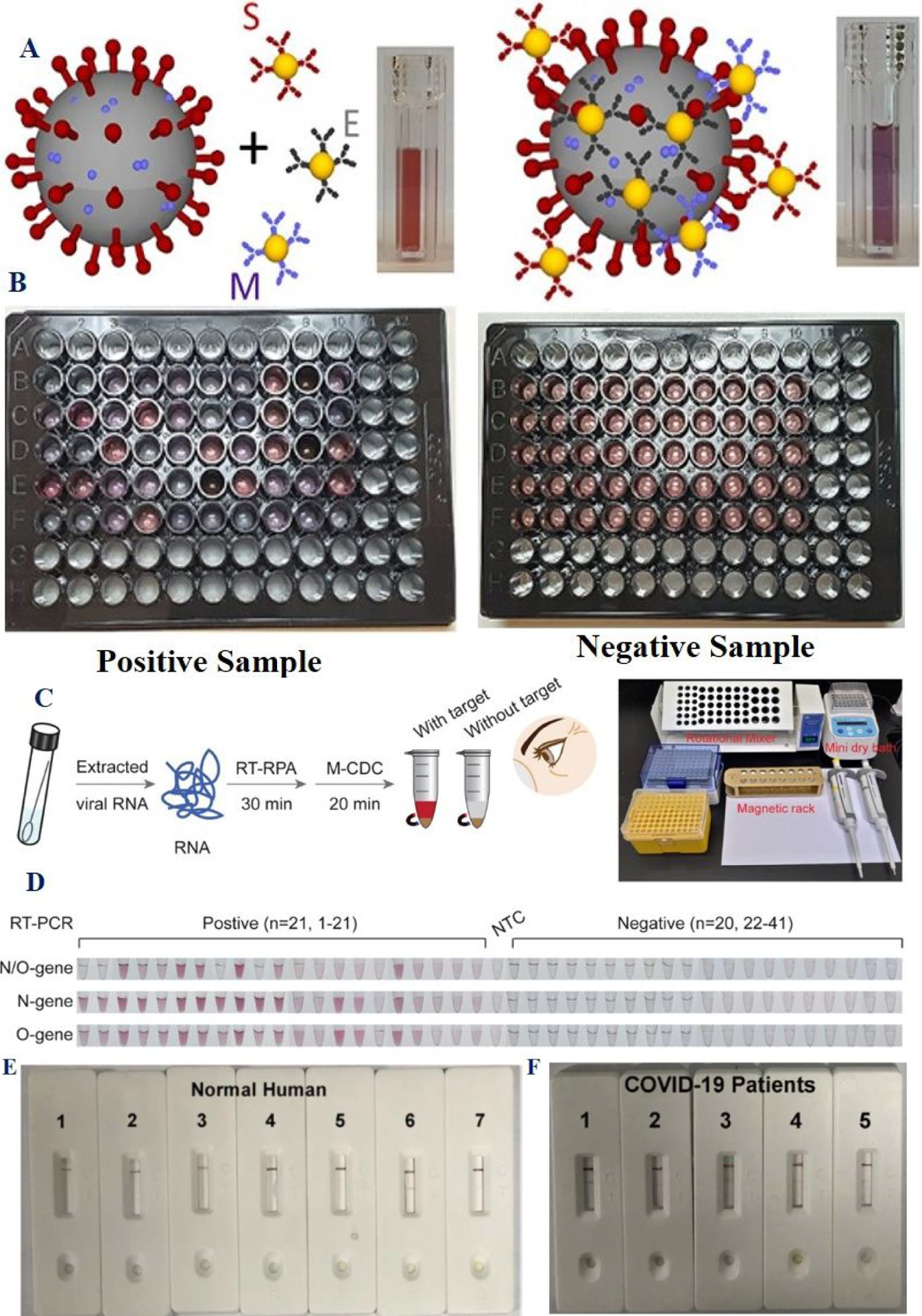
A) The scheme represents anti-spike, anti-membrane and anti-envelope antibody attached gold nanoparticles-based detection of viral particles. B) The photograph shows colorimetric test results for COVID-19 positive and negative clinical samples. Reproduced with permission from Ref (21). Copyright 2020 American Chemical Society. C) Schematics shows gold nanoparticle-CRISPR/Cas12a system-based diagnosis of clinical sample. D) Assessment of clinical sample detection using gold nanoparticle-CRISPR/Cas12a system-based. Reproduced with permission from Ref (22). Copyright 2021 American Chemical Society. E-F) Performance of gold nanoparticle based LFA for human serum sample from clinical sample, E) without COVID-19 infection and F) with COVID-19 infected patients. Reproduced with permission from Ref (27). Copyright 2020 American Chemical Society.
Similarly, recently researchers have used gold nanoparticle-CRISPR-Cas12a system to analyze clinical SARS-CoV-2 RNA (Figures 6C–6D)22. Reported data shows 95.12% consistency with clinically approved PCR data, which indicates that gold nanoparticle-CRISPR-Cas12a has potential for SARS-CoV-2 screening in clinics22.
Due to the enormous advantages of portability, the lateral flow assay (LFA) using bio-conjugated gold nanoparticles has been developed27–32. It is now well documented that antibody against SARS-CoV-2 will be produced by the human body after infection, which is the primary immune response to fight against COVID-1927–32. To design a possible point of care device for SARS-CoV-2, gold nanoparticle-based lateral-flow strips have been developed for rapid identification of the IgM antibody against the SARS-CoV-2 virus (Figure 5G)27. Reported positive and negative SARS-CoV-2 serum samples data demonstrated a good consistency with RT-PCR data, and specificity is 93.3% (Figure 5G)27. It should be noted that since it takes 7–14 days to produce antibodies after an infection, gold nanoparticles based LFA cannot be used for early detection. All reported clinical sample data discussed here indicates that bio-conjugated nanomaterial-based naked eye assay can be used for the design of inexpensive platforms for virus diagnosis on a mass scale for countries where health systems are lacking state-of-the-art laboratory infrastructure.
Although bio-conjugated GNP-based colorimetric assay is very rapid and requires minimal equipment, due to the low sensitivity, use of the assay for an early stage of the SARS-CoV-2 infection is limited. To overcome the above problem, researchers are working to develop nanomaterial-based ultra-sensitive tools for SARS-CoV-2.
3. Bio-conjugated graphene and plasmonic nanomaterial-based SERS for highly sensitive diagnosis of coronavirus
A paradigmatic example of the highly sensitive diagnostic method is nanomaterial-based surface-enhanced Raman scattering (SERS) spectroscopy, as it has the capability for very low-level specific identification of virus, bacteria, and finger-print identification of pathogens23–24,45–46,51–54. After the discovery of SERS in 1974, it is known to be one of the most sensitive detection techniques for bio-molecular sensing (Figure 7A–7F)51–54. It is well understood that placing the analyte within nanometer-sized gaps between two plasmonic nanoparticles is very important to enable massive enhancement of the Raman signals51–54 (Figure 7B). Along these lines, we have reported bio-conjugated nanomaterial-based SERS for the diagnosis of COVID-19 antigen, SARS-CoV-2 virus, rotavirus, dengue virus, West Nile virus, different superbugs, and cancer biomarkers23–24,45–46,53.
Figure 7:
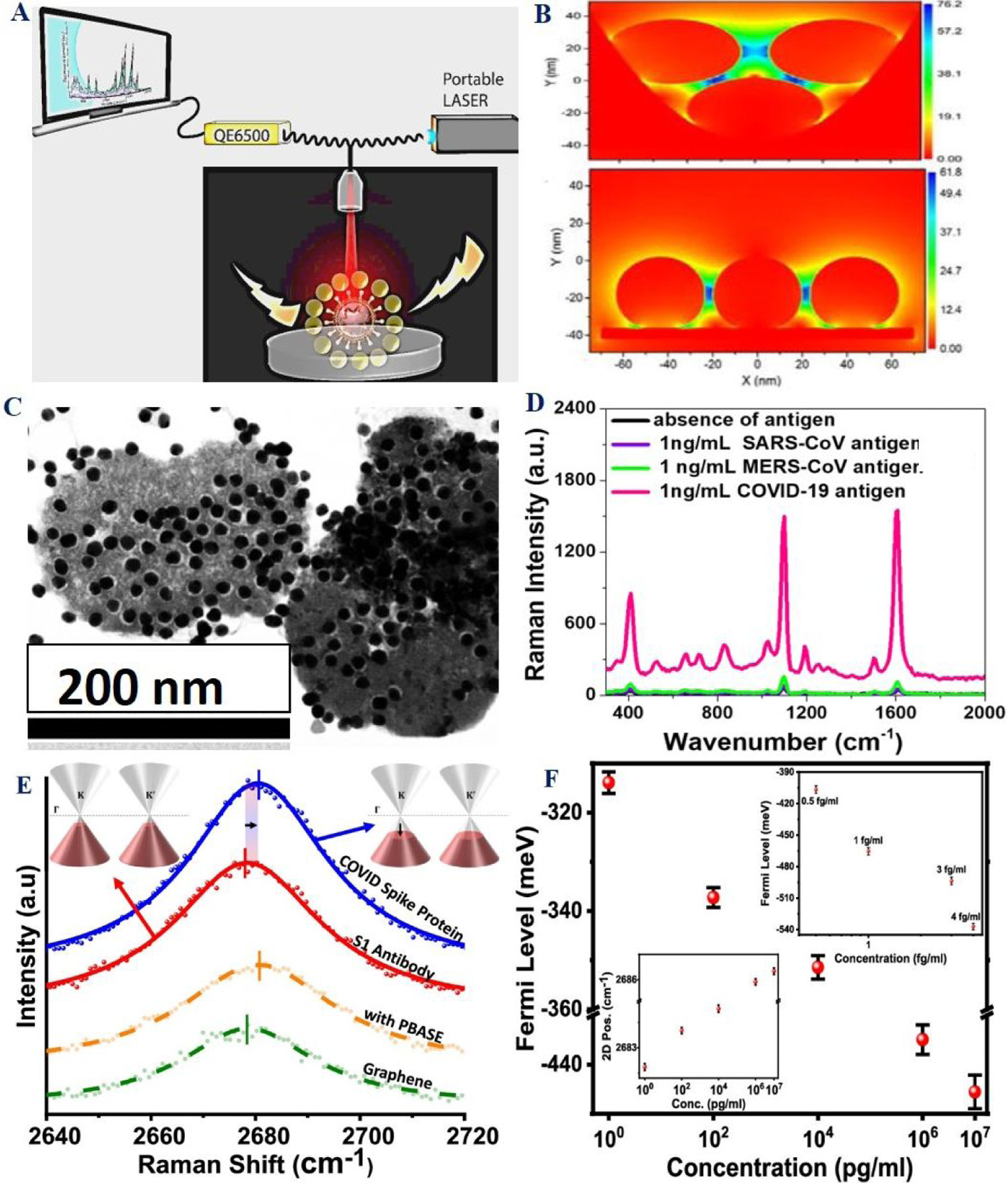
A) Schematic representation of portable SERS probe used for biomolecule identification. B) FDTD simulation data show how electric field enhancement varies for three GNPs in bent and straight assembly structure. Reproduced with permission from Ref (53). Copyright 2016 Royal Society of Chemistry. C) TEM data represent the aggregation of the antibody attached gold nanoparticles on the surface of virus via antigen-antibody interaction. D) Selectivity of SERS assay for COVID-19 antigens. Reproduced with permission from Ref (23). Copyright 2021 Royal Society of Chemistry. E) Plot shows the detection of the COVID spike protein using graphene 2D Raman band. F) Plot shows the variation of the 2D peak position and the Fermi level of graphene with the concentration of the COVID spike protein. Reproduced with permission from Ref (38). Copyright 2021 American Chemical Society.
3.1. Fundamentals of nanomaterial-based SERS assays.
SERS enhancement is based on electromagnetic as well as chemical enhancement mechanisms51–53. SERS exploits the strong electromagnetic fields generated at the inter-particle junctions of plasmonic nanostructures, known as “plasmonic hot spots” (Figure 7B–7C)51–54. The strong plasmon coupling in the “hot spots” allows Raman signals to be enhanced by 108-1010 orders of magnitude, which is sufficient to detect analytes at single molecular level single molecular level 51–54. Since SERS electromagnetic enhancement factor is proportional to the fourth power of the plasmonic enhancement electric field ([E]4), FDTD simulation data indicate that the enhancement factor can be 6–8 orders magnitude higher in the “hot spot” position (Figure 7B)51–54. Similarly, SERS chemical enhancement depends on the charge transfer between analytes and nanostructures51–54.
3.2. Targeted SERS diagnosis of SARS-CoV-2 antigen and virus
Due to the excellent sensitivity and potable sensor development capability, we have developed a SERS assay for COVID-19 antigen and virus identification using bio-conjugated gold nanoparticles (Table 1–2 and Figure 7–8)23. For this purpose, 4- amino thiophenol (4-ATP) SERS reporter and anti-spike antibody attached GNP have been developed 23. Due to the strong interaction via plasmon-excitation coupling, the SERS assay has the capability to diagnose 1pg/mL antigen (Figure 7D and Table 1)23. Selectivity for SERS assay has been demonstrated using severe acute respiratory syndrome coronaviruses (SARS-CoV) nucleoprotein antigen and Middle East respiratory syndrome coronavirus (MERS-CoV) nucleoprotein antigen separately (Figure 7D)23.
Table 2:
Major Raman bands observed in SERS spectra from dengue, West Nile Virus, rotavirus and coronavirus
| Corona virus (cm−1) | Rotavirus (cm−1) | West Nile Virus (cm−1) | Dengue virus (cm−1) | Vibration mode |
|---|---|---|---|---|
| 1680 | Amide-I | |||
| 1610 | 1606 | 1610 | phenylalanine | |
| 1303 | v(NH2) stretch for adenine | |||
| 1550 | Amide-II | |||
| 1495 | 1495 | -C=C- stretch | ||
| 1482 | 1482 | Base stacking of guanine | ||
| 1468 | Lipid alkyl C-H bend | |||
| 1310 | 1310 | Amide-III | ||
| 1190 | 1190 | -CO2H deformation | ||
| 1070 | 1070 | -CN stretch | ||
| 1045 | Unsaturated lipids | |||
| 950 | 950 | -CH3 rocking | ||
| 712 | 712 | Adenine band | ||
| 550 | 550 | 550 | Guanine band |
Figure 8:
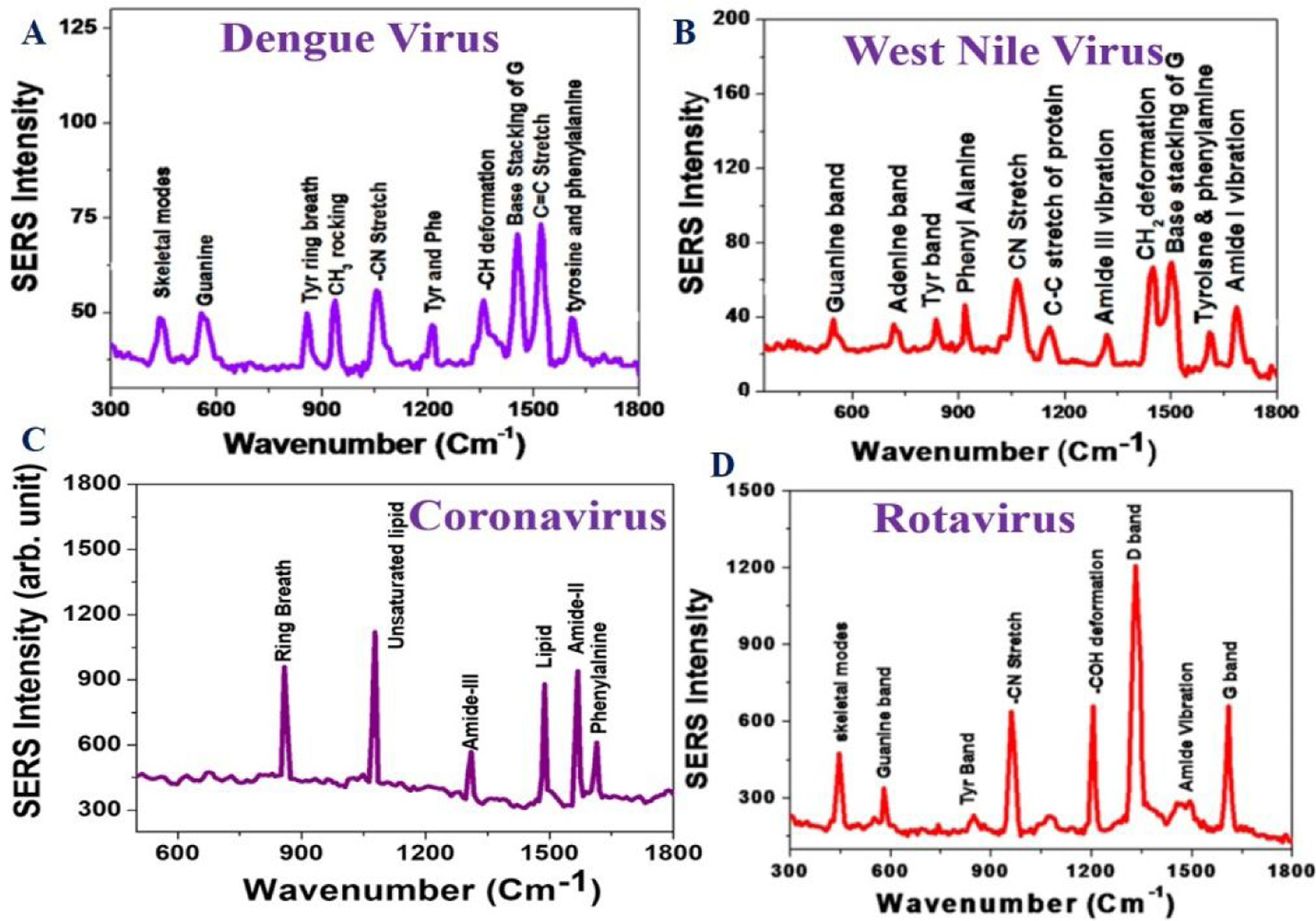
A) SERS spectra of Dengue virus in the presence of the anti-flavivirus antibody attached GNP. B) SERS spectra of West Nile virus in the presence of the anti-flavivirus antibody attached GNP. Reproduced with permission from Ref (45). Copyright 2015 American Chemical Society. C) SERS spectra of corona virus in the presence of the anti-spike antibody attached GNPs. D) SERS spectra of rotavirus in the presence of the anti-rotavirus antibody attached GNP. Reproduced with permission from Ref (46). Copyright 2014 American Chemical Society.
In an interesting study, recently, antibody attached graphene has been used as a Raman transducer for the detection of COVID-19 S-protein (Figure 7E–7F)38. Reported data shows no measurable cross-reactivity with MERS-CoV spike protein38. Experimental data demonstrated that antibody attached graphene-based Raman has the capability to detect SARS-CoV-2 spike protein as low as 3.75 fg/mL level in saliva38 (Table 1). Although Raman reporters-based SERS has very high sensitivity, the sensing assay will have huge benefits if the diagnostic technologies can be used as fingerprinting identification51–54. Last few decades, researchers have developed several sensing technologies to target the above goal51–54. Among them, label-free SERS has attracted considerable attention, which has the capability for unique fingerprint profiling of a biological sample51–54.
Taking the above advantage of Raman spectroscopy-based assay, we have demonstrated that antibody attached GNP-based SERS has the capability to be used for fingerprint identification of dengue, West Nile, rotavirus, and coronavirus, respectively (Figures 8A–8D, Table 2)23,45–46. Raman bands from pseudo-SARS-CoV-2 can be assigned to spike protein phenylalanine ring breath mode, amide II, and amide III modes (Table 2 and Figure 8C). Raman modes due to lipid and unsaturated lipids are also powerful (Table 2 and Figure 8C). From all the spectra, one can find that the Raman bands for lipid and phospholipids are unique for coronavirus spike protein which has not been observed for dengue, West Nile, or rotavirus23,45–46 (Table 2 and Figure 8).
3.3. SERS based diagnosis of SARS-CoV-2 antigen and antibody from clinical sample
Similarly, gold nanoparticle-based SERS assay has been used for SARS-CoV-2 spike antigen identification in a clinical sample using throat swabs from 102 healthy people and 30 confirmed COVID-19 patients (Figure 9A–9H).42 For this purpose, deep learning-based SERS has been developed for on-site detection of the SARS-CoV-2 antigen, using human throat swabs and sputum samples (Figures 9A–9C)42. Reported data shows that SERS can be used for SARS-CoV-2 antigen detection within 20 min time period with an accuracy of 87.7% from clinical sample42.
Figure 9:
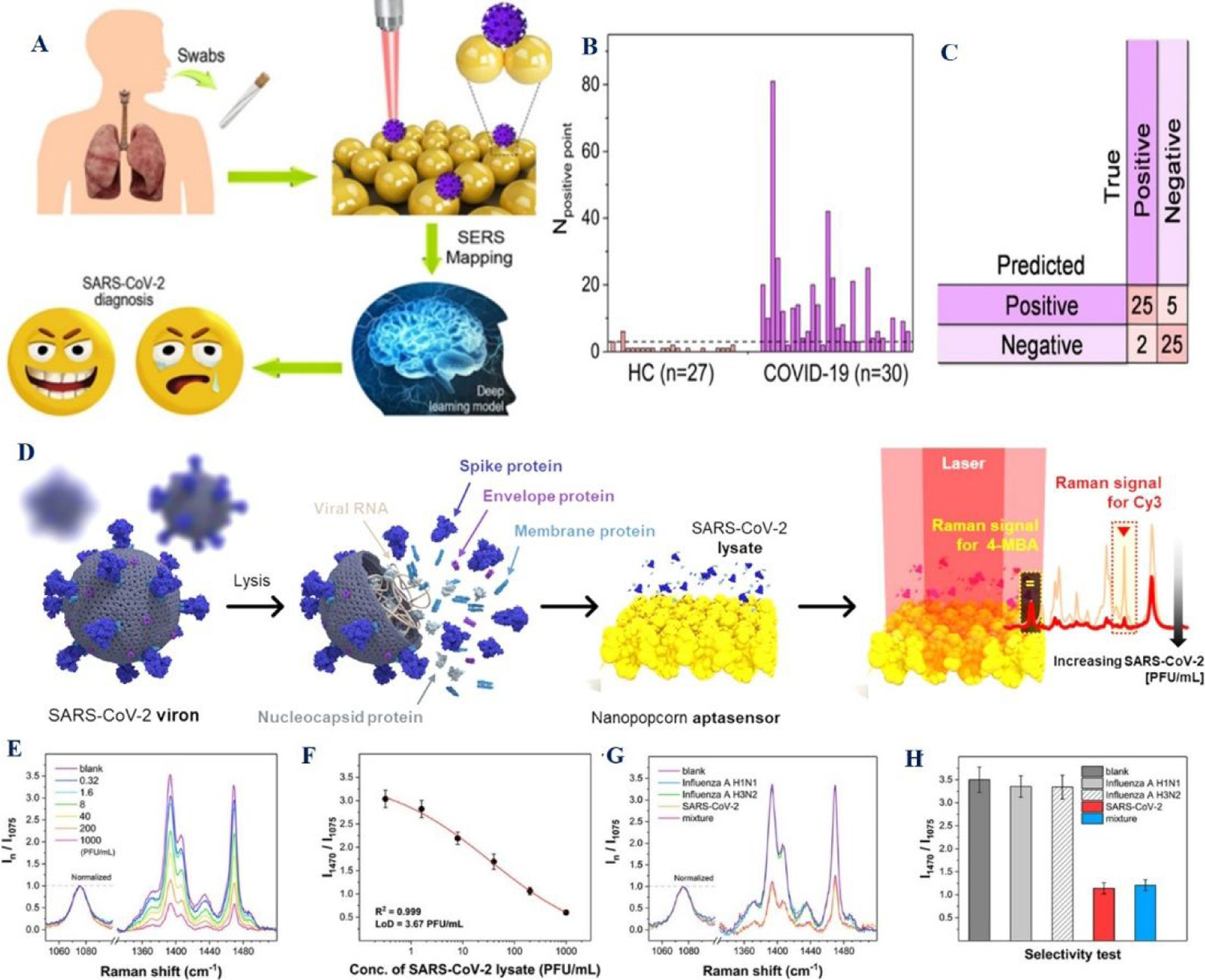
A) The scheme shows the detection process of the SARS-CoV-2 antigen from clinical sample using the deep learning-based SERS technique. B) The graph shows the number of the predicted positive points of clinical specimens. HC represents healthy controls. C) The plot shows the diagnostic results at the optimal cutoff value. Reproduced with permission from Ref (42). Copyright 2021 American Chemical Society. D) The scheme shows the quantitative evaluation of SARS-CoV-2 using the SERS-based aptasensor. E) The plot shows the Raman peak intensity variations with increasing SARS-CoV-2 concentration in real clinical negative respiratory specimens. F) The plot shows the variation of Raman intensity as a function of the SARS-CoV-2 lysate concentration. G) The plot shows the selectivity of the assay using influenza A/H1N1 (4500 PFU/mL), influenza A/H3N2 (15 000 PFU/mL), SARS-Cov-2 (200 PFU/mL), and their mixtures. H) The plot shows the histograms for the normalized Raman peak intensity ratios. Reproduced with permission from Ref (40). Copyright 2021 American Chemical Society.
Since LFA is the paper-based cheapest tool for achieving rapid screening, the researcher has developed SERS-based LFA for SARS-CoV-2 IgM/IgG detection. For this purpose, SERS tags were fabricated by coating a complete Ag shell on the SiO2 core.35 Reported result indicates that SERS-LFIA strips have the capability to detect SARS-CoV-2 IgM/IgG within 25 minutes from the clinical sample with 100% accuracy35. All the clinical sample data discussed here indicates that bio-conjugated graphene and GNP nanomaterial-based SERS has the capability to be used for the diagnosis of SARS-CoV-2 in-clinics.
Since genetic variants of SARS-CoV-2 such as B.1.1.7 (Alpha), B.1.351 (Beta), B.1.617.2 (Delta), and P.1 (Gamma) are circulating, whose infection rates are much higher than original version first detected in China, we anticipate that researchers will design many more innovative strategies in the coming years for the finger-print Raman sensing of different variants.
4. CNT and fluorescence nanoparticle-based sensor for SARS-CoV-2 sensing
In the last two centuries, light microscopy became the most used technique in clinical diagnosis55–57.. Last decade, we and others demonstrated that carbon dots (CDs), perovskite quantum dots (PQDs), semiconductor quantum dots, and lanthanum nanoparticles have been proven to be better fluorescent probes than conventional fluorescent dyes55–57. Since fluorescence nanodots exhibit a much higher quantum yield than organic dyes and nano-dots, and are resistant to photo-bleaching, researchers have used them in a variety of bio-detection and imaging purpose55–57.
4.1. CNT -based fluorescence sensor for SARSCoV-2 sensing
In an exciting study recently, single-walled carbon nanotubes (SWCNTs), which emits near-infrared emission, has been used to develop nano-sensor for the detection of SARS-CoV-2 (Figure 10A–10D) 37. For this purpose, SWCNTs were noncovalently functionalized with angiotensin-converting enzyme 2 (ACE2), which has a very high binding affinity for spike protein (Figure 10A)37.
Figure 10:
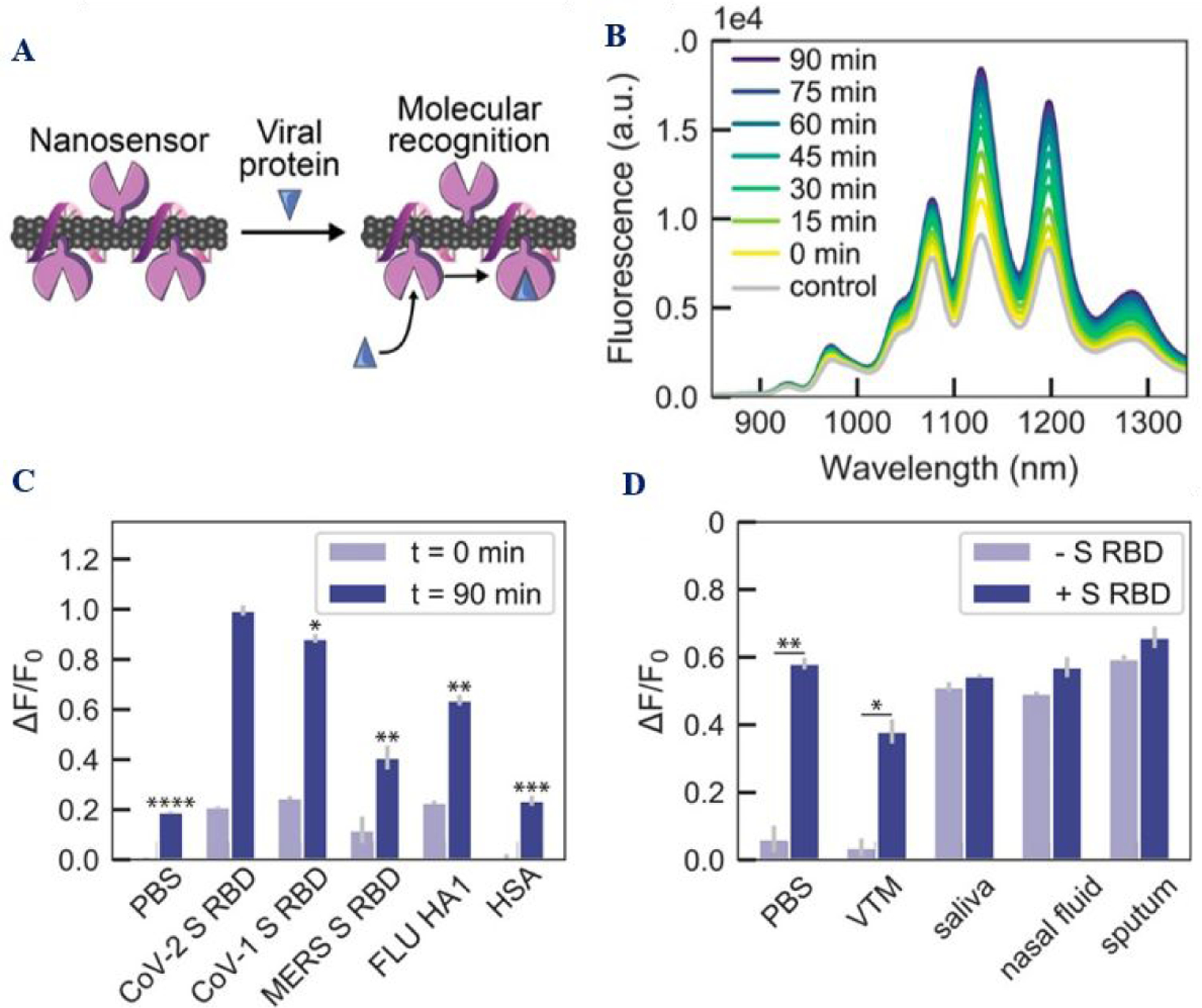
A) The scheme shows the ACE2-SWCNT nanosensor working principle. (B) The spectra show how the fluorescence spectrum of ACE2-SWCNT varies with time. C) The plot shows selectivity of the sensor for SARS-CoV-2 spike receptor-binding domain (S RBD) and SARS-CoV-1 S RBD with respect to MERS S RBD, and FLU hemagglutinin subunit (HA1). D) ACE2-SWCNT nanosensor response after exposure to 1 μM S RBD in the presence of 1% viral transport medium (VTM), saliva, nasal fluid, and sputum (treated with sputasol). Reproduced with permission from Ref (37). Copyright 2021 American Chemical Society.
Since CNT exhibits near-infrared (NIR) emission in biological II window, it has been used as signal transducers for spike protein detection (Figure 10B). Reported data indicates that when viral S proteins are present, the fluorescence signal from SWCNT changes due to the binding between ACE2 and spike protein (Figures 10B–10D)37. Reported data demonstrate that bio-conjugated SWCNT can be used for the identification of SARS-CoV-2 virus-like particles within 5 s of exposure time37. The detection limit was reported as 12.6 nM. (Table 1). Continuing innovations on the design of smart nanomaterial-based robust fluorescence assay will undoubtedly further expedite the development to achieve clinical testing performance.
4.2. Fundamentals of nanomaterial-based FRET assays
Although researchers have made huge advances in the design of the fluorescence microscope, it is now well documented that the fluorescence microscope resolution is not enough for understanding the interaction between biomolecules55–57. To overcome the above problem, researchers have developed a Förster or Fluorescence Resonance Energy Transfer (FRET) microscope which has capability to determine the spatial proximity of biomolecules55–57.
FRET was discovered by Theodor Förster in 1948, where the excited donor transfers energy to the ground state acceptor, which has been used routinely as a biosensor in the scientific community24, 55–57. In the last three decades, due to the advancement in state-of-the-art digital imaging techniques, FRET-based light microscopy imaging became very popular. It has been used to monitor the dynamics of biomolecules in vitro and in vivo24, 55–57. Although FRET has been used in biological research routinely due to the dipole-dipole coupling mechanism between donor and acceptor and the maximum distance limit for biomolecular interaction measurement is 10 nm for FRET24, 55–57. To overcome this 10 nm distance limit, we and other groups have reported a long-range nanoparticle-based surface energy transfer (NSET), where energy transfer can be monitored even above 20 nm distance (Figure 11A–11B)24, 55–57. Since the quenching efficiency of the gold nanoparticle is 9–10 orders of magnitude higher than that of the organic acceptor used for FRET, GNP-based NSET nanoprobe has much higher efficiency than FRET24, 55–57.
Figure 11:
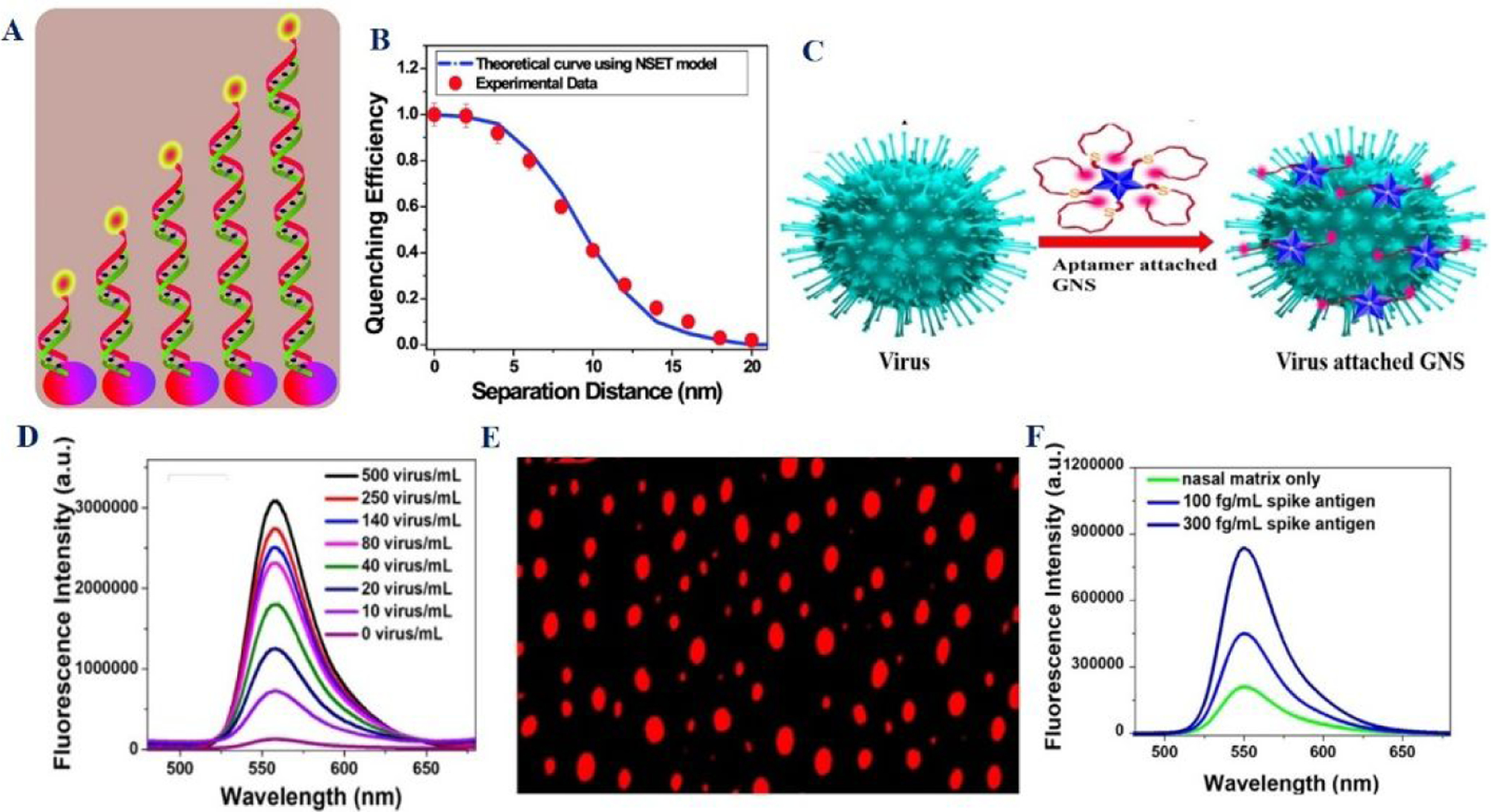
A) Schematic representation of NSET ruler with difference lengths. B) Variation of quenching efficiency with separation distance for NSET ruler. Reproduced with permission from Ref (47). Copyright 2012 American Chemical Society. C) Schematic indicates the designing of NSET assay using gold nanostar and COVID specific aptamer. D) Variation of NSET intensity with the concentration of pseudo-SARS-CoV-2 virus. E) Single photon luminescence image of pseudo baculovirus attached dye conjugated aptamer. Reproduced with permission from Ref (24). Copyright 2021 American Chemical Society.
4.3. Targeted FRET diagnosis of SARS-CoV-2 antigen and virus.
Using the above advantages of NSET and aptamer-based bio-recognition technology, we have developed rhodamine 6G (Rh-6G) dye conjugated COVID-19 spike protein-specific DNA aptamer attached gold nanostar-based NSET, which can be used for rapid diagnosis of COVID-19 antigen or virus rapidly (Figures 11D–11F)24.
Due to the strong interaction between aptamers and COVID-19 spike antigen via non-covalent interaction, the reported NSET assay has the capability to diagnose 130 fg/mL antigen (Table 1, Figures 11D–11F) 24. Similarly, using DNA aptamer-spike protein interaction, NSET has been used for the identification of virus with the sensitivity of 8 virus particles/mL (Table 1)24. For the possible application of the NSET assay for COVID-19 from clinical sample, NSET assay has been used to detect antigen and virus in infected nasal matrix 24. Interestingly, NSET has the capability to be used for the detection of COVID-19-specific antigen at a concentration of 100 fg/mL and virus at the concentration of 20 virus particles/mL (Figures 11F)24.
Using similar strategies, a FRET sensor based on the fluorescent QDs (green QD514, fluorescence maximum at 514 nm) and gold nanoparticles have been developed to monitor in vitro spike-ACE2 binding30 (Figures 12A–12B). Experimentally reported data indicates that QD fluorescence is quenched upon binding due to the change in distance between gold nanoparticles and QDs (Figures 12A–12B)30. Reported data demonstrated that FRET intensity could be disrupted by unlabeled ACE2 or by neutralizing SARS-CoV-2 antibodies (Figures 12A–12B)30. The in vitro data indicates that FRET platform can be used for rapid screening of SARS-CoV-2 infection30.
Figure 12:
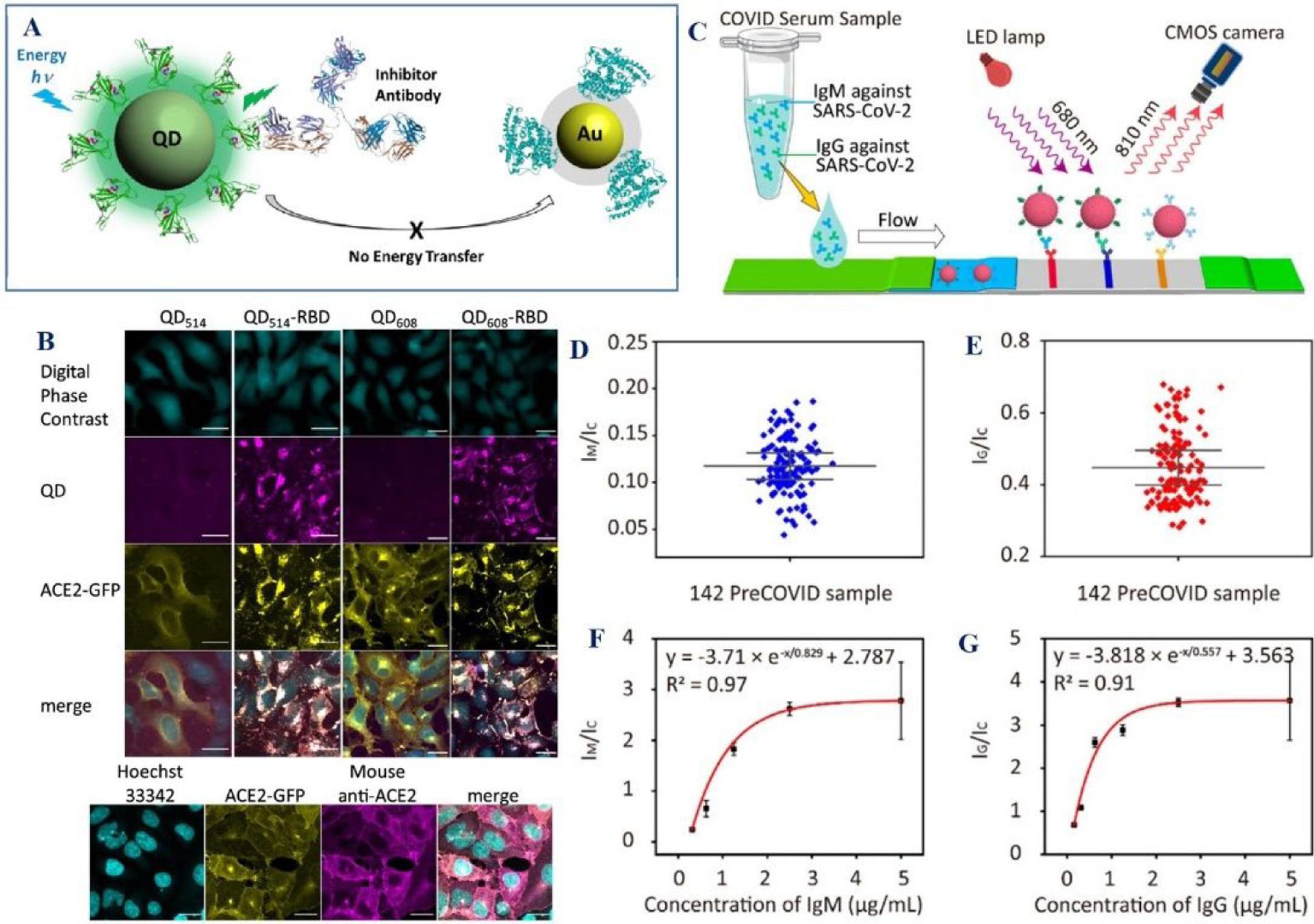
A) The scheme shows the design of QD and gold nanoparticles -based FRET for finding the interaction between RBD and ACE2. B) Fluorescence image shows the image of ACE2-GFP (yellow) HEK293T clone 2 treated with 100 nM QD514-RBD (magenta) and QD608-RBD (magenta). Reproduced with permission from ref (30). Copyright 2020 American Chemical Society. C) The scheme shows the NIR-Emissive AIE Nanoparticle-Labeled Lateral Flow Immunoassay for Detection of IgM and IgG. D-E) Clinical sample performance of test strip for the detection of IgM and IgG in 142 pre-COVID samples. The error bars represent the standard deviation of the values. The standard deviation is calculated from the results of 142 independent tests. F) Calibration curve indicates the LoD of IgM is 0.236 μg mL‒1. G) Calibration curve indicates the LoD of IgG is 0.125 μg mL‒1. Reproduced with permission from Ref (19). Copyright 2021 American Chemical Society.
4.4. FRET based diagnosis of SARS-CoV-2 antibody from clinical samples
Since Fluorescence-based LFA can be an easy-to-use point of care device for society, the researchers have developed high sensitivity, portable fluorescence lateral flow test strip for early detection of IgM and IgG in human serum (Figure 12C)19. For this purpose, aggregation-induced emission (AIE) dye-loaded nanoparticle-based LFA was designed (Figure 12C)19. Using 172 COVID samples from clinics, the reported data shows that the sensitivity of AIE810NP-based test strips are 78 and 95% in detecting IgM and IgG respectively. Fluorescence-based LFA data are comparable with the ELISA data, which shows that the sensitivities are 85 and 95% for IgM and IgG respectively19.
Reported clinical sample data indicates that bio-conjugated nanomaterials based NSET may pave new roads for SARS-CoV-2 detection in clinics. Although reported data are exciting, in future, researchers need to design robust, reusable and cheap substrates, so that it can be applied in inadequate clinical laboratories.
5. Conclusions and Outlook
In this Account, we introduced how bio-conjugated nanomaterials have been used in the last few months for the possible applications in SARS-CoV-2 diagnostics. We summarized recent efforts to harness the diversity of structures, surface chemistry, and plasmonic properties of nanomaterials for the development of naked-eye colorimetric identification of SERS-CoV-2 viral RNA, antigen, and antibody. The reported clinical sample shows that bio-conjugated nanomaterial-based naked eye assay can be used for the design of inexpensive platforms for virus diagnosis, which is critical for low- and middle-income countries. We have also discussed how bio-conjugated nanomaterial-based, highly sensitive SERS and NSET assays have been designed for the identification of SARS-CoV-2 virus, antigen, and antibody from a clinical sample. As material design for the diagnosis of SARS-CoV-2 is still in its infancy, which started less than two years before, the works presented here are the initial steps towards the new development. Since clear proof of concept exists, after continuing innovations on proper engineering design, the device will achieve clinical testing performance for rapid, accurate, and massive infection diagnosis. One smart way to design such a device is by combining nanomaterial-based devices with CRISPR recognition, which has the capability for unprecedented detection performance.
Despite the considerable achievements made in last year on the design of bio-conjugated nanomaterial for the major fundamental challenge is the development of cost-effective, biocompatible, and environmentally friendly nanomaterials, which have the capability to exhibit high selectivity, sensitivity, accuracy, and precision for real-life infectious disease marker sensing applications. With time it is now clear that slowly COVID-19 is becoming an endemic disease. As a result, researchers need to find out multifunctional material-based devices, which have the capability to test and to differentiate between different coronavirus variants.
To further promote this field to help survive from the current and future pandemics, the researchers need to explore the design of mass-producible synthetic process for bio-functionalized nanomaterials, where size, shape, defects and stability can be controlled. Future theoretical and experimental efforts are necessary for profound understanding on how to control the batch-to-batch design process, so that it will have higher potential for healthcare applications. Ultimately, the performance of the sensor in clinics will vary if the shelf life of bio-conjugated substrates is short and minimization of non-specific binding cannot be controlled in clinics. Further exploration on the design of a robust and stable immobilization technique is highly desired. For achieving best positive outcomes, a collaborative effort with clinical sector to receive their feedback on the novel bio-conjugated material-based sensor performance is very important for improving the accuracy of the diagnostic device and design of no or minimum toxic novel materials for health care industry. As history taught us that crisis could create new potential for discovery, the current pandemic could inspire all scientists to re-shape the future of the material field for complex biological systems.
ACKNOWLEDGEMENTS
Dr. Ray is supported by NSF-RAPID grant # DMR-2030439, NSF-PREM grant # DMR-1826886 and NIH-NIMHD grant # 1U54MD015929-01.
Biographies
Dr. Avijit Pramanik is currently a Senior Research Associate at Jackson State University. His current research involves the design of different anti-viral materials.
Ye Gao is currently a graduate student at Jackson State University. His current research involves the design of the nano-sensor for COVID-19.
Shamily Patibandla is currently a graduate student at Jackson State University. Her current research involves the design of the anti-viral material.
Kalein Gates was a graduate student at Jackson State University. His was working on the design of the novel nanomaterial.
Dr. Paresh C. Ray, Professor of Chemistry at Jackson State University, Jackson, MS, USA. His current research involves the design of nano-sensor and anti-microbial materials.
REFERENCES
- 1.Morens DM; Folkers GK; Fauci AS.; Emerging infections: a perpetual challenge. Lancet Infect Dis 2008, 8, 710–719. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Huremović D Brief History of Pandemics (Pandemics Throughout History). Psychiatry Pandemics 2019, 7–35 [Google Scholar]
- 3.Jones DS History in a Crisis—Lessons for Covid-19. N. Engl. J. Med 2020, 382, 1681–1683. [DOI] [PubMed] [Google Scholar]
- 4.Wang C; Horby PW; Hayden FG; Gao GF A novel coronavirus outbreak of global health concern. Lancet 2020, 395, 470–473 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Gates B Responding to Covid-19—A Once-in-a-Century Pandemic?. N. Engl. J. Med 2020, 382, 1677–1679 [DOI] [PubMed] [Google Scholar]
- 6.Phillips N The Coronavirus Will Become Endemic. Nature 2021, 590, 382–384 [DOI] [PubMed] [Google Scholar]
- 7.Vandenberg O; Martiny D; Rochas O; van Belkum A; Kozlakidis Z Considerations for Diagnostic COVID-19 Tests. Nat. Rev. Microbiol 2021, 19, 171–183 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Valera E,: Jankelow A; Lim J; Kindratenko V; Ganguli A; White K: Kumar J; Bashir R; COVID-19 Point-of-Care Diagnostics: Present and Future, ACS Nano 2021, 15, 5, 7899–7906 [DOI] [PubMed] [Google Scholar]
- 9.Derakhshan MA: Amani A; and Faridi-Majidi R; State-of-the-Art of Nanodiagnostics and Nanotherapeutics against SARS-CoV-2, ACS Appl. Mater. Interfaces 2021, 13, 13, 14816–14843 [DOI] [PubMed] [Google Scholar]
- 10.Zhang L; Lin D; Sun X; Curth U; Drosten C; Sauerhering L; Becker S; Rox K; Hilgenfeld R Crystal Structure of SARS-CoV-2 Main Protease Provides a Basis for Design of Improved α-Ketoamide Inhibitors. Science 2020, 368, 409–412 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Yue H; Huang M; Tian T; Xiong E; Zhou X; Advances in Clustered, Regularly Interspaced Short Palindromic Repeats (CRISPR)-Based Diagnostic Assays Assisted by Micro/ Nanotechnologies, ACS Nano 2021, 15, 5, 7848–7859 [DOI] [PubMed] [Google Scholar]
- 12.Tang Z; Kong N; Zhang X; Liu Y; Hu P; Mou S; Liljeström P; Shi J; Tan W; Kim JS; Cao Y; Langer R; Leong KW; Farokhzad OC; Tao W A Materials-Science Perspective on Tackling COVID-19. Nat. Rev. Mater 2020, 5, 847–860 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Wang C; Li W; Drabek D; Okba NMA; van Haperen R; Osterhaus ADME; van Kuppeveld FJM; Haagmans BL; Grosveld F; Bosch B-J A human monoclonal antibody blocking SARS-CoV-2 infection. Nat. Commun 2020, 11 (1), 1–6 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Huang HY; Fan CH; Li M; Nie HL; Wang FB; Wang H; Wang R; Xia JB; Zheng X; Zuo XL; Huang JX Covid-19: A Call for Physical Scientists and Engineers. ACS Nano 2020, 14, 3747–3754, [DOI] [PubMed] [Google Scholar]
- 15.Ding X; Yin K; Li Z; Lalla RV; Ballesteros E; Sfeir MM; Liu C Ultrasensitive and Visual Detection of SARS-CoV-2 Using All-in-One Dual CRISPR-Cas12a Assay. Nat. Commun 2020, 11, No. 4711. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Moitra P; Alafeef M; Dighe K; Frieman MB; Pan D Selective Naked-Eye Detection of SARS-CoV-2 Mediated by N Gene Targeted Antisense Oligonucleotide Capped Plasmonic Nanoparticles. ACSNano 2020, 14, 7617–7627 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Seo G; Lee G; Kim MJ; Baek SH; Choi M; Ku KB; Lee CS; Jun S; Park D; Kim SJ; Lee JO; Kim BT; Park EC; Kim S Rapid Detection of COVID-19 Causative Virus (SARS-CoV-2) in Human Nasopharyngeal Swab Specimens Using Field-Effect Transistor-Based Biosensor. ACS Nano 2020, 14, 5135–5142 [DOI] [PubMed] [Google Scholar]
- 18.Choi J; Lim J; Shin M; Paek S; Choi J CRISPR-Cas12aBased Nucleic Acid Amplification-Free DNA Biosensor via Au Nanoparticle-Assisted Metal-Enhanced Fluorescence and Colorimetric Analysis. Nano Lett. 2021, 21, 693–699 [DOI] [PubMed] [Google Scholar]
- 19.Chen R; Ren C; Liu M: Ge X; Qu M; Zhou; Liang; Lu; Li; Early Detection of SARS-CoV-2 Seroconversion in Humans with Aggregation-Induced Near-Infrared Emission Nanoparticle-Labeled Lateral Flow Immunoassay, ACS Nano 2021, 15,8996–9004 [DOI] [PubMed] [Google Scholar]
- 20.Yadav S; Sadique MA; Ranjan P; Kumar N; Singhal A; Srivastava AK; Khan R; SERS Based Lateral Flow Immunoassay for Point-of-Care Detection of SARS-CoV-2 in Clinical Samples. ACS Applied Bio Materials 2021, 4 (4) 2974–2995 [DOI] [PubMed] [Google Scholar]
- 21.Ventura B Della, Cennamo M, Minopoli A, Campanine R; Censi BS: Terracciano D; Portella G; Velotta R; Colorimetric Test for Fast Detection of SARS-CoV-2 in Nasal and Throat Swabs. ACS sensors 2020, 5, 3043–3048 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Jiang Y; Hu M; Liu AN; Lin Y: Liu L: Yu B: Zhou X: Pan DW: Detection of SARS-CoV-2 by CRISPR/Cas12a-Enhanced Colorimetry, ACS Sens. 2021, 6, 3, 1086–1093 [DOI] [PubMed] [Google Scholar]
- 23.Pramanik A; Gao Y; Patibandla S; Mitra D; McCandless MG; Fassero LA; Gates K; Tandon R; and Ray PC; Rapid Diagnosis and Effective Inhibition of Corona Virus Using Spike Antibody Attached Gold Nanoparticle, Nanoscale Advances, 2021, 3, 1588–1596 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Pramanik A; Gao Y; Patibandla S; Mitra D; McCandless MG; Fassero LA; Gates K; Tandon R; and Ray PC; Aptamer Conjugated Gold Nanostar-Based Distance-Dependent Nanoparticle Surface Energy Transfer Spectroscopy for Ultrasensitive Detection and Inactivation of Corona Virus, J. Phys. Chem. Lett 2021, 12, 2166–2171. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Alafeef M; Dighe K; Moitra P; Pan D; Rapid, Ultrasensitive, and Quantitative Detection of SARS-CoV-2 Using Antisense Oligonucleotides Directed Electrochemical Biosensor Chip. ACS Nano 2020, 14, 17028–17045 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Zhang Z, Tang Z, Farokhzad N, Chen T, Tao W; Sensitive, Rapid, Low-cost and Multiplexed COVID-19 Monitoring by the Wireless Telemedicine Platform, Matter 2020, 3, 1818–1820 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Huang C; Wen T; Shi F-J; Zeng XY; Jiao Y-J Rapid Detection of IgM Antibodies against the SARS-CoV-2 Virus via Colloidal Gold Nanoparticle-Based Lateral-Flow Assay. ACS Omega 2020, 5, 12550. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Wang C; Yang X; Gu B; Liu H; Zhou Z; Shi L; Cheng X; Wang S Sensitive and Simultaneous Detection of Sars-Cov-2-Specific Igm/Igg Using Lateral Flow Immunoassay Based on Dual-Mode Quantum Dot Nanobeads. Anal. Chem 2020, 92 (23), 15542–15549. [DOI] [PubMed] [Google Scholar]
- 29.Wang Z; Zheng Z; Hu H; Zhou Q; Liu W; Li X; Liu Z; Wang Y; Ma Y A Point-of-Care Selenium Nanoparticle-Based Test for the Combined Detection of Anti-Sars-Cov-2 Igm and Igg in Human Serum and Blood. Lab Chip 2020, 20 (22), 4255–4261 [DOI] [PubMed] [Google Scholar]
- 30.Gorshkov K; Susumu K; Chen J; Xu M; Pradhan M; Zhu W; Hu X; Breger JC; Wolak M; Oh E Quantum Dot-Conjugated Sars-Cov-2 Spike Pseudo-Virions Enable Tracking of Angiotensin Converting Enzyme 2 Binding and Endocytosis. ACS Nano 2020, 14 (9), 12234–12247 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Chen Z; Zhang Z; Zhai X; Li Y; Lin L; Zhao H; Bian L; Li P; Yu L; Wu Y; Lin G Rapid and Sensitive Detection of Anti-Sars-Cov-2 Igg, Using Lanthanide-Doped Nanoparticles-Based Lateral Flow Immunoassay. Anal.Chem 2020, 92 (10), 722 6–7231 [DOI] [PubMed] [Google Scholar]
- 32.Xiong H; Ye X; Li Y; Qi J; Fang X; Kong J; Efficient Microfluidic-Based Air Sampling/Monitoring Platform for Detection of Aerosol SARS-CoV-2 On-site. Analytical Chemistry 2021, 93, 4270–4276 [DOI] [PubMed] [Google Scholar]
- 33.Hristov D: Rijal H; Gomez-Marquez J; and Hamad-Schifferli K: Developing a Paper-Based Antigen Assay to Differentiate between Coronaviruses and SARS-CoV-2 Spike Variants, Anal. Chem 2021, 93, 7825–7832 [DOI] [PubMed] [Google Scholar]
- 34.Yao Z: Zhang Q; Zhu W; Galluzzi M: Zhou W: Li J: Zayats AV: Yu XF: Rapid detection of SARS-CoV-2 viral nucleic acids based on surface enhanced infrared absorption spectroscopy. Nanoscale 2021,13, 10133–10142 [DOI] [PubMed] [Google Scholar]
- 35.Liu HF; Dai EH; Xiao R; Zhou ZH; Zhang ML; Bai ZK; Shao Y; Qi KZ; Tu J; Wang CW; Wang SQ Development of a SERS-Based Lateral Flow Immunoassay for Rapid and Ultra-Sensitive Detection of Anti-SARS-CoV-2 IgM/IgG in Clinical Samples. Sens. Actuators, B 2021, 329, 129196. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Li Y: Peng Z; Holl NJ: Hassan MF: Pappas JM: Wei C: Izadi OH: Wang Y: Dong X: Wang C: Huang YW: Kim DY: Wu C: MXene–Graphene Field-Effect Transistor Sensing of Influenza Virus and SARS-CoV-2. ACS Omega 2021, 6 (10) , 6643–6653 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Pinals RL: Ledesma F; Yang D: Navarro N: Jeong S: Pak JE: Kuo L: Chuang YC: Cheng YW: Sun HY :. Landry MP: Rapid SARS-CoV-2 Spike Protein Detection by Carbon Nanotube-Based Near-Infrared Nanosensors. Nano Letters 2021, 21 , 2272–2280 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Nagueyn NH; Kim S; Lindemann G: Berry V: COVID-19 Spike Protein Induced Phononic Modification in Antibody-Coupled Graphene for Viral Detection Application, ACS Nano 2021, 15, 7, 11743–11752 [DOI] [PubMed] [Google Scholar]
- 39.Liu Y; Wang J; Xiong Q; Hornburg D; Tao W; Farokhzad OC Nano-Bio Interactions in Cancer: From Therapeutics Delivery to Early Detection. Acc. Chem. Res 2021, 54, 291–301 [DOI] [PubMed] [Google Scholar]
- 40.Chen H; Park SG; Choi N: Kwon HJ; Kang T: Lee MK: Choo J; Sensitive Detection of SARS-CoV-2 Using a SERS-Based Aptasensor, ACS Sens. 2021, 6, 2378–2385 [DOI] [PubMed] [Google Scholar]
- 41.Carlomagno C: Bertazioli D: Gualerzi A: Picciolini S; Banfi PI: Lax A; Messina E, Navarro J, Bianchi LA, Caronni F, Marenco S, Monteleone C, Arienti C: Bedoni M: COVID-19 salivary Raman fingerprint: innovative approach for the detection of current and past SARS-CoV-2 infections, Scientific Report, 2021, 11, 4943. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Huang J; Wen J: Zhou M; Ni S; Le W: Chen G: Wei L: Zeng Y; Qi D; Pan M; et al. On-Site Detection of SARS-CoV-2 Antigen by Deep Learning-Based Surface-Enhanced Raman Spectroscopy and Its Biochemical Foundations, Anal. Chem 2021, 93, 9174–9182. [DOI] [PubMed] [Google Scholar]
- 43.Song Y; Song J; Wei X; Huang M; Sun M; Zhu L; Lin B; Shen H; Zhu Z; Yang C Discovery of Aptamers Targeting the Receptor-Binding Domain of the SARS-CoV-2 Spike Glycoprotein. Anal. Chem 2020, 92, 9895–9900 [DOI] [PubMed] [Google Scholar]
- 44.Nellore BPV; Kanchanapally R; Pramanik A; Sinha SS; Chavva SR; Hamme A; Ray PC Aptamer-Conjugated Graphene Oxide Membranes for Highly Efficient Capture and Accurate Identification of Multiple Types of Circulating Tumor Cells Bioconjugate Chem. 2015, 26, 235–242 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Paul AM; Fan Z; Sinha SS; Shi Y; Le L; Bai F; Ray PC Bioconjugated Gold Nanoparticle Based SERS Probe for Ultrasensitive Identification of Mosquito-Borne Viruses Using Raman Fingerprinting. J. Phys. Chem. C 2015, 119, 23669–23675 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Fan Z; Yust B; Nellore BPV; Sinha SS; Kanchanapally R; Crouch RA; Pramanik A; Chavva SR; Sardar D; Ray PC Accurate Identification and Selective Removal of Rotavirus Using a Plasmonic-Magnetic 3D Graphene Oxide Architecture. J. Phys. Chem. Lett 2014, 5, 3216–3221 [DOI] [PubMed] [Google Scholar]
- 47.Cutler JI; Auyeung E; Mirkin CA Spherical nucleic acids. J. Am. Chem. Soc 2012, 134 (3), 1376–91 [DOI] [PubMed] [Google Scholar]
- 48.Rosi NL; Mirkin CA Nanostructures in Biodiagnostics. Chem. Rev 2005, 105, 1547–1562 [DOI] [PubMed] [Google Scholar]
- 49.Farokhzad N: Tao W; Materials chemistry-enabled platforms in detecting sexually transmitted infections: progress towards point-of-care tests. Trends in Chemistry 2021, 3, 589–602 [Google Scholar]
- 50.Mu Q; Jiang G; Chen L; Zhou H; Fourches D; Tropsha A; Yan B Chemical Basis of Interactions between Engineered Nanoparticles and Biological Systems. Chem. Rev 2014, 114, 7740–7781. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Sinha SS; Paul DK; Kanchanapally R; Pramanik A; Chavva SR; Nellore BPV; Jones SJ; Ray PC Long-range Two-photon Scattering Spectroscopy Ruler for Screening Prostate Cancer Cells, Chem. Sci 2015, 6, 2411–2418 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Sinha SS; Jones S; Pramanik A; Ray PC Nanoarchitecture Based SERS for Biomolecular Fingerprinting and Label-Free Disease Markers Diagnosis. Acc. Chem. Res 2016, 49, 2725–2735 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Jones S; Sinha SS; Pramanik A; Ray PC Three-dimensional (3D) plasmonic hot spots for label-free sensing and effective photothermal killing of multiple drug resistant-superbugs. Nanoscale 2016, 8, 18301–18308 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Singh AK; Khan SA; Fan Z; Demeritte T; Senapati D; Kanchanapally R; Ray PC Development of a Long-Range Surface-Enhanced Raman Spectroscopy Ruler. J. Am. Chem. Soc 2012, 134, 8662–8669. [DOI] [PubMed] [Google Scholar]
- 55.Gonçalves MST Fluorescent Labeling of Biomolecules with Organic Probes. Chem. Rev 2009, 109, 190–212 [DOI] [PubMed] [Google Scholar]
- 56.Pramanik A; Patibandla S, Gao Y; Gates K; and Ray PC, Water Triggered Synthesis of Highly Stable and Biocompatible 1D Nanowire, 2D Nanoplatelet, and 3D Nanocube CsPbBr3 Perovskites for Multicolor Two-Photon Cell Imaging, J. Am. Chem. Soc. Au 2021, 1, 1, 53–65. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Ou X; Liu Y; Lei X; Li P; Mi D; Ren L; Guo L; Guo R; Chen T; Hu J; Xiang Z; Mu Z; Chen X; Chen J; Hu K; Jin Q; Wang J; Qian Z Characterization of Spike Glycoprotein of SARS-CoV-2 on Virus Entry and Its Immune Cross-Reactivity with SARS-CoV. Nat. Commun 2020, 11, 1620. [DOI] [PMC free article] [PubMed] [Google Scholar]


