Summary:
Tumor data from the ABCGS5 trial of chemotherapy versus endocrine therapy for premenopausal ER+ breast cancer supports molecular subtyping by Ki-67 immunochemistry as a prognostic marker. But while this tissue was handled uniformly, Ki67 testing overall is unstandardized, complicating clinical utility. Increasing potential biomarkers herald more challenges in biomarker validation.
In this issue of Clinical Cancer Research, Bago-Horvath and colleagues (1) return to the seminal Austrian Breast and Colorectal Cancer Study Group Trial 5 (ABCSG5) to explore whether molecular subtyping by Ki-67 immunohistochemistry may provide prognostic or predictive information about benefit from chemotherapy in premenopausal women with early stage, hormone-receptor positive breast cancer. Until the turn of the last century, these women often received chemotherapy as part of their treatment course. Estrogen, however, had long been recognized as a powerful driver of breast cancer, raising the question of whether many could be spared the toxicities of chemotherapy. ABCSG5, first published in the Journal of Clinical Oncology in 2002, pitted five years of tamoxifen plus three years of goserelin against six cycles of cyclophosphamide, methotrexate and 5-flourouracil, and found superior outcomes for combined endocrine therapy for premenopausal women with stage I and II endocrine-responsive tumors.
ABCSG5 and similar studies underlie the substantial efforts (MINDACT, TAILORx, and many others) to find tools that can identify which patients should receive chemotherapy. Providers have relied on Ki-67 for this purpose in the past; even now, it is now sometimes viewed as a cheaper, simpler risk stratification device than the more widely used Oncotype Dx (a 21-gene transcriptomic assay which incorporates, among others, the gene encoding the Ki-67 protein(2)).
The authors found that while patients in ABCSG5 with higher Ki-67 suffered worse outcomes, they received little benefit from chemotherapy regardless of molecular (i.e., Ki-67) subtype. They conclude that the study demonstrates the clinical relevance of Ki-67, and that based on these results, many patients with low-risk luminal A tumors could safely be treated with endocrine therapy alone.
While it is indisputable that some premenopausal women will derive no benefit from chemotherapy, translating these results to present-day treatment of breast cancer is difficult. No patients in the ABCCSG 5 chemotherapy arm received estrogen-modulating therapy during the follow-up period, while patients in the tamoxifen + ovarian suppression arm received five years of what is now standard endocrine therapy. To confidently assess the predictive power of a biomarker like Ki-67, a comparator arm of patients who received chemotherapy followed by standard endocrine therapy would be needed.
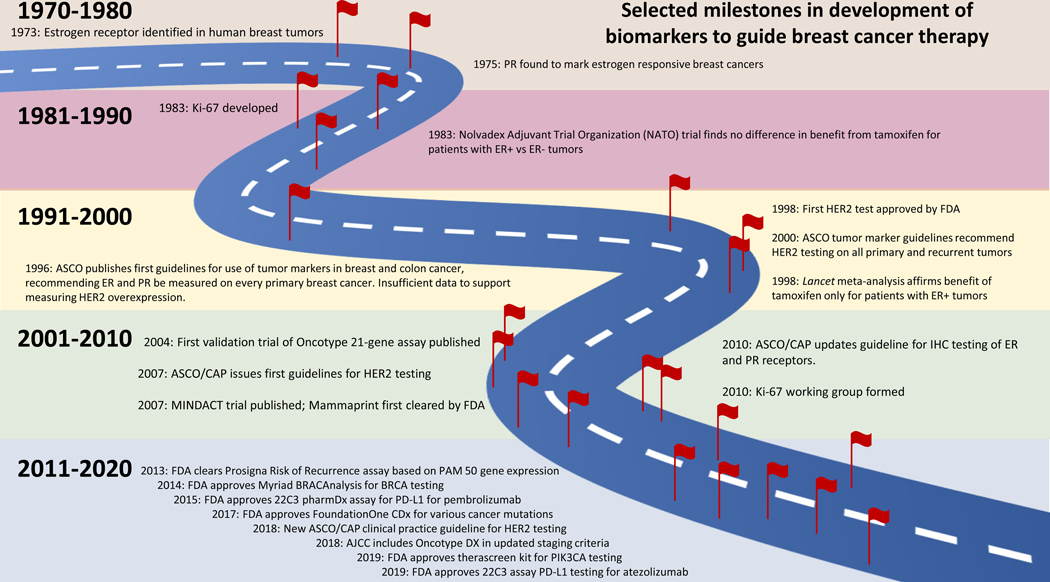
That said, molecular subtyping using the investigators’ centralized tissue handling process and their well-described, uniform methodology for categorizing luminal A and B and HER2+ tumors does support Ki-67 as a robust prognostic marker in this study. In a world with exponentially expanding arrays of complex predictive biomarkers for breast cancer therapies (Fig. 1), it is appealing to think of returning to a simpler and more cost-effective test that can be performed locally to stratify patients. But Ki-67 tells a story that illustrates the challenges in developing reliable biomarkers.
Figure 1. Selected milestones in development of biomarkers to guide breast cancer therapy.
First identified in the early 1980s and named after the German town, Kiel, where the test was developed, Ki-67 is an antigen expressed in cells in all phases of the cell cycle except G0, making it a promising proliferative marker. Numerous studies have demonstrated its prognostic value, but its predictive role in adjuvant, neoadjuvant or endocrine therapy has not been well established (3). In 2010, a multinational group of investigators launched a working group to review Ki-67 and potentially solidify its clinical utility, but to date, no standard method has been adopted. Indeed in 2014, the College of American Pathologists (CAP) reported “a lack of consensus on scoring, definition of low versus high expression, an appropriate cut point for positivity, or which part of the tumor should be scored (e.g., leading edge, hot spots, overall average).” The CAP statement adds that the American Society of Clinical Oncology (ASCO) and the National Comprehensive Cancer Network (NCCN) do not recommend routine testing of breast cancers for Ki-67 expression because of these issues (4).
In a 1991 commentary, Dr. William McGuire, a pioneer in breast cancer research, noted that identification of useful prognostic markers for breast cancer “is becoming more confusing because of a lack of guidelines for investigators to use to study new factors and for reviewers and readers to use to evaluate papers on this topic.” He proposed a set of minimal criteria to consider, including the biological hypothesis, the purpose of the study (pilot vs. definitive), the sample size, biases in patient population, method validation, and optimized cutoff values (5).
Thirty years later, we still struggle with the same challenges Dr. McGuire outlined. Ki-67 is one example, but other biomarkers have similarly complex histories. Investigators labored to refine Human Epidermal Growth Factor Receptor 2 (HER2) testing for nearly 20 years, despite the strong incentive to pair a suitable diagnostic test with a class of therapeutic agents. With the release of the ASCO/CAP 2018 clinical practice guideline, HER2 testing is becoming more consistently reproducible and interpretable, but questions still remain how these results correspond to therapy response. Meanwhile, new medications like trastuzumab deruxtecan, which seems to benefit patients with HER2-low disease, mandate investigations to define additional clinically relevant cut-off points. More recently, the drive to measure programmed death-ligand 1 (PDL1) by various companion diagnostics corresponding to different agents is complicated by substantial knowledge gaps regarding how to determine which patients may benefit from immunotherapy.
Ki-67, HER2 and PD-L1 are immunohistochemistry tests. But of course, numerous other gene-expression and next-generation sequencing (NGS) assays have been validated and are widely used for clinical decision making; many more are under development. Private and academic labs with competing interests develop different assays for the same marker, racing to ensure their place in an ever-evolving catalogue of proprietary and trademarked tests. For such proprietary assays, while similar samples may consistently produce similar results, classic reproducibility studies require different labs to confirm results, which necessitates test transparency. Comparing results across tests is also challenging, e.g., few head-to-head studies examined concordance between NGS platforms, and comparisons between assays like Oncotype Dx and Mammaprint are complicated by problems of normalizing the results that each test generates.
All these issues are drawing the attention of regulatory bodies. Since the implementation of the Medical Device Act of 1976, the U.S. Food and Drug Administration (FDA) has exercised “enforcement discretion” over laboratory developed tests (LDTs), i.e., a hands-off policy unless alerted to problems. But in 2010, citing radical changes in the technological and business landscape for assay development, the FDA announced that it will begin to exercise more active oversight. What this means for the growing array of diagnostic and prognostic tests entering the clinical space remains to be seen.
We are entering an era where access to even more personalized markers to guide therapy selection for breast cancer patients seems within reach. We can imagine highly sensitive and specific biomarker combinations that determine patient status at various time points, allowing de-escalation of care when appropriate, as well as re-escalation when a biomarker alerts us to reactivation of disease.
These truly are hopeful times for our patients, with ever more treatments available, and similarly burgeoning numbers of biomarkers to guide therapy. But reproducibility, dissemination, and proof of clinical utility must lie at the heart of all biomarker development. In the past these have required large trials and coordinated efforts. The future may bring novel approaches to validation, but the essential scientific principles required to confirm clinical utility must remain constant if we are to achieve the potential we envision.
Acknowledgments
This effort was partially supported by the Breast Cancer Research Foundation (N. E. Davidson) and NIH P30CA015704 (all authors).
Footnotes
The authors have no conflicts of interest.
Contributor Information
Natasha B. Hunter, Breast Medical Oncology, University of Washington.
Mark R. Kilgore, Breast and Gynecological Surgical Pathology, University of Washington.
Nancy E. Davidson, Head, Division of Medical Oncology, Department of Medicine, University of Washington; Director and Full Member, Clinical Research Division, Fred Hutchinson Cancer Research Center; Seattle Cancer Care Alliance.
References
- 1.Bago-Horvath Z, Rudas M, Singer CF, Greil R, Balic M, Lax SF, et al. Predictive value of molecular subtypes in premenopausal women with hormone receptor-positive early breast cancer: Results from the ABCSG Trial 5. Clin Can Res 2020. [DOI] [PubMed] [Google Scholar]
- 2.Sparano JA, Gray RJ, Makower DF, Pritchard KI, Albain KS, Hayes DF, et al. Adjuvant Chemotherapy Guided by a 21-Gene Expression Assay in Breast Cancer. N Engl J Med. 2018;379(2):111–21. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Yerushalmi R, Woods R, Ravdin PM, Hayes MM, Gelmon KA. Ki67 in breast cancer: prognostic and predictive potential. Lancet Oncol. 2010;11(2):174–83. [DOI] [PubMed] [Google Scholar]
- 4.Fitzgibbons PL, Dillon DA, Alsabeh R, Berman MA, Hayes DF, Hicks DG, et al. Template for reporting results of biomarker testing of specimens from patients with carcinoma of the breast. Arch Pathol Lab Med. 2014;138(5):595–601. [DOI] [PubMed] [Google Scholar]
- 5.McGuire WL. Breast cancer prognostic factors: evaluation guidelines. J Natl Cancer Inst. 1991;83(3):154–5 [DOI] [PubMed] [Google Scholar]