Abstract
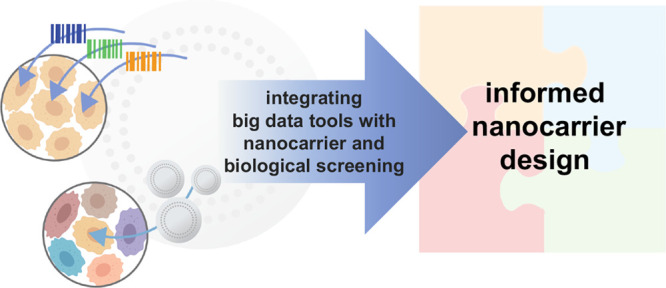
Nanocarriers have significant potential to advance personalized medicine through targeted drug delivery. However, to date, efforts to improve nanoparticle accumulation at target disease sites have largely failed to translate clinically, stemming from an incomplete understanding of nano–bio interactions. While progress has been made to evaluate the effects of specific physical and chemical nanoparticle properties on trafficking and uptake, there is much to be gained from controlling these properties singularly and in combination to determine their interactions with different cell types. We and others have recently begun leveraging library-based nanoparticle screens to study structure–function relationships of lipid- and polymer-based drug delivery systems to guide nanoparticle design. These combinatorial screening efforts are showing promise in leading to the successful identification of critical characteristics that yield improved and specific accumulation at target sites. However, there is a crucial need to equally consider the influence of biological complexity on nanoparticle delivery, particularly in the context of clinical translation. For example, tissue and cellular heterogeneity presents an additional dimension to nanoparticle trafficking, uptake, and accumulation; applying imaging and screening tools as well as bioinformatics may further expand our understanding of how nanoparticles engage with cells and tissues. Given recent advances in the fields of omics and machine learning, there is substantial promise to revolutionize nanocarrier development through the use of integrated screens, harnessing the combinatorial parameter space afforded both by nanoparticle libraries and clinically annotated biological data sets in combination with high throughput in vivo studies.
Keywords: drug delivery, nanocarriers, nanomedicine, DNA barcode, layer-by-layer, combinatorial screen, big data, omics, machine learning
Introduction
Nanomaterial-based drug delivery systems can be used to encapsulate therapeutic cargos, ranging from small molecule compounds to nucleic acids, preventing undesired cargo degradation and increasing circulation time.1−3 Recent advances in lipid nanoparticle (LNP) technology enabled the successful development of both the first FDA-approved siRNA therapy4 and also mRNA delivery platforms to create the first vaccines against COVID-19.5−7 While these LNPs and other nanoparticle systems can be effective at stabilizing and delivering cargo to cells in filtration organs such as the liver when delivered systemically, or locally but nonspecifically delivering to cells at the site of injection, targeted delivery to specific tissues and cells remains an unaddressed challenge.8,9 In fact, targeted nanomedicines often fail to translate to the clinic; of the nanoparticle-based drug delivery strategies that have been successfully approved for clinical use, the overwhelming majority are untargeted liposomal formulations.10 Further, both the translational and clinical successes of nanocarriers are currently much lower compared to other types of pharmaceutical technologies.11
The complexity and heterogeneity of both the biological environment and nanocarrier constructs pose a major translational hurdle, making it prohibitively challenging to deconvolute individual factors contributing to nanocarrier delivery. The field of nanomedicine currently suffers from a stark disconnection between fundamental science and translational advancement.12 However, we will only be able to meaningfully advance this field by bridging the gap between basic and clinical science and allowing each to inform the other. This can be achieved by developing the tools and methodologies that enable the fundamental study of nano–bio interactions in a clinically meaningful way (Figure 1). While protein interactions with nanocarriers are an integral part of delivery success, and protein corona effects have been reviewed extensively,13−16 this Perspective will mainly focus on the use of high throughput and pooled screening approaches to gain a new level of insight into nano–bio interactions.
Figure 1.
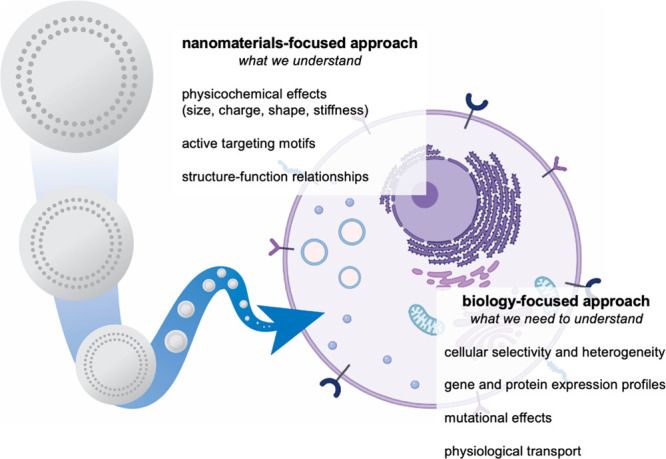
Development of nanomaterials systems for drug delivery is traditionally focused on the study and optimization of materials properties. To overcome biological delivery barriers, we must shift the focus to understanding the interactions of cells and tissues with nanocarriers. We can achieve this through integrated approaches, including the use of nanoparticle libraries, pooled screening, and omics characterization.
In recent years, the advent of high throughput sequencing techniques and the accessibility of these technologies have dramatically advanced the field of clinical genomics, such as through the use of DNA-encoded libraries. And in the area of small molecule drug design, advances in sequencing and barcoding technologies have enabled the development of potent targeted therapies at a rapidly accelerating pace through the generation of large biomedical data sets and use of omics analyses.17−23 These pivotal approaches are beginning to be applied to nanomedicine, providing the opportunity to advance the field in an unprecedented manner (Figure 2).
Figure 2.
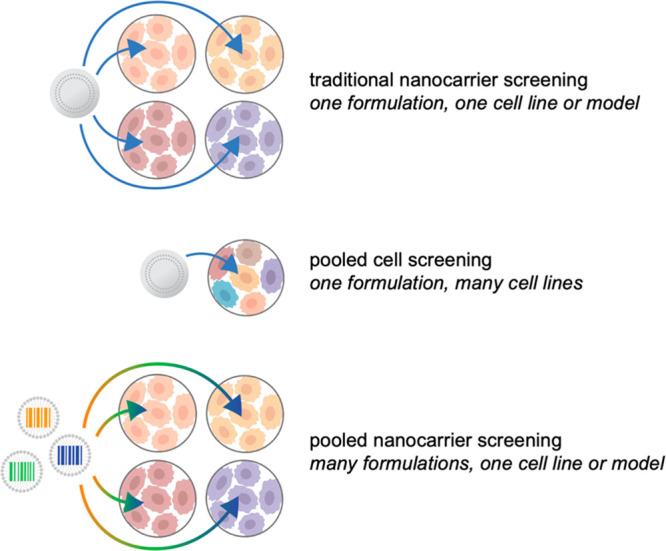
Illustrated examples of nanocarrier screening approaches. Traditionally, candidate formulations are tested iteratively, in one or two models at a time, with a focus on materials property testing. Through the use of pooled cell screening, the same formulations can be screened against hundreds of cell lines simultaneously, providing insight into the biological features mediating successful nanocarrier targeting and uptake. Alternatively, barcoding strategies can be implemented to pool nanocarriers for accelerated biological screening.
To leverage these approaches and maximize their impact on nanocarrier design and performance, barcoding and pooling strategies should ideally be compatible with a wide range of nanocarrier materials and formulations. Developing broadly applicable capabilities would enable the generation of large, nanocarrier-specific data sets to interface with existing biomedical and omics databases. Such integrated data sets could further amplify our understanding of drug delivery through the development and application of predictive machine learning algorithms. If successful, these strategies would pave the way to a holistic understanding of both the materials properties and biological features mediating successful drug delivery, ultimately allowing us, as a field, to realize the full potential of nanomedicine.
Combinatorial Nanocarrier Libraries
Advances in nanocarrier fabrication have led to large, combinatorial libraries of lipid and polymer-based nanocarriers for drug and gene delivery applications, including lipid nanoparticles for RNA delivery, chemically diverse core–shell NPs, and lipocationic polyesters.24−28 More recently, nanocarrier library screens have been coupled with machine learning algorithms to identify materials properties needed for efficient uptake, gene silencing, and biocompatibility. For example, Reineke and co-workers employed random forest algorithms to identify physicochemical properties required for ribonucleoprotein (RNP) delivery and efficient gene editing using a library of 43 chemically diverse copolymers.29 Using this approach, the researchers found that there are three main components contributing to successful transfection and that each of these components is mediated by distinct physicochemical properties of the nanocarrier: editing efficiency (hydrophobicity), RNP uptake (protonation state), and cellular toxicity (polyplex diameter).
The parameter space and statistical power afforded by these libraries have enabled detailed structure–function investigations to probe and decouple nanocarrier features leading to effective and efficient delivery. However, a majority of these screening efforts have focused predominantly on the materials properties leading to successful delivery, testing these combinatorial libraries only in limited in vitro settings. This is due to the large combination of nanocarriers to trial, as both the scale and cost of expanding, or even implementing combinatorial screens in vivo, present a significant barrier to further study. As it is well established that in vitro and in vivo results often do not correlate due to the additional complexities presented by the body, new methods to screen nanocarrier performance in the context of diverse biological settings must be considered.30,31 In addition to biological complexity stemming from both cellular heterogeneity within a given tumor and patient-to-patient heterogeneity, protein and serum interactions (opsonization), detection and clearance by the reticuloendothelial system (RES), and transport barriers (tissue penetration, blood vessel transport) must also be accurately taken into account. This again underscores the critical need to develop tools, such as high throughput pooling strategies that enable simultaneous screening of multiple nanocarriers in a single system, to study structure–function relationships of nanocarriers in vivo.
Pooled Nanoparticle Libraries and DNA Barcodes
Barcoded nanocarriers are one approach to rapidly screen large numbers of drug delivery vehicles in a single system. Nanocarrier barcoding strategies have come a long way from the first reports of barcoded nanoparticles that relied on imaging-based decoding approaches with limited biological applications.32−36 In recent years, advances in sequencing technologies and high throughput screening have inspired the use of nucleic-acid-based barcodes to profile nanocarrier performance and function as an alternative to relying on the nanocarrier itself for barcode identity.
Specifically, DNA barcodes, which leverage both the low limits of detection and almost limitless combinations of nonfunctional nucleic acid sequences, have enabled the pooling and screening of multi-thousand member LNP libraries for in vivo identification of successful lipid formulations with desired properties. These structure–function studies have enabled the identification of lipid structures for tissue-specific targeting without the use of additional targeting ligands.37−40 These approaches have since been leveraged in pooled settings to rapidly evaluate thousands of lipid NP formulations in biological contexts. Machine learning and artificial intelligence are being incorporated to further guide and predict successful nanocarrier design.41 Even though these pooled screens have rapidly accelerated the rate at which successful LNP candidates are identified for their clinical potential, the majority of reported efforts have focused on materials properties. There have been significantly fewer investigations into the biological components contributing to LNP success, a key parameter for overcoming the translational hurdle of patient-to-patient heterogeneity. Given the availability of these LNP data sets and possibility for multidimensional analyses,42 there is significant potential to interface them with existing biological information in an effort to understand the cellular phenotypes that dictate successful LNP uptake (for specific, targeted delivery) and those that result in low LNP uptake (for reduction of nonspecific delivery).
DNA barcodes have also been incorporated into liposomes in conjunction with small molecule therapeutics to evaluate both delivery success and therapeutic efficacy in a pooled fashion. Using this approach, the efficacy of small molecule delivery can be evaluated by coupling therapeutic response (e.g., viability) with DNA barcode count per cell. Specifically, the delivery efficacy of liposomal formulations of three small molecule anticancer agents, gemcitabine, cisplatin, and doxorubicin, has been probed in a pooled fashion. In vitro and in vivo profiling of the murine breast cancer cell line 4T1 revealed that the highest therapeutic efficacy was observed in cells with greatest DNA barcode presence, indicating high liposome uptake.43 While this study provides an elegant means to correlate liposome delivery and therapeutic efficacy, it also highlights drawbacks of nucleic-acid-based barcodes. One such drawback is the need to incorporate nontherapeutic DNA (barcodes) into each tested formulation, adding significant cost and complexity to nanocarrier design. DNA barcodes are also noncovalently incorporated into nanocarrier constructs. Therefore, direct tracking of the nanocarrier is not possible, just the nucleic acid cargo. This makes it challenging to decouple therapeutic efficacy and only provides incomplete information on the fate of the nanocarrier, including tissue accumulation, degradation, and clearance, after dissociation from the barcode.
DNA barcodes have begun to demonstrate the power pooled screening provides to accelerate nanocarrier development. However, because the use of nucleic-acid-based barcodes is limited to nanocarrier formulations compatible with nucleic acid delivery, formulations engineered to deliver different classes of therapeutics, including small molecules and peptide- and protein-based drugs, have thus far been predominantly excluded from such screens. To gain a holistic understanding of structure–function relationships mediating successful drug delivery, these approaches should be expanded to additional drug delivery systems. We urge the field to consider alternative barcoding strategies, especially approaches that allow for direct nanocarrier tracking in vivo and are compatible with wide ranges of drug delivery vehicles.
Layer-by-Layer Assembly: A Modular Approach to Biology-Focused Nanocarrier Design
Layer-by-layer (LbL) assembly can be used to deposit layers of functional polyelectrolytes on a charged surface. We have used this highly versatile approach to create multifunctional, colloidal drug delivery vehicles.44 Beyond therapeutic delivery applications, the modular nature of LbL assembly lends itself to fundamental studies on the effects of individual nanocarrier parameters (Figure 3). For example, we have previously employed LbL coatings to study the impact of core identity for theranostic applications as well as to investigate the benefits of tumor targeting surface coatings.45−47
Figure 3.
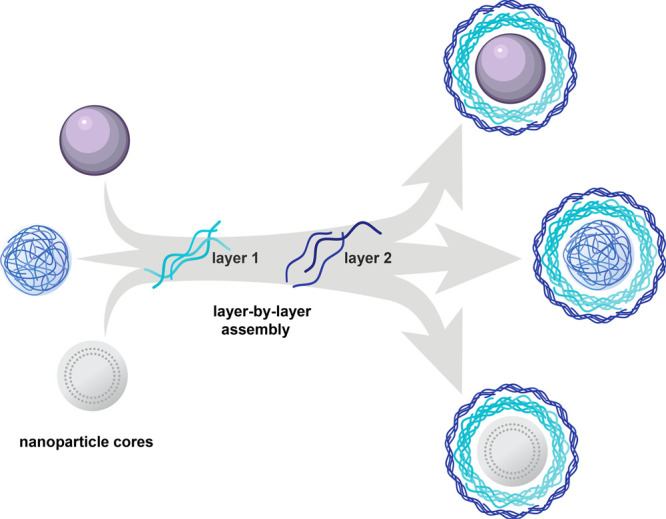
Layer-by-layer assembly can be used to electrostatically coat a wide range of nanoparticle cores with functional polyelectrolytes. The approach enables complete coating of the carrier core and thus decouples the outer layer functionality from physical or chemical characteristics of the core.
We recently pioneered the use of LbL assembly to generate NP libraries to screen new architectures with tumor targeting properties (Figure 4). Using this powerful NP screening approach, we identified novel surface chemistries with unique subcellular trafficking features and exquisite affinity for ovarian cancer cells over non-neoplastic cells.48 Specifically, we took advantage of the modularity provided by LbL assembly to generate a NP library comprising ten different LbL outer layers to investigate the role of NP surface chemistry on ovarian cancer cell interactions. The LbL NPs were prepared on 100 nm carboxylate modified latex cores, containing both natural and synthetic sulfated and carboxylated polyanions as the outermost layer. We probed interactions of these LbL NPs with panels of human ovarian cancer cell lines and noncancerous immune cells using flow cytometry. We found that carboxylated LbL NPs had greater NP–cell association in the tested ovarian cancer cell lines. Notably, while hyaluronic acid (HA) is both carboxylated and provides a well-characterized receptor–ligand interaction with CD44,49−51 HA coated NPs lagged behind NPs coated with carboxylated polyions with no prior established tumor targeting properties, including poly-l-aspartic acid (PLD) and poly-l-glutamic acid (PLE). This carboxylate-dependent trend was not observed in the screened noncancerous cells. Subsequent study by fluorescence microscopy showed marked differences in subcellular trafficking in human ovarian cancer cells (OVCAR8), particularly between PLD and PLE coated NPs. These results indicate the potential for larger NP screens to identify further NP parameters resulting in improved and controlled NP-cell association and uptake.
Figure 4.
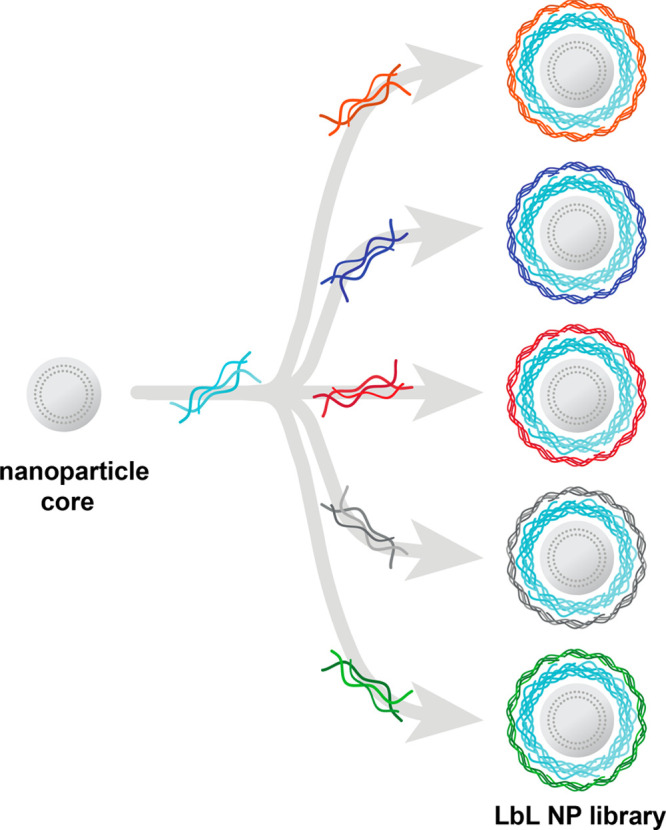
Layer-by-layer assembly enables the generation of nanocarrier libraries wherein one component is varied while all others are kept constant. Illustrated here is an example of a common nanoparticle core and polyelectrolyte layer being separately coated with a range of polyanions to generate a nanocarrier library focused on evaluating surface chemistry effects.
We have found that the in vitro surface chemistry-dependent trends also correlated with their in vivo performance; carboxylated LbL nanoparticles successfully target ovarian cancer cells both in vitro and in vivo. In a metastatic mouse model of ovarian cancer, we observed that carboxylated LbL NPs had significant and improved accumulation in neoplastic tissue over time. Additionally, these formulations were able to target not only the main tumors but also metastases, resulting in greater than 80% NP accumulation in tumor tissue following intraperitoneal (IP) administration. These findings underscore the potential of expanding the capacity of NP screening technologies, and using modular platforms that enable systematic screening of individual NP parameters, to enable the development of new nanocarriers targeted to specific cell populations.
We have since applied the newly identified tumor targeting properties of PLD and PLE-coated LbL NPs. We generated a new class of targeted theranostic LbL nanoparticles consisting of a liposomal core, poly-l-arginine (PLR) and siRNA inner layers, and PLD as the tumor targeting surface coating.52 We observed efficient tumor targeting not only in ovarian cancer mouse models but also in murine models of colorectal and pancreatic cancer. Taking further advantage of the unique trafficking features afforded by our newly discovered LbL NP surface chemistries, we recently employed PLE coatings to create a potent and safe cytokine delivery platform.53
Expanding on the modularity of LbL, we have taken advantage of this charge-based assembly to develop a new electrostatic conjugation strategy that enables simultaneous screening of nanocarrier surface chemistry and active tumor targeting ligand effects on nanocarrier uptake and trafficking. We functionalized anionic nanocarrier surfaces with cationic tumor penetrating peptides while retaining the particle’s negative charge characteristics and stability as well as peptide bioactivity. Through this approach, we investigated the interplay of carboxylated, sulfated, natural, and synthetic polyelectrolyte outer layers with both cyclic and linear tumor penetrating peptides.54
In addition to investigating the role of surface chemistry through a library-based approach, we have also begun to evaluate nanoparticle core parameters for drug delivery applications in an analogous manner. Specifically, we have generated LbL libraries wherein liposomal formulations were generated with ranging cholesterol content to modulate stiffness. These liposomes were then coated with the same polyelectrolyte coatings, composed of PLR and HA.55 This approach decouples the characteristics of the core composition from the nanoparticle surface properties, enabling study of core-specific effects. We observed that the liposomal cores with lower mechanical stiffness accumulated in higher amounts and penetrated more deeply into tumors using a murine model of ovarian cancer.
Colloidal LbL assembly provides a unique breadth of parameter space. One can explore and decouple features across many nanocarrier platforms, including the effects of core composition, surface chemistry, charge, size, and targeting features. The modularity of this approach uniquely enables us to interface LbL NP libraries with high throughput screening approaches to begin understanding the components mediating nanocarrier binding and uptake from a cellular perspective.
Addressing Additional Delivery Barriers
Effectively addressing biological heterogeneity as a translational barrier in the context of nanomedicine becomes even more challenging due to the added complexities of nanocarrier constructs, which are often incompatible with high throughput screens. However, the field of nanomedicine stands to benefit significantly from the advances being made toward understanding biological heterogeneity and how these potentially influence nanocarrier recognition, binding, and uptake. We envision that incorporating both existing and new biological data sets to overcome current hurdles could be achieved via two synergistic approaches: (1) comprehensive in vitro screens to better understand key cellular characteristics leading to therapeutic success of nanocarriers and (2) the development and use of appropriate, relevant models to relate cellular and tissue-specific nanocarrier selectivity with effective circulation and trafficking properties.
In Vitro Screens: Learning from Drug Discovery to Address Biological Heterogeneity
In the field of small molecule drug discovery, there are numerous barriers to the successful translation of new therapeutic agents stemming from biological heterogeneity. These include a lack of reproducible findings and failure to recapitulate efficacy beyond simplistic models, often attributed to limited preclinical screens that fail to capture the variability and complexity of human patients. To address these challenges, there are ongoing efforts to more effectively model biological systems in early stage, in vitro screens, including through the use of organoids and model organisms.56 To better capture patient heterogeneity, options to test compounds for safety and efficacy using “clinical trials in a dish”, composed of patient-specific induced pluripotent stem cells, are also becoming more widespread.57,58 These provide the ability to test drug candidates in a more comprehensive manner prior to first-in-human trials. There are also major, ongoing efforts to profile patient phenotypes and therapeutic response using molecular biology tools. Particularly in the context of cancer biology, extensive genetic and molecular profiling have been carried out to understand tumor origin and progression, as well as therapy response and resistance mechanisms, both in patient samples and established cell lines.23 The results of these efforts are available to the scientific community through data sets such as the Cancer Cell Line Encyclopedia (CCLE) and the Cancer Dependency Map (DepMap).59−61 These, in turn, have enabled in-depth studies into cancer cell metabolism62 and pediatric cancers63 and the potential for repurposing existing drugs for oncology applications.64 Computational tools are also being developed to factor genomic and expression profiles of tumors and cancer cells into choice of preclinical models. As these tools, such as TumorComparer,65 are available as interactive resources via the web and R packages, they are easy to incorporate into existing experimental workflows and can help provide a new level of information with respect to the role of cellular phenotype on nanocarrier performance. Overall, integrating preclinical screening tools and annotated data sets to study nanomedicine, possible through the generation of nanocarrier libraries, barcoding strategies, and modular design elements, could provide new insight into nanocarrier performance in heterogeneous biological settings.
It is additionally possible to augment the level of information obtained from high throughput screens via these data sets. For example, human cancer cell lines have been transfected with DNA barcodes, enabling pooling and multiplexed viability screening in hundreds of cell lines, spanning greater than 20 lineages, simultaneously. This method, referred to as Profiling Relative Inhibition Simultaneously in Mixtures (PRISM), has been used for both in vitro and in vivo applications. Resulting viability data can be combined with correlative genomics databases to elucidate the mechanism of action of unknown compounds.66 PRISM has further enabled the development of a pan-cancer metastasis map (MetMap), enabling large-scale, in vivo study of tumor progression.67 Application of such multiplexed pooled screening approaches to the study of nanocarrier-cell interactions could provide key information to genomic mediators of nanocarrier trafficking.
Realizing the potential of pooled, pan-cancer screens to provide a new level of insight, as well as the necessary statistical power, into understanding the mechanisms mediating drug delivery, we designed and implemented a competition assay to screen nanocarrier–cell interactions of a 35-member nanoparticle library across 488 pooled cancer cell lines from the PRISM platform.68 Using fluorescence-activated cell sorting (FACS), we generated interaction profiles across nanocarrier formulations of varying size, core composition, and surface chemistry. We found that core identity was the predominant materials parameter driving cell interactions. By coupling our findings with existing omics data sets and machine learning algorithms, we identified expression of a highly interconnected network of genes, or biomarkers, to play a significant role in nanocarrier–cell interactions. These genes provide a roadmap, and also indicate the complexity, of the biological components driving nanocarrier recognition and uptake. These findings suggest that gene expression profiles, or cellular phenotypes more generally, need to be taken into consideration when engineering nanocarriers for targeted delivery to a specific cell population.
Pooled, high throughput screens have the potential to dramatically advance nanomedicine. However, to fully harness the potential of omics and big data, we must also develop the toolsets to enable meaningful study of a wide range of nanomaterials in these settings. Moreover, while efforts have been made to comprehensively address patient heterogeneity in vitro, we must also consider the biological barriers stemming from circulation, transport, and immune system clearance.
Addressing Transport Barriers: The Right Tools for the Job
It has been established that there is a lack of appropriate preclinical models to fully evaluate the therapeutic value of nanomedicine; models should be able to effectively represent human complexity and disease as well as provide predictive response to tested treatments.69 As there is currently a disconnect between in vitro and in vivo nanocarrier performance due to limited mechanistic insight into the origin of these failures, there is also a need to develop and implement physiologically representative models that enable translation of promising candidates identified from in vitro screens. We envision this can be accomplished through the use of models with increasing complexity, including complex cell cocultures (e.g., organoids, organ-on-a-chip) and mouse models. Successful application of these models would enable the decoupling and identification of both the physicochemical nanocarrier properties and physiological features mediating successful drug delivery.
There are significant limitations to traditional, two-dimensional cell screens, which are often the starting place for evaluating biological functionality of newly designed nanocarriers in preclinical settings. To circumvent these drawbacks, three-dimensional multicellular constructs, referred to as organoids, are being developed to better mimic in vivo tissue.70 A recent editorial highlights the advances that have been made to develop such multicellular tools to further study development, pathology, and physiology.71 In addition to serving as tissue mimics, organoids can be used to recapitulate disease physiology. For example, in the context of cancer medicine, organoids are routinely generated from patient samples (tumor spheroids) for the study of tumor progression and therapy response.72 Methods have also been developed for organoid-based high throughput drug screens, and through omics profiling relationships between therapy response and cell phenotype can be drawn.73 As the immune system plays a key role in tumor development and response, there are also efforts to develop complex tumor organoids capable of sustaining tumor-associated immune cell populations in addition to tumor cells.74
Organoids present a unique opportunity to study nanocarrier trafficking in a three-dimensional in vitro setting. In the simplest system, organoids composed of a single cell type could be used to study nanocarrier penetration and uptake. Moving into multicellular tissue mimics, nanocarrier trafficking could be investigated through the use of flow cytometry- and imaging-based tools, useful for studying trafficking of nanocarriers to various cell populations in addition to evaluating therapeutic response of various nanocarrier cargo needing to be trafficked to multiple cell types or intracellular components. As organoid technologies progress, they are positioned to become a very meaningful and also accessible platform for preclinical nanocarrier screening, particularly in the context of the immune system.
While organoids and spheroids are able to recapitulate some of the cellular heterogeneity and spatial organization observed in tissue, their development has also been faced with low throughput production, high variability, and size restrictions stemming from poor nutrient supply. Microfluidics and other microfabrication techniques have been implemented to address these, particularly in the context of nutrient delivery and spatially controlled structures, both of which are also critical parameters for nanocarrier delivery.75 Microfabricated tissue mimics, such as organs-on-a-chip, present another promising avenue for evaluating both nanocarrier properties and cellular characteristics needed for successful drug delivery in the context of biological transport barriers, including blood vessel endothelial barriers, mucus membranes, and extracellular matrices.76,77 As the ultimate goal is to engineer systems capable of accurately predicting patient response to tested therapies in a controlled setting, efforts to develop three-dimensional culture systems that more completely model the complexity of the human body are ongoing. These include platforms that sustain immune cell populations and enable comparisons of healthy and diseased tissues.78−81 As we seek to understand how cellular and tissue-specific characteristics affect drug delivery success, application of these tools will become key to shedding light on the interplay of nanocarrier-cell interactions in the context of physiologic transport barriers.
Mouse models remain one of the most accessible preclinical animal models to evaluate efficacy of newly engineered drug delivery systems. However, their use is often criticized due to their simplicity and the failure to recapitulate immune responses of humans in mice. Moreover, modeling disease, such as tumors generated from human cancer cells, requires the use of immunocompromised models.82 As the immune system is responsible for clearing the vast majority of administered nanoparticles,83,84 models without intact immune systems cannot adequately predict the success of nanocarriers. On the other hand, immunodeficient mice can be used to recreate the human immune system through implantation of human stem and lymphoid cells. These models are particularly effective in studying autoimmune diseases, infectious diseases, as well as cancers,85,86 and have been used to evaluate nanoparticle immunogenicity and efficacy.87−89 While humanized models are well positioned to provide a lot of valuable information surrounding nanocarrier–immune cell interactions, for both targeted delivery and immune evasion applications, it is important to also note their limitations. This includes the inability to fully recapitulate the complete functional immune response observed in humans, which stems from incompatibilities between the murine immune system and human cells.90 Moreover, in the context of cancer research, it is important to bear in mind that the human immune systems of humanized mouse models are typically of a different origin, or allogeneic, from implanted tumor cells, which can affect communication between immune and tumor cells as well as tumor progression.
Alternatively, genetically engineered mouse models (GEMMs) provide a different and perhaps complementary means to study nanocarrier performance in a fully immunocompetent setting. GEMMs have induced mutations, introduced through biological (e.g., transgenes, targeted mutations) or chemical means, that lead to disease. The use of GEMMs enables the study of relationships between mutations and disease phenotype as well as human disease modeling, including spontaneous tumor formation and progression. Particularly in the context of cancer research, large numbers of models that mimic the histopathological and molecular characteristics of human disease have been developed and tested.91,92 We believe that utilizing GEMMs to evaluate nanocarrier performance in vivo would provide valuable insight into nanocarrier targeting, circulation, and immune evasion, particularly in the context of disease progression. One should consider the use of these models to study nanocarriers in both spatial and temporal contexts (i.e., how does nanocarrier performance vary in early versus late-stage disease?). However, as the immune makeup of GEMMs is murine, it is important to keep in mind that these models do not recapitulate human immune cells. Therefore, it is important to consider a range of models for comprehensive nanocarrier evaluation.
We recognize that the development and testing of specialized mouse models often falls outside the capabilities of nanotechnology-focused research groups. Therefore, we encourage researchers to seek out collaborations with biological and clinical researchers to leverage their expertise in disease-focused, preclinical testing. Such interdisciplinary approaches also open the door to new, mutually beneficial collaborative relationships by providing biologists and clinicians with the tools afforded by nanomedicine to tackle previously unmet challenges.
Outlook and Future Directions
The development of nanocarriers for therapeutic delivery applications has already led to significant clinical advancements by protecting encapsulated cargo from degradation, extending circulation time, and mitigating toxic side effects. However, for targeted drug delivery, translational successes have been more modest. This stems from an incomplete understanding of the biological features mediating successful nanocarrier trafficking, particularly challenging considering the patient and disease heterogeneity. In recent years, high throughput screening approaches have started to be applied to advance nanomedicine. This includes the use of DNA barcodes to pool and rapidly assay lipid nanoparticles in vivo, electrostatic layer-by-layer assembly to create modular nanoparticle libraries that enable the decoupling and identification of individual parameters modulating delivery success, and the combination of nanoparticle libraries with pooled cell assays to identify biomarkers predictive of nanoparticle uptake. Given the success of these early efforts, there is significant potential to develop further toolsets and methodologies that enable high throughput interrogation of broad ranges of nanomaterials and provide key insight into the biological features mediating selective and successful drug targeting. We believe that developing additional nanoparticle barcoding approaches that go beyond the constraints of DNA will yield pooled screening methodologies that significantly advance our understanding of nanocarrier performance. The field should prioritize strategies that enable direct tracking of nanocarrier fate in multiple biological contexts and allow for multiplexing with multiple classes of therapeutics.
As omics and machine learning tools are rapidly being implemented to gain mechanistic insight into patient variability and disease progression from a molecular biology perspective, nanomedicine also stands to benefit from applying these tools to understand drug delivery failures and successes. There is a plethora of biological data readily available for the scientific community to apply to the study of nanomedicine. Specifically, we believe that these data sets should be applied to account for discrepancies in nanocarrier delivery efficacy arising from biological heterogeneity. For example, investigators should take into account heterogeneity of gene and protein expression levels within targeted cells and how those might relate to successful nanocarrier accumulation. Moreover, these studies should be carried out in an unbiased way and examine a range of particle surface chemistries and materials systems while moving beyond the traditional cell surface receptors often leveraged in current targeted drug delivery platforms. In order for the field to leverage this level of biological information, we must first develop the methodology to interface nanocarrier performance metrics with cellular, tissue, and organism-level characteristics. We envision this can be accomplished through the implementation of machine learning algorithms and predictive models that accurately account for nanocarrier performance within this biological context. Additionally, as new tools, such as spatial omics, become more accessible to the general research community, we should begin to leverage these as a means to profile nanocarrier accumulation in target tissues, relating nanocarrier penetration with transcriptional and expression level changes on the tissue level as they relate to targeted microenvironments.
Coupling new screening and big data approaches to advance our understanding of the nano–bio interface will also require the continued development and use of relevant preclinical models, including multicellular organoids and mouse models capable of mimicking human immune responses. Ultimately, by combining these approaches in a comprehensive manner, we will be able to holistically identify the key parameters mediating successful cell targeting and uptake, from both materials and biology perspectives (Figure 5). As many of these tools and models exist at the interface of biology, engineering, and medicine, we believe there are also boundless possibilities for new cross-field collaborations to tackle the challenges facing targeted drug delivery in an entirely new manner.
Figure 5.
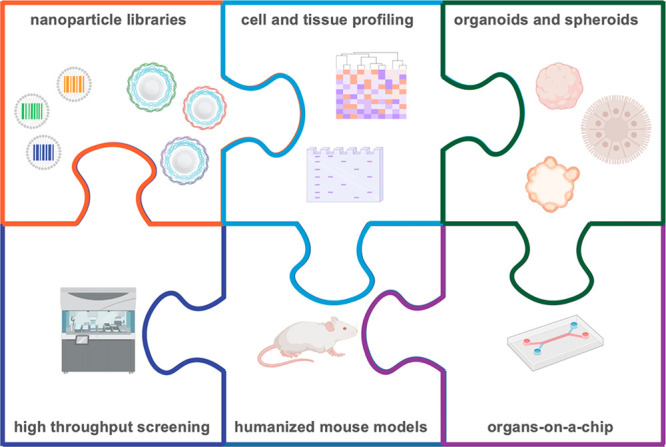
Informed nanocarrier design is possible through the use of nanoparticle barcodes, pools, and libraries, which can be interfaced with high throughput screening, biological profiling, and physiological in vitro and in vivo models. Through these integrated approaches, we can gain a more thorough understanding of the biological characteristics necessary for successful delivery of therapeutics.
Acknowledgments
Biorender was used in part for the figure design.
This work was supported by a grant from the Koch Institute’s Marble Center for Cancer Nanomedicine. N.B. also gratefully acknowledges funding support from the NIH-NCI (K99CA255844).
The authors declare no competing financial interest.
References
- Shi J. J.; Kantoff P. W.; Wooster R.; Farokhzad O. C. Cancer nanomedicine: progress, challenges and opportunities. Nat. Rev. Cancer 2017, 17 (1), 20–37. 10.1038/nrc.2016.108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mitchell M. J.; Billingsley M. M.; Haley R. M.; Wechsler M. E.; Peppas N. A.; Langer R. Engineering precision nanoparticles for drug delivery. Nat. Rev. Drug Discovery 2021, 20, 101–124. 10.1038/s41573-020-0090-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tran S.; DeGiovanni P. J.; Piel B.; Rai P. Cancer nanomedicine: a review of recent success in drug delivery. Clin. Transl. Med. 2017, 6, e44. 10.1186/s40169-017-0175-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Akinc A.; Maier M. A.; Manoharan M.; Fitzgerald K.; Jayaraman M.; Barros S.; Ansell S.; Du X. Y.; Hope M. J.; Madden T. D.; Mui B. L.; Semple S. C.; Tam Y. K.; Ciufolini M.; Witzigmann D.; Kulkarni J. A.; van der Meel R.; Cullis P. R. The Onpattro story and the clinical translation of nanomedicines containing nucleic acid-based drugs. Nat. Nanotechnol. 2019, 14 (12), 1084–1087. 10.1038/s41565-019-0591-y. [DOI] [PubMed] [Google Scholar]
- Chung Y. H.; Beiss V.; Fiering S. N.; Steinmetz N. F. COVID-19 Vaccine Frontrunners and Their Nanotechnology Design. ACS Nano 2020, 14 (10), 12522–12537. 10.1021/acsnano.0c07197. [DOI] [PubMed] [Google Scholar]
- Walsh E. E.; Frenck R. W.; Falsey A. R.; Kitchin N.; Absalon J.; Gurtman A.; Lockhart S.; Neuzil K.; Mulligan M. J.; Bailey R.; Swanson K. A.; Li P.; Koury K.; Kalina W.; Cooper D.; Fontes-Garfias C.; Shi P. Y.; Tureci O.; Tompkins K. R.; Lyke K. E.; Raabe V.; Dormitzer P. R.; Jansen K. U.; Sahin U.; Gruber W. C. Safety and Immunogenicity of Two RNA-Based Covid-19 Vaccine Candidates. N. Engl. J. Med. 2020, 383 (25), 2439–2450. 10.1056/NEJMoa2027906. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Baden L. R.; El Sahly H. M.; Essink B.; Kotloff K.; Frey S.; Novak R.; Diemert D.; Spector S. A.; Rouphael N.; Creech C. B.; McGettigan J.; Khetan S.; Segall N.; Solis J.; Brosz A.; Fierro C.; Schwartz H.; Neuzil K.; Corey L.; Gilbert P.; Janes H.; Follmann D.; Marovich M.; Mascola J.; Polakowski L.; Ledgerwood J.; Graham B. S.; Bennett H.; Pajon R.; Knightly C.; Leav B.; Deng W. P.; Zhou H. H.; Han S.; Ivarsson M.; Miller J.; Zaks T. Efficacy and Safety of the mRNA-1273 SARS-CoV-2 Vaccine. N. Engl. J. Med. 2021, 384 (5), 403–416. 10.1056/NEJMoa2035389. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wilhelm S.; Tavares A. J.; Dai Q.; Ohta S.; Audet J.; Dvorak H. F.; Chan W. C. W. Analysis of nanoparticle delivery to tumours. Nat. Rev. Mater. 2016, 1 (5), 16014. 10.1038/natrevmats.2016.14. [DOI] [Google Scholar]
- Cheng Y. H.; He C. L.; Riviere J. E.; Monteiro-Riviere N. A.; Lin Z. M. Meta-Analysis of Nanoparticle Delivery to Tumors Using a Physiologically Based Pharmacokinetic Modeling and Simulation Approach. ACS Nano 2020, 14 (3), 3075–3095. 10.1021/acsnano.9b08142. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Anselmo A. C.; Mitragotri S. Nanoparticles in the clinic: An update. Bioeng. Transl. Med. 2019, 4, e10143 10.1002/btm2.10143. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhong H.; Chan G.; Hu Y. J.; Hu H.; Ouyang D. F. A Comprehensive Map of FDA-Approved Pharmaceutical Products. Pharmaceutics 2018, 10 (4), 263. 10.3390/pharmaceutics10040263. [DOI] [PMC free article] [PubMed] [Google Scholar]
- The two directions of cancer nanomedicine. Nat. Nanotechnol. 2019, 14 ( (12), ), 1083–1083. 10.1038/s41565-019-0597-5 [DOI] [PubMed] [Google Scholar]
- Corbo C.; Molinaro R.; Parodi A.; Furman N. E. T.; Salvatore F.; Tasciotti E. The impact of nanoparticle protein corona on cytotoxicity, immunotoxicity and target drug delivery. Nanomedicine 2016, 11 (1), 81–100. 10.2217/nnm.15.188. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Caracciolo G.; Farokhzad O. C.; Mahmoudi M. Biological Identity of Nanoparticles In Vivo: Clinical Implications of the Protein Corona. Trends Biotechnol. 2017, 35 (3), 257–264. 10.1016/j.tibtech.2016.08.011. [DOI] [PubMed] [Google Scholar]
- Donahue N. D.; Acar H.; Wilhelm S. Concepts of nanoparticle cellular uptake, intracellular trafficking, and kinetics in nanomedicine. Adv. Drug Delivery Rev. 2019, 143, 68–96. 10.1016/j.addr.2019.04.008. [DOI] [PubMed] [Google Scholar]
- Rampado R.; Crotti S.; Caliceti P.; Pucciarelli S.; Agostini M. Recent Advances in Understanding the Protein Corona of Nanoparticles and in the Formulation of “Stealthy” Nanomaterials. Front. Bioeng. Biotechnol. 2020, 8, 166. 10.3389/fbioe.2020.00166. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brenner S.; Lerner R. A. Encoded Combinatorial Chemistry. Proc. Natl. Acad. Sci. U. S. A. 1992, 89 (12), 5381–5383. 10.1073/pnas.89.12.5381. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Song M.; Hwang G. T. DNA-Encoded Library Screening as Core Platform Technology in Drug Discovery: Its Synthetic Method Development and Applications in DEL Synthesis. J. Med. Chem. 2020, 63 (13), 6578–6599. 10.1021/acs.jmedchem.9b01782. [DOI] [PubMed] [Google Scholar]
- Reiher C. A.; Schuman D. P.; Simmons N.; Wolkenberg S. E. Trends in Hit-to-Lead Optimization Following DNA-Encoded Library Screens. ACS Med. Chem. Lett. 2021, 12 (3), 343–350. 10.1021/acsmedchemlett.0c00615. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen B.; Garmire L.; Calvisi D. F.; Chua M. S.; Kelley R. K.; Chen X. Harnessing big ‘omics’ data and AI for drug discovery in hepatocellular carcinoma. Nat. Rev. Gastroenterol. Hepatol. 2020, 17 (4), 238–251. 10.1038/s41575-019-0240-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Matthews H.; Hanison J.; Nirmalan N. Omics“-Informed Drug and Biomarker Discovery: Opportunities, Challenges and Future Perspectives. Proteomes 2016, 4 (3), 28. 10.3390/proteomes4030028. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Das T.; Andrieux G.; Ahmed M.; Chakraborty S. Integration of Online Omics-Data Resources for Cancer Research. Front. Genet. 2020, 11, 578345. 10.3389/fgene.2020.578345. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Conesa A.; Beck S. Making multi-omics data accessible to researchers. Sci. Data 2019, 6, 251. 10.1038/s41597-019-0258-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen D. L.; Love K. T.; Chen Y.; Eltoukhy A. A.; Kastrup C.; Sahay G.; Jeon A.; Dong Y. Z.; Whitehead K. A.; Anderson D. G. Rapid Discovery of Potent siRNA-Containing Lipid Nanoparticles Enabled by Controlled Microfluidic Formulation. J. Am. Chem. Soc. 2012, 134 (16), 6948–6951. 10.1021/ja301621z. [DOI] [PubMed] [Google Scholar]
- Hajj K. A.; Whitehead K. A. Tools for translation: non-viral materials for therapeutic mRNA delivery. Nat. Rev. Mater. 2017, 2 (10), 17056. 10.1038/natrevmats.2017.56. [DOI] [Google Scholar]
- Kauffman K. J.; Dorkin J. R.; Yang J. H.; Heartlein M. W.; DeRosa F.; Mir F. F.; Fenton O. S.; Anderson D. G. Optimization of Lipid Nanoparticle Formulations for mRNA Delivery in Vivo with Fractional Factorial and Definitive Screening Designs. Nano Lett. 2015, 15 (11), 7300–7306. 10.1021/acs.nanolett.5b02497. [DOI] [PubMed] [Google Scholar]
- Siegwart D. J.; Whitehead K. A.; Nuhn L.; Sahay G.; Cheng H.; Jiang S.; Ma M. L.; Lytton-Jean A.; Vegas A.; Fenton P.; Levins C. G.; Love K. T.; Lee H.; Cortez C.; Collins S. P.; Li Y. F.; Jang J.; Querbes W.; Zurenko C.; Novobrantseva T.; Langer R.; Anderson D. G. Combinatorial synthesis of chemically diverse core-shell nanoparticles for intracellular delivery. Proc. Natl. Acad. Sci. U. S. A. 2011, 108 (32), 12996–13001. 10.1073/pnas.1106379108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hao J.; Kos P.; Zhou K. J.; Miller J. B.; Xue L.; Yan Y. F.; Xiong H.; Elkassih S.; Siegwart D. J. Rapid Synthesis of a Lipocationic Polyester Library via Ring-Opening Polymerization of Functional Valerolactones for Efficacious siRNA Delivery. J. Am. Chem. Soc. 2015, 137 (29), 9206–9209. 10.1021/jacs.5b03429. [DOI] [PubMed] [Google Scholar]
- Kumar R.; Le N.; Tan Z.; Brown M. E.; Jiang S.; Reineke T. M. Efficient Polymer-Mediated Delivery of Gene-Editing Ribonucleoprotein Payloads through Combinatorial Design, Parallelized Experimentation, and Machine Learning. ACS Nano 2020, 14 (12), 17626–17639. 10.1021/acsnano.0c08549. [DOI] [PubMed] [Google Scholar]
- Whitehead K. A.; Matthews J.; Chang P. H.; Niroui F.; Dorkin J. R.; Severgnini M.; Anderson D. G. In Vitro-In Vivo Translation of Lipid Nanoparticles for Hepatocellular siRNA Delivery. ACS Nano 2012, 6 (8), 6922–6929. 10.1021/nn301922x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jain P.; Pawar R. S.; Pandey R. S.; Madan J.; Pawar S.; Lakshmi P. K.; Sudheesh M. S. In-vitro in-vivo correlation (IVIVC) in nanomedicine: Is protein corona the missing link?. Biotechnol. Adv. 2017, 35 (7), 889–904. 10.1016/j.biotechadv.2017.08.003. [DOI] [PubMed] [Google Scholar]
- Bruchez M.; Moronne M.; Gin P.; Weiss S.; Alivisatos A. P. Semiconductor nanocrystals as fluorescent biological labels. Science 1998, 281 (5385), 2013–2016. 10.1126/science.281.5385.2013. [DOI] [PubMed] [Google Scholar]
- Chan W. C. W.; Nie S. M. Quantum dot bioconjugates for ultrasensitive nonisotopic detection. Science 1998, 281 (5385), 2016–2018. 10.1126/science.281.5385.2016. [DOI] [PubMed] [Google Scholar]
- Mitchell G. P.; Mirkin C. A.; Letsinger R. L. Programmed assembly of DNA functionalized quantum dots. J. Am. Chem. Soc. 1999, 121 (35), 8122–8123. 10.1021/ja991662v. [DOI] [Google Scholar]
- Han M. Y.; Gao X. H.; Su J. Z.; Nie S. Quantum-dot-tagged microbeads for multiplexed optical coding of biomolecules. Nat. Biotechnol. 2001, 19 (7), 631–635. 10.1038/90228. [DOI] [PubMed] [Google Scholar]
- Nicewarner-Pena S. R.; Freeman R. G.; Reiss B. D.; He L.; Pena D. J.; Walton I. D.; Cromer R.; Keating C. D.; Natan M. J. Submicrometer metallic barcodes. Science 2001, 294 (5540), 137–141. 10.1126/science.294.5540.137. [DOI] [PubMed] [Google Scholar]
- Paunovska K.; Sago C. D.; Monaco C. M.; Hudson W. H.; Castro M. G.; Rudoltz T. G.; Kalathoor S.; Vanover D. A.; Santangelo P. J.; Ahmed R.; Bryksin A. V.; Dahlman J. E. A Direct Comparison of in Vitro and in Vivo Nucleic Acid Delivery Mediated by Hundreds of Nanoparticles Reveals a Weak Correlation. Nano Lett. 2018, 18 (3), 2148–2157. 10.1021/acs.nanolett.8b00432. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dahlman J. E.; Kauffman K. J.; Xing Y. P.; Shaw T. E.; Mir F. F.; Dlott C. C.; Langer R.; Anderson D. G.; Wang E. T. Barcoded nanoparticles for high throughput in vivo discovery of targeted therapeutics. Proc. Natl. Acad. Sci. U. S. A. 2017, 114 (8), 2060–2065. 10.1073/pnas.1620874114. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gan Z. B.; Lokugamage M. P.; Hatit M. Z. C.; Loughrey D.; Paunovska K.; Sato M.; Cristian A.; Dahlman J. E. Nanoparticles containing constrained phospholipids deliver mRNA to liver immune cells in vivo without targeting ligands. Bioeng. Transl. Med. 2020, 5 (3), e10161. 10.1002/btm2.10161. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lokugamage M. P.; Sago C. D.; Gan Z. B.; Krupczak B. R.; Dahlman J. E. Constrained Nanoparticles Deliver siRNA and sgRNA to T Cells In Vivo without Targeting Ligands. Adv. Mater. 2019, 31 (41), 1902251. 10.1002/adma.201902251. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Adir O.; Poley M.; Chen G.; Froim S.; Krinsky N.; Shklover J.; Shainsky-Roitman J.; Lammers T.; Schroeder A. Integrating Artificial Intelligence and Nanotechnology for Precision Cancer Medicine. Adv. Mater. 2020, 32 (13), 1901989. 10.1002/adma.201901989. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Paunovska K.; Loughrey D.; Sago C. D.; Langer R.; Dahlman J. E. Using Large Datasets to Understand Nanotechnology. Adv. Mater. 2019, 31 (43), 1902798. 10.1002/adma.201902798. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yaari Z.; da Silva D.; Zinger A.; Goldman E.; Kajal A.; Tshuva R.; Barak E.; Dahan N.; Hershkovitz D.; Goldfeder M.; Roitman J. S.; Schroeder A. Theranostic barcoded nanoparticles for personalized cancer medicine. Nat. Commun. 2016, 7, 13325. 10.1038/ncomms13325. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Deng Z. J.; Morton S. W.; Ben-Akiva E.; Dreaden E. C.; Shopsowitz K. E.; Hammond P. T. Layer-by-Layer Nanoparticles for Systemic Codelivery of an Anticancer Drug and siRNA for Potential Triple-Negative Breast Cancer Treatment. ACS Nano 2013, 7 (11), 9571–9584. 10.1021/nn4047925. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Poon Z.; Lee J. B.; Morton S. W.; Hammond P. T. Controlling in Vivo Stability and Biodistribution in Electrostatically Assembled Nanoparticles for Systemic Delivery. Nano Lett. 2011, 11 (5), 2096–2103. 10.1021/nl200636r. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Morton S. W.; Poon Z. Y.; Hammond P. T. The architecture and biological performance of drug-loaded LbL nanoparticles. Biomaterials 2013, 34 (21), 5328–5335. 10.1016/j.biomaterials.2013.03.059. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dang X. N.; Gu L.; Qi J. F.; Correa S.; Zhang G.; Belcher A. M.; Hammond P. T. Layer-by-layer assembled fluorescent probes in the second near-infrared window for systemic delivery and detection of ovarian cancer. Proc. Natl. Acad. Sci. U. S. A. 2016, 113 (19), 5179–5184. 10.1073/pnas.1521175113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Correa S.; Boehnke N.; Barberio A. E.; Deiss-Yehiely E.; Shi A.; Oberlton B.; Smith S. G.; Zervantonakis I.; Dreaden E. C.; Hammond P. T. Tuning Nanoparticle Interactions with Ovarian Cancer through Layer-by-Layer Modification of Surface Chemistry. ACS Nano 2020, 14 (2), 2224–2237. 10.1021/acsnano.9b09213. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang S.; Balch C.; Chan M. W.; Lai H. C.; Matei D.; Schilder J. M.; Yan P. S.; Huang T. H. M.; Nephew K. P. Identification and characterization of ovarian cancer-initiating cells from primary human tumors. Cancer Res. 2008, 68 (11), 4311–4320. 10.1158/0008-5472.CAN-08-0364. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ricardo S.; Vieira A. F.; Gerhard R.; Leitao D.; Pinto R.; Cameselle-Teijeiro J. F.; Milanezi F.; Schmitt F.; Paredes J. Breast cancer stem cell markers CD44, CD24 and ALDH1: expression distribution within intrinsic molecular subtype. J. Clin. Pathol. 2011, 64 (11), 937–946. 10.1136/jcp.2011.090456. [DOI] [PubMed] [Google Scholar]
- Choi K. Y.; Yoon H. Y.; Kim J. H.; Bae S. M.; Park R. W.; Kang Y. M.; Kim I. S.; Kwon I. C.; Choi K.; Jeong S. Y.; Kim K.; Park J. H. Smart Nanocarrier Based on PEGylated Hyaluronic Acid for Cancer Therapy. ACS Nano 2011, 5 (11), 8591–8599. 10.1021/nn202070n. [DOI] [PubMed] [Google Scholar]
- Boehnke N.; Correa S.; Hao L.; Wang W.; Straehla J. P.; Bhatia S. N.; Hammond P. T. Theranostic Layer-by-Layer Nanoparticles for Simultaneous Tumor Detection and Gene Silencing. Angew. Chem., Int. Ed. 2020, 59 (7), 2776–2783. 10.1002/anie.201911762. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Barberio A. E.; Smith S. G.; Correa S.; Nguyen C.; Nhan B.; Melo M.; Tokatlian T.; Suh H.; Irvine D. J.; Hammond P. T. Cancer Cell Coating Nanoparticles for Optimal Tumor-Specific Cytokine Delivery. ACS Nano 2020, 14, 11238. 10.1021/acsnano.0c03109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Boehnke N.; Dolph K. J.; Juarez V. M.; Lanoha J. M.; Hammond P. T. Electrostatic Conjugation of Nanoparticle Surfaces with Functional Peptide Motifs. Bioconjugate Chem. 2020, 31 (9), 2211–2219. 10.1021/acs.bioconjchem.0c00384. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kong S. M.; Costa D. F.; Jagielska A.; Van Vliet K. J.; Hammond P. T. Stiffness of targeted layer-by-layer nanoparticles impacts elimination half-life, tumor accumulation and tumor penetration. Proc. Natl. Acad. Sci. U. S. A. 2021, 118, e2104826118. 10.1073/pnas.2104826118. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Seyhan A. A. Lost in translation: the valley of death across preclinical and clinical divide - identification of problems and overcoming obstacles. Transl. Med. Commun. 2019, 4, 18. 10.1186/s41231-019-0050-7. [DOI] [Google Scholar]
- Sayed N.; Liu C.; Ameen M.; Himmati F.; Zhang J. Z.; Khanamiri S.; Moonen J. R.; Wnorowski A.; Cheng L. L.; Rhee J. W.; Gaddam S.; Wang K. C.; Sallam K.; Boyd J. H.; Woo Y. J.; Rabinovitch M.; Wu J. C. Clinical trial in a dish using iPSCs shows lovastatin improves endothelial dysfunction and cellular cross-talk in LMNA cardiomyopathy. Sci. Transl. Med. 2020, 12 (554), eaax9276. 10.1126/scitranslmed.aax9276. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lam C. K.; Wu J. C. Clinical Trial in a Dish Using Patient-Derived Induced Pluripotent Stem Cells to Identify Risks of Drug-Induced Cardiotoxicity. Arterioscler., Thromb., Vasc. Biol. 2021, 41 (3), 1019–1031. 10.1161/ATVBAHA.120.314695. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Barretina J.; Caponigro G.; Stransky N.; Venkatesan K.; Margolin A. A.; Kim S.; Wilson C. J.; Lehar J.; Kryukov G. V.; Sonkin D.; Reddy A.; Liu M. W.; Murray L.; Berger M. F.; Monahan J. E.; Morais P.; Meltzer J.; Korejwa A.; Jane-Valbuena J.; Mapa F. A.; Thibault J.; Bric-Furlong E.; Raman P.; Shipway A.; Engels I. H.; Cheng J.; Yu G. Y. K.; Yu J. J.; Aspesi P.; de Silva M.; Jagtap K.; Jones M. D.; Wang L.; Hatton C.; Palescandolo E.; Gupta S.; Mahan S.; Sougnez C.; Onofrio R. C.; Liefeld T.; MacConaill L.; Winckler W.; Reich M.; Li N. X.; Mesirov J. P.; Gabriel S. B.; Getz G.; Ardlie K.; Chan V.; Myer V. E.; Weber B. L.; Porter J.; Warmuth M.; Finan P.; Harris J. L.; Meyerson M.; Golub T. R.; Morrissey M. P.; Sellers W. R.; Schlegel R.; Garraway L. A. The Cancer Cell Line Encyclopedia enables predictive modelling of anticancer drug sensitivity (vol 483, pg 603, 2012). Nature 2019, 565 (7738), E5–E6. 10.1038/s41586-018-0722-x. [DOI] [PubMed] [Google Scholar]
- Barretina J.; Caponigro G.; Stransky N.; Venkatesan K.; Margolin A. A.; Kim S.; Wilson C. J.; Lehar J.; Kryukov G. V.; Sonkin D.; Reddy A.; Liu M.; Murray L.; Berger M. F.; Monahan J. E.; Morais P.; Meltzer J.; Korejwa A.; Jane-Valbuena J.; Mapa F. A.; Thibault J.; Bric-Furlong E.; Raman P.; Shipway A.; Engels I. H.; Cheng J.; Yu G. K.; Yu J. J.; Aspesi P.; de Silva M.; Jagtap K.; Jones M. D.; Wang L.; Hatton C.; Palescandolo E.; Gupta S.; Mahan S.; Sougnez C.; Onofrio R. C.; Liefeld T.; MacConaill L.; Winckler W.; Reich M.; Li N. X.; Mesirov J. P.; Gabriel S. B.; Getz G.; Ardlie K.; Chan V.; Myer V. E.; Weber B. L.; Porter J.; Warmuth M.; Finan P.; Harris J. L.; Meyerson M.; Golub T. R.; Morrissey M. P.; Sellers W. R.; Schlegel R.; Garraway L. A. The Cancer Cell Line Encyclopedia enables predictive modelling of anticancer drug sensitivity. Nature 2012, 483, 603–607. 10.1038/nature11003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ghandi M.; Huang F. W.; Jane-Valbuena J.; Kryukov G. V.; Lo C. C.; McDonald E. R.; Barretina J.; Gelfand E. T.; Bielski C. M.; Li H.; Hu K.; Andreev-Drakhlin A. Y.; Kim J.; Hess J. M.; Haas B. J.; Aguet F.; Weir B. A.; Rothberg M. V.; Paolella B. R.; Lawrence M. S.; Akbani R.; Lu Y.; Tiv H. L.; Gokhale P. C.; De Weck A.; Mansour A. A.; Oh C.; Shih J.; Hadi K.; Rosen Y.; Bistline J.; Venkatesan K.; Reddy A.; Sonkin D.; Liu M.; Lehar J.; Korn J. M.; Porter D. A.; Jones M. D.; Golji J.; Caponigro G.; Taylor J. E.; Dunning C. M.; Creech A. L.; Warren A. C.; McFarland J. M.; Zamanighomi M.; Kauffmann A.; Stransky N.; Imielinski M.; Maruvka Y. E.; Cherniack A. D.; Tsherniak A.; Vazquez F.; Jaffe J. D.; Lane A. A.; Weinstock D. M.; Johannessen C. M.; Morrissey M. P.; Stegmeier F.; Schlegel R.; Hahn W. C.; Getz G.; Mills G. B.; Boehm J. S.; Golub T. R.; Garraway L. A.; Sellers W. R. Next-generation characterization of the Cancer Cell Line Encyclopedia. Nature 2019, 569 (7757), 503–508. 10.1038/s41586-019-1186-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li H. X.; Ning S. Y.; Ghandi M.; Kryukov G. V.; Gopal S.; Deik A.; Souza A.; Pierce K.; Keskula P.; Hernandez D.; Ann J.; Shkoza D.; Apfel V.; Zou Y. L.; Vazquez F.; Barretina J.; Pagliarini R. A.; Galli G. G.; Root D. E.; Hahn W. C.; Tsherniak A.; Giannakis M.; Schreiber S. L.; Clish C. B.; Garraway L. A.; Sellers W. R. The landscape of cancer cell line metabolism. Nat. Med. 2019, 25 (5), 850–860. 10.1038/s41591-019-0404-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dharia N. V.; Kugener G.; Guenther L. M.; Malone C. F.; Durbin A. D.; Hong A. L.; Howard T. P.; Bandopadhayay P.; Wechsler C. S.; Fung I.; Warren A. C.; Dempster J. M.; Krill-Burger J. M.; Paolella B. R.; Moh P.; Jha N.; Tang A.; Montgomery P.; Boehm J. S.; Hahn W. C.; Roberts C. W. M.; McFarland J. M.; Tsherniak A.; Golub T. R.; Vazquez F.; Stegmaier K. A first-generation pediatric cancer dependency map. Nat. Genet. 2021, 53 (4), 529–538. 10.1038/s41588-021-00819-w. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Corsello S. M.; Nagari R. T.; Spangler R. D.; Rossen J.; Kocak M.; Bryan J. G.; Humeidi R.; Peck D.; Wu X.; Tang A. A.; Wang V. M.; Bender S.; Lemire E.; Narayan R.; Montgomery P.; Ben-David U.; Garvie C. W.; Chen Y.; Rees M. G.; Lyons N. J.; McFarland J. M.; Wong B. T.; Wang L.; Dumont N.; O’Hearn P. J.; Stefan E. J.; Doench J. G.; Harrington C.; Greulich H.; Meyerson M.; Vazquez F.; Subramaniam A.; Roth J. A.; Bittker J. A.; Boehm J. S.; Mader C. C.; Tsherniak A.; Golub T. R. Discovering the anticancer potential of non-oncology drugs by systematic viability profiling. Nat. Cancer 2020, 1, 235. 10.1038/s43018-019-0018-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sinha R.; Luna A.; Schultz N.; Sander C. A pan-cancer survey of cell line tumor similarity by feature-weighted molecular profiles. Cell Rep. Methods 2021, 1, 100039. 10.1016/j.crmeth.2021.100039. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yu C. N.; Mannan A. M.; Yvone G. M.; Ross K. N.; Zhang Y. L.; Marton M. A.; Taylor B. R.; Crenshaw A.; Gould J. Z.; Tamayo P.; Weir B. A.; Tsherniak A.; Wong B.; Garraway L. A.; Shamji A. F.; Palmer M. A.; Foley M. A.; Winckler W.; Schreiber S. L.; Kung A. L.; Golub T. R. High-throughput identification of genotype-specific cancer vulnerabilities in mixtures of barcoded tumor cell lines. Nat. Biotechnol. 2016, 34 (4), 419–423. 10.1038/nbt.3460. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jin X.; Demere Z.; Nair K.; Ali A.; Ferraro G. B.; Natoli T.; Deik A.; Petronio L.; Tang A. A.; Zhu C.; Wang L.; Rosenberg D.; Mangena V.; Roth J.; Chung K.; Jain R. K.; Clish C. B.; Heiden M. G. V.; Golub T. R. A metastasis map of human cancer cell lines. Nature 2020, 588 (7837), 331–336. 10.1038/s41586-020-2969-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Boehnke N.; Straehla J. P.; Safford H. C.; Kocak M.; Rees M. G.; Ronan M.; Adelmann C. H.; Chivukula R. R.; Cheah J.; Li H.; Sabatini D. M.; Roth J. A.; Koehler A. N.; Hammond P. T.. Massively parallel pooled screening reveals genomic determinants of nanoparticle-cell interactions. BioRXiv, 2021, 10.1101/2021.04.05.438521. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lazzari G.; Couvreur P.; Mura S. Multicellular tumor spheroids: a relevant 3D model for the in vitro preclinical investigation of polymer nanomedicines. Polym. Chem. 2017, 8 (34), 4947–4969. 10.1039/C7PY00559H. [DOI] [Google Scholar]
- Method of the Year 2017: Organoids. Nat. Methods 2018, 15, 1. 10.1038/nmeth.4575. [DOI] [Google Scholar]
- The promise of organoids and embryoids. Nat. Mater. 2021, 20 ( (2), ), 121. 10.1038/s41563-021-00926-3 [DOI] [PubMed] [Google Scholar]
- Drost J.; Clevers H. Organoids in cancer research. Nat. Rev. Cancer 2018, 18 (7), 407–418. 10.1038/s41568-018-0007-6. [DOI] [PubMed] [Google Scholar]
- Pauli C.; Hopkins B. D.; Prandi D.; Shaw R.; Fedrizzi T.; Sboner A.; Sailer V.; Augello M.; Puca L.; Rosati R.; McNary T. J.; Churakova Y.; Cheung C.; Triscott J.; Pisapia D.; Rao R.; Mosquera J. M.; Robinson B.; Faltas B. M.; Emerling B. E.; Gadi V. K.; Bernard B.; Elemento O.; Beltran H.; Demichelis F.; Kemp C. J.; Grandori C.; Cantley L. C.; Rubin M. A. Personalized In Vitro and In Vivo Cancer Models to Guide Precision Medicine. Cancer Discovery 2017, 7 (5), 462–477. 10.1158/2159-8290.CD-16-1154. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jenkins R. W.; Aref A. R.; Lizotte P. H.; Ivanova E.; Stinson S.; Zhou C. W.; Bowden M.; Deng J. H.; Liu H. Y.; Miao D. N.; He M. X.; Walker W.; Zhang G.; Tian T.; Cheng C. R.; Wei Z.; Palakurthi S.; Bittinger M.; Vitzthum H.; Kim J. W.; Merlino A.; Quinn M.; Venkataramani C.; Kaplan J. A.; Portell A.; Gokhale P. C.; Phillips B.; Smart A.; Rotem A.; Jones R. E.; Keogh L.; Anguiano M.; Stapleton L.; Jia Z. H.; Barzily-Rokni M.; Canadas I.; Thai T. C.; Hammond M. R.; Vlahos R.; Wang E. S.; Zhang H.; Li S.; Hanna G. J.; Huang W.; Hoang M. P.; Piris A.; Eliane J. P.; Stemmer-Rachamimov A. O.; Cameron L.; Su M. J.; Shah P.; Izar B.; Thakuria M.; LeBoeuf N. R.; Rabinowits G.; Gunda V.; Parangi S.; Cleary J. M.; Miller B. C.; Kitajima S.; Thummalapalli R.; Miao B. C.; Barbie T. U.; Sivathanu V.; Wong J. H.; Richards W. G.; Bueno R.; Yoon C. H.; Miret J.; Herlyn M.; Garraway L. A.; Van Allen E. M.; Freeman G. J.; Kirschmeier P. T.; Lorch J. H.; Ott P. A.; Hodi F. S.; Flaherty K. T.; Kamm R. D.; Boland G. M.; Wong K. K.; Dornan D.; Paweletz C. P.; Barbie D. A. Ex Vivo Profiling of PD-1 Blockade Using Organotypic Tumor Spheroids. Cancer Discovery 2018, 8 (2), 196–215. 10.1158/2159-8290.CD-17-0833. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Velasco V.; Shariati S. A.; Esfandyarpour R. Microtechnology-based methods for organoid models. Microsyst. Nanoeng. 2020, 6 (1), 76. 10.1038/s41378-020-00185-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen X.; Zhang Y. S.; Zhang X. P.; Liu C. S. Organ-on-a-chip platforms for accelerating the evaluation of nanomedicine. Bioact. Mater. 2021, 6 (4), 1012–1027. 10.1016/j.bioactmat.2020.09.022. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jia Z.; Guo Z.; Yang C.-T.; Prestidge C.; Thierry B. “Mucus-on-Chip”: A new tool to study the dynamic penetration of nanoparticulate drug carriers into mucus. Int. J. Pharm. 2021, 598, 120391. 10.1016/j.ijpharm.2021.120391. [DOI] [PubMed] [Google Scholar]
- Shelton S. E.; Nguyen H. T.; Barbie D. A.; Kamm R. D. Engineering approaches for studying immune-tumor cell interactions and immunotherapy. iScience 2021, 24 (1), 101985. 10.1016/j.isci.2020.101985. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang S.; Wan Z. P.; Kamm R. D. Vascularized organoids on a chip: strategies for engineering organoids with functional vasculature. Lab Chip 2021, 21 (3), 473–488. 10.1039/D0LC01186J. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wu J. D.; Dong M. L.; Rigatto C.; Liu Y.; Lin F. Lab-on-chip technology for chronic disease diagnosis. npj Digital Med. 2018, 1, 7. 10.1038/s41746-017-0014-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hajal C.; Le Roi B.; Kamm R. D.; Maoz B. M. Biology and Models of the Blood-Brain Barrier. Annu. Rev. Biomed. Eng. 2021, 23, 359–384. 10.1146/annurev-bioeng-082120-042814. [DOI] [PubMed] [Google Scholar]
- Perlman R. L. Mouse models of human disease An evolutionary perspective. Evol. Med. Public Health 2016, (1), 170–176. 10.1093/emph/eow014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Alexis F.; Pridgen E.; Molnar L. K.; Farokhzad O. C. Factors affecting the clearance and biodistribution of polymeric nanoparticles. Mol. Pharmaceutics 2008, 5 (4), 505–515. 10.1021/mp800051m. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Longmire M.; Choyke P. L.; Kobayashi H. Clearance properties of nano-sized particles and molecules as imaging agents: considerations and caveats. Nanomedicine 2008, 3 (5), 703–717. 10.2217/17435889.3.5.703. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brehm M. A.; Wiles M. V.; Greiner D. L.; Shultz L. D. Generation of improved humanized mouse models for human infectious diseases. J. Immunol. Methods 2014, 410, 3–17. 10.1016/j.jim.2014.02.011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brehm M. A.; Shultz L. D.; Greiner D. L. Humanized mouse models to study human diseases. Curr. Opin. Endocrinol., Diabetes Obes. 2010, 17 (2), 120–125. 10.1097/MED.0b013e328337282f. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Robbins G. R.; Roberts R. A.; Guo H. T.; Reuter K.; Shen T.; Sempowski G. D.; McKinnon K. P.; Su L. S.; DeSimone J. M.; Ting J. P. Y. Analysis of human innate immune responses to PRINT fabricated nanoparticles with cross validation using a humanized mouse model. Nanomedicine 2015, 11 (3), 589–599. 10.1016/j.nano.2014.11.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dou H.; Destache C. J.; Morehead J. R.; Mosley R. L.; Boska M. D.; Kingsley J.; Gorantla S.; Poluektova L.; Nelson J. A.; Chaubal M.; Werling J.; Kipp J.; Rabinow B. E.; Gendelman H. E. Development of a macrophage-based nanoparticle platform for antiretroviral drug delivery. Blood 2006, 108 (8), 2827–2835. 10.1182/blood-2006-03-012534. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhou J. H.; Neff C. P.; Liu X. X.; Zhang J.; Li H. T.; Smith D. D.; Swiderski P.; Aboellail T.; Huang Y. Y.; Du Q.; Liang Z. C.; Peng L.; Akkina R.; Rossi J. J. Systemic Administration of Combinatorial dsiRNAs via Nanoparticles Efficiently Suppresses HIV-1 Infection in Humanized Mice. Mol. Ther. 2011, 19 (12), 2228–2238. 10.1038/mt.2011.207. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tian H. M.; Lyu Y. N.; Yang Y. G.; Hu Z. Humanized Rodent Models for Cancer Research. Front. Oncol. 2020, 10, 1696. 10.3389/fonc.2020.01696. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kersten K.; de Visser K. E.; van Miltenburg M. H.; Jonkers J. Genetically engineered mouse models in oncology research and cancer medicine. EMBO Mol. Med. 2017, 9 (2), 137–153. 10.15252/emmm.201606857. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lampreht Tratar U.; Horvat S.; Cemazar M. Transgenic Mouse Models in Cancer Research. Front. Oncol. 2018, 8, 268. 10.3389/fonc.2018.00268. [DOI] [PMC free article] [PubMed] [Google Scholar]


