ABSTRACT
Aspergillus flavus aflR, a gene encoding a Zn(II)2Cys6 DNA-binding domain, is an important transcriptional regulator of the aflatoxin biosynthesis gene cluster. Our previous results of Gene ontology (GO) analysis for the binding sites of AflR in A. flavus suggest that AflR may play an integrative regulatory role. In this study the ΔaflR and overexpression (OE) strains based on the well-established double-crossover recombinational technique were constructed to investigate the integrative function of the aflR gene in A. flavus. The disruption of aflR severely affected the aflatoxin biosynthetic pathway, resulting in a significant decrease in aflatoxin production. The aflatoxin B1 (AFB1) of the ΔaflR strain was 180 ng/mL and aflatoxin B2 (AFB2) was 2.95 ng/mL on YES medium for 5 days, which was 1/1,000 of that produced by the wild-type strain (WT). In addition, the ΔaflR strain produced relatively sparse conidia and a very small number of sclerotia. On the seventh day, the sclerotia yield on each plate of the WT and OE strains exceeded 1,000, while the sclerotial formation of the ΔaflR strain was not detected until 14 days. However, the biosynthesis of cyclopiazonic acid (CPA) was not affected by aflR gene disruption. Transcriptomic analysis of the ΔaflR strain grown on potato dextrose agar (PDA) plates at 0 h, 24 h, and 72 h showed that expression of clustering genes involved in the biosynthesis of aflatoxin was significantly downregulated. Meanwhile, the ΔaflR strain compared with the WT strain showed significant expression differences in genes involved in spore germination, sclerotial development, and carbohydrate metabolism compared to the WT. The results demonstrated that the A. flavus aflR gene also played a positive role in the fungal growth and development in addition to aflatoxin biosynthesis.
IMPORTANCE Past studies of the A. flavus aflR gene and its orthologues in related Aspergillus species were solely focused on their roles in secondary metabolism. In this study, we used the ΔaflR and OE strains to demonstrate the role of aflR in growth and development of A. flavus. For the first time, we confirmed that the ΔaflR strain also was defective in production of conidia and sclerotia, asexual propagules of A. flavus. Our transcriptomic analysis further showed that genes involved in spore germination, sclerotial development, aflatoxin biosynssssthesis, and carbohydrate metabolism exhibited significant differences in the ΔaflR strain compared with the WT strain. Our study indicates that AflR not only plays an important role in regulating aflatoxin synthesis but also in playing a positive role in the conidial formation and sclerotial development in A. flavus. This study reveals the critical and positive role of the aflR gene in fungal growth and development, and provides a theoretical basis for the genetic studies of other aspergilli.
KEYWORDS: Aspergillus flavus, aflatoxin, aflR, asexual reproduction, sclerotial development
INTRODUCTION
Zinc cluster proteins are one of the largest families of transcriptional regulators in eukaryotes and play multiple functions in transcription (1, 2). Based on their unique and highly conserved amino acid sequences, eukaryotic zinc cluster proteins are classified into three major classes: Cys2His2 (C2H2), Cys4 (C4), and Cys6 (C6) (3). The protein AflR encoded by aflR is a binuclear cluster protein Zn(II)2Cys6 (C6) transcription factor of the GAL4-type family (4, 5). C6 proteins commonly contain two functional domains, the DNA-binding domain which includes the Zn(II)2Cys6 motif, the linker region, and downstream basic dimerization region, and the regulatory domain which is a specific transcription factor (TF) domain (6). The dimerization region, which is usually located at the C-terminus of the linker, consists of a heptapeptide repeat sequence similar to that found in the leucine zipper. The heptapeptide repeat sequence forms a highly conserved convoluted helix structure that most likely leads to dimerization and protein–protein interactions (7).
C6 proteins are mainly associated with genes of the utilization of carbon sources, nitrogen sources, secondary metabolism, growth and development, and play a global regulatory role in fungi (6). In Aspergillus flavus, disruption of aswA renders abnormalities in sclerotial development and biosynthesis of secondary metabolites (8). In Aspergillus nidulans, the zinc cluster protein SfgA negatively regulates the activator of FluG in asexual development (9). The C6 protein ADA-6 plays a vital role in conidial formation, sexual development, and oxidative stress in Neurospora crassa (10). C6 transcription factors have an important role in fungal growth, development, and secondary metabolism-related gene expression or repression. Therefore, unraveling the integrated regulatory role and mechanism of important transcription factors is essential to gain insight into the fungal growth and development and metabolic pathways research.
In A. flavus, the AflR protein binds to at least 17 genes in the aflatoxin biosynthetic cluster, leading to the activation of an enzymatic cascade reaction that results in aflatoxin biosynthesis (11–15). When the aflR gene was overexpressed, it resulted in higher transcript accumulation and increased aflatoxin production (16). Promoter regions of several biosynthesis genes are bound by AflR in a dimeric form with a 5′-TCG(N5)CGA-3′ binding motif. AflR also recognizes 5′-TTAGGCCTAA-3′ and 5′-TCGCAGCCCGG-3′ binding sequences (17). In A. parasiticus, AFLR transcription factor binding sites for the genes nadA, hlyC, and niiA were found outside the aflatoxin gene cluster providing the first evidence that genes outside the aflatoxin gene cluster may be AflR regulation (18). Kong et al. reported that the binding motif of AflR in A. flavus is 5′-CSSGGGWTCGAWCCCSSG-3′, an 18 bp palindrome sequence (19). Their ChIP-seq analysis showed that the consensus AflR binding sequences are present in 5′-NTR of 540 genes that were outside the aflatoxin biosynthesis gene cluster (19). To date, research on AflR has focused on the effects of AflR on aflatoxin biosynthesis, but the role of AflR in fungal growth and development and on genome-wide gene expression remains to be explored. This study aimed to investigate the integrated regulatory role of aflR in A. flavus.
We verified the effect of aflR gene on A. flavus conidial formation, sclerotial development, and biosynthesis of aflatoxin and cyclopiazonic acid (CPA) by examining aflR gene disrupted strains under different media. In addition to the effect on aflatoxin biosynthesis, aflR knockout significantly affected sclerotial development of A. flavus. Then, transcriptome profiles generated by RNA-seq of the ΔaflR mutant and wild type at 0 h, 24 h, and 72 h on potato dextrose agar (PDA) media was measured to study the effect of aflR on A. flavus.
RESULTS
Structural prediction and phylogenetic analysis of AflR in A. flavus.
A. flavus NRRL3357 aflR gene is 1,335 bp in length. AflR contains 444 amino acids. Amino acid residues 29 to 56 of AflR form a GAL4-type binuclear zinc cluster Zn(II)2Cys6 (C6) structure (Fig. 1A). AflR and its homologs from closely related Aspergillus species were analyzed by using MEGA X, and an evolutionary tree was constructed (Fig. 1B) (20). AflR shared 99.10% and 99.32% sequence identity with those of A. oryzae and A. transmontanensis, respectively. According to the phylogenetic relationship, A. flavus is more closely related to A. oryzae than to other Aspergillus species. The aflS gene is adjacent to the aflR gene and encodes an important transcriptional regulator, which in combination with the aflR gene plays a role in regulating aflatoxin biosynthesis (21, 22). Therefore, in this study, we compared the full gene sequence of aflS by whole gene sequencing, and the sequencing comparison. Our sequencing results showed that showed that the full-length sequence of the aflS gene of the ΔaflR strain was identical to the full sequence of aflS gene to that of the wild-type (WT) strain (Fig. S3).
FIG 1.
Conserved domains of the AflR protein (A) and phylogenetic analysis of the AflR proteins (B).
Effect of aflR on mycelial growth and conidial formation.
Colony morphologies of A. flavus WT, ΔaflR, and overexpression (OE) strains on the three media (glucose minimal medium [GMM], PDA, and YES) were significantly different (Fig. 2A). The WT, ΔaflR, and OE strains on YES medium, with dense mycelia, produced large numbers of yellow-green spores. On PDA medium, they also produced dense mycelia. Their radial growth on GMM was slow and the colony diameter was small, but the OE strain showed the largest colony diameter. Compared with the WT strains, the ΔaflR strains had defective conidial head development. The WT strains began to form conidial heads and strong stalks at 24 h, while the ΔaflR strains did not begin to form scattered conidial heads and slender stalks until 30 h (Fig. S4). The WT strains had round-shaped conidial heads full of well-developed conidia, while the ΔaflR strain showed broom-like, elongated conidial heads with smaller conidia (Fig. 2B).
FIG 2.
Colony morphology of the WT, ΔaflR, and OE strains on different media (GMM, PDA, and YES) (A) and microscopic observation of the WT and ΔaflR strains conidiophores at day 3 on GMM medium (10× eyepiece, 40× objective) (B).
To further investigate the effect of aflR on conidia formation, the conidial count was determined at different times (3, 5, and 7 days) on GMM, PDA, and YES. The ΔaflR strain, in general, showed a small but significant decrease in spore count compared with the WT strain especially at day 7 on all three media. Conidia of the ΔaflR strain were restricted, but conidial production was not completely lost. On day 3 (Fig. 3A), conidial formation of A. flavus on GMM and PDA media were at an early stage of development. Thus, there was no significant difference in conidial production between the ΔaflR and WT strains on GMM and PDA media, while the ΔaflR strain showed significantly lower spore production compared with the WT strain on YES media. The WT strain conidia on GMM and PDA media started to develop rapidly from the fifth day, while the ΔaflR strain conidia were relatively slow, resulting in a significant difference in conidial numbers (Fig. 3B). Compared with the WT strain, significant differences were observed on all media for the ΔaflR strain from day 7 (Fig. 3C). The smallest difference was observed on the GMM medium, where spore production was similar, while the largest difference was observed on the YES medium, reaching a difference of about 5-fold. Compared with the WT strain, the conidia of the OE strain increased in different degrees on three media (Fig. S5). The ΔaflR and WT strains showed significant differences in colony diameters on GMM and PDA media (Fig. 3D), while at 5 days on YES medium, the ΔaflR and WT strains had grown all over the YES plates and colony diameters could not be determined.
FIG 3.
Conidial production of the WT and ΔaflR strains on different media at day 3 (A), day 5 (B), and day 7 (C). Plates were incubated at 30°C and circular agar plugs (1.5 cm in diameter) in triplicate were sampled from each plate and the number of conidia was counted with a hemocytometer. At day 5, the colony diameter was different between the WT and ΔaflR strains (D). * P < 0.05; ** P < 0.01.
Effect of aflR on the development of sclerotia of A. flavus.
The Wickerham medium was used to research sclerotial development in the WT, ΔaflR, and OE strains. The growth of sclerotia of the WT and ΔaflR strains was observed after 7 days and 14 days. The WT strain produced many immature sclerotia on the seventh day and that appeared brown in color, while the ΔaflR strain had no sclerotia on the seventh day (Fig. 4A). After 14 days, the WT and OE strains produced a large number of mature sclerotia, while the ΔaflR strain produced only a much smaller number of sclerotia. On the seventh day the sclerotia produced by the WT strain exceeded 1,000 on each plate. On the 14th day, the average yield of the ΔaflR strain sclerotia was 22.5 per plate, and the average yield of the WT strain sclerotia reached 1,458 per plate, which was about 65 times that of the ΔaflR strain (Fig. 4C). The average yield of sclerotia of the OE strain was about 3,733 per plate, which was significantly higher than that of the WT strain. (Fig. S6).
FIG 4.
Sclerotial development and production of the WT and ΔaflR strains. (A) Top panels: colony morphology of the WT strain (left side) and the ΔaflR strain (right side) grown on WKM medium at 30°C for a week in the dark. Bottom panels: closeup of the central area that shows sclerotia produced by the ΔaflR strain in comparison to that of the WT strain. (B) Sclerotial development of the WT and ΔaflR strains after 14 days. (C) Sclerotial production of the WT and ΔaflR strains at day 7 and day 14.
Effect of the aflR deletion on aflatoxin and CPA biosynthesis.
Production of AFB1 and AFB2 by the WT and ΔaflR strains were determined using thin-layer chromatography (TLC) after 5 and 7 days of growth on different media (Fig. 5), and the highest yield of the WT and ΔaflR strains was obtained on YES medium. The AFB1 and AFB2 production of the WT and ΔaflR strains cultured on YES medium for 5 days was also determined using HPLC. The AFB1 of ΔaflR strain was 180 ng/ml and the AFB2 was 2.95 ng/mL (Table 1). The AFB1 and AFB2 content of OE strains were determined after 7 days of growth on YES medium. TLC showed that the AFB1 and AFB2 content of OE strain were higher than that of the WT strain (Fig. S7). The production of CPA was determined using TLC for the WT and ΔaflR strains (Fig. 5G) where CPA was colored as purple spots by the chromogenic agent. There was no significant difference in CPA production between the ΔaflR and WT strains.
FIG 5.
AFB production of the WT and ΔaflR strains after 5 days (A, C, E) and 7 days (B, D, F) and CPA production of the WT and ΔaflR strains after 7 days (G). AFB yields on GMM (A, B), PDA (C, D), and YES (E, F) were determined by TLC.
TABLE 1.
AFB produced by the WT and ΔaflR strains
Data results were the average of three parallel experiments. Error range of all values are between ± 3% −8%. The actual error values were omitted in the table.
Transcriptome analysis of WT and ΔaflR strains.
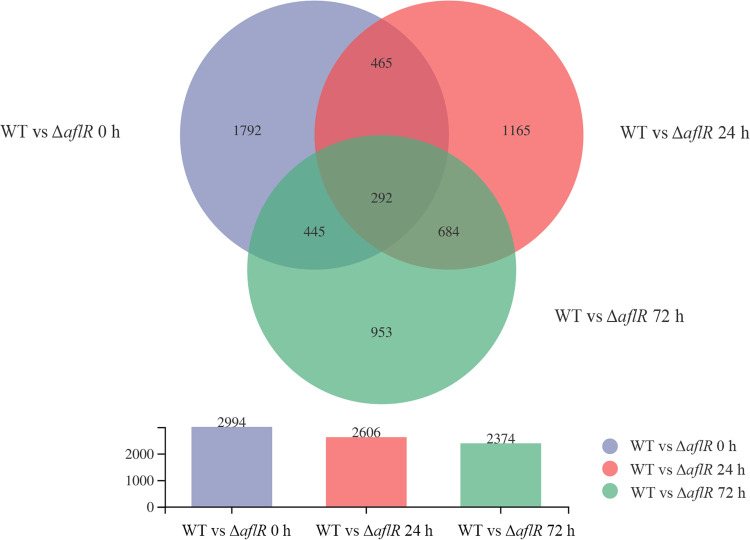
In this study, 2,994 differentially expressed genes (DEGs) were identified at 0 h, of which 1,606 were downregulated DEGs and 1,388 were upregulated DEGs (Data set S1). At 24 h, 2,606 DEGs were identified, of which 1,268 were downregulated and 1,338 were upregulated (Data set S2). At 72 h, 2,374 DEGs were identified, of which 1,201 were downregulated and 1,173 were upregulated (Data set S3). The WT and ΔaflR strains shared 757 common DEGs at 0 h and 24 h, 737 common DEGs at 0 h and 72 h, 976 common DEGs at 24 h and 72 h, and 292 common DEGs at 0 h, 24 h, and 72 h (Fig. 6).
FIG 6.
Venn analysis for the WT and ΔaflR strains.
DEGs identified at 0 h, 24 h, and 72 h were subjected to Gene ontology (GO) enrichment analysis (Fig. 7A). Enriched GO terms for DEGs of the ΔaflR strain at 0 h included maturation of LSU-rRNA from tricistronic rRNA transcript, ribosomal large subunit biogenesis, and RNA 5′-end processing. At 24 h, enriched GO terms for DEGs of the ΔaflR strain included mycotoxin metabolic process, mycotoxin biosynthetic process, aflatoxin metabolic process, and aflatoxin biosynthetic process (Fig. 7B). At 72 h, the GO function enrichment of DEGs for the ΔaflR strain showed significant changes including transmembrane transport, membrane components, transport activity, toxin biosynthesis process, toxin metabolism process, mycotoxin metabolism process, and mycotoxin biosynthesis process, etc. (Fig. 7C). At 24 h and 72 h, GO terms for the WT and ΔaflR strains were mainly enriched in processes related to toxin biosynthesis and toxin metabolism, including aflatoxin biosynthesis and metabolism.
FIG 7.
GO enrichment analysis of the WT and ΔaflR strains (A, 0 h; B, 24 h; C, 72 h). The rich factor represents the ratio of the number of DEGs annotated as GO functional classes to the number of all identified genes annotated as GO functional classes.
KEGG pathway enrichment analysis further performed on DEGs of the WT and ΔaflR strains. The significantly different metabolic pathways of the ΔaflR strain compared with the WT strain at 0 h, 24 h, and 72 h (Fig. 8). Secondary metabolism especially aflatoxin biosynthesis was significantly affected by the aflR deletion at 24 h, especially aflatoxin biosynthesis. Additionally, the common 292 DEGs obtained by Veen analysis at 0 h, 24 h, and 72 h were analyzed to KEGG pathway enrichment analysis to find metabolic pathways significantly associated with aflR. The pathways identified were those involved in pyruvate metabolism, methane metabolism, aflatoxin biosynthesis, starch and sucrose metabolism, galactose metabolism, interconversions of pentose and glucuronate interconversions, biosynthesis of unsaturated fatty acids, and fatty acid biosynthesis pathways (Fig. 8D).
FIG 8.
KEGG enrichment analysis of the WT and ΔaflR strains (A, 0 h; B, 24 h; C, 72 h) and KEGG enrichment analysis of common DEGs of the WT and ΔaflR strains at 0 h, 24 h, and 72 h (D). The rich factor indicates the ratio of the number of DEGs involved in the KEGG pathway to the number of genes involved in the pathway among all identified genes.
DISCUSSION
Impacts of aflR on mycelial growth and conidial formation.
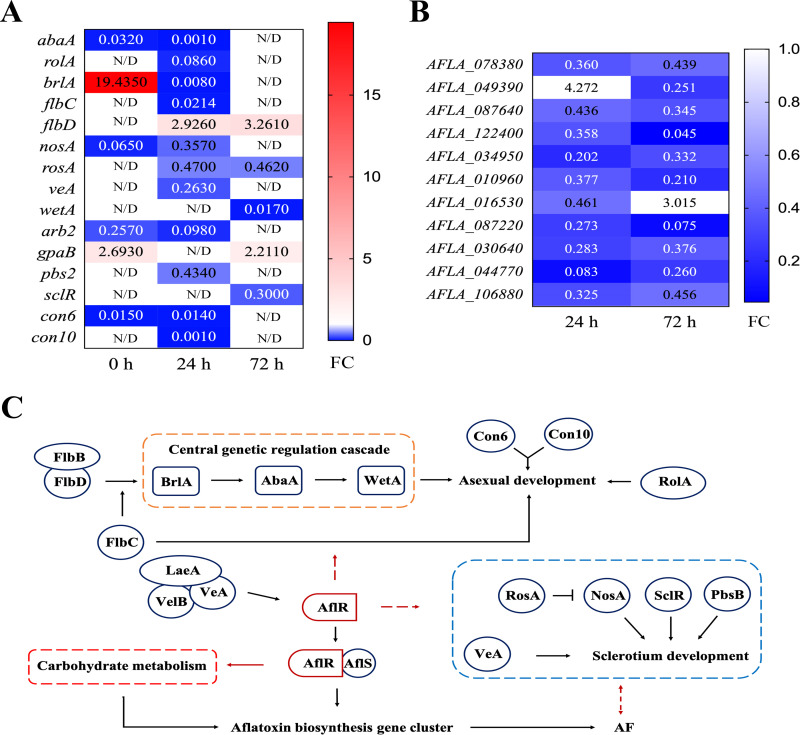
Growth and development are the two phases of asexual reproduction that Aspergillus undergoes. The growth period includes conidial germination and mycelial formation. When nutrition is limited, mycelial cells stop growth by forming complex structures (becoming conidia) and begin asexual reproduction (23). In the present study, we found significant differences in the expression of many genes related to the growth and development of A. flavus in the ΔaflR strain (Fig. 9A) and their associated functions are summarized as follows (Table 2). The abaA gene plays a crucial role in the differentiation of phialides, the hyphal cells necessary for the formation of conidia (24). The abaA gene also affects secondary metabolism by regulating the expression of veA, velB, and velC (25). The brlA gene encodes a C2H2 zinc finger transcription factor (TF), a key regulator of Aspergillus conidia production, which mediates vesicle formation and budding cell growth during the early stages of asexual development (26, 27). The wetA gene plays a key role in the coordinated control of Aspergillus spore production, though the exact molecular mechanism of wetA is not known, deletion of wetA results in defects in conidiophore development (23). The wetA gene activated by abaA completes the developmental role in the late stage of conidiation (28). The wetA gene at 72 h in the ΔaflR strain was only 0.017-fold of the WT strain. The significant decreased expression of the wet gene in the ΔaflR strain, likely caused the related defects in conidial formation. The central genetic regulatory cascade formed by brlA→abaA→wetA acts in concert with other genes to control gene expression in spore-specific cells and to determine the sequence of gene activation in spores during cellular and chemical development (28).
FIG 9.
Effect of aflR knockout on the expression of growth and development-related genes (A) and carbohydrate metabolism (B) of A. flavus compared with the WT strain, and the putative model of the comprehensive regulation of A. flavus (C).
TABLE 2.
Growth and development related genes and their functions affected by aflR gene knockout
| Gene | Associated function |
|---|---|
| abaA | Transcription factor for conidia formation |
| rolA | Encoding hydrophobic protein |
| brlA | C2H2 zinc finger protein transcriptional activator of conidiophore |
| flbC | Putative C2H2 conidiation transcription factor |
| flbD | MYB family conidiophore development |
| nosA | Necessary for the induction of gene transcription in filamentous fungi in sexual reproduction |
| rosA | Zn2-Cys6 binuclear cluster domain-containing protein |
| veA | Global regulator |
| wetA | Developmental regulatory protein |
| arb2 | Conidial pigment biosynthesis oxidase |
| gpaB | Asexual sporulation, AF biosynthesis, and virulence by regulating cAMP signaling |
| pbs2 | Involved in the growth of mycelium, conidiation, and sclerotia formation |
| sclR | Involved in mycelial morphology, conidial development, and sclerotial formation |
| con6 | Conidiation protein |
| con10 | Conidiation-specific protein |
The flbC and flbD genes regulate the expression of the brlA gene upstream to control the asexual development of Aspergillus (29). Disruption of the flbC gene resulted in delayed spore germination and reduced expression of the brlA gene, while FlbC bound to abaA and vosA play a regulatory role (30). FlbD is a c-Myb transcription factor, and deletion of the flbD gene results in a fluffy phenotype. Overexpression of flbD leads to sporulation in a liquid culture environment, which is caused by a failure of brlA activation (31). The FlbB/FlbE complex is required for the expression of flbD, and mechanisms exist for both FlbB/FlbE and FlbD to activate brlA gene expression (32). However, the activation of the brlA gene seems to be the result of the accumulation of multiple pathways (30). In this study, the expression of flbC in the ΔaflR strain was significantly reduced at 24 h, which was only 0.021 times that of the WT strain, while the expression of flbD was significantly upregulated at 24 h and 72 h to 2.926 and 3.261 times that of the WT strain, respectively. This may be due to the presence of multiple pathways activating brlA gene expression, and when one of the pathways is repressed, the repressive effect is offset by an increase in expression of another pathway. However, in the ΔaflR strain, the extent of flbC downregulation fold exceeded that of flbD, which is one of the possible reasons why brlA was not effectively activated.
Suzuki et al. found that a single deletion of con genes in A. nidulans did not cause significant phenotypic changes, while simultaneous inactivation of two or three con genes would result in delayed spore germination (33). Among them, con6 and con10 were representatives of con genes preferentially expressed during conidiation (34, 35). In this study, con6 and con10 were significantly downregulated in ΔaflR strain, which seems to be the reason for the limited spore formation in the ΔaflR strain compared with the WT strain.
Hydrophobin proteins are effective regulators that play a structural or enzymatic role in the formation of asexual developmental structures (36). The A. flavus hydrophobin protein is encoded by the rolA (rodA) gene, and deletion of the rolA gene results in reduced hydrophobicity of conidia (37). The expression of rolA gene may affect aflatoxin biosynthesis. For example, the inhibition of aflatoxin biosynthesis by Bacillus megaterium was related to significant inhibition of rolA gene expression (38). The results showed that the aflR gene could affect conidial development, but the aflR gene deletion did not cause complete impairment of conidial development, indicating that the aflR gene was not a key gene for the growth and development of A. flavus colonies and spores.
Regulation of sclerotial formation by aflR.
Sclerotium is another way of Aspergillus reproduction. Aspergillus hypha branches aggregate into a dense network to form the initial white immature sclerotia, and then the immature sclerotia with pigment deposition form mature sclerotia (8, 39). Mature mycorrhizal sclerotia contain a large number of metabolites such as aflatoxins and indoles. The sclerotium, as an important asexual reproduction structure for the spread of A. flavus in the soil and its growth in adverse conditions, is related to secondary metabolites. The gene nosA is one of the genes necessary for the induction of gene transcription in filamentous fungi in sexual reproduction (40). In A. fumigatus, nosA knockout led to impaired sexual reproduction in the fungus, and nosA expression was found to correlate with virulence (40). Inactivation of RosA did not affect asexual and sexual reproduction when VeA was present, indicating that the function of the rosA gene in asexual and sexual reproduction depends on veA level (41). In this study, both nosA and rosA were significantly downregulated in the ΔaflR strain. However, rosA is a developmental repressor gene whose downregulation should be positively regulating sexual reproduction. This may be because the rosA gene functions under the influence of veA, and significant downregulation of the veA gene prevents rosA from functioning properly. In A. oryzae, the sclR gene plays an important role in mycelial morphology, conidial development, and sclerotial formation, and overexpression of sclR promotes high mycelial aggregation to initiate sclerotia formation (42). The sclR gene was significantly downregulated in the ΔaflR strain at 72 h, which was only 0.300-fold of the WT strain. The veA gene functions as a global regulator of a variety of morphogenetic and secondary metabolic genes in fungi and is one of the genes essential for fungal growth and development (43). In the nucleus, three proteins, VeA, LaeA, and VelB, form a heterotrimeric velvet complex to coordinate and control the development and secondary metabolism of the fungi (25). In this study, a significant downregulation of veA at 24 h was observed in the ΔaflR strain, which was only 0.263 times that of the WT strain. In this study, veA of the ΔaflR strain was significantly downregulated to 0.263 times that of the WT strain at 24 h, which may affect its morphogenesis and secondary metabolism.
Effect of aflR on metabolic pathways.
The mitogen-activated protein (MAP) kinase cascade reaction is one of the mechanisms by which eukaryotic cells transfer information from the extracellular environment to target gene expression in the nucleus via plasma membrane-associated receptors, and the MAP kinase signaling pathway regulates development and toxin biosynthesis (44). The pbs2 gene was significantly downregulated in the ΔaflR strain at 24 h. pbs2 (pbsB), as MAPKK, is critical in the growth and virulence of A. flavus (45).
Carbohydrate metabolism plays a key role in fungal secondary metabolism and fungal infection, and fungal adaptation to changes in the extracellular environment; it is critical to their growth and development under stress conditions (46, 47). Pyruvate metabolism, starch, and sucrose metabolism, and the interconversion of pentose and glucuronide are parts of carbohydrate metabolism, and genes associated with these metabolic pathways were significantly downregulated in the ΔaflR strain. Therefore, aflR apparently affects carbohydrate metabolism. Kong et al. found that AfAflR can bind to genes encoding carbohydrate transporters, which suggests that aflR is involved in carbohydrate metabolism and there may regulate carbohydrate metabolism (19). Carbohydrate-related metabolic pathways were significantly altered after aflR knockdown, consistent with the findings of Kong et al. (Fig. 9B).
To sum up, Fig. 9C shows a presumptive model map on the full regulation of growth and development of A. flavus. AflR may regulate the process of conidial germination by regulating the central genetic regulatory cascade brlA→abaA→wetA genes and other genes related to conidial development, such as flbD, flbC, rolA, con6, and con10. It also may regulate the formation and development of sclerotia by regulating the expression of rosA, nosA, pbsB, and sclR.
aflR gene regulates gene expression of aflatoxin biosynthesis gene cluster.
The aflatoxin biosynthetic pathway has long been regarded as one of the most complex metabolic pathways of natural secondary metabolites, involving at least 27 enzymatic reactions (11, 48). In this study, the transcriptomic analysis revealed that genes of the aflatoxin biosynthesis gene cluster were significantly downregulated (Fig. 10). At 0 h, most aflatoxin biosynthesis genes were not expressed by both WT and ΔaflR strains. In particular, all 27 genes on the aflatoxin biosynthetic pathway were significantly downregulated at 24 h (P < 0.01). A total of 13 genes, hypC, aflG, aflH, aflK, aflV, aflJ, aflN, aflM, aflP, aflQ, hypE, aflE, and aflS, were not expressed in the ΔaflR strain at 24 h. In contrast, at 24 h, all genes on the aflatoxin biosynthetic gene cluster of the A. flavus were normally expressed. The aflatoxin biosynthetic pathway of the ΔaflR strains is disrupted, indicating that the aflR gene, as an important transcriptional regulator in the aflatoxin biosynthetic gene cluster, is one of the indispensable genes in the aflatoxin biosynthetic pathway. AflR binding sites were present in at least 17 genes in the aflatoxin biosynthesis pathway (13). Interestingly, the aflS gene was not expressed after aflR knockdown at 0 h, 24 h, or 72 h. In addition, the knockout of the aflR gene resulted in a small amount of aflatoxin production. Although genes related to the aflatoxin biosynthetic pathway were significantly downregulated, these genes were still expressed at low levels. These likely resulted from basal expression of those aflatoxin pathway structural genes after the aflR gene was deleted, resulting in the eventual possibility of still producing low levels of aflatoxin. Combined phenotypic and transcriptomic data analysis showed that the aflR gene not only affected the aflatoxin biosynthesis but also played a positive role in asexual reproduction, sclerotial development, and growth of A. flavus.
FIG 10.
Effect of aflR knockdown on the aflatoxin biosynthesis pathway (A) and on the expression profile of genes in the pathway (B). FC, fold change; N/D, not detected.
MATERIALS AND METHODS
Strains and culture conditions.
Fungal strains used in this study included A. flavus NRRL3357 (WT strain), TJES19.1 (ΔpyrG, ΔKu70) (49), ΔaflR strain (ΔaflR, ΔKu70, pyrG+), and OE strain (gpdA+, aflR+,ΔKu70, pyrG+). All strains were cultured at 30°C and stored short-term at −20°C (long-term, −80°C) in a glycerol stock solution. WT, ΔaflR, and OE strains stored at −20°C were spotted with 3 μL on V8 juice media for activation and incubated at 30°C for 5 days. The spores at 108 spores/mL were harvested from V8 juice medium (5% V8 juice, 2% agar, pH 5.2) using 0.1% Triton X-100 solution and inoculated in 100 mL Czepak-Dox Broth (supplemented with 0.5% casein amino acids, 10 mM ammonium sulfate, 0.2% uracil and 1% glucose) after resuspension with sterilized water.
Construction of strains.
Gene replacement by double crossover recombination was carried out in strains lacking the non-homologous end joining (NHEJ) (50). The ΔaflR strain were constructed by a deletion cassette. The two flanking fragments of the aflR gene and the screening marker gene pyrG were amplified using PrimeSTAR HS DNA polymerase (TaKaRa Bio Co., Ltd., Japan). The deletion cassette was generated by the fusion PCR (Fig. S1A) (51). The aflR gene was overexpressed via Gibson Assembly Cloning Kit (New England Bio Co., Ltd., USA). 5′-UTR, pyrG, gpdA, aflR, and puC19 were amplified and overlapped with particular primers. A molar ratio of 1:1 between vector and each fragment should be used. In a final volume of 20 μL at 50°C, 10 μL of 2X Gibson Assembly Master Mix was incubated with five fragments (0.05 pmol each) for 60 min. All primers used in the study were shown in Table S1.
TJES19.1 was transformed as described by Chang et al. (50). The obtained transformants were cultured in a regeneration medium (Czepak-Dox Broth supplemented with 0.6 M KCl, 5 mM ammonium sulfate) for 7 to 10 days and then validated by PCR and sequencing (Fig. S1B, Fig. S2). There is an intergenic region (491 bp) between aflR and aflS, and the primers FAflS and RAflS were used to amplify and check the intact aflS gene band in the aflR knockout positive transformants, and the selected PCR-positive transformants were purified (E.Z.N.A Cycle Pure Kit D6492, Omega Bio-tek Ltd., USA) and verified by sequencing (commissioned by RuiBiotech, Beijing, China) (Fig. S3).
Mycelial growth and conidial formation.
Approximately 3 μL 106 spores/mL of WT and ΔaflR strains were spotted in the center of GMM (52), PDA (Beijing Solarbio Science & Technology Co., Ltd., Beijing, China), and YES media and incubated at 30°C for 5 days. After 3 days in GMM medium, a small piece of fungal culture was removed, placed on a microscope slide, and a drop of 0.85% NaCl solution was added to the fungal culture for microscopic observation. The morphology of conidial were also recorded at 24 h, 30 h, 36 h, 42 h, and 48 h.
Conidia were determined as follows. A spore suspension of approximately 3 μL 108 spores/mL was spotted in the center of GMM, PDA, and YES media and incubated at 30°C for 3, 5, and 7 days. Agar plugs of 1.5 cm in diameter media (three samples per plate, three replicates per strain) were sampled and transferred to a 10-mL centrifuge tube, 5 mL of 0.1% Tween 80 solution was added and vortexed for 5 min. Then the agar plugs were removed, and spores were counted using a hemocytometer.
Sclerotial formation analysis.
Sclerotial formation analysis was performed as follows. Approximately 3 μL 108/mL WT and ΔaflR spore suspensions were inoculated in the center of the Wickerham medium (8). After grown at 30°C for 7 and 14 days, fungal cultures were sprayed with 95% ethanol, and the sclerotia were exposed and counted.
Determination of aflatoxin and semi-quantitative determination of cyclopiazonic acid.
The TLC analytical method for aflatoxins was modified (53). Strains were incubated on GMM, PDA, and YES for 5 and 7 days at 30°C. Three agar plugs (1.5-cm diameter), 1 cm from each inoculation site, were cored with transfer tubes and placed in 10 mL centrifuge tubes and extracted twice, each time with 5 mL of methanol (three replicates per strain). The extracts were vortexed for 3 to 5 min at 30°C and shaken at 200 rpm for 45 min. The supernatant was transferred, centrifuged at 4,000 g for 15 min, blown dry with nitrogen at 55°C, and resuspended with 1 mL CHCl3. Then, 10 microliters (2.5 μL/time) of WT and ΔaflR aflatoxin extracts were spotted onto a TLC plate. The plate (105554, Merck, Germany) was developed in methanol:ethyl acetate:acetic acid (96:3:1 vol/vol) and air-dried. Aflatoxin was visible under 365 nm UV light. High-performance liquid chromatography (HPLC) determination of aflatoxin produced by 5-day-old cultures on YES at 30°C was entrusted to Pony Testing International Group (Beijing, China). HPLC method for AFB1 and AFB2 was referenced to national standards of P. R. China (GB 5009.22-2016).
To determine the CPA production of the WT and ΔaflR mutants, approximately 3 μL of 108 spores/mL spore suspension was spotted in the center of each Wickerham medium plate and incubated at 30°C for 7 days. Agar plugs of 1.5 cm in diameter media (three samples per plate, three replicates per strain) were sampled and transferred to a 5-mL centrifuge tube; 3 mL CHCl3 was added, vortexed for 5 min, and shaken at 30°C for 45 min. The supernatant was removed and determined by TLC following the method of Chang et al. (54).
Transcriptome analysis of the WT and ΔaflR strains.
(i) Preparation of transcriptome samples and RNA extraction. Approximately 108 spores/mL of the WT and ΔaflR strains were inoculated in the PDB medium and grown for 24 h at 30°C 180 rpm. Mycelia were collected through sterilized Miracloth and washed with sterile water. The water on the surface of the mycelia was blotted out. The mycelia were spread flat on PDA plates at 30°C and sampled at 0 h, 24 h, and 72 h. The removed samples were quickly snap-frozen in liquid nitrogen for 1 h and stored at −80°C. Total RNA was extracted using the TRIzolTM kit (Thermo Fisher Scientific, USA) and DNA was removed using DNase I (TaKaRa, Japan). RNA integrity was examined using RNA electrophoresis and RNA quality was determined using an Agilent 2100 Nano (Agilent Technologies, Palo Alto, CA, United States). The concentration of RNA was determined using NanoDrop 2000 (Agilent Technologies, Palo Alto, CA, USA).
(ii) RNA-seq and enrichment analysis of differentially expressed genes. The total RNA of three biological replicates for the ΔaflR and WT strains were sequenced. RNA libraries were generated using the TruSeqTM RNA sample preparation kit (Illumina, USA). Raw sequencing data were converted to sequence data (clean data) by base calling and stored in fastq format. The clean data were blasted to the reference genome of the A. flavus genome sequence, and the mapped data (reads) were obtained for subsequent analysis. Cufflinks counts mapped clean reads for each gene and normalizes them to fragment per thousand base pairs of transcribed fragments mapped reads per million (FPKM) values (55). Expression profiles in genes and transcripts were quantified by Salmon expression quantification software (56). A gene was treated as significantly DEGs when |log2(fold change)| ≥ 2 with an adjusted P value ≤ 0.05 (Data set S1, Data set S2, Data set S3). GO terms (including cellular component, molecular function, and biological process) and Kyoto Encyclopedia of Genes and Genomes (KEGG) pathways were classified as significantly enriched among differentially expressed genes (P < 0.05).
Statistical analysis.
The data were processed by SPSS 25 software and significance analysis (P < 0.05) was performed.
Data availability.
The results of the RNA-seq data were submitted to NCBI's GEO database and assigned accession no. GSE179978.
ACKNOWLEDGMENTS
We thank Nancy Keller of the University of Wisconsin-Madison and Fuguo Xing of the Institute of Food Science and Technology, CAAS for providing the necessary TJES19.1 strain and plasmids in construction of knockout strains. This research is supported by the National Natural Science Foundation of China (32072328).
Conceptualization, Q.K., and P.W.; methodology, Q.K., P.W., and J.X.; formal analysis, Q.K., P.W., and P.K.C.; investigation, P.W., J.X., and Z.L.; writing—original draft preparation, Q.K., and P.W.; writing—review and editing; all authors contributed to writing and reviewing the paper.
Footnotes
Supplemental material is available online only.
Contributor Information
Qing Kong, Email: kongqing@ouc.edu.cn.
Gustavo H. Goldman, Universidade de Sao Paulo
REFERENCES
- 1.Zhang C, Huang H, Deng W, Li T. 2019. Genome-wide analysis of the Zn(II)2Cys6 zinc cluster-encoding gene family in tolypocladium guangdongense and its light-induced expression. Genes 10:179. doi: 10.3390/genes10030179. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Akache B, Wu K, Turcotte B. 2001. Phenotypic analysis of genes encoding yeast zinc cluster proteins. Nucleic Acids Res 29:2181–2190. doi: 10.1093/nar/29.10.2181. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.MacPherson S, Larochelle M, Turcotte B. 2006. A fungal family of transcriptional regulators: the zinc cluster proteins. Microbiol Mol Biol Rev 70:583–604. doi: 10.1128/MMBR.00015-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Shimizu K, Hicks JK, Huang T-P, Keller NP. 2003. Pka, Ras and RGS protein interactions regulate activity of AflR, a Zn(II)2Cys6 transcription factor in Aspergillus nidulans. Genetics 165:1095–1104. doi: 10.1093/genetics/165.3.1095. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Pan T, Coleman JE. 1989. Structure and function of the Zn(II) binding site within the DNA-binding domain of the GAL4 transcription factor. Proc Natl Acad Sci USA 86:3145–3149. doi: 10.1073/pnas.86.9.3145. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Chang P-K, Ehrlich KC. 2013. Genome-wide analysis of the Zn(II)2Cys6 zinc cluster-encoding gene family in Aspergillus flavus. Appl Microbiol Biotechnol 97:4289–4300. doi: 10.1007/s00253-013-4865-2. [DOI] [PubMed] [Google Scholar]
- 7.Schjerling P, Holmberg S. 1996. Comparative amino acid sequence analysis of the C6 zinc cluster family of transcriptional regulators. Nucleic Acids Res 24:4599–4607. doi: 10.1093/nar/24.23.4599. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Chang P-K, Scharfenstein LL, Li RW, Arroyo-Manzanares N, De Saeger S, Diana Di Mavungu J. 2017. Aspergillus flavus aswA, a gene homolog of Aspergillus nidulans oefC, regulates sclerotial development and biosynthesis of sclerotium-associated secondary metabolites. Fungal Genet Biol 104:29–37. doi: 10.1016/j.fgb.2017.04.006. [DOI] [PubMed] [Google Scholar]
- 9.Seo J-A, Guan Y, Yu J-H. 2006. FluG-dependent asexual development in Aspergillus nidulans occurs via derepression. Genetics 172:1535–1544. doi: 10.1534/genetics.105.052258. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Sun X, Wang F, Lan N, Liu B, Hu C, Xue W, Zhang Z, Li S. 2019. The Zn(II)2Cys6-type transcription factor ADA-6 regulates conidiation, sexual development, and oxidative stress response in Neurospora crassa. Front Microbiol 10:750. doi: 10.3389/fmicb.2019.00750. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Caceres I, Khoury AA, Khoury RE, Lorber S, Oswald IP, Khoury AE, Atoui A, Puel O, Bailly J-D. 2020. Aflatoxin biosynthesis and genetic regulation: a review. Toxins 12:150. doi: 10.3390/toxins12030150. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Woloshuk C, Foutz K, Brewer J, Bhatnagar D, Cleveland T, Payne G. 1994. Molecular characterization of aflR, a regulatory locus for aflatoxin biosynthesis. Appl Environ Microbiol 60:2408–2414. doi: 10.1128/aem.60.7.2408-2414.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Yu J-H, Keller N. 2005. Regulation of secondary metabolism in filamentous fungi. Annu Rev Phytopathol 43:437–458. doi: 10.1146/annurev.phyto.43.040204.140214. [DOI] [PubMed] [Google Scholar]
- 14.Ehrlich KC, Montalbano BG, Cotty PJ. 2003. Sequence comparison of aflR from different Aspergillus species provides evidence for variability in regulation of aflatoxin production. Fungal Genet Biol 38:63–74. doi: 10.1016/s1087-1845(02)00509-1. [DOI] [PubMed] [Google Scholar]
- 15.Chang P-K, Cary J, Bhatnagar D, Cleveland T, Bennett J, Linz J, Woloshuk C, Payne G. 1993. Cloning of the Aspergillus parasiticus apa-2 gene associated with the regulation of aflatoxin biosynthesis. Appl Environ Microbiol 59:3273–3279. doi: 10.1128/aem.59.10.3273-3279.1993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Flaherty JE, Payne G. 1997. Overexpression of aflR leads to upregulation of pathway gene transcription and increased aflatoxin production in Aspergillus flavus. Appl Environ Microbiol 63:3995–4000. doi: 10.1128/aem.63.10.3995-4000.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Ehrlich K, Montalbano B, Cary J. 1999. Binding of the C6-zinc cluster protein, AFLR, to the promoters of aflatoxin pathway biosynthesis genes in Aspergillus parasiticus. Gene 230:249–257. doi: 10.1016/S0378-1119(99)00075-X. [DOI] [PubMed] [Google Scholar]
- 18.Price MS, Yu J, Nierman WC, Kim HS, Pritchard B, Jacobus CA, Bhatnagar D, Cleveland TE, Payne GA. 2006. The aflatoxin pathway regulator AflR induces gene transcription inside and outside of the aflatoxin biosynthetic cluster. FEMS Microbiol Lett 255:275–279. doi: 10.1111/j.1574-6968.2005.00084.x. [DOI] [PubMed] [Google Scholar]
- 19.Kong Q, Chang P-K, Li C, Hu Z, Zheng M, Sun Q, Shan S. 2020. Identification of AflR binding sites in the genome of Aspergillus flavus by ChIP-Seq. JoF 6:52. doi: 10.3390/jof6020052. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Kumar S, Stecher G, Li M, Knyaz C, Tamura K. 2018. MEGA X: molecular evolutionary genetics analysis across computing platforms. Mol Biol Evol 35:1547–1549. doi: 10.1093/molbev/msy096. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Chang P-K. 2003. The Aspergillus parasiticus protein AFLJ interacts with the aflatoxin pathway-specific regulator AFLR. Mol Genet Genomics 268:711–719. doi: 10.1007/s00438-003-0809-3. [DOI] [PubMed] [Google Scholar]
- 22.Du W, Obrian G, Payne G. 2007. Function and regulation of aflJ in the accumulation of aflatoxin early pathway intermediate in Aspergillus flavus. Food Addit Contam 24:1043–1050. doi: 10.1080/02652030701513826. [DOI] [PubMed] [Google Scholar]
- 23.Wu M-Y, Mead ME, Kim S-C, Rokas A, Yu J-H. 2017. WetA bridges cellular and chemical development in Aspergillus flavus. PLoS One 12:e0179571. doi: 10.1371/journal.pone.0179571. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Shin K-S, Kim YH, Yu J-H. 2015. Proteomic analyses reveal the key roles of BrlA and AbaA in biogenesis of gliotoxin in Aspergillus fumigatus. Biochem Biophys Res Commun 463:428–433. doi: 10.1016/j.bbrc.2015.05.090. [DOI] [PubMed] [Google Scholar]
- 25.Bayram O, Krappmann S, Ni M, Bok JW, Helmstaedt K, Valerius O, Braus-Stromeyer S, Kwon N-J, Keller NP, Yu J-H, Braus GH. 2008. VelB/VeA/LaeA complex coordinates light signal with fungal development and secondary metabolism. Science 320:1504–1506. doi: 10.1126/science.1155888. [DOI] [PubMed] [Google Scholar]
- 26.Adams TH, Boylan MT, Timberlake WE. 1988. brlA is necessary and sufficient to direct conidiophore development in Aspergillus nidulans. Cell 54:353–362. doi: 10.1016/0092-8674(88)90198-5. [DOI] [PubMed] [Google Scholar]
- 27.Han S, Adams T. 2001. Complex control of the developmental regulatory locus brlA in Aspergillus nidulans. Mol Genet Genomics 266:260–270. doi: 10.1007/s004380100552. [DOI] [PubMed] [Google Scholar]
- 28.Wu M-Y, Mead ME, Lee M-K, Loss EMO, Kim S-C, Rokas A, Yu J-H. 2018. Systematic dissection of the evolutionarily conserved WetA developmental regulator across a genus of filamentous fungi. mBio 9:e01130-18. doi: 10.1128/mBio.01130-18. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Kwon N-J, Shin K-S, Yu J-H. 2010. Characterization of the developmental regulator FlbE in Aspergillus fumigatus and Aspergillus nidulans. Fungal Genet Biol 47:981–993. doi: 10.1016/j.fgb.2010.08.009. [DOI] [PubMed] [Google Scholar]
- 30.Kwon NJ, Garzia A, Espeso EA, Ugalde U, Yu JH. 2010. FlbC is a putative nuclear C2H2 transcription factor regulating development in Aspergillus nidulans. Mol Microbiol 77:1203–1219. doi: 10.1111/j.1365-2958.2010.07282.x. [DOI] [PubMed] [Google Scholar]
- 31.Wieser J, Adams TH. 1995. flbD encodes a Myb-like DNA-binding protein that coordinates initiation of Aspergillus nidulans conidiophore development. Genes Dev 9:491–502. doi: 10.1101/gad.9.4.491. [DOI] [PubMed] [Google Scholar]
- 32.Garzia A, Etxebeste O, Herrero‐García E, Ugalde U, Espeso EA. 2010. The concerted action of bZip and cMyb transcription factors FlbB and FlbD induces brlA expression and asexual development in Aspergillus nidulans. Mol Microbiol 75:1314–1324. doi: 10.1111/j.1365-2958.2010.07063.x. [DOI] [PubMed] [Google Scholar]
- 33.Suzuki S, Bayram ÖS, Bayram Ö, Braus GH. 2013. conF and conJ contribute to conidia germination and stress response in the filamentous fungus Aspergillus nidulans. Fungal Genet Biol 56:42–53. doi: 10.1016/j.fgb.2013.04.008. [DOI] [PubMed] [Google Scholar]
- 34.Berlin V, Yanofsky C. 1985. Isolation and characterization of genes differentially expressed during conidiation of Neurospora crassa. Mol Cell Biol 5:849–855. doi: 10.1128/mcb.5.4.849-855.1985. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Roberts A, Berlin V, Hager K, Yanofsky C. 1988. Molecular analysis of a Neurospora crassa gene expressed during conidiation. Mol Cell Biol 8:2411–2418. doi: 10.1128/mcb.8.6.2411-2418.1988. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Krijgsheld P, Bleichrodt R, Van Veluw G, Wang F, Müller W, Dijksterhuis J, Wösten H. 2013. Development in aspergillus. Stud Mycol 74:1–29. doi: 10.3114/sim0006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Chang P-K, Scharfenstein LL, Ehrlich KC, Wei Q, Bhatnagar D, Ingber BF. 2012. Effects of laeA deletion on Aspergillus flavus conidial development and hydrophobicity may contribute to loss of aflatoxin production. Fungal Biol 116:298–307. doi: 10.1016/j.funbio.2011.12.003. [DOI] [PubMed] [Google Scholar]
- 38.Kong Q, Chi C, Yu J, Shan S, Li Q, Li Q, Guan B, Nierman WC, Bennett JW. 2014. The inhibitory effect of Bacillus megaterium on aflatoxin and cyclopiazonic acid biosynthetic pathway gene expression in Aspergillus flavus. Appl Microbiol Biotechnol 98:5161–5172. doi: 10.1007/s00253-014-5632-8. [DOI] [PubMed] [Google Scholar]
- 39.Pfannenstiel BT, Greco C, Sukowaty AT, Keller NP. 2018. The epigenetic reader SntB regulates secondary metabolism, development and global histone modifications in Aspergillus flavus. Fungal Genet Biol 120:9–18. doi: 10.1016/j.fgb.2018.08.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Soukup AA, Farnoodian M, Berthier E, Keller NP. 2012. NosA, a transcription factor important in Aspergillus fumigatus stress and developmental response, rescues the germination defect of a laeA deletion. Fungal Genet Biol 49:857–865. doi: 10.1016/j.fgb.2012.09.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Vienken K, Fischer R. 2006. The Zn (II) 2Cys6 putative transcription factor NosA controls fruiting body formation in Aspergillus nidulans. Mol Microbiol 61:544–554. doi: 10.1111/j.1365-2958.2006.05257.x. [DOI] [PubMed] [Google Scholar]
- 42.Jin FJ, Takahashi T, Matsushima K-i, Hara S, Shinohara Y, Maruyama J-i, Kitamoto K, Koyama Y. 2011. SclR, a basic helix-loop-helix transcription factor, regulates hyphal morphology and promotes sclerotial formation in Aspergillus oryzae. Eukaryot Cell 10:945–955. doi: 10.1128/EC.00013-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Duran RM, Gregersen S, Smith TD, Bhetariya PJ, Cary JW, Harris-Coward PY, Mattison CP, Grimm C, Calvo AM. 2014. The role of Aspergillus flavus veA in the production of extracellular proteins during growth on starch substrates. Appl Microbiol Biotechnol 98:5081–5094. doi: 10.1007/s00253-014-5598-6. [DOI] [PubMed] [Google Scholar]
- 44.Kawasaki L, Castañeda-Bueno M, Sánchez-Paredes E, Velázquez-Zavala N, Torres-Quiroz F, Ongay-Larios L, Coria R. 2008. Protein kinases involved in mating and osmotic stress in the yeast Kluyveromyces lactis. Eukaryot Cell 7:78–85. doi: 10.1128/EC.00362-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Yuan J, Chen Z, Guo Z, Li D, Zhang F, Shen J, Zhang Y, Wang S, Zhuang Z. 2018. PbsB regulates morphogenesis, aflatoxin B1 biosynthesis, and pathogenicity of Aspergillus flavus. Front Cell Infect Microbiol 8:162. doi: 10.3389/fcimb.2018.00162. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Abad A, Fernández-Molina JV, Bikandi J, Ramírez A, Margareto J, Sendino J, Hernando FL, Pontón J, Garaizar J, Rementeria A. 2010. What makes Aspergillus fumigatus a successful pathogen? Genes and molecules involved in invasive aspergillosis. Rev Iberoam Micol 27:155–182. doi: 10.1016/j.riam.2010.10.003. [DOI] [PubMed] [Google Scholar]
- 47.Mellon JE, Dowd MK, Cotty PJ. 2005. Substrate utilization by Aspergillus flavus in inoculated whole corn kernels and isolated tissues. J Agric Food Chem 53:2351–2357. doi: 10.1021/jf040276g. [DOI] [PubMed] [Google Scholar]
- 48.Yu J. 2012. Current understanding on aflatoxin biosynthesis and future perspective in reducing aflatoxin contamination. Toxins (Basel) 4:1024–1057. doi: 10.3390/toxins4111024. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Zhao X, Spraker JE, Bok JW, Velk T, He Z-M, Keller NP. 2017. A cellular fusion cascade regulated by LaeA is required for sclerotial development in Aspergillus flavus. Front Microbiol 8:1925. doi: 10.3389/fmicb.2017.01925. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Chang P-K, Scharfenstein LL, Wei Q, Bhatnagar D. 2010. Development and refinement of a high-efficiency gene-targeting system for Aspergillus flavus. J Microbiol Methods 81:240–246. doi: 10.1016/j.mimet.2010.03.010. [DOI] [PubMed] [Google Scholar]
- 51.Szewczyk E, Nayak T, Oakley CE, Edgerton H, Xiong Y, Taheri-Talesh N, Osmani SA, Oakley BR, Oakley B. 2006. Fusion PCR and gene targeting in Aspergillus nidulans. Nat Protoc 1:3111–3120. doi: 10.1038/nprot.2006.405. [DOI] [PubMed] [Google Scholar]
- 52.Shimizu K, Keller NP. 2001. Genetic involvement of a cAMP-dependent protein kinase in a G protein signaling pathway regulating morphological and chemical transitions in Aspergillus nidulans. Genetics 157:591–600. doi: 10.1093/genetics/157.2.591. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Chang P-K, Zhang Q, Scharfenstein L, Mack B, Yoshimi A, Miyazawa K, Abe K. 2018. Aspergillus flavus GPI-anchored protein-encoding ecm33 has a role in growth, development, aflatoxin biosynthesis, and maize infection. Appl Microbiol Biotechnol 102:5209–5220. doi: 10.1007/s00253-018-9012-7. [DOI] [PubMed] [Google Scholar]
- 54.Chang P-K, Horn BW, Dorner JW. 2009. Clustered genes involved in cyclopiazonic acid production are next to the aflatoxin biosynthesis gene cluster in Aspergillus flavus. Fungal Genet Biol 46:176–182. doi: 10.1016/j.fgb.2008.11.002. [DOI] [PubMed] [Google Scholar]
- 55.Trapnell C, Williams BA, Pertea G, Mortazavi A, Kwan G, Van Baren MJ, Salzberg SL, Wold BJ, Pachter L. 2010. Transcript assembly and quantification by RNA-Seq reveals unannotated transcripts and isoform switching during cell differentiation. Nat Biotechnol 28:511–515. doi: 10.1038/nbt.1621. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Patro R, Duggal G, Love MI, Irizarry RA, Kingsford C. 2017. Salmon provides fast and bias-aware quantification of transcript expression. Nat Methods 14:417–419. doi: 10.1038/nmeth.4197. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Supplemental material. Download SPECTRUM00791-21_Supp_1_seq15.xls, XLS file, 0.4 MB (447.7KB, xls)
Supplemental material. Download SPECTRUM00791-21_Supp_2_seq16.xls, XLS file, 0.4 MB (409KB, xls)
Supplemental material. Download SPECTRUM00791-21_Supp_3_seq17.xls, XLS file, 0.3 MB (363.2KB, xls)
Supplemental material. Download SPECTRUM00791-21_Supp_4_seq14.pdf, PDF file, 0.7 MB (715.2KB, pdf)
Data Availability Statement
The results of the RNA-seq data were submitted to NCBI's GEO database and assigned accession no. GSE179978.