Abstract
Epstein-Barr virus (EBV) DNA load monitoring in peripheral blood has been shown to be a useful tool for the diagnosis of aberrant EBV infections. In the present study we compared the relative diagnostic values of EBV DNA load monitoring in unfractionated whole blood and simultaneously obtained serum or plasma samples from Burkitt's lymphoma (BL) patients, transplant recipients, human immunodeficiency virus (HIV)-infected individuals, and infectious mononucleosis (IM) patients by a quantitative competitive PCR (Q-PCR). The EBV DNA load in BL patients was mainly situated in the cellular blood compartment (up to 4.5 × 106 copies/ml). EBV DNA loads in unfractionated whole blood and parallel serum samples showed no correlation. In transplant recipients, IM patients, and HIV-infected patients, the EBV burden in the circulation was almost exclusively restricted to the cellular blood compartment, because serum or plasma samples from these patients yielded negative results by Q-PCR, despite high viral loads in corresponding whole-blood samples. A 10-fold more sensitive but qualitative BamHI-W-repeat PCR occasionally revealed the presence of EBV at <2,000 copies of EBV DNA per ml of serum. Spiking of 100 copies of EBV DNA in samples with negative Q-PCR results excluded the presence of inhibitory factors in serum or plasma that influenced the Q-PCR result. Serum samples from all populations were often positive for β-globin DNA, indicating cell damage in vivo or during serum preparation. We conclude that serum is an undesirable clinical specimen for EBV DNA load monitoring because it omits the presence of cell-associated virus and uncontrolled cell lysis may give irreproducible results or overestimation of the DNA load. Unfractionated whole blood is strongly preferred since it combines all blood compartments that may harbor EBV and it best reflects the absolute viral burden in the patient's circulation.
Epstein-Barr virus (EBV), a widespread human γ1-herpesvirus and the etiological agent of infectious mononucleosis, is associated with a broad spectrum of epithelio- and lymphoproliferative disorders. In immunocompetent hosts these include Burkitt's lymphoma, Hodgkin's disease, T- or NK-cell lymphoma, B-cell non-Hodgkin's lymphoma, nasopharyngeal carcinoma, and gastric carcinoma. In immunocompromised individuals, EBV is strongly associated with oral hairy leukoplakia, AIDS-related lymphoma, and both early- and late-onset posttransplantation lymphoproliferative disease (PTLD) (34).
In order to define individuals at high risk for the development of EBV-linked disease, a variety of nucleic acid diagnostic methods for detection and quantification of the virus in peripheral blood have been developed. These methods include semiquantitative PCR (12, 21, 26, 28), end-point dilution PCR (14), quantitative competitive PCR (Q-PCR) (2, 3, 27, 30), and real-time PCR (15, 19, 23). These studies revealed the clinical relevance of EBV load monitoring for the diagnosis of EBV-associated disorders and assessment of the efficacy of therapeutic intervention. However, standardization between different methods and institutes and definition of EBV load cutoff levels in different patient populations are required (24).
Although elevated peripheral blood EBV loads are observed in both immunocompetent and immunocompromised patients with epithelio- or lymphoproliferative disease, virtually nothing is known about the physical state of EBV detected in the peripheral blood of these patients or in healthy carriers. Three kinds of viral infection are thought to exist (25) and include a “silent” latent state with little or even no viral gene expression activity, a latent, growth-transforming infection of B cells with transcription of a limited number of latency genes, and a lytic infection in which infectious virus is produced.
Several clinical specimens have been used to determine EBV loads in the blood compartment including unfractionated whole (2, 30), serum (4, 9, 10, 17, 19), plasma (20, 35), and isolated peripheral blood leukocytes and mononuclear cells (14, 21, 26, 27, 28). However, no study thus far has analyzed the amount of EBV DNA in whole blood and simultaneously obtained serum or plasma by a standardized Q-PCR. Such a comparative study could provide insight into the type of EBV replication that occurs in different patient populations, either immunocompetent or immunocompromised patient populations. In addition, it might provide important clues about the type of clinical specimen that should be used for viral load monitoring in these patients.
The aim of the study described here was to compare EBV DNA loads in whole blood and serum or plasma simultaneously obtained from patients with different EBV-associated disorders. For this, a highly reproducible single-copy EBV gene-based Q-PCR and a sensitive multicopy target PCR for the BamHI-W repeat region of the EBV genome were used. We found that the EBV DNA loads in most patients with EBV-associated disorders are restricted to the cellular compartment, with no elevated EBV DNA loads in serum or plasma. When serum was found to be EBV DNA positive, this was associated with parallel detectable cellular DNA signals, suggesting cell death or damage as the source of viral DNA in the serum. We conclude that serum and plasma are unsuitable clinical specimens for monitoring in of EBV loads Burkitt's lymphoma patients, transplant recipients, human immunodeficiency virus (HIV)-infected individuals, and infectious mononucleosis patients. Whole blood is the preferred clinical sample type, as it is simple, is more standardized and directly reflects the overall dynamic changes in EBV DNA load in the circulation.
MATERIALS AND METHODS
Healthy donors.
Whole-blood samples (n = 18) and simultaneously obtained plasma samples (n = 18) were obtained from healthy laboratory volunteers (n = 11). All donors were EBV seropositive with no serological signs of viral reactivation, as determined by standard EBV serology and immunoblot analysis (33).
Patients with EBV-associated proliferative disorders.
Twelve whole-blood samples and corresponding serum samples were obtained from juvenile patients from Malawi with Burkitt's lymphoma. All samples were taken at the time of diagnosis, before the start of therapeutic intervention. As a control population, family members, mostly mothers of the Burkitt's lymphoma patients, were selected (African controls; n = 12). A whole-blood sample and a simultaneous serum sample were obtained from all individuals (kindly provided by L. Molineux and R. Broadhead, University of Malawi Medical School, Blantyre, Malawi).
Prospectively collected follow-up whole-blood samples (n = 35) and parallel serum samples (n = 35) were obtained from lung transplant recipients (n = 4) diagnosed with PTLD. Samples were obtained before, during, and after PTLD diagnosis and at periods of either increased, decreased, or maintenance immune suppression levels (samples were kindly provided by Erik Verschuuren and Hauw The, Department of Clinical Immunology, University Hospital Groningen, The Netherlands).
Nine plasma samples and two corresponding whole-blood samples were obtained from two HIV-infected individuals with increased EBV DNA loads (samples were kindly provided by Paul Smits, Slotervaart Hospital, Amsterdam, The Netherlands). These patients were receiving highly active anti-retroviral therapy but were not receiving any antiherpesvirus therapy at the time of sample collection.
Whole-blood and corresponding serum samples were obtained from five patients with infectious mononucleosis (all kindly provided by Nadja Prang, Medizinische Immunologische Laboratorien, Munich, Germany). Samples were taken at the time of diagnosis, which was done by routine serology.
Isolation of DNA from whole blood, serum, and plasma.
One milliliter of freshly obtained whole blood was diluted in 9 ml of NASBA lysis buffer (5 M guanidine thiocyanate, 0.1 M Tris · HCl [pH 6.4], 1.2% Triton X-100, 20 mM EDTA, Organon Teknika, Boxtel, The Netherlands) and stored at −80°C until use. DNA was isolated from 1 ml of blood lysate (equivalent to 0.1 ml of whole blood) by silica-based extraction essentially as described by Boom et al. (6). Whole blood from African Burkitt's lymphoma patients and African controls was directly frozen in liquid nitrogen, stored at −80°C, and lysed by thawing in NASBA lysis buffer at the time of use. DNA was subsequently isolated as described above.
Serum or plasma samples were stored at −20°C until use. For DNA isolation they were diluted 1:9 in NASBA lysis buffer, and DNA was isolated as described above for whole blood.
As a positive control, healthy donor serum was spiked with defined amounts of DNA isolated from EBV-positive JY cells. As a negative control, serum from an EBV-negative donor and water were included in all isolations.
Qualitative PCR and Q-PCR.
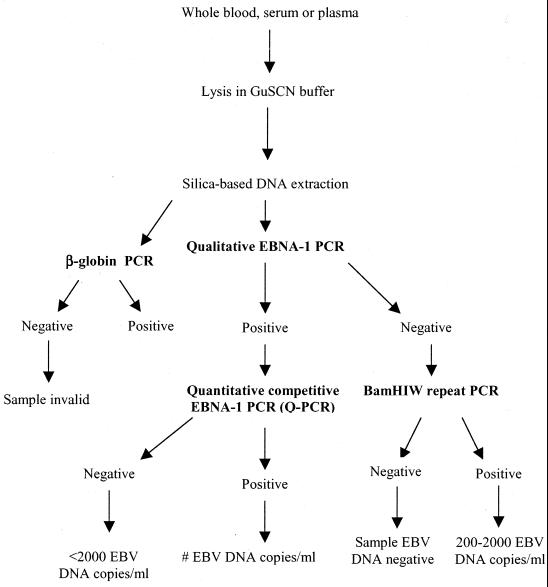
An outline of the experimental approach is depicted in Fig. 1. First, the EBV DNA status of whole blood, serum, or plasma samples was determined by qualitative EBNA-1 PCR, which has a sensitivity of approximately 10 genomic EBV DNA copies or approximately 1 EBV-infected cell (30). The cutoff value of this PCR was shown to be clinically relevant since the values for all healthy EBV-positive donors were below this value (31).
FIG. 1.
Experimental approach for EBV DNA load monitoring in whole-blood, serum, and plasma samples. After DNA isolation by silica-based DNA extraction, samples were prescreened by qualitative EBNA-1 PCR. The EBV DNA load in positive samples was subsequently quantified by Q-PCR. Samples negative by the qualitative EBNA-1 PCR were additionally amplified by the BamHI-W-repeat PCR in order to detect putative small (trace) amounts of EBV. DNA quality was assessed by β-globin PCR.
The EBV DNA loads in samples positive by qualitative EBNA-1 PCR were subsequently determined by Q-PCR with EBNA-1 combined with enzyme immunoassay detection, precisely as described previously (30). An equivalent of 5 μl of a whole-blood, serum, or plasma sample was used as input in the PCR, with each sample spiked with three different amounts of internal standard plasmid DNA in three different reaction mixtures (10, 100 or 1,000 copies per reaction mixture). A fixed amount of plasmid containing the wild-type amplicon served as a control for quantification in all Q-PCR runs. To ensure the validity of the results, several precautions were taken to avoid product carryover and PCR contamination (16). In all experiments appropriate negative controls were included and all clinical samples were screened blindly.
The clinically relevant cutoff value of the Q-PCR is 2,000 EBV DNA copies/ml, which is based on the low EBV DNA loads normally detected in the blood of healthy EBV-seropositive donors (1, 30, 31).
BamHI-W-repeat PCR.
Samples that were negative by the EBNA-1 PCR were additionally tested by a PCR for the large internal BamHI-W repeat, which was performed as described previously (13). This was done in order to assess qualitatively the EBV DNA status of these samples by using the most sensitive EBV PCR available and detect putative small (trace) amounts of EBV in the range of 200 to 2,000 copies/ml. Although this PCR in theory is more sensitive than the EBNA-1 PCR, the BamHI-W-repeat PCR is not suitable for quantitation since the number of BamHI-W repeats differs between clinical EBV isolates. PCR products were detected by enzyme immunoassay by the same protocol used for the EBNA-1 PCR (30) but with 1× SSC (1× SSC is 0.15 M Nacl plus 0.015 M sodium nitrate)–0.5% Tween 20 instead of 4× SSC–0.5% Tween 20 as the wash buffer and with pEBV (5′-AATCTGACACTTTAGAGCTCTGGAGGACTT-3′) as the digoxigenin-labeled oligonucleotide. The BamHI-W-repeat PCR has a sensitivity of at least 10 plasmid targets, containing a single BamHI-W insert, which in theory equals approximately 1 EBV genome (13).
β-Globin PCR.
To check for the presence of cellular DNA in whole blood, serum, or plasma, β-globin PCR was performed as described previously (7). The positivity of the samples was assessed by standard agarose gel analysis.
RESULTS
The results of the qualitative and quantitative PCRs with whole blood and serum or plasma from various patient and control groups are summarized in Table 1.
TABLE 1.
PCR results for EBV and β-globin with whole-blood, serum, and plasma samples from healthy donors and patients with EBV-related disease
| Population | Qualitative EBNA-1 PCR (no. of positive samples/no. of samples tested)
|
Quantitative EBNA-1 PCR (no. of samples with values above cutoff value/no. of samples tested)a
|
No. of serum or plasma samples/no. of samples tested additionally positive by BamHI-W-repeat PCR | β-Globin PCR result (no. of positive samples/no. of samples tested)
|
Range of EBV DNA load (no. of copies/ml)
|
||||
|---|---|---|---|---|---|---|---|---|---|
| Whole blood | Serum or plasma | Whole blood | Serum or plasma | Whole blood | Serum or plasma | Whole blood | Serum or plasma | ||
| Healthy EBV carriers (n = 11) | 1/18 | 1/18 | 0/1 | 0/1 | 0/18 | 18/18 | 13/18 | ||
| Burkitt's lymphoma patients (n = 12) | 12/12 | 12/12 | 12/12 | 8/12 | 12/12 | 12/12 | 14,600–4,591,900 | 4,400–108,600 | |
| African controls (n = 12) | 12/12 | 7/12 | 3/12 | 0/12 | 12/12 | 12/12 | 3,800–17,500 | ||
| PTLD patients (n = 4) | 35/35 | 15/35 | 30/35 | 0/15 | 2/35 | 35/35 | 35/35 | 2,600–308,000 | |
| HIV infected patients (n = 2) | 2/2 | 0/9 | 2/2 | 0/9 | 0/9 | 2/2 | 0/9 | 15,600–89,400 | |
| Infectious mononucleosis patients (n = 5) | 4/5 | 0/5 | 4/5 | 0/5 | 4/5 | 5/5 | 4/5 | 34,000–600,000 | |
Quantitative EBNA-1 PCR cut off value, 2,000 EBV DNA copies/ml of blood, serum, or plasma.
Healthy donors.
Of 18 whole-blood samples from healthy donors tested, 1 sample was positive by the qualitative EBNA-1 PCR, but the amount of EBV DNA in the sample was below the cutoff value of 2,000 copies/ml in the Q-PCR. The same results were found for the corresponding serum sample from this healthy donor. All other whole-blood and serum samples were also negative by the qualitative BamHI-W-repeat PCR, confirming the very low EBV DNA levels in the circulation of healthy donors (1, 2, 27, 30). Thirteen of 18 serum samples and all 18 whole-blood samples were positive by the β-globin PCR.
Burkitt's lymphoma patients.
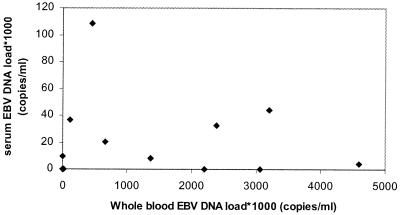
Twelve of 12 whole-blood samples from Burkitt's lymphoma patients had EBV DNA loads above the limit of detection of the Q-PCR. The EBV DNA loads in these samples ranged from 14,600 to 4,591,900 copies/ml of blood. Eight of the 12 corresponding serum samples had EBV DNA loads above the cutoff level of the Q-PCR, with EBV DNA loads in the range of 4,400 to 108,600 copies/ml of serum. However, there was a poor correlation between whole-blood EBV DNA loads and serum EBV DNA loads (Fig. 2). Furthermore, all serum samples contained β-globin DNA, indicating cell damage either in vivo or ex vivo during serum preparation.
FIG. 2.
Comparison of EBV DNA load in unfractionated whole blood and simultaneously obtained serum of Burkitt's lymphoma patients and African controls.
African controls.
All 12 whole-blood samples from African control donors were positive for EBV by qualitative EBNA-1 PCR. Only 3 of 12 whole-blood samples from African controls had EBV DNA loads above the cutoff value by the Q-PCR, with values ranging from 3,800 to 17,500 EBV DNA copies/ml of blood. All corresponding serum samples had EBV DNA loads below the cutoff value of the Q-PCR, although EBV DNA could be detected by qualitative EBNA-1 PCR in 7 of 12 of the samples. Again, all serum samples also contained cellular DNA, as they were found to be positive by the β-globin PCR.
PTLD patients.
All 35 whole-blood samples from PTLD patients tested were positive by the qualitative EBNA-1 PCR. Thirty of 35 samples had EBV DNA loads above the limit of detection of the Q-PCR, with EBV DNA loads ranging from 2,600 to 308,000 EBV DNA copies/ml of blood. Fifteen of 35 serum samples were positive only by the qualitative EBNA-1 PCR, while an additional 2 serum samples were positive by the BamHI-W-repeat PCR. All EBV DNA-positive serum samples had EBV DNA loads below the cutoff value of the Q-PCR, indicating that most, if not all, of the EBV burden in these patients could be attributed to the cellular blood compartment. All 35 serum samples were positive for β-globin, indicating the presence of cellular DNA and the absence of sample-related inhibition of the PCR.
HIV-infected individuals.
Samples from two HIV-infected patients were included in the study. Both had highly elevated EBV DNA loads in their circulations (15,600 and 89,400 EBV DNA copies/ml of blood, respectively). No EBV DNA could be detected in the corresponding plasma samples by EBNA-1 PCR or by the more sensitive BamHI-W-repeat PCR, indicating the complete absence of any detectable cell-free EBV DNA in the blood of these patients. EBV DNA was also absent from plasma samples (n = 7) that were obtained at several time points in the year before the whole-blood samples were taken. None of the plasma samples were positive by the β-globin PCR.
Infectious mononucleosis patients.
Four of five infectious mononucleosis patients had high whole-blood EBV DNA loads (34,000, 600,000, 74,000, and 37,000 copies/ml of blood, respectively). All simultaneously obtained serum samples had EBV DNA loads below the limit of detection by the EBNA-1 PCR; but EBV DNA could be detected in four of five serum samples by the BamHI-W-repeat PCR, and these four samples were also positive for β-globin DNA. This indicates that very low levels of EBV DNA may be present in the serum or, more likely, that EBV DNA is released from damaged EBV-positive cells during serum preparation.
Control for putative PCR inhibition by serum or plasma.
In order to exclude the possibility of putative PCR inhibition as a reason for the absence of detectable EBV DNA, eluates of serum and plasma samples, which tested negative by both the Q-PCR and the BamHI-W-repeat PCR, were spiked with 100 copies of plasmid containing the wild-type EBNA-1 target and amplified (30). All samples became positive by PCR, indicating the absence of PCR inhibitors in serum and plasma samples.
DISCUSSION
Monitoring of the EBV DNA load in the circulation has been shown to be a useful parameter for the diagnosis of EBV infection in various patient populations (3, 9, 26, 31). However, interlaboratory differences in sample type and EBV DNA detection assays have thus far limited appropriate standardization of EBV DNA load determinations and prevented direct comparison of different patient populations (24).
In the present study we evaluated the diagnostic value of unfractionated whole blood and serum or plasma as clinical specimens for determination of the EBV DNA loads in the blood compartment of patients with different EBV-associated proliferative disorders. We propose the use of unfractionated whole blood as the clinical specimen in quantitative EBV DNA assays, which is a step toward standardization of such assays.
In Burkitt's lymphoma patients, elevated EBV DNA loads were detected in both serum and whole blood samples at the time of primary diagnosis, indicating that the EBV burden may be present in both the cellular and fluid blood compartments of these patients. The major part of the EBV burden, however, is situated in the cellular fraction since the serum EBV levels are much lower than the whole-blood EBV levels. In addition, serum EBV DNA loads do not correlate with the levels in whole blood (Fig. 2). Therefore, serum is undesirable as a clinical specimen for viral load monitoring in Burkitt's lymphoma patients. The presence of EBV DNA in the serum of these patients could be due to lytic EBV replication, viral DNA release from damaged apoptotic or necrotic cells into the serum in vivo, or damage of the abundantly present EBV-positive B cells ex vivo as an artifact of blood coagulation during serum preparation. The positive result for β-globin DNA in serum samples from all Burkitt's lymphoma patients and African control donors and the absence of lytic replication in Burkitt's lymphoma patients are strongly suggestive of the latter.
We could not detect significantly elevated EBV DNA loads in the serum of transplant recipients, despite highly elevated EBV DNA loads in simultaneously obtained whole-blood speciments. This indicates that the elevated EBV DNA loads in these patients can be attributed mainly to the cellular compartment of the blood, directly reflecting proliferation of latently infected B cells, with very little or no lytic viral replication and associated release of virion DNA. Although viral DNA was detected by qualitative PCR in some serum samples from lung transplant recipients, the absence of significantly elevated viral DNA levels may be a result of the extensive use of antiviral drugs such as acyclovir, which was given both prophylactically (low dose) and as PTLD therapy (high dose), and ganciclovir and foscarnet, which were given for the treatment of active cytomegalovirus infection. These drugs inhibit lytic viral replication (36) and thus limit elevation of EBV levels in serum, which would be expected to originate from virions produced during lytic replication of the virus. However, a larger, more comprehensive study is required to further substantiate this hypothesis.
Our results indicate the poor diagnostic and predictive values of tests with serum and plasma samples as clinical specimens. In a previous study we showed that the whole-blood EBV load in lung transplant recipients can be used to both predict and diagnose PTLD and strongly correlates with changes in the immune status of the patients (31). The absence of any significantly elevated EBV DNA levels in the serum of the tested patients marks this specimen type as unsuitable for quantitative monitoring of EBV in lung transplant patients.
Similar to the findings in for Burkitt's lymphoma patients and transplant recipients, we found a complete lack of correlation between whole-blood EBV DNA levels and plasma EBV DNA levels in HIV-infected individuals. This is also the case for whole blood and serum samples from infectious mononucleosis patients. It is noteworthy that none of these patients was receiving acyclovir or ganciclovir-foscarnet as antiherpesvirus treatment at the time of sample collection. Our results suggest that the EBV DNA in these patients is mostly linked to the cellular blood compartment, i.e., proliferation of EBV-positive B cells.
Several studies indicated that the serum EBV DNA load could be used as a tumor marker for nasopharyngeal carcinoma (19, 20, 22, 29). Possibly, the EBV DNA present in the serum of patients with nasopharyngeal carcinoma may reflect lytic viral replication and virus secretion in some differentiating epithelial cells in the nasopharyngeal carcinoma-derived tumors, which is also reflected by the characteristic presence of immunoglobulin A antibodies directed to early and late structural antigens (37). It was recently shown that irradiation can induce EBV lytic gene expression in nasopharyngeal carcinoma-derived tumor cells (18). Whether the increased EBV DNA levels in nasopharyngeal carcinoma patients reflect (treatment-induced) release of virion DNA or tumor cell-derived DNA remains to be established, however.
One theoretical reason for the absence of EBV DNA in the majority of serum and plasma samples in the present study could be inhibition of the PCR. However, this is very unlikely since parallel whole-blood samples did contain amplifiable viral DNA and whole blood contains more putative inhibitors of PCR. The fact that a small amount of EBV plasmid DNA spiked into EBV DNA-negative serum and plasma eluates could be amplified very easily and the fact that nearly all serum samples contained β-globin as detected by PCR indicates the absence of PCR inhibition. We conclude that PCR negativity for EBV targets in serum and plasma samples represent the true absence of elevated levels of cell-free virus or viral DNA fragments in a patient's circulation.
The use of unfractionated whole blood has several additional advantages over the use of plasma or serum. It combines all compartments that may harbor EBV and is therefore the best reflection of the absolute EBV burden in the circulation, in contrast to isolated cell fractions. Although a fixed amount of leukocytes is frequently used as input in EBV PCR assays (14, 26, 27, 28), cell numbers may vary considerably in a patient over time and between different patients due to immunomodulation and underlying disease, thereby complicating interpretation. In addition, the isolation of leukocytes may introduce uncontrolled cell loss due to technical failure and inappropriate handling. Especially in patient populations with fluctuating B- and T-cell counts, such as transplant recipients and AIDS patients, the amount of EBV DNA per milliliter of blood gives a better indication of the absolute viral burden in the circulatory compartment than the amount of EBV DNA per, for example, 105 B cells or peripheral blood mononuclear cells. Whole blood represents a simple and uniform sample type.
We frequently observe DNA of cellular origin in serum samples as determined by β-globin PCR. DNA of cellular origin is generally absent from plasma, indicating that preparation of plasma leads to less cell damage than preparation of serum, as expected. Cell lysis is probably inevitable during blood coagulation. This again indicates that serum is an unfavorable clinical specimen, as in patients with high cell-associated EBV loads (e.g., Burkitt's lymphoma patients), and that preparation artifacts that appear during sample preparation could cause irreproducible results, putative overestimation of EBV loads in serum, and poor standardization of tests for EBV DNA load monitoring. One milliliter of unfractionated whole blood is a simple and reliable sample type which represents a suitable unit for EBV DNA load determination, irrespective of latent or lytic replication or the occurence of cell damage. Whole blood may be stored as such upon immediate freezing in liquid nitrogen, as we showed for the whole blood from Burkitt's lymphoma patients. Simple guanidine thiocyanate lysis of whole blood enables long-term storage and omits the need for serum, plasma, or cell preparation techniques which are laborious and complicated and which increase the risk of sample carryover. Furthermore, only a small volume of unfractionated whole blood (0.1 to 1 ml) is needed for silica-based DNA extraction, making this method much less invasive for the patient than cell separation procedures, for which relatively large volumes of blood are needed. The use of unfractionated whole blood enables the simultaneous screening for other microorganisms (e.g., cytomegalovirus and HIV) and cellular targets. Finally, simultaneous DNA and RNA isolation from lysed whole blood by silica extraction allows the analysis of viral and cellular transcription profiles (5, 11, 32).
In conclusion, when only serum or plasma is used, a (small) part of the viral burden in a patient's circulation is quantified and cell-associated virus is omitted. Use of a serum preparation may lead to the nonreproducible release of EBV DNA from the cellular compartment as a consequence of cell death. Also, plasma DNA could originate from cell death or from remaining variable amounts of leukocytes (8). Therefore, both serum and plasma are considered undesirable clinical specimens for use in EBV DNA load monitoring. Unfractionated whole blood is the prefered clinical specimen in lung transplant recipients,. HIV-infected individuals, infectious mononucleosis patients, and Burkitt's lymphoma patients. The viral loads in these patients are mainly present in the cellular blood compartment. The use of unfractionated whole blood in diagnostic settings is an important step toward standardization of EBV DNA load monitoring in patients at risk for the development of EBV-related disease.
ACKNOWLEDGMENTS
We are grateful to all collaborators who kindly provided the clinical specimens that were used in this study.
This work was supported by European Community grant 1C-18-CT96-0132.
REFERENCES
- 1.Babcock G J, Decker L L, Freeman R B, Thorley-Lawson D A. Epstein-Barr virus-infected resting memory B cells, not proliferating lymphoblasts, accumulate in the peripheral blood of immunosuppressed patients. J Exp Med. 1999;4:567–576. doi: 10.1084/jem.190.4.567. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Bai X, Hosler G, Rogers B, Dawson D B, Scheuermann R H. Quantitative polymerase chain reaction for human herpesvirus diagnosis and measurement of Epstein-Barr virus burden in posttransplant lymphoproliferative disorder. Clin Chem. 1997;43:1843–1849. [PubMed] [Google Scholar]
- 3.Baldanti F, Grossi P, Furione M, Simoncini L, Sarasini A, Comoli P, Maccario R, Fiocchi R, Gerna G. High levels of Epstein-Barr virus DNA in blood of solid-organ transplant recipients and their value in predicting posttransplant lymphoproliferative disorders. J Clin Microbiol. 2000;38:613–619. doi: 10.1128/jcm.38.2.613-619.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Barkholt L M, Dahl H, Enbom M, Linde A. Epstein-Barr virus DNA in serum after liver transplantation—surveillance of viral activity during treatment with different immunosuppressive agents. Transplant Int. 1998;9:439–445. doi: 10.1007/BF00336820. [DOI] [PubMed] [Google Scholar]
- 5.Blok M J, Christiaans M H, Goossens V J, van Hooff J P, Sillekens P, Middeldorp J M, Bruggeman C A. Early detection of human cytomegalovirus infection after kidney transplantation by nucleic acid sequence-based amplification. Transplantation. 1999;67:1274–1277. doi: 10.1097/00007890-199905150-00013. [DOI] [PubMed] [Google Scholar]
- 6.Boom R, Sol C J A, Salimans M M M, Jansen C L, Wertheim-van Dillen P M E, van der Noordaa J. Rapid and simple method for purification of nucleic acids. J Clin Microbiol. 1990;28:495–503. doi: 10.1128/jcm.28.3.495-503.1990. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.de Roda Husman A M, Walboomers J M M, van den Brule A J C, Meijer C J L M, Snijders P J F. The use of general primer GP5 and GP6 elongated at their 3′ ends with adjacent highly conserved sequences improves human papillomavirus detection by polymerase chain reaction. J Gen Virol. 1995;76:412–417. doi: 10.1099/0022-1317-76-4-1057. [DOI] [PubMed] [Google Scholar]
- 8.Fournie G J, Martres F, Pourrat J P, Alary C, Rumeau M. Plasma DNA as cell death marker in elderly patients. Gerontology. 1993;39:215–221. doi: 10.1159/000213536. [DOI] [PubMed] [Google Scholar]
- 9.Gallagher A, Armstrong A A, MacKenzie J, Shield L, Khan G, Lake A, Proctor S, Taylor P, Clements G B, Jarrett R F. Detection of Epstein-Barr virus (EBV) genomes in the serum of patients with EBV-associated Hodgkin's disease. Int J Cancer. 1999;84:442–448. doi: 10.1002/(sici)1097-0215(19990820)84:4<442::aid-ijc20>3.0.co;2-j. [DOI] [PubMed] [Google Scholar]
- 10.Gan Y, Sullivan J L, Sixbey J W. Detection of cell-free Epstein-Barr virus DNA in serum during acute infectious mononucleosis. J Infect Dis. 1994;170:436–439. doi: 10.1093/infdis/170.2.436. [DOI] [PubMed] [Google Scholar]
- 11.Gerna G, Baldanti F, Lilleri D, Parea M, Alessandrino E, Pagani A, Locatelli F, Middeldorp J M, Revello M G. Human cytomegalovirus immediate-early mRNA detection by nucleic acid sequence-based amplification as a new parameter for preemptive therapy in bone marrow transplant recipients. J Clin Microbiol. 2000;38:1845–1853. doi: 10.1128/jcm.38.5.1845-1853.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Gustafsson A, Levitsky V, Zou J, Frisan T, Dalianis T, Ljungman P, Ringden O, Winiarski J, Ernberg I, Masucci M G. Epstein-Barr virus (EBV) load in bone marrow transplant recipients at risk to develop posttransplant lymphoproliferative disease: prophylactic infusion of EBV-specific cytotoxic T cells. Blood. 2000;95:807–813. [PubMed] [Google Scholar]
- 13.Jiwa N M, Kanavaros P, van der Valk P, Walboomers J M M, Horstman A, Vos W, Mullink H, Meijer C J. Expression of c-myc and bcl-2 oncogene products in Reed-Sternberg cells independent of presence of Epstein-Barr virus. J Clin Pathol. 1993;46:211–217. doi: 10.1136/jcp.46.3.211. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Kenagy D N, Schlesinger Y, Weck K, Ritter J H, Gaudreault-Keener M M, Storch G A. Epstein-Barr virus DNA in peripheral blood leukocytes of patients with posttransplant lymphoproliferative disease. Transplantation. 1995;60:547–554. doi: 10.1097/00007890-199509270-00005. [DOI] [PubMed] [Google Scholar]
- 15.Kimura H, Morita M, Yabuta Y. Quantitative analysis of Epstein-Barr virus load by using a real-time PCR assay. J Clin Microbiol. 1999;37:132–136. doi: 10.1128/jcm.37.1.132-136.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Kwok S, Higuchi R. Avoiding false negatives with PCR. Nature. 1989;339:237–238. doi: 10.1038/339237a0. [DOI] [PubMed] [Google Scholar]
- 17.Limaye A P, Huang M, Atienza E E, Ferrenberg J M, Corey L. Detection of Epstein-Barr virus DNA in sera from transplant recipients with lymphoproliferative disorders. J Clin Microbiol. 1999;37:1113–1116. doi: 10.1128/jcm.37.4.1113-1116.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Liu J H, Huang D, Sun B F, Zhang X, Middeldorp J M, Klamut H, Liu F F. Efficacy of ionizing radiation combined with adenoviral p53 therapy in EBV-positive nasopharyngeal carcinoma. Int J Cancer. 2000;87:606–610. [PubMed] [Google Scholar]
- 19.Lo Y M, Chan L Y S, Lo K, Leung S, Zhang J, Chan A T C, Lee J C K, Hjelm N M, Johnson P J, Huang D P. Quantitative analysis of cell-free Epstein-Barr virus DNA in plasma of patients with nasopharyngeal carcinoma. Cancer Res. 1999;59:1188–1191. [PubMed] [Google Scholar]
- 20.Lo Y M, Chan L Y S, Chan A T C, Leung S, Lo K, Zhang J, Lee J C K, Hjelm N M, Johnson P J, Huang D P. Quantitative and temporal correlation between circulating cell-free Epstein-Barr virus DNA and tumor recurrence in nasopharyngeal carcinoma. Cancer Res. 1999;59:5452–5455. [PubMed] [Google Scholar]
- 21.Lucas K G, Burton R L, Zimmerman S E, Wang J, Cornetta K G, Robertson K A, Lee C H, Emanuel D J. Semiquantitative Epstein-Barr virus (EBV) polymerase chain reaction for the determination of patients at risk for EBV-induced lymphoproliferative disease after stem cell transplantation. Blood. 1998;91:3654–3661. [PubMed] [Google Scholar]
- 22.Mutirangura A, Porthanakasem W, Theamboonlers A, Sriurangpong V, Lertsanguansinchi P, Yenrudi S, Voravud N, Supiyaphun P, Poovorowan Y. Epstein-Barr virus DNA in serum of patients with nasopharyngeal carcinoma. Clin Cancer Res. 1998;4:665–669. [PubMed] [Google Scholar]
- 23.Niesters H G, van Esser J, Fries E, Wolthers K C, Cornelissen J, Osterhaus A D. Development of a real-time quantitative assay for detection of Epstein-Barr virus. J Clin Microbiol. 2000;38:712–715. doi: 10.1128/jcm.38.2.712-715.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Paya C V, Fung J J, Nalesnik M, A, Kieff E, Green M, Gores G. Epstein-Barr virus-induced posttransplant lymphoproliferative disorders. Transplantation. 1999;10:1517–1525. doi: 10.1097/00007890-199911270-00015. [DOI] [PubMed] [Google Scholar]
- 25.Rickinson A B, Lane P J L. Epstein-Barr virus: co-opting B-cell memory and migration. Curr Biol. 2000;10:R120–R123. doi: 10.1016/s0960-9822(00)00308-0. [DOI] [PubMed] [Google Scholar]
- 26.Riddler S A, Breinig M C, McKnight J L C. Increased levels of circulating Epstein-Barr virus (EBV)-infected lymphocytes and decreased EBV nuclear antigen antibody responses are associated with the development of posttransplant lymphoproliferative disease in solid-organ transplant recipients. Blood. 1994;84:972–984. [PubMed] [Google Scholar]
- 27.Rowe D T, Qu L, Reyes J, Jabbour N, Yunis E, Putnam P, Todo S, Green M. Use of quantitative competitive PCR to measure Epstein-Barr virus genome load in the peripheral blood of pediatric transplant recipients with lymphoproliferative disorders. J Clin Microbiol. 1997;35:1612–1615. doi: 10.1128/jcm.35.6.1612-1615.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Savoie A, Perpete C, Carpentier L, Joncas J, Alfieri C. Direct correlation between the load of Epstein-Barr virus-infected lymphocytes in the peripheral blood of pediatric transplant recipients and risk of lymphoproliferative disease. Blood. 1994;83:2715–2722. [PubMed] [Google Scholar]
- 29.Shotelersuk K, Khorprasert C, Sakdikul S, Pornthanakasem W, Voravud N, Mutirangura A. Epstein-Barr virus DNA in serum/plasma as a tumor marker for nasopharyngeal cancer. Clin Canc Res. 2000;6:1046–1051. [PubMed] [Google Scholar]
- 30.Stevens S J C, Vervoort M B H J, van den Brule A J C, Meenhorst P L, Meijer C J L M, Middeldorp J M. Monitoring of Epstein-Barr virus DNA load in peripheral blood by quantitative competitive PCR. J Clin Microbiol. 1999;37:2852–2857. doi: 10.1128/jcm.37.9.2852-2857.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Stevens S J C, Verschuuren E A M, Pronk I, van der Bij W, Harmsen M C, The T H, Meijer C J L M, van den Brule A J C, Middeldorp J M. Frequent monitoring of Epstein-Barr virus DNA load in unfractionated whole blood is essential for early detection of posttransplant lymphoproliferative disease in high-risk patients. Blood. 2001;97:1165–1171. doi: 10.1182/blood.v97.5.1165. [DOI] [PubMed] [Google Scholar]
- 32.van Deursen P B, Gunther A W, van Riel C C, van der Eijnden M M, Vos H L, van Gemen B, van Strijp D A, Tacken N M, Bertina R M. A novel quantitative multiplex NASBA method: application to measuring tissue factor and CD14 mRNA levels in human monocytes. Nucleic Acids Res. 1999;27:e15. doi: 10.1093/nar/27.17.e15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Van Grunsven W M J, Nabbe A, Middeldorp J M. Identification and molecular characterization of two diagnostically relevant marker proteins of the Epstein-Barr virus capsid antigen complex. J Med Virol. 1993;40:161–169. doi: 10.1002/jmv.1890400215. [DOI] [PubMed] [Google Scholar]
- 34.World Health Organization International Agency for Research on Cancer. IARC monographs on the evaluation of carcinogenic risks to humans. Vol. 70. Lyon, France: International Agency for Research on Cancer; 1997. Epstein-Barr virus and Kaposi's sarcoma herpesvirus/human herpesvirus 8. [Google Scholar]
- 35.Yamamoto M, Kimura H, Hironaka T, Hirai K, Hisegawa S, Kuzushima K, Shibata M, Morishima T. Detection and quantification of virus DNA in plasma of patients with Epstein-Barr virus-associated disease. J Clin Microbiol. 1995;33:1765–1768. doi: 10.1128/jcm.33.7.1765-1768.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Yao Q Y, Ogan P, Rowe M, Wood M, Rickinson A B. Epstein-Barr virus-infected B cells persist in the circulation of acyclovir-treated virus carriers. Int J Cancer. 1989;43:67–71. doi: 10.1002/ijc.2910430115. [DOI] [PubMed] [Google Scholar]
- 37.Zhang J X, Chen H L, Zong Y S, Chan K H, Nicholls J, Middeldorp J M, Sham J S, Griffin B E, Ng M H. Epstein-Barr virus expression within keratinizing nasopharyngeal carcinoma. J Med Virol. 1998;55:227–233. doi: 10.1002/(sici)1096-9071(199807)55:3<227::aid-jmv8>3.0.co;2-3. [DOI] [PubMed] [Google Scholar]