Graphical abstract
Abstract
Objective
The large-scale collection of observational data and digital technologies could help curb the COVID-19 pandemic. However, the coexistence of multiple Common Data Models (CDMs) and the lack of data extract, transform, and load (ETL) tool between different CDMs causes potential interoperability issue between different data systems. The objective of this study is to design, develop, and evaluate an ETL tool that transforms the PCORnet CDM format data into the OMOP CDM.
Methods
We developed an open-source ETL tool to facilitate the data conversion from the PCORnet CDM and the OMOP CDM. The ETL tool was evaluated using a dataset with 1000 patients randomly selected from the PCORnet CDM at Mayo Clinic. Information loss, data mapping accuracy, and gap analysis approaches were conducted to assess the performance of the ETL tool. We designed an experiment to conduct a real-world COVID-19 surveillance task to assess the feasibility of the ETL tool. We also assessed the capacity of the ETL tool for the COVID-19 data surveillance using data collection criteria of the MN EHR Consortium COVID-19 project.
Results
After the ETL process, all the records of 1000 patients from 18 PCORnet CDM tables were successfully transformed into 12 OMOP CDM tables. The information loss for all the concept mapping was less than 0.61%. The string mapping process for the unit concepts lost 2.84% records. Almost all the fields in the manual mapping process achieved 0% information loss, except the specialty concept mapping. Moreover, the mapping accuracy for all the fields were 100%. The COVID-19 surveillance task collected almost the same set of cases (99.3% overlaps) from the original PCORnet CDM and target OMOP CDM separately. Finally, all the data elements for MN EHR Consortium COVID-19 project could be captured from both the PCORnet CDM and the OMOP CDM.
Conclusion
We demonstrated that our ETL tool could satisfy the data conversion requirements between the PCORnet CDM and the OMOP CDM. The outcome of the work would facilitate the data retrieval, communication, sharing, and analysis between different institutions for not only COVID-19 related project, but also other real-world evidence-based observational studies.
1. Introduction
Widespread enthusiasm for big data approaches has led to increasing popularity in conducting large observational studies in the assessment of new medical technologies in recent years [1]. There is a growing consensus that the large-scale collection of observational data and digital technologies may be beneficial in advancing studies to address major public health concerns, such as the recent COVID-19 pandemic [2], [3]. As compared with other medical experimental studies such as randomized controlled trials (RCT), observational studies have several potential advantages including lower costs, better generalizability across a wider population, and a more rapid turnaround time [4], [5], [6]; in the context of the COVID-19 pandemic, the ability to conduct observational studies would drastically facilitate COVID-19 surveillance and research. The most commonly used data sources for real-world evidence-based observational studies include patient registries, electronic health record (EHR) data, administrative health insurance databases, pharmaceutical databases, and regulatory databases [7]. These databases differ however in both purpose and design, and a substantial issue in the context of utilizing this observational data to facilitate medical research that we are confronted with is how these disparate heterogeneous data sources can be integrated together.
Common data models (CDM) are an important tool that make it possible to integrate data from multiple sources. As each observational database may have a distinct physical format and use different medical terminologies or coding standards, CDMs are implemented utilizing a common data format, apply standardized data transformation rules and assumptions to the data, and develop common definitions and terminology during the data preparation process to help minimize variability and promote a common interpretation of the underlying data source [8]. CDM implementations aim to “standardize and facilitate the exchange, pooling, sharing, or storing of data from multiple sources [9]”. Within the last decade, several CDMs have been developed and widely deployed in medical observational studies for multiple sources/sites. Several CDMs currently deployed in support of medical studies include the National Patient-Centered Clinical Research Network (PCORnet) CDM [10], the Observational Medical Outcomes Partnership (OMOP) CDM [11], the Sentinel CDM [12], and the Informatics for Integrating Biology and the Bedside (i2b2) CDM [13]. As data standardization and data integration help improve data quality and facilitate data sharing [2], these CDMs have also been widely used in building data repositories in support of COVID-19 research.
The coexistence of multiple CDMs causes a potential issue that may make interoperability difficult between different institutions. Healthcare institutions typically do not support all of these different CDMs, but rather some specific subset (if any), and the choice of CDMs at any given institution often depends on the specific national initiatives for which the institution participates. Due to different design philosophies, each of these CDMs have their own data table format, value sets, terminologies, and value representations, and the data in different CDMs cannot be easily shared among different networks and institutions [14], [15]. On the other hand, many healthcare institutions have recently used different CDMs to standardize the COVID-19 data and build data warehouses to monitor the pandemic. It follows that building ETL (extract, transform and load) tools to promote the data transformation between different CDMs can accelerate the COVID-19 data retrieval, communication, sharing, and analysis in multi-institutional settings.
It is therefore critically important to develop tools to facilitate data transformation and evaluate the data consistency between different CDMs. Recently, there have been several studies that attempt to contribute to this topic. Klann et al. [14], [16], [17] conducted a series of studies that focus on developing tools that could convert i2b2 data into the OMOP CDM and the PCORnet CDM format. In 2017, FDA led a “Common Data Model Harmonization” (CDMH) project to attempt to facilitate broader access to data from different CDMs. The group developed an intermediary common data architecture named “BRIDG” (Biomedical Research Integrated Domain Group) between four CDMs (Sentinel, PCORNET, i2b2, OMOP) and the FHIR (Fast Healthcare Interoperability Resources) standard, to harmonize different CDMs and to support research and analyses across multiple data networks [15], [18]. In harmonizing between FHIR and OHDSI CDM, a Georgia Tech team [19], [20] developed an OMOP on FHIR Platform to map OMOP CDM to FHIR resources. For the transformation tool between the PCORnet CDM into the OMOP CDM, although there have been some efforts in coordinating the standard concepts between the two CDMs [21], to the best of our knowledge, no tooling has been designed to convert the PCORnet CDM into the OMOP CDM directly.
In this study, we focus on the development, evaluation, and validation of such an ETL tool that could transform the PCORnet CDM data into the OMOP CDM. To this end, we collaborate with two EHR data-driven COVID-19 surveillance projects, the National COVID Cohort Collaborative (N3C) [22] and the Minnesota EHR Consortium COVID-19 Project (MN EHR Consortium COVID-19) [23]. N3C is a partnership among the National Center for Advancing Translational Sciences (NCATS), the National Center for Data to Health (CD2H), and National Institute of General Medical Sciences (NIGMS), with the management by NCATS [22]. Key aims of the N3C include rapid collection of standardized clinical data to build a centralized national data resource for COVID-19 research. It follows that harmonization of data communication and analysis across the disparate data sites participating in the collaborative is one of the key objectives of N3C [24]. Similarly, the MN EHR Consortium COVID-19 Project, funded by the Minnesota Department of Health, attempts to identify COVID-19 related data through data-driven collaboration among members of Minnesota's health care community [25]. Due to the multi-institutional nature of this consortium, utilization of CDMs to standardize the COVID-19 data is of key interest. This work aims to help address both of these use cases. Specifically, the main contributions of this work are:
-
1)
We collaborate with N3C to develop an open-source ETL tool to facilitate the data conversion between the PCORnet CDM and the OMOP CDM, two widely used CDMs.
-
2)
We implement the ETL tool to the PCORnet CDM at Mayo Clinic and conduct several analyses to evaluate the feasibility of our ETL tool and identify the gaps between the PCORnet CDM and the OMOP CDM.
-
3)
We assess the capacity of data collection for COVID-19 surveillance in both the PCORnet CDM and the OMOP CDM by using data collection template of the MN EHR Consortium COVID-19 Project, and evaluate whether our ETL tool could completely support the COVID-19 data collection.
2. Materials
2.1. The PCORnet CDM
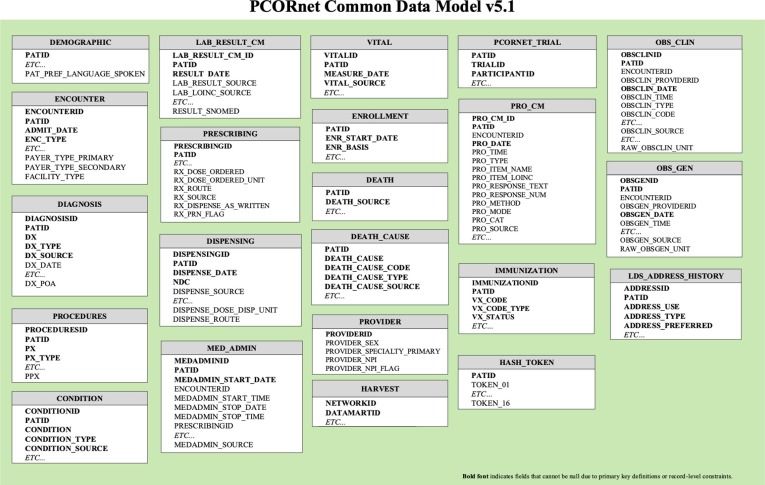
In 2013, Patient-Centered Outcomes Research Institute (PCORI) established PCORnet, a national Distributed Research Network (DRN). PCORnet focuses on building a national infrastructure that will enable the conduct of observational research and clinical trials while allowing each participating organization to maintain physical and operational control over its data [26], [27], [28]. The PCORnet CDM, a key component of PCORnet DRN infrastructure, was designed to provide an organization and representation for the data in 2014 [26]. Currently, the PCORnet CDM has released version 6.0, which contains 23 tables to represent all the EHR-related data. In this study, we use the PCORnet CDM v5.1 as a source database model to develop the ETL tool (Fig. 1 ).
Fig. 1.
Overview of tables in the PCORnet CDM version 5.1 [27].
2.2. The OMOP CDM
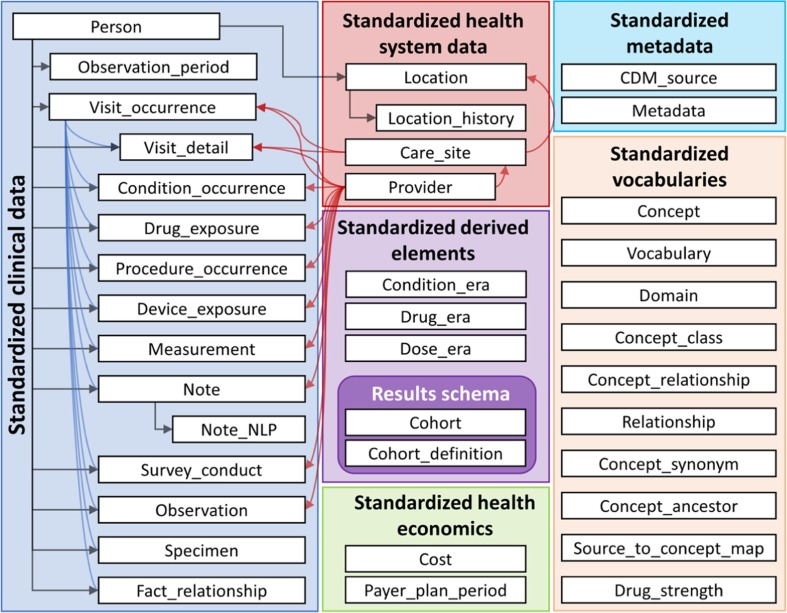
The OMOP CDM was developed by the Observational Medical Outcomes Partnership (OMOP), a project formerly chaired by the US Food and Drug Administration (FDA), administered by the Foundation for the National Institutes of Health (NIH), and funded by a consortium of pharmaceutical companies [2], [29]. Currently, the CDM is maintained by an open-science community, Observational Health Data Sciences and Informatics (OHDSI). The OMOP CDM also aims to represent healthcare data from diverse sources in a consistent and standardized manner. As compared with the PCORnet CDM, the OMOP CDM is distinguished in that it possesses a comprehensive vocabulary component which contains hundreds of medical terminologies and maps them to a common coding system. Moreover, the encoding and relationships among distinct medical concepts are explicitly and formally specified [30]. In the current OMOP CDM version 6.0, there are 38 tables distributed across 6 domains (Fig. 2 ). We use the OMOP CDM v6.0 as a destination data model for the ETL processing in this study.
Fig. 2.
Overview of all tables in the OMOP CDM [29].
2.3. The PCORnet CDM at Mayo Clinic
Mayo Clinic has an enterprise-level PCORnet CDM containing patient data starting in 2010, which is updated on a quarterly basis. In this study, in order to evaluate the ETL performance of our transformation tool and assess the feasibility of COVID-19 data collection for both the PCORnet CDM and the OMOP CDM, we randomly selected data associated with 1000 distinct patients who had at least one new encounter between 01/01/2020 and 04/28/2021 from the CDM. We then use our ETL tool to convert all PCORnet-format patient data into the OMOP COM format.
3. Methods
3.1. ETL tool development for converting the PCORnet CDM to the OMOP CDM
3.1.1. CDM structure mapping
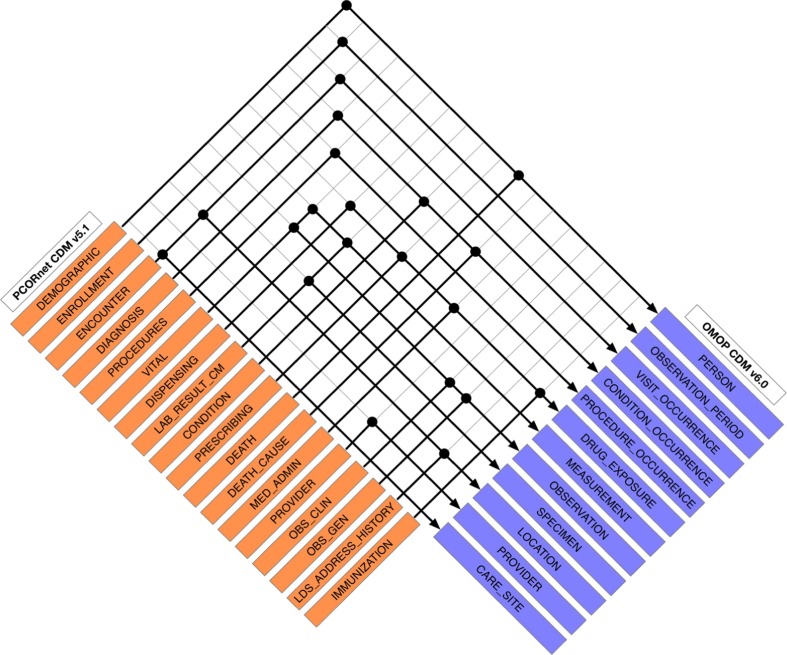
To provide a mapping definition to follow for our ETL tool, we first developed a CDM structure mapping by selecting corresponding tables/fields from the OMOP CDM (v6.0) that correspond to existing tables/fields in the PCORnet CDM (v5.1) utilizing the PCORnet to OMOP mapping dictionary created by N3C [31]. A total of 18 PCORnet tables were mapped to 12 OMOP CDM tables. We have outlined the mappings at a tabular level between the PCORnet and OMOP definitions in Fig. 3 . For each table, individual fields (columns) were also mapped, which we have provided in full detail in Supplementary File S1.
Fig. 3.
Table level mapping between the PCORnet CDM and the OMOP CDM. Each dot in the figure indicates a corresponding table mapping relationship.
3.1.2. ETL tool development and data transformation
The ETL tool was built by using SQL scripts that follow the mapping definitions that were previously created. The ETL tool was designed to achieve three data transformation tasks: value transformation, rule-based transformation, and concept mapping.
For the value transformation task, we designed a value transformation module to account for inconsistencies in data format, such as the data type and any null constraints on table columns, which may be different for the data elements between the two CDMs despite the field themselves being semantically equivalent between the two CDMs. For the rule-based transformation task, we established several rules to handle some special fields that were not directly transferrable from related fields in the PCORnet CDM. For example, several “Datetime” fields in the OMOP CDM are mapped to a combination of a date field and a time field in the PCORnet CDM, and rules to combine the two fields and map to a datetime equivalent while still accounting for null values is needed. For the standard concept code mapping task, codified values are transformed as appropriate into the CDM-preferred coding system. Codes derived from general medical terminologies such as ICD (International Classification of Diseases), LOINC (Logical Observation Identifiers Names and Codes), RxNorm, CPT (Current Procedural Terminology), SNOMED CT (Systematized Nomenclature of Medicine - Clinical Terms) as used in the PCORnet database were mapped to the corresponding preferred concept code in the OMOP vocabulary by parsing the concept/concept_relationship table of the OMOP CDM. For those concepts which are not from shared medical terminologies (i.e. not a source vocabulary for OMOP) such as unit and route concepts, we employed regular expressions to conduct string-matching so as to find the best mapping between the PCORnet CDM concept and the OMOP standard vocabulary. If these codes were specific codes defined in the PCORnet CDM value sets such as status, type or source information, manual definition of a corresponding OMOP concept was required. To ensure the manual mapping accuracy, a two-round mapping approach was conducted by two experts with medical terminology knowledge. The manual code mapping dictionary can be found in Supplementary Table S1. The ETL tool scripts are available at https://github.com/yuey11/PCORnet2OMOP_ETL_tool.
3.2. ETL result evaluation
To evaluate the performance of our ETL pipeline, we randomly sampled 1000 cases amongst all the patients in Mayo Clinic’s PCORnet CDM. The data corresponding to these 1000 patients was then extracted from the tables in the PCORnet CDM and transformed into the OMOP CDM format through our ETL tool.
To achieve a comprehensive assessment of the ETL performance, we designed several different approaches to conduct our evaluation. Firstly, we performed a record counts comparison between the original PCORnet data tables and corresponding target OMOP data tables. Secondly, we evaluated the data transformation and standardization quality through the use of two descriptive indicators: information loss and data mapping accuracy. Thirdly, we investigated the difference between two CDMs to understand the gap during the ETL process. Finally, several COVID-19 surveillance queries were deployed to both CDMs as a simulation to validate the ETL performance for real-world practical tasks.
3.2.1. Overall ETL result evaluation
To ensure there weren’t any records missing during the ETL process. We implemented a record counts comparison across the original PCORnet data tables and corresponding target OMOP data tables. Specifically, after the ETL process, we calculated the record number statistic for both 18 PCORnet CDM tables and 12 OMOP CDM tables. Then, we compared the record counts table by table according to the table-level mapping relationship to make sure all the data from the original database were successfully converted into the target database.
3.2.2. Information loss and data mapping accuracy
Differences in data definitions between the PCORnet CDM and the OMOP CDM may cause data quality issues: firstly, mapping approach may lead to information loss due to differences in information granularity; secondly, differences in preferred medical terminologies between the two CDMs may induce concept mismatch problems during the standard concept code mapping process. In this study, we define the information loss rate as the number of unmapped values in a field of the target OMOP CDM-based database divided by the number of original values from the source field of the PCORnet CDM-based database (Eq. (1)). We also perform a manual review to assess the accuracy of the concept code mappings (Eq. (2)).
| (1) |
| (2) |
3.2.3. Gap analysis between the PCORnet CDM and the OMOP CDM
Although the design philosophy and principle are not the same between the PCORnet CDM and the OMOP CDM, a significant portion of the two CDMs' contents overlap in the clinical domain. There are however still several gaps in terms of information content between these two CDMs. In this study, we try to conduct gap analysis by investigating which tables could not be successfully transferred during the ETL process to discover the data domain differences between the PCORnet CDM and the OMOP CDM.
3.2.4. ETL evaluation by COVID-19 surveillance task
To evaluate ETL performance from real-world practical perspective, we designed a utility evaluation experiment through a COVID-19 surveillance task. Specifically, N3C-defined phenotypic patient cohort acquisition queries [32] (shown in Supplementary File S2) were run on both the source PCORnet CDM and the target OMOP CDM to identify lab-confirmed, suspected, and possible cases of COVID-19, and COVID-19 test negative/equivocal controls. Table 1 below demonstrates the inclusion criteria of these queries (see Supplementary Table S2 for the specific data identification codes in Table 1). We then compared the resultant patient cohorts across the two CDMs to assess the ETL performance in the real-world practical applications.
Table 1.
Inclusion criteria of N3C COVID-19 surveillance queries [31].
| Case group inclusion criteria | Control group inclusion criteria |
|---|---|
|
|
3.3. COVID-19 data collection capacity evaluation for the PCORnet CDM and the OMOP CDM
We utilized the data collection template designed by the MN EHR Consortium COVID-19 Project to further illustrate the data collection capacity of the PCORnet CDM and the OMOP CDM for the COVID-19 surveillance task. Table 2 shows the data elements of the template. We compared the data elements in the template with the data coverage scope in both CDMs. Whether conversion across the different CDMs for each data element of interest is possible/successful was also noted.
Table 2.
Data elements in the data collection template of the MN EHR Consortium COVID-19 project.
| Domain | Data Elements | Identification Method |
|---|---|---|
| COVID test data | COVID PCR test positive/negative Symptomatic/Asymptomatic |
Lab test codes Flag |
| Flu test data | Flu test positive/negative | Lab test codes |
| Viral case data | Influenza-like illness Pneumonia More specific pneumonia COVID-19, virus identified Coronavirus COVID-19, virus not identified COVID Exposure Cough Acute respiratory failure Shortness of breath Fever Sore Throat Muscle aches Headache Diarrhea Loss of smell Fatigue/malaise |
ICD codes |
| Vaccine | Vaccination Dose Manufacturer |
Drug/Procedure Codes Value Value |
| Demographics | Age Race/Ethnicity Gender BMI Language/Interpreter service |
Value |
| Comorbidities | Smoking Asthma COPD HIV Cancer Heart disease Diabetes Mellitus Hypertension Chronic Kidney Disease Substance use (opioids, amphetamines, cocaine, alcohol) |
ICD codes |
| Geography | Zip | Value |
4. Results
4.1. Overall evaluation
1000 patients were randomly sampled from the Mayo Clinic CDM. Of the 1000 cases, female patients (53.5%, n = 535) were in the majority compared to the male patients (46.5%, n = 465). The age group containing most of the COVID-19 cases is 45–64 (24.9%, n = 249), followed by 75 and older (23.1%, n = 231), 65–74 (19.7%, n = 197), 25–44 (19.3%, n = 193), 0–14 (7.7%, n = 77), and 15–24 (5.3%, n = 53). With respect to race, the majority of our sample is white (87.5%, n = 875), followed by Black or African American (5.5%, n = 55), Asian (2.0%, n = 20), and American Indian or Alaska Native (0.8%, n = 8). Most of the patients (96.6%, n = 966) choose English as their preferred language. A detailed demographic breakdown is presented in Table 3 .
Table 3.
Demographic profiles distribution of the 1000 patients.
| Demographic | Value | Frequency | % |
|---|---|---|---|
| Sex | Female Male |
535 465 |
53.5 46.5 |
| Age | 0–14 15-24 25-44 45-64 65-74 75 and older |
77 53 193 249 197 231 |
7.7 5.3 19.3 24.9 19.7 23.1 |
| Race | American Indian or Alaska Native Asian Black or African American White Refuse to answer No information Unknown Other |
8 20 55 875 4 6 9 23 |
0.8 2.0 5.5 87.5 0.4 0.6 0.9 2.3 |
| Ethnicity | Hispanic Not Hispanic Refuse to answer No information Unknown |
39 933 12 6 10 |
3.9 93.3 1.2 0.6 1.0 |
| Language | English Spanish Unknown Other |
966 14 3 17 |
96.6 1.4 0.3 1.7 |
The data from 18 PCORnet CDM tables was mapped into 12 OMOP CDM tables as part of our ETL process. Table 4 shows a comparison of the records counts of tables from both the original PCORnet CDM-based dataset and the OMOP CDM-based database. Note that there are 5 PCORnet tables that were not captured by the CDM (Shown as N/A in Table 4). Amongst the final 12 OMOP CDM tables used as destination tables for the ETL process, 5 tables (PERSON, OBSERVATION_PERIOD, VISIT_OCCURRENCE, PROCEDURE_OCCURRENCE, PROVIDER) have the same number of records as their corresponding source tables in the PCORnet CDM (DEMOGRAPHIC, ENROLLMENT, ENCOUNTER, PROCEDURES, PROVIDER). The records in the CONDITION_OCCURRENCE table and DRUG_EXPOSURE table of OMOP CDM are transformed from multiple PCORnet CDM tables. Thus, records in these two tables are equivalent to the sum of the source tables (CONDITION_OCCURRENCE (364,816) = DIAGNOSIS (323,418) + CONDITION (41,284) + DEATH_CAUSE (1 1 4); DRUG_EXPOSURE (753,716) = PRESCRIBING (402,424) + MED_ADMIN (351,292)). Similarly, 1:1 mapping of the VITAL table of the PCORnet CDM was not possible as there are different types of the records in the VITAL table of PCORnet CDM as compared to the OMOP CDM equivalent. Specifically, the height, weight, BMI, and blood pressure data are mapped into the OMOP MEASUREMENT table, whereas smoking, tobacco, and tobacco type information are mapped into the OBSERVATION of the OMOP CDM. As a result, the records in the MEASUREMENT table (1,630,981) can be considered to be equivalent to the sum of the records in the PCORnet LAB_RESULT_CM table (1,055,436), and the height (38,199), weight (42,567), BMI (54,059), diastolic blood pressure (220,335), systolic blood pressure (220,385) records in the PCORnet VITAL table. Similarly, the record count in the OMOP CDM OBSERVATION table (76,113) is equal to the record count for smoking (25,371) + tobacco (25,371) + tobacco type (25,371) in the PCORnet table. For the SPECIMEN table in the target OMOP CDM, we collected 213,873 records from the LAB_RESULT_CM table of the source PCORnet CDM. Finally, 30 and 31 records are transferred into the OMOP LOCATION and CARE_SITE tables, respectively, from the PCORnet ENCOUNTER table.
Table 4.
Basic Statistics of the PCORnet CDM tables and the OHDSI CDM tables.
|
PCORnet CDM Table name |
Records |
OMOP CDM Table name |
Records |
ETL completeness to OMOP CDM |
|---|---|---|---|---|
| DEMOGRAPHIC | 1,000 | PERSON | 1,000 | 100% |
| ENROLLMENT | 997 | OBSERVATION_PERIOD | 977 | 100% |
| ENCOUNTER | 85,977 | VISIT_OCCURRENCE | 85,977 | 100% |
| DIAGNOSIS | 323,418 | CONDITION_OCCURRENCE | 364,816 | 100% |
| PROCEDURES | 485,385 | PROCEDURE_OCCURRENCE | 485,385 | 100% |
| VITAL | 380,614 | DRUG_EXPOSURE | 753,716 | 100% |
| DISPENSING* | N/A | MEASUREMENT | 1,630,981 | 100% |
| LAB_RESULT_CM | 1,055,436 | OBSERVATION | 76,113 | 100% |
| CONDITION | 41,284 | SPECIMEN | 213,873 | 100% |
| PRESCRIBING | 402,424 | LOCATION | 30 | 100% |
| DEATH | 56 | PROVIDER | 33,787 | 100% |
| DEATH_CAUSE | 114 | CARE_SITE | 31 | 100% |
| MED_ADMIN | 351,292 | |||
| PROVIDER | 33,787 | |||
| OBS_CLIN* | N/A | |||
| OBS_GEN* | N/A | |||
| LDS_ADDRESS_HISTORY* | N/A | |||
| IMMUNIZATION* | N/A |
*This table is not captured by the PCORnet CDM-based database at Mayo Clinic.
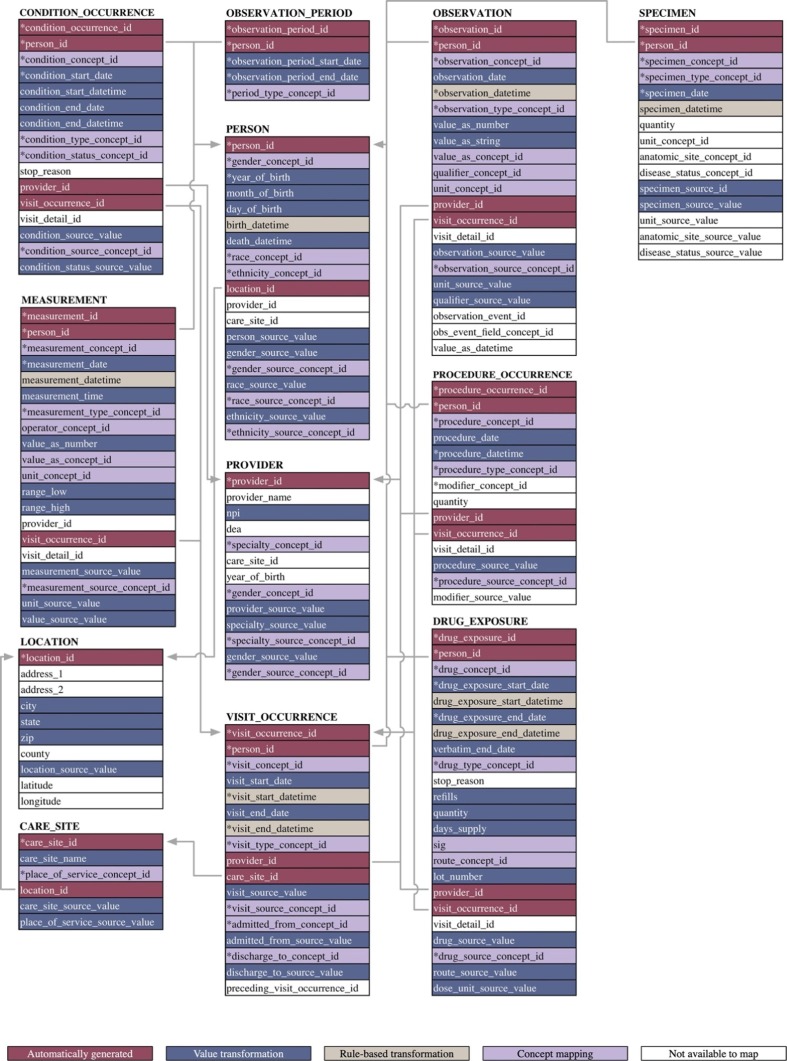
Fig. 4 presents an overview of the mapping quality of the PCORnet CDM into the OMOP CDM at the field level. Several data transformation strategies are performed for the ETL process. For the “id” fields, which are primary keys in some of the OMOP CDM tables, the “id” value is automatically generated by the system. For the other fields, we performed the value transformation, rule-based transformation, and concept mapping process as previously described to convert this data into the OMOP CDM format. Note that there is one required field, modifier_concept_id in the PROCEDURE_OCCURRENCE table of the OMOP CDM, for which we could not identify a corresponding mapping from the PCORnet CDM. So, we inputted “0” for this field to comply the requirement of the OMOP CDM.
Fig. 4.
Database mapping quality evaluation map. * represents the required fields.
4.2. Information loss
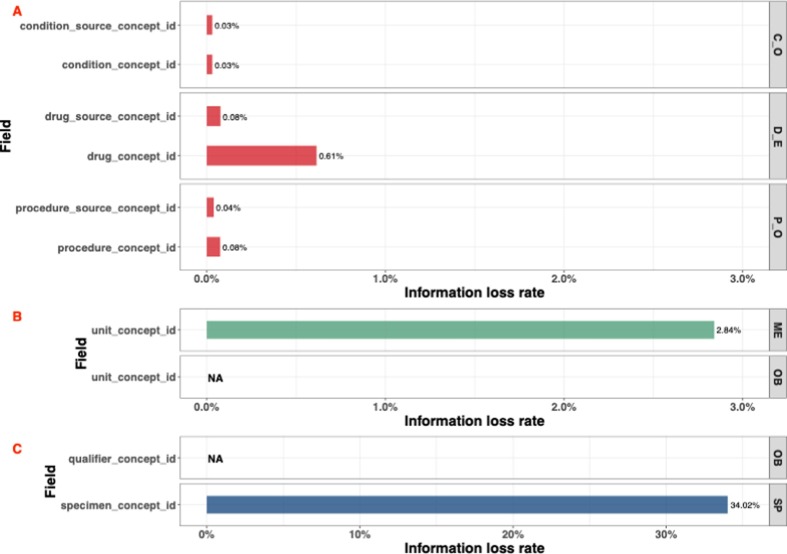
To evaluate the data transformation and standardization performance of our ETL tool, we further investigated information loss. As shown in Fig. 4 three distinct transformation strategies exist as part of our pipeline: value transformation, rule-based transformation, and concept code mapping. Due to the data quality control has been performed by the original PCORnet CDM and all the data has been formatted into the standard data type. All source data in the value transformation process and the rule-based transformation process could be successfully converted into the OMOP CDM by our ETL process. However, the data from concept mapping is not. We therefore analyzed the information loss during the concept code mapping process. Fig. 5 shows the information loss with respect to the three concept mapping approaches that were adopted as part of this study: vocabulary-based, regular-expression based, and manual. Note that we don’t show the 33 fields (6 fields for the concept mapping process, 1 field for string mapping process, and 26 fields for the manual mapping process) that information loss is equal to 0% in this figure. Please see Supplementary Table S3 view these no information loss fields. The largest information loss rate for vocabulary-based concept mapping is 0.61%, which appears in the drug_concept_id mapping, indicating that 0.61% (4,630 records) of the drug codes existing in the PCORnet source database could not be mapped with OMOP CDM standard concept IDs. The information loss rate of all the other fields is less than 0.1%. We also note that there are six fields, measurement_source_concept_id, measurement_concept_id, observation_source_concept_id, observation_concept_id, specialty_source_concept_id, and specialty_concept_id, for which the information loss rate during the ETL process is 0%. For the regular-expression-based string concept mapping process, the information loss rate of the route_concept_id field in the OMOP DRUG_EXPOSURE table is 0%. Conversely, unit_concept_id in the MEASUREMENT table lost 2.84% of unit data (46,332 records). The information loss of unit_concept_id in the OBSERVATION table couldn’t be calculated because this data did not exist in our source PCORnet CDM-based database. Finally, information loss during the manual concept mapping approach was 0% for 26 out of 28 fields, with the exception of two fields: qualifier_concept_id and specimen_concept_id. The information loss of qualifier_concept_id is also not available because no data is collected and converted from the source database. A relatively high information loss rate was observed for specimen_concept_id, which was 34.02%. We will further analyze and discuss the reason why there is some information loss for the unit_concept_id and specimen_concept_id in the discussion section.
Fig. 5.
Information loss during the ETL process. A. Concept mapping information loss. B. String mapping information loss. C. Manual mapping information loss. The Table name abbreviation: C_O, CONDITION_OCCURRENCE; D_E, DRUG_EXPOSURE; ME, MEASUREMENT; OB, OBSERVATION; P_O, PROCEDURE_OCCURRENCE; SP, SPECIMEN. NA in the figure means we didn’t capture related data from our original PCORnet CDM based database at Mayo Clinic.
4.3. Mapping accuracy
The concept mapping accuracy was conducted by a medical terminology expert to further evaluate the data standardization performance. Note that for the manual mapping process, the mapping accuracy is considered as “100%” due to the mapping having been confirmed through expert validation. We therefore only validated the mapping accuracy of the regex-based string mapping and the concept mapping approaches. We randomly selected 50 mappings for each concept mapping field to perform the evaluation. Table 5 shows the mapping accuracy review results. The results indicate that all the original concept codes from the PCORnet CDM are successfully mapped to the OMOP CDM concept IDs through the concept mapping process.
Table 5.
Concept mapping accuracy.
| Mapping Process | Table | Field | Mapping Accuracy (%) |
|---|---|---|---|
| Concept mapping accuracy | CONDITION_OCCURRENCE | condition_concept_id | 100% |
| CONDITION_OCCURRENCE | condition_source_concept_id | ||
| DRUG_EXPOSURE | procedure_concept_id | ||
| DRUG_EXPOSURE | procedure_source_concept_id | ||
| MEASUREMENT | drug_concept_id | ||
| MEASUREMENT | drug_source_concept_id | ||
| OBSERVATION | measurement_concept_id | ||
| OBSERVATION | measurement_source_concept_id | ||
| PROCEDURE_OCCURRENCE | observation_concept_id | ||
| PROCEDURE_OCCURRENCE | observation_source_concept_id | ||
| PROVIDER | specialty_concept_id | ||
| PROVIDER | specialty_source_concept_id | ||
| String mapping accuracy | DRUG_EXPOSURE | route_concept_id | |
| MEASUREMENT | unit_concept_id | ||
| OBSERVATION | unit_concept_id | NA* |
* NA means we didn’t capture related data from our original PCORnet CDM based database at Mayo clinic.
4.4. Transformation gap analysis
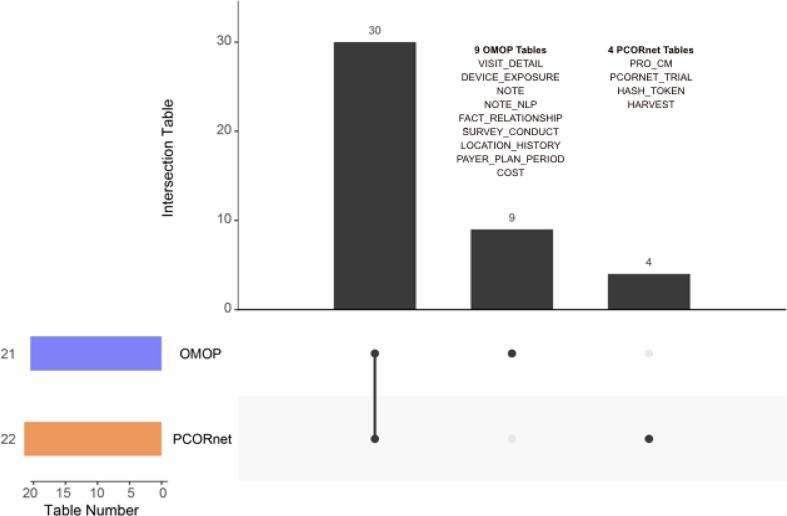
To analyze the data transformation gap between the two CDMs, we investigated which tables could not be converted from the PCORnet CDM into the OMOP CDM during the ETL process, the results of which we have summarized in Fig. 6 . Amongst the 22 PCORnet CDM tables and 21 clinical data related OMOP CDM tables (15 Clinical Data Tables, 4 Health System Data Tables, and 2 Health Economics Data Tables), our ETL tool could transform the data from 18 PCORnet CDM tables into 12 OMOP CDM tables. That indicates there remains 4 PCORnet CDM tables that could not be converted into the OMOP CDM, and 9 OMOP CDM tables that did not have source mappings from the PCORnet CDM-based database. In addition, we also found that in the mapped tables (18 PCORnet CDM tables and 12 OMOP CDM tables), there are some fields that were not involved in the ETL process (indicated by white cells in Fig. 4). Furthermore, all these un-transferred fields in both the PCORnet CDM and the OMOP CDM are listed in Supplementary Table S4.
Fig. 6.
Upset figure of table level data transformation between the PCORnet CDM and the OMOP CDM.
4.5. COVID-19 surveillance task evaluation
A real-world COVID-19 surveillance task based on phenotypic patient cohort identification was run to evaluate the ETL performance. We used N3C’s COVID-19 inclusion criteria and modified their phenotype acquisition codes to identify a COVID-19 case/control cohort from both the original PCORnet CDM and the target OMOP CDM. Table 6 shows the cohort identification results across the two CDMs that are loaded with data from the randomly selected 1000 patients. In total, we found 726 patients from the original PCORnet CDM-based database and 731 patients from the OMOP CDM-based database. All 726 patients derived from the phenotyping query in the original PCORnet database were also in the results for the OMOP CDM query. For case group identification, we identified 119 and 120 COVID-19 cases from the PCORnet CDM and the OMOP CDM respectively. Specifically, when we investigate the subsets of the case group, the “Strong Case” subgroup and “Lab Test Positive Case” groups were identical across both CDM results. As for the “Weak Case” subgroup, we identified 2 additional cases from the OMOP CDM compared to the PCORnet CDM (Note that there are overlaps between Weak Case group and the other two case groups. So, for the total number of case group, only 1 more case was identified by the OMOP CDM). Similarly, we identified 611 cases for the control group in the OMOP CDM cohort, 4 more cases than the control group derived from the PCORnet CDM cohort.
Table 6.
The patient number of Case/Control group identification result across the PCORnet CDM and the OMOP CDM.
| PCORnet CDM | OMOP CDM | Overlap | ||
|---|---|---|---|---|
| Case group |
Total Strong Case Weak Case Lab Test Positive Case |
119 43 64 55 |
120 43 66 55 |
119 43 64 55 |
| Control group | Total | 607 | 611 | 607 |
| All cases | Total | 726 | 731 | 726 |
4.6. COVID-19 data collection capacity evaluation
Table 7 shows the data collection capacity of the two CDMs according to the data collection template of the MN EHR Consortium COVID-19 project. We found that all the data elements required to support this COVID-19 surveillance project could be captured from both the PCORnet CDM and the OMOP CDM. Additionally, all the specific data elements used could be successfully converted during the ETL process.
Table 7.
The data collection capacity of the PCORnet CDM and OMOP CDM for the domain in the MN EHR Consortium COVID-19 Project.
| Domain | PCORnet CDMCapacity | OMOP CDMCapacity |
|---|---|---|
| COVID test data | Yes | Yes |
| Flu test data | Yes | Yes |
| Viral case data | Yes | Yes |
| Vaccine | Yes | Yes |
| Demographics | Yes | Yes |
| Comorbidities | Yes | Yes |
| Geography | Yes | Yes |
5. Discussions
This study developed a comprehensive ETL tool to facilitate data transformation from the PCORnet CDM to the OMOP CDM. This tool makes it possible to rapidly deploy the OMOP CDM for any institution that only has the PCORnet CDM. The ETL evaluation from different perspectives demonstrates that our tool has a satisfactory data transformation performance. The COVID-19 surveillance study simulation also suggests that the ETL performance is sufficient to support a real-world study.
5.1. Overall evaluation
The overall mapping evaluation suggests that we achieved a high-quality ETL result. At a table level, the record counts are the same before/after the ETL process, which proves we do not lose any records at the patient level. We also note that several records in some OMOP CDM tables are transformed from multiple PCORnet CDM tables in the same domain. Since the original PCORnet CDM tables typically indicate different types of data and may be incorporated as part of a many-to-one mapping to an equivalent OMOP CDM table, it is essential to distinguish these records in the OMOP CDM. This issue was addressed by supplying different values to the “concept_type_id” field of the OMOP CDM table after mapping. For example, the DRUG_EXPOSURE table in the OMOP CDM received drug-related data from the DISPENSING, PRESCRIBING, and MED_ADMIN table of the PCORnet CDM. To ensure that the users could identify different sources in the OMOP CDM, the “drug_type_concept_id” field was populated with the OMOP CDM concept ID “32838 (EHR prescription)”, “32825 (EHR dispensing record)”, “32830 (EHR medication list)” depending on the source table as appropriate. A similar strategy was applied in our mappings to the OMOP CONDITION_OCCURENCE table. At the field level, 146 out of the 179 fields from the 12 target OMOP CDM tables successfully loaded converted data from the PCORnet CDM. Only one required field, “modifier_concept_id” in the PROCEDURE_OCCURRENCE table of OMOP CDM, did not have an equivalent in the original PCORnet CDM. To satisfy the constraints of the OMOP CDM, we input a pseudo value “0” into this field. The data conversion performance at the field level further demonstrates our ETL tool's excellent capability for data transformation between the OMOP CDM and the PCORnet CDM.
To develop and evaluate the ETL tool, it takes a lot of effort from our team members with various backgrounds from different institutions. Specifically, 1 month was used for developing a specific manual concept mapping table for 2 experts with medical terminology domain knowledge. Then, two developers who were familiar with the CDM knowledge spent around 3 months developing the ETL tool. The data transformation and ETL performance evaluation cost about 2 months’ part-time work for 6 team members.
In addition, one of the main advantages to use the CDM-based approach to standardize the EHR data for observational studies is that it enables the distributed analytics for research collaborations. That means as long as each institution has a CDM established, the analytic scripts and results (rather than sensitive clinical data) can be shared among research collaborators across institutions. This distributed analytic approach is a common process used by the CDM-based research consortia (e.g., OHDSI or PCORI). On the other hand, some consortia (e.g., N3C) aimed to create a centralized CDM research repository for the research collaborations. In this situation, a data use agreement (DUA)-based process would be required for a separate institution to access deidentified EHR data.
5.2. Information loss and mapping accuracy
The information loss and mapping accuracy results demonstrate an outstanding data standardization performance. The information loss mainly occurs in the concept mapping approach. There are two significant reasons why the original concept codes in the PCORnet CDM could not be matched with the standard OMOP CDM concept IDs. Firstly, several error codes exist in the original PCORnet CDM data for which there is no equivalent OMOP mapping. For example, condition codes “****” and “Error” exist in our original database, which could not be mapped. 2) During the concept mapping process, PCORnet CDM concept codes are first mapped to the “XX_source_concept_id” and then map it to the “XX_concept_id”. However, for some cases, the OMOP CDM vocabulary may not provide a standard “XX_concept_id” for a “XX_source_concept_id”. This is reflected in our results, where the information loss of the “XX_source_concept_id” is usually lower than the “XX_concept_id”. For example, due to that the code “D0160” being retired in the original HCPCS (Healthcare Common Procedure Coding System) terminology, this code was marked as “invalid”, and a related standard concept ID was not found in the OMOP CDM vocabulary. We also analyzed reasons why there is information loss in the unit_concept_id field and the specimen_concept_id field. As for regular expression-based string mapping, all the route terms in our original PCORnet CDM-based database could be matched with a corresponding OMOP CDM concept IDs. Although we used a regular expression to normalize some unit terms, some units in the PCORnet CDM still could not be mapped to an appropriate OMOP CDM concept IDs. We found that some of unmatched units may not be collected by the OMOP CDM vocabulary, such as “nmol/mL/h”, or that the format of the unit is different in the two CDMs, which makes it difficult to find the related mapping through the string mapping process. For example, for the unit “m[IU]/L” in the PCORnet CDM, the corresponding concept in the OMOP CDM is “10*-3.[iU]/L”. We used manual mapping to deal with the unmatched concepts to decrease the information loss. Due to extensive manual work, most of the fields in the manual mapping process achieve 100% mapping rate between the two CDMs. For the “specimen_concept_id” field however, more than 30% of records could not be converted because we could not find a mapping between the vocabulary of the two CDMs, such as the “^MOTHER”, “URINE + SER_PLAS”, “BREAST_TUMOR”, etc. concepts in the PCORnet CDM, even if we performed a manual mapping approach. We also noticed that some of the PCORnet CDM-specific concepts have already been collected by the OMOP CDM vocabulary, which could facilitate our manual mapping. With recent updates to the PCORnet CDM, more specific concepts are however included in its vocabulary, and current related concepts in the OMOP CDM vocabulary only cover a small part of these PCORnet CDM specific concepts. We plan to contact the OMOP CDM maintenance community to consider extending the PCORnet CDM-related concepts to enhance the data transformation support in the future.
As our concept mapping and string mapping approach is based on the concepts/relationships preset in the OMOP CDM vocabulary, a mapping accuracy of 100% was achieved. The results show that the OMOP CDM provides an integrated vocabulary and comprehensive relationships between the concepts for multiple mapping purposes. However, we found a potential mapping issue caused by the concept mapping of the condition_concept_id in the CONDITION_OCCURRENCE table. The original condition concept codes in the PCORnet CDM are primarily from ICD to 9/10, whereas SNOMED CT is the preferred terminology for the condition concepts in the OMOP CDM. We must therefore first use the OMOP CONCEPT_RELATIONSHIP table to map the ICD codes to the SNOMED CT and then transform the codes into the OMOP CDM. In most cases, the mapping between the ICD codes and SNOMED CT codes is a one-to-one mapping. On rare occasions however, this is not the case. On one hand, some ICD codes with a more specific meaning would be mapped to the semantic hypernym terms in the SNOMED CT by the OMOP CDM vocabulary, such as the ICD-10 code “N60.31, Fibrosclerosis of right breast” being mapped to a more generic SNOMED CT term “29070004, Fibrosclerosis of breast”. On the other hand, one ICD code could be matched with multiple SNOMED CT codes. For example, the ICD-10 code “O09.892, Supervision of other high risk pregnancies, second trimester” is mapped to three SNOMED CT terms “47200007, High risk pregnancy”, “59466002, Second trimester pregnancy”, and “702738006, Supervision of high risk pregnancy” in the OMOP CDM. Because we can only input one OMOP CDM concept ID for each record, we usually choose the first concept which could be used in the condition domain to ensure the mapping consistency of these concepts. This concept mapping issue may slightly impact the real-world analysis result in some situations. The effect is also illustrated in the following discussion about the real-world COVID-19 surveillance task.
Furthermore, although there are only 119 COVID-19 positive patients in our evaluation cohort may not cover all the COVID-19 related clinical data. We still believe our ETL tool could successfully accomplish the COVID-19 related clinical data transformation between the PCORnet CDM and OMOP CDM. Specifically, the concept code mapping results indicated that with the help of the comprehensive vocabulary of the OMOP CDM, most of the clinical concept codes such as diagnosis, lab test, and medication codes from PCORnet could be successfully converted into OMOP CDM standard concepts. Moreover, thanks to the hard work of the PCORnet team and the OMOP community, most of the COVID-19 related concepts were added to the two CDMs once they were released. So, as long as the standard COVID-19 related concept codes appear in the two CDMs, our ETL tool could achieve high-quality transformation work.
5.3. Transformation gap
We investigated the tables that were not included in the ETL process to analyze the gap between the PCORnet CDM and the OMOP CDM. We realize that most of the data domains in the two CDMs overlap, but information gaps still exist. From the perspective of ensuring all data in the PCORnet database is converted, four PCORnet CDM tables are not convertible into the OMOP CDM. The PRO_CM table of PCORnet CDM is designed to store responses to patient-reported outcome measures (PROs) or questionnaires. This table may be converted into the SURVEY_CONDUCT table and OBSERVATION table of the OMOP CDM from a semantic content perspective. However, many concepts in this table, such as clinical questionnaires, require manual conversion to the OMOP CDM format. This conversion has been delegated to future work. The PCORNET_TRIAL table is used to record patients enrolled in PCORnet clinical trials. Although there is no corresponding table to store the trial-related information in the OMOP CDM directly, the OMOP CDM provides an application named “ATLAS”[33] to help users build clinical cohorts and store the cohort at the patient level. The HASH_TOKEN table of the PCORnet CDM stores encrypted, keyed secure hash tokens to match patient records across data marts. This encryption is not included in the OMOP CDM due to differing information-sharing strategies for intra-network collaboration. For the HARVEST table, it records the information associated with the specific PCORnet data mart implementation such as the PCORnet network name/ID. In the OMOP CDM, we can also abstract and record ETL information into the CDM_SOURCE table, but not directly collect those data from the PCORnet HARVEST table. From the perspective of ensuring that all OMOP CDM information is captured, the information gap against the PCORnet CDM can be categorized into three types. Firstly, the PCORnet CDM does not have tables relating to medical device, clinical notes, and health economics data. Secondly, the VISIT_DETAIL and the LOCATION_HISTORY table in the OMOP CDM are used to record some additional details for the VISIT_OCCURRENCE table and the LOCATION table. While the PCORnet CDM does provide the encounter and location data, the granularity of the data captured is insufficient to fully populate these additional details captured by OMOP CDM. Thirdly, although the data in the FACT_RELATIONSHIP table is not directly transformed from the related table of the PCORnet CDM, we can develop some algorithms to derive the relationship data from the original database. Moreover, another advantage of the OMOP CDM is its robust OBSERVATION table, which could capture any clinical facts that are not captured by any other domains about a patient. The patient data such as social and lifestyle facts, medical history, family history, etc., can be all recorded here.
5.4. COVID-19 surveillance task
The ETL performance was also evaluated by a real-world COVID-19 cohort identification task. In general, the consistency of identification results indicates a satisfactory ETL performance across the two CDMs. The N3C’s surveillance queries collected slightly more cases/controls from the target OMOP CDM-based database than the source PCORnet CDM-based database. We further investigated the reason of the inconsistency. 1) For the “Weak Case” subgroup of the case-cohort, 2 more cases were identified due to the different usage of the phenotype vocabularies. In the PCORnet CDM, the diagnosis and lab test data were directly collected by the ICD-10, SNOMED CT, and LOINC concept codes. In the OMOP CDM, said ICD-10, SNOMED CT, and LOINC codes need to first be converted to the standard OMOP CDM concept IDs, which are then used to collect the phenotype data. In most cases, the OMOP CDM concept ID and the phenotype concept are one-to-one mappings, leading to a consistent result for the “Strong Case” and “Lab Test Positive Case” identification across the two CDMs. However, in some cases, some of the standard OMOP CDM concept IDs may correspond to multiple phenotype concepts. For example, the OMOP CDM concept ID “320136” could be matched with several ICD-10 codes such as “J98.8”, “J98.9”, “J95.7”, etc. But only “J98.8” was used to identify the “Weak Case”. As a result, utilizing the OMOP CDM concept ID to retrieve the phenotype data may cause our phenotyping query to retrieve more cases due to differing concept granularity as opposed to using the concept code as defined in the phenotype itself. 2) Due to the differing lab test data collection strategy, we collected 4 more controls in the OMOP CDM than the PCORnet CDM. Specifically, in addition to the LOINC code and OMOP CDM concept ID, we also used some other strategies to collect the additional COVID-19 lab test data, which could not be captured by the concepts in the code list. In the PCORnet CDM, we utilized string mapping to search “COVID-19” and “SARS-COV-2” related data that is not captured by the LOINC codes. As a comparison, in the OMOP CDM, we searched all the descendant concepts of OMOP CDM concept ID “756055” (Measurement of Severe acute respiratory syndrome coronavirus 2 (SARS-CoV-2)) to collect the additional COVID-19 lab test data.
5.5. COVID-19 data collection capacity
The COVID-19 data collection capacity assessment shows the two CDMs could provide sufficient support for the COVID-19 surveillance project. In addition, it also confirmed that our ETL tool could facilitate data transformation for the perspective of a real-world use case. With the exception of symptomatic/asymptomatic COVID-19 infection and vaccine manufacture information, all other data elements in the MN EHR Consortium COVID-19 Project could be directly collected or transformed between the PCORnet CDM and the OMOP CDM. While neither CDMs had a specific field to collect the symptomatic/asymptomatic information, asymptomatic-related diagnosis code, such as the SNOMED CT code “189486241000119100, Asymptomatic SARS-CoV-2” could be used in the condition tables of both the PCORnet CDM and the OMOP CDM to identify the asymptomatic COVID-19 infection information. The vaccine manufacture information would be captured by the “VX_MANUFACTURER” field in the IMMUNIZATION table of the PCORnet CDM. Although OMOP CDM doesn’t provide such a field to store the vaccine manufacture information, such information can be derived by specific vaccine related OMOP CDM concept IDs, such as “1202358, JANSSEN COVID-19 VACCINE”, “36371349, MODERNA COVID-19 VACCINE”, “42794278, PFIZER-BIONTECH COVID-19 VACCINE”. Note that because we didn’t get all vaccine data in our original PCORnet CDM-based database, we could not evaluate whether the manufacture information could be directly converted into the OMOP CDM format. It is likely that such a task would need to be delegated to a rule-based transformation to combine the vaccine code and the “VX_MANUFACTURER” data together.
5.6. Limitations and future work
This study has several limitations. Firstly, our research protocol approved by the Institutional Review Boards (IRB) at Mayo only supports us to conduct a sampling-based ETL process for the study purpose. So, we only randomly selected 1,000 patients from our PCORnet CDM-based EHR database to perform our ETL experiment, other than transforming the entire PCORnet database at Mayo into the OMOP CDM. Secondly, as there isn’t any data for five tables (DISPENSING, OBS_CLIN, OBS_GEN, LDS_ADDRESS_HISTORY, IMMUNIZATION) in our original PCORnet-based database, we could not evaluate the ETL performance regarding these tables for our tool. Thirdly, our current COVID-19 data collection capacity evaluation results only show the data capturing performance in the COVID-19 surveillance field due to the data collection template table we used. Since the PCORnet CDM and/or the OMOP CDM don’t cover some other important COVID-19 event related data such as diagnostic imaging, ICU admissions, and long-term oxygen support data, etc., the two CDM may not support the needs of each specific COVID-19 research use case. Fourthly, by using the ETL tool we developed in this study, we assume the source CDM (i.e., PCORnet CDM) does not have any data quality issue. However, to establish multiple different CDMs in an institution, it may potentially cause some project management issues. One of significant challenges would be the “source of truth” issue. [34] In a previous study, we found that different CDMs in an institution provided different results for a set of identical queries. To resolve this issue, an enterprise level project management strategy should be established to ensure the source data consistently populated to different CDMs. Finally, we designed current concept mapping in our ETL tool based on the PCORnet CDM vocabulary published in 2020. As the PCORnet CDM vocabulary is continuously updated with new concept additions, our tool may not cover all the concepts in the latest version of the PCORnet CDM. To address these limitations, in future work, we will try to collaborate with the IT department at Mayo to deploy enterprise-level database transformation and perform a more comprehensive ETL evaluation. We will also work on extending the PCORnet CDM at Mayo to include more data such as immunization data and questionnaire data. And we would like to check the data collection requirement for more COVID-19 projects to conduct some more comprehensive capacity evaluation. In addition, we would like to collaborate with the CDM-based observational study research communities to conduct more evaluations for our ETL tool, and we will further update our tool to keep up with the latest version of the PCORnet CDM. Furthermore, we will use our ETL tool to conduct more cross-institution evaluation and real-world data-based study in future.
6. Conclusions
In this study, we developed an ETL tool to support data transformation from the PCORnet CDM to the OMOP CDM. The outcome of the work would facilitate the data retrieval, communication, sharing, and analysis between different institutions for not only COVID-19 related projects, but also other real-world evidence-based observational studies.
Declaration of Competing Interest
The authors declare that they have no known competing financial interests or personal relationships that could have appeared to influence the work reported in this paper.
Acknowledgments
Acknowledgements
This study is supported in part by the NCATS CD2H Project (U24 TR002306) and the NIH FHIRCat Projects (R56 EB028101 and R01 EB030529).
Author contribution
Y.Y., A.M.C, E.P., C.G.C, N.S., and G.J. contributed to the study design. D.G., and C.G.C contributed to design the mapping dictionary between the PCORnet CDM and the OMOP CDM. Y.Y. and A.W. contributed to develop the ETL tool. Y.Y., N.Z., A.W., S.L., D.S., D.K. contributed to conduct the ETL process and evaluate the ETL performance. Y.Y., A.W., A.M.C, and G.J. contributed to draft the manuscript. All the authors contributed to the manuscript review and editing.
Footnotes
Supplementary data to this article can be found online at https://doi.org/10.1016/j.jbi.2022.104002.
Appendix A. Supplementary material
The following are the Supplementary data to this article:
References
- 1.Gill J., Prasad V. Improving observational studies in the era of big data. Lancet. 2018;392(10149):716–717. doi: 10.1016/S0140-6736(18)31619-2. [DOI] [PubMed] [Google Scholar]
- 2.Ienca M., Vayena E. On the responsible use of digital data to tackle the COVID-19 pandemic. Nat Med. 2020;26(4):463–464. doi: 10.1038/s41591-020-0832-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Budd J., Miller B.S., Manning E.M., Lampos V., Zhuang M., Edelstein M., Rees G., Emery V.C., Stevens M.M., Keegan N., Short M.J., Pillay D., Manley E.d., Cox I.J., Heymann D., Johnson A.M., McKendry R.A. Digital technologies in the public-health response to COVID-19. Nat Med. 2020;26(8):1183–1192. doi: 10.1038/s41591-020-1011-4. [DOI] [PubMed] [Google Scholar]
- 4.Overhage J.M., Ryan P.B., Reich C.G., Hartzema A.G., Stang P.E. Validation of a common data model for active safety surveillance research. J Am Med Inform Assoc. 2012;19:54–60. doi: 10.1136/amiajnl-2011-000376. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Benson K., Hartz A.J. A comparison of observational studies and randomized, controlled trials. N Engl J Med. 2000;342(25):1878–1886. doi: 10.1056/NEJM200006223422506. [DOI] [PubMed] [Google Scholar]
- 6.Concato J., Shah N., Horwitz R.I. Randomized, controlled trials, observational studies, and the hierarchy of research designs. N Engl J Med. 2000;342(25):1887–1892. doi: 10.1056/NEJM200006223422507. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.P. Velentgas, N.A. Dreyer, P. Nourjah, et al., editors. Developing a Protocol for Observational Comparative Effectiveness Research: A User's Guide. Rockville (MD): Agency for Healthcare Research and Quality (US); (2013) Jan. Available from: https://www.ncbi.nlm.nih.gov/books/NBK126190/. [PubMed]
- 8.R.E. Gliklich, M.B. Leavy, N.A. Dreyer, (Eds.), Tools and Technologies for Registry Interoperability, Registries for Evaluating Patient Outcomes: A User’s Guide, 3rd Edition, Addendum 2 [Internet]. Rockville (MD): Agency for Healthcare Research and Quality (US); (2019) Oct. Report No.: 19(20)-EHC017-EF. PMID: 31891455. [PubMed]
- 9.Garza M., Del Fiol G., Tenenbaum J., Walden A., Zozus M.N. Evaluating common data models for use with a longitudinal community registry. J Biomed Inform. 2016;64:333–341. doi: 10.1016/j.jbi.2016.10.016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.PCORnet Common Data Model (CDM), https://pcornet.org/data/. Accessed Jan. 25, 2022.
- 11.Observational Health DataSciences and Informatics (OHDSI), https://www.ohdsi.org/. Accessed Jan. 25, 2022.
- 12.Sentinel Common Data Model, https://www.sentinelinitiative.org/methods-data-tools/sentinel-common-data-model. Accessed Jan. 25, 2022.
- 13.i2b2Research Data Warehouse, https://community.i2b2.org/wiki/display/BUN/i2b2+Common+Data+Model+Documentation. Accessed Jan. 25, 2022.
- 14.Klann J.G., Joss M.A.H., Embree K., Murphy S.N., Lovis C. Data model harmonization for the All Of Us Research Program: Transforming i2b2 data into the OMOP common data model. PLoS ONE. 2019;14(2):e0212463. doi: 10.1371/journal.pone.0212463. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Food and Drug Administration (FDA), National Institutes of Health's National Library of Medicine (NLM), National Cancer Institute (NCI) and National Center for Advancing Translational Sciences (NCATS), Office of the National Coordinator for Health Information Technology (ONC). Common Data Model Harmonization (CDMH) and Open Standards for Evidence Generation: Final Report. (2020). Available from: https://aspe.hhs.gov/sites/default/files/private/pdf/259016/CDMH-Final-Report-14August2020.pdf.
- 16.Klann J.G., et al. Web services for data warehouses: OMOP and PCORnet on i2b2. J Am Med Inform Assoc. 2018;25:1331–1338. doi: 10.1093/jamia/ocy093. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Klann J.G., Abend A., Raghavan V.A., Mandl K.D., Murphy S.N. Data interchange using i2b2. J Am Med Inform Assn. 2016;23:909–915. doi: 10.1093/jamia/ocv188. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Common Data Models Harmonization, https://build.fhir.org/ig/HL7/cdmh/index.html. Accessed Aug. 13, 2020.
- 19.M. Choi, R. Starr, M. Braunstein, J. Duke, OHDSI on FHIR platform development with OMOP CDM mapping to FHIR Resources. In OHDSI Symposium, Observational Health Data Sciences and Informatics, Washington, DC (2016).
- 20.The OMOPonFHIR Project at Georgia Tech, http://omoponfhir.org/ . Accessed Aug. 13, 2020.
- 21.Belenkaya R., Mirhaji P., Khayter M., Torok D., Khare R., Ong T., Schilling L. In OHDSI workshop. 2015. Establishing Interoperability Standards between OMOP CDM v4, v5, and PCORnet CDM v1. [Google Scholar]
- 22.National COVID Cohort Collaborative (N3C), https://ncats.nih.gov/n3c. Accessed Aug. 13, 2020.
- 23.MN EHR Consortium, https://www.hennepinhealthcare.org/ehrconsortium/. Accessed Aug. 13, 2020.
- 24.Haendel M.A., et al. The National COVID Cohort Collaborative (N3C): Rationale, design, infrastructure, and deployment. J Am Med Inform Assoc. 2021;28:427–443. doi: 10.1093/jamia/ocaa196. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Winkelman T., et al. Minnesota Electronic Health Record Consortium COVID-19 Project: Informing Pandemic Response Through Statewide Collaboration Using Observational Data. Public Health Rep. 2022 doi: 10.1177/00333549211061317. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Fleurence R.L., Curtis L.H., Califf R.M., Platt R., Selby J.V., Brown J.S. Launching PCORnet, a national patient-centered clinical research network. J Am Med Inform Assoc. 2014;21(4):578–582. doi: 10.1136/amiajnl-2014-002747. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Common Data Model (CDM) Specification, Version 5.1, https://cprn.org/data-submission/PCORnet-Common-Data-Model-v51-2019_09_12.pdf. Accessed Aug. 13, 2020.
- 28.Forrest C.B., et al. PCORnet(R) 2020: current state, accomplishments, and future directions. J Clin Epidemiol. 2021;129:60–67. doi: 10.1016/j.jclinepi.2020.09.036. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.The Book of OHDSI, https://ohdsi.github.io/TheBookOfOhdsi/CommonDataModel.html . Accessed Aug. 13, 2020.
- 30.Hripcsak G., et al. Observational Health Data Sciences and Informatics (OHDSI): Opportunities for Observational Researchers. Stud Health Technol Inform. 2015;216:574–578. [PMC free article] [PubMed] [Google Scholar]
- 31.N3C Data Ingestion and Harmonization, https://github.com/National-COVID-Cohort-Collaborative/Data-Ingestion-and-Harmonization/tree/master/CDMDataMaps/PCORNet2OMOP . Accessed Aug. 13, 2020.
- 32.N3C Phenotype Data Acquisition, https://github.com/National-COVID-Cohort-Collaborative/Phenotype_Data_Acquisition/wiki/Latest-Phenotype. Accessed Aug. 13, 2020.
- 33.ATLAS, https://atlas.ohdsi.org/. Accessed Aug. 13, 2020.
- 34.W.L. Schulz, H.P. Young, K.J. Ruddy, N.D. Shah, J.S. Ross, S. Gordon, M. Rocca, G. Jiang, A Multi-Institutional Review and Validation of Federated Query Results in Multiple Common Data Models. In AMIA (2018).
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.