LETTER
As 2021 comes to a close, the advances in vaccination against COVID-19 allow the world to glimpse an end to the pandemic. In Brazil, the disease has cost more than 600,000 lives and affected more than 21 million people. When the second wave of COVID-19 hit in early 2021, the country saw more than 3,500 daily deaths. As Brazil started to recover from this number, the first reports of infection by the Delta (B.1.617.2) variant of concern (VoC) in the country were emerging. The first confirmed case of this variant occurred on 26 April 2021, with five states registering infections by it in the following 3 months. At the time, these cases were considered isolated or contained imported events. Here, we describe the early phase of the first large-scale community transmission of the Delta variant in Brazil and the associated interstate dispersal.
Through biweekly sequencing of severe acute respiratory syndrome coronavirus 2 (SARS-CoV-2) randomized sampling, the Corona-ômica-RJ Network (http://www.corona-omica.rj.lncc.br) has been tracking lineage dynamics of the pandemic in the state of Rio de Janeiro, Brazil (1). We first detected the Delta VoC in two samples collected on 16 and 17 June 2021, which represented 0.57% (n = 2/353) of genomes sequenced at the time. Both originated from citizens without prior contact or history of travel in the 14 days preceding symptom onset. By tracking direct contacts with both cases, three more active infections with the variant were promptly identified. No hospital admission was necessary for any of these cases. As monitoring of contacts progressed, we conducted the next randomized sequencing round of positive COVID-19 samples from the state. We identified 60 additional samples carrying the Delta lineage, an astounding increase to 16% of samples (n = 60/377) in 15 days. Because of this fast growth, we consider Delta to have already jumped from individual to community transmission in Rio de Janeiro in June 2021.
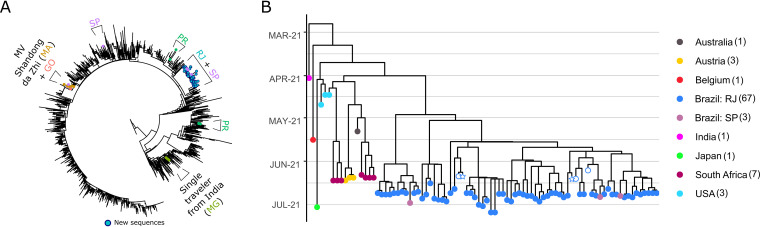
The evolutionary relationship between all SARS-CoV-2 sequences from Brazil available in mid-July 2021 indicates that the spread of Delta in the country can be described by at least six introductory events (Fig. 1A). The first reported cases, from the MV Shandong Da Zhi carrier ship in the state of Maranhão (2), are possibly related to the origin of the variant in the state of Goiás, more than 1,600 km away. Two transmission clusters in the state of Paraná have different origins, and one isolated case was found in the states of Minas Gerais and São Paulo. The outbreak in Rio de Janeiro originated with a single entrance, though it has spread to the state of São Paulo independently at least three times (Fig. 1B). We estimate the introduction in Rio de Janeiro to have happened in the last days of April or the beginning of May 2021, and most of the spread has occurred around mid-June. The Rio de Janeiro clade has four mutations in ORF1ab pervasive to all samples sequenced as follows: I1091V, T4087I, syn5373A→G and syn12951T→C. Indeed, all independent introductions have characteristic mutations when compared to one another, which can be a helpful resource to rapidly assign new cases to each of these events (see Table S1 in the supplemental material).
FIG 1.
Evolutionary relationship between samples of Delta variant collected in Brazil. (A) Maximum likelihood tree including 1,409 SARS-CoV-2 genomes shows that at least six independent introductions of the variant occurred in Brazil. Colored points indicate samples from different Brazilian states (MG, Minas Gerais; MA, Maranhão; GO, Goiás; PR, Paraná; RJ, Rio de Janeiro; SP, São Paulo). All samples from Rio de Janeiro were sequenced by the Corona-ômica-RJ Network. (B) Bayesian time tree of the outbreak in Rio de Janeiro. Color of points corresponds to sampling locality. Empty stars represent the first two cases detected in Rio de Janeiro, and empty circles represent their primary contacts.
Recent studies have demonstrated that Delta has higher transmissibility (3) and suggest that it can evade the immune response induced by previous infections or incomplete vaccination (4–6). Even though the variant has now become dominant in Brazil, a new surge of COVID-19 cases in the country was not observed. We attribute the current success in subduing the pandemic to the advances in vaccination coverage: as of 19 October 2021, at least 70% of the population had taken the first vaccine dose and 49.7% have completed the immunization chronogram. We warn that prematurely relaxing nonpharmacological interventions, such as social distancing and use of masks, can threaten the current trend of decline in cases. This work provides a first view of how Delta has repeatedly entered Brazil. Still, we note the possibility of other importing events passing undetected in genomic surveillance programs across the country. Enhancing comprehensive genomic surveillance programs in all Brazilian states can help track and monitor the introduction and spread of lineages across the country. The proper financing of these genomic surveys is of paramount importance to Brazil.
Data availability.
Next-generation sequencing (NGS) data generated in our study are publicly available in SRA-NCBI under BioProject accession no. PRJNA774631. Genome sequences were also deposited in GISAID (https://www.gisaid.org/) and are fully accessible for registered users within the “browse” option of the EpiCoV database (see Table S1 in the supplemental material).
ACKNOWLEDGMENTS
We thank all of the authors and administrators of the GISAID database, which allowed this study of genomic epidemiology to be conducted properly.
We would like to specially thank the following Brazilian institutes for the collective effort taken in quickly generating and analyzing the SARS-CoV-2 sequences used here: Instituto Butantan, Instituto Adolfo Lutz, Instituto Oswaldo Cruz, Universidade Federal de Goiás, Coordenação Geral de Laboratórios de Saúde Pública, Secretaria Municipal de Saúde de Guaratinguetá, Laboratório Central de Saúde Pública Noel Nutels (MA, RJ, and PR), Laboratório de Imunologia de Transplantes de Goiás, Inside Diagnósticos, Laboratório Dr. Paulo Emílio D' Alessandro, and Hospital Israelita Albert Einstein. A full list acknowledging the authors of the data used in this study can be found in Table S2 in the supplemental material.
This work was developed in the frameworks of Corona-ômica-RJ (FAPERJ E-26/210.179/2020). A.T.R.V. is supported by CNPq (303170/2017-4) and FAPERJ (E-26/202.903/20), and A.T. is supported by FAPERJ E-26/010.002434/2019 and E-26/210.178/2020. R.D.S.F., Jr., is a recipient of a graduate fellowship from CNPq, and A.P.L. was granted a postdoctoral scholarship (DTI-A) from CNPq.
We acknowledge support from the Rede Corona-ômica BR MCTI/FINEP, affiliated with RedeVírus/MCTI (FINEP 01.20.0029.000462/20, CNPq 404096/2020-4).
Footnotes
Supplemental material is available online only.
Contributor Information
Ana Tereza R. Vasconcelos, Email: atrv@lncc.br.
Kanta Subbarao, The Peter Doherty Institute for Infection and Immunity.
REFERENCES
- 1.Francisco Junior RDS, Lamarca AP, de Almeida LGP, Cavalcante L, Machado DT, Martins Y, Brustolini O, Gerber AL, Guimarães APDC, Gonçalves RB, Alves C, Mariani D, Cruz TF, de Souza IV, de Carvalho EM, Ribeiro MS, Carvalho S, da Silva FD, Garcia MHDO, de Souza LM, da Silva CG, Ribeiro CLP, Cavalcanti AC, de Mello CMB, Struchiner CJ, Tanuri A, de Vasconcelos ATR. 2021. Turnover of SARS-CoV-2 lineages shaped the pandemic and enabled the emergence of new variants in the state of Rio de Janeiro, Brazil. Viruses 13:2013. 10.3390/v13102013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Santos MCD, Sousa Júnior EC, Ferreira JDA, Barbagelata LS, da Silva SP, Silva AM, Cardoso J, Bedran RL, Chagas Junior WD, Bezerra DAM, da Pinheiro C, Monteiro DT, Lobo P, Muniz GM, Ferreira JL, Lobato LFL, Maramaldo CEC, Ferreira HL, Lima Neto LG, Vallinoto ACR, Cruz ACR, Medeiros BDA, Soares LS. 2021. First reported cases of SARS-CoV-2 sub-lineage B.1.617.2 in Brazil: an outbreak in a ship and alert for spread. Virological https://virological.org/t/first-reported-cases-of-sars-cov-2-sub-lineage-b-1-617-2-in-brazil-an-outbreak-in-a-ship-and-alert-for-spread/706. [Google Scholar]
- 3.Campbell F, Archer B, Laurenson-Schafer H, Jinnai Y, Konings F, Batra N, Pavlin B, Vandemaele K, Van Kerkhove MD, Jombart T, Morgan O, Le Polain de Waroux O. 2021. Increased transmissibility and global spread of SARS-CoV-2 variants of concern as at June 2021. Euro Surveill 26(24):pii=2100509. 10.2807/1560-7917.ES.2021.26.24.2100509. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Farinholt T, Doddapaneni H, Qin X, Menon V, Meng Q, Metcalf G, Chao H, Gingras M-C, Farinholt P, Agrawal C, Muzny DM, Piedra PA, Gibbs RA, Petrosino J. 2021. Transmission event of SARS-CoV-2 Delta variant reveals multiple vaccine breakthrough infections. medRxiv 10.1101/2021.06.28.21258780. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Williams SV, Vusirikala A, Ladhani SN, De Olano EFR, Iyanger N, Aiano F, Stoker K, Rao GG, John L, Patel B, Andrews N, Dabrera G, Ramsay M, Brown KE, Bernal JL, Saliba V. 2021. An outbreak caused by the SARS-CoV-2 Delta (B.1.617.2) variant in a care home after partial vaccination with a single dose of the COVID-19 vaccine Vaxzevria, London, England, April 2021. Euro Surveill 26(27):pii=2100626. 10.2807/1560-7917.ES.2021.26.27.2100626. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Planas D, Veyer D, Baidaliuk A, Staropoli I, Guivel-Benhassine F, Rajah MM, Planchais C, Porrot F, Robillard N, Puech J, Prot M, Gallais F, Gantner P, Velay A, Le Guen J, Kassis-Chikhani N, Edriss D, Belec L, Seve A, Courtellemont L, Péré H, Hocqueloux L, Fafi-Kremer S, Prazuck T, Mouquet H, Bruel T, Simon-Lorière E, Rey FA, Schwartz O. 2021. Reduced sensitivity of SARS-CoV-2 variant Delta to antibody neutralization. Nature 596:276–280. 10.1038/s41586-021-03777-9. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Table S1<br>. Download JVI.01228-21-s0001.xlsx, XLSX file, 0.02 MB (19.5KB, xlsx)
Table S2<br>. Download JVI.01228-21-s0002.pdf, PDF file, 0.2 MB (163.5KB, pdf)
Data Availability Statement
Next-generation sequencing (NGS) data generated in our study are publicly available in SRA-NCBI under BioProject accession no. PRJNA774631. Genome sequences were also deposited in GISAID (https://www.gisaid.org/) and are fully accessible for registered users within the “browse” option of the EpiCoV database (see Table S1 in the supplemental material).