ABSTRACT
Antibody-dependent cellular cytotoxicity (ADCC) has been correlated with reduced risk of human immunodeficiency virus type 1 (HIV-1) infection in several preclinical vaccine trials and in the RV144 clinical trial, indicating that this is a relevant antibody function to study. Given the diversity of HIV-1, the breadth of vaccine-induced antibody responses is a critical parameter to understand if a universal vaccine is to be realized. Moreover, the breadth of ADCC responses can be influenced by different vaccine strategies and regimens, including adjuvants. Therefore, to accurately evaluate ADCC and to compare vaccine regimens, it is important to understand the range of HIV Envelope (Env) susceptibility to these responses. These evaluations have been limited because of the complexity of the assay and the lack of a comprehensive panel of viruses for the assessment of these humoral responses. Here, we used 29 HIV-1 infectious molecular clones (IMCs) representing different Envelope subtypes and circulating recombinant forms to characterize susceptibility to ADCC from antibodies in plasma from infected individuals, including 13 viremic individuals, 10 controllers, and six with broadly neutralizing antibody responses. We found in our panel that ADCC susceptibility of the IMCs in our panel did not cluster by subtype, infectivity, level of CD4 downregulation, level of shedding, or neutralization sensitivity. Using partitioning around medoids (PAM) clustering to distinguish smaller groups of IMCs with similar ADCC susceptibility, we identified nested panels of four to eight IMCs that broadly represent the ADCC susceptibility of the entire 29-IMC panel. These panels, together with reagents developed to specifically accommodate circulating viruses at the geographical sites of vaccine trials, will provide a powerful tool to harmonize ADCC data generated across different studies and to detect common themes of ADCC responses elicited by various vaccines.
IMPORTANCE Antibody-dependent cellular cytotoxicity (ADCC) responses were found to correlate with reduced risk of infection in the RV144 trial of the only human HIV-1 vaccine to show any efficacy to date. However, reagents to understand the breadth and magnitude of these responses across preclinical and clinical vaccine trials remain underdeveloped. In this study, we characterize HIV-1 infectious molecular clones encoding 29 distinct Envelope strains (Env-IMCs) to understand factors that impact virus susceptibility to ADCC and use statistical methods to identify smaller nested panels of four to eight Env-IMCs that accurately represent the full set. These reagents can be used as standardized reagents across studies to fully understand how ADCC may affect efficacy of future vaccine studies and how studies differ in the breadth of responses developed.
KEYWORDS: antibody dependent cellular cytotoxicity, human immunodeficiency virus
INTRODUCTION
Antibody-dependent cellular cytotoxicity (ADCC) responses have been associated with lower viremia in human immunodeficiency virus type 1 (HIV-1)-infected individuals (1, 2), observed to be enriched in HIV-1 controllers (3, 4), and associated with curbing early simian immunodeficiency virus (SIV) viral replication in nonhuman primates (NHPs) (5–7). In addition, ADCC-mediating antibodies in breast milk have been correlated with reduced vertical transmission from viremic mothers (8). Consequently, there is substantial evidence to suggest that ADCC plays an important role in HIV-1 infection.
This is further supported by the results of the RV144 clinical trial, the only HIV-1 vaccine trial to date to show modest efficacy, which identified ADCC responses as a correlate of reduced risk of infection (9). Several preclinical trials support these observations. A pentavalent vaccine designed to improve antibody responses and enhance the protection observed in the RV144 trial and tested against a simian-human immunodeficiency virus (SHIV) challenge in NHPs again identified ADCC responses as a correlate of protection (10), and recently, ADCC responses elicited by a 6-valent vaccine were associated with decreased SHIV transmission risk (11). Conversely, the recent finding of lack of efficacy in the HVTN702 clinical trial brings into question the breadth of coverage that this vaccine could provide against circulating endemic HIV-1. As a result, ADCC responses elicited in future HIV-1 vaccine trials need to be methodically investigated.
Similarly to neutralization, assessments of HIV-1 vaccine-elicited ADCC-mediating antibody responses need to adequately address viral genetic and phenotypic diversity. Several large panels of pseudoviruses inclusive of numerous subtypes have been constructed and neutralization profiles comprehensively characterized using plasma from infected individuals and broadly neutralizing monoclonal antibodies (12–14). These studies revealed that variants have a range of neutralization susceptibility, referred to as tiers, with viruses clustering into one of four subgroups, as follows: very high (tier 1a), above average (tier 1b), moderate (tier 2), or low sensitivity (tier 3) to neutralization (15).
Assessment of ADCC requires de novo expression of Envelope (Env) in target cells, ideally from provirus in the context of infection, and thus cannot be done with pseudovirus. Given that utilization of replication-competent infectious molecular clones of HIV-1 strains of choice adds significant complexity to ADCC assays, the magnitude of testing described above for neutralization in TZM-bl cells would be prohibitive to implement for ADCC. Therefore, we investigated the possibility of implementing a panel of HIV-1 reference strains that reflects the global epidemic in order to facilitate testing and comparisons of ADCC responses across studies, especially for vaccine preclinical and clinical trials, which rely heavily on comparative immunogenicity data. We chose to test Envs from diverse strains (n = 29) encoded by infectious molecular clones with an isogenic backbone (Env-IMCs), and characterized ADCC responses against these Env-IMCs using plasma from nonmatched infected individuals (n = 29) in an attempt to develop standardized panels for assessment of vaccine-elicited ADCC-mediating antibody responses.
RESULTS
Env-IMCs broadly represent env diversity.
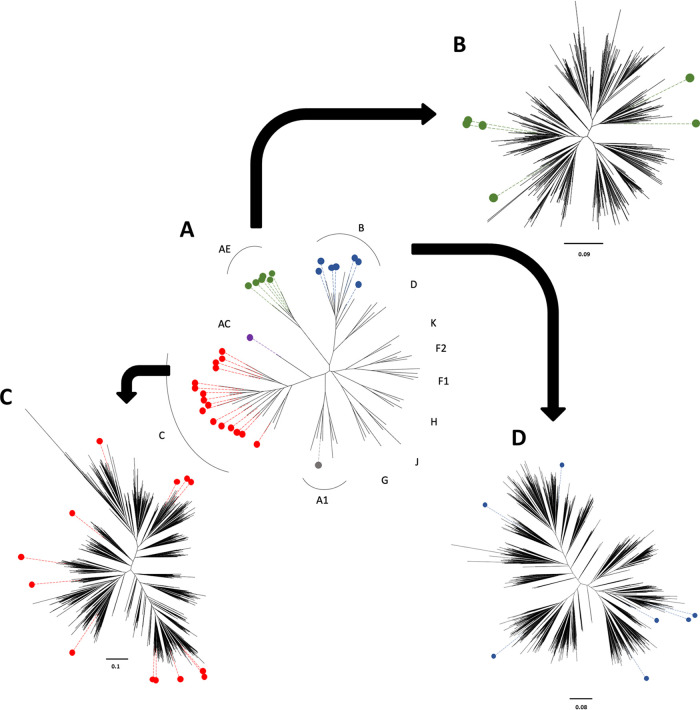
We used 29 infectious molecular clones, each designed using an isogenic proviral backbone and encoding the ectodomain of Envs of different subtypes (Env-IMCs) for this study (Fig. 1; characteristics described in Table 1), essentially as described before (16). Phylogenetic tree analysis (Fig. 1A) illustrates that these respective env genes represent five different subtypes or recombinants that are responsible for the majority (∼65%) of the HIV-1 epidemic (17), namely, A1 (n = 1), A1C (n = 1), CRF01_AE (n = 6), B (n = 7), and C (n = 14).
FIG 1.
Phylogenetic trees showing the genetic diversity covered by 29 Env-infectious molecular clones (IMCs). (A) env ectodomains from the 29 Env-IMCs we had available were aligned with sequences representing each major subtype (four sequences per subtype), and a maximum-likelihood tree was constructed. Six CRF01 A/E sequences (green) were then aligned with 549 superfiltered subtype A/E env sequences (B), 16 subtype C sequences (red) were aligned with 1,315 superfiltered subtype C env sequences (C), and seven subtype B sequences (blue) were aligned with 1,939 superfiltered subtype B env sequences (D) available in the Los Alamos database, and maximum-likelihood trees were constructed to show the genetic diversity covered by the three panels.
TABLE 1.
List of 29 Env-IMCs
| Virus | Full Env-IMC name | Subtype | Yr isolated | Accession no. for env gene sequence |
|---|---|---|---|---|
| BaL | NL-LucR.T2A-BaL.ecto | B | 1985 | DQ318211 |
| MW96.5 | NL-LucR.T2A-MW96.5.ecto | C | 1993 | U08455 |
| Q23.17 | NL-LucR.T2A-Q23.17.ecto | A1 | 1994 | AF004885 |
| SF162 | NL-LucR.T2A-SF162.ecto | B | 1988 | EU123924 |
| TH023 | NL-LucR.T2A-TH023.ecto | AE | 1992 | KU562843 |
| 1086.C | NL-LucR.T2A-Ce1086_B2.ecto | C | 2004 | FJ444395 |
| 246-F3.C10 | NL-LucR.T2A-246-F3_C10_2.ecto | A1C | 2000 | HM215279 |
| 427299 | NL-LucR.T2A-427299.c12.ecto | AE | 2006 | JN944655 |
| 816763 | NL-LucR.T2A-816763.c2.ecto | AE | 2006 | JN944659 |
| C1080.C03 | NL-LucR.T2A-C1080.c03.ecto | AE | 1999 | JN944660 |
| CH0505 | NL-LucR.T2A-CH0505.ecto | C | 2008 | KC247557 |
| CH058 | NL-LucR.T2A-CH58.ecto | B | 2006 | EU289194 |
| CH040 | NL-LucR.T2A-CH40.ecto | B | 2006 | EU289193 |
| CM235 | NL-LucR.T2A-CM235-2.ecto | AE | 1990 | JN944662 |
| CM244 | NL-LucR.T2A-CM244.c01-ETH2220.ecto | AE | 1990 | KC822429 |
| Du151 | NL-LucR.T2A-Du151.2.ecto | C | 1998 | DQ411851 |
| Du422 | NL-LucR.T2A-Du422.1.ecto | C | 1999 | DQ411854 |
| SUMA | NL-LucR.T2A-SUMA.ecto | B | 1991 | EU577073 |
| TV1 | NL-LucR.T2A-TV1.21.ecto | C | 2002 | HM215437 |
| WITO | NL-LucR.T2A-WITO.ecto | B | 2000 | AY835451 |
| YU2 | NL-LucR.T2A-Yu2.ecto | B | 1992 | M93258 |
| CAP210 | NL-LucR.T2A-CAP210.TF.ecto | C | 2005 | KC894136 |
| CAP228 | NL-LucR.T2A-CAP228.2.00.18.ecto | C | 2005 | EF203969 |
| CAP239 | NL-LucR.T2A-CAP239.TF.ecto | C | 2005 | FJ443353 |
| CAP255 | NL-LucR.T2A-CAP255.2.00.16J.ecto | C | 2005 | EF203982 |
| CAP256 | NL-LucR.T2A-CAP256.2.00.C7.ecto | C | 2005 | EF203981 |
| CAP45 | NL-LucR.T2A-CAP45.2.00.G3.ecto | C | 2005 | DQ435682 |
| CAP8 | NL-LucR.T2A-CAP8.TF.ecto | C | 2005 | FJ443492 |
| CAP88 | NL-LucR.T2A-CAP88.2.00.17_5A.ecto | C | 2005 | EF203970 |
These Env-IMCs, furthermore, represented the genetic diversity among three of the major subtypes (B, C, and CRF01_AE), assessed by using subtype-specific alignments consisting of superfiltered sequences available from the Los Alamos National Laboratory database. By aligning env genes of the 6 CRF01_AE Env-IMCs with 549 available sequences, we found the env sequences of the Env-IMCs broadly represented the genetic diversity of CRF01_AE env genes (Fig. 1B). Similarly, the env genes from the 16 subtype C Env-IMCs were distributed around the tree, indicating that they represented a large degree of subtype C diversity (Fig. 1C). Last, the env genes of the 7 subtype B Env-IMCs we had available fell into the half of the tree generated with 1,939 subtype B sequences (Fig. 1D).
Envelope susceptibility to ADCC and neutralization.
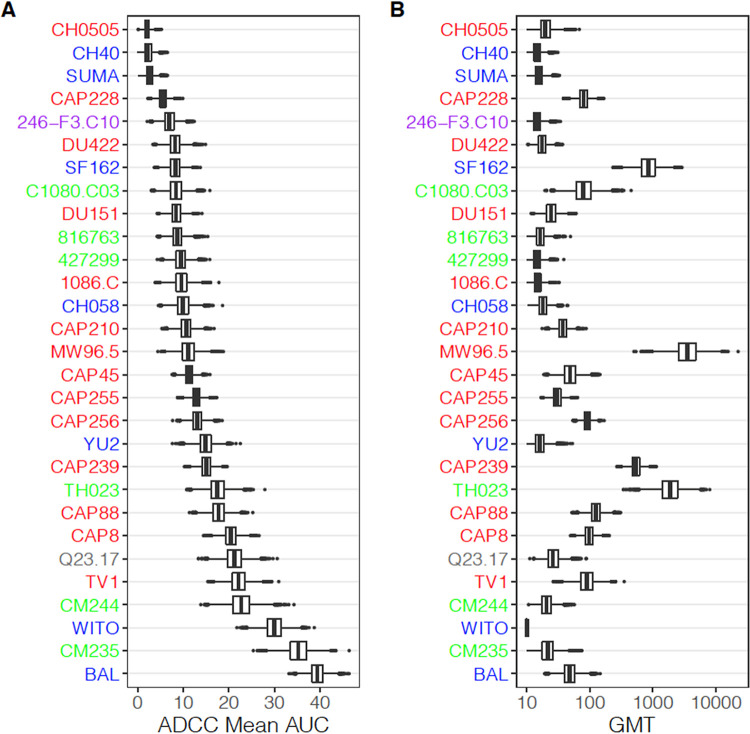
To assess the susceptibility of these viruses to ADCC, we used nonmatching plasma from 29 HIV-1-seropositive individuals infected with subtype B (n = 13), C (n = 10), or unknown subtype (n = 6) viruses and representing different clinical status with regard to their neutralization capacity and viral replication control, as well as plasma from three uninfected individuals as negative controls (Table 2). When we evaluated average virus susceptibility to ADCC, we observed differential recognition of viruses by ADCC-mediating plasma antibodies (Fig. 2A). For example, the subtype C Env-IMC CH0505 was extremely resistant to ADCC mediated by antibodies in plasma from the 29 individuals, with a mean ADCC area under the curve (AUC) of 3, while the subtype B Env-IMC BaL was highly susceptible to ADCC, with a mean AUC of 40.
TABLE 2.
Plasma samples used to assess ADCC susceptibility
| Neutralization profile | No. of samples of infecting strain of: |
||
|---|---|---|---|
| Subtype B | Subtype C | Unknown subtype | |
| Weakly neutralizing | 7 | 3 | 3 |
| Controller | 5 | 4 | 1 |
| Broadly neutralizing | 1 | 3 | 2 |
FIG 2.
Box plots showing distribution of mean area under the curve (AUC) of ADCC and geometrical mean neutralization titer of each virus over 10,000 bootstrap samples. Viruses are sorted according to mean AUC of ADCC.
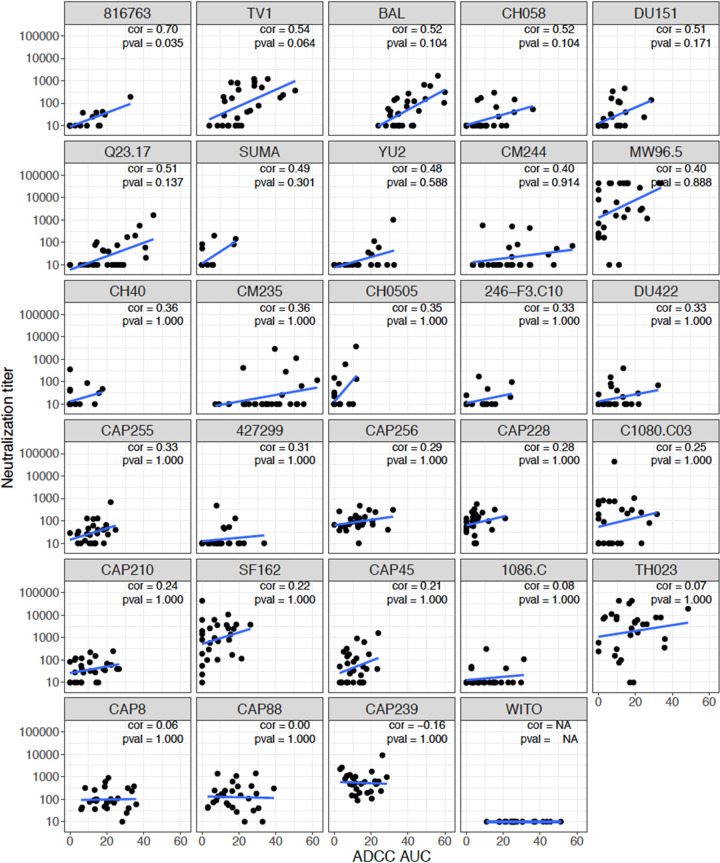
We then compared average virus susceptibility to ADCC with average virus susceptibility to neutralization using the TZM-bl neutralizing assay, shown by geometrical mean titer (GMT) for each virus in the same 29-virus panel across the set of plasma samples. Interestingly, the Env-IMCs exhibited differential susceptibility to ADCC and neutralization. The hierarchy of Env-IMC susceptibility to ADCC was not recapitulated when the same Env-IMCs were used in the TZM-bl neutralization assay and the same panel of plasma samples were used (Fig. 2). This observation was supported by a correlation analysis of the average virus susceptibility to ADCC (AUC) against average virus susceptibility to neutralization (GMTs) of the full 29-virus panel. We observed a nonsignificant positive Spearman correlation of 0.25. When looking at viruses individually, most viruses did not show a correlation between ADCC AUC and neutralization titer (Fig. 3). Responses against the CRF01_AE Env-IMC 816763 showed the highest, although moderate, Spearman correlation (rs = 0.70, P = 0.035), while correlations for all other Env-IMCs were less than 0.55 and tended to be positive, although none were significant after multiplicity correction.
FIG 3.
Scatterplots of neutralization titers versus ADCC (AUC) for 29 Env-IMCs. Spearman’s rank-order correlation and Bonferroni-adjusted P value are shown for each virus. Plots are ordered by Spearman’s rank-order correlation.
Effect of virus characteristics on ADCC susceptibility.
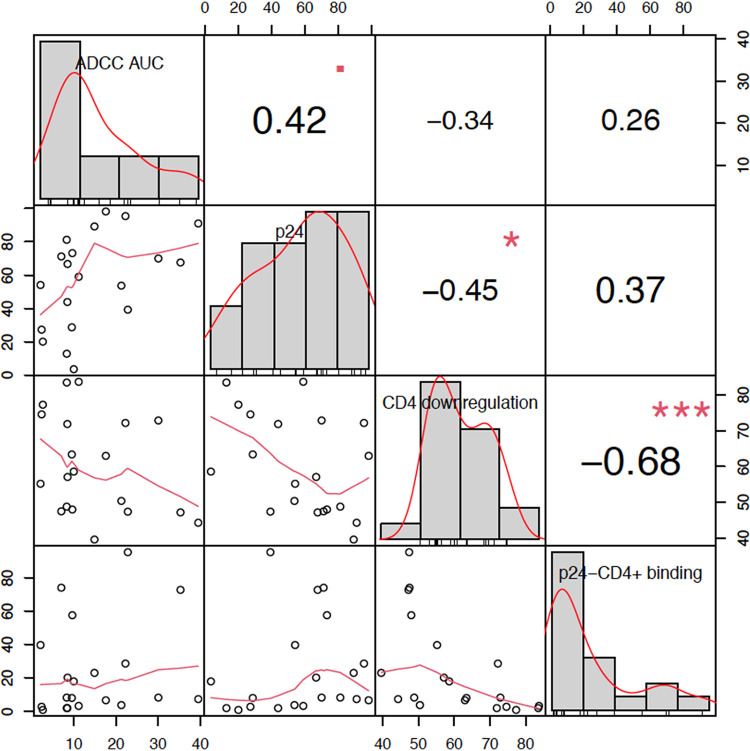
Based upon data from 21 of the Env-IMCs, we determined the impact of several characteristics of virus-infected cells on ADCC AUC, including the degree of target cell infection (percentage of p24+ cells), proportion of CD4 downregulated p24+ cells, and level of gp120 shedding (evaluated by the percentage of A32 binding to CD4+ p24− cells), using pairwise correlation. A modest relationship (rs = 0.42, P = 0.06) was observed between ADCC mean AUC and the percentage of p24+ cells (Fig. 4), while there was a nonsignificant moderate negative relationship between levels of CD4 downregulation and mean ADCC AUC (rs = −0.34, P = 0.1371). A moderate negative relationship was observed between the level of CD4 downregulation and gp120 shedding (rs = −0.68, P = 0.0009), indicating that as CD4 downregulation increased the level of shedding decreased, possibly due to the fact that as CD4 downregulation increases, more stable trimers are formed on the surface of the infected cell. Consequently, there are fewer unstable gp120 monomers to shed from the cell surface.
FIG 4.
Pairwise correlation plot showing correlations between average virus susceptibility to ADCC AUC, %p24 positivity at time of ADCC assay, %CD4 downregulation, and %A32 binding to p24− CD4+ cells for Env-IMCs in the global panel. The distribution of each variable (ADCC AUC, %p24 positivity, %CD4 downregulation, and %A32 binding to p24− CD4+ cells) is shown on the diagonal. The bivariate scatterplots indicate the exact value for each virus with respect to the variable above in the column and the variable in the row to the left, with a fitted line displayed below the diagonal. Correlation values with the significance level represented by stars (the greater the number of stars, the higher the significance) are displayed above the diagonal.
Downselection of Env-IMCs for assessment of ADCC responses in clinical trials.
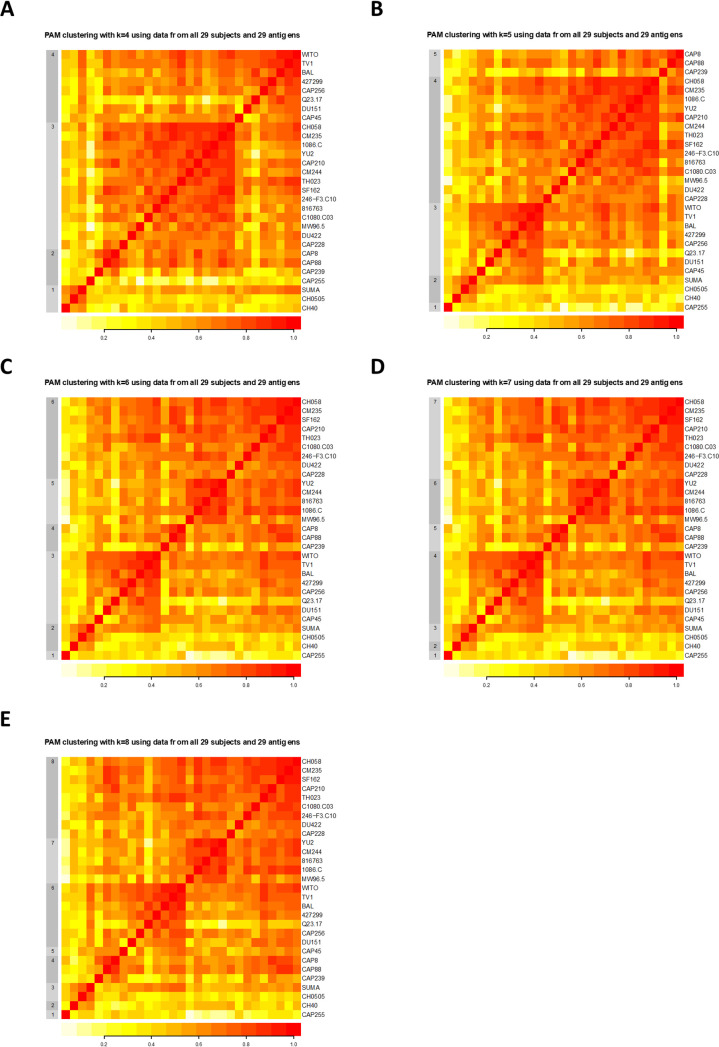
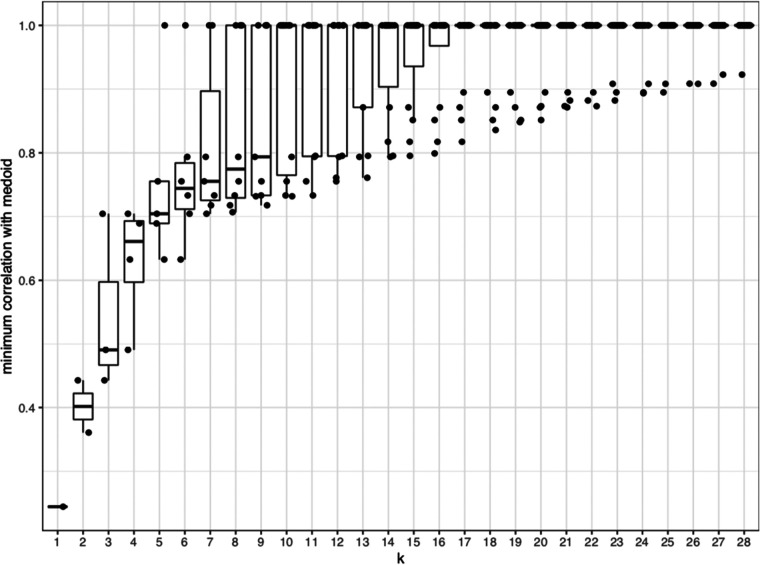
After establishing the profile of susceptibility of the full Env-IMC panel to the plasma antibody (Ab) ADCC responses, we next sought to determine whether or not smaller subsets of IMCs could be used to evaluate ADCC breadth in large clinical studies. In order to achieve this goal, partitioning around medoids (PAM) clustering was used to identify groups of viruses with similar ADCC susceptibility, defined by correlation-based distances (18). PAM identifies the viruses at the center of each cluster (termed “medoids”) for a given number (k) of clusters, which are used to construct panels of k viruses representing the different clusters. Taking the need for a balanced panel into consideration, together with pragmatic factors such as the number of viruses that could realistically be tested in vaccine immunogenicity studies given the cost of the assay, we chose to focus on panels ranging between k = 4 and k = 8 viruses. We were able to identify different panels based on the number of k-medoids (Fig. 5). As expected, the minimum correlation of viruses within a particular cluster to the medoid increased as k increased (Fig. 6), such that when k = 4, the minimum correlation was approximately 0.5, whereas when k = 7, the minimum correlation was greater than 0.7. This suggested that as the number of clusters increased, each medoid better represented the ADCC susceptibility or resistance of the other viruses in the cluster. In addition, once the viruses were split into clusters of five or more medoids, we observed at least one cluster of a single virus (minimum correlation = 1), indicating that the ADCC susceptibility profile of those singleton medoids was unique enough to not cluster with any other viruses.
FIG 5.
Partitioning around medoids (PAM) clustering using k = 4 (A), k = 5 (B), k = 6 (C), k = 7 (D), and k = 8 (E). The color scale represents the Spearman rank correlation of ADCC mediated by the 29 plasma samples against any two viruses.
FIG 6.
Box plots showing distribution of minimum correlation with medoid for each k.
When dividing the 29 viruses into four clusters, the four medoids were the subtype B Env-IMCs CH058, WITO, and SUMA and the subtype C Env-IMC CAP8 (Table 3). As the number of clusters was increased to eight, these four Env-IMCs remained the same, while other Env-IMCs were added as medoids, indicating that there was a nesting of medoids as k increased. These data indicate that a minimum of a four Env-IMC panel could be useful for study comparison purposes, even when different numbers of viruses have been used.
TABLE 3.
Nesting of medoids for k = 4 to k = 8 using all 29 Env-IMCs
| k = 4 | k = 5 | k = 6 | k = 7 | k = 8 |
|---|---|---|---|---|
| CH058 | CH058 | CH058 | CH058 | CH058 |
| WITO | WITO | WITO | WITO | WITO |
| SUMA | SUMA | SUMA | SUMA | SUMA |
| CAP8 | CAP8 | CAP8 | CAP8 | CAP8 |
| CAP255 | CAP255 | CAP255 | CAP255 | |
| YU2 | YU2 | YU2 | ||
| CH40 | CH40 | |||
| CAP45 |
The full panel and the seven-medoid panel exhibit similar susceptibility to ADCC.
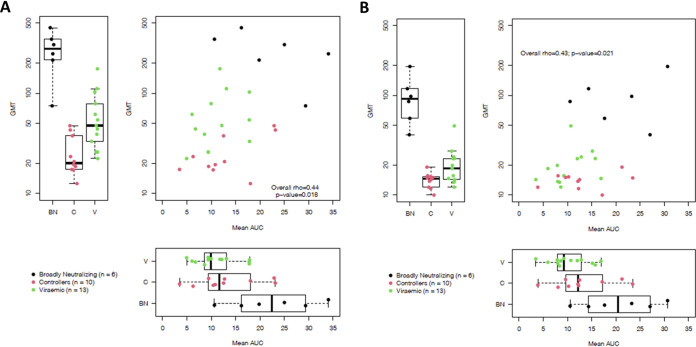
To ensure that the medoids accurately represented overall patterns of ADCC activity mediated by plasma antibodies from the 29 individuals, we compared these responses against the entire 29-virus panel and the Env-IMCs representing the seven-medoid panel. We performed these analyses by simultaneously using data from both neutralization and ADCC assays to make sure that functional responses were guiding our interpretation of the panels’ comparison. When we considered the correlation of average serum neutralization potency and average serum ADCC potency against the full 29-virus panel, we observed a low positive correlation between neutralization GMT and ADCC mean AUC (rs = 0.44, P = 0.018) (Fig. 7A). When we investigated neutralization alone (Fig. 7A, left), we observed that broadly neutralizing samples (black) exhibited the greatest neutralization, as expected, followed by plasma from viremic individuals and then controllers. While broadly neutralizing samples also mediated the highest levels of ADCC against the 29-virus panel (Fig. 7A, bottom), ADCC mediated by antibodies in controller sample plasma was higher than that mediated by antibodies in viremic plasma samples.
FIG 7.
ADCC and neutralization activity of 29 plasma samples using all 29 Env-IMCs (A) and data from the 7 medoid Env-IMCs (k = 7) obtained using PAM (B).
These relationships remained remarkably similar when assessing the correlation of average serum-neutralizing and ADCC activity against the seven medoid viruses, where we observed a Spearman correlation of 0.43, compared to 0.44 for the full panel (P = 0.021 versus 0.018) (Fig. 7B). While the medoid viruses became more resistant to neutralization (Fig. 7B, left), they maintained the same susceptibility to ADCC (P = 0.688; Fig. 7B, bottom) suggesting an accurate representation of the full panel of 29 viruses.
DISCUSSION
To understand how efficacious proposed HIV-1 vaccines may be, an understanding of the breadth of HIV-1 variant recognition elicited by these vaccines is crucial. In addition, it is important to be able to directly compare immune responses elicited by different vaccines when making decisions on which vaccine candidates to move forward. This requires a better understanding of the susceptibility of the Env to antibody responses to facilitate the selection of reagents dedicated to these tasks. Previously, robust statistical methods have been applied to identify a small panel of pseudoviruses that accurately represented the global epidemic for use in assessing the breadth of neutralizing antibody responses elicited by vaccine trials (19). However, there are currently no such panels of viruses for standardized assessment of vaccine-elicited ADCC-mediating antibody responses. Here, we used 29 Env-IMCs of different subtypes and sera from 29 nonmatched HIV-1-infected individuals to evaluate susceptibility of viruses produced from these IMCs to ADCC-mediating antibodies in a luciferase-based ADCC assay. Using PAM clustering, we were able to identify more economical panels of 4 to 8 Env-IMCs that accurately represented the susceptibility of the full panel of 29 Env-IMCs to ADCC responses. These panels will be essential to facilitating the standardized assessment of the breadth of vaccine-induced ADCC responses across past and future vaccine trials, in addition to the evaluation of responses to vaccine-matched reagents.
The PAM methodology used to determine appropriate panels of Env-IMCs resulted in clustering of Env-IMCs that nested for panels of size k = 4 to k = 8, such that each panel of size k + 1 contains all Env-IMCs in the panel of size k. This allows the use of 8 Env-IMCs where budget allows, while still permitting the comparison of vaccine-induced responses in other studies where fewer viruses were used. Panels derived by PAM clustering consisted entirely of subtype B and C Env-IMCs, suggesting that subtype is not a major factor in ADCC susceptibility. For example, this study suggests that little to no difference would be introduced by replacing the subtype B Env-IMC CH058 with CRF01_AE CM235, which had similar susceptibility to ADCC and correlation to other Env-IMCs in the cluster. Nevertheless, because of the relatively limited diversity of the panel, while the panel can be used for any HIV-1 vaccine, it will be most relevant for vaccines targeting regions dominated by subtype B and C and CRF01_AE strains, and it can be used to directly compare the diversity of ADCC responses across vaccines targeting the same geographic region.
The breadth of ADCC-mediating monoclonal antibodies has been reported to various degrees in numerous previous studies using both infected cells and Env-coated cells (20–24). Furthermore, we have previously characterized the breadth of ADCC responses in sera from vaccinees in HVTN100 (used as the go/no-go criteria for HVTN702 [25]) against both subtype C and CRF01_AE Env-IMCs (26). These studies suggest that using a standardized panel of viruses to characterize magnitude and breadth will improve evaluation of ADCC responses in clinical trials and provide novel insights into requirements for protection.
It is well established that neutralizing antibodies can mediate ADCC (21, 27), and a previous study found, for monoclonal antibodies, that neutralization and ADCC were generally correlated (28). While this had not previously been found using sera from infected individuals (29, 30), this may be due to the limited number of viruses used in these studies. We observed an overall positive but limited relationship between antibody neutralization and ADCC in our panel of sera, suggesting there is an overlap in neutralization and ADCC function of antibodies in polyclonal antibody responses. In addition, similarly to previous studies, we observed that plasma from broad neutralizers tended to have the highest ADCC activity (31).
However, when we ranked viruses by their susceptibility to ADCC or neutralization, we found no relationship. Viruses in this panel could be resistant to neutralization and sensitive to ADCC or vice versa, but were rarely sensitive or resistant to both responses (the highest correlation we observed between neutralization and ADCC was rs = 0.7 for Env-IMC 816763). This observation suggests there are different requirements for neutralization of virions and ADCC of virus-infected cells, which may be largely due to differences in Env epitopes targeted by neutralizing and nonneutralizing antibodies in serum. In addition, a further difference may be the presentation of Env epitopes on the membrane of virions and infected cells. Env from some viruses may spend more or less time in an “open” conformation, impacting their susceptibility to antibodies targeting CD4-induced epitopes (which are typically nonneutralizing antibodies that mediate potent ADCC) (32).
We previously reported that large portions of the HIV-1 Envelope trimer are present on the surface of target cells in a non-CD4i conformation (33), despite the luciferase gene included in our IMCs interfering with Nef function, resulting in incomplete downregulation of CD4 from the cell surface (34). In addition, it is important to note that the ability of Nef to downregulate CD4 differs between virus isolates (35), as does the intrinsic closed or open status of Env trimers (36, 37), making it unlikely that single-virus infection, assayed at a single time postinfection, will reflect the complexity of HIV infections in vivo. Furthermore, the importance of recognizing both conformations has been reported by different groups in the context of preclinical (10, 11) and clinical studies (32, 38, 39). Previous studies have identified CRF01_AE viruses as being generally more sensitive to ADCC due to the presence of a histidine residue at position 375 of the Env that fills in a phenylalanine cavity at position 43 (40). While all six CRF01_AE Env-IMCs in our panel contained histidine 375, we did not observe this association between virus subtype and ADCC susceptibility, which may be due to the characteristics of the Envs included in our panel.
Last, while subtypes B, C, and CRF01_AE account for most of the 29-Env–IMC panel, we observed no impact of subtype on susceptibility to ADCC in our analyses. In addition, we hope to construct additional Env-IMCs consisting of env genes from subtypes A, D, and G to more accurately represent HIV-1 genetic diversity. We intend to use the panels designed above, together with sequencing of breakthrough infections in past (including RV144 and the recently completed HVTN702) and ongoing or future vaccine trials, to fully understand the breadth and magnitude of ADCC responses being elicited by vaccines and the impact on protection.
In summary, we report here the most extensive effort to-date, using robust statistical analyses, to provide standard panels of reagents for assessing vaccine-elicited ADCC responses. We have identified panels of 4 to 8 Env strains that, expressed in the context of Env-IMCs, may be used to assess past, present, and future trials to different extents, dependent on budget, and that include env genes from three different subtypes (B, C, and CRF01_AE). We believe that these panels, together with reagents developed to specifically accommodate circulating viruses at the geographical sites of vaccine trials, will provide a powerful tool to harmonize generated data and detect common themes of ADCC responses elicited by various vaccines.
MATERIALS AND METHODS
Ethics statement.
The studies were reviewed and approved by the Duke University Medical Center Institutional Review Board, and all participants provided written informed consent. All research was performed in accordance with the Duke University School of Medicine guidelines and regulations.
Plasma samples.
Twenty-six samples were obtained from the CHAVI 001 cohort, and three samples were obtained from the Duke controller cohort, infected by either subtype B HIV-1 (n = 13), subtype C HIV-1 (n = 10), or unknown (n = 6). Controllers were defined as having a viral load of <50 copies/mL, and broadly neutralizing samples were chosen from broadly neutralizing sera identified in Hraber et al. (41).
Cell lines.
TZM-bl cells (42) were obtained from the NIH AIDS Research and Reference Reagent Program (NIH ARRRP, catalogue number 8129; contributed by John Kappes and Xiaoyun Wu). The HEK293T cell line was obtained from George Shaw (University of Pennsylvania, Philadelphia, PA). All adherent cell lines were cultured at 37°C and 5% CO2 in Dulbecco’s modified Eagle’s medium (DMEM) containing 10% heat-inactivated fetal calf serum (FCS; Biochrom) with 50 μg/mL gentamicin (Lonza, Basel, Switzerland) and disrupted at confluence by treatment with 0.25% trypsin in 1 mM EDTA (Lonza). CEM.NKRCCR5 cells were obtained through the NIH AIDS Research and Reference Reagent Program (NIH ARRRP, catalogue number 4376; contributed by Alexandra Trkola).
The CEM.NKRCCR5 cell line was used in ADCC assays to reduce variability in infections and to provide target cells that can be reproducibly used in combinations with effector cells from different donors.
Peripheral blood mononuclear cells.
Peripheral blood mononuclear cells (PBMCs) obtained by leukapheresis from an HIV-seronegative individual (Fc gamma receptor IIIA [FcγRIIIA] 158 V/F heterozygous) and previously assessed for FcyRIIIA genotype, frequency of NK cells, and ADCC activity using control sera and monoclonal antibodies under GCLP guidelines and W oversight were used as a source of effector cells.
Construction of HIV-1 infectious molecular clones and virus preparation.
HIV-1 infectious molecular clones (IMCs) carrying the env genes for different transmitted/founder viruses in an isogenic proviral backbone were constructed as previously described (16); these also expressed the Renilla luciferase (LucR) reporter gene under the control of the HIV-1 Tat protein. Replication-competent reporter viruses like NL-LucR.T2A-CAP239.TF.ecto and other env-carrying strains are collectively referred to here as Env-IMC-LucR viruses. These Env-IMC-LucR viruses were originally designed to preserve all nine viral open reading frames linking Nef expression to LucR via a ribosome-skipping T2A peptide, but we later found that Nef expression of these viruses was reduced in primary CD4 T cells (43). Replication-competent viruses were obtained by transfecting the Env-IMC-LucR plasmid in HEK293T cells using PolyFect transfection reagent (Qiagen). Virus-containing supernatants were harvested 48 h following transfection, clarified by 0.45-μm filtration, and adjusted to 10% FCS (Biochrom). The 50% tissue culture infectious dose (TCID50) for each IMC preparation was determined by infection of TZM-bl cells as previously described (44, 45).
Viruses were then used to infect CEM.NKRCCR5 cells, which were characterized for suitability in a Renilla luciferase ADCC assay using criteria previously determined in assay qualification for use in clinical trial immunogenicity assessments. These criteria included ensuring >5% p24+ (infected) cells after infection and assessing the level of CD4 downregulation induced by infection of cells (Fig. 4).
Infection of the CEM.NKRCCR5 cell line with HIV-1 Env-IMC-LucR.
A total of 1 × 106 CEM.NKRCCR5 cells (NIH AIDS Reagent Program, Division of AIDS, NIAID, NIH; from Alexandra Trkola) were infected with virus inoculum at a titer determined to achieve at least 5% infection in the presence of DEAE-dextran (7.5 μg/mL). The cells were subsequently resuspended at 0.3 × 106/mL and cultured for 3 days in complete medium containing 7.5 μg/mL DEAE-dextran. On the day of assay, the infection was monitored by measuring the frequency of cells expressing intracellular p24. Assays performed using the Env-IMC-LucR-infected target cells were considered reliable if the percentage of viable p24+ target cells on the day of assay was ≥5%. Following 72 h of infection, cells were washed in phosphate-buffered saline (PBS), dispensed in 96-well V-bottomed plates at 1 × 105 viable cells per well, and stained with a vital dye (Live/Dead Fixable Aqua dead cell stain; Invitrogen) to exclude nonviable cells from subsequent analyses. The cells were then washed with washing buffer (WB; PBS plus 1% FBS) and incubated with an anti-CD4 antibody (clone OKT4-PerCP-cyanine 5.5; BioLegend) at 1:40 for 20 min at room temperature. Cells were then washed with WB again, incubated with Cytofix/Cytoperm (BD Bioscience) for 20 min at 4°C, and washed with 1% Cytoperm washing buffer (BD Biosciences). After the final wash, the anti-p24 antibody (clone KC57-RD1; Beckman Coulter) was added to a final dilution of 1:400, and the plates were incubated for 30 min at 4°C. The plates were washed twice with Cytoperm washing buffer, and the cells were resuspended in 1% formaldehyde-PBS. The samples were acquired within 24 h using a BD Fortessa flow cytometer. A minimum of 10,000 total singlet events was acquired for each analysis. Gates were set to include singlet and live events. The appropriate compensation beads were used to compensate the spillover signal for the three fluorophores. Data analysis was performed using FlowJo 10.2 software (BD Biosciences). Uninfected CEM.NKRCCR5 and chronically infected A1953 cell lines were used to titer the vital dye and anti-p24 antibody and appropriately determine the position of the CD4+/CD4− gate and were used as negative and positive controls, respectively, for the described staining procedure.
Neutralization assay.
Neutralization was measured, as previously described (44), by a reduction in luciferase gene expression after single round infection of TZM-bl cells with Env pseudoviruses or infectious viruses. Titers were calculated as the reciprocal plasma dilution that resulted in a 50% inhibitory dose (ID50) of the relative light units (RLU).
Renilla luciferase-based ADCC assay.
The LucR-based ADCC assay was conducted as described by Pollara et al. (46). The day prior to the ADCC assay, cryopreserved PBMCs to be used as effectors in the assay were thawed in R10, counted and assessed for viability, and resuspended in R10 overnight. On the day of the assay, infected CEM.NKRCCR5 cells were counted, assessed for viability (viability ≥ 80%), and the concentration was adjusted to 2 × 105 viable cells/mL (5 × 103 cells/well). PBMCs were then counted, assessed for viability, pelleted, and resuspended in the infected CEM.NKRCCR5 cells at a concentration of 6 × 106 PBMCs/mL (1.5 × 105 PBMCs/well) (effector:target cell ratio of 30:1). Heat-inactivated autologous plasma was serially diluted. The effector/target cell mix and antibody dilutions were plated in opaque 96-well half-area plates, centrifuged at 300 × g for 1 min after 30 min incubation at room temperature, and then incubated for 5.5 h at 37°C and 5.5% CO2 to allow ADCC-mediated cell lysis to proceed. After 5.5 h, ViviRen substrate (Promega) was diluted 1:500 in R10 and added 1:1 to the assay wells. The substrate generates luminescence only in live, infected cells, not in dead or lysed cells. The final readout was the luminescence intensity generated by the presence of residual intact target cells that had not been lysed by the effector population in the presence of ADCC-mediating antibodies. The percent specific killing was calculated using the following formula:
In the analysis, the RLU of the target and effector wells represent lysis by NK cells in the absence of any source of antibody. Plasma from three seronegative donors were used as a negative control, and plasma from a chronically infected individual was used as a positive control. Titers were calculated as the reciprocal of the highest plasma dilution that resulted in a percentage of specific killing above the background signal.
Statistical analyses.
(i) ADCC AUC. The specific killing activities in the ADCC assay were summarized for each subject and antigen by computing the area under the dilution curve (AUC) as the mean of the specific killing activities across the dilution series. Specific killing activities less than 15 were assigned a value of 0 prior to AUC calculation.
(ii) NAb ID50 titers.
ID50 titers from the neutralization assay that were lower than the limit of detection of 20 were assigned a value of 10.
(iii) Bootstrapped confidence intervals.
Confidence intervals (95%) for the antigen-specific mean ADCC AUCs and geometrical mean neutralizing antibody (NAb) ID50 titers (GMTs) across the 29 seropositive plasma samples were computed from 10,000 bootstrapped samples generated with replacement.
(iv) Correlation analysis.
Spearman’s rank-order correlations (rs) were computed between ADCC AUCs and NAb GMTs for each antigen using only pairwise complete observations. The significance of correlation coefficients was adjusted using Bonferroni correction. Spearman’s rank-order correlations were also computed between mean AUCs, percent p24 positivity at time of ADCC assay, percent CD4 downregulation, and percent A32 binding to p24− CD4+ cells for antigens in the global panel.
(v) Clustering analysis.
Using ADCC AUC data from 29 plasma samples, a matrix of Spearman rank correlations was generated between all pairs of 29 Env-IMCs, which was subsequently used to create a distance matrix using 1 minus the correlation. Based on the distance matrix, k clusters were formed in which k ranged from 1 to 28, using the method of partitioning around medoids (PAM) from the cluster package in R (M. Maechler, P. Rousseeuw, A. Struyf, M. Hubert, and K. Hornik, “Cluster analysis basics and extensions.” R package version 2.0.5). For each k, the proposed panel of size k was defined as the k medoids identified by the PAM algorithm. The distance matrix was also used to create heatmaps to visually depict the clusters and the medoids identified by PAM. Within each k cluster, the minimum correlation between antigens and between antigens and medoid were used to identify the optimal k medoids, or the minimal subset of antigens that could be used to represent the complete panel.
Data availability.
Envelope sequences of all Env-IMCs are accessible through GenBank; accession numbers are provided in Table 1. Other data and reagents are available upon request.
ACKNOWLEDGMENTS
This study was supported in part by the Bill & Melinda Gates Foundation’s Collaboration for AIDS Vaccine Discovery (CAVD) grants to the Comprehensive Antibody Vaccine Immune Monitoring Consortium (CAVIMC; grant OPP1146996) and the Vaccine Immunology Statistical Center (VISC; grant OPP1151646). We thank Coleen Cunningham and John Bartlett of the Duke CFAR Clinical Core, Clinical Core staff (Stuart Carr and Sunita Patil), and the patients, physicians, and staff of the Duke Adult Infectious Diseases Clinic (Gary Cox, Nathan Thielman, Cameron Wolfe, Elizabeth Livingston, Brianna Norton, Kristen Dicks, Mehri McKellar, Vivian Chu, Jason Stout, and Ann Mosher) for virus controller patient recruitment, as well as Myron Cohen and the CHAVI cohort clinical staff and patients.
Author contributions were as follows: D. Mielke, S.S.O., B.B., L.F., D. Montefiori, J.P., A.D.C., G.F.: conceptualization and methodology. D. Mielke, S.S.O., B.B., K.F., M.T.: investigation. K.G., H.G., C.W., L.M., C.O., B.H., G.T., A.D.C., G.F.: resources. D. Mielke, S.S.O., B.B., J.P., A.D.C., G.F.: writing—original draft and visualization. K.G., H.G., A.D.C., G.F.: project administration and supervision. D. Montefiori, G.F.: funding acquisition.
We declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Contributor Information
Dieter Mielke, Email: dieter.mielke@duke.edu.
Guido Ferrari, Email: gflmp@duke.edu.
Viviana Simon, Icahn School of Medicine at Mount Sinai.
REFERENCES
- 1.Ahmad R, Sindhu STAK, Toma E, Morisset R, Vincelette J, Menezes J, Ahmad A. 2001. Evidence for a correlation between antibody-dependent cellular cytotoxicity-mediating anti-HIV-1 antibodies and prognostic predictors of HIV infection. J Clin Immunol 21:227–233. 10.1023/A:1011087132180. [DOI] [PubMed] [Google Scholar]
- 2.Johansson SE, Rollman E, Chung AW, Center RJ, Hejdeman B, Stratov I, Hinkula J, Wahren B, Kärre K, Kent SJ, Berg L. 2011. NK cell function and antibodies mediating ADCC in HIV-1-infected viremic and controller patients. Viral Immunol 24:359–368. 10.1089/vim.2011.0025. [DOI] [PubMed] [Google Scholar]
- 3.Lambotte O, Ferrari G, Moog C, Yates NL, Liao H-X, Parks RJ, Hicks CB, Owzar K, Tomaras GD, Montefiori DC, Haynes BF, Delfraissy J-F. 2009. Heterogeneous neutralizing antibody and antibody-dependent cell cytotoxicity responses in HIV-1 elite controllers. AIDS 23:897–906. 10.1097/QAD.0b013e328329f97d. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Lambotte O, Pollara J, Boufassa F, Moog C, Venet A, Haynes BF, Delfraissy J-F, Saez-Cirion A, Ferrari G. 2013. High antibody-dependent cellular cytotoxicity responses are correlated with strong CD8 T cell viral suppressive activity but not with B57 status in HIV-1 elite controllers. PLoS One 8:e74855. 10.1371/journal.pone.0074855. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Xiao P, Zhao J, Patterson LJ, Brocca-Cofano E, Venzon D, Kozlowski PA, Hidajat R, Demberg T, Robert-Guroff M. 2010. Multiple vaccine-elicited nonneutralizing antienvelope antibody activities contribute to protective efficacy by reducing both acute and chronic viremia following simian/human immunodeficiency virus SHIV89.6P challenge in rhesus macaques. J Virol 84:7161–7173. 10.1128/JVI.00410-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Hidajat R, Xiao P, Zhou Q, Venzon D, Summers LE, Kalyanaraman VS, Montefiori DC, Robert-Guroff M. 2009. Correlation of vaccine-elicited systemic and mucosal nonneutralizing antibody activities with reduced acute viremia following intrarectal simian immunodeficiency virus SIVmac251 challenge of rhesus macaques. J Virol 83:791–801. 10.1128/JVI.01672-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Xiao P, Patterson LJ, Kuate S, Brocca-Cofano E, Thomas MA, Venzon D, Zhao J, DiPasquale J, Fenizia C, Lee EM, Kalisz I, Kalyanaraman VS, Pal R, Montefiori D, Keele BF, Robert-Guroff M. 2012. Replicating adenovirus-simian immunodeficiency virus (SIV) recombinant priming and envelope protein boosting elicits localized, mucosal IgA immunity in rhesus macaques correlated with delayed acquisition following a repeated low-dose rectal SIVmac251 challenge. J Virol 86:4644–4657. 10.1128/JVI.06812-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Mabuka J, Nduati R, Odem-Davis K, Peterson D, Overbaugh J. 2012. HIV-specific antibodies capable of ADCC are common in breastmilk and are associated with reduced risk of transmission in women with high viral loads. PLoS Pathog 8:e1002739-13. 10.1371/journal.ppat.1002739. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Haynes BF, Gilbert PB, McElrath MJ, Zolla-Pazner S, Tomaras GD, Alam SM, Evans DT, Montefiori DC, Karnasuta C, Sutthent R, Liao H-X, DeVico AL, Lewis GK, Williams C, Pinter A, Fong Y, Janes H, DeCamp A, Huang Y, Rao M, Billings E, Karasavvas N, Robb ML, Ngauy V, de Souza MS, Paris R, Ferrari G, Bailer RT, Soderberg KA, Andrews C, Berman PW, Frahm N, De Rosa SC, Alpert MD, Yates NL, Shen X, Koup RA, Pitisuttithum P, Kaewkungwal J, Nitayaphan S, Rerks-Ngarm S, Michael NL, Kim JH. 2012. Immune-correlates analysis of an HIV-1 vaccine efficacy trial. N Engl J Med 366:1275–1286. 10.1056/NEJMoa1113425. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Bradley T, Pollara J, Santra S, Vandergrift N, Pittala S, Bailey-Kellogg C, Shen X, Parks R, Goodman D, Eaton A, Balachandran H, Mach LV, Saunders KO, Weiner JA, Scearce R, Sutherland LL, Phogat S, Tartaglia J, Reed SG, Hu S-L, Theis JF, Pinter A, Montefiori DC, Kepler TB, Peachman KK, Rao M, Michael NL, Suscovich TJ, Alter G, Ackerman ME, Moody MA, Liao H-X, Tomaras G, Ferrari G, Korber BT, Haynes BF. 2017. Pentavalent HIV-1 vaccine protects against simian-human immunodeficiency virus challenge. Nat Commun 8:15711. 10.1038/ncomms15711. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Felber BK, Lu Z, Hu X, Valentin A, Rosati M, Remmel CAL, Weiner JA, Carpenter MC, Faircloth K, Stanfield-Oakley S, Williams WB, Shen X, Tomaras GD, LaBranche CC, Montefiori D, Trinh HV, Rao M, Alam MS, Vandergrift NA, Saunders KO, Wang Y, Rountree W, Das J, Alter G, Reed SG, Aye PP, Schiro F, Pahar B, Dufour JP, Veazey RS, Marx PA, Venzon DJ, Shaw GM, Ferrari G, Ackerman ME, Haynes BF, Pavlakis GN. 2020. Co-immunization of DNA and protein in the same anatomical sites induces superior protective immune responses against SHIV challenge. Cell Rep 31:107624. 10.1016/j.celrep.2020.107624. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Hraber P, Rademeyer C, Williamson C, Seaman MS, Gottardo R, Tang H, Greene K, Gao H, LaBranche C, Mascola JR, Morris L, Montefiori DC, Korber B. 2017. Panels of HIV-1 subtype C Env reference strains for standardized neutralization assessments. J Virol 91:e00991-17. 10.1128/JVI.00991-17. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Rademeyer C, Korber B, Seaman MS, Giorgi EE, Thebus R, Robles A, Sheward DJ, Wagh K, Garrity J, Carey BR, Gao H, Greene KM, Tang H, Bandawe GP, Marais JC, Diphoko TE, Hraber P, Tumba N, Moore PL, Gray GE, Kublin J, McElrath MJ, Vermeulen M, Middelkoop K, Bekker LG, Hoelscher M, Maboko L, Makhema J, Robb ML, Abdool Karim SS, Abdool Karim Q, Kim JH, Hahn BH, Gao F, Swanstrom R, Morris L, Montefiori DC, Williamson C. 2016. Features of recently transmitted HIV-1 clade C viruses that impact antibody recognition: implications for active and passive immunization. PLoS Pathog 12:e1005742-29. 10.1371/journal.ppat.1005742. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Doria-Rose NA, Louder MK, Yang Z, O’Dell S, Nason M, Schmidt SD, McKee K, Seaman MS, Bailer RT, Mascola JR. 2012. HIV-1 neutralization coverage is improved by combining monoclonal antibodies that target independent epitopes. J Virol 86:3393–3397. 10.1128/JVI.06745-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Seaman MS, Janes H, Hawkins N, Grandpre LE, Devoy C, Giri A, Coffey RT, Harris L, Wood B, Daniels MG, Bhattacharya T, Lapedes A, Polonis VR, McCutchan FE, Gilbert PB, Self SG, Korber BT, Montefiori DC, Mascola JR. 2010. Tiered categorization of a diverse panel of HIV-1 Env pseudoviruses for assessment of neutralizing antibodies. J Virol 84:1439–1452. 10.1128/JVI.02108-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Edmonds TG, Ding H, Yuan X, Wei Q, Smith KS, Conway JA, Wieczorek L, Brown B, Polonis V, West JT, Montefiori DC, Kappes JC, Ochsenbauer C. 2010. Replication competent molecular clones of HIV-1 expressing Renilla luciferase facilitate the analysis of antibody inhibition in PBMC. Virology 408:1–13. 10.1016/j.virol.2010.08.028. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Hemelaar J, Gouws E, Ghys PD, Osmanov S, WHO-UNAIDS Network for HIV Isolation and Characterisation. 2011. Global trends in molecular epidemiology of HIV-1 during 2000–2007. AIDS 25:679–689. 10.1097/QAD.0b013e328342ff93. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Rousseeuw P, Struyf A, Hubert M. 1990. Partitioning around medoids (program PAM), p 68–125. In Kaufman L, Rousseeuw PJ (ed), Finding groups in data. John Wiley & Sons, Inc., Hoboken, NJ. [Google Scholar]
- 19.deCamp A, Hraber P, Bailer RT, Seaman MS, Ochsenbauer C, Kappes J, Gottardo R, Edlefsen P, Self S, Tang H, Greene K, Gao H, Daniell X, Sarzotti-Kelsoe M, Gorny MK, Zolla-Pazner S, LaBranche CC, Mascola JR, Korber BT, Montefiori DC. 2014. Global panel of HIV-1 Env reference strains for standardized assessments of vaccine-elicited neutralizing antibodies. J Virol 88:2489–2507. 10.1128/JVI.02853-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Bruel T, Guivel-Benhassine F, Lorin V, Lortat-Jacob H, Baleux F, Bourdic K, Noël N, Lambotte O, Mouquet H, Schwartz O. 2017. Lack of ADCC breadth of human nonneutralizing anti-HIV-1 antibodies. J Virol 91:e02440-16. 10.1128/JVI.02440-16. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Bruel T, Guivel-Benhassine F, Amraoui S, Malbec M, Richard L, Bourdic K, Donahue DA, Lorin V, Casartelli N, Noël N, Lambotte O, Mouquet H, Schwartz O. 2016. Elimination of HIV-1-infected cells by broadly neutralizing antibodies. Nat Commun 7:10844. 10.1038/ncomms10844. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Williams KL, Cortez V, Dingens AS, Gach JS, Rainwater S, Weis JF, Chen X, Spearman P, Forthal DN, Overbaugh J. 2015. HIV-specific CD4-induced antibodies mediate broad and potent antibody-dependent cellular cytotoxicity activity and are commonly detected in plasma from HIV-infected humans. EBioMedicine 2:1464–1477. 10.1016/j.ebiom.2015.09.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Mayr LM, Decoville T, Schmidt S, Laumond G, Klingler J, Ducloy C, Bahram S, Zolla-Pazner S, Moog C. 2017. Non-neutralizing antibodies targeting the V1V2 domain of HIV exhibit strong antibody-dependent cell-mediated cytotoxic activity. Sci Rep 7:12655. 10.1038/s41598-017-12883-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Easterhoff D, Pollara J, Luo K, Tolbert WD, Young B, Mielke D, Jha S, O’Connell RJ, Vasan S, Kim J, Michael NL, Excler J-L, Robb ML, Rerks-Ngarm S, Kaewkungwal J, Pitisuttithum P, Nitayaphan S, Sinangil F, Tartaglia J, Phogat S, Kepler TB, Alam SM, Wiehe K, Saunders KO, Montefiori DC, Tomaras GD, Moody MA, Pazgier M, Haynes BF, Ferrari G. 2019. Boosting with AIDSVAX B/E enhances Env constant region 1 and 2 antibody-dependent cellular cytotoxicity breadth and potency. J Virol 94:e01120-19. 10.1128/JVI.01120-19. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Bekker L-G, Moodie Z, Grunenberg N, Laher F, Tomaras GD, Cohen KW, Allen M, Malahleha M, Mngadi K, Daniels B, Innes C, Bentley C, Frahm N, Morris DE, Morris L, Mkhize NN, Montefiori DC, Sarzotti-Kelsoe M, Grant S, Yu C, Mehra VL, Pensiero MN, Phogat S, DiazGranados CA, Barnett SW, Kanesa-Thasan N, Koutsoukos M, Michael NL, Robb ML, Kublin JG, Gilbert PB, Corey L, Gray GE, McElrath MJ. 2018. Subtype C ALVAC-HIV and bivalent subtype C gp120/MF59 HIV-1 vaccine in low-risk, HIV-uninfected, South African adults: a phase 1/2 trial. Lancet HIV 5:e366–e378. 10.1016/S2352-3018(18)30071-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Fisher L, Zinter M, Stanfield-Oakley S, Carpp LN, Edwards RW, Denny T, Moodie Z, Laher F, Bekker L-G, McElrath MJ, Gilbert PB, Corey L, Tomaras G, Pollara J, Ferrari G. 2019. Vaccine-induced antibodies mediate higher antibody-dependent cellular cytotoxicity after interleukin-15 pretreatment of natural killer effector cells. Front Immunol 10:2741. 10.3389/fimmu.2019.02741. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Tuyishime M, Garrido C, Jha S, Moeser M, Mielke D, LaBranche C, Montefiori D, Haynes BF, Joseph S, Margolis DM, Ferrari G. 2020. Improved killing of HIV-infected cells using three neutralizing and non-neutralizing antibodies. J Clin Invest 130:5157–5170. 10.1172/JCI135557. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.von Bredow B, Arias JF, Heyer LN, Moldt B, Le K, Robinson JE, Zolla-Pazner S, Burton DR, Evans DT. 2016. Comparison of antibody-dependent cell-mediated cytotoxicity and virus neutralization by HIV-1 Env-specific monoclonal antibodies. J Virol 90:6127–6139. 10.1128/JVI.00347-16. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Smalls-Mantey A, Doria-Rose N, Klein R, Patamawenu A, Migueles SA, Ko S-Y, Hallahan CW, Wong H, Liu B, You L, Scheid J, Kappes JC, Ochsenbauer C, Nabel GJ, Mascola JR, Connors M. 2012. Antibody-dependent cellular cytotoxicity against primary HIV-Infected CD4+ T cells is directly associated with the magnitude of surface IgG binding. J Virol 86:8672–8680. 10.1128/JVI.00287-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Borggren M, Jensen SS, Heyndrickx L, Palm AA, Gerstoft J, Kronborg G, Hønge BL, Jespersen S, da Silva ZJ, Karlsson I, Fomsgaard A. 2016. Neutralizing antibody response and antibody-dependent cellular cytotoxicity in HIV-1-infected individuals from Guinea-Bissau and Denmark. AIDS Res Hum Retroviruses 32:434–442. 10.1089/AID.2015.0118. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Richardson SI, Chung AW, Natarajan H, Mabvakure B, Mkhize NN, Garrett N, Abdool Karim S, Moore PL, Ackerman ME, Alter G, Morris L. 2018. HIV-specific Fc effector function early in infection predicts the development of broadly neutralizing antibodies. PLoS Pathog 14:e1006987. 10.1371/journal.ppat.1006987. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Alsahafi N, Bakouche N, Kazemi M, Richard J, Ding S, Bhattacharyya S, Das D, Anand SP, Prévost J, Tolbert WD, Lu H, Medjahed H, Gendron-Lepage G, Ortega Delgado GG, Kirk S, Melillo B, Mothes W, Sodroski J, Smith AB, Kaufmann DE, Wu X, Pazgier M, Rouiller I, Finzi A, Munro JB. 2019. An asymmetric opening of HIV-1 envelope mediates antibody-dependent cellular cytotoxicity. Cell Host Microbe 25:578–587.e5. 10.1016/j.chom.2019.03.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Pollara J, Jones DI, Huffman T, Edwards RW, Dennis M, Li H, Jha S, Goodman D, Kumar A, Labranche CC, Montefiori DC, Fouda GG, Hope TJ, Tomaras GD, Staats HF, Ferrari G, Permar R. 2019. Bridging vaccine-induced HIV-1 neutralizing and effector antibody responses in rabbit and rhesus macaque animal models. J Virol 93:1–22. 10.1128/JVI.02119-18. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Prévost J, Richard J, Medjahed H, Alexander A, Jones J, Kappes JC, Ochsenbauer C, Finzi A. 2018. Incomplete downregulation of CD4 expression affects HIV-1 Env conformation and antibody-dependent cellular cytotoxicity responses. J Virol 92:e00484-18. 10.1128/JVI.00484-18. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Alsahafi N, Ding S, Richard J, Markle T, Brassard N, Walker B, Lewis GK, Kaufmann DE, Brockman MA, Finzi A. 2015. Nef proteins from HIV-1 elite controllers are inefficient at preventing antibody-dependent cellular cytotoxicity. J Virol 90:2993–3002. 10.1128/JVI.02973-15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Munro JB, Gorman J, Ma X, Zhou Z, Arthos J, Burton DR, Koff WC, Courter JR, Smith AB, Kwong PD, Blanchard SC, Mothes W. 2014. Conformational dynamics of single HIV-1 envelope trimers on the surface of native virions. Science 346:759–763. 10.1126/science.1254426. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.de Taeye SW, Ozorowski G, Torrents de la Peña A, Guttman M, Julien J-P, van den Kerkhof TLGM, Burger JA, Pritchard LK, Pugach P, Yasmeen A, Crampton J, Hu J, Bontjer I, Torres JL, Arendt H, DeStefano J, Koff WC, Schuitemaker H, Eggink D, Berkhout B, Dean H, LaBranche C, Crotty S, Crispin M, Montefiori DC, Klasse PJ, Lee KK, Moore JP, Wilson IA, Ward AB, Sanders RW. 2015. Immunogenicity of stabilized HIV-1 envelope trimers with reduced exposure of non-neutralizing epitopes. Cell 163:1702–1715. 10.1016/j.cell.2015.11.056. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Bonsignori M, Pollara J, Moody MA, Alpert MD, Chen X, Hwang K-K, Gilbert PB, Huang Y, Gurley TC, Kozink DM, Marshall DJ, Whitesides JF, Tsao C-Y, Kaewkungwal J, Nitayaphan S, Pitisuttithum P, Rerks-Ngarm S, Kim JH, Michael NL, Tomaras GD, Montefiori DC, Lewis GK, DeVico A, Evans DT, Ferrari G, Liao H-X, Haynes BF. 2012. Antibody-dependent cellular cytotoxicity-mediating antibodies from an HIV-1 vaccine efficacy trial target multiple epitopes and preferentially use the VH1 gene family. J Virol 86:11521–11532. 10.1128/JVI.01023-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Tuen M, Bimela JS, Banin AN, Ding S, Harkins GW, Weiss S, Itri V, Durham AR, Porcella SF, Soni S, Mayr L, Meli J, Torimiro JN, Tongo M, Wang X, Kong X-P, Nádas A, Kaufmann DE, Brumme ZL, Nanfack AJ, Quinn TC, Zolla-Pazner S, Redd AD, Finzi A, Gorny MK, Nyambi PN, Duerr R. 2019. Immune correlates of disease progression in linked HIV-1 infection. Front Immunol 10:1062. 10.3389/fimmu.2019.01062. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Prévost J, Zoubchenok D, Richard J, Veillette M, Pacheco B, Coutu M, Brassard N, Parsons MS, Ruxrungtham K, Bunupuradah T, Tovanabutra S, Hwang K-K, Moody MA, Haynes BF, Bonsignori M, Sodroski J, Kaufmann DE, Shaw GM, Chenine AL, Finzi A. 2017. Influence of the envelope gp120 Phe 43 cavity on HIV-1 sensitivity to antibody-dependent cell-mediated cytotoxicity responses. J Virol 91:e02452-16. 10.1128/JVI.02452-16. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Hraber P, Seaman MS, Bailer RT, Mascola JR, Montefiori DC, Korber BT. 2014. Prevalence of broadly neutralizing antibody responses during chronic HIV-1 infection. AIDS 28:163–169. 10.1097/QAD.0000000000000106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Wei X, Decker JM, Wang S, Hui H, Kappes JC, Wu X, Salazar-Gonzalez JF, Salazar MG, Kilby JM, Saag MS, Komarova NL, Nowak MA, Hahn BH, Kwong PD, Shaw GM. 2003. Antibody neutralization and escape by HIV-1. Nature 422:307–312. 10.1038/nature01470. [DOI] [PubMed] [Google Scholar]
- 43.Alberti MO, Jones JJ, Miglietta R, Ding H, Bakshi RK, Edmonds TG, Kappes JC, Ochsenbauer C. 2015. Optimized replicating Renilla luciferase reporter HIV-1 utilizing novel internal ribosome entry site elements for native Nef expression and function. AIDS Res Hum Retroviruses 31:1278–1296. 10.1089/aid.2015.0074. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Gray ES, Moore PL, Choge IA, Decker JM, Bibollet-Ruche F, Li H, Leseka N, Treurnicht F, Mlisana K, Shaw GM, Karim SSA, Williamson C, Morris L, CAPRISA 002 Study Team. 2007. Neutralizing antibody responses in acute human immunodeficiency virus type 1 subtype C infection. J Virol 81:6187–6196. 10.1128/JVI.00239-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Li M, Gao F, Mascola JR, Stamatatos L, Polonis VR, Koutsoukos M, Voss G, Goepfert P, Gilbert P, Greene KM, Bilska M, Kothe DL, Salazar-Gonzalez JF, Wei X, Decker JM, Hahn BH, Montefiori DC. 2005. Human immunodeficiency virus type 1 env clones from acute and early subtype B infections for standardized assessments of vaccine-elicited neutralizing antibodies. J Virol 79:10108–10125. 10.1128/JVI.79.16.10108-10125.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Pollara J, Bonsignori M, Moody MA, Liu P, Alam SM, Hwang K-K, Gurley TC, Kozink DM, Armand LC, Marshall DJ, Whitesides JF, Kaewkungwal J, Nitayaphan S, Pitisuttithum P, Rerks-Ngarm S, Robb ML, O’Connell RJ, Kim JH, Michael NL, Montefiori DC, Tomaras GD, Liao H-X, Haynes BF, Ferrari G. 2014. HIV-1 vaccine-induced C1 and V2 Env-specific antibodies synergize for increased antiviral activities. J Virol 88:7715–7726. 10.1128/JVI.00156-14. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Data Availability Statement
Envelope sequences of all Env-IMCs are accessible through GenBank; accession numbers are provided in Table 1. Other data and reagents are available upon request.