ABSTRACT
The positive-sense, single-stranded RNA genome SARS-CoV-2 harbors functionally important cis-acting elements governing critical aspects of viral gene expression. However, insights on how these elements sense various signals from the host cell and regulate viral protein synthesis are lacking. Here, we identified two novel cis-regulatory elements in SARS-CoV-2 ORF1a and S RNAs and describe their role in translational control of SARS-CoV-2. These elements are sequence-unrelated but form conserved hairpin structures (validated by NMR) resembling gamma activated inhibitor of translation (GAIT) elements that are found in a cohort of human mRNAs directing translational suppression in myeloid cells in response to IFN-γ. Our studies show that treatment of human lung cells with receptor-binding S1 subunit, S protein pseudotyped lentivirus, and S protein-containing virus-like particles triggers a signaling pathway involving DAP-kinase1 that leads to phosphorylation and release of the ribosomal protein L13a from the large ribosomal subunit. Released L13a forms a virus activated inhibitor of translation (VAIT) complex that binds to ORF1a and S VAIT elements, causing translational silencing. Translational silencing requires extracellular S protein (and its interaction with host ACE2 receptor), but not its intracellular synthesis. RNA-protein interaction analyses and in vitro translation experiments showed that GAIT and VAIT elements do not compete with each other, highlighting differences between the two pathways. Sequence alignments of SARS-CoV-2 genomes showed a high level of conservation of VAIT elements, suggesting their functional importance. This VAIT-mediated translational control mechanism of SARS-CoV-2 may provide novel targets for small molecule intervention and/or facilitate development of more effective mRNA vaccines.
IMPORTANCE Specific RNA elements in the genomes of RNA viruses play important roles in host-virus interaction. For SARS-CoV-2, the mechanistic insights on how these RNA elements could sense the signals from the host cell are lacking. Here we report a novel relationship between the GAIT-like SARS-CoV-2 RNA element (called VAITs) and the signal generated from the host cell. We show that for SARS-CoV-2, the interaction of spike protein with ACE2 not only serves the purpose for viral entry into the host cell, but also transduces signals that culminate into the phosphorylation and the release of L13a from the large ribosomal subunit. We also show that this event leads to the translational arrest of ORF1a and S mRNAs in a manner dependent on the structure of the RNA elements. Translational control of viral mRNA by a host-cell generated signal triggered by viral protein is a new paradigm in the host-virus relationship.
KEYWORDS: RNA virus, ribosomal loading, viral RNA element, cell signaling, ribosomal proteins, translational control, virus-host interactions
INTRODUCTION
The coronavirus disease 2019 (COVID-19) pandemic that resulted from the emergence of severe acute respiratory syndrome coronavirus 2 (SARS-CoV-2) is proving to be one of the most challenging scientific and public health crises in modern times. SARS-CoV-2 infection has caused more than 4.5 million deaths worldwide and often leaves survivors with serious multisystem long-term health issues. Infection of lung cells via the ACE2 receptor leading to damage of the respiratory system represents a major aspect of the pathogenesis of SARS-CoV-2; however, it is clear that numerous other organ systems are impacted as well. The approved prophylactic or therapeutic interventions for COVID-19 are very limited and only under the category of emergency use authorization thus far.
SARS-CoV-2 is a member of the highly diverse family of Coronaviridae with an enveloped positive-sense single-stranded RNA genome of approximately 30,000 nucleotides (1). Upon infection, the ORF1a and ORF1b regions of the SARS-CoV-2 genomic RNA are translated into a continuous polypeptide using the host cell’s ribosome. This polypeptide is then cleaved into 16 nonstructural proteins, NSP1 to NSP16 (1). Subsequently, four structural proteins, spike (S), envelope (E), membrane (M), and nucleocapsid (N), as well as several other accessory proteins, are produced from a nested set of subgenomic mRNAs transcribed from the 3′ one-third of the genome (1). Importantly, SARS-CoV-2 has successfully adopted a multipronged strategy to repress cellular translation, which results in robust downregulation of retinoic acid-inducible gene 1 (RIG-1), a cytosolic pattern recognition receptor that plays a key role in type-1 interferon (IFN1) response (2–5). This severely compromises IFN-dependent immune responses that would be the host’s first line of defense against the virus. Mechanisms of translation suppression utilized by SARS-CoV-2 include (i) binding of the viral NSP1 protein to the mRNA entry channel of the host 40S ribosomal subunit (2, 3); (ii) impairment of the translation of cellular transcripts encoding cytokines and other factors involved in innate immune responses (4, 5); and (iii) accelerated degradation of cytosolic cellular mRNAs, preventing newly transcribed host mRNAs from accessing ribosomes (4). Apart from direct inhibition of translation of host proteins as mentioned above, NSP8 and NSP9 of SARS-CoV-2 disrupt protein trafficking through targeting the 7SL RNA component of the signal recognition particle (SRP) (2).
It must be noted that the spike protein (S) of SARS-CoV-2 is well known to play a critical role in the process of cellular entry of the virus (6). Interaction between the receptor-binding domain (RBD) of the S1 subunit of the S protein and ACE2 receptor creates a conformational lock and increases the susceptibility to multiple cellular proteases pivotal for entry of the virus within the host cell (7–9). Work from SARS-CoV showed that S-ACE2 interaction is not only crucial for viral entry, but also transduces signals from cell surface to the nucleus through the CK2-Ras-ERK-AP-1 pathway, thus regulating the expression of host chemokine CCL2 (10).
Previously, we identified and characterized a novel translational silencing-dependent mechanism of controlling inflammation in myeloid cells. We found that IFN-γ-induced signaling in macrophages and monocytes leads to phosphorylation of ribosomal protein L13a and its subsequent release from the 60S ribosomal subunit (11). The released phospho-L13a incorporates into a large multiprotein complex that binds to structurally conserved “gamma-activated inhibitor of translation” (GAIT) elements found in the 3′ untranslated regions (3’UTRs) of certain mRNAs and silence their translation (12, 13). Phospho-L13a as a part of GAIT complex directly interacts with eIF4G subunit of the eIF4F translation initiation complex, thereby intercepts the recruitment of 43S pre-initiation complex by blocking the interaction between eIF4G and eIF3 (14). Notably, GAIT element-containing mRNAs typically encode proinflammatory factors such as chemokines and chemokine receptors. Thus, GAIT-mediated silencing of translation provides a feedback mechanism that effectively resolves inflammatory response (15, 16). The physiological importance of this translational silencing mechanism as an endogenous defense mechanism against uncontrolled inflammation was illustrated by our findings that induced endotoxemia (17), colitis (18, 19), and high-fat diet-induced atherosclerosis were all more severe in myeloid-specific L13a-knockout (KO) mice than in controls (20, 21).
A role for a similar translational regulatory mechanism(s) in cellular responses to coronaviruses and other respiratory pathogens has started to emerge from work done by others (22) and us (23). The Almazan laboratory identified a GAIT element in the 3′ end of the positive-strand RNA genome of transmissible gastroenteritis coronavirus (TGEV) that modulates the innate immune response to the virus (22). Similarly, our work has identified an L13a-dependent translation inhibitory element in the M gene of human respiratory syncytial virus (RSV) with significant structural (but not sequence) homology to the GAIT element and showed that it is responsible for inhibition of viral replication in human lung cells upon virus entry (23). Moreover, cis-acting RNA elements located in the 5′ and 3′ termini and in the open reading frames of the 3′-proximal one-third of the coronavirus genomes have been implicated in the viral genome replication and packaging of the virus (24). The functional relevance of cis-acting elements is supported by bioinformatics analyses showing a significant level of conservation of RNA secondary structures among different coronavirus genera (24). In addition, in silico analysis of SARS-CoV-2 genomes using the structRNAfinder tool predicted the existence of several structured cis-acting RNA elements that could play roles in the discontinuous transcription of subgenomic mRNAs, translation of viral proteins, and/or virus replication (25, 26). Despite these indications of a possible GAIT-like system of translational control in viruses a complete picture of the mechanism(s) by which RNA elements and associated proteins may regulate the final stage of viral gene expression - protein synthesis from genomic RNA and subgenomic mRNAs remains lacking. An interesting consideration in this context is the potential role of virus-induced signals in activating translational silencing similar to the role of IFN-γ induced signals in the GAIT system.
In the current study, we aimed to determine whether SARS-CoV-2 is subject to translation control mediated by the structurally conserved “GAIT-like” RNA elements within its genomic RNA or subgenomic mRNAs. We identified two novel RNA elements in the ORF1a RNA and S mRNA of SARS-CoV-2 that were predicted and subsequently shown by NMR to form stem-loop secondary structures similar to a canonical GAIT element. We found that treatment of human lung cells with the SARS-CoV2 S protein (in the form of recombinant S1 subunit harboring the RBD domain, S protein pseudotyped lentivirus, or S protein-containing virus-like particles [VLP]) activates a signaling mechanism that leads to specific translational silencing of reporter mRNAs containing these elements. This led us to name these elements virus activated inhibitor of translation (VAIT) elements. We also demonstrated VAIT element-dependent translation inhibition through analysis of polysome loading of the full length SARS-CoV-2 S mRNA (Wuhan MN988668) in S protein-treated human lung cells, a system more closely resembling viral infection than in in vitro translation experiments. Our results indicate that S protein-induced VAIT element-mediated translational silencing involves S protein-ACE2 receptor interaction on the cell surface (a condition that mimics viral entry), activation of DAP kinase1, phosphorylation and ribosomal release of L13a and formation of L13a-dependent RNA-binding complexes on VAIT elements. Despite the structural homology of VAIT and GAIT elements and the critical involvement of L13a in both translational control mechanisms, the two systems also have significant distinctions. We found that these two elements do not compete with each other and the activating signal and cell type is specific to each. Interestingly, the ORF1a and S VAIT elements are highly conserved among all SARS-CoV-2 genomes published to date, suggesting their function may be important for virus fitness.
Together, these results establish a new paradigm of potential self-regulation of SARS-CoV-2 virus gene expression in which engagement of the viral spike protein with its receptor on host cells triggers a novel signaling mechanism leading to L13a-dependent translation suppression specifically for RNAs containing structurally conserved VAIT elements such as SARS-CoV-2 ORF1a RNA and S mRNA. This regulatory mechanism could have important implications for understanding the consequences of SARS-CoV-2 infection and designing new vaccines and treatments.
RESULTS
Sequence-unrelated RNA elements of SARS-CoV-2 S mRNA and ORF1a RNA are predicted to fold into stem-loop structures resembling canonical Cp GAIT elements.
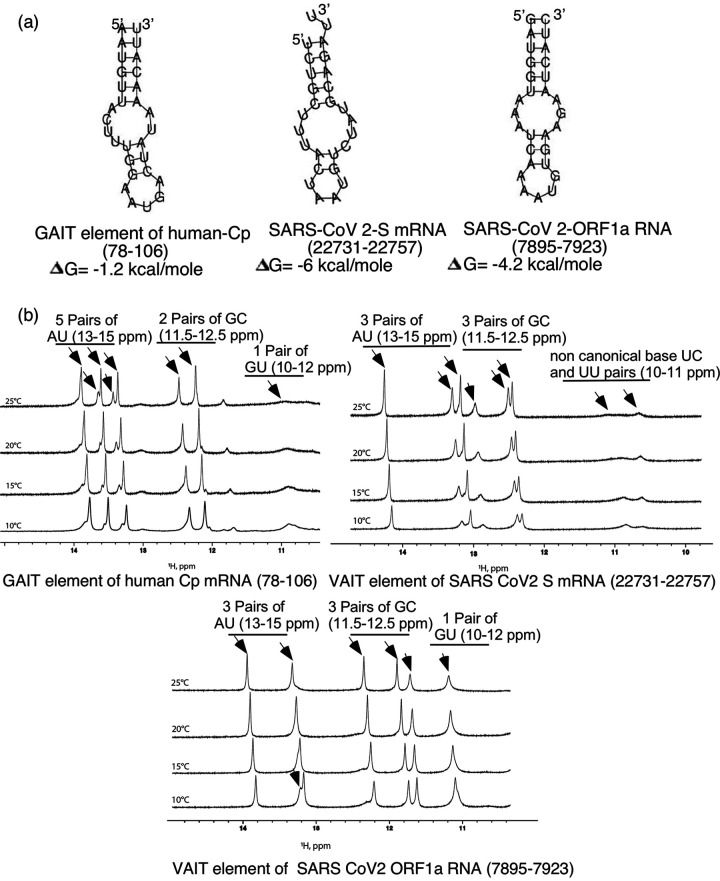
Based on our previous work on translational silencing in myeloid cells mediated by structurally conserved GAIT RNA elements (15, 16) and emerging evidence of a similar translation control mechanism in some viruses (22, 23) we hypothesized that such sequence-unrelated but structurally conserved GAIT like RNA elements (13) might be present in the positive-strand RNA genome of SARS-CoV-2. Using the canonical GAIT element in the human ceruloplasmin (Cp) mRNA as a reference, we employed “Foldalign” (http://foldalign.ku.dk) (27–29) and “RNAstructure version 6.3” (https://rna.urmc.rochester.edu/RNAstructureWeb/) programs to search the sequence of SARS-CoV-2 (Wuhan MN988668.1) genome for elements predicted to form GAIT-like secondary structures. Using these programs to perform pairwise structural RNA alignments and mutual motif searches, we identified several stem-loop structures in SARS-CoV-2 ORF1a RNA and S mRNA sequences homologous to the structure formed by the Cp GAIT element. In the subsequent studies reported here, two of these predicted elements, one in the S mRNA (genomic nucleotide [n.t.] position 22731 to 22757) and another in ORF1a of the SARS-CoV-2 genomic RNA (genomic n.t. position 7895 to 7923) (Fig. 1a) were structurally and functionally validated using 1D-1H-NMR and translation inhibition assays, respectively (see below).
FIG 1.
Identification of SARS-CoV-2 RNA elements with structural similarity to the canonical Cp GAIT element. (a) Bioinformatics-predicted secondary structures of the Cp GAIT element and SARS-CoV-2 S and ORF1a VAIT elements. VAIT sequences were identified using the Foldalign program and secondary structure and free energy predictions were made using RNA Folding server RNAstructure version 6.3 (see Materials and Methods). For the Cp GAIT element, the numbers in parentheses indicate nucleotide position in the 3′ UTR (counting from the first base after the stop codon); for the SARS-CoV-2 VAIT elements, the nucleotide position in the virus genome is given. (b) 1D-1H-NMR spectroscopic analysis of Cp GAIT and SARS-CoV-2 S and ORF1a VAIT elements. The imino region of 1D-1H-NMR spectra obtained for chemically synthesized and HPLC-purified RNA elements at four different temperatures are shown. Sharp resonances between 10 and 14.5 ppm (typical for canonical Watson–Crick or G-U wobble base pairing) indicate that all three RNA elements adopt structures consistent with in silico predictions shown in (a).
1D-1H-NMR analyses confirm that the in silico-predicted “GAIT-like” structures of the SARS-CoV-2 ORF1a and S RNA elements are formed in solution.
To test our in silico folding predictions, we analyzed each identified “GAIT-like” SARS-CoV-2 RNA element using 1D-1H-NMR spectroscopy in which imino protons show diagnostic chemical shifts for canonical and noncanonical base pairs. The obtained spectra showed that all tested RNA elements form stable helical regions over a broad temperature range (Fig. 1b). Imino protons for canonical A-U base pairs resonate in the region between 13 ppm to 15 ppm, whereas those for canonical G-C, and G-U base pairs typically resonate in the range of 12–13.5 ppm and 10–12 ppm, respectively (30). Noncanonical base pairs, which are often, found in loop regions like those predicted for GAIT-like elements, typically have imino chemical shifts that resonate between 9.5 ppm and 14 ppm.
The predicted human Cp GAIT element consists of six AU and two GC Watson-Crick base pairs, and one GU wobble pair. With the exception of terminal- or loop-proximal base pairs that are prone to rapid solvent exchange, we expected to observe at least eight imino signals consistent with the predicted structure in the 1D 1H spectrum of the Cp GAIT construct used for NMR. Evidence for five sharp and well-resolved signals were seen at each temperature in which NMR spectra were collected (Fig. 1b). The chemical shifts (13 to 15 ppm) of three of the imino signals occurred within the range expected for AU base pairs, and the remaining two imino signals exhibited chemical shifts (11.5 to 12.5 ppm) consistent with GC base pairs. As the temperature was elevated from 10° to 25°C, two additional sharp imino signals (13 to 14 ppm) appeared more resolved. Conversely, two broad imino signals (10 to 11 ppm) became more solvent exchanged at from 10° to 25°C. The chemical shifts and solvent exchange behavior of these NMR signals are consistent with that expected for a GU wobble pair. Taken together, the 1D-1H-NMR spectra of Cp GAIT demonstrate that it is stably folded with base-pair composition in agreement with its predicted secondary structure.
The base pair composition of the SARS-CoV-2 S mRNA GAIT-like element (n.t. 22731 to 22757) is predicted to consist of three AU and four GC Watson-Crick base pairs. Excluding terminal- or loop-proximal pairs, we expected to observe at least five sharp imino signals in the 1D 1H spectrum of the SARS-CoV-2 S mRNA construct used for NMR. Indeed, three imino signals within the chemical shift range (13 to 14.5 ppm) expected for AU base pairs, and three other imino signals (12 to 13 ppm), consistent with the predicted GC composition, are seen over the entire temperature range (Fig. 1b). However, it is not possible to unambiguously assign the sharp signal at ∼13 ppm to a GC or AU pair using chemical shifts alone, but we note that both base pair types are proximal to the predicted internal loop (Fig. 1b). Lastly, two broad NMR signals were observed between 10 and 11 ppm at low temperature (10° C) but become exchanged broadened as the temperature is elevated 25° C. These additional NMR signals likely correspond to imino protons engaged in noncanonical base pairs within the large internal loop.
Like the other elements, the composition of the predicted SARS-CoV-2 ORF1a RNA (n.t. 7895 to 7923) consists of three GC base pairs, five AU base pairs, and one GU wobble pair. Excluding terminal- or loop-proximal pairs, we expected to observe at least seven imino signals in the 1D 1H spectrum of the SARS-CoV-2 ORF1a construct used for NMR. At 10° C, the 1D 1H spectrum of SARS-CoV-2 ORF1a RNA element showed a well-resolved imino signal at ∼11 ppm with an obvious shoulder on its upfield side (Fig. 1b). As the temperature is elevated, the two peaks coalesce into one signal. The chemical shifts and temperature profile of these imino signals are consistent with that expected for a relatively stable GU base pair. At each temperature where data were collected, three sharp and well-resolved imino signals are observed at ∼11.5 to 12 ppm in the range consistent with GC base pairs. By comparison, two well-resolved and sharp signals are seen between 13 and 14 ppm at 25° C, and as the temperature is lowered to 10° C, a shoulder appears on the downfield side of the imino signal at ∼13 ppm (Fig. 1b).
Taken together, the NMR chemical shifts and temperature dependent properties of the imino protons of each RNA element evaluated here are consistent with their predicted secondary structure. However, additional NMR experiments, which are under way, are needed to unambiguously assign the chemical shifts to specific base pair types and to determine the overall tertiary folds of these RNAs. Evidence for stable tertiary contacts within the various loops can be gleaned from these preliminary studies.
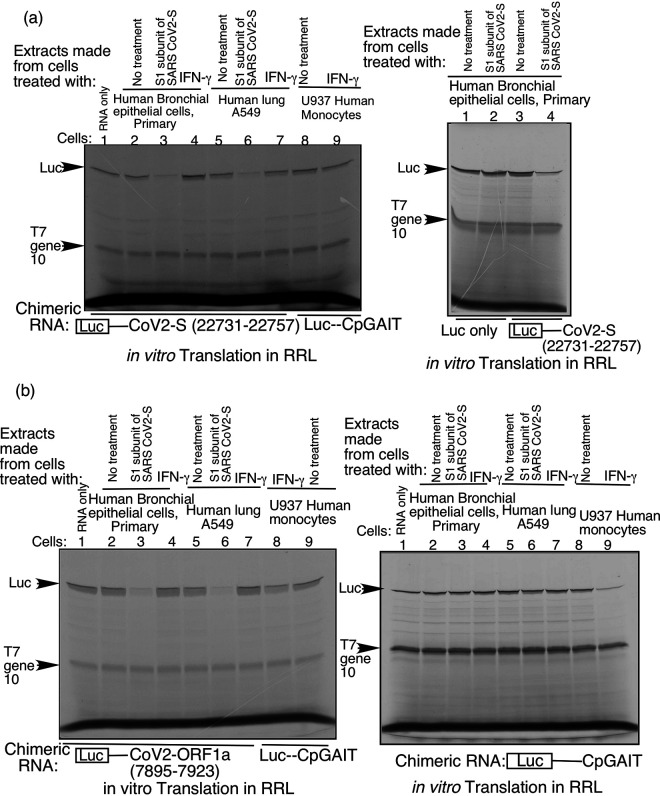
Two RNA elements in SARS-CoV-2 S mRNA and ORF1a RNA support translational silencing of a heterologous reporter in response to treatment of human lung cells with SARS-CoV-2 S protein.
To test the potential translation regulatory activity of predicted GAIT-like SARS-CoV-2 RNA elements, we created reporter constructs with four such elements [S (n.t. 22019 to 22047), S (n.t. 22384 to 22408), S (n.t. 22731 to 22757), and ORF1a (n.t. 7895 to 7923)] inserted individually into the 3’UTR of the luciferase mRNA. These chimeric mRNAs were translated in cell-free translation system with rabbit reticulocyte lysate (RRL) as described previously (16) with or without addition of cell extracts prepared from human primary bronchial/tracheal epithelial cells (HBTEC) or lung carcinoma A549 cells treated with SARS-CoV-2 S protein. For these experiments, we used recombinant purified protein corresponding to the S1 subunit of the S protein, which contains the ACE2 RBD. Addition of extracts from S protein-treated lung cells to the system resulted in specific inhibition of translation of the chimeric luciferase reporter mRNAs containing two of the four tested GAIT-like elements, those located in the S mRNA at nucleotide position 22731 to 22757 of the SARS-CoV-2 genome (Fig. 2a, left panel) and in the ORF1a RNA at nucleotide position 7895 to 7923 (Fig. 2b, left panel). Translation from reporters containing the other two tested RNA elements (S n.t. 22019 to 22047 and S n.t. 22384-22408) was not changed by addition of cell extracts made from S protein-treated cells (data not shown) and these elements were not investigated further in the current work. As expected, translation from the internal control mRNA of T7 gene 10 present in all tested reporters (Fig. 2) and from reporter luciferase lacking an added RNA element in 3’UTR (lanes 1 to 2 of Fig. 2a right panel) was not affected by addition of extract from S protein-treated lung cells. Our finding that specific SARS-CoV-2 sequences with GAIT-like secondary structure mediate S protein-induced translation suppression led us to name them VAIT elements.
FIG 2.
In vitro translation of reporter mRNAs containing S and ORF1a VAIT elements is suppressed by extracts from SARS-CoV-2 spike protein-treated lung cells. (a) Chimeric luciferase reporter mRNAs harboring either the S VAIT element or the Cp GAIT element in their 3′UTRS were translated in vitro in rabbit reticulocyte lysates in the presence of [35S]-Methionine. Cell extracts prepared from SARS-CoV-2 spike protein (S1 subunit) or IFN-γ -treated (for 24 h) human bronchial epithelial cells (HBTEC), A549 cells or U937 cells were added to the translation reactions. [35S]-Methionine-labeled translation products from the luciferase reporter and phage T7 gene 10 cRNA (included in the reaction as an internal control, no GAIT or VAIT element present) were resolved by SDS-PAGE (indicated by arrows). The experiment shown in the right panel includes a “luciferase-only” reporter (no GAIT or VAIT element, first two lanes) to illustrate that repression of luciferase translation by extracts of S protein-treated lung cells requires the VAIT element. (b) Translational control assays were performed as described in (a) with luciferase reporters containing either the SARS-CoV-2 ORF1a VAIT element (left panel) or the Cp GAIT element (right panel) in their 3’UTRs.
Functionality of the S and ORF1a VAIT elements in directing translational silencing in S protein-treated lung epithelial cells is particularly interesting since they were predicted based on structural homology with GAIT elements (Fig. 1) which our previous work showed mediate translation control specifically in cells of myeloid origin (31). To clarify how distinct the two mechanisms are, we tested whether IFN-γ, a classical activator of GAIT-mediated translation inhibition (32), can also activate translational silencing of mRNAs containing SARS-CoV-2 VAIT elements. As shown in Fig. 2a and b (left panels, lanes 4 and 7), in vitro translation of reporter mRNAs containing SARS-CoV-2 VAIT elements was not inhibited by addition of extracts from U937 cells treated with IFN-γ even though the same extracts effectively reduced translation of a reporter containing the canonical Cp GAIT element (lanes 8 and 9) and even though the translation of the VAIT element containing reporters was blocked by extracts from the S protein-treated lung cells (lanes 3 and 6). Next, we performed the opposite experiment, testing whether SARS-CoV-2 S protein can activate GAIT element-mediated translational silencing. As shown in Fig. 2b (right panel, lane 3 and 6), extracts from S protein-treated lung cells failed to silence translation of a chimeric luciferase reporter containing the canonical Cp GAIT element (13). Similarly, IFN-γ did not activate GAIT element-mediated translational silencing in lung cells (lanes 4 and 7), although as expected, it did trigger the GAIT pathway in human monocytes (lane 9). Together, these results suggest that, while fundamentally similar, VAIT and GAIT element-mediated translational silencing likely have substantial mechanistic differences.
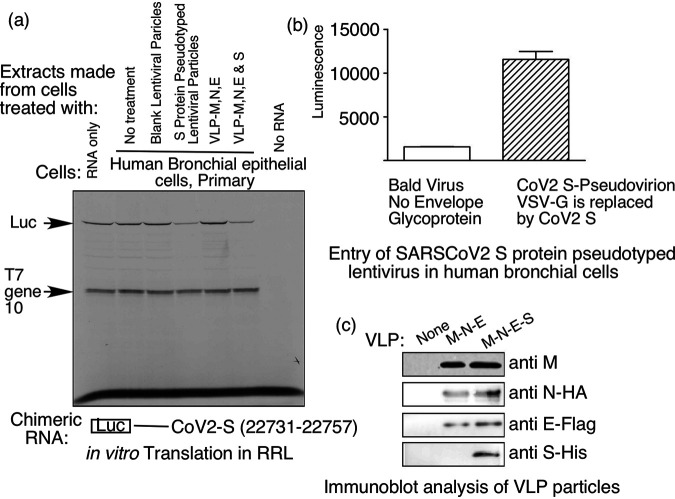
Treatment of human lung cells with SARS-CoV-2 pseudovirus or virus like particles stimulates VAIT element-mediated translational silencing.
To demonstrate the virological relevance of our results obtained with recombinant S protein (Fig. 2), we next tested whether VAIT element-mediated translational silencing could also be induced by treatment of lung cells with more natural forms of SARS-CoV-2 S protein, namely, S protein-pseudotyped lentivirus and S protein-containing VLPs. Both of these tools recapitulate critical mechanistic features of the native virus’s interaction with its cellular receptor. However, since they lack genetic material and are therefore, replication-incompetent and non-infectious, they can be handled safely in BSL2 facilities (33–35). For lentiviral treatment of cells, we used S protein-pseudotyped lentivirus expressing luciferase purchased from Genecopoeia, (Rockville, MD) and as control, “bald” lentivirus expressing luciferase without S protein purchased from BPS Bioscence (San Diego, CA). VLPs were produced in our BSL2 facility from A549 cells transfected with expression constructs for the four structural proteins M, N, E, and S of SARS-CoV-2. VLPs lacking S protein were used as controls. As shown in Figure 3a, treatment of HBTEC with S pseudotyped lentivirus or S protein-containing VLPs resulted in effective translational silencing of a luciferase RNA reporter with the S VAIT element (n.t. 22731 to 22757) in its 3’UTR without any effect on translation of the internal control T7 gene10 RNA. Control reagents (bald lentivirus and VLPs lacking S protein) did not induce translational silencing. Transduction of the S protein-pseudotyped recombinant lentivirus (but not the S protein-negative bald lentivirus) in lung cells was confirmed by measuring the luciferase expression (Fig. 3b) and presence of viral M, N, E, and S proteins in VLPs was evaluated by immunobloting (Fig. 3c).
FIG 3.
SARS-CoV-2 spike protein pseudotyped lentivirus or virus like particles (VLPs) containing S protein can induce VAIT element-mediated translational control in human bronchial epithelial cells. (a) Translational control assay performed as in Fig. 2 with a luciferase reporter mRNA containing the SARS-CoV-2 S VAIT element in the 3’UTR. Cell extracts added to in vitro translation reactions were prepared from HBTEC cells left untreated or treated for 72 h with bald (“blank”) lentivirus, SARS-CoV-2 spike protein pseudotyped lentivirus, or VLPs (containing structural proteins M, N, and E only or M, N, E, and S). (b) Transduction of SARS-CoV-2 spike protein pseudotyped lentivirus in human bronchial epithelial cells was confirmed by luciferase expression. (c) Presence of SARS-CoV-2 structural proteins in VLPs purified from the culture medium of A549 cells 48 h after transfection with plasmids directing expression of SARS-CoV-2 M, N, and E, or M, N, E, and S proteins. Viral structural proteins were detected in VLPs by Western blotting using antibodies against either the native protein (M) or tags (HA, FLAG, and His) attached to the viral N, E, and S proteins.
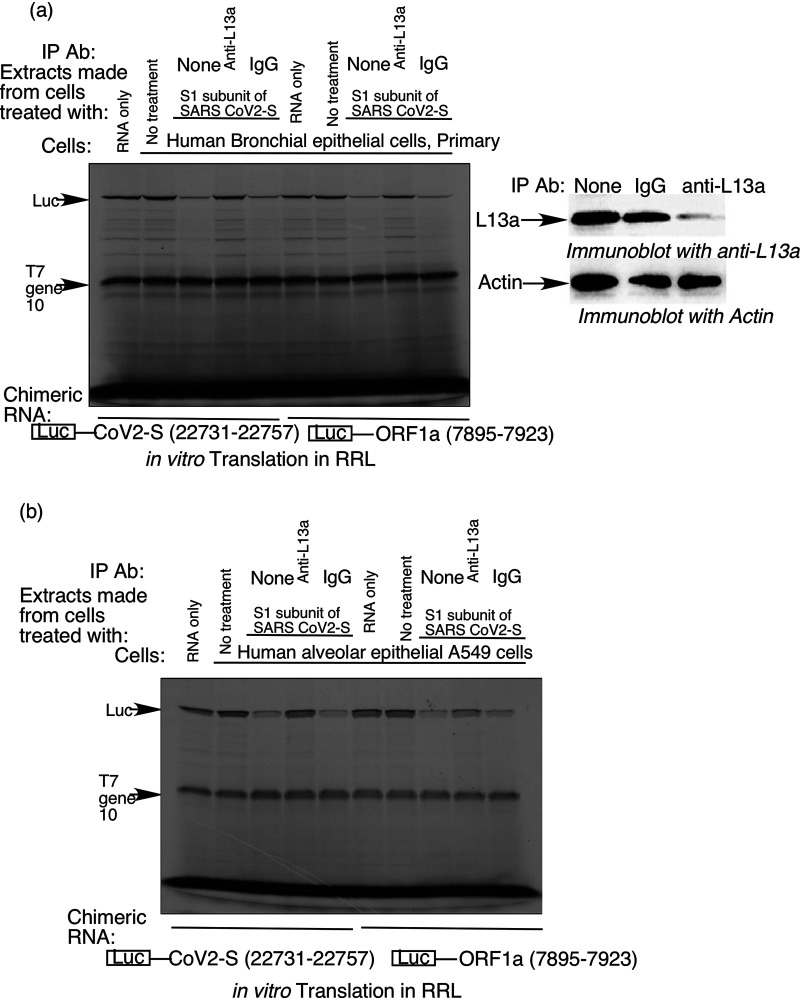
S protein-induced VAIT element-mediated translational silencing requires ribosomal protein L13a.
Although the specificity of VAIT and GAIT element-mediated translational silencing with respect to cell type and inducing signal (see Fig. 2) suggest differences in their underlying mechanisms, we hypothesized that they might share a requirement for ribosomal protein L13a, a critical component of the GAIT pathway (11). Due to the structural homology between VAIT and GAIT elements (Fig. 1), we felt it was reasonable to test this possibility. To accomplish this, we used anti-L13a antibody/protein A agarose beads to immunodeplete L13a from extracts prepared from HBTEC and A549 cells treated with recombinant S protein. Extracts with and without L13a immunodepletion were then tested for their effect on in vitro translation of chimeric luciferase RNA reporters containing either SARS-CoV2 S (n.t. 22731 to 22757) or ORF1a (n.t. 7895 to 7923) VAIT elements. We found that immunodepletion of L13a (confirmed by immunobloting, see Fig. 4a right panel) specifically abrogated translational silencing of the chimeric RNAs while “mock” immunodepletion with control IgG antibody had no effect (Fig. 4). Similar results were obtained for both HBTEC (Fig. 4a) and A549 cells (Fig. 4b) and for both the S and ORF1a VAIT elements. Thus, as in the GAIT system, L13a plays an essential role in virus-activated VAIT element-mediated translational silencing.
FIG 4.
Ribosomal protein L13a is required for SARS-CoV-2 S protein-induced suppression of translation of VAIT element-containing mRNAs. Extracts were prepared from human bronchial epithelial cells (HBTEC) (a) and human alveolar epithelial cells (A549) (b) left untreated or treated with recombinant spike protein (S1 subunit). The immunodepletion of L13a was confirmed by immunoblot (a, right panel). Extracts were added to in vitro translation reactions performed as described for Fig. 2 using the same luciferase reporter cRNAs harboring S or ORF1a VAIT elements (as shown below the panels). To test for involvement of L13a, L13a protein was immunodepleted from cell extracts by preincubation with anti-L13a antibody. IgG antibody was used as a negative control for immunodepletion.
The L13a-dependent translation-inhibitory RNA-protein complex that forms on SARS-CoV-2 S and ORF1a VAIT elements is different from the GAIT protein complex.
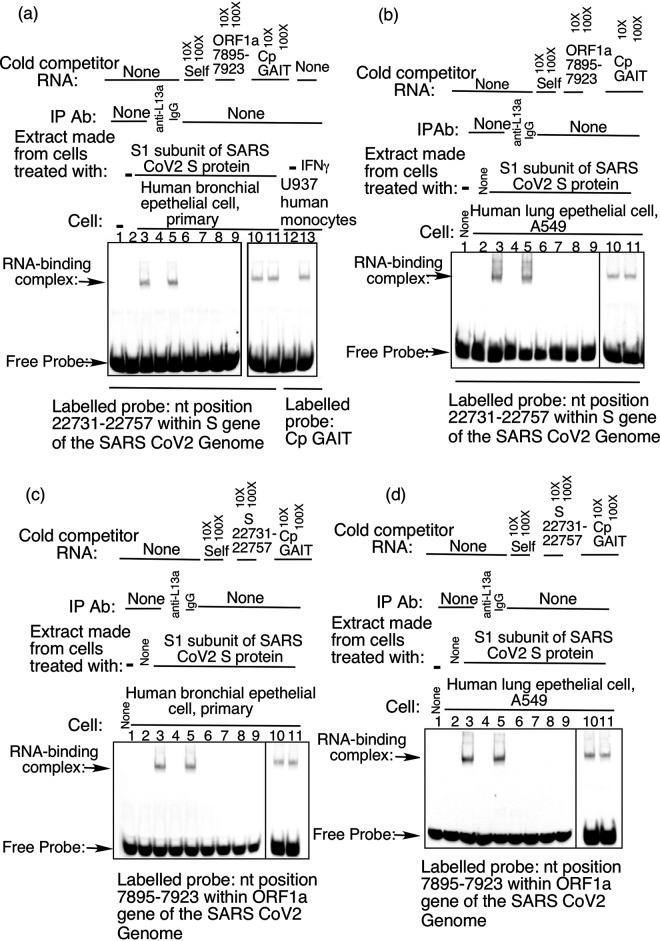
To gain further insight into the mechanism of SARS-CoV-2 S protein-induced translation control and how it may differ from the GAIT system we performed RNA electrophoretic mobility shift assay (RNA-EMSA) using RNA oligonucleotide probes corresponding to the SARS-CoV-2 S and ORF1a VAIT elements. As a positive control for the GAIT complex, we relied on our previous findings (13, 31) showing formation of a RNP complex on GAIT element probes upon incubation with extracts made from U937 monocytic cells treated with IFN-γ for 16 h, which were reproduced here (Fig. 5a, left panel, right two lanes). For VAIT element probes, we used extracts made from S protein treated HBTEC and A549 cells as a source of RNA-binding proteins. We observed formation of specific RNA-protein complexes on both S (Fig. 5a and b) and ORF1a (Fig. 5c and d) VAIT probes upon incubation with these extracts, but not with extracts from untreated lung cells. Immunodepletion of L13a from S protein-treated lung cell extracts using anti-L13a antibody abrogated RNP formation (lane 4 in Fig. 5a to c and d) while immunodepletion with control IgG antibody (Cell Signaling Technology #2729) had not effect (lane 5 in Fig. 5a to c and d). Therefore, formation of the RNA-protein complex on VAIT elements in response to S protein treatment requires L13a, similar to the GAIT system.
FIG 5.
Formation of L13a-dependent, SARS-CoV-2 VAIT element-specific RNA-protein complexes in spike protein-treated human bronchial epithelial cells (HBTEC) and A549 cells. Complexes were detected by RNA-EMSA in which biotin-labeled VAIT or GAIT element probes were incubated with extracts prepared from cells treated with spike protein or IFN-γ and then subjected to native gel electrophoresis. (a) RNA-EMSA analysis of the S VAIT element with cell extracts from S protein-treated HBTEC and of the Cp GAIT element with extracts from IFN-γ-treated U937 cells as indicated below (RNA element/probe) and above (cell extract) each lane. (b) RNA-EMSA analysis of the S VAIT element with extracts prepared from S protein-treated A549 cells. (c, d) RNA-EMSA analysis of the ORF1a VAIT element using extracts prepared from S protein-treated HBTEC (c) and A549 cells (d). L13a dependency was shown by immunodepleting L13a protein from cell extracts with anti-L13a antibody as in specificity of the complexes formed on S (a, b) and ORF1a (c, d) VAIT elements was determined by competition EMSA using 10- and 100-fold molar excess of unlabeled (“cold”) RNA oligos corresponding to the S VAIT, ORF1a VAIT, and Cp GAIT elements.
To test for differences between the RNP complexes that form on GAIT and VAIT elements in response to their respective inducing signals we performed RNA-EMSA competition experiments in which an excess of unlabeled “competitor” probe was included in binding reactions along the labeled probe of interest. Complex formation on S (n.t. 22731 to 22757) and ORF1a (n.t. 7895 to 7923) VAIT element probes was effectively competed by just a 10-fold excess of unlabeled probe consisting of the same (“self”) sequence or that of the other VAIT element (Fig. 5). In contrast, unlabeled GAIT element probe failed to compete against formation of RNP complexes on both VAIT elements even at 100X-fold excess (Fig. 5). Together, these results demonstrate that S protein-activated translational silencing of VAIT element-containing RNAs, while sharing the requirement for L13a is in fact mechanistically distinct from IFN-γ- activated GAIT element-mediated silencing. Details underlying this distinction, including whether RNA-protein complexes formed on VAIT elements contain any GAIT complex components other than L13a remains to be determined.
S protein-induced VAIT element-mediated translation control in lung cells is ACE2-dependent and requires extracellular S protein but not intracellular synthesis.
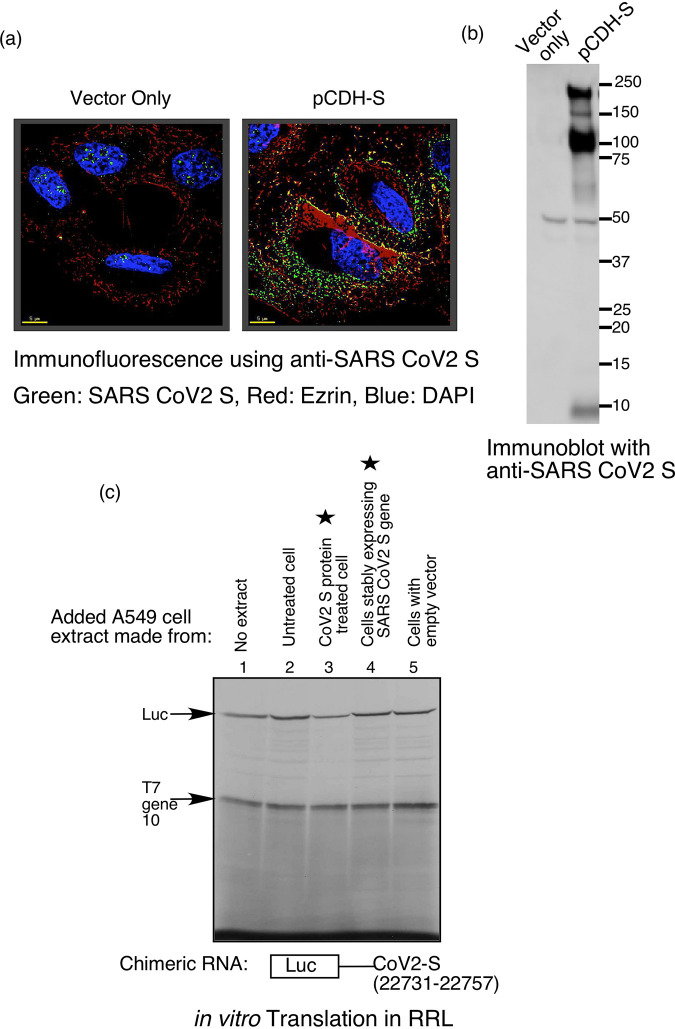
Productive infection of host cells by SARS-CoV-2 requires interaction of S protein of the invading virion with the cell surface receptor ACE2 followed by intracellular synthesis of S protein from the viral subgenomic mRNA (1, 9). This led us to test whether intracellular synthesis of S protein could induce VAIT element-mediated translation control similar to exposure of cells to extracellular S protein as described above. As a model of intracellular synthesis, we generated a stably transfected A549 cell line constitutively expressing humanized S protein based on data from an earlier study (9). Expression of S protein in these cells was confirmed by immunofluorescence (Fig. 6a, left two panels) and immunoblot (Fig. 6a, right panel) with anti-SARS-CoV-2 S antibodies (antibodies-online.com #ABIN1030641); however, extracts prepared from them did not inhibit in vitro translation of the chimeric luciferase reporter RNA containing the SARS-CoV-2 S VAIT element (Fig. 6c, lane 4). In contrast, as shown in our earlier experiments (Fig. 2 to 4), extracts prepared from A549 cells treated with extracellular S protein (recombinant S1 subunit) effectively suppressed translation of the VAIT element-containing construct (Fig. 6c, lane 3). Control extracts prepared from untreated cells or from cells stably transfected with the empty vector did not inhibit translation (Fig. 6c lane 2 and 5), thus ruling out a nonspecific effect.
FIG 6.
While exogenous addition of S protein induces VAIT element-mediated translation suppression, intracellular S protein production does not. (a) Expression of SARS-CoV-2 S protein in A549 cells stably transfected with either an empty vector (left panel) or a plasmid directing expression of S protein (pCDH-S, right panel) was detected by immunofluorescence. Cells were co-stained with antibodies against SARS-CoV-2 S protein (green) (antibodies-online.com #ABIN1030641), the membrane protein ezrin, (red) (Thermofisher #MA5-13862), and DAPI (blue; to visualize DNA). (b) Western blot analysis of extracts made from the same cells as in (a) using an antibody specific for SARS-CoV-2 S protein (antibodies-online.com #ABIN1030641). (c) Translational control assay performed as in Fig. 2 using the luciferase reporter mRNA harboring the S VAIT element and cell extracts made from A549 cells left untreated, treated exogenously treated with recombinant S protein, or stably transfected with pCDH-S (directing intracellular synthesis of S protein) or empty vector (as a negative control). For comparison, two lanes corresponding to the exogenous treatment and intracellular production (by stable expression) are marked with a star.
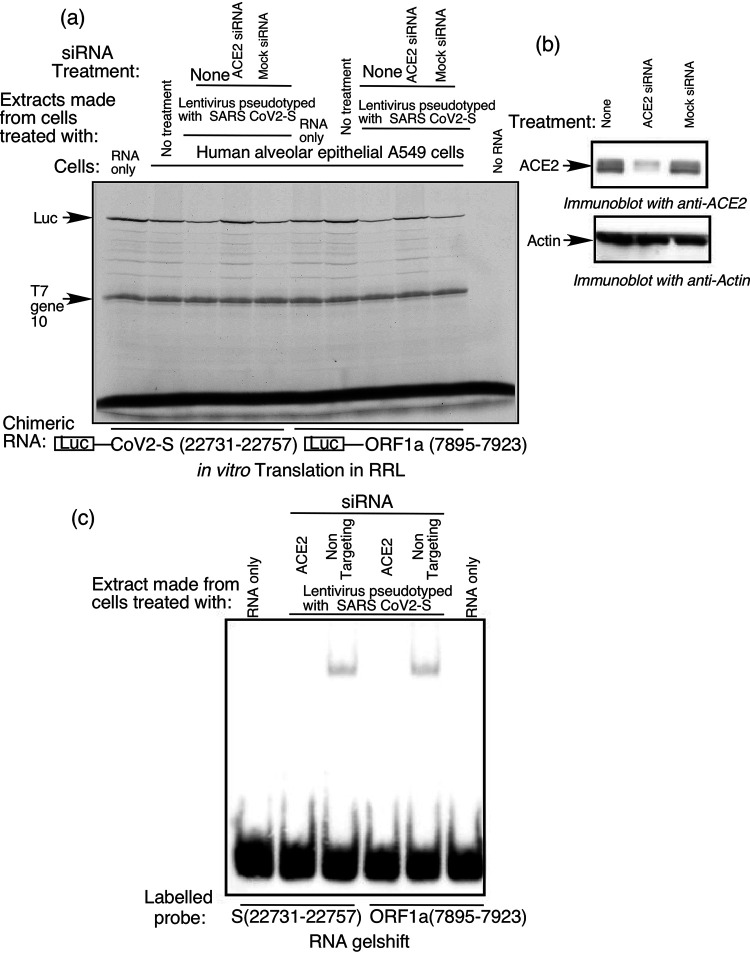
Our finding that VAIT-mediated translation control in lung cells requires extracellular S protein suggests that interaction of S protein with ACE2 on the host cell surface provides the initiating signal of the pathway. To test this possibility we used ACE2-specific siRNA to deplete ACE2 from A549 cells and nontargeting siRNA to generate mock-treated control cells. Effective depletion of ACE2 specifically in ACE2 siRNA-treated cells was confirmed by immunoblot analysis with an anti-ACE2 antibody (Fig. 7a, right panel). We then exposed ACE2-depleted cells and mock-treated cells to S protein-pseudotyped lentivirus and prepared extracts to test for translational silencing activity. As shown in Fig. 7a (left panel), in vitro translation of luciferase reporter RNAs containing either the S or ORF1a VAIT element was suppressed by extracts from cells treated with nontargeting mock siRNA, but not by those from cells depleted of ACE2 using ACE2-specific siRNA. In addition, because mRNA-specific translation control is generally driven by formation of RNA-protein complexes bound to cis-acting RNA elements in target mRNAs (36, 37), we further tested the impact of ACE2 depletion on VAIT element-mediated translation silencing using RNA-EMSA. This method showed formation of RNA-protein complexes on S and ORF1a VAIT element probes upon incubation with extract from cells treated with nontargeting mock siRNA followed by S protein-pseudotyped lentivirus, but not with extract from ACE2-depleted cells treated with the same lentivirus (Fig. 7b). Thus, absence of ACE2 abrogated RNA-protein complex formation on VAIT elements as well as inhibition of translation of reporter RNAs containing VAIT elements. Together, these results suggest that interaction of SARS-CoV-2 S protein with ACE2 on the extracellular side of the host cell membrane is required for initiating the signal(s) that leads to formation of translation-inhibitory complexes on VAIT elements, and ultimately, reduced production of proteins encoded by viral RNAs containing VAIT elements.
FIG 7.
The ACE2 receptor is required for SARS-CoV-2 S protein-induced translation control in lung cells. (a) RNAi-mediated depletion of ACE2 protein from A549 cells abrogates VAIT element-mediated translational control. In vitro translation reactions were performed as described for Fig. 2 with luciferase reporter cRNAs harboring S or ORF1a VAIT elements as indicated below lanes. Cell extracts added to in vitro translation reactions were prepared from A549 cells that were transfected with a mock (nontargeting) siRNA pool or a specific ACE2 siRNA pool and then transduced with SARS-CoV-2 spike protein pseudotyped lentivirus as indicated above lanes. (b) Reduced expression of ACE2 protein in A549 cells transfected with ACE2-specific siRNA (but not in A549 cells transfected with a nontargeting “mock” siRNA pool) was confirmed by Western blotting with anti-ACE2 antibody. Actin was used as a specificity/loading control. (c) RNA-EMSA performed with biotin-labeled S and OFR1a VAIT element RNA probes and extracts prepared from A549 cells treated as in (a) (transfected with ACE2-targeting or nontargeting siRNA pools and transduced with S pseudotyped lentivirus).
The S protein-induced VAIT pathway involves phosphorylation and release of L13a from the ribosome in a DAP kinase-dependent manner.
Our previous work showed that treatment of the myeloid cells with IFN-γ induces phosphorylation and release of L13a from the 60S ribosomal subunit culminating in GAIT-element mediated translational silencing of a cohort of chemokine and chemokine receptors mRNAs (11, 15). In this pathway, a DAP kinase-1/ZiP kinase cascade was found to be responsible for phosphorylation of L13a at Ser77 leading to release of the protein from the ribosome (38). While our work described above indicates that S protein-induced VAIT element-mediated translational silencing has some mechanistic differences compared with the IFN-γ-induced GAIT system, its shared requirement for L13a led us to investigate the involvement of DAP kinase-dependent phosphorylation and ribosomal release of L13a.
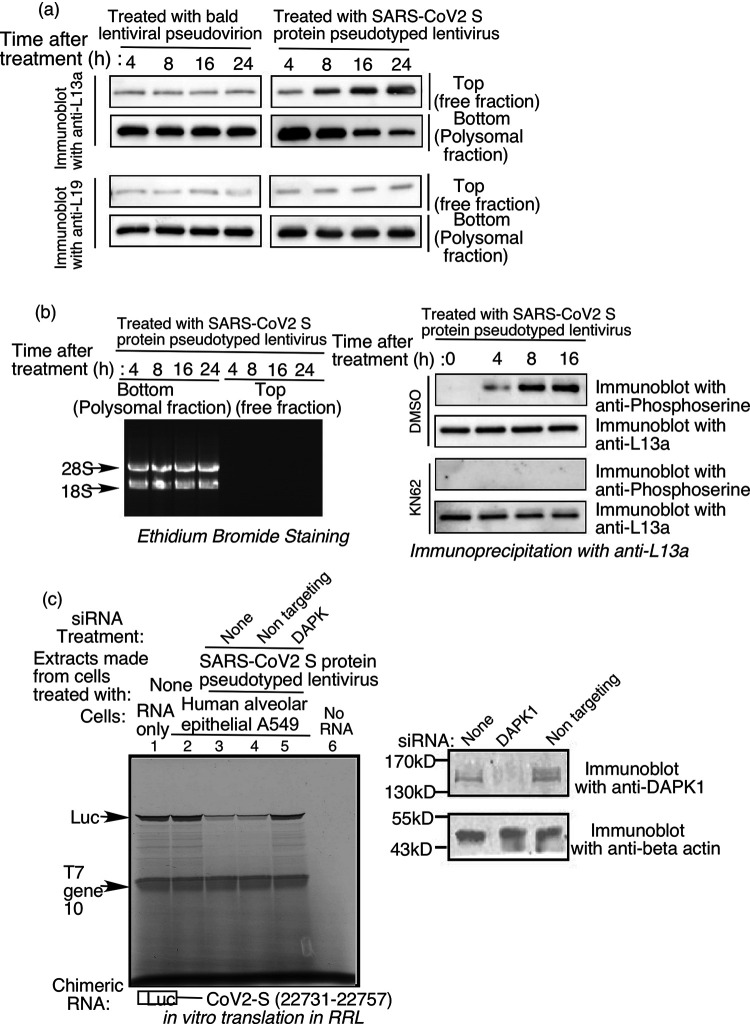
To test for these events in the context of the VAIT translational control system, extracts were made from A549 cells treated with SARS-CoV-2 S protein- pseudotyped lentivirus (or “bald” lentivirus lacking S protein as a negative control) for various amounts of time and centrifuged through a 20% sucrose cushion to separate ribosomal and non-ribosomal fractions. Presence of L13a in the two fractions was determined by immunobloting with anti-L13a antibodies. We previously used this method to demonstrate time-dependent release of L13a from 60S ribosomal subunit in IFN-γ activated human myeloid cells (11, 31). Here, we observed time-dependent release of L13a from the ribosome as early as 8 h after treatment of A549 cells with SARS-CoV-2 S protein pseudotyped lentivirus (Fig. 8a, upper panel). In contrast, treatment of cells with the bald control lentivirus did not have any effect on the distribution of L13a between ribosome-associated and non-ribosomal (free) pools (Fig. 8a, upper panel). Under the same conditions of S protein pseudotyped virus treatment, another ribosomal protein, L19, was not released from the ribosome (Fig. 8a, lower panel), thus showing that S protein-induced release was specific for L13a. Efficient removal of all intact ribosomes from the top fraction (the supernatant following ultracentrifugation) in this experiment was verified by the absence of 28S and 18S rRNA (Fig. 8b, left panel).
FIG 8.
Treatment of A549 lung cells with SARS-CoV-2 S pseudotyped lentivirus triggers phosphorylation and release of L13a from the ribosome. (a) Detection of ribosome-associated and free L13a in A549 cells exposed to S protein. Extracts from A549 cells treated with “bald” (negative control lacking S protein) or S protein pseudotyped lentivirus for the indicated amounts of time were separated into polysome (bottom) and ribosome-free cytosolic (top) fractions, which were then immunoblotted with anti-L13a or anti-L19 antibodies (Thermofisher #14701-1-AP). (b) (left panel), Confirmation of ribosomal and non-ribosomal fractions used in (a). RNA was extracted from the separated fractions using TRIzol, resolved on an agarose gel, and visualized by staining with ethidium bromide. 8b (right panel), Treatment with SARS-CoV-2 S pseudotyped lentivirus induces DAPK-dependent serine phosphorylation of L13a in A549 cells. A549 cells were pretreated with DAPK inhibitor KN62 (or DMSO solvent as a negative control) for 1 h before incubation with the lentivirus. After the indicated amounts of time, cell extracts were prepared and subjected to immunoprecipitation (IP) with anti-L13a antibody, followed by immunoblotting with anti-phosphoserine or anti-L13a antibodies. (c) Requirement of DAPK1 in SARS-CoV-2 S protein-induced and VAIT element-mediated translation control. A549 cells were either untreated or treated with a DAPK1 specific or a non-targeting (used as negative control) siRNA and the steady state level of DAPK1 and beta actin were monitored by immunoblot analysis with anti-DAPK1 and anti-beta actin antibodies. 48 h after siRNA transfection, cells were incubated with SARS-CoV-2 S protein pseudotyped lentivirus for 24 h. Cell lysates were then prepared and used in translation control assay as described earlier. Results showed DAPK1 knocked down cells failed to inhibit translation from chimeric RNA of luciferase and VAIT element (lanes 3 and 4 vs lane 5).
Next, we sought to determine whether the observed release of L13a from ribosomes in response to SARS-CoV-2 S protein treatment is due to L13a phosphorylation and whether such phosphorylation is mediated by DAP kinase. For this, extracts were prepared from A549 lung cells treated with SARS-CoV-2 S protein pseudotyped lentivirus for various amounts of time in the presence or absence of the DAP Kinase inhibitor, KN62. The extracts were subjected to immunoprecipitation with anti-L13a antibody and then immunoblotted using anti-phosphoserine and anti-L13a antibodies to determine relative amounts of phosphorylated and total L13a. To further confirm the role of DAP Kinase in SARS-CoV-2 S protein induced translational silencing, we depleted DAP kinase from human lung A549 cells by RNAi. We first confirmed the depletion of DAP Kinase by immunoblot analysis with anti-DAPK antibody (Fig. 8c, right panel). The DAPK siRNA and Mock siRNA (nontargeting) treated cells were treated with lentivirus particles pseudotyped with SARS-CoV-2 S protein. Cell extracts were made from these cells and tested for their ability to silence the translation of the chimeric luciferase reporter mRNA containing VAIT element from SARS-CoV-2 S mRNA (22731 to 22757). Our results show that depletion of DAP kinase by RNAi abrogates VAIT element-mediated translational silencing (Fig. 8c, left panel, lane 5). However, translational silencing was observed upon treatment with non-targeting mock siRNA similar to no siRNA treatment (Fig. 8c, left panel, lane 3 and 4), thus showing specificity. Together these results show that in consistence with IFN-γ-induced GAIT pathway in myeloid cells, the SARS-CoV-2 S protein-induced VAIT pathway in lung cells proceeds through DAP-kinase mediated phosphorylation of L13a and its subsequent release from the ribosome.
Regulation of S protein-induced polyribosomal association of full-length S mRNA in human lung cells relies on the VAIT element.
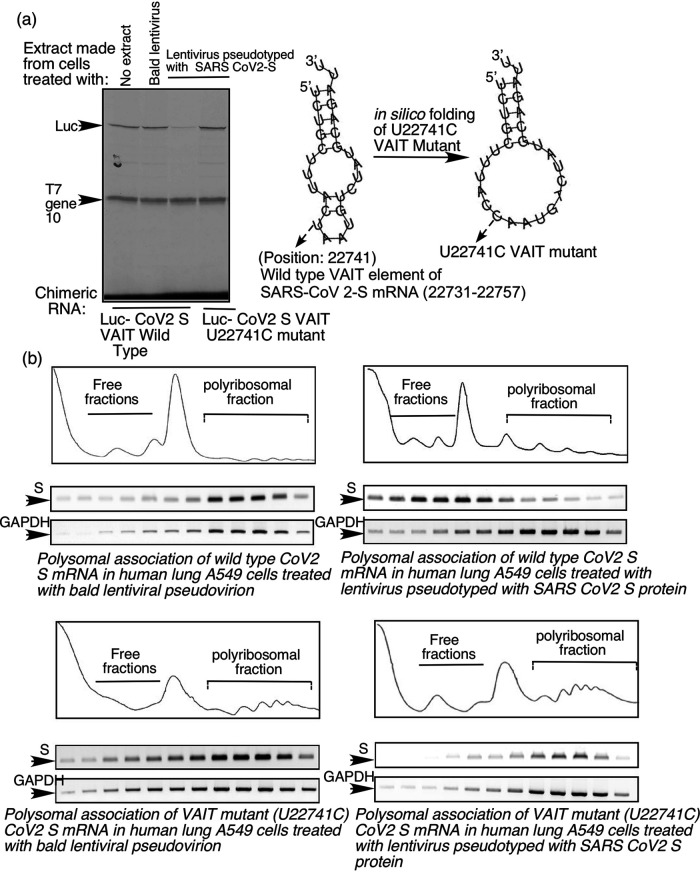
All of our experiments described above showing VAIT element-mediated translational control were performed with chimeric reporter RNAs (i.e., luciferase RNA with the SARS-CoV-2 ORF1a or S VAIT elements inserted in the 3’UTR). To test if this process can be observed in the context of a full-length, native viral RNA and assess the importance of VAIT element secondary structure for directing translational silencing, we focused on the SARS-CoV-2 S VAIT element. Unlike GAIT elements, which are located in the 3’UTRs, the VAIT element in the SARS-CoV-2 S mRNA is located at genomic positions 22731 to 22757, within the S protein-coding region. To test the importance of the structural context of this region, we introduced an “structure-inactivating” point mutation of U to C at position 22741 of the SARS-CoV-2 genome (U22741C). This is a silent mutation coding for threonine in both wild-type (ACU) and mutant (ACC) versions of the S protein. However, in silico folding of the U22741C mutant predicts substantial alteration of the secondary structure of the VAIT element in which the internal bulge and small terminal loop are converted into a single, comparatively large loop (Fig. 9a right panel). Initial testing of the ability of this mutant VAIT element to direct translational silencing was done using chimeric luciferase reporter constructs with VAIT elements inserted into the 3’UTR and a RRL cell-free translation system as in our earlier experiments (e.g., see Fig. 2). Our results show that unlike the wild-type VAIT element, the U22741C mutant VAIT element cannot confer translational silencing on the luciferase reporter RNA when incubated with cell extract prepared from A549 lung cells treated with SARS-CoV-2 S protein pseudotyped lentivirus (Fig. 9a, left panel). This finding confirmed our assumption that the GAIT-like secondary structure of SARS-CoV-2 VAIT elements is the key determinant of their cis-acting translational silencing activity.
FIG 9.
The SARS-CoV-2 S VAIT element controls translation of the full-length S mRNA in A549 cells. (a) A mutant S VAIT element with altered secondary structure fails to support S protein-induced translation control. Right panel: Structures of the wild type and U22741C mutant S VAIT elements predicted using m-fold software. Left panel: In vitro translation of chimeric luciferase reporter cRNAs containing wild type or U22741C mutant S VAIT elements (as indicated below lanes) in the presence of extracts prepared from A549 lung cells treated with bald or S pseudotyped lentivirus (as indicated above lanes). Arrows show luciferase and T7 gene 10 (internal control) translation products. (b) VAIT element structure determines the polysomal association of full-length S mRNA in A549 cells in response to S protein treatment. Polyribosomal and free ribosomal fractions were prepared from A549 cells transfected with plasmids expressing full-length native S cDNA harboring either the wild type (upper panels) or U22741C mutant (lower panels) S VAIT element (and treated with either bald (left panels) or S pseudotyped (right panels) lentivirus). S mRNA and GAPDH mRNA (control) in each fraction was determined by RT-PCR. For each panel, a plot of the A254 values of the fractions (1 through 12) is provided above agarose gels showing ethidium bromide-stained RT-PCR products in the corresponding fractions.
Next, we introduced the U22741C mutation in the full-length mRNA of the native SARS-CoV-2 S sequence (Wuhan MN988668) and used polysome analysis to determine its impact on the efficiency of translation of the full-length S mRNA in cells. A549 cells were transfected with plasmids containing cDNAs corresponding to the unmodified (wild-type) full-length S sequence or the U22741C mutant S sequence, resulting in constitutive, endogenous production of full-length wild type or mutant S mRNA. The transfected cells were then treated with SARS-CoV-2 S protein pseudotyped lentivirus (or “bald” lentivirus lacking S protein as a negative control) for 24 h. Cell extracts were prepared and subjected to sucrose gradient centrifugation, which allows separation of “polyribosomal” and “ribosome-free” fractions containing actively translated (polyribosome-associated) mRNAs and undertranslated (free) mRNAs, respectively. Polyribosomal loading of plasmid-derived wild type and mutant S mRNAs was compared by RT-PCR to quantify the amount of S mRNA and GAPDH mRNA (as a control) in polysomal and free ribosomal fractions. This experiment showed reduced polyribosomal association of the wild type S mRNA in cells treated with SARS-CoV-2 S protein pseudotyped lentivirus compared to cells treated with bald lentivirus lacking S protein (Fig. 9b, upper right and left panels). However, there was not a similar reduction in polyribosomal association for the mutant (U22741C) S RNA upon treatment of cells with S protein pseudotyped lentivirus (Fig. 9b, lower right and left panels). These results demonstrate that the SARS-CoV-2 S VAIT element controls translational efficiency of the full-length S mRNA in human lung cells in response to the treatment with SARS-CoV-2 S protein and that this control is dependent on the structure of the VAIT element.
VAIT elements in the S mRNA and ORF1a RNA of SARS-CoV-2 show high conservation among all evolved genomes of the virus.
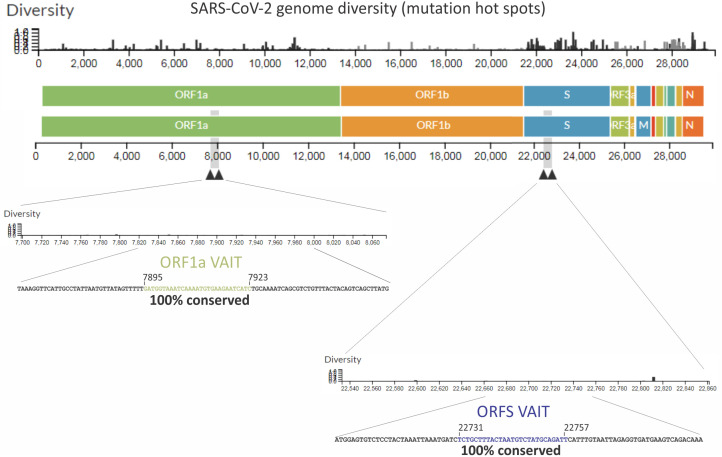
Complete genomic sequences are available for a large number of SARS-CoV-2 isolates, with more becoming available every day. Since emerging from Wuhan, China, in December of 2019, SARS-CoV-2 has been constantly mutating, and thousands of variants have been identified (39–41). Analysis of the genomic locations of mutations observed among SARS-CoV-2 variants showed that the majority of mutations lie in the sequence encoding the RBD of the spike (S) protein (39–41) (Fig. 10). The prevalence of such mutations might be due to their potential to improve receptor binding or cell entry by the virus. Mutations are also found in many other locations across the SARS-CoV-2 genome. Surprisingly, however, we found that the ORF1a and S VAIT elements are extremely well conserved, with no mutations in these regions among all currently available sequenced genomes (Fig. 10). The striking degree of sequence conservation of the S and ORF1a VAIT elements suggests that they have a function that is important for fitness of the virus.
FIG 10.
Sequence conservation of SARS-CoV-2 VAIT elements. The colored bar provides a schematic representation of the SARS-CoV-2 genome structure spanning nucleotides 0 to ∼30,000. The black histogram above the colored bar shows the degree of diversity among currently available SARS-CoV-2 sequences at each position of the genome (data from https://nextstrain.org/ncov/), thus illustrating mutation host spots. The positions and sequence diversity of VAIT elements in ORF1a and ORF S are shown in expanded form below the colored bar.
DISCUSSION
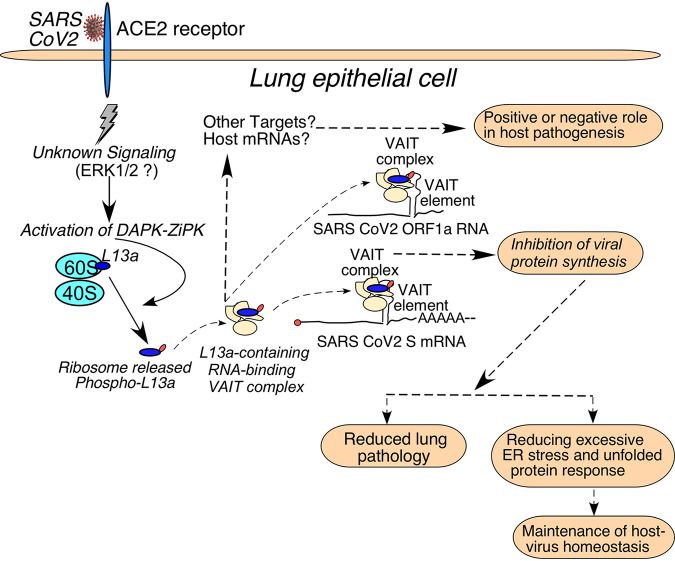
The primary finding of this study is the identification of a novel signaling pathway in human lung cells that is activated by SARS-CoV-2 viral S protein treatment and leads to translational silencing of two SARS-CoV-2 viral mRNAs, ORF1a and S. Our results suggest that this signaling is initiated by interaction of S protein with the ACE2 receptor on host cells, which activates a pathway involving DAP kinase-mediated phosphorylation of ribosomal protein L13a and its release from the large ribosomal unit. The released L13a forms an RNA-binding VAIT complex (likely involving other proteins as well) that binds to structurally conserved VAIT elements present in the S mRNA and ORF1a RNA and suppresses their translation.
To our knowledge, this is the first report of self-regulated translation of SARS-CoV-2 viral transcripts triggered by S protein-ACE2 receptor interaction. For SARS-CoV, it is known that S protein-ACE2 interaction in human lung cells activates ERK1/2 leading to transcriptional upregulation of CCL2 (10). Other studies showed that ERK1/2 could act as an upstream kinase activating DAP kinase (42, 43). This suggests that ERK1/2 may be involved in the VAIT translation control pathway providing the link between ACE2 engagement and DAP kinase-mediated phosphorylation of L13a, but that remains to be directly tested.
Our initial observation of S protein-induced VAIT element-mediated translational silencing was made in a cell-free translation system using extracts from lung cells treated with recombinant S1 subunit of SARS-CoV-2 S protein, which binds ACE2, and chimeric luciferase reporter RNAs containing ORF1a and S VAIT elements in their 3’UTRs (Fig. 2). To address the possibility that recombinant S1 interaction with ACE2 may not faithfully recapitulate the first step in natural viral infection, we performed similar experiments in which lung cells were treated with either S protein pseudotyped lentivirus or S protein-containing VLPs. Both of these treatments also produced extracts that suppressed translation of VAIT element-containing RNAs (Fig. 3). Notably, however, translational silencing was not observed in response to intracellular synthesis of S protein using lung cells stably transfected with the cDNA encoding S protein (Fig. 6). The demonstrated requirement for both extracellular S protein (Fig. 6) and ACE2 (Fig. 7) supports our belief that the VAIT translation control system is activated in vivo by the initial S-ACE2 interaction of invading virions with host lung cells. We note that the VAIT sequences are located in the open reading frames (ORFs) of ORF1a RNA and S mRNA of SARS-CoV-2. However, we have tested the translational silencing activity of these VAIT sequences by positioning them in the 3’UTR of the reporter luciferase. We were unsure if placing a foreign sequence within the ORF of luciferase may affect luciferase activity and thus initially tested the activity of the VAIT elements within 3’UTR. However, we have also found that, like luciferase reporter constructs with inserted VAIT elements at the 3’UTR, the full-length S mRNA of the native Wuhan strain of SARS-CoV-2 is also a target of translational silencing in S protein-treated lung cells (Fig. 9), thus supporting the functional activity of the VAIT element within the ORF of native S mRNA. Importantly, the same full-length S mRNA harboring a point mutation in the VAIT element that changes its structure but does not alter the encoded protein sequence (U22741C) was not translationally silenced (Fig. 9). We believe that these studies clearly demonstrate that S protein-induced translational silencing of VAIT element-containing transcripts is a virologically relevant phenomenon with potentially important consequences for host-virus interaction.
It is important to note that GAIT-element mediated translational silencing in immune cells and VAIT-element mediated translational silencing in lung epithelial cells both rely on conserved RNA element secondary structure rather than sequence motifs. This was shown in our previous work on GAIT elements (16) and in the current study by testing a structure-altering point mutation in the VAIT element of the full-length SARS-CoV-2 S (U22741C; Fig. 9). Our initial bioinformatics-based identification of sequence-unrelated VAIT elements in SARS-CoV-2 RNAs was based on their structural homology with the canonical Cp GAIT element. Our subsequent experiments revealed additional similarities between the GAIT and VAIT pathways, including (i) both require ribosomal protein L13a and its phosphorylation-mediated release from the 60S ribosomal subunit (11) (Fig. 8a); (ii) both require the presence of a cis-acting RNA hairpin structure within the target mRNAs (11, 13) (Fig. 2 and 5); and (iii) both require activation of death-associated protein kinase (DAPK) (38) (Fig. 8b and c). However, there are also substantial differences between the two pathways: (i) the GAIT pathway is triggered by IFN-γ treatment of the cells but not by SARS-CoV-2 S protein interaction with ACE2 receptor; conversely, the VAIT pathway is insensitive to IFN-γ treatment but is activated by viral S protein treatment of cells (Fig. 2); (ii) the GAIT pathway occurs exclusively in cells of myeloid origin (13, 31) while the VAIT pathway occurs in nonimmune cells, e.g., lung epithelial cells (shown herein; see Fig., 2, 3, 4, and 5); (iii) GAIT RNA elements are located in the 3’UTRs of target messages while the identified SARS-CoV-2 VAIT elements are located within the open reading frames of the S mRNA and ORF1a RNA; and (iv) the RNA-protein complexes that form on GAIT and VAIT elements, while both involving L13a, are distinct (Fig. 5). The latter difference was shown by failure of GAIT and VAIT elements to compete with one another in our RNA-EMSA experiments.
There are several potential explanations for our finding that complexes formed on GAIT and VAIT elements are different despite the facts that they both require L13a and that the RNA elements have similar structures. While ribosome-released phosphorylated L13a may be an essential component of both complexes, other binding proteins may be different in the two cases. The canonical GAIT complex is comprised of L13a, glutamyl-prolyl tRNA synthetase (EPRS), NS1-associated protein 1 (NSAP1), and glyceraldehyde-3-phosphate dehydrogenase (GAPDH) (12). Whether EPRS, NSAP1, and/or GAPDH are present in complexes formed on SARS-CoV-2 VAIT elements remains to be established. It is also possible that differences in complex formation may be due to changes in the conformation/structure of VAIT and GAIT hairpins within the RNA-bound protein complex. In addition, VAIT and GAIT elements could potentially recruit a specific additional protein(s) that might impact the specificity of the complex. This possibility is consistent with our previous work showing differential affinities of the GAIT protein complex for the conserved structures of different GAIT elements (16). Future studies will be aimed at identifying the individual protein components and structure of RNA-protein complexes formed on VAIT elements, which should provide insight into the reasons underlying the differences between VAIT and GAIT pathways.
Currently, the consequences of S protein-induced VAIT element-mediated translational control of SARS-CoV-2 viral protein expression for the virus and for the host are far from clear. Prior studies with SARS-CoV-2 and SARS-CoV showed that the flux of S protein through the endoplasmic reticulum (ER) during a productive infection might cause ER stress and activation of the unfolded protein response (UPR) (44, 45) as part of the host’s antiviral response. Therefore, it is tempting to speculate that controlling the synthesis of viral proteins through the VAIT system could serve to promote host-virus homeostasis by reducing the ER burden and ER stress and might act as a rheostat of viral growth regulated by a ribosomal protein of the host. It is also possible that the SARS-CoV-2 S protein-induced VAIT complex reported here could have additional targets beyond the S and ORF1a transcripts, including host mRNAs, which could have further implications for host-virus interaction. Existence of such additional targets remains to be explored. Our current view of the SARS-CoV-2 S protein-induced VAIT element-mediated translational control mechanism(s) is summarized in the schematic model presented in Fig. 11. Another important point to be noted is A549 cells do not significantly support SARS-CoV-2 infection without ACE2 over-expression because of low endogenous level of ACE2. Our work clearly shows that the endogenous level of ACE2 is required and sufficient to mediate S protein induced signaling. This result is consistent with the previous work by others using S protein of SARS-CoV-1and A549 cells (10). At present, it is not known to what extent the S protein induced signaling is correlated with the level of ACE2. However, future experiment involving live virus infection model of SARS-CoV-2 using A549 cells with or without ACE2 over expression will unfold new information on the relationship between infection and the signaling leading to translational silencing.
FIG 11.
Model for VAIT element-driven regulation of SARS-CoV-2 S and ORF1a protein synthesis in host cells. S protein-mediated interaction of SARS-CoV-2 virus with ACE2 at the cell surface leads to phosphorylation of L13a and its release from ribosomes through a novel signaling pathway that involves DAPK but is otherwise not yet defined. The released extra-ribosomal phosphorylated L13a joins with other (currently unknown) proteins to form complexes on VAIT elements in viral RNAs, which suppresses their translation. Inhibition of viral protein synthesis via this mechanism might benefit the host by reducing lung damage and/or promoting host-virus homeostasis through the reduction of ER stress. Whether any host mRNAs are regulated by this translational control mechanism is currently unknown.
For RNA viruses, selection pressure often drives rapid emergence of mutations with the potential to improve virus fitness (46, 47). It is not currently known whether the cellular mechanism of L13a-dependent translational suppression of VAIT element-containing viral transcripts (e.g., S and ORF1a) compromises viral replication. If it does, then one would expect to observe emergence of mutations in the SARS-CoV-2 S and ORF1a VAIT elements that could allow the virus to evade this cellular antiviral defense mechanism. However, our analysis of 3,782 SARS-CoV-2 genomes of curated variants (Nextrain, dated August 3, 2021) and 417,595 noncurated genomes (NCBI SARS-CoV-2 resource portal, dated August 12, 2021) showed no mutations in the S and ORF1a VAIT elements, despite the presence of thousands of changes in other parts of the viral genome (Fig. 10). This extremely high level of sequence conservation among SARS-CoV-2 variants suggests that the S and ORF1a VAIT elements do not negatively impact virus fitness, but rather have a role of critical importance for the virus.
It is far from clear whether the translation inhibitory VAIT sequence presents in the open reading frame of ORF1a RNA and S mRNA of SARS-CoV-2 is the weakness or strength of the virus. Translational inhibition of the two viral proteins mediated by the elements of the viral RNA is consistent with the notion that these elements could be the potential weakness. However, conservation of the sequence among the evolving genomes of SARS-CoV-2 argues against it. Loss or gain of function studies through manipulating these sequences in the context of the infectious virus could give the ultimate answer. However, we understand that these kinds of studies must face stringent regulatory burdens. We envision that the recently developed replicon model of SARS-CoV-2 (48–50) may potentially allow the safe testing of the consequences of the VAIT pathway both for the virus and the host. Together, in these studies, we have provided evidence for a novel paradigm of host-virus relationship for SARS-CoV-2. A potential host-innate immune mechanism involving L13a targets specific viral RNA sequences, thus inhibiting viral protein synthesis. Complete understanding of the mechanistic insights of this process may have significant impact on the development of novel antiviral drugs and or the mRNA vaccine design.
MATERIALS AND METHODS
Cells and culture condition.
A549 human alveolar basal epithelial cells (ATCC; #CCL-185) were cultured in Ham's F12K medium supplemented with 10% heat-inactivated fetal bovine serum, 20 mM HEPES pH 7.5, 100 U penicillin, and 100 mg/mL streptomycin at 37°C with 5% CO2. HBTEC human primary bronchial/tracheal epithelial cells were purchased from Lifeline cell technology (Frederick, MD; #FC-0035) and cultured in Lifeline BronchiaLife Medium (#LL-0023) according to the vendor’s protocol.
Plasmids and Lentiviruses.
Expression constructs (plasmids) containing codon-optimized cDNA clones encoding SARS-CoV-2 proteins M, N, S, and E were purchased from Sino Biological (#VG40608-UT, #VG40588-CY, #VG40589-CH, and #VG40609-CF, respectively). SARS-CoV-2 S gene with the native sequence (Wuhan MN988668) was purchased from Origene (Rockville, MD; #VC102566). Lentiviral particles pseudotyped with full-length SARS-CoV-2 spike glycoprotein were purchased from Genecopoeia (Rockville, MD; #SP101-100; 6.62 × 105 TU [(transduction unit)/mL]) and bald lentiviral pseudovirion particles with no VSV-G or SARS-CoV-2 spike protein were purchased from BPS Bioscience (San Diego, CA; #79943).
Bioinformatic prediction of “GAIT-like” elements in SARS-CoV-2.
SARS-CoV-2 genomic sequences corresponding to ORF1ab, S, ORF3a, E, M, ORF6, ORF7a, ORF8, N, ORF10, and 3′ UTR were separately examined for GAIT-like structural elements. We used the “FOLDALIGN” algorithm (foldalign.ku.dk/server/index.html) to analyze candidate SARS-CoV-2 mRNA sequences and canonical ceruloplasmin (Cp) GAIT element which were subjected to pairwise local structural alignment and prediction. The resulting hits were then separately folded using the “RNAstructure version 6.3” webserver (https://rna.urmc.rochester.edu/RNAstructureWeb/). Hits showing structural similarity with the Cp GAIT element were further tested by RNA-EMSA and translational silencing assay (see below).
Analysis of SARS-CoV-2 genome diversity.
The NCBI SARS-CoV-2 resources portal (https://www.ncbi.nlm.nih.gov/sars-cov-2/) containing (as of August 12, 2021) 417,595 SARS-CoV-2 genome records was the source for SARS-CoV-2 sequence analysis. The SARS-CoV-2 ORF1a VAIT sequence 5′-GATGGTAAATCAAAATGTGAAGAATCATC-3′ spanning nucleotides 7895–7923 and ORF S VAIT sequence 5′-TCTGCTTTACTAATGTCTATGCAGATT-3′ spanning nucleotides 22731–22757 of the Wuhan strain RefSeq genome (NC_045512) were used to analyze conservation of these sequences using BLAST. The Nextstrain (51) SARS-CoV-2 resource (https://nextstrain.org/ncov/) was used for graphic visualization of the data. As of August 3, 2021, this resource contained 3,782 genomes sampled from December 20, 2019 to July 25, 2021. The resource subsamples approximately 600 genomes per continental region: Africa, Asia, Europe, North America, Oceania, and South America.
1D-1H-NMR spectroscopic analysis of RNA elements.
Chemically synthesized HPLC-purified RNA sequences corresponding to Cp GAIT and SARS-CoV-2 VAIT elements were purchased from IDT DNA technologies. The lyophilized RNA samples were dissolved in 50 μL RNase-free water immediately prior to analysis. RNA folding reactions were performed by heating samples to 95°C for 2 min followed by flash cooling for 20 min on ice. The buffer was then exchanged to NMR buffer (10 mM K2HPO4 and 10 mM KCl, pH 6.0) in 90% H2O/10% D2O. Concentrations were determined using theoretical molar extinction coefficients and were typically in the range of ∼0.3 μM at 200 μL sample volume. The 1D-1H-NMR spectra were recorded at 10°C, 15°C, 20°C, and 25°C on a Bruker 900-MHz high-field NMR spectrometer using a cryogenically cooled H-C-N triple-resonance probe. The WATERGATE (p3919gp) pulse sequence was applied with 256 scans (NS), 32,000 points in time domain (TD), and a relaxation delay (d1) of 1.0 s. After collection, all NMR data were processed and analyzed using Bruker Topspin 4.1.1 software.
Production of SARS-CoV-2 VLPs.
SARS-CoV2 VLPs were generated in A549 cells by cotransfection of plasmids directing constitutive expression of SARS-CoV-2 viral proteins (M, N, S, and E) following a previously published protocol with minor modifications (33). 2 × 106 A549 cells were plated in a 100-mm culture dish overnight to achieve ∼70% confluence on the day of transfection. Cells were transfected using 6 μg of each of the four SARS-CoV-2 cDNA expression plasmids (M, N, S, and E) and 36 μl Fugene 6 transfection reagent (Promega, # E2691). After 48 h of transfection, the cell medium was cleared by low-speed centrifugation (1,000 × g for 10 min) followed by a passage through a 0.45-μm-pore-size filter. The cleared medium was then layered on top of a 20% sucrose cushion and ultra-centrifuged for 3 h at 28,000 rpm using a Beckman SW32.1 Ti rotor at 4°C. VLP-containing pellets were resuspended in TNE buffer (50 mM Tris-HCl, 100 mM NaCl, 0.5 mM EDTA, [pH 7.4]) and stored at −80°C. 20 μL of purified SARS-CoV-2 VLPs were examined by Western blotting analysis. VLPs prepared from A549 cells transfected with only M, N, and E (no S) were used as a negative control.
Lentiviral transduction and Luciferase assay.
To obtain 70% to 80% confluence at the time of infection, 5 × 104 A549 cells per well were plated in a 24-well plate 24 h before viral infection. Cells were infected by incubating with 0.5 mL of complete medium containing 1 μL virus particle (662 TU) and 8 μg/mL Polybrene. After 72 h of incubation at 37°C, cells were analyzed for Firefly luciferase expression using the Brightglo luciferase assay system (Promega, #E2610). Mock-infected cells were used to normalize data.
ACE2 and DAPK1 knockdown.
A549 cells (3 × 105 cells/well) were seeded into 6-well plates 1 day before transfection in order to achieve ∼70% confluence at transfection. Three ON-TARGETplus siRNA pools were obtained from Dharmacon: (i) control nontargeting siRNA pool (#D-001810-10-05); (ii) human ACE2 specific siRNA-SMARTpool (#L-005755-00-0005); and (iii) human DAPK1 specific siRNA-SMARTpool (#L-004417-00-005). Cells were transfected using 30 pmol siRNA and DharmaFECT 2 transfection reagent (#T-2002-01) according to the vendor’s protocol. At 48 h posttransfection, cells were treated with lentivirus particles pseudotyped with full-length SARS-CoV-2 spike protein (Genecopoeia, Rockville, MD #SP101-100) at a concentration of 6.62 × 104 TU/106 cells for 24 h. Expression of ACE2, DAPK1 and beta-actin was monitored by immunoblotting of whole cell extracts with corresponding specific antibodies. (anti-ACE2, Cell signaling technology #4355; anti-DAPK1, Thermofisher #MA1-24696 and anti-beta-actin, Cell signaling technology #3700).
Preparation of cell extracts for use in translation silencing assay.
Cytosolic extracts were prepared from A549 or HBTEC cells either left untreated or treated with SARS CoV2 S protein. S protein treatment was delivered as (i) recombinant purified protein (S1 subunit, 1 μg/106 cells); (ii) VLP particles containing M, N, E, and S proteins (50 μg VLP/5 × 104 cells); or (iii) S protein pseudotyped lentivirus (6.62 × 104 TU/106 cells) as described above. Exponentially growing cells were harvested after treatment, washed three times with cold PBS, resuspended in lysis buffer (50 mM Tris [pH 7.6], 50 mM NaCl, 1 mM PMSF, 1 mM DTT, and proteinase inhibitor cocktail) and kept on ice for 15 min. Cells were then lysed by repeated freeze-thaw cycles followed by centrifugation at 10,000 × g for 30 min. Supernatants were collected and used in RNA-EMSA and in vitro translation assays.
Immunodepletion of L13a.
L13a protein was immunodepleted from A549 and HBTEC cell lysates following a protocol previously described protocol (16). Briefly, cell lysates were incubated with mouse anti-ribosomal protein L13A antibody (C-11) (Santa Cruz Biotech, #sc-390131) and complexes were immunoprecipitated using Protein A Agarose beads. Depletion of L13a was confirmed by Western blotting with ribosomal protein L13a antibody (Cell Signaling Technology, #2765).
3′ biotinylation of RNA.
Oligonucleotides corresponding to SARS-CoV-2 VAIT RNA elements were synthesized by Integrated DNA Technologies (Coralville, IA) and biotinylated using the Thermo Scientific Pierce RNA biotinylation kit (#20160) according to the manufacturer’s protocol.
RNA electromobility shift assay.
RNA electromobility shift assay (RNA-EMSA) was performed as described previously (16). In brief, 15 fmol of biotinylated RNA oligonucleotide probe and 5 μg cell lysates were incubated in REMSA buffer (10 mM HEPES [pH 7.3], 20 mM KCl, 1 mM MgCl2, 1 mM DTT, 5% glycerol, 10 μg yeast tRNA, and 40 U RNasin [Promega]) for 30 min on ice. Specificity of binding was tested by competition experiments in which nonbiotinylated “competitor” RNA oligonucleotides were added to the reaction mixture along with the biotinylated RNA probe. Native gel electrophoresis (5% polyacrylamide in 0.5× Tris Borate EDTA [TBE] buffer) was performed to separate free probe and RNA-protein complexes. After transfer to a nylon membrane, the biotinylated probe was detected using the Pierce Lightshift chemiluminescent RNA EMSA kit (Thermo Scientific, # 20158).
In vitro translation.
Annealed DNA oligonucleotides corresponding to the SARS-CoV-2 VAIT elements were cloned in the SpeI-HindIII sites of the pMIR-REPORT Luciferase vector (Thermofisher Scientific, # AM5795), downstream of a luciferase reporter gene under the control of the T7 promoter. VAIT element containing luciferase cRNAs were then synthesized from these constructs by in vitro transcription. We used 100 ng of cRNA for in vitro translation using rabbit reticulocyte lysates (Promega, #L4960) in the presence of [35S]-methionine. To test for translational silencing activity of cell extracts (from e.g., S protein-treated human lung cells, see above), 4 μg of lysates was added per reaction. To detect translation products, a 10-μl aliquot of each translation reaction mixture was resolved by SDS-PAGE (10% polyacrylamide), followed by autoradiography. T7 gene 10 cRNA was used as a control for nonspecific inhibition of translation in rabbit reticulocyte lysate as described previously (11). This RNA was prepared by in vitro transcription of pGEMEX-2 (Promega) vector using T7 RNA polymerase (mMessage mMachine T7 transcription kit, Thermofisher, #AM1344).
Site-directed mutagenesis.
To disrupt the native secondary structure of the S VAIT element without altering the encoded S protein amino acid sequence, a single silent mutation (T to C) was made in the S gene at position 22741 by site-directed mutagenesis using the QuikChange II site-directed mutagenesis system (Agilent, # 200523) and primers SDMFwd (5′- GATCTCTGCTTTACCAATGTCTATGCAG-3′) and SDMRev (5′- CTGCATAGACATTGGTAAAGCAGAGATC-3′).
Polysomal analysis.
A549 cells were transiently transfected with plasmids containing the native SARS-CoV-2 S mRNA sequence, either wild type or mutated (U22741C) in the S VAIT element. Cells were treated with lentivirus particles pseudotyped with full-length SARS-CoV-2 spike protein (Genecopoeia, Rockville, MD #SP101-100) 48 h after transfection at a concentration of 6.62 × 104 TU/106 cells for 24 h. Bald lentiviral pseudovirion particles without the S protein were used as a control. Following lentivirus treatment, cytoplasmic extracts were prepared and resolved by centrifugation for 18 h at 17,000 rpm using 5% to 50% linear sucrose density gradients in polysome buffer (10 mM HEPES [pH 7.5], 100 mM KCl, 2.5 mM MgCl2, 1 mM DTT, 50 U recombinant RNasin [Promega, Madison, WI], and 0.1% NP-40). Fractions were collected using an ISCO gradient fractionation system equipped with a UA-6 detector. Total RNA from these fractions was isolated by TRIzol extraction (Invitrogen) and used for reverse transcription (RT)-PCR with primers specific for the SARS-CoV-2 S gene (SRTF [5′-CGCTTGTTAAACAACTTAGCTC-3′] and SRTR [5′- CCCTTTCCACAAAAATCAACTC-3′], 256 bp product) and the human GAPDH gene (GAPDHRTF [5′- GACCACAGTCCATGCCATCACTGC-3′] and GAPDHRTR [5′- TGTTGCTGTAGCCAAATTCGTTG-3′], 443 bp product).
Ribosomal protein L13a release and phosphorylation.
To detect SARS-CoV-2 S protein-induced ribosomal release and phosphorylation of Ribosomal protein L13a (RPL13a), A549 cells were incubated with lentivirus particles pseudotyped with full-length SARS-CoV-2 S protein (Genecopoeia, Rockville, MD #SP101-100) at a concentration of 6.62 × 104 TU/106 cells for 24 h. A cytoplasmic extract was then prepared from the cells, layered on a 20% sucrose cushion, and centrifuged as described above. The top portion of the layer (containing proteins released from ribosomes) and the precipitated ribosome at the bottom were collected and analyzed by immunoblotting with appropriate antibodies. To detect L13a phosphorylation, cell lysates from S protein-treated A549 cells were subjected to immunoprecipitation by L13a-specific antibody followed by Western blotting with anti-phospho-serine/threonine antibody (Thermofisher, #61-8300). In some experiments, DAPK kinase inhibitor KN62 (Sigma, # I2142) was added to the culture medium during S protein treatment.
ACKNOWLEDGMENTS
This work was supported by Public Health Service grants NIH HL79164 (B.M.), GM128981 (A.A.K.), and GM101979 (B.S.T.) and a gift from the Nord Family Foundation (J.K.). We thank Patricia Stanhope Baker for assistance with manuscript editing.
Contributor Information
Barsanjit Mazumder, Email: b.mazumder@csuohio.edu.
Tom Gallagher, Loyola University Chicago.
REFERENCES
- 1.V’Kovski P, Kratzel A, Steiner S, Stalder H, Thiel V. 2021. Coronavirus biology and replication: implications for SARS-CoV-2. Nat Rev Microbiol 19:155–170. 10.1038/s41579-020-00468-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Banerjee AK, Blanco MR, Bruce EA, Honson DD, Chen LM, Chow A, Bhat P, Ollikainen N, Quinodoz SA, Loney C, Thai J, Miller ZD, Lin AE, Schmidt MM, Stewart DG, Goldfarb D, De Lorenzo G, Rihn SJ, Voorhees RM, Botten JW, Majumdar D, Guttman M. 2020. SARS-CoV-2 disrupts splicing, translation, and protein trafficking to suppress host defenses. Cell 183:1325–1339. 10.1016/j.cell.2020.10.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Thoms M, Buschauer R, Ameismeier M, Koepke L, Denk T, Hirschenberger M, Kratzat H, Hayn M, Mackens-Kiani T, Cheng J, Straub JH, Sturzel CM, Frohlich T, Berninghausen O, Becker T, Kirchhoff F, Sparrer KMJ, Beckmann R. 2020. Structural basis for translational shutdown and immune evasion by the Nsp1 protein of SARS-CoV-2. Science 369:1249–1255. 10.1126/science.abc8665. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Finkel Y, Gluck A, Nachshon A, Winkler R, Fisher T, Rozman B, Mizrahi O, Lubelsky Y, Zuckerman B, Slobodin B, Yahalom-Ronen Y, Tamir H, Ulitsky I, Israely T, Paran N, Schwartz M, Stern-Ginossar N. 2021. SARS-CoV-2 uses a multipronged strategy to impede host protein synthesis. Nature 594:240–245. 10.1038/s41586-021-03610-3. [DOI] [PubMed] [Google Scholar]
- 5.Alexander MR, Brice AM, Jansen van Vuren P, Rootes CL, Tribolet L, Cowled C, Bean AGD, Stewart CR. 2021. Ribosome-profiling reveals restricted post transcriptional expression of antiviral cytokines and transcription factors during SARS-CoV-2 infection. Int J Mol Sci 22. 10.3390/ijms22073392. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Li F. 2016. Structure, function, and evolution of coronavirus spike proteins. Annu Rev Virol 3:237–261. 10.1146/annurev-virology-110615-042301. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Yan R, Zhang Y, Li Y, Xia L, Guo Y, Zhou Q. 2020. Structural basis for the recognition of SARS-CoV-2 by full-length human ACE2. Science 367:1444–1448. 10.1126/science.abb2762. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Hoffmann M, Kleine-Weber H, Pohlmann S. 2020. A multibasic cleavage site in the spike protein of SARS-CoV-2 is essential for infection of human lung cells. Mol Cell 78:779–784. 10.1016/j.molcel.2020.04.022. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Hoffmann M, Kleine-Weber H, Schroeder S, Kruger N, Herrler T, Erichsen S, Schiergens TS, Herrler G, Wu NH, Nitsche A, Muller MA, Drosten C, Pohlmann S. 2020. SARS-CoV-2 cell entry depends on ACE2 and TMPRSS2 and is blocked by a clinically proven protease inhibitor. Cell 181:271–280. 10.1016/j.cell.2020.02.052. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Chen IY, Chang SC, Wu HY, Yu TC, Wei WC, Lin S, Chien CL, Chang MF. 2010. Upregulation of the chemokine (C-C motif) ligand 2 via a severe acute respiratory syndrome coronavirus spike-ACE2 signaling pathway. J Virol 84:7703–7712. 10.1128/JVI.02560-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Mazumder B, Sampath P, Seshadri V, Maitra RK, DiCorleto PE, Fox PL. 2003. Regulated release of L13a from the 60S ribosomal subunit as a mechanism of transcript-specific translational control. Cell 115:187–198. 10.1016/s0092-8674(03)00773-6. [DOI] [PubMed] [Google Scholar]
- 12.Sampath P, Mazumder B, Seshadri V, Gerber CA, Chavatte L, Kinter M, Ting SM, Dignam JD, Kim S, Driscoll DM, Fox PL. 2004. Noncanonical function of glutamyl-prolyl-tRNA synthetase: gene-specific silencing of translation. Cell 119:195–208. 10.1016/j.cell.2004.09.030. [DOI] [PubMed] [Google Scholar]
- 13.Sampath P, Mazumder B, Seshadri V, Fox PL. 2003. Transcript-selective translational silencing by gamma interferon is directed by a novel structural element in the ceruloplasmin mRNA 3' untranslated region. Mol Cell Biol 23:1509–1519. 10.1128/MCB.23.5.1509-1519.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Kapasi P, Chaudhuri S, Vyas K, Baus D, Komar AA, Fox PL, Merrick WC, Mazumder B. 2007. L13a blocks 48S assembly: role of a general initiation factor in mRNA-specific translational control. Mol Cell 25:113–126. 10.1016/j.molcel.2006.11.028. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Vyas K, Chaudhuri S, Leaman DW, Komar AA, Musiyenko A, Barik S, Mazumder B. 2009. Genome-wide polysome profiling reveals an inflammation-responsive posttranscriptional operon in gamma interferon-activated monocytes. Mol Cell Biol 29:458–470. 10.1128/MCB.00824-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Basu A, Jain N, Tolbert BS, Komar AA, Mazumder B. 2017. Conserved structures formed by heterogeneous RNA sequences drive silencing of an inflammation responsive post-transcriptional operon. Nucleic Acids Res 45:12987–13003. 10.1093/nar/gkx979. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Poddar D, Basu A, Baldwin WM, 3rd, Kondratov RV, Barik S, Mazumder B. 2013. An extraribosomal function of ribosomal protein L13a in macrophages resolves inflammation. J Immunol 190:3600–3612. 10.4049/jimmunol.1201933. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Poddar D, Kaur R, Baldwin WM, 3rd, Mazumder B. 2016. L13a-dependent translational control in macrophages limits the pathogenesis of colitis. Cell Mol Immunol 13:816–827. 10.1038/cmi.2015.53. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Mazumder B. 2018. GAITing the gut. Cell Mol Immunol 15:1082–1084. 10.1038/s41423-018-0039-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Basu A, Poddar D, Robinet P, Smith JD, Febbraio M, Baldwin WM, 3rd, Mazumder B. 2014. Ribosomal protein L13a deficiency in macrophages promotes atherosclerosis by limiting translation control-dependent retardation of inflammation. Arterioscler Thromb Vasc Biol 34:533–542. 10.1161/ATVBAHA.113.302573. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Basu A, Dvorina N, Baldwin WM, 3rd, Mazumder B. 2020. High-fat diet-induced GAIT element-mediated translational silencing of mRNAs encoding inflammatory proteins in macrophage protects against atherosclerosis. FASEB J 34:6888–6906. 10.1096/fj.201903119R. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Marquez-Jurado S, Nogales A, Zuniga S, Enjuanes L, Almazan F. 2015. Identification of a gamma interferon-activated inhibitor of translation-like RNA motif at the 3' end of the transmissible gastroenteritis coronavirus genome modulating innate immune response. mBio 6:e00105. 10.1128/mBio.00105-15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Mazumder B, Poddar D, Basu A, Kour R, Verbovetskaya V, Barik S. 2014. Extraribosomal l13a is a specific innate immune factor for antiviral defense. J Virol 88:9100–9110. 10.1128/JVI.01129-14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Madhugiri R, Fricke M, Marz M, Ziebuhr J. 2016. Coronavirus cis-acting RNA elements. Adv Virus Res 96:127–163. 10.1016/bs.aivir.2016.08.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Ahmed F, Sharma M, Al-Ghamdi AA, Al-Yami SM, Al-Salami AM, Refai MY, Warsi MK, Howladar SM, Baeshen MN. 2020. A comprehensive analysis of cis-acting RNA elements in the SARS-CoV-2 genome by a bioinformatics approach. Front Genet 11:572702. 10.3389/fgene.2020.572702. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Manfredonia I, Incarnato D. 2021. Structure and regulation of coronavirus genomes: state-of-the-art and novel insights from SARS-CoV-2 studies. Biochem Soc Trans 49:341–352. 10.1042/BST20200670. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Havgaard JH, Lyngso RB, Stormo GD, Gorodkin J. 2005. Pairwise local structural alignment of RNA sequences with sequence similarity less than 40%. Bioinformatics 21:1815–1824. 10.1093/bioinformatics/bti279. [DOI] [PubMed] [Google Scholar]
- 28.Havgaard JH, Torarinsson E, Gorodkin J. 2007. Fast pairwise structural RNA alignments by pruning of the dynamical programming matrix. PLoS Comput Biol 3:1896–1908. 10.1371/journal.pcbi.0030193. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Torarinsson E, Havgaard JH, Gorodkin J. 2007. Multiple structural alignment and clustering of RNA sequences. Bioinformatics 23:926–932. 10.1093/bioinformatics/btm049. [DOI] [PubMed] [Google Scholar]
- 30.Furtig B, Richter C, Wohnert J, Schwalbe H. 2003. NMR spectroscopy of RNA. Chembiochem 4:936–962. 10.1002/cbic.200300700. [DOI] [PubMed] [Google Scholar]
- 31.Mazumder B, Fox PL. 1999. Delayed translational silencing of ceruloplasmin transcript in gamma interferon-activated U937 monocytic cells: role of the 3' untranslated region. Mol Cell Biol 19:6898–6905. 10.1128/MCB.19.10.6898. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Mazumder B, Li X, Barik S. 2010. Translation control: a multifaceted regulator of inflammatory response. J Immunol 184:3311–3319. 10.4049/jimmunol.0903778. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Siu YL, Teoh KT, Lo J, Chan CM, Kien F, Escriou N, Tsao SW, Nicholls JM, Altmeyer R, Peiris JS, Bruzzone R, Nal B. 2008. The M, E, and N structural proteins of the severe acute respiratory syndrome coronavirus are required for efficient assembly, trafficking, and release of virus-like particles. J Virol 82:11318–11330. 10.1128/JVI.01052-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Chen M, Zhang XE. 2021. Construction and applications of SARS-CoV-2 pseudoviruses: a mini review. Int J Biol Sci 17:1574–1580. 10.7150/ijbs.59184. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Yu J, Li Z, He X, Gebre MS, Bondzie EA, Wan H, Jacob-Dolan C, Martinez DR, Nkolola JP, Baric RS, Barouch DH. 2021. Deletion of the SARS-CoV-2 spike cytoplasmic tail increases infectivity in pseudovirus neutralization assays. J Virol 95:00044. 10.1128/JVI.00044-21. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Standart N, Jackson RJ. 1994. Regulation of translation by specific protein/mRNA interactions. Biochimie 76:867–879. 10.1016/0300-9084(94)90189-9. [DOI] [PubMed] [Google Scholar]
- 37.Mazumder B, Seshadri V, Fox PL. 2003. Translational control by the 3'-UTR: the ends specify the means. Trends Biochem Sci 28:91–98. 10.1016/S0968-0004(03)00002-1. [DOI] [PubMed] [Google Scholar]
- 38.Mukhopadhyay R, Ray PS, Arif A, Brady AK, Kinter M, Fox PL. 2008. DAPK-ZIPK-L13a axis constitutes a negative-feedback module regulating inflammatory gene expression. Mol Cell 32:371–382. 10.1016/j.molcel.2008.09.019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Koyama T, Platt D, Parida L. 2020. Variant analysis of SARS-CoV-2 genomes. Bull World Health Organ 98:495–504. 10.2471/BLT.20.253591. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Srivastava S, Banu S, Singh P, Sowpati DT, Mishra RK. 2021. SARS-CoV-2 genomics: An Indian perspective on sequencing viral variants. J Biosci 46:22. 10.1007/s12038-021-00145-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Sanyaolu A, Okorie C, Marinkovic A, Haider N, Abbasi AF, Jaferi U, Prakash S, Balendra V. 2021. The emerging SARS-CoV-2 variants of concern. Ther Adv Infect Dis 8:20499361211024372. 10.1177/20499361211024372. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Mebratu Y, Tesfaigzi Y. 2009. How ERK1/2 activation controls cell proliferation and cell death: is subcellular localization the answer? Cell Cycle 8:1168–1175. 10.4161/cc.8.8.8147. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Houle F, Poirier A, Dumaresq J, Huot J. 2007. DAP kinase mediates the phosphorylation of tropomyosin-1 downstream of the ERK pathway, which regulates the formation of stress fibers in response to oxidative stress. J Cell Sci 120:3666–3677. 10.1242/jcs.003251. [DOI] [PubMed] [Google Scholar]
- 44.Versteeg GA, van de Nes PS, Bredenbeek PJ, Spaan WJ. 2007. The coronavirus spike protein induces endoplasmic reticulum stress and upregulation of intracellular chemokine mRNA concentrations. J Virol 81:10981–10990. 10.1128/JVI.01033-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Sicari D, Chatziioannou A, Koutsandreas T, Sitia R, Chevet E. 2020. Role of the early secretory pathway in SARS-CoV-2 infection. J Cell Biol 219. 10.1083/jcb.202006005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Domingo E, Holland JJ. 1997. RNA virus mutations and fitness for survival. Annu Rev Microbiol 51:151–178. 10.1146/annurev.micro.51.1.151. [DOI] [PubMed] [Google Scholar]
- 47.Novella IS, Quer J, Domingo E, Holland JJ. 1999. Exponential fitness gains of RNA virus populations are limited by bottleneck effects. J Virol 73:1668–1671. 10.1128/JVI.73.2.1668-1671.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.He X, Quan S, Xu M, Rodriguez S, Goh SL, Wei J, Fridman A, Koeplinger KA, Carroll SS, Grobler JA, Espeseth AS, Olsen DB, Hazuda DJ, Wang D. 2021. Generation of SARS-CoV-2 reporter replicon for high-throughput antiviral screening and testing. Proc Natl Acad Sci USA 118:e2025866118. 10.1073/pnas.2025866118. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Kotaki T, Xie X, Shi PY, Kameoka M. 2021. A PCR amplicon-based SARS-CoV-2 replicon for antiviral evaluation. Sci Rep 11:2229. 10.1038/s41598-021-82055-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Luo Y, Yu F, Zhou M, Liu Y, Xia B, Zhang X, Liu J, Zhang J, Du Y, Li R, Wu L, Zhang X, Pan T, Guo D, Peng T, Zhang H. 2021. Engineering a reliable and convenient SARS-CoV-2 replicon system for analysis of viral RNA synthesis and screening of antiviral inhibitors. mBio 12 10.1128/mBio.02754-20. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Hadfield J, Megill C, Bell SM, Huddleston J, Potter B, Callender C, Sagulenko P, Bedford T, Neher RA. 2018. Nextstrain: real-time tracking of pathogen evolution. Bioinformatics 34:4121–4123. 10.1093/bioinformatics/bty407. [DOI] [PMC free article] [PubMed] [Google Scholar]