Abstract
A novel PCR detection assay that amplifies the Helicobacter pylori-specific vacuolating cytotoxin gene (vacA) and thus enables rapid diagnosis of infection is described. Additionally, a real-time probe hybridization melting point analysis assay to detect all three mutations in the 23S rRNA gene associated with clarithromycin resistance was applied directly to antral gastric biopsy samples. Comparison with culture and an alternative PCR assay targeting the 16S rrn gene showed that the vacA assay was sensitive and specific when tested on biopsy samples from 121 patients. Clarithromycin susceptibilities could be determined in the majority (92.3%) of culture-positive gastric biopsy samples analyzed, four of which generated melting peaks indicative of clarithromycin resistance by either an A→G or A→C mutation. The presence of the mutations correlated with the clarithromycin disk diffusion sensitivities of matched cultures. This PCR-based system was simple to perform and could be completed in 3 to 4 h, thereby overcoming the delays associated with conventional culture methods for H. pylori identification and susceptibility testing.
Helicobacter pylori, a major cause of chronic gastritis, is strongly associated with the development of gastric and duodenal ulcers and has been linked with gastric adenocarcinoma and B-cell mucosa-associated lymphoid tissue lymphoma (15, 17, 18). Infection can be eradicated in up to 90% of patients using current combination triple therapies, of which the macrolide antibiotic clarithromycin is a key component (5). Rates of resistance to clarithromycin of 1 to 9% have been reported in several European countries and the United States, with even higher rates in some countries, such as France and Belgium (24). The development of clarithromycin resistance in H. pylori is recognized as a significant contributing factor in treatment failure (8, 14), and the mechanism is attributed to single point mutations in the peptidyltransferase region of the 23S rRNA gene (25). Adenine residues at either position 2143 or 2144 can mutate. Transition to guanine (A2143G and A2144G) is the most common mutation type, with the transversion mutation to cytosine (A2143C) less common (16, 21, 22, 25). Although culture of gastric biopsy samples allows further H. pylori strain analysis, including determination of antibiotic susceptibility, tests can take up to 2 weeks to complete. The aim of the present study was to develop a PCR-based system using conventional and real-time techniques enabling same-day diagnosis of H. pylori infection and determination of clarithromycin resistance.
MATERIALS AND METHODS
Gastric biopsy samples and strain isolation.
Two sets of gastric biopsy samples were used in this study. First, we examined a series of preserved (−20°C) gastric biopsy samples from 39 dyspeptic patients attending an open-access endoscopy clinic in Chelmsford during 1995 and 1996. These biopsy samples were confirmed positive for H. pylori by both culture and histology, and the patients were also confirmed seropositive for H. pylori (19). Second, we performed a prospective study to assess the clinical applicability of the assays in which two biopsy samples per patient were obtained over a 3-month period in 1999 from the gastric antra of 121 dyspeptic patients referred for endoscopy by general practitioners. One biopsy sample was used directly for culture of H. pylori on Columbia base agar containing 10% (vol/vol) defibrinated horse blood at 36°C under microaerobic conditions (90% CO2, 4% O2, 4% N2, 2% H2). For antibiotic susceptibility testing, approximately 107 CFU of H. pylori and a disk containing 2 μg of clarithromycin were applied to a blood agar plate. Clarithromycin susceptibility was recorded after microaerobic incubation for 48 h at 36°C. The second biopsy sample was stored at −20°C until required for DNA extraction.
Genomic DNA was extracted from both sets of gastric biopsy samples using a modification of a previously described method (12). Briefly, the thawed biopsy samples were homogenized in Griffith's tubes containing 400 μl of sterile saline (0.85% [wt/vol]), transferred to 1.5-ml screw-cap Eppendorf tubes, and centrifuged (10,000 × g for 2 min) and the supernatant was discarded. Pellets were resuspended in extraction buffer (20 mM Tris-HCl [pH 8.0], 0.5% [vol/vol] Tween 20) and proteinase K (0.5 mg/ml), vortexed for 2 to 5 s, and incubated at 56°C for 1 h and then 100°C for 10 min. DNA extracts were stored at −20°C until required.
Conventional PCR assays.
A PCR assay targeted at the 16S rRNA gene of H. pylori was performed with primer pair Hp1 and Hp2 (Table 1) by a modification of the protocol described previously (9). Briefly, the 50-μl PCR mixture, containing 5 μl of extracted DNA, 200 μM (each) deoxynucleoside trisphosphates (dNTPs) (Gibco BRL, Paisley, Scotland), 0.4 μM (each) primer (MWG Biotech, Milton Keynes, England), 1.5 mM MgCl2, and 1 U of Taq polymerase (Gibco BRL) in PCR buffer (20 mM Tris-HCl [pH 8.4], 50 mM KCl, 0.2% [vol/vol] glycerol), was held for 5 min at a denaturation temperature of 95°C, followed by 35 cycles of 30 s each at a denaturation temperature of 95°C, an annealing temperature of 60°C, and an elongation temperature of 72°C and by 5 min at 72°C.
TABLE 1.
Details of primers and probe system used in PCR-based assays for H. pylori detection and mutation identification
| Primer or probe | Target (reference) | Primer or probe sequence (5′–3′) |
|---|---|---|
| Conventional PCR | ||
| VAC3624F | vacA | GAG CGA GCT ATG GTT ATG AC |
| VAC3853R | vacA | ACT CCA GCA TTC ATA TAG A |
| Hp1 | 16S rRNA (16) | CTG GAG AGA CTA AGC CCT CC |
| Hp2 | 16S rRNA (16) | ATT ACT GAC GCT GAT TGT GC |
| LightCycler PCR | ||
| 23F | 23S rRNA (14) | CAA CCA GAG ATT CAG TGA AA |
| 23R | 23S rRNA (14) | GTG CTA AGT TGT AGT AAA GGT |
| 23Pr | 23S rRNA (14) | Cy5-GGC AAG ACG GAA AGA CCC-biotin |
A novel PCR assay to detect the vacA gene was developed using primers designed from in silico analysis of the 30 complete sequences of vacA currently deposited in GenBank. All alignments were performed with GeneBase, version 1 (Applied Maths, Kortrijk, Belgium). A 229-bp fragment of vacA at the 3′ end of the gene was amplified with the primer pair VAC3624F and VAC3853R (Table 1) in a 50-μl reaction mixture containing PCR buffer, 2.0 mM MgCl2, 200 μM (each) dNTP, 0.3 μM (each) primer, 1 U of Taq polymerase (Gibco BRL), and 5 μl of extracted DNA. Following denaturation at 95°C (5 min), the vacA fragment was amplified through 35 cycles as follows: 95, 53, and 72°C for 30 s each; extension was continued at 72°C for 5 min. Aliquots of each PCR product were electrophoresed in a 1% (wt/vol) agarose gel (Ultra Pure; Gibco BRL) in Tris-borate-EDTA buffer (90 mM Tris-HCl, 90 mM boric acid, 0.002 M EDTA), and stained in ethidium bromide at 0.5 μg/ml.
Real-time PCR assay and probe melting point hybridization analysis.
Point mutations in the 23S rRNA gene associated with the acquisition of clarithromycin resistance were detected using a modification of the previously described assay developed for the LightCycler (BioGene Ltd., Kimbolton, England) (7). The reaction mixture was prepared as follows: 200 μM (each) dNTP, 0.4 U of platinum Taq polymerase (Gibco BRL), Idaho Technology buffer (50 mM Tris-HCl [pH 8.3], 2 mM MgCl2) (Biogene) adjusted to give a final MgCl2 concentration of 3 mM, SYBR Green 1 (BioGene) diluted 1/10,000, 5 pmol of each primer (Table 1), and 5 pmol of probe (Table 1). DNA (1 μl) was added to a 9-μl reaction mixture, and the PCR was performed as described previously (7), but with the modification that the amplification stage was increased to include 75 cycles.
RESULTS
Detection of H. pylori directly in gastric biopsy samples.
Our results showed that the majority (36 of 39) of the DNA samples derived from stored gastric biopsy samples (set 1) from confirmed H. pylori-positive patients were positive by the diagnostic PCR assays for both the 16S rrn and the vacA targets (Table 2). Both assays were of equal sensitivity (92.3%) in this biopsy set. DNA samples obtained from biopsy samples received during the prospective study (set 2) showed that 17 (14.0%) of the 121 antral gastric biopsy samples were confirmed both culture positive and histology positive for H. pylori. The conventional PCR assay targeting vacA amplified the specific product of 229 bp in 15 of 17 (88.2%) of the culture-positive biopsy samples, as well in one sample from a patient who was culture negative. The 16S rRNA PCR assay confirmed the presence of H. pylori in 14 of 17 (82.3%) of these biopsy samples, although three culture-negative biopsy samples also generated the expected PCR product of 109 bp. The sensitivities of the vacA and 16S rRNA assays were 89.5, and 85.0%, respectively, and their specificities were 99.0 and 98.1%, respectively (Table 2).
TABLE 2.
Comparison of performances of PCR-based detection assays targeting the H. pylori vacA and 16S rRNA genes with culture from the two sets of gastric biopsy samples
| Culture result | No. of samples with indicated result in biopsy set:
|
|||||||
|---|---|---|---|---|---|---|---|---|
| 1a(n = 39) by test for:
|
2b(n = 121) by test for:
|
|||||||
|
vacA
|
16S rRNA
|
vacA
|
16S rRNA
|
|||||
| + | − | + | − | + | − | + | − | |
| + | 36 | 3 | 36 | 3 | 15 | 2 | 14 | 3 |
| − | nac | na | na | na | 1d | 103 | 3d | 101 |
H. pylori-positive status confirmed prior to analyses.
H. pylori status unknown at time of analyses.
na, not applicable.
One culture-negative biopsy sample was positive for both PCR assays.
Direct detection of mutations conferring clarithromycin resistance.
The LightCycler PCR assay to detect point mutations in the 23S rRNA gene was applied to determine clarithromycin susceptibilities on DNA from 142 gastric biopsy samples from both sets of patients, comprising 56 culture-positive and 86 culture-negative biopsy samples. Analysis of DNA from 48 of the 56 culture-positive biopsy samples and from one culture-negative biopsy sample produced melting peaks characteristic of a clarithromycin-sensitive (wild-type) genotype (Table 3). Repeat PCR analyses were necessary on five of these samples to generate sufficient product to allow mutation detection. Antibiotic susceptibility testing by disk diffusion of matched cultures of H. pylori confirmed that strains isolated from 47 of 48 of these biopsy samples were clarithromycin sensitive, while one was a mixed population of clarithromycin-sensitive and -resistant phenotypes. DNA from three biopsy samples generated melting peaks indicative of a clarithromycin-resistant phenotype with an A→G mutation (Table 3). Disk diffusion results on the two available matched cultures of H. pylori confirmed that one of these was clarithromycin resistant. The other was sensitive to clarithromycin, which suggested that the in vivo infection might be a mixture of both clarithromycin-resistant and -sensitive forms even though only the latter form was isolated on culture. The remaining culture-positive biopsy sample produced a melting peak indicative of clarithromycin resistance due to an A→C mutation. Disk diffusion testing of the matched culture confirmed clarithromycin resistance, and the LightCycler assay when applied to this culture generated a melting peak (A→C mutation) identical to that of the corresponding biopsy sample. Finally, LightCycler PCR was not successful for the four remaining culture-positive biopsy specimens (PCR negative), and therefore 23S rRNA genotypes could not be determined (Table 3).
TABLE 3.
Performance of PCR-based probe melting point hybridization assay to determine clarithromycin susceptibility of H. pylori directly from 142 gastric biopsy samples compared with that of the culture-based disk diffusion test
| Result of 23S rRNA mutation assay | No. of samples having indicated:
|
|||
|---|---|---|---|---|
| Biopsy culture status
|
Clarithromycin susceptibilitya
|
|||
| + (n = 56) | − (n = 86) | Sensitive | Resistant | |
| Wild type | 48 | 1 | 47 | 1b |
| A→G | 3 | 0 | 1 | 1c |
| A→C | 1 | 0 | 0 | 1 |
| PCR negative | 4 | 85 | 3 | 1 |
Determined by disk diffusion test on matched cultures (n = 55) from positive biopsy samples.
Mixed population of clarithromycin-sensitive and -resistant variants and strains.
One matched culture not available.
DISCUSSION
The results of the study showed that H. pylori DNA could be detected by PCR directly in human gastric biopsy samples after frozen storage with high specificity and sensitivity. Our approach was to use two target genes for detection to reduce uncertainties associated with use of a single target. The 16S rrn primers have been applied extensively to a range of clinical samples and are established as sensitive and specific (6, 9, 11). But as such DNA sequences encoding rRNA are highly ubiquitous, there is a possible risk of nonspecific products, particularly when analyzing DNA extracted from mammalian tissue (3). We therefore developed a novel PCR assay that targeted the vacA gene, which is a species-specific and highly conserved locus in H. pylori (1, 4). Although vacA has a mosaic structure and some regions such as the midregion are highly diverse, the assay was designed to target a conserved region based on in silico comparisons of vacA sequences in GenBank.
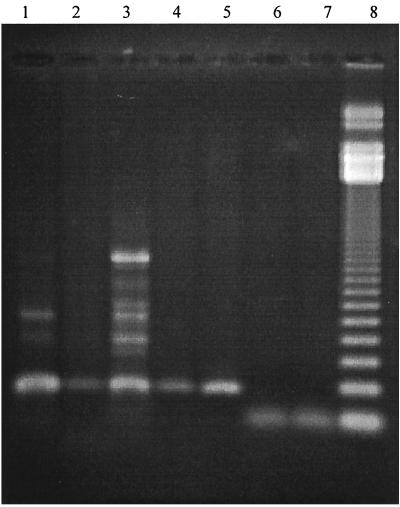
The performance of both diagnostic PCR assays in relation to culture and histology was established in an analysis of 39 known culture-positive biopsy samples, and we found that H. pylori could be detected in both assays with comparably high sensitivity (92.3%). Further analyses based on 121 biopsy samples (comprising culture-positive and -negative samples) demonstrated that both assays were highly specific, with the vacA assay giving a specificity of 99.0% compared with 98.1% for the 16S rrn assay. There were minor differences between the sensitivity results for the two sets of biopsy samples, with the vacA assay being the most sensitive (89.5 versus 85.0%). The exact reason for the lower sensitivities observed is unclear although for this set of biopsy samples culture was performed on one biopsy sample at the primary diagnostic laboratory and a second biopsy sample was frozen and subsequently transported to our laboratory for molecular analyses. Reduced sensitivities could have been due to differences in transport conditions that occurred with some specimens. We have evidence (data not shown) suggesting that H. pylori DNA in gastric biopsy samples could be rapidly degraded if transport conditions were suboptimal due to delays before freezing, repeated freezing and thawing, and use of an inappropriate suspending medium. Alternatively H. pylori colonization may not be uniformly distributed across the gastric mucosa and thus biopsy samples taken from different sites may occasionally give conflicting results. Overall, however, the new vacA PCR assay generally performed better than the 16S rrn assay. Nonspecific bands were amplified occasionally from some gastric biopsy DNA extracts (Fig. 1), but these were significantly larger than the expected specific 229-bp product of the vacA assay, and therefore reporting a false-positive result was unlikely. Generation of nonspecific DNA bands could theoretically reduce the sensitivity of the vacA assay due to competitive coamplification of specific and nonspecific products. However, this did not appear to be a problem, as strong specific signals were observed regardless of any nonspecific amplification. The exact origin of these bands is not known. They were generated occasionally in both culture-positive and culture-negative biopsy samples but not from DNA extracted from a pure culture of the infecting strain of H. pylori (data not shown). It is therefore most likely that human DNA was amplified. A PCR-based approach to diagnostic testing has the advantage over existing non-culture-based tests in that it is simple to perform and can provide additional genotypic information about the infecting strain, including markers associated with antibiotic susceptibilities. In addition, although the initial cost of equipment is high, the cost of reagents and consumables for each test is extremely low in comparison with corresponding costs for other methods such as the urea breath test and stool antigen test.
FIG. 1.
Examples to illustrate the quality of vacA and 16S rRNA PCR products from H. pylori culture-positive biopsy samples. Shown are PCR products generated by assays targeting vacA (lanes 1 to 5) and 16S rrn (lanes 6 and 7). Lane 8, 123-bp molecular weight marker.
Clarithromycin is a key component of most current triple-therapy regimens for treatment of H. pylori infection; however, resistance can dramatically decrease the chances of successful eradication. The identification of specific point mutations (namely, A2143G, A2144G, or A2143C) in the 23S rRNA gene (16, 21, 22, 25) has enabled development of molecular tests that allow determination of clarithromycin resistance directly from biopsies (2, 10, 12, 13, 20). The main limitations are that most assays could not detect the A2143C mutation (2, 13, 20) and some were of low sensitivity (20), while others required multiple PCRs (10, 12, 23). The LightCycler assay we describe is simpler and more rapid. All three common mutations were detectable directly from a gastric biopsy sample in a single reaction tube in under 1 h, so avoiding the associated delay of culture-based susceptibility testing. Most assays provided information on the clarithromycin susceptibility of the infecting strain that corresponded accurately with culture-based disk diffusion results. Nevertheless, DNA from one biopsy sample generated a melting peak suggesting an A→G mutation while the matched culture was clarithromycin sensitive. That was possibly indicative of a mixed infection, as a similar discrepancy between mutation detection and phenotypic susceptibility testing was reported previously (13). Three culture-positive biopsy samples that were also positive by the diagnostic PCR assays were negative for the clarithromycin susceptibility assay. The precise reason for this was unclear, but the sensitivity may have been affected by sampling error, as the target copy number contained in a volume as small as 1 μl of DNA could be extremely low in some biopsy samples, particularly if degradation of specific DNA has indeed occurred. Sensitivity may be improved by modification of the assay to a nested-PCR format. Alternatively, given that the method of DNA extraction was extremely simple, we speculate that the differences in sensitivity may be due to the presence of low levels of substances inhibitory to the PCR, which may affect the efficiency of the LightCycler assay to a greater extent than that of the conventional PCR assays.
In conclusion, application of our PCR-based approach not only allowed same-day diagnosis of H. pylori infection but also supplied further strain information concerning clarithromycin susceptibility. Rapid provision of such information could have a significant impact on patient management in terms of turnaround times, appropriate antibiotic prescription, and ultimately treatment outcome.
REFERENCES
- 1.Atherton J C, Cao P, Peek R M, Jr, Tummuru M K, Blaser M J, Cover T L. Mosaicism in vacuolating cytotoxin alleles of Helicobacter pylori. Association of specific vacA types with cytotoxin production and peptic ulceration. J Biol Chem. 1995;270:17771–17777. doi: 10.1074/jbc.270.30.17771. [DOI] [PubMed] [Google Scholar]
- 2.Bjorkholm B, Befrits R, Jaup B, Engstrand L. Rapid PCR detection of Helicobacter pylori-associated virulence and resistance genes directly from gastric biopsy material. J Clin Microbiol. 1998;36:3689–3690. doi: 10.1128/jcm.36.12.3689-3690.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Chong S K, Lou Q, Fitzgerald J F, Lee C H. Evaluation of 16S rRNA gene PCR with primers Hp1 and Hp2 for detection of Helicobacter pylori. J Clin Microbiol. 1996;34:2728–2730. doi: 10.1128/jcm.34.11.2728-2730.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Cover T L. The vacuolating cytotoxin of Helicobacter pylori. Mol Microbiol. 1996;20:241–246. doi: 10.1111/j.1365-2958.1996.tb02612.x. [DOI] [PubMed] [Google Scholar]
- 5.de Boer W A, Tytgat G N. Regular review: treatment of Helicobacter pylori infection. BMJ. 2000;320:31–34. doi: 10.1136/bmj.320.7226.31. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Dowsett S A, Archila L, Segreto V A, Gonzalez C R, Silva A, Vastola K A, Bartizek R D, Kowolik M J. Helicobacter pylori infection in indigenous families of Central America: serostatus and oral and fingernail carriage. J Clin Microbiol. 1999;37:2456–2460. doi: 10.1128/jcm.37.8.2456-2460.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Gibson J R, Saunders N A, Burke B, Owen R J. Novel method for rapid determination of clarithromycin sensitivity in Helicobacter pylori. J Clin Microbiol. 1999;37:3746–3748. doi: 10.1128/jcm.37.11.3746-3748.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Goodwin C S. Antimicrobial treatment of Helicobacter pylori infection. Clin Infect Dis. 1997;25:1023–1026. doi: 10.1086/516078. [DOI] [PubMed] [Google Scholar]
- 9.Ho S A, Hoyle J A, Lewis F A, Secker A D, Cross D, Mapstone N P, Dixon M F, Wyatt J I, Tompkins D S, Taylor G R. Direct polymerase chain reaction test for detection of Helicobacter pylori in humans and animals. J Clin Microbiol. 1991;29:2543–2549. doi: 10.1128/jcm.29.11.2543-2549.1991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Maeda S, Yoshida H, Matsunaga H, Ogura K, Kawamata O, Shiratori Y, Omata M. Detection of clarithromycin-resistant Helicobacter pylori strains by a preferential homoduplex formation assay. J Clin Microbiol. 2000;38:210–214. doi: 10.1128/jcm.38.1.210-214.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Mapstone N P, Lynch D A, Lewis F A, Axon A T, Tompkins D S, Dixon M F, Quirke P. Identification of Helicobacter pylori DNA in the mouths and stomachs of patients with gastritis using PCR. J Clin Pathol. 1993;46:540–543. doi: 10.1136/jcp.46.6.540. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Marais A, Monteiro L, Occhialini A, Pina M, Lamouliatte H, Megraud F. Direct detection of Helicobacter pylori resistance to macrolides by a polymerase chain reaction/DNA enzyme immunoassay in gastric biopsy specimens. Gut. 1999;44:463–467. doi: 10.1136/gut.44.4.463. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Matsuoka M, Yoshida Y, Hayakawa K, Fukuchi S, Sugano K. Simultaneous colonisation of Helicobacter pylori with and without mutations in the 23S rRNA gene in patients with no history of clarithromycin exposure. Gut. 1999;45:503–507. doi: 10.1136/gut.45.4.503. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Megraud F, Doermann H P. Clinical relevance of resistant strains of Helicobacter pylori: a review of current data. Gut. 1998;43(Suppl. 1):S61–S65. doi: 10.1136/gut.43.2008.s61. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.NIH Consensus Development Panel on Helicobacter pylori in Peptic Ulcer Disease. Helicobacter pylori in peptic ulcer disease. JAMA. 1994;272:65–69. [PubMed] [Google Scholar]
- 16.Occhialini A, Urdaci M, Doucet-Populaire F, Bebear C M, Lamouliatte H, Megraud F. Macrolide resistance in Helicobacter pylori: rapid detection of point mutations and assays of macrolide binding to ribosomes. Antimicrob Agents Chemother. 1997;41:2724–2728. doi: 10.1128/aac.41.12.2724. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Parsonnet J. Gastric adenocarcinoma and Helicobacter pylori infection. West J Med. 1994;161:60. [PMC free article] [PubMed] [Google Scholar]
- 18.Parsonnet J, Hansen S, Rodriguez L, Gelb A B, Warnke R A, Jellum E, Orentreich N, Vogelman J H, Friedman G D. Helicobacter pylori infection and gastric lymphoma. N Engl J Med. 1994;330:1267–1271. doi: 10.1056/NEJM199405053301803. [DOI] [PubMed] [Google Scholar]
- 19.Peters T M, Owen R J, Teare E L, Goodbourn C. Detection of Helicobacter pylori by PCR on culture- and histology-negative gastric biopsies from seropositive patients. PHLS Microbiol Dig. 1997;14:227–229. [Google Scholar]
- 20.Sevin E, Lamarque D, Delchier J C, Soussy C J, Tankovic J. Co-detection of Helicobacter pylori and of its resistance to clarithromycin by PCR. Microbiol Lett. 1998;165:369–372. doi: 10.1111/j.1574-6968.1998.tb13172.x. [DOI] [PubMed] [Google Scholar]
- 21.Stone G G, Shortridge D, Versalovic J, Beyer J, Flamm R K, Graham D Y, Ghoneim A T, Tanaka S K. A PCR-oligonucleotide ligation assay to determine the prevalence of 23S rRNA gene mutations in clarithromycin-resistant Helicobacter pylori. Antimicrob Agents Chemother. 1997;41:712–714. doi: 10.1128/aac.41.3.712. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Taylor D E, Ge Z, Purych D, Lo T, Hiratsuka K. Cloning and sequence analysis of two copies of a 23S rRNA gene from Helicobacter pylori and association of clarithromycin resistance with 23S rRNA mutations. Antimicrob Agents Chemother. 1997;41:2621–2628. doi: 10.1128/aac.41.12.2621. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Trebesius K, Panthel K, Strobel S, Vogt K, Faller G, Kirchner T, Kist M, Heesemann J, Haas R. Rapid and specific detection of Helicobacter pylori macrolide resistance in gastric tissue by fluorescent in situ hybridisation. Gut. 2000;46:608–614. doi: 10.1136/gut.46.5.608. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Tytgat G N. Antimicrobial therapy for Helicobacter pylori infection. Helicobacter. 1997;2(Suppl. 1):S81–S88. doi: 10.1111/j.1523-5378.1997.06b01.x. [DOI] [PubMed] [Google Scholar]
- 25.Versalovic J, Shortridge D, Kibler K, Griffy M V, Beyer J, Flamm R K, Tanaka S K, Graham D Y, Go M F. Mutations in 23S rRNA are associated with clarithromycin resistance in Helicobacter pylori. Antimicrob Agents Chemother. 1996;40:477–480. doi: 10.1128/aac.40.2.477. [DOI] [PMC free article] [PubMed] [Google Scholar]