ABSTRACT
The host restriction factor APOBEC3G (A3G) inhibits an extensive variety of viruses, including retroviruses, DNA viruses, and RNA viruses. Our study shows that A3G inhibits enterovirus 71 (EV71) and coxsackievirus A16 (CA16) via competitively binding the 5′ untranslated region (UTR) with the host protein poly(C)-binding protein 1 (PCBP1), which is required for the replication of multiple EVs. However, whether A3G inhibits other EVs in addition to EV71 and CA16 has not been investigated. Here, we demonstrate that A3G could inhibit the replication of EVD68, which requires PCBP1 for its replication, but not CA6, which does not require PCBP1 for replication. Further investigation revealed that the nucleic-acid-binding activity of A3G is required for EVD68 restriction, similar to the mechanism presented for EV71 restriction. Mechanistically, A3G competitively binds to the cloverleaf (1 to 123 nucleotides [nt]) and the stem-loop IV (234 to 446 nt) domains of the EVD68 5′ UTR with PCBP1, thereby inhibiting the 5′ UTR activity of EVD68; by contrast, A3G does not interact with CA6 5′ UTR, resulting in no effect on CA6 replication. Moreover, the nonstructural protein 2C, encoded by EVD68, overcomes A3G suppression by inducing A3G degradation via the autophagy-lysosome pathway. Our findings revealed that A3G might have broad-spectrum antiviral activity against multiple EVs through this general mechanism, and they might provide important information for the development of an anti-EV strategy.
IMPORTANCE As the two major pathogens causing hand, foot, and mouth disease (HFMD), enterovirus 71 (EV71) and coxsackievirus A16 (CA16) attract a lot of attention for the study of their pathogenesis, their involvement with cellular proteins, and so on. However, other EVs such as CA6 and EVD68 constantly occur sporadically or have spread worldwide in recent years. Therefore, more information related to these EVs is needed in order to develop a broad-spectrum anti-EV inhibitor. In this study, we first reveal that the protein poly(C)-binding protein 1 (PCBP1), involved in PV- and EV71 virus replication, is also required for the replication of EVD68, but not for the replication of CA6. Next, we found that the host-restriction factor A3G specifically inhibits the replication of EVD68, but not the replication of CA6, by competitively binding to the 5′ UTR of EVD68 along with PCBP1. Our findings broaden knowledge related to EV replication and the interplay between EVs and host factors.
KEYWORDS: A3G, EVD68, CA6, inhibition, PCBP1
INTRODUCTION
The Enterovirus genus of the Picornaviridae family contains 13 species, of which 7 infect humans and 6 are animal pathogens. The human pathogens are composed of enteroviruses A to D and rhinoviruses A to C. Enteroviruses A to D contain more than 100 subtypes, including EV71, enterovirus D68 (EVD68), coxsackieviruses A and B, and poliovirus (PV) (1). EV71 and CA16 are the two major pathogens responsible for hand, foot, and mouth disease (HFMD) in children under 5 years old, which has become a severe public health problem. However, other EVs, such as CA6 and CA10, constantly occur sporadically in some provinces of China (2–5). EVD68 was initially isolated in 1962 from children with respiratory infections (California, USA). In recent years, an unprecedented epidemic of respiratory disease, temporally associated with acute flaccid myelitis (AFM), was thought to have been caused by EVD68, which is different from historical reports on EVD68 (6–8). In China, EVD68 is thought to have spread widely in recent years based on neutralizing antibody (NAb) detection (9).
EVs are single-stranded positive RNA molecules of 7,000 to 8,000 nucleotides, with a single open reading frame (ORF) encoding a polyprotein that is flanked by untranslated regions (UTR) at the 5′ and 3′ ends. An EV 5′ UTR contains an RNA structure formed by the cloverleaf and the internal ribosome entry site (IRES), which is important for viral RNA synthesis and translation. The poly(rC)-binding protein 2 (PCBP2) was originally identified as a host factor that binds the IRES on the PV genome (10). Subsequent studies revealed that PCBP2 and its paralog PCBP1 bind the cloverleaf-like (stem-loop I) domain of the PV genome for viral replication, and the IRES region of the PV genome for viral translation, respectively, with different affinities (11–13). Moreover, these PCBPs are also involved in the translation of other EVs, such as CVB3 (14, 15). In summary, PCBP2 and PCBP1 are host factors for the initiation of both EV translation and RNA synthesis, either by binding to the stem-loop IV of the IRES or by binding to the cloverleaf in concert with viral 3CD, respectively (16–19). Other host proteins, such as hnRNP A1, hnRNP K, far-upstream element-binding protein (FBP), and others have been found to interact with the 5′ UTR of EVs, enhancing viral translation and viral replication (20–24).
Several intrinsic host factors, such as APOBEC3G (A3G), SAMHD1, and Mx2, were discovered one after another and identified as inhibitors of HIV infection (25). A3G has been extensively investigated and found to possess broad-spectrum antiviral activity against retroviruses, DNA viruses, single-stranded RNA viruses, and endogenous retro-elements (26–30). Our recent study showed that A3G also suppresses EV71 replication (31). Classical inhibition by A3G is mediated by its cytidine-deaminase activity, which deaminates viral cDNA cytidines to uridines, thereby affecting viral reverse transcription or integration (32–34). In addition, the nucleic-acid-binding activity of A3G also contributes its viral restriction (35, 36). We have previously shown that A3G is independent of its cytidine-deaminase activity, and dependent on its nucleic-acid-binding activity, to inhibit EV71 (31). Viruses have developed sophisticated strategies to escape restriction, such as using its encoded proteins or its host’s proteins to replicate efficiently (25, 31, 37–40). The ubiquitin-proteasome pathway is extensively employed by HIV proteins such as Vif and Vpx to antagonize the restriction of APOBEC3 family members and SAMHD1 (39–43), while the autophagy-lysosome pathway is utilized by viral proteins such as HIV-1 Vpu and EV71 2C to degrade the host restrictive factors BST-2 and A3G (31, 44, 45). Although A3G is an inhibitor of EV71, whether it inhibits other EVs such EVD68 or CA6 has not been investigated.
In this study, we demonstrate that not all EVs require PCBP1 to facilitate viral translation. A3G could potently inhibit the replication of EVD68, which needs PCBP1 for replication, but it has no effect on CA6, which does not need PCBP1 for replication. Mechanistically, the nucleic-acid-binding characteristic of A3G is essential for inhibiting EVD68; together with PCBP1, it competitively binds to the EVD68 5′ UTR, which is important for viral RNA synthetic and translational activities. Similar to EV71, EVD68 utilizes 2C protein to induce A3G degradation through an autophagy-lysosome pathway, thereby blocking A3G restriction. Our study provides an attractive target for the development of a novel anti-EV D68 inhibitor.
RESULTS
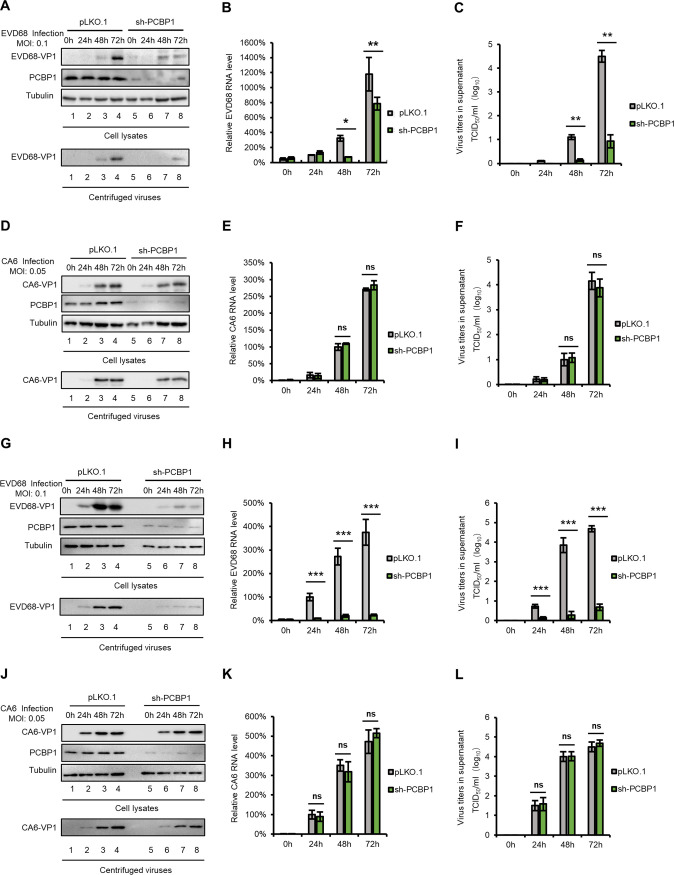
PCBP1 is required for EVD68 replication but not for CA6 replication.
Previous studies have reported that picornavirus utilizes some host factors to finish an infectious cycle. For instance, the cellular proteins PCBP1 and PCBP2 are required for the replication of PV, EV71, CA16, and CVB3 (11, 12, 15, 31, 46–48), interacting with the 5′ UTR to enhance viral RNA synthesis or viral translation. To investigate whether PCBP1 is required for the replication of other EVs, we tested the replication of EVD68 and CA6 in PCBP1-silencing HEK293T cells and in negative-control cells; these were infected by EVD68 and CA6 at a multiplicity of infection (MOI) of 0.1 and 0.05, respectively. VP1 protein expression levels (Fig. 1A and D), mRNA levels (Fig. 1B and E), and viral titers in supernatants (Fig. 1C and F) were detected by immunoblotting (IB), real-time quantitative PCR (RT-qPCR) and the cytopathic effect (CPE) method, respectively. The results showed that EVD68, but not CA6, had reduced replication in PCBP1-silencing cells. Similar results were obtained in another cell line, RD (Fig. 1G to L), which was sensitive to EV infection.
FIG 1.
PCBP1 is required for EVD68 replication but not for CA6 replication. PCBP1-silencing cell line or pLKO.1 negative-control cell line were constructed in HEK293T cells using sh-PCBP1 or a pLKO.1 vector, then infected with EVD68 (A to C) and CA6 (D to F) at a MOIs of 0.1 and 0.05, respectively. The cells and culture supernatants were harvested at the indicated time points. Immunoblotting (IB) analysis of EVD68 (A) or CA6 (D) VP1 in cells was performed, with tubulin as a loading control. The VP1 protein in the supernatants was detected after ultracentrifugation. EVD68 (B) or CA6 (E) viral RNA levels in cell lysates was detected by RT-qPCR and normalized to GAPDH. Then, the EVD68-RNA levels in 293T cells at 24 h postinfection and the CA6-RNA levels in 293T cells at 48 h postinfection were set as 100%. Viral titers of EVD68 (C) or CA6 (F) in the supernatants were measured by the cytopathic effect (CPE) method. Similar experiments were performed in another cell line, RD (G to L). Mean values are presented from three independent repeats (±SD, *, P < 0.05, **, P < 0.01, ***, P < 0.001, ns stands for no significance, paired t test).
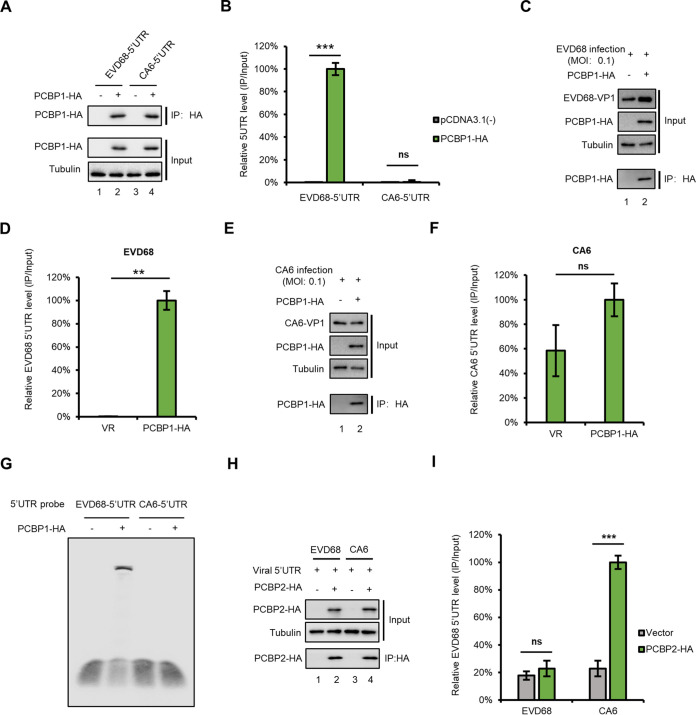
PCBP1 specifically binds with EVD68 5′ UTR but not with CA6 5′ UTR.
To examine whether PCBP1 interacts with either the EVD68 5′ UTR or the CA6 5′ UTR, cells were transfected with PCBP1 plus either EVD68-5′-UTR- or CA6-5′-UTR expression vectors as indicated in Fig. 2A, and then immunoprecipitation (IP) and RT-qPCR assays were employed. The results showed that PCBP1 strongly interacts with EVD68 5′ UTR but not with CA6 5′ UTR (Fig. 2B); the expression of PCBP1 was confirmed by IB analysis (Fig. 2A). We also examined the binding ability of PCBP1 with viral genome by using EVD68- or CA6 virus infection. HEK293T cells were transfected with PCBP1 for 24 h, then infected with either EVD68 or CA6 virus at a MOI of 0.1. As expected, PCBP1 specifically interacted with the EVD68 5′ UTR but not with the CA6 5′ UTR (Fig. 2C to F). An electrophoretic mobility shift assay (EMSA) showed similar results (Fig. 2G). The homologue of PCBP1, PCBP2, was initially identified as a host factor that binds with the PV genome, although PCBP1 itself was not (10). Thus, we examined the binding capacities of PCBP2 with the EVD68 5′ UTR and with the CA6 5′ UTR (Fig. 2H and I). IP and RT-qPCR assays showed that the CA6 5′ UTR, but not the EVD68 5′ UTR, specifically interacts with PCBP2, indicating that the 5′ UTR of different EVs have different affinities with PCBPs. These results suggested that various PCBPs are selectively required for the replication of different EVs.
FIG 2.
PCBP1 specifically interacts with the EVD68 5′ UTR but not with the CA6 5′ UTR. (A and B) Interaction between PCBP1 with transfected 5′ UTR expression vector. PCBP1 or the control vector was cotransfected with either EVD68 5′ UTR or CA6 5′ UTR, respectively; cells were then harvested, and cell lysates were immunoprecipitated with anti-HA agarose beads. Elutions were analyzed by IB (A) and RT-qPCR (B), respectively. The eluted EVD68 5′ UTR by PCBP1 was set as 100%. (C to F) Interaction between PCBP1 and viral 5′ UTR in the context of a viral infection. HEK293T cells were transfected with PCBP1-HA for 24 h, then infected with EVD68 (C and D) and CA6 (E and F) at a MOI of 0.1. Cells were harvested and treated as in panels A and B. (G) Interaction between PCBP1 and viral 5′ UTR by EMSA. RNA of the EVD68 5′ UTR and the CA6 5′ UTR was transcribed by a MEGAscript T7 kit. PCBP1 was purified from HEK293T transfected with PCBP1-HA using HA-beads. An EMSA of PCBP1 and either EVD68 5′ UTR or CA6 5′ UTR was then performed using a LightShift Chemiluminescent EMSA kit. (H and I) Interaction between PCBP2 and either the EVD68 5′ UTR or the CA6 5′ UTR in the context of a viral infection. HEK293T cells were transfected with PCBP2-HA for 24 h and then infected with either EVD68 or CA6, respectively. Cells were harvested and treated as in panels A and B. (D, F, I) Mean values are shown as repeats (±SD, n = 3 *, P < 0.05, **, P < 0.01, ***, P < 0.001, ns stands for no significance, paired t test) in the presence of PCBPs relative those obtained in the presence of the control vector (100%).
A3G restricts EVD68 replication, but not CA6 replication, through interfering with 5′ UTR activity.
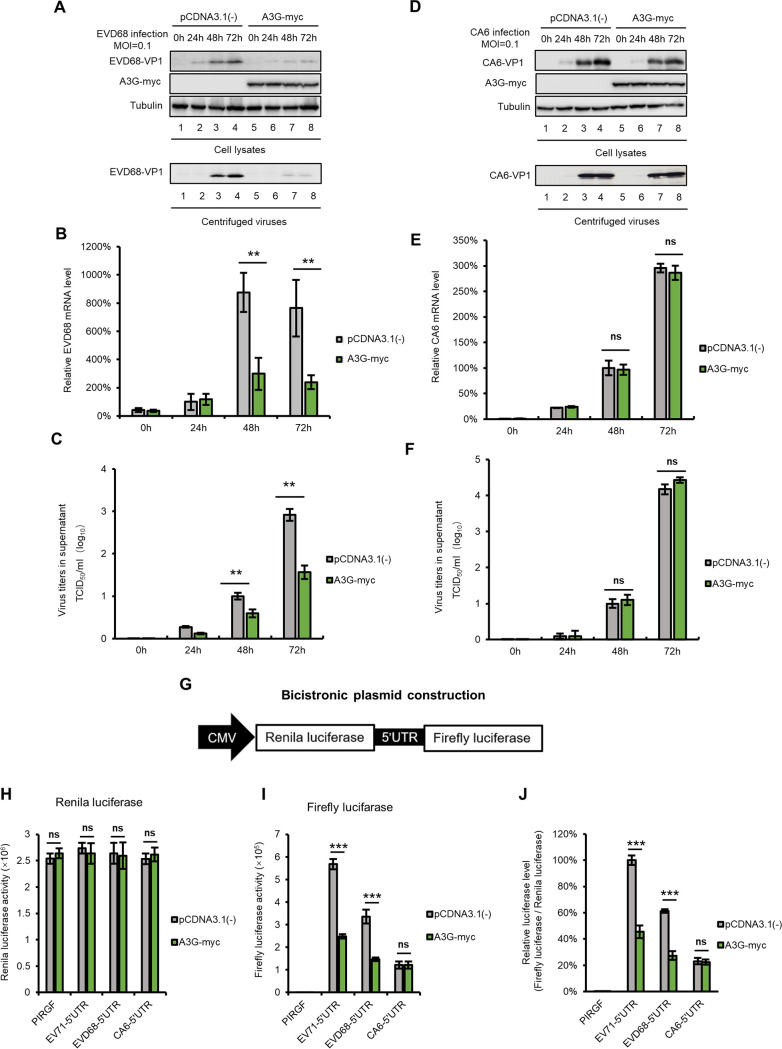
Having proven that A3G inhibits the replication of EV71 and CA16 depending on PCBP1 (31), we next tested whether A3G inhibits the replication of EVD68 and CA6. We transfected either the negative-control expression vector VR1012, or A3G, into HEK293T cells for 24 h, after which the cells were infected with either EVD68 or CA6 at a MOI of 0.1. We found that overexpressing A3G in HEK293T cells restricted EVD68 replication, but not CA6 replication, as measured by VP1-IB-viral-mRNA analyses in intracellular and viral titers in the supernatant at 24 h, 48 h, and 72 h postinfection (Fig. 3A to F).
FIG 3.
A3G suppresses EVD68 replication but not CA6 replication. HEK293T cells transfected with either A3G-myc or the control vector were infected with EVD68 (A to C) and CA6 (D to F) at a MOI of 0.1. Cells and culture supernatants were harvested at the indicated time points. IB analysis of EVD68 (A) or CA6 (D) VP1 in cells was performed, with tubulin as a loading control. The VP1 protein in the supernatants was detected after ultracentrifugation. EVD68 (B) or CA6 (E) viral RNA levels in cell lysates were detected by RT-qPCR and normalized to GAPDH; EVD68-RNA levels in 293T cells at 24h postinfection, and CA6-RNA levels in 293T cells at 48h postinfection, were set as 100%. Viral titers of EVD68 (C) or CA6 (F) in the supernatants were measured by CPE method. (G) Bicistronic plasmid construction. (H and I) A3G-myc or the control vector was cotransfected with either EV71-5′ UTR-luc, EVD68-5′ UTR-luc, or CA6-5′ UTR-luc, into HEK293T cells. Cells were harvested at 48h posttransfection and tested using a Dual-Luciferase Reporter Assay System. Raw data for Renilla are shown in panel H, and the relative luciferase activity was normalized to Renilla (I). The activity of luciferase in cells cotransfected with EV71-5′ UTR and pCDNA3.1(–) was set as 100%. Mean data are shown as previously (±SD, n = 3, **, P < 0.01, ***, P < 0.001, ns stands for no significance, paired t test).
The picornavirus 5′ UTR contains an IRES, which is required for translation initiation. Next, we tested whether A3G inhibits 5′ UTR activity in EV68 and CA6. We constructed and employed bicistronic plasmids with a fluorescent reporter downstream on both the EVD68 5′ UTR and the CA6 5′ UTR (Fig. 3G). The raw data for Renilla luciferase and firefly luciferase are shown in Fig. 3H and I, and the relative activity of the 5′ UTR from EV71, EVD68, and CA6 are shown in Fig. 3J. The luciferase assay showed that A3G decreased the luciferase activity of the EV71 and EVD68 5′ UTR, but not the activity of the CA6 5′ UTR.
A3G restricts 5′ UTR activity by competitively binding to the EVD68 5′ UTR with PCBP1.
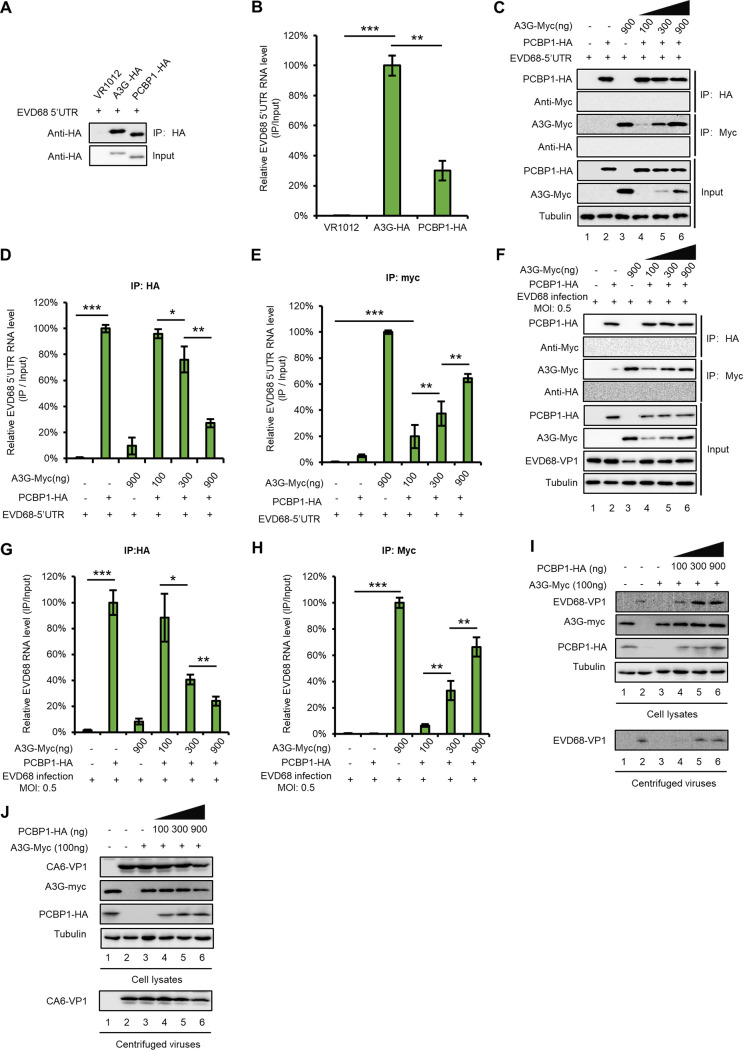
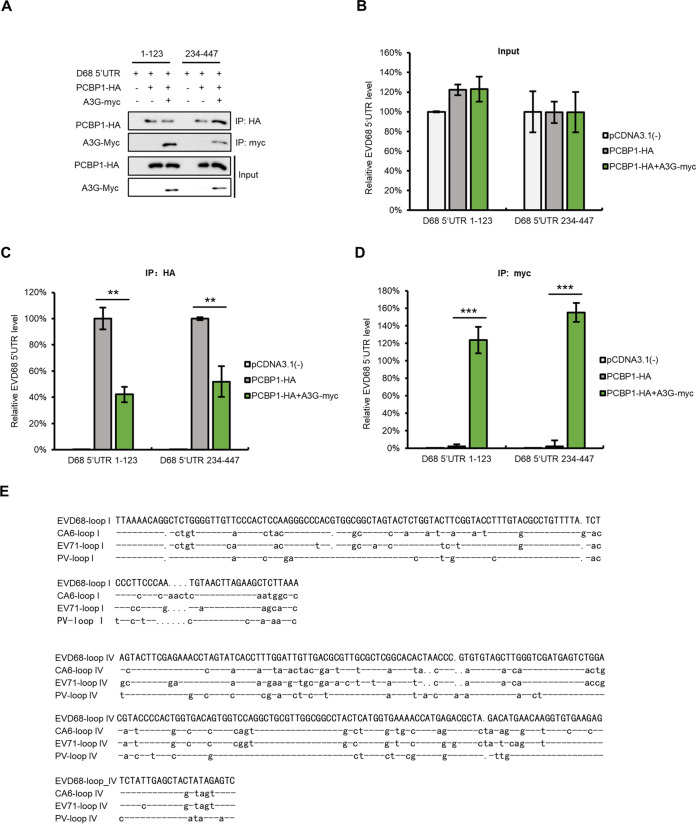
Since both PCBP1 and A3G interact with the 5′ UTR of EVD68, we further investigated and compared their respective abilities to bind to the 5′ UTR. IP and RT-qPCR assays showed that A3G has a stronger ability to bind to the EVD68 5′ UTR compared with that of PCBP1 (Fig. 4B), while the number of proteins immunoprecipitated by HA beads was almost equal (Fig. 4A). We speculated that A3G may competitively bind to the 5′ UTR of EVD68, impairing the interaction between the EVD68 5′ UTR and PCBP1. To verify this hypothesis, we cotransfected an EVD68-5′-UTR expression vector with either pcDNA3.1 or with increasing doses of A3G-myc-tagged and PCBP1-HA-tagged expression vectors into HEK293T cells as indicated in Fig. 4C. As expected, A3G-myc and PCBP1-HA were expressed efficiently and could be immunoprecipitated from cell lysates (Fig. 4C), The 5′-UTR-RNA levels for EVD68 in the cell lysates from all samples were similar (data not shown). However, the interaction of PCBP1 with the 5′ UTR of EVD68 was profoundly reduced with increasing A3G protein levels (Fig. 4D), whereas the increasing doses of A3G bound increasing amounts of EVD68 5′ UTR (Fig. 4E). As expected, the 5′ UTR did not coprecipitate with the sample in the absence of either A3G-myc or PCBP1-HA, indicating that the interaction between the 5′ UTR and the protein was specific (Fig. 4D and E). To further confirm the competitive binding capacity of A3G with PCBP1 in the context of viral infection, HEK293T cells were transfected with PCBP1-HA and increasing doses of A3G-myc for 24 h, and then infected with EVD68 at a MOI of 0.5. We also observed that an increase in A3G reduced the binding of PCBP1 with EVD68 5′ UTR (Fig. 4F to H). To confirm the relevance of A3G- and PCBP1 expression levels and their effect on viral replication, we designed an experiment to examine whether PCBP1 overexpression can rescue the inhibition of EVD68 by A3G. The results showed that even in the presence of A3G, increased doses of PCBP1 gradually restored the replication of EVD68 that was inhibited by A3G, whereas CA6 replication was not affected by either A3G- or PCBP1 expression (Fig. 4I and J). These results suggested that A3G restricted EVD68, but not CA6, by competitively interacting with the EVD68 5′ UTR, interfering with the interaction between PCBP1 and the EVD68 5′ UTR and then suppressing 5′-UTR activity.
FIG 4.
A3G competes with PCBP1 to bind to the EVD68 5′ UTR. (A and B) The binding capacity of A3G or PCBP1 with the EVD68 5′ UTR. HEK293T cells were transfected with either the control vector, A3G-HA, or PCBP1-HA, plus the EVD68 5′ UTR expression vector. At 48 h posttransfection, cells were harvested and immunoprecipitated with anti-HA agarose beads. (A) Cell lysates and immunoprecipitated products were analyzed by IB. (B) The 5′-UTR-RNA levels in the immunoprecipitated products were detected by RT-qPCR. (C to E) A3G competitively binds EVD68 5′ UTR with PCBP1 by using the 5′ UTR expression vector. HEK293T cells were transfected with increasing doses of A3G-myc and 1 μg of PCBP1-HA plus the 5′ UTR of EVD68. At 48 h posttransfection, half of the cells were harvested and immunoprecipitated with anti-HA agarose beads, and the other cells were precipitated with anti-myc agarose beads. The cell lysates and immunoprecipitated products were analyzed by IB (C). RNA levels in elution from anti-HA agarose beads (D) and anti-myc agarose beads (E) were extracted by TRIzol reagent and tested by RT-qPCR. (F to H) Competitive binding assays for A3G were performed using EVD68 virus infection. HEK293T cells transfected with the increasing doses of A3G-myc and 1 μg of PCBP1-HA were infected with EVD68 at a MOI of 0.5, then treated as in panels C to E. (I to J) Overexpression of PCBP1 rescued the replication of EVD68 but not CA6 in the presence of A3G. HEK293T cells transfected with A3G and increasing doses of PCBP1 were infected with either EVD68 or CA6. At 48 h postinfection, the cells and the supernatants were harvested for IB. Means with SD from at least three independent experiments are shown. (*, P < 0.05, **, P < 0.01, ***, P < 0.001, paired t test).
The nucleic-acid-binding activity, but not the cytidine-deaminase activity, of A3G is essential for EVD68 restriction.
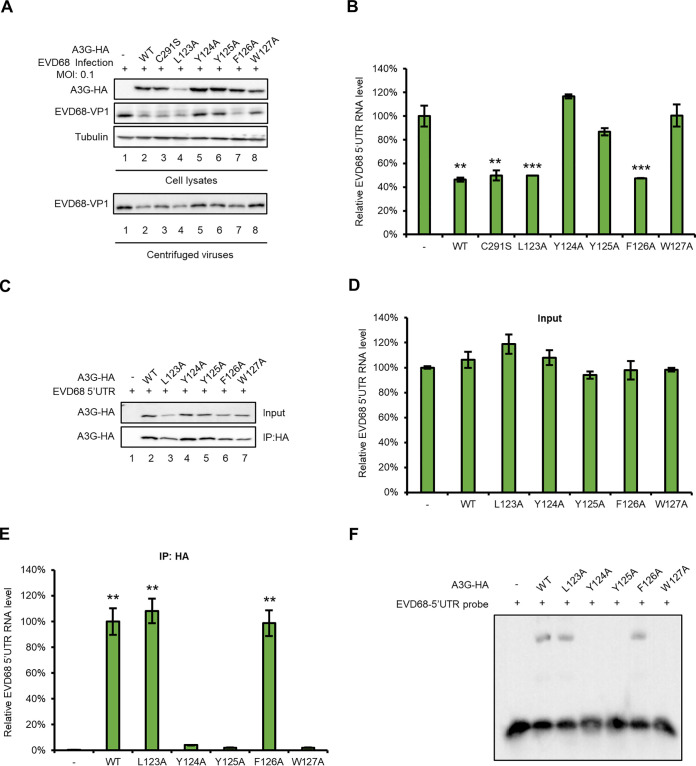
It is well known that A3G has two characteristics: the cytidine-deaminase activity mediated by the carboxy-terminal domain, and the nucleic-acid-binding property mediated by the amino-terminal domain, which is responsible for incorporating A3G into viral particles (35, 49–53). Previous studies have reported that the 123-to-127 region contributes to the A3G virion package by binding with nucleic acid (54, 55), while C291 is one of three key residues in the cytidine deaminase active site of A3G (56). To investigate whether either the cytidine-deaminase activity or the nucleic-acid-binding property is required for binding to the EVD68 5′ UTR, we tested a series of A3G mutants to examine their effect on EVD68 replication. A3G C291S with a mutation at the C terminus showed similar inhibitory effects on EVD68 replication compared to wild-type (WT) A3G (Fig. 5A, lanes 2 and 3). A3G C291S also inhibited virus production, as indicated by the supernatant IB analysis (Fig. 5A, lower panel, lanes 2 and 3); and viral RNA synthesis, as shown by using RT-qPCR to detect viral (Fig. 5B). These results showed that A3G inhibits EVD68 replication without requiring cytidine deaminase. However, we found that A3G mutants Y124A, Y125A, and W127A, located at the amino terminus of A3G, which is responsible for the nucleic-acid-binding activity of A3G, could not inhibit EVD68 replication compared to WT A3G (Fig. 5A and B). Immunoprecipitation and RNA EMSA further confirmed that these three mutants lost the ability to bind the EVD68 5′ UTR (Fig. 5C to F). These data demonstrated that the nucleic-acid-binding property of A3G is closely associated with its inhibition of EVD68.
FIG 5.
The nucleic-acid-binding activity of A3G is essential for suppressing EVD68 replication. HEK293T cells were transfected with A3G, and its mutants or the control vector were infected with EVD68, at a of MOI 0.1, respectively (A and B). Cells and culture supernatants were harvested 48 h after transfection. (A) Immunoblotting (IB) analysis of EVD68VP1 in cells was performed, with tubulin as a loading control. The VP1 protein in the supernatants was tested after ultracentrifugation. (B) RNA levels of EVD68 in cell lysates were detected by RT-qPCR and normalized to GAPDH, and the EVD68-RNA level in 293T cells transfected with the control vector was set as 100%. HEK293T cells cotransfected with EVD68 5′ UTR and A3G-myc, its mutants, or the control vector, respectively, were harvested at 48 h after transfection, and IP and RT-qPCR assays were performed (C to E). (C) A3G expression in cell lysates was tested by IB. (D) RNA levels of EVD68 5′ UTR were tested in inputs by RT-qPCR. (E) RNA levels of EVD68 5′ UTR were tested in elution by RT-qPCR. Mean values from three independent repeats are shown (±SD, **, P < 0.01, ***, P < 0.001, paired t test). (F) RNA of EVD68 5′ UTR was transcribed by a MEGAscript T7 kit. A3G-HA and its mutants were purified from HEK293T transfected with either A3G-HA or its mutants, respectively, using HA-beads. Finally, an EMSA for EVD68 5′ UTR and either A3G-HA or its mutants was performed using a LightShift Chemiluminescent EMSA kit.
A3G competes with PCBP1 to bind the cloverleaf and stem-loop IV regions of the EVD68 5′ UTR.
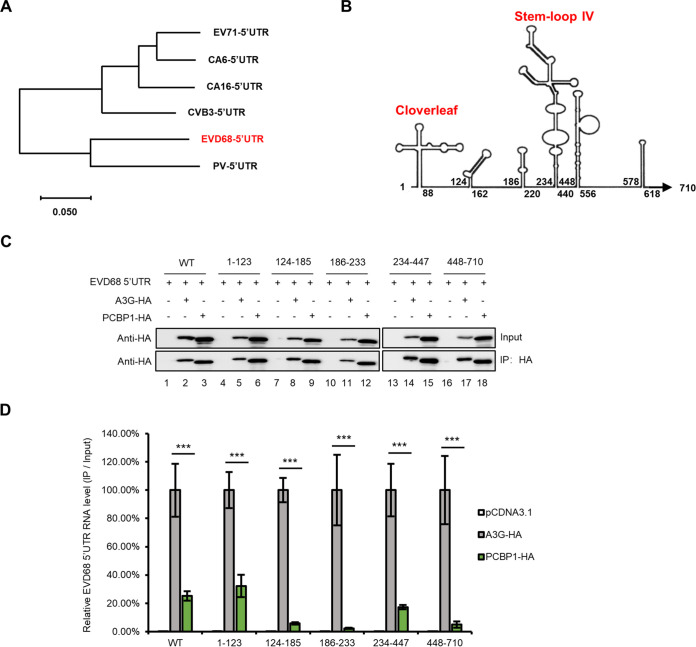
To investigate the competitive binding site of A3G on the 5′ UTR of EVD68, we aligned the sequences of 5′ UTRs of diverse EVs, including EV71, CA16, CVB3, CA6, EVD68, and PV, and analyzed the homology between them. We found that the EVD68 5′ UTR had high homology with the PV 5′ UTR (Fig. 6A). Based on the secondary structure predicted with Mfold, we simulated the secondary structure of the EVD68 5′ UTR (Fig. 6B) and constructed a series of truncated mutants. We first examined the binding abilities of A3G and PCBP1 with EVD68-5′-UTR-truncated mutants using IP and RT-qPCR, respectively (Fig. 6C and D). The results showed that PCBP1 interacts with the cloverleaf and the stem-loop IV regions of the EVD68 5′ UTR, which is consistent with previous reports (12, 46, 57), while A3G showed stronger binding ability with any region of the 5′ UTR compared to PCBP1 (Fig. 6D) (12, 46, 57). We next examined whether A3G competitively binds to the cloverleaf (1 to 123) and stem-loop IV (234 to 447) regions of the EVD68 5′ UTR with PCBP1 (Fig. 7A). We observed that PCBP1 showed reduced binding capacity with the cloverleaf and stem-loop IV regions of the EVD68 5′ UTR in the presence of A3G (Fig. 7C), while A3G showed higher binding capacity with the 5′ UTR (Fig. 7D). As expected, the amounts of proteins immunoprecipitated by HA and myc beads were almost equal (Fig. 7B). We also aligned the sequences of the cloverleaf and stem-loop IV regions of PV, EV71, EVD68, and CA6 (Fig. 7E). For the cloverleaf and stem-loop IV regions, EVD68 showed higher homology with PV (∼82% and 80%) and medium homology with EV71 (∼73% and 65%), while CA6 showed lower homology with PV (68% and 72%) and relatively high homology with EV71 (∼77% and 88%). These results suggest that not only is sequence homology determinant for 5′-UTR- and host-protein-binding, but that spatial confirmation might also contribute binding ability.
FIG 6.
The binding ability of A3G or PCBP1 with various regions of the EVD68 5′ UTR. (A) The phylogenetic tree of multiple EV 5′ UTRs was built using MEGA 7. Information on 5′ UTR sequences was described in a previous study (77) and human PV1 Mahoney (GenBank: V01149.1) was adopted for this analysis. (B) The secondary structure of the 5′ UTR was predicted by MFold. (C to D) Interaction of A3G and PCBP1 with various truncated mutants of EVD68 5′ UTR. HEK293T cotransfected with EVD68 5′ UTR truncated mutants, plus PCBP1-HA or A3G-HA as indicated, was harvested at 48 h posttransfection, and IP was performed using anti-HA agarose and RT-qPCR. Results are presented as means with SD from at least three independent experiments (±SD, **, P < 0.01, ***, P < 0.001).
FIG 7.
A3G competes with PCBP1 to bind to the cloverleaf and the stem-loop IV of EVD68 5′ UTR. (A to D) HEK293T cells cotransfected with EVD68-5′-UTR-truncated mutants, PCBP1-HA, and A3G-myc, as shown in the figure, were harvested at 48 h posttransfection, and IP and RT-qPCR assays were performed . PCBP1-HA and A3G-myc were tested by IB (A); 5′ UTR in inputs (B), and elution from anti-HA agarose beads (C) or anti-myc agarose beads (D) was extracted by TRIzol reagent and tested by RT-qPCR. Results are presented as means with SD from at least three independent experiments (±SD, **, P < 0.01, ***, P < 0.001). (E) Alignment of the cloverleaf and the stem-loop IV regions of various EVs.
2C protein of EVD68 triggers A3G degradation through the autophagy-lysosome pathway.
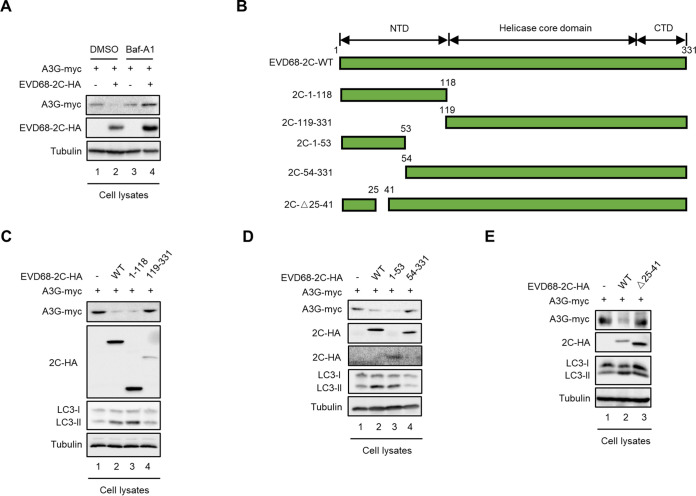
Although host restrictive factors such as A3G, SAMHD1, and BST-2 exhibit potential antiviral activity, viruses evolve diverse strategies to evade this restriction (25, 42). We had shown that EV71 antagonizes A3G inhibition using its 2C protein, via the autophagy-lysosome pathway (31). We next examined whether EVD68 2C could antagonize A3G restriction by inducing A3G degradation. By aligning the 2C sequences of EV71 and EVD68, we found that the sequence homology of the two proteins is 62.8% (data not shown). However, EVD68 2C also caused reduced A3G expression, while the autophagy-lysosome inhibitor Baf-A1 could rescue A3G expression in the presence of 2C (Fig. 8A).
FIG 8.
Amino acids 25 to 41 in 2C of EVD68 are required for triggering A3G degradation through the autophagy lysosomal pathway. (A) HEK293T cells cotransfected with A3G-myc and EVD68-2C-HA, or the control vector, were treated with DMSO or 100 nM Baf-A1 at 12 h prior to harvest. A3G-myc and EVD68-2C-HA were tested by IB, with tubulin as a loading control. (B) Schematic diagram of EVD68 2C truncations. (C to E) HEK293T cells cotransfected with A3G-myc and EVD68-2C-HA, truncations of EVD68-2C-HA, or the control vector were harvested at 48 h after transfection. A3G-myc, EVD68-2C-HA and truncations of EVD68-2C-HA (1 to 118 and 119 to 331 in panel C; 1 to 53 and 54 to 331 in panel D; Δ25 to 41 in panel E) were tested in cell lysates, with tubulin as a loading control.
According to the sequence alignment of EV71 2C and EVD68 2C, a series of truncated EVD68 2C mutants were constructed based on computation-generated structural modeling of 2C (Fig. 8B). To identify the functional domain in EVD68 2C that is required for A3G degradation, we performed IB assays which showed that amino acids 1 to 118 at the N terminus of EVD68 2C were sufficient for A3G degradation, while amino acids 119 to 331 of EVD68 2C were not required for A3G degradation (Fig. 8C, lanes 3 and 4). Further investigation showed that the truncated EVD68 2C mutant amino acids 54 to 331 lost the ability to degrade A3G, while amino acids 1 to 53 induced A3G degradation, indicating that amino acids 1 to 53 of EVD68 are sufficient for A3G degradation (Fig. 7D, lane 3). Finally, we determined that amino acids 25 to 41 of EVD68 2C are critical for A3G degradation (Fig. 8E, lane 3). During autophagy, cytosolic LC3 I is converted to its lipidated form, LC3 II; thus, we investigated the ratio of LC2 II to LC3 I. Accordingly, the functional domains of EVD68 2C responsible for A3G degradation are also required for 2C-induced autophagy: amino acids 1 to 118, 1 to 53, and 25 to 41 could increase the ratio of LC3 II to LC3 I in a manner similar to that of WT 2C, but amino acids 118 to 331 and 53 to 331 could not (Fig. 8C to E). In summary, we determined that the N terminal of EVD68 2C, especially amino acids 25 to 41, is essential for A3G degradation. These results indicate that the 2C protein of EVD68 triggers A3G degradation through the autophagy-lysosome pathway, similarly to the 2C protein of EV71.
DISCUSSION
PCBP1 consists of three K-homology (KH) domains and belongs to the KH superfamily, the members of which are characterized by their poly(C)-binding specificity. The KH superfamily contains five proteins: hnRNP K, and PCBP1 through PCBP4. PCBP1 and PCBP2 have been well studied and are also referred to as αCP-1 and αCP-2, or hnRNP-E1 and hnRNP-E2 (58, 59). Accumulating evidence has shown that PCBPs are involved in multiple biological processes, including mRNA stabilization, translational silencing, translational enhancement, transcriptional controls, and apoptotic pathways (11, 57, 59–62). The involvement of PCBPs in viral infection was initially identified in PV by stimulating the translation of IRES-dependent mRNAs (11, 12, 47). PCBPs were then shown to be required for CVB3 replication, binding to the extended cloverleaf RNA and domain IV RNA of the IRES in the CVB3 5′ UTR (15, 48). Subsequently, a series of IRES-specific trans-acting factors (ITAFs) required for IRES-dependent translation were identified and characterized. For instance, hnRNP A1, which interacts with conserved RNA structural elements, is required for EV71 translation and replication (24, 63). In addition to their function in stimulating translation, PCBP1 and PCBP2 are utilized by porcine reproductive and respiratory syndrome virus (PRRSV) to interact with its encoded nonstructural protein 1, resulting in the colocalization of PCBP1 and PCBP2 with viral replication-transcription complexes and the regulation of PRRSV RNA synthesis (64). Li et al. identified PCBP1 as a novel mediator of antiviral innate immunity and demonstrated that PCBP1 enhances the replication of classical swine fever virus (CSFV) by interacting with the Npro protein (65). Due to a wide outbreak of EV71 and CA16 in the Asia-Pacific region, most researchers focus on the pathogenesis of these viruses, with only a few studies illustrating the involvement of cellular proteins in the viral replication of other EVs such as EVD68 and CA6. Our recent study showed that A3G competitively binds with the 5′ UTRs of EV71 and CA16 together with PCBP1, inhibiting EV71 and CA16; this indicates that PCBP1 is required for EV71 and CA16 replication. Here, we further demonstrated that PCBP1 is required for the replication of EVD68 but not the replication of CA6 (Fig. 1).
As a broad-spectrum antiviral factor, A3G has two important characteristics for its antiviral activity. Our previous research (31) and current studies have demonstrated that A3G inhibits the replication of EVD68, EV71, and CA16 using its nucleic-acid-binding activity but not its classical cytidine-deaminase activity (Fig. 3 and 5). Thus, A3G might present antiviral activity for other RNA viruses, if A3G can specifically interact with some viral RNAs that are required for viral translation or for viral RNA replication. Since A3G is a promising target against HIV, small molecular compounds targeting it have been developed, either by specifically stabilizing A3G or by the interrupting its interaction with HIV-1 Vif (66, 67). Our findings provide important information on the common mechanism or agent which might be utilized to antagonize diverse viruses.
Approximately 490 nucleotides (nt) of IRES elements exist in the 5′ UTRs of different groups of viruses, including picornaviral mRNAs and cellular mRNAs (68–70). Most picornaviral IRESs have been divided into four types, with PV, coxsackievirus, rhinovirus, and other enteroviruses belonging to type I. Based on PV, viruses with a type I IRES contain five major stem-loop regions. The 5′-terminal cloverleaf is mainly responsible for RNA stability or replication, and might also influence translation (71, 72). Domains IV (stem-loop IV) and V are essential for viral translation (73, 74). Previous research and our own studies have found that the cloverleaf and stem-loop IV regions of most EV 5′ UTRs, such as those of PV, EV71, and EVD68, are essential for PCBP1 binding, while A3G has strong binding capacity with the cloverleaf and the stem-loop II regions of EV71 and stronger binding ability with any regions of the EVD68 5′ UTR. Due to the strong binding capacity of A3G with the 5′ UTR, A3G competitively binds to the 5′ UTR along with PCBP1, thereby inhibiting EV replication (Fig. 6 and 7).
To replicate successfully, viruses develop various strategies to antagonize host restriction factors, such as ubiquitin proteasomal degradation. The 2C protein encoded by EVs can induce autophagy (31, 75). We found that the 2C protein of multiple EVs can induce A3G degradation via the autophagy-lysosomal pathway. The complicated interplay between viruses and host also provides an attractive target for the development of an anti-EV inhibitor.
MATERIALS AND METHODS
Plasmid construction.
RNAs of EVD68, CA6, and HEK293T cells were extracted with TRIzol (Invitrogen, Carlsbad, CA, USA) and reverse-transcripted with oligonucleotide (dT) primers and M-MLV reverse transcriptase (Invitrogen) according to the manufacturer’s instructions. The resulting cDNA was used for amplification of the EVD68 5′ UTR, EVD68 2C, PCBP1, and PCBP2; then, an amplified fragment of 5′ UTR was inserted into the NsiI/SalI sites of pIRIGF (Addgene, catalog no. 101139) or the SalI/BamHI sites of the VR1012 vector to generate the plasmid constructs 5′ UTR-luciferase-pIRIGF and 5′ UTR-VR1012. Various truncated 5′ UTRs were constructed by amplification and inserted into VR1012. The fragment of EVD68-2C followed by an HA tag was inserted into the SalI/BamHI sites of the VR1012 vector. Various truncated 5′ UTR were constructed by amplification and inserted into VR1012 by replacing EVD68-2C WT or by standard site-directed mutagenesis. Fragment of PCBP1 followed by an HA tag were inserted into the SalI/BamHI sites of the VR1012 vector. A3G-myc-pCDNA3.1(–), A3G-HA-pCDNA3.1(–), and mutants of A3G-HA were described in a previous study (31).
Cell culture and viruses.
HEK293T (ATCC, catalog no. CRL-11268) and HeLa (ATCC, catalog no. CCL-2) cells were cultured as monolayers in Dulbecco’s modified Eagle’s medium and minimum essential medium (HyClone, Logan, UT, USA) supplemented with 10% heat-inactivated (56°C, 30 min) fetal bovine serum (Gibco/Brl, Grand Island, NY, USA) and maintained at 37°C with 5% CO2 in a humidified atmosphere. EV71 CC063 was deposited in the China General Microbiological Culture Collection Center (CGMCC No.7753), which was isolated from HFMD patients in 2010 (76). CA6 (GenBank accession number: KT779410.1) and EVD68 (GenBank accession number: KM851231.1) were described in a previous study (77). PCBP1-specific shRNAs with the following target sites were cloned in the lentiretroviral vector pLKO.1-puro (Addgene, Cambridge, MA, USA): 5′-CCGGAAGGGAGAGTCATGACCATTCCTCGAGGAATGGTCATGACTCTCCCTTTTTTTG-3′ and 5′-AATTCAAAAAAAGGGAGAGTCATGACCATTCCTCGAGGAATGGTCATGACTCTCCCTT-3′. pLKO.1 was used as a negative control.
Transfection and infection.
HEK293T cells were transfected with Lipofectamine 2000 (Invitrogen) according to the manufacturer's instructions. The DNA transfection efficiency for the HEK293T cells was about 80%. For viral infections, cells were grown to 80% confluence in a 6-well plate, washed twice with phosphate-buffered saline (PBS), and incubated with virus at 37°C for 1 h. During adsorption, the plate was gently agitated at 15-min intervals. Following adsorption, the virus-containing medium was replaced with fresh medium containing 2% FCS, followed by incubation at 37°C in 5% CO2 for indicated time points.
Western blotting and antibodies.
In brief, transfected or infected HEK293T cells were harvested and boiled in 1× loading buffer (0.08 M Tris [pH 6.8] with 2.0% SDS, 10% glycerol, 0.1 M dithiothreitol, and 0.2% bromophenol blue) followed by separation on a 12% polyacrylamide gel. Proteins were transferred onto a polyvinylidene difluoride membrane for Western blotting analysis. The membranes were incubated with primary antibodies, followed by a corresponding horseradish peroxidase (HRP)-conjugated secondary antibody (Jackson ImmunoResearch, Suffolk, United Kingdom) diluted 1:10,000, respectively. Proteins incubated with HRP-conjugated secondary antibody were visualized using an ultrasensitive ECL Chemiluminescence Detection kit (Proteintech, Rosemont, IL, USA, catalog no. B500024) and then imaged and scanned by laser imaging system (Azure Biosystem). For membranes in Fig. 4A and I, a column defect is due to long-term use of CCD camera pixels which needed to be periodically corrected.
The following antibodies were used in this study: anti-EVD68-VP1 polyclonal antibody (pAb, GeneTex, Irvine, CA, USA, catalog no. GTX132313), anti-CA6-VP1 pAb (GeneTex, catalog no. GTX78102), anti-PCBP1 monoclonal antibody (MAb; Sangon Biotech, Shanghai, China, catalog no. D198382), anti-LC3 pAb (Proteintech, catalog no. 14600-1-AP), anti-hemagglutinin (anti-HA) MAb (Covance, Princeton, NJ, USA, catalog no. MMS-101R-10000), anti-HA pAb (Invitrogen, catalog no. 71-5500), anti-myc MAb (Millipore, Billerica, MA, USA, catalog no. 05-724), and anti-tubulin MAb (Abcam, Cambridge, MA, USA, catalog no. ab11323).
RNA extraction and RT-qPCR.
For RT-qPCR, viral RNA was extracted from HEK293T cells, RD cells transfected with APOBEC3-expression vector, or cells infected with virus by TRIzol reagent (Invitrogen), diethyl pyrocarbonate-treated water, and RNase inhibitor (New England BioLabs, Ipswich, MA, USA). The cDNA was generated with a High-Capacity cDNA Reverse Transcription kit (Applied Biosystems, Carlsbad, CA, USA) and oligonucleotide d(T)18 primers according to the supplier’s instructions. To avoid contamination, DNase was used to digest DNA (Promega, M6101). RT-qPCR was carried out on an Mx3005P instrument (Agilent Technologies, Stratagene, La Jolla, CA, USA) with the RealMaster Mix (Sybr Green Kit, Takara Bio, Inc., Shiga, Japan) and primers designed by conserved sequences of human A3G (hA3G). The RT-qPCR assay was carried out in a 20-μL volume consisting of 9 μL of 2.5× RealMaster Mix/20× Sybr Green solution containing HotMaster Taq DNA polymerase, 1 μL of 5 μmol/liter of each oligonucleotide primer, and 2 μg of cDNA template. Amplification of the target fragment was carried out as follows: initial activation of HotMaster Taq DNA polymerase at 95°C for 2 min was followed by 45 cycles of 95°C for 15 s, 57°C for 15 s, and 68°C for 20 s. All primers used in this study are listed in Table 1.
TABLE 1.
Primers used in this study
| Primer | Sequence (5′ to 3′) |
|---|---|
| SalI-HA-PCBP1-F | GCGTCGACATGTACCCTTACGACGTCCCAGATTACGCGATGGATGCCGGTGTGACTG |
| PCBP1-BglII-R | GAAGATCTCTAGCTGCACCCCATGCC |
| SalI-HA-PCBP2-F | GCGTCGACATGTACCCTTACGACGTCCCAGATTACGCGATGGACACCGGTGTGATTG |
| PCBP2-BamHI-R | CGCGGATCCTCAGCTGCTCCCCATGCCAC |
| GAPDH-RT-F | 5TGCACCACCAACTGCTTAGC |
| GAPDH-RT-R | GGCATGGACTGTGGTCATGAG |
| EVD68 VP1-RT-F | CAGTCACAGCCACACTAGC |
| EVD68 VP1-RT-R | CAATCTAAACCCCTGAGAGC |
| CA6 VP1-RT-F | AATGAGGCGAGTGTGGAAC |
| CA6 VP1-RT-R | AGGTTGGACACAAAAGTGAACT |
| EVD68 5′ UTR-RT-F | GAGACGCTAAACATGAACAAG |
| EVD68 5′ UTR-RT-R | CATTACGACAAGCAACTCACT |
| CA6 5′ UTR-RT-F | TGTCTCCCCGGATTGAGTAT |
| CA6 5′ UTR-RT-R | TGGGGAATGCAGTGACTCAT |
| CA6 5′ UTR-Xhol-F | CCCTCGAGTTAAAACAGCCTGTGGGTTG |
| CA6 5′ UTR-BamHI-R | CTGGATCCTTTCTATTATTCAG |
| EVD68 5′ UTR-1-XholI-F | CCCTCGAGTTAAAACAGGCTCTGGGGTTG |
| EVD68 5′ UTR-123-BamHI-R | CGGGATCCGATTTGAAAGCTTCTAAGTTACA |
| EVD68 5′ UTR-124-XholI-F | CCCTCGAGTCAAAGCTCAATAGGTGGAG |
| EVD68 5′ UTR-186-BamHI-R | CGGGATCCCACCGGGGAAACAGGAGTGCTTGC |
| EVD68 5′ UTR-163-XholI-F | CCCTCGAGGCAAGCACTCCTGTTTCCC |
| EVD68 5′ UTR-233-BamHI-R | CGGGATCCTAACGGATAGGTTGTTTTCAAC |
| EVD68 5′ UTR-234-XholI-F | CCCTCGAGGTTATCCGCTATAGTACTTCG |
| EVD68 5′ UTR-446-BamHI-R | CGGGATCCGAGGACTCTATAGTAGCTCAA |
| EVD68 5′ UTR-447-XholI-F | CCCTCGAGTCCTCCGGCCCCTGAATG |
| EVD68 5′ UTR-710-BamHI-R | GGATCCCTTTGTAAGTAGGTAT |
| EVD68 2C-1-SalI-F | GCGTCGACATGGGTGAATCGTGGCTTAAG |
| EVD68 2C-53-BamHI-R | CGGGATCCTTATAACTGTTTGAGCCTTTGCAC |
| EVD68 2C-54-SalI-R | GCGTCGACATGCCGGTGATAGAAAACCAAG |
| EVD68 2C-331-BamHI-R | CGGGATCCTTACTGAAACAGAGCTTCCAGC |
| EVD68 2C△25-41-F | ATTGGTTGTCACAAAAGGAGAAATATGAATTTGTG |
| EVD68 2C△25-41-R | CCTTTTGTGACAACCAATCTAAACCTCTG |
| EVD68 5′UTR-123-Biotin | GATTTGAAAGCTTCTAAGTTACA |
| EVD68 5′UTR-Biotin | CTTTGTAAGTAGGTATAAAGTC |
| CA6 5′UTR-Biotin | TTTCTATTATTCAGGATTAAAATG |
Luciferase assays.
HEK293T cells in 12-well plates were cotransfected with 0.5 μg of bicistronic pIRIGF-5′-UTR expression vector and 0.2 μg of either A3G-myc expression vector or pCDNA3.1(-) for 48 h before harvesting. Luciferase activity in the cells was detected with Fluoroskan Ascent FL (Thermo Fisher) with Dual-Luciferase Reporter Assay System (Promega).
Co-immunoprecipitation.
Co-immunoprecipitation (co-IP) experiments were performed as previously reported (40, 55). For A3G, PCBP1, or PCBP2 with 5′ UTR IP, HEK293T cells were transfected either with 5′-UTR-VR1012 and hA3G-myc or with PCBP1-HA. The cells were then harvested and washed twice with cold PBS, followed by disruption with lysis buffer (PBS containing 1% Triton X-100 and complete protease inhibitor cocktail [Roche]) at 4°C for 1 h. Cell lysates were clarified by centrifugation at 10,000 × g for 30 min at 4°C. Anti-HA agarose beads (Roche, Basel, Switzerland, catalog no. 190-119) or anti-myc agarose beads (Sigma, St. Louis, MO, USA, catalog no. A7470) were mixed with the pre-cleared cell lysates and incubated at 4°C for 4 h on an end-over-end rocker. The reaction mixtures were then washed eight times with cold wash buffer (20 mM Tris-HCl [pH 7.5] 100 mM NaCl, 0.1 mM EDTA, 0.05% Tween 20) and subsequently analyzed either by immunoblotting or by extracting RNA for RT-qPCR analysis.
Electrophoretic mobility shift assay.
RNA of 5′ UTRs from EVD68 and CA6 was transcribed with a MEGAscriptTM T7 kit (Ambion, Austin, TX, USA), and RNAs purified with a MEGA clear kit (Ambion) were annealed with the DNA probe (5′-CTTTGTAAGTAGGTATAAAGTC-3′ for EVD68 and 5′-TTTCTATTATTCAGGATTAAAATG-3′ for CA6)-labeled biotin by a Biotin 3′ End DNA Labeling kit (Thermo Fisher). For EMSA, A3G and its mutants with HA tags were purified by co-IP with anti-HA agarose beads from 5 × 106 HEK293T cells transfected with either A3G or a mutant expression plasmid for 48 h, and then incubated with 4 μg of biotinylated RNA for detecting the interaction between the 5′ UTRs and either A3G or its mutants with a LightShift Chemiluminescent EMSA kit (Thermo Fisher), according to the manufacturer's instructions. In brief, proteins with or without DNAs were separated by a 4 to 6% native polyacrylamide gel, then transferred onto nylon membranes by wet transfer (Bio-Rad, Hercules, CA) at 100 V for 30 min. When the transfer was complete, we proceeded to cross-link at 120 mJ/cm2 using a commercial UV-light cross-linking instrument equipped with 254-nm bulbs. The membrane was blocked and washed, and then biotin-labeled DNAs were detected by Chemiluminescence.
ACKNOWLEDGMENTS
We thank C. Y. Dai for the critical reagents.
This work was supported in part by funding from the National Natural Science Foundation of China (no. 81930062 and 81672004 to W.Y.Z. and no. 81701987 to Z.L.L.), the Science and Technology Department of Jilin Province (20190101003JH and 20190201272JC), the China Postdoctoral Science Foundation (2020M670826), the Key Laboratory of Molecular Virology, Jilin Province (20102209), the Education Department of Jilin Province (JJKH20211141KJ), the Health Commission of Jilin Province (2020J059), and the Fundamental Research Funds for the Central Universities.
The authors declare no conflicts of interest.
Contributor Information
Wenyan Zhang, Email: zhangwenyan@jlu.edu.cn.
Susana López, Instituto de Biotecnologia (UNAM).
REFERENCES
- 1.Baggen J, Thibaut HJ, Strating J, van Kuppeveld FJM. 2018. The life cycle of non-polio enteroviruses and how to target it. Nat Rev Microbiol 16:368–381. 10.1038/s41579-018-0005-4. [DOI] [PubMed] [Google Scholar]
- 2.Bian L, Wang Y, Yao X, Mao Q, Xu M, Liang Z. 2015. Coxsackievirus A6: a new emerging pathogen causing hand, foot and mouth disease outbreaks worldwide. Expert Rev Anti Infect Ther 13:1061–1071. 10.1586/14787210.2015.1058156. [DOI] [PubMed] [Google Scholar]
- 3.Fu X, Wan Z, Li Y, Hu Y, Jin X, Zhang C. 2020. National epidemiology and evolutionary history of four hand, foot and mouth disease-related enteroviruses in China from 2008 to 2016. Virol Sin 35:21–33. 10.1007/s12250-019-00169-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Zhang C, Zhu R, Yang Y, Chi Y, Yin J, Tang X, Yu L, Zhang C, Huang Z, Zhou D. 2015. Phylogenetic analysis of the major causative agents of hand, foot and mouth disease in Suzhou City, Jiangsu province, China, in 2012–2013. Emerg Microbes Infect 4:e12. 10.1038/emi.2015.12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Zhao TS, Du J, Sun DP, Zhu QR, Chen LY, Ye C, Wang S, Liu YQ, Cui F, Lu QB. 2020. A review and meta-analysis of the epidemiology and clinical presentation of coxsackievirus A6 causing hand-foot-mouth disease in China and global implications. Rev Med Virol 30:e2087. 10.1002/rmv.2087. [DOI] [PubMed] [Google Scholar]
- 6.Dyda A, Stelzer-Braid S, Adam D, Chughtai AA, MacIntyre CR. 2018. The association between acute flaccid myelitis (AFM) and Enterovirus D68 (EV-D68) - what is the evidence for causation? Euro Surveill 23:17-00310. 10.2807/1560-7917.ES.2018.23.3.17-00310. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Hixon AM, Frost J, Rudy MJ, Messacar K, Clarke P, Tyler KL. 2019. Understanding Enterovirus D68-induced neurologic disease: a basic science review. Viruses 11:821 10.3390/v11090821. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Holm-Hansen CC, Midgley SE, Fischer TK. 2016. Global emergence of enterovirus D68: a systematic review. Lancet Infect Dis 16:e64–e75. 10.1016/S1473-3099(15)00543-5. [DOI] [PubMed] [Google Scholar]
- 9.Xiang Z, Li L, Ren L, Guo L, Xie Z, Liu C, Li T, Luo M, Paranhos-Baccala G, Xu W, Wang J. 2017. Seroepidemiology of enterovirus D68 infection in China. Emerg Microbes Infect 6:e32. 10.1038/emi.2017.14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Blyn LB, Swiderek KM, Richards O, Stahl DC, Semler BL, Ehrenfeld E. 1996. Poly(rC) binding protein 2 binds to stem-loop IV of the poliovirus RNA 5′ noncoding region: identification by automated liquid chromatography-tandem mass spectrometry. Proc Natl Acad Sci USA 93:11115–11120. 10.1073/pnas.93.20.11115. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Blyn LB, Towner JS, Semler BL, Ehrenfeld E. 1997. Requirement of poly(rC) binding protein 2 for translation of poliovirus RNA. J Virol 71:6243–6246. 10.1128/JVI.71.8.6243-6246.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Gamarnik AV, Andino R. 1997. Two functional complexes formed by KH domain containing proteins with the 5′ noncoding region of poliovirus RNA. RNA 3:882–892. [PMC free article] [PubMed] [Google Scholar]
- 13.Gamarnik AV, Andino R. 1998. Switch from translation to RNA replication in a positive-stranded RNA virus. Genes Dev 12:2293–2304. 10.1101/gad.12.15.2293. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Sharma N, Ogram SA, Morasco BJ, Spear A, Chapman NM, Flanegan JB. 2009. Functional role of the 5′ terminal cloverleaf in Coxsackievirus RNA replication. Virology 393:238–249. 10.1016/j.virol.2009.07.039. [DOI] [PubMed] [Google Scholar]
- 15.Zell R, Ihle Y, Seitz S, Gundel U, Wutzler P, Gorlach M. 2008. Poly(rC)-binding protein 2 interacts with the oligo(rC) tract of coxsackievirus B3. Biochem Biophys Res Commun 366:917–921. 10.1016/j.bbrc.2007.12.038. [DOI] [PubMed] [Google Scholar]
- 16.Perera R, Daijogo S, Walter BL, Nguyen JH, Semler BL. 2007. Cellular protein modification by poliovirus: the two faces of poly(rC)-binding protein. J Virol 81:8919–8932. 10.1128/JVI.01013-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Toyoda H, Franco D, Fujita K, Paul AV, Wimmer E. 2007. Replication of poliovirus requires binding of the poly(rC) binding protein to the cloverleaf as well as to the adjacent C-rich spacer sequence between the cloverleaf and the internal ribosomal entry site. J Virol 81:10017–10028. 10.1128/JVI.00516-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Vogt DA, Andino R. 2010. An RNA element at the 5′-end of the poliovirus genome functions as a general promoter for RNA synthesis. PLoS Pathog 6:e1000936. 10.1371/journal.ppat.1000936. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Walter BL, Parsley TB, Ehrenfeld E, Semler BL. 2002. Distinct poly(rC) binding protein KH domain determinants for poliovirus translation initiation and viral RNA replication. J Virol 76:12008–12022. 10.1128/jvi.76.23.12008-12022.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Huang PN, Lin JY, Locker N, Kung YA, Hung CT, Lin JY, Huang HI, Li ML, Shih SR. 2011. Far upstream element binding protein 1 binds the internal ribosomal entry site of enterovirus 71 and enhances viral translation and viral growth. Nucleic Acids Res 39:9633–9648. 10.1093/nar/gkr682. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Lin JY, Li ML, Huang PN, Chien KY, Horng JT, Shih SR. 2008. Heterogeneous nuclear ribonuclear protein K interacts with the enterovirus 71 5′ untranslated region and participates in virus replication. J Gen Virol 89:2540–2549. 10.1099/vir.0.2008/003673-0. [DOI] [PubMed] [Google Scholar]
- 22.Lin JY, Shih SR, Pan M, Li C, Lue CF, Stollar V, Li ML. 2009. hnRNP A1 interacts with the 5′ untranslated regions of enterovirus 71 and Sindbis virus RNA and is required for viral replication. J Virol 83:6106–6114. 10.1128/JVI.02476-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Shih SR, Stollar V, Li ML. 2011. Host factors in enterovirus 71 replication. J Virol 85:9658–9666. 10.1128/JVI.05063-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Tolbert M, Morgan CE, Pollum M, Crespo-Hernandez CE, Li ML, Brewer G, Tolbert BS. 2017. HnRNP A1 alters the structure of a conserved Enterovirus IRES domain to stimulate viral translation. J Mol Biol 429:2841–2858. 10.1016/j.jmb.2017.06.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Simon V, Bloch N, Landau NR. 2015. Intrinsic host restrictions to HIV-1 and mechanisms of viral escape. Nat Immunol 16:546–553. 10.1038/ni.3156. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Bogerd HP, Wiegand HL, Hulme AE, Garcia-Perez JL, O'Shea KS, Moran JV, Cullen BR. 2006. Cellular inhibitors of long interspersed element 1 and Alu retrotransposition. Proc Natl Acad Sci USA 103:8780–8785. 10.1073/pnas.0603313103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Fehrholz M, Kendl S, Prifert C, Weissbrich B, Lemon K, Rennick L, Duprex PW, Rima BK, Koning FA, Holmes RK, Malim MH, Schneider-Schaulies J. 2012. The innate antiviral factor APOBEC3G targets replication of measles, mumps and respiratory syncytial viruses. J Gen Virol 93:565–576. 10.1099/vir.0.038919-0. [DOI] [PubMed] [Google Scholar]
- 28.Seppen J. 2004. Unedited inhibition of HBV replication by APOBEC3G. J Hepatol 41:1068–1069. 10.1016/j.jhep.2004.10.008. [DOI] [PubMed] [Google Scholar]
- 29.Sheehy AM, Gaddis NC, Choi JD, Malim MH. 2002. Isolation of a human gene that inhibits HIV-1 infection and is suppressed by the viral Vif protein. Nature 418:646–650. 10.1038/nature00939. [DOI] [PubMed] [Google Scholar]
- 30.Turelli P, Mangeat B, Jost S, Vianin S, Trono D. 2004. Inhibition of hepatitis B virus replication by APOBEC3G. Science 303:1829. 10.1126/science.1092066. [DOI] [PubMed] [Google Scholar]
- 31.Li Z, Ning S, Su X, Liu X, Wang H, Liu Y, Zheng W, Zheng B, Yu XF, Zhang W. 2018. Enterovirus 71 antagonizes the inhibition of the host intrinsic antiviral factor A3G. Nucleic Acids Res 46:11514–11527. 10.1093/nar/gky840. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Lecossier D, Bouchonnet F, Clavel F, Hance AJ. 2003. Hypermutation of HIV-1 DNA in the absence of the Vif protein. Science 300:1112. 10.1126/science.1083338. [DOI] [PubMed] [Google Scholar]
- 33.Vartanian JP, Guetard D, Henry M, Wain-Hobson S. 2008. Evidence for editing of human papillomavirus DNA by APOBEC3 in benign and precancerous lesions. Science 320:230–233. 10.1126/science.1153201. [DOI] [PubMed] [Google Scholar]
- 34.Zhang H, Yang B, Pomerantz RJ, Zhang C, Arunachalam SC, Gao L. 2003. The cytidine deaminase CEM15 induces hypermutation in newly synthesized HIV-1 DNA. Nature 424:94–98. 10.1038/nature01707. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Apolonia L, Schulz R, Curk T, Rocha P, Swanson CM, Schaller T, Ule J, Malim MH. 2015. Promiscuous RNA binding ensures effective encapsidation of APOBEC3 proteins by HIV-1. PLoS Pathog 11:e1004609. 10.1371/journal.ppat.1004609. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Polevoda B, McDougall WM, Tun BN, Cheung M, Salter JD, Friedman AE, Smith HC. 2015. RNA binding to APOBEC3G induces the disassembly of functional deaminase complexes by displacing single-stranded DNA substrates. Nucleic Acids Res 43:9434–9445. 10.1093/nar/gkv970. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Li Z, Huan C, Wang H, Liu Y, Liu X, Su X, Yu J, Zhao Z, Yu XF, Zheng B, Zhang W. 2020. TRIM21-mediated proteasomal degradation of SAMHD1 regulates its antiviral activity. EMBO Rep 21:e47528. 10.15252/embr.201847528. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Malim MH, Bieniasz PD. 2012. HIV restriction factors and mechanisms of evasion. Cold Spring Harb Perspect Med 2:a006940. 10.1101/cshperspect.a006940. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Yu X, Yu Y, Liu B, Luo K, Kong W, Mao P, Yu XF. 2003. Induction of APOBEC3G ubiquitination and degradation by an HIV-1 Vif-Cul5-SCF complex. Science 302:1056–1060. 10.1126/science.1089591. [DOI] [PubMed] [Google Scholar]
- 40.Zhang W, Du J, Evans SL, Yu Y, Yu XF. 2011. T-cell differentiation factor CBF-beta regulates HIV-1 Vif-mediated evasion of host restriction. Nature 481:376–379. 10.1038/nature10718. [DOI] [PubMed] [Google Scholar]
- 41.Goldstone DC, Ennis-Adeniran V, Hedden JJ, Groom HC, Rice GI, Christodoulou E, Walker PA, Kelly G, Haire LF, Yap MW, de Carvalho LP, Stoye JP, Crow YJ, Taylor IA, Webb M. 2011. HIV-1 restriction factor SAMHD1 is a deoxynucleoside triphosphate triphosphohydrolase. Nature 480:379–382. 10.1038/nature10623. [DOI] [PubMed] [Google Scholar]
- 42.Hrecka K, Hao C, Gierszewska M, Swanson SK, Kesik-Brodacka M, Srivastava S, Florens L, Washburn MP, Skowronski J. 2011. Vpx relieves inhibition of HIV-1 infection of macrophages mediated by the SAMHD1 protein. Nature 474:658–661. 10.1038/nature10195. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Marin M, Rose KM, Kozak SL, Kabat D. 2003. HIV-1 Vif protein binds the editing enzyme APOBEC3G and induces its degradation. Nat Med 9:1398–1403. 10.1038/nm946. [DOI] [PubMed] [Google Scholar]
- 44.Mitchell RS, Katsura C, Skasko MA, Fitzpatrick K, Lau D, Ruiz A, Stephens EB, Margottin-Goguet F, Benarous R, Guatelli JC. 2009. Vpu antagonizes BST-2-mediated restriction of HIV-1 release via beta-TrCP and endo-lysosomal trafficking. PLoS Pathog 5:e1000450. 10.1371/journal.ppat.1000450. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Shi J, Xiong R, Zhou T, Su P, Zhang X, Qiu X, Li H, Li S, Yu C, Wang B, Ding C, Smithgall TE, Zheng YH. 2018. HIV-1 Nef antagonizes SERINC5 restriction by downregulation of SERINC5 via the endosome/lysosome system. J Virol 92 10.1128/JVI.00196-18. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Luo Z, Dong X, Li Y, Zhang Q, Kim C, Song Y, Kang L, Liu Y, Wu K, Wu J. 2014. PolyC-binding protein 1 interacts with 5′-untranslated region of enterovirus 71 RNA in membrane-associated complex to facilitate viral replication. PLoS One 9:e87491. 10.1371/journal.pone.0087491. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Parsley TB, Towner JS, Blyn LB, Ehrenfeld E, Semler BL. 1997. Poly (rC) binding protein 2 forms a ternary complex with the 5′-terminal sequences of poliovirus RNA and the viral 3CD proteinase. RNA 3:1124–1134. [PMC free article] [PubMed] [Google Scholar]
- 48.Souii A, M'Hadheb-Gharbi MB, Gharbi J. 2015. Cellular proteins act as bridge between 5′ and 3' ends of the coxsackievirus B3 mediating genome circularization during RNA translation. Curr Microbiol 71:387–395. 10.1007/s00284-015-0866-y. [DOI] [PubMed] [Google Scholar]
- 49.Bogerd HP, Cullen BR. 2008. Single-stranded RNA facilitates nucleocapsid: APOBEC3G complex formation. RNA 14:1228–1236. 10.1261/rna.964708. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Hakata Y, Landau NR. 2006. Reversed functional organization of mouse and human APOBEC3 cytidine deaminase domains. J Biol Chem 281:36624–36631. 10.1074/jbc.M604980200. [DOI] [PubMed] [Google Scholar]
- 51.Khan MA, Kao S, Miyagi E, Takeuchi H, Goila-Gaur R, Opi S, Gipson CL, Parslow TG, Ly H, Strebel K. 2005. Viral RNA is required for the association of APOBEC3G with human immunodeficiency virus type 1 nucleoprotein complexes. J Virol 79:5870–5874. 10.1128/JVI.79.9.5870-5874.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Tian C, Wang T, Zhang W, Yu XF. 2007. Virion packaging determinants and reverse transcription of SRP RNA in HIV-1 particles. Nucleic Acids Res 35:7288–7302. 10.1093/nar/gkm816. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Wang T, Tian C, Zhang W, Luo K, Sarkis PT, Yu L, Liu B, Yu Y, Yu XF. 2007. 7SL RNA mediates virion packaging of the antiviral cytidine deaminase APOBEC3G. J Virol 81:13112–13124. 10.1128/JVI.00892-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Huthoff H, Malim MH. 2007. Identification of amino acid residues in APOBEC3G required for regulation by human immunodeficiency virus type 1 Vif and virion encapsidation. J Virol 81:3807–3815. 10.1128/JVI.02795-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Zhang W, Du J, Yu K, Wang T, Yong X, Yu XF. 2010. Association of potent human antiviral cytidine deaminases with 7SL RNA and viral RNP in HIV-1 virions. J Virol 84:12903–12913. 10.1128/JVI.01632-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Holden LG, Prochnow C, Chang YP, Bransteitter R, Chelico L, Sen U, Stevens RC, Goodman MF, Chen XS. 2008. Crystal structure of the anti-viral APOBEC3G catalytic domain and functional implications. Nature 456:121–124. 10.1038/nature07357. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Gamarnik AV, Andino R. 2000. Interactions of viral protein 3CD and poly(rC) binding protein with the 5′ untranslated region of the poliovirus genome. J Virol 74:2219–2226. 10.1128/jvi.74.5.2219-2226.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Kiledjian M, Wang X, Liebhaber SA. 1995. Identification of two KH domain proteins in the alpha-globin mRNP stability complex. EMBO J 14:4357–4364. 10.1002/j.1460-2075.1995.tb00110.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Makeyev AV, Liebhaber SA. 2002. The poly(C)-binding proteins: a multiplicity of functions and a search for mechanisms. RNA 8:265–278. 10.1017/s1355838202024627. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Charroux B, Angelats C, Fasano L, Kerridge S, Vola C. 1999. The levels of the bancal product, a Drosophila homologue of vertebrate hnRNP K protein, affect cell proliferation and apoptosis in imaginal disc cells. Mol Cell Biol 19:7846–7856. 10.1128/MCB.19.11.7846. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Ostareck-Lederer A, Ostareck DH. 2004. Control of mRNA translation and stability in haematopoietic cells: the function of hnRNPs K and E1/E2. Biol Cell 96:407–411. 10.1016/j.biolcel.2004.03.010. [DOI] [PubMed] [Google Scholar]
- 62.Ostareck-Lederer A, Ostareck DH, Hentze MW. 1998. Cytoplasmic regulatory functions of the KH-domain proteins hnRNPs K and E1/E2. Trends Biochem Sci 23:409–411. 10.1016/s0968-0004(98)01301-2. [DOI] [PubMed] [Google Scholar]
- 63.Levengood JD, Tolbert M, Li ML, Tolbert BS. 2013. High-affinity interaction of hnRNP A1 with conserved RNA structural elements is required for translation and replication of enterovirus 71. RNA Biol 10:1136–1145. 10.4161/rna.25107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Beura LK, Dinh PX, Osorio FA, Pattnaik AK. 2011. Cellular poly(c) binding proteins 1 and 2 interact with porcine reproductive and respiratory syndrome virus nonstructural protein 1beta and support viral replication. J Virol 85:12939–12949. 10.1128/JVI.05177-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Li D, Li S, Sun Y, Dong H, Li Y, Zhao B, Guo D, Weng C, Qiu HJ. 2013. Poly(C)-binding protein 1, a novel N(pro)-interacting protein involved in classical swine fever virus growth. J Virol 87:2072–2080. 10.1128/JVI.02807-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Cen S, Peng ZG, Li XY, Li ZR, Ma J, Wang YM, Fan B, You XF, Wang YP, Liu F, Shao RG, Zhao LX, Yu L, Jiang JD. 2010. Small molecular compounds inhibit HIV-1 replication through specifically stabilizing APOBEC3G. J Biol Chem 285:16546–16552. 10.1074/jbc.M109.085308. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Ma L, Zhang Z, Liu Z, Pan Q, Wang J, Li X, Guo F, Liang C, Hu L, Zhou J, Cen S. 2018. Identification of small molecule compounds targeting the interaction of HIV-1 Vif and human APOBEC3G by virtual screening and biological evaluation. Sci Rep 8:8067. 10.1038/s41598-018-26318-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Hellen CU, Sarnow P. 2001. Internal ribosome entry sites in eukaryotic mRNA molecules. Genes Dev 15:1593–1612. 10.1101/gad.891101. [DOI] [PubMed] [Google Scholar]
- 69.Jang SK, Krausslich HG, Nicklin MJ, Duke GM, Palmenberg AC, Wimmer E. 1988. A segment of the 5′ nontranslated region of encephalomyocarditis virus RNA directs internal entry of ribosomes during in vitro translation. J Virol 62:2636–2643. 10.1128/JVI.62.8.2636-2643.1988. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Pelletier J, Sonenberg N. 1988. Internal initiation of translation of eukaryotic mRNA directed by a sequence derived from poliovirus RNA. Nature 334:320–325. 10.1038/334320a0. [DOI] [PubMed] [Google Scholar]
- 71.Bonderoff JM, Lloyd RE. 2008. CVB translation: lessons from the polioviruses. Curr Top Microbiol Immunol 323:123–147. 10.1007/978-3-540-75546-3_6. [DOI] [PubMed] [Google Scholar]
- 72.Simoes EA, Sarnow P. 1991. An RNA hairpin at the extreme 5′ end of the poliovirus RNA genome modulates viral translation in human cells. J Virol 65:913–921. 10.1128/JVI.65.2.913-921.1991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Dildine SL, Semler BL. 1989. The deletion of 41 proximal nucleotides reverts a poliovirus mutant containing a temperature-sensitive lesion in the 5′ noncoding region of genomic RNA. J Virol 63:847–862. 10.1128/JVI.63.2.847-862.1989. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Percy N, Barclay WS, Sullivan M, Almond JW. 1992. A poliovirus replicon containing the chloramphenicol acetyltransferase gene can be used to study the replication and encapsidation of poliovirus RNA. J Virol 66:5040–5046. 10.1128/JVI.66.8.5040-5046.1992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Lee YR, Wang PS, Wang JR, Liu HS. 2014. Enterovirus 71-induced autophagy increases viral replication and pathogenesis in a suckling mouse model. J Biomed Sci 21:80. 10.1186/s12929-014-0080-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.Wang X, Zhu C, Bao W, Zhao K, Niu J, Yu XF, Zhang W. 2012. Characterization of full-length enterovirus 71 strains from severe and mild disease patients in northeastern China. PLoS One 7:e32405. 10.1371/journal.pone.0032405. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77.Xu N, Yang J, Zheng B, Zhang Y, Cao Y, Huan C, Wang S, Chang J, Zhang W. 2020. The pyrimidine analog FNC potently inhibits the replication of multiple enteroviruses. J Virol 94 10.1128/JVI.00204-20. [DOI] [PMC free article] [PubMed] [Google Scholar]