Visual Abstract
Keywords: acute kidney injury and ICU nephrology, body fluids, coronavirus, COVID-19, enzyme-linked immunosorbent assay, SARS-CoV-2, spike glycoprotein, spike protein, viral envelope proteins
Key Points
Using an antigen capture assay to detect severe acute respiratory syndrome coronavirus 2 (SARS-CoV-2) spike S1 protein, we found that the protein is present in the urine of 25% of patients with coronavirus disease 2019 (COVID-19).
Further, we found that 24% and 21% of adult patients with COVID-19 have high levels of urine albumin and cystatin C, respectively.
The presence of SARS-CoV-2 spike protein in the urine suggests renal abnormalities resulting from COVID-19.
Abstract
Background
SARS-CoV-2 infection has, as of April 2021, affected >133 million people worldwide, causing >2.5 million deaths. Because the large majority of individuals infected with SARS-CoV-2 are asymptomatic, major concerns have been raised about possible long-term consequences of the infection.
Methods
Wedeveloped an antigen capture assay to detect SARS-CoV-2 spike protein in urine samples from patients with COVID-19whose diagnosis was confirmed by positive PCR results from nasopharyngeal swabs (NP-PCR+) forSARS-CoV-2. We used a collection of 233 urine samples from 132 participants from Yale New Haven Hospital and the Children’s Hospital of Philadelphia that were obtained during the pandemic (106 NP-PCR+ and 26 NP-PCR−), and a collection of 20 urine samples from 20 individuals collected before the pandemic.
Results
Our analysis identified 23 out of 91 (25%) NP-PCR+ adult participants with SARS-CoV-2 spike S1 protein in urine (Ur-S+). Interestingly, although all NP-PCR+ children were Ur-S−, one child who was NP-PCR− was found to be positive for spike protein in their urine. Of the 23 adults who were Ur-S+, only one individual showed detectable viral RNA in urine. Our analysis further showed that 24% and 21% of adults who were NP-PCR+ had high levels of albumin and cystatin C, respectively, in their urine. Among individuals with albuminuria (>0.3 mg/mg of creatinine), statistical correlation could be found between albumin and spike protein in urine.
Conclusions
Together, our data showed that one of four individuals infected with SARS-CoV-2 develop renal abnormalities, such as albuminuria. Awareness about the long-term effect of these findings is warranted.
Introduction
Coronavirus disease 2019 (COVID-19) is a highly contagious disease caused by severe acute respiratory syndrome coronavirus 2 (SARS-CoV-2). The disease was first reported in December 2019 by health authorities in Wuhan, China, where a cluster of patients with symptoms of unidentified pneumonia had characteristics reminiscent of viral pneumonia (1–4). By March 12, 2020, the virus spread across the globe, having infected 145,200 individuals, and leading the World Health Organization to declare COVID-19 a global pandemic (5). As of early April 2021, >133 million cases and >2.5 million deaths have been recorded worldwide. Epidemiologic estimates suggest that approximately 70% of the world’s population could become infected with COVID-19, with a global case fatality rate between 0.1% and >25% (6).
Coronavirus-related illnesses in humans are caused by seven viruses, four of which (human coronavirus 229E [HCoV-229E], HCoV-OC43, HCoV-NL63, and HCoV-HKU1) cause common cold symptoms, and the remaining three (SARS-CoV, SARS-CoV-2, and Middle East respiratory syndrome–CoV [MERS-CoV]) cause SARS and MERS, respectively. SARS-CoV and MERS-CoV are known to have fatal outcomes, as occurred during the two outbreaks reported in 2002/2003 and 2012, respectively (7,8). SARS-CoV-2, the agent of the COVID-19 pandemic, is a member of β-coronaviruses, which mostly infect bats (9). However, this enveloped, positive-stranded RNA virus is also capable of infecting humans (10). Genetic characterization of SARS-CoV-2 shows it is closely related to bat virus RaTG13 (11).
Similar to the extensively studied SARS-CoV (12,13), the spike protein of SARS-CoV-2 plays a crucial role in viral attachment to the angiotensin-converting enzyme 2 receptor of the human cell membrane and entry into the target cell (14,15). This process is accompanied by proteolytic activation of the SARS-CoV-2 spike protein at the spike 1 (S1)/S2 site, where host proteases cleave the S1 from the S2 protein (15). Therefore, screening for the SARS-CoV-2 spike protein could be an excellent strategy for monitoring active and recent COVID-19. Analysis of the tissue distribution of angiotensin-converting enzyme 2 showed high expression of the receptor in the epithelial cells of the intestine, kidneys, alveoli, heart, arteries, and the gastrointestinal system (16); this suggests that, in addition to the lungs and the upper respiratory tract, SARS-CoV-2 could also invade other important organs, including the kidneys, and cause inflammation with possibly long-lasting injuries (17). To date, the effect of SARS-CoV-2 infection on renal function has been controversial. Analysis of a cohort of 193 adult patients with laboratory-confirmed SARS-CoV-2 infection from three hospitals in and around Wuhan, China showed kidney abnormalities (computed-tomography scan), with 63% of the participants showing proteinuria (Z. Li et al., unpublished observations). In the United States, a study by Hirsch et al. (18), showed that 37% of patients with COVID-19 developed AKI during hospitalization, most of whom were on mechanical ventilation. However, another study by Wang et al. (19), on 116 patients with confirmed COVID-19 (from the Renmin Hospital in Wuhan, China), found that Acute Kidney Infection (AKI) was uncommon, and that SARS-CoV-2 infection did not result in AKI or exacerbate Chronic Kidney Disease (CKD). Another indication for renal abnormality in patients with COVID-19 was observed by Diao et al. (unpublished observations), who found SARS-CoV-2 nucleocapsid protein in almost three quarters of urine samples collected from patients with COVID-19.
In this study, we developed an antigen capture assay that detects the presence of SARS-CoV-2 spike protein in biologic specimens. We used this assay to evaluate the presence of the antigen in urine samples collected during the COVID-19 pandemic from adults and children with PCR results from nasopharyngeal swabs that were positive (NP-PCR+) and negative (NP-PCR−) for SARS-CoV-2. Our study population also included urine samples collected 3–5 years before the pandemic. We found that, in our study population, 25% of adult patients with COVID-19 had the SARS-CoV- 2 S1 protein in their urine at least once during the course of infection. Urine protein analyses further revealed proteinuria with elevated albumin and cystatin C in 24% and 21% of NP-PCR+ individuals, respectively.
Methods
Definitions and Calculations
A urine sample was considered positive for the SARS-CoV-2 spike protein if its OD value was greater than the OD value obtained from a NP-PCR− urine sample spiked with 5 ng/ml of the SARS-CoV-2 spike protein. Urinary levels of SARS-CoV-2 spike protein, albumin, and cystatin C were normalized to urine creatinine levels and expressed as milligram per milligram of urine creatinine (20,21). The fractional excretion of sodium (FENa) was calculated using the formula (22–23):
The fractional excretion of urea (FEUrea) was calculated using the formula (22–23):
Study Population
In this study, we analyzed 253 urine samples from 152 participants. Of these, 233 urine samples were collected between March and August 2020 (COVID-19 period; N=132) and 20 urine samples were collected before December 2019 (pre–COVID-19 period; N=20). Participants from the COVID-19 period were recruited from Yale New Haven Hospital (YNHH; NP-PCR+ inpatients, N=91; NP-PCR− healthcare workers [HCWs], N=13) and Children’s Hospital of Philadelphia (CHOP; NP-PCR+ children, N=12 (outpatients); NP-PCR− healthy children, N=14). In addition, we included two adult participants who were NP-PCR−. A participant was considered positive for COVID-19 by performing quantitative RT-PCR (RT-qPCR) for SARS-CoV-2 on nasopharyngeal swabs (NP-PCR), as previously described (24).
The samples from the pre–COVID-19 period that were used as controls included Yale Kidney BioBank participants (N=10), patients with heart failure (N=5), and healthy participants (N=5). These samples were collected between 2015 and 2018. We used the convenience sampling technique, where no statistical methods were used to predetermine sample size. A flowchart describing the study population is shown in Figure 1A. In addition, we also screened 49 serum samples from 38 NP-PCR+ individuals for the SARS-CoV-2 spike protein using spike capture ELISA. These samples were collected on the same day urine samples were collected from the individual (not shown in Figure 1A).
Figure 1.
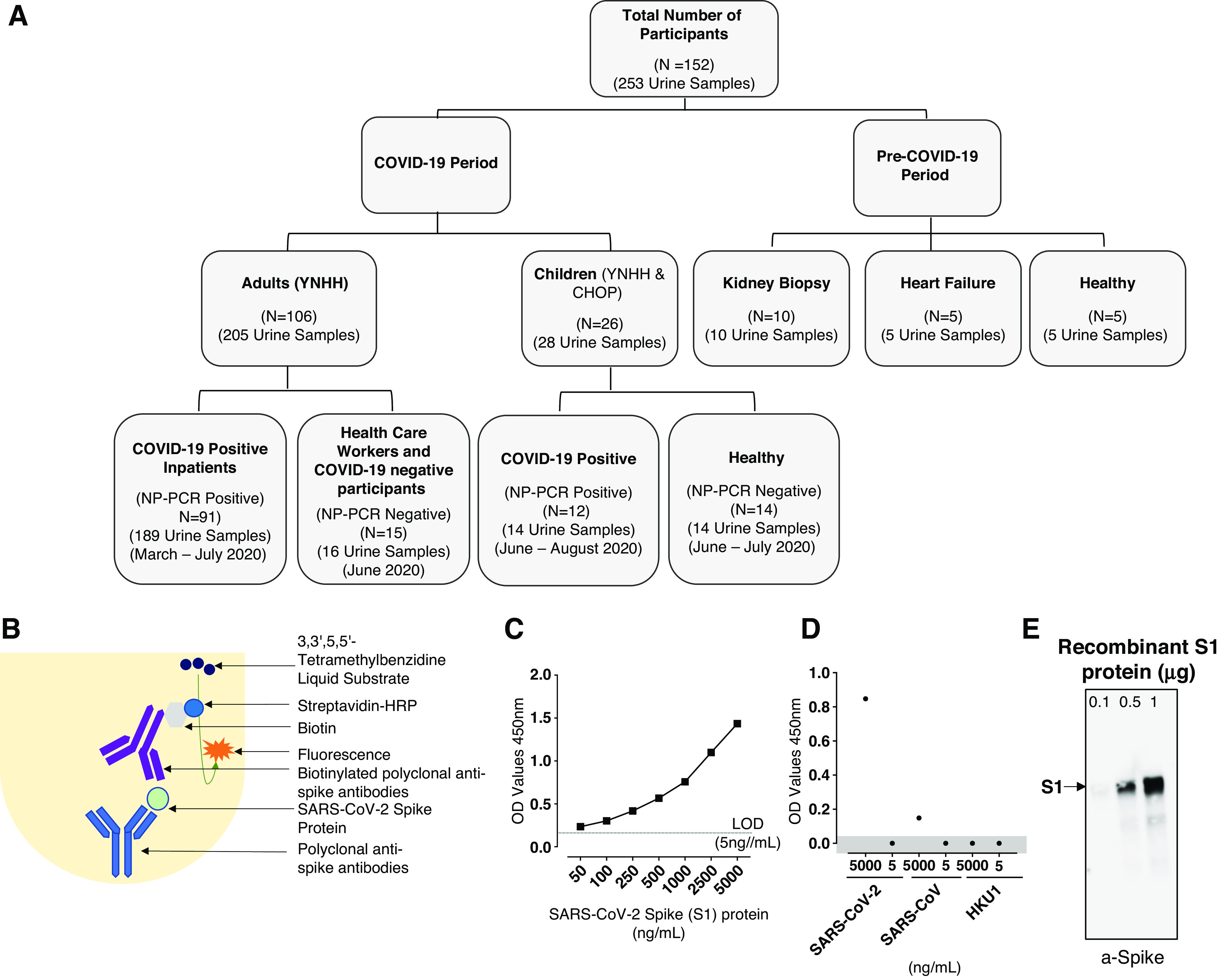
Consolidated summary of study population, assay chemistry, and sensitivity and specificity of the severe acute respiratory syndrome coronavirus 2 (SARS-CoV-2) spike protein using capture ELISA. (A) Flowchart describing the study population. Samples used in this study were collected both before and during the coronavirus disease 2019 (COVID-19) pandemic. (B) Schematic representation of the capture ELISA assay chemistry. (C) Representative standard curve generated using 5 µg/ml SARS-CoV-2 polyclonal anti-spike antibodies. LOD, limit of detection (D) Assay used to define the specificity of the SARS-CoV-2 capture ELISA. Two different concentrations (5 µg/ml and 5 ng/ml) of different human-infecting coronaviruses (SARS-CoV-2, SARS-CoV, and human coronavirus HKU1 [HCoV-HKU1]) were assessed to determine the specificity of the polyclonal anti-spike SARS-CoV-2 antibodies. Data points in the shaded area are below the limit of detection. (E) Sensitivity of the polyclonal antibodies to detect SARS-CoV-2 spike S1 protein using Western blot. SARS-CoV-2 in three different concentrations was measured: 0.1 µg, 0.5 µg, and 1 µg. CHOP, Children’s Hospital of Philadelphia; HRP, horseradish peroxidase; NP-PCR, PCR of nasopharyngeal swab; YNHH, Yale New Haven Hospital.
Sample Processing, RNA Extraction, and RT-qPCR Detection
Urine samples were centrifuged at 2000 × g for 10 minutes at room temperature, and immediately used for RNA extraction or stored frozen at −80°C. RNA extraction and RT-qPCR detection for SARS-CoV-2 in urine followed the procedures detailed in Kaplan and Kohn (24). All urine samples from CHOP were treated with nonidet P-40 (NP-40) for viral inactivation before shipping to Yale School of Medicine. Our comparative analysis using capture ELISA on NP-40–treated versus –nontreated urine samples revealed that NP-40 had no effect on the sensitivity of the assay. The processing of nasopharyngeal samples to detect the presence of SARS-CoV-2 RNA has been described in detail elsewhere (25).
Detection of SARS-CoV-2 Spike Protein Using Urine Capture ELISA and Serum Capture ELISA
Rabbit polyclonal anti-spike protein antibody, purified using protein G immunoaffinity chromatography, was purchased from MyBioSource (MBS434243; 2 µg/µl) and used as the capture antibody in the capture ELISA. This antibody was also biotinylated using the EZ-Link Micro Sulfo-NHS-Biotinylation Kit (catalog number 21925; Thermo Fisher) and used as the detection antibody. In addition, the SARS-CoV-2 S1 protein (78.3 kD), obtained from GenScript (catalog number Z03501), was used as a positive control (Figure 1B). For the urine capture ELISA (UELISA), urine samples from participants who were NP-PCR− were spiked with different concentrations of SARS-CoV-2 S1 protein (5000 ng/ml, 2500 ng/ml, 1000 ng/ml, 500 ng/ml, 250 ng/ml, 100 ng/ml, 50 ng/ml, 5 ng/ml, and 0.5 ng/ml; Figure 1C) and used as a standard in every plate. For the serum capture ELISA (SELISA), human serum sample from a healthy individual was spiked with different concentrations of the SARS-CoV-2 S1 protein (5000 ng/ml, 2500 ng/ml, 1000 ng/ml, 500 ng/ml, 250 ng/ml, 100 ng/ml, 50 ng/ml, and 5 ng/ml; Supplemental Figures 1 and 2). A 96-well ELISA plate (Nunc MaxiSorp Plate, catalog number 442404; Thermo Fisher) was coated with 5 µg/ml polyclonal anti-spike antibody diluted in carbonate coating buffer (0.848 g sodium carbonate, 1.428 g sodium bicarbonate, 500 ml distilled water) and incubated at room temperature for 2 hours. Unbound antibodies were removed, and 300 µl of blocking solution (PBS with 0.05% Tween 20 [PBST] and 2% BSA) was added per well. The plate was then incubated for an hour at room temperature. For UELISA, this was followed by an addition of 100 µl of urine sample per well to screen for the presence of SARS-CoV-2 spike protein, and the plate was incubated overnight at 4°C. For SELISA, incubation was followed by the addition of 20 µl of serum sample and 80 µl of PBS per well to screen for the presence of SARS-CoV-2 spike protein, and the plate was incubated overnight at 4°C. After 15–16 hours, the wells were washed four times with PBST; biotinylated polyclonal anti–COVID-19 antibodies (spike protein) were added at a concentration of 5µg/ml; and plates were incubated for 1 hour at room temperature. After four washes with PBST, horseradish peroxidase–streptavidin conjugate (catalog number KPL 474-3000; Seracare Life Sciences Inc.) was added at a dilution of 1:10000 in PBST, and plates were incubated for 1 hour at room temperature. A final wash (four times) with PBST was then carried out, before adding 100 µl of 3,3′,5,5′-tetramethylbenzidine liquid substrate (SureBlue Reserve TMB 1-Component Microwell Peroxidase Substrate, catalog number KPL 5120-0083; Seracare Life Sciences Inc.) to each well. The OD was measured with a BioTek FLx800 fluorescence plate reader at 450 nm. A schematic of the antigen capture ELISA is shown in Figure 1B.
In addition, to determine the specificity of polyclonal SARS-CoV-2 anti-spike antibodies (2 µg/µl, MBS434243; MyBioSource), different concentrations (0.1 µg, 0.5 µg, and 1 µg) of purified recombinant SARS-CoV-2 S1 (catalog number Z03501; GenScript) were probed using a Western blot (Figure 1E).
Evaluation of the Specificity of the Detection of SARS-CoV-2 Spike Protein by UELISA
Two recombinant proteins, SARS-CoV spike protein (NR-722, lot number 660P029) and HCoV-HKU1 spike glycoprotein (NR-53713, lot number 70037425), were obtained from BEI Resources, National Institute of Allergy and Infectious Diseases (NIAID), National Institutes of Health (NIH). These proteins (at 5 µg/ml and 5 ng/ml) were used to spike NP-PCR− urine sample in the UELISA. In addition, two NP-PCR+ urine samples were spiked with SARS-CoV-2 S1 protein at the same concentrations as stated above. The crossreactivity of the polyclonal anti-spike antibody was measured at an OD of 450 nm.
Urine Electrolyte, Albumin, and Cystatin C Analyses
Urine electrolytes were measured using ion-sensitive electrodes on the Randox Imola clinical chemistry analyzer (Randox Laboratories, Crumlin, Northern Ireland). Urine creatinine was determined using a modified Jaffe method. Creatinine measurements were standardized to National Institute of Standards and Technology Standard Reference Materials (SRM 967). Cystatin C and microalbumin were used in accordance with the manufacturer’s instructions (Randox Laboratories).
Determining the Integrity of SARS-CoV-2 S1 Protein Using Western Blot
To determine the integrity of SARS-CoV-2 spike protein in urine, 10 µl urine samples from NP-PCR+, NP-PCR−, and kidney biopsy (KB) participants were analyzed on a 4%–20% Mini-Protean TGX gel (catalog number 4568096; Bio-Rad). The gel was analyzed by Western blot after transfer to a nitrocellulose membrane (catalog number 1620214; Bio-Rad). The membrane was blocked with 5% milk (catalog number AB10109-00100; American Bio) in PBST, followed by treatment with polyclonal antibody against SARS-CoV-2 S1 and S2 proteins raised in rabbit (MBS434243; MyBioSource) and used at 1:1000 dilution in PBST. Goat anti-rabbit IgG conjugated with horse radish peroxidase (catalog number 31466; Thermo Fisher Scientific) was used as the secondary antibody at a dilution of 1:5000. The membrane was then treated with SuperSignal West Pico PLUS Chemiluminescent Substrate (catalog number 34577; Thermo Fisher Scientific), and scanned and imaged using an Odyssey Fc system (catalog number 2800-03; LI-COR Biosciences).
Statistical Analyses
Continuous variables are expressed as either mean (95% CI) or median (interquartile range [IQR]). Statistical analyses were performed using a two-tailed, unpaired t test, or one-way ANOVA in case of multiple variables. Categoric variables are expressed as numbers (%). Differences were considered statistically significant when P<0.05.
Ethics Statement
This study was approved by the Yale Human Research Protection Program Institutional Review Boards (FWA00002571, protocol ID 2000027690). Urine collection from children who were positive and negative for COVID-19 was conducted under protocol IRB20-017503, approved by the CHOP Institutional Review Board (FWA00000459). Informed consent was obtained from all enrolled participants. The pre–COVID-19 urine samples from KB participants were provided by Yale BioBank, and the study was approved by the Yale Human Investigation Committee (HIC) (HIC number 11110009286). The pre–COVID-19 urine samples from the heart failure cohort and healthy individuals were provided by J.T. (HIC number 1311013065).
Results
Identification of SARS-CoV-2 Spike Protein in Urine and Demographic Characteristics
Large-scale screening of individuals positive for SARS-CoV-2 infection, to identify both symptomatic and asymptomatic individuals, is a major priority in the control of COVID-19 transmission worldwide. This screening may be facilitated by the use of easy-to-collect biospecimens over several days during the course of an infection or after suspected exposure. To detect the presence of SARS-CoV-2 spike protein in biologic specimens from YNHH and CHOP (Figure 1A, Table 1), we developed a capture ELISA using anti-spike polyclonal antibodies for antigen capture, and biotinylated antibodies for detection of the antigen-antibody complex (Figure 1B). To use this sandwich ELISA for detection of SARS-CoV-2 spike protein in a collection of urine samples, the assay was optimized using purified recombinant S1 antigen in PBS (data not shown) and urine samples from individuals who were NP-PCR−, and standard curves were generated (Figure 1C).
Table 1.
Clinical and demographic characteristics of the study participants
| Characteristics | COVID-19 Period | Pre–COVID-19 Period | ||||||
| Adults | Children | Kidney Biopsy | Heart Failure | Healthy | ||||
| Inpatients (NP-PCR Positive; YNHH) | Healthcare Workers (NP-PCR Negative; YNHH) (COVID-19 negative; NP-PCR) |
COVID-19 positive (CHOP; YNHH) | Healthy (CHOP) | |||||
| UELISA SARS-CoV-2 S1 Positive | UELISA SARS-CoV-2 S1 Negative | |||||||
| Demographic characteristics | ||||||||
| Age | 68.3 (61.27–75.43) | 63.72 (59.16–68.29) | 44.36 (37.02–51.69) | 12.18 (9.062–15.30) | 13.93 (12.33–15.53) | 57.5 (57–71) | 66.5 (62.7–68.7) | NA |
| Sex | NA | |||||||
| Male | 10 | 24 | 3 | 5 | 6 | 6 | 2 | |
| Female | 10 | 34 | 12 | 6 | 9 | 4 | 2 | |
| Unknown | 3 | 10 | 0 | 0 | 1 | |||
| Race | NA | NA | ||||||
| Asian | 1 | 1 | 0 | 0 | ||||
| Black | 9 | 21 | 4 | 0 | ||||
| White | 9 | 29 | 6 | 0 | ||||
| Hispanic or Latino | 1 | 6 | 2 | 2 | 0 | 0 | ||
| Other/not listed | 3 | 11 | 9 | 13 | 0 | 4 | ||
| Clinical characteristics | NA | NA | NA | |||||
| Duration of hospitalization (days) | 21.6 (13.06–30.24) | 19.07 (14.43–23.71) | NA | |||||
| Discharge disposition | NA | |||||||
| Alive | 20 | 47 | 10 | |||||
| Died | 0 | 11 | 0 | |||||
| Unknown | 3 | 10 | 0 | |||||
| BMI | 36.2 (31.33–41.09) | 32.4 (29.31–35.46) | 24.87 (21.65–28.09) | 21.70 (15.73–27.66) | 30.57 (19.56–41.57) | 27.83 (26.57–41.89) | 35.97(31.41–38.97) | NA |
| Blood (7 day mean) | NA | NA | NA | NA | ||||
| WBC (1000/μl) | 7.724 (5.39–10.06) | 7.966 (7.013–8.918) | 9.6 (6.6–9.9) | NA | ||||
| Hemoglobin (g/dl) | 11.77 (10.82–12.71) | 11.42 (10.78–12.05) | 9.6 (8.9–9.6) | NA | ||||
| Platelet (1000/μl) | 245.8 (198.9–292.8) | 223.4 (198.2–292.8) | 309 (289–325) | NA | ||||
| Glucose (mg/dl) | 146.7 (121.4–172.0) | 141.4 (127.3–155.6) | NA | 103 (85.25–124) | ||||
| BUN (mg/dl) | 25.83 (18.96–32.71) | 30.59 (23.55–37.63) | 21 (21–38) | 31 (11.75–45) | ||||
| Creatinine (mg/dl) | 1.320 (0.8765–1.763) | 1.527 (1.090–1.964) | 2.45 (2.3–2.8) | 1.35(0.75–2.25) | ||||
| Total protein (g/dl) | 6.610 (6.301–6.919) | 6.408 (6.230–6.585) | NA | NA | ||||
| D-dimer | 3.000 (1.728–4.272) | 3.288 (2.381–4.195) | NA | NA | ||||
| Chloride (mmol/L) | 102.9 (100.6–105.1) | 103.7 (102.3–105.0) | 101 (101–104) | 100 (92.75–102.75) | ||||
| Potassium (mmol/L) | 4.102 (3.941–4.263) | 4.074 (3.970–4.178) | 3.8 (3.6–4.1) | 4(3.9–4.1) | ||||
| Urine | ||||||||
| Total protein (g/L) | 3.423 (1.139–5.708) | 3.254 (2.006–4.502) | 0.6277 (0.3905–0.8648) | 3.628 (0.2663–6.989) | 0.7950 (0.4618–1.128) | NA | NA | NA |
| Creatinine (mg/dl) | 105.5 (71.20–139.9) | 80.99 (66.14–95.85) | 66.94 (42.47–91.42) | 67.01 (28.69–105.3) | 137.2 (60.91–213.4) | 60.89 (48.56–85.39) | 68.19 (37.37–168.28) | 55.16 (49.44–253.57) |
| Microalbumin (mg/dl) | 7.548 (4.294–10.80) | 12.93 (6.038–19.83) | 0.9923 (0.05204–1.933) | 16.60 (-17.45–50.64) | 0.9110 (0.2679–1.554) | 42.84 (1.46–110.88) | 3.03 (0.5–10.85) | 0.5 (0.5–1.56) |
| Cystatin C (mg/L) | 0.2152 (0.044960–0.3809) | 1.179 (0.4372–1.921) | 0.04154 (0.02656–0.05651) | 0.09125 (-0.004743–0.1872) | 0.07100 (0.04589–0.09611) | NA | NA | NA |
| Sodium (mmol/L) | 66.11 (48.68–83.55) | 77.23 (64.76–89.69) | 109.3 (68.54–150.0) | 124.7 (38.62–210.9) | 158.9 (119.4–198.3) | NA | 49 (24–83.5) | 120 (71.5–153) |
| Urea (mmol/L) | 287.7 (220.0–355.5) | 250.3 (215.3–285.2) | 275.2 (169.1–381.3) | 245.4 (110.4–380.3) | 28.2 (152.4–404.0) | NA | 152.83 (91.31–399.28) | 220.89 (126.06–333.49) |
| Potassium (mol/L) | 28.87 (20.51–37.23) | 25.00 (21.73–28.26) | 62.62 (36.12–89.11) | 18.25 (7.036–29.46) | 38.00 (10.01–65.99) | NA | 36 (22–54.6) | 49.6 (48.15–63.35) |
COVID-19, coronavirus disease 2019; NP-PCR, PCR result from nasopharyngeal swab; YNHH, Yale New Haven Hospital; CHOP, Children’s Hospital of Philadelphia; UELISA, urine capture ELISA; SARS-CoV-2, severe acute respiratory syndrome coronavirus 2; NA, not available; BMI, body mass index; WBC, white blood cells.
Using this assay, we determined the lower limit of detection (LOD) to be 5 ng/ml of protein in the urine (Figure 1C). The specificity of the antibodies was further evaluated by ELISA using available recombinant spike proteins from SARS-CoV and HCoV-HKU1. Although no crossreactivity could be detected with HCoV-HKU1 spike protein at concentrations as high as 5 µg/ml, or with SARS-CoV spike protein at 5 ng/ml, a weak signal could be detected by ELISA with the SARS-CoV spike protein at 5 µg/ml (Figure 1D). Western blot analysis confirmed the specificity of the antibodies to the SARS-CoV-2 spike protein (Figure 1E and Supplemental Figure 1)
Having determined sensitivity and specificity, the assay was then used to examine a repository of 253 urine samples collected from 152 participants for the presence of SARS-CoV-2 spike protein (Figure 1A; Table 1). This repository included 203 urine samples from NP-PCR+ adults (189 urine samples; N=91) and children (14 urine samples; N=12) from YNHH and CHOP. Samples from NP-PCR− HCWs (N=13) and children (N=12) from both hospitals, adult participants negative for COVID-19 (N=2), and urine samples collected before the COVID-19 pandemic (N=20) were used as controls (Figure 1A). Creatinine levels in all of the urine samples were measured and used to calculate the urine protein-creatinine ratio to standardize measurements. Of the 203 urine samples from 103 NP-PCR+ adults and children analyzed in this study, 29 (N=23; 25%) were found to contain the SARS-CoV-2 spike protein in the urine (Figure 2A). Interestingly, one child (with no respiratory symptoms) who was NP-PCR− (tested for preadmission screening) appeared to have high levels of the SARS-CoV-2 spike protein in their urine (Figure 2A). Overall, the mean concentration of the SARS-CoV-2 spike protein in adults was 0.033 (95% CI, 0.01 to 0.06) mg/mg of urine creatinine, and the child had 0.0083 mg/mg of urine creatinine. None of the urine samples from adult HCWs (N=15; YNHH), NP-PCR+ children (N=12), or pre–COVID-19 participants (N=20) showed the presence of the SARS-CoV-2 spike protein (Figure 2A). No correlation between the presence of the SARS-CoV-2 spike protein in the urine and the sex of patients with COVID-19 could be found (P=0.34; Figure 2B) among 20 of the 23 participants with the SARS-CoV-2 spike protein in their urine (information on sex for three individuals was not available). Similarly, no significant association between the presence of the SARS-CoV-2 spike protein and factors such as body mass index (BMI; P=0.16), age (P=0.29), and duration of hospitalization (P=0.49) could be found (Figure 2C). In this cohort, we also examined possible correlations between the levels of albumin and cystatin C in the urine samples and BMI, age, and duration of hospitalization of the patient. However, no significant association was observed between these confounding factors and elevated levels of albumin (BMI, P=0.33; age, P=0.06; duration of hospitalization, P=0.12) and cystatin C (BMI, P=0.36; age, P=0.88; duration of hospitalization, P=0.11) (Supplemental Figure 4, A and B). Among the adults who were NP-PCR+, no correlation between nasopharyngeal SARS-CoV-2 viral load and presence of the SARS-CoV-2 spike protein in urine could be found (P=0.47; Supplemental Figure 5). For individuals who were negative for the urine spike (Ur-S−), the mean±SD viral titer was 2.4 × 107±6.4 × 107; the mean±SD viral titer for individuals positive for the urine spike (Ur-S+) was 4.4 × 106±1.1 × 107.
Figure 2.
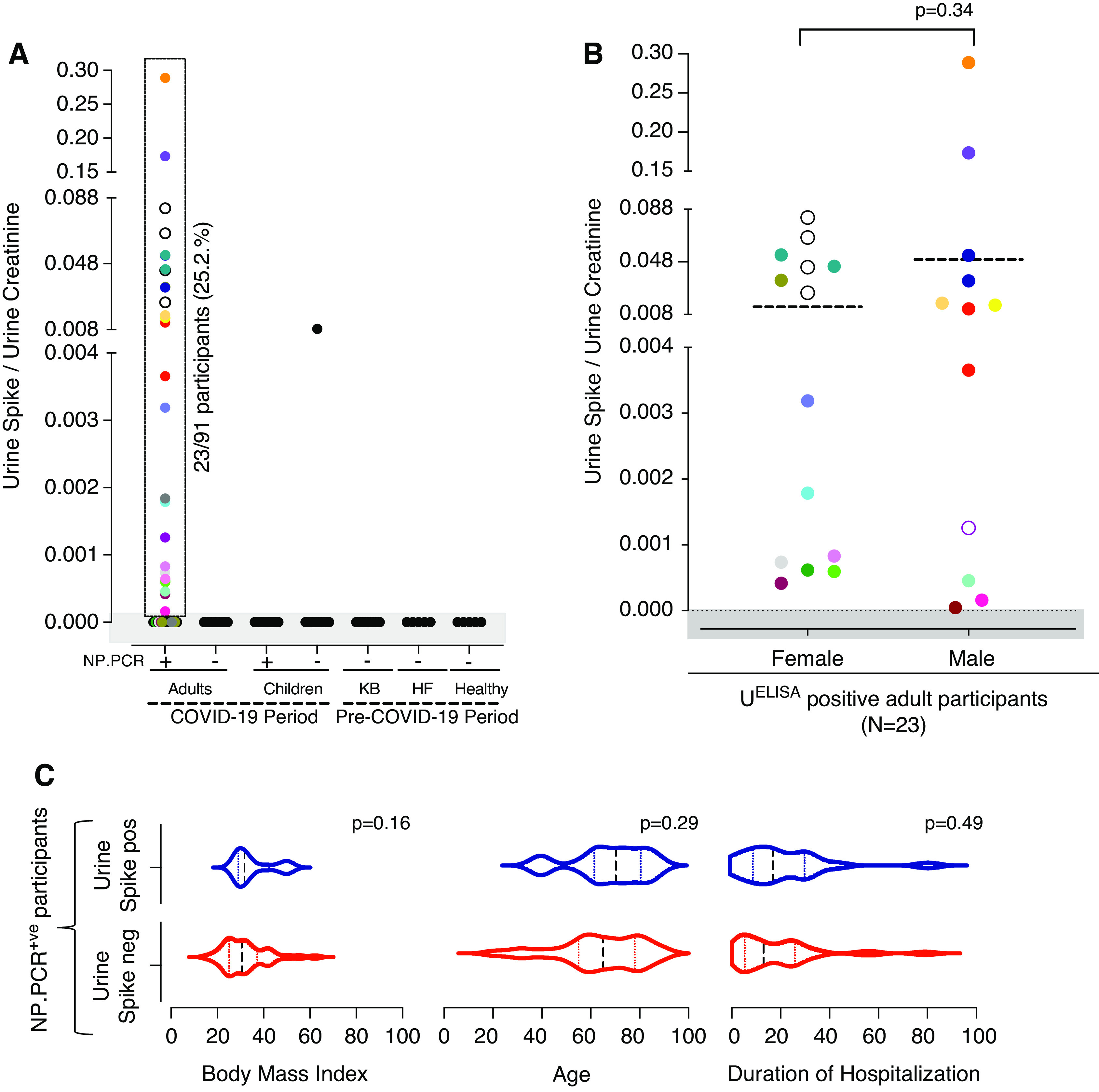
Detection of SARS-CoV-2 in urine and role of confounding factors. (A) Concentration of SARS-CoV-2 spike protein in urine, measured using capture ELISA. Multiple urine samples from the same individual are color matched. Multiple urine samples from the same individual are color matched. (B–C) Effect of sex, body mass index, and duration of hospitalization on the presence of SARS-CoV-2 spike protein in urine. Dotted lines indicate (B) mean and (C) median (interquartile range). HF, heart failure; KB, kidney biopsy; UELISA, urine capture ELISA.
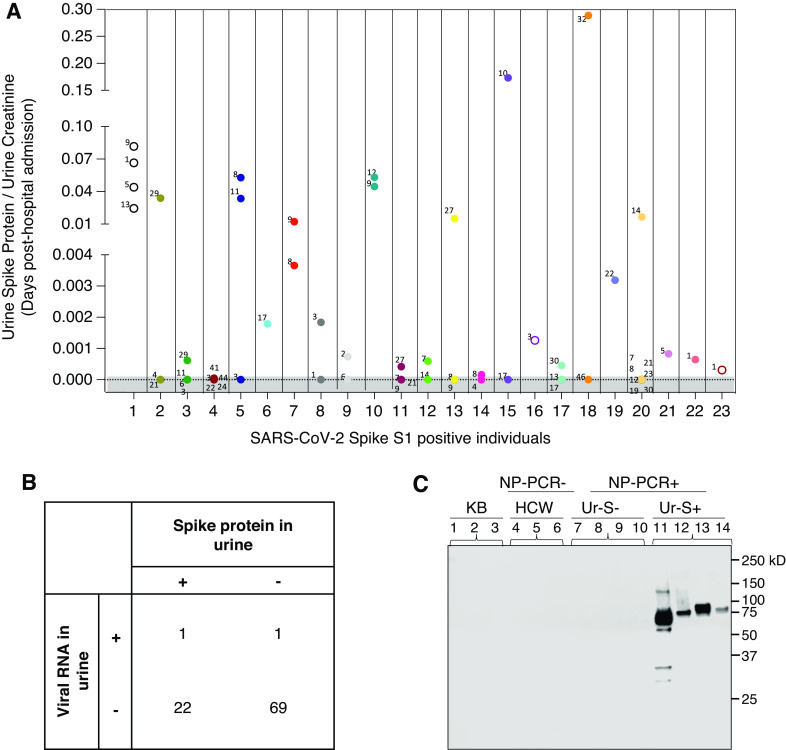
Of the 23 patients with COVID-19 with the spike protein in their urine, 17 provided at least two urine samples during the course of hospitalization (Figure 3A). The SARS-CoV-2 spike protein could be detected in urine from day 1 to day 44 post–hospital admission (Figure 3A). However, no correlation could be found between the concentration of the SARS-CoV-2 spike protein in the urine and the day urine sample was collected post–hospital admission. To assess whether the presence of the spike protein in the urine of a subset of individuals with COVID-19 is due to the presence SARS-CoV-2–infected cells in this biospecimen, RT-qPCR analysis was conducted on all urine samples from NP-PCR+ individuals using two primer pairs, as previously reported (24). Of 93 patients who were NP-PCR+, only two individuals were positive for viral RNA in their urine (approximately 2%; Figure 3B). Of these two positive individuals (one male and one female), only one (female) was positive for both the spike protein and viral RNA in urine.
Figure 3.
Detection of SARS-CoV-2 in urine samples. (A) Assessment on the concentration of SARS-CoV-2 spike protein in urine using capture ELISA on samples from the same individuals collected at different time points (shown in days since hospitalization). Multiple urine samples from the same individual are color matched. Data points in the shaded area are below the limit of detection. (B) Detection of viral RNA in urine to determine if the presence of spike protein is due to kidney infection. (C) Detection of SARS-CoV-2 spike protein in urine by Western blot. Lanes 1–3 are samples from KB participants, lanes 4–6 are from NP-PCR− healthcare workers (HCW), lanes 7–10 are from NP-PCR+ individuals who are negative for SARS-CoV-2 spike protein in urine (Ur-S−), and lanes 11–14 are NP-PCR+ individuals who are positive for the SARS-CoV-2 spike protein in urine (Ur-S+). A protein fragment size of 78.3 kD, corresponding to the SARS-CoV-2 spike S1 protein, is seen in individuals positive for the urine spike.
More importantly, considering the importance of protein size in renal filtration, we detected the spike protein in urine samples by Western blot. As shown in Figure 3C, a band of 78.3 kD that comigrates with the recombinant S1 antigen was detected in the urine of adults positive for the SARS-CoV-2 spike. No S1 protein could be detected in the Ur-S− adults positive for SARS-CoV-2, HCWs, or individuals whose urine samples were collected before the pandemic.
Evidence of Proteinuria in Individuals Infected with SARS-CoV-2
To assess whether the presence of the spike protein in the urine of individuals with COVID-19 may indicate renal abnormalities caused or exacerbated by viral infection, we analyzed the link between SARS-CoV-2 infection and renal filtration of the human proteins albumin and cystatin C. Urine samples from ten patients who underwent KB, which were collected before the pandemic (2015–2018), were included as controls. Our analysis revealed that the median (IQR) concentration of urine albumin among all NP-PCR+ individuals in our cohort was 0.073 (0.019–0.276) mg/mg of urine creatinine, whereas urine cystatin C was 0.00014 (0.00008–0.00106) mg/mg of urine creatinine. Among individuals who were Ur-S+, the median (IQR) urine albumin concentration was 0.089 (0.016–0.299) mg/mg of urine creatinine), whereas urine cystatin C concentration was 0.00012 (0.00007–0.00029) mg/mg of urine creatinine. The median (IQR) concentration of urine albumin in individuals who were NP-PCR+ was 0.061 (0.019–0.158) mg/mg of urine creatinine, whereas that for urine cystatin C was 0.00012 (0.000007–0.00050) mg/mg of urine creatinine. In contrast, the median (IQR) concentration of urine albumin in HCWs who were NP-PCR− was 0.011 (0.007–0.024) mg/mg of urine creatinine, whereas that for urine cystatin C was 0.00007 (0.00006–0.00008) mg/mg of urine creatinine. Eight urine samples from six individuals who were Ur-S+ had an albumin concentration >0.3 mg/mg of urine creatinine (Figure 4A, Supplemental Figure 3A). In total, 22 adult participants with COVID-19 (24%; 31 urine samples) had urine albumin levels >0.3 mg/mg of urine creatinine (Figure 4A, Supplemental Figure 3A). Similarly, 18 adult participants with COVID-19 (21%; 26 samples) had a median (IQR) urine cystatin C concentration of >0.0022 (0.001–0.003) mg/mg of urine creatinine (Figure 4C, Supplemental Figure 3B). Interestingly, using a cutoff for albumin of 0.3 mg/mg of urine creatinine, which is considered a marker for AKI (26), we found significant association between elevated urine albumin-creatinine ratio and concentration of the SARS-CoV-2 spike protein in urine (P=0.02; Figure 4B). There were six individuals with an elevated urine albumin-creatinine ratio >0.3 mg/mg who had spike protein in their urine, with a median (IQR) concentration of 0.0086 (0.00019–0.1341) mg/mg of urine creatinine. As shown in Figure 4D, for two patients who were NP-PCR+ (INP.1.019 and INP.1.095) with three or four urine samples testing positive for the SARS-CoV-2 spike protein, our analysis showed no correlation between the levels of urine albumin and cystatin C (normalized to creatinine) and the levels of urine SARS-CoV-2 spike protein (Figure 4D).
Figure 4.
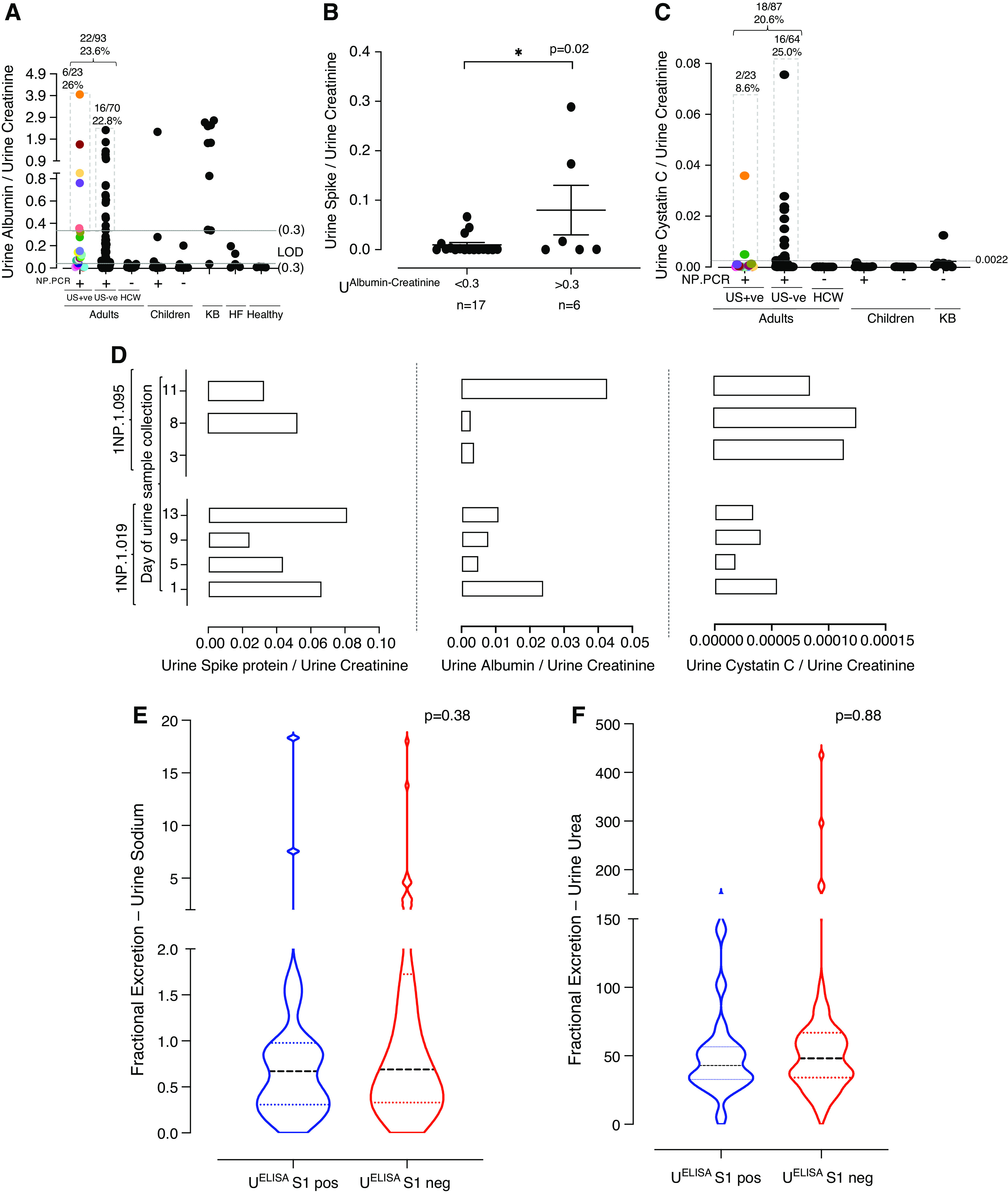
Urine proteomics to determine presence of SARS-CoV-2 spike protein as a biomarker for kidney injury in patients with COVID-19. (A) Concentration of urine albumin (in milligram per milligram of urine creatinine) across one urine sample collected per study participant. Multiple urine samples from the same individual are color matched. (B) Comparison of the concentration of urine spike to albumin concentration (normalized to urine creatinine) >0.3 and <0.3 mg/mg of urine creatinine. One sample per Ur-S+ participant was considered. The line represents mean±SEM. (C) Concentration of urine cystatin C (in milligrams per milligram of urine creatinine) across urine samples, with one collected per study participant. Mean value of cystatin C for KB individuals was determined (0.0022 mg/mg of urine creatinine) and used as a cutoff across our study population. Multiple urine samples from the same individual are color matched. (D) Determining the correlation between the concentrations of SARS-CoV-2 spike protein, albumin, and cystatin C in multiple urine samples from two representative adult NP-PCR+ participants. (E) Fractional excretion of urine sodium. Dashed lines represent median (interquartile range [IQR]). (F) Fractional excretion of urine urea. Dashed lines represent median (IQR). HF, heart failure; LOD, limit of detection; neg, negative; pos, positive.
Previous studies demonstrated an increased level of blood creatinine (>0.3 mg/dl) in individuals with AKI (27–29). To determine whether the presence of spike protein in urine was due to AKI, we measured the concentration of creatinine (in milligrams per deciliter) in sera of individuals that were either Ur-S+ or Ur-S−. No significant difference was found between the levels of serum creatinine and the presence or absence of SARS-CoV-2 spike protein in urine among these individuals (P=0.70; Supplemental Figure 2B). Among the individuals who were Ur-S+, the mean concentration of serum creatinine was 1.319 (95% CI, 0.88 to 1.76) mg/dl, whereas the mean concentration of individuals who were Ur-S− was 1.526 (95% CI, 1.09 to 1.96) mg/dl. In addition, we analyzed the FENa and FEUrea levels between samples from individuals who were Ur-S+ and Ur-S− by considering one urine sample per participant (adults, N=89; children, N=2). We found no statistically significant difference for both FENa (P=0.38) and FEUrea (P=0.88) between Ur-S+ and Ur-S− samples (Figure 4, E and F). The mean FENa for Ur-S+ samples was 1.86 (95% CI, −0.09 to 3.82), whereas that for Ur-S− samples was 1.73 (95% CI, 0.93 to 2.54). The mean FEUrea for Ur-S+ samples was 48.98 (95% CI, 35.06 to 62.90), whereas that for Ur-S− samples was 62.13 (95% CI, 45.25 to 79.00).
The Presence of Spike Protein in Urine Does Not Correlate with Increased Levels of Viral Protein in Serum
To assess whether the presence of high levels of SARS-CoV-2 spike protein in the urine samples from patients with COVID-19 may be due to high levels of the protein in the serum, we screened 49 available serum samples from the cohort of 38 patients with COVID-19 using SELISA (48 urine samples; N=38). Only four samples from three individuals showed levels of spike protein in serum above the lower LOD (5 ng/ml), and none of these were positive in the ELISA assay performed on the urine samples from patients with COVID-19 (Figure 5). Together, these data suggest there is no correlation between high levels of spike protein in urine and serum concentrations of the protein.
Figure 5.
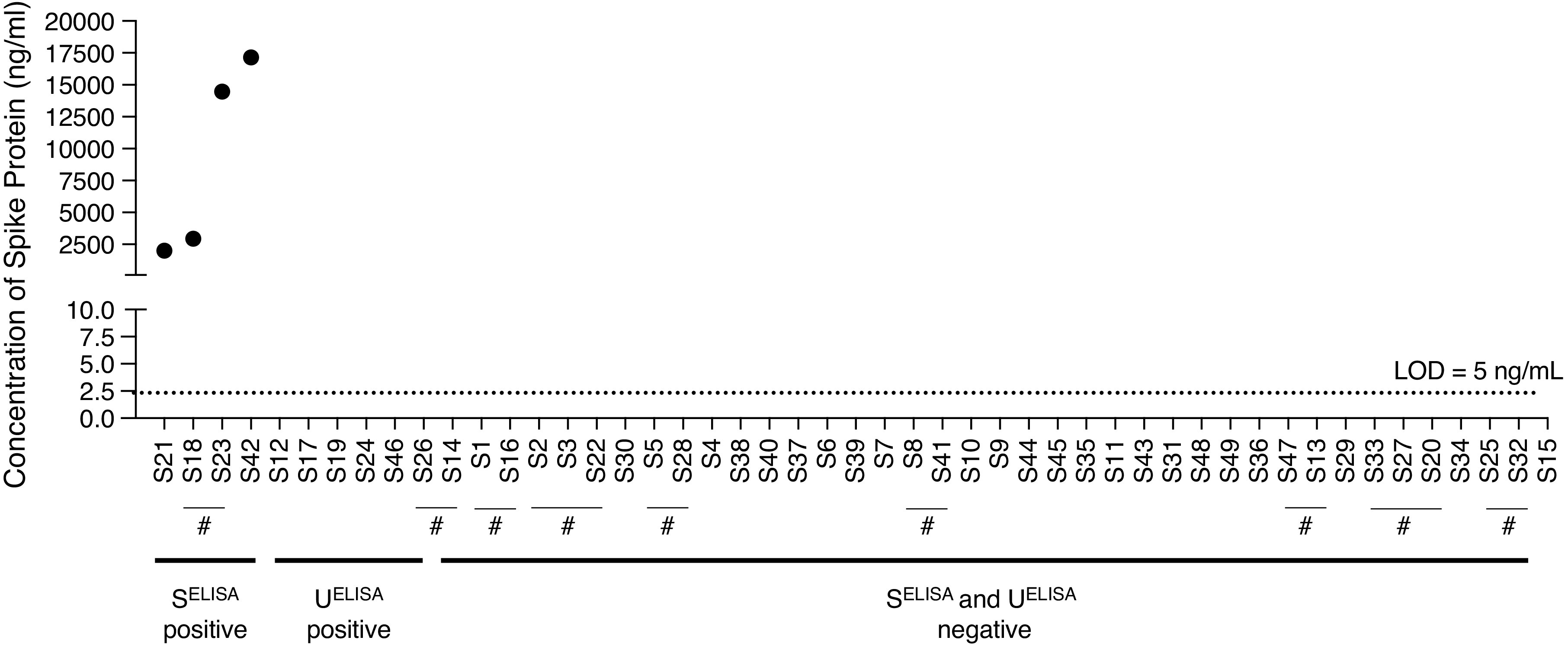
Serum capture ELISA (SELISA) to determine the presence of SARS-CoV-2 spike protein in patients with COVID-19. ELISA assay performed on 49 serum samples collected from 38 patients with COVID-19. Multiple serum samples from the same individual is indicated with #. Dashed line represents the LOD, i.e., 5 ng/ml of spike protein. “SELISA positive” indicates the serum samples positive for the spike protein in SELISA. “UELISA positive” indicates the serum samples that are positive for the spike protein in UELISA, but are negative in SELISA. “SELISA and in UELISA negative” samples indicate the serum samples that are negative for the spike protein in both SELISA and UELISA.
Discussion
In this study, we developed an antigen capture ELISA assay to detect SARS-CoV-2 spike protein and used it to analyze urine samples from a cohort of 152 individuals, including adults and children who were NP-PCR+. Although antigen-based detection assays have been reported (30,31), to the best of our knowledge, this is the first report using an antigen capture assay to detect spike protein in the urine of patients with COVID-19 and asymptomatic individuals.
Analysis of our urine collection revealed that approximately 25% of patients who were NP-PCR+ shed the SARS-CoV-2 spike protein in their urine. In addition, the urine from one NP-PCR− child was found to be positive for SARS-CoV-2 spike protein. In our study, the overall mean ratio of SARS-CoV-2 spike protein/urine creatinine in adults was 0.033 (95% CI, 0.01 to –0.06) mg/mg of urine creatinine, whereas that of the positive child was 0.0083 mg/mg of urine creatinine. None of the other urine samples used in our study showed the presence of the SARS-CoV-2 spike protein. There was also no correlation between the presence of the SARS-CoV-2 spike protein and the confounding factors of BMI, age, sex, and duration of hospitalization. We further assessed whether the presence of spike protein in urine was due to kidney infection, injury, or dysfunction in patients with COVID-19. No correlation was observed between the presence of the SARS-CoV-2 spike protein in urine and markers of kidney dysfunction, including serum creatinine, FENa, FEUrea, or cystatin C. However, we noted the level of the SARS-CoV-2 spike protein in urine was higher in patients with albuminuria (Figure 4B). A 2003 study by Chu et al. (32) suggested the development of AKI in patients with SARS-CoV was likely to be due to multiorgan failure rather than kidney tropism of the virus. Interestingly, our Western blot analysis of the urine samples showed the presence of a protein fragment that was the expected size (78.3 kD) of theS1 fragment of the spike protein (Figure 3C). In addition, we also observed additional fragments, suggesting proteolysis (Figure 3C). Considering that both the spike protein and albumin have molecular masses >60 kD, it is likely that their release may be the result of similar filtration abnormalities (33). Altogether, our data suggest that the presence of the spike protein in urine samples of some patients with COVID-19 may still be indicative of an unknown or unpredicted kidney injury, most likely involving the spilling of spike protein from serum. Notably, the presence of proteinuria and microscopic hematuria has been associated with greater clinical severity of COVID-19 (19). The predominant form of kidney injury in COVID-19 seems to be acute tubular injury, which might be secondary to cytokine storm or shock. Direct viral infection, when present, may only occur in the most severe cases, as noted in autopsy studies (34–36).
Another important finding of this study is the lack of viral RNA in the urine of most individuals who were NP-PCR+. Of 93 patients who were NP-PCR+, only two individuals were positive for viral RNA in their urine (approximately 2%; Figure 3B). Of these two positive individuals (one male and one female), only one (female) was positive for both spike protein and viral RNA in urine. This suggests that the SARS-CoV-2 spike protein detected in the urine is a direct result of a filtration abnormality, rather than a viral infection of the kidney. Whether the presence of viral RNA in the urine of a small percentage of patients with COVID-19 is a result of viral shedding, or simply due to contamination during urine collection, remains to be further elucidated. However, our analysis of urine samples collected at different times from the same individuals during hospitalization do not show a pattern of viral RNA detection that would be consistent with viral RNA shedding in the urine. More importantly, it cannot be ruled out that the urine samples, in general, have not gone through as rigorous accuracy verification or matrix equivalency studies for RT-qPCR as have the nasopharyngeal or nasal swab samples.
A significant finding in this study is that one child was found to have the SARS-CoV-2 spike protein in their urine, despite being NP-PCR−. Notably, this urine sample was collected on the same day the nasopharyngeal swab was performed for RT-qPCR analysis. One possible explanation is that the child was previously infected and then tested negative due to viral clearance, but continued to shed the viral spike protein in their urine. Another possibility is that the PCR result in this patient was a false negative. The ability of the antigen capture assay to detect spike protein in an individual who is NP-PCR− highlights the need for the development of assays that are not intrusive, are rapid, and can be deployed for large-scale detection of active infection in the general population, and at different times, to prevent continued propagation of the virus. Although urine ELISA-based tests are particularly suitable for large-scale, repeat, and rapid diagnostic campaigns, the fact that only 25% of infected individuals in our study were found to have the spike protein in their urine suggests that the sensitivity of the current urine spike capture assay is not sufficient for population-based screening. Efforts to evaluate the usefulness of this antigen-based assay to detect SARS-CoV-2 infection in other biospecimens, such as saliva, are warranted.
In conclusion, our data demonstrate that 25% of individuals infected with SARS-CoV-2 shed spike protein in their urine. RT-qPCR on urine samples demonstrated that this shedding is neither due to the presence of infected cells in this specimen, nor to high levels of this viral protein in the serum. However, it does not preclude the possibility of kidney infection by the virus. Nevertheless, our data highlight possible kidney abnormalities resulting from SARS-CoV-2 infection. Considering the possible long-term implications of these findings, longitudinal studies aimed at understanding the long-term effects of SARS-CoV-2 infection on renal filtration and injury are warranted.
Disclosures
A.I. Ko reportsreceiving research funding from Bristol Myers Squibb, Regeneron, Serimmune, and Tata Medical and Diagnostics; and having consultancy agreements with Tata Medical and Diagnostics. D.G. Moledina reports receiving honoraria from the British Medical Journal, National Kidney Foundation, and Remedy Health Media; serving as an editorial board member for Kidney360; receiving research funding from National Institute of Diabetes and Digestive and Kidney Diseases (grants K23DK117065, R01DK12681, UH3DK114866, and P30DK079310), outside the submitted work; and being a coinventor of the pending patent application “Methods and Systems for Diagnosis of Acute Interstitial Nephritis,” which is subject to an option for a license agreement with RenalytixAI Inc. A.R. John reports receiving research funding from the Burroughs Wellcome Fund and National Institutes of Health (NIH); being a scientific advisor for, or member of, Pluton Biosciences; and having patents and inventions involving antimalarials and malaria and SARS-CoV-2 biomarkers. V. Rao reports patents and inventions with Corvidia Therapeutics, and having consultancy agreements with Translational Catalyst. J. Testani reports receiving research funding from Abbott, Boehringer Ingelheim, Bristol Myers Squibb, 3ive Labs, Merck, NIH, Otsuka, Sanofi, Sequana Medical, The Foundry, and the US Food and Drug Administration; having consultancy agreements with, and receiving honoraria from, AstraZeneca, Bayer, Boehringer Ingelheim, Bristol Myers Squibb, Cardionomic, FIRE1, Lexicon, Magenta Med, Merck, Novartis, Regeneron, Reprieve, Sanofi, Sequana Medical, Windtree Therapeutics, and W.L. Gore; having patents and inventions with Corvidia, Reprieve Inc., and Yale University; and having ownership interest in Reprive Inc. All remaining authors have nothing to disclose.
Funding
C.B. Mamoun’s research is supported by NIHhttps://doi.org/10.13039/100000002 grants and the Steven and Alexandra Cohen Foundation. As an Investigator in the Pathogenesis of Infectious Diseases of the Burroughs Wellcome Fund, A.R. John is supported by NIH/NIAID grants R01AI103280, R21AI144472, and R21AI154370. J. Testani was supported by National Heart, Lung, and Blood Institute https://doi.org/10.13039/100000050 awards K23HL114868, L30HL115790, R01HL139629, R21HL143092, R01HL128973, and R01HL148354. IMPACT received support from the Yale COVID-19 Research Resource Fund.
Acknowledgments
This manuscript previously appeared on MedRxiv at https://doi.org/10.1101/2021.01.27.21250637.
Author Contributions
E.H. Akaho, M. Campbell, A. Casanovas-Massana, J.E. Chiu, C.S. Dela Cruz, S.F. Farhadian, S. George, N.D. Grubaugh, C.A. Harden, A.R. John, A.I. Ko, M. Ledizet, C.B. Mamoun, D.G. Moledina, I.M. Ott, M. Tokuyama, C.B.F. Vogels, A.E. Watkins, and A.L. Wyllie were responsible for investigation; A.Z. Berna Perez, J.E. Chiu, S.F. Farhadian, J. Gagnon, S. George, A.R. John, M. Ledizet, C.B. Mamoun, D.G. Moledina, A.C. Pal, I. Renard, P. Singh, J. Testani, P. Vydyam, and A.L. Wyllie reviewed and edited the manuscript; A.Z. Berna, J. Gagnon, S. George, A.R. John, M. Ledizet, P. Lu, A.C. Pal, V. Rao, I. Renard, P. Singh, J. Testani, S. Timalsina, M. Tokuyama, A. Venkataraman, C.B.F. Vogels, and P. Vydyam were responsible for methodology;
S. George was responsible for validation and visualization; S. George, M. Ledizet, C.B. Mamoun conceptualized the study; S. George, D.G. Moledina, and P. Singh were responsible for data curation; S. George and C.B. Mamoun wrote the original draft; S. George, M. Munshi, and A.C. Pal were responsible for formal analysis; A.R. John and C.B. Mamoun were responsible for funding acquisition; and C.B. Mamoun provided supervision and was responsible for project administration and resources.
Supplemental Material
This article contains the following supplemental material online at http://kidney360.asnjournals.org/lookup/suppl/doi:10.34067/KID.0002172021/-/DCSupplemental.
Specificity of the polyclonal antibodies to detect SARS-CoV-2 spike S1 protein using Western blot. Download Supplemental Figure 1, PDF file, 1.4 MB (1.4MB, pdf)
Concentration of urine and serum creatinine across the study population. Download Supplemental Figure 2, PDF file, 1.4 MB (1.4MB, pdf)
Urine proteomics to determine presence of SARS-CoV-2 spike protein as a biomarker for kidney injury in COVID-19 patients across all the urine samples. Download Supplemental Figure 3, PDF file, 1.4 MB (1.4MB, pdf)
(A) Comparison on the role of confounding factors in the elevated levels of albumin in urine. (B) Comparison of role of confounding factors in the elevated levels of cystatin C in urine. Download Supplemental Figure 4, PDF file, 1.4 MB (1.4MB, pdf)
Correlation of SARS-CoV-2 viral RNA load to the presence of spike protein in urine among COVID-19 NP-PCR+ adults. Download Supplemental Figure 5, PDF file, 1.4 MB (1.4MB, pdf)
Representative standard curve generated using 5μg/mL SARS-CoV-2 polyclonal anti-spike antibodies. Download Supplemental Figure 6, PDF file, 1.4 MB (1.4MB, pdf)
References
- 1.Chan JF, Yuan S, Kok KH, To KK, Chu H, Yang J, Xing F, Liu J, Yip CC, Poon RW, Tsoi HW, Lo SK, Chan KH, Poon VK, Chan WM, Ip JD, Cai JP, Cheng VC, Chen H, Hui CK, Yuen KY: A familial cluster of pneumonia associated with the 2019 novel coronavirus indicating person-to-person transmission: a study of a family cluster. Lancet 395: 514–523, 2020. 10.1016/S0140-6736(20)30154-9 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Huang C, Wang Y, Li X, Ren L, Zhao J, Hu Y, Zhang L, Fan G, Xu J, Gu X, Cheng Z, Yu T, Xia J, Wei Y, Wu W, Xie X, Yin W, Li H, Liu M, Xiao Y, Gao H, Guo L, Xie J, Wang G, Jiang R, Gao Z, Jin Q, Wang J, Cao B: Clinical features of patients infected with 2019 novel coronavirus in Wuhan, China. Lancet 395: 497–506, 2020. 10.1016/S0140-6736(20)30183-5 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Wang C, Horby PW, Hayden FG, Gao GF: A novel coronavirus outbreak of global health concern. Lancet 395: 470–473, 2020. 10.1016/S0140-6736(20)30185-9 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Zhu N, Zhang D, Wang W, Li X, Yang B, Song J, Zhao X, Huang B, Shi W, Lu R, Niu P, Zhan F, Ma X, Wang D, Xu W, Wu G, Gao GF, Tan W; China Novel Coronavirus Investigating and Research Team: A novel coronavirus from patients with pneumonia in China, 2019. N Engl J Med 382: 727–733, 2020. 10.1056/NEJMoa2001017 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.World Health Organization: Coronavirus disease 2019 (COVID-19) situation report–57, Geneva, Switzerland, World Health Organization, 2020 [Google Scholar]
- 6.World Health Organization (WHO): Estimating mortality from COVID-19: Scientific brief, 2020. Available at: https://www.who.int/publications/i/item/WHO-2019-nCoV-Sci-Brief-Mortality-2020.1Accessed April 27, 2021
- 7.Zhong NS, Zheng BJ, Li YM, Poon, Xie ZH, Chan KH, Li PH, Tan SY, Chang Q, Xie JP, Liu XQ, Xu J, Li DX, Yuen KY, Peiris, Guan Y: Epidemiology and cause of severe acute respiratory syndrome (SARS) in Guangdong, People’s Republic of China, in February, 2003. Lancet 362: 1353–1358, 2003. 10.1016/S0140-6736(03)14630-2 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Pollack MP, Pringle C, Madoff LC, Memish ZA: Latest outbreak news from ProMED-mail: Novel coronavirus – Middle East. Int J Infect Dis 17: e143–e144, 2013. 10.1016/j.ijid.2012.12.001 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Malaiyan J, Arumugam S, Mohan K, Radhakrishnan GG: An update on the origin of SARS-CoV- 2: Despite closest identity, bat (RaTG13) and pangolin derived coronaviruses varied in the critical binding site and O-linked glycan residues. J Med Virol 93: 499–505, 2021 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Andersen KG, Rambaut A, Lipkin WI, Holmes EC, Garry RF: The proximal origin of SARS-CoV-2. Nat Med 26: 450–452, 2020. 10.1038/s41591-020-0820-9 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Zhou P, Yang XL, Wang XG, Hu B, Zhang L, Zhang W, Si HR, Zhu Y, Li B, Huang CL, Chen HD, Chen J, Luo Y, Guo H, Jiang RD, Liu MQ, Chen Y, Shen XR, Wang X, Zheng XS, Zhao K, Chen QJ, Deng F, Liu LL, Yan B, Zhan FX, Wang YY, Xiao GF, Shi ZL: A pneumonia outbreak associated with a new coronavirus of probable bat origin. Nature 579: 270–273, 2020. 10.1038/s41586-020-2012-7 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Belouzard S, Millet JK, Licitra BN, Whittaker GR: Mechanisms of coronavirus cell entry mediated by the viral spike protein. Viruses 4: 1011–1033, 2012. 10.3390/v4061011 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Heald-Sargent T, Gallagher T: Ready, set, fuse! The coronavirus spike protein and acquisition of fusion competence. Viruses 4: 557–580, 2012. 10.3390/v4040557 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Walls AC, Park YJ, Tortorici MA, Wall A, McGuire AT, Veesler D: Structure, function, and antigenicity of the SARS-CoV-2 spike glycoprotein. Cell 181: 281–292.e6, 2020. 10.1016/j.cell.2020.02.058 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Letko M, Marzi A, Munster V: Functional assessment of cell entry and receptor usage for SARS-CoV-2 and other lineage B betacoronaviruses. Nat Microbiol 5: 562–569, 2020. 10.1038/s41564-020-0688-y [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Zou X, Chen K, Zou J, Han P, Hao J, Han Z: Single-cell RNA-seq data analysis on the receptor ACE2 expression reveals the potential risk of different human organs vulnerable to 2019-nCoV infection. Front Med 14: 185–192, 2020. 10.1007/s11684-020-0754-0 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Chong WH, Saha B: Severe acute respiratory syndrome coronavirus 2 (SARS-CoV-2) associated with rhabdomyolysis and acute kidney injury (AKI). Am J Med Sci 360: 738–739, 2020. 10.1016/j.amjms.2020.07.032 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Hirsch JS, Ng JH, Ross DW, Sharma P, Shah HH, Barnett RL, Hazzan AD, Fishbane S, Jhaveri KD; Northwell COVID-19 Research Consortium; Northwell Nephrology COVID-19 Research Consortium: Acute kidney injury in patients hospitalized with COVID-19. Kidney Int 98: 209–218, 2020. 10.1016/j.kint.2020.05.006 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Wang L, Li X, Chen H, Yan S, Li D, Li Y, Gong Z: Coronavirus disease 19 infection does not result in acute kidney injury: An analysis of 116 hospitalized patients from Wuhan, China. Am J Nephrol 51: 343–348, 2020. 10.1159/000507471 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Jensen JS, Clausen P, Borch-Johnsen K, Jensen G, Feldt-Rasmussen B: Detecting microalbuminuria by urinary albumin/creatinine concentration ratio. Nephrol Dial Transplant 12[Suppl 2]: 6–9, 1997 [PubMed] [Google Scholar]
- 21.Keane WF, Eknoyan G: Proteinuria, albuminuria, risk, assessment, detection, elimination (PARADE): A position paper of the National Kidney Foundation. Am J Kidney Dis 33: 1004–1010, 1999. 10.1016/S0272-6386(99)70442-7 [DOI] [PubMed] [Google Scholar]
- 22.Steiner RW: Interpreting the fractional excretion of sodium. Am J Med 77: 699–702, 1984. 10.1016/0002-9343(84)90368-1 [DOI] [PubMed] [Google Scholar]
- 23.Park R, Rabinowitz L: Effect of reduced glomerular filtration rate on the fractional excretion of urea in the dg. Proc Soc Exp Biol Med 132: 27–29, 1969. 10.3181/00379727-132-34139 [DOI] [PubMed] [Google Scholar]
- 24.Kaplan AA, Kohn OF: Fractional excretion of urea as a guide to renal dysfunction. Am J Nephrol 12: 49–54, 1992. 10.1159/000168417 [DOI] [PubMed] [Google Scholar]
- 25.Wyllie AL, Fournier J, Casanovas-Massana A, Campbell M, Tokuyama M, Vijayakumar P, Warren JL, Geng B, Muenker MC, Moore AJ, Vogels CBF, Petrone ME, Ott IM, Lu P, Venkataraman A, Lu-Culligan A, Klein J, Earnest R, Simonov M, Datta R, Handoko R, Naushad N, Sewanan LR, Valdez J, White EB, Lapidus S, Kalinich CC, Jiang X, Kim DJ, Kudo E, Linehan M, Mao T, Moriyama M, Oh JE, Park A, Silva J, Song E, Takahashi T, Taura M, Weizman OE, Wong P, Yang Y, Bermejo S, Odio CD, Omer SB, Dela Cruz CS, Farhadian S, Martinello RA, Iwasaki A, Grubaugh ND, Ko AI: Saliva or nasopharyngeal swab specimens for detection of SARS-CoV-2. N Engl J Med 383: 1283–1286, 2020. 10.1056/NEJMc2016359 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Chancharoenthana W, Leelahavanichkul A: Acute kidney injury spectrum in patients with chronic liver disease: Where do we stand? World J Gastroenterol 25: 3684–3703, 2019. 10.3748/wjg.v25.i28.3684 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Ostermann M, Joannidis M: Acute kidney injury 2016: Diagnosis and diagnostic workup. Crit Care 20: 299, 2016. 10.1186/s13054-016-1478-z [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Garrido JM, Candela-Toha AM, Parise-Roux D, Tenorio M, Abraira V, Del Rey JM, Prada B, Ferreiro A, Liaño F: Impact of a new definition of acute kidney injury based on creatinine kinetics in cardiac surgery patients: A comparison with the RIFLE classification. Interact Cardiovasc Thorac Surg 20: 338–344, 2015. 10.1093/icvts/ivu393 [DOI] [PubMed] [Google Scholar]
- 29.Waikar SS, Bonventre JV: Creatinine kinetics and the definition of acute kidney injury. J Am Soc Nephrol 20: 672–679, 2009. 10.1681/ASN.2008070669 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Mak GC, Cheng PK, Lau SS, Wong KK, Lau CS, Lam ET, Chan RC, Tsang DN: Evaluation of rapid antigen test for detection of SARS-CoV-2 virus. J Clin Virol 129: 104500, 2020. 10.1016/j.jcv.2020.104500 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Porte L, Legarraga P, Vollrath V, Aguilera X, Munita JM, Araos R, Pizarro G, Vial P, Iruretagoyena M, Dittrich S, Weitzel T: Evaluation of a novel antigen-based rapid detection test for the diagnosis of SARS-CoV-2 in respiratory samples. Int J Infect Dis 99: 328–333, 2020. 10.1016/j.ijid.2020.05.098 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Chu KH, Tsang WK, Tang CS, Lam MF, Lai FM, To KF, Fung KS, Tang HL, Yan WW, Chan HW, Lai TS, Tong KL, Lai KN: Acute renal impairment in coronavirus-associated severe acute respiratory syndrome. Kidney Int 67: 698–705, 2005. 10.1111/j.1523-1755.2005.67130.x [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Tojo A, Kinugasa S: Mechanisms of glomerular albumin filtration and tubular reabsorption. Int J Nephrol 2012: 481520, 2012. 10.1155/2012/481520 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Gupta RK, Bhargava R, Shaukat AA, Albert E, Leggat J: Spectrum of podocytopathies in new-onset nephrotic syndrome following COVID-19 disease: A report of 2 cases. BMC Nephrol 21: 326, 2020. 10.1186/s12882-020-01970-y [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Batlle D, Soler MJ, Sparks MA, Hiremath S, South AM, Welling PA, Swaminathan S; COVID-19 and ACE2 in Cardiovascular, Lung, and Kidney Working Group: Acute kidney injury in COVID-19: Emerging evidence of a distinct pathophysiology. J Am Soc Nephrol 31: 1380–1383, 2020. 10.1681/ASN.2020040419 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Ahmadian E, Hosseiniyan Khatibi SM, Razi Soofiyani S, Abediazar S, Shoja MM, Ardalan M, Zununi Vahed S: Covid-19 and kidney injury: Pathophysiology and molecular mechanisms [published online ahead of print October 6, 2020]. Rev Med Virol 10.1002/rmv.2176 [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Specificity of the polyclonal antibodies to detect SARS-CoV-2 spike S1 protein using Western blot. Download Supplemental Figure 1, PDF file, 1.4 MB (1.4MB, pdf)
Concentration of urine and serum creatinine across the study population. Download Supplemental Figure 2, PDF file, 1.4 MB (1.4MB, pdf)
Urine proteomics to determine presence of SARS-CoV-2 spike protein as a biomarker for kidney injury in COVID-19 patients across all the urine samples. Download Supplemental Figure 3, PDF file, 1.4 MB (1.4MB, pdf)
(A) Comparison on the role of confounding factors in the elevated levels of albumin in urine. (B) Comparison of role of confounding factors in the elevated levels of cystatin C in urine. Download Supplemental Figure 4, PDF file, 1.4 MB (1.4MB, pdf)
Correlation of SARS-CoV-2 viral RNA load to the presence of spike protein in urine among COVID-19 NP-PCR+ adults. Download Supplemental Figure 5, PDF file, 1.4 MB (1.4MB, pdf)
Representative standard curve generated using 5μg/mL SARS-CoV-2 polyclonal anti-spike antibodies. Download Supplemental Figure 6, PDF file, 1.4 MB (1.4MB, pdf)