Abstract
The last two decades have witnessed a tremendous growth in probiotics and in the numbers of publications on their potential health benefits. Owing to their distinguishing beneficial effects and long history of safe use, species belonging to the Lactobacillus genus are among the most widely used probiotic species in human food and dietary supplements and are finding increased use in animal feed. Here, we isolated, identified, and evaluated the safety of two novel Limosilactobacillus reuteri (L. reuteri) isolates, ATCC PTA-126787 & ATCC PTA-126788. More specifically, we sequenced the genomes of these two L. reuteri strains using the PacBio sequencing platform. Using a combination of biochemical and genetic methods, we identified the two strains as belonging to L. reuteri species. Detailed in silico analyses showed that the two strains do not encode for any known genetic sequences of concern for human or animal health. In vitro assays confirmed that the strains are susceptible to clinically relevant antibiotics and do not produce potentially harmful by-products such as biogenic amines. In vitro bile and acid tolerance studies demonstrated that the two strains have similar survival profiles as the commercial L. reuteri probiotic strain DSM 17938. Most importantly, daily administration of the two probiotic strains to broiler chickens in drinking water for 26 days did not induce any adverse effect, clinical disease, or histopathological lesions, supporting the safety of the strains in an in vivo avian model. All together, these data provide in silico, in vitro and in vivo evidence of the safety of the two novel candidates for potential probiotic applications in humans as well as animals.
Introduction
The term “probiotic” was derived from “pro” (Latin, means “for”) and “bios” (Greek, means “life”) and thereby means “for life”. Probiotics are defined as “live microorganisms that, when administered in adequate amounts, confer a benefit on the host” [1]. In recent years, there has been an unprecedented growth in the application of probiotics to support health and well-being. Often consumed as dietary supplements, nutraceuticals or as part of functional foods, probiotics are associated with many health benefits in the form of promoting gut barrier function, including studies on their potential to prevent and/or treat gastrointestinal diseases, inhibit pathogenic bacteria, and favourably modulate gut bacteria, the immune system, and host metabolism [2–5]. Several species of microorganisms are used as probiotics and the lactic acid bacteria belonging to the Lactobacillus genus, first described in 1901 [6], are among the most commonly used and well-studied probiotic bacteria with a long history of safe use [7].
Limosilactobacillus reuteri (L. reuteri), a member of the Lactobacillus genus, are Gram positive, non-spore forming, non-motile bacteria, which are naturally adapted to survive under low pH, bile-rich, and microaerophilic to strictly anaerobic gastrointestinal environments [8]. German microbiologist Gerhard Reuter first isolated L. reuteri from human fecal and intestinal samples and classified it as L. fermentum biotype II [9]; later, Kandler et al., (1980) identified L. reuteri as a distinct species [10]. L. reuteri is considered one of the few true autochthonous lactobacilli present frequently in the gastrointestinal tract of all vertebrates, including humans, monkeys, chicken, turkeys, doves, pigs, dogs, lambs, cattle and rodents [11, 12]. L. reuteri strains are often known to produce reuterin (a bacteriocin with antimicrobial properties), cobalamin and folate, exclude or inhibit pathogens, modulate immune response, and enhance gut barrier function [13–17]. Several clinical studies have been published on the efficacy of L. reuteri in treating gastrointestinal disorders such as infantile colic, regurgitation, functional constipation, abdominal pain, and necrotizing enterocolitis [17–22]. L. reuteri was used in sourdough bread in 1980, and was introduced into human functional foods as a starter in the production of a special drink called “BRA (stands for Bifidobacterium, Reuteri and Acidophilus)” and a fermented milk called “BRA fil” in 1991 in Sweden [23]. Since then, L. reuteri strains, such as DSM 17938 and RC-14, have been widely used as a part of many commercially available dietary supplements and functional foods [17].
Lactic acid bacteria are known for their safety and are one of the probiotic microbial types with the longest history of safe use [7]. The L. reuteri species is usually considered safe for human and animal consumption due to the facts that they have been used as part of fermented foods for more than 30 years, they are normal inhabitants of the human and animal gut microflora, and they have regulatory stature in the United States and the European Union. Indeed, several strains belonging to the L. reuteri species were notified to the United States Food and Drug Administration (FDA), including three strains as “Generally Regarded As Safe (GRAS)” for use in specific foods and two strains as new dietary ingredients; the FDA cited no objections to such strains [24–28]. Moreover the L. reuteri strains was granted “Qualified Presumption of Safety (QPS)” status and is considered safe for use in, or as a source of food for, human and animal consumption by European Food Safety Agency (EFSA) [24–26, 29, 30]. A growing number of clinical studies have repeatedly confirmed the safety of L. reuteri not only in healthy individuals but also in immunocompromised individuals such as those positive for HIV [31, 32].
Despite the prior safe use of a probiotic genus and species, the survival properties, efficacy, and safety of probiotics are evaluated on a strain-specific basis. Hence, screening for such properties for every new strain is required before any new probiotic candidate is accepted for human and animal consumption. Hence, various regulatory agencies and experts have established comprehensive recommended guidelines for efficacy and safety assessment of new probiotic candidates [33–35]. These guidelines encompass a series of in vitro, in silico and in vivo studies, of which genomics is considered a powerful tool for rapid screening of probiotic candidates for safety.
Genomic characterizations are instrumental in selecting a safe and efficacious probiotic strain. Safety assessment begins with the correct identification of the probiotic candidate and this is important for both scientific and regulatory reasons. Genomic approaches offer high resolution identification of strains by comparing those with other well-characterized, safe, and efficacious probiotic strains. Comparative genomics studies further help to understand the molecular basis of probiotic efficacy, as well as the survival and adaptation of these probiotic strains in the gastrointestinal tract. Most importantly, genomic analyses allow for rapid screening of probiotic candidates for genes encoding antimicrobial resistance, virulence factors, toxins, and biogenic amines, facilitating better understanding of the safety of the probiotic strain of interest. Finally, genome-based analyses also help to investigate the stability of probiotic strains.
The goal of this study was to provide in silico, in vitro and in vivo evidence to support the safety of L. reuteri ATCC PTA-126787 & ATCC PTA-126788 (hereafter referred to as PTA-126787 and PTA-126788) for their use as probiotics in humans as well as animals. More specifically, the strains were identified using a combination of biochemical, 16S rRNA and whole-genome sequencing analyses. The genomes were screened for potential genes encoding antimicrobial resistance, toxins, virulence factors and other harmful metabolites. In silico data were further confirmed using in vitro experiments. The strains were finally analysed for safety using the broiler chicken as an in vivo model.
Materials and methods
Ethics statement
All chickens were housed and cared for under the Guide for the Care and Use of Agricultural Animals in Research and Teaching and all local standard operating procedures. The study was reviewed and approved by the Animal Care and Use Committee of the institution performing the study (Assigned ACUP# 1399).
Bacterial strains and culture conditions
The L. reuteri strains described in this study were routinely propagated on Lactobacilli de Man Rogosa Sharpe (MRS, BD Difco) medium anaerobically at 37°C. L. reuteri strain DSM 17938 was used as a reference strain for biochemical identification, D- and L-lactate production, autoaggregation, resistance to bile salts and acidic pH assays. L. reuteri strain ATCC 23272 was used as a reference strain for growth kinetics, autoaggregation, biogenic amine production and D- and L-lactate production assays. L. acidophilus strain ATCC 4356 was used as a reference strain for growth kinetics assay.
Molecular identification
The strains were identified using 16S rRNA sequencing. Briefly, L. reuteri strains were grown in Lactobacilli MRS broth overnight for 14–16 hours under anaerobic conditions at 37°C. One hundred microliters of the culture were pelleted by centrifugation and resuspended in 50 μL of nuclease-free water. The resuspended culture was heated at 98°C for 10 minutes. The debris was pelleted by brief centrifugation and the supernatant was used as a template for PCR. The 16S rRNA gene was amplified by PCR using 3 μL of the DNA template and universal primers 16S rRNA gene F, 5’-AGAGTTTGATCCTGGCTCAG-3’ and 16S rRNA gene R, 5’-CTTGTGCGGGCCCCCGTCAATTC-3’. The amplicons were PCR purified using the QIAquick PCR Purification Kit (Qiagen Inc.) following manufacturer’s instructions and sequenced by Sanger sequencing by GenScript. The sequences were then searched against the NCBI nucleotide collection (nr/nt) database using the BLAST algorithm.
Biochemical identification
The strains were profiled for enzymatic activity and carbohydrate fermentation using API 50 CHL strips (bioMérieux), following the manufacturer’s instructions. The L. reuteri strain DSM 17938 was used as a positive control.
Enzyme profiling
The enzymatic profiles of L. reuteri strains were determined using the APIZym test strips (bioMérieux), following manufacturer’s instructions.
Growth kinetics
The L. reuteri strains were grown in MRS broth overnight for 14–16 hours under anaerobic conditions at 37°C. The next morning, the cultures were adjusted to an OD600 of 0.1 and monitored for growth by plating on Lactobacilli MRS agar at 0, 1, 2, 4, and 8 hours. Human L. reuteri strain 23272 and L. acidophilus strain ATCC 4356 were used as controls.
Isolation of high molecular weight DNA
High molecular weight DNA for PacBio sequencing was isolated using the phenol:chloroform method. Briefly, L. reuteri strains were grown in Lactobacilli MRS broth overnight under anaerobic conditions for 14–16 hours. The cells were harvested by centrifugation at 4,000g RCF for 10 minutes at 4°C. The pellet was washed once by 1 mL of TE buffer (10 mM Tris-HCl and 1 mM EDTA, pH 8.0) and resuspended in 0.5 mL of TE containing 1.2% Triton X-100 and 10 mg/mL of lysozyme (Sigma Aldrich) and incubated at 37°C for 1 hour. After incubation, 20 μL of proteinase K was added, mixed several times, and incubated at 55°C for 1 hour. Twenty microliters of RNase was then added and incubated at 37°C for an additional 30 minutes. Approximately, 600 μL of phenol:chloroform:isoamyl alcohol (25:24:1; ThermoFisher Scientific) mixture (pH 8.0) was added, the tubes were inverted several times and centrifuged at 11,200 × g for 10 minutes at 4°C. The upper aqueous phase was carefully transferred to a new 1.5 mL centrifuge tube. The above phenol:chloroform step was repeated one more time. 0.5 mL of chloroform was added, the tubes were inverted several times and centrifuged at 11,200g RCF for 10 minutes. The upper aqueous phase was carefully transferred to a new centrifuge tube. 0.45 mL of isopropanol was layered onto the aqueous phase containing genomic DNA and the tubes were gently shaken to precipitate high molecular weight genomic DNA. The precipitated DNA was removed with a sterile loop and transferred to a new tube containing 1 mL 70% ethanol. The tubes were centrifuged at 11,200g RCF for 5 minutes at 4°C. The DNA pellet was briefly air-dried and 300 μL of nuclease free water was added, and then allowed to dissolve overnight at 4°C. The dissolved DNA was gently mixed with a big-bore tip and stored at -20°C. The isolated DNA was analyzed for quantity using Qubit (Thermo Fisher Scientific, Inc.).
Whole genome sequencing and assembly
The bacterial genomic DNA samples were shipped on dry-ice to DNA Link, Inc (San Diego, CA; https://www.dnalink.com/english/) for whole genome sequencing using PacBio RSII platform. Briefly, 20 kb DNA fragments were generated by shearing genomic DNA using the Covaris G-tube according to the manufacturer’s recommended protocol (Covaris). Smaller fragments were purified by the AMpureXP bead purification system (Beckman Coulter). For library preparation, 5μg of genomic DNA was used. The SMRTbell library was constructed using SMRTbell™ Template Prep Kit 1.0 (PacBio®). Small fragments were removed using the BluePippin Size selection system (Sage Science). The remaining DNA sample was used for large-insert library preparation. A sequencing primer was annealed to the SMRTbell template and DNA polymerase was bound to the complex using DNA/Polymerase Binding kit P6 (PacBio®). Following the polymerase binding reaction, the MagBead was bound to the library complex with MagBeads Kit (PacBio®). This polymerase-SMRTbell-adaptor complex was loaded into zero-mode waveguides. The SMRTbell library was sequenced by 2 PacBio® SMRT cells (PacBio®) using the DNA sequencing kit 4.0 with C4 chemistry (PacBio®). A 1×240-minute movie was captured for each SMRT cell using the PacBio® RS sequencing platform. The genome was further assembled by DNA link, Inc with HGAP.3 protocol.
Genome annotation and feature prediction
Genome annotation was carried out using a custom annotation pipeline by combining several prediction tools. Coding sequences, transfer RNA and transmembrane RNA were predicted and annotated using Prokka v 1.14.5 [36–38]. Ribosomal binding site (RBS) prediction was carried out using RBSFinder [39]. TranstermHP v2.08 was used to predict Rho-independent transcription terminators (TTS) [40]. Ribosomal RNA and other functional RNAs such as riboswitches and non-coding RNA was annotated with Infernal v1.1.2 [41]. Operons were predicted based on primary genome sequence information with Rockhopper v2.0.3 using default parameters [42].
Data deposition
The raw sequencing reads, genome assemblies and annotations in this study were deposited in the NCBI BioProject under project PRJNA675717.
Accession numbers:
| Serial No. | Sample | BioSample | Accession number | SRA number |
|---|---|---|---|---|
| 1. | ATCC PTA-126787 | SAMN16712075 | CP065330-CP065334 | SRX9689306 |
| 2. | ATCC PTA-126788 | SAMN16712076 | CP065849-CP065855 | SRX9689307 |
Phylogenetic analyses
Phylogenetic relationships of the genomes were explored with UBCG v3.0 using default settings [43]. This software tool employs a set of 92 single-copy core genes commonly present in all bacterial genomes. These genes then were aligned and concatenated within UBCG using default parameters. The estimation of robustness of the nodes is done through the gene support index (GSI), defined as the number of individual gene trees, out of the total genes used, that present the same node. A maximum-likelihood phylogenetic tree was inferred using FastTree v.2.1.10 with the GTR+CAT model [44].
Comparative genomic analyses
OrthoFinder v2.3.11 [45] was used to determine orthologous relationships between protein sequences inferred from PTA-126787 and PTA-126788 with protein sequences of strains ATCC 53608, CF48-3A, DSM20016 and SD2112 (the parent strain of DSM17938) downloaded from GenBank [46]. Pairwise Average Nucleotide Identities (ANI) values were calculated all-against-all, using FastANI v 1.32 [47].
Identification of prophages, transposases and other insertion sequences (IS)
Insertion sequence prediction was done using ISEscan v.1.7.2.1 [48]. Prophage prediction was done using PhiSpy v4.2.6 which combines similarity‐ and composition‐based strategies [49].
Identification of CRISPR-Cas sequences
Coding sequences for Clustered Regularly Interspaced Short Palindromic Repeats (CRISPR) and CRISPR-associated genes (Cas) were searched using CRISPRDetect version 2.2 [50]. However, no CRISPR sequences were identified in both the genomes.
Identification of virulence determinants and antimicrobial resistance genes
Protein-encoding genes related to virulence were searched manually based on functional annotation of the genomes. Automated screening of whole genome sequences of both strains against the Virulence Factor Database (VFDB), a comprehensive repository of known bacterial virulence factors and other putative adverse metabolites [51], ARG-ANNOT [52], ResFinder [53] and NCBI-AMR databases (2020-Jun-15) was performed using Abricate version 0.9.9 [54].
Identification of genes encoding toxic metabolites
Analysis was performed on the genomes manually to identify homologs of histidine decarboxylase, tyrosine decarboxylase, lysine decarboxylase, ornithine decarboxylase, agmatine deiminase, agmatine::putrescine antiporter, multicopper oxidase and other potential genes involved in the production of biogenic amines.
Genes involved in lactic acid production and other beneficial metabolites
Sequences encoding putative genes involved in lactic acid production and other metabolites were identified by manual search of functional annotations.
Antimicrobial susceptibility profiling
Antimicrobial susceptibility testing was performed using broth microdilution method, using LSB medium (Mueller Hinton broth containing 5% horse blood) following Clinical and Laboratory Standards Institute (CLSI, 28th edition) guidelines. Two-fold dilutions of the clinically relevant antibiotics (Clindamycin, Chloramphenicol, Erythromycin, Gentamicin, Kanamycin, Streptomycin, Tetracycline and Ampicillin, all purchased from Sigma Aldrich) were prepared in LSB medium. Approximately, 50 μL of 1 × 105 CFU/mL of the L. reuteri cells were added into each well. “No antibiotic” and “medium” alone controls were included. Escherichia coli ATCC 25923, Pseudomonas aeruginosa ATCC 27853, Staphylococcus aureus ATCC 29213, Enterococcus faecalis ATCC 29212, Streptococcus pneumonia ATCC 49619 and Lacticaseibacillus paracasei ATCC 334 were used as quality control organisms. The plates were incubated for 24–48 hours under microaerophilic conditions. Minimum inhibitory concentration (MIC) was defined as the lowest concentration of antibiotic that showed complete inhibition of L. reuteri growth. The strains were classified as susceptible or resistant using the microbiological cut offs established by EFSA [35].
Biogenic amine production
The ability of L. reuteri strains to produce biogenic amines was determined as previously described [55]. Briefly, L. reuteri cultures were grown in MRS broth supplemented with L-tyrosine (0.1% m/v), L-histidine (0.1% m/v), L-arginine (0.1% m/v) or L-lysine (0.1% m/v) and pyridoxal-5-phosphate (0.005% m/v) under anaerobic conditions at 37°C overnight. The cultures were then plated on supplemented decarboxylase broth base as described by Bover-Cid and Holzapfel [56] and colour development was recorded after 48 hours of incubation under anaerobic conditions at 37°C.
D- and L-lactate production
The amounts of D- and L-lactate produced were quantified using D-/L-Lactic Acid (D-/L-Lactate) (Rapid) Assay Kit (Megazyme), following manufacturer’s instructions. Briefly, L. reuteri strains were grown in MRS broth incubated at 37°C for 14–16 hours. The cultures were centrifuged at 4,000g RCF for 10 minutes at 4°C and the supernatant was collected into a 1.5 mL Eppendorf tube and filter sterilized. 1.0 mL of the filter sterilized supernatant was used for lactic acid quantification as described in the manual provided by the manufacturer.
Autoaggregation
The ability of L. reuteri strains to autoaggregate was assayed as follows. L. reuteri strains were grown in MRS broth overnight for 14–16 hours under anaerobic conditions at 37°C. The cultures were adjusted to an OD600 of 0.1 and allowed to grow for another 14–16 hours and observed for aggregate formation. Autoaggregation was quantified as described previously with some minor modifications [57]. L. reuteri strains were grown in MRS broth overnight for 14–16 hours under anaerobic conditions at 37°C. The cultures were washed twice with PBS (pH 7.2) by centrifuging at 11,200g RCF for 10 minutes at 4°C. The washed cell pellets were then resuspended in PBS (pH 7.2) and adjusted to an OD600 of 0.5 to standardize the number of bacterial cells (107−108 CFU/ml). The suspensions were incubated as 1 ml aliquots under anaerobic conditions at 37°C for 5 hours. The OD600 was recorded after 5 hours. Autoaggregation percentage was calculated as follows: [1- (Absorbance at 5 hours/ Absorbance at 0 hour)] x 100.
Hydrogen peroxide production
The ability of L. reuteri strains to produce hydrogen peroxide was assessed as previously described [58]. Briefly, MRS agar plates were prepared with 0.25 mg/mL of tetramethylbenzidine and 0.01 mg/mL of horseradish peroxidase. L. reuteri strains were streaked on the supplemented MRS agar plates and incubated for 24 hours and coloration of the colonies/culture was recorded. White bacterial colonies/culture indicates no hydrogen peroxide production, pale blue colonies/culture indicates poor production and dark blue colonies/culture indicates high production.
Resistance to bile salts
The ability of cultures to tolerate bile salts was determined as follows: L. reuteri strains were grown under anaerobic conditions for 14–16 hours at 37°C. The culture was inoculated into fresh MRS broth (pH adjusted to 6.4, optimal pH for L. reuteri growth) containing 0.3% bile salts (Oxoid, USA) at a rate of 1% inoculum and incubated under anaerobic conditions at 37°C for 4 hours. Samples were collected at 0 and 4 hours after incubation and analysed for CFU counts.
Resistance to acidic pH
The tolerance of L. reuteri strains to low pH was determined as described below. L. reuteri strains were grown under anaerobic conditions for 14–16 hours at 37°C. The cells were harvested by centrifugation at 4,000g RCF for 10 minutes at 4°C and the pellet was resuspended in sterile PBS (adjusted to a pH of 2.5) to an OD600 of 0.5. The cultures were then incubated at 37°C for 3 hours under anaerobic conditions. Aliquots were collected at time 0 and 3 hours, respectively after incubation, serially diluted in PBS and plated on MRS to determine the CFU counts.
In vivo safety assessment
The safety of L. reuteri strains was tested using Specific Pathogen Free (SPF) broiler chickens. Forty-two White Leghorn, mixed sex chicks of day-old age were purchased from Valo BioMedia. At arrival, the chicks were tagged via wing web. The birds were fed commercially available non-medicated feed ad libitum. Briefly, day-old chicks were randomly grouped into three groups with 14 birds in each group. Group 1 was administered with 1 × 107 CFU/bird/day of L. reuteri PTA-126787 in drinking water from day 1 to day 26. Group 2 was administered with 1 × 107 CFU/bird/day of L. reuteri PTA-126788 in drinking water from day 1 to day 26. Group 3 served as no treatment control. The birds were examined for adverse events, morbidity, and mortality on a daily basis. On day 31, the birds were euthanized, and observed for any gross lesions, indicative of health issues. More specifically, lungs, trachea, liver, spleen, kidneys, intestine were observed for gross lesions and scored as normal or abnormal. Gut (2 cm of cecal-rectal junction), lung (dime size) and tracheal (2 cm long) samples were collected from 5 birds per group in buffered formalin, analysed for histopathology and scored as described in S1 File.
Results
L. reuteri isolation and molecular identification
A library of seven L. reuteri strains along with the two strains described in this study were isolated from the cecum of older broiler chickens at Elanco Animal Health, Cuxhaven, (Germany). Based on the 16S rRNA amplicon sequencing and respective BLAST search comparison results, all the seven strains, including PTA-126787 and PTA-126788 showed closest homology to published L. reuteri sequences, suggesting that our strains belong to the L. reuteri species (Fig 1A and 1B).
Fig 1. Identification of L. reuteri strains by 16S rRNA amplicon sequencing.
L. reuteri strains were identified by PCR amplification and sequencing of the 16S rRNA variable region. A. Agarose gel electrophoresis of the 16S rRNA PCR product. B. Phylogenetic analysis of the 16S rRNA sequence along with other L. reuteri sequences. Streptococcus pyogenes was included as an outgroup.
Biochemical identification
When tested with API 50 CHL, the final two L. reuteri candidates, PTA-126787 and PTA-126788, were identified as Limosilactobacillus fermentum (previously Lactobacillus fermentum) with 92.3% identity (Table 1). The positive control L. reuteri DSM 17938 was also identified as L. fermentum with 92.3% identity (Table 1). The fermentation profile of L. reuteri is similar to that of L. fermentum and the APIwebTM software version 5.0 does not have the capability to distinguish between the 2 species.
Table 1. Carbohydrate fermentation profile of L. reuteri strains PTA-126787 and PTA-126788 by API 50 CHL.
| Substrate | PTA-126787 | PTA-126788 | DSM 17938 | Substrate | PTA-126787 | PTA-126788 | DSM 17938 |
|---|---|---|---|---|---|---|---|
| Negative control | - | - | - | Esculin ferric citrate | + | + | + |
| Glycerol | - | - | - | Salicin | - | - | - |
| Erythritol | - | - | - | D-Cellobiose | - | - | - |
| D-Arabinose | - | - | - | D-Maltose | + | + | + |
| L-Arabinose | + | + | + | D-Lactose | + | + | + |
| D-Ribose | + | + | + | D-Melibiose | + | + | + |
| D-Xylose | - | - | - | D-Saccharose | + | + | + |
| L-Xylose | - | - | - | D-Trehalose | - | - | - |
| D-Adonitol | - | - | - | Inulin | - | - | - |
| Methyl-βD-xylopyranoside | - | - | - | D-Melezitose | - | - | - |
| D-Galactose | + | + | + | D-Raffinose | + | + | + |
| D-Glucose | + | + | + | Amidon | - | - | - |
| D-Fructose | - | - | - | Glycogen | - | - | - |
| D-Mannose | - | - | - | Xylitol | - | - | - |
| L-Sorbose | - | - | - | Gentibiose | - | - | - |
| L-Rhamnose | - | - | - | D-Turanose | - | - | - |
| Dulcitol | - | - | - | D-Lyxose | - | - | - |
| Inositol | - | - | - | D-Tagatose | - | - | - |
| D-Mannitol | - | - | - | D-Fucose | - | - | - |
| D-Sorbitol | - | - | - | L-Fucose | - | - | - |
| Methyl-αD-mannopyroside | - | - | - | D-Arabitol | - | - | - |
| Methyl-αD-glucopyranoside | - | - | - | L-Arabitol | - | - | - |
| N-Acetylglucosamine | - | - | - | Potassium gluconate | + | + | + |
| Amygdalin | - | - | - | Potassium 2-ketogluconate | - | - | - |
| Arbutin | - | - | - | Potassium 5-ketogluconate | - | - | - |
+, positive reaction; -, negative reaction
Enzyme profile
Enzyme profile is a good indicator of both the probiotic function as well as safety. APIZym test is a rapid semiquantitative assay to detect 19 enzymatic reactions. Unlike API 50 CHL, no databases exist to identify bacteria based on APIZym profiles. As shown in Table 2, both L. reuteri strains showed similar enzyme profiles to that of L. reuteri strain DSM 17938. The two strains showed strong leucine arylamidase, valine arylamidase, acid phosphatase, α-galactosidase and β-galactosidase activities, while both were negative for alkaline phosphatase, lipase, trypsin, α-chymotrypsin, β-glucosidase, α-mannosidase and α-fucosidase activities. In general, the enzymatic reactions from APIZym testing were in good agreement with carbohydrate fermentation by API 50 CHL.
Table 2. Enzymatic profiles of L. reuteri strains PTA-126787 and PTA-126788 by APIZym.
| Enzyme assayed | Substrate | PTA-126787 | PTA-126788 | DSM 17938 |
|---|---|---|---|---|
| Alkaline phosphatase | 2-naphthyl phosphate | - | - | - |
| Esterase (C 4) | 2-naphthyl butyrate | + | + | + |
| Esterase Lipase (C 8) | 2-naphthyl capylate | +/- | +/- | - |
| Lipase (C 14) | 2-naphthyl myristate | - | - | - |
| Leucine arylamidase | L-leucyl-2-naphthylamide | +++ | +++ | ++ |
| Valine arylamidase | L-valyl-2-naphthylamide | ++ | ++ | + |
| Cystine arylamidase | L-cystyl-2-naphthylamide | + | + | + |
| Trypsin | N-benzoyl-DL-argine-2-naphthylamide | - | - | - |
| α-chymotrypsin | N-glutaryl-phenylalanine-2-naphthylamide | - | - | - |
| Acid phosphatase | 2-naphthyl phosphate | +++ | +++ | ++ |
| Naphthol-AS BI-phosphohydrolase | Naphthol-AS-BI-phosphate | + | + | - |
| α-galactosidase | 6-Br-2-naphthyl-αD-galactopyranoside | +++ | +++ | +++ |
| β-galactosidase | 2-naphthyl-βD-galactopyranoside | +++ | +++ | +++ |
| β-glucuronidase | Naphthol-AS-BI-βD-glucuronide | + | + | + |
| α-glucosidase | 2-naphthyl-αD-glucopyranoside | + | + | + |
| β-glucosidase | 6-Br-2-naphthyl-βD-glucopyranoside | - | - | - |
| N-acetyl-β-glucosaminidase | 1-naphthyl-N-acetyl-βD-glucosaminide | +/- | +/- | - |
| α-mannosidase | 6-Br-2-naphthyl-αD-mannopyranoside | - | - | - |
| α-fucosidase | 2-naphthyl-αL-fucopyranoside | - | - | - |
+++, very strong positive enzymatic reaction; ++, strong positive enzymatic reaction; +, positive enzymatic reaction; -, negative enzymatic reaction; +/-, inconclusive
Growth profiles
All the L. reuteri strains had similar growth profiles, including PTA-126787 and PTA-126788 (Fig 2), and the profiles were comparable to that of human L. reuteri strain ATCC 23272 and Lactobacillus acidophilus strain ATCC 4356.
Fig 2. Growth profiles of L. reuteri strains in MRS broth.
Growth profiles were assessed by growing the strains in MRS broth and determining the CFU counts at different time points. The data shown is representative of 3 independent experiments.
In silico analyses
A. Genomic characterization
The genomes of L. reuteri strains PTA-126787 and PTA-126788 were sequenced by PacBio sequencing platform. Strain PTA-126787 contains 5 contigs yielding a total estimated genome size of 2.4 Mb and strain PTA-126788 contains 7 contigs yielding an estimated genome size of 2.4 Mb. The genome properties, prediction and annotation of different features are summarized in Table 3. The circular representation of the complete genomes of both strains is shown in Fig 3. The whole-genome sequencing project was deposited at DDBJ/ENA/GenBank under BioProject number PRJNA675717.
Table 3. Genomic properties of L. reuteri strains PTA-126787 and PTA-126788.
| Feature | PTA-126787 | PTA-126788 |
|---|---|---|
| Contigs | 5 | 7 |
| Coding sequence | 2427 | 2495 |
| Prophages | 6 | 8 |
| Mobile Element | 86 | 88 |
| Non-coding RNA | 21 | 17 |
| Operons | 501 | 541 |
| Ribosomal RNA | 18 | 17 |
| Ribosomal binding site | 2359 | 2396 |
| Transcription terminator | 1182 | 1241 |
| Riboswitch | 26 | 25 |
| Transfer RNA | 73 | 74 |
| Transfer-messenger RNA | 1 | 1 |
Fig 3. Chromosomal map of L. reuteri strains PTA-126787 and PTA-126788.
The concentric circles show, reading outwards: GC skew, GC content, AT skew, AT content, COG classification of proteins, CDS on reverse strand, ORFs on three frames in reverse strand, ORFs on three frames in forward strand, CDS on forward strand and COG classification of proteins on forward strand.
B. Phylogenetic analysis
Phylogenetic relationships of the genomes were explored with UBCG v3.0 which employs a set of 92 single-copy core genes commonly present in all bacterial genomes. These genes then were aligned and concatenated within UBCG using default parameters. The estimation of robustness of the nodes is done through the gene support index (GSI), defined as the number of individual gene trees, out of the total genes used, that present the same node. As shown in Fig 4, both strains PTA-126787 and PTA-126788 showed closest relationship to L. reuteri. Average Nucleotide Identities were calculated between closely related genomes and is shown in Table 4.
Fig 4. Phylogenetic relationship of L. reuteri strains PTA-126788 and PTA-126787 to other known human L. reuteri strains using 92 core genes.
The phylogenetic relationship was explored using UBCG v3.0 and a maximum likelihood tree was inferred using GTR+CAT model. Streptococcus thermophilus and Enterococcus faecalis were used as outgroups.
Table 4. Average Nucleotide Identity (ANI) of L. reuteri PTA-126787 and PTA-126788 with closely related human probiotic strains.
| Query genome | Reference genome | %ANI | Orthologous matches | Sequence fragments |
|---|---|---|---|---|
| PTA-126787 | CF48-3A | 98.1 | 560 | 797 |
| PTA-126787 | RC-14 | 98.0 | 537 | 797 |
| PTA-126787 | RC-18 | 98.0 | 537 | 797 |
| PTA-126787 | SD2112 | 98.0 | 590 | 797 |
| PTA-126787 | DSM17938 | 98.0 | 591 | 797 |
| PTA-126787 | DSM20016 | 95.4 | 523 | 797 |
| PTA-126787 | ATCC53608 | 95.1 | 532 | 797 |
| PTA-126788 | CF48-3A | 98.2 | 553 | 825 |
| PTA-126788 | SD2112 | 98.1 | 581 | 825 |
| PTA-126788 | DSM17938 | 98.1 | 582 | 825 |
| PTA-126788 | RC-14 | 98.0 | 532 | 825 |
| PTA-126788 | RC-18 | 98.0 | 532 | 825 |
| PTA-126788 | DSM20016 | 95.6 | 516 | 825 |
| PTA-126788 | ATCC53608 | 95.0 | 545 | 825 |
C. Comparative genomics analyses
Ortholog analysis was performed to identify paralogous and/or orthologous relationships between genomes of L. reuteri strains PTA-126787 and PTA-126788 against L. reuteri strains ATCC 53608, CF48-3A, DSM20016 and SD2112 (the parent strain of DSM17938) using OrthoFinder (S1 and S2 Tables). Genes unique to strains PTA-126787 and PTA-126788 are presented in S3 Table. L. reuteri strains PTA-126787 and PTA-126788 shared the highest number of orthologs amongst the strains compared in the analysis with 2264 and 2242 shared genes among them, respectively (S1 Table).
D. Screening for prophages, insertion sequences and transposases
Both strains were scanned for the presence of mobile genetic elements such as prophages, insertion sequences (IS) and transposases. Six prophage regions in strain PTA-126787 and eight regions in PTA-126788 were identified (S1 Fig). However, there were 12 phage genes (all coding for Tyrosine recombinase protein) in PTA-126788 that were outside of prophage regions. Putative IS and associated proteins predicted by ISEscan reveal 86 coding sequences in 10 IS families in strain PTA-126787 and 88 coding sequences in 18 IS families in strain PTA-126788 (Fig 5; S4 Table).
Fig 5.
Distribution of predicted IS family within the genomes of Lactobacillus reuteri strains PTA-126787 (A) and PTA-126788 (B) using ISEScan.
E. Absence of virulence factors and toxins
Both L. reuteri PTA-126787 (5 contigs) and PTA-126788 (7 contigs) strains were confirmed to be free of known virulence factors and/or toxins by comparing against virulence factor database (VFDB; search parameters of ≥80% identity and ≥80% alignment length/coverage), which is an integrated comprehensive online resource database for curating information about bacterial virulence factors and/or toxins [51].
F. Absence of acquired antimicrobial resistance genes
The Pariza et al. [34] decision tree and the EFSA Panel on Additives and Products or Substances used in Animal Feed [35] recommend that microbial strains used in food applications must not harbor acquired antimicrobial resistance genes to clinically relevant antimicrobials. Search for antimicrobial resistance genes was carried out for both L. reuteri strains by comparing the genomes against multiple AMR databases including NCBI-AMR, Resfinder DB and ARG-ANNOT using Abricate. The screening identified tetracycline-resistant ribosomal protection protein (tetW) that confers resistance to tetracycline as one potential gene of health concern (S5 Table) [34].
G. Screening for genes involved in biogenic amines and toxins
Functional annotation of the entire genomes of L. reuteri strains PTA-126787 and PTA-126788 revealed that they do not contain any known protein-encoding genes involved in the production of biogenic amines with the exception of CDS encoding for arginine deiminase. No other toxins were identified (S6 Table).
H. Genes involved in the production of lactic acid and other beneficial metabolites
Both strains, PTA-126787 and PTA-126788, contain genes responsible for production of lactic acids. A total of four coding sequences (CDS) were predicted to encode for D-lactate dehydrogenase (EC 1.1.1.28) and four CDS for L-lactate dehydrogenase (EC 1.1.1.27) were found on different loci within the genome (S7 Table). However, IVR12_00498 gene in strain PTA-126788 is a pseudogene due to a frameshift mutation. The coding sequence putative for a therapeutically useful peptide, S-ribosylhomocysteinelyase (EC 4.4.1.21; IU404_00512 and IVR12_00964) was also present in the genomes of strains PTA-126787 and PTA-126788, respectively.
Several coding sequences involved in adhesion of Lactobacilli to intestinal epithelium were identified in the genome (S8 Table). Some of the genes involved in adhesion to host found in both strains are sortase A, epsilon subunit related 3’-5’ exonuclease, exopolysaccharide biosynthesis protein and ATP synthase epsilon subunit (S8 Table). Search for desired stress tolerance features in both L. reuteri strains revealed the presence of CDS predictably encoding for DNA protection during starvation protein (S8 Table). Another stress resistant gene putatively encoding for Phosphate starvation-inducible PhoH-like protein, predicted ATPase was also found in both strains (S8 Table).
Antimicrobial susceptibility
Minimum inhibitory concentrations were analyzed against relevant antibiotics according to EFSA guidelines (EFSA Panel on Additives and Products or Substances used in Animal Feed) [35], including Ampicillin, Vancomycin, Gentamicin, Kanamycin, Streptomycin, Erythromycin, Clindamycin, Tetracycline and Chloramphenicol. L. reuteri PTA-126788 and PTA-126787 strains were determined to be sensitive to all relevant tested antibiotics according to EFSA guidelines [35], with MIC values at or below the reported species characteristic cut-off values (Table 5), except for tetracycline. For tetracycline, the MIC values for our strains were two-fold dilution above the EFSA microbiological cut off value, in one of the two biological replicates. However, this is considered acceptable due to the technical variation of the phenotypic method as recognized previously [59].
Table 5. Susceptibility of L. reuteri PTA-126787 and PTA-126788 to EFSA critically important antibiotics.
| L. reuteri PTA-126788 | L. reuteri PTA-126787 | EFSA microbiological cut off values for L. reuteri | |
|---|---|---|---|
| Clindamycin | ≤0.06 | ≤0.06 | 4 |
| Chloramphenicol | 2 | 2 | 4 |
| Erythromycin | 0.12 | 0.12 | 1 |
| Gentamicin | 1 | 1 | 8 |
| Kanamycin | 16 | 16 | 64 |
| Streptomycin | 8 | 8 | 64 |
| Tetracycline | 32/64 | 32/64 | 32 |
| Ampicillin | 1 | 1 | 2 |
Biogenic amine production
Many lactic acid bacteria produce biogenic amines such as histamine, tyramine, putrescine and/or cadaverine by amino acid decarboxylation of histidine, tyrosine, ornithine and/or lysine, respectively. The few instances of toxicity cases are associated with histamine and to some extent tyramine. Consistent with the bioinformatics results, neither of the subject L. reuteri strains were able to produce the major biogenic amines histamine, tyramine, putrescine or cadaverine. As expected, L. reuteri ATCC 23272 produced a positive reaction in the area of bacterial growth on the decarboxylase base media supplemented with L-histidine. Control plates lacking these amino acids showed no positive reaction for any of the strains tested.
D- and L-Lactate production
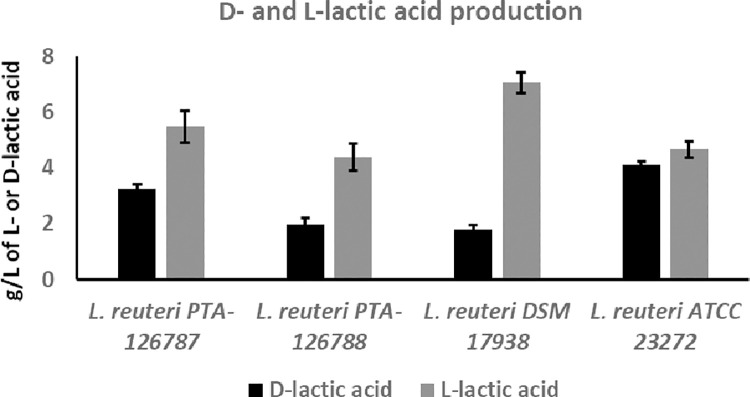
Quantitative determination of lactic acid production showed that the two L. reuteri strains produce both L- and D-lactic acids but predominantly L-lactic acid (Fig 6). Similarly, L. reuteri DSM 17938 also produced L- and D-lactic acids but predominantly L-lactic acid (Fig 6). L. reuteri strain ATCC 23272 produced approximately equal amounts of L- and D-lactic acids (Fig 6).
Fig 6. Production of D- and L-lactic acid by L. reuteri strains.
L- and D-lactic acids were quantified using D-/L-lactic acid (D-L-lactate) Rapid Assay Kit (Megazyme). The data represent the mean ± SD from 3 independent experiments.
Autoaggregation
Autoaggregation is a phenomenon where bacteria form fibrous-like aggregates after overnight growth and settle to the bottom of the tube. Once the bacteria are aggregated, they generally do not redisperse unless vigorously mixed manually. Autoaggregation appears to be one of the key properties needed for probiotic strains to attach to the epithelial cells in the gastrointestinal tract. The ability to aggregate has also been suggested to play a role in preventing pathogen colonization. As shown in Fig 7A, L. reuteri PTA-126788 showed excellent ability to autoaggregate, while the other L. reuteri strains PTA-126787 and DSM 17938 showed no ability to form aggregates. Similar to L. reuteri PTA-126788, the positive control, L. reuteri ATCC 23272 also showed ability to autoaggregate (Fig 7A). Quantification of autoaggregation showed that L. reuteri PTA-126788 exhibited the highest ability to autoaggregate, while L. reuteri strains PTA-126787 and DSM 17938 had the least ability to form aggregates (Fig 7B). The positive control L. reuteri ATCC 23272 showed moderate ability to autoaggregate (Fig 7B).
Fig 7. Ability of L. reuteri strains to undergo autoaggregation.
A. Ability to undergo autoaggregation was determined by growing the strains overnight in MRS broth and observing for aggregate formation. B. Autoaggregation was quantified by measuring the OD600 in PBS after incubation for 5 hours and calculating the autoaggregation % as described in the methods section. The data represents the mean ± SE of 3 independent experiments.
Hydrogen peroxide production
The ability of probiotic strains to produce hydrogen peroxide at physiological levels is highly desirable. Hydrogen peroxide production by Lactobacillus johnsonii NCC533 has been attributed to inducing recovery of the epithelial barrier and remission in inflammatory bowel disease [60]. Similarly, an L. reuteri probiotic strain ATCC PTA 5289 producing hydrogen peroxide was able to significantly reduce proinflammatory response and improved clinical outcomes in human patients with chronic periodontitis [61]. All the L. reuteri strains including PTA-126787 and PTA-126788 strains showed moderate to high ability to produce hydrogen peroxide as shown in Fig 8.
Fig 8. Ability of L. reuteri strains to produce hydrogen peroxide.
Hydrogen peroxide production was assessed by growing the strains on MRS agar supplemented with 0.25mg/ml of tetramethylbenzidine and 0.01mg/ml of horseradish peroxidase and observing for color change. Dark blue coloration indicates high production of hydrogen peroxide. The data are representative of 3 independent experiments.
Resistance to bile
The ability of the two L. reuteri strains to tolerate bile salts was also assessed by incubating the strains in the presence of 0.3% bile salts. The viability of L. reuteri PTA-126787, PTA-126788 and DSM 17938 did not change after incubation with 0.3% bile salts for 4 hours, suggesting that our strains are resistant to 0.3% bile salts similar to DSM 17938 (Fig 9).
Fig 9. Tolerance of L. reuteri strains to 0.3% bile.
The ability of L. reuteri strains to tolerate bile salts was assessed by growing the strains in the presence of 0.3% bile salts for 4 hours and determining the CFU counts at 0 hours and 4 hours after incubation with bile salts. The data represent the mean ± SD from 3 independent experiments.
Resistance to acidic pH
The viability of L. reuteri PTA-126787 decreased from 2.81 x 108 to 2.75 x 106 after incubation at pH 2.5 for 3 hours (Fig 10). Similarly, the viability of L. reuteri PTA-126788 decreased from 1.22 x 108 to 6.67 x 105 after 3 hours incubation at pH 2.5 (Fig 10). Overall, there was approximately 2-log reduction in viability for PTA-126787 and approximately 2.5-log reduction in viability for PTA-126788 after incubation at pH 2.5 for 3 hours (Fig 10). The control strain DSM 17938 also showed approximately 2.5-log reduction in CFU counts from 5.51 x 108 to 1.57 x 106 after 3 hours incubation at pH 2.5 (Fig 10).
Fig 10. Tolerance of L. reuteri strains to acidic pH.
The ability of L. reuteri strains to tolerate acidic pH was assessed by growing the strains at pH 2.5 for 3 hours and determining the CFU counts at 0 hours and 3 hours after incubation. The data represent the mean ± SD from 3 independent experiments.
In vivo safety in broilers
Compared to untreated control, members of groups treated with L. reuteri PTA-126787 and PTA-126788 daily in drinking water had no mortality, morbidity or adverse events in broiler chickens. Necropsy on day 26 showed no gross lesions indicative of health issues. Compared to the control group, histopathological analysis of trachea, lung and cecal tonsils from the groups treated with L. reuteri PTA-126787 or PTA-126788 showed no evidence of inflammation or abnormal pathology compared to untreated group (S2 Fig).
Discussion
Our understanding of the microbiome and its impact on human and animal health is rapidly evolving, leading to the identification of innovative ways to impact human and animal diseases. Undoubtedly, the past two decades have witnessed a tremendous progress in the area of probiotics highlighting their key role in supporting general health, enhancing immune function and showing the potential to reduce specific diseases. Research has repeatedly shown that the survival, safety, and efficacy properties of probiotic candidates are strain specific and cannot be generalized. In the present study, we isolated two novel Lactobacillus strains from chicken cecum, identified them as L. reuteri and established their safety using various genomic, in vitro, and in vivo studies, supporting their application as potential probiotics for human and animal health.
Identification is the first step in establishing the safety of a probiotic candidate and regulatory agencies recommend that at least two state-of-the-art methods be used to correctly identify a probiotic candidate [35, 62]. API 50 CHL analysis identified our strains as L. fermentum. L. reuteri is a subtype of L. fermentum and the two species are indistinguishable at the biochemical level [63]. 16S rRNA identification confirmed that our strains have closest homology to L. reuteri strains. Whole-genome sequencing coupled with phylogenetic analyses further confirmed that our strains have closest relatedness to L. reuteri and that our strains genetically cluster with DSM 17938, SD2112 (parent strain of DSM 17938) and RC-14. DSM 17938 and RC-14 are widely used as part of several commercially marketed dietary supplements and functional foods and there exists a plethora of clinical evidence supporting their safety and efficacy for different disease indications in humans. Consistent with our findings, several previous whole-genome phylogenetic studies also showed that the parent strain of DSM 17938, SD2112 indeed clusters with poultry isolates under poultry/human lineage IV and the authors from these studies hypothesized that SD2112 may have indeed originated from poultry [64–67]. All together, these findings clearly establish that our strains belong to L. reuteri species and that our strains have closest homology to the two commercially marketed probiotic candidates with proven human clinical safety, DSM 17938 and RC-14.
Long read sequencing technology enabled complete genome characterization with each chromosome and plasmid represented by large, nearly complete contigs. Comprehensive functional annotation of the L. reuteri strains PTA-126787 and PTA-126788 revealed presence of several genes important for probiotic efficacy. Probiotic bacteria are known to contain bioactive secondary metabolites that interact with other pathogenic bacteria to attenuate virulence [68–71]. For instance, lactic acids produced by lactic acid bacteria inhibit the growth and survival of nearby pathogens and inactivate human immunodeficiency virus by increasing acidity of the surrounding environment [72]. Both PTA-126787 and PTA-126788 strains contain four coding sequences encoding D-lactate dehydrogenase (EC 1.1.1.28) and four encoding L-lactate dehydrogenase (EC 1.1.1.27) which are responsible for lactic acid production. However, IVR12_00498 from strain PTA-126787 is a pseudogene due to a frameshift mutation.
One of the key desirable traits in a probiotic candidate is the ability to adhere to epithelial cells. The genes identified in both strains of L. reuteri putatively encode proteins involved in adhesion, providing stability to the strains and the ability to compete with other undesirable resident gut bacteria, thereby enabling effective colonization of the gut and exclusion of pathogens [73, 74]. Sortase-dependent proteins are an important group of cell surface proteins in Lactobacillus spp. and are responsible for sorting various kinds of cell surface proteins, thus playing an important role in adhesion [75]. The genomes of both strains contain the gene encoding phosphate starvation-inducible protein PhoH, a member of both the Pho regulon and the sB-regulated general stress regulon. Pho regulon plays a key role in regulating phosphate homeostasis and is generally induced in response to phosphate starvation. The sB-dependent general stress proteins are predicted to provide cells with several kinds of non-specific stress tolerance [76].
While the diversity of phages in gut ecosystems is getting increasingly well-characterized, knowledge is limited on how phages contribute to the evolution and ecology of their host bacteria [77, 78]. Prophage analysis of L. reuteri strain DSM 17938 showed 5 prophage regions while the strains, PTA-126787 and PTA-126788 had 6 and 8 prophage regions, respectively (S9 Table). Prophages can be advantageous for gut symbionts like L. reuteri by increasing its competitiveness in the intestinal niche [77].
Genome analysis identified the presence of tetW in both L. reuteri strains. tetW was found to be present on the chromosome and no elements indicative of horizontal transfer (plasmids, phages, transposons, or conjugation elements) were identified in the 15-kb flanking regions on both sides of tetW (S2 File). Phenotypic analysis showed that the two strains are susceptible to all clinically relevant antimicrobials with MICs below the EFSA recommended microbiological cut offs, except for tetracycline. For tetracycline, both strains showed a marginal 2-fold increase in MIC than the recommended microbiological cut off and a 2-fold variation in the MIC is considered acceptable due to technical variation in the MIC assay and hence the strains can be considered phenotypically susceptible [59, 79]. Together, these data suggest that the presence of tetW in our L. reuteri poses minimal risk to human and animal health.
During carbohydrate fermentation, Lactobacillus species are known to produce either exclusively L-lactic acid, exclusively D-lactic acid or a racemic mix of L- and D-lactic acid [80]. Many commercially used Lactobacillus species produce a racemic mix of L- and D-lactic acid, including the most widely used L. reuteri probiotic strains DSM 17938 and NCIMB 30242 [81–84]. Screening for D-lactic acid has gained much attention due to D-lactic acidosis and encephalopathy reported in individuals with short bowel syndrome and intestinal failure [85–87]. However, such illnesses have not yet been reported in healthy individuals. Quantification of L- and D-lactic acid showed that our strains produce a racemic mix of L- and D-lactic acid with predominance of L-lactic acid. Consistent with the previous reports, DSM 17938 also produced a racemic mix of D- and L-lactic acid [81]. NCIMB 30242, another widely used L. reuteri probiotic strain, also produces a racemic mix of D- and L-lactic acid in a ratio of 9:11 [55]. Many clinical studies conducted on DSM 17938 and NCIMB 30242 in infants, children and adults showed no evidence of adverse effects from D-lactic acidosis [26].
Lactobacillus species also possess amino acid decarboxylase activity, which results in production of toxic metabolites such as histamine, tyramine, cadaverine and putrescine. Toxicity from biogenic amines are rare but when reported is mostly associated with histamine and less commonly with tyramine [88–90]. Genome analysis showed that our strains do not encode for any known genes encoding for histamine or tyramine production. Analysis of the strains for their ability to produce biogenic amines using decarboxylase media developed by Bover-Cid and Holzapfei [56] showed that our strains are not capable of producing histamine or tyramine. The data clearly suggest that our strains do not produce the two major biogenic amines associated with toxicity in humans—histamine, and tyramine.
Our bioinformatic search identified a CDS predicted to encode arginine deiminase in both L. reuteri PTA-126787 and PTA-126788. Arginine deiminase is a common enzyme present in most lactic acid bacteria and is used to convert arginine into ornithine via citrulline and allows bacteria to adapt to non-optimal stress conditions such as acid, osmotic and temperature stresses [91]. Expectedly, a gene encoding arginine deiminase was also present in the genome of the commercially marketed L. reuteri strain DSM 17938 (Accession no. WP_003670382.1). Our bioinformatics analysis showed that the downstream gene ornithine decarboxylase required for putrescine production is absent in the genomes of L. reuteri PTA-126787 and PTA-126788. Consistent with this, in vitro analysis of biogenic amines using decarboxylase media showed that our strains are not capable of producing putrescine using L-ornithine as a substrate. Thus, the presence of arginine deiminase may not result in production of the harmful biogenic amine putrescine.
Studies on the survival properties of probiotic candidates in simulated gastrointestinal conditions are key to our understanding of their safety and efficacy. Probiotic candidates are exposed to a variety of harsh extremes in the gastrointestinal tract, but acidic pH of the stomach and bile salts appear to be the dominant factors determining the survival and growth of probiotic candidates in the gastrointestinal tract. The chicken duodenum has a typical bile salt concentration of 0.175% and likewise, the human duodenum has a bile salt concentration of around 0.3% [92, 93]. Both of our L. reuteri strains showed a similar survival profile to that of commercially marketed probiotic L. reuteri DSM 17938 in the presence of 0.3% bile salts. The human and chicken (proventriculus) stomachs have a pH of around 1.5–4.0 [94, 95]. Both of our strains showed similar survival at pH 2.5, similar to DSM 17938. Together, these data suggest that the two probiotic candidates possess desirable survival properties in a simulated gastrointestinal environment.
In the present study, broiler chickens (SPF White Leghorn chickens) were used as a model for preliminary screening of L. reuteri PTA-126787 and PTA-126788 strains for gross safety parameters. Our data showed that daily administration of the two L. reuteri strains for 26 days to chickens was safe and did not induce any adverse events. The data provided here serves as a preliminary safety evidence of the two probiotic candidates for potential animal health applications. Future studies will focus on further safety evaluation of the two strains in a rat toxicity model for potential human health applications.
In conclusion, we provide comprehensive genomic, in vitro, and in vivo evidence to support the safety of two novel L. reuteri candidates, PTA-126787 and PTA-126788. These findings would serve as the basis for designing future studies to establish efficacy in humans as well as animals.
Supporting information
(TIF)
(TIF)
(DOCX)
(DOCX)
(DOCX)
(DOCX)
(DOCX)
(DOCX)
(DOCX)
(DOCX)
(DOCX)
(XLSX)
(PDF)
(PDF)
Data Availability
The raw sequencing reads, genome assemblies and annotations in this study were deposited in the NCBI BioProject under project PRJNA675717. Accession numbers: 1. ATCC PTA-126787 SAMN16712075 CP065330-CP065334 2. ATCC PTA-126788 SAMN16712076 CP065849-CP065855.
Funding Statement
Elanco Animal Health, Inc. provided support in the form of salaries for authors [DG, SPM, NRT, NL, VR, AV, DS, MP, AA, AK, EBH], but did not have any additional role in the study design, data collection and analysis, decision to publish, or preparation of the manuscript. The specific roles of these authors are articulated in the ‘author contributions’ section.
References
- 1.FAO/WHO. Guidelines for the evaluation of probiotics in food report of a joint FAO/WHO working group on drafting guidelines for the evaluation of probiotics in food. London Ontario, Canada: World Health Organization. 2002. [Google Scholar]
- 2.Hill C, Guarner F, Reid G, Gibson GR, Merenstein DJ, Pot B, et al. Expert consensus document. The international scientific association for probiotics and prebiotics consensus statement on the scope and appropriate use of the term probiotic. Nat Rev Gastroenterol Hepatol. 2014;11(8):506–14. doi: 10.1038/nrgastro.2014.66 [DOI] [PubMed] [Google Scholar]
- 3.Vandenplas Y, Huys G, Daube G. Probiotics: An update. J Pediatr (Rio J). 2015;91(1):6–21. doi: 10.1016/j.jped.2014.08.005 [DOI] [PubMed] [Google Scholar]
- 4.Aggarwal J, Swami G, Kumar M. Probiotics and their effects on metabolic diseases: An update. J Clin Diagn Res. 2013;7(1):173–7. doi: 10.7860/JCDR/2012/5004.2701 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Zoumpopoulou G, Tzouvanou A, Mavrogonatou E, Alexandraki V, Georgalaki M, Anastasiou R, et al. Probiotic features of lactic acid bacteria isolated from a diverse pool of traditional greek dairy products regarding specific strain-host interactions. Probiotics Antimicrob Proteins. 2018;10(2):313–22. doi: 10.1007/s12602-017-9311-9 [DOI] [PubMed] [Google Scholar]
- 6.Beijerinck MW. Sur les ferments lactiques de l’industrie. Archives Neerlandaises des Sciences Exactes et Naturelles. 1901(6):212–43. [Google Scholar]
- 7.Bourdichon F, Casaregola S, Farrokh C, Frisvad JC, Gerds ML, Hammes WP, et al. Food fermentations: Microorganisms with technological beneficial use. Int J Food Microbiol. 2012;154(3):87–97. doi: 10.1016/j.ijfoodmicro.2011.12.030 [DOI] [PubMed] [Google Scholar]
- 8.Fuller R. Probiotics in man and animals. J Appl Bacteriol. 1989;66(5):365–78. [PubMed] [Google Scholar]
- 9.Reuter G. Das vorkommen von laktobazillen in lebensmitteln und ihr verhalten im menschlichen intestinaltrakt. ZBL Bak Parasit Infec Hyg I Orig. 1965;197:468–87. [Google Scholar]
- 10.Kandler O, Stetter K-O, Köhl R. Lactobacillus reuteri sp. nov., a new species of heterofermentative lactobacilli. Zentralblatt für Bakteriologie: I Abt Originale C: Allgemeine, angewandte und ökologische Mikrobiologie. 1980;1(3):264–9. [Google Scholar]
- 11.Casas IA, Dobrogosz WJ. Validation of the probiotic concept: Lactobacillus reuteri confers broad-spectrum protection against disease in humans and animals. Microbial Ecology in Health and Disease. 2000;12(4):247–85. [Google Scholar]
- 12.Mitsuoka T. The Human Gastrointestinal Tract. In: B.J.B. W, editor. The lactic acid bacteria. Boston, MA: Springer; 1992. [Google Scholar]
- 13.Valeur N, Engel P, Carbajal N, Connolly E, Ladefoged K. Colonization and immunomodulation by Lactobacillus reuteri ATCC 55730 in the human gastrointestinal tract. Appl Environ Microbiol. 2004;70(2):1176–81. doi: 10.1128/AEM.70.2.1176-1181.2004 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Talarico TL, Dobrogosz WJ. Chemical characterization of an antimicrobial substance produced by Lactobacillus reuteri. Antimicrob Agents Chemother. 1989;33(5):674–9. doi: 10.1128/AAC.33.5.674 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Weizman Z, Asli G, Alsheikh A. Effect of a probiotic infant formula on infections in child care centers: comparison of two probiotic agents. Pediatrics. 2005;115(1):5–9. doi: 10.1542/peds.2004-1815 [DOI] [PubMed] [Google Scholar]
- 16.Kolodziej M, Szajewska H. Lactobacillus reuteri DSM 17938 in the prevention of antibiotic-associated diarrhoea in children: Protocol of a randomised controlled trial. BMJ Open. 2017;7(1):e013928. doi: 10.1136/bmjopen-2016-013928 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Mu Q, Tavella VJ, Luo XM. Role of Lactobacillus reuteri in human health and diseases. Front Microbiol. 2018;9:757. doi: 10.3389/fmicb.2018.00757 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Indrio F, Riezzo G, Raimondi F, Bisceglia M, Cavallo L, Francavilla R. The effects of probiotics on feeding tolerance, bowel habits, and gastrointestinal motility in preterm newborns. J Pediatr. 2008;152(6):801–6. doi: 10.1016/j.jpeds.2007.11.005 [DOI] [PubMed] [Google Scholar]
- 19.Jones ML, Martoni CJ, Parent M, Prakash S. Cholesterol-lowering efficacy of a microencapsulated bile salt hydrolase-active Lactobacillus reuteri NCIMB 30242 yoghurt formulation in hypercholesterolaemic adults. Br J Nutr. 2012;107(10):1505–13. doi: 10.1017/S0007114511004703 [DOI] [PubMed] [Google Scholar]
- 20.Shornikova AV, Casas IA, Mykkanen H, Salo E, Vesikari T. Bacteriotherapy with Lactobacillus reuteri in rotavirus gastroenteritis. Pediatr Infect Dis J. 1997;16(12):1103–7. doi: 10.1097/00006454-199712000-00002 [DOI] [PubMed] [Google Scholar]
- 21.Abrahamsson TR, Sinkiewicz G, Jakobsson T, Fredrikson M, Bjorksten B. Probiotic lactobacilli in breast milk and infant stool in relation to oral intake during the first year of life. J Pediatr Gastroenterol Nutr. 2009;49(3):349–54. doi: 10.1097/MPG.0b013e31818f091b [DOI] [PubMed] [Google Scholar]
- 22.Schreiber O, Petersson J, Phillipson M, Perry M, Roos S, Holm L. Lactobacillus reuteri prevents colitis by reducing P-selectin-associated leukocyte- and platelet-endothelial cell interactions. Am J Physiol Gastrointest Liver Physiol. 2009;296(3):G534–42. doi: 10.1152/ajpgi.90470.2008 [DOI] [PubMed] [Google Scholar]
- 23.Klantschitsch T, Spillmann H, Puhan Z, editors. Lactobacillus reuteri: A Newcomer in Dairy Technology; 1996. [Google Scholar]
- 24.US-FDA. GRAS exemption claim and exemption notification for Lactobacillus reuteri DSM 17938. In: FDA U, editor.: GRAS Notice (GRN) No. 254; 2008. [Google Scholar]
- 25.US-FDA. GRAS exemption claim and exemption notification for Lactobacillus reuteri DSM 17938. GRAS Notice (GRN) No. 410; 2012. [Google Scholar]
- 26.US-FDA. GRAS exemption claim and exemption notification for Lactobacillus reuteri NCIMB 30242. GRAS Notice (GRN) No. 440.; 2013. [Google Scholar]
- 27.US-NDIN. NDI 78—Lactobacillus reuteri—Original NDI Notification. In: 78 NN, editor. 2000. [Google Scholar]
- 28.US-NDIN. NDI Notification 460. RepHresh Pro-B (Lactobacillus reuteri RC-14™ and Lactobacillus rhamnosus GR-1™). NDI Notification 4602000. [Google Scholar]
- 29.EFSA panel on Biological Hazards, Ricci A, Allende A, Bolton D, Chemaly M, Davies R, et al. Scientific opinion on the update of the list of QPS-recommended biological agents intentionally added to food or feed as notified to EFSA. EFSA Journal. 2017;15(3):e04664. doi: 10.2903/j.efsa.2017.4664 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.US-FDA. GRAS exemption claim and exemption notification for Lactobacillus reuteri NCIMB 30242. GRAS Notice (GRN) No. 4092012. [Google Scholar]
- 31.Wolf BW, Garleb KA, Ataya DG, Casas IA. Safety and tolerance of Lactobacillus reuteri in healthy adult male subjects. Microbial Ecology in Health and Disease. 1995;8(2):41–50. [Google Scholar]
- 32.Wolf BW, Wheeler KB, Ataya DG, Garleb KA. Safety and tolerance of Lactobacillus reuteri supplementation to a population infected with the human immunodeficiency virus. Food Chem Toxicol. 1998;36(12):1085–94. doi: 10.1016/s0278-6915(98)00090-8 [DOI] [PubMed] [Google Scholar]
- 33.Pradhan D, Mallappa RH, Grover S. Comprehensive approaches for assessing the safety of probiotic bacteria. Food Control. 2020;108:106872. [Google Scholar]
- 34.Pariza MW, Gillies KO, Kraak-Ripple SF, Leyer G, Smith AB. Determining the safety of microbial cultures for consumption by humans and animals. Regul Toxicol Pharmacol. 2015;73(1):164–71. doi: 10.1016/j.yrtph.2015.07.003 [DOI] [PubMed] [Google Scholar]
- 35.Rychen G, Aquilina G, Azimonti G, Bampidis V, Bastos MdL, Bories G, et al. Guidance on the characterisation of microorganisms used as feed additives or as production organisms. EFSA Journal. 2018;16(3):e05206. doi: 10.2903/j.efsa.2018.5206 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Seemann T. Prokka: Rapid prokaryotic genome annotation. Bioinformatics. 2014;30(14):2068–9. doi: 10.1093/bioinformatics/btu153 [DOI] [PubMed] [Google Scholar]
- 37.Hyatt D, Chen G-L, LoCascio PF, Land ML, Larimer FW, Hauser LJ. Prodigal: Prokaryotic gene recognition and translation initiation site identification. BMC Bioinformatics. 2010;11(1):119. doi: 10.1186/1471-2105-11-119 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Laslett D, Canback B. ARAGORN, a program to detect tRNA genes and tmRNA genes in nucleotide sequences. Nucleic Acids Res. 2004;32(1):11–6. doi: 10.1093/nar/gkh152 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Suzek BE, Ermolaeva MD, Schreiber M, Salzberg SL. A probabilistic method for identifying start codons in bacterial genomes. Bioinformatics. 2001;17(12):1123–30. doi: 10.1093/bioinformatics/17.12.1123 [DOI] [PubMed] [Google Scholar]
- 40.Kingsford CL, Ayanbule K, Salzberg SL. Rapid, accurate, computational discovery of Rho-independent transcription terminators illuminates their relationship to DNA uptake. Genome Biol. 2007;8(2):R22. doi: 10.1186/gb-2007-8-2-r22 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Nawrocki EP, Kolbe DL, Eddy SR. Infernal 1.0: Inference of RNA alignments. Bioinformatics. 2009;25(10):1335–7. doi: 10.1093/bioinformatics/btp157 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Tjaden B. A computational system for identifying operons based on RNA-seq data. Methods. 2020;176:62–70. doi: 10.1016/j.ymeth.2019.03.026 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Na SI, Kim YO, Yoon SH, Ha SM, Baek I, Chun J. UBCG: Up-to-date bacterial core gene set and pipeline for phylogenomic tree reconstruction. J Microbiol. 2018;56(4):280–5. doi: 10.1007/s12275-018-8014-6 [DOI] [PubMed] [Google Scholar]
- 44.Price MN, Dehal PS, Arkin AP. FastTree 2—approximately maximum-likelihood trees for large alignments. PLoS One. 2010;5(3):e9490. doi: 10.1371/journal.pone.0009490 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Emms DM, Kelly S. OrthoFinder: Phylogenetic orthology inference for comparative genomics. Genome Biology. 2019;20(1):238. doi: 10.1186/s13059-019-1832-y [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Sayers EW, Beck J, Bolton EE, Bourexis D, Brister JR, Canese K, et al. Database resources of the National Center for Biotechnology Information. Nucleic Acids Res. 2021;49(D1):D10–d7. doi: 10.1093/nar/gkaa892 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Jain C, Rodriguez-R LM, Phillippy AM, Konstantinidis KT, Aluru S. High throughput ANI analysis of 90K prokaryotic genomes reveals clear species boundaries. Nature Communications. 2018;9(1):5114. doi: 10.1038/s41467-018-07641-9 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Xie Z, Tang H. ISEScan: Automated identification of insertion sequence elements in prokaryotic genomes. Bioinformatics. 2017;33(21):3340–7. doi: 10.1093/bioinformatics/btx433 [DOI] [PubMed] [Google Scholar]
- 49.Akhter S, Aziz RK, Edwards RA. PhiSpy: A novel algorithm for finding prophages in bacterial genomes that combines similarity- and composition-based strategies. Nucleic Acids Res. 2012;40(16):e126. doi: 10.1093/nar/gks406 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Biswas A, Staals RH, Morales SE, Fineran PC, Brown CM. CRISPRDetect: A flexible algorithm to define CRISPR arrays. BMC Genomics. 2016;17:356. doi: 10.1186/s12864-016-2627-0 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Liu B, Zheng D, Jin Q, Chen L, Yang J. VFDB 2019: a comparative pathogenomic platform with an interactive web interface. Nucleic Acids Res. 2019;47(D1):D687–D92. doi: 10.1093/nar/gky1080 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Gupta SK, Padmanabhan BR, Diene SM, Lopez-Rojas R, Kempf M, Landraud L, et al. ARG-ANNOT, a new bioinformatic tool to discover antibiotic resistance genes in bacterial genomes. Antimicrob Agents Chemother. 2014;58(1):212–20. doi: 10.1128/AAC.01310-13 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Bortolaia V, Kaas RS, Ruppe E, Roberts MC, Schwarz S, Cattoir V, et al. ResFinder 4.0 for predictions of phenotypes from genotypes. J Antimicrob Chemother. 2020;75(12):3491–500. doi: 10.1093/jac/dkaa345 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Seemann T. Abricate: mass screening of contigs for antimicrobial and virulence genes. [cited 2018. Available from: https://github.com/tseemann/abricate]. [Google Scholar]
- 55.Branton WB, Jones ML, Tomaro-Duchesneau C, Martoni CJ, Prakash S. In vitro characterization and safety of the probiotic strain Lactobacillus reuteri cardioviva NCIMB 30242. International Journal of Probiotics & Prebiotics. 2011;6(1):1–12. [Google Scholar]
- 56.Bover-Cid S, Holzapfel WH. Improved screening procedure for biogenic amine production by lactic acid bacteria. Int J Food Microbiol. 1999;53(1):33–41. doi: 10.1016/s0168-1605(99)00152-x [DOI] [PubMed] [Google Scholar]
- 57.Ferreira CL, Grzeskowiak L, Collado MC, Salminen S. In vitro evaluation of Lactobacillus gasseri strains of infant origin on adhesion and aggregation of specific pathogens. J Food Prot. 2011;74(9):1482–7. doi: 10.4315/0362-028X.JFP-11-074 [DOI] [PubMed] [Google Scholar]
- 58.Eschenbach DA, Davick PR, Williams BL, Klebanoff SJ, Young-Smith K, Critchlow CM, et al. Prevalence of hydrogen peroxide-producing Lactobacillus species in normal women and women with bacterial vaginosis. J Clin Microbiol. 1989;27(2):251–6. doi: 10.1128/jcm.27.2.251-256.1989 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.NCCLS. Development of in vitro susceptibility testing criteria and quality control parameters; approved 579 guideline 2nd ed. Wayne, PA, USA.: NCCLS documents M23 -A2 NCCLS; 2001. [Google Scholar]
- 60.Singh AK, Hertzberger RY, Knaus UG. Hydrogen peroxide production by lactobacilli promotes epithelial restitution during colitis. Redox Biol. 2018;16:11–20. doi: 10.1016/j.redox.2018.02.003 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Szkaradkiewicz AK, Stopa J, Karpiński TM. Effect of oral administration involving a probiotic strain of Lactobacillus reuteri on pro-inflammatory cytokine response in patients with chronic periodontitis. Arch Immunol Ther Exp (Warsz). 2014;62(6):495–500. doi: 10.1007/s00005-014-0277-y [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.FAO/WHO. Guidelines for the evaluation of probiotics in food report of a joint FAO/WHO working group on drafting guidelines for the evaluation of probiotics in food. London Ontario, Canada: World Health Organization; May 1, 2002. [Google Scholar]
- 63.Cadieux P, Wind A, Sommer P, Schaefer L, Crowley K, Britton RA, et al. Evaluation of reuterin production in urogenital probiotic Lactobacillus reuteri RC-14. Appl Environ Microbiol. 2008;74(15):4645–9. doi: 10.1128/AEM.00139-08 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Greppi A, Asare PT, Schwab C, Zemp N, Stephan R, Lacroix C. Isolation and comparative genomic analysis of reuterin-producing Lactobacillus reuteri from the chicken gastrointestinal tract. Front Microbiol. 2020;11:1166. doi: 10.3389/fmicb.2020.01166 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Lee JY, Han GG, Choi J, Jin GD, Kang SK, Chae BJ, et al. Pan-genomic approaches in Lactobacillus reuteri as a porcine probiotic: Investigation of host adaptation and antipathogenic activity. Microb Ecol. 2017;74(3):709–21. doi: 10.1007/s00248-017-0977-z [DOI] [PubMed] [Google Scholar]
- 66.Duar RM, Frese SA, Lin XB, Fernando SC, Burkey TE, Tasseva G, et al. Experimental evaluation of host adaptation of Lactobacillus reuteri to different vertebrate species. Appl Environ Microbiol. 2017;83(12). doi: 10.1128/AEM.00132-17 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Oh PL, Benson AK, Peterson DA, Patil PB, Moriyama EN, Roos S, et al. Diversification of the gut symbiont Lactobacillus reuteri as a result of host-driven evolution. The ISME Journal. 2010;4(3):377–87. doi: 10.1038/ismej.2009.123 [DOI] [PubMed] [Google Scholar]
- 68.Choi H-J, Lee N-K, Paik H-D. Health benefits of lactic acid bacteria isolated from kimchi, with respect to immunomodulatory effects. Food Science and Biotechnology. 2015;24(3):783–9. [Google Scholar]
- 69.Oliveira LC, Saraiva TDL, Silva WM, Pereira UP, Campos BC, Benevides LJ, et al. Analyses of the probiotic property and stress resistance-related genes of Lactococcus lactis subsp. lactis NCDO 2118 through comparative genomics and in vitro assays. PLoS ONE. 2017;12(4):e0175116. doi: 10.1371/journal.pone.0175116 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Sandes S, Alvim L, Silva B, Acurcio L, Santos C, Campos M, et al. Selection of new lactic acid bacteria strains bearing probiotic features from mucosal microbiota of healthy calves: Looking for immunobiotics through in vitro and in vivo approaches for immunoprophylaxis applications. Microbiological Research. 2017;200:1–13. doi: 10.1016/j.micres.2017.03.008 [DOI] [PubMed] [Google Scholar]
- 71.Dlamini ZC, Langa RLS, Aiyegoro OA, Okoh AI. Safety evaluation and colonisation abilities of four lactic acid bacteria as future probiotics. Probiotics Antimicrob Proteins. 2019;11(2):397–402. doi: 10.1007/s12602-018-9430-y [DOI] [PubMed] [Google Scholar]
- 72.Nordeste R, Tessema A, Sharma S, Kovač Z, Wang C, Morales R, et al. Molecules produced by probiotics prevent enteric colibacillosis in pigs. BMC Vet Res. 2017;13(1):335. doi: 10.1186/s12917-017-1246-6 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Salas-Jara MJ, Ilabaca A, Vega M, García A. Biofilm forming Lactobacillus: New challenges for the development of probiotics. Microorganisms. 2016;4(3):35. doi: 10.3390/microorganisms4030035 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Granato D, Bergonzelli GE, Pridmore RD, Marvin L, Rouvet M, Corthésy-Theulaz IE. Cell surface-associated elongation factor Tu mediates the attachment of Lactobacillus johnsonii NCC533 (La1) to human intestinal cells and mucins. Infect Immun. 2004;72(4):2160–9. doi: 10.1128/IAI.72.4.2160-2169.2004 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Boekhorst J, de Been MW, Kleerebezem M, Siezen RJ. Genome-wide detection and analysis of cell wall-bound proteins with LPxTG-like sorting motifs. J Bacteriol. 2005;187(14):4928–34. doi: 10.1128/JB.187.14.4928-4934.2005 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.Antelmann H, Scharf C, Hecker M. Phosphate starvation-inducible proteins of Bacillus subtilis: Proteomics and transcriptional analysis. Journal of Bacteriology. 2000;182(16):4478. doi: 10.1128/JB.182.16.4478-4490.2000 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77.Oh J-H, Lin XB, Zhang S, Tollenaar SL, Özçam M, Dunphy C, et al. Prophages in Lactobacillus reuteri are associated with fitness trade-offs but can increase competitiveness in the gut ecosystem. Applied and Environmental Microbiology. 2019;86(1):e01922–19. doi: 10.1128/AEM.01922-19 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78.Lennon JT, Khatana SA, Marston MF, Martiny JB. Is there a cost of virus resistance in marine cyanobacteria? Isme j. 2007;1(4):300–12. doi: 10.1038/ismej.2007.37 [DOI] [PubMed] [Google Scholar]
- 79.US-FDA. GRAS Exemption Claim and Exemption Notification for Lactobacillus paracasei ssp. paracasei strain Fl 9.: GRAS Notice (GRN) No. 840.; 2018. [Google Scholar]
- 80.Axelsson L. Lactic acid bacteria: Classification and physiology. In: Salminen S, Wright A.V. and Ouwehand A., editor. Lactic Acid Bacteria: Microbiological and Functional Aspects. 3 ed. New York: Marcel Dekker; 2004. p. 1–67. [Google Scholar]
- 81.Papagaroufalis K, Fotiou A, Egli D, Tran LA, Steenhout P. A Randomized double blind controlled safety trial evaluating d-lactic acid production in healthy infants fed a Lactobacillus reuteri-containing formula. Nutr Metab Insights. 2014;7:19–27. doi: 10.4137/NMI.S14113 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 82.Iino T, Manome A, Okada S, Uchimura T, Komagata K. Effects of sodium acetate on the production of stereoisomers of lactic acid by Lactobacillus sakei and other lactic acid bacteria. J Gen Appl Microbiol. 2001;47(5):223–39. doi: 10.2323/jgam.47.223 [DOI] [PubMed] [Google Scholar]
- 83.Saulnier DM, Santos F, Roos S, Mistretta TA, Spinler JK, Molenaar D, et al. Exploring metabolic pathway reconstruction and genome-wide expression profiling in Lactobacillus reuteri to define functional probiotic features. PLoS One. 2011;6(4):e18783. doi: 10.1371/journal.pone.0018783 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 84.Kyla-Nikkila K, Hujanen M, Leisola M, Palva A. Metabolic engineering of Lactobacillus helveticus CNRZ32 for production of pure L-(+)-lactic acid. Appl Environ Microbiol. 2000;66(9):3835–41. doi: 10.1128/AEM.66.9.3835-3841.2000 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 85.Karton M, Rettmer RL, Lipkin EW. Effect of parenteral nutrition and enteral feeding on D-lactic acidosis in a patient with short bowel. JPEN J Parenter Enteral Nutr. 1987;11(6):586–9. doi: 10.1177/0148607187011006586 [DOI] [PubMed] [Google Scholar]
- 86.Scully TB, Kraft SC, Carr WC, Harig JM. D-lactate-associated encephalopathy after massive small-bowel resection. J Clin Gastroenterol. 1989;11(4):448–51. doi: 10.1097/00004836-198908000-00020 [DOI] [PubMed] [Google Scholar]
- 87.Hudson M, Pocknee R, Mowat NA. D-lactic acidosis in short bowel syndrome—an examination of possible mechanisms. Q J Med. 1990;74(274):157–63. [PubMed] [Google Scholar]
- 88.Becker K, Southwick K, Reardon J, Berg R, MacCormack JN. Histamine poisoning associated with eating tuna burgers. JAMA. 2001;285(10):1327–30. doi: 10.1001/jama.285.10.1327 [DOI] [PubMed] [Google Scholar]
- 89.Ohnuma S, Higa M, Hamanaka S, Matsushima K, Yamamuro W. An outbreak of allergy-like food poisoning. Intern Med. 2001;40(8):833–5. doi: 10.2169/internalmedicine.40.833 [DOI] [PubMed] [Google Scholar]
- 90.Miki M, Ishikawa T, Okayama H. An outbreak of histamine poisoning after ingestion of the ground saury paste in eight patients taking isoniazid in tuberculous ward. Intern Med. 2005;44(11):1133–6. doi: 10.2169/internalmedicine.44.1133 [DOI] [PubMed] [Google Scholar]
- 91.Vrancken G, Rimaux T, Wouters D, Leroy F, De Vuyst L. The arginine deiminase pathway of Lactobacillus fermentum IMDO 130101 responds to growth under stress conditions of both temperature and salt. Food Microbiol. 2009;26(7):720–7. doi: 10.1016/j.fm.2009.07.006 [DOI] [PubMed] [Google Scholar]
- 92.Van Deest BW, Fordtran JS, Morawski SG, Wilson JD. Bile salt and micellar fat concentration in proximal small bowel contents of ileectomy patients. J Clin Invest. 1968;47(6):1314–24. doi: 10.1172/JCI105823 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 93.Lin J, Sahin O, Michel LO, Zhang Q. Critical role of multidrug efflux pump CmeABC in bile resistance and in vivo colonization of Campylobacter jejuni. Infect Immun. 2003;71(8):4250–9. doi: 10.1128/IAI.71.8.4250-4259.2003 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 94.Musikasang H, Tani A, H-kittikun A, Maneerat S. Probiotic potential of lactic acid bacteria isolated from chicken gastrointestinal digestive tract. World Journal of Microbiology and Biotechnology. 2009;25(8):1337–45. [Google Scholar]
- 95.Kararli TT. Comparison of the gastrointestinal anatomy, physiology, and biochemistry of humans and commonly used laboratory animals. Biopharm Drug Dispos. 1995;16(5):351–80. doi: 10.1002/bdd.2510160502 [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
(TIF)
(TIF)
(DOCX)
(DOCX)
(DOCX)
(DOCX)
(DOCX)
(DOCX)
(DOCX)
(DOCX)
(DOCX)
(XLSX)
(PDF)
(PDF)
Data Availability Statement
The raw sequencing reads, genome assemblies and annotations in this study were deposited in the NCBI BioProject under project PRJNA675717. Accession numbers: 1. ATCC PTA-126787 SAMN16712075 CP065330-CP065334 2. ATCC PTA-126788 SAMN16712076 CP065849-CP065855.