TO THE EDITOR:
Like many others, I was very interested to see the recent 2021 guidelines by the American Society of Hematology, International Society on Thrombosis and Haemostasis, National Hemophilia Foundation, and World Federation of Hemophilia on the diagnosis1 and management2 of von Willebrand disease (VWD). These now add to the many prior guidance documents, in particular several key publications3-5; nevertheless, the new guidelines1,2 are destined to become those most closely followed. This commentary is in relation to the diagnostic guidelines1 and focuses on 3 main items related to laboratory testing. First, the “panel suggests newer assays that measure the platelet-binding activity of von Willebrand factor (VWF) (eg, VWF:GPIbM [glycoprotein Ib binding assay for VWF using recombinant mutated GPIb (no ristocetin)], VWF:GPIbR [glycoprotein Ib binding assay for VWF using recombinant GPIb (and ristocetin)]) over the VWF ristocetin cofactor assay (VWF:RCo) (automated or nonautomated assay) for the diagnosis of VWD.”1(p283) This is because classical VWF:RCo is diagnostically problematic as a result of high assay variability and poor low VWF level quantification limits, leading to a high diagnostic error rate.6,7 Moreover, VWF:GPIbM seems to be favored over VWF:GPIbR,1 because assays based on ristocetin may falsely identify type 2 VWD as a result of exon 28 polymorphisms that may reduce ristocetin binding.8 In particular, for 1 polymorphism (D1472H), 35% of the Black controls were homozygous for the H allele, whereas all the White controls with D1472H were heterozygous.8 Second, for type 2 VWD, the “panel suggests against a platelet-dependent VWF activity/VWF:Ag [VWF antigen] ratio <0.5 cutoff, and rather using a higher cutoff of <0.7 to confirm type 2 VWD (2A, 2B, or 2M) for patients with an abnormal initial VWD screen.”1(p283) Third, although not a specific recommendation, the panel supports, within its published algorithm, a core test panel comprising VWF:Ag, platelet-dependent VWF activity, and factor VIII coagulant assay (FVIII:C) and relegates collagen binding activity (VWF:CB) to a supplementary assay, as a potential alternative to multimer analysis, to further characterize type 2 VWD, but notably relies on using the prior platelet-dependent VWF activity/Ag ratio of <0.7. Many of the recommendations build upon one another; for example, poor initial selection of a platelet-dependent VWF activity assay may lead to failure to appropriately characterize type 2 VWD for further testing, including application of VWF:CB or multimer analysis. Also important to note, the term “suggests” indicates a conditional recommendation that is likely to be strengthened (for future updates or adaptation) by further research. Therefore, in this commentary, I provide some additional data to help inform future iterations of such guidance. For this, I largely highlight some recently published work.6,7,9-13
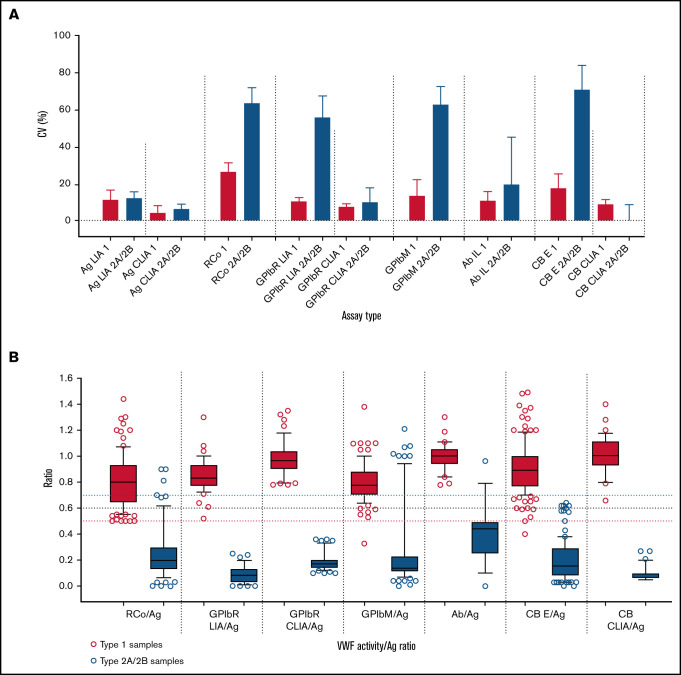
First, in our most recent external quality assessment (EQA) publication,9 highlighting VWD diagnostic errors as linked to test types/test panels, we found, as detailed in Table 1, (1) VWF:RCo showed the highest assay variability and poorest quantification limit among all platelet-dependent VWF activity assays; (2) VWF:GPIbR performed by CLIA was the least variable (with the lowest quantification limit) platelet-dependent VWF activity assay, followed in turn by VWF:GPIbR by LIA, VWF:GPIbM (LIA), and VWF:RCo (Figure 1A); (3) CLIA methodology was the least variable method overall (all available assays) (Figure 1A); (4) for associated diagnostic errors of type 1 (quantitative VWF deficiency) vs type 2A/2B (high molecular weight [HMW] VWF deficiency) samples, the most problematic platelet-dependent VWF activity assay was VWF:RCo, followed by VWF:GPIbM, whereas VWF:GPIbR and VWF:CB were associated with few errors; (5) error rates associated with a standard 3-test panel (ie, as per the guidelines1: FVIII:C, VWF:Ag, and platelet-dependent VWF activity assay) were twice those of a 4-test panel including the VWF:CB; (6) no single universal cutoff value (eg, 0.5 or 0.7) for activity/Ag was sufficiently robust to effectively identify/exclude type 2A/B VWD; indeed, ideal cutoff values are method specific (Figure 1B), but if a universal cutoff is required, perhaps 0.6, in line with recommendations by the UK Haemophilia Doctors organization,4 could be favored (supplemental Figure 1); and (7) the best VWD type discrimination was achieved using CLIA methods (ie, GPIbR/Ag or CB/Ag), followed by GPIbR/Ag (LIA); other methods, including RCo/Ag and GPIbM/Ag, showed substantial ratio overlap between type 1 vs 2A/2B VWD (Figure 1B).
Table 1.
Summary of main findings through recent EQA exercises6,9
| Main finding |
|---|
| The least variable platelet-dependent VWF activity assay (with the lowest quantification limit) was VWF:GPIbR performed by CLIA procedure on the ACL AcuStar instrument, followed by VWF:GPIbR performed by LIA, VWF:GPIbM (LIA), and VWF:RCo (Figure 1A) |
| CLIA was by far the least variable method overall, which was true for VWF:Ag, VWF:GPIbR, and VWF:CB, as compared with other comparative methodologies (Figure 1A) |
| In terms of associated diagnostic errors between identification of type 1 vs type 2A/B VWD samples, the most problematic platelet-dependent VWF activity assay was VWF:RCo (responsible for 26.7% of such errors), followed in order by VWF:GPIbM (22.2%) and VWF:GPIbR (LIA; 3.7%) |
| The least problematic VWF activity assay overall was VWF:CB (1.5% of errors), especially if performed by CLIA (no errors) |
| A standard 3-test panel, as recommended in the recent guidelines,1 was associated with a twofold error rate compared with a 4-test panel incorporating VWF:CB, as recommended by UK guidelines4 |
| No single universal cutoff value (eg, 0.5 or 0.7) for activity/Ag is sufficiently robust to effectively identify/exclude type 2 VWD, with cutoff values being somewhat method specific (Figure 1B); if a universal cutoff is required, perhaps 0.6, in line with the recommendations of the UK Haemophilia Doctors organization,4 could be favored (supplemental Figure 1) |
| The best type 1 vs 2A/2B discrimination was seen using CLIA methods (ie, GPIbR/Ag or CB/Ag), followed by GPIbR/Ag using LIA; other methods, including RCo/Ag and GPIbM/Ag, showed substantial overlap in ratios between type 1 vs 2A/2B VWD (Figure 1B) |
| The composite of these data favors CLIA methodology over all other methodologies; for the platelet-binding activity assays, VWF:GPIbR is favored over both VWF:GPIbM and VWF:RCo |
CLIA, chemiluminescence immunoassay; LIA, latex immunoassay.
Figure 1.
Summary data from the Royal College of Pathologists Quality Assurance Program (RCPAQAP) for the years 2014 to 2021 inclusive. (A) Summary data for VWF assay variability. Data shown as median and interquartile range of the coefficient of variation (CV; %) on the y-axis. Assay is type listed along the x-axis in the following order: (1) VWF:Ag as LIA- and CLIA-based assays; (2) VWF:RCo; (3) VWF:GPIbR by LIA and CLIA; (4) VWF:GPIbM (LIA only); (5) VWF:Ab; and (6) VWF:CB by enzyme-linked immunosorbent assay (ELISA) and CLIA. Data compare type 1 plasma (n = 7; red) vs type 2A plasma (n = 5)/type 2B plasma (n = 5) as composite (blue) and as sent to participants over the period of analysis. There are fewer data points for CLIAs, because this method emerged in 2016; however, this method showed the overall least variability. Note that higher CVs are expected for functional VWF tests in type 2 VWD as test values approach 0. (B) Summary data for VWF activity/Ag ratios. Data respectively shown as box plots of ratios (y-axis) of VWF:RCo/VWF:Ag, VWF:GPIbR/VWF:Ag (LIA then CLIA based), VWF:GPIbM/VWF:Ag, VWF:Ab/VWF:Ag, and VWF:CB/VWF:Ag (ELISA then CLIA based) (x-axis) for data using type 1 plasma (n = 7; red) and type 2A/2B VWD patient plasma (n = 5; blue), as tested by RCPAQAP participants over the period of 2014 to 2021. Box plots show median and 10th to 90th percentile, with outliers shown as dots. Long horizontal dashed lines indicate 0.5 (red), 0.6 (black), and 0.7 (blue) cutoff values. Note that a cutoff of 0.7 is more inclusive and, for some assays, ensures greater capture of type 2 VWD cases; however, this comes at the cost of specificity, because the higher cutoff also inappropriately captures more cases of type 1 VWD.
In summary, the composite of these EQA data favors CLIA methodology over all other methodologies, and for platelet-binding activity assays, VWF:GPIbR is favored over both VWF:GPIbM and VWF:RCo. Moreover, the data confirm the findings reported in an earlier article,6 which stated that initial inclusion of VWF:CB, using a 4-test panel, is most diagnostically accurate, as supported by the UK Haemophilia Doctors guidance.4 The data also favor a cutoff of 0.6 instead of 0.7, also in line with the UK guidance.4 A cutoff of 0.7, being more inclusive, may identify more cases of type 2 VWD for some assays, but this comes at the cost of specificity and will also inappropriately capture cases of type 1 VWD. This places a high subsequent test burden on laboratories and may also lead to additional errors. For example, if multimer or VWF:CB (ELISA) assays are performed, then falsely captured type 1 cases may ultimately be incorrectly identified as 2M (no loss of HMW VWF or normal CB/Ag) or 2A/2B (false loss of HMW7 or CB/Ag ratio <0.7).
Naturally, there are limitations to the use of EQA data to inform diagnostic practice. First, testing is undertaken by real-world laboratories, not just expert laboratories. However, that is also a strength, because these laboratories assess most cases of VWD. Second, samples may comprise both true patient samples and in vitro prepared samples that mimic VWD (either type 1 or type 2A).10,11 However, in our experience, data using true patient samples are strikingly similar to those using prepared samples, and in Figure 1B, only true patient data are shown for 2A/2B samples. Third, such studies cannot address the issue of polymorphisms affecting ristocetin binding and influencing the panel’s recommendation of VWF:GPIbM over VWF:GPIbR; however, although apparently common in Black populations, homozygous changes are uncommon in White populations,8 and my own experience indicates low incidence in my geographic locality of Australia.12 Indeed, my laboratory has only ever identified a single known case of such a polymorphism causing a false type 2M VWD diagnosis.12 Therefore, using a VWF:GPIbR assay rather than VWF:GPIbM could be considered appropriate in areas where few Black individuals live, especially given the comparatively favorable findings shown here. Lastly, data are influenced by the assays available to EQA participants, as well as by samples tested. In regard to VWF:GPIbM, only the automated commercial method is used, and so the data may not hold true for the ELISA-based VWF:GPIbM potentially used in expert laboratories.13 Also, findings represent a sampling, including, for example, only 5× 2A/2B patient samples (Figure 1B), and thus may not translate to all 2A/2B VWD cases investigated by laboratories.
In conclusion, access to the best tests is not always guaranteed. The United States, for example, only has access to 2 classes of US Food and Drug Administration (FDA)–cleared VWF activity assays (VWF:RCo and VWF:Ab).14 There is no FDA-cleared VWF:GPIbR, VWF:GPIbM, or VWF:CB assay. In contrast, in Australia and Europe, all such assays are widely available. I therefore request that future iterations of the guidelines consider these findings. I also urge US laboratories to support manufacturers of the best VWF test assays to gain FDA clearance, thereby ultimately improving the diagnosis of VWD around the world.
Supplementary Material
The full-text version of this article contains a data supplement.
Acknowledgments
Acknowledgments: The author thanks staff members and laboratory participants of Royal College of Pathologists Quality Assurance Program (RCPAQAP) Haematology, as well as past and present members of the Westmead ICPMR laboratory.
The opinions expressed in this article are those of the author and not necessarily those of NSW Health Pathology or the RCPAQAP.
Contribution: E.J.F. wrote the paper.
Conflict-of-interest disclosure: The author declares no competing financial interests.
Correspondence: Emmanuel J. Favaloro, Department of Haematology, Institute of Clinical Pathology and Medical Research, Westmead, NSW, Australia 2145; e-mail: emmanuel.favaloro@health.nsw.gov.au.
References
- 1.James PD, Connell NT, Ameer B, et al. ASH ISTH NHF WFH 2021 guidelines on the diagnosis of von Willebrand disease. Blood Adv. 2021;5(1):280-300. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Connell NT, Flood VH, Brignardello-Petersen R, et al. ASH ISTH NHF WFH 2021 guidelines on the management of von Willebrand disease. Blood Adv. 2021;5(1):301-325. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Sadler JE, Budde U, Eikenboom JC, et al. ; Working Party on von Willebrand Disease Classification . Update on the pathophysiology and classification of von Willebrand disease: a report of the Subcommittee on von Willebrand Factor. J Thromb Haemost. 2006;4(10):2103-2114. [DOI] [PubMed] [Google Scholar]
- 4.Laffan MA, Lester W, O’Donnell JS, et al. The diagnosis and management of von Willebrand disease: a United Kingdom Haemophilia Centre Doctors Organization guideline approved by the British Committee for Standards in Haematology. Br J Haematol. 2014;167(4):453-465. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Nichols WL, Hultin MB, James AH, et al. von Willebrand disease (VWD): evidence-based diagnosis and management guidelines, the National Heart, Lung, and Blood Institute (NHLBI) Expert Panel report (USA). Haemophilia. 2008;14(2):171-232. [DOI] [PubMed] [Google Scholar]
- 6.Favaloro EJ, Bonar RA, Meiring M, et al. Evaluating errors in the laboratory identification of von Willebrand disease in the real world. Thromb Res. 2014;134(2):393-403. [DOI] [PubMed] [Google Scholar]
- 7.Chandler WL, Peerschke EI, Castellone DD, Meijer P; NASCOLA Proficiency Testing Committee . Von Willebrand factor assay proficiency testing. The North American Specialized Coagulation Laboratory Association experience. Am J Clin Pathol. 2011;135(6):862-869. [DOI] [PubMed] [Google Scholar]
- 8.Flood VH, Gill JC, Morateck PA, et al. Common VWF exon 28 polymorphisms in African Americans affecting the VWF activity assay by ristocetin cofactor. Blood. 2010;116(2):280-286. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Favaloro EJ, Dean E, Arunachalam S, Vong R, Mohammed S. Evaluating errors in the laboratory identification of von Willebrand disease using contemporary von Willebrand factor assays [published online ahead of print 20 September 2021]. Pathology. doi: 10.1016/j.pathol.2021.07.001. [DOI] [PubMed] [Google Scholar]
- 10.Favaloro EJ. Rethinking internal quality control and external quality assessment for laboratory diagnostics of von Willebrand disease. Ann Blood. 2019;4:4. [Google Scholar]
- 11.Favaloro EJ, Bonar R, Hollestelle MJ, et al. Differential sensitivity of von Willebrand factor activity assays to reduced VWF molecular weight forms: a large international cross-laboratory study. Thromb Res. 2018;166:96-105. [DOI] [PubMed] [Google Scholar]
- 12.Favaloro EJ, Mohammed S, Vong R, et al. How we diagnose 2M von Willebrand disease (VWD): use of a strategic algorithmic approach to distinguish 2M VWD from other VWD types. Haemophilia. 2021;27(1):137-148. [DOI] [PubMed] [Google Scholar]
- 13.Flood VH, Gill JC, Morateck PA, et al. Gain-of-function GPIb ELISA assay for VWF activity in the Zimmerman Program for the Molecular and Clinical Biology of VWD. Blood. 2011;117(6):e67-e74. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Favaloro EJ, Lippi G. Recent advances in mainstream hemostasis diagnostics and coagulation testing. Semin Thromb Hemost. 2019;45(3):228-246. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.