ABSTRACT
Monoclonal antibodies are susceptible to chemical and enzymatic modifications during manufacturing, storage, and shipping. Deamidation, isomerization, and oxidation can compromise the potency, efficacy, and safety of therapeutic antibodies. Recently, in silico tools have been used to identify liable residues and engineer antibodies with better chemical stability. Computational approaches for predicting deamidation, isomerization, oxidation, glycation, carbonylation, sulfation, and hydroxylation are reviewed here. Although liable motifs have been used to improve the chemical stability of antibodies, the accuracy of in silico predictions can be improved using machine learning and molecular dynamic simulations. In addition, there are opportunities to improve predictions for specific stress conditions, develop in silico prediction of novel modifications in antibodies, and predict the impact of modifications on physical stability and antigen-binding.
KEYWORDS: In silico prediction, post-translational modifications, chemical stability, developability, therapeutic antibody development
Introduction
Monoclonal antibodies (mAbs) have become one of the predominant classes of therapeutic proteins and they are used to treat various diseases, such as breast cancer, multiple sclerosis, and asthma.1 Therapeutic antibodies can be engineered to have high specificity and affinity for their targets.2 However, physical and chemical instabilities can have a negative impact on the manufacturability, safety, and efficacy of therapeutic antibodies.3 Like all proteins, mAbs are susceptible to chemical degradation (e.g., oxidation)4 and enzymatic modifications (e.g., sulfation) during cell culture.5 Chemical and enzymatic modifications contribute to heterogeneity. For example, asparagine (Asn) deamidation can generate charge variants; tryptophan (Trp) oxidation can generate hydrophilic or hydrophobic variants.6 In addition, chemical modifications can affect the physical stability and biological activity of antibodies. For example, isomerization within the Fab region can reduce conformational stability,7 whereas deamidation within complementary-determining region (CDR) loops can reduce binding affinity.8 Therefore, identifying liable sites for post-translational modifications (PTMs) has become a critical step in assessing the developability of therapeutic candidates.
Developability assessments aim to identify candidates with long-term stability, manufacturability, and low heterogeneity.9 Forced degradation with thermal, pH, and light stress has been used to accelerate chemical degradation and identify liable residues.10 Peptide mapping can identify the specific sites for chemical modifications after forced degradation. In contrast, chromatographic techniques such as cation exchange chromatography and hydrophobic interaction chromatography can monitor the overall change in charge and hydrophilic variants, respectively.11 However, experimental approaches for identifying PTM liabilities are time-consuming and require high quantities of the purified protein.12 The sample preparation and data analysis for peptide mapping is incredibly labor-intensive.13 At earlier stages of drug development, the number of forced degradation conditions is limited by the low availability of the purified protein.10 Computational tools are becoming more common during developability assessments due to the low cost, lack of sample consumption, and high speed. In the past decade, computational tools have been used to predict PTM liable sites and engineer antibodies with better chemical stability.14
Computational approaches for predicting PTMs
Computational approaches for predicting PTMs can be divided into three categories: sequence-based, structure-based, and physics-based. Sequence-based approaches either flag individual residues prone to chemical degradation (e.g., methionine oxidation) or liable motifs (e.g., NG, NS, and NT for deamidation).15 Liable motifs for deamidation and isomerization were identified by investigating the effects of protein sequence on deamidation16 and isomerization17 rates for model peptides. Model peptides are suitable for assessing the effects of protein sequence on chemical degradation due to the generation of substantial chemical degradation in a short time.17
Structure-based approaches predict PTM liabilities by using structural features correlated with enzymatic and chemical modifications.18 Common structural features include, but are not limited to, secondary structure, water coordination number (WCN), and solvent-accessible surface area (SASA).9 WCN represents the average number of water molecules within the radius of an atom;19 SASA represents the surface area of the protein that interacts with the solvent.20 Structural features such as SASA are typically extracted from crystal21 or predicted structures for antibodies.22 Alternatively, machine learning algorithms (e.g., NetSurfP) can also be used to predict structural features such as secondary structure, solvent exposure, and structural disorder for protein sequences.23
Physics-based approaches are based on physical principles: for example, molecular dynamic (MD) simulations use Newtonian physics to predict the spatial position of atoms over time.24 Physics-based approaches can predict the free energy barriers for chemical modifications25,26 and probe protein dynamics.27 MD simulations can be used to estimate averaged SASA, which captures changes in solvent exposure due to conformational changes. In addition, MD simulation can provide the root-mean-square fluctuations (RMSF) for Cα atoms, which captures structural flexibility.28
Comparison of computational approaches
Sequence-based approaches are simple and easy to implement. Once the protein sequence is available, candidates with poor chemical stability can be eliminated by checking for liable motifs.15 However, using liable motifs alone can overestimate the number of liable residues or miss potential degradation hotspots.12 The rate of chemical degradation for model peptides does not represent chemical degradation in native proteins, where the tertiary structure plays a role in chemical and enzymatic modifications.16 For example, high solvent exposure can increase the risk of chemical modifications due to the increased exposure to degrading agents such as water, peroxides, and light.27
Structure-based approaches have better accuracy than sequence-based approaches because the in silico prediction includes the impact of secondary and tertiary structure on PTMs.29 The accuracy of structure-based approaches is dependent on the quality of crystal or predicted structures and the selection of structural features. Knowledge of the chemical mechanisms for modifications (e.g., deamidation) can improve the accuracy of structure- and physics-based approaches. For example, some studies use the nucleophilic attack (Cγ – Nn+1) distance to predict the risk of deamidation due to the nucleophilic attack on the side chain by the backbone nitrogen during succinimide formation.30,31
Although physics-based approaches provide valuable insight into the dynamics and energetics of chemical degradation, physics-based approaches are more computationally demanding compared to sequence- and structure-based approaches. It is challenging and time-consuming to predict free energy barriers and conduct MD simulations for larger proteins like antibodies.32 Improvements in the speed and accuracy of MD simulations and quantum chemical calculations are required for the broader adoption of physics-based approaches. Moreover, coarse-grained models can potentially improve the efficiency of MD simulations for larger proteins such as antibodies.33
More recent studies combine these three computational approaches for in silico prediction of PTMs. Protein sequence and structural features have been incorporated into machine learning algorithms (e.g., random forest) to classify cold and hot spots for chemical modifications.34 This review discusses current computational approaches for predicting isomerization, deamidation, oxidation, and lysine glycation in mAbs. In silico prediction of novel PTMs in mAbs, such as carbonylation, sulfation, and hydroxylation, is also considered.
Aspartic acid isomerization
Aspartic acid (Asp) residues in mAbs can undergo isomerization to form IsoAsp. During isomerization, there is a nucleophilic attack on the carbonyl group of Asp by the ionized amine group at the n + 1 residue. The Asp side chain requires a proton (H+) to form the succinimide intermediate and leaving water molecule, and thus isomerization is more favorable at low pH.35 Finally, the succinimide intermediate is hydrolyzed at the n + 1 peptide bond to form isoAsp.17 At pH 4–7, Asp isomerization forms succinimide, which can cause a change in net charge. In contrast, IsoAsp formation can cause conformational changes because of the methyl group added to the peptide backbone. These conformational changes can cause changes in surface charge distribution or surface hydrophobicity.5 Depending on the location of IsoAsp formation, isomerization can generate basic,8 acidic,36 hydrophilic, or hydrophobic variants.37 Aspartate isomerization at the CDR loops can decrease the antigen-binding affinity.38
Asp isomerization is sensitive to temperature and the dielectric constant of the solvent. At neutral pH, a high dielectric constant for solvents increases the pKa of Asp, which increases the concentration of the carboxylic acid (COOH) form of the Asp side chain. The COOH form is more reactive and prone to isomerization than the carboxylate form (COO−).39 High temperatures can also accelerate the rate of isomerization reactions. Moreover, flanking residues, ionization state, and higher-order structure also influences isomerization.40 For example, the risk of isomerization is highest for Asp residues within random coils due to structural flexibility and higher solvent exposure.41 The risk of isomerization also differs for different CDR loops. Asp isomerization at liable motifs is more likely with the CDR H3, H2, and L1 loops.42
Sequence-based approaches
Sequence-based approaches for predicting Asp isomerization flag liable motifs for Asp isomerization such as DG, DS, DD, DT, and DH.42 Flanking glycine (Gly) residues make succinimide formation more favorable due to the lower steric hindrance. Serine and threonine residues can act as proton donors during isomerization.40 Positively charged residues at n + 1 or n-1 position can accelerate Asp isomerization due to electrostatic effects.35 Moreover, flanking residues that act as proton donors and acceptors can make Asp residues more reactive and prone to isomerization. For example, histidine residues can act as proton donors (H+) to OH− ions at low pH during succinimide formation.43
During the developability stage, protein engineering can be used to remove PTM liabilities and improve chemical stability.14 The risk of isomerization can be reduced by substituting Asp with glutamic acid, which is less prone to isomerization. Patel et al. identified a liable DS motif within the CDR loop that was prone to isomerization: substituting Asp with glutamic acid led to a loss in bioactivity. However, engineering the n + 1 residue (i.e., serine) retained the bioactivity while reducing the risk of Asp isomerization.44
Structure-based approaches
The higher-order structure of antibodies also plays a crucial role in the risk for Asp isomerization. Dihedral angles can influence the reactivity of Asp residues and affect the risk of isomerization.26 Diepold and colleagues observed isomerization for liable motifs within the solvent-exposed and flexible CDR loops. However, isomerization did not occur at the DG motif in the conserved region of the antibody.45 Asp isomerization is the least favorable within beta-sheets due to structural rigidity. Sreedhara and colleagues observed succinimide formation for Asp74, which was solvent-exposed and part of a loop, whereas the Asp73 within beta-helix was less prone to isomerization.40 In addition, hydrogen bonding for peptide backbone and side chains can make succinimide formation less favorable.46
Structure-based approaches for predicting Asp isomerization incorporate solvent exposure, secondary structure, nucleophilic attack (Cγ – Nn+1) distance, and hydrogen bonding.46 Sydow and colleagues used the root mean square deviation of Asp Cα atoms, size of the n + 1 residue, and secondary structure to develop a decision tree for classifying liable Asp residues.46 They reported that Asp residues with high conformational flexibility and small size for n + 1 residue were more prone to isomerization. Although, a smaller side chain leads to lower steric hindrance for forming succinimide;26 using the size of n + 1 residue for predicting isomerization neglects potential electrostatic effects from flanking residues on isomerization.35
Physics-based approaches
MD simulations can supplement structure-based approaches for predicting Asp isomerization. Sharma et al. developed a logistic regression model to predict Asp isomerization at liable motifs (i.e., DG, DS, DT, DD, DH). 29 The averaged SASA for Asp residues, RMSF for Cα atoms, and the averaged SASA for the n + 1 residue correlated well with Asp isomerization liability. Sharma and colleagues’ regression model successfully predicted five out of six liable Asp residues and all non-liable sites. Most of the parameters used in the study were derived from MD simulations. Thus, MD simulations can provide insight into how conformational flexibility plays a role in chemical degradation.
Recent studies have used MD simulations to predict chemical modifications; however, few studies attempt to predict the free energy barriers for chemical modifications. Plotnikov et al. used quantum mechanical/molecular mechanics (QM/MM) MD simulations to identify isomerization and deamidation hotspots. They estimated the free-energy barriers at different steps for isomerization and deamidation. The steric hindrance for succinimide formation is captured by the free energy barriers for adopting more reactive conformations to form succinimide.26 In addition, Plotnikov et al. included long- and short-range electrostatic interactions in their QM/MM model. Compared to structure-based approaches, free energy predictions can incorporate electrostatic effects on Asp isomerization.26
Therapeutic candidates with high and low risk for isomerization can be classified based on liable motifs and structural predictors such as SASA. In addition, knowledge of the antigen-binding sites, either from crystal structures or in silico predictions, can guide protein engineering to improve chemical stability. For example, if a liable Asp residue is involved in antigen binding, then the n + 1 residue can be substituted to improve chemical stability without affecting the bioactivity of the antibody.44
Asparagine deamidation
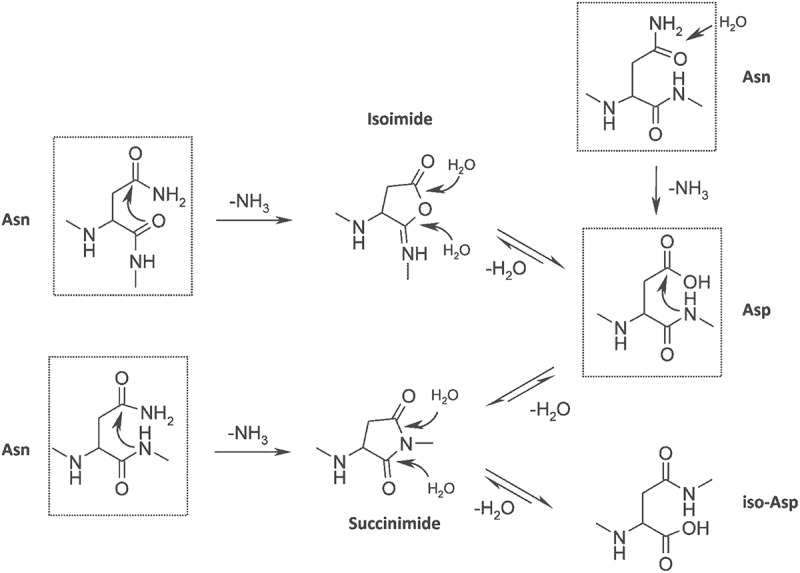
Asn residues can undergo deamidation to form Asp or IsoAsp. There are three possible pathways for Asn deamidation. A nucleophilic attack on the backbone nitrogen at the n + 1 residue forms succinimide, which is hydrolyzed to form Asp. 46 Alternatively, the nucleophilic attack by the backbone carbonyl group can form isoimide, which is hydrolyzed to IsoAsp and Asp (Figure 1). At acidic pH (pH ≤ 4), direct hydrolysis of Asn can also form Asp (Figure 1).47 The direct hydrolysis pathway competes with succinimide-mediated deamidation.48 Overall, deamidation is more favorable at pH ≥ 6. Forced degradation by thermal and high pH stress are used to confirm liable Asn residues.10 Flanking residues, secondary and tertiary structure, solvent exposure, and structural flexibility can affect deamidation.49
Figure 1.
Asparagine deamidation and aspartic acid isomerisation pathway. Reproduced from Sydow JF et al. (2014).46 (Creative Commons Attribution License).
Alt Text: The top half of the figure shows the removal of amine group from asparagine to form isoimide. Isoimide is hydrolysed by two water molecules hydrolysing isoimide to form aspartic acid in a reversible reaction. The top right-hand side of the figure shows the loss of an amine group from asparagine to form aspartic acid. The bottom half of the figure shows the loss of an amine group from asparagine to succinimide. The succinimide is hydrolysed by two water molecules to form aspartic acid and isoaspartic acid.
Sequence-based approaches
Liable motifs for deamidation include NG, NS, NN, NT, and NH:42 the NG motif has the highest deamidation rate, followed by NS, NT, and NH.16 The high deamidation rate for the NG motif can be attributed to lower steric hindrance for forming succinimide. Alternatively, Gly residues at the n + 1 position can provide more conformational flexibility, allowing Asn residues to adopt more reactive conformations to form succinimide.41 The n + 1 flanking residues can also play a role in deamidation. Chelius et al. reported the highest level of deamidation at SNG, ENN, LNG, and LNN.50 Lu et al. investigated the deamidation for 131 clinical-stage antibodies and reported deamidation at liable motifs was more common at CDR-H2 and CDR-L1 loops.42 Deamidation at NT, NF, and NY only occurred within the CDR-H3 loop,14 which could be due to the length and flexibility of the CDR-H3 loop, allowing it to adopt more conformations.51 Excluding the CDR-H3 loop, the remaining CDR loops are clustered into canonical structures, where a few critical amino acids at specific positions can determine the CDR conformation.52 Non-H3 CDR loops are clustered based on structural similarity: dihedral angles and root mean square deviation have been used to compare and cluster structures for the same type of CDR loops (e.g., CDR-L1).53 For example, the L1-11-A cluster refers to CDR-L1 loops with a length of 11 residues, which fall into the first cluster of CDR-L1 structures. 53 Future studies could investigate how differences in conformations for different CDR-L1 and H2 clusters affect the risk of deamidation.
Structure-based approaches
The higher-order structure also influences the rate of Asn deamidation.45 The risk of deamidation is low for Asn within beta-sheets due to structural rigidity and an extensive network of hydrogen bonds. Structure-based approaches for predicting deamidation incorporate secondary structure, hydrogen bonding, SASA, dihedral angles, and nucleophilic attack (Cγ – N) distance.30 Random forest and decision trees are the most common machine learning algorithms for classifying hot and cold spots (Table 1). Delmar et al. developed a random forest model to identify liable Asn residue and predict the deamidation rate.30 The backbone and side-chain dihedral angles are used to assess if the Asn side chain and backbone alignment are favorable for forming succinimide. However, the backbone dihedral angles are more critical than side-chain dihedral for predicting deamidation hot spots.30,54
Table 1.
Summary of parameters used for machine learning methods for predicting asparagine deamidation in mAbs. Abbreviations: RMSD, root mean square deviation; SASA, solvent-accessible surface area
| Authors | Machine learning algorithm | Parameters | Data Set | Accuracy and precision1 | Reference |
|---|---|---|---|---|---|
| Sydow, J. F. et al. (2014). | Decision tree | RMSD of asparagine Cα-atoms, C-terminal amino acid size (>102.7 Å), SASA (>89.4 Å2), and phi angles (>75.2º) | Data set consisted of 55 deamidation hotspots and 940 non-hotspots from forced degradation data for 37 mAbs. | Accuracy: 95.7 % Precision: 100.0 % |
46 |
| Yan, Q., Huang, M., Lewis, M. J. & Hu, P. (2018)2 | Decision tree | Secondary structure, SASA (> 10 Å2), Cγ-Nn+1distance (> 3.4 Å), motifs (NS, NT, NG). |
Data set consisted of deamidation sites for five IgG1 and five IgG4 antibodies. | Accuracy: 83.8% Precision: 41.7% |
54 |
| Delmar, J. A., Wang, J., Choi, S. W., Martins, J. A. & Mikhail, J. P. (2019).2 | Random forest | Residue at n + 1 position, half-life of pentapeptide, backbone dihedral angles, Cγ-Nn+1 distance, side-chain dihedral angles, and hydrogen bonding within side chains and secondary structures. | Combination of in-house data and deamidation data from Lu et al.39 | Accuracy: 95.6 % Precision: 100.0 % |
30 |
1Accuracy is determined by the following formula: and precision was determined by the following formula: . TP = true positive, TN = true negative, FN = false negative, and FP = false positive.
2The accuracy and precision is reported from Delmar et al. (2019)30
There are contradicting results for the impact of Cγ – Nn+1 distance on Asn deamidation. Yan et al. reported that a Cγ – Nn+1 distance > 3.4 Å was unfavorable for Asn deamidation, 54 whereas Sydow et al. found no correlation between deamidation risk and Cγ – Nn+1 distance.46 The discrepancy in results could be due to different stress conditions used to identify liable Asn residues for both studies. Yan et al. used high pH stress to identify liable residues, while Sydow et al. thermally stressed their samples at the formulation pH of 6.0. At basic pH, deamidation by forming succinimide is more favorable, and the Cγ – Nn+1 distance is a useful predictor for succinimide formation.48 Therefore, the impact of structural features such as Cγ – Nn+1 distance on deamidation can differ between thermal stress at acidic and basic pH. Future studies could select different structural features to develop machine learning models to predict the risk of deamidation for different types of stress conditions.
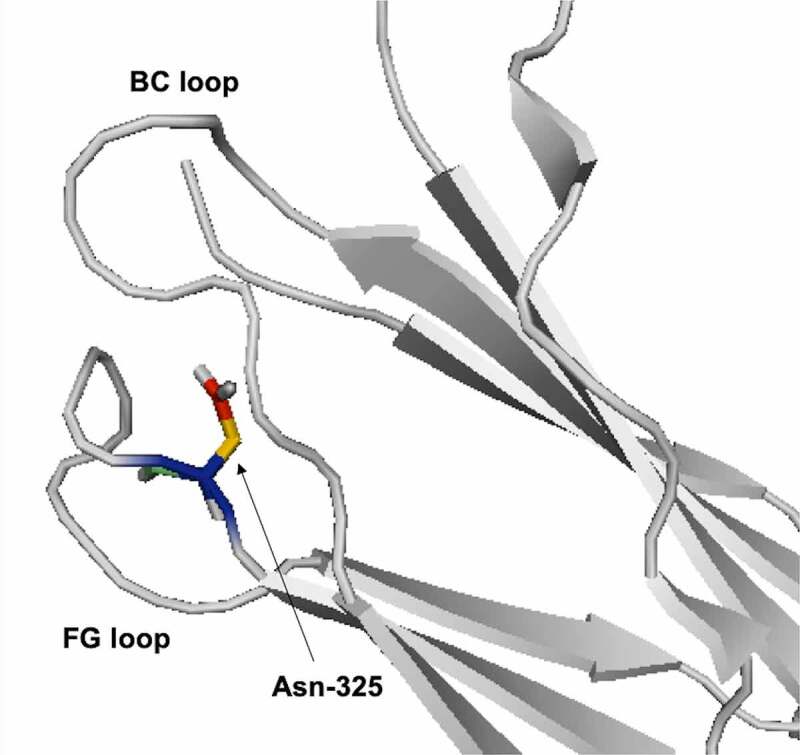
Antibodies can also change conformations at different pH, which can cause a change in the solvent exposure for Asn residues. At high pH, heavy chain (HC)-Asn325 at the FG loop of the CH2 domain is buried between the FG and BC loop (Figure 2). There is a conformational change in the BC loop at low pH, which causes HC-Asn325 to become exposed and susceptible to deamidation.54 The CDR-H3 loop can adopt multiple conformations due to its length and flexibility. Lan et al. observed different conformations for the CDR-H3 loop at different pH. At neutral pH, the CDR-H3 loop is extended and solvent-exposed. In contrast, at low pH, the CDR-H3 loop is bent. 55 Future studies could use MD simulations to identify more reactive conformations for deamidation at different pH.
Figure 2.
Ribbon diagram representation for HC-Asn325 between BC and FG loop (PDB: 4BYH).
[Alt Text: Zoom-in on three loops within the CH2 domain. Each loop is connected to two beta-sheets. Two loops are labelled as FG and BC loop. Both loops are facing each other and the BC loop is above the FG loop. A stick representation for the asparagine-325 side chain is shown at bottom right side of the BC loop.].
Oxidation
Oxidation of antibodies can occur due to peroxides within formulations, exposure to trace metals during manufacturing, and light exposure. Aromatic amino acids and the sulfur groups of methionine and cysteine residues are the most susceptible to oxidation.56 Methionine (Met) oxidation generates sulfoxides and sulfone. Histidine (His) oxidation generates oxo-His. For tryptophan (Trp) oxidation, singlet oxygen reacts with the indole group to form kynurenine and N-formylkynurenine.4,49 Met is the most prone to oxidation due to the reactive sulfur atom.57
Sequence-based approaches flag all Met and Trp residues in the variable domains as liable residues.58 However, sequence-based approaches can overestimate the risk of oxidation due to the impact of higher-order structure on oxidation. Exposed Met, Trp, and His are more prone to oxidation. SASA is a common predictor for identifying liable Met and Trp residues.9 More recent studies have successfully incorporated MD simulations and machine learning to predict Met and Trp oxidation.29,59However, specific factors that affect oxidation risk can vary depending on the type of liable residue and the type of oxidative stress.60 The subsequent sections outline in silico prediction for Met, Trp, and His oxidation in more depth.
Methionine oxidation
Met is oxidized by peroxides, light stress, and trace metal ions. Oxidation by peroxides involves transferring oxygen from the peroxide to Met through nucleophilic substitution.61 Water molecules stabilize the transition state near the sulfur atom.62 Alternatively, Met can also react with singlet oxygen to form Met sulfoxide. Met oxidation in mAbs can reduce conformational stability,63 generate hydrophilic variants,5 cause structural changes,64 and affect antigen binding.65 Met residues within CDR loops are more prone to oxidation than Met residues within framework regions. For example, Yan et al. reported oxidation of Met-56 in the solvent-exposed CDR-H2 loops.7
SASA and WCN are useful predictors when water molecules play a role in stabilizing the transition state during oxidation by H2O2.62 WCN represents the average number of water molecules within the radius of an atom.19 The accuracy of WCN as a predictor depends on the selected radius. If the radius is too small, the risk of Met oxidation is underestimated; if the radius is too big, the risk of Met oxidation is overestimated. A radius of 5 Å- 6 Å for WCN is sufficient for predicting Met oxidation. 19,66 Recent studies use the averaged SASA values of Met residues from MD simulations. MD simulations capture changes in solvent exposure for Met residues due to the flexibility of flanking residues and conformational changes.23 In addition, the SASA of the sulfur atom can be a better predictor than overall SASA for Met residues.19
Machine learning models have been used to identify liable Met residues.22,59 Yang and colleagues used the SASA of Met side chains derived from a random forest model to identify liable Met residues for 121 antibodies.67 They observed a strong correlation between side-chain solvent exposure and experimental Met oxidation, but there were some false negatives. The study used SASA values from static crystal structures, which fails to capture changes in solvent exposure of Met residues due to conformational changes.67 Sankar et al. developed a random forest model incorporating SASA, WCN, and distance between sulfur and aromatic residues, to predict Met oxidation by 2,2ʹ-azobis(2-amidinopropane) dihydrochloride (AAPH). Most of the incorrect predictions for Met oxidation were for the CDR-H3 loop. 22 Predicting the structure of CDR-H3 is challenging due to the longer length and high structural variation of the CDR-H3 loops.68 Thus, the accuracy of SASA values is limited by the quality of the crystal or predicted structures.
Although MD simulations have provided more accurate SASA values, most studies conduct MD simulations with water. Excipients and buffers impact the risk of Met oxidation and the hydrodynamic radius of antibodies.69 Changes in hydrodynamic radius of IgG can lead to changes in solvent exposure for Met residues. Conducting MD simulations with the formulation buffer can further bridge the gap between in silico predictions and experimental data. In addition, in silico tools can be used to identify formulations with a lower risk of Met oxidation.70
In silico prediction of Met oxidation by peroxides is very well established. More recent studies have explored in silico prediction for Met oxidation under light stress and forced oxidation by AAPH. 22,59 The mechanism for Met oxidation by peroxides and photolysis is different. Hydrogen peroxide oxidizes Met residues through nucleophilic substitution, whereas light exposure oxidizes Met by generating free radicals.4 In addition to using solvent exposure, Delmar and colleagues also used the closest atomic distance between phenylalanine and Met residues as a predictor for Met oxidation under light stress. Neighboring aromatic residues can prevent Met oxidation by acting as scavengers for free radicals.59 Forced oxidation by H2O2 mimics exposure to trace peroxides from polysorbates in formulations, whereas photolysis is meant to mimic degradation due to light exposure during shipping and storage.71 Therefore, different predictors for Met oxidation (e.g., WCN) should be selected for different types of forced oxidation (e.g., peroxides and photolysis).19
Tryptophan oxidation
Trp oxidation can cause color changes due to kynurenine formation,72 reduce physical stability,73 and oxidation of Trp residues in CDR loops can reduce binding affinity.65 Structural flexibility, solvent exposure, secondary structure, and side-chain and backbone conformations affect Trp oxidation.59 Most aromatic residues get buried within the hydrophobic core during protein folding;74 however, the CDR loops are enriched with tyrosine and Trp residues, which play a role in antigen binding.75
Trp oxidation is more common in the solvent-exposed and flexible CDR loops:9 Trp oxidation in CDR-H3,76 CDR-L1,77 CDR-L3,78,79 CDR-H2, and CDR-L2 loops60 has been reported. Neighboring aromatic residues and disulfide bonds can prevent photooxidation of Trp residues by acting as a light sink.80 IgG1 antibodies have a conserved Cys-Cys-Trp triad, where the disulfide bond protects the neighboring Trp residue from photooxidation. Electron transfer from the Trp residue to the disulfide bond occurs as the disulfide bond dissipates the UV energy absorbed by Trp residues. 80 Solvent exposure is the predominant predictor for Trp oxidation. SASA from static structures does not capture fluctuations in the exposure of Trp residues due to conformational changes. An averaged SASA sampled from MD simulations can provide a more accurate prediction for Trp oxidation. Solvent exposure for Trp side chains is a good predictor because the oxidation of the indole ring is a critical step for forming kynurenine.27 In addition, SASA for Trp side chains can be an accurate predictor for oxidation in cases where the Trp backbone is buried, but the side chain is exposed. Sharma et al. observed that a Trp side chain SASA of > 80 Å2 correlated well with Trp oxidation after incubation with AAPH.29
Other structural factors also influence the oxidation rate and liability for different Trp residues (e.g., side chain and backbone dihedral angles) and the electrostatic environment.60 Recently, Delmar and colleagues developed a random forest model to predict Trp oxidation under light stress, which incorporated secondary structure and psi and phi angles.59 Future studies could investigate the role of other structural factors on Trp oxidation for a broader panel of antibodies.
The risk of Trp oxidation can also differ under long-term thermal stress compared to forced oxidation by AAPH and photolysis. For example, Jacobitz et al. reported that solvent exposure of Trp residues correlated better with Trp oxidation under long-term thermal stress than oxidation by AAPH and light stress.60 Future studies can continue to tailor in silico prediction of Trp oxidation to specific stress conditions, such as thermal stress, photolysis, and forced oxidation by AAPH and peroxides combined with trace metals.
Histidine oxidation
Photo-oxidation and metal-catalyzed oxidation (MCO) are the primary mechanisms for His oxidation. During light stress or MCO, a singlet oxygen reacts with the imidazole ring of His to form peroxide intermediates that then yield oxo-His or cross-linked products.4 Recently, His oxidation has been reported for mAbs: Luo et al. observed oxidation at His-304 and His-428 for IgG2 antibodies after incubation with Cu2+/ascorbate,81 and Amano et al. reported His oxidation in the CH2 domain of an IgG1 antibody under light stress.82 Studies have also reported cross-linking of mAbs caused by His oxidation, with cross-linking being more favorable at higher pH.83 For IgG1 antibodies, oxidized His residues in the Fc and hinge region can cross-link with His, cysteine, and lysine residues after light stress.84 Interestingly, His-His cross-linking has also occurred between the His side chain of an IgG4 antibody and the free His in the formulation.85 Therefore, in silico tools can be used to select formulations for antibodies with a higher risk of His oxidation.
Solvent exposure,85 conformational flexibility,84 surrounding residues, and pKa influence His oxidation. Not all solvent-exposed His residues will get oxidized,82 but solvent exposure is still a prerequisite for His oxidation. His residues within highly flexible and solvent-exposed regions are more prone to oxidation. For example, the oxidized HC-His440 and HC-His292 were located at the CH2-CH3 interface within loops,84 whereas the oxidized His23184 and His22083 were within the hinge region. Surrounding residues can also enhance or inhibit His oxidation. Neighboring Trp residues can act as photosensitizers and increase the risk of His oxidation. Under light stress, Trp residues can generate singlet oxygen or superoxide anions, which can oxidize neighboring His residues.86,87
Studies on the impact of pKa on the risk of His oxidation have yielded inconsistent results. For example, Amano et al. reported no correlation between pKa of His residues and His oxidation,82 whereas Miyahara et al. reported His residues with lower pKa values were more prone to photooxidation.88 The discrepancy in the reported role of pKa on His oxidation could be due to the different stress conditions (i.e., MCO versus UVC light exposure) used in these studies and the different by-products formed after the stress condition (i.e., oxo-His versus Asn and Asp residues). Compared to Met and Trp oxidation, in silico prediction of His oxidation is understudied.49 Future studies could investigate the impact of pKa on His oxidation for different types of oxidative stress (e.g., photooxidation versus MCO).
Lysine glycation
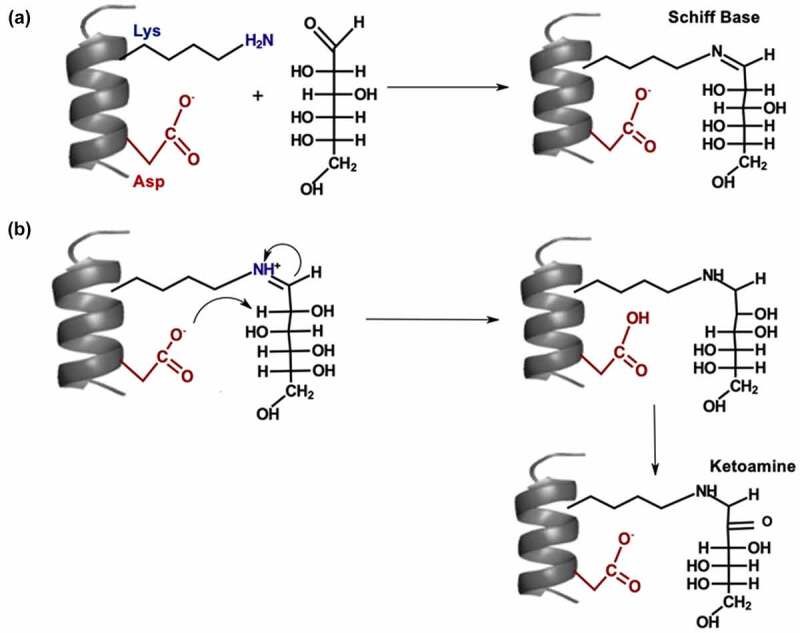
Glycation is a non-enzymatic modification, where amino groups at lysine (Lys) and arginine residues or the N-terminal are glycated by reducing sugars such as glucose.89 The Schiff base is formed after a condensation reaction between the aldehydes of the reducing sugars and the amine groups of Lys residues (Figure 3(a)). Schiff base formation is reversible; however, the multistep Amadori rearrangement can generate more stable ketoamines (Figure 3(b)).90 Glycated Lys residues can further degrade to form advanced glycation end products,91 which can cause an immunogenic response. Lys glycation typically occurs during long-term storage in formulations, cell culture, and in vivo.92 Disaccharides in formulations can degrade to form reducing sugars, leading to glycation during storage.92 During cell culture, a high glucose concentration can accelerate Lys glycation and reduce protein yield.89 The effects of Lys glycation on antigen binding varies from decreased binding affinity93 to minimal change in binding affinity.94 Lys glycation can increase protein aggregation by affecting the net surface charge, reducing the electrostatic repulsion between antibodies.95 Forced glycation is used to identify liable Lys residues by incubating the IgG with high concentrations of reducing sugars (e.g., glucose) at high temperatures.92 Forced glycation in citrate buffers correlates well with glycation during cell culture.96
Figure 3.
(a) Mechanism for Schiff base formation. (b) Mechanism for Amadori rearrangement on glycated lysine.
[Alt Text: (A) The side chains of lysine and aspartic are shown within a coil. There is a condensation reaction between lysine side chain and first carbon atom of glucose to form Schiff base. (B) The reaction mechanism starts with nucleophilic attack on the NH+ group of the lysine side chain by the first carbon atom of glucose and a nucleophilic attack on the proton attached to the second carbon by the carboxylate ion of the aspartic acid. Two hydrogen atoms are removed from the second carbon of glucose to form ketoamine.].
The use of SASA to identify liable residues is a popular approach.27 High solvent exposure is a prerequisite for Lys glycation, but not all exposed Lys residues are glycated.96 Lys glycation is also influenced by flanking residues (i.e., polar, 97 acidic, and basic residues92), three-dimensional structure, and pKa of Lys residues.90 Neighboring Asp and His residues can slow Schiff base formation; however, flanking His and Asp residues can promote Amadori rearrangement by acting as proton donors and acceptors (Figure 3(b)).90,97 Liable motifs for glycation include KD, KXD, KXK, and KXE.96 Structure-based approaches can identify acidic residues that are not part of motifs, but are close enough to catalyze glycation.98 For example, Zhang et al. reported glycation at light chain (LC)-Lys49, which was 11 Å apart from LC-Asp31.97 They suggested that the flexibility of Lys and Asp side-chains promoted Amadori rearrangement by reducing the distance between LC-Lys49 and LC-Asp31.97
Basic residues such as arginine can promote Schiff base formation by lowering the pKa of the Lys side chain.90,95 Lys side chains with lower pKa values are more likely to adopt the deprotonated form (NH2), which is more favorable for forming the Schiff base. The deprotonated form of the Lys side chain can perform a nucleophilic attack on the carbonyl carbon of the sugar to form the Schiff base (Figure 3(a)). Miller et al. suggested that neighboring arginine and His residues increased reactivity of HC-Lys98 by influencing the pKa of HC-Lys98.95 However, Zhang et al. found no correlation between Lys side chain pKa and glycation risk.97 They inferred that Amadori rearrangement (Figure 3(b)) was the rate-limiting step for the glycation of a recombinant humanized mAb. Thus, the impact of the Lys side chain pKa on glycation may depend on the rate-limiting step for Lys glycation. QM/MM simulations of free energy calculations could be used to help identify the rate-limiting step for the glycation of different antibodies. Moreover, future studies could investigate if different backbone and side-chain conformations are more favorable for glycation.
Carbonylation
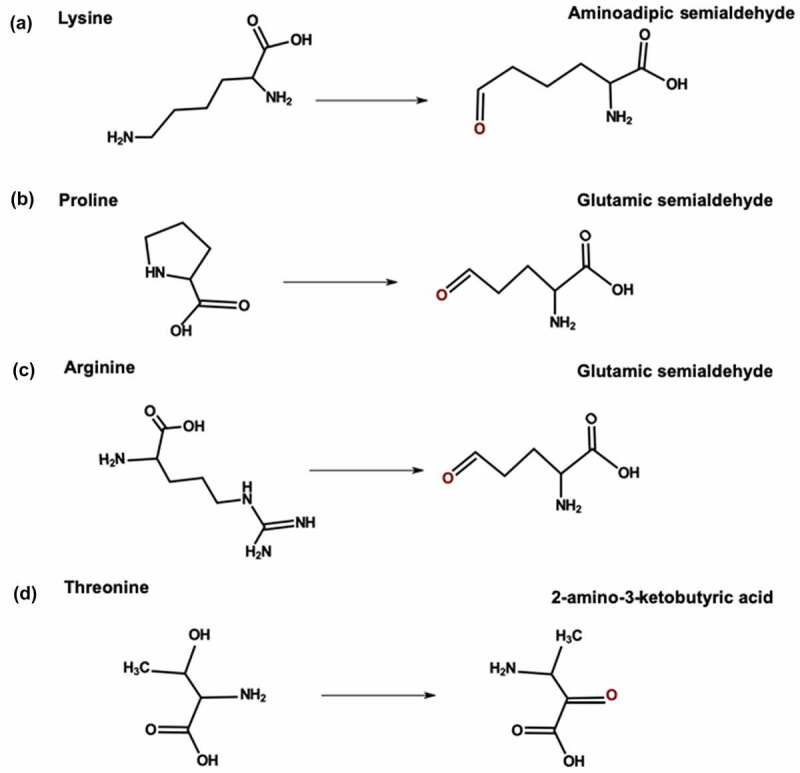
Metal ions can catalyze oxidative carbonylation of arginine, Lys, proline (Pro), and threonine residues. Transition metals such as iron and copper can convert oxygen (O2) to superoxide radical anions (O2–·).99 During carbonylation, free radicals attack the side chain and add an amine or ketone group. Arginine and Pro are converted to glutamic semialdehyde; Lys is converted to aminoadipic semialdehyde, and threonine is converted to 2-amino-3-ketobutyric acid (Figure 4).100 Exposure to trace metals from stainless steel surfaces and glass vials can cause carbonylation of mAbs during manufacturing and storage.101 Carbonylation of arginine and Lys residues leads to a loss in positive charge, which generates acidic variants. For example, Yang et al. reported increased acidic variants after forced oxidation with ferrous sulfate and hydrogen peroxide.102 Oxidative carbonylation of mAbs can also increase protein aggregation.103
Figure 4.
Carbonylation pathway for (a) Lysine (b) Proline (c) Arginine and (d) Threonine.
[Alt Text: (a) The amine group of lysine is converted into a carbonyl group to form aminoadipic semialdehyde. (b) The proline ring opens up and hydroxyl group is converted into a carbonyl group to form glutamic semialdehyde. (c) The guanidino group of the arginine side chain is converted to form glutamic semialdehyde. (d) The hydroxyl group of threonine side chain is converted to a carbonyl group to form 2-amino-3-ketobutyric acid.].
Lys is most susceptible to carbonylation, followed by Pro as the next most susceptible residue, and arginine and threonine are the least susceptible residues. Pro residues tend to form kinks that make Pro residues more exposed and increases the risk of carbonylation.104 Carbonylation sites are more likely to occur in regions enriched with arginine, Lys, Pro, and threonine residues. Regions at the N- and C-terminal of the carbonylation sites tend to be enriched with positively charged residues.105,106 Solvent exposure and structural flexibility also increase the risk of carbonylation.105 Currently, there is no clear association between carbonylation sites and different types of secondary structures.104,107 Multiple carbonylation predictors such as CarSPred,106 iCar-PseCp,108 and iCarPS,109 have been developed using random forest and support vector machine (SVM) algorithms (Table 2). The most common features of machine algorithms for predicting carbonylation include the position-specific propensity of amino acid, hydrophilicity, SASA, side-chain interaction parameter, and positive charge (Table 2). 106,108,109
Table 2.
Comparison of online carbonylation predictors used for machine learning methods. Abbreviations: KNN, k-nearest neighbor; ROC, Receiver Operating Characteristic
| Carbonylation predictor | Machine learning algorithm | Parameters | Data Set | Area under the ROC1 | Reference |
|---|---|---|---|---|---|
| CarSPred | Weighted support vector machine | Position-specific propensity of amino acid, k-spaced amino acid pair, KNN scores, physicochemical properties (electric properties, hydrophobicity, alpha and turn propensities, etc.) |
331 lysine, 131 arginine, 128 threonine, and 129 proline carbonylation sites were extracted from 230 carbonylated human proteins. In addition, 22 lysine, 3 arginine, 6 threonine, and 15 proline carbonylation sites were extracted from carbonylated mouse, rabbit and bovine proteins. |
Lysine: 0.6704 Arginine: 0.5345 Threonine: 0.6800 Proline: 0.7873 |
106 |
| iCar-PseCp | Random forest | pseudo amino acid composition | Data was derived from 230 human carbonylated protein sequences and 20 carbonylated proteins from Photobacterium and Escherichia coli. | Lysine: 0.8728 Arginine: 0.8668 Threonine: 0.8603 Proline: 0.8484 |
108 |
| iCarPS | Random forest | 3-D conical coordinates and physicochemical properties (hydrophobicity, hydrophilicity, mass, pK1, pK2, pI, rigidity, flexibility, and irreplaceability) | Same benchmark dataset as Lv, et al. (2014).109 | Lysine: 0.789 Arginine: 0.726 Threonine: 0.790 Proline: 0.814 |
109 |
1Area under the curve was derived for the ROC. The ROC plots the sensitivity (i.e., true positive rate) versus selectivity.
More precise, sensitive, and site-specific analytical methods for detecting carbonylation in antibodies have been developed in the past decade.5,99,100,110 Joshi et al. identified 27 carbonylation sites within trastuzumab. Most carbonylation sites were within the framework region; they reported no carbonylation sites within the CH3 domain and CDR loops.101 The abundance of carbonylation sites within the structurally rigid framework regions100,101 is surprising because previous studies have suggested that carbonylation is more common in structurally flexible or disordered regions. 104,107 However, residues within the framework region can have a high solvent exposure and undergo carbonylation. 100 Future studies could investigate the impact of the structure of antibodies on carbonylation. In addition, carbonylation sites from a wide panel of antibodies could be used to develop machine learning models for specifically predicting carbonylation sites for therapeutic antibodies.
Tyrosine sulfation
Tyrosine sulfation is an enzymatic modification that is catalyzed by tyrosylprotein sulfotransferases (TPSTs). During sulfation, a sulfate group is attached to the hydroxyl group of tyrosine residues.111 Tyrosine sulfation has been reported for a few monoclonal and bispecific antibodies.111–114 Sulfation of mAbs can occur in Chinese hamster ovary (CHO) cells during the cell culture. The degree of tyrosine sulfation in CHO cells varies due to differential expression of phosphoadenosine-5ʹ-phosphosulfate (PAPS) synthetase and TPST. PAPS synthetase converts ATP to PAPS; TPST transfers the sulfo group from PAPs to tyrosine residues.115 Sulfation in mAbs generates acidic variants,111 but the impact of tyrosine sulfation on the safety and efficacy of therapeutic antibodies has not yet been established.5
Common predictors of tyrosine sulfation include secondary structure, flanking residues, structural flexibility,101, and SASA.116 Sulfation is more favorable for structurally flexible or disordered regions that can fit into the cleft of TPSTs. Tyrosine residues flanked by acidic residues are also more likely to get sulfated (Figure 5(a)).111 During sulfation, the flanking acidic residues within the IgG form electrotactic interactions with arginine and Lys residues of TPST.117 The role of structural flexibility in sulfation suggests that sulfation is more likely to occur in the solvent-exposed and flexible CDR loops than in the framework regions.51 Zhao et al. reported tyrosine sulfation in the CDR-L1 loop,111 and Tyshchuk et al. reported sulfation in the CDR-L2 loop.112
Figure 5.
Sequence logos showing the most conserved amino acids around (a) sulfotyrosine (sTyr). (b) Sequence logos showing the most conserved amino acids around 4-hydroxyproline (4Hyp) residues. Reproduced from Tyshchuk, O. et al.112 (Creative Commons Attribution License).
[Alt Text: (a) The x-axis in the sequence logo shows the amino acid position relative to the sulfated tyrosine. The y-axis indicates the depletion or enrichment of different amino acids surrounding sulfotyrosine. Aspartic acid and glutamic acid are larger in size to indicate the enrichment of acidic residues surrounding sulfotyrosine. (b) The x-axis in shows the amino acid position relative to the 4-hydroxyproline. The y-axis indicates the depletion or enrichment of different amino acids surrounding sulfotyrosine. Glycine is larger in size at −8, −5, −2, 1, 4, 7, and 10 position and proline is larger in size at −7, −4 and +2 from 4-hydroxyproline.].
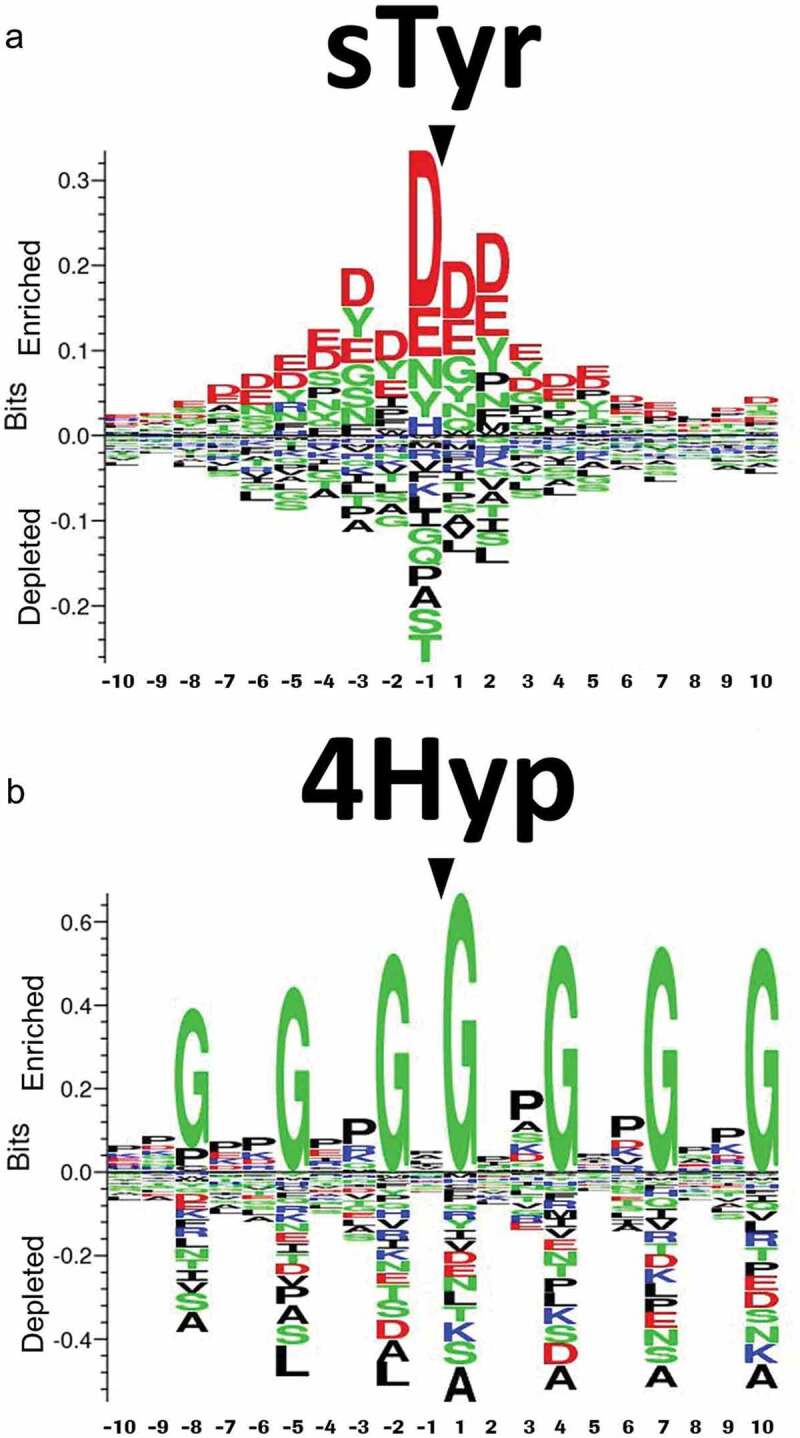
There are multiple online tools for predicting sulfation, such as Sulfinator,118 PredSulSite,119 and Sulfotyrosine.120 However, the accuracy of online tools is limited by the training set used to develop machine learning algorithms to predict PTMs.34 The training sets for most online predictors include experimentally identified tyrosine sulfation sites from databases like Swiss Prot. However, mAbs are underrepresented, and an overtrained machine learning algorithm may fail to identify sulfation sites in mAbs. Hence, sulfation predictors have to be used with caution for assessing the risk of tyrosine sulfation in mAbs. Tyrosine sulfation needs to be identified in a broader panel of antibodies to improve tyrosine sulfation prediction for mAbs. Future studies could also explore the impact of structural features such as dihedral angles, pKa of surrounding residues, and position within IgG (i.e., CDR or framework region) on tyrosine sulfation.
Hydroxylation
Hydroxylation is an enzymatic modification that is catalyzed by hydroxylases. Hydroxylation can occur at arginine, tyrosine, Trp, and phenylalanine, but it is more common for Pro and Lys.121 During hydroxylation, a hydroxyl (OH) group is added to Pro or Lys residues. As a result, Pro is converted to 3-hydroxyPro or 4-hydroxyPro, whereas Lys is converted to 5-hydroxyLys.122 Hydroxylation of Lys and Pro residues at consensus motifs (Xaa-Lys-Gly or Xaa-Pro-Gly) is common in collagen, where hydroxylation helps stabilize the collagen triple-helix.123 Common parameters for designing hydroxylation predictors include solvent exposure, intrinsic disorder, hydrophilicity, and sequence.122,123 Hydroxylation sites tend to be disordered, exposed, and enriched with Pro and Gly residues.124
There are multiple online predictors for hydroxylation sites such as PredHydroxy, HydPred, and ModPred.123 Online hydroxylation predictors are suitable for collagen. However, most hydroxylation predictors have limited training sets, which leads to a high number of false positives and false negatives for non-collagen proteins and uncommon hydroxylation motifs. To date, two studies have reported hydroxylation in antibodies. Xie et al. reported Lys hydroxylation for an IgG1 antibody,125 and Tyshchu et al. observed Pro hydroxylation for a bispecific antibody.112 Using consensus motifs can lead to the overestimation of hydroxylation sites. For example, Xie et al. did not report Lys hydroxylation at all of the consensus motifs.125 Most hydroxylation predictors are not specific to mammalian and human proteins, limiting their utility for therapeutic antibodies.123 Tyshchuk and colleagues used hydroxylation sites for mammalian proteins to generate sequence logos (Figure 5(b)) and develop a kNN model:112 the in silico prediction correlated well with the experimental data for the bispecific antibody. Huang et al. used a similar approach to create HydLoc, which had a training set composed of human proteins and had better accuracy for predicting hydroxylation of human proteins.124 Machine learning methods for predicting hydroxylation can be improved with more experimental data for uncommon hydroxylation sites, as well as hydroxylation sites for mammalian proteins, human proteins, and antibodies.123
Perspectives and future directions
In the past decade, in silico prediction of PTMs has evolved from simple sequence-based approaches to more sophisticated approaches incorporating antibody modeling, MD, and machine learning.12 Although the structure- and physics-based approaches have better accuracy than sequence-based approaches, there is still a gap between the in silico prediction and experimental data from forced degradation studies. External factors such as formulation and cell culture conditions can contribute to the gap between in silico predictions and forced degradation results.3 The quality of predicted structures is a limiting factor for structure- and physics-based approaches. Homology modeling is suitable for modeling the conserved framework regions. Recent advances in loop modeling have provided more accurate predictions for the L1, L2, L3, H1, and H2 loops.126 However, modeling the CDR-H3 loop is still challenging due to the length and structural variation.127 Future advancements in antibody modeling will improve the accuracy in silico prediction of PTMs. In addition, MD simulations can provide more representative SASA values for predicted and crystal structures and refine the quality of predicted structures.
Machine learning algorithms such as random forest and decision trees have been used to predict the risk of deamidation, isomerization, and oxidation. However, machine learning models are limited by the training set and selection of parameters. Building a training set can require a lot of in-house experimental data. Experimentally confirmed liable sites for publicly available antibody sequences are essential for developing better machine learning models in the future. Selecting the best parameters is complicated by contradictory findings for the impact of structural features (e.g., pKa values) on PTMs. Studies investigating the effect of different structural features on PTMs for a broad panel of antibodies are crucial for improving in silico prediction of PTMs. 42,67 Moreover, the selection parameters for machine learning models should account for different mechanisms for chemical modifications (e.g., oxidation) under different types of stress conditions, such as forced oxidation by peroxides66 and photolysis.
Therapeutic antibodies can also undergo in vivo chemical modifications (e.g., deamidation and oxidation) due to rapid temperature and pH changes after drug administration.128 In vivo chemical modifications can impact the safety and efficacy of therapeutic antibodies. For example, in vivo deamidation can lead to a loss in activity and cause immunogenicity. 57 Few studies have investigated in vivo modifications by monitoring PTMs for IgG in serum or by using in vitro conditions to mimic in vivo degradation.57 Monitoring in vivo modifications is difficult because the human serum is a mixture of endogenous proteins and the administered therapeutic antibodies. It is challenging to separate and distinguish therapeutic antibodies from endogenous proteins. Fluorescence labeling techniques or affinity purification with anti-Fc antibodies are required for mass spectrometry (MS) analysis of in vivo samples. The harsh purification conditions (e.g., low pH elution) can introduce artifacts in liquid chromatography-MS analysis.129 Computational approaches for improving the stability of therapeutic antibodies focus on preventing chemical degradation under the types of stresses (e.g., thermal stress) encountered during manufacturing and storage.27 In silico prediction of in vivo modifications remains relatively unexplored. Developing computational approaches to predict in vivo chemical modifications provides opportunities for improving in vivo stability of therapeutic candidates.
The impact of PTMs on heterogeneity, safety, and efficacy can vary depending on the location of the liable residue. A more holistic approach to engineering mAbs could identify PTM hotspots and predict the impact of PTMs on binding affinity and physical stability. Removing liable sites is not always feasible, especially if the amino acid residue plays a role in antigen binding. Paratope prediction and molecular docking of IgG-antigen complex can guide antibody engineering for developability.130 The surface charge distribution and surface hydrophobicity131 could be used to predict the impact of modifications of liable residues on colloidal stability.
Substantial strides have been made in silico prediction of deamidation, isomerization, and Met oxidation.22,30,46 However, the in silico prediction of glycation, sulfation, hydroxylation, and carbonylation mAbs is underdeveloped compared to other PTMs. Another understudied area is in silico prediction of in vivo chemical modifications.57 Advances in analytical characterization of novel modifications (e.g., sulfation) and in vivo modifications are crucial for developing computational approaches for predicting PTMs.
After two decades of establishing a solid foundation for in silico prediction of chemical modifications, future studies can focus on tailoring in silico predictions to more specific stress conditions and formulations.
Funding Statement
The author(s) reported there is no funding associated with the work featured in this article.
Abbreviations
| AAPH | 2,2ʹ-Azobis(2-amidinopropane) dihydrochloride |
| Asn | asparagine |
| Asp | aspartic acid |
| CDR | complementary-determining region |
| CHO | Chinese hamster ovary |
| Gly | glycine |
| HC | heavy chain |
| His | histidine |
| kNN | K-nearest neighbor |
| LC | light chain |
| Lys | Lysine |
| mAbs | monoclonal antibodies |
| MCO | metal-catalyzed oxidation |
| Met | methionine |
| MD | molecular dynamics |
| PAPS | phosphoadenosine-5ʹ-phosphosulfate |
| Pro | proline |
| PTM | post-translational modification |
| QM/MM | quantum mechanics/molecular mechanics |
| RMSF | root-mean-square fluctuations |
| SASA | solvent-accessible surface area |
| SVM | support vector machine |
| TPST | tyrosylprotein sulfotransferases |
| Trp | tryptophan |
| WCN | water coordination number. |
Disclosure statement
No potential conflict of interest was reported by the author(s).
References
- 1.Chames P, Van Regenmortel M, Weiss E, Baty D.. Therapeutic antibodies: successes, limitations and hopes for the future. Br J Pharmacol. 2009;157(2):220–15. PMID: 19459844. doi: 10.1111/j.1476-5381.2009.00190.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Kapingidza AB, Kowal K, Chruszcz M.. Antigen-antibody complexes. Subcell Biochem. 2020;94:465–97. PMID: 32189312. doi: 10.1007/978-3-030-41769-7_19. [DOI] [PubMed] [Google Scholar]
- 3.Xu Y, Wang D, Mason B, Rossomando T, Li N, Liu D, Cheung JK, Xu W, Raghava S, Katiyar A, et al. Structure, heterogeneity and developability assessment of therapeutic antibodies. MAbs. 2019;11(2):239–64. PMID: 30543482. doi: 10.1080/19420862.2018.1553476. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Grassi L, Cabrele C. Susceptibility of protein therapeutics to spontaneous chemical modifications by oxidation, cyclization, and elimination reactions. Amino Acids. 2019;51(10–12):1409–31. PMID: 31576455. doi: 10.1007/s00726-019-02787-2. [DOI] [PubMed] [Google Scholar]
- 5.Beck A, Liu H. Macro- and micro-heterogeneity of natural and recombinant IgG antibodies. Antibodies. 2019;8(1):18. PMID: 31544824. doi: 10.3390/antib8010018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Barton C, Spencer D, Levitskaya S, Feng J, Harris R, Schenerman MA. Heterogeneity of IgGs: role of production, processing, and storage on structure and function [Internet]. In: ACS Symposium Series. American Chemical Society, State-of-the-Art and Emerging Technologies for Therapeutic Monoclonal Antibody Characterization Volume 1. Monoclonal Antibody Therapeutics: Structure, Function, and Regulatory Space; 2014. 69–98. doi: 10.1021/bk-2014-1176.ch003 [DOI] [Google Scholar]
- 7.Yan Y, Wei H, Fu Y, Jusuf S, Zeng M, Ludwig R, Krystek SR, Chen G, Tao L, Das TK. Isomerization and oxidation in the complementarity-determining regions of a monoclonal antibody: a study of the modification-structure-function correlations by hydrogen-deuterium exchange mass spectrometry. Anal Chem. 2016;88(4):2041–50. PMID: 26824491. doi: 10.1021/acs.analchem.5b02800. [DOI] [PubMed] [Google Scholar]
- 8.Du Y, Walsh A, Ehrick R, Xu W, May K, Liu H. Chromatographic analysis of the acidic and basic species of recombinant monoclonal antibodies. MAbs. 2012;4(5):578–85. PMID: 22820257. doi: 10.4161/mabs.21328. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Jarasch A, Koll H, Regula JT, Bader M, Papadimitriou A, Kettenberger H. Developability assessment during the selection of novel therapeutic antibodies. J Pharm Sci. 2015;104(6):1885–98. PMID: 25821140. doi: 10.1002/jps.24430. [DOI] [PubMed] [Google Scholar]
- 10.Nowak CK, Cheung JM, Dellatore S, Katiyar A, Bhat R, Sun J, Ponniah G, Neill A, Mason B, Beck A, et al. Forced degradation of recombinant monoclonal antibodies: a practical guide. MAbs. 2017;9(8):1217–30. PMID: 28853987. doi: 10.1080/19420862.2017.1368602. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Michels DA, Ip AY, Dillon TM, Brorson K, Lute S, Chavez B, Prentice KM, Brady LJ, Miller KJ. Separation Methods and Orthogonal Techniques [Internet]. In: State-of-the-art and emerging technologies for therapeutic monoclonal antibody characterization Volume 2. Biopharmaceutical characterization: the NISTmAb case study. American Chemical Society; 2015, p. 237–84. DOI: 10.1021/bk-2015-1201.ch005. [DOI] [Google Scholar]
- 12.Kuroda D, Tsumoto K. Engineering stability, viscosity, and immunogenicity of antibodies by computational design. J Pharm Sci. 2020;109(5):1631–51. PMID: 31958430. doi: 10.1016/j.xphs.2020.01.011. [DOI] [PubMed] [Google Scholar]
- 13.Temel DB, Kinderman F, Eryilmaz E. Developability in biophysical characterization. In: Houde, Damian J and Berkowitz, Steven A, editors. Biophysical Characterization of proteins in developing biopharmaceuticals. Elsevier; 2019. 505–26. DOI: 10.1016/B978-0-444-64173-1.00017-2. [DOI] [Google Scholar]
- 14.Zurdo J, Arnell A, Obrezanova O, Smith N, Gómez De La Cuesta R, Gallagher TRA, Michael R, Stallwood Y, Ekblad C, Abrahmsén L, et al. Early implementation of QbD in biopharmaceutical development: a practical example. Biomed Res Int. 2015;2015:1–19. PMID: 26075248. doi: 10.1155/2015/605427. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Strohl WR, Strohl LM. Development issues: antibody stability, developability, immunogenicity, and comparability [Internet. In: Therapeutic antibody engineering. Elsevier: Woodhead Publishing; 2012. 377–595. DOI: 10.1533/9781908818096.377. [DOI] [Google Scholar]
- 16.Robinson NE, Robinson AB. Prediction of protein deamidation rates from primary and three-dimensional structure. Proc Natl Acad Sci U S A. 2001;98(8):4367–72. PMID: 11296285. doi: 10.1073/pnas.071066498. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Wakankar AA, Borchardt RT. Formulation considerations for proteins susceptible to asparagine deamidation and aspartate isomerization. J Pharm Sci. 2006;95(11):2321–36. PMID: 16960822. doi: 10.1002/jps.20740. [DOI] [PubMed] [Google Scholar]
- 18.Norman RA, Ambrosetti F, Bonvin AMJJ, Colwell LJ, Kelm S, Kumar S, Krawczyk K. Computational approaches to therapeutic antibody design: established methods and emerging trends. Brief Bioinform. 2020;21(5):1549–67. PMID: 31626279. doi: 10.1093/bib/bbz095. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Agrawal NJ, Dykstra A, Yang J, Yue H, Nguyen X, Kolvenbach C, Angell N. Prediction of the hydrogen peroxide–induced methionine oxidation propensity in monoclonal antibodies. J Pharm Sci. 2018;107(5):1282–89. PMID: 29325924. doi: 10.1016/j.xphs.2018.01.002. [DOI] [PubMed] [Google Scholar]
- 20.Ali S, Hassan M, Islam A, Ahmad F. A review of methods available to estimate solvent-accessible surface areas of soluble proteins in the folded and unfolded states. Curr Protein Pept Sci. 2014;15(5):456–76. PMID: 24678666. doi: 10.2174/1389203715666140327114232. [DOI] [PubMed] [Google Scholar]
- 21.Yang Y, Zhao J, Geng S, Hou C, Li X, Lang X, Qiao C, Li Y, Feng J, Lv M, et al. Improving Trastuzumab’s stability profile by removing the two degradation hotspots. J Pharm Sci. 2015;104(6):1960–70. PMID: 25820189. doi: 10.1002/jps.24435. [DOI] [PubMed] [Google Scholar]
- 22.Sankar K, Hoi KH, Yin Y, Ramachandran P, Andersen N, Hilderbrand A, McDonald P, Spiess C, Zhang Q. Prediction of methionine oxidation risk in monoclonal antibodies using a machine learning method. MAbs. 2018;10(8):1281–90. PMID: 30252602. doi: 10.1080/19420862.2018.1518887. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Xu KN, Uversky V, Xue B. Local flexibility facilitates oxidization of buried methionine residues. Protein Pept Lett. 2012;19(6):688–97. PMID: 22519542. doi: 10.2174/092986612800494084. [DOI] [PubMed] [Google Scholar]
- 24.Hollingsworth SA, Dror RO. Molecular dynamics simulation for all. Neuron. 2018;99(6):1129–43. PMID: 30236283. doi: 10.1016/j.neuron.2018.08.011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Kaliman I, Nemukhin A, Varfolomeev S. Free energy barriers for the N-terminal asparagine to succinimide conversion: quantum molecular dynamics simulations for the fully solvated model. J Chem Theory Comput. 2010;6(1):184–89. PMID: 26614331. doi: 10.1021/ct900398a. [DOI] [PubMed] [Google Scholar]
- 26.Plotnikov NV, Singh SK, Rouse JC, Kumar S. Quantifying the risks of asparagine deamidation and aspartate isomerization in biopharmaceuticals by computing reaction free-energy surfaces. J Phys Chem B. 2017;121(4):719–30. PMID: 28051868. doi: 10.1021/acs.jpcb.6b11614. [DOI] [PubMed] [Google Scholar]
- 27.Kumar S, Plotnikov NV, Rouse JC, Singh SK. Biopharmaceutical informatics: supporting biologic drug development via molecular modelling and informatics. J Pharm Pharmacol. 2018;70(5):595–608. PMID: 28155992. doi: 10.1111/jphp.12700. [DOI] [PubMed] [Google Scholar]
- 28.Salsbury FR. Molecular dynamics simulations of protein dynamics and their relevance to drug discovery. Curr Opin Pharmacol. 2010;10(6):738–44. PMID: 20971684. doi: 10.1016/j.coph.2010.09.016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Sharma VK, Patapoff TW, Kabakoff B, Pai S, Hilario E, Zhang B, Li C, Borisov O, Kelley RF, Chorny I, et al. In silico selection of therapeutic antibodies for development: viscosity, clearance, and chemical stability. Proc Natl Acad Sci U S A. 2014;111(52):18601–06. PMID: 25512516. doi: 10.1073/pnas.1421779112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Delmar JA, Wang J, Choi SW, Martins JA, Mikhail JP. Machine learning enables accurate prediction of asparagine deamidation probability and rate. Mol Ther Methods Clin Dev. 2019;15:264–74. PMID: 31890727. doi: 10.1016/j.omtm.2019.09.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Jia L, Sun Y, de Brevern AG. Protein asparagine deamidation prediction based on structures with machine learning methods. PLoS One. 2017;12(7):1–17. PMID: 28732052. doi: 10.1371/journal.pone.0181347. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Koehl P, Delarue M. Combined approaches from physics, statistics, and computer science for ab initio protein structure prediction: ex unitate vires (unity is strength)? F1000Research. 2018;7:11. PMID: 30079234. doi: 10.12688/f1000research.14870.1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Lai PK, Swan JW, Trout BL. Calculation of therapeutic antibody viscosity with coarse-grained models, hydrodynamic calculations and machine learning-based parameters. MAbs. 2021;13(1). PMID: 33834944. doi: 10.1080/19420862.2021.1907882. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Tatjewski M, Kierczak M, Plewczynski D. Predicting post-translational modifications from local sequence fragments using machine learning algorithms: overview and best practices. Methods Mol Biol. 2017;1484:275–300. PMID: 27787833. doi: 10.1007/978-1-4939-6406-2_19. [DOI] [PubMed] [Google Scholar]
- 35.Yi L, Beckley N, Gikanga B, Zhang J, Wang YJ, Chih HW, Sharma VK. Isomerization of Asp-Asp motif in model peptides and a monoclonal antibody fab fragment. J Pharm Sci. 2013;102(3):947–59. PMID: 23280575. doi: 10.1002/jps.23423. [DOI] [PubMed] [Google Scholar]
- 36.Miao S, Xie P, Zou M, Fan L, Liu X, Zhou Y, Zhao L, Ding D, Wang H, Tan WS. Identification of multiple sources of the acidic charge variants in an IgG1 monoclonal antibody. Appl Microbiol Biotechnol. 2017;101(14):5627–38. PMID: 28439623. doi: 10.1007/s00253-017-8301-x. [DOI] [PubMed] [Google Scholar]
- 37.Haverick M, Mengisen S, Shameem M, Ambrogelly A. Separation of mAbs molecular variants by analytical hydrophobic interaction chromatography HPLC: overview and applications. MAbs. 2014;6(4):852–58. PMID: 24751784. doi: 10.4161/mabs.28693. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Harris RJ, Kabakoff B, Macchi FD, Shen FJ, Kwong M, Andya JD, Shire SJ, Bjork N, Totpal K, Chen AB. Identification of multiple sources of charge heterogeneity in a recombinant antibody. J Chromatogr B Biomed Sci Appl. 2001;752(2):233–45. PMID: 11270864. doi: 10.1016/S0378-4347(00)00548-X. [DOI] [PubMed] [Google Scholar]
- 39.Wakankar AA, Liu J, Vandervelde D, Wang YJ, Shire SJ, Borchardt RT. The effect of cosolutes on the isomerization of aspartic acid residues and conformational stability in a monoclonal antibody. J Pharm Sci. 2007;96(7):1708–18. PMID: 17238195. doi: 10.1002/jps.20823. [DOI] [PubMed] [Google Scholar]
- 40.Sreedhara A, Cordoba A, Zhu Q, Kwong J, Liu J. Characterization of the isomerization products of aspartate residues at two different sites in a monoclonal antibody. Pharm Res. 2012;29(1):187–97. PMID: 21809161. doi: 10.1007/s11095-011-0534-2. [DOI] [PubMed] [Google Scholar]
- 41.Radkiewicz JL, Zipse H, Clarke S, Houk KN. Neighboring side chain effects on asparaginyl and aspartyl degradation: an Ab initio study of the relationship between peptide conformation and backbone NH acidity. J Am Chem Soc. 2001;123(15):3499–506. PMID: 11472122. doi: 10.1021/ja0026814. [DOI] [PubMed] [Google Scholar]
- 42.Lu X, Nobrega RP, Lynaugh H, Jain T, Barlow K, Boland T, Sivasubramanian A, Vásquez M, Xu Y. Deamidation and isomerization liability analysis of 131 clinical-stage antibodies. MAbs. 2019;11(1):45–57. PMID: 30526254. doi: 10.1080/19420862.2018.1548233. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Brennan TV, Clarke S. Effect of adjacent histidine and cysteine residues on the spontaneous degradation of asparaginyl- and aspartyl-containing peptides. Int J Pept Protein Res. 2009;45(6):547–53. PMID: 7558585. doi: 10.1111/j.1399-3011.1995.tb01318.x. [DOI] [PubMed] [Google Scholar]
- 44.Patel CN, Bauer SP, Davies J, Durbin JD, Shiyanova TL, Zhang K, Tang JX. N+1 engineering of an aspartate isomerization hotspot in the complementarity-determining region of a monoclonal antibody. J Pharm Sci. 2016;105(2):512–18. PMID: 26869414. doi: 10.1016/S0022-3549(15)00185-9. [DOI] [PubMed] [Google Scholar]
- 45.Diepold K, Bomans K, Wiedmann M, Zimmermann B, Petzold A, Schlothauer T, Mueller R, Moritz B, Stracke JO, Mølhøj M, et al. Simultaneous assessment of Asp isomerization and Asn deamidation in recombinant antibodies by LC-MS following incubation at elevated temperatures. PLoS One. 2012;7(1):1–11. PMID: 22272329. doi: 10.1371/journal.pone.0030295. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Sydow JF, Lipsmeier F, Larraillet V, Hilger M, Mautz B, Mølhøj M, Kuentzer J, Klostermann S, Schoch J, Voelger HR, et al. Structure-based prediction of asparagine and aspartate degradation sites in antibody variable regions. PLoS One. 2014;9(6):e100736. PMID: 24959685. doi: 10.1371/journal.pone.0100736. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Pace AL, Wong RL, Zhang YT, Kao YH, Wang YJ. Asparagine deamidation dependence on buffer type, pH, and temperature. J Pharm Sci. 2013;102(6):1712–23. PMID: 23568760. doi: 10.1002/jps.23529. [DOI] [PubMed] [Google Scholar]
- 48.Catak S, Monard G, Aviyente V, Ruiz-López MF. Deamidation of asparagine residues: direct hydrolysis versus succinimide-mediated deamidation mechanisms. J Phys Chem A. 2009;113(6):1111–20. PMID: 19152321. doi: 10.1021/jp808597v. [DOI] [PubMed] [Google Scholar]
- 49.Shire SJ. Stability of monoclonal antibodies (mAbs). In: Monoclonal antibodies. Elsevier: Woodhead Publishing; 2015, 45–92. DOI: 10.1016/b978-0-08-100296-4.00003-8. [DOI] [Google Scholar]
- 50.Chelius D, Render DS, Bondarenko PV. Identification and characterization of deamidation sites in the conserved regions of human immunoglobulin gamma antibodies. Anal Chem. 2005;77(18):6004–11. PMID: 16159134. doi: 10.1021/ac050672d. [DOI] [PubMed] [Google Scholar]
- 51.Chiu ML, Goulet DR, Teplyakov A, Gilliland GL. Antibody structure and function: the basis for engineering therapeutics. Antibodies. 2019;8(4):55. PMID: 31816964. doi: 10.3390/antib8040055. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.North B, Lehmann A, Dunbrack RL. A new clustering of antibody CDR loop conformations. J Mol Biol. 2011;406(2):228–56. PMID: 21035459. doi: 10.1016/j.jmb.2010.10.030. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Nowak J, Baker T, Georges G, Kelm S, Klostermann S, Shi J, Sridharan S, Deane CM. Length-independent structural similarities enrich the antibody CDR canonical class model. MAbs. 2016;8(4):751–60. PMID: 26963563. doi: 10.1080/19420862.2016.1158370. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Yan Q, Huang M, Lewis MJ, Hu P. Structure based prediction of asparagine deamidation propensity in monoclonal antibodies. MAbs. 2018;10(6):901–12. PMID: 29958069. doi: 10.1080/19420862.2018.1478646. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Lan W, Valente JJ, Ilott A, Chennamsetty N, Liu Z, Rizzo JM, Yamniuk AP, Qiu D, Shackman HM, Bolgar MS. Investigation of anomalous charge variant profile reveals discrete pH-dependent conformations and conformation-dependent charge states within the CDR3 loop of a therapeutic mAb. MAbs. 2020;12(1):1763138. PMID: 32432964. doi: 10.1080/19420862.2020.1763138. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Le Basle Y, Chennell P, Tokhadze N, Astier A, Sautou V. Physicochemical stability of monoclonal antibodies: a review. J Pharm Sci. 2020;109(1):169–90. PMID: 31465737. doi: 10.1016/j.xphs.2019.08.009. [DOI] [PubMed] [Google Scholar]
- 57.Liu H, Ponniah G, Zhang HM, Nowak C, Neill A, Gonzalez-Lopez N, Patel R, Cheng G, Kita AZ, Andrien B. In vitro and in vivo modifications of recombinant and human IgG antibodies. MAbs. 2014;6(5):1145–54. PMID: 25517300. doi: 10.4161/mabs.29883. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Xu A, Kim HS, Estee S, Viajar S, Galush WJ, Gill A, Hötzel I, Lazar GA, McDonald P, Andersen N, et al. Susceptibility of antibody CDR residues to chemical modifications can be revealed prior to antibody humanization and aid in the lead selection process. Mol Pharm. 2018;15(10):4529–37. PMID: 30118239. doi: 10.1021/acs.molpharmaceut.8b00536. [DOI] [PubMed] [Google Scholar]
- 59.Delmar JA, Buehler E, Chetty AK, Das A, Quesada GM, Wang J, Chen X. Machine learning prediction of methionine and tryptophan photooxidation susceptibility. Mol Ther Methods Clin Dev. 2021;21:466–77. PMID: 33898635. doi: 10.1016/j.omtm.2021.03.023. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Jacobitz AW, Liu Q, Suravajjala S, Agrawal NJ. Tryptophan oxidation of a monoclonal antibody under diverse oxidative stress conditions: distinct oxidative pathways favor specific tryptophan residues. J Pharm Sci. 2021;110(2):719–26. PMID: 33198947. doi: 10.1016/j.xphs.2020.10.039. [DOI] [PubMed] [Google Scholar]
- 61.Shah DD, Zhang J, Hsieh MC, Sundaram S, Maity H, Mallela KMG. Effect of peroxide- versus alkoxyl-induced chemical oxidation on the structure, stability, aggregation, and function of a therapeutic monoclonal antibody. J Pharm Sci. 2018;107(11):2789–803. PMID: 30075161. doi: 10.1016/j.xphs.2018.07.024. [DOI] [PubMed] [Google Scholar]
- 62.Chu JW, Brooks BR, Trout BL. Oxidation of methionine residues in aqueous solutions: free methionine and methionine in granulocyte colony-stimulating factor. J Am Chem Soc. 2004;126(50):16601–07. PMID: 15600366. doi: 10.1021/ja0467059. [DOI] [PubMed] [Google Scholar]
- 63.Liu D, Ren D, Huang H, Dankberg J, Rosenfeld R, Cocco MJ, Li L, Brems DN, Remmele RL. Structure and stability changes of human IgG1 Fc as a consequence of methionine oxidation. Biochemistry. 2008;47(18):5088–100. PMID: 18407665. doi: 10.1021/bi702238b. [DOI] [PubMed] [Google Scholar]
- 64.Tang L, Sundaram S, Zhang J, Carlson P, Matathia A, Parekh B, Zhou Q, Hsieh MC. Conformational characterization of the charge variants of a human IgG1 monoclonal antibody using H/D exchange mass spectrometry. MAbs. 2013;5(1):114–25. PMID: 23222183. doi: 10.4161/mabs.22695. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Torosantucci R, Schöneich C, Jiskoot W. Oxidation of therapeutic proteins and peptides: structural and biological consequences. Pharm Res. 2014;31(3):541–53. PMID: 24065593. doi: 10.1007/s11095-013-1199-9. [DOI] [PubMed] [Google Scholar]
- 66.Chennamsetty N, Quan Y, Nashine V, Sadineni V, Lyngberg O, Krystek S. Modeling the oxidation of methionine residues by peroxides in proteins. J Pharm Sci. 2015;104(4):1246–55. PMID: 25641333. doi: 10.1002/jps.24340. [DOI] [PubMed] [Google Scholar]
- 67.Yang R, Jain T, Lynaugh H, Nobrega RP, Lu X, Boland T, Burnina I, Sun T, Caffry I, Brown M, et al. Rapid assessment of oxidation via middle-down LCMS correlates with methionine side-chain solvent-accessible surface area for 121 clinical stage monoclonal antibodies. MAbs. 2017;9(4):646–53. PMID: 28281887. doi: 10.1080/19420862.2017.1290753. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Regep C, Georges G, Shi J, Popovic B, Deane CM. The H3 loop of antibodies shows unique structural characteristics. Proteins Struct Funct Bioinf. 2017;85(7):1311–18. PMID: 28342222. doi: 10.1002/prot.25291. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Baek Y, Singh N, Arunkumar A, Zydney AL. Effects of histidine and sucrose on the biophysical properties of a monoclonal antibody. Pharm Res. 2017;34(3):629–39. PMID: 28035628. doi: 10.1007/s11095-016-2092-0. [DOI] [PubMed] [Google Scholar]
- 70.Razinkov VI, Treuheit MJ, Becker GW. Accelerated formulation development of monoclonal antibodies (MABS) and mab-based modalities: review of methods and tools. J Biomol Screen. 2015;20(4):468–83. PMID: 25576149. doi: 10.1177/1087057114565593. [DOI] [PubMed] [Google Scholar]
- 71.Hawe A, Wiggenhorn M, van de Weert M, Garbe JHO, Mahler H, Jiskoot W. Forced degradation of therapeutic proteins. J Pharm Sci. 2012;101(3):895–913. PMID: 22083792. doi: 10.1002/jps.22812. [DOI] [PubMed] [Google Scholar]
- 72.Li Y, Polozova A, Gruia F, Feng J. Characterization of the degradation products of a color-changed monoclonal antibody: tryptophan-derived chromophores. Anal Chem. 2014;86(14):6850–57. PMID: 24937252. doi: 10.1021/ac404218t. [DOI] [PubMed] [Google Scholar]
- 73.Alam ME, Slaney TR, Wu L, Das TK, Kar S, Barnett GV, Leone A, Tessier PM. Unique impacts of methionine oxidation, tryptophan oxidation, and asparagine deamidation on antibody stability and aggregation. J Pharm Sci. 2020;109(1):656–69. PMID: 31678251. doi: 10.1016/j.xphs.2019.10.051. [DOI] [PubMed] [Google Scholar]
- 74.Feige MJ, Hendershot LM, Buchner J. How antibodies fold. Trends Biochem. 2010;35(4):139–48. PMID: 20022755. doi: 10.1016/j.physbeh.2017.03.040. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Dalkas GA, Teheux F, Kwasigroch JM, Rooman M. Cation–π, amino–π, π–π, and H‐bond interactions stabilize antigen–antibody interfaces. Proteins Struct Funct Bioinf. 2014;82(9):1734–46. PMID: 24488795. doi: 10.1002/prot.24527. [DOI] [PubMed] [Google Scholar]
- 76.Wei Z, Feng J, Lin H-Y-Y, Mullapudi S, Bishop E, Tous GI, Casas-Finet J, Hakki F, Strouse R, Schenerman MA. Identification of a single tryptophan residue as critical for binding activity in a humanized monoclonal antibody against respiratory syncytial virus. Anal Chem. 2007;79(7):2797–805. PMID: 17319649. doi: 10.1021/ac062311j. [DOI] [PubMed] [Google Scholar]
- 77.Hensel M, Steurer R, Fichtl J, Elger C, Wedekind F, Petzold A, Schlothauer T, Molhoj M, Reusch D, Bulau P. Identification of potential sites for tryptophan oxidation in recombinant antibodies using tert-butylhydroperoxide and quantitative LC-MS. PLoS One. 2011;6(3):e17708. PMID: 21390239. doi: 10.1371/journal.pone.0017708. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78.Qi PP, Volkin DB, Zhao H, Nedved ML, Hughes R, Bass R, Yi SC, Panek ME, Wang D, Dalmonte P, et al. Characterization of the photodegradation of a human IgG1 monoclonal antibody formulated as a high-concentration liquid dosage form. J Pharm Sci. 2009;98(9):3117–30. PMID: 19009595. doi: 10.1002/jps.21617. [DOI] [PubMed] [Google Scholar]
- 79.Dashivets T, Stracke J, Dengl S, Knaupp A, Pollmann J, Buchner J, Schlothauer T. Oxidation in the complementarity-determining regions differentially influences the properties of therapeutic antibodies. MAbs. 2016;8(8):1525–35. PMID: 27612038. doi: 10.1080/19420862.2016.1231277. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80.Wecksler AT, Yin J, Lee Tao P, Kabakoff B, Sreedhara A, Deperalta G. Photodisruption of the structurally conserved Cys-Cys-Trp triads leads to reduction-resistant scrambled intrachain disulfides in an IgG1 monoclonal antibody. Mol Pharm. 2018;15(4):1598–606. PMID: 29502420. doi: 10.1021/acs.molpharmaceut.7b01128. [DOI] [PubMed] [Google Scholar]
- 81.Luo Q, Joubert MK, Stevenson R, Ketchem RR, Narhi LO, Wypych J. Chemical modifications in therapeutic protein aggregates generated under different stress conditions. J Biol Chem. 2011;286(28):25134–44. PMID: 21518762. doi: 10.1074/jbc.M110.160440. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 82.Amano M, Kobayashi N, Yabuta M, Uchiyama S, Fukui K. Detection of histidine oxidation in a monoclonal immunoglobulin gamma (IgG) 1 antibody. Anal Chem. 2014;86(15):7536–43. PMID: 24940720. doi: 10.1021/ac501300m. [DOI] [PubMed] [Google Scholar]
- 83.Liu M, Zhang Z, Cheetham J, Ren D, Zhou ZS. Discovery and characterization of a photo-oxidative histidine-histidine cross-link in IgG1 antibody utilizing 18O-labeling and mass spectrometry. Anal Chem. 2014;86(10):4940–48. PMID: 24738698. doi: 10.1021/ac500334k. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 84.Xu CF, Chen Y, Yi L, Brantley T, Stanley B, Sosic Z, Zang L. Discovery and characterization of histidine oxidation initiated cross-links in an IgG1 monoclonal antibody. Anal Chem. 2017;89(15):7915–23. PMID: 28635253. doi: 10.1021/acs.analchem.7b00860. [DOI] [PubMed] [Google Scholar]
- 85.Powell T, Knight MJ, O’Hara J, Burkitt W. Discovery of a photoinduced histidine-histidine cross-link in an IgG4 antibody. J Am Soc Mass Spectrom. 2020;31(6):1233–40. PMID: 32392057. doi: 10.1021/jasms.0c00076. [DOI] [PubMed] [Google Scholar]
- 86.Sreedhara A, Lau K, Li C, Hosken B, MacChi F, Zhan D, Shen A, Steinmann D, Schöneich C, Lentz Y. Role of surface exposed tryptophan as substrate generators for the antibody catalyzed water oxidation pathway. Mol Pharm. 2013;10(1):278–88. PMID: 23136850. doi: 10.1021/mp300418r. [DOI] [PubMed] [Google Scholar]
- 87.Bane J, Mozziconacci O, Yi L, Wang YJ, Sreedhara A, Schöneich C. Photo-oxidation of IgG1 and model peptides: detection and analysis of triply oxidized his and trp side chain cleavage products. Pharm Res. 2017;34(1):229–42. PMID: 27800571. doi: 10.1007/s11095-016-2058-2. [DOI] [PubMed] [Google Scholar]
- 88.Miyahara Y, Shintani K, Hayashihara-Kakuhou K, Zukawa T, Morita Y, Nakazawa T, Yoshida T, Ohkubo T, Uchiyama S. Effect of UVC irradiation on the oxidation of histidine in monoclonal antibodies. Sci Rep. 2020;10(1). PMID: 32286391. doi: 10.1038/s41598-020-63078-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 89.Quan C, Alcala E, Petkovska I, Matthews D, Canova-Davis E, Taticek R, Ma S. A study in glycation of a therapeutic recombinant humanized monoclonal antibody: where it is, how it got there, and how it affects charge-based behavior. Anal Biochem. 2008;373(2):179–91. PMID: 18158144. doi: 10.1016/j.ab.2007.09.027. [DOI] [PubMed] [Google Scholar]
- 90.Venkatraman J, Aggarwal K, Balaram P. Helical peptide models for protein glycation: proximity effects in catalysis of the Amadori rearrangement. Chem Biol. 2001;8(7):611–25. PMID: 11451663. doi: 10.1016/S1074-5521(01)00036-9. [DOI] [PubMed] [Google Scholar]
- 91.Goetze AM, Liu YD, Arroll T, Chu L, Flynn GC. Rates and impact of human antibody glycation in vivo. Glycobiology. 2012;22(2):221–34. PMID: 21930650. doi: 10.1093/glycob/cwr141. [DOI] [PubMed] [Google Scholar]
- 92.Wei B, Berning K, Quan C, Zhang YT. Glycation of antibodies: modification, methods and potential effects on biological functions. MAbs. 2017;9(4):586–94. PMID: 28272973. doi: 10.1080/19420862.2017.1300214. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 93.Kennedy DM, Skillen AW, Self CH. Glycation of monoclonal antibodies impairs their ability to bind antigen. Clin Exp Immunol. 1994;98(2):245–51. PMID: 7955529. doi: 10.1111/j.1365-2249.1994.tb06133.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 94.Vrdoljak A, Trescec A, Benko B, Hecimovic D, Simic M. In vitro glycation of human immunoglobulin G. Clin Chim Acta. 2004;345(1–2):105–11. PMID: 15193984. doi: 10.1016/j.cccn.2004.03.026. [DOI] [PubMed] [Google Scholar]
- 95.Miller AK, Hambly DM, Kerwin BA, Treuheit MJ, Gadgil HS. Characterization of site-specific glycation during process development of a human therapeutic monoclonal antibody. J Pharm Sci. 2011;100(7):2543–50. PMID: 21287557. doi: 10.1002/jps.22504. [DOI] [PubMed] [Google Scholar]
- 96.Jacobitz AW, Dykstra AB, Spahr C, Agrawal NJ. Effects of buffer composition on site-specific glycation of lysine residues in monoclonal antibodies. J Pharm Sci. 2020;109(1):293–300. PMID: 31150698. doi: 10.1016/j.xphs.2019.05.025. [DOI] [PubMed] [Google Scholar]
- 97.Zhang B, Yang Y, Yuk I, Pai R, McKay P, Eigenbrot C, Dennis M, Katta V, Francissen KC. Unveiling a glycation hot spot in a recombinant humanized monoclonal antibody. Anal Chem. 2008;80(7):2379–90. PMID: 18307322. doi: 10.1021/ac701810q. [DOI] [PubMed] [Google Scholar]
- 98.Saleem RA, Affholter BR, Deng S, Campbell PC, Matthies K, Eakin CM, Wallace A. A chemical and computational approach to comprehensive glycation characterization on antibodies. MAbs. 2015;7(4):719–31. PMID: 26030340. doi: 10.1080/19420862.2015.1046663. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 99.Ros J, Ashraf G., Madian, Fred E., Regnier, Ao, Zeng. Protein carbonylation: principles, analysis, and biological implications. In Editor Joaquim Ros. John Wiley & Sons, Inc; 2017. p. 24–47. doi: 10.1002/9781119374947. [DOI] [Google Scholar]
- 100.Yang Y, Stella C, Wang W, Schöneich C, Gennaro L. Characterization of oxidative carbonylation on recombinant monoclonal antibodies. Anal Chem. 2014;86(10):4799–806. PMID: 24731230. doi: 10.1021/ac4039866. [DOI] [PubMed] [Google Scholar]
- 101.Joshi S, Kumari S, Rathore AS. Identification and characterization of carbonylation sites in trastuzumab biosimilars. Int J Biol Macromol. 2021;169:95–102. PMID: 33338527. doi: 10.1016/j.ijbiomac.2020.12.095. [DOI] [PubMed] [Google Scholar]
- 102.Yang Y, Mah A, Yuk IH, Grewal PS, Pynn A, Cole W, Gao D, Zhang F, Chen J, Gennaro L, et al. Investigation of metal-catalyzed antibody carbonylation with an improved protein carbonylation assay. J Pharm Sci. 2018;107(10):2570–80. PMID: 29935298. doi: 10.1016/j.xphs.2018.06.015. [DOI] [PubMed] [Google Scholar]
- 103.Bee JS, Davis M, Freund E, Carpenter JF, Randolph TW. Aggregation of a monoclonal antibody induced by adsorption to stainless steel. Biotechnol Bioeng. 2010;105(1):121–29. PMID: 19725039. doi: 10.1002/bit.22525. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 104.Shyama Prasad Rao R, Møller IM. Pattern of occurrence and occupancy of carbonylation sites in proteins. Proteomics. 2011;11(21):4166–73. PMID: 21919202. doi: 10.1002/pmic.201100223. [DOI] [PubMed] [Google Scholar]
- 105.Weng SL, Huang KY, Kaunang FJ, Huang CH, Kao HJ, Chang TH, Wang HY, Lu JJ, Lee TY. Investigation and identification of protein carbonylation sites based on position-specific amino acid composition and physicochemical features. BMC Bioinform. 2017;18(S3). PMID: 28361707. doi: 10.1186/s12859-017-1472-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 106.Lv H, Han J, Liu J, Zheng J, Liu R, Zhong D, Lisacek F. CarSPred: a computational tool for predicting carbonylation sites of human proteins. PLoS One. 2014;9(10):1–8. PMID: 25347395. doi: 10.1371/journal.pone.0111478. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 107.Maisonneuve E, Ducret A, Khoueiry P, Lignon S, Longhi S, Talla E, Dukan S, Hofmann A. Rules governing selective protein carbonylation. PLoS One. 2009;4(10):1–10. PMID: 19802390. doi: 10.1371/journal.pone.0007269. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 108.Jia J, Liu Z, Xiao X, Liu B, Chou KC. ICar-PseCp: identify carbonylation sites in proteins by Monto Carlo sampling and incorporating sequence coupled effects into general PseAAC. Oncotarget. 2016;7(23):34558–70. PMID: 27153555. doi: 10.18632/oncotarget.9148. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 109.Zhang D, Xu ZC, Su W, Yang YH, Lv H, Yang H, Lin H, Xu J. ICarPS: a computational tool for identifying protein carbonylation sites by novel encoded features. Bioinformatics. 2021;37(2):171–77. PMID: 32766811. doi: 10.1093/bioinformatics/btaa702. [DOI] [PubMed] [Google Scholar]
- 110.Yi Y, Zang L. High-throughput carbonyl content method of therapeutic mAb using size-exclusion chromatography with ultraviolet and fluorescence detection. Anal Biochem. 2019;571:25–36. PMID: 30653944. doi: 10.1016/j.ab.2018.12.025. [DOI] [PubMed] [Google Scholar]
- 111.Zhao J, Saunders J, Schussler SD, Rios S, Insaidoo FK, Fridman AL, Li H, Liu YH. Characterization of a novel modification of a CHO-produced mAb: evidence for the presence of tyrosine sulfation. MAbs. 2017;9(6):985–95. PMID: 28590151. doi: 10.1080/19420862.2017.1332552. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 112.Tyshchuk O, Gstöttner C, Funk D, Nicolardi S, Frost S, Klostermann S, Becker T, Jolkver E, Schumacher F, Koller CF, et al. Characterization and prediction of positional 4-hydroxyproline and sulfotyrosine, two post-translational modifications that can occur at substantial levels in CHO cells-expressed biotherapeutics. MAbs. 2019;11(7):1219–32. PMID: 31339437. doi: 10.1080/19420862.2019.1635865. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 113.Jiang H, Xu W, Liu R, Gupta B, Kilgore B, Du Z, Yang X. Characterization of bispecific antibody production in cell cultures by unique mixed mode size exclusion chromatography. Anal Chem. 2020;92(13):9312–21. PMID: 32497423. doi: 10.1021/acs.analchem.0c01641. [DOI] [PubMed] [Google Scholar]
- 114.Gomez N, Lull J, Yang X, Wang Y, Zhang X, Wieczorek A, Harrahy J, Pritchard M, Cano DM, Shearer M, et al. Improving product quality and productivity of bispecific molecules through the application of continuous perfusion principles. Biotechnol Prog. 2020;36(4). PMID: 31991523. doi: 10.1002/btpr.2973. [DOI] [PubMed] [Google Scholar]
- 115.Liu R, Zhang Y, Kumar A, Huhn S, Hullinger L, Du Z. Modulating tyrosine sulfation of recombinant antibodies in CHO cell culture by host selection and sodium chlorate supplementation. Biotechnol J. 2021;16(9):2100142. PMID: 34081410. doi: 10.1002/biot.202100142. [DOI] [PubMed] [Google Scholar]
- 116.Chang WC, Lee TY, Shien DM, Hsu JBK, Horng JT, Hsu PC, Wang TY, Huang HD, Pan RL. Incorporating support vector machine for identifying protein tyrosine sulfation sites. J Comput Chem. 2009;30(15):2526–37. PMID: 19373826. doi: 10.1002/jcc.21258. [DOI] [PubMed] [Google Scholar]
- 117.Teramoto T, Fujikawa Y, Kawaguchi Y, Kurogi K, Soejima M, Adachi R, Nakanishi Y, Mishiro-Sato E, Liu MC, Sakakibara Y, et al. Crystal structure of human tyrosylprotein sulfotransferase-2 reveals the mechanism of protein tyrosine sulfation reaction. Nat Commun. 2013;4(1). PMID: 23481380. doi: 10.1038/ncomms2593. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 118.Monigatti F, Gasteiger E, Bairoch A, Jung E. The Sulfinator: predicting tyrosine sulfation sites in protein sequences. Bioinformatics. 2002;18(5):769–70. PMID: 12050077. doi: 10.1093/bioinformatics/18.5.769. [DOI] [PubMed] [Google Scholar]
- 119.Huang SY, Shi SP, Qiu JD, Sun XY, Suo SB, Liang RP. PredSulSite: prediction of protein tyrosine sulfation sites with multiple features and analysis. Anal Biochem. 2012;428(1):16–23. PMID: 22691961. doi: 10.1016/j.ab.2012.06.003. [DOI] [PubMed] [Google Scholar]
- 120.Yang ZR. Predicting sulfotyrosine sites using the random forest algorithm with significantly improved prediction accuracy. BMC Bioinform. 2009;10(1):361. PMID: 19874585. doi: 10.1186/1471-2105-10-361. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 121.Raju TS. Hydroxylation of proteins. In: Co- and post-translational modifications of therapeutic antibodies and proteins. John Wiley & Sons Inc; 2019. p. 119–31. doi: 10.1002/9781119053354.ch10. [DOI] [Google Scholar]
- 122.Hu LL, Niu S, Huang T, Wang K, Shi XH, Cai YD, Uversky VN. Prediction and analysis of protein hydroxyproline and hydroxylysine. PLoS One. 2010;5(12):1–8. PMID: 21209839. doi: 10.1371/journal.pone.0015917. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 123.Piovesan D, Hatos A, Minervini G, Quaglia F, Monzon AM, Tosatto SCE, Iakoucheva LM. Assessing predictors for new post translational modification sites: a case study on hydroxylation. PLoS Comput Biol. 2020;16(6):1–15. PMID: 32569263. doi: 10.1371/journal.pcbi.1007967. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 124.Huang Q, Chen X, Wang Y, Li J, Liu H, Xie Y, Dai Z, Zou X, Li Z. HydLoc: a tool for hydroxyproline and hydroxylysine sites prediction in the human proteome. Chemom Intell Lab Syst. 2020;202:104035. doi: 10.1016/j.chemolab.2020.104035. [DOI] [Google Scholar]
- 125.Xie Q, Moore B, Beardsley RL. Discovery and characterization of hydroxylysine in recombinant monoclonal antibodies. MAbs. 2016;8(2):371–78. PMID: 26651858. doi: 10.1080/19420862.2015.1122148. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 126.Choi Y, Deane CM. Predicting antibody complementarity determining region structures without classification. Mol Biosyst. 2011;7(12):3327–34. PMID: 22011953. doi: 10.1039/c1mb05223c. [DOI] [PubMed] [Google Scholar]
- 127.Almagro JC, Teplyakov A, Luo J, Sweet RW, Kodangattil S, Hernandez-Guzman F, Gilliland GL. Second Antibody Modeling Assessment (AMA-II). Proteins Struct Funct Bioinforma. 2014;82(8):1553–62. PMID: 24668560. doi: 10.1002/prot.24567. [DOI] [PubMed] [Google Scholar]
- 128.Schuster J, Koulov A, Mahler HC, Detampel P, Huwyler J, Singh S, Mathaes R. In vivo stability of therapeutic proteins. Pharm Res. 2020;37. PMID: 31900680. doi: 10.1007/s11095-019-2689-1. [DOI] [PubMed] [Google Scholar]
- 129.Xu X, Huang Y, Pan H, Molden R, Qiu H, Daly TJ, Li N, Popoff MR. Quantitation and modeling of posttranslational modifications in a therapeutic monoclonal antibody from single- And multiple-dose monkey pharmacokinetic studies using mass spectrometry. PLoS One. 2019;14(10):e0223899. PMID: 31618250. doi: 10.1371/journal.pone.0223899. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 130.Qiu H, Wei R, Jaworski J, Boudanova E, Hughes H, VanPatten S, Lund A, Day J, Zhou Y, McSherry T, et al. Engineering an anti-CD52 antibody for enhanced deamidation stability. MAbs. 2019;11(7):1266–75. PMID: 31199181. doi: 10.1080/19420862.2019.1631117. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 131.Hebditch M, Warwicker J. Web-based display of protein surface and pH-dependent properties for assessing the developability of biotherapeutics. Sci Rep. 2019;9(1):1–9. PMID: 30760735. doi: 10.1038/s41598-018-36950-8. [DOI] [PMC free article] [PubMed] [Google Scholar]


