Abstract
Circulating tumor DNA (ctDNA) has emerged as a biomarker with wide-ranging applications in cancer management. While its role in guiding precision medicine in certain tumors via noninvasive detection of susceptibility and resistance alterations is now well established, recent evidence has pointed to more generalizable use in treatment monitoring. Quantitative changes in ctDNA levels over time (i.e., ctDNA kinetics) have shown potential as an early indicator of therapeutic efficacy and could enable treatment adaptation. However, ctDNA kinetics are complex and heterogeneous, affected by tumor biology, host physiology, and treatment factors. This review outlines the current preclinical and clinical knowledge of ctDNA kinetics in cancer and how early on-treatment changes in ctDNA levels could be applied in clinical research to collect evidence to support implementation in daily practice.
Understanding the rationale of early changes in circulating tumor DNA in cancer patients may tailor treatment in the near future.
INTRODUCTION
Every cancer carries a unique complement of molecular alterations acquired stepwise through tumorigenesis. Through next-generation sequencing (NGS), the unique molecular fingerprint of each cancer is more accessible than ever (1). There is a growing body of research on the detectability of circulating tumor DNA (ctDNA) at cross-sectional time points such as the end of definitive treatment and detection of molecular/minimal residual disease (MRD), which has prognostic implications in numerous cancer types (2). However, the understanding of ctDNA kinetics early on during the treatment course remains nascent. This is an area with great potential to guide clinical decision-making, especially with the increasing use of immune checkpoint inhibitors, where imaging at the usual decision time points may not accurately reflect treatment response (3).
ctDNA is the component of fragmented cell-free DNA (cfDNA) that is derived from tumor cells. Quantitative and digital polymerase chain reactions (PCRs) were the first ctDNA detection approaches for small numbers of targets and remain useful in tumors with highly frequent driver mutations (i.e., KRAS in colorectal cancer) (4, 5). The advent of NGS allows high-throughput analysis of multiple targets, expanding both the scope and accuracy of ctDNA methods. Advancements in both sequencing accuracy and analytical error correction (6, 7) have markedly improved the sensitivity of ctDNA sequencing (8). Meanwhile, the increasing availability and affordability of NGS, including targeted panels, exome, and whole genome sequencing (WGS), expands the number of available targets that can improve the precision of ctDNA concentration estimates. The use of bespoke panels has also rapidly emerged, wherein a personalized panel is constructed based on selected mutations captured during tissue sequencing and then applied to detect these tumor-specific mutations in plasma.
The kinetics of ctDNA depends on various biological, clinical, and treatment-related factors. ctDNA concentration varies by tumor type (9) and is broadly correlated with tumor volume (6); however, this correlation is confounded by tumor heterogeneity (10–12) and ctDNA release mechanisms (13). Having established a baseline, variations in ctDNA concentration are well correlated with changing tumor burden and may outperform conventional tumor markers (14). With effective treatment, ctDNA levels fall rapidly with complex and variable decay patterns (15).
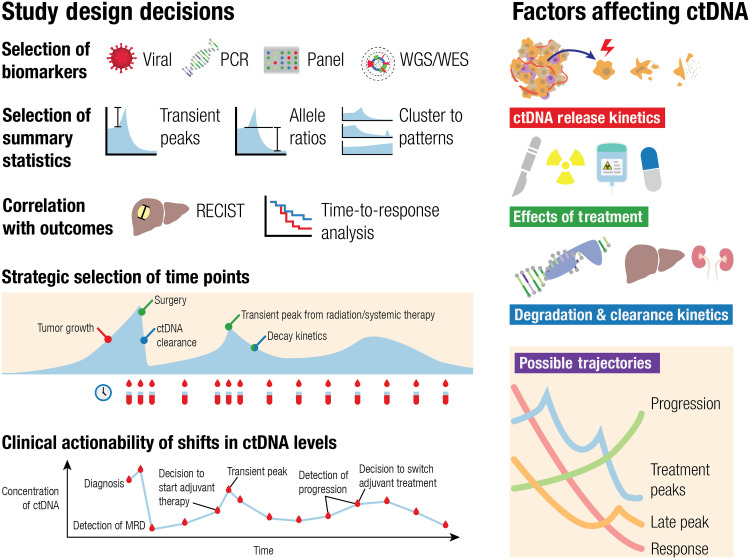
Here, we review the current state of knowledge regarding ctDNA kinetics during treatment. We will begin with a brief overview of the biology of ctDNA release as it pertains to treatment monitoring. More comprehensive reviews on ctDNA biology and technology are available elsewhere (2, 16). We will next survey the landscape of studies on clinical application of ctDNA kinetics, first outlining the wide array of differing approaches used in such studies (Fig. 1). We will limit our attention to studies that track ctDNA at multiple time points during treatment and attempt to use these on-treatment measurements to predict clinical endpoints. Our goal is to identify opportunities for clinical translation of ctDNA kinetics as an early on-treatment biomarker.
Fig. 1. ctDNA kinetics vary with kinetics of release and degradation/clearance and interact closely with treatment effects.
The interplay of these factors governs the various potential applications of ctDNA along the cancer treatment timeline. Both early transient changes in ctDNA and gradual decay kinetics may be informative of response to treatment. Understanding the implications of these kinetics may inform treatment decisions, allowing for early adjustments. Studies examining ctDNA kinetics vary by how they quantify ctDNA, which time points they sample, what metrics are used to summarize ctDNA kinetics, and which clinical outcomes they collect. WES, whole exome sequencing.
Mechanisms of ctDNA release and clearance in cancer
Our understanding of the mechanisms of ctDNA release stems from fundamental studies on cellular death and turnover in the body. Under normal circumstances, the vast majority of cfDNA in the circulation is derived from nucleated blood cells (17). This is thought to be explained by the accessibility of marrow-resident and circulating blood cells to the vascular space as well as rapid cellular turnover (18). Natural within-subject variability of cfDNA levels day to day has been estimated at 25%, with levels typically declining over the course of the day (19). Statistical modeling suggests that a shift of over 70% would be required to denote a substantial shift above normal background variation (19). Regular activities such as exercise can result in a marked increase in cfDNA levels (20, 21). In pathological states including cancer, as well as sepsis, inflammatory conditions, or organ injury, other tissue-specific circulating cfDNA is found in greater abundance (17, 22, 23). ctDNA levels are influenced by rates of cell turnover and mechanisms of cell death. In lung cancer, for example, mathematical modeling suggests that tumor cells shed 0.014% of their DNA into the bloodstream per cell death (24). In this model, the tumor size necessary for detectability is inversely proportional to the rate of ctDNA shedding. Induction of necrosis and apoptosis stimulates ctDNA release (25, 26), while senescence prevents ctDNA release (13).
Once cfDNA/ctDNA enters the circulation, it is rapidly cleared within a matter of minutes (min). This was first demonstrated in the setting of fetal cfDNA in the maternal circulation where the mean cfDNA half-life was 16.3 min (range, 4 to 30 min) (27). Tumor resection produces a similarly rapid decline in ctDNA concentration (28–30), albeit with a slower decay rate in patients with residual cancer (15). This finding highlights how careful elucidation of ctDNA kinetics could be a powerful indicator of recurrence risk.
The mechanisms of cfDNA clearance are incompletely characterized. Renal excretion plays a role (31–33) but may account for less than 20% of total cfDNA turnover (34, 35). Moreover, renal clearance appears to be variable and has been observed to yield cyclical peaks and troughs when monitored daily (32). Mouse studies showed that a sizeable fraction of nucleic acids was sequestered into the liver and spleen, suggesting immune-mediated clearance through the reticuloendothelial system (35, 36). Delayed clearance of fetal DNA from maternal plasma is observed during preeclampsia, implicating the relevance of hepatic or renal dysfunction in the clearance process (37). It has also been postulated that circulating endonucleases and hepatocyte-associated exonucleases contribute to cfDNA degradation (38). Together, there are multiple phases of cfDNA metabolism and clearance, which can result in complex decay kinetics (34, 36).
Study methodologies are variable and evolving
Coincident with investigations into the biological mechanisms dictating ctDNA kinetics has been a plethora of observational studies describing these kinetics across a range of clinical settings. Synthesizing findings across studies is challenging because of variability in study design, patient populations under evaluation, and techniques used. One source of variability unique to the study of ctDNA kinetics arises from differences in time points for blood collection. These variables allow studies to address different scientific or clinical questions. Another source of variability across studies stems from the steady progression of ctDNA detection techniques. This includes advancements in DNA capture, sequencing, and downstream bioinformatics analysis, which have had the combined effect of improving detection sensitivity through signal enrichment and error suppression. What follows is a brief discussion of variables in study design and techniques, with mention of trends observed across the literature.
Biomarkers
NGS has expanded the pool of available ctDNA biomarkers. While nearly all studies included in this review estimate ctDNA concentration, the specific biomarkers examined vary. In virally driven tumors such as human papilloma virus–driven cervical or head and neck cancers and Epstein-Barr virus (EBV)–driven nasopharyngeal cancer (NPC) and lymphoma, measurement of viral DNA provides a robust cancer-specific biomarker (39–41). In cancer types with characteristic driver mutations, droplet digital PCR (ddPCR)–based assays were used to detect mutations in genes such as EGFR, BRAF, KRAS, or EWSR1 fusions (42–45). Although there can be variability in concentration between different mutations stemming from copy number variation or tumor heterogeneity, ctDNA allele fractions are often well correlated with cancer tissue sequencing (46). Inference of copy number profiles and sequencing multiple genes using NGS to identify “trunk” mutations can help mitigate these sources of variability. Another emerging method is the detection of methylated ctDNA, which has characteristic patterns in distinct tumor types; notably, there are numerous recurrent cancer-specific DNA methylation aberrations, and these can be measured using PCR- and NGS-based assays (47–51). The strength of methylated DNA as a biomarker is that it does not rely on the presence of specific mutations.
Collection time points
Studies varied substantially in their temporal resolution; many studies sampled only a few on-treatment time points (52, 53), whereas others collected an abundance of time points, even daily in the case of one study of cfDNA in urine (32). The sampling strategy generally reflected the clinical or biological phenomenon under study. For example, studies examining rapid shifts in ctDNA triggered by treatment performed frequent blood draws shortly after treatment, followed by more widely spaced collections thereafter (15, 54, 55). One common strategy for systemic therapy studies was to collect samples at baseline and at various predetermined endpoints such as specific treatment cycles or upon disease progression (56).
Summary metrics
Although studies can report mutant allele counts or frequencies directly, there has been a desire for normalized scores to quantify ctDNA kinetics. Mutation allele ratio in therapy (MART) (57) is defined as the ratio of the plasma mutation score (number of reads containing a mutation in cfDNA) after the initiation of therapy to that before therapy. The circulating DNA ratio (CDR) score, which has also been called the molecular response ratio, is defined similarly as the variant allele fraction (VAF, the number of reads matching a specific DNA variant divided by the total number of reads covering the genomic locus) during therapy divided by the baseline VAF (58–60). A potential weakness of VAF-based metrics is that their denominator can be confounded by the natural background variability of nontumor cfDNA and by exercise, infection, or trauma each of which can raise cfDNA levels, as discussed above. This can cause artificial shifts in ctDNA allele frequencies, which can obfuscate kinetics. Therefore, concentration-based metrics such as mutations per milliliter of plasma are preferable for clinical reporting purposes. It is worth noting that, while concentration-based metrics reduce clinical and biological uncertainty, a recent analysis has shown that they may be more susceptible to certain technical artifacts such as coverage bias, especially when using NGS rather than ddPCR (61). Therefore, care is always advised in technical validation of laboratory methods and analytical workflows.
The choice of time points also affects summary metrics. Studies with few time points, such as one baseline and one mid-treatment time point, are limited to only one comparison to establish ctDNA kinetics and are almost certain to miss transient shifts. However, studies that capture multiple time points produce an abundance of data that can be challenging to interpret, especially if there is variability in the timing of collections between participants. A common strategy of such studies is to classify participants into a limited number of typical clearance profiles (54, 62). These characteristic classes can be identified statistically using clustering-based approaches (62). Treatment outcomes are then compared between participants falling into these classes, allowing all time points to be used in the discovery of dimensionally reduced model simple enough for clinical application. Another strategy to account for variability in sampling times is to assess the slope of ctDNA changes, defined as the change in ctDNA levels divided by the difference in time between each pair of consecutive samples. Each of these strategies requires further validation.
Clinical endpoints
Given the time-dependent nature of ctDNA kinetics metrics, there is substantial temporal dependency of outcome measures. Radiographic response is a commonly used outcome measure. Studies also examined various time-to-response or survival-based endpoints based on the study population and the nature of treatment. Clinical studies often attempt to predict patients with higher chances of durable response based on ctDNA change or clearance. Many also investigate whether ctDNA kinetics can predict relapse with meaningful lead time.
ctDNA detection and analysis
Most of the studies have used a form of quantitative/digital PCR to measure ctDNA bearing a specific variant of interest. However, application of NGS-based techniques is growing increasingly common and includes a wide spectrum of approaches ranging from small targeted panels to WGS (63, 64). Panel-based NGS approaches have varied in their breadth of targeted sequences from single genes to many hundreds (65). Most panels target a small number of genes for deep sequencing; these are designed for target detection rather than ctDNA tracking and are thus limited in their detection sensitivity and the ability to reveal emerging subclones. Larger panels that allow for aggregation of multiple mutations per patient are potentially better suited for high sensitivity ctDNA tracking (6). On the other end of the spectrum, WGS can profile all tumor-derived genomic aberrations but does not generally have sufficient depth for tracking individual mutations; for this reason, WGS has not yet been widely explored for ctDNA tracking (66, 67). The aforementioned NGS methods can been applied in either a “tumor-informed” manner (i.e., tissue analysis is conducted to verify tumor specificity of aberrations observed in plasma) or a “tumor-agnostic” manner (6, 67). While tumor-informed approaches introduce logistical complexity into assay workflow, they have the advantage of boosting confidence in the origin of putative cancer-derived aberrations through analysis of concordant mutations (68). Both tumor-informed and tumor-agnostic NGS methods are susceptible to spurious signals as a result of clonal hematopoiesis (69). For this reason, peripheral blood leukocytes are often sequenced alongside plasma cfDNA, and aberrations derived from clonal hematopoiesis are filtered.
To further ensure specificity of putative ctDNA signals, many studies are now using individualized (i.e., bespoke) NGS panels. This approach may improve specificity and standardization by tracking a fixed number of robust tumor-specific variants (54). Bespoke ctDNA assays are best suited to ensure that the tracked mutations are truly tumor-derived as opposed to arising from noncancerous tissues. One potential limitation of bespoke ctDNA assays is that its performance can be affected by spatial and temporal tumor heterogeneity; panels could be constructed from nonrepresentative biopsies and are not designed to detect emerging mutations in resistant subclones. Furthermore, the process of designing individualized NGS panels can also lengthen turnaround time of the assay compared with fixed-panel NGS or WGS.
CLINICAL APPLICATIONS OF ctDNA KINETICS
Surgery
Levels of nonspecific cfDNA increase after trauma and surgery regardless of the procedure or its indication (70, 71). Substantial increases have been observed after resection of malignant tumors (72, 73), as well as hip replacement (72) and endoscopic decompression (74). The level of cfDNA increase was as much as eightfold, varied depending on the procedure, and persisted for up to 4 weeks.
By contrast, in the case of oncologic surgery, ctDNA shows the opposite kinetics. For instance, in patients who underwent complete resection (R0) of colorectal carcinoma (CRC), a sharp drop in ctDNA level was observed by the day of discharge (2 to 10 days after surgery) with a 99% median decrease in ctDNA (30). Most of this decrease occurred by 24 hours after surgery with an estimated half-life of 114 min. Recently, the DYNAMIC study examined rapid postoperative shifts in ctDNA levels in patients undergoing lung cancer resection, collecting samples at 5 min, 30 min, 2 hours, 24 hours, 3 days, and 30 days after surgery (15). Median ctDNA half-life was estimated to be shorter than the aforementioned CRC study (35 min), possibly due to finer temporal resolution. A transient ctDNA peak 5 min after surgery was observed in patients who underwent lobectomy as opposed to wedge resection (15). On the other hand, incomplete resection is correlated with delayed clearance of ctDNA. Patients with CRC with incomplete resections displayed slower decrease or even elevation of ctDNA within the first 24 hours after surgery, perhaps due to injury of remnant tumor tissue during the surgery. In DYNAMIC, although all patients had complete resections, patients who were later found to have MRD had significantly longer ctDNA decay half-life (103.2 min versus 29.7 min). Moreover, all patients without ctDNA clearance by day 3 after surgery recurred or died later during follow-up, suggesting that rapid ctDNA clearance may have prognostic implications.
MRD after surgery detected by ctDNA predicts worse disease-free survival and overall survival (OS) across many tumor types such as colorectal (30, 75, 76), pancreatic (77), breast (78–80), and lung (81) carcinoma and may precede radiological recurrence. However, few studies have focused on detecting changes in ctDNA within a short period after surgery (i.e., 2 months) (15, 78, 82–88). Early findings suggest that slower ctDNA decay rate is associated with incomplete resection and MRD, which may enable earlier prediction of recurrence risk and stratify patients to receive short-interval postoperative therapy. Most of these studies are retrospective with variable postsurgery collection time points, and prospective trials are needed to establish optimal sampling. Thus, early postsurgical ctDNA kinetics are a promising potential predictor of recurrence risk that requires further study before it can be used clinically.
Use in radiotherapy
A subset of patients exhibit a transient peak in ctDNA levels in the days following initiation of radiotherapy (89). Subsequently, most patients who undergo definitive radiotherapy demonstrate a steady decline in ctDNA concentration over the course of treatment, measurable by weeks 2 and 3. The median half-life of this decline has been estimated at 3 to 8 days (89, 90). Most of the patients clear ctDNA completely, and residual ctDNA at the end of treatment is a poor prognostic marker (52, 53, 91, 92). However, even some patients without detectable end-of-treatment ctDNA nevertheless recur. This raises the question of whether monitoring ctDNA kinetics during radiotherapy may enable early detection of high-risk disease, granting a window of opportunity for treatment intensification, for example, by expanding the treatment field or adding a concurrent systemic therapy such as an immune checkpoint inhibitor. Conversely, rapid ctDNA clearance may allow for de-escalation by withdrawing radiosensitizing chemotherapy and/or shortening the treatment course. Both the transient peak and the decay phase have been examined for treatment monitoring and may inform patient stratification for early treatment modification.
The transient peak has been hypothesized to result from a brief period of rapid cell death and is a potential indicator of treatment response. The timing of peaks was predictive of treatment response in patients with NPC who underwent induction chemotherapy followed by chemoradiotherapy. This study of EBV DNA in 673 patients with nonmetastatic NPC undergoing induction chemotherapy followed by chemoradiotherapy described eight distinct ctDNA kinetics patterns. Samples with transient peaks during the earlier induction chemotherapy phase had significantly better outcomes than those with transient peaks during the later chemoradiotherapy phase. Allowing for differences in treatment modality, the authors postulated that late transient peaks may have been indicative of chemotherapy-resistant subclones and thus predictive of increased recurrence risk (62).
The decay phase varies in its rate and completeness and has been studied at various temporal resolutions. The simplest approach is to sample ctDNA at baseline, end of treatment, and one mid-treatment time point. Mid-treatment ctDNA was first investigated in 107 patients with locally advanced NPC, where persistent mid-treatment EBV DNA was associated with worse rates of distant recurrence, progression-free survival (PFS), and OS (53). Increasing sampling allows for more granular observation of ctDNA kinetics and more precise estimation of the decay half-life. The importance of half-life was illustrated in a study of nonmetastatic NPC receiving radical intensity modulated radiotherapy with concurrent chemotherapy wherein clearance half-life was predictive of PFS (90). While postradiotherapy EBV DNA levels represent a more specific poor prognostic marker than mid-radiotherapy levels (93), persistent mid-radiotherapy EBV DNA could be a sensitive early predictor of recurrence risk. Delayed ctDNA clearance may result from radioresistance, incomplete tumor coverage, or occult regional/distant tumor extension.
Use in chemotherapy
The most consistent finding regarding ctDNA kinetics in chemotherapy is that ctDNA levels decline within the first few days of treatment regardless of clinical response (4, 94), and a greater decline is often associated with improved outcomes (4, 55, 95–97). Less consistent, however, is the observation of transient ctDNA peaks after chemotherapy being noted by some (4, 96), while others show an immediate decrease after treatment (55, 95). Some studies have suggested that such peaks may predict eventual radiological response. In a prospective trial of 53 patients with CRC, four showed a transient spike on day 3, and three of these four patients had an excellent response to standard first line chemotherapy (96).
Determining the timing of ctDNA nadir could also provide early clues of chemotherapy response and help to optimize the timing of ctDNA sampling. Although different studies have addressed this question, there is no clear conclusion due to disparity in the timing of blood collection and methods for ctDNA detection. Estimate nadir has varied from 2 weeks after the first dose of gemcitabine-based chemotherapy in pancreatic cancer (4) to 3 weeks in Ewing Sarcoma treated with induction chemotherapy (94) or to 37 days in high-grade serous ovarian cancer (97).
Clearance of ctDNA has been suggested as the most robust biomarker for response and survival in patients treated with chemotherapy across tumor types (98, 99). Moreover, there is evidence in selected tumor types that absence of early clearance within the first weeks of chemotherapy could predict relapse (94). Nevertheless, ctDNA clearance may be delayed or incomplete (100), and studies that examine earlier relative ctDNA changes compared to baseline are needed to predict treatment outcome before standard radiological assessments. Table 1 summarizes several studies and their varying approaches to answer this question (65, 66, 96, 97, 101–104). A limitation that many of these studies share is that they target only a single mutation in ctDNA, such as KRAS in pancreatic cancer (104) or TP53 in ovarian cancer (97) or a single gene fusion such as EWRS1 in Ewing Sarcoma (94). While these mutations may be prevalent within these tumor types and the technology used (ddPCR) is per se highly sensitive, this can limit generalizability to tumors lacking these mutations. The remaining studies used fixed NGS panels (i.e., small cell lung, colorectal, and pancreas cancers) (65, 66, 103), cancer personalized profiling by deep sequencing (CAPP-seq) (i.e., lymphoma) (102), or tumor-plasma intersection bespoke panels (i.e., gastrointestinal malignancies) (101), with limitations of each of these platforms as discussed earlier in this review.
Table 1. Studies that analyze early ctDNA with treatment outcome in patients treated with chemotherapy.
C, cycle; DFS, disease-free survival; dPCR, digital polymerase chain reaction; GI, gastrointestinal; HR, hazard ratio; PDAC, pancreatic ductal adenocarcinoma; pts, patients; PR, partial response; Safe-SeqS, safe sequencing system; SCLC, small cell lung carcinoma; SD, stable disease; TTP, time to progression.
| Author | N | Tumor | ctDNA method | Time point | Conclusions |
| stage | |||||
| Parikh et al. (101) | 101 pts | GI tumors stage IV | ddPCR for mutations found in NGS on tumor |
Baseline and 4 weeks | Percent change of ctDNA by 4 weeks predicted PR and clinical benefit (PR and SD). |
| A decrease by 4 weeks ≥30% predicted a longer PFS. | |||||
| Tie et al. (96) | 53 pts | CRC stage IV | Safe-SeqS | Baseline and C2 (2 weeks after first dose) |
Fold reduction in ctDNA predicts better radiological response than the absolute level of ctDNA. |
| 74% of patients who had a ≥10-fold reduction in ctDNA levels had a radiological response at the first radiological measurement. | |||||
| Wei et al. (65) | 17 pts | Pancreatic adeno- carcinoma stage IV |
560-gene panel NGS | Baseline and C2 (2 weeks after first dose) |
Relative changes in ctDNA prior cycle 2 predict radiological response as all patients with PD as best response had an increase in ctDNA, whereas 91% of patients with at least SD at first radiological assessment had a decrease in ctDNA. |
| Parkinson et al. (97) | 32 pts | Relapsed high-grade serous ovarian carcinoma |
dPCR for TP53 mutation | Baseline and C2 (21 to 28 days after first dose) |
A percentage ctDNA decrease of ≥60% between baseline and cycle 2 predicted a longer TTP compared to those with a decrease of <60% irrespective of disease volume. |
| Kurtz et al. (102) | 217 pts | Diffuse large B cell lymphoma |
CAPP-seq | Baseline, mid cycle, cycle 2 (28 days), and cycle 3 |
ctDNA drop by midpoint of first cycle (6 to 16 days) predicts responders versus nonresponders. |
| A 100-fold decrease (log2) drop by the start of cycle 2 and a log2.5 drop by cycle 3 were also predictive for a better event-free survival and OS irrespective other prognosis factors. | |||||
| Osumi et al. (103) | 29 pts | CRC stage IV | 14-gene panel NGS | Baseline, weeks 2 and 8 | Change in ctDNA levels at 2 weeks could be a possible predictor of PFS, while change in ctDNA levels at 8 weeks predicts independently PFS and OS. |
| Almodovar et al. (66) | 25 pts | SCLC stage IV | 14-gene panel NGS | Baseline, cycles 2 and 3 | ctDNA decrease from baseline to cycle 2 or 3 predicts radiological response but not PFS or OS. |
| Perets et al. (104) | 5 pts | PDAC stage IV | KRAS dPCR | Baseline and 4 weeks later | A significant negative correlation between the ctDNA slopes and survival times was found, suggesting that a deep fall in ctDNA over a short time correlates with longer OS, whereas a fast and marked rise in ctDNA predicted a shorter OS. |
One of the main hurdles to using ctDNA kinetics as a predictive factor is to identify the optimal time point to measure changes, which may vary according to tumor biology or drug mechanism. Findings of Osumi et al. (103) highlight an important trade-off: Earlier time points could support more prompt clinical decisions but at the cost of poorer sensitivity in predicting eventual treatment response or progression. The BEECH study (a randomized phase 2 study evaluating paclitaxel ± AKT inhibitor in advanced estrogen receptor–positive breast cancer) provides an innovative approach for selecting the best time point for ctDNA analysis (58). This study has a discovery cohort where intensive plasma analysis was performed weekly for 4 weeks, and day 28 after treatment was identified as the optimal time point for predicting PFS, which was subsequently confirmed in the validation cohort.
ctDNA kinetics may assist in predicting response in potentially curative settings such as neoadjuvant chemotherapy. The I-SPY-2 clinical trial evaluated ctDNA kinetics in 58 patients with early stage breast carcinoma treated with neoadjuvant chemotherapy (anthracyclines followed by paclitaxel) ± AKT inhibitor (99). Bespoke ctDNA positivity decreased from 73% of patients at baseline to 35% after 3 weeks, 14% at the end of anthracyclines, and 9% at the end of all chemotherapy. At the early time point of 3 weeks, patients with undetectable ctDNA were more likely to have pathologic complete response (48% versus 17%) and had longer relapse-free survival even if they did not achieve pathologic complete response. Detectable ctDNA predicted metastatic recurrence regardless of the direction of change. These findings suggest an opportunity to modify treatment or introduce additional therapies if ctDNA does not decrease significantly during the early phase of treatment.
In conclusion, early ctDNA kinetics from mutation-based ctDNA seems to predict outcome of chemotherapy. Moreover, there are other alternative ctDNA markers that have also been suggested as treatment response predictors such as methylated ctDNA kinetics in patients with CRC (47) or viral-related DNA kinetics in certain lymphomas (41) and NPC.
Use in targeted therapy
The most relevant studies evaluating ctDNA changes in relation to targeted therapy have been performed in patients with non–small cell lung carcinoma (NSCLC) treated with epidermal growth factor receptor (EGFR) inhibitors. ctDNA kinetics share many similar features to those described during chemotherapy. Intensive ctDNA analysis of EGFR mutations at multiple time points within the first week of treatment demonstrated an 11-fold peak at 26 hours in one responder followed by a ctDNA reduction during the next 2 days. By contrast, a nonresponder showed a steep rise of EGFR mutation levels at day 5 followed by a later decline, but levels remained above baseline (43). Similar kinetics were reported in patients with BRAF mutant melanoma who were treated with BRAF and MEK inhibitors (105). Early decrease in ctDNA (2 to 4 weeks) after the start of EGFR inhibitor therapy (erlotinib and gefitinib) was correlated with an improvement in median OS by around 7 months (106) or with a favorable RECIST (response evaluation criteria in solid tumors) radiological response (44). However, these correlations have only been shown with specific EGFR mutations, such as L858R, but validation in other EGFR mutations is needed (107).
Normalized metrics of ctDNA change have been used in targeted therapy tracking such as the previously described MART and CDR scores (which capture the ratio of on-treatment to pretreatment mutant allele count or fraction). In EGFR mutant NSCLC, all cases with disease progression showed MART > 0.1, while this result was only seen in 22% of patients without disease progression (57). Furthermore, among all patients with MART > 0.1, those with disease progression had higher plasma mutation score. Thus, combining plasma mutation score and MART may be a more comprehensive approach to predict benefit from EGFR inhibitors in EGFR mutant NSCLC. Similarly, CDR at day 15 was evaluated in hormone receptor–positive breast cancer patients enrolled in the PALOMA-3 trial that evaluated fulvestrant and palbociclib (59). Samples at baseline and day 15 were analyzed for PI3KCA and ESR1 mutations using ddPCR. PI3KCA and ESR1 CDR at day 15 value was lower in patients treated with palbociclib, indicating a larger reduction in ctDNA abundance. Moreover, PI3KCA CDR at day 15 correlated with PFS on palbociclib and fulvestrant.
Again, many of these studies use highly targeted approaches that cannot detect emergence of previously unidentified mutations. This limitation carries particular relevance to targeted therapies in which failure is often related to specific well-characterized resistance mutations. This limitation can be mitigated by including common resistance mutations in testing, such as T790M in EGFR-mutated lung cancers, as has been extensively reviewed previously (108). However, this is more challenging when the resistance mechanism is not well known.
Use in immunotherapy
Early changes in ctDNA have become a relevant topic in patients treated with immunotherapy, especially checkpoint inhibitors. Different patterns of response to immunotherapy according to ctDNA kinetics have been proposed. For instance, patients with NSCLC have been observed to follow one of the three trends: rapid complete clearance (responders), fluctuation or rise in 3 to 16 weeks (nonresponders), and initial reduction followed by an increase in ctDNA (responders with acquired resistance) (68). It is also well known that immunotherapy requires more time to produce cell death than other cancer therapies (i.e., targeted therapies), and therefore, decrease in ctDNA may take place later (105, 109, 110). Similarly, median time to significant response or decrease (>50% from baseline) in ctDNA is 24.5 days in a cohort of patients with metastatic NSCLC treated with checkpoint inhibitors (110). Unlike with other therapies, peaks in ctDNA have not been widely described with immunotherapy.
Clearance of ctDNA has again been suggested as the most robust predictive ctDNA biomarker. For instance, clearance within 12 weeks of immune checkpoint inhibitors predicted a 2-year OS rate of 90% in patients with advanced melanoma (111). Recently, the INSPIRE study (a pan-cancer investigator-initiated study) showed that 12 of 73 patients who achieved ctDNA clearance at any time during treatment with pembrolizumab (typically, between weeks 9 and 18) were alive at median follow-up of 25 months (54). Nevertheless, clearance is not seen in most of the patients, and time to clearance can be lengthy (up to 12 cycles).
Therefore, earlier quantitative changes in ctDNA have been studied as potential predictive biomarkers for specific tumor types. For instance, a decrease in ctDNA within 4 weeks of checkpoint inhibitors predicted longer OS and PFS in patients with metastatic melanoma (109, 112). Similar findings were observed in patients with metastatic NSCLC within the first month of checkpoint inhibitor treatment (68, 110, 113), including a cohort of patients where a very early decrease in ctDNA (median time 2.4 weeks) predicted treatment outcome (114). Moreover, multiparameter models combining early ctDNA kinetics and immune profiling in this cohort improved treatment outcome prediction (114). Similar results have been observed in patients with metastatic gastric adenocarcinoma treated with pembrolizumab within a clinical trial (115) where a decrease in ctDNA at 6 weeks predicted better response and PFS. Kinetics of ctDNA have also been studied in earlier disease stages such as patients with stage IIII NSCLC. For instance, patients with rising ctDNA during consolidation immunotherapy after chemoradiation have a poorer outcome than those with decreasing ctDNA (116), suggesting that those with no response by ctDNA could potentially benefit from treatment escalation. Although the above studies showed consistent findings, heterogeneity in their approaches to detect ctDNA should be highlighted to understand such limitations. The ctDNA platforms used in these studies ranged from focused sequencing of a few alterations (109, 111) to broader NGS multigene panels (110, 113, 115), to CAPP-seq (114, 116), and to bespoke ctDNA evaluations (68, 112).
ctDNA kinetics may be predictive of response to immunotherapy in pan-cancer settings, as reported by two comprehensive studies addressing this issue (Table 2). Within the prospective multitumor INSPIRE trial, ctDNA variations from baseline to week 6 have been suggested as a predictive marker of clinical benefit and improved OS and PFS in patients treated with pembrolizumab, on the basis of multivariable analysis that also accounted for programmed death-ligand 1 (PD-L1) status and tumor mutational burden (54). Second, a pooled analysis has correlated ctDNA changes with response rate, OS, and PFS in patients with advanced cancers treated with durvalumab and/or tremelimumab (54), validating a prior analysis of two trials with durvalumab (60). In this study, on-treatment VAF and a change between baseline and on-treatment VAF at week 6 were complementary in predicting PFS and OS. Both studies showed robust results despite different methodologies for tracking ctDNA. The INSPIRE trial uses a highly sensitive personalized approach based on tracking in plasma variants found in primary tumor using whole exome sequencing. On the other hand, the pooled analysis assay used a fixed 72-gene panel, which may have a lower sensitivity and specificity but does not rely on tissue availability.
Table 2. ctDNA kinetics in main pan-tumor studies with checkpoint inhibitors.
AUC, area under the curve; CI, confidence interval; CR, complete response; HNSCC, head and neck squamous cell carcinoma; RECIST, response evaluation criteria in solid tumors.
| INSPIRE | Study 1108, ATLANTIC, and Study 10 | |
| [Bratman et al. (54)] | [Zhang et al. (60] | |
| Patients | 73 pts in a single study | 171 pts in three different studies |
| Number of tumor types | 25 tumor types | 16 tumor types (mostly NSCLC and urothelial) |
| (five cohorts HNSCC, triple-negative breast cancer, ovarian cancer, melanoma and mixed tumor types) | ||
| Drugs | Pembrolizumab | Durvalumab ± tremelimumab |
| ctDNA analysis | Bespoke ctDNA (Signatera) | 72-gene panel NGS (Guardant 360) |
| ctDNA kinetics metrics | ΔctDNAC3: Relative change in ctDNA levels from baseline to C3 |
Delta VAF: Mean change in VAF |
| Molecular response: Ratio between on-treatment VAF and pretreatment VAF | ||
| Correlation with response | 42% pts with negative ΔctDNAC3 achieved an objective response whereas 2% with positive ΔctDNAC3 had an objective response |
Ratio-based molecular response has a strong association with RECIST response (AUC = 0.82; 95% CI, 0.71–0.93) |
| OR: 28.74 (95% CI, 3.51 to 253.04) | Molecular response (ratio < 50%) is associated with higher overall response rate in each study |
|
| ΔctDNAC3 was also associated with higher clinical benefit rate (CR, PR, and SD ≥ 6 cycles) | ||
| Correlation with PFS | Favorable PFS (adjusted HR: 0.33; 95% CI, 0.19 to 0.58) for negative ΔctDNAC3 |
pts with delta-VAF > 0 had the worst PFS, those with decreased but not completely cleared ctDNA had intermediate PFS, and those without on-treatment ctDNA had the best PFS (P < 0.0001) |
| Consistent in all cohorts | Molecular response (ratio < 50%) is associated with improved HR for OS in each study (HR: 0.28, 0.3, and 0.11, respectively) |
|
| Correlation with OS | Favorable OS (adjusted HR: 0.36; 95% CI 0.18 to 0.71) for negative ΔctDNAC3. |
pts with delta-VAF > 0 had the worst OS, those with decreased but not completely cleared ctDNA had intermediate OS, and those without on-treatment ctDNA had the best OS (P < 0.0001) |
| Consistent in all cohorts. | Molecular response (ratio < 50%) is associated with improved HR for OS in each study (HR: 0.29, 0.29, and 0.12, respectively) |
Therefore, current evidence suggests that decrease or clearance of ctDNA within the first 6 weeks of treatment may predict outcome. However, there is no published evidence on whether earlier time points (i.e., 2 to 3 weeks after first dose) may correlate with outcome despite observations that significant ctDNA decreases occur in the first 2 to 4 weeks. These earlier time points may spare clinical and financial toxicities in nonresponders. Another potential application of ctDNA kinetics in patients treated with immunotherapy could be the ability to distinguish between true progressors and pseudoprogressors (when tumor size can initially increase due to immune infiltration before tumor shrinkage) (117). This hypothesis has been studied in patients with melanoma, where a decrease in ctDNA of over 10-fold within 12 weeks of checkpoint inhibitors was able to predict pseudoprogression with 90% sensitivity and 100% specificity (118). Similar findings were also observed in a small cohort of patients with metastatic NSCLC treated with nivolumab, where patients with pseudoprogression had a marked decrease of ctDNA at 30 days, while true progressors did not (119). Although prospective trials are needed, these findings provide a first step to validate ctDNA as a predictive biomarker for immune checkpoint inhibitors.
While most existing studies examine checkpoint inhibitors (i.e., anti–PD-L1, PD-1, and CTLA-4), ctDNA kinetics have also been evaluated in other immunotherapies such as adoptive cell therapies, specifically tumor-infiltrating lymphocyte transfer. Three different ctDNA dynamic patterns have been seen in a cohort of 39 BRAF mutant melanoma patients: early peak with clearance, early peak without clearance, and no peak with or without clearance (120). Early peak was observed before day 9 of treatment in more than two-thirds of the patients, suggesting on-target recognition and subsequent killing of malignant cells. Moreover, ctDNA clearance was observed in a median of 27 days. Interestingly, patients with early ctDNA peak and clearance had radiological response and better OS compared to the other patterns.
SUMMARY AND FUTURE DIRECTIONS
Early ctDNA kinetics are emerging as a promising predictive biomarker of treatment outcomes. However, further development and validation are required before these approaches can be used clinically.
First, a deeper knowledge of how early ctDNA kinetics relate to therapy and response is needed. To date, studies have been heterogeneous in the ctDNA methods used, summary statistics collected, and time points analyzed. Moreover, most studies examining these questions have had small sample sizes, limiting power and generalizability. It is necessary to establish standards and best practices to better systematize the evaluation of ctDNA kinetics to improve comparability between studies. Initiatives are now being developed to standardize ctDNA evaluations such as the ctMoniTR Project by Friends of Cancer Research Initiative that is collecting and integrating data from multiple clinical trials to better understand ctDNA patterns (42). Moreover, there are ongoing studies using systematic intensive ctDNA analysis (every 3 to 7 days within 4 weeks of the first infusion) to better characterize early shifts, for instance, in patients with metastatic head and neck squamous cell carcinoma (NCT04606940 and NCT03540563) or with metastatic NSCLC (NCT03926260). Moreover, there is no clear threshold to distinguish whether ctDNA changes are clinically significant or a validated optimal time point for analysis. These questions will require resolution before early ctDNA kinetics can be used in clinical practice.
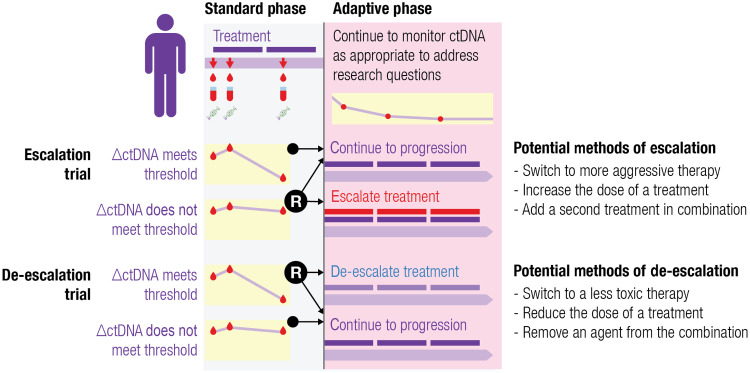
Second, there is an unmet need for prospective clinical trials to address whether modifying therapy due to ctDNA changes before radiological progression improves survival outcomes. To this end, prior trials using other liquid biopsy approaches such as circulating tumor cells (CTCs) have not shown benefit. SWOG S0500 study evaluated 595 patients with metastatic breast cancer receiving systemic therapy, using persistent high CTCs at 21 days to decide between treatment continuation or switch to a different systemic agent. Unfortunately, primary endpoint was not met, as OS was not improved for patients who changed therapy when no response in CTCs was seen. One criticism of this study was that treatment was switched to ineffective second line chemotherapy, which may account for the negative result. Moreover, CTCs may not be as sensitive as ctDNA to reflect microscopic changes at an earlier time point of molecular progression. Since this study, novel treatment options and improvements in ctDNA analysis provide hope for successful implementation of adaptive treatment paradigms. Several adaptive trial designs are summarized in Fig. 2. Many ongoing trials provide examples of treatment escalation designs and are summarized in Table 3. The variety of ongoing trials is notable, involving the full range of available therapies in creative combinations to achieve rational escalation or de-escalation in response to ctDNA results.
Fig. 2. Adaptive clinical trials are a potential strategy for evaluating treatment optimizations guided by early ctDNA kinetics.
In an escalation trial, patients with below threshold ctDNA levels (suggesting poor biochemical response) could be randomized to a predetermined treatment escalation with the goal of improved disease control over continuing standard management. Conversely, in a de-escalation trial, patients whose ctDNA levels exceed threshold (excellent biochemical response) could be randomized to a predetermined treatment de-escalation with hope of sparing toxicity while achieving a noninferior outcome.
Table 3. Examples of ongoing adaptive trials using ctDNA kinetics to modify treatment regimens.
| Trial | Population | Initial therapy | Adaptation |
| NCT04093167 | Metastatic NSCLC |
Immune checkpoint inhibitor (pembrolizumab) |
ctDNA after 1 to 3 cycles determines whether patients continue or switch to next standard line of therapy |
| NCT04166487 | Metastatic NSCLC |
Immune checkpoint inhibitor (pembrolizumab) |
Cycle 2 ctDNA determines candidates for addition of chemotherapy |
| NCT04358562 | EGFR mutant NSCLC | Targeted therapy (gefitinib) | Lack of ctDNA clearance at 8 weeks prompts addition of second targeted agent (anlotinib) |
|
NCT04680260 (OPTIMISE) |
Oligometastatic CRC | Radical-intent resection or ablative therapy |
Randomization to standard of care or ctDNA-guided intensified adjuvant chemotherapy regimen |
| NCT04567420 (DARE) | High-risk stage II-III estrogen receptor–positive breast cancer |
Adjuvant hormone therapy (letrozole or tamoxifen) |
Increase in ctDNA prompts switch to combination hormone therapy and targeted agent (fulvestrant and palbociclib) |
| NCT03808441 (CACTUS) | BRAF mutant melanoma |
Targeted therapy (dabrafenib + trametinib) |
ctDNA decrease prompts switch to immunotherapy (nivolumab + ipilimumab) |
Another relevant issue to be addressed in future studies is the threshold for ctDNA detection, as this may vary by both the method used and the time point. Therefore, defining ctDNA shifts or clearance across two or more consecutive time points may be more accurate than at a single time point. While most trials examine ctDNA at a single time point, in the future, more sophisticated models could stratify patients to one of several characteristic temporal profiles using multiple samples drawn at arbitrary time points. This approach has been demonstrated with EBV DNA (62), in which rich temporal profiles across definitive chemoradiation and consolidative chemotherapy were clustered into distinct patterns with diverging clinical outcomes. Previously described studies involving intensive sampling may help to establish such patterns, to which more sparse real-world clinical data could be fit against. These promising approaches will need to overcome a few practical challenges. First, it can be challenging to define clear endpoints in the context of varying time points. Second, while robust and repeatable, machine-fitted models often lack intuitive interpretations, complicating clinical decision-making. Last, intensive sampling may hamper recruitment as many patients may not wish to be involved in trials that require frequent, possibly superfluous, blood collections.
Apart from adaptive trials, ctDNA kinetics may have other applications in clinical research. In early phase clinical trials (when treatment effectiveness is not known), dynamic biomarkers can help to optimize the recommended phase 2 dose in combination with adverse events and pharmacokinetics. Moreover, we may be able to identify patients unlikely to respond based on early ctDNA assessments, limiting exposure to potential drug toxicity. As detection sensitivity improves, we expect to see ctDNA analysis being used earlier after initial diagnosis. Window-of-opportunity trials would be an ideal environment to study rapid shifts in ctDNA during treatment with experimental therapies and will help to identify the treatments for which ctDNA kinetics are predictive. Last, ctDNA can be combined with other accessible biomarkers such as peripheral and tumor-infiltrating immune cell counts, PD-L1 immunohistochemistry, and tumor mutation burden, which have been shown to have orthogonal and complementary predictive effects for immune checkpoint inhibition (62).
Third, an improvement in the sensitivity of current methods as well as a consensus on definitions for ctDNA clearance are needed to implement ctDNA kinetics in clinical practice. These definitions will necessary vary depending on the clinical context, type of cancer being treated, and sensitivity of the method under use. Ultimately, such definitions will require evidence from clinical trials to define practical treatment goals with measurable impact on prognosis and will evolve with improving methodology. Most ctDNA approaches described above only detect mutations (and copy number alterations and structural variants in CAPP-seq assays) that can be quantified and monitored in plasma. However, a proliferation of resistant clones may go undetected by many ctDNA panels. Genome analysis methods with broader coverage such as genome-wide cfDNA analysis (121) and methylation DNA analysis (67) could enable quantification of ctDNA levels beyond mutant allele fraction levels in plasma and help to advance discovery of previously unknown biology and clinically relevant biomarkers. The analysis of orthogonal cancer-associated molecular alterations such as epigenetics could improve ctDNA sensitivity and even identify tissue-specific molecular signatures associated with treatment response or resistance.
The promise of ctDNA analysis is to make cancer biomolecular testing noninvasive. Treatment monitoring is one of the most exciting potential clinical applications of this advancing technology. The totality of evidence from published and ongoing studies shows a steady progression in the ability to glean early indicators of treatment effectiveness from ctDNA. This field has been long dominated by small studies with heterogeneous technologies, time points, and treatment metrics. However, the advent of novel therapies coupled with increasing accessibility of cancer genome sequencing has given rise to an array of innovative trial designs well suited to assessing the clinical utility of ctDNA kinetics. The use of ctDNA kinetics as a predictive biomarker may enable rapid adjustment early in cancer treatment, with the promise of improved outcomes while patients are still well enough to alter therapy. To realize this promise, we urge clinical trials in oncology to capture ctDNA-based metrics at early treatment time points. This should be paired with the development of computational analysis models and machine learning algorithms to maximize the utility of this powerful tool. Collectively, these efforts will establish an evidence base of standards and best practices for clinical monitoring and early treatment optimization using ctDNA analysis.
Acknowledgments
L. L. Siu holds the BMO Chair in Precision Cancer Genomics. S. V. Bratman holds the Dr. Mariano Antonio Elia Chair in Head and Neck Cancer Research.
Funding: The authors acknowledge that they received no funding in support of this article.
Author contributions: E.S.-G., E.Z., S.V.B., and L.L.S. conceived the idea. E.S.-G. and E.Z. wrote the original manuscript and performed the review of the literature. L.L.S. and S.V.B. supervised the manuscript.
Competing interests: E.S.-G.: None. E.Z.: None. S.V.B.: Stock ownership in Adela; leadership position in Adela; patents licensed to Roche, Adela; and royalties from Roche. L.L.S.: Consulting/advisory arrangements with Merck, Pfizer, Celgene, AstraZeneca, Morphosys, Roche, Oncorus, Symphogen, Seattle Genetics, GlaxoSmithKline, Voronoi, Arvinas, Tessa, Navire, Relay, Rubius, Janpix, and Daiichi Sanyko; stock ownership of Agios (spouse); leadership position in Treadwell Therapeutics (spouse); and institution receives clinical trials support from Novartis, Bristol-Myers Squibb, Pfizer, Boerhinger-Ingelheim, GlaxoSmithKline, Roche/Genentech, Karyopharm, AstraZeneca, Merck, Celgene, Astellas, Bayer, Abbvie, Amgen, Symphogen, Intensity Therapeutics, Mirati Therapeutics, Shattucks, and Avid.
Data and materials availability: All data needed to evaluate the conclusions in the paper are present in the paper and/or the materials cited herein.
REFERENCES AND NOTES
- 1.Mardis E. R., DNA sequencing technologies: 2006-2016. Nat. Protoc. 12, 213–218 (2017). [DOI] [PubMed] [Google Scholar]
- 2.Cescon D. W., Bratman S. V., Chan S. M., Siu L. L., Circulating tumor DNA and liquid biopsy in oncology. Nat. Cancer 1, 276–290 (2020). [DOI] [PubMed] [Google Scholar]
- 3.Anagnostou V., Yarchoan M., Hansen A. R., Wang H., Verde F., Sharon E., Collyar D., Chow L. Q. M., Forde P. M., Immuno-oncology trial endpoints: Capturing clinically meaningful activity. Clin. Cancer Res. 23, 4959–4969 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Kruger S., Heinemann V., Ross C., Diehl F., Nagel D., Ormanns S., Liebmann S., Prinz-Bravin I., Westphalen C. B., Haas M., Jung A., Kirchner T., von Bergwelt-Baildon M., Boeck S., Holdenrieder S., Repeated mutKRAS ctDNA measurements represent a novel and promising tool for early response prediction and therapy monitoring in advanced pancreatic cancer. Ann. Oncol. 29, 2348–2355 (2018). [DOI] [PubMed] [Google Scholar]
- 5.Elez E., Chianese C., Sanz-García E., Martinelli E., Noguerido A., Mancuso F. M., Caratù G., Matito J., Grasselli J., Cardone C., Esposito Abate R., Martini G., Santos C., Macarulla T., Argilés G., Capdevila J., Garcia A., Mulet N., Maiello E., Normanno N., Jones F., Tabernero J., Ciardello F., Salazar R., Vivancos A., Impact of circulating tumor DNA mutant allele fraction on prognosis in RAS-mutant metastatic colorectal cancer. Mol. Oncol. 13, 1827–1835 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Newman A. M., Bratman S. V., To J., Wynne J. F., Eclov N. C., Modlin L. A., Liu C. L., Neal J. W., Wakelee H. A., Merritt R. E., Shrager J. B., Loo B. W. Jr., Alizadeh A. A., Diehn M., An ultrasensitive method for quantitating circulating tumor DNA with broad patient coverage. Nat. Med. 20, 548–554 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Newman A. M., Lovejoy A. F., Klass D. M., Kurtz D. M., Chabon J. J., Scherer F., Stehr H., Liu C. L., Bratman S. V., Say C., Zhou L., Carter J. N., West R. B., Sledge G. W., Shrager J. B., Loo B. W. Jr., Neal J. W., Wakelee H. A., Diehn M., Alizadeh A. A., Integrated digital error suppression for improved detection of circulating tumor DNA. Nat. Biotechnol. 34, 547–555 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Zhao E. Y., Bratman S. V., Emerging precision oncology applications of liquid biopsy using circulating tumour DNA and methylome profiling. Clin. Oncol. 32, 626–631 (2020). [DOI] [PubMed] [Google Scholar]
- 9.Bettegowda C., Sausen M., Leary R. J., Kinde I., Wang Y., Agrawal N., Bartlett B. R., Wang H., Luber B., Alani R. M., Antonarakis E. S., Azad N. S., Bardelli A., Brem H., Cameron J. L., Lee C. C., Fecher L. A., Gallia G. L., Gibbs P., Le D., Giuntoli R. L., Goggins M., Hogarty M. D., Holdhoff M., Hong S. M., Jiao Y., Juhl H. H., Kim J. J., Siravegna G., Laheru D. A., Lauricella C., Lim M., Lipson E. J., Marie S. K., Netto G. J., Oliner K. S., Olivi A., Olsson L., Riggins G. J., Sartore-Bianchi A., Schmidt K., Shihl M., Oba-Shinjo S. M., Siena S., Theodorescu D., Tie J., Harkins T. T., Veronese S., Wang T. L., Weingart J. D., Wolfgang C. L., Wood L. D., Xing D., Hruban R. H., Wu J., Allen P. J., Schmidt C. M., Choti M. A., Velculescu V. E., Kinzler K. W., Vogelstein B., Papadopoulos N., Diaz L. A. Jr., Detection of circulating tumor DNA in early- and late-stage human malignancies. Sci. Transl. Med. 6, 224ra224 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Nygaard A. D., Holdgaard P. C., Spindler K. L., Pallisgaard N., Jakobsen A., The correlation between cell-free DNA and tumour burden was estimated by PET/CT in patients with advanced NSCLC. Br. J. Cancer 110, 363–368 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Heidary M., Auer M., Ulz P., Heitzer E., Petru E., Gasch C., Riethdorf S., Mauermann O., Lafer I., Pristauz G., Lax S., Pantel K., Geigl J. B., Speicher M. R., The dynamic range of circulating tumor DNA in metastatic breast cancer. Breast Cancer Res. 16, 421 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.García-Saenz J. A., Ayllón P., Laig M., Acosta-Eyzaguirre D., García-Esquinas M., Montes M., Sanz J., Barquín M., Moreno F., Garcia-Barberan V., Díaz-Rubio E., Caldes T., Romero A., Tumor burden monitoring using cell-free tumor DNA could be limited by tumor heterogeneity in advanced breast cancer and should be evaluated together with radiographic imaging. BMC Cancer 17, 210 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Rostami A., Lambie M., Yu C. W., Stambolic V., Waldron J. N., Bratman S. V., Senescence, necrosis, and apoptosis govern circulating cell-free DNA release kinetics. Cell Rep. 31, 107830 (2020). [DOI] [PubMed] [Google Scholar]
- 14.Dawson S. J., Tsui D. W., Murtaza M., Biggs H., Rueda O. M., Chin S. F., Dunning M. J., Gale D., Forshew T., Mahler-Araujo B., Rajan S., Humphray S., Becq J., Halsall D., Wallis M., Bentley D., Caldas C., Rosenfeld N., Analysis of circulating tumor DNA to monitor metastatic breast cancer. N. Engl. J. Med. 368, 1199–1209 (2013). [DOI] [PubMed] [Google Scholar]
- 15.Chen K., Zhao H., Shi Y., Yang F., Wang L. T., Kang G., Nie Y., Wang J., Perioperative dynamic changes in circulating tumor DNA in patients with lung cancer (DYNAMIC). Clin. Cancer Res. 25, 7058–7067 (2019). [DOI] [PubMed] [Google Scholar]
- 16.Wan J. C. M., Massie C., Garcia-Corbacho J., Mouliere F., Brenton J. D., Caldas C., Pacey S., Baird R., Rosenfeld N., Liquid biopsies come of age: Towards implementation of circulating tumour DNA. Nat. Rev. Cancer 17, 223–238 (2017). [DOI] [PubMed] [Google Scholar]
- 17.Sun K., Jiang P., Chan K. C. A., Wong J., Cheng Y. K. Y., Liang R. H. S., Chan W.-k., Ma E. S. K., Chan S. L., Cheng S. H., Chan R. W. Y., Tong Y. K., Ng S. S. M., Wong R. S. M., Hui D. S. C., Leung T. N., Leung T. Y., Lai P. B. S., Chiu R. W. K., Lo Y. M. D., Plasma DNA tissue mapping by genome-wide methylation sequencing for noninvasive prenatal, cancer, and transplantation assessments. Proc. Natl. Acad. Sci. 112, E5503–E5512 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Sender R., Milo R., The distribution of cellular turnover in the human body. Nat. Med. 27, 45–48 (2021). [DOI] [PubMed] [Google Scholar]
- 19.Madsen A. T., Hojbjerg J. A., Sorensen B. S., Winther-Larsen A., Day-to-day and within-day biological variation of cell-free DNA. EBioMedicine 49, 284–290 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Atamaniuk J., Vidotto C., Kinzlbauer M., Bachl N., Tiran B., Tschan H., Cell-free plasma DNA and purine nucleotide degradation markers following weightlifting exercise. Eur. J. Appl. Physiol. 110, 695–701 (2010). [DOI] [PubMed] [Google Scholar]
- 21.Fatouros I. G., Destouni A., Margonis K., Jamurtas A. Z., Vrettou C., Kouretas D., Mastorakos G., Mitrakou A., Taxildaris K., Kanavakis E., Papassotiriou I., Cell-free plasma DNA as a novel marker of aseptic inflammation severity related to exercise overtraining. Clin. Chem. 52, 1820–1824 (2006). [DOI] [PubMed] [Google Scholar]
- 22.Moss J., Magenheim J., Neiman D., Zemmour H., Loyfer N., Korach A., Samet Y., Maoz M., Druid H., Arner P., Fu K.-Y., Kiss E., Spalding K. L., Landesberg G., Zick A., Grinshpun A., Shapiro A. M. J., Grompe M., Wittenberg A. D., Glaser B., Shemer R., Kaplan T., Dor Y., Comprehensive human cell-type methylation atlas reveals origins of circulating cell-free DNA in health and disease. Nat. Commun. 9, 5068 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Lehmann-Werman R., Neiman D., Zemmour H., Moss J., Magenheim J., Vaknin-Dembinsky A., Rubertsson S., Nellgård B., Blennow K., Zetterberg H., Spalding K., Haller M. J., Wasserfall C. H., Schatz D. A., Greenbaum C. J., Dorrell C., Grompe M., Zick A., Hubert A., Maoz M., Fendrich V., Bartsch D. K., Golan T., Sasson S. A. B., Zamir G., Razin A., Cedar H., Shapiro A. M. J., Glaser B., Shemer R., Dor Y., Identification of tissue-specific cell death using methylation patterns of circulating DNA. Proc. Natl. Acad. Sci. 113, E1826–E1834 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Avanzini S., Kurtz D. M., Chabon J. J., Moding E. J., Hori S. S., Gambhir S. S., Alizadeh A. A., Diehn M., Reiter J. G., A mathematical model of ctDNA shedding predicts tumor detection size. Sci. Adv. 6, eabc4308 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Stroun M., Lyautey J., Lederrey C., Olson-Sand A., Anker P., About the possible origin and mechanism of circulating DNA apoptosis and active DNA release. Clin. Chim. Acta 313, 139–142 (2001). [DOI] [PubMed] [Google Scholar]
- 26.Jahr S., Hentze H., Englisch S., Hardt D., Fackelmayer F. O., Hesch R.-D., Knippers R., DNA fragments in the blood plasma of cancer patients: Quantitations and evidence for their origin from apoptotic and necrotic cells. Cancer Res. 61, 1659–1665 (2001). [PubMed] [Google Scholar]
- 27.Lo Y. M., Zhang J., Leung T. N., Lau T. K., Chang A. M., Hjelm N. M., Rapid clearance of fetal DNA from maternal plasma. Am. J. Hum. Genet. 64, 218–224 (1999). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Muhanna N., Di Grappa M. A., Chan H. H. L., Khan T., Jin C. S., Zheng Y., Irish J. C., Bratman S. V., Cell-free DNA kinetics in a pre-clinical model of head and neck cancer. Sci. Rep. 7, 16723 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.To E. W., Chan K. C., Leung S. F., Chan L. Y., To K. F., Chan A. T., Johnson P. J., Lo Y. M., Rapid clearance of plasma Epstein-Barr virus DNA after surgical treatment of nasopharyngeal carcinoma. Clin. Cancer Res. 9, 3254–3259 (2003). [PubMed] [Google Scholar]
- 30.Diehl F., Schmidt K., Choti M. A., Romans K., Goodman S., Li M., Thornton K., Agrawal N., Sokoll L., Szabo S. A., Kinzler K. W., Vogelstein B., Diaz L. A. Jr., Circulating mutant DNA to assess tumor dynamics. Nat. Med. 14, 985–990 (2008). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Shi X.-Q., Xue W.-H., Zhao S.-F., Zhang X.-J., Sun W., Dynamic tracing for epidermal growth factor receptor mutations in urinary circulating DNA in gastric cancer patients. Tumour Biol. 39, 1010428317691681 (2017). [DOI] [PubMed] [Google Scholar]
- 32.Husain H., Melnikova V. O., Kosco K., Woodward B., More S., Pingle S. C., Weihe E., Park B. H., Tewari M., Erlander M. G., Cohen E., Lippman S. M., Kurzrock R., Monitoring daily dynamics of early tumor response to targeted therapy by detecting circulating tumor DNA in urine. Clin. Cancer Res. 23, 4716–4723 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Markus H., Zhao J., Contente-Cuomo T., Stephens M. D., Raupach E., Odenheimer-Bergman A., Connor S., McDonald B. R., Moore B., Hutchins E., McGilvrey M., de la Maza M. C., Van Keuren-Jensen K., Pirrotte P., Goel A., Becerra C., Von Hoff D. D., Celinski S. A., Hingorani P., Murtaza M., Analysis of recurrently protected genomic regions in cell-free DNA found in urine. Sci. Transl. Med. 13, eaaz3088 (2021). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Yu S. C., Lee S. W., Jiang P., Leung T. Y., Chan K. A., Chiu R. W., Lo Y. D., High-resolution profiling of fetal DNA clearance from maternal plasma by massively parallel sequencing. Clin. Chem. 59, 1228–1237 (2013). [DOI] [PubMed] [Google Scholar]
- 35.Chused T. M., Steinberg A. D., Talal N., The clearance and localization of nucleic acids by New Zealand and normal mice. Clin. Exp. Immunol. 12, 465–476 (1972). [PMC free article] [PubMed] [Google Scholar]
- 36.Tsumita T., Iwanaga M., Fate of injected deoxyribonucleic acid in mice. Nature 198, 1088–1089 (1963). [DOI] [PubMed] [Google Scholar]
- 37.Lau T.-W., Leung T. N., Chan L. Y., Lau T. K., Chan K. A., Tam W. H., Lo Y. D., Fetal DNA clearance from maternal plasma is impaired in preeclampsia. Clin. Chem. 48, 2141–2146 (2002). [PubMed] [Google Scholar]
- 38.Emlen W., Mannik M., Kinetics and mechanisms for removal of circulating single-stranded DNA in mice. J. Exp. Med. 147, 684–699 (1978). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Cabel L., Bidard F. C., Servois V., Cacheux W., Mariani P., Romano E., Minsat M., Bieche I., Farkhondeh F., Jeannot E., Buecher B., HPV circulating tumor DNA to monitor the efficacy of anti-PD-1 therapy in metastatic squamous cell carcinoma of the anal canal: A case report. Int. J. Cancer 141, 1667–1670 (2017). [DOI] [PubMed] [Google Scholar]
- 40.Jones K., Nourse J. P., Keane C., Crooks P., Gottlieb D., Ritchie D. S., Gill D., Gandhi M. K., Tumor-specific but not nonspecific cell-free circulating DNA can be used to monitor disease response in lymphoma. Am. J. Hematol. 87, 258–265 (2012). [DOI] [PubMed] [Google Scholar]
- 41.Kwong Y.-L., Pang A. W., Leung A. Y., Chim C.-S., Tse E., Quantification of circulating Epstein-Barr virus DNA in NK/T-cell lymphoma treated with the SMILE protocol: Diagnostic and prognostic significance. Leukemia 28, 865–870 (2014). [DOI] [PubMed] [Google Scholar]
- 42.Xi L., Pham T. H., Payabyab E. C., Sherry R. M., Rosenberg S. A., Raffeld M., Circulating tumor DNA as an early indicator of response to T-cell transfer immunotherapy in metastatic melanoma. Clin. Cancer Res. 22, 5480–5486 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Riediger A. L., Dietz S., Schirmer U., Meister M., Heinzmann-Groth I., Schneider M., Muley T., Thomas M., Sültmann H., Mutation analysis of circulating plasma DNA to determine response to EGFR tyrosine kinase inhibitor therapy of lung adenocarcinoma patients. Sci. Rep. 6, 33505 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Taus Á., Camacho L., Rocha P., Hardy-Werbin M., Pijuan L., Piquer G., López E., Dalmases A., Longarón R., Clavé S., Salido M., Albanell J., Bellosillo B., Arriola E., Dynamics of EGFR mutation load in plasma for prediction of treatment response and disease progression in patients with EGFR-mutant lung adenocarcinoma. Clin. Lung Cancer 19, 387–394.e2 (2018). [DOI] [PubMed] [Google Scholar]
- 45.Taus Á., Camacho L., Rocha P., Hernández A., Longarón R., Clavé S., Fernández-Ibarrondo L., Salido M., Hardy-Werbin M., Fernández-Rodríguez C., Albanell J., Bellosillo B., Arriola E., Plasmatic KRAS kinetics for the prediction of treatment response and progression in patients with KRAS-mutant lung adenocarcinoma. Arch. Bronconeumol. 57, 323–329 (2021). [DOI] [PubMed] [Google Scholar]
- 46.Wyatt A. W., Annala M., Aggarwal R., Beja K., Feng F., Youngren J., Foye A., Lloyd P., Nykter M., Beer T. M., Alumkal J. J., Thomas G. V., Reiter R. E., Rettig M. B., Evans C. P., Gao A. C., Chi K. N., Small E. J., Gleave M. E., Concordance of circulating tumor DNA and matched metastatic tissue biopsy in prostate cancer. J. Natl. Cancer Inst. 109, djx118 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Thomsen C. B., Hansen T. F., Andersen R. F., Lindebjerg J., Jensen L. H., Jakobsen A., Early identification of treatment benefit by methylated circulating tumor DNA in metastatic colorectal cancer. Ther. Adv. Med. Oncol. 12, 175883592091847 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Visvanathan K., Fackler M. S., Zhang Z., Lopez-Bujanda Z. A., Jeter S. C., Sokoll L. J., Garrett-Mayer E., Cope L. M., Umbricht C. B., Euhus D. M., Forero A., Storniolo A. M., Nanda R., Lin N. U., Carey L. A., Ingle J. N., Sukumar S., Wolff A. C., Monitoring of serum DNA methylation as an early independent marker of response and survival in metastatic breast cancer: TBCRC 005 prospective biomarker study. J. Clin. Oncol. 35, 751–758 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Shen S. Y., Singhania R., Fehringer G., Chakravarthy A., Roehrl M. H. A., Chadwick D., Zuzarte P. C., Borgida A., Wang T. T., Li T., Kis O., Zhao Z., Spreafico A., Medina T. D. S., Wang Y., Roulois D., Ettayebi I., Chen Z., Chow S., Murphy T., Arruda A., O’Kane G. M., Liu J., Mansour M., McPherson J. D., O’Brien C., Leighl N., Bedard P. L., Fleshner N., Liu G., Minden M. D., Gallinger S., Goldenberg A., Pugh T. J., Hoffman M. M., Bratman S. V., Hung R. J., De Carvalho D. D., Sensitive tumour detection and classification using plasma cell-free DNA methylomes. Nature 563, 579–583 (2018). [DOI] [PubMed] [Google Scholar]
- 50.Liu M. C., Oxnard G. R., Klein E. A., Swanton C., Seiden M. V.; CCGA Consortium , Sensitive and specific multi-cancer detection and localization using methylation signatures in cell-free DNA. Ann. Oncol. 31, 745–759 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Burgener J. M., Zou J., Zhao Z., Zheng Y., Shen S. Y., Huang S. H., Keshavarzi S., Xu W., Liu F. F., Liu G., Waldron J. N., Weinreb I., Spreafico A., Siu L. L., de Almeida J. R., Goldstein D. P., Hoffman M. M., De Carvalho D. D., Bratman S. V., Tumor-naïve multimodal profiling of circulating tumor DNA in head and neck squamous cell carcinoma. Clin. Cancer Res. 27, 4230–4244 (2021). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.He S. S., Wang Y., Bao Y., Cai X. Y., Yang X. L., Chen D. M., Chen Y., Lu L. X., Dynamic changes in plasma Epstein-Barr virus DNA load during treatment have prognostic value in nasopharyngeal carcinoma: A retrospective study. Cancer Med. 7, 1110–1117 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Leung S. F., Chan K. C., Ma B. B., Hui E. P., Mo F., Chow K. C., Leung L., Chu K. W., Zee B., Lo Y. M., Chan A. T., Plasma Epstein-Barr viral DNA load at midpoint of radiotherapy course predicts outcome in advanced-stage nasopharyngeal carcinoma. Ann. Oncol. 25, 1204–1208 (2014). [DOI] [PubMed] [Google Scholar]
- 54.Bratman S. V., Yang S. Y. C., Iafolla M. A. J., Liu Z., Hansen A. R., Bedard P. L., Lheureux S., Spreafico A., Razak A. A., Shchegrova S., Louie M., Billings P., Zimmermann B., Sethi H., Aleshin A., Torti D., Marsh K., Eagles J., Cirlan I., Hanna Y., Clouthier D. L., Lien S. C., Ohashi P. S., Xu W., Siu L. L., Pugh T. J., Personalized circulating tumor DNA analysis as a predictive biomarker in solid tumor patients treated with pembrolizumab. Nat. Cancer 1, 873–881 (2020). [DOI] [PubMed] [Google Scholar]
- 55.Patsch K., Matasci N., Soundararajan A., Diaz P., Agus D. B., Ruderman D., Gross M. E., Monitoring dynamic cytotoxic chemotherapy response in castration-resistant prostate cancer using plasma cell-free DNA (cfDNA). BMC. Res. Notes 12, 275 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Uchida J., Imamura F., Kukita Y., Oba S., Kumagai T., Nishino K., Inoue T., Kimura M., Kato K., Dynamics of circulating tumor DNA represented by the activating and resistant mutations in epidermal growth factor receptor tyrosine kinase inhibitor treatment. Cancer Sci. 107, 353–358 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Kato K., Uchida J., Kukita Y., Kumagai T., Nishino K., Inoue T., Kimura M., Oba S., Imamura F., Numerical indices based on circulating tumor DNA for the evaluation of therapeutic response and disease progression in lung cancer patients. Sci. Rep. 6, 29093 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Hrebien S., Citi V., Garcia-Murillas I., Cutts R., Fenwick K., Kozarewa I., McEwen R., Ratnayake J., Maudsley R., Carr T. H., de Bruin E. C., Schiavon G., Oliveira M., Turner N., Early ctDNA dynamics as a surrogate for progression-free survival in advanced breast cancer in the BEECH trial. Ann. Oncol. 30, 945–952 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.O’Leary B., Hrebien S., Morden J. P., Beaney M., Fribbens C., Huang X., Liu Y., Bartlett C. H., Koehler M., Cristofanilli M., Garcia-Murillas I., Bliss J. M., Turner N. C., Early circulating tumor DNA dynamics and clonal selection with palbociclib and fulvestrant for breast cancer. Nat. Commun. 9, 896 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Zhang Q., Luo J., Wu S., Si H., Gao C., Xu W., Abdullah S. E., Higgs B. W., Dennis P. A., van der Heijden M. S., Segal N. H., Chaft J. E., Hembrough T., Barrett J. C., Hellmann M. D., Prognostic and predictive impact of circulating tumor DNA in patients with advanced cancers treated with immune checkpoint blockade. Cancer Discov. 10, 1842–1853 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Bos M. K., Nasserinejad K., Jansen M. P. H. M., Steendam C. M. J., Angus L., Atmodimedjo P. N., de Jonge E., Dinjens W. N. M., van Schaik R. H. N., Del Re M., Dubbink H. J., Sleijfer S., Martens J. W. M., Comparison of variant allele frequency and number of mutant molecules as units of measurement for circulating tumor DNA. Mol. Oncol. 15, 57–66 (2021). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Lv J., Chen Y., Zhou G., Qi Z., Tan K. R. L., Wang H., Lin L., Chen F., Zhang L., Huang X., Liu R., Xu S., Chen Y., Ma J., Chua M. L. K., Sun Y., Liquid biopsy tracking during sequential chemo-radiotherapy identifies distinct prognostic phenotypes in nasopharyngeal carcinoma. Nat. Commun. 10, 3941 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Cai Z., Chen G., Zeng Y., Dong X., Li Z., Huang Y., Xin F., Qiu L., Xu H., Zhang W., Su X., Liu X., Liu J., Comprehensive liquid profiling of circulating tumor DNA and Protein biomarkers in long-term follow-up patients with hepatocellular carcinoma. Clin. Cancer Res. 25, 5284–5294 (2019). [DOI] [PubMed] [Google Scholar]
- 64.Nygård L., Ahlborn L. B., Persson G. F., Chandrananda D., Langer J. W., Fischer B. M., Langer S. W., Gabrielaite M., Kjær A., Rosenfeld N., Mouliere F., Østrup O., Vogelius I. R., Bentzen S. M., Circulating cell free DNA during definitive chemo-radiotherapy in non-small cell lung cancer patients - initial observations. PLOS ONE 15, e0231884 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Wei T., Zhang Q., Li X., Su W., Li G., Ma T., Gao S., Lou J., Que R., Zheng L., Bai X., Liang T., Monitoring tumor burden in response to FOLFIRINOX chemotherapy via profiling circulating cell-free DNA in pancreatic cancer. Mol. Cancer Ther. 18, 196–203 (2019). [DOI] [PubMed] [Google Scholar]
- 66.Almodovar K., Iams W. T., Meador C. B., Zhao Z., York S., Horn L., Yan Y., Hernandez J., Chen H., Shyr Y., Lim L. P., Raymond C. K., Lovly C. M., Longitudinal cell-free DNA analysis in patients with small cell lung cancer reveals dynamic insights into treatment efficacy and disease relapse. J. Thorac. Oncol. 13, 112–123 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Zviran A., Schulman R. C., Shah M., Hill S. T. K., Deochand S., Khamnei C. C., Maloney D., Patel K., Liao W., Widman A. J., Wong P., Callahan M. K., Ha G., Reed S., Rotem D., Frederick D., Sharova T., Miao B., Kim T., Gydush G., Rhoades J., Huang K. Y., Omans N. D., Bolan P. O., Lipsky A. H., Ang C., Malbari M., Spinelli C. F., Kazancioglu S., Runnels A. M., Fennessey S., Stolte C., Gaiti F., Inghirami G. G., Adalsteinsson V., Houck-Loomis B., Ishii J., Wolchok J. D., Boland G., Robine N., Altorki N. K., Landau D. A., Genome-wide cell-free DNA mutational integration enables ultra-sensitive cancer monitoring. Nat. Med. 26, 1114–1124 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Anagnostou V., Forde P. M., White J. R., Niknafs N., Hruban C., Naidoo J., Marrone K., Sivakumar I. K. A., Bruhm D. C., Rosner S., Phallen J., Leal A., Adleff V., Smith K. N., Cottrell T. R., Rhymee L., Palsgrove D. N., Hann C. L., Levy B., Feliciano J., Georgiades C., Verde F., Illei P., Li Q. K., Gabrielson E., Brock M. V., Isbell J. M., Sauter J. L., Taube J., Scharpf R. B., Karchin R., Pardoll D. M., Chaft J. E., Hellmann M. D., Brahmer J. R., Velculescu V. E., Dynamics of tumor and immune responses during immune checkpoint blockade in non-small cell lung cancer. Cancer Res. 79, 1214–1225 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Razavi P., Li B. T., Brown D. N., Jung B., Hubbell E., Shen R., Abida W., Juluru K., De Bruijn I., Hou C., Venn O., Lim R., Anand A., Maddala T., Gnerre S., Satya R. V., Liu Q., Shen L., Eattock N., Yue J., Blocker A. W., Lee M., Sehnert A., Xu H., Hall M. P., Santiago-Zayas A., Novotny W. F., Isbell J. M., Rusch V. W., Plitas G., Heerdt A. S., Ladanyi M., Hyman D. M., Jones D. R., Morrow M., Riely G. J., Scher H. I., Rudin C. M., Robson M. E., Diaz L. A. Jr., Solit D. B., Aravanis A. M., High-intensity sequencing reveals the sources of plasma circulating cell-free DNA variants. Nat. Med. 25, 1928–1937 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Gögenur M., Burcharth J., Gögenur I., The role of total cell-free DNA in predicting outcomes among trauma patients in the intensive care unit: A systematic review. Crit. Care 21, 14 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Jackson Chornenki N. L., Coke R., Kwong A. C., Dwivedi D. J., Xu M. K., McDonald E., Marshall J. C., Fox-Robichaud A. E., Charbonney E., Liaw P. C., Comparison of the source and prognostic utility of cfDNA in trauma and sepsis. Intensive Care Med. Exp. 7, 29 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Szpechcinski A., Chorostowska-Wynimko J., Kupis W., Maszkowska-Kopij K., Dancewicz M., Kowalewski J., Orlowski T., Quantitative analysis of free-circulating DNA in plasma of patients with resectable NSCLC. Expert Opin. Biol. Ther. 12 ( Suppl. 1), S3–S9 (2012). [DOI] [PubMed] [Google Scholar]
- 73.Henriksen T. V., Reinert T., Christensen E., Sethi H., Birkenkamp-Demtröder K., Gögenur M., Gögenur I., Zimmermann B. G., Dyrskjøt L., Andersen C. L., The effect of surgical trauma on circulating free DNA levels in cancer patients-implications for studies of circulating tumor DNA. Mol. Oncol. 14, 1670–1679 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Takahashi G., Yamada T., Iwai T., Takeda K., Koizumi M., Shinji S., Uchida E., Oncological assessment of stent placement for obstructive colorectal cancer from circulating cell-free DNA and circulating tumor DNA dynamics. Ann. Surg. Oncol. 25, 737–744 (2018). [DOI] [PubMed] [Google Scholar]
- 75.Reinert T., Schøler L. V., Thomsen R., Tobiasen H., Vang S., Nordentoft I., Lamy P., Kannerup A.-S., Mortensen F. V., Stribolt K., Hamilton-Dutoit S., Nielsen H. J., Laurberg S., Pallisgaard N., Pedersen J. S., Ørntoft T. F., Andersen C. L., Analysis of circulating tumour DNA to monitor disease burden following colorectal cancer surgery. Gut 65, 625–634 (2016). [DOI] [PubMed] [Google Scholar]
- 76.Tie J., Wang Y., Tomasetti C., Li L., Springer S., Kinde I., Silliman N., Tacey M., Wong H.-L., Christie M., Kosmider S., Skinner I., Wong R., Steel M., Tran B., Desai J., Jones I., Haydon A., Hayes T., Price T. J., Strausberg R. L., Diaz L. A., Papadopoulos N., Kinzler K. W., Vogelstein B., Gibbs P., Circulating tumor DNA analysis detects minimal residual disease and predicts recurrence in patients with stage II colon cancer. Sci. Transl. Med. 8, 346ra392 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77.Pietrasz D., Pécuchet N., Garlan F., Didelot A., Dubreuil O., Doat S., Imbert-Bismut F., Karoui M., Vaillant J.-C., Taly V., Laurent-Puig P., Bachet J.-B., Plasma circulating tumor DNA in pancreatic cancer patients is a prognostic marker. Clin. Cancer Res. 23, 116–123 (2017). [DOI] [PubMed] [Google Scholar]
- 78.Garcia-Murillas I., Schiavon G., Weigelt B., Ng C., Hrebien S., Cutts R. J., Cheang M., Osin P., Nerurkar A., Kozarewa I., Garrido J. A., Dowsett M., Reis-Filho J. S., Smith I. E., Turner N. C., Mutation tracking in circulating tumor DNA predicts relapse in early breast cancer. Sci. Transl. Med. 7, 302ra133 (2015). [DOI] [PubMed] [Google Scholar]
- 79.Olsson E., Winter C., George A., Chen Y., Howlin J., Tang M.-H. E., Dahlgren M., Schulz R., Grabau D., van Westen D., Fernö M., Ingvar C., Rose C., Bendahl P.-O., Rydén L., Borg Å., Gruvberger-Saal S. K., Jernström H., Saal L. H., Serial monitoring of circulating tumor DNA in patients with primary breast cancer for detection of occult metastatic disease. EMBO Mol. Med. 7, 1034–1047 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80.McDonald B. R., Contente-Cuomo T., Sammut S.-J., Odenheimer-Bergman A., Ernst B., Perdigones N., Chin S.-F., Farooq M., Mejia R., Cronin P. A., Anderson K. S., Kosiorek H. E., Northfelt D. W., McCullough A. E., Patel B. K., Weitzel J. N., Slavin T. P., Caldas C., Pockaj B. A., Murtaza M., Personalized circulating tumor DNA analysis to detect residual disease after neoadjuvant therapy in breast cancer. Sci. Transl. Med. 11, eaax7392 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 81.Abbosh C., Birkbak N. J., Wilson G. A., Jamal-Hanjani M., Constantin T., Salari R., Le Quesne J., Moore D. A., Veeriah S., Rosenthal R., Marafioti T., Kirkizlar E., Watkins T. B. K., McGranahan N., Ward S., Martinson L., Riley J., Fraioli F., Al Bakir M., Grönroos E., Zambrana F., Endozo R., Bi W. L., Fennessy F. M., Sponer N., Johnson D., Laycock J., Shafi S., Czyzewska-Khan J., Rowan A., Chambers T., Matthews N., Turajlic S., Hiley C., Lee S. M., Forster M. D., Ahmad T., Falzon M., Borg E., Lawrence D., Hayward M., Kolvekar S., Panagiotopoulos N., Janes S. M., Thakrar R., Ahmed A., Blackhall F., Summers Y., Hafez D., Naik A., Ganguly A., Kareht S., Shah R., Joseph L., Quinn A. M., Crosbie P. A., Naidu B., Middleton G., Langman G., Trotter S., Nicolson M., Remmen H., Kerr K., Chetty M., Gomersall L., Fennell D. A., Nakas A., Rathinam S., Anand G., Khan S., Russell P., Ezhil V., Ismail B., Irvin-Sellers M., Prakash V., Lester J. F., Kornaszewska M., Attanoos R., Adams H., Davies H., Oukrif D., Akarca A. U., Hartley J. A., Lowe H. L., Lock S., Iles N., Bell H., Ngai Y., Elgar G., Szallasi Z., Schwarz R. F., Herrero J., Stewart A., Quezada S. A., Peggs K. S., Van Loo P., Dive C., Lin C. J., Rabinowitz M., Aerts H. J. W. L., Hackshaw A., Shaw J. A., Zimmermann B. G.; TRACERx cosnsortium; PEACE consortium, Swanton C., Phylogenetic ctDNA analysis depicts early-stage lung cancer evolution. Nature 545, 446–451 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 82.Ohara S., Suda K., Sakai K., Nishino M., Chiba M., Shimoji M., Takemoto T., Fujino T., Koga T., Hamada A., Soh J., Nishio K., Mitsudomi T., Prognostic implications of preoperative versus postoperative circulating tumor DNA in surgically resected lung cancer patients: A pilot study. Trans. Lung Cancer Res. 9, 1915–1923 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 83.Yang J., Gong Y., Lam V. K., Shi Y., Guan Y., Zhang Y., Ji L., Chen Y., Zhao Y., Qian F., Chen J., Li P., Zhang F., Wang J., Zhang X., Yang L., Kopetz S., Futreal P. A., Zhang J., Yi X., Xia X., Yu P., Deep sequencing of circulating tumor DNA detects molecular residual disease and predicts recurrence in gastric cancer. Cell Death Dis. 11, 346 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 84.Tie J., Cohen J. D., Lo S. N., Wang Y., Li L., Christie M., Lee M., Wong R., Kosmider S., Skinner I., Wong H. L., Lee B., Burge M. E., Yip D., Karapetis C. S., Price T. J., Tebbutt N. C., Haydon A. M., Ptak J., Schaeffer M. J., Silliman N., Dobbyn L., Popoli M., Tomasetti C., Papadopoulos N., Kinzler K. W., Vogelstein B., Gibbs P., Prognostic significance of postsurgery circulating tumor DNA in nonmetastatic colorectal cancer: Individual patient pooled analysis of three cohort studies. Int. J. Cancer 148, 1014–1026 (2021). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 85.Tarazona N., Gimeno-Valiente F., Gambardella V., Zuñiga S., Rentero-Garrido P., Huerta M., Roselló S., Martinez-Ciarpaglini C., Carbonell-Asins J. A., Carrasco F., Ferrer-Martínez A., Bruixola G., Fleitas T., Martín J., Tébar-Martínez R., Moro D., Castillo J., Espí A., Roda D., Cervantes A., Targeted next-generation sequencing of circulating-tumor DNA for tracking minimal residual disease in localized colon cancer. Ann. Oncol. 30, 1804–1812 (2019). [DOI] [PubMed] [Google Scholar]
- 86.Jiang J., Ye S., Xu Y., Chang L., Hu X., Ru G., Guo Y., Yi X., Yang L., Huang D., Circulating tumor DNA as a potential marker to detect minimal residual disease and predict recurrence in pancreatic cancer. Front. Oncol. 10, 1220–1220 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 87.Lee B., Lipton L., Cohen J., Tie J., Javed A. A., Li L., Goldstein D., Burge M., Cooray P., Nagrial A., Tebbutt N. C., Thomson B., Nikfarjam M., Harris M., Haydon A., Lawrence B., Tai D. W. M., Simons K., Lennon A. M., Wolfgang C. L., Tomasetti C., Papadopoulos N., Kinzler K. W., Vogelstein B., Gibbs P., Circulating tumor DNA as a potential marker of adjuvant chemotherapy benefit following surgery for localized pancreatic cancer. Ann. Oncol. 30, 1472–1478 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 88.Tan L., Sandhu S., Lee R. J., Li J., Callahan J., Ftouni S., Dhomen N., Middlehurst P., Wallace A., Raleigh J., Hatzimihalis A., Henderson M. A., Shackleton M., Haydon A., Mar V., Gyorki D. E., Oudit D., Dawson M. A., Hicks R. J., Lorigan P., McArthur G. A., Marais R., Wong S. Q., Dawson S. J., Prediction and monitoring of relapse in stage III melanoma using circulating tumor DNA. Ann. Oncol. 30, 804–814 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 89.Lo Y. M., Leung S. F., Chan L. Y., Chan A. T., Lo K. W., Johnson P. J., Huang D. P., Kinetics of plasma Epstein-Barr virus DNA during radiation therapy for nasopharyngeal carcinoma. Cancer Res. 60, 2351–2355 (2000). [PubMed] [Google Scholar]
- 90.Chan S.-K., Chan S.-Y., Choi H. C.-W., Tong C.-C., Lam K.-O., Kwong D. L.-W., Vardhanabhuti V., Leung T.-W., Luk M.-Y., Lee A. W.-M., Lee V. H.-F., Prognostication of half-life clearance of plasma EBV DNA in previously untreated non-metastatic nasopharyngeal carcinoma treated with radical intensity-modulated radiation therapy. Front. Oncol. 10, 1417 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 91.Han K., Leung E., Barbera L., Barnes E., Croke J., Grappa M. A. D., Fyles A., Metser U., Milosevic M., Pintilie M., Wolfson R., Zhao Z., Bratman S. V., Circulating human papillomavirus DNA as a biomarker of response in patients with locally advanced cervical cancer treated with definitive chemoradiation. JCO Precis. Oncol. 1–8 (2018). [DOI] [PubMed] [Google Scholar]
- 92.Leung E., Han K., Zou J., Zhao Z., Zheng Y., Wang T. T., Rostami A., Siu L. L., Pugh T. J., Bratman S. V., HPV Sequencing facilitates ultrasensitive detection of HPV circulating tumor DNA. Clin. Cancer Res. 27, 5857–5868 (2021). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 93.Chan A. T. C., Hui E. P., Ngan R. K. C., Tung S. Y., Cheng A. C. K., Ng W. T., Lee V. H. F., Ma B. B. Y., Cheng H. C., Wong F. C. S., Loong H. H. F., Tong M., Poon D. M. C., Ahuja A. T., King A. D., Wang K., Mo F., Zee B. C. Y., Chan K. C. A., Lo Y. M. D., Analysis of plasma epstein-barr virus DNA in nasopharyngeal cancer after chemoradiation to identify high-risk patients for adjuvant chemotherapy: A randomized controlled trial. J. Clin. Oncol. Jco2018777847 (2018). [DOI] [PubMed] [Google Scholar]
- 94.Krumbholz M., Hellberg J., Steif B., Bäuerle T., Gillmann C., Fritscher T., Agaimy A., Frey B., Juengert J., Wardelmann E., Hartmann W., Juergens H., Dirksen U., Metzler M., Genomic EWSR1 fusion sequence as highly sensitive and dynamic plasma tumor marker in ewing sarcoma. Clin. Cancer Res. 22, 4356–4365 (2016). [DOI] [PubMed] [Google Scholar]
- 95.Moser T., Waldispuehl-Geigl J., Belic J., Weber S., Zhou Q., Hasenleithner S. O., Graf R., Terzic J. A., Posch F., Sill H., Lax S., Kashofer K., Hoefler G., Schoellnast H., Heitzer E., Geigl J. B., Bauernhofer T., Speicher M. R., On-treatment measurements of circulating tumor DNA during FOLFOX therapy in patients with colorectal cancer. npj Precision Oncol. 4, 30 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 96.Tie J., Kinde I., Wang Y., Wong H. L., Roebert J., Christie M., Tacey M., Wong R., Singh M., Karapetis C. S., Desai J., Tran B., Strausberg R. L., Diaz L. A. Jr., Papadopoulos N., Kinzler K. W., Vogelstein B., Gibbs P., Circulating tumor DNA as an early marker of therapeutic response in patients with metastatic colorectal cancer. Ann. Oncol. 26, 1715–1722 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 97.Parkinson C. A., Gale D., Piskorz A. M., Biggs H., Hodgkin C., Addley H., Freeman S., Moyle P., Sala E., Sayal K., Hosking K., Gounaris I., Jimenez-Linan M., Earl H. M., Qian W., Rosenfeld N., Brenton J. D., Exploratory analysis of TP53 mutations in circulating tumour DNA as biomarkers of treatment response for patients with relapsed high-grade serous ovarian carcinoma: A retrospective study. PLOS Med. 13, e1002198 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 98.Song Y., Hu C., Xie Z., Wu L., Zhu Z., Rao C., Liu L., Chen Y., Liang N., Chen J., Hu C., Yang N., Hu J., Zhao W., Tong G., Dong X., Zheng D., Jin M., Chen J., Huang M., He Y., Rosell R., Lippi G., Mino-Kenudson M., Han-Zhang H., Mao X., Zhang L., Liu H., Field J. K., Chuai S., Ye J., Han Y., Lu S.; Written o behalf of AME Lung Cancer Collaborative Group , Circulating tumor DNA clearance predicts prognosis across treatment regimen in a large real-world longitudinally monitored advanced non-small cell lung cancer cohort. Transl. Lung Cancer Res. 9, 269–279 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 99.Magbanua M. J. M., Swigart L. B., Wu H. T., Hirst G. L., Yau C., Wolf D. M., Tin A., Salari R., Shchegrova S., Pawar H., Delson A. L., DeMichele A., Liu M. C., Chien A. J., Tripathy D., Asare S., Lin C. H. J., Billings P., Aleshin A., Sethi H., Louie M., Zimmermann B., Esserman L. J., van’t Veer L. J., Circulating tumor DNA in neoadjuvant-treated breast cancer reflects response and survival. Ann. Oncol. 32, 229–239 (2021). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 100.Moati E., Blons H., Taly V., Garlan F., Wang-Renault S.-F., Pietrasz D., Didelot A., Garrigou S., Saint A., Pernot S., Taieb J., Laurent-Puig P., Zaanan A., Plasma clearance of RAS mutation under therapeutic pressure is a rare event in metastatic colorectal cancer. Int. J. Cancer 147, 1185–1189 (2020). [DOI] [PubMed] [Google Scholar]
- 101.Parikh A. R., Mojtahed A., Schneider J. L., Kanter K., Van Seventer E. E., Fetter I. J., Thabet A., Fish M. G., Teshome B., Fosbenner K., Nadres B., Shahzade H. A., Allen J. N., Blaszkowsky L. S., Ryan D. P., Giantonio B., Goyal L., Nipp R. D., Roeland E., Weekes C. D., Wo J. Y., Zhu A. X., Dias-Santagata D., Iafrate A. J., Lennerz J. K., Hong T. S., Siravegna G., Horick N., Clark J. W., Corcoran R. B., Serial ctDNA monitoring to predict response to systemic therapy in metastatic gastrointestinal cancers. Clin. Cancer Res. 26, 1877–1885 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 102.Kurtz D. M., Scherer F., Jin M. C., Soo J., Craig A. F. M., Esfahani M. S., Chabon J. J., Stehr H., Liu C. L., Tibshirani R., Maeda L. S., Gupta N. K., Khodadoust M. S., Advani R. H., Levy R., Newman A. M., Dührsen U., Hüttmann A., Meignan M., Casasnovas R. O., Westin J. R., Roschewski M., Wilson W. H., Gaidano G., Rossi D., Diehn M., Alizadeh A. A., Circulating tumor DNA measurements as early outcome predictors in diffuse large B-cell lymphoma. J. Clin. Oncol. 36, 2845–2853 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 103.Osumi H., Shinozaki E., Yamaguchi K., Zembutsu H., Early change in circulating tumor DNA as a potential predictor of response to chemotherapy in patients with metastatic colorectal cancer. Sci. Rep. 9, 17358 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 104.Perets R., Greenberg O., Shentzer T., Semenisty V., Epelbaum R., Bick T., Sarji S., Ben-Izhak O., Sabo E., Hershkovitz D., Mutant KRAS circulating tumor DNA is an accurate tool for pancreatic cancer monitoring. Oncologist 23, 566–572 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 105.Schreuer M., Meersseman G., van Den Herrewegen S., Jansen Y., Seremet T., Bott A., Chevolet I., Wilgenhof S., Maertens G., Neyns B., Applications for quantitative measurement of BRAF V600 mutant cell-free tumor DNA in the plasma of patients with metastatic melanoma. Melanoma Res. 26, 157–163 (2016). [DOI] [PubMed] [Google Scholar]
- 106.He J., Tan W., Tang X., Ma J., Variations in EGFR ctDNA correlates to the clinical efficacy of afatinib in non small cell lung cancer with acquired resistance. Pathol. Oncol. Res. 23, 307–315 (2017). [DOI] [PubMed] [Google Scholar]
- 107.Zhou Q., Yang J. J., Chen Z. H., Zhang X. C., Yan H. H., Xu C. R., Su J., Chen H. J., Tu H. Y., Zhong W. Z., Yang X. N., Wu Y. L., Serial cfDNA assessment of response and resistance to EGFR-TKI for patients with EGFR-L858R mutant lung cancer from a prospective clinical trial. J. Hematol. Oncol. 9, 86 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 108.Passiglia F., Rizzo S., Di Maio M., Galvano A., Badalamenti G., Listì A., Gulotta L., Castiglia M., Fulfaro F., Bazan V., Russo A., The diagnostic accuracy of circulating tumor DNA for the detection of EGFR-T790M mutation in NSCLC: A systematic review and meta-analysis. Sci. Rep. 8, 13379 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 109.Ashida A., Sakaizawa K., Uhara H., Okuyama R., Circulating tumour DNA for monitoring treatment response to anti-PD-1 immunotherapy in melanoma patients. Acta Derm. Venereol. 97, 1212–1218 (2017). [DOI] [PubMed] [Google Scholar]
- 110.Goldberg S. B., Narayan A., Kole A. J., Decker R. H., Teysir J., Carriero N. J., Lee A., Nemati R., Nath S. K., Mane S. M., Deng Y., Sukumar N., Zelterman D., Boffa D. J., Politi K., Gettinger S. N., Wilson L. D., Herbst R. S., Patel A. A., Early assessment of lung cancer immunotherapy response via circulating tumor DNA. Clin. Cancer Res. 24, 1872–1880 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 111.Lee J. H., Long G. V., Boyd S., Lo S., Menzies A. M., Tembe V., Guminski A., Jakrot V., Scolyer R. A., Mann G. J., Kefford R. F., Carlino M. S., Rizos H., Circulating tumour DNA predicts response to anti-PD1 antibodies in metastatic melanoma. Ann. Oncol. 28, 1130–1136 (2017). [DOI] [PubMed] [Google Scholar]
- 112.Forschner A., Battke F., Hadaschik D., Schulze M., Weißgraeber S., Han C. T., Kopp M., Frick M., Klumpp B., Tietze N., Amaral T., Martus P., Sinnberg T., Eigentler T., Keim U., Garbe C., Döcker D., Biskup S., Tumor mutation burden and circulating tumor DNA in combined CTLA-4 and PD-1 antibody therapy in metastatic melanoma - results of a prospective biomarker study. J. Immunother. Cancer 7, 180 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 113.Guibert N., Jones G., Beeler J. F., Plagnol V., Morris C., Mourlanette J., Delaunay M., Keller L., Rouquette I., Favre G., Pradines A., Mazieres J., Targeted sequencing of plasma cell-free DNA to predict response to PD1 inhibitors in advanced non-small cell lung cancer. Lung Cancer 137, 1–6 (2019). [DOI] [PubMed] [Google Scholar]
- 114.Nabet B. Y., Esfahani M. S., Moding E. J., Hamilton E. G., Chabon J. J., Rizvi H., Steen C. B., Chaudhuri A. A., Liu C. L., Hui A. B., Almanza D., Stehr H., Gojenola L., Bonilla R. F., Jin M. C., Jeon Y. J., Tseng D., Liu C., Merghoub T., Neal J. W., Wakelee H. A., Padda S. K., Ramchandran K. J., Das M., Plodkowski A. J., Yoo C., Chen E. L., Ko R. B., Newman A. M., Hellmann M. D., Alizadeh A. A., Diehn M., Noninvasive early identification of therapeutic benefit from immune checkpoint inhibition. Cell 183, 363–376.e13 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 115.Kim S. T., Cristescu R., Bass A. J., Kim K.-M., Odegaard J. I., Kim K., Liu X. Q., Sher X., Jung H., Lee M., Lee S., Park S. H., Park J. O., Park Y. S., Lim H. Y., Lee H., Choi M., Talasaz A., Kang P. S., Cheng J., Loboda A., Lee J., Kang W. K., Comprehensive molecular characterization of clinical responses to PD-1 inhibition in metastatic gastric cancer. Nat. Med. 24, 1449–1458 (2018). [DOI] [PubMed] [Google Scholar]
- 116.Moding E. J., Liu Y., Nabet B. Y., Chabon J. J., Chaudhuri A. A., Hui A. B., Bonilla R. F., Ko R. B., Yoo C. H., Gojenola L., Jones C. D., He J., Qiao Y., Xu T., Heymach J. V., Tsao A., Liao Z., Gomez D. R., Das M., Padda S. K., Ramchandran K. J., Neal J. W., Wakelee H. A., Loo B. W., Lin S. H., Alizadeh A. A., Diehn M., Circulating tumor DNA dynamics predict benefit from consolidation immunotherapy in locally advanced non-small-cell lung cancer. Nat. Cancer 1, 176–183 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 117.Raja R., Kuziora M., Brohawn P. Z., Higgs B. W., Gupta A., Dennis P. A., Ranade K., Early reduction in ctDNA predicts survival in patients with lung and bladder cancer treated with durvalumab. Clin. Cancer Res. 24, 6212–6222 (2018). [DOI] [PubMed] [Google Scholar]
- 118.Borcoman E., Kanjanapan Y., Champiat S., Kato S., Servois V., Kurzrock R., Goel S., Bedard P., Le Tourneau C., Novel patterns of response under immunotherapy. Ann. Oncol. 30, 385–396 (2019). [DOI] [PubMed] [Google Scholar]
- 119.Lee J. H., Long G. V., Menzies A. M., Lo S., Guminski A., Whitbourne K., Peranec M., Scolyer R., Kefford R. F., Rizos H., Carlino M. S., Association between circulating tumor DNA and pseudoprogression in patients with metastatic melanoma treated with anti–programmed cell death 1 antibodies. JAMA Oncol. 4, 717–721 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 120.Guibert N., Mazieres J., Delaunay M., Casanova A., Farella M., Keller L., Favre G., Pradines A., Monitoring of KRAS-mutated ctDNA to discriminate pseudo-progression from true progression during anti-PD-1 treatment of lung adenocarcinoma. Oncotarget 8, 38056–38060 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 121.Chen X., Chang C. W., Spoerke J. M., Yoh K. E., Kapoor V., Baudo C., Aimi J., Yu M., Liang-Chu M. M. Y., Suttmann R., Huw L. Y., Gendreau S., Cummings C., Lackner M. R., Low-pass whole-genome sequencing of circulating cell-free DNA demonstrates dynamic changes in genomic copy number in a squamous lung cancer clinical cohort. Clin. Cancer Res. 25, 2254–2263 (2019). [DOI] [PubMed] [Google Scholar]