Abstract
Whether induced tissue-resident memory T (TRM) cells in nonlymphoid organs alone can mediate allograft rejection is unknown. By grafting alloskin or heart into severe combined immunodeficient or Rag2KO mice in which a piece of induced CD4+ and/or CD8+ TRM cell–containing MHC-matched or syngeneic skin was transplanted in advance, we addressed this issue. The induced CD4+ TRM cells in the skin alone acutely rejected alloskin or heart grafts. RNA-seq analysis showed that induced CD4+ TRM cells in skin favorably differentiated into TH17-like polarization during the secondary immune response. Inhibition of the key TH17 signaling molecule RORγt attenuated TRM cell–mediated graft rejection. Thus, we offer a unique mouse model to specifically study TRM cell–mediated allograft rejection without the involvement of lymphocytes in lymphoid organs and tissues. Our study provides strong evidence supporting the hypothesis that long-lived alloreactive TRM cells resident in other organs/tissues substantially contribute to organ allograft rejection.
The induced tissue-resident memory T cells in the skin alone could efficiently mediate acute allogeneic skin or heart rejection.
INTRODUCTION
Tissue-resident memory T (TRM) cells are a distinct population of lymphocytes, which reside permanently in tissues and organs of mice and humans without recirculating and are recognized as contributing significantly to enhanced local immune protection in nonlymphoid tissues (1–3). Increasingly, studies have revealed CD69+CD103+TRM cells located mainly in barrier tissues, such as the intestines, lungs, skin, and reproductive tract, whereas CD69+CD103−TRM cells are located in both barrier and nonbarrier tissues, such as the pancreas, kidneys, and brain (4–7). However, a series of studies showed nicely that CD4+ or CD8+ TRM cells had different migration patterns during the first and secondary activation and that these formed TRM cells could rejoin the circulating pool after reactivation in mice and humans (8–11). So far, many more studies have been performed on CD8+ TRM cells than on CD4+TRM cells. CD4+TRM cells and CD8+TRM cells display some similarities as they share a set of common key markers, such as CD103 and CD49a-based adhesion molecules as well as CXCR6 and CX3CR1-based chemokine receptors (12). However, there are obvious differences between CD4+ and CD8+TRM cells, such as different mechanisms for CD4+TRM and CD8+TRM cell homeostasis in different locations and different patterns of peripheral migration (2, 8). It is recognized that CD4+ TRM and/or CD8+ TRM cells play important roles in infectious diseases, chronic inflammatory diseases, and tumors [reviewed in (13–16)].
The involvement of CD4+TRM and/or CD8+TRM cells in organ transplant rejection has been elucidated in recent years. In human lymphocyte antigen (HLA)–mismatched liver transplantation, the infiltrated recipient cells form long-lived CD8+TRM populations, and a small pool of donor CXCR3high TRM cells have persisted in allografts for more than a decade (17). It is reported that the donor resident CD4+ and CD8+TRM cells in human small intestine allografts have survived for >1 year (18, 19). The donor and host resident T cell turnover kinetics in human intestinal allografts are correlated with clinical outcomes (20). In the absence of rejection, donor-derived graft-versus-host (GvH)–reactive T cells have been enriched and have persisted long term in human intestinal grafts (20). Early expansion of GvH clones in grafts was correlated with rapid replacement of donor antigen-presenting cells by the recipient (20). Graft rejection was associated with transient infiltration of the recipient host-versus-graft–reactive CD28+NKG2DhighCD8+T cells, which could acquire a steady-state TRM phenotype (20). In human lung transplantation, Snyder et al. (21) reported that the persistence of donor lung TRM cells was associated with the decreased incidence of clinical primary graft dysfunction and acute cellular rejection in clinics. Abou-Daya et al. (22) recently demonstrated that antigen-specific CD8+ TRM cells indeed form in allografts and have greatly contributed to the following chronic graft rejection in a mouse kidney transplantation model in which kidney allografts undergo delayed chronic rejection.
Whether the formerly induced alloreactive CD4+TRM cells and/or CD8+TRM cells in recipient nonlymphoid organs or tissues such as skin alone could mediate allogeneic organ graft rejection has never been investigated. Here, we reported that the induced long-lived alloreactive TRM cells, especially CD4+TRM cells in recipient skin tissue alone, could be sufficient to rapidly reject allogeneic skin and heart grafts in immunodeficient severe combined immunodeficient (SCID) mice that received a piece of the alloreactive TRM cell–containing major histocompatibility complex (MHC)–matched or syngeneic skin tissue. Our present studies indicate that long-lived alloreactive TRM cells resident in recipient nonlymphoid organs or tissues may contribute significantly to organ allograft rejection even in the absence of lymphocytes in lymphoid organs and tissues.
RESULTS
Long-lived CD103+CD69+TRM cells in skin induced by allogenic skin grafting
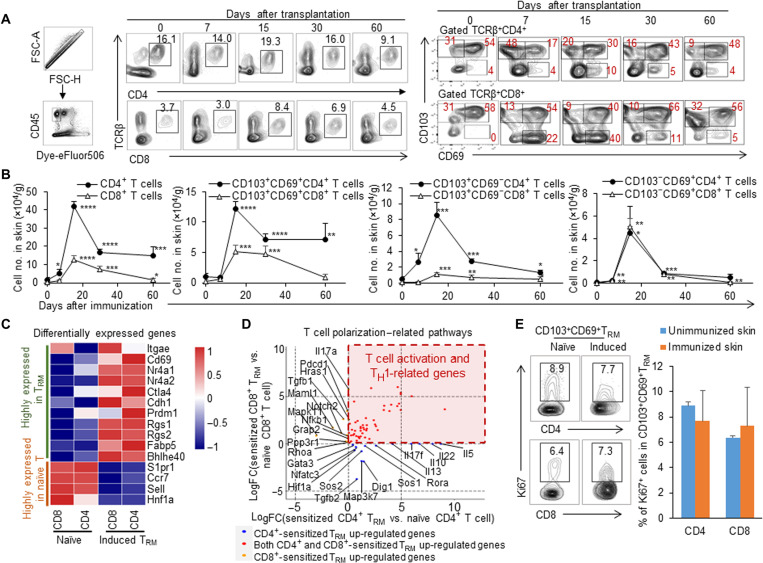
To investigate whether more alloreactive TRM cells in the recipient skin tissue could be induced by immunization with allogeneic skin tissue grafting, we immunized BALB/c mice, grafting an MHC-mismatched C57BL/6 (B6) mouse tail skin, and analyzed the cell numbers and phenotypes of the accumulated T cells in the local recipient skin tissue at 0, 7, 15, 30, and 60 days after immunization (Fig. 1A). The mice at time point day 0 after transplantation were not immunized and served as controls. The allogeneic skin grafts were rejected by B6 or BALB/c recipient mice around 10 days after transplantation (fig. S1), as reported previously (23). We found that the cell numbers of CD4+ and CD8+ T cells in recipient skin rapidly increased from day 7 and reached the summit at day 15. They then began to decrease and persisted at high levels (3 to 10 orders of magnitude more than the unimmunized mice) for the long term, as detected at 60 days after immunization (Fig. 1B). Consistently, these accumulated T cells increasingly expressed the key classical TRM cell surface makers CD103 and CD69; the percentage of CD103+CD69+T cells in TCRβ+CD4+T cells significantly increased from days 7 to 60 (n = 4, P < 0.01) (Fig. 1A). The predominate cell subpopulation of the long-lived alloreactive CD4+ T cells in the skin tissue after allogeneic graft challenging consists of CD103+CD69+CD4+ T cells. The numbers of CD103+CD69+CD4+T cells in skin tissue of immunized mice were about seven times that of the numbers of these cells in the skin of the unimmunized control mice 60 days after immunization (Fig. 1B). Similarly, a high percentage of CD103+CD69+ T cells in TCRβ+CD8+ T cells was observed in the skin tissue of the immunized mice 15 days after skin grafting (Fig. 1A). The numbers of CD103+CD69+CD8+ T cells in the skin of immunized mice were around four times those of the numbers of these cells in the unimmunized control mice 60 days after immunization (Fig. 1B). The numbers of CD103+CD69+CD4+ T cells, significantly higher than those of CD103+CD69+CD8+ T cells resident in the local skin of the immunized mice, indicated that the predominant subpopulation of the long-term–surviving T cells in recipient skin tissue was CD4+ T cells, specifically CD103+CD69+CD4+ T cells, in this immunized mouse model. To explore whether TRM cells exist in the long-distance skin tissue of the immunized mice, we detected levels of CD4+ or CD8+TRM cells in the skin tissues at around 0 to 1, 1 to 2, and 2 to 3 cm from the immunized point (fig. S2A). We found that most CD103+CD69+, CD103+CD69−, and CD103−CD69+ cells of CD4+ or CD8+ T cells were located within 2 cm of the immunized site. There was no significant change in the levels of these TRM cells in skin tissues located between 0 to 1 and 1 to 2 cm, while there were few induced TRM cells in the distal skin tissue (>2 cm from the immunized site), which were similar to those in the unimmunized mice (fig. S2B). On the other hand, to see whether donor-derived T cells were present in the recipient skin tissue, we detected donor and recipient MHC I+ cells in T cell receptor–positive (TCR+) cells in the immunized local BALB/c skin tissue 30 days after B6 skin grafting. There were no donor-derived TCR+ cells in the local recipient skin tissue of the immunized mice (fig. S3), indicating the efficient allograft rejection by immunocompetent recipient mice without any immunosuppression.
Fig. 1. Long-lived CD103+CD69+TRM cells in recipient skin tissue induced by allogenic skin grafting.
BALB/c mice were immunized with C57BL/6 mice tail skin. The percentages and cell numbers of CD4 and CD8 T or TRM cells in recipient skin tissue were analyzed 0, 7, 15, 30, and 60 days after immunization; the 2 cm–by–2 cm recipient skin samples were taken around the immunized site. The skin samples on day 0 represent skin tissues from the unimmunized mice. (A) Flow cytometric analysis of CD45+, CD4+TCRβ+, CD8+TCRβ+, CD103+, CD69+, and CD103+CD69+ T cells in the recipient skin of the immunized BALB/c mice from days 0 to 60. (B) Quantification of cell number of CD4+TCRβ+, CD8+TCRβ+, CD103+CD69+, CD103+CD69−, and CD103−CD69+cells in recipient skin of the immunized BALB/c mice. (C) The heatmap of highly expressed genes in T effector cells and TRM cells. q < 0.05. (D) Dot plot showing the differentially expressed genes in T cell polarization–related pathways in the induced TRM. The blue, red, and orange colors represent the up-regulated genes in induced CD4+ TRM cells, up-regulated genes in both induced CD4+ and CD8+ TRM cells, and up-regulated genes in induced CD8+ TRM cells, respectively. (E) Flow cytometric analysis of Ki67 expression in CD4+/CD8+CD69+ T cells from TRM of unimmunized mice and induced TRM of immunized mice at 60 days after immunization. Data are representative of independent experiments (n ≥ 3). Data are means ± SD. *P < 0.05, **P < 0.01, ***P < 0.001 , and ****P < 0.0001.
To determine whether these long-term–surviving alloreactive T cells in recipient skin tissue induced by allogeneic skin grafting are indeed the classically defined TRM cells, we sorted CD4+/CD8+CD62L+CD44low T cells from the spleen and CD4+/CD8+CD69+ T cells from the skin of BALB/c mice at days 0 and 100 after immunization with allogeneic B6 skin and performed RNA sequencing (RNA-seq) assays. The DEGseq was used to analyze the differentially expressed genes. To confirm the characteristics of induced TRM, we compared the TRM-related genes previously reported in various mouse models (24–29). The result showed that the expressions of Itgae, Cd69, Nr4a1 (Nur77), Nr4a2, Ctla4, Cdh1, Cd244, Prdm1, Rgs, Fabp5, and Bhlhe40 are up-regulated, and S1pr1, Ccr7, Sell, and Hnf1a were down-regulated in induced CD8+ TRM (Fig. 1C and fig. S4). These up- and down-regulated genes in the induced CD8+ TRM in our transplant mouse model were well in line with the reported results of induced CD8+ TRM in an infection mouse model, compared with naïve, central memory, and effector memory CD8+ T cells (fig. S5A) (30). Because the reported marker gene expression profile for the skin-resident CD4+ TRM was absent, we used the marker genes identified in CD8+ TRM cells to see their expression patterns in the induced CD4+ TRM in the immunized skin. Most of these genes were similarly regulated in the induced CD4+ TRM as those in CD8+ TRM except for CD244. The up- and down-regulated genes in induced CD4+ TRM in our model were identical with the reported results with that of human CD4+CD69+ TRM cells in lungs, which were compared with splenic CD4+CD69− T cells (fig. S5B) (12). Thus, most of these long-term–surviving CD4+ and CD8+ T cells displayed the TRM phenotype and the recognized marker gene expression. Compared with naïve T cells, the T cell polarization analysis showed that both induced CD4+ and CD8+ TRM up-regulate T cell activation and T helper 1 (TH1)–related genes. In addition, the induced CD4+ TRM cells highly expressed the TH2-type cytokines such as interleukin-10 (IL-10) and IL-5 and the TH17-type cytokines such as IL-22 and IL-17f (Fig. 1D). Furthermore, the percentage of Ki67+ cells in long-term–living CD103+CD69+CD4+ CD8+ T cells (60 days after immunization) was similar to that of naïve CD103+CD69+CD4+ CD8+ T cells in unimmunized mice as detected by intracellular flow cytometry (Fig. 1E), thus supporting the self-renewal potentiality of these surviving T cells. Therefore, immunization with allogeneic skin grafts resulted in a significant accumulation of long-lived TRM cells in skin. These induced CD4+ and CD8+ TRM cells had tissue-resident memory characteristics in terms of cell surface phenotypes, transcriptional profiles, and potential self-renewal ability.
Immunized skin tissue endowed immunodeficient mice to reject skin allografts with low specificity
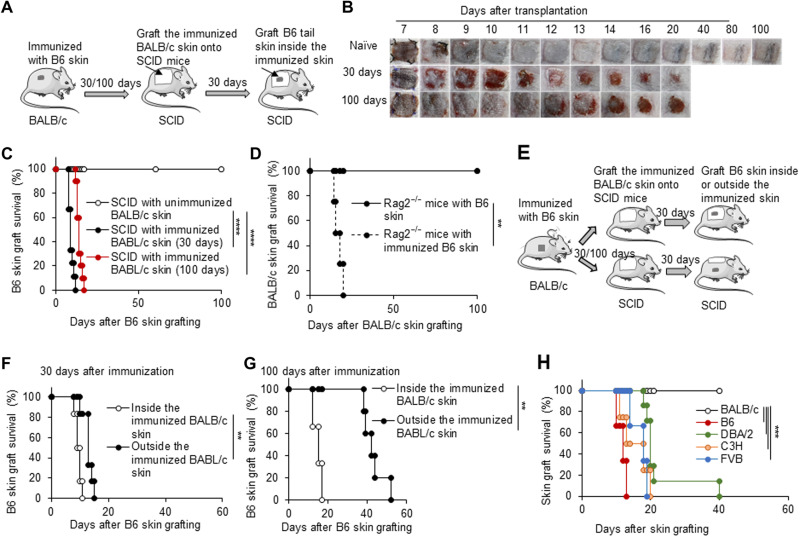
To explore the biofunction of the induced alloreactive TRM cells resident in recipient skin tissue during the secondary anti-alloantigen immune response, we established a novel mouse model that contains only alloreactive TRM cells in skin tissue without T and B lymphocytes in lymphoid tissues and organs by grafting a piece of the skin tissue from the alloskin-immunized mice (immunized skin) to MHC-matched or syngeneic immunodeficient SCID or Rag2 knockout (KO) mice (Fig. 2A). Thirty or 100 days after BALB/c mice had been immunized with B6 skin, BALB/c skin (2 cm by 2 cm) from either the unimmunized or immunized mice was transplanted onto MHC-matched SCID mice. After another 30 days or so, allogeneic B6 or other strain tail skin grafts were grafted onto these SCID mice, respectively (Fig. 2A). When B6 tail skin was transplanted inside the BALB/c skin grafts in SCID mice, allogeneic B6 skin grafts were rapidly rejected (within 17 days) by SCID mice that had received the immunized BALB/c skin, regardless of whether the BALB/c skin was gained from immunized mice 30 or 100 days after immunization, but B6 skin grafts were accepted for the long term by SCID mice onto which was transplanted the unimmunized BALB/c skin (P < 0.0001; Fig. 2, B and C). Conversely, after the B6 mice were immunized with allogeneic BALB/c skin, we then transplanted the immunized B6 skin onto syngeneic Rag2KO mice. Another 30 days later, allogeneic BALB/c tail skin was transplanted onto these Rag2KO mice. The Rag2KO mice that received the immunized B6 skin tissue also rejected the allogeneic BALB/c skin rapidly, but Rag2KO mice that received the unimmunized B6 skin failed to do so (P < 0.01; Fig. 2D).
Fig. 2. Immunized skin tissue endowed immunodeficient mice to reject skin allografts with low specificity.
(A) Animal model: (i) By 30 or 100 days after BALB/c mice were immunized with C57BL/6 tail skin, one piece of the immunized BALB/c skin (2 cm by 2 cm) was harvested and grafted onto MHC-matched SCID mice; (ii) after SCID mice were recovered for about 30 days, a C57BL/6 tail skin was transplanted inside the immunized skin in SCID mice. (B) The photos of C57BL/6 skin grafts in SCID recipients with the unimmunized or immunized BALB/c skin tissue. (C) C57BL/6 skin graft survival in SCID recipients with the unimmunized or immunized BALB/c skin tissue (n = 8 to 10 per group). (D) BALB/c skin graft survival on Rag2KO recipients with syngeneic unimmunized or immunized C57BL/6 skin tissue (n = 4 per group). (E) The animal model was the same as in (A), except the C57BL/6 tail skin was transplanted inside or outside the immunized skin. (F) The survival of C57BL/6 skin grafts that were grafted either inside or outside the immunized BALB/c skin tissue gained from the immunized BALB/c mice 30 days after immunization. (G) The survival of C57BL/6 skin grafts that were grafted either inside or outside the immunized BALB/c skin tissue gained from the immunized BALB/c mice 100 days after immunization. (H) The survival of C57BL/6, FvB, C3H, DBA/2, and BALB/c skin grafts that were grafted inside the immunized BALB/c skin tissue in SCID mice. n = 6 per group. **P < 0.01, ***P < 0.001, and ****P < 0.0001 between the indicated groups.
We compared the TRM cell–mediated rejection efficiency of allogeneic B6 skin grafts transplanted either inside or outside the immunized BALB/c skin grafts in SCID recipient mice (Fig. 2E). SCID mice that received the immunized BALB/c skin harvested 30 days after the initial immunization efficiently rejected B6 skin grafts within 15 days, whether or not B6 skin was grafted inside or outside the immunized BALB/c skin (Fig. 2F). However, SCID mice that received BALB/c skin harvested 100 days after the initial immunization quickly rejected B6 skin grafted inside the BALB/c skin within 20 days but rejected B6 skin grafted outside the BALB/c skin somewhat more slowly (within 50 days, P < 0.01; Fig. 2G), possibly because of the decreased cell abundance of TRM cells in the immunized BALB/c skin tissue 60 days after immunization. Regardless, immunodeficient SCID mice that received the previously immunized BALB/c skin tissue gained the ability to efficiently reject allogeneic skin grafts from the same donor used to immunize BALB/c mice. Thus, the MHC-matched or syngeneic immunized skin tissue endowed T/B cell–deficient recipient mice with the ability to reject subsequent allogeneic grafts, regardless of whether the allogeneic skin grafts were transplanted inside or outside the immunized skin tissue. On the other hand, when we grafted either B6(H-2b) or the third-party skin from FvB(H-2q), C3H(H-2k), and DBA/2(H-2d) mice inside the immunized BALB/c skin of the SCID recipients, respectively, the immunized skin tissue efficiently rejected the third-party skin grafts too, although they were in a slower rejection kinetics compared with B6 skin (P < 0.001; Fig. 2H), possibly because of the presence of the induced long-lived polyclonal and/or bystander T cells resident in the immunized skin tissue and/or the cross-reactivity of TRM cells (31, 32).
Induced TRM cells in skin proliferated and mediated skin allograft rejection
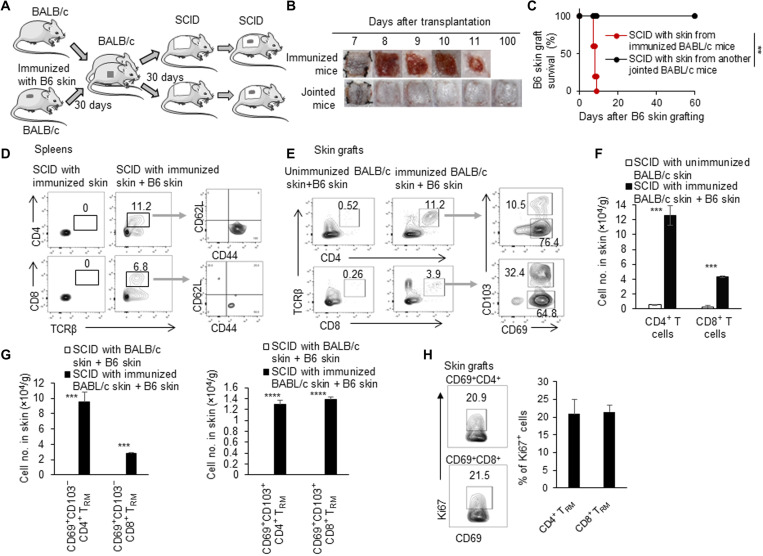
To determine whether it is the induced TRM cells resident in skin tissue that mediate allograft rejection in immunodeficient recipient mice, we carried out experiments with parabiosis mouse models as TRM cell–containing skin tissue donors (Fig. 3A). First, we connected a previously immunized BALB/c mouse and a normal BALB/c mouse for 30 days. We then transplanted the skin tissue (2 cm by 2 cm) from either the immunized mice or the jointed unimmunized mice onto MHC-matched SCID mice. Another 30 days later, these SCID mice were grafted with allogeneic B6 tail skin (Fig. 3A). SCID mice that received skin from the immunized mice quickly rejected B6 skin allografts, while SCID mice that received skin from the jointed unimmunized mice failed to do so (P < 0.01; Fig. 3, B and C), suggesting that the induced TRM cells reside in the skin tissue in the long term and tend not to circulate in the steady state, as indicated by the parabiosis mouse model.
Fig. 3. Induced TRM cells in skin proliferated and mediated skin allograft rejection.
(A) Animal model: (i) We connected a previously immunized BALB/c mouse and an unimmunized BALB/c mouse. (ii) Thirty days later, the skin (2 cm by 2 cm) tissues from both immunized mice and jointed mice were transplanted onto two other SCID mice, respectively. (iii) C57BL/6 tail skin was then grafted onto these SCID mice. (B) Photos of C57BL/6 skin grafts. (C) C57BL/6 skin graft survival in SCID recipients with the unimmunized and immunized BALB/c skin tissue (n = 9 to 10 per group). (D) Flow cytometric analysis of CD4+, CD8+, CD62L+, and CD44+ T cells in spleens in SCID mice that received immunized BALB/c skin with or without C57BL/6 skin. (E) Flow cytometric analysis of CD4+, CD8+, CD103−CD69+, and CD103+CD69+ T cells in C57BL/6 skin grafts in SCID mice that received unimmunized and immunized BALB/c skin. (F) Quantification of numbers of CD4+ and CD8+ T cells in C57BL/6 skin grafts in SCID mice that received unimmunized and immunized BALB/c skin. (G) Quantification of numbers of CD103−CD69+ and CD103+CD69+ T cells in C57BL/6 skin grafts in SCID mice that received unimmunized and immunized BALB/c skin. (H) Flow cytometric analysis of Ki67 in CD4+CD69+ and CD8+CD69+ T cells in C57BL/6 skin grafts in SCID mice that received immunized BALB/c skin. Data are representative of independent experiments (n ≥ 3). Data are means ± SD. **P < 0.01, ***P < 0.001, and ****P < 0.0001, compared between the indicated groups.
Next, to see whether the immunized skin tissue promoted repopulation of lymphocytes in peripheral lymphoid tissues or organs in steady state or immune response phase, we analyzed the T cell subsets in the spleens and skin grafts of SCID mice that received the immunized MHC-matched BALB/c skin with or without B6 skin grafts. There were no detectable CD4+ and CD8+ T cells in the spleens of SCID mice that received the immunized BALB/c skin for 30 days or so without allogeneic B6 skin challenging, further supporting that TRM cells in skin tissue did not egress and circulate in the peripheral immune system. However, SCID mice that received the immunized BALB/c skin with allogeneic B6 skin had significantly higher percentages of CD4+ and CD8+ T cells in the spleens 10 days after B6 skin grafting (6 to 12%; Fig. 3D), indicating that TRM cells in skin tissue recirculated in the peripheral immune system during the secondary immune response. Furthermore, most of these CD4+ and CD8+ T cells in spleens of the immunized BALB/c skin–transplanted SCID mice after allogeneic B6 skin grafting showed activated/memory CD62LlowCD44high T cell phenotype (Fig. 3D). There were about 11% CD4+ T cells and about 4% CD8+ T cells in B6 skin allografts in SCID mice with the immunized BALB/c skin tissue at 10 days after grafting but only about 0.5% CD4+ T cells and about 0.3% CD8+ T cells in B6 skin grafts in SCID mice that received unimmunized BALB/c skin tissue (Fig. 3E). The number of CD4+ T cells in B6 skin grafts was about 10 × 104 cells/g, and the number of CD8+ T cells in B6 skin grafts was about 0.4 × 104 cells/g (Fig. 3F). This indicated that most of the expanded and infiltrated T cells were CD4+ T cells in this transplantation mouse model. Almost all of these infiltrated T cells were CD69+ (Fig. 3, E and G), but a higher percentage of CD8+ T cells than CD4+ T cells expressed the CD103 molecule. Some of these T cells expressed the cell proliferation protein ki67 (Fig. 3H), which may explain the increased cell numbers in the grafts. Thus, the induced alloreactive TRM cells were mainly resident in the skin tissue in the physiological situation but significantly expanded, recirculated in the periphery lymphoid tissues, and infiltrated into the allograft during rejection.
Induced CD4+ TRM cells alone were sufficient to reject skin allografts
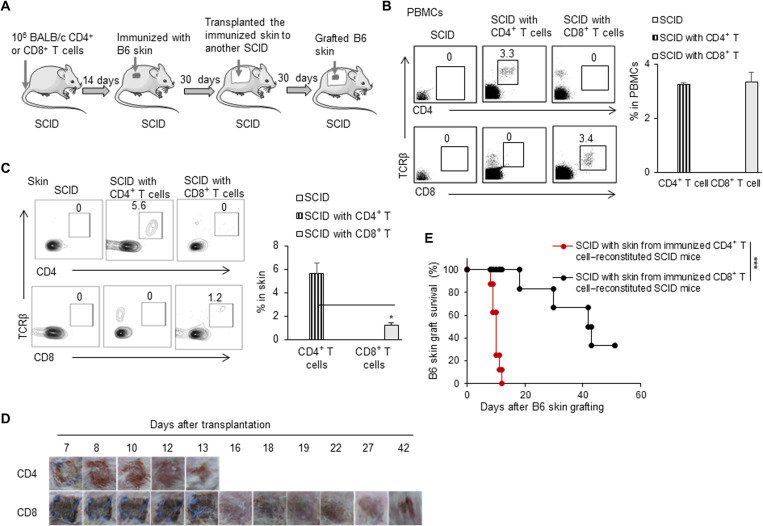
To explore the roles of the induced alloreactive CD4+ TRM and CD8+ TRM cells in skin tissues in mediating allograft rejection, we adoptively transferred either the sorted BALB/c CD4+ T cells or CD8+ T cells to MHC-matched SCID mice to reconstitute their peripheral CD4+ and CD8+ T cell pools, respectively (Fig. 4A). Before immunization, the reconstituted SCID mice showed about 3% CD4+ T cells or CD8+ T cells in the peripheral blood on a relatively equal cell level (Fig. 4B). Thirty days after immunization with allogeneic B6 skin tissue, the recipient skin tissue of the CD4+ T cell– or CD8+ T cell–reconstituted SCID mice had about 5% CD4+ TRM cells or 1% CD8+ TRM cells, respectively (Fig. 4C). The low levels of the induced CD8+ TRM cells in the skin tissue of the CD8+ T cell–reconstituted SCID mice likely result from the absence of the help of CD4+ T cells, as it was previously reported in other models that CD4+ T cells help the formation of lung-resident memory CD103+CD8+ T cells during influenza virus infection (33). Thirty days after immunization, we grafted these CD4+ TRM cell– or CD8+ TRM cell–reconstituted SCID mouse skin tissues to the secondary SCID mice; allogeneic B6 skin was transplanted onto these mice as shown in Fig. 4A. SCID mice with the induced CD4+ TRM–reconstituted skin tissue efficiently rejected allogeneic B6 skin grafts, but SCID mice with the induced CD8+ TRM–reconstituted skin tissue rejected allogeneic B6 skin grafts much more slowly (P < 0.001; Fig. 4, D and E). These results showed that long-lived alloreactive CD4+ TRM cells resident in the skin tissue were sufficient to mediate allograft rejection.
Fig. 4. Induced CD4+ TRM cells alone were sufficient to reject skin allografts.
(A) Animal model: (i) We transfused 1 × 106 CD4+ or CD8+ T cells into SCID mice, (ii) immunized these SCID with C57BL/6 tail skin, and (iii) transplanted the immunized skin onto another naïve SCID; after SCID mice recovery, C57BL/6 tail skin was transplanted onto those SCID mice inside the immunized skin. (B) Flow cytometric analysis of TCRβ+CD4+ and TCRβ+CD8+ T cells in peripheral blood mononuclear cells (PBMCs) of CD4+ T cell– or CD8+ T cell–reconstituted SCID. (C) Flow cytometric analysis of CD4+ TRM and CD8+ TRM in the skin tissue of CD4+ T cell– or CD8+ T cell–reconstituted SCID mice 30 days after C57BL/6 skin immunization. (D) Photos of C57BL/6 skin grafts in SCID mice that received skin tissues from either CD4+ T cell– or CD8+ T cell–reconstituted SCID mice 30 days or so after C57BL/6 skin immunization. (E) C57BL/6 skin graft survival in SCID mice that received skin tissues from either CD4+ T cell– or CD8+ T cell–reconstituted SCID mice 30 days or so after C57BL/6 skin immunization (n = 6 to 8 per group). Data are representative of two to three independent experiments. Data are means ± SD. *P < 0.05 and ***P < 0.001, compared between the indicated groups.
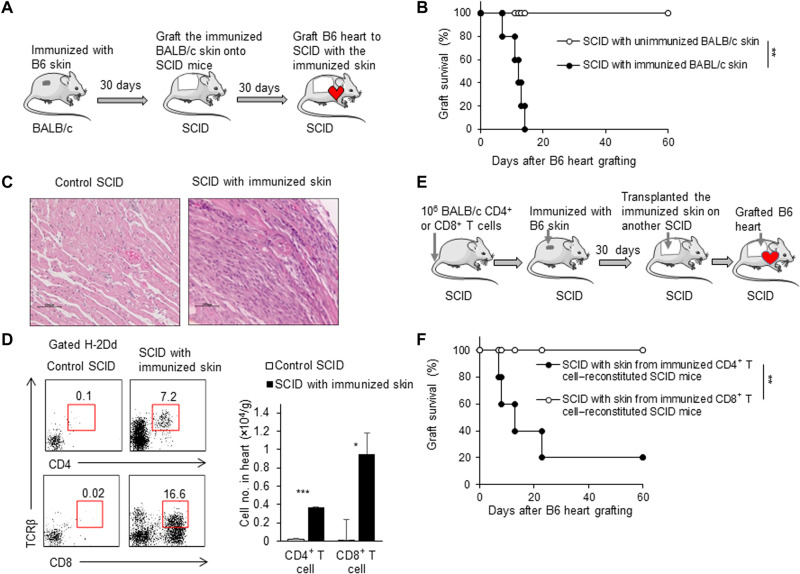
Induced TRM cells or CD4+ TRM cells in skin rejected heart allografts
It is interesting and important to understand whether alloreactive TRM cells resident in skin tissue could reject allogeneic heart or other organ grafts. We thus performed heterotopic mouse heart transplantation instead of skin transplantation in this mouse model to address this issue (Fig. 5A). Unexpectedly, SCID mice with a piece of the MHC-matched immunized BALB/c skin tissue rapidly rejected allogeneic heart grafts (within 14 days), but SCID mice with the unimmunized BALB/c skin failed to reject allogeneic heart grafts by the end point of the experiment (P < 0.01; Fig. 5B). Pathological examination showed severe leukocyte infiltration in heart grafts of SCID mice with the immunized BALB/c skin, whereas there was no obvious leukocyte infiltration in heart grafts of SCID mice with the unimmunized BALB/c skin (Fig. 5C). The severe leukocyte infiltration in heart grafts of SCID mice with the immunized BALB/c skin was further confirmed by flow cytometry assays showing a large number of CD4+ T and CD8+ T cells in heart grafts of SCID mice with the immunized BALB/c skin during rejection (Fig. 5D). In addition, we used the CD4+ T cell– or CD8+ T cell–reconstituted SCID mouse model to examine the effect of the induced CD4+ TRM or CD8+ TRM in the skin tissue to mediate heart allograft rejection (Fig. 5E). The result showed that SCID mice grafted with the immunized SCID skin tissue of the B6-immunized CD4+ T cell–reconstituted SCID mice could reject B6 heart grafts (four of five mice), while SCID mice grafted with the immunized SCID skin tissue of the B6 skin–immunized CD8+ T cell–reconstituted SCID mice failed to reject B6 heart grafts (P < 0.01; Fig. 5F). Therefore, the induced alloreactive TRM cells or CD4+ TRM cells resident in the skin tissue in the long term were sufficient to mediate acute heart allograft immune rejection without the involvement of preexisting lymphocytes in lymphoid organs and tissues.
Fig. 5. Induced TRM cells or CD4+ TRM cells in skin rejected heart allografts.
(A) Animal model: (i) BALB/c mice were immunized by C57BL/6 tail skin; (ii) after BALB/c mice were immunized for 30 days, the skin tissue (2 cm by 2 cm) was taken from immunized BALB/c mice and grafted onto SCID mice; (iii) after SCID mice recovery, C57BL/6 heart grafts were transplanted into SCID mice. (B) C57BL/6 heart graft survival (n = 6 per group). (C) Pathological examination of heart grafts in SCID mice with unimmunized and immunized skin tissues. (D) Flow cytometric analysis and the cell numbers of recipient CD4+ and CD8+T cells that infiltrated into the heart grafts. (E) Animal model: (i) We transfused 1 × 106 CD4+ or CD8+ T cells into SCID mice, (ii) immunized these SCID mice with C57BL/6 tail skin, and (iii) transplanted the immunized skin onto another naïve SCID mouse; (4) after SCID mice recovery for 30 days, C57BL/6 heart grafts were transplanted into those SCID mice. (F) C57BL/6 heart graft survival in SCID mice that received skin tissues from either CD4+ T cell– or CD8+ T cell–reconstituted SCID mice 30 days or so after C57BL/6 skin immunization (n = 5). Data are representative of three independent experiments (n ≥ 3). Data are means ± SD. *P < 0.05, **P < 0.01, and ***P < 0.001, compared between the indicated groups.
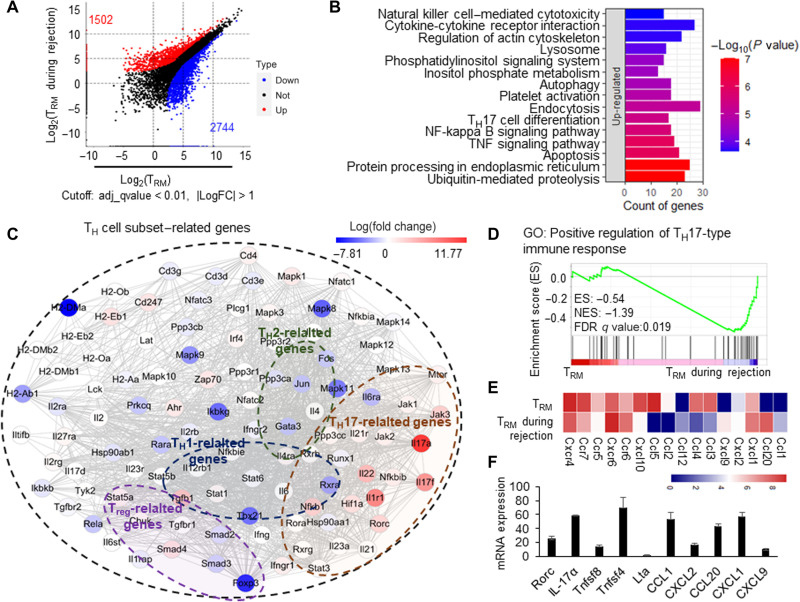
CD4+ TRM cells in skin acquired a TH17-like transcriptional profile during the secondary immune response
To explore the major molecular mechanisms for skin alloreactive CD4+ TRM cell–mediated allograft rejection, we sorted the quiescent CD4+ TRM cells in skin without allograft challenge and the activated CD4+ TRM cells during allograft rejection to perform RNA-seq assays. The differential gene analysis showed 1502 up-regulated and 2744 down-regulated genes in the activated CD4+ TRM cells during rejection (Fig. 6A). We found that the signaling pathways of protein processing in endoplasmic reticulum and cytokine-cytokine receptor interaction were up-regulated significantly, which indicated that the activated CD4+ TRM cells synthesize and secrete cytokines or other proteins in large quantities during allograft rejection (Fig. 6B and fig. S6A). In addition, the up-regulated genes in CD4+ TRM cells during rejection enriched the inflammatory pathways, including TH17 cell differentiation and nuclear factor kappa B signaling pathways (Fig. 6B and fig. S6A). When we further conducted network analysis of the genes relevant to TH cell subset polarizations, we found that TH17-related genes [IL-17a, IL-17f, IL-21, IL-22, and the key transcription factor retinoid-related orphan receptor c (RORc) (34, 35)] were up-regulated in CD4+ TRM cells during rejection, whereas the related genes of TH1 (T-bet and Stat3), TH2 (IL-13, IL-5, Gata3, Fos, and Jun), and regulatory T cells (Tregs) (Smad2, Foxp3, and Tgfbr1) were down-regulated in CD4+ TRM cells during rejection (Fig. 6C and fig. S6B). Furthermore, we performed gene set enrichment analysis (GSEA) on genes that were relevant to TH cell polarization and regulated in CD4+ TRM cells and found that the TH17-related genes were significantly enriched in CD4+ TRM cells during rejection (Fig. 6D). Figure 6E summarizes the major changes of cytokines, chemokines, and transcription factors in activating CD4+ TRM cells compared with the inactivated, induced CD4+ TRM cells. The changes of the major interesting genes in the activated CD4+ TRM cells during rejection were confirmed by real-time polymerase chain reaction analysis (Fig. 6F). To verify the TH17-like phenotype of CD4+ TRM cells in the skin during rejection, we performed flow cytometry analysis after intracellular cytokine staining. After the CD4+ TRM cells isolated from the immunized BALB/c skin of SCID mice or B6 skin grafts inside the immunized BALB/c skin of SCID mice were stimulated with phorbol 12-myristate 13-acetate/ionomycin and Golgi-plug in vitro for 6 hours, the intracellular staining of IL-17a or IL-17f was performed and analyzed using flow cytometry (36). The results showed that a considerable proportion of CD69+CD4+ TRM cells (about 20%) expressed IL-17a and IL-17f during allograft rejection, whereas CD69+CD4+ TRM cells in the immunized BALB/c skin of SCID mice without B6 skin grafting did not show detectable expression of IL-17a and IL-17f (Fig. 7A). These results collectively suggested that the induced alloreactive CD4+ TRM cells in the immunized skin favorably differentiated to TH17-like cells, likely in a RORγt pathway–dependent manner during their secondary immune response to allografts.
Fig. 6. CD4+ TRM cells in skin acquired a TH17-like transcriptional profile during the secondary immune response.
(A) Dot plot of induced CD4+ TRM cells and CD4+ TRM cells during rejection showing the differentially expressed genes. The red and blue colors represent the up- and down-regulated genes in CD4+ TRM during rejection, respectively. (B) Top 15 up-regulated pathways by differentially expressed genes of CD4+ TRM during rejection. KEGG database was used to map the signaling pathways. (C) Network summary of TH cell subset–related genes in CD4+ TRM during rejection; the color represents the log(fold change) of differentially expressed genes. (D) The GSEA enrichment of positive regulation of TH17-type immune response in CD4+ TRM during rejection. GO, gene ontology. (E) The heatmap of differentially expressed chemokines and chemokine receptors. (F) Relative expression of RORc, IL-17a, Tnfsf8, Tnfsf4, Lta, CCL1, CXCL2, CCL20, CXCL1, and CXCL9 in CD4+ TRM during rejection. The gene expressions in CD4+ TRM in skin tissues were used as control 1. Data are representative of three independent experiments (n ≥ 3). Data are means ± SD.
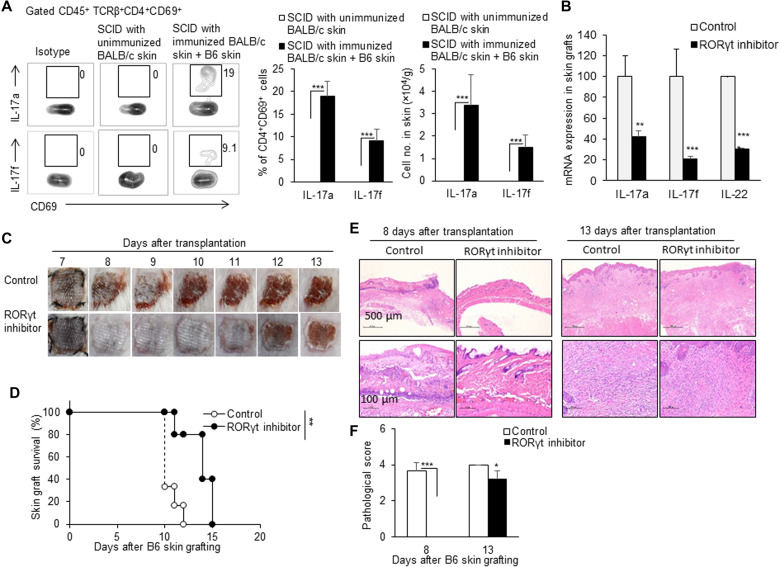
Fig. 7. RORγt inhibitor delayed TRM cell–mediated allograft rejection.
(A) Flow cytometric analysis of cytokines IL-17a and IL-17f in CD4+ TRM cells in skin grafts in SCID mice that received a piece of MHC-matched, immunized BALB/c skin tissue during the secondary allograft rejection. The percentages of IL-17a+ or IL-17f+ in CD4+CD69+ TRM cells and the cell number of these cells in skin are shown. (B) mRNA expressions of IL-17A, IL-17F, and IL-22 in skin grafts in the corn oil control group and RORγt inhibitor group. RORγt inhibitor (10 mg/kg) was given once every 2 days via oral gavage. (C) Photos of C57BL/6 skin grafts in the control group and RORγt inhibitor group. (D) C57BL/6 skin graft survival in control and RORγt inhibitor–treated SCID mice (n = 5 per group). (E) Pathological examination of the skin grafts in the control group and RORγt inhibitor group on days 8 and 13. (F) Quantification of H&E staining corresponding to the control group and RORγt inhibitor group on days 8 and 13. Data are representative of three independent experiments (n ≥ 3). Data are means ± SD. **P < 0.01 and ***P < 0.001.
RORγt inhibitor delayed TRM cell–mediated allograft rejection
Considering the involvement of IL-17, IL-21, and IL-22 in allograft rejection in other transplantation models [(37–39) and reviewed in (40, 41)] and the key role of TH17-specific transcription factor RORγt in mastering these cytokine expressions in T cells (42), we used a RORγt inhibitor to block the RORγt pathway in the immunized BALB/c skin–grafted SCID mouse model to test the roles of RORγt pathway in the induced TRM cells in allogeneic graft rejection. The treatment with RORγt inhibitor via oral gavage significantly reduced the RNA expressions of cytokines IL-17A, IL-17F, and IL-22 in B6 skin grafts in SCID mice with immunized BALB/c skin tissue, compared with mice that received corn oil gavage alone (Fig. 7B). The RORγt inhibitor–treated, immunized BALB/c skin–grafted SCID mice showed a significantly prolonged allogeneic B6 skin graft survival time, compared with the control immunized BALB/c skin–grafted SCID mice without the RORγt inhibitor treatment (P < 0.01; Fig. 7, C and D). Consistently, pathological examination showed that there was less leukocyte infiltration in allogeneic B6 skin grafts in the RORγt inhibitor–treated, immunized BALB/c skin–grafted SCID mice than those in the RORγt inhibitor–untreated control mice on both days 8 and 13 (Fig. 7E). The skin tissue inflammation scores of the control group were significantly higher than those of the RORγt inhibitor group (Fig. 7F). On the other hand, Rag2KO mice that received syngeneic immunized IL-17KO mouse skin rejected allogeneic grafts significantly more slowly than Rag2KO mice that received syngeneic immunized wild-type mouse skin (P < 0.01; fig. S7). Therefore, inhibition of RORγt pathway or IL-17 deficiency could significantly attenuate the allograft rejection mediated only by the induced alloreactive TRM cells residing in the immunized skin to a certain extent.
DISCUSSION
The formation and the biological roles of TRM cells resident within allografts have been elegantly studied (20–22, 43, 44). With polychromatic flow cytometry and high-throughput TCR sequencing technology, Zuber et al. (20) nicely demonstrated that the antidonor alloreactive recipient T cells entered human intestinal allografts, expanded during rejection, and acquired the TRM phenotype. Here, we established a novel mouse model in which allogeneic skin or heart was grafted to an immunodeficient SCID or Rag2KO mouse with a piece of immunized MHC-matched or syngeneic skin tissue harvested from wild-type mice that were immunized with allogeneic skin grafting 30 or 60 days earlier. Meanwhile, the immunodeficient SCID or Rag2KO mice with a piece of unimmunized MHC-matched or syngeneic skin tissue were used as a negative control. In this unique transplantation mouse model, we could study the roles of the induced long-lived alloreactive TRM cells in skin in allograft rejection without the bother or involvement of T/B lymphocytes in lymphoid organs and tissues. This transplantation mouse model will be of significance for studies on the specific roles of the long-lived alloreactive TRM cells in nonlymphoid organs or tissues or for the evaluation and identification of novel drugs specifically targeting TRM cell–mediated allograft rejection. This and other related studies have clearly indicated that the induced alloreactive TRM cells resident within recipient tissues and/or donor allografts could be important reservoirs for alloreactive T cells for the subsequent graft rejection episodes in mice and humans (17–21).
With this unique mouse model, we demonstrated that the long-lived alloreactive TRM cells resident in skin tissue alone could efficiently mediate acute allograft rejection, as determined by allogeneic skin or heart grafts that were rapidly rejected by the immunodeficient SCID or Rag2KO mice with an immunized MHC-matched or syngeneic skin tissue. However, allogeneic skin or heart grafts survived long term in immunodeficient SCID or Rag2KO mice with an unimmunized MHC-matched or syngeneic skin tissue. The efficient acute allograft rejection by the immunized MHC-matched or syngeneic skin tissue, but not by the unimmunized skin tissue, indicated that the induction and maintenance of more alloreactive TRM cells in skin tissue in wild-type mice were achieved by immunization with grafting of allogeneic skin tissue. The induced TRM cells in the skin tissue acutely rejected allogeneic heart grafts and skin grafts outside the immunized skin tissue. Furthermore, although the induced CD8+ TRM cells in the immunized skin tissue could reject allogeneic skin grafts, the induced alloreactive CD4+ TRM cells in the immunized skin tissue alone are sufficient to mediate allogeneic skin and heart graft rejection without the involvement of CD8+ T cells. This is because SCID mice with a piece of syngeneic skin tissue from the immunized CD4+ T cell–reconstituted SCID mice rapidly rejected the subsequent allogeneic skin and heart grafts within 20 days. Considering the high number of CD4+ TRM cells in nonlymphoid organs and tissues, their roles in allogeneic organ graft rejection should be emphasized.
It is true that significantly more CD4+ or CD8+ TRM cells including CD103+CD69+ T cells, CD103+CD69− T cells, and CD103−CD69+ T cells were detected in immunized skin tissue even at 30 or 60 days after immunization. This observation was consistent with the previous reports showing the formation of TRM cells in viral infection (45, 46), solid tumors (47–50), and autoimmune diseases (51). Significantly more long-lived CD4+ TRM cells in skin tissue were induced in our allogeneic skin–grafted wild-type mouse model than CD8+ TRM cells, which is in line with the results that the number of CD4+ TRM cells is more than two orders of magnitude that of CD8+ TRM cells in most barrier tissues (52). When SCID mice were reconstituted with either CD4+ T cells or CD8+ T cells equally in the peripheral blood and then immunized with allogeneic grafting, significantly more CD4+ TRM cells in skin tissue were induced in CD4+ T cell–reconstituted SCID mice than CD8+ TRM cells in skin tissue in CD8+ T cell–reconstituted SCID mice. It is reported that CD4+ TH cells were important for the development of functional CD103+CD8+ TRM cells in the lung airways following respiratory influenza virus infection (33). Whether the low number of CD8+ TRM cells in skin tissue in CD8+ T cell–reconstituted SCID mice is due to the less potential ability of CD8+ T cells to differentiate into CD8+ TRM cells and/or the induction of CD8+ TRM cells requires the help of CD4+ T cells in this model is unclear and needs to be clarified in the future.
Our results showed almost no detectable CD4+ or CD8+ T cells in the spleens of the immunized skin–grafted SCID mice without grafting allogeneic skin (in steady state) but that high levels of activated/memory CD62LlowCD44high CD4+ or CD8+ T cells were observed in the spleens of these mice after grafting allogeneic skin. These results together suggest that the induced long-lived TRM cells are mainly resident in local skin tissue in physiological situation (in the parabiosis mouse model and the immunized skin–grafted SCID mice without allogeneic skin grafting) but could egress from their resident skin tissue and migrate to allografts during immune response (in the immunized skin–grafted SCID mice with subsequent allogeneic skin grafting). This observation is consistent with a series of studies showing that the formed CD4+ or CD8+ TRM cells could rejoin the circulating pool after reactivation in mice and humans (8–11). However, it should be recognized that low levels of circulating CD103−CD69− T cells were present in the immunized and unimmunized mouse skin tissue. Whether these circulating T cells in skin tissue contributed to the allograft rejection in this model is unclear. Because there were similarly low levels of CD103−CD69− T cells in the immunized and unimmunized mouse skin tissue and because SCID mice with a piece of unimmunized skin tissue failed to reject allogeneic grafts, we speculated that these CD103−CD69− T cells in the skin tissue were unlikely to be key to mediating the allograft rejection or contributed at a weak degree, if any.
It is well known that naïve CD4+ T cells would proliferate and differentiate into functionally polarized subsets, including TH1, TH2, TH17, and Treg cells. We found that, in addition to a cell-proliferative ability during induction phase in the first immune response and the activation phase in the secondary immune response, the induced CD4+ TRM cells in the immunized skin tissue have the potential ability to differentiate into TH1 and TH17 cells. RNA-seq assays showed that the key genes and cytokines related to TH17 cells were significantly up-regulated in the activated CD4+ TRM cells in skin tissue during the following allograft rejection; meanwhile, the major gene clusters related to TH2, Treg, and TH1 cells were down-regulated in these cells. Consistently with the gene transcriptional expression, a significantly higher percentage of IL-17a+CD4+ and IL-17f+CD4+ TRM cells was observed during allogeneic skin graft rejection as detected by intracellular staining flow cytometry assays. Thus, the induced alloreactive CD4+ TRM cells in skin are more likely to differentiate into TH17-like cell type during subsequent allograft rejection. This observation is in line with studies showing that the pathogen-induced TRM cells in kidneys had a TH17 transcriptional signature and remarkably exacerbated renal pathogenesis upon experimental glomerulonephritis induction by producing IL-17A (53). Our RNA-seq analysis showed that one of the key TH17-relevant transcription factors, RORγt, was significantly up-regulated in the induced alloreactive CD4+ TRM cells during the following allograft rejection. Meanwhile, the key IL-6/IL-6R downstream and important signaling pathway in promoting the RORγt transcription and expression (54), Janus kinase–signal transducer and activator of transcription 3 (STAT3), was also significantly up-regulated in the activated CD4+ TRM cells. The IL-17 KO recipients show a delayed graft rejection, although some arguments on the roles of IL-17 in graft rejection still exist (37, 55, 56). The treatment with RORγt inhibitor or IL-17 deficiency significantly delayed the TRM cell–mediated skin allograft rejection and decreased the immune cell infiltration in skin allografts in the immunized SCID mouse model. Therefore, targeting RORγt or IL-17 might be a potential approach to inhibit TRM cell–mediated allograft rejection. It should be noticed that the allogeneic grafts were rejected quickly in mice with RORγt inhibitor treatment or IL-17 deficiency. Thus, other effector pathways were certainly involved in TRM-mediated allograft rejection in this model, which needs future study.
In summary, we established a useful mouse model specifically to investigate the allograft rejection mediated solely by TRM cells resident in skin tissue without the involvement of lymphocytes in lymphoid organs and tissues. Significantly higher levels of long-lived alloreactive TRM cells in recipient skin can be induced by grafting allogeneic skin tissue. The sole presence of the induced TRM or CD4+ TRM cells in skin tissue could efficiently mediate acute skin and heart allograft rejection with less antigen-recognizing specificity and high cross-reactivity. The TH17-like cell polarization of the induced CD4+ TRM cells is involved in allograft rejection by CD4+ TRM cells in the skin. Inhibition of RORγt significantly delayed TRM cell–mediated skin allograft rejection. Targeting the RORγt-TH17 signaling pathway might be a potential approach to block TRM cell–mediated allograft rejection and improve organ transplantation outcomes. Nevertheless, recognizing the roles of TRM cells resident in other nonlymphoid tissues in organ allograft rejection is fundamental and of clinical significance.
MATERIALS AND METHODS
Study design
We offered a mouse model to specifically investigate the allograft rejection mediated solely by the defined TRM cells resident in skin tissue without the involvement of lymphocytes in lymphoid organs and tissues. Significantly higher cell numbers of CD103+CD69+CD4+ and/or CD8+ TRM cells in BALB/c skin (termed as immunized skin) were induced by grafting allogeneic skin than by grafting skin from unimmunized BALB/c mice (unimmunized skin). SCID or Rag2KO mice with immunized skin tissue did not show detectable levels of T cells in peripheral blood and spleens in a steady state, but they acutely rejected the following allogenic skin or heart grafts (<17 days), whereas SCID or Rag2KO mice with the unimmunized skin tissue failed to do so. The immunodeficient SCID or Rag2KO mice that received a piece of preimmunized MHC-matched or syngeneic skin tissue could acutely reject the following allogeneic skin or heart grafts, but the immunodeficient SCID or Rag2KO mice that received a piece of unimmunized MHC-matched or syngeneic skin tissue could not reject the following allogeneic skin or heart grafts. With this established mouse model, we found that the sole presence of the induced TRM or CD4+ TRM cells in skin tissue could efficiently mediate acute skin and heart allograft rejection with less antigen-recognizing specificity and high cross-reactivity. The TH17-like cell polarization of the induced CD4+ TRM cells in the skin is involved in allograft rejection. Inhibition of RORγt significantly delayed TRM cell–mediated skin allograft rejection.
Mice
Six- to 8-week-old C57BL/6 (H-2b) and BALB/c (H-2d) mice were purchased from SPF (Beijing) Biotechnology Co. Ltd. (Beijing, China). Six- to 8-week-old SCID (H-2d) and Rag2KO (H-2b) mice were purchased from Beijing HFK Bioscience Co. Ltd. (Beijing, China). Six- to 8-week-old FvB (H-2q), C3H (H-2k), and DBA/2 (H-2d) mice were purchased from Vital River Laboratories (Beijing, China). All mice were maintained in a specific pathogen–free facility and were housed in microisolator cages containing sterilized feed, autoclaved bedding, and water. All experimental manipulations were undertaken in accordance with the Institutional Guidelines for the Care and Use of Laboratory Animals by the Institute of Zoology, CAS (Beijing, China).
Skin transplantation
For immunization, tail skin grafts from C57BL/6 (H-2b) mice were transplanted onto BALB/c (H-2d) mice as described previously (23). For skin transfer, previously immunized BALB/c (H-2d) skin (2 cm by 2 cm) was transplanted on SCID (H-2d) mice. The hair was shaved with a clipper before skin transfer. The direction of the BALB/c (H-2d) hair needed to be easily distinguishable from the direction of the SCID hair, reverse or upward, with three to four stitches on each side. For rejection, tail skin grafts from C57BL/6, FvB (H-2q), C3H (H-2k), and DBA/2 (H-2d) mice were transplanted onto SCID mice as described previously (23). For skin transplantation, grafts were followed by daily visual and tactile inspections. The healthy skin graft was soft and pliable to touch, and the hairs on the graft regrew. For visual inspection, grafts were considered to be rejected when less than 10% remained viable. For visual and tactile inspection, skin grafts were considered as rejected at the time of complete sloughing or when they formed a dry scar. We combined the above two criteria to define the end point of rejection. Survival was expressed as the mean survival time ± SD. Seven days after transplantation, skin graft photos were taken every 1 to 2 days with a digital camera until the graft was rejected. The skin grafts were removed at the indicated time points. Sections (4 to 6 mm) were fixed in 4% paraformaldehyde and stained with hematoxylin and eosin (H&E) for assessment of cellular infiltration. The H&E experiment was carried out by Beijing Brightshines Technology Co. Ltd. (Beijing, China).
Heart transplantation
Cardiac grafts from C57BL/6 (H-2b) mice were heterotopically transplanted into SCID mice with cuff technique (57). Graft loss was defined as no palpable beat. In some experiments, grafts were excised, fixed in 4% paraformaldehyde, and stained with H&E for assessment of cellular infiltration. The H&E experiment was carried out by Beijing Brightshines Technology Co. Ltd. (Beijing, China).
Gavage
RORγt inhibitor (catalog no. S6767) and corn oil (catalog no. S6701) were purchased from Selleckchem (Houston, TX, USA). The RORγt inhibitor was dissolved and stored in dimethyl sulfoxide (DMSO; Sigma-Aldrich) at a storage concentration of 50 mg/ml. It was suspended with corn oil (2% DMSO and 98% corn oil) and used at 10 mg/kg of body weight by a gastric applicator to inject corn oil suspension directly into the mouse’s stomach from the mouse’s mouth.
RNA-seq analysis
Naïve CD4+T cells (CD4+CD62L+CD44low) and CD8+T cells (CD8+CD62L+CD44low) from C57BL/6 mice were sorted using a MoFlo XDP cell sorter (Beckman Coulter, Brea, CA, USA). Total RNA was extracted using TRIzol agent (Thermo Fisher Scientific, 15596018). The CD4+ TRM and CD8+ TRM cells were sorted by CD4+CD69+ and CD8+CD69+ cells in the skin from BALB/c mice at 100 days after the first allogeneic skin transplantation. The activating TRM cells in skin during rejection were sorted by CD4+CD69+ cells during the skin transplant rejection. All samples were collected in tubes containing lysis components and ribonuclease inhibitors. For specific sequencing steps, refer to the Supplementary Materials. We used the mapping software HISAT2 to map the reads to the mouse mm10 reference genome and StringTie to construct transcripts independently for each cell (32). DEGseq was used to identify the differentially expressed genes. We set q < 0.01 and |log2(fold change)| > 1 as a significant difference in the activating TRM versus induced TRM comparison thresholds and set q < 0.05 and |log2(fold change)| > 0 to a significant difference in the naïve T versus induced TRM comparison thresholds.
Function and pathway enrichment
Gene ontology functional annotation analysis was performed on all cell differential genes using the DAVID Bioinformatics Resources 6.8 online search tool (https://david.ncifcrf.gov/) (58). Kyoto Encyclopedia of Genes and Genomes (KEGG) pathway analysis was performed on all the different genes of each cell using the KOBAS online search tool (http://bioinfo.org/kobas) (59, 60). The protein interaction network was obtained by STRING (https://string-db.org/) and visualized by Cytoscape (61).
Statistical analysis
All data are presented as the means ± SD. T test was used for comparison between groups. The log-rank (Mantel-Cox) test was used for graft survival curve comparisons. Hypergeometric test/Fisher’s exact test was used for KEGG pathway, and MA-plot–based method with random sampling model (62) was used for analysis of differentially expressed genes. *P < 0.05, **P < 0.01, ***P < 0.001, and ****P < 0.0001 were used to show statistical difference; ns indicates no significance achieved.
Acknowledgments
We acknowledge Q. Meng and X. Yang for the assistance in cell sorting and L. Li for excellent laboratory assistance. We acknowledge Q. Li from the pathology department of Capital Medical University Beijing Chaoyang Hospital for pathological grading.
Funding: This work is funded by the National Key Research and Development Program of China (2017YFA0105002, 2017YFA0104401, and 2017YFA0104402 to Y.Z.), National Natural Science Foundation for General and Key Programs (31930041 to Y.Z.; 81771720 and 82070764 to W.W.), and Knowledge Innovation Program of Chinese Academy of Sciences (XDA16030301 to Y.Z.);
Author contributions: Conceptualization: F.Y. and Y.Z. Methodology: Q.T., F.Y., L.T., and Z.Z. Investigation: Q.T., F.Y., L.T., Y.X., Z.Z., D.W., P.C., J.J., C.W., and Y.G. Visualization: Q.T., F.Y., and L.T. Funding acquisition: Y.Z. Project administration: Q.T. and Y.Z. Supervision: W.W., X.X., and Y.Z. Writing—original draft: Q.T., F.Y., Y.G., and Y.Z. Writing—review and editing: W.W., X.X., and Y.Z.
Competing interests: The authors declare that they have no competing interests.
Data and materials availability: The RNA-seq data presented in the study are deposited in the NCBI database (https://ncbi.nlm.nih.gov/bioproject/), accession number PRJNA758612. All other data needed to evaluate the conclusions in the paper are present in the paper and/or the Supplementary Materials.
Supplementary Materials
This PDF file includes:
Supplementary Materials and Methods
Figs. S1 to S7
Other Supplementary Material for this manuscript includes the following:
Data files S1 and S2
REFERENCES AND NOTES
- 1.Gebhardt T., Wakim L. M., Eidsmo L., Reading P. C., Heath W. R., Carbone F. R., Memory T cells in nonlymphoid tissue that provide enhanced local immunity during infection with herpes simplex virus. Nat. Immunol. 10, 524–530 (2009). [DOI] [PubMed] [Google Scholar]
- 2.Thome J. J., Yudanin N., Ohmura Y., Kubota M., Grinshpun B., Sathaliyawala T., Kato T., Lerner H., Shen Y., Farber D. L., Spatial map of human T cell compartmentalization and maintenance over decades of life. Cell 159, 814–828 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Steinert E. M., Schenkel J. M., Fraser K. A., Beura L. K., Manlove L. S., Igyártó B. Z., Southern P. J., Masopust D., Quantifying memory CD8 T cells reveals regionalization of immunosurveillance. Cell 161, 737–749 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Masopust D., Choo D., Vezys V., Wherry E. J., Duraiswamy J., Akondy R., Wang J., Casey K. A., Barber D. L., Kawamura K. S., Fraser K. A., Webby R. J., Brinkmann V., Butcher E. C., Newell K. A., Ahmed R., Dynamic T cell migration program provides resident memory within intestinal epithelium. J. Exp. Med. 207, 553–564 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Teijaro J. R., Turner D., Pham Q., Wherry E. J., Lefrançois L., Farber D. L., Cutting edge: Tissue-retentive lung memory CD4 T cells mediate optimal protection to respiratory virus infection. J. Immunol. 187, 5510–5514 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Park S. L., Buzzai A., Rautela J., Hor J. L., Hochheiser K., Effern M., Bain N. M., Wagner T., Edwards J., Conville R. M., Wilmott J. S., Scolyer R. A., Tüting T., Palendira U., Gyorki D., Mueller S. N., Huntington N. D., Bedoui S., Hölzel M., Mackay L. K., Waithman J., Gebhardt T., Tissue-resident memory CD8+ T cells promote melanoma–immune equilibrium in skin. Nature 565, 366–371 (2019). [DOI] [PubMed] [Google Scholar]
- 7.Iijima N., Iwasaki A., T cell memory. A local macrophage chemokine network sustains protective tissue-resident memory CD4 T cells. Science 346, 93–98 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Gebhardt T., Whitney P. G., Zaid A., Mackay L. K., Brooks A. G., Heath W. R., Carbone F. R., Mueller S. N., Different patterns of peripheral migration by memory CD4+ and CD8+ T cells. Nature 477, 216–219 (2011). [DOI] [PubMed] [Google Scholar]
- 9.Fonseca R., Beura L. K., Quarnstrom C. F., Ghoneim H. E., Fan Y., Zebley C. C., Scott M. C., Fares-Frederickson N. J., Wijeyesinghe S., Thompson E. A., Borges da Silva H., Vezys V., Youngblood B., Masopust D., Developmental plasticity allows outside-in immune responses by resident memory T cells. Nat. Immunol. 21, 412–421 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Collins N., Jiang X., Zaid A., Macleod B. L., Li J., Park C. O., Haque A., Bedoui S., Heath W. R., Mueller S. N., Kupper T. S., Gebhardt T., Carbone F. R., Skin CD4(+) memory T cells exhibit combined cluster-mediated retention and equilibration with the circulation. Nat. Commun. 7, 11514 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Klicznik M. M., Morawski P. A., Höllbacher B., Varkhande S. R., Motley S. J., Kuri-Cervantes L., Goodwin E., Rosenblum M. D., Long S. A., Brachtl G., Duhen T., Betts M. R., Campbell D. J., Gratz I. K., Human CD4+CD103+ cutaneous resident memory T cells are found in the circulation of healthy individuals. Sci. Immunol. 4, eaav8995 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Kumar B. V., Ma W., Miron M., Granot T., Guyer R. S., Carpenter D. J., Senda T., Sun X., Ho S. H., Lerner H., Friedman A. L., Shen Y., Farber D. L., Human tissue-resident memory T cells are defined by core transcriptional and functional signatures in lymphoid and mucosal sites. Cell Rep. 20, 2921–2934 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Paik D. H., Farber D. L., Anti-viral protective capacity of tissue resident memory T cells. Curr. Opin. Virol. 46, 20–26 (2021). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Samat A. A. K., van der Geest J., Vastert S. J., van Loosdregt J., van Wijk F., Tissue-resident memory T cells in chronic inflammation-local cells with systemic effects? Cell 10, 409 (2021). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Okła K., Farber D. L., Zou W., Tissue-resident memory T cells in tumor immunity and immunotherapy. J. Exp. Med. 218, (2021). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Chen L., Shen Z., Tissue-resident memory T cells and their biological characteristics in the recurrence of inflammatory skin disorders. Cell. Mol. Immunol. 17, 64–75 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Pallett L. J., Burton A. R., Amin O. E., Rodriguez-Tajes S., Patel A. A., Zakeri N., Jeffery-Smith A., Swadling L., Schmidt N. M., Baiges A., Gander A., Yu D., Nasralla D., Froghi F., Iype S., Davidson B. R., Thorburn D., Yona S., Forns X., Maini M. K., Longevity and replenishment of human liver-resident memory T cells and mononuclear phagocytes. J. Exp. Med. 217, (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Bartolomé-Casado R., Landsverk O. J. B., Chauhan S. K., Sætre F., Hagen K. T., Yaqub S., Øyen O., Horneland R., Aandahl E. M., Aabakken L., Bækkevold E. S., Jahnsen F. L., CD4+ T cells persist for years in the human small intestine and display a TH1 cytokine profile. Mucosal Immunol. 14, 402–410 (2021). [DOI] [PubMed] [Google Scholar]
- 19.Bartolomé-Casado R., Landsverk O. J. B., Chauhan S. K., Richter L., Phung D., Greiff V., Risnes L. F., Yao Y., Neumann R. S., Yaqub S., Øyen O., Horneland R., Aandahl E. M., Paulsen V., Sollid L. M., Qiao S. W., Baekkevold E. S., Jahnsen F. L., Resident memory CD8 T cells persist for years in human small intestine. J. Exp. Med. 216, 2412–2426 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Zuber J., Shonts B., Lau S. P., Obradovic A., Fu J., Yang S., Lambert M., Coley S., Weiner J., Thome J., DeWolf S., Farber D. L., Shen Y., Caillat-Zucman S., Bhagat G., Griesemer A., Martinez M., Kato T., Sykes M., Bidirectional intragraft alloreactivity drives the repopulation of human intestinal allografts and correlates with clinical outcome. Science Immunol. 1, eaah3732 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Snyder M. E., Finlayson M. O., Connors T. J., Dogra P., Senda T., Bush E., Carpenter D., Marboe C., Benvenuto L., Shah L., Robbins H., Hook J. L., Sykes M., D’Ovidio F., Bacchetta M., Sonett J. R., Lederer D. J., Arcasoy S., Sims P. A., Farber D. L., Generation and persistence of human tissue-resident memory T cells in lung transplantation. Science Immunol. 4, eaav5581 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Abou-Daya K. I., Tieu R., Zhao D., Rammal R., Sacirbegovic F., Williams A. L., Shlomchik W. D., Oberbarnscheidt M. H., Lakkis F. G., Resident memory T cells form during persistent antigen exposure leading to allograft rejection. Science Immunol. 6, eabc8122 (2021). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Wu T., Sun C., Chen Z., Zhen Y., Peng J., Qi Z., Yang X., Zhao Y., Smad3-deficient CD11b+Gr1+ myeloid-derived suppressor cells prevent allograft rejection via the nitric oxide pathway. J. Immunol. 189, 4989–5000 (2012). [DOI] [PubMed] [Google Scholar]
- 24.Boddupalli C. S., Nair S., Gray S. M., Nowyhed H. N., Verma R., Gibson J. A., Abraham C., Narayan D., Vasquez J., Hedrick C. C., Flavell R. A., Dhodapkar K. M., Kaech S. M., Dhodapkar M. V., ABC transporters and NR4A1 identify a quiescent subset of tissue-resident memory T cells. J. Clin. Invest. 126, 3905–3916 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Hombrink P., Helbig C., Backer R. A., Piet B., Oja A. E., Stark R., Brasser G., Jongejan A., Jonkers R. E., Nota B., Basak O., Clevers H. C., Moerland P. D., Amsen D., van Lier R. A. W., Programs for the persistence, vigilance and control of human CD8+ lung-resident memory T cells. Nat. Immunol. 17, 1467–1478 (2016). [DOI] [PubMed] [Google Scholar]
- 26.Milner J. J., Toma C., Yu B., Zhang K., Omilusik K., Phan A. T., Wang D., Getzler A. J., Nguyen T., Crotty S., Wang W., Pipkin M. E., Goldrath A. W., Erratum: Runx3 programs CD8+ T cell residency in non-lymphoid tissues and tumours. Nature 554, 392 (2018). [DOI] [PubMed] [Google Scholar]
- 27.Mueller S. N., Mackay L. K., Tissue-resident memory T cells: Local specialists in immune defence. Nat. Rev. Immunol. 16, 79–89 (2016). [DOI] [PubMed] [Google Scholar]
- 28.Kallies A., Xin A., Belz G. T., Nutt S. L., Blimp-1 transcription factor is required for the differentiation of effector CD8+ T cells and memory responses. Immunity 31, 283–295 (2009). [DOI] [PubMed] [Google Scholar]
- 29.van Gisbergen K. P. J. M., Kragten N. A. M., Hertoghs K. M. L., Wensveen F. M., Jonjic S., Hamann J., Nolte M. A., van Lier R. A. W., Mouse Hobit is a homolog of the transcriptional repressor Blimp-1 that regulates NKT cell effector differentiation. Nat. Immunol. 13, 864–871 (2012). [DOI] [PubMed] [Google Scholar]
- 30.Pan Y., Tian T., Park C. O., Lofftus S. Y., Mei S., Liu X., Luo C., O’Malley J. T., Gehad A., Teague J. E., Divito S. J., Fuhlbrigge R., Puigserver P., Krueger J. G., Hotamisligil G. S., Clark R. A., Kupper T. S., Survival of tissue-resident memory T cells requires exogenous lipid uptake and metabolism. Nature 543, 252–256 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Halloran P. F., T cell-mediated rejection of kidney transplants: A personal viewpoint. Am. J. Transplant. 10, 1126–1134 (2010). [DOI] [PubMed] [Google Scholar]
- 32.Pertea M., Kim D., Pertea G. M., Leek J. T., Salzberg S. L., Transcript-level expression analysis of RNA-seq experiments with HISAT, StringTie and Ballgown. Nat. Protoc. 11, 1650–1667 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Laidlaw B. J., Zhang N., Marshall H. D., Staron M. M., Guan T., Hu Y., Cauley L. S., Craft J., Kaech S. M., CD4+ T cell help guides formation of CD103+ lung-resident memory CD8+ T cells during influenza viral infection. Immunity 41, 633–645 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.O’Shea J. J., Paul W. E., Mechanisms underlying lineage commitment and plasticity of helper CD4+ T cells. Science 327, 1098–1102 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Korn T., Bettelli E., Oukka M., Kuchroo V. K., IL-17 and Th17 cells. Annu. Rev. Immunol. 27, 485–517 (2009). [DOI] [PubMed] [Google Scholar]
- 36.Li Y., Zhu L., Chu Z., Yang T., Sun H. X., Yang F., Wang W., Hou Y., Wang P., Zhao Q., Tao Y., Zhang L., Zhang X., Zhao Y., Characterization and biological significance of IL-23-induced neutrophil polarization. Cell. Mol. Immunol. 15, 518–530 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Gorbacheva V., Fan R., Li X., Valujskikh A., Interleukin-17 promotes early allograft inflammation. Am. J. Pathol. 177, 1265–1273 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Yuan X., Paez-Cortez J., Schmitt-Knosalla I., D’Addio F., Mfarrej B., Donnarumma M., Habicht A., Clarkson M. R., Iacomini J., Glimcher L. H., Sayegh M. H., Ansari M. J., A novel role of CD4 Th17 cells in mediating cardiac allograft rejection and vasculopathy. J. Exp. Med. 205, 3133–3144 (2008). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Agorogiannis E. I., Regateiro F. S., Howie D., Waldmann H., Cobbold S. P., Th17 cells induce a distinct graft rejection response that does not require IL-17A. Am. J. Transplant. 12, 835–845 (2012). [DOI] [PubMed] [Google Scholar]
- 40.Abadja F., Sarraj B., Ansari M. J., Significance of T helper 17 immunity in transplantation. Curr. Opin. Organ Transplant. 17, 8–14 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Chen Y., Wood K. J., Interleukin-23 and TH17 cells in transplantation immunity: Does 23+17 equal rejection? Transplantation 84, 1071–1074 (2007). [DOI] [PubMed] [Google Scholar]
- 42.Ivanov I. I., McKenzie B. S., Zhou L., Tadokoro C. E., Lepelley A., Lafaille J. J., Cua D. J., Littman D. R., The orphan nuclear receptor RORgammat directs the differentiation program of proinflammatory IL-17+ T helper cells. Cell 126, 1121–1133 (2006). [DOI] [PubMed] [Google Scholar]
- 43.de Leur K., Dieterich M., Hesselink D. A., Corneth O. B. J., Dor F. J. M. F., de Graav G. N., Peeters A. M. A., Mulder A., Kimenai H. J. A. N., Claas F. H. J., Clahsen-van Groningen M. C., van der Laan L. J. W., Hendriks R. W., Baan C. C., Characterization of donor and recipient CD8+ tissue-resident memory T cells in transplant nephrectomies. Sci. Rep. 9, 5984 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.de Leur K., Dieterich M., Corneth O., de Graav G. D., Mulder A., Dor F., Kimenai H., Claas F., Hesselink D., van Groningen M. V., van der Laan L. J., Hendriks R., Baan C., Tissue-resident memory T cells of donor origin are short-lived in renal allografts after transplantation. Transplantation 102, S146 (2018). [Google Scholar]
- 45.Shin H., Formation and function of tissue-resident memory T cells during viral infection. Curr. Opin. Virol. 28, 61–67 (2018). [DOI] [PubMed] [Google Scholar]
- 46.Buggert M., Japp A. S., Betts M. R., Everything in its right place: Resident memory CD8(+) T cell immunosurveillance of HIV infection. Curr. Opin. HIV AIDS 14, 93–99 (2019). [DOI] [PubMed] [Google Scholar]
- 47.Amsen D., van Gisbergen K., Hombrink P., van Lier R. A. W., Tissue-resident memory T cells at the center of immunity to solid tumors. Nat. Immunol. 19, 538–546 (2018). [DOI] [PubMed] [Google Scholar]
- 48.Mami-Chouaib F., Blanc C., Corgnac S., Hans S., Malenica I., Granier C., Tihy I., Tartour E., Resident memory T cells, critical components in tumor immunology. J. Immunother. Cancer 6, 87 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Park S. L., Gebhardt T., Mackay L. K., Tissue-resident memory T cells in cancer immunosurveillance. Trends Immunol. 40, 735–747 (2019). [DOI] [PubMed] [Google Scholar]
- 50.Byrne A., Savas P., Sant S., Li R., Virassamy B., Luen S. J., Beavis P. A., Mackay L. K., Neeson P. J., Loi S., Tissue-resident memory T cells in breast cancer control and immunotherapy responses. Nat. Rev. Clin. Oncol. 17, 341–348 (2020). [DOI] [PubMed] [Google Scholar]
- 51.Wu H., Liao W., Li Q., Long H., Yin H., Zhao M., Chan V., Lau C. S., Lu Q., Pathogenic role of tissue-resident memory T cells in autoimmune diseases. Autoimmun. Rev. 17, 906–911 (2018). [DOI] [PubMed] [Google Scholar]
- 52.Schreiner D., King C. G., CD4+ memory T cells at home in the tissue: Mechanisms for health and disease. Front. Immunol. 9, 2394 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Krebs C. F., Reimers D., Zhao Y., Paust H. J., Bartsch P., Nuñez S., Rosemblatt M. V., Hellmig M., Kilian C., Borchers A., Enk L. U. B., Zinke M., Becker M., Schmid J., Klinge S., Wong M. N., Puelles V. G., Schmidt C., Bertram T., Stumpf N., Hoxha E., Meyer-Schwesinger C., Lindenmeyer M. T., Cohen C. D., Rink M., Kurts C., Franzenburg S., Koch-Nolte F., Turner J. E., Riedel J. H., Huber S., Gagliani N., Huber T. B., Wiech T., Rohde H., Bono M. R., Bonn S., Panzer U., Mittrücker H. W., Pathogen-induced tissue-resident memory TH17 (TRM17) cells amplify autoimmune kidney disease. Sci. Immunol. 5, (2020). [DOI] [PubMed] [Google Scholar]
- 54.Zhou L., Ivanov I. I., Spolski R., Min R., Shenderov K., Egawa T., Levy D. E., Leonard W. J., Littman D. R., IL-6 programs T(H)-17 cell differentiation by promoting sequential engagement of the IL-21 and IL-23 pathways. Nat. Immunol. 8, 967–974 (2007). [DOI] [PubMed] [Google Scholar]
- 55.Burrell B. E., Csencsits K., Lu G., Grabauskiene S., Bishop D. K., CD8+ Th17 mediate costimulation blockade-resistant allograft rejection in T-bet-deficient mice. J. Immunol. 181, 3906–3914 (2008). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Rao D. A., Eid R. E., Qin L., Yi T., Kirkiles-Smith N. C., Tellides G., Pober J. S., Interleukin (IL)-1 promotes allogeneic T cell intimal infiltration and IL-17 production in a model of human artery rejection. J. Exp. Med. 205, 3145–3158 (2008). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Tan L., Xie X., Xu Y., Tian Q., Zhang Q., Lan G., Wang H., Zhao Y., Peng L., Skills to perform vessel eversion in mouse cervical cardiac transplantation with cuff technique. Braz. J. Cardiovasc. Surg. 36, 318–322 (2021). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Huang D. W., Sherman B. T., Lempicki R. A., Systematic and integrative analysis of large gene lists using DAVID bioinformatics resources. Nat. Protoc. 4, 44–57 (2009). [DOI] [PubMed] [Google Scholar]
- 59.Wu J., Mao X., Cai T., Luo J., Wei L., KOBAS server: A web-based platform for automated annotation and pathway identification. Nucleic Acids Res. 34, W720–W724 (2006). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Xie C., Mao X., Huang J., Ding Y., Wu J., Dong S., Kong L., Gao G., Li C.-Y., Wei L., KOBAS 2.0: A web server for annotation and identification of enriched pathways and diseases. Nucleic Acids Res. 39, W316–W322 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Shannon P., Markiel A., Ozier O., Baliga N. S., Wang J. T., Ramage D., Amin N., Schwikowski B., Ideker T., Cytoscape: A software environment for integrated models of biomolecular interaction networks. Genome Res. 13, 2498–2504 (2003). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Jiang H., Wong W. H., Statistical inferences for isoform expression in RNA-seq. Bioinformatics 25, 1026–1032 (2009). [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Supplementary Materials and Methods
Figs. S1 to S7
Data files S1 and S2