Abstract
T-cell acute lymphoblastic leukemias (T-ALL) represent 15% of pediatric and 25% of adult ALL. Since they have a particularly poor outcome in relapsed/refractory cases, identifying prognosis factors at diagnosis is crucial to adapting treatment for high-risk patients. Unlike acute myeloid leukemia and BCP ALL, chromosomal rearrangements leading to chimeric fusion-proteins with strong prognosis impact are sparsely reported in T-ALL. To address this issue an RT-MPLA assay was applied to a consecutive series of 522 adult and pediatric T-ALLs and identified a fusion transcript in 20% of cases. PICALM-MLLT10 (4%, n = 23), NUP214-ABL1 (3%, n = 19) and SET-NUP214 (3%, n = 18) were the most frequent. The clinico-biological characteristics linked to fusion transcripts in a subset of 235 patients (138 adults in the GRAALL2003/05 trials and 97 children from the FRALLE2000 trial) were analyzed to identify their prognosis impact. Patients with HOXA trans-deregulated T-ALLs with MLLT10, KMT2A and SET fusion transcripts (17%, 39/235) had a worse prognosis with a 5-year EFS of 35.7% vs 63.7% (HR = 1.63; p = 0.04) and a trend for a higher cumulative incidence of relapse (5-year CIR = 45.7% vs 25.2%, HR = 1.6; p = 0.11). Fusion transcripts status in T-ALL can be robustly identified by RT-MLPA, facilitating risk adapted treatment strategies for high-risk patients.
Subject terms: Acute lymphocytic leukaemia, Cancer genetics
Introduction
T-cell acute lymphoblastic leukemia (T-ALL) is an aggressive hematological cancer arising from the transformation of T cell precursors arrested at specific stages of differentiation [1, 2]. T-ALL represent 15% of pediatric and 25% of adult ALL. Despite modern poly-chemotherapy protocols, followed by allogeneic hematopoietic stem cell transplantation (HSCT) in high-risk cases, outcome of pediatric and adult patients with T-ALL remains poor, particularly in primary refractory and relapsed cases. After a transient initial response, about 20–30% of pediatric [3–5] and 40% of adult T-ALL patients relapse [6] with a 5-years overall survival (OS) below 20%. Understanding the mechanisms leading to treatment failure is therefore crucial to identify patients at high risk and adapt treatment in order to improve long term prognosis. Cytogenetic and global transcriptomic analyses led to the classification of T-ALL into molecular subgroups characterized by the abnormal expression of specific transcription factors (TF) (TAL1; LMO1/2; TLX1/3; LYL1; HOXA; MEF2C) and their blocked differentiation at specific stages of maturation [1, 7, 8]. Aberrant TF expression can be due to their juxtaposition to T Cell Receptor (TCR) loci or to somatic mutations in their enhancer regions. A number of additional, recurrent genetic abnormalities are found, including the loss of major tumor suppressive pathways (e.g., inactivating mutations of PTEN and of the CDKN2A tumor suppressor locus) and activation of oncogenic pathways (e.g., activating mutations in NOTCH1/FBXW7, IL7R/JAK pathway, epigenetic regulators, cell cycle, PI3K, and RAS signaling) [9–12]. Acquisition and selection of these molecular alterations entails the complex clonal evolution at the cellular level that occurs during T-ALL progression. Unlike acute myeloid leukemia (AML) and B-Cell Precursor acute lymphoblastic leukemia (BCP-ALL), which harbor numerous translocations leading to chimeric protein fusions [13, 14] such abnormalities are more rarely reported in large series of T-ALLs [15–20]. Recurrent chimeric protein fusions in T-ALL include rearrangements of KMT2A (AFDN (AF6), MLLT1, ELL), SET-NUP214, ABL1 (NUP214-ABL1, BCR-ABL1), MLLT10 (PICALM, DDX3X, NAP1L1, XPO1), and the ETS family (SPI and ETV6). Given their individual low frequency, the clinico-biological features of T-ALLs harboring chimeric fusions within a comprehensive series remains elusive.
To address this issue, we designed and developed an RT-MPLA assay allowing identification of the majority of known fusion transcripts leading to chimeric proteins in T-ALLs. Applying this panel to a comprehensive, consecutive series of 522 adult and pediatric T-ALLs, we here report an unexpected overall incidence of 20% of fusion transcript. Their mutational landscape, associated clinico-biological features and prognostic impact on patients enrolled in the French GRAALL protocol for adult patients and the FRALLE 2000T protocol for pediatric patients are described.
Methods
T-ALL samples and clinical trials
Diagnostic peripheral blood (PB) or bone marrow (BM) samples from a consecutive series of 522 T-ALL patients, with RNA available, 206 children and 316 adults, were screened for fusion transcripts (fig. S1). Sample collection and analyses were obtained with informed consent in accordance with the Declaration of Helsinki with approval from the institutional review boards of institutions that participated in this study. Diagnosis of T-ALL was based on the World Health Organization 2008 criteria revised in 2016, defined by expression of cytoplasmic and/or surface CD3, and negativity of CD19 and MPO [21].
Adult patients aged from 16 to 59 years (n = 138) were included in the GRAALL03/05 trials (GRAALL-2003, #NCT00222027; GRAALL-2005, #NCT00327678) and pediatric patients aged from 1 to 19 years (n = 97) were treated according to FRALLE 2000 T guidelines in ten French centers members of the Société Française de lutte contre les Cancers et les leucémies de l’Enfant et de l’adolescent (SFCE) centers.
Definitions: Corticosensitivity was defined as circulating blasts <1 G/L on day 8. Complete remission (CR) was defined as: absence of physical signs of leukemia, BM with active hematopoiesis and <5% leukemic blast cells (identified morphologically), and normal cerebrospinal fluid.
Biological analysis of T-ALL samples
Diagnostic PB or BM T-ALL samples were analyzed for immunophenotype and classified as follows: Immature T-ALL are TCR-and cTCRβ-. αβ-lineage T-ALL are TCRαβ or preαβ (TCR-) but with cTCRβ + . γδ-lineage T-ALL are TCRγδ, SIL-TAL1 and PICALM-MLLT10 detection was performed by RT-PCR as previously described [2, 22]). Oncogenic transcripts TLX1 and TLX3 quantification was performed by RQ-PCR (primer and probe sequences as previously described [23, 24]). Quantification of HOXA9 expression was performed by qRT-PCR as previously described [25]. All methods are detailed in supplementary methods.
Diagnostic available DNA samples from 273 cases were also analyzed using an 80-gene pan-exon next-generation sequencing capture-panel (details included in supplementary methods and Table S1).
RT-MLPA
Using previously published methods [26], we created a mix of 268 RT-MLPA probes to target 106 different genes (Table S2; Fig. S2). The procedure is detailed in supplementary data. All 141 5′ probes have a GTGCCAGCAAGATCCAATCTAGA tail at their 5′ ends and all 127 3′ probes a TCCAACCCTTAGGGAACCC tail at their 3′ ends to allow for final PCR amplification.
Data analysis
Fisher’s exact and Mann–Whitney tests were used for clinical, biological and mutational analyses. Overall survival (OS) was calculated from the date of pre-phase initiation to the last follow-up date. The cumulative incidence of relapse (CIR) was calculated from CR to relapse date, censoring patients alive without relapse at the last follow-up date. The Event-Free-Survival was calculated from diagnosis date to first event among: induction failure, first hematologic relapse and death from any cause in first CR. Relapse and death in CR were considered as competitive events. Univariate analyses assessing the impact of categorical and continuous variables were performed with a Cox model. Statistical analyses were performed with STATA software (STATA 12.0 Corporation, College Station, TX) for survival and R software for others (v4.0.2). All p values were two-sided, with p < 0.05 denoting statistical significance.
Results
RT-MLPA detect an unexpected 20% incidence of fusion proteins in T-ALLs
A series of 522 T-ALL, all systematically screened for PICALM-MLLT10, SET-NUP214, and NUP214-ABL1 by RT/qPCR, were evaluated for fusion transcripts by RT-MLPA. Fusion transcripts were detected in 104/522 (20%) cases (Table 1, S3A, B), leading to the identification of 99/104 fusion partners by pyrosequencing (Fig. 1A). Among the 5 unresolved cases 2 PICALM-MLLT10, 1 NUP214-ABL1 and 1 KMT2A-MLLT1 were identified by RT/qPCR. Only one t(11;16) involving KMT2A identified by cytogenetics and called “KMT2A-?” remained only partially identified. Importantly, among fusion transcripts systematically screened by RT/qPCR, no false negative or false positive RT-MLPA case was observed.
Table 1.
Biological characteristics of T acute lymphoblastic leukemia according to fusion transcripts status.
| RT-MLPA | Positive | Negative | All | ||||
|---|---|---|---|---|---|---|---|
| 104 | 418 | 522 | |||||
| Fusion groups | MLLT10 n = 37 (7%) | KMT2A/SET n = 34 (7%) | ABL1 n = 22 (4%) | ETS n = 6 (1%) | Other n = 5 (1%) | 80% | |
| Transcripts |
PICALM-MLLT10 (23) DDX3X-MLLT10 (8) NAP1L1-MLLT10 (4) XPO1-MLLT10 (2) |
SET-NUP214 (18) KMT2A-AFDN (10) KMT2A-MLLT1 (4) KMT2A-ELL (1) KMT2A-? (1) |
NUP214-ABL1 (19) BCR-ABL1 (2) ETV6-ABL1 (1) |
ETV6-NCOA2 (3) STMN1-SPI1 (2) TCF7-SPI1 (1) |
NUP98-RAP1GDS1 (3) PCM1-FLT3 (1) P2RY8-CRLF2 (1) |
||
| Population | |||||||
| Age mean (range) | 23.4 (3–44) | 27.9 (8–63) | 23.1 (4–61) | 4.5 (1–8) | 18.8 (8–34) | 25.9 (1–78) | 25.4 (1–78) |
| Pediatric cases | 41% (n = 15) | 24% (n = 8) | 55% (n = 12) | 100 % (n = 6) | 60% (n = 3) | 39% (n = 162) | 39% (n = 206) |
| Phenotype | |||||||
| Immature | 3% (n = 1) | 4% (n = 1) | 7% (n = 1) | 33% (n = 1) | 33% (n = 1) | 10% (n = 34) | 9% (n = 39) |
| γδ-lineage | 84% (n = 27) | 81% (n = 22) | 27% (n = 4) | 0 | 33% (n = 1) | 26% (n = 85) | 34% (n = 139) |
| αβ-lineage | 12% (n = 4) | 15% (n = 4) | 67% (n = 10) | 67 % (n = 2) | 33% (n = 1) | 64% (n = 214) | 57% (n = 235) |
| Not available | 14% (n = 5) | 21% (n = 7) | 32% (n = 7) | 50% (n = 3) | 40% (n = 2) | 20% (n = 85) | 21% (n = 109) |
| Homeobox Genes Deregulation | |||||||
| HOXA9 | 95% (n = 35) | 100% (n = 34) | 5% (n = 1) | 0 | 60% (n = 3) | 20% (n = 82) | 30% (n = 155) |
| TLX1 | 0 | 0 | 14% (n = 3) | 0 | 0 | 14% (n = 60) | 12% (n = 63) |
| TLX3 | 0 | 0 | 73% (n = 16) | 0 | 0 | 14% (n = 57) | 14% (n = 73) |
| Negative | 5% (n = 2) | 0 | 14% (n = 3) | 100% (n = 6) | 40% (n = 2) | 55% (n = 231) | 47% (n = 244) |
All means are compared against the Negative group with Wilcoxon test or Student test depending on the Shapiro test result. All proportions are compared against the Negative group with Pearson’s χ2 test or for small values with the Fisher exact test. Results with p ≤ 0.05 are displayed in bold. Immature T-ALL are TCR- and cTCRβ-. αβ-lineage T-ALL are TCRαβ or preαβ (TCR-) but with cTCRβ+ . γδ-lineage T-ALL are TCRγδ.
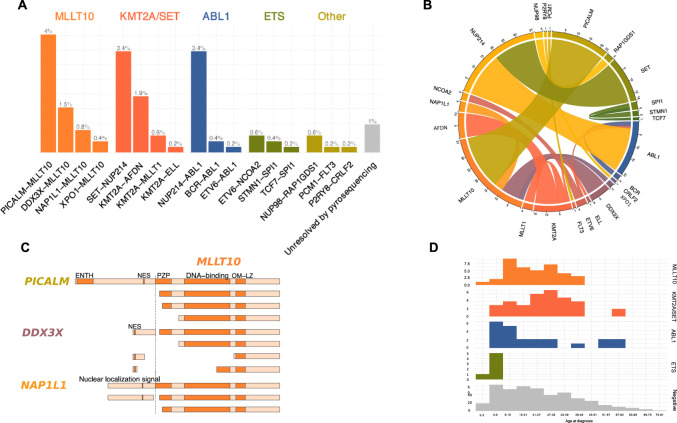
Fig. 1. Identified fusion transcripts and incidence.
A Histogram of identified transcripts, assigned to fusion groups. On top of each bars the percentage of all cases. PICALM-MLLT10 (4%), SET-NUP214 (3.4%), and NUP214-ABL1 (3.4%) are the most recurrent. B Circos plot representing the fusion transcripts. C Schematic representation of MLLT10 fused polypeptide chains with relevant domains. OM-LZ and nuclear addressing signals are always conserved. D Histograms of incidence of cases by age classes and fusion groups (absolute count on y axis). ETS fusion group is constituted with only pediatric cases. ENTH epsin N-terminal homology, NES nuclear export domain, PZP pregnancy zone protein.
Fusion transcripts are individually rare in T-ALL
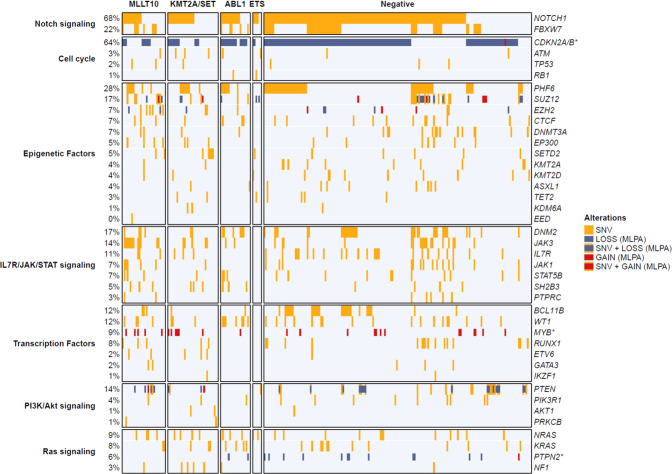
Among identified transcripts PICALM-MLLT10 (4%, n = 23), (Fig. 1A, B), NUP214-ABL1 (3%, n = 19) and SET-NUP214 (3%, n = 18) were the most frequently detected. All others fusion transcripts were rare (<2%). In order to facilitate analysis, RT-MPLA tested T-ALL were assigned into six groups: MLLT10 recombinome (7%, n = 37) (Table 1); KMT2A/SET (grouped together due to their HOXA overexpression) (7%, n = 34); ABL1 (4%, n = 22); ETS (1%, n = 6); other (1%, n = 5) and negative (80%, n = 418). Their available phenotype (n = 413), targeted DNA mutations (n = 273), HOXA9 and TLX1/3 overexpression data are resumed in Table 1 and Fig. 2.
Fig. 2. Mutational landscape according to fusion transcript group.
Oncoplot depicting the genetic anomalies observed in each fusion transcript group. Genes are classified by functional groups. Each potent mutation is represented in yellow. Each line is a sequenced gene, each column a case.
MLLT10 recombinome
MLLT10 recombinome was the most frequent chimeric protein (36% of identified transcripts). By order of occurrence MLLT10 partners were PICALM (11q14, n = 23), DDX3X (Xp11, n = 8), NAP1L1 (12q21, n = 4) and XPO1 (2p15, n = 2) (Table 1). In keeping with the fact that OM-LZ domains are a key for immortalization [27], all MLLT10 breakpoints conserved this domain (Fig. 1C).
The MLLT10 cases were more frequently of TCRγ/δ lineage (p < 0.05) and HOXA9 deregulated (Table 1). They were less likely to have NOTCH1 signaling mutations (p = 0.03) and cell cycle mutation/deletion (p < 0.001) and showed an higher frequency of RAS, IL7R/JAK/STAT and epigenetic regulator mutations compared to negative samples (Table 2; Fig. 2).
Table 2.
Genetic profile according to fusion transcripts status.
| Fusion Group | Notch signaling | Cell cycle | Epigenetic factors | IL7R/JAK/STAT signaling | Transcription factors | PI3K/Akt signaling | Ras signaling | Total |
|---|---|---|---|---|---|---|---|---|
| MLLT10 | 59% (n = 17) | 41% (n = 12) | 76% (n = 22) | 3% (n = 1) | 52% (n = 15) | 24% (n = 7) | 24% (n = 7) | 29 |
| KMT2A/SET | 53% (n = 18) | 41% (n = 14) | 59% (n = 20) | 3% (n = 1) | 35% (n = 12) | 21% (n = 7) | 32% (n = 11) | 34 |
| ABL1 | 75% (n = 15) | 80% (n = 16) | 60% (n = 12) | 5% (n = 1) | 45% (n = 9) | 5% (n = 1) | 50% (n = 10) | 20 |
| ETS | 67% (n = 4) | 67% (n = 4) | 67% (n = 4) | 17% (n = 1) | 17% (n = 1) | 17% (n = 1) | 33% (n = 2) | 6 |
| Negative | 79% (n = 143) | 76% (n = 136) | 57% (n = 102) | 1% (n = 1) | 37% (n = 67) | 19% (n = 35) | 20% (n = 36) | 180 |
All proportions are compared against the Negative group with Pearson’s χ2 test or for small values with the Fisher exact test. Results with p ≤ 0.05 are displayed in bold.
KMT2A recombinome and SET-NUP214 fusions
Seven percent of T-ALLs (34/522) (Table 1) demonstrated KMT2A or SET fusion transcripts, leading to HOXA overexpression without implication of MLLT10.
SET-NUP214 gene fusions were identified in 18 cases, with uniform breakpoint positions. KMT2A 3′ partners were, by decreasing incidence, AFDN (n = 10), MLLT1 (n = 4) and ELL (n = 1). As for the MLLT10 group, and according to previous reports [20], significantly more of these T-ALL were from the γδ lineage (p < 0.05) and overexpressed HOXA transcripts (Table 1). They were significantly associated with less cell cycle genes mutations/deletion (p < 0.001) and fewer NOTCH1 signaling genes mutations (53%, p = 0.002) compared to the negative group (Table 2; Fig. 2).
ABL1 recombinome
This group represented 4% of all T-ALLs and was dominated by NUP214-ABL1 (19/22) transcripts. As expected [28], most of them were TLX3 deregulated (p < 0.05), and a minority were TLX1 deregulated [29]. This group harbored significantly more RAS signaling genes mutations/deletions (50%, p < 0.01) (Table 2; Fig. 2). Among the two BCR-ABL1 transcripts, both were p190, one e6a2 and one e1a2.
ETS family recombinome
ETS fusion transcripts were detected only in 1% of T-ALLs, all of which were pediatric cases, (Fig. 1D) with an incidence of 3% in the pediatric cohort. They included ETV6-NCOA2 (n = 3), STMN1-SPI1 (n = 2), and TCF7-SPI1 (n = 1) transcripts. All SPI1 breakpoints involved exon 3 [17] and all ETS domains were conserved in the fusion transcripts. No specific phenotype or transcription factor (HOXA9, TLX1 nor TLX3) overexpression was observed within this subgroup (Table 1). Targeted sequencing identified significantly more cell cycle gene mutations (33%, p = 0.03) (Fig. 2; Table 2).
Clinico-biological characteristics of fusion transcripts in GRAALL and FRALLE treated T-ALL
We then investigated the clinico-biological characteristics linked to fusion transcripts in a subset of 235 patients, including 138 adults enrolled in the GRAALL-2003/05 trials and 97 children enrolled in the FRALLE-2000T trial. A fusion transcript was observed in 27% of adults and 22% of children and the overall incidence of fusion transcripts in these cohorts was 25% (58/235).
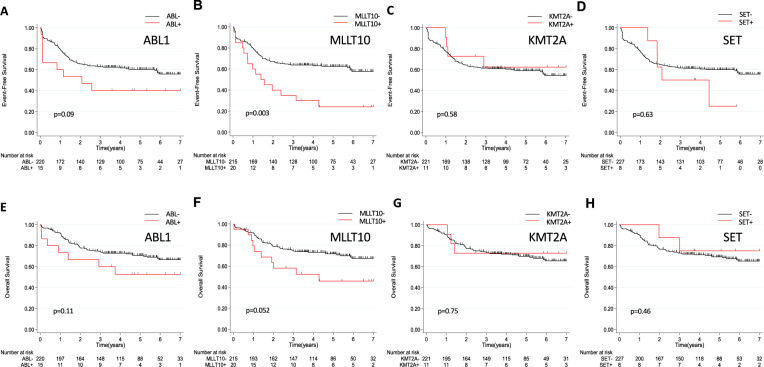
The incidence of ABL1 fusion was 6% (15/235). Patient outcome in this group did not differ significantly from negative cases (Table 3). Despite lower CR rates (66.7%) at the end of induction patients did not receive more frequent allogeneic HSCT and prognosis was not significantly (Table S4) different, with only a trend for shorter 5-year EFS (40% vs 60.2%, HR = 1.81; 95% CI [0.9–3.6]; p = 0.09) (Fig. 3).
Table 3.
Clinical characteristics and treatment response in GRAALL and FRALLE treated patients according to fusion transcript status.
| Fusion groups | Negative n = 177 (75%) | ABL1 n = 15 (6%) | KMT2A/SET n = 19 (8%) | MLLT10 n = 20 (9%) | ETS n = 2 (1%) | other n = 2 (1%) | All n = 235 |
|---|---|---|---|---|---|---|---|
| Transcripts | NUP214-ABL1 (14) | SET-NUP214 (8) | PICALM-MLLT10 (16) | ETV6-NCOA2 (1) | NUP98-RAP1GDS1 (2) | ||
| BCR-ABL1 (1) | KMT2A-AFDN (5) | NAP1L1-MLLT10 (1) | TCF7-SPI1 (1) | ||||
| KMT2A-MLLT1 (4) | DDX3X-MLLT10 (2) | ||||||
| KMT2A-ELL (1) | XPO-MLLT10 (1) | ||||||
| KMT2A-? (1) | |||||||
| Clinical subsets | |||||||
| Age, median (range), y | 19.9 (1.1–59) | 15.9 (4.9–40.4) | 27.5 (8.5–44.6) | 29.8 (5.8–45.6) | 4.6–4.2 | 12–12.8 | 21 (1.1–58.9) |
| WBC, median (range) | 84.9 (2.8–788) | 98 (12–980) | 29.2 (2.8–195) | 28 (1.2–352) | 16–641 | 24.3–49 | 73.2 (1.2–980) |
| CNS involvement (%) | 20 (11) | 3 (20) | 1 (5.3) | 4 (20) | 0 | 0 | 28 (11.9) |
| Male, n (%) | 134 (76) | 12 (80) | 15 (79) | 12 (60) | 1 (50) | 2 (100) | 176 (74.9) |
| Treatment response, n (%) | |||||||
| Poor steroid response | 84 (47.4) | 11 (73.3) | 16 (84.2) | 14 (70.0) | 1 (50) | 0 | 126 (53.6) |
| CR | 161 (91) | 10 (66.7) | 19 (100) | 17 (85.0) | 2 (100) | 2 (100) | 211 (89.8) |
| MRD > 10–4 | 38 (29) | 5 (55.6) | 8 (53.3) | 7 (58.3) | 0 | 2 (100) | 59 (34.3) |
| Allo HSCT | 37 (22) | 2 (20) | 7 (36.8) | 6 (35.3) | 0 | 1 (50) | 59 (28) |
All subgroups characteristics are compared with the negative group. All results with p value ≤ 0.05 are displayed in bold.
allo HSCT allogeneic hematopoietic stem cell transplantation, CNS central nervous system, CR complete remission, MRD minimal residual disease, WBC white blood cell.
Fig. 3. Clinical impact of the fusion transcripts group.
A–D Event-Free Survival (EFS) and (E–H) Overall Survival (OS) according to fusion transcripts status in GRAALL and FRALLE treated patients.
The incidence of HOXA trans-deregulated T-ALL with identified fusion transcripts was 17% (39/235) distributed as follows: MLLT10 fusions in 20 patients (9%), KMT2A fusions in 11 patients (5%), SET fusions in 8 patients (3%). Compared to the negative group, patients with one of these 3 fusion transcripts were older with lower white blood cell counts (WBC). Overall, these patients have worse prognosis with a 5-year EFS of 35.7% vs 63.7% (HR = 1.63; 95% CI [1.02–2.6]; p = 0.04) and a trend for higher CIR (5-year CIR = 45.7% vs 25.2%, HR = 1.6; 95% CI [0.9–2.9]; p = 0.11) (Fig. S3). However, 5-year OS was not significantly different (59.3% vs 71.4%, HR1.26; 95% CI [0.7–2.2]; p = 0.43). This outcome cannot be explained by a significantly increased rate of allogeneic HSCT in this subgroup (36.1% vs 26.3%, p = 0.23).
When comparing each of these three fusions groups with their respective negative group, different patterns of treatment response and prognosis were identified. KMT2A fusions have no impact on treatment response and prognosis. On the contrary, patients with SET fusions were all corticosteroid-resistant with a higher rate of MRD positivity (71.4%) at the end of induction (data not shown) and higher 5-year CIR (75% vs 27.2%, p = 0.17). Despite good initial treatment response, patients with MLLT10 fusions demonstrated a worse outcome, with shorter 5-year EFS (24% vs 62.4%, HR = 2.33; 95% CI [1.3–4.1]; p = 0.003) and shorter 5-year OS (45.7% vs 71.6%, HR = 1.94; 95% CI [0.99–3.8]; p = 0.052) (Fig. 3; table S4). This poor outcome remained on EFS and OS analysis of adult patients only (HR = 2.18; 95% CI [1.14–4.18]; p = 0.02 and HR = 2.19; 95%CI [1.07–4.51]; p = 0.03 respectively) (Fig. S4) and persists as a trend in children only in terms of EFS (HR = 2.81; 95% CI [0.99–8]; p = 0.052).
ETS-family (n = 2) and NUP98 (n = 2) fusions were only identified in pediatric patients. Despite high MRD at the end of induction (>10−4) for both patients with NUP98 rearrangements, only 1 patient required allogeneic HSCT. In contrast to previous report [17], all four patients had favorable outcome and remained in CR with a median follow up of 4.5 years (R, 3.4–6.7).
Discussion
Last decade transcriptomic and genomic studies identified biological subgroups of T-ALL and uncovered major oncogenic and tumor suppressor pathways [1]. This molecular characterization provided a strong rationale for targeted therapies in T-ALL, such as drugs directed against JAK, NOTCH1, BCL-2 or PI3-AKT signaling pathways. However, contrary to BCP -ALL or AML, prognostic biomarkers identified in large prospective studies are yet lacking or debated to stratify patients at first line and adapt treatment.
In 522 consecutive adult and pediatric T-ALL RT-MLPA identified an unexpected 20% incidence of chimeric fusion protein. Fusion transcripts were correlated with their respective immunophenotypic, transcriptional and mutational landscapes, resulting in an unprecedented global overview.
MLLT10 (10p12) (previously AF10) is a frequent 5′ and 3′ partner in chimeric fusion proteins harbored by T-ALL (8–10%) [30]. Published fusion partners include: PICALM (11q14) [31], XPO1 (2p15) [32], NAP1L1 (12q21) [11], NUP98 (11p15) [33], DDX3X (Xp11) [19], and HNRNPH1 (5q35) [19]. Within T-ALL, the most frequent 5′ partner is PICALM, with an incidence of 4–9% [2, 34, 35]. MLLT10 translocations are associated with HOXA overexpression [36] and TCRγ/δ lineage orientation [2]. MLLT10 contains a OM-LZ domain, known to bind to the epigenetic factor histone methyltransferase DOT1L [37, 38]. Direct fusion of DOT1L to MLLT10 results in leukemic transformation and upregulation of HOXA9 [37]. The outcome of cases harboring MLLT10 fusion proteins is insufficiently described. Published studies are limited to children, with a trend for a pejorative outcome [35, 39]. In our series this unfavorable outcome was confirmed, with a shorter EFS and OS in the entire cohort and in the adult cohort highlighting the requirement alternative treatment in these cases. Same trends were observed in children but not reached significance because of low number of patients. Preclinical data showed an antiproliferative effect of demethylating agents via DOT1L in a model of transformed PICALM-MLLT10 cells [40].
Rearrangements involving KMT2A (11q23) [41] (previously MLL) are also recurrent in T-ALL with reported occurrence of 5–8% [7, 42]. Consistent with previous reports, the most frequent 3′ partners in our T-ALL cohort were: AFDN (6q27) [43] (previously AF6) and MLLT1 (19p13, previously ENL) [44]. Of note, none of our 34 cases with KMT2A fusion demonstrated KMT2A-AFF1 (previously MLL-AF4), or KMT2A-MLLT3 (previously MLL-AF9) fusions, commonly observed in BCP-ALL and AML respectively. KMT2A rearranged T-ALLs over-express HOXA and are arrested in an early differentiation step after commitment to the γδ-lineage [1, 45]. The prognosis of KMT2A- rearrangements in T-ALL was unclear. Contrary to BCP-ALL, KMT2A-MLLT1 in T-ALL has been reported to be favorable in children [46]. In keeping with this, in our series, patients with KMT2A rearrangements do not demonstrate worse outcome neither in terms of initial response to treatment neither in terms of relapse.
SET-NUP214 (previously TAF1-CAN) is a recurrent chimeric protein found in 3–10% of T-ALLs [18, 47]. Most SET-NUP214 chimeric proteins result from a cryptic deletion at chromosomic 9q34, leading to loss of one ABL1 allele [48]. SET-NUP214 induces HOXA deregulation by interacting with XPO1, DOT1L and the HOXA promoter [18, 49, 50]. As confirmed here, this recombination occurs in T-ALL from the γδ-lineage that are associated with a poor outcome [20]. Patients with SET-NUP214 chimeric protein have significantly poor initial treatment response and a trend for a higher risk of relapse without significant prognostic impact on EFS or OS. Thus, the need of intensifying treatment protocol in these patients is still matter of debate.
In T-ALL, ABL1 (9q34) is predominantly rearranged with NUP214 (4–6%, 9q34) by episomal amplification in association with TLX1/3 deregulation [51, 52]. Other, rare reported, 3′ partners include BCR (22q11), EML1 (14q32) [53], and ETV6 (12p13) [54]. All confer constitutive activation of the tyrosine kinase domain of ABL1, with downstream LCK mediated proliferation [55] which could be targeted by tyrosine kinase inhibitors (TKI) such as dasatinib or bosutinib [56, 57]. The clinical impact of these translocations is debated, with a trend for a relatively pejorative impact [58]. In our series no prognosis impact of this rearrangement was demonstrated. However, due to this potentially targetable rearrangement these patients could benefit from the opportunity to add tyrosine kinase inhibitors to standard therapy [29].
The ETS family of proteins are TF containing an ETS DNA-binding domain [59] which are detected in pediatric T-ALL, fused in-frame to different 3′ or 5′ partners. SPI1 (11p11.2 encoding PU.1) is fused in the 3′ position with STMN1 (1p36) or TCF7 (5q31) and ETV6 (12p13) is fused in 5′ with NCOA2 (8q13). SPI1 fusions transcripts have been described in 4% of one pediatric T-ALL series, when they demonstrated a distinct gene expression profile and a poor outcome [17]. In contrast to this, all 4 FRALLE 2000T treated patients in our series had a favorable outcome.
RT-MLPA has the capacity to easily detect many rearrangements and its fast turnaround time and superior exhaustivity and reduced cDNA consumption could make it an interesting alternative to RT-PCR. Such a screening strategy will also identify patients with targetable rearrangement such as ABL1 [57] (with tyrosine kinase inhibitor) or KMT2A (with Dot1l or menin inhibitor) [40, 60] or who have poor prognosis and require treatment adaptation. However, RT-MLPA only allows detection of known fusion transcripts. New fusion transcripts detected by T-ALL RNA sequencing can easily be added to the RT-MLPA, thus providing a complement and extension to RNA-Seq approaches. In addition, the excellent specificity of this method was already demonstrated on a previous cohort of 540 patients with acute leukemia, confirming virtually all (98%) fusion transcripts detected by RT-PCR and Sanger sequencing [26]. As such, it is well adapted to routine clinical screening in Acute Leukemia.
Supplementary information
Acknowledgements
The authors would like to thank all participants in the GRAALL-2003 and GRAALL-2005 study groups, the SFCE and the investigators of the 16 SFCE centers involved in collection and provision of data and patient samples, and V. Lheritier for collection of clinical data. The GRAALL was supported by grants P0200701 and P030425/AOM03081 from the Programme Hospitalier de Recherche Clinique, Ministère de l’Emploi et de la Solidarité, France and the Swiss Federal Government in Switzerland. Samples were collected and processed by the AP-HP “Direction de Recherche Clinique” Tumor Bank at Necker-Enfants Malades. MED was supported by “CARPEM”. MS was supported by « Action Leucémie » and « Soutien pour la formation à la recherche translationnelle en cancérologie dans le cadre du Plan cancer 2009–2013 ». This work was supported by grants to Necker laboratory from the “Association Laurette Fugain”, Association pour la Recherche contre le Cancer (Equipe labellisée), Institut National du Cancer PRT-K 18–071 and the Fédération Leucémie espoir and Horizon Hemato.
Author contributions
VA, TS, PR, MA, and MED conceived the study and oversaw the project; MA, MS, AT, JM, LL, JB, CG, NG, JMC, IA, VG, NI, HD, AB, AP, PR, and NB provided study materials or patients; TS, MA, and VA performed molecular analyses; TS, MED, and VA collected and assembled data; NB, TS, and MED performed statistical analysis; TS, MED, NB, and VA analyzed and interpreted data; TS, MED, EM, NB, and VA wrote the paper; all authors approved the paper.
Competing interests
The authors declare no competing interests.
Footnotes
Publisher’s note Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
These authors contributed equally: Thomas Steimlé, Marie-Emilie Dourthe.
Supplementary information
The online version contains supplementary material available at 10.1038/s41408-022-00613-9.
References
- 1.Ferrando AA, Neuberg DS, Staunton J, Loh ML, Huard C, Raimondi SC, et al. Gene expression signatures define novel oncogenic pathways in T cell acute lymphoblastic leukemia. Cancer Cell. 2002;1:75–87. doi: 10.1016/s1535-6108(02)00018-1. [DOI] [PubMed] [Google Scholar]
- 2.Asnafi V, Radford-Weiss I, Dastugue N, Bayle C, Leboeuf D, Charrin C, et al. CALM-AF10 is a common fusion transcript in T-ALL and is specific to the TCRgammadelta lineage. Blood. 2003;102:1000–6. doi: 10.1182/blood-2002-09-2913. [DOI] [PubMed] [Google Scholar]
- 3.Pui C-H, Yang JJ, Hunger SP, Pieters R, Schrappe M, Biondi A, et al. Childhood Acute Lymphoblastic Leukemia: Progress Through Collaboration. J Clin Oncol. 2015;33:2938–48. doi: 10.1200/JCO.2014.59.1636. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Pui C-H, Robison LL, Look AT. Acute lymphoblastic leukaemia. Lancet. 2008;371:1030–43. doi: 10.1016/S0140-6736(08)60457-2. [DOI] [PubMed] [Google Scholar]
- 5.Hocking J, Schwarer AP, Gasiorowski R, Patil S, Avery S, Gibson J, et al. Excellent outcomes for adolescents and adults with acute lymphoblastic leukemia and lymphoma without allogeneic stem cell transplant: the FRALLE-93 pediatric protocol. Leuk Lymphoma. 2014;55:2801–7. doi: 10.3109/10428194.2014.894191. [DOI] [PubMed] [Google Scholar]
- 6.Huguet F, Leguay T, Raffoux E, Thomas X, Beldjord K, Delabesse E, et al. Pediatric-Inspired Therapy in Adults With Philadelphia Chromosome–Negative Acute Lymphoblastic Leukemia: The GRAALL-2003 Study. JCO. 2009;27:911–8. doi: 10.1200/JCO.2008.18.6916. [DOI] [PubMed] [Google Scholar]
- 7.Graux C, Cools J, Michaux L, Vandenberghe P, Hagemeijer A. Cytogenetics and molecular genetics of T-cell acute lymphoblastic leukemia: from thymocyte to lymphoblast. Leukemia. 2006;20:1496–510. doi: 10.1038/sj.leu.2404302. [DOI] [PubMed] [Google Scholar]
- 8.Van Vlierberghe P, Pieters R, Beverloo HB, Meijerink JP. Molecular-genetic insights in paediatric T-cell acute lymphoblastic leukaemia. Br J Haematol. 2008;143:153–68. doi: 10.1111/j.1365-2141.2008.07314.x. [DOI] [PubMed] [Google Scholar]
- 9.Fattizzo B, Rosa J, Giannotta JA, Baldini L, Fracchiolla NS. The Physiopathology of T- Cell Acute Lymphoblastic Leukemia: focus on Molecular Aspects. Front Oncol. 2020;10. 10.3389/fonc.2020.00273. [DOI] [PMC free article] [PubMed]
- 10.Liu Y, Easton J, Shao Y, Maciaszek J, Wang Z, Wilkinson MR, et al. The genomic landscape of pediatric and young adult T-lineage acute lymphoblastic leukemia. Nat Genet. 2017;49:1211–8. doi: 10.1038/ng.3909. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Zhang J, Ding L, Holmfeldt L, Wu G, Heatley SL, Payne-Turner D, et al. The genetic basis of early T-cell precursor acute lymphoblastic leukaemia. Nature. 2012;481:157–63. doi: 10.1038/nature10725. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Belver L, Ferrando A. The genetics and mechanisms of T cell acute lymphoblastic leukaemia. Nat Rev Cancer. 2016;16:494–507. doi: 10.1038/nrc.2016.63. [DOI] [PubMed] [Google Scholar]
- 13.Yang JJ, Park TS, Wan TSK. Recurrent Cytogenetic Abnormalities in Acute Myeloid Leukemia. In: Wan TSK, editors. Cancer Cytogenetics: Methods and Protocols. New York, NY:Springer; 2017. p. 223–45. [DOI] [PubMed]
- 14.Schwab C, Harrison CJ. Advances in B-cell Precursor Acute Lymphoblastic Leukemia Genomics. Hemasphere. 2018;2:e53. doi: 10.1097/HS9.0000000000000053. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Chen B, Jiang L, Zhong M-L, Li J-F, Li B-S, Peng L-J, et al. Identification of fusion genes and characterization of transcriptome features in T-cell acute lymphoblastic leukemia. PNAS. 2018;115:373–8. doi: 10.1073/pnas.1717125115. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Matlawska-Wasowska K, Kang H, Devidas M, Wen J, Harvey RC, Nickl CK, et al. MLL rearrangements impact outcome in HOXA-deregulated T-lineage acute lymphoblastic leukemia: a Children’s Oncology Group Study. Leukemia. 2016;30:1909–12. doi: 10.1038/leu.2016.60. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Seki M, Kimura S, Isobe T, Yoshida K, Ueno H, Nakajima-Takagi Y, et al. Recurrent SPI1 (PU.1) fusions in high-risk pediatric T cell acute lymphoblastic leukemia. Nat Genet. 2017;49:1274–81. doi: 10.1038/ng.3900. [DOI] [PubMed] [Google Scholar]
- 18.Van Vlierberghe P, van Grotel M, Tchinda J, Lee C, Beverloo HB, van der Spek PJ, et al. The recurrent SET-NUP214 fusion as a new HOXA activation mechanism in pediatric T-cell acute lymphoblastic leukemia. Blood. 2008;111:4668–80. doi: 10.1182/blood-2007-09-111872. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Brandimarte L, Pierini V, Di Giacomo D, Borga C, Nozza F, Gorello P, et al. New MLLT10 gene recombinations in pediatric T-acute lymphoblastic leukemia. Blood. 2013;121:5064–7. doi: 10.1182/blood-2013-02-487256. [DOI] [PubMed] [Google Scholar]
- 20.Ben Abdelali R, Roggy A, Leguay T, Cieslak A, Renneville A, Touzart A, et al. SET-NUP214 is a recurrent γδ lineage-specific fusion transcript associated with corticosteroid/chemotherapy resistance in adult T-ALL. Blood. 2014;123:1860–3. doi: 10.1182/blood-2013-08-521518. [DOI] [PubMed] [Google Scholar]
- 21.Asnafi V, Beldjord K, Boulanger E, Comba B, Le Tutour P, Estienne MH, et al. Analysis of TCR, pT alpha, and RAG-1 in T-acute lymphoblastic leukemias improves understanding of early human T-lymphoid lineage commitment. Blood. 2003;101:2693–703. doi: 10.1182/blood-2002-08-2438. [DOI] [PubMed] [Google Scholar]
- 22.Delabesse E, Bernard M, Landman-Parker J, Davi F, Leboeuf D, Varet B, et al. Simultaneous SIL-TAL1 RT-PCR detection of all tal(d) deletions and identification of novel tal(d) variants. Br J Haematol. 1997;99:901–7. doi: 10.1046/j.1365-2141.1997.4833286.x. [DOI] [PubMed] [Google Scholar]
- 23.Asnafi V, Beldjord K, Libura M, Villarese P, Millien C, Ballerini P, et al. Age-related phenotypic and oncogenic differences in T-cell acute lymphoblastic leukemias may reflect thymic atrophy. Blood. 2004;104:4173–80. doi: 10.1182/blood-2003-11-3944. [DOI] [PubMed] [Google Scholar]
- 24.Ballerini P, Blaise A, Busson-Le Coniat M, Su XY, Zucman-Rossi J, Adam M, et al. HOX11L2 expression defines a clinical subtype of pediatric T-ALL associated with poor prognosis. Blood. 2002;100:991–7. doi: 10.1182/blood-2001-11-0093. [DOI] [PubMed] [Google Scholar]
- 25.Bond J, Marchand T, Touzart A, Cieslak A, Trinquand A, Sutton L, et al. An early thymic precursor phenotype predicts outcome exclusively in HOXA-overexpressing adult T-cell acute lymphoblastic leukemia: a Group for Research in Adult Acute Lymphoblastic Leukemia study. Haematologica. 2016;101:732–40. doi: 10.3324/haematol.2015.141218. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Ruminy P, Marchand V, Buchbinder N, Larson T, Joly B, Penther D, et al. Multiplexed targeted sequencing of recurrent fusion genes in acute leukaemia. Leukemia. 2016;30:757–60. doi: 10.1038/leu.2015.177. [DOI] [PubMed] [Google Scholar]
- 27.Deshpande AJ, Rouhi A, Lin Y, Stadler C, Greif PA, Arseni N, et al. The clathrin-binding domain of CALM and the OM-LZ domain of AF10 are sufficient to induce acute myeloid leukemia in mice. Leukemia. 2011;25:1718–27. doi: 10.1038/leu.2011.153. [DOI] [PubMed] [Google Scholar]
- 28.Graux C, Stevens-Kroef M, Lafage M, Dastugue N, Harrison CJ, Mugneret F, et al. Heterogeneous patterns of amplification of the NUP214-ABL1 fusion gene in T-cell acute lymphoblastic leukemia. Leukemia. 2009;23:125–33. doi: 10.1038/leu.2008.278. [DOI] [PubMed] [Google Scholar]
- 29.Hagemeijer A, Graux C. ABL1 rearrangements in T-Cell acute lymphoblastic leukemia. Genes Chromosom Cancer. 2010;49:299–308. doi: 10.1002/gcc.20743. [DOI] [PubMed] [Google Scholar]
- 30.Forgione MO, McClure BJ, Yeung DT, Eadie LN, White DL. MLLT10 rearranged acute leukemia: incidence, prognosis, and possible therapeutic strategies. Genes, Chromosomes Cancer. 2020;59:709–21. doi: 10.1002/gcc.22887. [DOI] [PubMed] [Google Scholar]
- 31.t(10;11)(p13–14;q14–21): A New Recurrent Translocation in T-Cell Acute Lymphoblastic Leukemias. Groupe Français de Cytogénétique Hématologique (GFCH). Genes Chromosom Cancer.1991;3:411–5. [PubMed]
- 32.Bond J, Bergon A, Durand A, Tigaud I, Thomas X, Asnafi V, et al. Cryptic XPO1-MLLT10 translocation is associated with HOXA locus deregulation in T-ALL. Blood. 2014;124:3023–5. doi: 10.1182/blood-2014-04-567636. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Romana SP, Radford-Weiss I, Ben Abdelali R, Schluth C, Petit A, Dastugue N, et al. NUP98 rearrangements in hematopoietic malignancies: a study of the Groupe Francophone de Cytogénétique Hématologique. Leukemia. 2006;20:696–706. doi: 10.1038/sj.leu.2404130. [DOI] [PubMed] [Google Scholar]
- 34.Ben Abdelali R, Asnafi V, Petit A, Micol J-B, Callens C, Villarese P, et al. The prognosis of CALM-AF10-positive adult T-cell acute lymphoblastic leukemias depends on the stage of maturation arrest. Haematologica. 2013;98:1711–7. doi: 10.3324/haematol.2013.086082. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Lo Nigro L, Mirabile E, Tumino M, Caserta C, Cazzaniga G, Rizzari C, et al. Detection of PICALM-MLLT10 (CALM-AF10) and outcome in children with T-lineage acute lymphoblastic leukemia. Leukemia. 2013;27:2419–21. doi: 10.1038/leu.2013.149. [DOI] [PubMed] [Google Scholar]
- 36.Dik WA, Brahim W, Braun C, Asnafi V, Dastugue N, Bernard OA, et al. CALM-AF10+ T-ALL expression profiles are characterized by overexpression of HOXA and BMI1 oncogenes. Leukemia. 2005;19:1948–57. doi: 10.1038/sj.leu.2403891. [DOI] [PubMed] [Google Scholar]
- 37.Okada Y, Feng Q, Lin Y, Jiang Q, Li Y, Coffield VM, et al. hDOT1L Links Histone Methylation to Leukemogenesis. Cell. 2005;121:167–78. doi: 10.1016/j.cell.2005.02.020. [DOI] [PubMed] [Google Scholar]
- 38.Okada Y, Jiang Q, Lemieux M, Jeannotte L, Su L, Zhang Y. Leukaemic transformation by CALM-AF10 involves upregulation of Hoxa5 by hDOT1L. Nat Cell Biol. 2006;8:1017–24. doi: 10.1038/ncb1464. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.van Grotel M, Meijerink JP, Beverloo HB, Langerak AW, Buys-Gladdines JG, Schneider P, et al. The outcome of molecular-cytogenetic subgroups in pediatric T-cell acute lymphoblastic leukemia: a retrospective study of patients treated according to DCOG or COALL protocols. Haematologica. 2006;91:1212–21. [PubMed] [Google Scholar]
- 40.Chen L, Deshpande AJ, Banka D, Bernt KM, Dias S, Buske C, et al. Abrogation of MLL–AF10 and CALM–AF10-mediated transformation through genetic inactivation or pharmacological inhibition of the H3K79 methyltransferase Dot1l. Leukemia. 2013;27:813–22. doi: 10.1038/leu.2012.327. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.McCabe NR, Kipiniak M, Kobayashi H, Thirman M, Gill H, Rowley JD, et al. DNA rearrangements and altered transcripts of the MLL gene in a human T-all cell line karpas 45 with a t(X;II) (q13;q23) translocation. Genes Chromosom Cancer. 1994;9:221–4. doi: 10.1002/gcc.2870090311. [DOI] [PubMed] [Google Scholar]
- 42.Hayette S, Tigaud I, Maguer-Satta V, Bartholin L, Thomas X, Charrin C, et al. Recurrent involvement of the MLL gene in adult T-lineage acute lymphoblastic leukemia. Blood. 2002;99:4647–9. doi: 10.1182/blood-2002-03-0717. [DOI] [PubMed] [Google Scholar]
- 43.Tanabe S, Zeleznik-Le NJ, Kobayashi H, Vignon C, Espinosa R, LeBeau MM, et al. Analysis of the t(6;11)(q27;q23) in leukemia shows a consistent breakpoint in AF6 in three patients and in the ML-2 cell line. Genes Chromosom Cancer. 1996;15:206–16. doi: 10.1002/(SICI)1098-2264(199604)15:4<206::AID-GCC2>3.0.CO;2-5. [DOI] [PubMed] [Google Scholar]
- 44.Peterson JF, Baughn LB, Pearce KE, Williamson CM, Benevides Demasi JC, Olson RM, et al. KMT2A (MLL) rearrangements observed in pediatric/young adult T-lymphoblastic leukemia/lymphoma: a 10-year review from a single cytogenetic laboratory. Genes Chromosom Cancer. 2018;57:541–6. doi: 10.1002/gcc.22666. [DOI] [PubMed] [Google Scholar]
- 45.Ferrando AA, Armstrong SA, Neuberg DS, Sallan SE, Silverman LB, Korsmeyer SJ, et al. Gene expression signatures in MLL-rearranged T-lineage and B-precursor acute leukemias: dominance of HOX dysregulation. Blood. 2003;102:262–8. doi: 10.1182/blood-2002-10-3221. [DOI] [PubMed] [Google Scholar]
- 46.Rubnitz JE, Camitta BM, Mahmoud H, Raimondi SC, Carroll AJ, Borowitz MJ, et al. Childhood acute lymphoblastic leukemia with the MLL-ENL fusion and t(11;19)(q23;p13.3) translocation. J Clin Oncol. 1999;17:191–6. doi: 10.1200/JCO.1999.17.1.191. [DOI] [PubMed] [Google Scholar]
- 47.Chae H, Lim J, Kim M, Park J, Kim Y, Han K, et al. Phenotypic and genetic characterization of adult T-cell acute lymphoblastic leukemia with del(9)(q34);SET-NUP214 rearrangement. Ann Hematol. 2012;91:193–201. doi: 10.1007/s00277-011-1289-x. [DOI] [PubMed] [Google Scholar]
- 48.Quentmeier H, Schneider B, Röhrs S, Romani J, Zaborski M, MacLeod RA, et al. SET-NUP214 fusion in acute myeloid leukemia- and T-cell acute lymphoblastic leukemia-derived cell lines. BioMed Central. 2009;2:1–5. doi: 10.1186/1756-8722-2-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Oka M, Mura S, Otani M, Miyamoto Y, Nogami J, Maehara K, et al. Chromatin-bound CRM1 recruits SET-Nup214 and NPM1c onto HOX clusters causing aberrant HOX expression in leukemia cells. eLife. 2019;8:e46667. doi: 10.7554/eLife.46667. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Saito S, Cigdem S, Okuwaki M, Nagata K. Leukemia-Associated Nup214 Fusion Proteins Disturb the XPO1-Mediated Nuclear-Cytoplasmic Transport Pathway and Thereby the NF-κB Signaling Pathway. Mol Cell Biol. 2016;36:1820–35. doi: 10.1128/MCB.00158-16. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Vanden Bempt M, Demeyer S, Broux M, De Bie J, Bornschein S, Mentens N, et al. Cooperative Enhancer Activation by TLX1 and STAT5 Drives Development of NUP214-ABL1/TLX1-Positive T Cell Acute Lymphoblastic Leukemia. Cancer Cell. 2018;34:271–.e7. doi: 10.1016/j.ccell.2018.07.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Burmeister T, Gökbuget N, Reinhardt R, Rieder H, Hoelzer D, Schwartz S. NUP214-ABL1 in adult T-ALL: the GMALL study group experience. Blood. 2006;108:3556–9. doi: 10.1182/blood-2006-04-014514. [DOI] [PubMed] [Google Scholar]
- 53.De Keersmaecker K, Graux C, Odero MD, Mentens N, Somers R, Maertens J, et al. Fusion of EML1 to ABL1 in T-cell acute lymphoblastic leukemia with cryptic t(9;14)(q34;q32) Blood. 2005;105:4849–52. doi: 10.1182/blood-2004-12-4897. [DOI] [PubMed] [Google Scholar]
- 54.Schlieben S, Borkhardt A, Reinisch I, Ritterbach J, Janssen JW, Ratei R, et al. Incidence and clinical outcome of children with BCR/ABL-positive acute lymphoblastic leukemia (ALL). A prospective RT-PCR study based on 673 patients enrolled in the German pediatric multicenter therapy trials ALL-BFM-90 and CoALL-05-92. Leukemia. 1996;10:957–63. [PubMed] [Google Scholar]
- 55.Keersmaecker KD, Porcu M, Cox L, Girardi T, Vandepoel R, Beeck JO de et al. NUP214-ABL1-mediated cell proliferation in T-cell acute lymphoblastic leukemia is dependent on the LCK kinase and various interacting proteins. 2014;99:85–93. [DOI] [PMC free article] [PubMed]
- 56.Graux C, Cools J, Melotte C, Quentmeier H, Ferrando A, Levine R, et al. Fusion of NUP214 to ABL1 on amplified episomes in T-cell acute lymphoblastic leukemia. Nat Genet. 2004;36:1084–9. doi: 10.1038/ng1425. [DOI] [PubMed] [Google Scholar]
- 57.Quintás-Cardama A, Tong W, Manshouri T, Vega F, Lennon PA, Cools J, et al. Activity of tyrosine kinase inhibitors against human NUP214-ABL1-positive T cell malignancies. Leukemia. 2008;22:1117–24. doi: 10.1038/leu.2008.80. [DOI] [PubMed] [Google Scholar]
- 58.Ballerini P, Landman-Parker J, Cayuela JM, Asnafi V, Labopin M, Gandemer V, et al. Impact of genotype on survival of children with T-cell acute lymphoblastic leukemia treated according to the French protocol FRALLE-93: the effect of TLX3/HOX11L2 gene expression on outcome. Haematologica. 2008;93:1658–65. doi: 10.3324/haematol.13291. [DOI] [PubMed] [Google Scholar]
- 59.Wasylyk B, Hahn SL, Giovane A. The Ets family of transcription factors. In: Christen P, Hofmann E, editors. EJB Reviews 1993. Berlin, Heidelberg:Springer;1994. p. 7–18. [DOI] [PubMed]
- 60.Klossowski S, Miao H, Kempinska K, Wu T, Purohit T, Kim E, et al. Menin inhibitor MI-3454 induces remission in MLL1-rearranged and NPM1-mutated models of leukemia. J Clin Investig. 2020;130:981–97. doi: 10.1172/JCI129126. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.