Abstract
The ViroSeq HIV-1 Genotyping System is a commercially available, integrated sequence-based system for analysis of human immunodeficiency virus type 1 (HIV-1) drug resistance. We evaluated the performance of this system by analyzing HIV-1 in pediatric plasma samples. Plasma samples from children 4 months to 17 years of age were obtained from a clinical trial protocol (PACTG 377). Children in PACTG 377 were randomized to four treatment arms, including different combinations of antiretroviral drugs. HIV-1 genotyping was performed using samples collected prior to antiretroviral therapy (baseline) and at the time of virologic failure. Performance of the genotyping system was compared in three university laboratories. A total of 196 samples were analyzed, including 135 baseline and 61 failure samples. Plasma volumes ranged from 0.05 to 0.5 ml, and viral loads ranged from 1,084 to 3,484,991 copies/ml. PCR products suitable for sequencing were obtained for 192 of the 196 samples. Complete sequences for protease and reverse transcriptase were obtained for all of these 192 samples. For 180 samples, data were obtained from both DNA strands for the entire region analyzed. There was no evidence of sample cross-contamination based on phylogenetic analysis of HIV-1 sequences. Performance of the genotyping system was similar in three laboratories. This genotyping system performs well for analysis of HIV-1 in pediatric plasma samples, including those with low volume and low viral load. The availability of this system should facilitate studies of HIV-1 drug resistance.
Antiretroviral therapy can prolong the lives and improve the health of patients with human immunodeficiency virus type 1 (HIV-1) infection. However, selection of HIV-1 with resistance to antiretroviral drugs can limit the efficacy of antiretroviral treatment regimens and limit a patient's treatment options. Drug-resistant HIV-1 can be transmitted from one adult to another (4, 13) and from women to their children by vertical transmission (1, 3). Therefore, an individual may have drug-resistant HIV-1 even prior to the initiation of antiretroviral therapy (6).
Currently licensed antiretroviral drugs include inhibitors of HIV-1 protease and reverse transcriptase (RT). Resistance to these drugs is associated with mutations in the genes encoding HIV-1 protease and RT which can be detected by genotyping assays (5). Recent studies suggest that HIV-1 genotypic resistance testing may be useful for patients with acute HIV-1 infection to assist in selection of an initial treatment regimen. Testing may also be useful in treatment failure in order to determine whether resistant HIV-1 has been selected and to help guide changes in therapy (5, 9).
A major challenge in the design of HIV-1 genotypic resistance assays is that HIV-1 viruses have a high level of genetic diversity (6). HIV-1 viruses differ not only from one geographic region to another, and from person to person but also within a given individual. Genotyping assays typically require the use of DNA primers to detect resistance mutations. Recent studies illustrate the problems that genetic heterogeneity can cause when genotypic assays are based on hybridization of primers to HIV-1 templates (8, 10, 11). Sequencing-based genotypic resistance assays typically require the use of several primers for reverse transcription, PCR, and sequencing. Each of these primers must be directed toward a highly conserved region of the HIV-1 genome in order for the analysis to be successful. Genotypic analysis is further complicated in a pediatric setting, since the plasma volumes available for analysis may be small. This problem is frequently encountered when stored samples collected in the context of clinical trials are analyzed in retrospective studies.
An integrated, sequence-based system for HIV-1 genotyping has been developed by Applied Biosystems (Foster City, Calif.). The ViroSeq version of this system is now commercially available for research use. This system utilizes a total of 10 DNA primers for genotypic analysis (1 for reverse transcription, 2 for PCR amplification, and 7 for cycle sequencing). We evaluated the performance of the Applied Biosystems ViroSeq HIV-1 Genotyping System by analyzing 196 plasma samples from pediatric patients with HIV-1 infection. Plasma samples were obtained as part of the Pediatric AIDS Clinical Trials Group (PACTG) Protocol 377 (12). Children ages 4 months to 17 years were enrolled in to PACTG 377 between December 1997 and September 1999. Prior treatment with zidovudine (AZT), zalcitabine (ddC), or didanosine (ddI) was acceptable. Plasma samples were obtained prior to starting antiretroviral therapy (baseline samples). Children were randomized to four treatment arms, including different combinations of the following antiretroviral drugs: stavudine (d4T), lamivudine (3TC), nevirapine (NVP), nelfinavir (NFV), and ritonavir (RTV) (12). Children were monitored for up to 48 weeks for analysis of virologic response to treatment. Some children did not achieve satisfactory viral suppression by 12 weeks or had elevated HIV-1 RNA levels at later time points (12 to 48 weeks). Plasma samples were obtained from these patients at the time of virologic failure before study treatment was changed or discontinued (failure samples). This study included a large number of samples with plasma volumes of <0.5 ml. In this analysis, we focused on the ability of this genotyping system to successfully amplify DNA for sequencing and to provide complete DNA sequences of the regions of interest.
MATERIALS AND METHODS
Samples used for analysis.
Plasma samples were obtained as part of PACTG 377 (see above) (12). Plasma was isolated from EDTA-anticoagulated whole blood within 6 h of sample collection. HIV-1 RNA levels (viral loads) were measured in PACTG 377 using the Amplicor HIV-1 Monitor Test kit (Roche Diagnostic Systems, Inc., Branchburg, N.J.) with a quantitative range from 400 to 750,000 copies/ml. Samples with viral loads of >750,000 copies/ml were analyzed following 1:100 dilution. Plasma samples were stored frozen at −80°C prior to genotypic analysis. Baseline and failure samples were distributed among three different laboratories for analysis. Each laboratory had a unique sample set.
HIV-1 genotyping.
HIV-1 genotyping was performed using the Applied Biosystems ViroSeq HIV-1 Genotyping System (Applied Biosystems) according to the manufacturer's instructions, with the following exception. Where indicated, less than the recommended 0.5 ml of plasma was used for analysis. In this system, HIV-1 RNA is extracted from plasma samples, and one-fifth of the extracted RNA is reverse transcribed with murine Moloney virus RT. A 1.8-kb DNA fragment is then amplified in the same tube in a single 40-cycle PCR with AmpliTaq gold polymerase and uracil N-deglycosylase decontamination control. PCR products are purified using spin columns and analyzed by agarose gel electrophoresis. The PCR products are sequenced with premixed BigDye sequencing reagents in seven separate reactions. BigDye terminator chemistry provides 98% accuracy at 550 bases for the ABI Prism 377 DNA Sequencer and 98.5% accuracy at 600 bases for the ABI Prism 310 Genetic Analyzer according to product bulletins issued by the manufacturer. The resulting sequences are assembled and analyzed using HIV-1 Genotyping System software.
Phylogenetic analysis.
Protease and RT nucleotide sequences (297 and 972 nucleotides, respectively) were aligned. Phylogenetic reconstructions were performed with PHYLIP 3.572c, by using the neighbor-joining method. Reference sequences were obtained from the 1999 HIV-1 Subtype Reference Alignments of the Los Alamos National Laboratory web site (http://hiv-web.lanl.gov/) and included subtypes A to K, as well as recombinant sequences and the laboratory strains pNL4-3 and HXB2.
RESULTS
The performance of the Applied Biosystems ViroSeq HIV-1 Genotyping System was evaluated in laboratories at The Johns Hopkins University School of Medicine, The University of California at Los Angeles School of Medicine, and the University of Medicine and Dentistry of New Jersey (laboratories 1 to 3, respectively in Table 1). Each laboratory analyzed baseline samples obtained prior to antiretroviral therapy, as well as failure samples obtained at defined time points while study subjects were still on antiretroviral therapy. A total of 196 plasma samples were analyzed, including 135 baseline samples and 61 failure samples (Table 1).
TABLE 1.
Samples used for analysis
| Laboratory | No. of samples
|
Mean volume (ml [range]) | No. (%) of samples with <0.5 ml | Mean viral load (copies/ml [range]) | No. (%) of samples with <10,000 copies/ml | No. of samples with <1,000 input RNA copies | ||
|---|---|---|---|---|---|---|---|---|
| Baseline | Failure | Total | ||||||
| 1 | 46 | 19 | 65 | 0.42 (0.1–0.5) | 29 (45) | 143,180 (1,084–1,794,664) | 12 (18) | 3 |
| 2 | 45 | 21 | 66 | 0.4 (0.1–0.5) | 30 (45) | 106,711 (1,205–750,000) | 16 (24) | 1 |
| 3 | 44 | 21 | 65 | 0.43 (0.05–0.5) | 24 (37) | 121,028 (1,370–3,484,991) | 19 (29) | 4 |
| Total | 135 | 61 | 196 | 0.42 (0.05–0.5) | 84 (43) | 123,533 (1,084–3,484,991) | 47 (24) | 8 |
Applied Biosystems recommends use of the ViroSeq system for 0.5-ml plasma samples with viral loads of >2,000 copies/ml. This amount corresponds to an input RNA copy number (i.e., RNA copies/milliliter × milliliters of plasma) of 1,000 copies. In this study, the volume of plasma available for analysis was <0.5 ml for 43% of the samples. The range of plasma volumes, mean plasma volume, and number of samples with plasma volumes of <0.5 ml analyzed in each laboratory are shown in Table 1. In addition, 47 (24%) of the samples had viral loads of <10,000 copies/ml. The ranges of viral loads and mean viral loads for samples analyzed in each laboratory are shown in Table 1. Of note, the sample set included eight samples with input RNA copy numbers of <1,000 (median 742 copies; range 480 to 899 copies).
RNA isolation, reverse transcription, and PCR amplification were performed for all 196 samples. The quality and quantity of the PCR products were analyzed by agarose gel electrophoresis. PCR yielded sufficient DNA for sequencing for 98% of the samples tested. This included seven of eight samples for which the input RNA copy number was <1,000 copies. The four samples for which a suitable product was not obtained had the following viral loads and plasma volumes: 1,205 copies/ml and 0.4 ml (482 copies), 17,738 copies/ml and 0.15 ml (2,661 copies), 6,003 copies/ml and 0.5 ml (3,002 copies), and 142,753 copies/ml and 0.33 ml (47,108 copies). The failure to obtain a suitable PCR product for the latter sample was attributed to loss of the viral pellet during viral RNA isolation. PCR products suitable for sequence analysis were obtained from samples collected at other times from all four of these individuals.
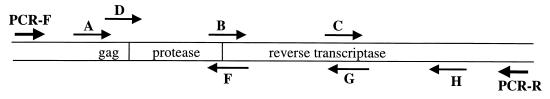
DNA sequencing was performed using premixed reagents with seven sequencing primers. Figure 1 shows the position and orientation of the sequencing primers on the PCR product. Sequencing reactions were analyzed on either an ABI Prism 377 DNA Sequencer or an ABI Prism 310 Genetic Analyzer. One of the three laboratories used a core facility for sequence analysis. We analyzed the performance of the seven sequencing primers. For this analysis, we considered a primer to have failed if the resulting sequence did not assemble with the other sequences from the same sample using the Applied Biosystems ViroSeq HIV-1 Genotyping System software package. In this system, the A and D primers are alternate sequencing primers that bind to the genetically heterogeneous gag region. Either of these primers can be used to generate a consensus sequence for analysis. In this study, sequencing with primer A was successful for 176 (92%) of the 192 samples, and sequencing with primer D was successful for 169 (88%) of the 192 samples. Both primers A and D failed in only 3 (1.6%) of 192 samples. When an individual primer fails on the first sequencing attempt, it is sometimes possible to obtain a usable sequence if the analysis with that primer is repeated. In this study, sequencing reactions and/or analysis of sequencing products was repeated if (i) both primers A and D failed (in this case, analysis with both primers was repeated) or (ii) any other primer (B, C, F, G, or H) failed. Using this strategy, each of the three laboratories performed repeat analysis for <4% of the primer reactions. Sequencing analysis was successful for >70% of the repeated reactions in all three laboratories (Table 2).
FIG. 1.
Orientation and position of PCR and sequencing primers. The position and orientation of primers in the ViroSeq HIV-1 Genotyping System are shown with respect to the protease and RT coding regions. The two PCR primers (PCR-F and PCR-R) and the seven sequencing primers are shown.
TABLE 2.
Assay performance
| Laboratory | No. (%) of samples with PCR product sufficient for sequencing | No. of primer reactions performed | No. (%) of primer reactions repeated | No. (%) of repeat primer reactions successful | No. (%) of genotypes obtained | No. (%) of genotypes based on full double-stranded sequences |
|---|---|---|---|---|---|---|
| 1 | 64/65 (98) | 448 | 17 (3.8) | 12/17 (71) | 64/65 (98.5) | 56/65 (86) |
| 2 | 64/66 (97) | 448 | 15 (3.3) | 12/15 (80) | 64/66 (97) | 62/66 (95) |
| 3 | 64/65 (98) | 448 | 11 (2.4) | 9/11 (82) | 64/65 (98.5) | 62/65 (95) |
| Total | 192/196 (98) | 1,344 | 43 (3.2) | 33/43 (77) | 192/196 (98) | 180/196 (92) |
We considered genotyping to be successful if the analysis yielded a complete sequence for HIV-1 protease amino acids 1 to 99 and HIV-1 RT amino acids 1 to 320. In this report, genotyping was successful for all 192 samples for which adequate PCR products were obtained (Table 2). For 180 samples, sequence data from both DNA strands was obtained for the entire region analyzed. For the remaining samples, a small region of the consensus sequence was generated using data from only one DNA strand. This reflected repeated failure of both primers A and D (three samples), the F primer (two samples), or the G primer (two samples). For five additional samples, sequences obtained with the H primer assembled successfully with the other sequences but had a small internal region of sequence ambiguity.
Mutations associated with resistance to the PACTG 377 study drugs (3TC, d4T, NVP, RTV, and NFV), as well as mutations associated with resistance to other drugs (e.g., ddI, ddC, and AZT) were detected in this cohort. Detailed analyses of the mutations detected and their relationship to outcome in protocol PACTG 377 will be reported elsewhere (S. H. Eshleman et al., unpublished results).
For quality control, we used phylogenetic analysis to compare the nucleotide sequences obtained for each plasma sample to one another and to those of laboratory strains of HIV-1. None of the sequences resembled those of strains pNL4-3 or HXB2, which were present in some of the genotyping laboratories. Furthermore, sequences from individual children derived from samples collected at different times clustered more closely with one another than with sequences from other children in the study, and no two sequences were identical. Therefore, it is unlikely that samples were cross-contaminated (e.g., during PCR) or misidentified.
DISCUSSION
This study demonstrates successful use of the Applied Biosystems ViroSeq HIV-1 Genotyping System for analysis of HIV-1 protease and RT in pediatric plasma samples. This system uses direct sequence analysis of PCR products to determine the predominant sequence in a mixed population. Plasma samples from HIV-1-infected individuals often contain mixtures of HIV-1 with or without drug resistance mutations. Drug resistance mutations can be detected as amino acid mixtures using direct sequencing. However, site-specific PCR or mutation-specific hybridization assays may be more sensitive for the detection of specific mutations that are present at low levels. It is usually necessary to sequence cloned cDNAs to determine the linkage of specific mutations in individual HIV-1 viruses.
Performance of the ViroSeq genotyping system was similar in three different laboratories. Genotyping was successful for 98% of the 196 samples analyzed in this study, even though 43% of the samples had plasma volumes below the 0.5-ml volume recommended for analysis. This is important, since samples with small volumes are often obtained from pediatric patients and since the volume of plasma available for genotyping from samples obtained in clinical trials is often well below the volume required by most commercial laboratories for genotypic resistance testing. Furthermore, genotyping was successful for seven of eight samples with <1,000 input RNA copies.
Genotypic analysis of HIV-1 is complicated by several factors. First, the concentration of viral RNA used as a template for reverse transcription and PCR amplification (viral load) varies significantly from sample to sample. In this study, the viral load varied by >3 orders of magnitude (from 1,084 to 3,484,991 copies/ml). Second, because HIV-1 has an RNA genome, special care must be taken to avoid degradation of the viral template, and reverse transcription must be performed prior to PCR. Finally, analysis is complicated by the high degree of genetic diversity of the virus. Viral sequences vary significantly from individual to individual, and viruses in a single plasma sample are also genetically diverse. Primers used for reverse transcription, PCR amplification, and DNA sequencing must be directed to conserved regions of the virus to maximize homology between the template and primer sequences. In this study, genotyping was unsuccessful for only 4 of 196 samples. In all four cases, RNA isolation, reverse transcription, or PCR amplification resulted in insufficient DNA for sequencing. In each case, samples from the same individuals collected at different times were successfully analyzed. This suggests that genetic differences between the primers used for analysis and the viral sequence were unlikely to be the cause of the analytic failures. Genetic differences among different HIV-1 subtypes can also complicate genotyping analysis. The ViroSeq HIV-1 Genotyping System was developed for analysis of subtype B HIV-1, which is the most common subtype in the United States. However, the system has been used successfully for analysis of diverse HIV-1 subtypes (2, 7).
For sequence-based assays, it is desirable to analyze sequences from both DNA strands. In this study, complete double-stranded sequence information was obtained for 94% of the samples that were successfully genotyped. In the remaining samples, a small region (typically encoding fewer than 100 amino acids) was sequenced in one direction only. In this study, single-stranded regions were obtained most frequently due to ambiguities in a region of the sequence obtained with the H primer.
A concern using any PCR-based assay is contamination (e.g., sample cross-contamination or contamination with reference strains used in the laboratory). One advantage of the ViroSeq system is that reagents and protocols are sufficiently optimized so that nested PCR is not required. This minimizes sample handling and lowers the risk of sample contamination. Another advantage of this system is the inclusion of a uracil N-deglycosylase contamination control system, which serves to degrade contaminating DNA generated in previous reactions. In this study, phylogenetic analysis confirmed the absence of sample cross-contamination or contamination with laboratory strains of HIV-1 in the three testing laboratories.
This report demonstrates successful use of a commercial system for HIV-1 drug resistance genotyping in three University laboratories. This system is currently available for research use only. Validation studies of this system are currently under way to assess its suitability for clinical use.
ACKNOWLEDGMENTS
This study was supported by the Adult and Pediatric AIDS Clinical Trials Groups (NIH-NIAID). P.K. is an Elizabeth Glaser Scientist supported by the Pediatric AIDS Foundation. S.H.E. was also supported by the NIH HIV-1 Prevention Trials Network (U01-AI-46745-01), the NIH-CH/HD (R29 34348), and the Elizabeth Glaser Pediatric AIDS Foundation. D.L., W.L., and P.P. were also supported by the NIH-NIAID (2U01-AI25883 and contract 97PVCL08).
We thank the following investigators: Andy Wiznia (Jacobi Medical Center, Bronx, N.Y.), Sharon Nachman (State University of New York at Stony Brook), Ken Stanley (Harvard School of Public Health), George Johnson (Medical University of South Carolina), and the PACTG 377 Study Team for providing plasma samples and clinical and laboratory data for this study. We also thank Eric Shulse, Craig Lindquist, and the HIV-1 Genotyping Team (Applied Biosystems) for helpful discussions and for providing reagents and supplies for this study and Yvonne Bryson (University of California, Los Angeles School of Medicine) for facilitating the study at UCLA.
REFERENCES
- 1.Colgrove R C, Pitt J, Chung P H, Welles S L, Japour A J. Selective vertical transmission of HIV-1 antiretroviral resistance mutations. AIDS. 1998;12:2281–2288. doi: 10.1097/00002030-199817000-00009. [DOI] [PubMed] [Google Scholar]
- 2.Dileanis J, Marlowe N, Hoo B, Brown R C, Bulmer M, Huang D, Palumbo P, Schuurman R, Van Laethem K, Vandamme A-M, Elbeik T. 5th International Congress on Drug Therapy in HIV Infection, Glasgow, United Kingdom. 2000. Performance of the new ViroSeq HIV-1 Genotyping System (version 2) with group M subtype panel and with subtype B clinical research samples at test sites. [Google Scholar]
- 3.Frenkel L M, Wagner L E, Demeter L M, Dewhurst S, Coombs R W, Murante B L, Reichman R C. Effects of zidovudine use during pregnancy on resistance and vertical transmission of human immunodeficiency virus type 1. Clin Infect Dis. 1995;20:1321–1326. doi: 10.1093/clinids/20.5.1321. [DOI] [PubMed] [Google Scholar]
- 4.Hecht F M, Grant R M, Petropoulos C J, Dillon B, Chesney M A, Hellmann N S, Bandrapalli N I, Digilio L, Branson B, Kahn J O. Sexual transmission of an HIV-1 variant resistant to multiple reverse transcriptase and protease inhibitors. N Engl J Med. 1998;339:307–311. doi: 10.1056/NEJM199807303390504. [DOI] [PubMed] [Google Scholar]
- 5.Hirsch M S, Brun-Vezinet F, D'Aquila R, Hammer S M, Johnson V A, Kuritzkes D R, Loveday C, Mellors J W, Clotet B, Conway B, Demeter L M, Vella S, Jacobsen D M, Richman D D. Antiretroviral drug resistance testing in adult HIV-1 infection: recommendations of an international AIDS society–USA panel. JAMA. 2000;283:2417–2426. doi: 10.1001/jama.283.18.2417. [DOI] [PubMed] [Google Scholar]
- 6.Hirsch M S, Conway B, D'Aquila R T, Johnson V A, Brun-Vezinet F, Clotet B, Demeter L M, Hammer S M, Jacobsen D M, Kuritzkes D R, Loveday C, Mellors J W, Vella S, Richman D D. Antiretroviral drug resistance testing in adults with HIV infection: implications for clinical management. JAMA. 1998;279:1984–1991. doi: 10.1001/jama.279.24.1984. [DOI] [PubMed] [Google Scholar]
- 7.Jackson J B, Mracna M, Guay L, Dileanis J, Musoke P, Mmiro F, Eshleman S H. XIII International AIDS Conference, Durban, South Africa. 2000. Selection of nevirapine (NVP) resistance mutations in Ugandan women and infants receiving NVP prophylaxis to prevent HIV-1 vertical transmission (HIVNET-012) [Google Scholar]
- 8.Koch N, Yahi N, Colson P, Fantini J, Tamalet C. Genetic polymorphism near HIV-1 reverse transcriptase resistance-associated codons is a major obstacle for the line probe assay as an alternative method to sequence analysis. J Virol Methods. 1999;80:25–31. doi: 10.1016/s0166-0934(99)00030-0. [DOI] [PubMed] [Google Scholar]
- 9.Panel on Clinical Practices for Treatment of HIV Infection. Guidelines for the use of antiretroviral agents in HIV-1-infected adults and adolescents. Washington D.C. http://www.hivatis.org 1/28/2000. Department of Health and Human Services; 2000. [DOI] [PubMed] [Google Scholar]
- 10.Puchhammer-Stockl E, Schmied B, Mandl C W, Vetter N, Heinz F X. Comparison of line probe assay (LIPA) and sequence analysis for detection of HIV-1 drug resistance. J Med Virol. 1999;57:283–289. doi: 10.1002/(sici)1096-9071(199903)57:3<283::aid-jmv12>3.0.co;2-2. [DOI] [PubMed] [Google Scholar]
- 11.Vahey M, Nau M E, Barrick S, Cooley J D, Sawyer R, Sleeker A A, Vickerman P, Bloor S, Larder B, Michael N L, Wegner S A. Performance of the Affymetrix GeneChip HIV PRT 440 platform for antiretroviral drug resistance genotyping of human immunodeficiency virus type 1 clades and viral isolates with length polymorphisms. J Clin Microbiol. 1999;37:2533–2537. doi: 10.1128/jcm.37.8.2533-2537.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Wiznia A, Stanley K, Krogstad P, Johnson G, Lee S, McNamara J, Moye J, Jackson J B, Mendez H, Aguayo R, Dieudonne A, Kovacs A, Bamji M, Abrams E, Rana S, Sever J, Nachman S. Pediatric AIDS Clinical Trials Group 377 Study Team. Combination nucleoside analog reverse transcriptase inhibitor(s) plus nevirapine, nelfinavir or ritonavir in stable antiretroviral therapy-experienced HIV-infected children: week 24 results of a randomized controlled trial–PACTG 377. AIDS Res Hum Retrovir. 2000;16:1113–1121. doi: 10.1089/088922200414956. [DOI] [PubMed] [Google Scholar]
- 13.Yerly, Kaiser S L, Race E, Bru J P, Clavel F, Perrin L. Transmission of antiretroviral-drug-resistant HIV-1 variants. Lancet. 1999;354:729–733. doi: 10.1016/S0140-6736(98)12262-6. [DOI] [PubMed] [Google Scholar]