Abstract
Purpose:
Immunotherapies targeting PD-1/L1 enhance pathologic complete response (pCR) rates when added to standard neoadjuvant chemotherapy (NAC) regimens in early-stage triple-negative, and possibly high-risk estrogen receptor–positive breast cancer. However, immunotherapy has been associated with significant toxicity, and most patients treated with NAC do not require immunotherapy to achieve pCR. Biomarkers discerning patients benefitting from the addition of immunotherapy from those who would achieve pCR to NAC alone are clearly needed. In this study, we tested the ability of MHC-II expression on tumor cells, to predict immunotherapy-specific benefit in the neoadjuvant breast cancer setting.
Patients and Methods:
This was a retrospective tissue-based analysis of 3 cohorts of patients with breast cancer: (i) primary nonimmunotherapy-treated breast cancers (n = 381), (ii) triple-negative breast cancers (TNBC) treated with durvalumab and standard NAC (n = 48), and (iii) HER2-negative patients treated with standard NAC (n = 87) or NAC and pembrolizumab (n = 66).
Results:
HLA-DR positivity on ≥5% of tumor cells, defined a priori, was observed in 10% and 15% of primary non-immunotherapy–treated hormone receptor–positive and triple-negative breast cancers, respectively. Quantitative assessment of MHC-II on tumor cells was predictive of durvalumab + NAC and pembrolizumab + NAC (ROC AUC, 0.71; P = 0.01 and AUC, 0.73; P = 0.001, respectively), but not NAC alone (AUC, 0.5; P = 0.99).
Conclusions:
Tumor-specific MHC-II has a strong candidacy as a specific biomarker of anti–PD-1/L1 immunotherapy benefit when added to standard NAC in HER2-negative breast cancer. Combined with previous studies in melanoma, MHC-II has the potential to be a pan-cancer biomarker. Validation is warranted in existing and future phase II/III clinical trials in this setting.
Translational Relevance.
Immunotherapies targeting the PD-1/L1 axis benefit a fraction of patients with breast cancer, and predictive biomarkers identifying these patients are lacking. Using two orthogonal detection approaches in two early-phase clinical trials, we demonstrate MHC-II expression on tumor cells is a clinical predictor of anti–PD-1/L1 benefit in the neoadjuvant setting.
Introduction
Addition of immune checkpoint drugs targeting the PD-1/L1 axis to chemotherapy provides clinical benefit in patients with breast cancer in the metastatic (1) and neoadjuvant (2) settings. Based on IMPassion130 (1) and KEYNOTE-355 (3), atezolizumab or pembrolizumab in combination with chemotherapy is now standard of care for locally advanced/metastatic PD-L1+ triple-negative breast cancer (TNBC). The addition of PD-1/L1 inhibitors also show varying levels of benefit in patients with metastatic (4, 5) and early-stage high-risk (6) hormone receptor–positive (HR+) HER2− breast cancer in phase Ib/II trials. Nonetheless, these agents do not benefit all patients, can cause severe and permanent toxicities, and come at a high financial cost. Thus, a priori identification of patients likely to benefit from anti–PD-1/L1 is needed. The most assessed biomarkers for immunotherapy outcome prediction in breast cancer are PD-L1 expression and stromal tumor infiltrating lymphocytes (sTIL).
IHC-based detection of PD-L1 expression on immune cells is the current companion diagnostic test for treatment with atezolizumab or pembrolizumab in advanced TNBC. However, the two PD-L1 assays approved as companion diagnostics in the United States show different positive rates due to differences in staining patterns and scoring systems (7). Additionally, concerns have been raised about interpathologist reproducibility for PD-L1 scoring on immune cells (8). In the neoadjuvant setting, increased pathologic complete response (pCR) was observed in both PD-L1–positive and -negative groups in the randomized controlled phase III trials KEYNOTE-522 (2) and IMPassion031 (9). Moreover, PD-L1 expression is not specific to immunotherapy in that it also predicts response to chemotherapy alone, complicating its usage to specifically define patients who require immunotherapy to achieve pCR (10).
Likewise, sTILs have been associated with response to PD-1/L1 inhibitors plus neoadjuvant chemotherapy (NAC) in published correlative studies (11). Assessed in a standardized manner, sTILs are a robust and reproducible biomarker (12), but strongly predict chemotherapy benefit in the neoadjuvant (13) and adjuvant settings (14). Moreover, sTILs were predictive of pCR in the durvalumab+NAC and NAC-alone arms of the GeparNuevo study (10), complicating usage of this marker as a specific biomarker of immunotherapy benefit, like PD-L1. Thus, an optimal biomarker in this setting is one that specifically identifies patients who require the addition of immunotherapy to NAC to achieve pCR, while not identifying patients who have high pCR rates to chemotherapy alone.
MHC-II expression on tumor cells is a predictive biomarker to single-agent anti–PD-1 therapy in melanoma (15) and is also associated with response in Hodgkin's lymphoma (16). Based on those results, and that the association of MHC-II expression with NAC response has not been studied yet, we evaluated the ability of MHC-II expression to predict the benefit of adding immunotherapy to NAC in breast cancer. We evaluated pCR rates as a primary outcome measure and secondarily as event-free survival (EFS).
Patients and Methods
Study approval and patient tissues
Primary nonimmunotherapy-treated breast cancers
Human breast cancer tissue microarrays were constructed from clinical surgical specimens collected at Vanderbilt University Medical Center in Nashville, and Instituto Nacional de Enfermedades Neoplásicas in Lima, Peru under institutionally approved protocols [Institutional Review Board (IRB) 030747, IRB130916, INEN 10–018]. All patients signed written informed consent prior to participation. Eight tissue microarrays (TMA) were built with 1 to 3 1 mm tumor cores per surgical specimen. Tumors were identified as TNBC or HR+ by IHC for estrogen receptor (ER), progesterone receptor (PR), and HER2 performed in a Clinical Laboratory Improvement Amendments (CLIA)–approved laboratory. CK/HLA-DR multiplex immunofluorescence (mIF) was performed on 8 TMAs, containing a total of 144 stage I–III TNBCs, of which 90 had received prior NAC, and 237 stage I–IV HR+/HER2− breast cancers, 9 of which had received prior NAC and 121 who had received letrozole for 10 to 21 days prior to surgery.
Standard NAC+ durvalumab cohort
NCT02489448 was an interventional single-arm phase I/II clinical trial in newly diagnosed histologically confirmed stage I to III TNBC. Further details of the clinical trial including a patient eligibility and a patients' characteristics table have been previously published (17). Participants received nab-paclitaxel 100 mg/m2 given weekly for 12 weeks, followed by doxorubicin 60 mg/m2 and cyclophosphamide 600 mg/m2, given every other week from 13 to 19 weeks as standard care for all patients. Seven patients were included in the Phase I part of the study, 4 at 3 mg/kg and 3 at 10 mg/kg dose. The patients in the Phase II part of the study were dosed at 10 mg/kg. Durvalumab was given every 2 weeks up until 20 weeks, at which point patients underwent surgery within 4 weeks of completion of treatment. Ethical approval was obtained from the Yale Human Investigations Committee (Yale University, HIC# 1409014537) in accordance with the Declaration of Helsinki. The study was approved and was annually reviewed by the internal IRB and all patients signed written informed consent prior to participation.
Residual cancer burden (RCB) was evaluated by a pathologist on surgical samples. CK/HLA-DR mIF was performed on 48 out of 57 formalin-fixed paraffin-embedded (FFPE) tissue sections pretreatment biopsy samples stored at −20°C, with loss of sample size due primarily to lack of tissue content in the biopsy sections. CONSORT diagram: Supplementary Fig. S1A.
Standard NAC± pembrolizumab cohort
The I-SPY2 study is an adaptively randomized phase II multicenter trial for stage II/III breast cancer with high risk of recurrence (NCT01042379) evaluating multiple investigational arms in parallel, each consisting of standard NACT (serving as the common control arm) plus an investigational agent/combination. HR, HER2 (ERBB2), and MammaPrint status performed at baseline are used to classify patients for adaptive randomization. Only ERBB2-negative patients were eligible for randomization to the pembrolizumab arm. Participants in the control arm received standard NAC consisting of 80 mg/m2 i.v. paclitaxel weekly for 12 weeks, followed by 4 cycles of 60 mg/m2 doxorubicin with 600 mg/m2 i.v. cyclophosphamide every 2 to 3 weeks (AC). Those participants in the pembrolizumab arm received standard NAC as above, with the addition of 200 mg i.v. pembrolizumab every 3 weeks for 4 cycles (weeks 1, 4, 7, and 10) concurrently with paclitaxel. EFS follow-up was up to 1,946 days. All participating sites received IRB approval, and patients provided written informed consent. The study was conducted in accordance with the Declaration of Helsinki. The I-SPY2 DSMB meets monthly to review patient safety and study progress. RCB was evaluated by a pathologist on surgical samples. Further details of this trial have been published elsewhere (6). Laser capture microdissection (LCM) and reverse-phase protein microarray (RPPA) were performed on fresh frozen tissue biopsies stored at −80°C from the T0 pretreatment (baseline) time-point from both control (NAC; n = 89) and pembrolizumab-treated (NAC+P; n = 67) arms. Analysis was performed for the whole cohort and according to HR status. CONSORT diagram: Supplementary Fig. S1B.
mIF
FFPE tissue sections were cut at 4 μm and deparaffinized. Antigen retrieval was performed with citrate buffer pH 6. Endogen peroxidase were blocked and protein block was applied. Sections were then incubated with the primary antibody (HLA-DR TAL1B5 Santa Cruz at 1:4000, panCK AE1/AE3 Biocare at 1:400, HLA-DR/DP/DQ/DX sc-53302 at 1:2000) overnight at 4°C. Sections were then incubated with the secondary antibody and tyramide signal amplification reagent (Tyramide Superboost Kit; catalog no. B40912, Invitrogen) applied according to manufacturer's recommendations. The procedure was repeated 1 or 2 more times with the subsequent different primary antibodies and then counterstained with DAPI for nuclei identification. Tonsil was used as a positive control. A detailed protocol can be found in Supplementary Data.
Image analysis and quantification
Whole-slide images were digitally acquired using an AxioScan Z1 slide scanner (Carl Zeiss) at 20x. Automated quantitative scoring was performed by a pathologist blinded to sample characteristics, using QuPath software v0.2.0. A CK mask was defined with the pixel classifier to outline tumor and nontumoral compartments. Out-of-focus areas, tissue folds, necrosis, normal breast, and in situ carcinoma were excluded from the analysis. For cases with patchy or null CK expression, tumor areas were manually annotated. Cell segmentation was determined on DAPI. Object classifiers were trained for HLA-DR and HLA-DR/DP/DQ/DX. Each core was visually assessed for correct performance of the quantification algorithm. HLA-DR expression on tumor cells was considered as ≥5% of tumor cells based on prior experience in melanoma (15).
LCM and reverse-phase protein microarray
Enriched epithelial-cell subpopulations were isolated from 8 μM cryosections (>95% purity) using an Arcturus Pixcell IIe LCM system (Arcturus). Resultant cell lysates were printed in triplicate spots (approx. 10nL per spot) onto nitrocellulose-coated slides (Grace Biolabs) using an Aushon 2470 Arrayer (Aushon Biosystems) as described (18). Antibodies used on the arrays were extensively validated before use and are listed in Supplementary Table S1. RPPA immunostaining was performed as previously described (19).
For the calibration study, 20 breast cancers with varying degrees of known HLA-DR or HLA-DR/DP/DQ/DX expression from the nonimmunotherapy-treated TMA cohort were analyzed by identical methods to the above and a linear correlation was established against mIF percent positivity, yielding a cut-off point of 17,000 units comparable with 5% tumor-cell positivity.
Statistical analysis
The statistical analyses included herein were not preplanned per study protocols but were preplanned per individual requests for existing tissues as secondary or exploratory correlative endpoints. Statistical analyses were performed in R or Graphpad Prism. For RPPA analysis, nominal p values were calculated using a two-sided t test, and then an adjusted p value (q value) was calculated (Benjamini–Hochberg). For 2-group quantitative comparisons, a one-sided t test was used in cases where a specific directionality of change was hypothesized based on previous data on melanoma. For response prediction based on defined biomarker subgroups, a one-sided χ2 test was used. To test interaction effects, a logistic regression model was used to test the interaction between MHC-II status and treatment arm on response (pCR rate) and survival, after adjusting for the main effects. ROC analysis was performed in Graphpad Prism to evaluate the overall response prediction performance of MHC-II expression across multiple cut-off points beyond the predefined 5%. Kaplan–Meier and log–rank tests were calculated with a cut-off point chosen based on the calibration study. Graphs show mean values ± SEM. P value cutoffs displayed on plots correspond to “ns” - P > 0.05, * P ≤ 0.05, ** P ≤ 0.01, *** P ≤ 0.001, and **** P ≤ 0.0001.
Results
MHC-II is expressed in a subgroup of primary TNBC and HR± breast cancers
Twenty percent of all breast cancers from the primary nonimmunotherapy-treated breast cancers expressed ≥1% HLA-DR in tumor cells by mIF (Supplementary Fig. S2; Supplementary Table S2). Using a predefined cutoff of 5% tumor-cell positivity that predicted benefit to single-agent anti–PD-1 in melanoma (15), 12% of all breast cancers assessed expressed ≥5% HLA-DR. Fifteen percent of TNBCs showed ≥5% HLA-DR compared with 10% of HR+ tumors. Six percent of treatment-naïve TNBCs and HR+ patients expressed ≥5% HLA-DR. Twenty-one percent of patients with TNBC who received neoadjuvant chemotherapy had ≥5% HLA-DR. Tumor-specific HLA-DR expression was not associated with sTILs (Spearman r = 0; P = 0.87; n = 292 accessible pairs), a common predictive marker for response to NAC alone.
Tumor-specific MHC-II is associated with pCR to durvalumab ± NAC
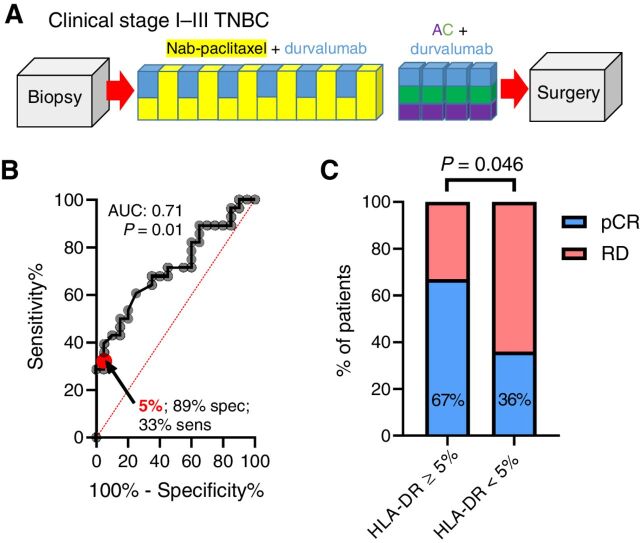
Fifteen percent of patients from the standard NAC+ durvalumab cohort (Fig. 1A) showed ≥5% HLA-DR expression on tumor cells. ROC analysis yielded an AUC of 0.71 (P = 0.01; Figure 1B). The predefined cut-off point of 5% resulted in 33% sensitivity, 89% specificity, 67% positive predictive value (PPV), and 64% negative predictive value (NPV). Patients with ≥5% HLA-DR+ tumor cells demonstrated higher pCR rates (67% versus 36%; one-sided χ2 P = 0.046, Fig. 1C and Table 1). HLA-DR expression on nontumor cells was not predictive of response (Supplementary Fig. S3A–S3C).
Figure 1.
Tumor-specific MHC-II expression is associated with pCR to NAC and anti–PD-L1 inhibition. A, NCT02489448 clinical trial schema depicting NAC (nab-paclitaxel in yellow, doxorubicin in purple, and cyclophosphamide in green) + durvalumab (blue) single arm. B, ROC curve for predictive capacity of tumor-specific MHC-II (≥5% tumor-cell positivity cut-off point) on pCR. C, pCR rates by HLA-DR biomarker group; P value represents the result of a one-sided χ2 test.
Table 1.
Association of tumor-specific MHC-II/HLA-DR expression with outcome to durvalumab and NAC.
| RCB | ||||
|---|---|---|---|---|
| pCR (0) | I | II | III | |
| n (%) | n (%) | n (%) | n (%) | |
| Total (n = 48) | 20 (42%) | 6 (13%) | 16 (33%) | 6 (13%) |
| HLA-DR ≥ 5% (n = 9) | 6 (67%) | 1 (11%) | 1 (11%) | 1 (11%) |
| HLA-DR < 5% (n = 39) | 14 (36%) | 5 (13%) | 15 (38%) | 5 (13%) |
| χ2 p value (1-tailed): | pCR vs. RD: P = 0.046‡ | RCB 0/I vs. II/III: P = 0.057 | ||
| Odds ratio for response (95% CI): | 3.57 (0.87–14.23) | 3.68 (0.81–18.81) | ||
Abbreviation: CI, confidence interval.
‡Trend for significance.
Eighty-nine percent of tumors expressing ≥5% HLA-DR were also PD-L1+, defined as PD-L1 (Ventana SP263) expression in ≥1% on immune or tumor cells and previously reported in Foldi and colleagues (Supplementary Fig. S3D; ref. 17). Interestingly, the 1 patient with a PD-L1-negative HLA-DR ≥5% tumor had a pCR (Supplementary Fig. S3E).
High tumor-specific MHC-II expression is associated with pembrolizumab benefit
As many markers of antitumor immunity are associated with pCR to NAC, a control arm of NAC alone is required to establish predictive capacity for anti–PD-1/L1 benefit. Using existing data from the control (NAC) and pembrolizumab arms of the I-SPY2 TRIAL in high-risk HER2-negative patients (Fig. 2A; ref. 6) we tested the association of tumor MHC-II expression with pCR by RPPA. The RPPA panel utilized was part of a planned correlative analysis and contained 32 protein-based biomarkers, including MHC-II (HLA-DR and pan-MHC-II; HLA-DR/DP/DQ/DX).
Figure 2.
![Figure 2. Tumor-specific MHC-II expression specifically predicts benefit to anti–PD-1 inhibition with NAC in high-risk HER2− patients. A, I-SPY2 clinical trial schema depicting NAC [paclitaxel (T) in yellow, doxorubicin in purple, and cyclophosphamide in green] + pembrolizumab (P; blue) arm and NAC-only control arm. B, HLADR/DP/DQ/DX values by outcome for NAC-only and NAC+ pembrolizumab arms. The dotted line shows the cut-off point (17,000 units) chosen based on a calibration study comparing RPPA with a 5% tumor-specific MHC-II cut-off point. C, ROC curve for predictive capacity of tumor-specific MHC-II on pCR in NAC-alone (T) arm. D, ROC curve for predictive capacity of tumor-specific MHC-II on pCR in pembrolizumab (T+P) arm. E, Interaction plot of biomarker group (stratified at 50,000 units) and treatment arm on pCR rate. P value represents interaction term in a logistic regression model after adjusting for main effects (treatment arm and MHC-II status).](https://cdn.ncbi.nlm.nih.gov/pmc/blobs/6191/9401480/8add6811871e/5299fig2.jpg)
Tumor-specific MHC-II expression specifically predicts benefit to anti–PD-1 inhibition with NAC in high-risk HER2− patients. A, I-SPY2 clinical trial schema depicting NAC [paclitaxel (T) in yellow, doxorubicin in purple, and cyclophosphamide in green] + pembrolizumab (P; blue) arm and NAC-only control arm. B, HLADR/DP/DQ/DX values by outcome for NAC-only and NAC+ pembrolizumab arms. The dotted line shows the cut-off point (17,000 units) chosen based on a calibration study comparing RPPA with a 5% tumor-specific MHC-II cut-off point. C, ROC curve for predictive capacity of tumor-specific MHC-II on pCR in NAC-alone (T) arm. D, ROC curve for predictive capacity of tumor-specific MHC-II on pCR in pembrolizumab (T+P) arm. E, Interaction plot of biomarker group (stratified at 50,000 units) and treatment arm on pCR rate. P value represents interaction term in a logistic regression model after adjusting for main effects (treatment arm and MHC-II status).
Multiple protein markers were quantitatively higher in patients achieving pCR at a nominal (uncorrected) p value in each of the subgroups tested (all patients, HR+/HER2−, and TNBC), including known correlates of NAC outcome, such as PD-L1 (SP142) and CD3 epsilon, in the control NAC arm for high-risk HR+ and all patients (Supplementary Fig. S4). Only pan–MHC-II was significantly higher in patients achieving pCR after correcting for multiple comparisons across all patients and high-risk HR+ subgroup (Fig. 2B; Supplementary Figs. S4 and S5A-S5B). ROC analysis was performed, yielding an AUC of 0.5 (P = 0.99; Figure 2C) in the NAC-alone arm and an AUC of 0.73 (P = 0.001; Fig. 2D) in the pembrolizumab arm. Since the RPPA analysis did not have a prior-defined cut-off point for pCR prediction, a calibration study was performed including 20 breast tumors with known MHC-II IF status by performing RPPA analysis for pan–MHC-II on serial sections (r = 0.5429; P = 0.0134) which yielded a comparable cut-off point of 17,000 normalized units, representing approximately 5% tumor-cell positivity by mIF. This cut-off point yielded 95% specificity, 38% sensitivity, 80% PPV, and 67% NPV (Fig. 2D), which was also comparable with that identified in the durvalumab study (Fig. 1B). Application of this cut-off point demonstrated predictive capacity in all pembrolizumab-treated patients and in the HR+ subgroups (one-tailed χ2 test; P = 0.0002 and P < 0.0001, respectively), but was not predictive of pCR in the control arm (NAC-alone; Table 2 and Fig. 2B). Moreover, testing the interaction between MHC-II status and treatment arm on pCR demonstrated a significant interaction term (P = 0.05; Figure 2E). While some clinical and tumor characteristic showed an association with pCR, only pan–MHC-II remained significant on multivariate analysis (Supplementary Table S3).
Table 2.
Association of tumor-specific MHC-II/HLA-DR expression with outcome to pembrolizumab and NAC or chemotherapy alone.
| pCR | RD | ||||||
|---|---|---|---|---|---|---|---|
| HLA-DR/DP/DQ/DX intensity units: | ≥17,000 (RPPA-high) n (%) | <17,000 (RPPA-low) n (%) | ≥17,000 (RPPA-high) n (%) | <17,000 (RPPA-low) n (%) | OR (95% CI)≥17,000 (RPPA-high) | χ2 P value (1-tailed) | |
| All patients | Paclitaxel (n = 87) | 2 (2%) | 11 (13%) | 12 (14%) | 62 (71%) | 1.063 (0.23–5.3) | 0.47 |
| Paclitaxel + pembrolizumab (n = 66) | 12 (18%) | 17 (26%) | 3 (2%) | 34 (55%) | 8 (1.938–28.42) | 0.0007 | |
| HR+/HER2− | Paclitaxel (n = 49) | 2 (4%) | 5 (10%) | 7 (14%) | 35 (71%) | 2 (0.3381–10.37) | 0.22 |
| Paclitaxel + pembrolizumab (n = 40) | 8 (20%) | 4 (10%) | 2 (5%) | 26 (65%) | 26 (4.253–136.3) | <0.0001 | |
| TNBC | Paclitaxel (n = 38) | 0 (0%) | 6 (16%) | 5 (13%) | 27 (71%) | 0 (0.000–4.53) | 0.15 |
| Paclitaxel + pembrolizumab (n = 26) | 4 (15%) | 13 (50%) | 1 (4%) | 8 (31%) | 2.46 (0.28–33.43) | 0.22 | |
Note: Values in bold are statistically significant.
Tumor-specific MHC-II expression is associated with improved survival in high-risk HER2-negative patients with the addition of anti–PD-1 inhibition to NAC, but not with NAC alone.
Patients with pan–MHC-II-positive tumors who received neoadjuvant pembrolizumab had an improved EFS over those patients with MHC-II–negative tumors (P = 0.079; Figure 3A). However, the same trend was not observed in the NAC-alone arm (P = 0.22; Figure 3B). Comparing only patients with MHC-II–positive tumors across treatment arms showed that this population had improved EFS when treated with pembrolizumab versus standard NAC alone (P = 0.042; Figure 3C). Interestingly, a trend toward worse outcomes was observed for the addition of pembrolizumab in patients with MHC-II–negative tumors (P = 0.063; Figure 3D). Cox proportional hazard ratio analyses including clinical and tumor characteristics showed that only pan–MHC-II was significantly associated with survival (Supplementary Table S4).
Figure 3.
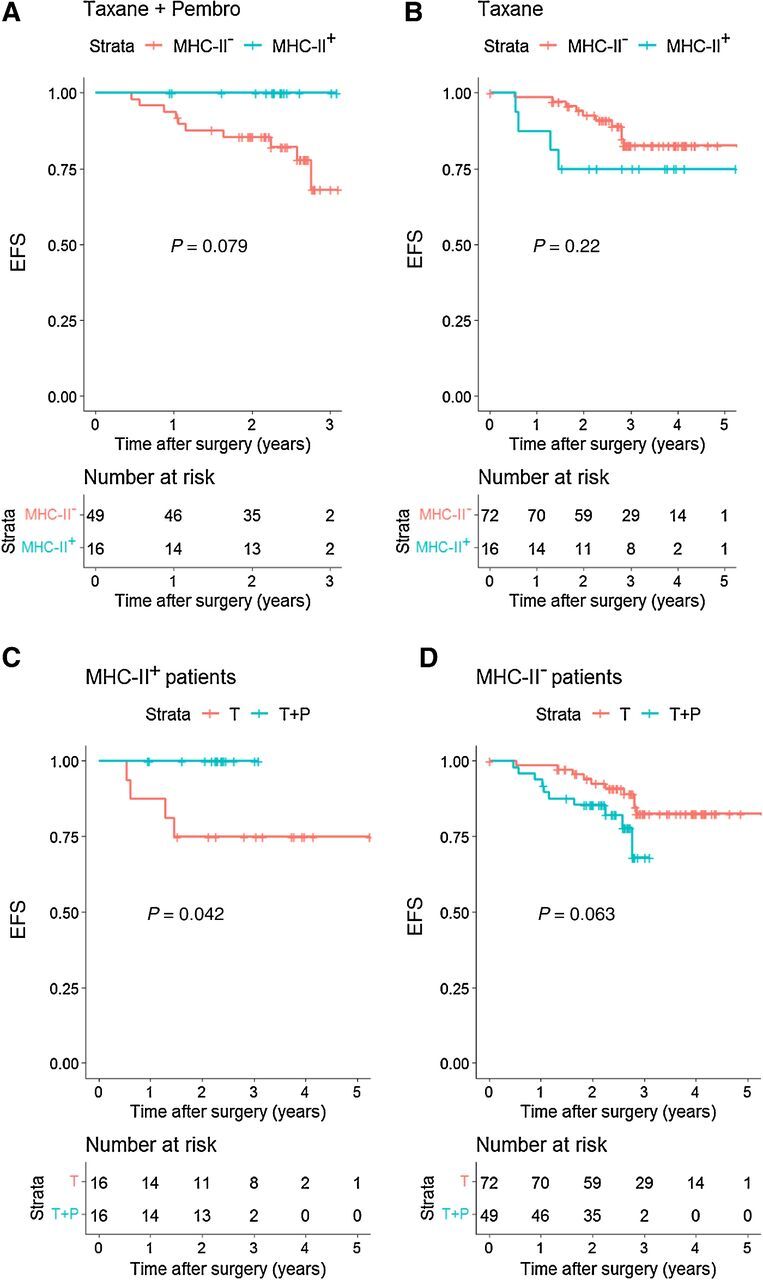
Tumor-specific MHC-II expression is associated with improved EFS in high-risk HER2− patients with the addition of anti–PD-1 inhibition to NAC, but not with NAC alone. A–D, Kaplan–Meier analysis of EFS for the NAC + pembrolizumab and NAC-alone arms of the I-SPY2 trial according to HLADR/DP/DQ/DX expression on tumor cells by RPPA. MHC-II+: tumors expressing ≥17,000 normalized units of pan–MHC-II. MHC-II−: <17,000 normalized units. The 17,000 units cut-off point was chosen based on a calibration study comparing RPPA with a 5% tumor-specific MHC-II cut-off point by mIF. P value represents the log–rank test.
Discussion
Although immunotherapies targeting the PD-1/L1 axis appear to provide benefit in combination with NAC in some patients, not all patients require these agents to achieve pCR. Moreover, these agents are not without toxicity. Thus, biomarker approaches to prioritize patients likely to derive benefit from immunotherapy in the neoadjuvant setting are clearly needed. TILs and PD-L1 expression cannot adequately define patients who need additional PD-1/L1–targeted therapy on top of chemotherapy, or at the least, use of such a biomarker would be complicated given that these biomarkers paradoxically define the patients who respond to NAC alone with the highest rates.
The factors driving tumor cell–specific MHC-II expression are not entirely clear, although its expression can be driven by inflammatory signals, such as interferons in the tumor microenvironment. Interestingly, there are two variations on this theme that extend the importance of MHC-II expression beyond simply being an interferon-responsive marker. First, there are a wide variety of tumor cell lines that will not upregulate MHC-II with interferon treatment, but will upregulate PD-L1, suggesting an intact interferon-driven JAK STAT-signaling pathway (15). Thus, many tumors may lose the ability to express MHC-II, which is thought to be an epigenetic event (20). Second, many cell lines constitutively express MHC-II without interferon stimulation. We hypothesize that this occurs due to innate inflammatory signaling resulting from DNA- and RNA-sensing pathways. Overall, the mechanisms and functionality of tumor-specific MHC-II are intriguing and an important subject of study and have recently been reviewed elsewhere (21).
In this study, we demonstrate that tumor epithelium expressed MHC-II protein has the potential to be a bona fide predictive biomarker for immunotherapy benefit in high-risk HR+ and TNBCs when added to standard NAC regimens and was not associated with pCR to NAC alone. The predefined cut-off point of 5% used in melanoma proved to be useful for breast cancer in the neoadjuvant setting. In the KEYNOTE-522 study, the addition of pembrolizumab to NAC yielded an improvement in pCR rate from 54.9% to 68.9%, or a 14% change, while in the IMPassion031 trial, the addition of atezolizumab improved pCR rates from 41% to 58%, or a 17% change. Thus, in a biomarker assessment of immunotherapy benefit in a single-arm trial, approximately 20% to 30% of responding patients should be identified by the biomarker (sensitivity), while most biomarker-negative patients should be predicted to have RD (specificity). The 5% cut-off point approximated these sensitivity and specificity values.
We found pan–MHC-II to be an independent specific predictor of response and EFS to the addition of PD-1/PD-L1 inhibitor to standard NAC. A previous publication on the standard NAC+ durvalumab cohort showed that age [Objective response (OR) 1.00 (0.96–1.04); P = 0.94], stage [T1 vs. T2 0.47 (0.15–1.44); P = 0.19 and N− vs. N+ 1.38 (0.49–4.00); P = 0.54], PD-L1 [≥1% vs. <1% 2.63 (0.82–9.21); P = 0.11], and TILs [0.99 (0.98–1.01); P = 0.56] were not predictive of pCR in univariate analysis (17). Additionally, PD-L1 was not associated with response to the addition of pembrolizumab in high-risk HR+ and TNBC subpopulations, while showing an association with pCR in the HR+ control (NAC alone) arm. Moreover, while the majority of patients with ≥5% HLA-DR+ tumors represent a subset of PD-L1+ (≥1%) tumors, HLA-DR also captured PD-L1− patients that responded to immunotherapy and standard chemotherapy combination.
There are clear limitations to the present study. The sample sizes in both studies contained less than 100 patients per arm, and there was no control arm in the durvalumab trial. Moreover, the data should be interpreted with caution due to the retrospective nature of the analyses. The use of 2 assays (mIF and RPPA) could be considered both a strength (e.g., rigor in 2 methods of assessing MHC-II expression demonstrate similar findings) and a limitation (e.g., the 2 trial results cannot be directly compared with one another). Importantly, we ensured that the RPPA cut-off point was calibrated to mIF in order to approximate the 5% cut-off point. Application of the cut-off points, at 5% positive tumor cells (IHC approaches) or 17,000 normalized units (RPPA approach), must be validated in a large randomized controlled trial to confirm these results. Where possible, REMARK guidelines were adhered to with exceptions primarily related to the retrospective (not preplanned per the study protocols) nature of the analysis.
Nonetheless, to our knowledge, this is the first study to evaluate and demonstrate the predictive capacity of tumor MHC-II for immunotherapy benefit in patients with breast cancer. The utility of tumor-specific MHC-II was verified by 2 independent approaches, in 2 trials comprising different populations of patients, slightly different NAC regimens, and antibodies (anti–PD-1; anti–PD-L1) targeting different sides of the anti–PD-1/L1 axis. These data suggest that tumor-specific MHC-II is an important, potentially pan-cancer biomarker, which can be measured easily by tissue analyses, including standard IHC, that should be included in correlative analyses in future breast cancer immunotherapy Phase II and III trials.
Authors' Disclosures
J.D. Wulfkhule reports grants from Gateway for Cancer Research during the conduct of the study, as well as other support from Theralink Technologies and personal fees and non-financial support from Baylor College of Medicine outside the submitted work; in addition, J.D. Wulfkhule has numerous patents pending, issued, licensed, and with royalties paid from George Mason University. R.I. Gallagher reports grants from Gateway Foundation for Cancer Research during the conduct of the study. M.L. Axelrod reports a patent for “Detection of immune signatures in a breast cancer subject” pending to Vanderbilt University. L. Pusztai reports grants and personal fees from Merck, Pfizer, Bristol Myers Squibb, and AstraZeneca and grants from Seagen outside the submitted work. E. Petricoin reports grants from Gateway for Cancer Research during the conduct of the study, as well as personal fees from Perthera, Inc. and Theralink Technologies, Inc. outside the submitted work. K.R.M. Blenman is on the scientific advisory board of CDI Labs (Puerto Rico). J.M. Balko reports grants from NIH during the conduct of the study, as well as grants from Genentech/Roche and Incyte outside the submitted work; in addition, J.M. Balko has a patent for “Immunotherapy biomarkers/HLA-DR” issued. No disclosures were reported by the other authors.
Supplementary Material
Supplementary Methods: detailed protocol. Supplementary Table 1: Antibodies used for RPPA. Supplementary Table 2: Rates of tumor-specific MHC-II/HLA-DR expression across HER2-negative primary breast cancers. Supplementary Table 3: Univariate and multivariate analysis for achieving pCR to pembrolizumab and neoadjuvant chemotherapy or chemotherapy alone. Supplementary Table 4: Cox proportional Hazard ratio univariate analysis for 3-year event-free survival associated with pembrolizumab and neoadjuvant chemotherapy or chemotherapy alone. Supplementary Figure legends.
Supplementary Figure 1: CONSORT diagrams for associated studies. Supplementary Figure 2: Tumor specific MHC-II pattern immunofluorescence in ER+ and TNBC. Supplementary Figure 3: Non-tumor MHC-II expression and comparison of PD-L1+ and HLA-DR+ patient populations. Supplementary Figure 4: RPPA associations with pCR for control and pembrolizumab-treated (Treat) arms, by clinical group. Supplementary Figure 5: RPPA associations with pCR for control and pembrolizumab-treated arms in high-risk ER+ patients.
Acknowledgments
This work was supported by Susan G. Komen Career Catalyst Grant CCRCR18552647 (to J.M. Balko), NIH/NCI SPORE 2P50CA098131–17 (to J.M. Balko), Department of Defense Era of Hope Award BC170037 (to J.M. Balko), and the Vanderbilt-Ingram Cancer Center Support Grant P30 CA68485. Additional funding was provided by NIH T32GM007347 (to M.L. Axelrod) and F30CA236157 (to M.L. Axelrod). The I-SPY 2 Trial and RPPA work was supported by the Gateway for Cancer Research (grant no. G-16–900), QuantumLeap Healthcare Collaborative, Foundation for the NIH, NCI Center for Biomedical Informatics and Information Technology (grant no. 28XS197), a Sta Op Tegen Kanker International Translational Cancer Research Grant, Safeway Foundation, the Bill Bowes Foundation, Quintiles Transnational, Johnson & Johnson, Genentech, Amgen, the San Francisco Foundation, Give Breast Cancer the Boot, Eli Lilly, Pfizer, Eisai, the Harlan family, the Avon Foundation for Women, and Alexandria Real Estate Equities. The Medimmune Durvalumab trial was partly supported by AstraZeneca and grants from the NCI (R01CA219647), the Breast Cancer Research Foundation, a Susan Komen Foundation Leadership Grant (SAC 160076), and an annual Investigator Award from the Breast Cancer Research Foundation (to L. Pusztai).
The costs of publication of this article were defrayed in part by the payment of page charges. This article must therefore be hereby marked advertisement in accordance with 18 U.S.C. Section 1734 solely to indicate this fact.
Footnotes
Note: Supplementary data for this article are available at Clinical Cancer Research Online (http://clincancerres.aacrjournals.org/).
Authors' Contributions
P.I. Gonzalez-Ericsson: Data curation, software, formal analysis, validation, investigation, visualization, methodology, writing–original draft, writing–review and editing. J.D. Wulfkhule: Data curation, formal analysis, writing–original draft, writing–review and editing. R.I. Gallagher: Data curation, formal analysis, writing–original draft. X. Sun: Data curation, writing–original draft. M.L. Axelrod: Data curation, writing–original draft. Q. Sheng: Data curation, writing–original draft. N. Luo: Data curation, writing–original draft. H. Gomez: Resources. V. Sanchez: Data curation, validation, methodology. M. Sanders: Resources, data curation, writing–original draft. L. Pusztai: Resources, data curation, supervision, funding acquisition, writing–original draft, writing–review and editing. E. Petricoin: Resources, data curation, supervision, funding acquisition, methodology, writing–original draft, writing–review and editing. K.R.M. Blenman: Resources, data curation, formal analysis, supervision, investigation, writing–original draft, writing–review and editing. J.M. Balko: Conceptualization, resources, data curation, formal analysis, supervision, investigation, writing–original draft, project administration, writing–review and editing. I-SPY2 Trial Team: Resources.
Collaborators
Investigation of Serial Studies to Predict Your Therapeutic Response With Imaging and Molecular Analysis 2 (I-SPY 2) trial investigators by site/affiliation: Avera Cancer Institute: B. Leyland-Jones; British Colombia Cancer Agency: S. Chia, R. Serpanchy, C. Yu; Emory University: S. McMillan, R. Mosley, K. Nguyen, E.C. Wood, A. Zelnak; Georgetown University: C. Dillis, R. Donnelly, T. Harrington, C. Isaacs, B. Kallakury, M. Liu, F. Lynce, B. Oppong, P. Pohlmann, E. Tousimis, R. Warren, S. Willey, J.E. Wong, J. Zeck; Loyola University Chicago Medical Center: K. Albain, M.B. Bartolotta, D. Bova, C. Brooks, B. Busby, K. Czaplicki, X. Duan, R. Gamez, K. Ganesh, E. Gaynor, C. Godellas, C. Grace-Louthen, T. Kuritza, S. Lo, A. Nagamine, C. Perez, P. Robinson, D. Rosi, F. Vaince, K. Ward; Inova Fairfax Hospital: K. Choquette, K. Edmiston, H. Gallimore, J. McGovern, K. Mokarem, M. Pajaniappan, S. Rassulova, K. Scott, K. Sherwood, J. Wright; Mayo Clinic, Arizona: K.S. Anderson, R.J. Gray, S.J. Myers, D.E. Northfelt, B.A. Pockaj, J. Roedig, N. Wasif; Mayo Clinic, Rochester: A.M. Arens, J.C. Boughey, K.R. Brandt, J.L. Carroll, B. Chen, A.L. Connors, A.C. Degnim, D.R. Farley, S.M. Greenlee, T.C. Haddad, T.J. Hieken, T.J. Hobday, J.W. Jakub, L.L. Liberte, M.C. Liu, C.L. Loprinzi, L.R. Menard, M.M. Moe, T.J. Moynihan, C. O'Sullivan, E.A. Olson, P.P. Peethambaram, K.J. Ruddy, B.A. Russell, A.L. Rynearson, D.R. Smith, D.W. Visscher, A.J. Windish; H. Lee Moffitt Cancer Center and Research Institute: K. Cox, K. Dawson, O. Newton, W. Ramirez; Oregon Health and Science University: H. Bengtson, J. Bucher, S. Chui, B. Gilbert-Ghormley, R. Hampton, K.A. Kemmer, D. Kurdyla, D. Nauman, J. Spear, A. Wilson; Swedish Cancer Institute: D. Beatty, P. Dawson, E.R. Ellis, M. Fer, J. Hanson, M.P. Goetz, T.C. Haddad, D. Iriarte, H.G. Kaplan, B. Porter, K. Rinn, H. Thomas, S. Thornton, R. Tickman, N. Varghis; University of Alabama at Birmingham: V. Caterinichia, J. Delos Santos, C. Falkson, A. Forero, H. Krontiras, C. Vaklavas, S. Wei; University of Arizona: A. Bauland, L. Inclan, D. Lewallen, A. Powell, C. Roney, K. Schmidt, R.K. Viscusi, H. Wright; University of California, San Diego: S. Blair, S. Boles, J. Bykowski, B. Datnow, L. Densley, M. Eghtedari, V. Genna, F. Hasteh, T. Helsten, P. Kormanik, H. Ojeda-Fournier, I. Onyeacholem, B. Parker, K. Podsada, R. Schwab, A. Wallace, C. Yashar; University of California, San Francisco: M.D. Alvarado, A. Au, R. Balassanian, C. Benz, M. Buxton, Y.Y. Chen, J. Chien, C. D'Andrea, S.E. Davis, L. Esserman, C. Ewing, A. Goga, G.L. Hirst, M. Hwang, N. Hylton, B. Joe, J. Lyandres, M. Kadafour, G. Krings, M. Melisko, M. Moasser, P. Munter, Z. Ngo, J. Park, E. Price, H. Rugo, L. van't Veer, J. Wong, C. Yau; University of Chicago: H. Abe, N.T. Jaskowiak, R. Nanda, F. Olopade, D.V. Schacht; University of Colorado, Denver: V. Borges, T. Colvin, J. Diamond, A.D. Elias, C. Finlayson, C. Fisher, L. Hardesty, P. Kabos, N. Kounalakis, J. Mayordomo, T. McSpadden, C. Murphy, R. Rabinovitch, S. Sams, E. Shagisultanova; University of Kansas: S. Baccaray, Q. Khan; University of Minnesota: H. Beckwith, A. Blaes, T. Emory, T.C. Haddad, J. Hui, M. Klein, J. Kuehn-Hajder, M. Nelson, D. Potter, T. Tuttle, D. Yee, R. Zera; University of Pennsylvania: L. Bayne, A. Bradbury, A. Clark, A. DeMichele, S. Domchek, C. Fisher, K. Fox, D. Frazee, M. Lackaye, J. Matro, E. McDonald, M. Rosen, P. Shah, J. Tchou, M. Volpe; University of Texas MD Anderson Cancer Center: R. Alvarez, C. Barcenas, D.A. Berry, D. Booser, A. Brewster, P. Brown, A. Gonzalez-Angulo, N. Ibrahim, M. Karuturi, K. Koenig, S. Moulder, J. Murray, R. Murthy, L. Pusztai, B. Saigal, W.F. Symmans, D. Tripathy, R. Theriault, N. Ueno, V. Valero; University of Southern California: M. Brown, M. Carranza, Y. Flores, J. Lang, A. Luna, N. Perez, D. Tripathy, K. Watkins; University of Texas Southwestern Medical Center: S. Armstrong, C. Boyd, L. Chen, V. Clark, A. Frankel, D.M. Euhus, T. Froehlich, S. Goudreau, B. Haley, A. Harker-Murray, D. Klemow, A.M. Leitch, R. Leon, H. Li, T. Morgan, N. Qureshi, R. Rao, M. Reeves, A. Rivers, N. Sadeghi, S. Seiler, B. Staves, V. Tagoe, G. Thomas, D. Tripathy, N. Unni, S. Weyandt, R. Wooldridge, J. Zuckerman; Universty of Washington: L. Korde, M. Griffin, B. Butler, A. Cundy, L. Rubinstein, C. Hixson
References
- 1. Schmid P, Adams S, Rugo HS, Schneeweiss A, Barrios CH, Iwata H, et al. Atezolizumab and Nab-Paclitaxel in advanced triple-negative breast cancer. N Engl J Med 2018;379:2108–21. [DOI] [PubMed] [Google Scholar]
- 2. Schmid P, Cortes J, Pusztai L, McArthur H, Kummel S, Bergh J, et al. Pembrolizumab for early triple-negative breast cancer. N Engl J Med 2020;382:810–21. [DOI] [PubMed] [Google Scholar]
- 3. Cortes J, Cescon DW, Rugo HS, Nowecki Z, Im SA, Yusof MM, et al. Pembrolizumab plus chemotherapy versus placebo plus chemotherapy for previously untreated locally recurrent inoperable or metastatic triple-negative breast cancer (KEYNOTE-355): a randomised, placebo-controlled, double-blind, phase 3 clinical trial. Lancet 2020;396:1817–28. [DOI] [PubMed] [Google Scholar]
- 4. Rugo HS, Delord J-P, Im S-A, Ott PA, Piha-Paul SA, Bedard PL, et al. Safety and antitumor activity of pembrolizumab in patients with estrogen receptor–positive/human epidermal growth factor receptor 2–negative advanced breast cancer. Clin Cancer Res 2018;24:2804–11. [DOI] [PubMed] [Google Scholar]
- 5. Dirix LY, Takacs I, Jerusalem G, Nikolinakos P, Arkenau H-T, Forero-Torres A, et al. Avelumab, an anti-PD-L1 antibody, in patients with locally advanced or metastatic breast cancer: a phase 1b JAVELIN Solid Tumor study. Breast Cancer Res Treat 2018;167:671–86. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6. Nanda R, Liu MC, Yau C, Shatsky R, Pusztai L, Wallace A, et al. Effect of pembrolizumab plus neoadjuvant chemotherapy on pathologic complete response in women with early-stage breast cancer: an analysis of the ongoing phase 2 adaptively randomized I-SPY2 trial. JAMA Oncol 2020;6:676–84. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7. Salgado R, Bellizzi AM, Rimm D, Bartlett JMS, Nielsen T, Holger M, et al. How current assay approval policies are leading to unintended imprecision medicine. The Lancet Oncol 2020;21:1399–401. [DOI] [PubMed] [Google Scholar]
- 8. Reisenbichler ES, Han G, Bellizzi A, Bossuyt V, Brock J, Cole K, et al. Prospective multi-institutional evaluation of pathologist assessment of PD-L1 assays for patient selection in triple negative breast cancer. Mod Pathol: 2020;33:1746–52. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9. Mittendorf EA, Zhang H, Barrios CH, Saji S, Jung KH, Hegg R, et al. Neoadjuvant atezolizumab in combination with sequential nab-paclitaxel and anthracycline-based chemotherapy versus placebo and chemotherapy in patients with early-stage triple-negative breast cancer (IMpassion031): a randomised, double-blind, phase 3 trial. Lancet 2020;396:1090–100. [DOI] [PubMed] [Google Scholar]
- 10. Loibl S, Untch M, Burchardi N, Huober J, Sinn BV, Blohmer JU, et al. A randomised phase II study investigating durvalumab in addition to an anthracycline taxane-based neoadjuvant therapy in early triple-negative breast cancer: clinical results and biomarker analysis of GeparNuevo study. Ann Oncol 2019;30:1279–88. [DOI] [PubMed] [Google Scholar]
- 11. Schmid P, Salgado R, Park Y, Muñoz-Couselo E, Kim S, Sohn J, et al. Pembrolizumab plus chemotherapy as neoadjuvant treatment of high-risk, early-stage triple-negative breast cancer: results from the phase 1b open-label, multicohort KEYNOTE-173 study. Ann Oncol 2020;31:569–81. [DOI] [PubMed] [Google Scholar]
- 12. Hendry S, Salgado R, Gevaert T, Russell PA, John T, Thapa B, et al. Assessing tumor-infiltrating lymphocytes in solid tumors: a practical review for pathologists and proposal for a standardized method from the international immuno-oncology biomarkers working group: Part 2: TILs in Melanoma, Gastrointestinal Tract Carcinomas, non-small cell lung carcinoma and mesothelioma, endometrial and ovarian carcinomas, squamous cell carcinoma of the head and neck, genitourinary carcinomas, and primary brain tumors. Adv Anat Pathol 2017;24:311–35. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13. Denkert C, von Minckwitz G, Brase JC, Sinn BV, Gade S, Kronenwett R, et al. Tumor-infiltrating lymphocytes and response to neoadjuvant chemotherapy with or without carboplatin in human epidermal growth factor receptor 2-positive and triple-negative primary breast cancers. J Clin Oncol 2015;33:983–91. [DOI] [PubMed] [Google Scholar]
- 14. Adams S, Gray RJ, Demaria S, Goldstein L, Perez EA, Shulman LN, et al. Prognostic value of tumor-infiltrating lymphocytes in triple-negative breast cancers from two phase III randomized adjuvant breast cancer trials: ECOG 2197 and ECOG 1199. J Clin Oncol 2014;32:2959. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15. Johnson DB, Estrada MV, Salgado R, Sanchez V, Doxie DB, Opalenik SR, et al. Melanoma-specific MHC-II expression represents a tumour-autonomous phenotype and predicts response to anti-PD-1/PD-L1 therapy. Nat Commun 2016;7:10582. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16. Roemer MGM, Redd RA, Cader FZ, Pak CJ, Abdelrahman S, Ouyang J, et al. Major histocompatibility complex class II and programmed death ligand 1 expression predict outcome after programmed death 1 blockade in classic hodgkin lymphoma. J Clin Oncol 2018;36:942–50. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17. Foldi J, Silber A, Reisenbichler E, Singh K, Fischbach N, Persico J, et al. Neoadjuvant durvalumab plus weekly nab-paclitaxel and dose-dense doxorubicin/cyclophosphamide in triple-negative breast cancer. NPJ breast cancer 2021;7:9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18. Wulfkuhle JD, Yau C, Wolf DM, Vis DJ, Gallagher RI, Brown-Swigart L, et al. Evaluation of the HER/PI3K/AKT family signaling network as a predictive biomarker of pathologic complete response for patients with breast cancer treated with Neratinib in the I-SPY 2 trial. JCO Precision Oncology 2018;2:PO.18.00024. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19. Pin E, Federici G, Petricoin III EF. Preparation and use of reverse protein microarrays. Curr Protoc Protein Sci 2014;75:27.27. 21–9. [DOI] [PubMed] [Google Scholar]
- 20. Mehta NT, Truax AD, Boyd NH, Greer SF. Early epigenetic events regulate the adaptive immune response gene CIITA. Epigenetics 2011;6:516–25. [DOI] [PubMed] [Google Scholar]
- 21. Axelrod ML, Cook RS, Johnson DB, Balko JM. Biological consequences of MHC-II expression by tumor cells in cancer. Clin Cancer Res 2019;25:2392–402. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Supplementary Methods: detailed protocol. Supplementary Table 1: Antibodies used for RPPA. Supplementary Table 2: Rates of tumor-specific MHC-II/HLA-DR expression across HER2-negative primary breast cancers. Supplementary Table 3: Univariate and multivariate analysis for achieving pCR to pembrolizumab and neoadjuvant chemotherapy or chemotherapy alone. Supplementary Table 4: Cox proportional Hazard ratio univariate analysis for 3-year event-free survival associated with pembrolizumab and neoadjuvant chemotherapy or chemotherapy alone. Supplementary Figure legends.
Supplementary Figure 1: CONSORT diagrams for associated studies. Supplementary Figure 2: Tumor specific MHC-II pattern immunofluorescence in ER+ and TNBC. Supplementary Figure 3: Non-tumor MHC-II expression and comparison of PD-L1+ and HLA-DR+ patient populations. Supplementary Figure 4: RPPA associations with pCR for control and pembrolizumab-treated (Treat) arms, by clinical group. Supplementary Figure 5: RPPA associations with pCR for control and pembrolizumab-treated arms in high-risk ER+ patients.