Abstract
Despite the dramatic underrepresentation of non-European populations in human genetics studies, researchers continue to exclude participants of non-European ancestry, as well as variants rare in European populations, even when these data are available. This practice perpetuates existing research disparities and can lead to important and large effect size associations being missed. Here, we conducted genome-wide association studies (GWAS) of 31 serum and urine biomarker quantitative traits in African (n=9354), East Asian (n=2559) and South Asian (n=9823) ancestry UK Biobank (UKBB) participants. We adjusted for all known GWAS catalog variants for each trait, as well as novel signals identified in a recent European ancestry focused analysis of UKBB participants. We identify 7 novel signals in African ancestry and 2 novel signals in South Asian ancestry participants (p < 1.61E-10). Many of these signals are highly plausible, including a cis pQTL for the gene encoding gamma-glutamyl transferase and PIEZO1 and G6PD variants with impacts on HbA1c through likely erythrocytic mechanisms. This work illustrates the importance of using the genetic data we already have in diverse populations, with novel discoveries possible in even modest sample sizes.
Introduction
Lack of representation of diverse global populations is a major problem in human genetics research. As recently reviewed, 78% of genome-wide association study (GWAS) participants are of European ancestry, with an additional 9% East Asian participants1. All other populations (as well as multi-ethnic studies) make up less than 13% of subjects, but account for 38% of significant associations in the GWAS catalog, demonstrating the scientific importance of including diverse populations for understanding the biology of complex traits. For example, only 2.4% of GWAS participants are of predominantly African ancestry, but 7% of GWAS catalog associations were found in these participants. Inclusion of diverse populations is also essential for risk prediction; polygenic risk score (PRS) instruments often perform poorly when trained using European only summary statistics and then applied to non-European populations2. As PRSs move into clinical use, this lack of representation risks perpetuating existing health disparities. Lack of inclusion of diverse populations could also result in missing many of the important insights into disease biology possible through human genetics.
However, as recently reviewed 3, we are still failing to use the data we have in ancestrally diverse populations. Even when non-European data are available, many researchers tend to focus only on large European sample sizes, and do not perform appropriate trans-ethnic or ancestry stratified analyses in participants with substantial non-European genetic ancestry. For example, the UK Biobank (UKBB) data, which is widely used due to its large sample size, broad data availability for qualified researchers, and variety of measured phenotypes and electronic health record data, includes >20,000 participants with non-European genetic ancestry. However, all 29 of the first papers indexed on the GWAS catalog that include UKBB participants included only the European ancestry sample (>400,000 individuals), likely for reasons of analytical convenience. Only recently, efforts such as the Pan-UK Biobank project 4 have made available summary statistics across UKBB participants with substantial non-European ancestry, and some efforts have been made to more extensively study whether additional variants and pathways are identified versus European ancestry participants only.
These existing studies support the value of including even small numbers of non-European ancestry participants, especially for biomarkers and endophenotypes for which a larger percentage of variance is often explained by a small number of genetic signals. Notably, in recent trans-ethnic analyses of blood cell traits including the UKBB data and other cohorts (total n=746,667), an IL7 coding variant associated with lymphocyte counts was identified in South Asian UKBB participants only (n=8189)5. The lymphocyte increasing allele of this variant increased secretion of IL7 by 83% in follow-up in vitro analyses. We here assess the genetic contributors to the UKBB serum and urine biomarker panel in non-European ancestry populations. We chose these quantitative traits based on a higher probability of previously undetected large effect size loci and improved statistical power versus dichotomous disease endpoints. Initial analyses of these serum and urine biomarkers have, similar to many other analyses in the UKBB, focused predominantly on European ancestry individuals 6; while all ancestry populations were included in the final published meta-analyses of these traits by Sinnott-Armstrong et al. (in contrast to the preprint version, which included European ancestry participants only 7), the posted meta-analysis results and subsequent follow-up analyses were limited to variants with a minor allele frequency (MAF) >1% in White British populations. This prior work on UKBB serum and urine biomarkers revealed important relationships, such as improved prediction of disease in the independent FinnGenn cohort for multi-biomarker PRS versus single-disease PRS, particularly for liver and renal disease, and novel signals, for example low frequency coding variants with impacts on kidney biomarkers and outcomes. However, we hypothesized that important novel variant-trait associations were missed by the focus only on variants common in British individuals. Mendelian randomization analyses suggest causal roles for a number of these biomarkers, including IGF-18, urine albumin9, urate10, so such ancestry differentiated variants may have important health consequences, as well as point to key genes and biological mechanisms relevant across populations and improve PRS prediction.
Materials and Methods
UK Biobank serum and urine biomarkers
The UK Biobank resource includes genetic and phenotypic data on nearly 500,000 individuals aged 40–69 at time of recruitment (2006–2010).11 All participants gave informed consent. UKBB released data on 34 serum and urine biomarkers, chosen based on their role as established risk factors or diagnostic measures for a wide range of diseases, with an emphasis on renal and liver health.12 We excluded three biomarkers with a high percentage of values below the reportable range (oestradiol, microalbumin in urine, and rheumatoid factor, with missingness>70%) and generated inverse normalized values for the remaining 31 biomarkers for genetic analysis (Table S1).
Derivation of Ancestry Clusters
We used a combination of self-reported ethnicity and k-means clustering of genetic principal components to derive lists of individuals to include in the African, South Asian, and East Asian clusters. First, we calculated principal components (PC) and their loadings for all 488,377 genotyped UKBB participants using high quality variants in the UKBB data set that overlapped with the participants in the 1000G Phase 3 v5 (1KG) reference panel (Figure S1). Reference ancestries used included 504 European (EUR), 347 American Admixed (AMR), 661 African (AFR), 504 East Asian (EAS), and 489 South Asian (SAS) samples (overall 2504). We projected the 1KG reference panel dataset on the calculated PC loadings from UKBB. We then used k-means clustering with four dimensions, defined by the first four PCs, to identify the individuals that clustered with the majority of individuals in each 1KG ancestry specific reference panel (PC1, PC2, PC3, and PC4 are displayed in Figure S1, those who are not in any k-means cluster (UKBB_other) are shown in grey).
We used self-reported ethnicity (variable “ethnic_background”, 21000–0.0 of the UKBB data, as reported by participants during the initial Assessment Centre visit) to assign individuals who fell outside of any 1KG cluster to a genetic analysis subset. For the African ancestry subset used in our analysis, we included all individuals that cluster with the 1KG AFR samples by k-means clustering, except n=7 individuals whose self-reported ethnicity was White, British, Irish, Any other White background, Indian, Pakistani, Bangladeshi, Any other Asian background, or Chinese. For individuals who did not cluster with the 1KG AFR population (or any other 1KG cluster) but self-reported White and Black Caribbean, White and Black African, Black or Black British, Caribbean, African, or Any other Black background, we assigned them to the African genetic ancestry analysis group (n=660). For the South Asian subset used in our analysis, we included all individuals that cluster with the 1KG SAS samples by k-means clustering, except 117 individuals with self-reported ethnicity as follows: White, British, Irish, Any other White background, White and Black Caribbean, White and Black African, Black or Black British, Black Caribbean, African, Any other Black background, or Chinese. 55 individuals with self-reported Indian, Pakistani, or Bangladeshi ethnicity (who did not cluster with any 1KG ancestry group) were also assigned to the South Asian subset (n=55). Finally, our East Asian ancestry subset is comprised of individuals that cluster with 1KG East Asians (EAS) by k-means clustering, removing 8 individuals with self-reported White, British, Irish, Any other White background, White and Black Caribbean, White and Black African, Indian, Pakistani, Bangladeshi, Black or Black British, Black Caribbean, African, or Any other Black background. 19 individuals with self-reported Chinese ethnicity (who did not cluster with any 1KG ancestry group) were also included in the East Asian subset. After clustering and exclusion of extreme outliers/potential sample swaps, we included n=9354 African, n=2559 East Asian, and n=9823 South Asian ancestry participants; these sample sizes are larger than those reported in Sinnott-Armstrong et al.6, largely due to inclusion of individuals which cluster based on principal components with a particular genetic ancestry group but have missing, “Mixed”, or “Other” for their self-report ethnicity data. For ease of comparison to reference allele frequencies (notably those from 1KG), we stratified analyses by these ancestry clusters.
Medication adjustment for lipids and diabetes traits
For subjects on lipid medications, we divided total cholesterol by 0.8 to approximate pre-medication values, and we divided directly assessed LDL by 0.7, as previously recommended.13 For analysis of both diabetes related traits (HbA1c and glucose), we excluded individuals with diabetes diagnosed by a doctor (UKBB variable 2443–0.0), those taking insulin (UKBB variable 6153–0.0), and those with HbA1c >=48 mmol/mol or glucose >=7 mmol/L.
Genotype imputation and association
Imputation was performed using 97,256 deeply sequenced reference genomes (freeze 8) from diverse populations from the National Heart, Lung, and Blood Institute’s Trans-Omics for Precision Medicine (TOPMed) Initiative (https://imputation.biodatacatalyst.nhlbi.nih.gov/#!), in order to better capture ancestry-specific rare variation (particularly in African ancestry populations) compared to the UK10K panel used for the public UKBB release. All listed positions are on build 38. We filtered to individuals and SNPs with a call rate >90% prior to imputation. For our analyses, we included only well-imputed variants in each cluster. For common (MAF > 0.5%) variants, we defined well-imputed as those with estimated r2 > 0.3, and for rare variants (MAF < 0.5%), those with estimated r2 > 0.8 were considered well-imputed. Association analyses were performed using the EMMAX test implemented in EPACTS 3.3.0, which accounts for population structure. Genotyped variants with MAF > 1% and missing rate < 1% were used in kinship matrix derivation. We removed variants with an estimated minor allele count (MAC) < 5 when running EPACTS to improve model stability. X chromosome analyses were conducted stratified by sex and then meta-analyzed using GWAMA, alleviating problems with inflation for some sex-differentiated biomarkers and allowing us to assess evidence of heterogeneity by sex. We assessed testosterone stratified by sex for both autosomes and the X chromosome due to the dramatically difference in trait distribution between males and females (see Table S1).
For our association analyses of serum and urine biomarkers, we first regressed out covariates (age, sex, first 10 PCs (provided by UKBB), genotyping array, centers) before inverse normalizing the resultant residuals. In our conditional GWAS analyses, we also included known variants from the GWAS catalog (accessed Spring 2020) as covariates in our association models (any variant previously identified on each tested chromosome, Table S2), as our primary aim was to identify novel signals missed in previous predominantly European analyses. For our identified signals, we checked if UKBB European focused analyses (as described in Sinnott-Armstrong et al.6, 7, Table S3) had identified genome-wide significant variants (p<5E-8) within 1MB of our sentinel signal. We then included these nearby associated variants as covariates in final conditional analyses reported here, to see whether our sentinel variants from non-European ancestry focused analyses were still genome-wide significant. Chromosome X was not included in previous European focused analyses from Sinnott-Armstrong et al., so this does not apply to those variants. We also assessed if any genome-wide significant signals remained after adjustment for significant novel variants in Table 1 to assess if there were multiple distinct novel signals at the locus.
Table 1: Novel Association Signals in African and South Asian Ancestry Participants in UK Biobank.
All biomarkers are measured in serum, except creatinine, potassium, and sodium, which were measured in urine. EAF, effect allele frequency, APOA, apolipoprotein A, APOB, apolipoprotein B, ALP, alkaline phosphatase, ALT, alanine aminotransferase, BRB, bilirubin, CysC, cystatin C, GGT, gamma glutamyltransferase, HbA1c, glycated hemoglobin, IGF-1, Insulin-like growth factor 1, LPA, Lipoprotein-A
| Unconditioned Results | Conditional Analysis | |||||||||
|---|---|---|---|---|---|---|---|---|---|---|
| rsID | Effect allele | Trait | Cohort | EAF | p-value | β | p-value | β | Nearest Gene | Annotation |
| rs28362286 | A | APOB | AFR | 0.9% | 3.14E-20 | −0.75 | 9.49E-16 | −0.85 | PCSK9 | coding, p.Cys679Ter |
| rs3211938 | G | ALP | AFR | 10.3% | 9.80E-15 | −0.20 | 8.00E-15 | −0.20 | CD36 | coding, p.Tyr325Ter |
| rs1050828 | T | BRB total | AFR female | 14.4% | 4.19E-38 | 0.31 | 1.61E-33 | 0.30 | G6PD | coding, p.Val98Met |
| AFR male | 7.6% | 3.91E-33 | 0.65 | 8.85E-32 | 0.66 | |||||
| AFR meta | 11.5% | 3.16E-63 | 0.36 | 8.52E-57 | 0.36 | |||||
| BRB direct | AFR female | 14.4% | 8.04E-19 | 0.23 | 4.13E-15 | 0.21 | ||||
| AFR male | 7.6% | 4.86E-20 | 0.50 | 3.31E-20 | 0.53 | |||||
| AFR meta | 11.5% | 5.76E-33 | 0.28 | 2.28E-28 | 0.27 | |||||
| rs334 | A | Creatinine | AFR | 6.3% | 2.62E-38 | −0.43 | 2.62E-38 | −0.43 | HBB | coding, p.Glu7Ala |
| Potassium | AFR | 2.84E-32 | −0.39 | 2.84E-32 | −0.39 | |||||
| Sodium | AFR | 5.43E-36 | −0.42 | 5.43E-36 | −0.42 | |||||
| rs112902560 | T | CysC | AFR | 4.7% | 1.92E-11 | 0.25 | 1.92E-11 | 0.25 | MMP26/HBB | noncoding |
| rs57719575 | C | GGT | AFR | 14.9% | 3.97E-38 | −0.28 | 9.18E-13 | −0.35 | GGT1 | noncoding |
| rs556126054 | G | HbA1c | SAS | 2.4% | 1.02E-27 | −0.59 | 2.40E-12 | −0.53 | PIEZO1 | noncoding |
| rs5030868 | A | HbA1c | SAS female | 1.7% | 6.98E-22 | −0.82 | 1.56E-21 | −0.82 | G6PD | coding, p.Ser218Phe |
| SAS male | 0.7% | 1.09E-33 | −1.77 | 8.79E-34 | −1.77 | |||||
| SAS meta | 1.2% | 7.51E-48 | −1.06 | 1.90E-47 | −1.06 | |||||
| rs115739169 | A | LPA | AFR | 1.27% | 6.15E-62 | 1.22 | 5.00E-12 | 1.05 | LPA | noncoding |
The conditional analysis p-value for our novel signals displayed is, along with GWAS catalog variants, adjusted for any variants within 1MB on each side of the sentinel variant which were genome-wide significant in analyses of serum and urine biomarkers in UK Biobank Europeans7, to ensure the signals we identify could not be found in European ancestry participants alone.
We adopted a significance threshold of 5E-9/31 traits, or p < 1.61E-10, based on reasonable estimates of the number of independent tests for testing all common and low frequency variants genome-wide14.
Inclusion of rs334
Our initial analyses identified several putative novel signals at the HBB locus; however, these results were difficult to interpret as the sickle cell trait variant rs334, which is known to have impacts on numerous traits including kidney function15 and HbA1c16, was excluded from the TOPMed freeze 8 reference panel. We extracted this variant from UK10K imputation provided by UKBB (imputation info score 0.899) for additional conditional analyses at these loci.
Replication Analyses
We conducted replication of our novel signals in African American women from the Women’s Health Initiative with Affymetrix 6.0 data from the WHI17 SHARe resource (dbGaP phs000386.v7.p3). Imputation was performed using TOPMed imputation server (https://imputation.biodatacatalyst.nhlbi.nih.gov) with TOPMed freeze 8 reference panel. We adopted the same analysis plan described above. Due to limited availability of serum and urine biomarkers with adequate sample sizes (>100 individuals with phenotype data), we only performed replication analyses for APOB (n=186) and LPA (n=1,599) associated variants in WHI (Table S5). Where adequate sample sizes were available, we also pursued replication analyses in African Americans from the BioVU biobank at Vanderbilt University Medical Center, which is comprised of >100,000 individuals who have DNA samples linked to their de-identified electronic health record (EHR) information18 and includes both cleaned and harmonized diagnosis codes and clinical laboratory values. Genotyping in BioVU was performed using the Multi-Ethnic Global (MEGA) array. Genetic ancestry clusters were determined using principal component analysis on the imputed data combined with 1KG reference panels, for a total of 15,123 African ancestry participants with at least some EHR based lab data. Imputation was performed using the TOPMed imputation server using TOPMed freeze 8 for rs1050828 and the Haplotype Reference Consortium (HRC) panel for rs334. Analyses for urine creatine (n=2522) and total bilirubin in serum or plasma (n=11960) were adjusted for age, sex, and 10 PCs; other biomarker trait associations in African ancestry individuals were not able to be replicated due to limited phenotype data.
Most of our serum and urine biomarker traits are not widely assayed in publicly available databases; however, multiple analyses of HbA1c are available as part of the AMP T2D portal (https://t2d.hugeamp.org/, accessed February 2, 2021). Replication results were also available from the Singapore Indian Eye Study (SINDI)20 population-based cohort of Indian ancestry individuals (n = 1512) with measured HbA1c and imputation to TOPMed freeze 8 using the TOPMed imputation server.
Results
All genome-wide significant variants are displayed in Table 1, Figure S2 (LocusZoom21 plots), Figure S3 (allele frequency spectrum plot for 1KG reference populations) and Table S5. We picked two traits as examples to show the genome-wide mirror Manhattan plots (Figure 1–2) and all the other plots are available in Figure S4, with unconditional results (bottom) and results conditioned on previously reported genome-wide significant variants (top). We did not observe evidence of significant genome-wide inflation (Table S4).
Figure 1.
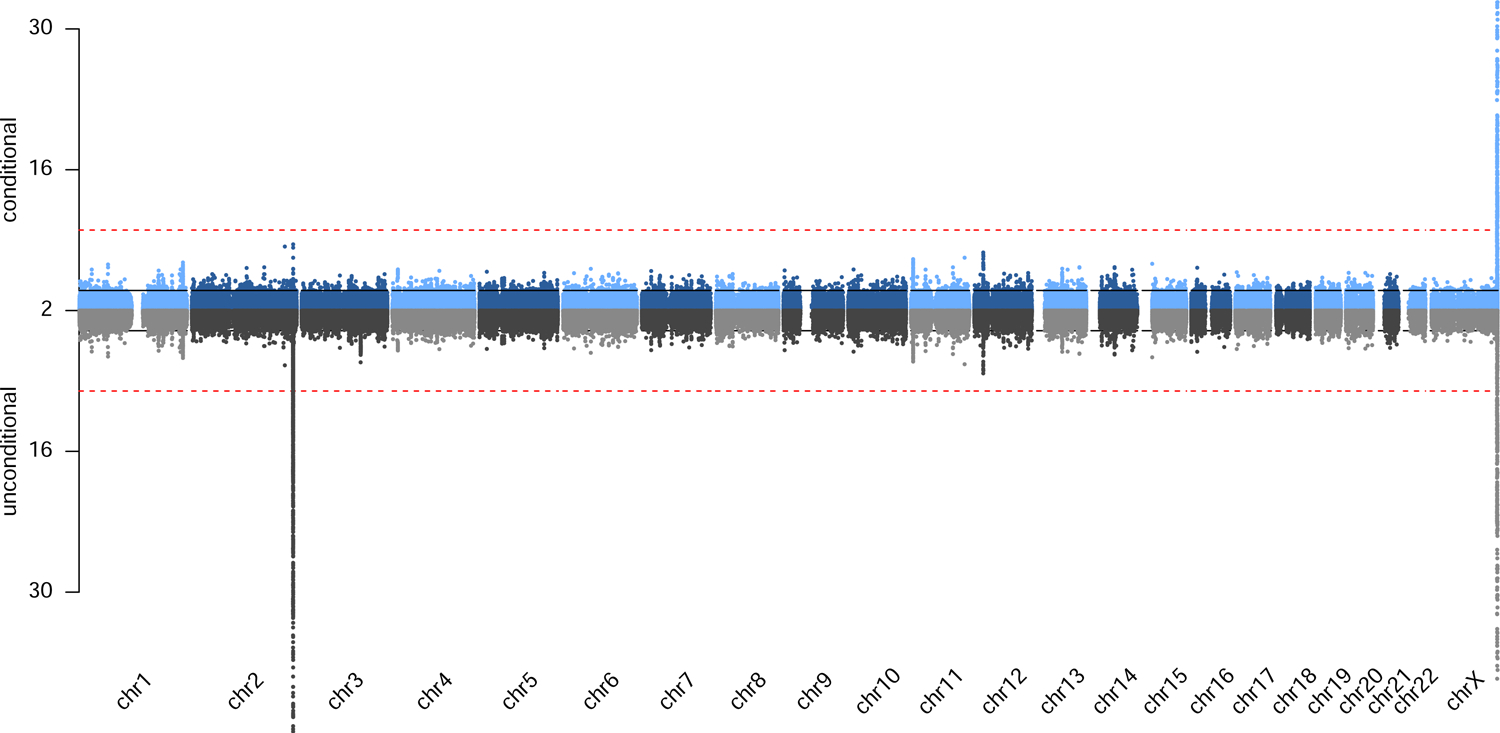
Genome-wide mirror Manhattan plot of association statistics for total bilirubin in African ancestry populations, with unconditional results (bottom) and results conditioned on previously reported genome-wide significant variants (top).
Figure 2.
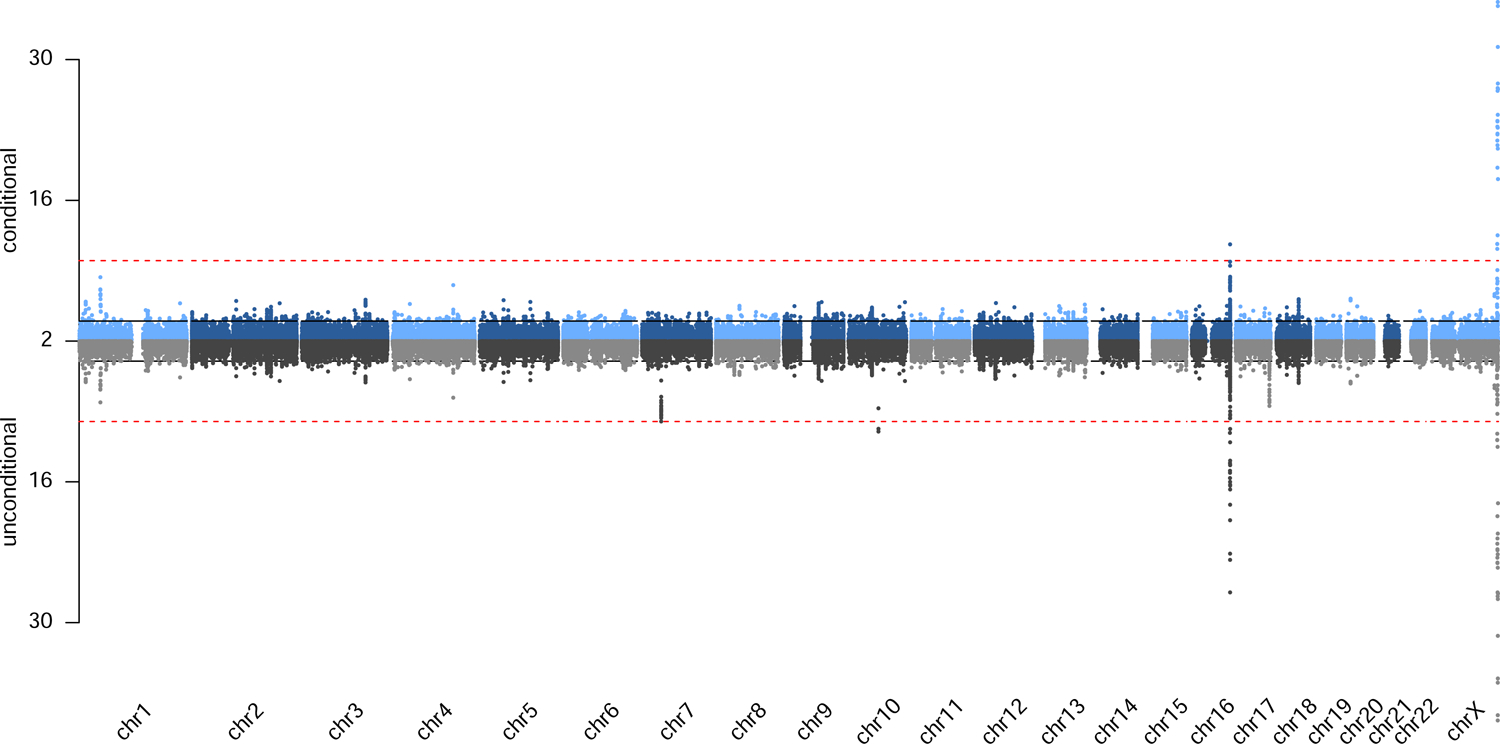
Genome-wide mirror Manhattan plot of association statistics for HbA1c in South Asian ancestry populations, with unconditional results (bottom) and results conditioned on previously reported genome-wide significant variants (top).
We identify two novel findings in South Asians (n=9823), both for HbA1c (a non-coding variant near PIEZO1 rs556126054 and a G6PD missense variant rs5030868). In the AMP T2D portal, rs5030868 was reported to be associated with lower HbA1c in 1774 multi-ethnic individuals from AMP T2D-GENES quantitative trait exome sequence analysis (p = 4.29E-8), a multi-ethnic meta-analysis of 10,338 individuals with whole genome sequencing data (p = 8.39E-6) and 7,159 European ancestry participants from the Exeter EXTEND Biobank (p = 0.04). rs556126054 near PIEZ01 was not available in the AMP T2D portal. For rs556126054, we replicated the association in SINDI (p = 1.84E-3, Table S5).
We identify 7 novel findings in African ancestry individuals (n=9354), including coding variants (for example, a CD36 loss of function variant, rs3211938, with ALP) and cis pQTLs (rs57719575 at GGT1 for liver enzyme gamma-glutamyl transferase (GGT)). We did not have access to appropriate replication datasets for all findings; however, all tested SNP-trait pairs did replicate. G6PD coding variant rs1050828’s association with bilirubin replicated in African Americans from BioVU (p = 2.24E-9), as did sickle cell trait (rs334) with creatinine in urine (p = 4.81E-18). We note that the rs334 associations could not be replicated in European ancestry individuals in BioVU due to very low allele frequency, which is consistent with exclusion of this variant from published European focused analysis results6. In WHI African Americans we replicated the association of noncoding LPA locus variant rs115739169 with LPA (p = 4.01E-32, Table S5) and stop variant rs28362286 at PCSK9 with APOB (p = 5.36E-3). We do not identify any novel findings in East Asians (n=2559), the smallest of the three samples. As shown in Table S5 and Figure S3, all novel variants are rare or low frequency in Europeans. At each locus, we also assessed if any genome-wide significant signals remained after adjusting for the sentinel variant in Table S5; none were identified, suggesting no additional novel distinct signals at these loci.
Novel G6PD locus associations with bilirubin (African ancestry) and HbA1c (South Asian ancestry)
The X chromosome is left out of the majority of GWAS analyses, with only around a third of GWAS including chromosome X 3, 22. We here identify a strong association of a G6PD coding variant (rs1050828), located on chromosome X, with total and direct bilirubin in African ancestry individuals, which has not yet been reported in the GWAS catalog despite the strong effect size. Direct bilirubin assesses bilirubin conjugated with glucuronic acid, which is secreted into bile. Indirect bilirubin (unconjugated) in plasma is usually low in healthy individuals, as this conjugation process is quite efficient, but can be elevated in many forms of hyperbilirubinemia, such as those caused by hemolysis, Gilbert syndrome, or in response to some medications23. This G6PD signal is also associated with indirect bilirubin (calculated as total minus direct bilirubin, βfemale = 0.22, pfemale = 1.39E-16; βmale = 0.58, pmale = 3.57E-24; βmeta = 0.29, pmeta =1.71E-32), concordant with the known risk of hemolytic anemia in those with G6PD deficiency. This association is concordant with existing literature that males with G6PD deficiency (including deficiency caused by rs1050828) are at elevated risk of neonatal hyperbilirubinemia and jaundice24, though the strong association with bilirubin in adults and in females as well as in males is less expected. Bilirubin is commonly measured in clinical settings to assess liver function or diagnose hemolytic anemia (which can occur upon exposure to triggers such as oxidative drugs or acute infections in individuals with G6PD deficiency); if used to assess liver function, it is possible that variation at G6PD, as well as alpha thalassemia copy number variation, which was recently reported to be associated with bilirubin25 and is also more common in African versus European ancestry populations, could interfere with accurate clinical inference.
We also identify a different G6PD coding variant strongly associated with HbA1c in South Asians (rs5030868, 1.1% MAF in UKBB South Asians, noted in ClinVar for G6PD deficiency, known as the G6PD Mediterranean variant in previous literature). Unlike the G6PD deficiency variant common in African Americans (rs1050828, reported here for bilirubin), which has been reported to strongly influence HbA1c,26 this variant is not previously reported in the GWAS catalog for HbA1c. Other G6PD coding variants (rs76723693 in African Americans,19 rs72554665 and rs72554664 in East Asians27) have also been reported to influence HbA1c. Our results are concordant with this previous literature, and add to concerns that use of HbA1c as a laboratory test in populations with a high prevalence of G6PD deficiency may lead to underdiagnosis of diabetes and poor management and prevention of complications in those with diagnosed diabetes.28 There is some literature to suggest that G6PD-deficient patients may have an increased risk of diabetes29 and its complications30; more study is needed to disentangle impacts of G6PD deficiency on diabetes diagnosis and monitoring (due to use of HbA1c) from potential impacts on disease pathogenesis.
PIEZO1 locus association with HbA1c in South Asian ancestry individuals
In addition to the signals described above at G6PD, we identify an additional novel signal for HbA1c which likely impedes accurate assessment of glycemic control in South Asians. A conserved noncoding variant near PIEZO1 (rs556126054, CADD score 9.72) more common in South Asian populations (4.7% in 1KG South Asians versus 0.8% in Europeans and 0.6% in admixed Americans, not found East Asian or African populations) was associated with HbA1c. PIEZO1 encodes an erythrocyte membrane protein, and African specific variants in this protein have been associated with red blood cell dehydration and lower malaria infection risk31. In recent analyses of UK Biobank blood cell trait data5, there is a strong signal in South Asians for PIEZO1 missense variant rs563555492 (p.Leu2277Met) for higher hematocrit (p = 6.09E-14), hemoglobin (p = 4.69E-22), and red blood cell count (p = 1.50E-11), suggesting this locus acts through an erythrocytic pathway on HbA1c. This variant is also significant in our results (p = 3.63E-21, LD r2=0.25 in UKBB South Asians) for HbA1c, but there is only one statistically distinct genome-wide significant signal at the locus upon iterative conditional analysis. Like the G6PD coding variants discussed above, this noncoding signal at PIEZO1 also likely acts through erythrocytic mechanisms (as suggested by prior red blood cell related trait associations for its LD buddy rs5635554925) and will interfere with how accurately HbA1c assess glycemic control, potentially leading to disparities in diabetes diagnosis and treatment.
Additional associations for known variants
We identified an association with ALP with African ancestry specific CD36 nonsense variant rs3211938, which has been previously associated with HDL cholesterol levels32, 33, ECG traits34, red cell distribution width35, platelet count36, and C-reactive protein37. This locus is under selective pressure38, potentially from malaria, though relationships are unclear, with this nonsense variant associated with risk of cerebral malaria and higher overall malaria incidence, but lower risk of severe anemia39. While this association with ALP was not anticipated from previous literature, our findings confirm evidence of pleiotropy at this locus. We also identified an association of an African ancestry specific PCSK9 stop variant already known to be associated with LDL and total cholesterol25, 32 with apolipoprotein B, an unsurprising extension of the existing literature.
We further extend the literature linking sickle cell trait (or rs334) to kidney function15, 40, including albumin to creatinine ratio in urine, with strong associations observed for urine potassium, sodium, and creatinine (Table 1, Table S5 and Figure S2). These associations are robust to adjustment for hemoglobin and estimated glomerular filtration rate (eGFR) (Table S6). A noncoding variant (rs112902560) in LD with rs334 (r2=0.41 in UKBB African ancestry participants) was also newly identified as associated with cystatin C, another kidney function measure.
Additional novel findings in African ancestry individuals
Our results also include two additional cis pQTL signals, or pQTLs near the encoding genes for our serum biomarkers. For example, we identify a novel cis pQTL, rs57719575, at GGT1, the encoding gene for liver enzyme GGT. Our results further include identification of a novel signal at the LPA locus for lipoprotein A, adding to the already extensive evidence of multiple distinct cis pQTL signals at this locus41–43. We were not able to adjust for KIV2-CN (copy number) in the Lp(a) region with our imputed single nucleotide variant data, which makes novel distinct signals somewhat difficult to interpret. Local ancestry has also been shown to be an important covariate at the LPA locus in analyses of African Americans and may be a confounder of results at this locus42. However, these highly interpretable and biologically relevant cis pQTL signals echo the results from recent focused analyses of urate, IGF-1, and testosterone in European populations.44 Many lead signals for these serum biomarkers were near genes involved in biosynthesis, transport, or signaling pathways relevant to the target trait, in contrast to the often difficult to interpret lead association signals for more complex phenotypes.
Discussion
Even in the relatively small number of African and South Asian ancestry individuals in UKBB, we identified novel and clinically relevant associations. These associations could not be found or tagged by any variants in close LD if we only restrict to European individuals for analysis, even in the very large UKBB sample size6. While it is possible these variants could be identified in even larger European ancestry cohorts, or cohorts recruited in countries other than the UK, identification of these signals in less than 10,000 individuals of African or South Asian ancestry demonstrates the importance of inclusion of non-European ancestry populations in genetic analysis of serum and urine biomarkers. Several associations replicate in external cohorts and biobanks such as WHI and BioVU. These novel findings also highlight the importance of X chromosome analysis, ancestry differentiated cis pQTLs and variants which impact HbA1c through likely erythrocytic mechanisms, and coding variant associations for urine and serum biomarker traits.
Some novel findings would not have been possible without TOPMed imputation, which has been demonstrated in previous analyses to have dramatically improved imputation quality for rare variants, particularly in Hispanic/Latino and African ancestry individuals45, including for identification of novel rare variant association signals in African45 and European ancestry46 UKBB participants. For many of our identified signals, imputation quality was similar to the Haplotype Reference Consortium (HRC) and UK10K haplotype imputation provided by UKBB. However, improvements were observed for most variants, with noticeable improvement particularly for G6PD coding variant rs5030868 (previously imputed with an info score < 0.3, imputed with an r2 of 0.86 using the TOPMed reference panel in South Asian ancestry individuals). We do note that due to stringent variant filtering in TOPMed some important known signals (like sickle cell trait) were not included in the reference panel; this is an important limitation for users of this reference panel.
Given the very large sample size now available for all of these biomarkers through the European focused analyses in UKBB 6, as well as in many cases other large GWAS meta-analyses, it is striking that a number of functionally plausible and novel signals could be identified in analyses of <10,000 African and South Asian individuals, a sample size much smaller than most current GWAS analyses. Our results highlight the potential impact of ancestry-differentiated results on the accuracy of clinical biomarker measures. Issues with the use of HbA1c in non-European populations due to G6PD variants, sickle cell trait, and other ancestry differentiated variants are recognized, but other clinical assays are also likely influenced by ancestry differentiated variants unrelated to disease risk. This bias may cause even more systematic problems as novel biomarkers and large-scale proteomics panels move into clinical risk prediction, as the largest training datasets for risk prediction and determination of reference ranges are composed of European ancestry individuals.
We note that a limitation of our results is our failure to provide replication for some of our putative novel findings, due to a lack of readily available replication datasets, especially for less frequently measured serum biomarkers (for example the association of the CD36 loss of function variant with ALP, which is not available in most cohort datasets). However, for the associations we have reasonable sized datasets to replicate findings in, we identified consistent replication. In addition, the number of variants identified with strong functional annotation near relevant genes suggests that these preliminary results include findings worthy of future exploration in larger datasets of diverse ancestry background, and clearly demonstrate the value of using genetic data from UKBB non-European ancestry participants.
Supplementary Material
Acknowledgements
This research has been conducted using the UK Biobank Resource under Application Number 25953.
LMR is supported by T32 HL129982, R01HG010297, and KL2 TR00249. NF is supported by the NIH DK117445, MD012765 and HL140385. MPL is supported by F32 HL149256. The content is solely the responsibility of the authors and does not necessarily represent the official views of the NIH.
We would like to acknowledge use of the Trans-Omics in Precision Medicine (TOPMed) program imputation panel (freeze 8 version) supported by the National Heart, Lung and Blood Institute (NHLBI); see www.nhlbiwgs.org. TOPMed study investigators contributed data to the reference panel, which was accessed through https://imputation.biodatacatalyst.nhlbi.nih.gov. The panel was constructed and implemented by the TOPMed Informatics Research Center at the University of Michigan (3R01HL-117626–02S1; contract HHSN268201800002I). The TOPMed Data Coordinating Center (R01HL-120393; U01HL-120393; contract HHSN268201800001I) provided additional data management, sample identity checks, and overall program coordination and support. We gratefully acknowledge the studies and participants who provided biological samples and data for TOPMed.
The BioVU projects at Vanderbilt University Medical Center are supported by numerous sources: institutional funding, private agencies, and federal grants. These include the NIH funded Shared Instrumentation Grant S10OD017985 and S10RR025141; CTSA grants UL1TR002243, UL1TR000445, and UL1RR024975 from the National Center for Advancing Translational Sciences. Its contents are solely the responsibility of the authors and do not necessarily represent official views of the National Center for Advancing Translational Sciences or the National Institutes of Health. Genomic data are also supported by investigator-led projects that include U01HG004798, R01NS032830, RC2GM092618, P50GM115305, U01HG006378, U19HL065962, R01HD074711; and additional funding sources listed at https://victr.vumc.org/biovu-funding/.
The PAGE Study is funded by the National Human Genome Research Institute (NHGRI) with co-funding from the National Institute on Minority Health and Health Disparities (NIMHD). The contents of this paper are solely the responsibility of the authors and do not necessarily represent the official views of the NIH. Genotyping services were provided by the Center for Inherited Disease Research (CIDR). CIDR is fully funded through a federal contract from the National Institutes of Health to The Johns Hopkins University. Genotype data quality control and quality assurance services were provided by the Genetic Analysis Center in the Biostatistics Department of the University of Washington, through support provided by the CIDR contract.
Funding support for the “Exonic variants and their relation to complex traits in minorities of the WHI” study is provided through the NHGRI PAGE program (NIH U01HG007376). The WHI program is funded by the National Heart, Lung, and Blood Institute, National Institutes of Health, U.S. Department of Health and Human Services through contracts HHSN268201100046C, HHSN268201100001C, HHSN268201100002C, HHSN268201100003C, HHSN268201100004C, and HHSN271201100004C. The authors thank the WHI investigators and staff for their dedication, and the study participants for making the program possible. A listing of WHI investigators can be found at: https://www.whi.org/researchers/Documents%20%20Write%20a%20Paper/WHI%20Investigator%20Long%20List.pdf.
Footnotes
Conflict of Interest Statement
The authors declare no competing interests.
Web Resources
Summary statistics are available at https://yunliweb.its.unc.edu/serum_biomarker/index.php
Data are available upon request from the UK Biobank https://www.ukbiobank.ac.uk/.
References
- 1.Gurdasani D, Barroso I, Zeggini E, Sandhu MS. Genomics of disease risk in globally diverse populations. Nature Reviews Genetics. 2019;20:520–35. [DOI] [PubMed] [Google Scholar]
- 2.Martin AR, Kanai M, Kamatani Y, Okada Y, Neale BM, Daly MJ. Clinical use of current polygenic risk scores may exacerbate health disparities. Nat Genet. 2019;51:584–91. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Manolio TA. Using the Data We Have: Improving Diversity in Genomic Research. Am J Hum Genet. 2019;105:233–36. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Pan-UKB team., (2020).
- 5.Chen M-H, Raffield LM, Mousas A, Sakaue S, Huffman JE, Jiang T, et al. Trans-ethnic and ancestry-specific blood-cell genetics in 746,667 individuals from 5 global populations. bioRxiv. 2020:2020.01.17.910497. [DOI] [PMC free article] [PubMed]
- 6.Sinnott-Armstrong N, Tanigawa Y, Amar D, Mars N, Benner C, Aguirre M, et al. Genetics of 35 blood and urine biomarkers in the UK Biobank. Nature Genetics. 2021;53:185–94. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Sinnott-Armstrong N, Tanigawa Y, Amar D, Mars NJ, Aguirre M, Venkataraman GR, et al. Genetics of 38 blood and urine biomarkers in the UK Biobank. bioRxiv. 2019:660506. [DOI] [PMC free article] [PubMed]
- 8.Larsson SC, Michaëlsson K, Burgess S. IGF-1 and cardiometabolic diseases: a Mendelian randomisation study. Diabetologia. 2020. [DOI] [PMC free article] [PubMed]
- 9.Haas ME, Aragam KG, Emdin CA, Bick AG, Hemani G, Davey Smith G, et al. Genetic Association of Albuminuria with Cardiometabolic Disease and Blood Pressure. Am J Hum Genet. 2018;103:461–73. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Li X, Meng X, He Y, Spiliopoulou A, Timofeeva M, Wei W-Q, et al. Genetically determined serum urate levels and cardiovascular and other diseases in UK Biobank cohort: A phenome-wide mendelian randomization study. PLOS Medicine. 2019;16:e1002937. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Bycroft C, Freeman C, Petkova D, Band G, Elliott LT, Sharp K, et al. The UK Biobank resource with deep phenotyping and genomic data. Nature. 2018;562:203–09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.UK Biobank., (2018).
- 13.Willer CJ, Schmidt EM, Sengupta S, Peloso GM, Gustafsson S, Kanoni S, et al. Discovery and refinement of loci associated with lipid levels. Nat Genet. 2013;45:1274–83. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Lin DY. A simple and accurate method to determine genomewide significance for association tests in sequencing studies. Genetic epidemiology. 2019;43:365–72. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Naik RP, Irvin MR, Judd S, Gutierrez OM, Zakai NA, Derebail VK, et al. Sickle Cell Trait and the Risk of ESRD in Blacks. Journal of the American Society of Nephrology : JASN. 2017;28:2180–87. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Lacy ME, Wellenius GA, Sumner AE, Correa A, Carnethon MR, Liem RI, et al. Association of Sickle Cell Trait With Hemoglobin A1c in African Americans. Jama. 2017;317:507–15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.The Women’s Health Initiative Study Group. Design of the Women’s Health Initiative clinical trial and observational study. Controlled clinical trials. 1998;19:61–109. [DOI] [PubMed] [Google Scholar]
- 18.Roden DM, Pulley JM, Basford MA, Bernard GR, Clayton EW, Balser JR, et al. Development of a large-scale de-identified DNA biobank to enable personalized medicine. Clinical pharmacology and therapeutics. 2008;84:362–9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Sarnowski C, Leong A, Raffield LM, Wu P, de Vries PS, DiCorpo D, et al. Impact of Rare and Common Genetic Variants on Diabetes Diagnosis by Hemoglobin A1c in Multi-Ancestry Cohorts: The Trans-Omics for Precision Medicine Program. Am J Hum Genet. 2019;105:706–18. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Lavanya R, Jeganathan VS, Zheng Y, Raju P, Cheung N, Tai ES, et al. Methodology of the Singapore Indian Chinese Cohort (SICC) eye study: quantifying ethnic variations in the epidemiology of eye diseases in Asians. Ophthalmic epidemiology. 2009;16:325–36. [DOI] [PubMed] [Google Scholar]
- 21.Pruim RJ, Welch RP, Sanna S, Teslovich TM, Chines PS, Gliedt TP, et al. LocusZoom: regional visualization of genome-wide association scan results. Bioinformatics. 2010;26:2336–7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Wise AL, Gyi L, Manolio TA. eXclusion: toward integrating the X chromosome in genome-wide association analyses. Am J Hum Genet. 2013;92:643–7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.VanWagner LB, Green RM. Evaluating elevated bilirubin levels in asymptomatic adults. Jama. 2015;313:516–17. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Frank JE. Diagnosis and management of G6PD deficiency. American family physician. 2005;72:1277–82. [PubMed] [Google Scholar]
- 25.Gurdasani D, Carstensen T, Fatumo S, Chen G, Franklin CS, Prado-Martinez J, et al. Uganda Genome Resource Enables Insights into Population History and Genomic Discovery in Africa. Cell. 2019;179:984–1002.e36. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Wheeler E, Leong A, Liu CT, Hivert MF, Strawbridge RJ, Podmore C, et al. Impact of common genetic determinants of Hemoglobin A1c on type 2 diabetes risk and diagnosis in ancestrally diverse populations: A transethnic genome-wide meta-analysis. PLoS Med. 2017;14:e1002383. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Leong A, Lim VJY, Wang C, Chai JF, Dorajoo R, Heng CK, et al. Association of G6PD variants with hemoglobin A1c and impact on diabetes diagnosis in East Asian individuals. BMJ open diabetes research & care. 2020;8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Paterson AD. HbA1c for type 2 diabetes diagnosis in Africans and African Americans: Personalized medicine NOW! PLoS Med. 2017;14:e1002384. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Lai YK, Lai NM, Lee SW. Glucose-6-phosphate dehydrogenase deficiency and risk of diabetes: a systematic review and meta-analysis. Annals of hematology. 2017;96:839–45. [DOI] [PubMed] [Google Scholar]
- 30.Cappai G, Songini M, Doria A, Cavallerano JD, Lorenzi M. Increased prevalence of proliferative retinopathy in patients with type 1 diabetes who are deficient in glucose-6-phosphate dehydrogenase. Diabetologia. 2011;54:1539–42. [DOI] [PubMed] [Google Scholar]
- 31.Ma S, Cahalan S, LaMonte G, Grubaugh ND, Zeng W, Murthy SE, et al. Common PIEZO1 Allele in African Populations Causes RBC Dehydration and Attenuates Plasmodium Infection. Cell. 2018;173:443–55.e12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Wojcik GL, Graff M, Nishimura KK, Tao R, Haessler J, Gignoux CR, et al. Genetic analyses of diverse populations improves discovery for complex traits. Nature. 2019;570:514–18. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Klarin D, Damrauer SM, Cho K, Sun YV, Teslovich TM, Honerlaw J, et al. Genetics of blood lipids among ~300,000 multi-ethnic participants of the Million Veteran Program. Nature genetics. 2018;50:1514–23. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Baldassari AR, Sitlani CM, Highland HM, Arking DE, Buyske S, Darbar D, et al. Multi-ethnic Genome-wide Association Study of Decomposed Cardioelectric Phenotypes Illustrates Strategies to Identify and Characterize Evidence of Shared Genetic Effects for Complex Traits. Circulation Genomic and precision medicine. 2020. [DOI] [PMC free article] [PubMed]
- 35.Chami N, Chen MH, Slater AJ, Eicher JD, Evangelou E, Tajuddin SM, et al. Exome Genotyping Identifies Pleiotropic Variants Associated with Red Blood Cell Traits. Am J Hum Genet. 2016. [DOI] [PMC free article] [PubMed]
- 36.Auer PL, Johnsen JM, Johnson AD, Logsdon BA, Lange LA, Nalls MA, et al. Imputation of exome sequence variants into population- based samples and blood-cell-trait-associated loci in African Americans: NHLBI GO Exome Sequencing Project. Am J Hum Genet. 2012;91:794–808. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Ellis J, Lange EM, Li J, Dupuis J, Baumert J, Walston JD, et al. Large multiethnic Candidate Gene Study for C-reactive protein levels: identification of a novel association at CD36 in African Americans. Human genetics. 2014;133:985–95. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Bhatia G, Patterson N, Pasaniuc B, Zaitlen N, Genovese G, Pollack S, et al. Genome-wide comparison of African-ancestry populations from CARe and other cohorts reveals signals of natural selection. Am J Hum Genet. 2011;89:368–81. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Penha-Gonçalves C Genetics of Malaria Inflammatory Responses: A Pathogenesis Perspective. Front Immunol. 2019;10:1771. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Naik RP, Derebail VK, Grams ME, Franceschini N, Auer PL, Peloso GM, et al. Association of sickle cell trait with chronic kidney disease and albuminuria in African Americans. Jama. 2014;312:2115–25. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Zekavat SM, Ruotsalainen S, Handsaker RE, Alver M, Bloom J, Poterba T, et al. Deep coverage whole genome sequences and plasma lipoprotein(a) in individuals of European and African ancestries. Nature communications. 2018;9:2606. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Li J, Lange LA, Sabourin J, Duan Q, Valdar W, Willis MS, et al. Genome- and exome-wide association study of serum lipoprotein (a) in the Jackson Heart Study. Journal of human genetics. 2015. [DOI] [PubMed]
- 43.Mack S, Coassin S, Rueedi R, Yousri NA, Seppälä I, Gieger C, et al. A genome-wide association meta-analysis on lipoprotein (a) concentrations adjusted for apolipoprotein (a) isoforms. J Lipid Res. 2017;58:1834–44. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Sinnott-Armstrong N, Naqvi S, Rivas M, Pritchard JK. GWAS of three molecular traits highlights core genes and pathways alongside a highly polygenic background. bioRxiv. 2020:2020.04.20.051631. [DOI] [PMC free article] [PubMed]
- 45.Kowalski MH, Qian H, Hou Z, Rosen JD, Tapia AL, Shan Y, et al. Use of >100,000 NHLBI Trans-Omics for Precision Medicine (TOPMed) Consortium whole genome sequences improves imputation quality and detection of rare variant associations in admixed African and Hispanic/Latino populations. PLoS genetics. 2019;15:e1008500. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Taliun D, Harris DN, Kessler MD, Carlson J, Szpiech ZA, Torres R, et al. Sequencing of 53,831 diverse genomes from the NHLBI TOPMed Program. bioRxiv. 2019:563866. [DOI] [PMC free article] [PubMed]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.


