Abstract
The recent outbreaks of West Nile virus (WNV) in the northeastern United States and other regions of the world have made it essential to develop an efficient protocol for surveillance of WNV. In the present report, we describe a high-throughput procedure that combines automated RNA extraction, amplification, and detection of WNV RNA. The procedure analyzed 96 samples in approximately 4.5 h. A robotic system, the ABI Prism 6700 Automated Nucleic Acid workstation, extracted RNA and set up reactions for real-time reverse transcription (RT)-PCR in a 96-well format. The robot extracted RNA with a recovery as efficient as that of a commercial RNA extraction kit. A real-time RT-PCR assay was used to detect and quantitate WNV RNA. Using in vitro transcribed RNA, we estimated the detection limit of the real-time RT-PCR to be approximately 40 copies of RNA. A standard RT-PCR assay was optimized to a sensitivity similar to that of the real-time RT-PCR. The standard assay can be reliably used to test a small number of samples or to confirm previous test results. Using internal primers in a nested RT-PCR, we increased the sensitivity by approximately 10-fold compared to that of the standard RT-PCR. The results of the study demonstrated for the first time that the use of an automated system for the purpose of large-scale viral RNA surveillance dramatically increased the speed and efficiency of sample throughput for diagnosis.
West Nile virus (WNV) belongs to the family Flaviviridae, which comprises over 70 viruses. Most flaviviruses are arthropod borne and are transmitted to vertebrates by infected mosquito or tick vectors. Many flaviviruses are significant human pathogens, including WNV, yellow fever virus, four serotypes of dengue virus, Japanese encephalitis virus, Murray Valley encephalitis virus, and tick-borne encephalitis virus (16). Like all flaviviruses, WNV is a single-stranded RNA virus with a positive-polarity RNA genome of approximately 11 kb. Both termini of the genomic RNA contain sequences that do not encode viral proteins, known as the 5′ untranslated region (UTR) and the 3′ UTR. The genomic RNA of flaviviruses contains a single, long open reading frame. The encoded polyprotein is translated and co- and posttranslationally processed by viral and cellular proteases into three structural (capsid [C], premembrane [prM] or membrane [M], and envelope [E]) proteins and seven nonstructural (NS1, NS2a, NS2b, NS3, NS4a, NS4b, and NS5) proteins (17). Flavivirus virions are spherical in shape with a diameter of 40 to 60 nm. The nucleocapsid of about 30 nm in diameter consists of capsid and genomic RNA and is surrounded by a lipid bilayer in which the viral envelope and membrane proteins are embedded (4).
WNV was originally isolated in 1937 from the blood of a febrile woman from the West Nile Province of Uganda (currently Nile Province) and was subsequently isolated from patients, birds, and mosquitoes in Egypt in the early 1950s (15, 19). The virus is widely distributed throughout Africa, the Middle East, part of Europe, Russia, India, and Indonesia. Transmission predominantly involves mosquitoes of the Culex genus and wild birds as the reservoir host. In regions of endemicity, many infections are often asymptomatic or cause mild disease (22). However, human epidemics of severe disease were reported in Israel (Centers for Disease Control and Prevention [CDC] web pages, accessed 29 October 2000), France (10), Romania (23), and Russia (7, 9).
In the summer of 1999, an outbreak of WNV in the northeastern United States sickened more than 60 people and caused 7 deaths (8). Extensive mortality in crows (Corvus species) and the deaths of several exotic birds at a zoologic park in the same geographic area were concurrently observed (20). This was the first evidence of WNV in North America (1, 12). The virus recurred during 2000 and spread to more states in the northeastern United States. Since the outbreak, intensive surveillance has been instituted to monitor the spread of WNV among mosquitoes, birds, and other vertebrates. The surveillance results provided direct evidence of WNV activity and greatly affected intervention decisions such as the spraying of mosquito pesticides to control the spread of virus. Accumulated data on viral infection among various species of mosquitoes and birds as well as other vertebrates (e.g., bats and horses) in an expanding geographic area during the transmission season provided valuable information for prevention and prediction of future outbreaks.
Confronted with increasing numbers of samples for surveillance, it was essential to establish a high-throughput detection procedure to analyze the specimens. Nucleic acid-based techniques, especially reverse transcription (RT)-PCR, have the advantages of speed, specificity, and sensitivity for detection of viral RNA. The procedure involves extraction of RNA from specimens, amplification by RT-PCR, and product detection. The recent development of real-time PCR has combined the steps of amplification and detection and thus has dramatically increased the throughput. In addition, real-time PCR has the ability to quantitate nucleic acid. For nucleic acid extraction, most commercial kits use a silica gel-based membrane to selectively bind RNA or DNA under differently formulated solutions. Although the standardized commercial kits are faster than the traditional phenol-chloroform-based extraction method, this protocol still requires manual operation and thus limits the number of samples that can be tested during the extensive surveillance season. A newly developed RNA extraction robot, the ABI Prism 6700 Automated Nucleic Acid workstation (Applied Biosystems, Foster City, Calif.), contains a novel chemistry for extraction (unpublished data) and dramatically improves the throughput for nucleic acid extraction, allowing the laboratory to increase the number of specimens for surveillance. In this report we describe a high-throughput detection procedure for WNV surveillance which can test 96 samples in approximately 4.5 h. The procedure combines automated RNA extraction, amplification, and detection of WNV RNA in a 96-well format. The results of the present study demonstrate for the first time that an automated system can be used for large-scale viral surveillance. In addition, we also report on protocols for reliable standard RT-PCR and sensitive nested RT-PCR for WNV RNA detection. The methodology described here should be applicable to the detection of viral or cellular RNA or DNA in other systems.
MATERIALS AND METHODS
Cells and viruses.
African green monkey kidney (Vero) cells were grown in minimal essential medium supplemented with 10% fetal calf serum, 2 mM l-glutamine, 0.3% sodium bicarbonate, 10 U of penicillin per ml, and 10 μg of streptomycin per ml. WNV 99-35262 flamingo was kindly provided by the National Veterinary Science Laboratory, Ames, Iowa. The virus was propagated and quantitated by plaque assay on Vero cells (17).
Mosquito and bird specimens.
Mosquitoes were collected with CDC light traps or gravid traps, identified and pooled by species, and transported to the Arbovirus Laboratory of the Wadsworth Center by county health departments. Dead birds were submitted to New York State county health departments and analyzed for pathologic signs of viral encephalitis at the Wildlife Pathology Unit of the New York State Department of Environmental Conservation. Tissues from birds with evidence suggestive of WNV infection (kidney, brain, liver, heart, and spleen) were delivered to the Arbovirus Laboratory for WNV detection. All specimens were kept on dry ice during transportation and stored at −70°C before being processed.
WNV RNA extraction.
Sample RNA was extracted with RNeasy (Qiagen, Valencia, Calif.) or an ABI Prism 6700 workstation (Applied Biosystems). For RNeasy extraction, pools of 10 to 50 individual mosquitoes were homogenized in diluent containing 20% fetal bovine serum, 50 μg of streptomycin per ml, 50 U of penicillin, and 2.5 μg of amphotericin B per ml in phosphate-buffered saline in a Spex CertiPrep (Metuchen, N.J.) 8000-D mixer mill for 3 min. RNA was extracted from 350 μl of homogenized samples. Approximately 50 mg (3 by 3 by 6 mm) of vertebrate tissues was homogenized in 700 μl of RNeasy lysis buffer in a Spex CertiPrep 8000-D mixer mill for 3 min. The homogenized tissue samples (350 μl) were subjected to RNeasy extraction. For testing of extraction with the ABI Prism 6700 workstation, uninfected tissues were spiked with a known amount of virus (in PFU) and homogenized as described above in 1 ml of 1× lysis buffer (Applied Biosystems) diluted with either phosphate-buffered saline (calcium- and magnesium-free) or minimal essential medium. A total of 250 μl of homogenized sample RNA was extracted by the robotic workstation according to the manufacturer's instructions. The extracted RNA was eluted in a total volume of 50 μl of RNase-free water for RNeasy extraction or 150 μl of elution buffer for extraction with the ABI Prism 6700 workstation.
To prepare the viral RNA standards, a large stock of WNV with a known amount of PFU was extracted by the RNeasy method. The extracted RNA was aliquoted in small amounts to avoid multiple rounds of freezing and thawing. This procedure ensured consistent standards throughout the surveillance season.
Plasmid construction.
WNV 99-35262 flamingo genomic RNA extracted with RNeasy was used as the template to synthesize cDNA with the One-Step RT-PCR kit (Qiagen, Valencia, Calif.). The RT-PCR product containing the genomic sequence from nucleotides (nt) 466 to 2666 included the coding regions of the complete prM or M and E proteins and the N-terminal region of the NS1 protein. The first-strand cDNA was generated by RT at 45°C for 30 min with reverse primer 1 (5′-GTATGGATCCTGATGCTCCAGTCTGGAAACTGATCGTA-3′; nt 2666 to 2639) containing a BamHI restriction site (underlined). Following RT, the reaction mixture was incubated at 95°C for 15 min to inactivate the reverse transcriptase and to activate the DNA Taq polymerase. The cDNA was then PCR amplified with reverse primer 1 and forward primer 2 (5′-CATCGAATTCGTTACCCTCTCTAACTTCCAAGGGAAGGTG-3′; nt 466 to 495) containing an EcoRI site (underlined). The thermal cycling consisted of 35 cycles of 94°C for 10 s, 64°C for 45 s, and 68°C for 3 min, with a final elongation at 72°C for 10 min. The cDNA was purified with a PCRpure kit (Qiagen), digested with BamHI and EcoRI, gel purified with a QIAquick gel extraction kit (Qiagen), and ligated to pGEM-3Zf(+) digested with the same restriction enzymes. The positive clone, pGEM/M/E, was verified by restriction enzyme digestion and DNA sequencing.
Preparation of RNA transcripts.
The pGEM/M/E plasmid was amplified in Escherichia coli strain DH5α, purified with a plasmid Maxi kit (Qiagen), linearized by BamHI digestion, and recovered by phenol-chloroform extraction followed by ethanol precipitation. The linearized plasmid was used as the template for in vitro transcription with T7 MEGAScript (Ambion, Austin, Tex.). Briefly, 1 μg of plasmid was used in a 20-μl reaction mixture. The transcription reaction was incubated at 37°C for 2 h, followed by DNase I digestion at 37°C for 30 min to remove the template DNA. The RNA was recovered by phenol-chloroform extraction and ethanol precipitation and quantitated with a spectrophotometer.
5′ Nuclease real-time RT-PCR.
Three sets of primers-probes (Table 1) targeting different regions of WNV RNA were tested in the 5′ nuclease real-time RT-PCR. The sequences of the primer-probe sets were generously provided by Robert Lanciotti (CDC, Fort Collins, Colo.). All probes contained a 5′ reporter, 6-carboxyfluorescein (FAM), and a 3′ quencher, 6-carboxy-N,N,N′,N′-tetramethylrhodamine (TAMRA). The assay was performed on an ABI Prism 7700 Sequence Detector with TaqMan One-Step RT-PCR master mixture (Applied Biosystems). The reaction mixture contained a total volume of 50 μl including each primer pair at a concentration of 1 μM and probe at a concentration of 0.2 μM. Because of the difference in elution volumes following extraction with RNeasy and the ABI 6700 workstation, as described above, a total of 5 and 20 μl of RNA eluate was used for each sample, respectively. The RNA samples were heated at 60°C for 3 min before being added to the reaction mixtures. The thermal cycling consisted of 48°C for 30 min for RT, 95°C for 10 min, and 40 cycles of 95°C for 15 s and 60°C for 1 min.
TABLE 1.
Primer-probe sets for 5′ nuclease real-time RT-PCR
| Set no. | Targetb | Sequencea
|
||
|---|---|---|---|---|
| Forward primer | Probe | Reverse primer | ||
| 1 | E | TCAGCGATCTCTCCACCAAAG (1160V) | TGCCCGACCATGGGAGAAGCTC (1186V) | GGGTCAGCACGTTTGTCATTG (1229C) |
| 2 | NS1 | GGCAGTTCTGGGTGAAGTCAA (3111V) | TGTACGTGGCCTGAGACGCATACCTTGT (3136V) | CTCCGATTGTGATTGCTTCGT (3239C) |
| 3 | 3′ UTR | CAGACCACGCTACGGCG (10668V) | TCTGCGGAGAGTGCAGTCTGCGAT (10691V) | CTAGGGCCGCGTGGG (10770C) |
Designations in parentheses are nucleotide number and polarity of the primer-probe. V, viral genomic sense; C, complementary sense. Nucleotide numbering is based on the sequence from Lanciotti et al. (14; GeneBank accession number AF196835).
The sequences for the primer-probe sets were designed by R. S. Lanciotti, CDC (14; Lanciotti, personal communication).
Standard RT-PCR assay.
A 408-bp fragment (nt 233 to 640) containing the C-terminal portion of the C gene and the N-terminal part of the prM gene was amplified to detect WNV RNA with the One-Step RT-PCR kit (Qiagen). The oligonucleotides for the standard RT-PCR were provided by Robert Lanciotti, CDC (14), and included forward primer 3 (5′-TTGTGTTGGCTCTCTTGGCGTTCTT-3′; nt 233 to 257) and reverse primer 4 (5′-CAGCCGACAGCACTGGACATTCATA-3′; nt 616 to 640). The amplification reaction was performed according to the manufacturer's instructions, with minor modifications. A 50-μl standard RT-PCR mixture contained 1× reaction buffer, 0.4 mM deoxynucleotide triphosphates, 0.6 μM primers 3 and 4, 1× Q solution, and 1 μl of reverse transcriptase-DNA Taq polymerase enzyme mix. A total of 5 and 20 μl of RNA eluate extracted with RNeasy and the ABI Prism 6700 workstation, respectively, was tested for each sample. The thermal cycling consisted of 50°C for 30 min to synthesize the first-strand cDNA; 95°C for 15 min to inactivate the reverse transcriptase and to activate DNA Taq polymerase; 35 cycles of 94°C for 45 s, 56°C for 45 s, and 72°C for 1 min for PCR amplification; and a final elongation at 72°C for 10 min. A total of 20 μl of RT-PCR products was analyzed on a 1.5% agarose gel with TAE (Tris-acetate-EDTA) running buffer and stained with 0.5 μg of ethidium bromide per ml.
Nested PCR assay.
The sample from the standard RT-PCR described above was diluted 10,000-fold for the nested PCR with the Taq PCR Core kit (Qiagen). The internal primer set included forward primer 5 (5′-CAGTGCTGGATCGATGGAGAGG-3′; nt 287 to 308) and reverse primer 6 (5′-CCGCCGATTGATAGCACTGGT-3′; nt 370 to 390). A 25-μl reaction mixture contained 1× reaction buffer, 0.2 mM deoxynucleoside triphosphates, 0.3 μM primers 5 and 6, 1 μl of diluted RT-PCR template, and 0.625 U of Taq DNA polymerase. The thermal cycling consisted of 94°C for 3 min; 22 cycles of 94°C for 45 s, 58°C for 45 s, and 72°C for 1 min; and a final elongation at 72°C for 10 min. A total of 15 μl of nested PCR products was analyzed on a 1.5% agarose gel, as described above.
DNA sequencing of RT-PCR products.
Products from selected assays performed by the standard RT-PCR protocol described above were sequenced by the Molecular Genetics Core at the Wadsworth Center. The RT-PCR mixtures were separated on a 1.5% agarose gel, and the 408-bp DNA product was gel purified with a QIAquick gel extraction kit (Qiagen) and sequenced with primers 3 and 4.
RESULTS
5′ Nuclease real-time RT-PCR is a high-throughput assay for detection of WNV RNA with high sensitivity.
Three primer-probe sets (Table 1) were provided by Robert Lanciotti, CDC (14; R. Lanciotti, personal communication). After optimization of the reaction conditions, we found that primer-probe set 1, which targeted the E gene, was the most sensitive. Therefore, it was used for the primary screening of all specimens. Primer-probe sets 2 and/or 3, targeting NS1 and 3′ UTR, respectively, were chosen for use in the confirmatory tests. The specificities of these primer-probe sets were tested against eastern equine encephalitis virus and St. Louis encephalitis virus, two common arboviruses in North America, and no signals were obtained (data not shown).
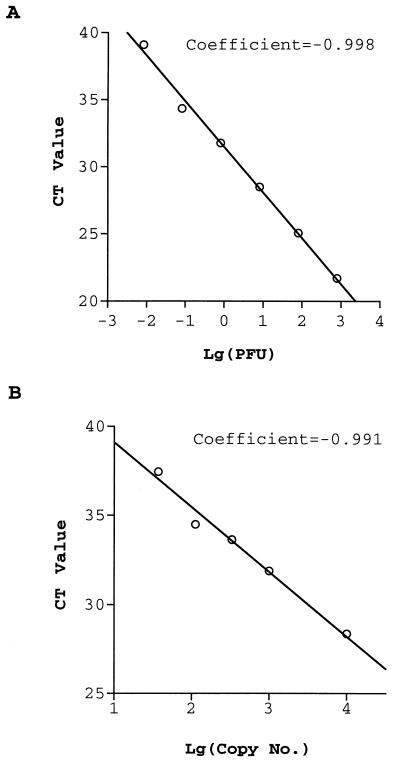
Two methods were used to quantitate the samples. One used RNA extracted from a virus stock with a known titer (in PFU). We routinely used 800, 80, 8, 0.8, 0.08, and 0.008 PFU of virus as our standards. During an authentic amplification, the multicomponent fluorescence of the reporter (FAM) and the quencher (TAMRA) increased and decreased, respectively. Therefore, for data analysis, the threshold line was set at the maximum fluorescence reached by a standard for which the fluorescences of both FAM and TAMRA minimally changed, usually with the 0.008 PFU standard. Figure 1A shows a plot of the threshold cycles (CT; the point at which the fluorescence rises appreciably above the background level) versus the log of the titrated amount of virus (in PFU) with primer-probe set 1. The CT values were 21.7, 25.04, 28.48, 31.77, 34.35, 39.09, and >40 for 800, 80, 8, 0.8, 0.08, 0.008, and 0.0008 PFU, respectively. These CT values were typically observed for the standard viral input. The linear dynamic range of the assay is more than 4 orders of magnitude from 800 to 0.08 PFU. We found that although robust and consistent curves were observed with input virus levels of 0.08 PFU or greater, the reproducibility decreased as the viral input was reduced to less than 0.08 PFU. This was expected since the low level of RNA input (less than 0.08 PFU of virus) was almost out of the linear range of the RT-PCR amplification. The unknown samples were analyzed and interpreted by comparing their CT values to those of the standards. These results indicate that real-time RT-PCR is a reliable method for detection of WNV RNA, with a detection limit of approximately 0.08 PFU.
FIG. 1.
Quantitation of WNV by real-time RT-PCR assay. CT values are plotted versus the log of a known amount of WNV (in PFU) (A) or in vitro transcribed RNA (B).
The method described above quantitated samples in terms of the number of infectious viral particles. It gave no direct quantitation of the numbers of copies of viral RNA because not all viral particles are infectious in a virus stock. In order to determine the detection limit in RNA copy number, the envelope gene of WNV was cloned, in vitro transcribed with T7 polymerase, and quantitated as described in Materials and Methods. Figure 1B is a plot of the CT values versus the log of the titrated copy numbers of the in vitro transcribed RNA obtained with primer-probe set 1. Inputs of 104, 103, 333, 111, 37, 12, and 4 RNA molecules yielded CT values of 28.35, 31.88, 33.64, 34.49, 37.45, >40, and >40, respectively. Similar results were obtained in repeat experiments, and for each experiment, tests with every input concentration were performed in duplicate. These results indicated that the detection limit of the real-time RT-PCR is approximately 40 copies of RNA.
Standard RT-PCR has similar sensitivity to 5′ nuclease real-time RT-PCR.
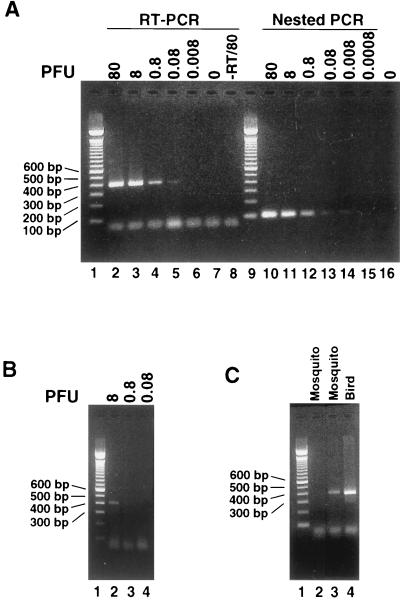
It was recently reported that the standard RT-PCR is less sensitive than the real-time RT-PCR (14). In order to improve the sensitivity, we used the same set of primers used in the previous study (14) to optimize the standard RT-PCR assay. The products of the expected 408-bp cDNA, which contained the 3′ region of the C gene and the 5′ region of the prM gene, were observed (Fig. 2A). The intensities of the bands decreased with decreasing amounts of input WNV RNA (Fig. 2A, lanes 2 to 6). A faint band was visible with input virus of 0.08 PFU, and no product was detected with 0.008 PFU (Fig. 2A, lanes 5 and 6). During the development of the protocol, we found that addition of Q solution, an additive reagent from the One-Step RT-PCR kit, improved the sensitivity by more than 10-fold since input virus at 0.8 PFU could not be detected in the absence of Q solution (Fig. 2B, lanes 2 to 4). Therefore, Q solution was added in all subsequent experiments. No DNA product was amplified from the reaction mixture without WNV RNA (Fig. 2A, lane 7), indicating that the observed products resulted from the amplification of the input viral RNA. The specificity of the RT-PCR was further tested with eastern equine encephalitis virus and St. Louis encephalitis virus, and no DNA products were observed, confirming the specificity of the assay (data not shown). These results suggested that the RT-PCR specifically amplified WNV RNA.
FIG. 2.
Standard RT-PCR and nested PCR for WNV RNA detection. (A) Comparison of sensitivities of RT-PCR and the nested PCR assay. A titrated amount of WNV (in PFU) was tested by standard RT-PCR (lanes 2 to 8) and nested RT-PCR (lanes 10 to 16). The amounts of viral RNA (in PFU) are indicated above each lane. Lane 8, 80 PFU of viral RNA was subjected to PCR without RT. All RT-PCR mixtures contained Q solution. Lanes 1 and 9, 100-bp marker. (B) Addition of Q solution to the RT-PCR mixture increases the sensitivity by more than 10-fold. In the absence of Q solution, a standard RT-PCR was performed with a titrated amount of virus, as indicated above each lane (lanes 2 to 4). Without Q solution, no product was detected with 0.8 PFU of virus (Lane 3). Lane 1, 100-bp marker. (C) Standard RT-PCR analysis of specimens. Lanes 2 and 3, mosquito specimens; lane 4, a bird specimen; lane 1, 100-bp marker.
It was possible that the observed product was from the amplification of contaminating DNA containing the WNV sequence rather than viral RNA. To eliminate this possibility, we performed a standard RT-PCR with 80 PFU of input viral RNA, except that the RT step was omitted. No product was observed from this reaction (Fig. 2A, lane 8), indicating that the 408-bp fragment was the result of amplification of viral RNA rather than contaminating DNA. To further demonstrate that the product originated from WNV RNA, the amplified DNA products were subjected to DNA sequencing. The sequencing results confirmed the identity of the amplicons as WNV. On the basis of the results described above, we chose 8, 0.8, and 0.08 PFU of WNV as positive controls and water as the negative control throughout our surveillance RT-PCR assays. Figure 2C presents the results of a representative assay and shows the results for one positive (lane 3) and one negative (lane 2) mosquito pool and one strongly positive bird sample (lane 4). Overall, the results demonstrated that the standard RT-PCR assay is specific for WNV RNA and has a sensitivity of approximately 0.08 PFU, similar to that of the 5′ nuclease real-time RT-PCR.
Nested PCR increases the sensitivity.
Although standard RT-PCR was reliable and consistent and was routinely used as a confirmatory assay, some samples yielded equivocal results by both the standard RT-PCR and the 5′ nuclease real-time RT-PCR. This was expected because the two assays had similar sensitivities (see above). A more sensitive assay was required to verify the results for samples with discrepancies. Therefore, a nested RT-PCR assay was developed. A set of internal primers was designed to perform nested PCR with the products from the standard RT-PCR as templates. As shown in Fig. 2A, a 104-bp DNA product from the nested RT-PCR was observed, and the intensities of the product decreased with decreasing amounts of input viral RNA for the standard RT-PCR (lanes 10 to 15). No DNA product was detected without the addition of viral RNA (lane 16). The results indicated that the nested RT-PCR could detect WNV at levels as as low as 0.008 PFU in the sample (lane 14). This was in contrast to the results obtained by the standard RT-PCR, with a sensitivity of about 0.08 PFU (see above and Fig. 2A, lane 5). These results demonstrate that nested RT-PCR increases the sensitivity by approximately 10-fold over those of both the standard RT-PCR and the 5′ nuclease real-time RT-PCR.
ABI Prism 6700 workstation dramatically increases the throughout of RNA extraction, with recovery as efficient as that by the RNeasy method.
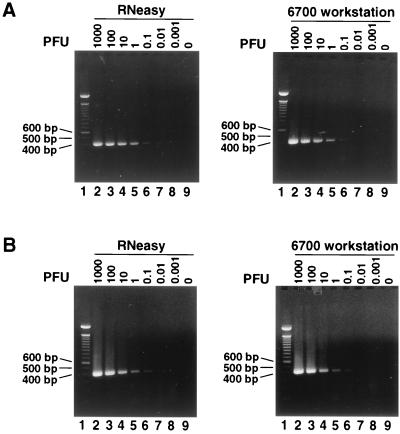
Although real-time RT-PCR significantly increased the throughout of the surveillance assays, the number of specimens that could be tested was limited by the RNA extraction step prior to RT-PCR. Commercial RNA extraction kits are efficient compared to the traditional phenol-chloroform extraction method, but they still require manual operation and thus remain a bottleneck for surveillance. Therefore, the use of an automated RNA extraction system was essential to increase sample throughput. We tested the ABI Prism 6700 workstation, a fully automated platform for purification of RNA or DNA as well as reaction setup for real-time RT-PCR assays. We spiked a known amount of virus into uninfected bird tissues, homogenized the specimens, and extracted the RNA using either RNeasy (Qiagen) or the ABI Prism 6700 workstation. The extracted RNA was then quantitated by the real-time RT-PCR assay. Table 2 compares the CT values for RNA from four bird organs (kidney, heart, brain, and spleen) extracted by use of either RNeasy or the ABI Prism 6700 workstation. For each tissue, we spiked various amounts of viruses ranging from 103 to 10−3 PFU. At each level of spiked virus, similar CT values were obtained for samples from which RNA was extracted by use of either the ABI Prism 6700 workstation or RNeasy, indicating similar recovery rates by the two extraction methods. No difference in viral RNA recovery was observed for the kidney, heart, brain, and spleen tissues. These results indicated that no tissue-specific factors affected the RNA extraction by either method. Similar results were obtained by two independent experiments. To confirm the results of the real-time RT-PCR, we performed the standard RT-PCR with the extracted RNA samples. Figure 3 shows the autographs of the amplified products analyzed on agarose gels stained with ethidium bromide. Similar intensities of the amplified products were observed by either extraction method at each level of spiked virus. Figures 3A and B show the results for samples extracted from kidney and heart tissues, respectively. Similar results were obtained with brain and spleen tissues (data not shown). These results demonstrated that the ABI Prism 6700 workstation extracted RNA as efficiently as RNeasy. Furthermore, the ABI Prism 6700 workstation dramatically increased the throughput of the procedure. Up to 96 samples could be processed in approximately 1 h by the robotic process, whereas it takes a trained technician approximately 4 h to process the same number of samples by the RNeasy method. When the ABI Prism 6700 workstation is programmed to proceed with plate setup for real-time RT-PCR, an additional hour of trained technician time can be saved.
TABLE 2.
Comparison of RNA extraction by ABI Prism 6700 workstation and with RNeasya
| No. of PFU spiked |
CT value
|
|||||||
|---|---|---|---|---|---|---|---|---|
| Kidney
|
Heart
|
Brain
|
Spleen
|
|||||
| 6700 | RNeasy | 6700 | RNeasy | 6700 | RNeasy | 6700 | RNeasy | |
| 103 | 21.4 | 21.44 | 21.36 | 21.4 | 20.74 | 20.75 | 21.39 | 21.29 |
| 102 | 24.7 | 24.89 | 24.89 | 25 | 24.85 | 24.88 | 24.77 | 24.8 |
| 10 | 28.22 | 28.13 | 28.22 | 28.15 | 28.14 | 28.19 | 28.18 | 28.18 |
| 1 | 31.14 | 31.48 | 31.4 | 31.52 | 30.59 | 31.37 | 31.38 | 31.34 |
| 10−1 | 34.86 | 35.23 | 34.89 | 35.18 | 35.73 | 34.32 | 34.84 | 34.66 |
| 10−2 | 36.93 | 37.08 | 37.46 | 36.28 | 36.96 | 38.81 | 37.98 | 37.67 |
| 10−3 | 40 | 40 | 40 | 40 | 40 | 40 | 40 | 40 |
| 0 | 40 | 40 | 40 | 40 | 40 | 40 | 40 | 40 |
Kidney, heart, brain, and spleen from an uninfected bird were spiked with a known amount of WNV, and the homogenized samples were subjected to RNA extraction by the RNeasy method and with the ABI Prism 6700 workstation (6700). The recovered RNA samples were analyzed by real-time RT-PCR. See details in Materials and Methods.
FIG. 3.
ABI Prism 6700 workstation extracts RNA as efficiently as the RNeasy method. Uninfected bird tissues spiked with titrated amounts of WNV were extracted by RNeasy methods (left panels) or with the ABI Prism 6700 workstation (right panels). The recovered RNA was subjected to standard RT-PCR amplification and analyzed on agarose gels stained with ethidium bromide. The amounts of WNV spiked into the sample (in PFU) are indicated above each lane. (A and B) Samples extracted from kidney and heart tissues, respectively. Lanes 1, 100-bp markers.
Next, we tested the potential cross-contamination among the 96 wells during the RNA extraction with the ABI Prism 6700 workstation. Samples with or without spiked viruses (1,000 PFU per sample) were processed in neighboring wells. The extracted RNA was analyzed for viral RNA by real-time RT-PCR. The results showed no cross-well contamination on the 96-well plate during the robotic extraction (data not shown). In addition, we tested the reproducibilities of tests with the robotic apparatus by extracting RNA from similar samples several times and quantitating the RNA recovery. The data showed consistent RNA recovery, with little variation (data not shown). In summary, the results presented above demonstrated that the ABI Prism 6700 workstation is a reliable and efficient RNA extraction system for high-throughput WNV surveillance.
Streamlined viral RNA detection procedure.
The earlier sections described two systems for RNA extraction (RNeasy and the ABI Prism 6700 workstation) and three methods of WNV RNA detection (real-time RT-PCR, standard RT-PCR, and nested PCR). The following is a streamlined protocol for our routine WNV RNA detection procedure. Once the RNA was extracted from the specimen, real-time RT-PCR was used as a primary screen to test for viral RNA. Because of its highest sensitivity, primer-probe set 1 (Table 1) was used in the initial assay to screen all specimens. For confirmatory purposes, the positive samples were then subjected to a secondary assay with primer-probe set 2 or 3 (Table 1). Samples that tested positive by both assays were considered confirmed positives. Since primer-probe sets 2 and 3 are less sensitive than set 1, it was expected that some samples with low levels of WNV-specific RNA from the initial screen would be negative by the confirmatory assay. To confirm the results for these samples, the RNA was tested by the standard RT-PCR. If the results of the standard RT-PCR were positive, the samples were considered positive. A portion of the specimens that were not confirmed were analyzed further by the nested PCR assay. During surveillance in the year 2000, all the samples were processed by the RNeasy procedure. However, with the introduction of the high-throughput ABI Prism 6700 workstation, laboratory capacity for future surveillance can be significantly increased.
DISCUSSION
The recent outbreaks of WNV in the northeastern United States and certain regions of Europe and the Middle East have made it essential to develop an efficient procedure for surveillance. High-throughput detection of WNV in specimens submitted to the laboratory is essential for surveillance because (i) the number of samples is very large and (ii) the diagnosis usually has a demanding timeline since the results will have an immediate effect on intervention decisions such as mosquito pesticide spraying. Nucleic acid-based techniques, especially RT-PCR, have the advantages of speed, specificity, and sensitivity. The combination of the ABI Prism 6700 workstation and real-time RT-PCR automated the complete procedure for nucleic acid extraction, amplification, and product detection and, therefore, dramatically increased the throughput and the capacity of diagnosis. This procedure allows testing of 96 samples in approximately 4.5 h, with a detection limit of about 40 copies of viral RNA. A standard RT-PCR was optimized to yield a sensitivity equal to that of real-time RT-PCR, which will be useful for the testing of small numbers of specimens or as a confirmatory assay. A nested PCR assay was developed to further improve the detection limit by approximately 10-fold. The combination of these protocols represents a high-throughput and sensitive procedure for WNV RNA detection.
A key component of high-throughput diagnostics is RNA extraction. The ABI Prism 6700 workstation alleviated the RNA extraction bottleneck and safely handled infectious virus. The robot extracts and sets up the real-time RT-PCR assays for 96 samples within 2 h. Two alternative systems are the Nuclisens RNA extractor from Organon Teknika (Durham, N.C.), which extracts RNA from 10 samples within approximately 1 h, and the BioRobot 9604 (Qiagen), which extracts RNA and sets up real-time RT-PCRs. These two systems are based on the silica membrane purification method; the ABI Prism 6700 workstation uses a novel extraction mechanism (unpublished data). The ABI Prism 6700 workstation preferentially captures RNA on a glass-fiber membrane with an electronically controlled vacuum filtration mechanism. Genomic DNA is retained in solution and passes through the glass-fiber membrane with virtually no retention. Although the throughput capacity of the BioRobot 9604 system is similar to that of the ABI Prism 6700 workstation, the BioRobot 9604 system is exposed to open air and thus needs to be installed in a safety cabinet to process infectious materials. In contrast, the ABI Prism 6700 workstation is enclosed in its own safety cabinet, thus preventing potential aerosol exposure and virus contamination.
A second key component of high-throughput nucleic acid detection is real-time RT-PCR, which incorporates amplification and detection into one step (11). We routinely tested positive specimens using two primer-probe sets in two separate reactions. To further improve the throughput and reduce the cost of the assay, multiplex real-time RT-PCR assays that combine the two primer-probe sets in one reaction tube are under development. Another advantage of real-time RT-PCR is the decreased risk of contamination of the laboratory with PCR product because the reaction mixture is discarded without opening the tube at the end of the amplification.
The sensitivity of our real-time RT-PCR assay was similar to those reported previously. Briese et al. (3) reported results for two primer-probe sets that targeted the NS3 and NS5 protein sequences and that was used to detect WNV in cerebrospinal fluid by real-time RT-PCR. The sensitivity of the primer-probe sets was 50 to 100 molecules. We estimated that the detection limit of the real-time RT-PCR assay was approximately 40 molecules. Lanciotti et al. (14) reported a real-time RT-PCR assay with primer-probe sets 1 and 3 and showed that it had a sensitivity of 0.1 PFU of viral RNA. We found a similar level of sensitivity (0.08 PFU of virus) using the same primer-probe set, which targeted the E gene. These assays, including the assay developed in the study described in this report, use the TaqMan technology in the real-time RT-PCR. An alternative technology, Molecular Beacon (24), exists, but there have been no reports on its use for WNV detection. A recent comparison of the TaqMan and Molecular Beacon technologies showed that they have similar sensitivities for the detection of DNA in clinical samples (21).
Lanciotti et al. (14) recently reported that a traditional RT-PCR assay was less sensitive than the 5′ real-time RT-PCR. However, using the same primer set, we optimized the standard RT-PCR so that it had a sensitivity similar to that of the real-time RT-PCR. Several factors could explain the improvement in sensitivity: (i) the use of different tissue processing protocols, (ii) the use of different RNA extraction kits, (iii) the use of different RT-PCR kits, or most likely (iv) optimization of assay conditions. We found that the addition of Q solution, an additive reagent from the One-Step RT-PCR kit, to the reaction mixture improved the sensitivity of the assay by more than 10-fold. This may be because Q solution changes the melting behavior of nucleic acids and thus increased the amplification efficiency. However, we found that the addition of Q solution did not increase the amplification efficiency in assays targeting other regions of the WNV genome (unpublished data). We also found that a high primer concentration (up to 0.6 μM) is important to maintain the high sensitivity. In order to maintain the consistency of the standard RT-PCR, we strictly controlled the conditions of the assay to minimize experimental variation. These included the use of freshly made gels, standard ethidium bromide concentrations, and standard electrophoresis times. Freshly thawed aliquots of control standards were used throughout the surveillance season. Caution should be taken when performing nested RT-PCR. Due to its high sensitivity, contamination can easily lead to false-positive results. In order to control the quality, reactions containing no WNV RNA should always be performed as negative controls. Both RT-PCR and nested PCR were performed in individual tubes rather than strip tubes because contamination can easily occur between the strip tubes during assembly of the reaction.
In addition to the methods described above, other sensitive nucleic acid-based techniques, including nucleic acid sequence-based amplification (NASBA) (6) and branched DNA (bDNA) methods, are available for development for WNV RNA detection. On the basis of the principles of NASBA and bDNA, the NucliSens (Organon Teknica) and Quantiplex (Bayer, Emeryville, Calif.), tests have been developed for quantitation of human immunodeficiency virus RNA copy numbers and are capable of detecting 80 and 50 copies of viral RNA per ml, respectively. NucliSens uses the electrochemiluminescence method for sensitive detection and yields clear-cut positive and negative signals. This is in contrast to the electrophoresis detection method, which frequently provides equivocal results when small amounts of amplified products are analyzed. Lanciotti et al. (13) recently reported a sensitive NASBA assay for WNV detection. The signal enhancement of bDNA is achieved by hybridization of viral RNA with bDNA probes (5, 25). A recent comparison of the NASBA and bDNA techniques for analysis of human immunodeficiency virus RNA showed that although the bDNA test exhibited a higher dynamic range and was more robust with low copy numbers, the interassay precision of the NASBA method is better (2). As we observed with the real-time RT-PCR assay, both the NASBA and the bDNA methods showed greater variations near the detection limit.
ACKNOWLEDGMENTS
We thank Robert Lanciotti, CDC, for providing sequence information on three primer-probe sets for real-time RT-PCR and primers for standard RT-PCR. We are very grateful to Paul Masters for help during the development of the assays. We thank the Molecular Genetics Core at the Wadsworth Center for sequencing.
This work was supported by the New York State Department of Health. J. H. Tai was supported by the Emerging Infectious Diseases Fellowship Program administered by the Association of Public Health Laboratories and funded by CDC.
REFERENCES
- 1.Anderson J F, Andreadis T G, Vossbrinck C R, Tirrell S, Wakem E M, French R A, Garmendia A E, Van Kruiningen H J. Isolation of West Nile virus from mosquitoes, crows, and a Cooper's hawk in Connecticut. Science. 1999;286:2331–2333. doi: 10.1126/science.286.5448.2331. [DOI] [PubMed] [Google Scholar]
- 2.Berndt C, Müllera U, Bergmannb F, Schmittc U, Kaiserd R, Müllera C. Comparison between a nucleic acid sequence-based amplification and branched DNA test for quantifying HIV RNA load in blood plasma J. Virol Methods. 2000;89:177–181. doi: 10.1016/s0166-0934(00)00225-1. [DOI] [PubMed] [Google Scholar]
- 3.Briese T, Glass W G, Lipkin W I. Detection of West Nile virus sequences in cerebrospinal fluid. Lancet. 2000;355:1614–1615. doi: 10.1016/s0140-6736(00)02220-0. [DOI] [PubMed] [Google Scholar]
- 4.Chambers T J, Hahn C S, Galler R, Rice C M. Flavivirus genome organization, expression, and replication. Annu Rev Microbiol. 1990;44:649–688. doi: 10.1146/annurev.mi.44.100190.003245. [DOI] [PubMed] [Google Scholar]
- 5.Collins M L, Irvine B, Tyner D, Fine E, Zayati C, Chang C, Horn T, Ahle D, Detmer J, Shen L, Kolberg J, Bushnell S, Urdea M, Ho D D. A branched DNA signal amplification assay for quantification of nucleic acid targets below 100 molecules/ml. Nucleic Acids Res. 1997;25:2979–2984. doi: 10.1093/nar/25.15.2979. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Compton J. Nucleic acid sequence-based amplification. Nature. 1990;350:91–92. doi: 10.1038/350091a0. [DOI] [PubMed] [Google Scholar]
- 7.Enserink M. New York's lethal virus came from Middle East, DNA suggests. Science. 1999;286:1450–1451. doi: 10.1126/science.286.5444.1450. [DOI] [PubMed] [Google Scholar]
- 8.Enserink M. New York's deadly virus may stage a comback. Science. 1999;287:2129–2130. doi: 10.1126/science.287.5461.2129a. [DOI] [PubMed] [Google Scholar]
- 9.Förster V T. Zwischenmolekulare energy-wanderung und fluoreszenz. Ann Physics. 1948;2:55–75. [Google Scholar]
- 10.Hannoun C, Panthier R, Corniou B. Epidemiology of West Nile infections in the South of France. In: Bardos V, editor. Arboviruses of the California complex and the Bunya-mwera group. Bratislava, Slovakia: Publishing House SAS; 1969. pp. 379–387. [Google Scholar]
- 11.Lakowicz J R. Principles of fluorescent spectroscopy. New York, N.Y: Plenum Press; 1983. Energy transfer; pp. 303–339. [Google Scholar]
- 12.Lanciotti R S, Roehrig J T, Deubel V, Smith J, Parker M, Steele K, Crise B, Volpe K E, Crabtree M B, Scherret J H, Hall R A, MacKenzie J S, Cropp C B, Panigrahy B, Ostlund E, Schmitt B, Malkinson M, Banet C, Weissman J, Komar N, Savage H M, Stone W, McNamara T, Gubler D J. Origin of the West Nile virus responsible for an outbreak of encephalitis in the northeastern United States. Science. 1999;286:2333–2337. doi: 10.1126/science.286.5448.2333. [DOI] [PubMed] [Google Scholar]
- 13.Lanciotti R S, Kerst A J, Allen B C. Development of NASBA based assay for the rapid detection of West Nile virus. Am J Trop Med Hyg. 2000;62(Suppl.):340. [Google Scholar]
- 14.Lanciotti R S, Kerst A J, Nasci R S, Godsey M S, Mitchell C J, Savage H M, Komar N, Panella N A, Allen B C, Volpe K E, Davis B S, Roehrig J T. Rapid detection of West Nile virus from human clinical specimens, field-collected mosquitoes, and avian samples by a TaqMan reverse transcriptase-PCR assay. J Clin Microbiol. 2000;38:4066–4071. doi: 10.1128/jcm.38.11.4066-4071.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Melnick J L, Paul J R, Riordan J T, Barnett V H H, Goldblum N, Zabin E. Isolation from human sera in Egypt of a virus apparently identical to West Nile virus. Proc Soc Exp Biol Med. 1951;77:661–665. doi: 10.3181/00379727-77-18884. [DOI] [PubMed] [Google Scholar]
- 16.Monath T P, Heinz F X. Flaviviruses. In: Fields B N, Knipe D M, Howley P M, et al., editors. Virology. New York, N.Y: Raven Press; 1996. pp. 961–1035. [Google Scholar]
- 17.Reisen, W. K., R. P. Meyer, S. B. Presser, and J. L. Hardy. 1993. Effect of temperature on the transmission of Western equine encephalomyelitis and St. Louis encephalitis viruses by Culex tarsalis (Diptera: Culcadea). J. Med. Entomol. 30:151–160. [DOI] [PubMed]
- 18.Rice C M. Flaviviruses. In: Fields B N, Knipe D M, Howley P M, et al., editors. Virology. New York, N.Y: Raven Press; 1996. pp. 931–959. [Google Scholar]
- 19.Smithburn K C, Hughes T P, Burke A W, Paul J H. A neurotropic virus isolated from the blood of a native of Uganda. Am J Trop Med. 1940;20:471–492. [Google Scholar]
- 20.Steele K E, Linn M J, Schoepp R J, Komar N, Geisbert T W, Manduca R M, Calle P P, Raphael B L, Clippinger T L, Larsen T, Smith J, Lanciotti R S, Panella N A, McNamara T S. Pathology of fatal West Nile Virus infections in native and exotic birds during the 1999 outbreak in New York City, New York. Vet Pathol. 2000;37:208–224. doi: 10.1354/vp.37-3-208. [DOI] [PubMed] [Google Scholar]
- 21.Tapp I, Malmberg L, Rennel E, Wik M, Syvanen A C. Homogeneous scoring of single-nucleotide polymorphisms: comparison of the 5′-nuclease TaqMan assay and Molecular Beacon probes. BioTechniques. 2000;28:732–738. doi: 10.2144/00284rr02. [DOI] [PubMed] [Google Scholar]
- 22.Taylor R M, Work T H, Hurlbut H S, Rizk F. A study of the ecology of West Nile virus in Egypt. Am J Trop Med Hyg. 1956;5:579–620. doi: 10.4269/ajtmh.1956.5.579. [DOI] [PubMed] [Google Scholar]
- 23.Tsai T F, Popovici F, Cernescu C, Campbell G L, Nedelcu N I. West Nile encephalitis epidemic in southeastern Romania. Lancet. 1998;352:767–771. doi: 10.1016/s0140-6736(98)03538-7. [DOI] [PubMed] [Google Scholar]
- 24.Tyagi S, Kramer F R. Molecular beacons: probes that fluoresce upon hybridization. Nat Biotechnol. 1996;14:303–308. doi: 10.1038/nbt0396-303. [DOI] [PubMed] [Google Scholar]
- 25.Urdea M S, Wilber J C, Yeghiazarian T, Todd J A, Kern D G, Fong S J, Besemer D, Hoo B, Sheridan P J, Kokka R, Neuwald P, Pachl C A. Direct and quantitative detection of HIV-1 RNA in human plasma with a branched DNA signal amplification assay. AIDS. 1993;7(Suppl. 2):S11–S14. doi: 10.1097/00002030-199311002-00004. [DOI] [PubMed] [Google Scholar]