Abstract
Clostridioides difficile produces toxins TcdA and TcdB during infection. Since the severity of the illness is directly correlated with the level of toxins produced, researchers have long been interested in the regulation mechanisms of toxin production. The advent of new genetics and mutagenesis technologies in C. difficile has allowed a slew of new investigations in the last decade, which considerably improved our understanding of this crucial regulatory network. The current body of work shows that the toxin regulatory network overlaps with the regulatory networks of sporulation, motility, and key metabolic pathways. This implies that toxin production is a complicated process initiated by bacteria in response to numerous host factors during infection. We summarize the existing knowledge about the toxin gene regulatory network here.
Keywords: C. difficile, Clostridioides difficile, toxin gene regulation, virulence gene regulation
Introduction
Clostridioides difficile has increasingly become one of the most clinically relevant human pathogens in the last decade. The frequency and severity of C. difficile infection (CDI) has increased worldwide to become one of the most common hospital-acquired infections, especially in industrialized countries [1]. CDI symptoms range from mild diarrhea to severe life-threatening pseudomembranous colitis and toxic megacolon [2]. These symptoms are essentially caused by the production of two major virulent factors, Toxin A (TcdA) and Toxin B (TcdB) [3]. Under laboratory conditions, production of TcdA and TcdB occurs in responce to various environmental conditions, such as availability of specific nutrients, temperature, cell density, redox potential changes, phage infection, and presence of antibiotics [4–11]. This review will provide a comprehensive discussion on the regulatory networks that control the production of TcdA and TcdB.
Regulators located in PaLoc (Pathogenicity Locus)
In C. difficile, the genes tcdA and tcdB are located in the pathogenicity locus (Paloc), a 19.5 kb chromosomal region found only in the toxigenic C. difficile strains. Besides the toxin genes, PaLoc consists of tcdR, tcdC and tcdE. TcdR and TcdC are involved in transcriptional regulation of the toxin genes [12,13], and TcdE might be needed for the efficient secretion of toxins [14,15]. While the function of TcdR as a positive regulator is well established, the roles of TcdC and TcdE are still under investigation.
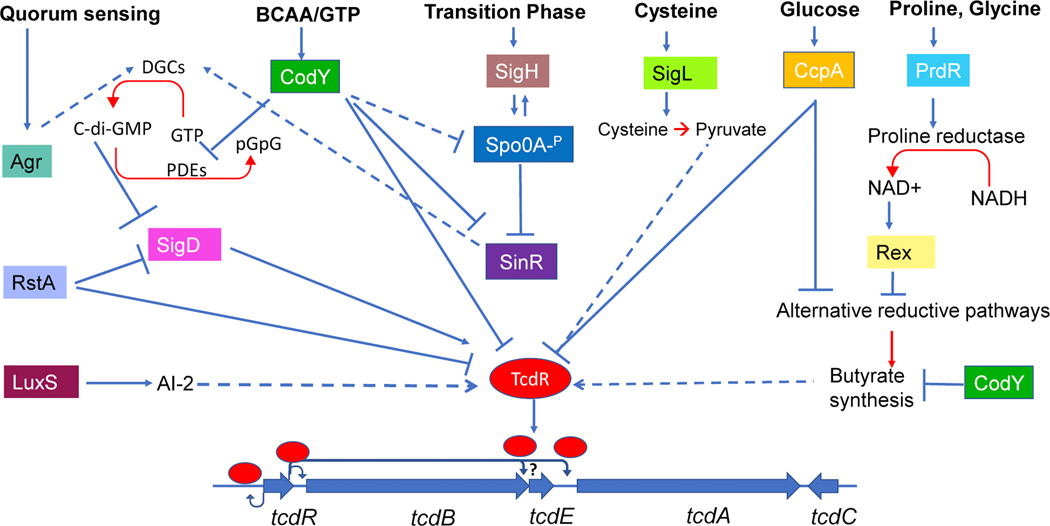
TcdR is a group V sigma factor homologous to toxin gene regulators of other clostridial pathogens, such as TetR of Clostridium tetani and BotR of Clostridium botulinum [reviewed in 16]. TcdR activates transcription by directing the RNAP from its own promoter and the promoters of tcdA and tcdB [12]. Utilizing single-cell analysis with a red fluorescent protein (RFP) as a reporter, Ransom et al. showed that TcdR accumulates to trigger toxin production only in a subset of cells in the population [17]. Thus, TcdR concentration appears to control a bistable switch that turns the toxin genes ON or OFF in a cell.It is important to note that TcdR is the only positive regulator capable of directly controlling the transcription of the toxin genes. Many known positive and negative regulators modulate tcdR expression and indirectly influence toxin gene transcription (Figure 1) [18–21].
Figure 1. Toxin gene regulatory network in response to various stimuli.
TcdR activates the expression of its gene (tcdR) and both toxin genes tcdA and tcdB, while SigD activates the expression of tcdR. The repressors CodY, CcpA, and RstA bind to the promoter-regulatory regions at tcdR. CodY and CcpA also repress tcdA, tcdB directly by binding to their promoter region (not shown). All the other regulatory proteins influence toxin gene transcription indirectly, by regulating tcdR expression. Blue arrowed lines indicate positive controls, while lines ending with a bar across correspond to negative controls. Blue dashed arrows indicate mechanisms that are not fully understood. Enzymatic reactions are marked in red arrows. Abbreviations used: AI-2 (Auto inducers), GTP (Guanosine triphosphate), pGpG (5′-phosphoguanylyl-(3′−5′)-guanosine), c-di-GMP (cyclic di-guanosyl-5′monophosphate), BCCAs (branch chain amino acids), NAD (Nicotinamide adenine dinucleotide), DGCs (Di-Guanylate cyclases), PDEs (Phosphodiesterases).
TcdC has long been thought to negatively affect toxin gene expression due to the inverse timing of its expression compared to the other PaLoc genes [22]. TcdC has been suggested to act as an anti-sigma factor by interfering with the capacity of the RNAP-TcdR complex to recognize the tcdA and tcdB promoters [13]. Carter et al. utilized a plasmid-borne copy of the tcdC gene to complement the C. difficile NAPI/027 strain (M7404) lacking the functional tcdC gene and showed that tcdC expression down-regulated toxin production and reduced pathogenicity [23]. Even though these studies suggested tcdC’s role in toxin gene regulation, other studies generated contradictory results [24,25]. TcdC, unlike any other known regulatory protein, does not have a DNA-binding domain. It is a membrane-associated protein with a transmembrane domain at the N-terminus [13,26]. Its C-terminal region was thought to be localized in the cytoplasm to interact with TcdR [13]. More recently, the HiBiT extracellular detection system demonstrated that the C- terminus of TcdC is located extracellularly [27]. This calls into question the present model of TcdC’s anti-sigma factor mechanism of action. More research is needed to fully understand the role of TcdC in C. difficile physiology and toxin gene regulation.
Toxin gene regulation in response to nutrients
Significant variations in the level of toxin production in C. difficile strains are seen in the presence of different nutrient sources. Glucose, certain amino acids and butyrate in the medium greatly influence toxin production [6,8,9]. Molecular pathways controlling C. difficile toxin gene expression in response to some of these nutrients involve major regulators, CcpA, CodY, PrdR, and Rex [18,19,28–30].
The effects of rapidly metabolizable sugars on toxin gene expression are examples of carbon catabolite repression (CCR). In this phenomenon where the presence of a carbon source in the medium represses the expression of certain genes and operons. CcpA, a Lac repressor family protein, is the main agent of CCR in low G+C Gram-positive bacteria [18]. CcpA reduces toxin gene expression in C. difficile by interacting directly with the PaLoc promoter regions, having the greatest affinity for the tcdR promoter region. [18]. In a ccpA mutant, toxin production remains the same irrespective of the presence or absence of glucose, however lower than in the parent strain grown without glucose [18]. This finding implies that CcpA also controls other regulators involved in toxin gene regulation. Transcriptomics analysis of the ccpA mutant indeed revealed CcpA directly or indirectly controlling several other global regulators, including CodY and Rex [18].
While CcpA controls toxin production in reaction to glucose, repression of toxin synthesis in response to amino acids is mediated by CodY. During the exponential growth phase, when the nutrients are abundant, CodY binds to branched-chain amino acids (BCAAs)/ or GTP and acts primarily as a repressor of various alternative metabolic pathways [30,31]. When nutrients become limited in the cell, CodY is no longer bound by the cofactors, and the transcriptional repression by CodY is alleviated. CodY controls toxin gene expression by binding to the tcdR promoter region at three distinct sites with variable affinities, two of which overlap with the TcdR binding sites. In addition to its direct action on the toxin gene locus, CodY also regulates other metabolic pathways that affect toxin production. For example, genes involved in the butyrate synthesis and c-di-GMP metabolism are direct targets of CodY and are repressed when BCAA and GTP are in excess in the medium [31–33]. CodY also regulates the master regulators SinR and Spo0A, which are known to govern toxin production (see below) (Figure 1).
Cysteine is one of the most potent amino acids that down-regulates toxin production in C. Difficile [8]. Through an unknown mechanism, SigL, which belongs to the σ54 family, has been identified to promote cysteine-dependent suppression of toxin synthesis, probably in response to the accumulation of by-products of cysteine degradation, such as pyruvate (Figure 1) [34]. Similar to cysteine, the amino acids proline and glycine also have a strong inhibitory effect on the production of toxins [9]. Both proline and glycine are part of Stickland reaction, where the coupled oxidation and reduction of these amino acids generate ATP and NAD+, respectively [29]. In the presence of proline and glycine, proline reductase (PR) and glycine reductase (GR), respectively are induced and reduce these amino acids. Among the two amino acids, C. difficile preferentially utilizes proline for NAD+ regeneration [29]. PrdR is a proline-responsive regulator that mediates proline-dependent PR activation and proline-dependent toxin gene suppression by an unknown mechanism (Figure 1) [29]. Analysis of the transcriptome of a PrdR mutant revealed that PrdR controls numerous additional reductive pathways, including glycine reductase, suggesting the direct regulator might be Rex, a sensor of redox status [28]. Rex is a DNA-binding protein that recognizes changes in the redox state and is only active when the intracellular NADH/NAD+ ratio is low. According to the current model [28], PrdR is activated and promotes PR expression when proline levels are high. As a result, the NADH/NAD+ ratio is low, and Rex acts as a repressor of alternative NAD+ synthesis pathways. When proline becomes scarce, on the other hand, the NADH/NAD+ ratio rises, and NADH inhibits Rex-dependent suppression of alternative pathways. When NAD+ is regenerated via these alternative reductive pathways, accumulation of butyrate occurs and stimulates toxin synthesis.
Role of c-di-GMP in toxin gene regulation
The signaling molecule cyclic di-guanosyl-5′monophosphate (c-di-GMP) is a second messenger in bacterial systems [35]. An increase in intracellular c-di-GMP concentration by ectopic expression of a c-di-GMP synthesis gene, dccA, has been shown to decrease the expression of toxin genes [33,36]. c-di-GMP has also been shown to negatively regulate the flgB flagellar operon by binding to the upstream riboswitch [36]. Along with flagellar genes, flgB operon also contains sigD, which positively regulates tcdR expression [20,21]. Therefore, c-di-GMP mediated regulation of toxin production is indirect and is mediated by SigD. It is important to note that a DNA inversion upstream of flgB, switch ON or OFF the expression of flagellar operon, including sigD. Thus, the phase variable expression of sigD, indirectly brings the toxin gene expression under the flgB regulatory switch [37]. In addition to SigD, other flagellar components, such as the flagellin FliC and FliD, also influence toxin production through unknown mechanisms [38,39].
Link between toxin production and sporulation
In sporulating bacteria, a complex regulatory network controls the transition between the stationary phase and the onset of sporulation. These regulatory mechanisms were extensively studied in Bacillus subtilis, where repressors such as AbrB, CodY, and SinR prevent the expression of genes that are expressed only during stationary phase or sporulation [40,41]. In B. subtilis, Spo0A is the master regulator that initiates sporulation after being phosphorylated through a phosphorelay system [40]. SigH, an alternative sigma factor, directs transcription of spo0A and the genes involved in the phosphorelay system [40]. C. difficile does not have orthologs of AbrB or the Spo0A-phosphorelay components, although it does have Spo0A, SigH, and SinR. At the onset of the stationary phase, SigH and Spo0A control toxin gene expression (Figure 1). In a sigH mutant, toxin genes and tcdR are overexpressed, implying that SigH represses toxin expression [42]. Spo0A effect on toxin gene expression was strain specific. In ribotype 027 strains, Spo0A acted as a repressor of toxin gene expression but had minimal impact in strains of ribotype 078 [43,44]. While it was clear that SigH and Spo0A indirectly affect toxin gene expression, the regulators involved were unknown. Results from the analysis of sin locus mutant suggested that SinR could be the link that connects toxin production with sporulation. The sin (sporulation inhibitor) locus in C. difficile encodes two proteins, SinR and SinI, and controls many genes involved in sporulation, motility, and biofilm formation [45–47]. SinR is a DNA-binding protein that inhibits target gene transcription, and SinI has an antagonistic interaction with SinR and inhibits its function by binding directly to the SinR protein. In the absence of SinR and SinI, decreased toxin production was observed [46]. The expression of sinR alone was sufficient to complement this phenotype, suggesting SinR is a positive regulator of toxin synthesis. SinR influences toxin gene transcription by controlling the sigD expression [46]. Spo0A suppresses sinR expression by directly binding to its upstream DNA region [48]. In the absence of Spo0A (in the spo0A mutant and the sigH mutant), SinR is produced more, increasing the SigD production, leading to more toxin synthesis [42,48]. Other than Spo0A, CodY also regulates sin locus expression [46].
RstA is a newly discovered regulator that promotes sporulation initiation while inhibiting toxin production [49–51]. An earlier study suggested that RstA regulates toxin production by repressing the transcription of SigD [50]. New data, however, indicates that this regulator binds to specific DNA sequences upstream of tcdR and controls its transcription directly [51]. The RstA is a member of the RNPP family of transcriptional regulators involved in quorum sensing [49]. While the link between RstA and quorum sensing is unknown, autoinducer (AI) like molecules have been found in C. difficile. All analyzed C. difficile genomes contain an incomplete agr1- locus that contains agrDB, which codes for components responsible for producing autoinducer peptides [52]. The two-component system agrAC that activates quorum sensing responsive genes in response to AI peptides is absent in the agr-1 locus [52]. When the agr-1 locus was deleted, the deletions strains produced no toxins, and the mutant was unable to colonize the mouse gut to initiate infection [53]. However, in a recent work, Ahmed et. al showed that deletion of agrDB locus in 630 Δ erm strain had little impact in toxin production, but accumulation of AgrD1 peptide in the cytoplasm in the absence of AgrB1 resulted in increased toxin gene transcription [54]. In addition to agr1 locus, C. difficile strains belonging to ribotype 027 carry a complete agrABCD operon (agr-2 locus) [52]. In strain R20291 (ribotype 027), the inactivation of agrA results in decreased toxin production (Figure 1) [53,55]. AgrA regulates the flagellar synthesis and the expression of genes involved in c-di-GMP metabolism [55], suggesting that AgrA’s impact on toxin gene transcription is indirect and mediated via SigD. While these initial studies highlight the importance of agr loci in C. difficile, questions regarding their mechanisms of action remain unanswered and warrant further studies. In addition to the agr system, C. difficile has been found to include the LuxS gene, which is known to produce a class of quorum sensing molecules known as AI-2 [56]. When added to the culture medium early during growth, AI-2 up-regulates the expression of PaLoc genes through an unknown mechanism.
Other regulators controlling toxin production
LexA is one of the major players in the bacterial SOS regulatory network. Absence of LexA influences the tcdA transcription negatively. Even though the interaction of LexA with tcdA promoter was not experimentally tested, potential LexA binding motifs were identified in the tcdA promoter region [57,58]. This might explain why antibiotics that are known to trigger SOS responses, such as levofloxacin and ciprofloxacin, enhanced toxin A production when added at sub-inhibitory doses [57,58]. Similar to LexA, Mfd is also a highly conserved protein and functions in the nucleotide excision repair pathway in response to DNA damage and is known to affect transcription elongation by removing the stalled RNA polymerases [59]. In C. difficile, inactivation of mfd resulted in enhanced transcription of tcdA and tcdB [59]. It was hypothesized that Mfd might affect the transcriptional regulation of toxin genes by relieving RNA polymerase stalled at roadblocks created by CodY and/or CcpA, the two major repressors of toxin gene expression. More recently, CelR, the repressor of cellobiose utilization operon was identified and characterized [60]. In the absence of CelR, C. difficile produced more toxins than in its presence. Potential CelR binding sites were identified in genes responsible for c-di-GMP turnover, indicating the effect of CelR on toxin gene transcription is indirect, possibly through the activation of SigD [60].
Highlights.
During infection, Clostridioides difficile produces the toxins TcdA and TcdB, which are directly linked to the severity of the disease.
Toxin gene expression is influenced by a number of positive and negative regulators in response to environmental cues.
Many of the same regulators that govern toxin gene expression also affect sporulation, biofilm formation, and motility.
Our present understanding of toxin gene regulation by various regulators is summarized in this review.
Concluding remarks.
Toxin gene expression in C. difficile is activated by a variety of environmental stimuli and regulated by several global regulators, demonstrating the intricacy of this regulatory network. Deeper insight into toxin genes regulations can help combat the rise in CDI incidence and may help identify alternative treatment strategies.
Acknowledgement
This paper is dedicated to the memory of Aritri Majumdar who lost her battle against cancer. She prepared part of this review during her rotation at the Division of Biology.
Funding Information
Grants from NIAID (1R15AI122173, 1R03AI135762-01A1) and Johnson Cancer Center-KSU, supported the preparation of this review.
Footnotes
Declaration of Interest
None
Conflict of Interest:
None exist
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
Reference
- 1.Gupta A, Ananthakrishnan AN: Economic burden and cost-effectiveness of therapies for Clostridiodes difficile infection: a narrative review. Therap Adv Gastroenterol 2021, 14:17562848211018654. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Mullish BH, Williams HR: Clostridium difficile infection and antibiotic-associated diarrhoea. Clin Med (Lond) 2018, 18:237–241. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Kuehne SA, Cartman ST, Heap JT, Kelly ML, Cockayne A, Minton NP: The role of toxin A and toxin B in Clostridium difficile infection. Nature 2010, 467:711–713. [DOI] [PubMed] [Google Scholar]
- 4.Aldape MJ, Packham AE, Nute DW, Bryant AE, Stevens DL: Effects of ciprofloxacin on the expression and production of exotoxins by Clostridium difficile. J Med Microbiol 2013, 62:741–747. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Chilton CH, Freeman J, Crowther GS, Todhunter SL, Nicholson S, Wilcox MH: Co-amoxiclav induces proliferation and cytotoxin production of Clostridium difficile ribotype 027 in a human gut model. J Antimicrob Chemother 2012, 67:951–954. [DOI] [PubMed] [Google Scholar]
- 6.Dupuy B, Sonenshein AL: Regulated transcription of Clostridium difficile toxin genes. Mol Microbiol 1998, 27:107–120. [DOI] [PubMed] [Google Scholar]
- 7.Govind R, Vediyappan G, Rolfe RD, Dupuy B, Fralick JA: Bacteriophage-mediated toxin gene regulation in Clostridium difficile. J Virol 2009, 83:12037–12045. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Karlsson S, Lindberg A, Norin E, Burman LG, Akerlund T: Toxins, butyric acid, and other short-chain fatty acids are coordinately expressed and down-regulated by cysteine in Clostridium difficile. Infect Immun 2000, 68:5881–5888. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Karlsson S, Burman LG, Åkerlund T: Suppression of toxin production in Clostridium difficile VPI 10463 by amino acids. Microbiology (Reading) 1999, 145 ( Pt 7):1683–1693. [DOI] [PubMed] [Google Scholar]
- 10.Sekulovic O, Meessen-Pinard M, Fortier L-C: Prophage-stimulated toxin production in Clostridium difficile NAP1/027 lysogens. J Bacteriol 2011, 193:2726–2734. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Karlsson S, Dupuy B, Mukherjee K, Norin E, Burman LG, Akerlund T: Expression of Clostridium difficile toxins A and B and their sigma factor TcdD is controlled by temperature. Infect Immun 2003, 71:1784–1793. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Mani N, Dupuy B: Regulation of toxin synthesis in Clostridium difficile by an alternative RNA polymerase sigma factor. Proc Natl Acad Sci U S A 2001, 98:5844–5849. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Matamouros S, England P, Dupuy B: Clostridium difficile toxin expression is inhibited by the novel regulator TcdC. Mol Microbiol 2007, 64:1274–1288. [DOI] [PubMed] [Google Scholar]
- 14.Govind R, Dupuy B: Secretion of Clostridium difficile toxins A and B requires the holin-like protein TcdE. PLoS Pathog 2012, 8:e1002727. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Olling A, Seehase S, Minton NP, Tatge H, Schröter S, Kohlscheen S, Pich A, Just I, Gerhard R: Release of TcdA and TcdB from Clostridium difficile cdi 630 is not affected by functional inactivation of the tcdE gene. Microb Pathog 2012, 52:92–100. [DOI] [PubMed] [Google Scholar]
- 16.Dupuy B, Matamouros S: Regulation of toxin and bacteriocin synthesis in Clostridium species by a new subgroup of RNA polymerase sigma-factors. Res Microbiol 2006, 157:201–205. [DOI] [PubMed] [Google Scholar]
- 17.Ransom EM, Kaus GM, Tran PM, Ellermeier CD, Weiss DS: Multiple factors contribute to bimodal toxin gene expression in Clostridioides (Clostridium) difficile.Mol Microbiol 2018, 110:533–549. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Antunes A, Camiade E, Monot M, Courtois E, Barbut F, Sernova NV, Rodionov DA, Martin-Verstraete I, Dupuy B: Global transcriptional control by glucose and carbon regulator CcpA in Clostridium difficile.Nucleic Acids Res 2012, 40:10701–10718. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Antunes A, Martin-Verstraete I, Dupuy B: CcpA-mediated repression of Clostridium difficile toxin gene expression. Mol Microbiol 2011, 79:882–899. [DOI] [PubMed] [Google Scholar]
- 20.El Meouche I, Peltier J, Monot M, Soutourina O, Pestel-Caron M, Dupuy B, Pons J-L: Characterization of the SigD regulon of C. difficile and its positive control of toxin production through the regulation of tcdR. PLoS One 2013, 8:e83748. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.McKee RW, Mangalea MR, Purcell EB, Borchardt EK, Tamayo R: The second messenger cyclic Di-GMP regulates Clostridium difficile toxin production by controlling expression of sigD. J Bacteriol 2013, 195:5174–5185. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Hundsberger T, Braun V, Weidmann M, Leukel P, Sauerborn M, von Eichel-Streiber C: Transcription analysis of the genes tcdA-E of the pathogenicity locus of Clostridium difficile. Eur J Biochem 1997, 244:735–742. [DOI] [PubMed] [Google Scholar]
- 23.Carter GP, Douce GR, Govind R, Howarth PM, Mackin KE, Spencer J, Buckley AM, Antunes A, Kotsanas D, Jenkin GA, et al. : The anti-sigma factor TcdC modulates hypervirulence in an epidemic BI/NAP1/027 clinical isolate of Clostridium difficile. PLoS Pathog 2011, 7:e1002317. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Cartman ST, Kelly ML, Heeg D, Heap JT, Minton NP: Precise manipulation of the Clostridium difficile chromosome reveals a lack of association between the tcdC genotype and toxin production. Appl Environ Microbiol 2012, 78:4683–4690. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Bakker D, Smits WK, Kuijper EJ, Corver J: TcdC does not significantly repress toxin expression in Clostridium difficile 630ΔErm. PLoS One 2012, 7:e43247. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Govind R, Vediyappan G, Rolfe RD, Fralick JA: Evidence that Clostridium difficile TcdC is a membrane-associated protein. J Bacteriol 2006, 188:3716–3720. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Oliveira Paiva AM, de Jong L, Friggen AH, Smits WK, Corver J: The C-Terminal Domain of Clostridioides difficile TcdC Is Exposed on the Bacterial Cell Surface. J Bacteriol 2020, 202:e00771–19. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Bouillaut L, Dubois T, Francis MB, Daou N, Monot M, Sorg JA, Sonenshein AL, Dupuy B: Role of the global regulator Rex in control of NAD+ -regeneration in Clostridioides (Clostridium) difficile. Mol Microbiol 2019, 111:1671–1688. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Bouillaut L, Self WT, Sonenshein AL: Proline-dependent regulation of Clostridium difficile Stickland metabolism. J Bacteriol 2013, 195:844–854. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Dineen SS, Villapakkam AC, Nordman JT, Sonenshein AL: Repression of Clostridium difficile toxin gene expression by CodY. Molecular Microbiology 2007, 66:206–219. [DOI] [PubMed] [Google Scholar]
- 31.Dineen SS, McBride SM, Sonenshein AL: Integration of metabolism and virulence by Clostridium difficile CodY. J Bacteriol 2010, 192:5350–5362. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Dhungel BA, Govind R: Phase-variable expression of pdcB, a phosphodiesterase, influences sporulation in Clostridioides difficile. Mol Microbiol 2021, doi: 10.1111/mmi.14828. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Purcell EB, McKee RW, Courson DS, Garrett EM, McBride SM, Cheney RE, Tamayo R: A Nutrient-Regulated Cyclic Diguanylate phosphodiesterase controls Clostridium difficile biofilm and toxin production during stationary Phase. Infect Immun 2017, 85:e00347–17. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Dubois T, Dancer-Thibonnier M, Monot M, Hamiot A, Bouillaut L, Soutourina O, Martin-Verstraete I, Dupuy B: Control of Clostridium difficile physiopathology in response to cysteine availability. Infect Immun 2016, 84:2389–2405. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Römling U, Galperin MY, Gomelsky M: Cyclic di-GMP: the first 25 years of a universal bacterial second messenger. Microbiol Mol Biol Rev 2013, 77:1–52. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.McKee RW, Harvest CK, Tamayo R: Cyclic diguanylate regulates virulence factor genes via multiple riboswitches in Clostridium difficile. mSphere 2018, 3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Anjuwon-Foster BR, Maldonado-Vazquez N, Tamayo R: Characterization of flagellum and toxin phase variation in Clostridioides difficile ribotype 012 isolates. J Bacteriol 2018, 200:e00056–18. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Dingle TC, Mulvey GL, Armstrong GD: Mutagenic analysis of the Clostridium difficile flagellar proteins, FliC and FliD, and their contribution to virulence in hamsters. Infect Immun 2011, 79:4061–4067. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Aubry A, Hussack G, Chen W, KuoLee R, Twine SM, Fulton KM, Foote S, Carrillo CD, Tanha J, Logan SM: Modulation of toxin production by the flagellar regulon in Clostridium difficile. Infect Immun 2012, 80:3521–3532. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Sonenshein AL: Control of sporulation initiation in Bacillus subtilis. Curr Opin Microbiol 2000, 3:561–566. [DOI] [PubMed] [Google Scholar]
- 41.Phillips ZEV, Strauch MA: Bacillus subtilis sporulation and stationary phase gene expression. Cell Mol Life Sci 2002, 59:392–402. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Saujet L, Monot M, Dupuy B, Soutourina O, Martin-Verstraete I: The Key Sigma Factor of transition phase, SigH, controls sporulation, metabolism, and virulence factor expression in Clostridium difficile. J Bacteriol 2011, 193:3186–3196. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Mackin KE, Carter GP, Howarth P, Rood JI, Lyras D: Spo0A differentially regulates toxin production in evolutionarily diverse strains of Clostridium difficile.PLoS ONE 2013, 8:e79666. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Rosenbusch KE, Bakker D, Kuijper EJ, Smits WK: C. difficile 630Δerm Spo0A Regulates Sporulation, but does not contribute to toxin production, by direct high-affinity binding to target DNA. PLoS One 2012, 7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Ciftci Y, Girinathan BP, Dhungel BA, Hasan MK, Govind R: Clostridioides difficile SinR’ regulates toxin, sporulation and motility through protein-protein interaction with SinR. Anaerobe 2019, 59:1–7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Girinathan BP, Ou J, Dupuy B, Govind R: Pleiotropic roles of Clostridium difficile sin locus. PLOS Pathogens 2018, 14:e1006940. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Poquet I, Saujet L, Canette A, Monot M, Mihajlovic J, Ghigo J- M, Soutourina O, Briandet R, Martin-Verstraete I, Dupuy B: Clostridium difficile Biofilm: Remodeling metabolism and cell surface to build a sparse and heterogeneously aggregated architecture. Front Microbiol 2018, 9:2084. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Dhungel BA, Govind R: Spo0A Suppresses sin locus expression in Clostridioides difficile. mSphere 2020, 5:e00963–20. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Edwards AN, Tamayo R, McBride SM: A novel regulator controls Clostridium difficile sporulation, motility and toxin production. Mol Microbiol 2016, 100:954–971. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Edwards AN, Anjuwon-Foster BR, McBride SM: RstA Is a Major Regulator of Clostridioides difficile toxin production and motility. mBio 2019, 10:e01991–18. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Edwards AN, Krall EG, McBride SM: Strain-Dependent RstA Regulation of Clostridioides difficile toxin production and sporulation. J Bacteriol 2020, 202:e00586–19. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Stabler RA, He M, Dawson L, Martin M, Valiente E, Corton C, Lawley TD, Sebaihia M, Quail MA, Rose G, et al. : Comparative genome and phenotypic analysis of Clostridium difficile 027 strains provide insight into the evolution of a hypervirulent bacterium. Genome Biol 2009, 10:R102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Darkoh C, DuPont HL, Norris SJ, Kaplan HB: Toxin synthesis by Clostridium difficile is regulated through quorum signaling. mBio 2015, 6:e02569. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Ahmed UKB, Shadid TM, Larabee JL, Ballard JD: Combined and distinct roles of Agr Proteins in Clostridioides difficile 630 Sporulation, motility, and toxin production. mBio 2020, 11:e03190–20. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Martin MJ, Clare S, Goulding D, Faulds-Pain A, Barquist L, Browne HP, Pettit L, Dougan G, Lawley TD, Wren BW: The agr locus regulates virulence and colonization genes in Clostridium difficile 027. J Bacteriol 2013, 195:3672–3681. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Lee ASY, Song KP: LuxS/autoinducer-2 quorum sensing molecule regulates transcriptional virulence gene expression in Clostridium difficile. Biochem Biophys Res Commun 2005, 335:659–666. [DOI] [PubMed] [Google Scholar]
- 57.Walter BM, Cartman ST, Minton NP, Butala M, Rupnik M: The SOS Response Master Regulator LexA is associated with sporulation, motility and biofilm formation in Clostridium difficile. PLoS One 2015, 10:e0144763. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Walter BM, Rupnik M, Hodnik V, Anderluh G, Dupuy B, Paulič N, Žgur-Bertok D, Butala M: The LexA regulated genes of the Clostridium difficile. BMC Microbiol 2014, 14:88. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Willing SE, Richards EJ, Sempere L, Dale AG, Cutting SM, Fairweather NF: Increased toxin expression in a Clostridium difficilemfd mutant. BMC Microbiol 2015, 15:280. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Hasan MK, Dhungel BA, Govind R: Characterization of an operon required for growth on cellobiose in Clostridioides difficile. Microbiology 2021, 16: 001079. doi: 10.1099/mic.0.001079. [DOI] [PMC free article] [PubMed] [Google Scholar]