Abstract
Metabolites are important biomarkers in human body fluids, conveying direct information of cellular activities and physical conditions. Metabolite detection has long been a research hotspot in the field of biology and medicine. Surface-enhanced Raman spectroscopy (SERS), based on the molecular “fingerprint” of Raman spectrum and the enormous signal enhancement (down to a single-molecule level) by plasmonic nanomaterials, has proven to be a novel and powerful tool for metabolite detection. SERS provides favorable properties such as ultra-sensitive, label-free, rapid, specific, and non-destructive detection processes. In this review, we summarized the progress in recent 10 years on SERS-based sensing of endogenous metabolites at the cellular level, in tissues, and in biofluids, as well as drug metabolites in biofluids. We made detailed discussions on the challenges and optimization methods of SERS technique in metabolite detection. The combination of SERS with modern biomedical technology were also anticipated.
Keywords: Surface enhanced Raman spectroscopy (SERS), Metabolite detection, Cellular metabolites, Bio-fluids, Drug metabolites
Graphical abstract
1. Introduction
Metabolites are small biomolecules that existed as precursors, intermediates and end products in the whole process of cellular activities [1,2]. They can initiate cell signaling cascades, regulate various biological procedures [3], and provide direct information of the state of cells [4]. Metabolomics data sets link cell activities with metabolic pathways as well as biological information in patients and healthy groups [5,6]. Therefore, the detection of metabolites shows significant potentials in the fundamental studies and clinical applications to investigate physiological activities [7].
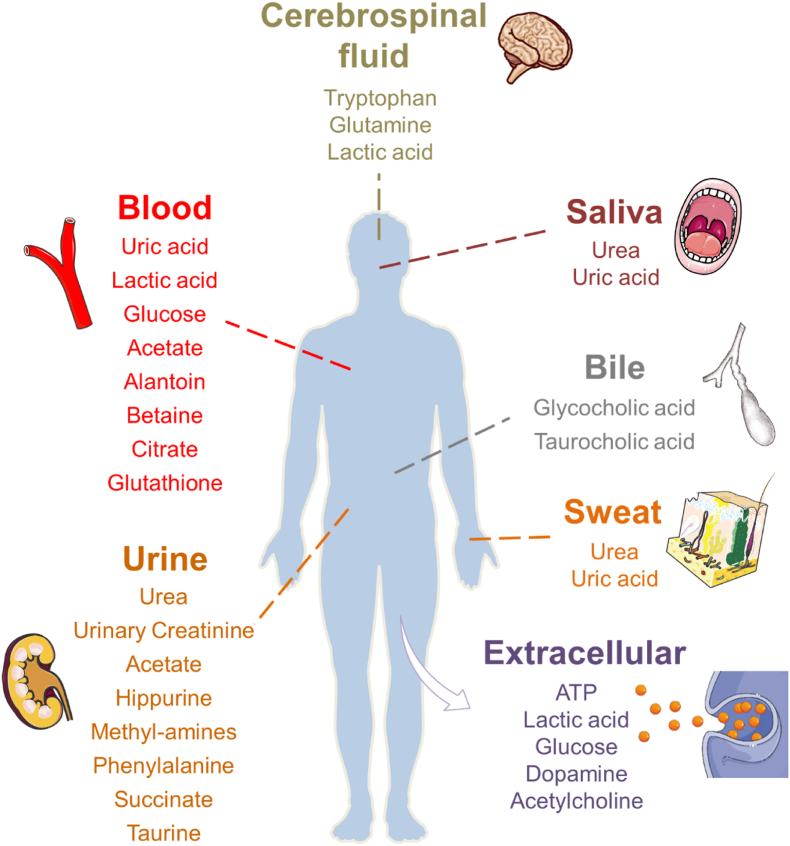
Generally, human metabolites are studied at the cellular level, in tissues, or in the body fluids, such as blood, saliva, bile, sweat, urine, and cerebrospinal fluid (Fig. 1). Cellular metabolism entails a series of complicated biological reactions, including the metabolic pathways of cell growth and reproduction as well as changes in cellular microenvironment [8]. The core set of metabolites includes amino acids, glucose, lactic acid, lactate dehydrogenase (LDH), pyruvate, adenosine triphosphate (ATP), reactive oxygen species (ROS), lipids, and nucleotides. They act as alternative fuels, signaling metabolites, stimulators, and inhibitors of enzymes during cell lifetime [[9], [10], [11], [12], [13]].
Fig. 1.
Common metabolites from biofluids and cells.
Metabolites extracted from body fluids have long been investigated for health assessment and clinical diagnosis. Neurotransmitters are one of the most commonly studied, since they are related to diseases such as schizophrenia and Parkinson's disease [14,15]. The main types of neurotransmitters are amino acids (e.g., glutamic acid), peptides (e.g., somatostatin), purine derivatives (e.g., ATP), and monoamines (e.g., dopamine) [16]. Amino acids metabolism is related to alkaline proteinuria, phenylketonuria and maple syrupuria [17], which are the components of proteins and precursors of neurotransmitters [18]. ATP is an important neurotransmitter that produces metabolites in the degradation process, including inosine (INO), hypoxanthine (HX) and uric acid (UA). INO and HX are key cardiac biomarkers, and UA can provide signals of heart failure and acute kidney injury [[19], [20], [21]]. Besides neurotransmitters, vitamins are a critical necessity for human body. Vitamin B complex is the precursor of a variety of coenzymes; vitamin B6 deficiency can cause depression and convulsions; scurvy may be due to a lack of vitamin C. The metabolites of vitamins provide a clue for the disease related to genetic dysfunction, dietary deficiency, and so on. Besides the above endogenous natural metabolites, drug metabolites have garnered much attention as they reflect in vivo drug dosage. The monitoring of drug metabolites helps to understand the working concentration of pills, potential side effects of overdose, and to control the use of illegal drugs. To sum up, extensive efforts have been made for the analysis of metabolites in biofluids, in order to fully understand the in vivo biological activities [[22], [23], [24], [25], [26], [27]].
Surface-enhanced Raman spectroscopy (SERS) is an ultra-sensitive and highly specific optical detection technique, benefited from the molecular “fingerprint” of Raman spectrum and the enormous signal enhancement (down to a single-molecule level) by plasmonic nanomaterials [28,29]. The applications of SERS in chemistry, biology and medicine have been widely reported [[30], [31], [32]]. For instance, SERS-based cancer diagnosis and in vivo tumor detection have been successfully proposed in vitro or in vivo [[33], [34], [35]]. SERS also holds great potential for the detection of metabolites because it is a sensitive, label-free, rapid, specific and non-destructive technique; particularly, the advance in nanotechnology provides abundant choices of plasmonic substrates for metabolite screening.
This review systematically summarizes SERS-based metabolite detection studies. Starting with the general introduction of different metabolites and their important functions, we then compared the conventional detection techniques including mass spectrometry (MS) and nuclear magnetic resonance (NMR), as well as the emerging SERS technique. In the main text, the progress on SERS-based metabolite detection in the recent decade was discussed, classified by the metabolic molecules at the cellular level and in body fluids. We then made perspectives on the prospects of metabolite SERS detection, by discussing the challenges and optimization methods of SERS technique. At last, the combinations of SERS with modern medical technology were anticipated. Other relevant reviews about SERS-based cancer diagnosis, biomarkers determination, and illicit drugs determination can be referred to literature [[36], [37], [38], [39], [40], [41]].
2. Main metabolic molecules detecting methods
MS, NMR, and the combination of them with chromatography have been widely applied for the separation and detection of metabolites [[42], [43], [44]]. SERS, as an emerging method for metabolite detection, has been developed with a standardized substrate preparation and detection procedure.
2.1. Traditional methods
MS is an analytical method for the measurement of the mass-to-charge ratio of ions. The basic principle of MS is to ionize each component in the sample to generate charged ions with different charge-to-mass ratios, then the ion beam enters the mass analyzer for mass determination. MS is widely applied to analyze biological samples through direct injection or chromatographic separation. The latest improvement in mass accuracy has greatly broadened the range of metabolites that MS can analyze with enhanced precision [45]. The quantification of varied metabolites could be achieved through MS with high sensitivity and resolution to obtain the information of metabolism [46]. For example, metabolites in urine were detected via matrix-assisted laser desorption/ionization MS (LDIMS) for kidney diseases subtypes characterization [47]. Similarly, with the help of LDIMS, the diagnosis of early-stage lung adenocarcinoma was achieved through serum metabolite detection [48].
Metabolite detection through NMR was based on the differences of spin motions from different nuclei. NMR is able to acquire the information of the whole metabolism, considered as one of the most successful analytical techniques in the analysis of bio-fluids [[49], [50], [51]]. The benefits and shortcomings of MS and NMR are listed in Table 1. Nevertheless, limited by relatively low sensitivity, NMR is only appliable for metabolites of adequate concentrations [52].
Table 1.
Comparison of MS, NMR and SERS in metabolite detection. The descriptions of MS and NMR are according to reference [58].
| MS | NMR | SERS | |
|---|---|---|---|
| Sensitivity | High and the limit of detection (LOD) reaches nM | Low but can be improved with high field intensity or low temperature | High and LOD reaches the range of nM - pM |
| Selectivity | Selective and non-selective analysis | Generally for non-selective analysis | Selective and non-selective analysis |
| Destruction | Destructive but a small number of samples required | Non-destructive and samples can be recycled after measurements | Non-destructive and a small number of samples required |
| Reproducibility | Moderate | High | Moderate, depending on substrate uniformity |
| Sample pretreatment | Demanding | Not necessary | Not necessary but recommended |
| Sample analysis time | Longer than NMR, and chromatography techniques required | Fast and the whole sample can be analyzed simultaneously | Fast and multiple analytes can be analyzed in one measurement |
| Targeted analysis | Relatively better | Not optimal | Not optimal |
Chromatography relies on the difference in forces (distribution, adsorption, ion exchange, etc.) between different solutes (in the sample) and stationary phase and mobile phase to separate different solutes from each other. Thus, as a powerful tool for sample separation, chromatography has been largely developed to be combined with MS or NMR, and occupies a prominent position in the science of identification and separation [53]. Advances in high performance liquid chromatography (HPLC) and MS have greatly promoted metabolomics analysis. MS and HPLC can be combined for unknown endogenous or exogenous metabolites characterization in complicated biological samples [[54], [55], [56]]. However, the combination of these technologies requires expensive equipment, complicated sample preparation and long test time [57].
2.2. SERS
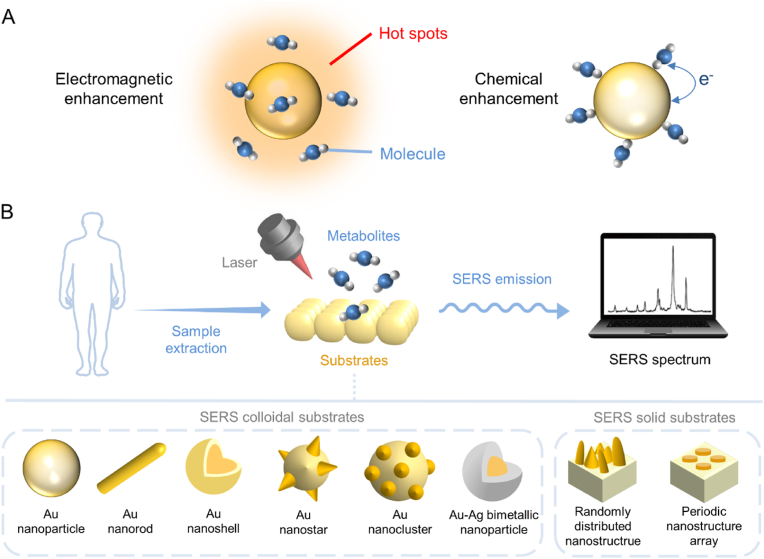
Raman spectroscopy, as an inelastic part of emission when light interacting with samples, exhibits high specificity with fingerprint feature and narrow bandwidths. It reveals the information of molecular vibrational, rotational, and transitional information [59,60]. Normal Raman signals can be largely enhanced when molecules are adsorbed on the surface of metallic nanostructures [61]. This is called surface-enhanced Raman scattering/spectroscopy (SERS). The mechanism of SERS enhancement is generally described in two aspects: electromagnetic (EM) and chemical (CM) enhancement (Fig. 2A) [62]. The localized surface plasmon resonance (LSPR) brings an enhanced local EM field, referred to as EM “hotspots”, which enhances Raman signals of molecules located in this region. In the narrow gaps between/in plasmonic structures, a much stronger EM enhancement can be generated [63]. Theoretically, EM enhancement takes effects for any molecules located in hotspot regions, and the direct attachment of molecule to the plasmonic surface is not necessary (Fig. 2A). CM mechanism, though not fully understood so far, often refers to (i) the increased polarizability of the molecules, (ii) molecular electronic excitation promoted by the photon excitation, or (iii) charge transfer transitions between the metal and molecules [64,65]. It is usually effective for a molecule that directly contacts nanostructure surface, especially via chemo-adsorption. Generally, EM mechanism dominates the total enhancement, even for the molecule directly adsorb on nanostructure surface [[66], [67], [68]].
Fig. 2.
(A) SERS enhancement mechanisms. (B) The direct SERS detection procedure of metabolites. The SERS substrates commonly utilized are presented.
SERS-based detection techniques could be recognized as direct (label-free) and indirect (with SERS labels). For the direct strategies, the target molecules are placed close to plasmonic nanoparticles (NPs) to generate enhanced molecular Raman spectra [69,70]. These Raman spectra contain abundant molecular information, reflecting the chemical and pathological state of cells or tissues [71]. So far, direct SERS has been applied for food safety testing, field testing, liquid biopsy and cancer diagnosis [67,[72], [73], [74]]. For the indirect strategies, NPs modified with Raman reporters are used as labels to specifically bind to targets, e.g., protein biomarkers. In this case, the detection of targets relies on the enhanced signals of Raman reporters on NPs. A variety of SERS NPs coated with different targeting ligands have been developed, benefiting SERS-based multiplexing analysis [75,76]. SERS NPs possesses great potentials in bioimaging, immunoassay, microtumor diagnosis, as well as intraoperative navigation [64,[77], [78], [79], [80]].
Despite the fact that indirect SERS has been widely used for detecting macro biomolecules and cells [[81], [82], [83]], the detection of metabolites, however, is commonly applied in a direct way. This can be attributed to two reasons. First, most metabolites, unlike immuno-biomolecules (e.g., antibodies), rarely show affinity toward plasmonic surfaces [84]. Second, the indirect SERS detection usually requires the targeting of SERS NPs to metabolites either through bio-recognition or by physical-chemo reactions. The targeting process is seriously hindered by the much smaller size of metabolite molecules than that of NPs. Therefore, it is more feasible to simply enhance metabolites by generated hotspots of substrates in a label-free approach. Fig. 2B presents the typical procedure of SERS metabolite detection: firstly, the metabolite samples are collected and added onto the SERS-active nanostructures for molecular adsorption; then, the exposure of samples to laser excitations induces high SERS signal, which can be detected by the Raman spectrometer; finally, the obtained SERS signals are processed and analyzed for quantification. The design of SERS substrates follows the guidelines of generating more plasmonic hotspots [[85], [86], [87], [88]]. As shown in Fig. 2B (bottom panel), the SERS substrates can be made in colloidal states (left), or as solid substrates such as randomly distributed or periodic array nanostructures (right).
SERS provides favorable properties in metabolite detections. First, the direct SERS technique has proven ultra-high sensitivity down to the single-molecule level, with the procedure being rapid and non-destructive. Second, the abundant choices of SERS active substrates make the detection flexible - in vitro, ex vivo or in vivo. For instance, the general substrates in Fig. 2B are feasible for in vitro or ex vivo applications; as for the in vivo studies, the proposed plasmonic needles are promising to be precisely locate near cells or in tissues for investigations on local biomolecules [72,89]. Third, the high specificity of Raman spectra benefits the high-throughput screening in metabolomic studies. Also, SERS allows the integration with other techniques (e.g., chromatography) on metabolites sensing to enhance the accuracy, multiplexing capability and specificity [90].
3. SERS detection of cellular metabolites
3.1. Cellular metabolite detection in physiological states
Cellular microenvironment is complicated, as the rapid metabolic kinetics leads to concentration variations, structural diversity and high complexity of analyte medium. Also, the relatively low concentrations make it challenging to study metabolites at the cellular level. To circumvent the above problems, a SERS-microfluidic droplet platform was reported [91]. The multi-functional magnetic SERS substrates were composed of Ag NPs modified Fe3O4 microspheres. Owing to efficient adsorption of analytes and aggregation-inducing ability of magnetic beads, numerous EM hotspots can be generated to obtain strong SERS signals. This study achieved the label-free, non-destructive, and synchronous quantification of three common cellular metabolites: pyruvate, ATP and lactic acid (Fig. 3A) [91]. The LOD value was 1.0, 0.1 and 0.01 pM, respectively. This SERS-microdroplet platform provided new insights for the design of SERS aggregates with dense hotspots.
Fig. 3.
(A) SERS detection based on Fe3O4@Ag NPs of normal cell metabolites, and the standard curves of SERS intensities versus ATP concentration (Reproduced with permission from Ref. [91]. Copyright 2019, American Chemical Society.). (B) SERS optophysiology distance curves experiments based on Au nanoraspberries and the extracellular ATP and urea events in different cell lines (Reproduced with permission from Ref. [92]. Copyright 2019, American Chemical Society.). (C) SERS detection based on Au nanospheres combined with a hydrogel-based 3D cancer model of metabolites in tumor extracellular media: adenine and hypoxanthine derivate metabolites, and the SERS mapping of hypoxanthine, followed by relative SERRS intensities after different incubation time with hemin (Reproduced with permission from Ref. [94]. Copyright 2020, John Wiley and Sons Inc.). (D) Dopant-functionalized 3D-metasensors and PCA from complete SERS spectra of different cell populations (Reproduced with permission from Ref. [96]. Copyright 2021, American Chemical Society.).
3.2. Cellular metabolite detection in disease/pathological states
By monitoring the variations in extracellular ATP concentration, it is potential to reveal chronic inflammatory diseases and cancers. Lussier et al. reported a non-invasive strategy to simultaneously monitor gradients of ATP and other metabolites of endothelial cells (HUVEC) and tumor cells (HeLa) [92]. In this work, the plasmonic nanosensors composed of Au nano-raspberries were located above cells. Then metabolites diffusion from the cells to the surface of nanosensors led to the enhancement in Raman spectra (Fig. 3B). SERS signals provided complex information about the cellular uptake and secretion of different metabolites. The results showed that the extracellular ATP of HUVEC and HeLa cells increased significantly, revealing the significance of extracellular ATP in tumor micro-environments.
Cancer-related metabolites can be specifically detected by SERS. In a demonstration work, lactic acid and thiocyanate in a hydrogel tumor phantom model were detected using Au nanostars and Ag-plated Au nanorods SERS biosensors [93]. Similarly, Plou et al. developed a SERS plasmonic substrate containing Au superlattices for cancer-associated metabolite detection [94]. The tumor cell culture supernatant was collected and deposited on the self-assembled SERS superlattice nanostructures. This is beneficial for nondestructive and multiplexing detection of extracellular metabolites relevant to tumor biology. The analysis of kynurenine (a secretory immune-modulating derivative of cancer metabolism) and related molecules tryptophan were achieved with a LOD of 1 μM. Other adenine and hypoxanthine derivate metabolites were also detected (Fig. 3C). This study demonstrated the feasibility of SERS detection on extracellular tumor metabolites under diverse cell culture conditions. Erzina et al. reported Au multi-branched SERS NPs (AuM) for the accurate identification of normal cells and tumor cells [95]. These NPs were modified with different functional groups for partially-selective capture of molecules from the cell culture medium. The convolutional neural network (CNN) was applied and the training results were evaluated using ad hoc feature selection methods. In this way, the SERS data sets of normal and tumor-related metabolites were recognized with 100% predicting accuracy. This work indicated that the introduction of functional groups could improve the specificity in capturing proteins, nucleic acids, and lipids, benefiting the distinguishment between normal cells and tumor cells.
SERS metabolite detection could also be applied in the prediction of cancer metastasis. Tumor metastasis generally originates from primary cancer cells, and in this evolution process, a specific population called metastatic cancer stem cell-like cells (MCSC) was considered to be related to cancer metastasis [96]. Dharmalingam et al. invented an ultra-sensitive 3D metasensor to effectively capture non-sticky MCSC and to amplify important pre-metastatic signals from individual cells. This 3D metasensor was made through the ultrafast laser ionization technology, which consisted of self-assembled 3D tissue nanoprobes and dopant (Fig. 3D). The Raman signal of trace-level MCSC could be amplified through the charge transfer and plasmon resonance via the introduction of dopant. As a result, the subsequent signals (metabolism, proliferation, and metastasis) in MCSC could be detected. By furthering applying principal component analysis (PCA), multispectral evaluation can accurately locate metabolic dynamics in MCSC and predict the occurrence of metastasis. This technique demonstrated the potential of SERS sensors in treatment prognosis and metastasis prediction.
3.3. Neurotransmitters
The neuronal activities are closely related to some key neurotransmitters (glutamate, γ-aminobutyric acid, and dopamine etc.). SERS can be used to simultaneously detect these neurotransmitters near neurons. Lussier et al. reported a dynamic SERS (D-SERS) nanosensor invented as a patch clamp nanopipette modified with Au nano-raspberries [97]. It could be accurately positioned in any areas containing analytes under a microscope, to measure ATP, glutamate, acetylcholine (ACh), GABA and dopamine at the same time. The SERS spectra of these neurotransmitters were further analyzed by barcode data processing methods. This D-SERS nanosensor could be a robust tool for the study on the secretion of other cell lines.
4. SERS detection towards metabolites in biofluids and tissues
4.1. SERS detection of metabolites in biofluids in physiological states
4.1.1. In cerebrospinal fluids
Accurate analysis of disease-related metabolites in human body fluids brings a new avenue to understand pathological mechanisms, enabling early diagnosis of underlying diseases. By taking advantages of highly specific and efficient enzymatic reactions, analytes can be detected optically or electrochemically for metabolism studies [98]. Using enzyme-based strategies, Liu et al. reported a biomimetic SERS-based nanoreactor for glucose quantification in cerebrospinal fluids. Briefly, borate probes and enzymes were loaded in mesoporous silica NPs. With the presence of enzyme, the reaction of glucose produced H2O2, which oxidized borate probes to its corresponding phenol form. As a result, SERS intensity at 882 cm−1 raised along with the increase of H2O2 concentration, while that at 998 cm−1 rarely changed, so the peak intensity ratio (I882/I998) was analyzed to determine the concentration of glucose (Fig. 4A). The linear dynamic range was 1–100 μM with the LOD of 0.47 μM. The results measured on the samples from two patients were consistent with that obtained using the clinical glucose detection kits, proving the clinic potential of this nanoreactor. This work showed that ratio metric measurement could be a reproducible and robust strategy for metabolite detection.
Fig. 4.
(A) Glucose detection in patient cerebrospinal fluids determined with nanoreactor based on ratiometric SERS response to the enzymatic by-product H2O2 (Reproduced with permission from Ref. [98]. Copyright 2020, John Wiley and Sons Inc.). (B) PCA and PLS analysis of pregnancy and tetrahydrocortisone in urine for miscarriage prediction (Reproduced with permission from Ref. [84]. Copyright 2020, American Chemical Society.). (C) PCA for metabolites in the urine of normal, pancreatic cancer, and prostate cancer patients based on the AgNW-GFF sensor (Reproduced with permission from Ref. [105]. Copyright 2021, American Chemical Society.). (D) SERS detection of sulfodibimane (representing polysulfides in tumor tissue) (Reproduced with permission from Ref. [107]. Copyright 2021, Elsevier.).
4.1.2. In blood
To reduce the interferences from large metabolites in the blood, a microdialysis-SERS method was used and improved the sensitivity in lactic acid detection [99]. In this work, a 13 kDa cut-off dialysis membrane was used. The dialysate was automatically blended with Ag colloidal NPs in the microfluidic chamber for SERS detection. The linear range of lactic acid measurement was 10−5–3 × 10−4 M with an R2 value of 0.99. This work demonstrated the advantages of microdialysis in improving the sensitivity of SERS metabolite detection.
4.1.3. In urine
The main components in urine are water, urea, creatinine and uric acid. For the SERS detection of urea biomolecules, Ag–Au compound SERS substrates were reported [100]. The Au porous nanostructure exhibited large surface area with more adsorption sites and plasmonic hotspots. Compared with Ag films, the SERS enhancement factor of this porous substrate was improved by 6 times, allowing the trace detection of urine metabolites. Its detection linear range covered a wide range of concentrations from 1 to 20 mM with the LOD of 1 mM. In another work on urea detection, SERS substrates comprised of closely spaced metal nanodomes were placed in a flexible tube used for intravenous administration and catheters [101]. The SERS peaks of promethazine and urea were used for individual testing and cross testing. This work aimed to improve the safety of intravenous administration through monitoring the concentration of promethazine and urea.
Creatinine is a protein produced by muscle and released into the blood. Its level in healthy people is relatively stable. Therefore, it is the most used urine component in renal clearance tests and provides details on the status of the kidneys. Wang et al. collected human urine samples from ten healthy people and detected creatinine in these samples by SERS [102]. The partial least squares cross-validation (PLSCV) method was then used to obtain the estimated creatinine concentration in the clinically relevant concentration range (55.9–208 mg/dl), and the root-mean square error of cross validation was 26.1 mg/dl. This work proved the viability of SERS detection towards human urine creatinine.
Similarly, with the use of partial least square (PLS), a “limit and capture” method was reported for multiple detection of two urinary metabolite families associated with the risk of miscarriage [84]. First, surface chemistry was applied on the self-assembled single-layer substrate grafted with Ag nanocubes, making it possible for the substrate to capture metabolites from the complicated urine matrix. As the SERS substrate was also hydrophobic, metabolites could be confined in the hotspots generated from the SERS nanomaterials (Fig. 4B). PCA was used to clarify the spectral changes that occur when urine metabolites were combined with the capture agent, to identify the molecular fingerprints corresponding to the target metabolites and eliminate background interferences. After that, PLS analysis converted all molecule fingerprints in the complete spectral window into quantifiable SERS readings, generating a detection range spanning 7 orders of magnitude in concentrations. Moreover, PLS analysis could also detect small changes in the concentration of metabolites at the level of 0.1 nM in urine, allowing rapid (within 30 min) quantitative screening of key metabolites in patient samples. This method brings new strategies of substrate design to improve the sensitivity and selectivity of SERS metabolite detection.
4.1.4. In fingerprints and tears
The most common indicator of kidney disease is the level of urea in blood or urine, and therefore, the urea detection was usually carried out in blood or urine samples. Early in 2012, an alternative method to detect urea by examining and comparing SERS spectra from different biological fluids (tears and fingerprints) was proposed, using a thin film of Au NPs as SERS substrates [103]. The results indicated the possibility of using other alternative biological fluids to monitor the urea content in the body. In contrast to the blood urea nitrogen test, the SERS analysis of tears and fingerprints produced by sebaceous and exocrine gland secretions was a rapid and non-invasive method for the early diagnosis of kidney disease.
4.2. SERS detection of metabolites in bio-fluids and tissues in disease/pathological states
4.2.1. Metabolites in bio-fluids in disease/pathological states
Metabolites are promising biomarkers for cancer diagnosis. The detection of the serum metabolic profile of hepatocellular carcinoma patients through SERS was firstly reported in 2016 [104]. It compared the SERS spectra of serum from hepatocellular carcinoma, breast cancer, and lung cancer patients to discover the changes in serum metabolism of patients. The Raman spectrum of the hepatocellular carcinoma serum biomarker alpha-fetoprotein was detected for the first time, and the significant peaks of hepatocellular carcinoma were analyzed. This work demonstrated the potential of hepatocellular carcinoma diagnosis based on the detection of patients’ serum through SERS.
For the diagnosis of pancreatic cancer and prostate cancer, the stacked Ag nanowires on glass fiber filters have been proved to be useful (Fig. 4D) [105]. In this work, calcium ions were added to enhance the absorption of metabolites to Ag nanowires. The detection of xanthopterin, inosine, hypoxanthine, xanthine and urea was achieved. Then, multivariate analysis of SERS spectra using unsupervised PCA and supervised orthogonal PLS discriminant analysis was applied. From the results, the discrimination between the pancreatic and prostate cancer group was rather effective (with a sensitivity of 100% and a specificity of 100%), so as the discrimination between the normal group and the combined cancer group. This study indicated that PCA and PLS improved the accuracy of diagnosis in a large number of patients’ statistics.
Except for blood and urine, saliva might be the ideal sample for non-invasive cancer (oral cancer) detection because it contains a large number of metabolites related to personal health conditions. Falamas et al. studied the SERS spectra of saliva using Au NPs [106]. The reproducible spectra were obtained at certain locations on the SERS map. Based on Raman bands of amino acids and proteins, the healthy and oral cancer saliva could be distinguished. This work indicated the possibility of SERS detection on saliva for oral cancer diagnosis.
4.2.2. Metabolites in tissues in disease/pathological states
Honda et al. reported that the detection of polysulfides on the tissue could predict the prognosis of cisplatin-based chemotherapy [107]. Au NPs based SERS substrate was utilized to enhance the electromagnetic field to sense a variety of sulfur-containing metabolites-S (Fig. 4E). As a result, SERS spectra of cancer regions of clear cell carcinoma (CCC) revealed that CCC showed a characteristic Raman peak at 480 cm−1 in cancer stromal regions. The CCC patients with higher SERS signal exhibit significantly shorter survival time than those with lower SERS signal, indicating that the SERS intensity at 480 cm−1 could be used to predict the overall survival time of CCC patients. The SERS signal further indicated the presence of polysulfides in cancer tissues. This work proved that the anti-tumor effects could be evaluated through detecting the specified metabolites change by SERS.
To distinguish different tumor models, SERS was reported to detect metabolites from tumor lysate with high sensitivity and chemical specificity [108]. In this work, liquid chromatography (LC) was performed, and this LC-SERS method presented a metabolite detection capability comparable to LC-MS, showing selectivity towards the detection of different metabolite subsets. Analysis of repeated LC-SERS experiments showed reproducible metabolite patterns, which could be converted into barcodes to distinguish different tumor models. This work demonstrated that SERS could be integrated with other techniques (chromatography, etc.) for higher selectivity in metabolite detection.
4.3. Metabolites of drugs
4.3.1. Anti-tumor drugs
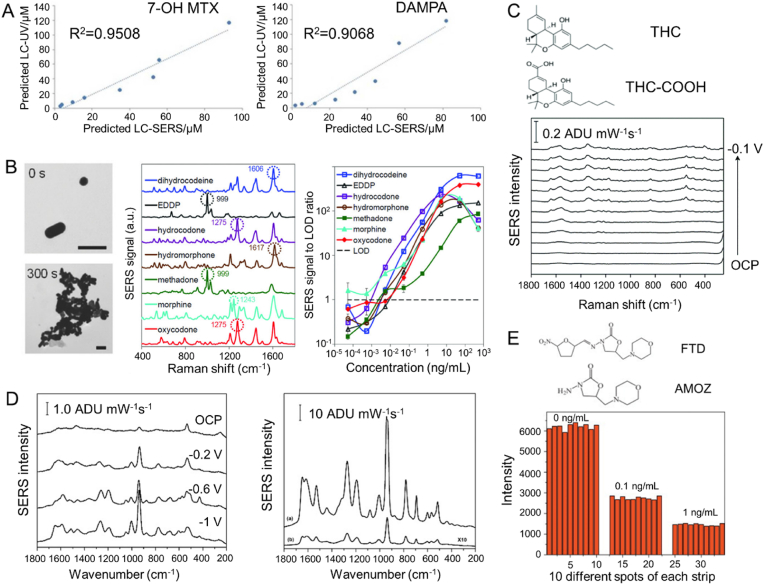
For the quantitative detection of the anti-tumor drug methotrexate and its main metabolites in urine, Subaihi et al. reported the combination of SERS with reversed-phase liquid chromatography (RP-LC) [109]. In this work, gradient elution was applied, and the composition of the mobile phase was gradually changed in the analytic process. The identification of methotrexate and its metabolites 7-hydroxy methotrexate (7-OH MTX) and 2,4-diamino-N (10)-methylpteroic acid (DAMPA) was based on their specific SERS spectra, providing LOD of 2.36, 1.84, and 3.26 μM, respectively (Fig. 5A). It proved that SERS was suitable for the detection of analytes without UV absorption or analytes not ionized for MS-based detection.
Fig. 5.
(A) Calibration plots of predicted LC-UV versus LC-SERS from 7-OH MTX and DAMPA with high R2 (Reproduced with permission from Ref. [109]. Copyright 2017, American Chemical Society.). (B) SERS spectra and LOD of various opioids metabolites with halide-conjugated gold nanoclusters (Reproduced with permission from Ref. [110]. Copyright 2018, John Wiley and Sons Inc.). (C) EC-SERS for THC-COOH detection in urine samples from 0 V to −1.0 V (Reproduced from Ref. [111] with permission from the Royal Society of Chemistry.). (D) EC-SERS spectra for 6-TUA at different potentials and the comparison of SERS signals in synthetic urine and NaF (Reproduced with permission from Ref. [118]. Copyright 2017, American Chemical Society.). (E) Ultrasensitive ICA based on SERS towards the metabolite of FTD AMOZ with high reproducibility (Reproduced with permission from Ref. [121]. Copyright 2015, Elsevier.).
4.3.2. Psychotropic drugs
With the popularization of anesthetics, the demand is increasing to detect anesthesia drugs and their metabolites in body using a rapid, non-invasive, and quantitative method. At present, opioid detection is mainly carried out on urine samples, including immunoassay screening, followed by confirmation analysis using gas chromatography MS (GC-MS). However, it may take several days to obtain results. Therefore, the rapid clinical identification of opioids is a challenge. In this case, a quantitative direct SERS strategy was reported [110]. In this work, citrate stabilized Au NPs were firstly mixed with opioids such as morphine. At this stage, NP aggregates were rare. The alkali metal halide was then added to replace the citrate ions on NP surface, allowing the adsorption of opioid molecules and leading to NP aggregates. This helped to generate more SERS hotspots. The obtained LOD in urine was equivalent to GC-MS (100 ng/mL in urine) (Fig. 5B). The work provided a novel strategy for nanocluster formation and SERS enhancement to improve the sensitivity of metabolite detection.
With the legalization of marijuana in several countries, more studies aim to develop advanced methods to monitor marijuana. The rapid detection of tetrahydrocannabinol (THC) and its main secondary metabolite carboxy-tetrahydrocannabinol (THC-COOH) via electrochemical SERS (EC-SERS) was firstly reported in 2020 [111]. In this study, the applied voltage was proved to be essential for effective SERS detection of trace cannabinoids in body fluids (Fig. 5C). What's more, it was predicted that the presence of carboxylic acid groups in THC-COOH brought stronger surface adsorption of the molecules compared to THC. Thus, it suggested that EC-SERS might be a more suitable method for detecting THC-COOH in urine, and could be used as a favorable strategy for cannabis monitoring.
The drug nicotine and its main xenogeneic metabolites cotinine and trans 3′-hydroxycotinine were also reported to be quantified by SERS [112]. In this work, SERS measurements could be conducted in the tertiary mixture containing nicotine, cotinine and trans 3′-hydroxycotinine with the concentration range of 10−7-10−5 M. Then, a stoichiometric analysis using kernel PLS (K-PLS) and artificial neural networks (ANN) was performed, and these models were proved using bootstrap resampling. As a result, the prediction of nicotine was most accurate, followed by that of cotinine. This work proved the advantages of SERS as the process was rapid without the use of time-consuming chromatography.
The abuse of psychiatric drugs is often associated with death. Therefore, the detection of psychiatric drugs is critical to clinical and forensic toxicology. Tricyclic antidepressants (TCA) are the first generation of antidepressants that use imipramine (Imi) as a precursor. The widely used method for TCA detection is immunoassay, which, however, could not identify the drug from its metabolites. The qualitative and semi-quantitative analysis of TCA could be achieved via SERS [113]. The SERS spectra of aqueous solution of Imi and its metabolite desipramine (Des) were collected and the LOD of Imi in Ag colloidal solution was 0.98 μM.
Besides TCA, caffeine is considered as a widely used psychoactive drug. It has been reported that caffeine as well as its major metabolite paraxanthine could be detected via EC-SERS [114]. Then, density functional theory (DFT) calculations were performed on the caffeine and paraxanthines in the saliva sample to further study the adsorption of biomolecules and assign each Raman band. The bands were determined to be related to paraxanthines (Fig. 5D), confirmed by the pharmacokinetic study of paraxanthine and caffeine in human saliva. In a similar work, SERS combined with machine learning achieved the quantification of caffeine and its two major metabolites (paraxanthine and theobromine) rapidly [115]. The above two studies were both carried out over the detection of caffeine and its metabolites, and both applied algorithms to improve the accuracy or the speed of the detection process.
To assess antipsychotic drug clozapine poisoning by detecting clozapine metabolites in urine samples, a SERS method based on the coffee ring effect was reported [116]. In coffee ring regions, more hotspots were formed, improving the detection sensitivity. The tested linear range was 0.5–50 μg/mL (covering the toxicity range) with adequate reproducibility. This strategy provides a feasible way to generate hotspots for sensitive detection.
Some weight-loss pills take effect by acting on the nervous system to suppress appetite. Sibutramine hydrochloride (SH) is an illegal drug for losing weight, but its banned addition into health products still remains, leading to the urgent demand for SH detection. Ouyang et al. reported the quantitative SERS detection of SH metabolites [117]. To overcome the weak affinity between SH and bare metal, β-cyclodextrin was covered in situ on the surface of Ag NPs to capture SH. Then the obtained Ag NPs were coated with polyvinyl alcohol (PVA) to prepare a SERS active hydrogel with excellent reproducibility. In this way, the quantification of trace amounts of SH was achieved, with a LOD as low as 3.0 g mL−1. This work implied that surface modification of SERS substrates could be applied for better metabolites capture ability.
4.3.3. Immunologic drugs
The most common type (75%) of childhood leukemia is acute lymphoblastic leukemia (ALL). There are many treatment options for ALL, including chemotherapy, radiation therapy, and bone marrow transplantation (BMT). One immunosuppressive drug commonly used in BMT is 6-mercaptopurine (6-MP). 6-thiouric acid (6-TUA) is one of the main metabolites of 6-MP, excreted through urine. Low-concentration quantitative detection of 6-TUA is regarded as a marker for the evaluation of immunosuppressive therapy effects for patients. EC-SERS was reported to study the adsorption and electrochemical behavior of 6-TUA on the surface of nanostructured Ag electrodes [118]. EC-SERS provided significant signals for 6-TUA in urine at a concentration as low as 1 μM, highlighting the huge potential of EC-SERS in detecting drug metabolites in urine (Fig. 5D).
6-thioguanine nucleotides (6-TGNs) and 6-methylmercaptopurine (6-MMP) are metabolites of two thiopurines (azathioprine, or its prodrug mercaptopurine), which can be used for inflammation bowel disease treatments. Both 6-TGN and 6-MMP may have side effects depending on their concentration. It is thus important to monitor their concentrations in body. Traditionally, their metabolites were quantified by HPLC. Through EC-SERS, 6-TGN and 6-MMP could also be quantified effectively and rapidly [119]. Compared with the conventional SERS detection with the LOD of 1 μM, EC-SERS helped to reduce the LOD of 6-TGN to 0.01 μM and the LOD of 6-MMP to 0.1 μM, well satisfying the clinical requirements. Moreover, through optimizing the SERS mapping process, the standard deviations were less than 10%. This work informed that the integration of EC-SERS and mapping was potential for sensitive and accurate quantitative analysis of trace concentrations, widely applicable to any EC-SERS active analyte.
4.3.4. Aspirin
Since aspirin is available as an over-the-counter drug, its overdose brings high risk and causes serious health symptoms. The application of SERS was introduced to detect salicylic acid (a metabolite of aspirin) in patient serum [120]. Under near-infrared (NIR) laser excitation, label-free colloidal SERS (spherical Ag NPs with citrate-terminated) could be used to detect salicylic acid at a concentration as low as 3 mM in serum, corresponding to the consumption of at least eight standard aspirin pills (mild toxicity). This diagnostic method based on SERS is more rapid than the conventional LC and MS, and can make a sensitive diagnosis of aspirin consumption in blood.
4.3.5. Illegal drugs
Furaltadone (FTD) is a nitrofuran antibiotic that has been banned due to its potential harm to human health. 3-amino-5-methylmorpholino-2-oxazolidinone (AMOZ) is a metabolite of FTD, which is stable in the tissue matrix and can be used as a marker of FTD use. A new ultra-sensitive competitive immunochromatography based on SERS has been developed for the direct detection of AMOZ in tissue and urine samples [121]. A monoclonal antibody (mAb) against AMOZ and Raman reporter 4-mercaptobenzoic acid (MBA) double-labeled colloidal Au NPs was prepared and used as an immune probe (Fig. 5E). The specific Raman scattering intensity of MBA on the immunochromatography test line was measured for the quantification of AMOZ, which could be finished in 15 min with high reproducibility. The IC50 value (when the binding rate is 50%, the corresponding concentration of the inhibitory substance is called IC50. Generally, when the value of IC50 is lower, the specificity of the antibody is higher) and LOD of AMOZ were 0.04 ng/mL and 0.28 pg/mL, respectively. The ICA strip could be used for 7 weeks without losing activity. The combination of immunochromatography with SERS improved the selectivity towards specified metabolites.
Among the illicit drugs, methamphetamine (MAMP) is the second widely abused. To evaluate the abuse of MAMP, various sample matrices, such as urine and blood, are required to be analyzed on-site. The detection of trace amounts of MAMP and its main metabolite amphetamine in urine via a nanostructured SERS chip was reported [122]. In this work, the SERS substrate were self-assembled vertically arranged Ag nanorods prepared by magnetron sputtering technology. Before the detection process, the urine samples were acidified to remove organic urea by-products. The remaining dissolved methamphetamine/amphetamine in the urine sample were used for SERS analysis with the LOD of 50 ng/mL, indicating the potential of SERS for the rapid detection of illicit drug in real samples. This work again reminded the importance of the pretreatment of real samples from patients to decrease the interferences of other biomolecules and improve the accuracy of SERS detection.
Here we summarized the metabolites, SERS substrates, assay types, LOD, and dynamic range for all the studies mentioned above (Table 2). It can be found that, over cellular level, the common LOD was at the level of 10−6 M, and the level of 10−12 M was possibly achieved in one study; in bio-fluids, the common LOD was at the level of 10−3 M; as for drug metabolites, the common LOD was at the level of 10−6 M, and the LOD could be low as 10−9 M in some work. For the fabrication of SERS substrate, Au and Ag are the most preferred with the form of NPs. For metabolites over cells and in bio-fluids, assays were mainly carried out on the solid medium; for drug metabolites, liquid assays were more common.
Table 2.
A summary of various metabolites and the corresponding SERS detection enhancement substrates, assay, limit of detection (LOD) and dynamic range using SERS technique.
| Metabolites | SERS Substrate | Assay Type |
LOD | Dynamic range | Ref. | |
|---|---|---|---|---|---|---|
| In Normal cells | Lactate | Fe3O4@Ag NPs | Droplet | 10−14 M | 10−14-5 × 10−12 M | [91] |
| Au@Ag NRs | Hydrogel | 10−7 M | 10−7-10−3 M | [93] | ||
| ATP | Fe3O4@Ag NPs | Droplet | 10−13 M | 10−13-5 × 10−11 M | [91] | |
| Pyruvate | Fe3O4@Ag NPs | Droplet | 10−12 M | 10−12-5 × 10−10 M | [91] | |
| In Cancer Cells | Kynurenine | Au NPs | Glass | 10−6 M | 10−6-10−4 M | [94] |
| Tryptophan | Au NPs | Glass | 10−6 M | 10−6-10−4 M | [94] | |
| In Cerebrospinal fluids | Glucose | Si foams with Raman reporters | Solution | 4.7 × 10−7 M | 4.7 × 10−7-10−4 M | [98] |
| In Blood | Lactic acid | Au NPs | Solution | 10−5 M | 10−5-3 × 10−4 M | [99] |
| In Urine | Urea | Ag film | Silicon wafer | 0.013 M | 0.013–0.17 M | [101] |
| Ag–Au compound NP film | Glass | 10−3 M | 10−3-0.02 M | [100] | ||
| Creatinine | Au NPs | Solution | 0.559 mg/mL | 0.559–2.08 mg/mL | [102] | |
| Pregnane | Ag layer | Silica bead | 10−14 M | 10−14-10−4 M | [84] | |
| In Fingerprints | Human IgG | Ag NPs | Glass | 10−7 mg/mL | 10−7-10−1 mg/mL | [123] |
| Cancer related metabolites | Pyrimidine and purine metabolites | Ag nanowires | Glass | 1.77 × 10−9 M | [105] | |
| Drug metabolites | 7-OH MTX | Ag NPs | Solution | 2.87 × 10−6 M | [109] | |
| DAMPA | Ag NPs | Solution | 1.66 × 10−6 M | [109] | ||
| Opiods | Au nanoclusters | Solution | 5 × 10−11 mg/mL | 5 × 10−11-5 × 10−6 mg/mL | [110] | |
| Paraxanthine | Ag NPs | Solution | 1.5 × 10−5 M | [114] | ||
| Theobromine | Au colloid | Solution | 5 × 10−7 M | 5 × 10−7-10−5 M | [115] | |
| 6-TUA | Ag colloid | Solution | 10−6 M | 10−6-10−3 M | [118] | |
| 6-TGNs | Au NPs | Solution | 10−8 M | [119] | ||
| 6-MMP | Au NPs | Solution | 10−7 M | [119] | ||
| Salicylic acid | Au NPs | Solution | 3 × 10−3 M | 3 × 10−3-0.01 M | [119] | |
| AMOZ | Au NPs | Nitrocellulose membrane | 2.8 × 10−10 mg/mL | 2.8 × 10−10-10−4 mg/mL | [121] | |
5. Challenges and optimization methods of SERS detection
SERS platforms are promising for the sensitive and multiplexed screening of metabolites. However, this technology is still under development for clinical translations. In this section, we elucidate the main concerns regarding the current challenges as well as possible methods to optimize SERS-based metabolite screening.
5.1. Toward high sensitivity
High sensitivity is the primary requirement for the analysis of a trace number of biological molecules. Although SERS allows the detection down to a single-molecule level, many metabolites show low affinity to plasmonic surfaces and exhibit small Raman cross-sections [84]. As mentioned before, SERS-based metabolite screening is commonly applied in a label-free approach, i.e., directly enhancing Raman signals of target molecules in plasmonic hotspots. In this process, the following factors should be considered.
-
(1)
The preparation of nanostructures with engineered hotspots. This can be fulfilled through forming particle aggregations, enlarging the surface area, and fabricating plasmon-coupling structures [[124], [125], [126]]. For example, colloidal aggregates are preferred in liquid phase, and nanopillar patterns [127], film layers [100] as well as porous structures [100] are frequently reported on a planar substrate. By dripping colloids on a substrate, the random agglomerates in “coffee ring” can be formed [116]. Additionally, SERS-magnetic complex can be introduced for the enrichment and separation of targets as well as the formation of additional hotspots [91].
-
(2)
The trap of metabolites into these hotspots. There are many types of metabolic molecules with varied structures, sizes, and concentrations, and corresponding strategies are therefore applied for different situations. Liquid analytes can be physically confined on planar SERS substrates. For example, super-hydrophobic SERS-active substrates were reported for the enrichment of droplets containing analytes and plasmonic colloids [84]. For solid substrates, the molecules with affinity to targets can be applied as surface-grafted agents to enhance the trapping ability. For instance, beta-cyclodextrin coated Ag substrate can enhance the capture of sibutramine hydrochloride [117].
-
(3)
The propagation of optical signals. For applications that integrated the primary sensors with other devices (e.g., flasks, microfluidic devices), the transmission of optical signals might be difficult. To overcome this spatial constraint, a waveguide could be considered to drive optical signals near plasmonic surfaces [128].
-
(4)
Pretreatment. We should also note that the direct detection of metabolites in complicated environments is still challenging, partly due to the competitive adsorption of other molecules. It is thus necessary to filter interfering molecules in the pretreatment step. For example, salt and water-soluble molecules can be removed through a silica column filtration [84]. Membrane filtration attached inside a micro-channel was also employed to filtrate and concentrate bacteria [129]. Micro-dialysis is useful for in vivo sampling of molecules in body fluids and has been applied for the SERS detection of lactic acids [99]. The removal of interfering molecules highly improves the LOD of SERS screening.
5.2. Quantitative SERS detection
Quantitativeness is an eternal theme in the SERS field. Quantitative SERS examination of metabolites from body fluidics remains problematic, which is mainly due to the poor signal reproducibility caused by random aggregations or nonuniform distribution of SERS NPs within the laser spot, or inhomogeneous adsorption of analytes. During past decades, a variety of methods has been adopted to circumvent this issue, and in the following we outlined three major approaches.
-
(1)
Uniform SERS substrates preparation and molecular distribution. The uniform substrate has proved to be feasibly achieved through micro-fabrication techniques. In contrast, it is challenging to keep uniform molecular distribution, particularly at ultralow concentration, due to the stochastic distributions of single molecules. Therefore, the concept of digital SERS measurements holds the promise to revolutionize the quantitative analysis at low concentrations. Its key point is to digitalize the signal generated by the molecule in each hotspot, and the number of SERS event counts is proportional to the solution concentration [127]. Using this methodology, the specific quantification was reported on cytokine (LOD of attomolar levels) and enrofloxacin and ciprofloxacin (LOD of 2.9 pM) [127].
-
(2)
Signal calibration using ratio-metric strategies. This calibration relies on the intensity of an internal-standard peak, to eliminate intensity fluctuations induced by the NP concentration variation. For example, to sense small molecule metabolites, SERS substrates are firstly modified with the internal-standard molecules, then functionalized with probe molecules that are sensitive to target analytes. Upon the exposure to targets, the probe molecules undergo chemical conversion and induce a change in Raman signals. The intensity of generated molecules can be calibrated by internal peak intensity to correct SERS intensity variations and estimate the concentrations of targets. This ratiometric strategy has been employed to measure the changes of H2O2, cholesterol, and cerebrospinal fluid glucose [98,130,131].
-
(3)
Spectral analysis methods. As an important procedure for Raman spectral information read-out, the spectral analysis helps to identify specific features from the mixed components (such as PLS, CLS) [112], and cluster similar spectra (such as PCA) [84]. The spectral analysis can be jointly used with the above two methods to improve the quantification capability.
5.3. Multiplexing and selectivity in mixed samples
Considering that a physiological pathway always relates more than one kind of metabolites, high throughput detection adds to the accuracy and efficacy of physiological state monitoring. Also, the multiplexing capability is preferred to be evaluated along with the selectivity, since they together represent the accuracy in capturing multiple target molecules from a complex sample matrix.
-
(1)
Combination of SERS with other analytical techniques. Different analytical techniques have respective analyte selection rules. In complex body fluids, there are various unknown substances that are hard to distinguish only using SERS. Also, Raman spectroscopy database on metabolites is yet to be populated. Therefore, for the high-throughput screening, it is promising to integrate SERS with other commonly used analytical techniques or devices, such as GC, MS, LC and micro-channel chips. For example, the LC-SERS approach was reported to have a selectivity for the detection on metabolites in complex tumor lysate samples and show comparable detection capabilities to the LC-MS [108].
-
(2)
Machine learning assisted analysis. As an advanced form of spectral analysis, the emerging machine learning has assisted faster and more accurate SERS multiplexing profiling. For example, slightly altered SERS spectra referring to different types of metabolite molecules on the SERS substrates can be recognized using a flexible supervised machine learning approach. This algorithm is adopted to extract various features and allocate multiple spectra to the same metabolites [92]. Such machine learning assisted analysis increased the number of sensed metabolites and provided better selectivity and sensitivity.
6. Outlook
Looking into the future, SERS technology is promising to contribute to a wide range of fundamental studies and clinical translations for analyzing metabolites at the cellular level, in tissues, and in body fluids. The most fascinating yet challenging application is the real-time SERS monitoring of in vivo metabolites.
At present, a few works that use endoscopes for intestinal metabolite detection were reported, which can conveniently, non-invasively, and visually obtain digestion-related metabolite information for disease diagnosis and evaluation [132]. Therefore, the endoscopic SERS probe can be expected as a powerful tool for early diagnosis of metabolic diseases and cancer with two advantages: (i) Better selectivity. The key information of the biochemical research of the target tissue could be obtained through reading the characteristic Raman peak intensity; (ii) High sensitivity. When the sample contacts with a rough metal surface, the usually weak Raman signal level will be significantly enhanced.
Smart homes have been used in metabolite detection, such as smartphones, smart toilets, and electronic noses [133]. SERS technology, as a mature detection method, can be introduced into smart homes. For instance, SERS sensors can be incorporated into the monitoring system of smart toilets for health assessment. Recently, a smartphone system was proposed with Raman spectrometer integrated into the phone backside [134]. The wireless communication function of the smartphone could be used to connect with the cloud server for spectral analysis. This type of system could be potential for the real-time detection of sweats metabolites.
Wearable devices based on SERS-active nanomaterials have also been reported, since integrating optically active plasmonic materials on flexible substrates allows the direct and sensitive recognition of metabolites [[135], [136], [137]]. SERS-active wearable devices, such as T-shirts made of flexible SERS fibers, can be designed as sensitive sweat sensors, which can monitor health by measuring sweat electrolytes. Similarly, toothbrushes made of SERS plasmonic fibers can be the choice for the detection of metabolites in saliva. Furthermore, compared to wearable devices that only get access to the metabolites on skin, mostly in sweats, the implantable devices can collect more molecular information from the lymphatic system and bloodstreams, offering a wider range of applications. The implantable sensor based on Au NPs and hydrogel was recently reported with good biosafety and extended lifetime of more than two months [138]. The implantable SERS sensors are thus expected, to be located within several centimeters deep inside the body surface; In this scenario, in vivo SERS signal is detected noninvasively through skin with a spatially-offset or a transmitted Raman spectroscopy setup.
Nevertheless, the biggest challenge to bring in vivo metabolite SERS sensors to the clinic would be the selective detection of appropriate analytes from the complex physiological environments. Also, the in vivo sensors would face the biosafety and efficiency issues before getting approval from the Food and Drug Administration. Still, SERS technique proves to be a powerful tool for metabolite detection. A bright future is thus anticipated!
Data availability
Data sharing is not applicable to this article as no new data were created or analyzed in this study.
Author contributions
Yao Lu: Literature collection and summary, Writing – original draft. Li Lin: Funding acquisition, Writing - original draft of section 5 and 2.2, Review & editing. Jian Ye: Supervision, Funding acquisition, Writing – review & editing.
Declaration of competing interest
The authors declare that they have no known competing financial interests or personal relationships that could have appeared to influence the work reported in this paper.
Acknowledgement
We acknowledge the financial support from the National Natural Science Foundation of China (No. 81871401 and 81901786), the Science and Technology Commission of Shanghai Municipality (Nos. 19441905300 and 21511102100), Shanghai Jiao Tong University (No. YG2019QNA28 and No. YG2022QN006), and Shanghai Key Laboratory of Gynecologic Oncology. We thank Dr. Fan Li for helping reviewing the biological terminology during the manuscript revision.
Contributor Information
Li Lin, Email: linli92@sjtu.edu.cn.
Jian Ye, Email: yejian78@sjtu.edu.cn.
References
- 1.Damiani C., Gaglio D., Sacco E., Alberghina L., Vanoni M. Systems metabolomics: from metabolomic snapshots to design principles. Curr. Opin. Biotechnol. 2020;63:190–199. doi: 10.1016/j.copbio.2020.02.013. [DOI] [PubMed] [Google Scholar]
- 2.Dufour-Rainfray D., Lamberioux M., Boulard P., Guidotti M., Delaye J.B., Ribeiro M.J., Gauchez A.S., Balageas A.C., Emond P., Agin A. Metabolomics - an overview. From basic principles to potential biomarkers (part 2) Med. Nucl. 2020;44(3):158–163. doi: 10.1016/j.mednuc.2020.02.004. [DOI] [Google Scholar]
- 3.Johnson C.H., Ivanisevic J., Siuzdak G. Metabolomics: beyond biomarkers and towards mechanisms. Nat. Rev. Mol. Cell Biol. 2016;17(7):451–459. doi: 10.1038/nrm.2016.25. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Duncan K.D., Fyrestam J., Lanekoff I. Advances in mass spectrometry based single-cell metabolomics. Analyst. 2019;144(3):782–793. doi: 10.1039/c8an01581c. [DOI] [PubMed] [Google Scholar]
- 5.Roque W., Romero F. Cellular metabolomics of pulmonary fibrosis, from amino acids to lipids. Am. J. Physiol. Cell Physiol. 2021;320(5):C689–C695. doi: 10.1152/ajpcell.00586.2020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Seyler L., Kujawinski E.B., Azua-Bustos A., Lee M.D., Marlow J., Perl S.M., Cleaves H.J. Metabolomics as an emerging tool in the search for astrobiologically relevant biomarkers. Astrobiology. 2020;20(10):1251–1261. doi: 10.1089/ast.2019.2135. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Patti G.J., Yanes O., Siuzdak G. Innovation: metabolomics: the apogee of the omics trilogy. Nat. Rev. Mol. Cell Biol. 2012;13(4):263–269. doi: 10.1038/nrm3314. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Panahi N., Arjmand B., Ostovar A., Kouhestani E., Heshmat R., Soltani A., Larijani B. Metabolomic biomarkers of low BMD: a systematic review. Osteoporos. Int. 2021;32(12):2407–2431. doi: 10.1007/s00198-021-06037-8. [DOI] [PubMed] [Google Scholar]
- 9.Yotter R.A., Wilson D.M. Sensor technologies for monitoring metabolic activity in single cells - Part II: nonoptical methods and applications. IEEE Sens. J. 2004;4(4):412–429. doi: 10.1109/Jsen.2004.830954. [DOI] [Google Scholar]
- 10.Zenobi R. Single-cell metabolomics: analytical and biological perspectives. Science. 2013;342(6163) doi: 10.1126/science.1243259. [DOI] [PubMed] [Google Scholar]
- 11.Vasdekis A.E., Stephanopoulos G. Review of methods to probe single cell metabolism and bioenergetics. Metab. Eng. 2015;27:115–135. doi: 10.1016/j.ymben.2014.09.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Kucherenko I.S., Topolnikova Y.V., Soldatkin O.O. Advances in the biosensors for lactate and pyruvate detection for medical applications: a review. Trac. Trends Anal. Chem. 2019;110:160–172. doi: 10.1016/j.trac.2018.11.004. [DOI] [Google Scholar]
- 13.Bispo D.S.C., Jesus C.S.H., Marques I.M.C., Romek K.M., Oliveira M.B., Mano J.F., Gil A.M. Metabolomic applications in stem cell research: a review. Stem Cell Rev. Rep. 2021;17(6):2003–2024. doi: 10.1007/s12015-021-10193-z. [DOI] [PubMed] [Google Scholar]
- 14.Moon J.M., Thapliyal N., Hussain K.K., Goyal R.N., Shim Y.B. Conducting polymer-based electrochemical biosensors for neurotransmitters: a review. Biosens. Bioelectron. 2018;102:540–552. doi: 10.1016/j.bios.2017.11.069. [DOI] [PubMed] [Google Scholar]
- 15.Arumugasamy S.K., Chellasamy G., Gopi S., Govindaraju S., Yun K. Current advances in the detection of neurotransmitters by nanomaterials: an update. Trac. Trends Anal. Chem. 2020;123 doi: 10.1016/j.trac.2019.115766. [DOI] [Google Scholar]
- 16.Mobed A., Hasanzadeh M., Ahmadalipour A., Fakhari A. Recent advances in the biosensing of neurotransmitters: material and method overviews towards the biomedical analysis of psychiatric disorders. Anal. Methods-Uk. 2020;12(4):557–575. doi: 10.1039/c9ay02390a. [DOI] [Google Scholar]
- 17.Gao P., Huang X., Fang X.Y., Zheng H., Cai S.L., Sun A.J., Zhao L., Zhang Y. Application of metabolomics in clinical and laboratory gastrointestinal oncology. World J. Gastrointest. Oncol. 2021;13(6):536–549. doi: 10.4251/wjgo.v13.i6.536. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Si Y., Lee H.J. Carbon nanomaterials and metallic nanoparticles- incorporated electrochemical sensors for small metabolites: detection methodologies and applications. Curr. Opin. Electroc. 2020;22:234–243. doi: 10.1016/j.coelec.2020.08.007. [DOI] [Google Scholar]
- 19.Farthing D.E., Farthing C.A., Xi L. Inosine and hypoxanthine as novel biomarkers for cardiac ischemia: from bench to point-of-care. Exp. Biol. Med. 2015;240(6):821–831. doi: 10.1177/1535370215584931. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Lisowska-Myjak B. Serum and urinary biomarkers of acute kidney injury. Blood Purif. 2010;29(4):357–365. doi: 10.1159/000309421. [DOI] [PubMed] [Google Scholar]
- 21.Rock K.L., Kataoka H., Lai J.J. Uric acid as a danger signal in gout and its comorbidities. Nat. Rev. Rheumatol. 2013;9(1):13–23. doi: 10.1038/nrrheum.2012.143. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Steckl A.J., Ray P. Stress biomarkers in biological fluids and their point-of-use detection. ACS Sens. 2018;3(10):2025–2044. doi: 10.1021/acssensors.8b00726. [DOI] [PubMed] [Google Scholar]
- 23.Smith L., Villaret-Cazadamont J., Claus S.P., Canlet C., Guillou H., Cabaton N.J., Ellero-Simatos S. Important considerations for sample collection in metabolomics studies with a special focus on applications to liver functions. Metabolites. 2020;10(3):e104. doi: 10.3390/metabo10030104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Lima A.R., Pinto J., Amaro F., Bastos M.D., Carvalho M., de Pinho P.G. Advances and perspectives in prostate cancer biomarker discovery in the last 5 Years through tissue and urine metabolomics. Metabolites. 2021;11(3):e181. doi: 10.3390/metabo11030181. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Araujo R., Bispo D., Helguero L.A., Gil A.M. Metabolomic studies of breast cancer in murine models: a review. Biochim. Biophys. Acta (BBA) - Mol. Basis Dis. 2020;1866(5) doi: 10.1016/j.bbadis.2020.165713. [DOI] [PubMed] [Google Scholar]
- 26.Dastmalchi F., Deleyrolle L.P., Karachi A., Mitchell D.A., Rahman M. Metabolomics monitoring of treatment response to brain tumor immunotherapy. Front. Oncol. 2021;11 doi: 10.3389/fonc.2021.691246. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Tang Z.Y., Xu Z.H., Zhu X., Zhang J.F. New insights into molecules and pathways of cancer metabolism and therapeutic implications. Cancer Commun. 2021;41(1):16–36. doi: 10.1002/cac2.12112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Yang B., Wang Y., Guo S., Jin S.L., Park E., Chen L., Jung Y.M. Charge transfer study for semiconductor and semiconductor/metal composites based on surface-enhanced Raman scattering. Bull. Kor. Chem. Soc. 2021;42(11):1411–1418. doi: 10.1002/bkcs.12387. [DOI] [Google Scholar]
- 29.Kim J., Sim K., Cha S., Oh J.W., Nam J.M. Single-particle analysis on plasmonic nanogap systems for quantitative SERS. J. Raman Spectrosc. 2021;52(2):375–385. doi: 10.1002/jrs.6030. [DOI] [Google Scholar]
- 30.Sun Y., Shi L., Mi L., Guo R., Li T. Recent progress of SERS optical nanosensors for miRNA analysis. J. Mater. Chem. B. 2020;8(24):5178–5183. doi: 10.1039/d0tb00280a. [DOI] [PubMed] [Google Scholar]
- 31.Fan M., Andrade G.F.S., Brolo A.G. A review on recent advances in the applications of surface-enhanced Raman scattering in analytical chemistry. Anal. Chim. Acta. 2020;1097:1–29. doi: 10.1016/j.aca.2019.11.049. [DOI] [PubMed] [Google Scholar]
- 32.Jiang L., Hassan M.M., Ali S., Li H.H., Sheng R., Chen Q.S. Evolving trends in SERS-based techniques for food quality and safety: a review. Trends Food Sci. Technol. 2021;112:225–240. doi: 10.1016/j.tifs.2021.04.006. [DOI] [Google Scholar]
- 33.Cialla-May D., Zheng X.S., Weber K., Popp J. Recent progress in surface-enhanced Raman spectroscopy for biological and biomedical applications: from cells to clinics. Chem. Soc. Rev. 2017;46(13):3945–3961. doi: 10.1039/c7cs00172j. [DOI] [PubMed] [Google Scholar]
- 34.Zhang Y., Liu Z., Thackray B.D., Bao Z., Yin X., Shi F., Wu J., Ye J., Di W. Intraoperative Raman-guided chemo-photothermal synergistic therapy of advanced disseminated ovarian cancers. Small. 2018 doi: 10.1002/smll.201801022. [DOI] [PubMed] [Google Scholar]
- 35.Bao Z., Zhang Y., Tan Z., Yin X., Di W., Ye J. Gap-enhanced Raman tags for high-contrast sentinel lymph node imaging. Biomaterials. 2018;163:105–115. doi: 10.1016/j.biomaterials.2018.02.020. [DOI] [PubMed] [Google Scholar]
- 36.Chakraborty A., Ghosh A., Barui A. Advances in surface-enhanced Raman spectroscopy for cancer diagnosis and staging. J. Raman Spectrosc. 2020;51(1):7–36. doi: 10.1002/jrs.5726. [DOI] [Google Scholar]
- 37.Reyes-Goddard J.M., Barr H., Stone N. Photodiagnosis using Raman and surface enhanced Raman scattering of bodily fluids. Photodiagn Photodyn. 2005;2(3):223–233. doi: 10.1016/S1572-1000(05)00066-9. [DOI] [PubMed] [Google Scholar]
- 38.Yu B., Ge M., Li P., Xie Q., Yang L. Development of surface-enhanced Raman spectroscopy application for determination of illicit drugs: towards a practical sensor. Talanta. 2019;191:1–10. doi: 10.1016/j.talanta.2018.08.032. [DOI] [PubMed] [Google Scholar]
- 39.Khlebtsov N.G., Lin L., Khlebtsov B.N., Ye J. Gap-enhanced Raman tags: fabrication, optical properties, and theranostic applications. Theranostics. 2020;10(5):2067–2094. doi: 10.7150/thno.39968. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Khlebtsov B., Khlebtsov N. Surface-enhanced Raman scattering-based lateral-flow immunoassay. Nanomaterials. 2020;10(11):e2228. doi: 10.3390/nano10112228. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Lussier F., Brule T., Vishwakarma M., Das T., Spatz J.P., Masson J.F. Dynamic-SERS optophysiology: a nanosensor for monitoring cell secretion events. Nano Lett. 2016;16(6):3866–3871. doi: 10.1021/acs.nanolett.6b01371. [DOI] [PubMed] [Google Scholar]
- 42.Zhang A., Sun H., Wang P., Han Y., Wang X. Modern analytical techniques in metabolomics analysis. Analyst. 2012;137(2):293–300. doi: 10.1039/c1an15605e. [DOI] [PubMed] [Google Scholar]
- 43.Feizi N., Hashemi-Nasab F.S., Golpelichi F., Saburouh N., Parastar H. Recent trends in application of chemometric methods for GC-MS and GCxGC-MS-based metabolomic studies. Trac. Trends Anal. Chem. 2021;138 doi: 10.1016/j.trac.2021.116239. [DOI] [Google Scholar]
- 44.Manzi M., Riquelme G., Zabalegui N., Monge M.E. Improving diagnosis of genitourinary cancers: biomarker discovery strategies through mass spectrometry-based metabolomics. J. Pharmaceut. Biomed. 2020;178 doi: 10.1016/j.jpba.2019.112905. [DOI] [PubMed] [Google Scholar]
- 45.Kulkarni A.S., Huang L., Qian K. Material-assisted mass spectrometric analysis of low molecular weight compounds for biomedical applications. J. Mater. Chem. B. 2021;9(17):3622–3639. doi: 10.1039/d1tb00289a. [DOI] [PubMed] [Google Scholar]
- 46.Gonzalez-Dominguez R., Gonzalez-Dominguez A., Sayago A., Gonzalez-Sanz J.D., Lechuga-Sancho A.M., Fernandez-Recamales A. Mechanistic insights into alzheimer's disease unveiled through the investigation of disturbances in central metabolites and metabolic pathways. Biomedicines. 2021;9(3):e298. doi: 10.3390/biomedicines9030298. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Yang J., Wang R., Huang L., Zhang M., Niu J., Bao C., Shen N., Dai M., Guo Q., Wang Q., Wang Q., Fu Q., Qian K. Urine metabolic fingerprints encode subtypes of kidney diseases. Angew Chem. Int. Ed. Engl. 2020;59(4):1703–1710. doi: 10.1002/anie.201913065. [DOI] [PubMed] [Google Scholar]
- 48.Huang L., Wang L., Hu X., Chen S., Tao Y., Su H., Yang J., Xu W., Vedarethinam V., Wu S., Liu B., Wan X., Lou J., Wang Q., Qian K. Machine learning of serum metabolic patterns encodes early-stage lung adenocarcinoma. Nat. Commun. 2020;11(1) doi: 10.1038/s41467-020-17347-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Wang R.C.C., Campbell D.A., Green J.R., Cuperlovic-Culf M. Automatic 1D H-1 NMR metabolite quantification for bioreactor monitoring. Metabolites. 2021;11(3):e157. doi: 10.3390/metabo11030157. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Lombo M., Ruiz-Diaz S., Gutierrez-Adan A., Sanchez-Calabuig M.J. Sperm metabolomics through nuclear magnetic resonance spectroscopy. Animals-Basel. 2021;11(6):e1669. doi: 10.3390/ani11061669. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Crook A.A., Powers R. Quantitative NMR-based biomedical metabolomics: current status and applications. Molecules. 2020;25(21):e5128. doi: 10.3390/molecules25215128. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Reimets N., Ausmees K., Vija S., Reile I. Developing analytical applications for parahydrogen hyperpolarization: urinary elimination pharmacokinetics of nicotine. Anal. Chem. 2021;93(27):9480–9485. doi: 10.1021/acs.analchem.1c01281. [DOI] [PubMed] [Google Scholar]
- 53.Dunn W.B., Wilson I.D., Nicholls A.W., Broadhurst D. The importance of experimental design and QC samples in large-scale and MS-driven untargeted metabolomic studies of humans. Bioanalysis. 2012;4(18):2249–2264. doi: 10.4155/bio.12.204. [DOI] [PubMed] [Google Scholar]
- 54.Sarbu M., Ica R., Zamfir A.D. Developments and applications of separation and microfluidics methods coupled to electrospray mass spectrometry in glycomics of nervous system gangliosides. Electrophoresis. 2021;42(4):429–449. doi: 10.1002/elps.202000236. [DOI] [PubMed] [Google Scholar]
- 55.Huang Y., Shi T., Luo X., Xiong H., Min F., Chen Y., Nie S., Xie M. Determination of multi-pesticide residues in green tea with a modified QuEChERS protocol coupled to HPLC-MS/MS. Food Chem. 2019;275:255–264. doi: 10.1016/j.foodchem.2018.09.094. [DOI] [PubMed] [Google Scholar]
- 56.Want E.J., Masson P., Michopoulos F., Wilson I.D., Theodoridis G., Plumb R.S., Shockcor J., Loftus N., Holmes E., Nicholson J.K. Global metabolic profiling of animal and human tissues via UPLC-MS. Nat. Protoc. 2013;8(1):17–32. doi: 10.1038/nprot.2012.135. [DOI] [PubMed] [Google Scholar]
- 57.La Nasa J., Modugno F., Degano I. Liquid chromatography and mass spectrometry for the analysis of acylglycerols in art and archeology. Mass Spectrom. Rev. 2021;40(4):381–407. doi: 10.1002/mas.21644. [DOI] [PubMed] [Google Scholar]
- 58.Emwas A.H. The strengths and weaknesses of NMR spectroscopy and mass spectrometry with particular focus on metabolomics research. Methods Mol. Biol. 2015;1277:161–193. doi: 10.1007/978-1-4939-2377-9_13. [DOI] [PubMed] [Google Scholar]
- 59.Langer J., Jimenez de Aberasturi D., Aizpurua J., Alvarez-Puebla R.A., Auguie B., Baumberg J.J., Bazan G.C., Bell S.E.J., Boisen A., Brolo A.G., Choo J., Cialla-May D., Deckert V., Fabris L., Faulds K., Garcia de Abajo F.J., Goodacre R., Graham D., Haes A.J., Haynes C.L., Huck C., Itoh T., Kall M., Kneipp J., Kotov N.A., Kuang H., Le Ru E.C., Lee H.K., Li J.F., Ling X.Y., Maier S.A., Mayerhofer T., Moskovits M., Murakoshi K., Nam J.M., Nie S., Ozaki Y., Pastoriza-Santos I., Perez-Juste J., Popp J., Pucci A., Reich S., Ren B., Schatz G.C., Shegai T., Schlucker S., Tay L.L., Thomas K.G., Tian Z.Q., Van Duyne R.P., Vo-Dinh T., Wang Y., Willets K.A., Xu C., Xu H., Xu Y., Yamamoto Y.S., Zhao B., Liz-Marzan L.M. Present and future of surface-enhanced Raman scattering. ACS Nano. 2020;14(1):28–117. doi: 10.1021/acsnano.9b04224. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Yu Y., Xiao T.H., Wu Y.Z., Li W.J., Zeng Q.G., Long L., Li Z.Y. Roadmap for single-molecule surface-enhanced Raman spectroscopy. Adv. Photon. 2020;2(1) doi: 10.1117/1.Ap.2.1.014002. [DOI] [Google Scholar]
- 61.Wang Y., Yan B., Chen L. SERS tags: novel optical nanoprobes for bioanalysis. Chem. Rev. 2013;113(3):1391–1428. doi: 10.1021/cr300120g. [DOI] [PubMed] [Google Scholar]
- 62.Yang Y.Q., Li J., Zhang M.X., Song P., Lu X.M., Ding Y. Universal and simple MoO3 substrate for identification of SERS enhancement mechanism. J. Raman Spectrosc. 2021;52(7):1275–1280. doi: 10.1002/jrs.6135. [DOI] [Google Scholar]
- 63.Ye J., Van Dorpe P. Plasmonic behaviors of gold dimers perturbed by a single nanoparticle in the gap. Nanoscale. 2012;4(22):7205–7211. doi: 10.1039/c2nr32353b. [DOI] [PubMed] [Google Scholar]
- 64.Lin L., Bi X.Y., Gu Y.Q., Wang F., Ye J. Surface-enhanced Raman scattering nanotags for bioimaging. J. Appl. Phys. 2021;129(19) doi: 10.1063/5.0047578. [DOI] [Google Scholar]
- 65.Lin L., Zhang Q., Li X., Qiu M., Jiang X., Jin W., Gu H., Lei D.Y., Ye J. Electron transport across plasmonic molecular nanogaps interrogated with surface-enhanced Raman scattering. ACS Nano. 2018;12(7):6492–6503. doi: 10.1021/acsnano.7b08224. [DOI] [PubMed] [Google Scholar]
- 66.Krajczewski J., Ambroziak R., Kudelski A. Substrates for surface-enhanced Raman scattering formed on nanostructured non-metallic materials: preparation and characterization. Nanomaterials-Basel. 2021;11(1):e75. doi: 10.3390/nano11010075. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Lin T., Song Y.L., Kuang P., Chen S., Mao Z., Zeng T.T. Nanostructure-based surface-enhanced Raman scattering for diagnosis of cancer. Nanomedicine. 2021;16(26):2389–2406. doi: 10.2217/nnm-2021-0298. [DOI] [PubMed] [Google Scholar]
- 68.Bell S.E.J., Charron G., Cortes E., Kneipp J., de la Chapelle M.L., Langer J., Prochazka M., Tran V., Schlucker S. Towards reliable and quantitative surface-enhanced Raman scattering (SERS): from key parameters to good analytical practice. Angew Chem. Int. Ed. Engl. 2020;59(14):5454–5462. doi: 10.1002/anie.201908154. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Lopez-Lorente A.I. Recent developments on gold nanostructures for surface enhanced Raman spectroscopy: particle shape, substrates and analytical applications. A review. Anal. Chim. Acta. 2021;1168 doi: 10.1016/j.aca.2021.338474. [DOI] [PubMed] [Google Scholar]
- 70.Sun J., Gong L., Wang W., Gong Z., Wang D., Fan M. Surface-enhanced Raman spectroscopy for on-site analysis: a review of recent developments. Luminescence. 2020;35(6):808–820. doi: 10.1002/bio.3796. [DOI] [PubMed] [Google Scholar]
- 71.Kim N., Thomas M.R., Bergholt M.S., Pence I.J., Seong H., Charchar P., Todorova N., Nagelkerke A., Belessiotis-Richards A., Payne D.J., Gelmi A., Yarovsky I., Stevens M.M. Surface enhanced Raman scattering artificial nose for high dimensionality fingerprinting. Nat. Commun. 2020;11(1) doi: 10.1038/s41467-019-13615-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Zhou B.B., Ou W.H., Zhao C.H., Shen J.D., Zhang G.B., Tang X.H., Deng Z.Q., Zhu G.Y., Li Y.Y., Lu J. Insertable and reusable SERS sensors for rapid on-site quality control of fish and meat products. Chem. Eng. J. 2021;426 doi: 10.1016/j.cej.2021.130733. [DOI] [Google Scholar]
- 73.Moisoiu V., Iancu S.D., Stefancu A., Moisoiu T., Pardini B., Dragomir M.P., Crisan N., Avram L., Crisan D., Andras I., Fodor D., Leopold L.F., Socaciu C., Balint Z., Tomuleasa C., Elec F., Leopold N. SERS liquid biopsy: an emerging tool for medical diagnosis. Colloids Surf. B Biointerfaces. 2021;208 doi: 10.1016/j.colsurfb.2021.112064. [DOI] [PubMed] [Google Scholar]
- 74.Shi R., Liu X., Ying Y. Facing challenges in real-life application of surface-enhanced Raman scattering: design and nanofabrication of surface-enhanced Raman scattering substrates for rapid field test of food contaminants. J. Agric. Food Chem. 2018;66(26):6525–6543. doi: 10.1021/acs.jafc.7b03075. [DOI] [PubMed] [Google Scholar]
- 75.Li C.C., Huang Y.M., Li X.Y., Zhang Y.R., Chen Q.L., Ye Z.W., Alqarni Z., Bell S.E.J., Xu Y.K. Towards practical and sustainable SERS: a review of recent developments in the construction of multifunctional enhancing substrates. J. Mater. Chem. C. 2021;9(35):11517–11552. doi: 10.1039/d1tc02134f. [DOI] [Google Scholar]
- 76.Ryu H.J., Lee W.K., Kim Y.H., Lee J.S. Interfacial interactions of SERS-active noble metal nanostructures with functional ligands for diagnostic analysis of protein cancer markers. Mikrochim. Acta. 2021;188(5):e164. doi: 10.1007/s00604-021-04807-z. [DOI] [PubMed] [Google Scholar]
- 77.Khlebtsov B.N., Bratashov D.N., Byzova N.A., Dzantiev B.B., Khlebtsov N.G. SERS-based lateral flow immunoassay of troponin I by using gap-enhanced Raman tags. Nano Res. 2019;12(2):413–420. doi: 10.1007/s12274-018-2232-4. [DOI] [Google Scholar]
- 78.Lin L., Ye J. Spontaneous Raman and surface-enhanced Raman scattering bioimaging. Adv. Exp. Med. Biol. 2021;3233:177–195. doi: 10.1007/978-981-15-7627-0_9. [DOI] [PubMed] [Google Scholar]
- 79.Qiu Y., Zhang Y., Li M., Chen G., Fan C., Cui K., Wan J.B., Han A., Ye J., Xiao Z. Intraoperative detection and eradication of residual microtumors with gap-enhanced Raman tags. ACS Nano. 2018;12(8):7974–7985. doi: 10.1021/acsnano.8b02681. [DOI] [PubMed] [Google Scholar]
- 80.Ye Z.X., Lin L., Tan Z.Y., Zeng Y.J., Ruan S.C., Ye J. Sub-100 nm multi-shell bimetallic gap-enhanced Raman tags. Appl. Surf. Sci. 2019;487:1058–1067. doi: 10.1016/j.apsusc.2019.05.200. [DOI] [Google Scholar]
- 81.Zhu W., Hutchison J.A., Dong M., Li M. Frequency shift surface-enhanced Raman spectroscopy sensing: an ultrasensitive multiplex assay for biomarkers in human health. ACS Sens. 2021;6(5):1704–1716. doi: 10.1021/acssensors.1c00393. [DOI] [PubMed] [Google Scholar]
- 82.Ji W., Li L.F., Zhang Y., Wang X.N., Ozaki Y. Recent advances in surface-enhanced Raman scattering-based sensors for the detection of inorganic ions: sensing mechanism and beyond. J. Raman Spectrosc. 2021;52(2):468–481. doi: 10.1002/jrs.5975. [DOI] [Google Scholar]
- 83.Chen J., Wang J., Geng Y., Yue J., Shi W., Liang C., Xu W., Xu S. Single-cell oxidative stress events revealed by a renewable SERS nanotip. ACS Sens. 2021;6(4):1663–1670. doi: 10.1021/acssensors.1c00395. [DOI] [PubMed] [Google Scholar]
- 84.Kao Y.C., Han X., Lee Y.H., Lee H.K., Phan-Quang G.C., Lay C.L., Sim H.Y.F., Phua V.J.X., Ng L.S., Ku C.W., Tan T.C., Phang I.Y., Tan N.S., Ling X.Y. Multiplex surface-enhanced Raman scattering identification and quantification of urine metabolites in patient samples within 30 min. ACS Nano. 2020;14(2):2542–2552. doi: 10.1021/acsnano.0c00515. [DOI] [PubMed] [Google Scholar]
- 85.Zheng T.T., Zhou Y., Feng E.D., Tian Y. Surface-enhanced Raman scattering on 2D nanomaterials: recent developments and applications dagger. Chin. J. Chem. 2021;39(3):745–756. doi: 10.1002/cjoc.202000453. [DOI] [Google Scholar]
- 86.Li D., Yao D.M., Li C.N., Luo Y.H., Liang A.H., Wen G.Q., Jiang Z.L. Nanosol SERS quantitative analytical method: a review. Trac. Trends Anal. Chem. 2020;127 doi: 10.1016/j.trac.2020.115885. [DOI] [Google Scholar]
- 87.Lai H.S., Li G.K., Xu F.G., Zhang Z.M. Metal-organic frameworks: opportunities and challenges for surface-enhanced Raman scattering - a review. J. Mater. Chem. C. 2020;8(9):2952–2963. doi: 10.1039/d0tc00040j. [DOI] [Google Scholar]
- 88.Feng E.D., Tian Y. Surface-enhanced Raman scattering of self-assembled superstructures. Chem. Res. Chin. Univ. 2021;37(5):989–1007. doi: 10.1007/s40242-021-1263-7. [DOI] [Google Scholar]
- 89.Liu J., Yin D., Wang S., Chen H.Y., Liu Z. Probing low-copy-number proteins in a single living cell. Angew Chem. Int. Ed. Engl. 2016;55(42):13215–13218. doi: 10.1002/anie.201608237. [DOI] [PubMed] [Google Scholar]
- 90.Plou J., Molina-Martinez B., Garcia-Astrain C., Langer J., Garcia I., Ercilla A., Perumal G., Carracedo A., Liz-Marzan L.M. Nanocomposite scaffolds for monitoring of drug diffusion in three-dimensional cell environments by surface-enhanced Raman spectroscopy. Nano Lett. 2021;21(20):8785–8793. doi: 10.1021/acs.nanolett.1c03070. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 91.Sun D., Cao F., Tian Y., Li A., Xu W., Chen Q., Shi W., Xu S. Label-free detection of multiplexed metabolites at single-cell level via a SERS-microfluidic droplet platform. Anal. Chem. 2019;91(24):15484–15490. doi: 10.1021/acs.analchem.9b03294. [DOI] [PubMed] [Google Scholar]
- 92.Lussier F., Missirlis D., Spatz J.P., Masson J.F. Machine-learning-Driven surface-enhanced Raman scattering optophysiology reveals multiplexed metabolite gradients near cells. ACS Nano. 2019;13(2):1403–1411. doi: 10.1021/acsnano.8b07024. [DOI] [PubMed] [Google Scholar]
- 93.Abalde-Cela S., Rebelo R., Wu L., Barbosa A.I., Rodriguez-Lorenzo L., Kant K., Reis R.L., Correlo V.M., Dieguez L. A SERS-based 3D nanobiosensor: towards cell metabolite monitoring. Mater. Adv. 2020;1(6):1613–1621. doi: 10.1039/d0ma00121j. [DOI] [Google Scholar]
- 94.Plou J., Garcia I., Charconnet M., Astobiza I., Garcia-Astrain C., Matricardi C., Mihi A., Carracedo A., Liz-Marzan L.M. Multiplex SERS detection of metabolic alterations in tumor extracellular media. Adv. Funct. Mater. 2020;30(17) doi: 10.1002/adfm.201910335. [DOI] [Google Scholar]
- 95.Erzina M., Trelin A., Guselnikova O., Dvorankova B., Strnadova K., Perminova A., Ulbrich P., Mares D., Jerabek V., Elashnikov R., Svorcik V., Lyutakov O. Precise cancer detection via the combination of functionalized SERS surfaces and convolutional neural network with independent inputs. Sensor. Actuator. B Chem. 2020;308 doi: 10.1016/j.snb.2020.127660. [DOI] [Google Scholar]
- 96.Dharmalingam P., Venkatakrishnan K., Tan B. Predicting metastasis from cues of metastatic cancer stem-like cells-3D-ultrasensitive metasensor at a single-cell level. ACS Nano. 2021;15(6):9967–9986. doi: 10.1021/acsnano.1c01436. [DOI] [PubMed] [Google Scholar]
- 97.Lussier F., Brule T., Bourque M.J., Ducrot C., Trudeau L.E., Masson J.F. Dynamic SERS nanosensor for neurotransmitter sensing near neurons. Faraday Discuss. 2017;205:387–407. doi: 10.1039/c7fd00131b. [DOI] [PubMed] [Google Scholar]
- 98.Liu J., Cai C., Wang Y., Liu Y., Huang L., Tian T., Yao Y., Wei J., Chen R., Zhang K., Liu B., Qian K. A biomimetic plasmonic nanoreactor for reliable metabolite detection. Adv. Sci. 2020;7(10) doi: 10.1002/advs.201903730. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 99.Heckman E.M., Hsu P.-H., Singh T.B., Tsai T.-H., Chiang H.K., Yoshida J. In vivo blood lactic acid monitoring using microdialysis and surface-enhanced Raman spectroscopy. Journal. 2008;7040 doi: 10.1117/12.794798. [DOI] [Google Scholar]
- 100.Li Y.L., Li Q.W., Sun C.B., Jin S.L., Park Y., Zhou T.L., Wang X., Zhao B., Ruan W.D., Jung Y.M. Fabrication of novel compound SERS substrates composed of silver nanoparticles and porous gold nanoclusters: a study on enrichment detection of urea. Appl. Surf. Sci. 2018;427:328–333. doi: 10.1016/j.apsusc.2017.08.230. [DOI] [Google Scholar]
- 101.Choi C.J., Wu H.Y., George S., Weyhenmeyer J., Cunningham B.T. Biochemical sensor tubing for point-of-care monitoring of intravenous drugs and metabolites. Lab Chip. 2012;12(3):574–581. doi: 10.1039/c2lc20586f. [DOI] [PubMed] [Google Scholar]
- 102.Wang T.L., Chiang H.K., Lu H., Hung Y. SERS quantitative urine creatinine measurement of human subject. Plasm. Biol. Med. Ii. 2005;5703:17–24. doi: 10.1117/12.591393. [DOI] [Google Scholar]
- 103.Leordean C., Canpean V., Astilean S. Surface-enhanced Raman scattering (SERS) analysis of urea trace in urine, fingerprint, and tear samples. Spectrosc. Lett. 2012;45(8):550–555. doi: 10.1080/00387010.2011.649439. [DOI] [Google Scholar]
- 104.Xiao R., Zhang X., Rong Z., Xiu B., Yang X., Wang C., Hao W., Zhang Q., Liu Z., Duan C., Zhao K., Guo X., Fan Y., Zhao Y., Johnson H., Huang Y., Feng X., Xu X., Zhang H., Wang S. Non-invasive detection of hepatocellular carcinoma serum metabolic profile through surface-enhanced Raman spectroscopy. Nanomedicine. 2016;12(8):2475–2484. doi: 10.1016/j.nano.2016.07.014. [DOI] [PubMed] [Google Scholar]
- 105.Phyo J.B., Woo A., Yu H.J., Lim K., Cho B.H., Jung H.S., Lee M.Y. Label-free SERS analysis of urine using a 3D-stacked AgNW-glass fiber filter sensor for the diagnosis of pancreatic cancer and prostate cancer. Anal. Chem. 2021;93(8):3778–3785. doi: 10.1021/acs.analchem.0c04200. [DOI] [PubMed] [Google Scholar]
- 106.Falamas A., Rotaru H., Hedesiu M. Surface-enhanced Raman spectroscopy (SERS) investigations of saliva for oral cancer diagnosis. Laser Med. Sci. 2020;35(6):1393–1401. doi: 10.1007/s10103-020-02988-2. [DOI] [PubMed] [Google Scholar]
- 107.Honda K., Hishiki T., Yamamoto S., Yamamoto T., Miura N., Kubo A., Itoh M., Chen W.Y., Takano M., Yoshikawa T., Kasamatsu T., Sonoda S., Yoshizawa H., Nakamura S., Itai Y., Shiota M., Koike D., Naya M., Hayakawa N., Naito Y., Matsuura T., Iwaisako K., Masui T., Uemoto S., Nagashima K., Hashimoto Y., Sakuma T., Matsubara O., Huang W., Ida T., Akaike T., Masugi Y., Sakamoto M., Kato T., Ino Y., Yoshida H., Tsuda H., Hiraoka N., Kabe Y., Suematsu M. On-tissue polysulfide visualization by surface-enhanced Raman spectroscopy benefits patients with ovarian cancer to predict post-operative chemosensitivity. Redox Biol. 2021;41 doi: 10.1016/j.redox.2021.101926. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 108.Xiao L., Wang C., Dai C., Littlepage L.E., Li J., Schultz Z.D. Untargeted tumor metabolomics with liquid chromatography-surface-enhanced Raman spectroscopy. Angew Chem. Int. Ed. Engl. 2020;59(9):3439–3443. doi: 10.1002/anie.201912387. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 109.Subaihi A., Trivedi D.K., Hollywood K.A., Bluett J., Xu Y., Muhamadali H., Ellis D.I., Goodacre R. Quantitative online liquid chromatography-surface-enhanced Raman scattering (LC-SERS) of methotrexate and its major metabolites. Anal. Chem. 2017;89(12):6702–6709. doi: 10.1021/acs.analchem.7b00916. [DOI] [PubMed] [Google Scholar]
- 110.Turzhitsky V., Zhang L., Horowitz G.L., Vitkin E., Khan U., Zakharov Y., Qiu L., Itzkan I., Perelman L.T. Picoanalysis of drugs in biofluids with quantitative label-free surface-enhanced Raman spectroscopy. Small. 2018;14(47) doi: 10.1002/smll.201802392. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 111.Bindesri S.D., Jebailey R., Albarghouthi N., Pye C.C., Brosseau C.L. Spectroelectrochemical and computational studies of tetrahydrocannabinol (THC) and carboxy-tetrahydrocannabinol (THC-COOH) Analyst. 2020;145(5):1849–1857. doi: 10.1039/c9an02173f. [DOI] [PubMed] [Google Scholar]
- 112.Alharbi O., Xu Y., Goodacre R. Simultaneous multiplexed quantification of nicotine and its metabolites using surface enhanced Raman scattering. Analyst. 2014;139(19):4820–4827. doi: 10.1039/c4an00879k. [DOI] [PubMed] [Google Scholar]
- 113.Jaworska A., Wietecha-Posluszny R., Wozniakiewicz M., Koscielniak P., Malek K. Evaluation of the potential of surface enhancement Raman spectroscopy for detection of tricyclic psychotropic drugs. Case studies on imipramine and its metabolite. Analyst. 2011;136(22):4704–4709. doi: 10.1039/c1an15598a. [DOI] [PubMed] [Google Scholar]
- 114.Velicka M., Zacharovas E., Adomaviciute S., Sablinskas V. Detection of caffeine intake by means of EC-SERS spectroscopy of human saliva. Spectrochim. Acta Mol. Biomol. Spectrosc. 2021;246 doi: 10.1016/j.saa.2020.118956. [DOI] [PubMed] [Google Scholar]
- 115.Alharbi O., Xu Y., Goodacre R. Simultaneous multiplexed quantification of caffeine and its major metabolites theobromine and paraxanthine using surface-enhanced Raman scattering. Anal. Bioanal. Chem. 2015;407(27):8253–8261. doi: 10.1007/s00216-015-9004-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 116.Zhu Q., Yu X., Wu Z., Lu F., Yuan Y. Antipsychotic drug poisoning monitoring of clozapine in urine by using coffee ring effect based surface-enhanced Raman spectroscopy. Anal. Chim. Acta. 2018;1014:64–70. doi: 10.1016/j.aca.2018.02.027. [DOI] [PubMed] [Google Scholar]
- 117.Ouyang L., Jiang Z., Wang N., Zhu L., Tang H. Rapid surface enhanced Raman scattering (SERS) detection of sibutramine hydrochloride in pharmaceutical capsules with a beta-cyclodextrin- Ag/polyvivnyl alcohol hydrogel substrate. Sensors. 2017;17(7) doi: 10.3390/s17071601. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 118.Greene B.H.C., Alhatab D.S., Pye C.C., Brosseau C.L. Electrochemical-surface enhanced Raman spectroscopic (EC-SERS) study of 6-thiouric acid: a metabolite of the chemotherapy drug azathioprine. J. Phys. Chem. C. 2017;121(14):8084–8090. doi: 10.1021/acs.jpcc.7b01179. [DOI] [Google Scholar]
- 119.Lin Y.K., Tai R.J., Wei S.C., Luo S.C. Electrochemical SERS on 2D mapping for metabolites detection. Langmuir. 2020;36(21):5990–5996. doi: 10.1021/acs.langmuir.0c00863. [DOI] [PubMed] [Google Scholar]
- 120.Adomavičiūtė S., Velička M., Šablinskas V. Detection of aspirin traces in blood by means of surface-enhanced Raman scattering spectroscopy. J. Raman Spectrosc. 2020;51(6):919–931. doi: 10.1002/jrs.5853. [DOI] [Google Scholar]
- 121.Li M.X., Yang H., Li S.Q., Liu C.W., Zhao K., Li J.G., Jiang D.N., Sun L.L., Wang H., Deng A.P. An ultrasensitive competitive immunochromatographic assay (ICA) based on surface-enhanced Raman scattering (SERS) for direct detection of 3-amino-5-methylmorpholino-2-oxazolidinone (AMOZ) in tissue and urine samples. Sensor. Actuator. B Chem. 2015;211:551–558. doi: 10.1016/j.snb.2014.12.135. [DOI] [Google Scholar]
- 122.Mao K., Zhou Z., Han S., Zhou X., Hu J., Li X., Yang Z. A novel biosensor based on Au@Ag core-shell nanoparticles for sensitive detection of methylamphetamine with surface enhanced Raman scattering. Talanta. 2018;190:263–268. doi: 10.1016/j.talanta.2018.07.071. [DOI] [PubMed] [Google Scholar]
- 123.Song W., Mao Z., Liu X., Lu Y., Li Z., Zhao B., Lu L. Detection of protein deposition within latent fingerprints by surface-enhanced Raman spectroscopy imaging. Nanoscale. 2012;4(7):2333–2338. doi: 10.1039/c2nr12030e. [DOI] [PubMed] [Google Scholar]
- 124.Masson J.F. The need for benchmarking surface-enhanced Raman scattering (SERS) sensors. ACS Sens. 2021;6(11):3822–3823. doi: 10.1021/acssensors.1c02275. [DOI] [PubMed] [Google Scholar]
- 125.Zhao X., Campbell S., Wallace G.Q., Claing A., Bazuin C.G., Masson J.F. Branched Au nanoparticles on nanofibers for surface-enhanced Raman scattering sensing of intracellular pH and extracellular pH gradients. ACS Sens. 2020;5(7):2155–2167. doi: 10.1021/acssensors.0c00784. [DOI] [PubMed] [Google Scholar]
- 126.Zhu H., Lussier F., Ducrot C., Bourque M.J., Spatz J.P., Cui W., Yu L., Peng W., Trudeau L.E., Bazuin C.G., Masson J.F. Block copolymer brush layer-templated gold nanoparticles on nanofibers for surface-enhanced Raman scattering optophysiology. ACS Appl. Mater. Interfaces. 2019;11(4):4373–4384. doi: 10.1021/acsami.8b19161. [DOI] [PubMed] [Google Scholar]
- 127.Li J., Wuethrich A., Sina A.A.I., Cheng H.H., Wang Y., Behren A., Mainwaring P.N., Trau M. A digital single-molecule nanopillar SERS platform for predicting and monitoring immune toxicities in immunotherapy. Nat. Commun. 2021;12(1) doi: 10.1038/s41467-021-21431-w. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 128.Valpapuram I., Candeloro P., Coluccio M.L., Parrotta E.I., Giugni A., Das G., Cuda G., Di Fabrizio E., Perozziello G. Waveguiding and SERS simplified Raman spectroscopy on biological samples. Biosensors. 2019;9(1):e37. doi: 10.3390/bios9010037. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 129.Chang K.W., Cheng H.W., Shiue J., Wang J.K., Wang Y.L., Huang N.T. Antibiotic susceptibility test with surface-enhanced Raman scattering in a microfluidic system. Anal. Chem. 2019;91(17):10988–10995. doi: 10.1021/acs.analchem.9b01027. [DOI] [PubMed] [Google Scholar]
- 130.Jiang X., Tan Z.Y., Lin L., He J., He C., Thackray B.D., Zhang Y.Q., Ye J. Surface-enhanced Raman nanoprobes with embedded standards for quantitative cholesterol detection. Small Methods. 2018;2(11) doi: 10.1002/smtd.201800182. [DOI] [Google Scholar]
- 131.Shen W., Lin X., Jiang C., Li C., Lin H., Huang J., Wang S., Liu G., Yan X., Zhong Q., Ren B. Reliable quantitative SERS analysis facilitated by core-shell nanoparticles with embedded internal standards. Angew Chem. Int. Ed. Engl. 2015;54(25):7308–7312. doi: 10.1002/anie.201502171. [DOI] [PubMed] [Google Scholar]
- 132.Ding Z., Wang W., Zhang K., Ming F., Yangdai T., Xu T., Shi H., Bao Y., Yao H., Peng H., Han C., Jiang W., Liu J., Hou X., Lin R. Novel scheme for non-invasive gut bioinformation acquisition with a magnetically controlled sampling capsule endoscope. Gut. 2021;70(12):2297–2306. doi: 10.1136/gutjnl-2020-322465. [DOI] [PubMed] [Google Scholar]
- 133.Yamamoto N., Kawashima N., Kitazaki T., Mori K., Kang H., Nishiyama A., Wada K., Ishimaru I. Ultrasonic standing wave preparation of a liquid cell for glucose measurements in urine by midinfrared spectroscopy and potential application to smart toilets. J. Biomed. Opt. 2018;23(5):1–4. doi: 10.1117/1.JBO.23.5.050503. [DOI] [PubMed] [Google Scholar]
- 134.Mu T.T., Li S., Feng H.H., Zhang C.C., Wang B., Ma X., Guo J.H., Huang B., Zhu L.Q. High-sensitive smartphone-based Raman system based on cloud network architecture. IEEE J. Sel. Top Quant. 2019;25(1) doi: 10.1109/Jstqe.2018.2832661. [DOI] [Google Scholar]
- 135.Xu K., Zhou R., Takei K., Hong M. Toward flexible surface-enhanced Raman scattering (SERS) sensors for point-of-care diagnostics. Adv. Sci. 2019;6(16) doi: 10.1002/advs.201900925. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 136.Koh E.H., Lee W.C., Choi Y.J., Moon J.I., Jang J., Park S.G., Choo J., Kim D.H., Jung H.S. A wearable surface-enhanced Raman scattering sensor for label-free molecular detection. ACS Appl. Mater. Interfaces. 2021;13(2):3024–3032. doi: 10.1021/acsami.0c18892. [DOI] [PubMed] [Google Scholar]
- 137.Wang Y., Zhao C., Wang J., Luo X., Xie L., Zhan S., Kim J., Wang X., Liu X., Ying Y. Wearable plasmonic-metasurface sensor for noninvasive and universal molecular fingerprint detection on biointerfaces. Sci. Adv. 2021;7(4) doi: 10.1126/sciadv.abe4553. eabe4553. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 138.Kaefer K., Kruger K., Schlapp F., Uzun H., Celiksoy S., Flietel B., Heimann A., Schroeder T., Kempski O., Sonnichsen C. Implantable sensors based on gold nanoparticles for continuous long-term concentration monitoring in the body. Nano Lett. 2021;21(7):3325–3330. doi: 10.1021/acs.nanolett.1c00887. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Data Availability Statement
Data sharing is not applicable to this article as no new data were created or analyzed in this study.