Abstract
Mycoplasma synoviae (MS), an important avian pathogen, can cause chronic respiratory disease, eggshell apex abnormalities, infectious synovitis, and arthritis in avian species, leading serious economic losses in the global poultry industry. To date, studies have shown significant different transcript profiles using various chicken cells after MS infection. However, in vitro cell models cannot fully represent the complex in vivo regulations after adventitious infection. The objective of this study was to explore the nature of the host-pathogen interaction during MS infection. The tracheal and spleen tissues of chickens were collected at d 0, 1, 3, and 5 postinoculation, and samples were analyzed for differential gene expression using Illumina RNA sequencing. A lot of significantly differentially expressed genes (DEGs) were observed in this analysis, and 861 DEGs were observed in trachea tissues and 753 DEGs were observed in spleen samples. Many of DEGs in trachea tissues participate in a variety of cellular activities, especially cellular metabolism. Immune-related DEGs were mainly enriched at d 3, and 5 postinfection in trachea tissues. While, DEGs in spleen tissues were significantly and mainly enriched into immune-related pathways. The results of this study show the direct interactions between MS and the chicken trachea and spleen for the first time. Early dysregulation of tissue-wide gene expression as observed here set the stage for persistent infection of MS.
Key words: Mycoplasma synoviae, transcriptional profiling, trachea, spleen
INTRODUCTION
Mycoplasma synoviae (MS) is an important avian pathogen. Although MS infection does not directly lead to death in poultry, it causes infectious synovitis, respiratory disease, growth retardation, decrease in egg production, and the production of deformed eggs, leading serious economic losses in the poultry industry (Dufour-Gesbert et al., 2006; Xu et al., 2021). Usually, MS co-infection with other infectious agents such as Mycoplasma gallisepticum (MG), Newcastle disease virus (NDV), Infectious bronchitis virus (IBV), Escherichia coli, and increases economic losses (Raviv et al., 2007; Xie et al., 2014; Hutton et al., 2017; Yadav et al., 2021). MS was reported in Australia, South America, Asia, Europe, and Africa (Gole et al., 2012; Hutton et al., 2017; Kreizinger et al., 2017; Sun et al., 2017; Reck et al., 2019) and have been listed as a notifiable mycoplasma by World Organization for Animal Health (OIE) (2018). Variability exists among MS strains, and subacute and chronic infections make the control and elimination of this pathogen particularly difficult. The effective measures to prevent and control MS infection include biosecurity, medication, and vaccination (Gong et al., 2020).
It has been shown that mycoplasmas interacted with the host's immune system on many levels, including modulating the host immune system and stimulating an inflammatory response, which enable mycoplasmas to establish a chronic, persistent infection (Razin et al., 1998). In vitro experimental studies showed that MS infection modulates several immune genes in chicken macrophages and apoptotic genes in chicken chondrocytes (Lavric et al., 2007; Lavric et al., 2008; Dusanic et al., 2012). Lavrič et al. (2008) reported that 14 chicken macrophage genetic elements, representing actually 11 distinct genes, shown to be significantly modulated consistently during all experimental models of macrophage exposure to MS. And the amino-terminal end of VlhA (Variable lipoprotein hemagglutinin), MSPB (Lipoprotein at N- terminal of VlhA) were able to induce NO (Nitric Oxide), IL-6 (Interleukin-6), and IL-1β (Interleukin-1β) (Lavric et al., 2007). The N-terminal diacylated lipopeptide (MDLP) of MS significantly induces the expression of TLR15 (Toll-like receptors 15) in chicken macrophages and chondrocytes, which increases the expression of transcription factor NF-κB (nuclear factor kappa-light-chain-enhancer of activated B cells) and NO production (Oven et al., 2013). And, MS strain HN01 infected chicken synovial fibroblasts (CSF) could alter the gene and protein expression, which consisting of proliferation- and apoptosis-related factors, inflammatory mediators, proangiogenic factors, antiangiogenic factors, and matrix metalloproteinases, as well as other arthritis-related proteins (Liu et al., 2020). The interaction of MS with chicken synovial sheath cells (SSC) contributes to the inflammatory response caused by the robust expression of related cytokines and macrophage chemotaxis (Xu et al., 2020). Besides, in the model of chicken chondrocytes (CCH), MS infection would change the metabolic and sensitivity profile of CCH (Dusanic et al., 2014), and induce upregulation of apoptotic genes, secretion of nitric oxide and appearance of an apoptotic phenotype of cells (Dusanic et al., 2012). Moreover, MS infection of embryos results in intensive upregulation of cytokine and chemokine genes, and the liver, spleen, bursa of fabricius, and thymus have different expression profiles (Bolha et al., 2013).
MS most frequently colonizes the upper respiratory tract, which may lead to the development of systemic infection and/or infectious synovitis. Although there have been many studies on the interaction between MS and host, the results were gotten based on in vitro model. The nature of the host-pathogen interaction during MS infection especially trachea-MS interaction has not been clarified. In this study, we have utilized RNA sequencing (RNA-Seq) of the host tracheal and spleen tissue transcripts subsequent to infection with virulent MS strain SD2 over a 5-d time course. This study will provide a comprehensive, unbiased assessment of the changes in gene expression.
MATERIALS AND METHODS
Animals and Infection
Ten-day-old Chinese yellow chickens with a negative cleft palate swab were selected and raised in HEPA-filtered avian isolators until 3-wk-old. The MS virulent strain SD2 was isolated from a sick yellow chicken with synovitis in the Shandong province, China, and maintained in our laboratory. MS was cultured in modified Frey's broth or solid medium at 37°C with 5% CO2. The titer of MS was determined by color-changing units (CCU) as previously described (Stemke and Robertson, 1982). MS was pelleted by centrifugation at 8,000 × g for 10 min and resuspended at 1 × 108 CCU/mL in phosphate-buffered saline solution (PBS). Chickens were challenged by intranasal inoculation with 200 μL each chicken. At 1, 3, 5 d postinfection (dpi.), 3 chickens were humanely euthanized respectively and then whole tracheas and whole spleens were collected and placed in tubes containing 2 mL of sterile PBS and stored at -80°C until processing for RNA extraction as described below. Before infection (at 0 dpi.), tissue samples were collected as negative control. The samples of Neg_T, d1_T, d3_T and d5_T means the samples of tracheas collected at 0, 1, 3 and 5 dpi., while Neg_S, d1_S, d3_S and d5_S means the samples of spleens collected at 0, 1, 3, and 5 dpi.
RNA Extraction
One sample consisted of 3 parallel whole tissues, and total 8 samples were subjected to RNA extraction. Total RNA was collected by TRIzol reagent (Invitrogen) according to the manufacturer's instructions. RNase-free DNase (Thermo Scientific) was used to remove contaminating genomic DNA. RNA degradation and contamination were monitored on 1% agarose gels. RNA purity was checked using the NanoPhotometer spectrophotometer (IMPLEN, CA). RNA integrity was assessed using the RNA Nano 6000 Assay Kit of the Bioanalyzer 2100 system (Agilent Technologies, CA).
Illumina Library Preparation and RNA Sequencing
A total amount of 1 μg RNA per sample was used as input material for the RNA sample preparations. Sequencing libraries were generated using NEBNext UltraTM RNA Library Prep Kit for Illumina (NEB) following manufacturer's recommendations and index codes were added to attribute sequences to each sample. Briefly, mRNA was purified from total RNA using poly-T oligo-attached magnetic beads. Fragmentation was carried out using divalent cations under elevated temperature in NEBNext First Strand Synthesis Reaction Buffer (5X). First strand cDNA was synthesized using random hexamer primer and M-MuLV Reverse Transcriptase (RNase H−). Second strand cDNA synthesis was subsequently performed using DNA Polymerase I and RNase H. Remaining overhangs were converted into blunt ends via exonuclease/polymerase activities. After adenylation of 3’ ends of DNA fragments, NEBNext Adaptor with hairpin loop structure were ligated to prepare for hybridization. In order to select cDNA fragments of preferentially 250 to 300 bp in length, the library fragments were purified with AMPure XP system (Beckman Coulter, Beverly, MA). Then 3 μL USER Enzyme (NEB) was used with size-selected, adaptor-ligated cDNA at 37°C for 15 m in followed by 5 min at 95°C before PCR. Then PCR was performed with Phusion High -Fidelity DNA polymerase, Universal PCR primers and Index (X) Primer. At last, PCR products were purified (AMPure XP system) and library quality was assessed on the Agilent Bioanalyzer 2100 system. The clustering of the index-coded samples was performed on a cBot Cluster Generation System using TruSeq PE Cluster Kit v3-cBot-HS (Illumina) according to the manufacturer's instructions. After cluster generation, the library preparations were sequenced on an Illumina Novaseq platform and 150 bp paired-end reads were generated.
Quality Control and Reads Mapping to the Reference Genome
Raw data (raw reads) of fastq format were firstly processed through in-house perl scripts. In this step, clean data (clean reads) were obtained by removing reads containing adapter, reads containing ploy-N and low-quality reads from raw data. At the same time, Q20 (percentage of total bases with a Phred value greater than 20), Q30 (percentage of total bases with a Phred value greater than 30), and GC content of the clean data were calculated. All the downstream analyses were based on the clean data with high quality. Reference genome and gene model annotation GRCg6a (Genome Reference Consortium Chicken Build 6a) files were downloaded from NCBI (https://www.ncbi.nlm.nih.gov/assembly/GCF_000002315.5/) directly. Index of the reference genome was built using Hisat2 v2.0.5 and paired-end clean reads were aligned to the reference genome using Hisat2 v2.0.5.
Quantification of Gene Expression Level and Differential Expression Analysis
FeatureCounts v1.5.0-p3 was used to count the reads numbers mapped to each gene. And then FPKM (expected number of Fragments Per Kilobase of transcript sequence per Millions base pairs sequenced) of each gene was calculated based on the length of the gene and reads count mapped to this gene. Differential expression analysis of 2 samples was performed using the DESeq2 R package (1.20.0). The resulting P-values indicated as padj (adjust P value) were adjusted using the Benjamini and Hochberg's approach for controlling the false discovery rate (Love et al., 2014). Genes with a padj <0.05 and |Log2(foldchange)|>1 were assigned as differentially expressed.
GO and KEGG Enrichment Analysis of Differentially Expressed Genes
Gene Ontology (GO) and KEGG (Kyoto encyclopedia of genes and genomes) pathways were used to classify the DEGs. GO annotations are divided into 3 categories: Molecular Function (MF), Biological Process (BP) and Cellular Components (CC). Through these 3 functional categories, the function of a gene can be defined and described in many aspects. KEGG is a comprehensive database integrating genomic, chemical and system function information. GO and KEGG pathway enrichment were performed using clusterProfiler R package (3.4.4), in which gene length bias was corrected. The GO term and KEGG pathway with a padj < 0.05 was significantly enriched.
Quantitative Real-Time PCR
To analyze mRNA expression, total RNA was used as templates to perform quantitative real-time PCR using the One Step TB Green PrimeScript RT-PCR Kit II (TAKARA, China) according the manufacturer's protocol. Quantitative real-time PCR was carried out with ABI 7500 Fast Real-Time PCR System (Applied Biosystems). The relative expression values of candidate mRNAs were normalized to that of β-actin in each sample using the 2-ΔΔCt method.
RESULTS
Global Differentially Expressed Genes Analysis of the Trachea and Spleen After MS Infection
In order to analyze the transcriptomic profile of the trachea and spleen of chickens after MS infection, a total of 8 sequencing libraries prepared at 0, 1, 3, 5 dpi. were sequenced, and about 348 million raw reads were obtained. About 334 million clean reads were obtained after filtering out the adapter, reads containing ploy-N and low-quality reads from raw data. The values of Q20 and Q30 were higher than 96.5 and 91.4% respectively, means the sequencing data can be used for further analysis (Figure 1A). From the results of Figure 1B, we can see that 4 trachea samples showed similar gene expression distributions, as well as 4 spleen samples, and different tissues have different gene expression distributions.
Figure 1.
Quantity analysis of the trachea and spleen after MS infection. (A) Quality summary of sequencing data. The raw_reads represent the number of reads in the raw data. The clean_reads represent the number of filtered reads. The error-rate represents the overall data sequencing error rate. The Q20 and Q30 represent the percentages of bases with Phred value greater than 20 and 30 in the total base, respectively. The GC_pct represents the percentages of G and C base in the clean reads. (B) Boxplot of gene expression distribution in samples. The horizontal axis is the samples and the vertical axis is the log2(FPKM+1), the statistics of each box are the maximum, upper quartile, median, lower quartile and minimum from top to bottom.
A total of 25,253 genes were compared, and among them, 1,412 genes were considered to be significantly differentially expressed genes (DEGs) in infected samples when compared to the uninfected samples under the padj value <0.05 and | log2 (fold change) | >1.0. Compared with the uninfected samples, the genes in each infected sample exhibiting the great expression pattern constitute ≥50% of the total DEGs identified, except the d1_T sample, and the DEGs number is different with the time of infection (Figure 2A). The precise numbers of DEGs at 1, 3, and 5 dpi. were 117, 353, and 224 great-expressed and 141, 226, and 138 less-expressed in the trachea and 340, 243, and 421 great-expressed and 111, 85, and 116 less-expressed in the spleen. Taking the union of all the DEGs in the comparison samples and drawing the heatmap according to the FPKM value of each gene. As shown in Figure 2B, genes had the similar expression profiles in each sample of spleens, as well as in each sample of tracheas, however, different expression trends were shown between spleens and tracheas. Taken together, these data demonstrate that MS infection induces widespread alterations in the expression pattern of chicken trachea and spleen, and different tissues have different expression patterns. Venn diagrams were generated to examine the unique and overlapping DEGs among subgroups with the infection at 3 infection time-points (Figures 2C and 2D). Eighty seven DEGs were common among 3 time-points in trachea and 44 DEGs were common in spleen.
Figure 2.
DEGs analysis of the trachea and spleen after MS infection. (A) The counts of DEGs among different infected samples compared with uninfected samples. The horizontal axis represents samples and the vertical axis represents the counts of DEGs. (B) Heatmap of the DEGs according to the FPKM value of each gene. The horizontal axis represents samples and different colors represent different FPKM values. (C, D) Venn diagrams of DEGs among trachea samples and spleen samples respectively. Abbreviation: DEGs, differentially expressed genes.
Pathway Enrichment Analysis of DEGs
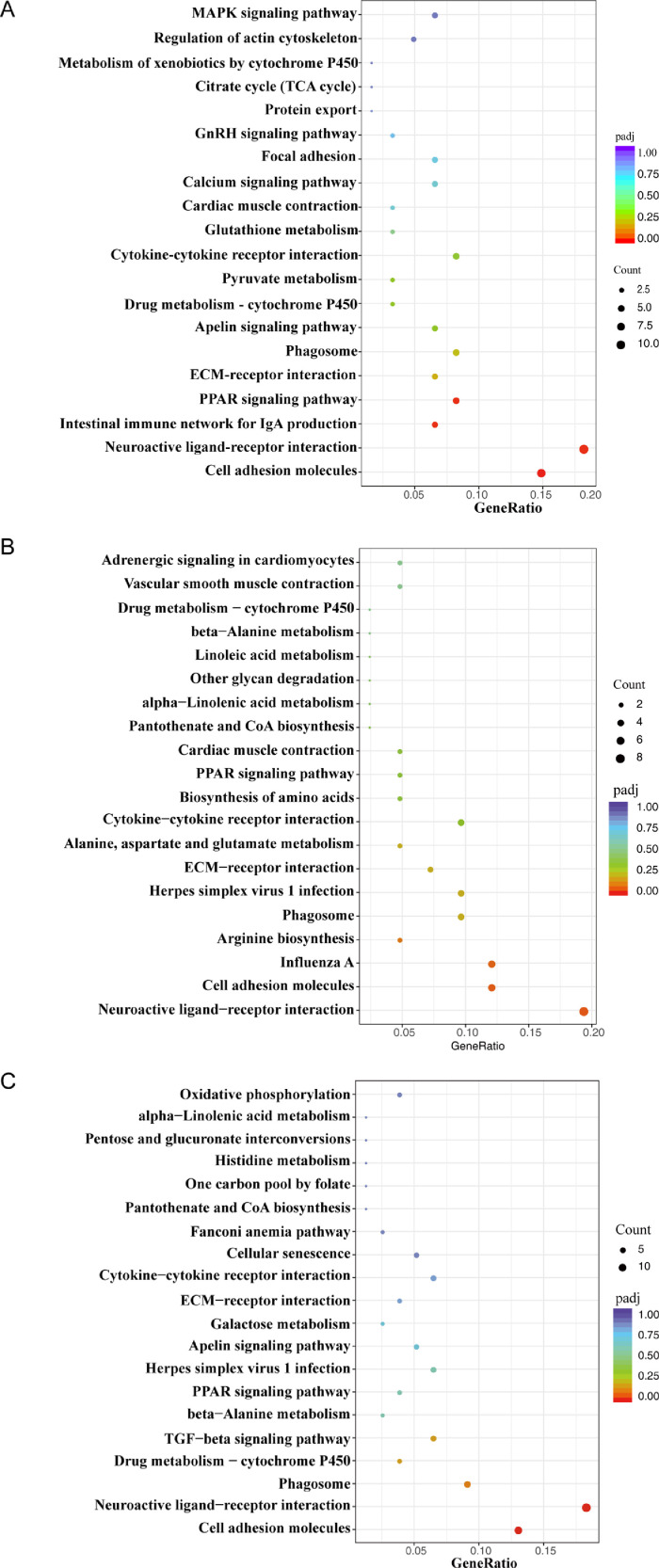
To identify the cellular pathways potentially involved during the MS infection, the KEGG was employed (Figure 3, Figure 4). The d 1 after MS infection, no KEGG pathways were significantly enriched in trachea sample (Figure 3A), while cell adhesion molecules (gga04514) pathway was most significantly enriched in spleen sample (Figure 4A), then were neuroactive ligand-receptor interaction (gga04080), intestinal immune network for IgA production (gga04672) and PPAR signaling pathway (gga03320). No pathways were significantly enriched in spleen sample at 3 dpi. (Figure 4B), while cytokine-cytokine receptor interaction (gga04060), cell adhesion molecules (gga04514) and ECM-receptor interaction (gga04512) were significantly enriched in trachea sample (Figure 3B). At 5 dpi., intestinal immune network for IgA production (gga04672), cytokine-cytokine receptor interaction (gga04060) and cell adhesion molecules (gga04514), neuroactive ligand-receptor interaction (gga04080) pathways were significantly enriched in trachea and spleen samples respectively (Figures 3C and 4C).
Figure 3.
Pathway enrichment of DEGs in trachea samples. (A) Pathway enrichment of DEGs of trachea at d 1 after MS infected. (B) Pathway enrichment of DEGs of trachea at d 3 after MS infected. (C) Pathway enrichment of DEGs of trachea at d 5 after MS infected. The different sizes of bubble mean the different gene counts, the different colors of bubble mean the different padj values. Abbreviation: DEGs, differentially expressed genes.
Figure 4.
Pathway enrichment of DEGs in spleen samples. (A) Pathway enrichment of DEGs of spleen at d 1 after MS infected. (B) Pathway enrichment of DEGs of spleen at d 3 after MS infected. (C) Pathway enrichment of DEGs of spleen at d 5 after MS infected. The different sizes of bubble mean the different gene counts, the different colors of bubble mean the different padj values. Abbreviation: DEGs, differentially expressed genes.
GO Enrichment Analysis of DEGs
To explore the biological functions of the DEGs, GO enrichment analysis was performed in 3 functional categories including MF, CC, and BP. The top 30 or less highly enriched GO terms (padj < 0.05) of each sample were considered (Figure 5, Figure 6). The categories of multiorganism process (GO:0051704), viral process (GO:0016032), symbiont process (GO:0044403), and interspecies interaction between organisms (GO:0044419) were highly enriched in trachea at d 1 after infection (Figure 5A). The categories of myosin complex (GO:0016459), actin cytoskeleton (GO:0015629), and cytoskeletal part (GO:0044430) were highly enriched in trachea at d 3 after infection (Figure 5B). The categories of immune response (GO:0006955), immune system process (GO:0002376) and multiorganism process (GO:0051704) were highly enriched in trachea at d 5 after infection (Figure 5C). While the immune response (GO:0006955) and immune system process (GO:0002376) categories were highly enriched in spleen samples throughout the infection process (Figure 6).
Figure 5.
GO enrichment analysis of DEGs in trachea samples. (A) GO enrichment of DEGs of trachea at d 1 after MS infected. (B) GO enrichment of DEGs of trachea at d 3 after MS infected. (C) GO enrichment of DEGs of trachea at d 5 after MS infected. The horizontal axis represents GO terms and different colors represent different GO category. The vertical axis represents –log10(padj). Abbreviations: DEGs, differentially expressed genes; GO, Gene Ontology.
Figure 6.
GO enrichment analysis of DEGs in spleen samples. (A) GO enrichment of DEGs of spleen at d 1 after MS infected. (B) GO enrichment of DEGs of spleen at d 3 after MS infected. (C) GO enrichment of DEGs of spleen at d 5 after MS infected. The horizontal axis represents GO terms and different colors represent different GO category. The vertical axis represents –log10(padj). Abbreviations: DEGs, differentially expressed genes; GO, Gene Ontology.
Trend of Gene Expression in Trachea After MS Infection
Trachea is the first important target after MS infection especially respiratory infection, and MS-trachea interaction is the key factor to determine whether MS can successfully colonize and complete the infection. Thus, after KEGG and GO enrichment, we constructed the directed acyclic graph (DAG) of DEGs to show the gene expression tendency of trachea and spleen at different time points. As shown in Figure S1, the biology process of less-expressed DEGs in trachea after MS infection were mainly associated with multiorganism process and the mainly function were effects nucleic acid metabolic process. However, the biology process of great-expressed DEGs was more abundant. As shown in Figure S2, on the first day after infection, the main work of MS is to regulate the growth of tracheal cells and change the process of cell metabolism. Then, the work focuses of MS changed to alter the lipid transport which is important for MS survival. And the implementation of a series of regulation measures would eventually lead to the recognition of the host immune system and the successful activation of the host immune system through a series of signal transduction pathways. Spleen is an important immune organ in the chicken. As we can see in Figure S3, the immune-related DAGs were downregulated in spleen from the d 1 to d 5 after infection. In the meanwhile, some immune-related DAGs were great-expressed at d 3 after infection (Figure S4), and most of these DAGs were cell adhesion molecules which act as receptor-ligand binding.
Immune-Related Genes With Greatest Differential Expression
Fold-change values for the top 15 immune-related DEGs over the course of infection were ranged (Table 1). The pathways of these genes involved in were cell adhesion molecules, cytokine-cytokine receptor interaction, TGF-beta signaling pathway, endocytosis, MAPK-signaling pathway, intestinal immune network for IgA production and so on.
Table 1.
Top 15 immune-related DEGs of trachea samples common to all time points.
| Gene_name | Log2 fold change on day p.i.: |
Tf_family | Pathways | ||
|---|---|---|---|---|---|
| 1 | 3 | 5 | |||
| ENSGALG00000014184 | −11.7363 | −5.10933 | −11.7528 | MH2 | TGF-beta signaling pathway |
| Smad7b | −9.14604 | −9.13345 | −9.16254 | MH2 | TGF-beta signaling pathway |
| ENSGALG00000031355 | −9.12371 | −9.11112 | −9.14021 | HECT | Endocytosis |
| ENSGALG00000048307 | −9.66206 | −5.65679 | −9.67857 | - | MAPK signaling pathway |
| ENSGALG00000049444 | 6.079679 | 10.55825 | – | MHC_I | Cell adhesion molecules |
| CACNG7 | −6.83426 | −2.27465 | −0.78468 | - | MAPK signaling pathway |
| TUBA8B | −5.47584 | −1.47083 | 3.385416 | Tubulin | Apoptosis |
| TLR1A | 3.709063 | 5.493616 | 7.066343 | TIR | Toll-like receptor signaling pathway |
| IL11 | 1.970566 | 5.06306 | 0.676594 | - | Cytokine-cytokine receptor interaction |
| CXCL13L2 | 1.877944 | 4.205412 | 6.802326 | - | Cytokine-cytokine receptor interaction |
| CXCR5 | 0.870156 | 3.370464 | 4.783849 | 7tm_1 | Cytokine-cytokine receptor interaction |
| TNFRSF13C | 0.335024 | 3.177463 | 4.333251 | - | Cytokine-cytokine receptor interaction &Intestinal immune network for IgA production |
| CLDN18 | – | 12.46919 | – | - | Cell adhesion molecules |
| ENSGALG00000052686 | – | 6.720185 | – | MHC_I | Cell adhesion molecules &Endocytosis |
| ENSGALG00000050544 | – | – | 8.049725 | - | NOD-like receptor signaling pathway |
Confirmation of DEGs by Quantitative Real-Time PCR
To confirm the differential expression in the sequencing data, we chose 4 DEGs (CYP2C23a, CDH22, KCNT1, and C3orf67) and then validated their expression in tissue samples by quantitative real-time PCR, primers used in our study are listed in Table S1. As shown in Figure S5, these DEGs exhibited similar expression signatures as were detected in the sequencing data, indicating the robustness of our experimental settings and bioinformatic analyses.
DISCUSSION
To better understand the pathogenesis of pathogens, profiling the omics changes is the most comprehensive and commonly used. There have many omics studies about MS infection, however, almost all of them were used cells as models, which could not give a completely accurate picture of the actual course of MS infection (Lavric et al., 2008; Oven et al., 2013; Liu et al., 2020; Xu et al., 2020). MS can be transmitted both vertically and horizontally (Ter Veen et al., 2020). Vertical transmission is more complex and will not be discussed here. MS mainly spreads horizontally through the respiratory tract and colonizes on the upper respiratory tract of chickens after infection, which was a key step in achieving a successful infection (Rebollo Couto et al., 2012). The colonization of MS in the respiratory tract and the transmission in chickens inevitably stimulate the immune system of the host (Gong et al., 2020). Spleen as one of the main immune organs may involve in the interaction between MS and chicken. Thus, in the present study, we collected the tracheas and spleens of infected or uninfected chickens at different time to profile the transcripts. The differential gene expression data observed in this study revealed the complexity of the host response to MS, especially at early time points. It is easy to envisage how the myriad differentially expressed inflammatory, signaling, and immune response genes could result in a profoundly dysregulated host response. Numerous genes were found to have significant differential expression over the course of infection. After preliminary analysis, we found that the gene expression profiles were similar in the same organs and different between trachea and spleen (Figures 1B and 2B), which was same with MS infection of chicken embryos produces different expression profiles in different organs (Bolha et al., 2013). Different expression profile may associate with infection degree and biological functions of organs.
Mycoplasma can directly affect cellular metabolism and physiology of the host organism by rewiring the process of nutrient consumption (Borchsenius et al., 2020). MS-infected chickens show lesions of the wing joints, intertarsal joints, digital flexor tendons, metatarsal extensor tendons, and footpads, manifested as the viscous to caseous exudation, tissue hyperplasia and hypertrophy, and the infiltration of inflammatory cells (Dufour-Gesbert et al., 2006; Xu et al., 2021). In the study of MS infection chicken synovial fibroblasts cells (Liu et al., 2020) and chicken synovial sheath cells (Xu et al., 2020), inflammatory response related cytokines were upregulated. And in Bolha et al. (2013) study, they tested the cytokine and chemokine gene expression in chicken embryos (Bolha et al., 2013), and resulted in intensive upregulation of IFN-γ (Interferon-γ), IL-1β, IL-6, IL-12β (Interleukin 12β), IL-16 (Interleukin 16), IL-18 (Interleukin 18), CCL4 (C-C motif chemokine ligand 4), iNOS (Inducible nitric oxide synthase), XCL1 (X-C motif chemokine ligand 1), and LITAF (Lipopolysaccharide induced TNF factor). In present study, we also found significant upregulation (|log2FoldChange|>2) of many pro-inflammatory cytokines, such as IL-11 (Interleukin 11), IL-22 (Interleukin 22), CSF-3 (Colony stimulating factor 3), and IFN-α (Interferon-α) in trachea samples and IL-1β, IL-2, IL-12β, IFN-β (Interferon-β), and CSF-3 in spleen samples. Significant differential expression of many cytokine and chemokine receptors was also observed over the course of infection (Figure 3). For example, on d 3 and d 5 after infection, cytokine/chemokine receptor genes, such as CXCR5, among DEGs in trachea samples were enriched significantly (Table 1 and Table 2). This indicates that MS infection results in increased cell sensitivity to inflammatory agonists by virtue of upregulating receptors for signaling molecules not present on mycoplasmas (Beaudet et al., 2017).
Table 2.
Top 15 immune-related DEGs of spleen samples common to all time points.
| Gene_name | Log2 fold change on day p.i.: |
Tf_family | Pathways | ||
|---|---|---|---|---|---|
| 1 | 2 | 3 | |||
| CLDN18 | 8.986176 | – | 8.397927 | - | Cell adhesion molecules |
| ENSGALG00000052686 | −8.99733 | −1.20433 | −8.89855 | MHC_I | Cell adhesion molecules &Endocytosis |
| ENSGALG00000054743 | 7.270238 | 6.409338 | 7.686966 | - | Cytokine-cytokine receptor interaction |
| NEGR1 | 6.759665 | 5.112648 | 7.628364 | I-set | Cell adhesion molecules |
| GREM2 | 5.962261 | 4.153755 | 6.855955 | - | TGF-beta signaling pathway |
| MPZ | 4.816639 | −4.02443 | 6.13273 | - | Cell adhesion molecules |
| ENSGALG00000034319 | −4.3485 | −3.18047 | −1.14045 | C1-set | Cell adhesion molecules &Intestinal immune network for IgA production |
| ENSGALG00000049342 | 3.355326 | −5.29216 | −2.12116 | - | Cytokine-cytokine receptor interaction &Intestinal immune network for IgA production |
| CXCL14 | −1.67338 | −4.22612 | −2.22088 | - | Cytokine-cytokine receptor interaction |
| ENSGALG00000049444 | −1.51517 | 2.996556 | −4.43861 | MHC_I | Cell adhesion molecules &Endocytosis |
| CLDN23 | −0.6478 | -1.93639 | −6.67937 | - | Cell adhesion molecules |
| ENSGALG00000014184 | – | – | 11.50011 | MH2 | TGF-beta signaling pathway & Endocytosis |
| ENSGALG00000031355 | – | – | 9.267254 | HECT | Endocytosis |
| ENSGALG00000048307 | – | – | 8.945911 | - | MAPK signaling pathway |
| Smad7b | – | – | 8.041578 | MH2 | TGF-beta signaling pathway |
In the study of MS infection chicken macrophages, chicken chondrocytes, and chicken synovial sheath cells, TLR15 was significantly induced. The expression quantity of TLR15 was related with infective dose and collection time (Xu et al., 2020). However, in this study, no significant difference in TLR15 expression was occurred. Different results may be attributed to different experimental models, different infection doses and different sampling times. In this study, both toll-like, such as TLR1 and NOD-like receptor signaling pathway were enriched in trachea and spleen samples, besides, RIG-I-like receptor signaling pathway was enriched in trachea samples only.
MS infection would change the cellular metabolism and cell cycle. MS infection chicken chondrocytes induces a wide range of metabolic and sensitivity modifications of cells which can contribute to pathological processes in the development of infectious synovitis (Dusanic et al., 2014). And MS induces upregulation of apoptotic genes, secretion of NO, and appearance of an apoptotic phenotype in infected chicken chondrocytes (Dusanic et al., 2012). MS also can induce the expression of serum amyloid A (Liu et al., 2021) and Nos2 (Nitric oxide synthase 2) (Liu et al., 2020) to promote proliferation and apoptosis of chicken synovial fibroblast cell. In this study, many genes associated with cell metabolism were enriched, but cell cycle-related genes were enriched little.
In conclusion, this study has provided global profiles of the chicken tracheal and splenic responses to MS over the course of early infection. A better understanding of these molecular events is critical to understand the pathogenic mechanisms of pathogenic microorganisms important in the development of future therapeutics and vaccines.
DISCLOSURES
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
ACKNOWLEDGMENTS
This work was supported by the China Postdoctoral Science Foundation (2021M692455).
Data availability statement: The BioSample accessions of datasets generated for this study were: SAMN22310569, SAMN22310570, SAMN22310571, SAMN22310572, SAMN22310573, SAMN22310574, SAMN22310575, SAMN22310576.
Ethics statement: The animal experiment was carried out in Sun Yat-Sen University with the approval of the Committee on the Ethics of Animal Experiments of (SYSU-IACUC-2021-B0507).
Author contributions: XNW conceived and designed the experiments, carried out the experiments, analyzed the data, and wrote the manuscript. WC and QJS participated in performing experiments. ZQY participated in revising the manuscript. FC and YCC participated in analyzing and interpreting the data. QFZ supervised and guided this work. All the authors have read and approved the final version of the manuscript.
Footnotes
Supplementary material associated with this article can be found in the online version at doi:10.1016/j.psj.2021.101660.
Contributor Information
Feng Chen, Email: fengch@scau.edu.cn.
Xiaona Wei, Email: weixiaona_na@126.com.
Appendix. Supplementary materials
Figure S1. Directed acyclic graph of the biology process of less-expressed DEGs of trachea samples. Each node represents a GO term, and the box represents the TOP5 enrichment degree of GO. The color depth represents the enrichment degree, and the darker the color, the higher the enrichment degree. The name of the term and padj of enrichment analysis are displayed on each node. The red, yellow and blue means the DEGs were obtained respectively at day 1, day 3 and day 5 after MS infection, and the green means the DEGs were obtained at both day3 and day 5.
Figure S2. Directed acyclic graph of the biology process and molecular function of great-expressed DEGs of trachea samples. A. Directed acyclic graph of the biology process of great-expressed DEGs. B. Directed acyclic graph of the molecular function of great-expressed DEGs. Each node represents a GO term, and the box represents the TOP5 enrichment degree of GO. The color depth represents the enrichment degree, and the darker the color, the higher the enrichment degree. The name of the term and padj of enrichment analysis are displayed on each node. The red, yellow and blue means the DEGs were obtained respectively at day 1, day 3 and day 5 after MS infection, and the green means the DEGs were obtained at both day3 and day 5, the orange means the DEGs were obtained at both day 1 and day 3.
Figure S3. Directed acyclic graph of the biology process of less-expressed DEGs of spleen samples. Each node represents a GO term, and the box represents the TOP5 enrichment degree of GO. The color depth represents the enrichment degree, and the darker the color, the higher the enrichment degree. The name of the term and padj of enrichment analysis are displayed on each node. The red, yellow and blue means the DEGs were obtained respectively at day 1, day 3 and day 5 after MS infection, and the black means the DEGs were obtained at all of the day.
Figure S4. Directed acyclic graph of the biology process of great-expressed DEGs of spleen samples. Each node represents a GO term, and the box represents the TOP5 enrichment degree of GO. The color depth represents the enrichment degree, and the darker the color, the higher the enrichment degree. The name of the term and padj of enrichment analysis are displayed on each node. The red, yellow and blue means the DEGs were obtained respectively at day 1, day 3 and day 5 after MS infection.
Figure S5. Validation of RNA-sequencing data by quantitative real-time PCR. Trachea and spleen samples collected from chicken at days of 0, 1, 3 and 5 after MS infection were subject to RNA extraction and relative mRNA expression levels of indicated genes (CYP2C23a, CDH22, KCNT1, and C3orf67) were measured by quantitative real-time PCR. A-D. Relative mRNA expression levels of indicated genes in trachea samples. E-H. Relative mRNA expression levels of indicated genes in spleen samples.
Table S1. Primers used in this study.
REFERENCES
- Beaudet J., Tulman E.R., Pflaum K., Liao X., Kutish G.F., Szczepanek S.M., Silbart L.K., Geary S.J. Transcriptional profiling of the chicken tracheal response to virulent Mycoplasma gallisepticum strain rlow. Infect Immun. 2017;85 doi: 10.1128/IAI.00343-17. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bolha L., Bencina D., Cizelj I., Oven I., Slavec B., Rojs O.Z., Narat M. Effect of Mycoplasma synoviae and lentogenic Newcastle disease virus coinfection on cytokine and chemokine gene expression in chicken embryos. Poult. Sci. 2013;92:3134–3143. doi: 10.3382/ps.2013-03332. [DOI] [PubMed] [Google Scholar]
- Borchsenius S.N., Vishnyakov I.E., Chernova O.A., Chernov V.M., Barlev N.A. Effects of Mycoplasmas on the host cell signaling pathways. Pathogens. 2020;9 doi: 10.3390/pathogens9040308. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dufour-Gesbert F., Dheilly A., Marois C., Kempf I. Epidemiological study on Mycoplasma synoviae infection in layers. Vet. Microbiol. 2006;114:148–154. doi: 10.1016/j.vetmic.2005.10.040. [DOI] [PubMed] [Google Scholar]
- Dusanic D., Bencina D., Narat M., Oven I. Phenotypic characterization of Mycoplasma synoviae induced changes in the metabolic and sensitivity profile of in vitro infected chicken chondrocytes. Biomed. Res. Int. 2014;2014 doi: 10.1155/2014/613730. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dusanic D., Bencina D., Oven I., Cizelj I., Bencina M., Narat M. Mycoplasma synoviae induces upregulation of apoptotic genes, secretion of nitric oxide and appearance of an apoptotic phenotype in infected chicken chondrocytes. Vet. Res. 2012;43:7. doi: 10.1186/1297-9716-43-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gole V.C., Chousalkar K.K., Roberts J.R. Prevalence of antibodies to Mycoplasma synoviae in laying hens and possible effects on egg shell quality. Prev. Vet. Med. 2012;106:75–78. doi: 10.1016/j.prevetmed.2012.02.018. [DOI] [PubMed] [Google Scholar]
- Gong X., Chen Q., Ferguson-Noel N., Stipkovits L., Szathmary S., Liu Y., et al. Evaluation of protective efficacy of inactivated Mycoplasma synoviae vaccine with different adjuvants. Vet. Immunol. Immunopathol. 2020;220 doi: 10.1016/j.vetimm.2019.109995. [DOI] [PubMed] [Google Scholar]
- Hutton S., Bettridge J., Christley R., Habte T., Ganapathy K. Detection of infectious bronchitis virus 793B, avian metapneumovirus, Mycoplasma gallisepticum and Mycoplasma synoviae in poultry in Ethiopia. Trop. Anim. Health Prod. 2017;49:317–322. doi: 10.1007/s11250-016-1195-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kreizinger Z., Grozner D., Sulyok K.M., Nilsson K., Hrivnak V., Bencina D., Gyuranecz M. Antibiotic susceptibility profiles of Mycoplasma synoviae strains originating from Central and Eastern Europe. BMC Vet. Res. 2017;13:342. doi: 10.1186/s12917-017-1266-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lavric M., Bencina D., Kothlow S., Kaspers B., Narat M. Mycoplasma synoviae lipoprotein MSPB, the N-terminal part of VlhA haemagglutinin, induces secretion of nitric oxide, IL-6 and IL-1beta in chicken macrophages. Vet. Microbiol. 2007;121:278–287. doi: 10.1016/j.vetmic.2006.12.005. [DOI] [PubMed] [Google Scholar]
- Lavric M., Maughan M.N., Bliss T.W., Dohms J.E., Bencina D., Keeler C.L., Jr., Narat M. Gene expression modulation in chicken macrophages exposed to Mycoplasma synoviae or Escherichia coli. Vet. Microbiol. 2008;126:111–121. doi: 10.1016/j.vetmic.2007.06.011. [DOI] [PubMed] [Google Scholar]
- Liu R., Xu B., Yu S., Zhang J., Sun H., Liu C., Lu F., Pan Q., Zhang X. Integrated transcriptomic and proteomic analyses of the interaction between chicken synovial fibroblasts and Mycoplasma synoviae. Front. Microbiol. 2020;11:576. doi: 10.3389/fmicb.2020.00576. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu R., Xu B., Zhang J., Sun H., Liu C., Lu F., Pan Q., Zhang X. Mycoplasma synoviae induces serum amyloid A upregulation and promotes chicken synovial fibroblast cell proliferation. Microb. Pathog. 2021;154 doi: 10.1016/j.micpath.2021.104829. [DOI] [PubMed] [Google Scholar]
- Love M.I., Huber W., Anders S. Moderated estimation of fold change and dispersion for RNA-seq data with DESeq2. Genome. Biol. 2014;15:550. doi: 10.1186/s13059-014-0550-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Oven I., Resman Rus K., Dusanic D., Bencina D., Keeler C.L., Jr., Narat M. Diacylated lipopeptide from Mycoplasma synoviae mediates TLR15 induced innate immune responses. Vet. Res. 2013;44:99. doi: 10.1186/1297-9716-44-99. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Raviv Z., Ferguson-Noel N., Laibinis V., Wooten R., Kleven S.H. Role of Mycoplasma synoviae in commercial layer Escherichia coli peritonitis syndrome. Avian. Dis. 2007;51:685–690. doi: 10.1637/0005-2086(2007)51[685:ROMSIC]2.0.CO;2. [DOI] [PubMed] [Google Scholar]
- Razin S., Yogev D., Naot Y. Molecular biology and pathogenicity of mycoplasmas. Microbiol. Mol. Biol. Rev. 1998;62:1094–1156. doi: 10.1128/mmbr.62.4.1094-1156.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rebollo Couto M.S., Klein C.S., Voss-Rech D., Terenzi H. Extracellular Proteins of Mycoplasma synoviae. ISRN Vet. Sci. 2012;2012 doi: 10.5402/2012/802308. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Reck C., Menin A., Canever M.F., Pilatic C., Miletti L.C. Molecular detection of Mycoplasma synoviae and avian reovirus infection in arthritis and tenosynovitis lesions of broiler and breeder chickens in Santa Catarina State, Brazil. J S Afr. Vet. Assoc. 2019;90:e1–e5. doi: 10.4102/jsava.v90i0.1970. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stemke G.W., Robertson J.A. Comparison of two methods for enumeration of mycoplasmas. J. Clin. Microbiol. 1982;16:959–961. doi: 10.1128/jcm.16.5.959-961.1982. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sun S.K., Lin X., Chen F., Wang D.A., Lu J.P., Qin J.P., Luo T.R. Epidemiological investigation of Mycoplasma Synoviae in native chicken breeds in China. BMC Vet. Res. 2017;13:115. doi: 10.1186/s12917-017-1029-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ter Veen C., de Wit J.J., Feberwee A. Relative contribution of vertical, within-farm and between-farm transmission of Mycoplasma synoviae in layer pullet flocks. Avian. Pathol. 2020;49:56–61. doi: 10.1080/03079457.2019.1664725. [DOI] [PubMed] [Google Scholar]
- Xie Z., Luo S., Xie L., Liu J., Pang Y., Deng X., Xie Z., Fan Q., Khan M.I. Simultaneous typing of nine avian respiratory pathogens using a novel GeXP analyzer-based multiplex PCR assay. J. Virol. Methods. 2014;207:188–195. doi: 10.1016/j.jviromet.2014.07.007. [DOI] [PubMed] [Google Scholar]
- Xu B., Chen X., Lu F., Sun Y., Sun H., Zhang J., Shen L., Pan Q., Liu C., Zhang X. Comparative genomics of Mycoplasma synoviae and new targets for molecular diagnostics. Front. Vet. Sci. 2021;8 doi: 10.3389/fvets.2021.640067. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xu B., Liu R., Ding M., Zhang J., Sun H., Liu C., Lu F., Zhao S., Pan Q., Zhang X. Interaction of Mycoplasma synoviae with chicken synovial sheath cells contributes to macrophage recruitment and inflammation. Poult. Sci. 2020;99:5366–5377. doi: 10.1016/j.psj.2020.08.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yadav J.P., Tomar P., Singh Y., Khurana S.K. Insights on Mycoplasma gallisepticum and Mycoplasma synoviae infection in poultry: a systematic review. Anim. Biotechnol. 2021:1–10. doi: 10.1080/10495398.2021.1908316. 1908316. [DOI] [PubMed] [Google Scholar]
- OIE. (2018). Avian mycoplasmosis (Mycoplasma gallisepticum, Mycoplasma synoviae) In OIE Terrestrial Manual, Ch. Paris: 3.3.5. OIE; 2018. pp. 844–859.
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Figure S1. Directed acyclic graph of the biology process of less-expressed DEGs of trachea samples. Each node represents a GO term, and the box represents the TOP5 enrichment degree of GO. The color depth represents the enrichment degree, and the darker the color, the higher the enrichment degree. The name of the term and padj of enrichment analysis are displayed on each node. The red, yellow and blue means the DEGs were obtained respectively at day 1, day 3 and day 5 after MS infection, and the green means the DEGs were obtained at both day3 and day 5.
Figure S2. Directed acyclic graph of the biology process and molecular function of great-expressed DEGs of trachea samples. A. Directed acyclic graph of the biology process of great-expressed DEGs. B. Directed acyclic graph of the molecular function of great-expressed DEGs. Each node represents a GO term, and the box represents the TOP5 enrichment degree of GO. The color depth represents the enrichment degree, and the darker the color, the higher the enrichment degree. The name of the term and padj of enrichment analysis are displayed on each node. The red, yellow and blue means the DEGs were obtained respectively at day 1, day 3 and day 5 after MS infection, and the green means the DEGs were obtained at both day3 and day 5, the orange means the DEGs were obtained at both day 1 and day 3.
Figure S3. Directed acyclic graph of the biology process of less-expressed DEGs of spleen samples. Each node represents a GO term, and the box represents the TOP5 enrichment degree of GO. The color depth represents the enrichment degree, and the darker the color, the higher the enrichment degree. The name of the term and padj of enrichment analysis are displayed on each node. The red, yellow and blue means the DEGs were obtained respectively at day 1, day 3 and day 5 after MS infection, and the black means the DEGs were obtained at all of the day.
Figure S4. Directed acyclic graph of the biology process of great-expressed DEGs of spleen samples. Each node represents a GO term, and the box represents the TOP5 enrichment degree of GO. The color depth represents the enrichment degree, and the darker the color, the higher the enrichment degree. The name of the term and padj of enrichment analysis are displayed on each node. The red, yellow and blue means the DEGs were obtained respectively at day 1, day 3 and day 5 after MS infection.
Figure S5. Validation of RNA-sequencing data by quantitative real-time PCR. Trachea and spleen samples collected from chicken at days of 0, 1, 3 and 5 after MS infection were subject to RNA extraction and relative mRNA expression levels of indicated genes (CYP2C23a, CDH22, KCNT1, and C3orf67) were measured by quantitative real-time PCR. A-D. Relative mRNA expression levels of indicated genes in trachea samples. E-H. Relative mRNA expression levels of indicated genes in spleen samples.
Table S1. Primers used in this study.