Summary
The role of Processing bodies (P-bodies), key sites of post-transcriptional control, in adult stem cells remains poorly understood. Here, we report that adult Drosophila intestinal stem cells, but not surrounding differentiated cells such as absorptive Enterocytes (ECs), harbor P-bodies that contain Drosophila orthologs of mammalian P-body components DDX6, EDC3, EDC4 and LSM14A/B. A targeted RNAi screen in intestinal progenitor cells identified 39 previously known and 64 novel P-body regulators, including Patr-1, a gene necessary for P-body assembly. Loss of Patr-1-dependent P-bodies leads to a loss of stem cells that is associated with inappropriate expression of EC-fate gene nubbin. Transcriptomic analysis of progenitor cells identifies a cadre of such weakly transcribed pro-differentiation transcripts that are elevated after P-body loss. Altogether, this study identifies a P-body-dependent repression activity that coordinates with known transcriptional repression programs to maintain a population of in vivo stem cells in a state primed for differentiation.
Graphical Abstract
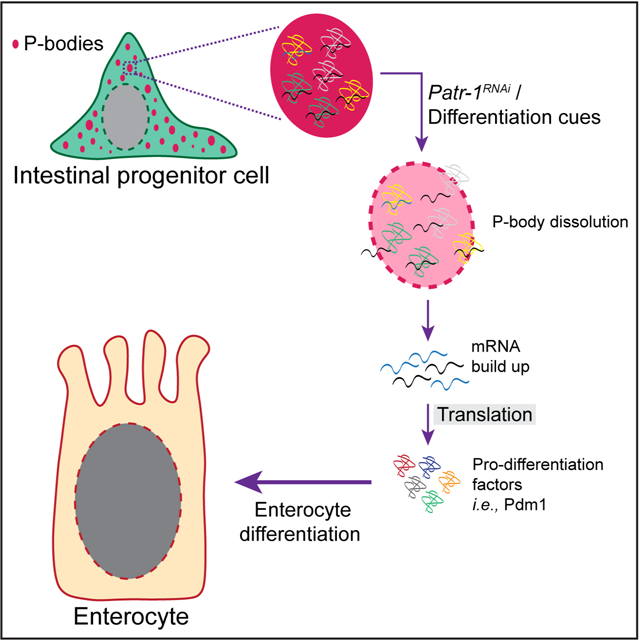
eTOC Blurb
How stem cells are maintained in adult tissues is a fundamental question. In a new study, Buddika et al., show that Drosophila intestinal stem cells contain P-bodies that repress the expression of weakly transcribed pro-differentiation genes to repress precocious differentiation and thereby ensure the maintenance of this cell.
Introduction
In eukaryotic cells, translationally inactive messenger RNAs (mRNAs) can assemble to form a distinct population of cytoplasmic messenger ribonucleoprotein (mRNP) particles known as Processing bodies (P-bodies).1–3 Like stress granules, P-bodies lack a limiting membrane and are visible via conventional microscopic techniques,1 are enhanced or induced by blocking translation initiation,4–8 are disassembled when treated with cycloheximide, a chemical that traps mRNAs on ribosomes,4–6,9,10 and are conserved from yeast to humans.11,12 Unlike stress granules, however, P-bodies are constitutive and contain proteins involved in mRNA decay.1,2,5,9,13–17 While stress granules have been identified in stem cells of adult tissues,18 the presence and role of P-bodies in adult stem cells remains unclear.
Biochemical profiling experiments in human HEK293 cells identified different classes of P-body residents that include proteins involved in mRNA translation and decay.19 In addition, the population of mRNAs recruited to these P-bodies is large but specific.19,20 Although proteins implicated in mRNA degradation localize to P-bodies,1,2 mRNAs can leave P-bodies to re-enter translation in the cytopasm.21,22 Therefore, current models propose that P-bodies coordinate the storage and protein production of mRNAs.20 Genetically tractable in vivo P-body models are necessary to better understand the exact role, organization, and dynamics of P-bodies in eukaryotic tissues including the identification and tracking of P-body regulated mRNAs in vivo.
Although some post-transcriptional processes such as alternative splicing, polyadenylation, and RNA modifications have been extensively studied in pluripotent stem cells,25 the roles of these and other RNA processes in the potency of adult somatic stem cells remain poorly understood. For example, from recent functional analysis of the P-body protein DDX6, it remains unclear why P-bodies promote differentiation in some in vitro derived stem cell lineages and repress it in others.26 To address this gap, tissue-based stem cell models will be critical for delineating P-body function in dynamic, in vivo contexts. The intestinal epithelium of adult Drosophila offers such a model to study post-transcriptional gene regulatory mechanisms.18,27,28 This actively regenerating tissue hosts a population of progenitor cells, which is composed of two main cell types: mitotic intestinal stem cells (ISCs) and their transient and non-mitotic daughters, enteroblasts (EBs).29,30 In this study we used the Drosophila midgut as a tissue-based stem cell model to delineate the biological roles of P-bodies in vivo.
Results
P-body components are enriched in intestinal progenitor cells
To test whether intestinal progenitors contained P-bodies, we stained intestines dissected from 7-day old, adult female Drosophila with four antibodies. These detected the Drosophila orthologs of proteins known to localize to P-bodies in other species: EDC3, GE-1 (known in metazoans as EDC4), Me31B (known in metazoans as DDX6), and TRAL (known in metazoans as LSM14A).1,17,19,31 Using previously verified antibodies for GE-1 and Me31B32–34 and newly validated antibodies for EDC3 and TRAL (Figure S1A–B), we found that all of these antibodies displayed elevated signal in progenitor cells (Figure 1A–D), which were labeled by horseradish peroxidase (HRP).35,36 Limited to the cytoplasm, confocal microscopy detected these proteins in clearly separated cytoplasmic granules that were similar in level and organization in both ISCs and 3Xgbe-smGFP::V5::nls positive EBs (Figure 1A’–D’). TRAL and Me31B staining perfectly overlapped, indicating colocalization (Figure 1E–E’). The absence of staining in neighboring polyploid ECs indicated that these proteins were downregulated during EC differentiation. Such downregulation also occurred during EE differentiation, since the majority of cells stained for Prospero (PROS), an EE-marker,30 showed very low-to-no detectable Me31B or TRAL, although ~20% of PROS+ EEs displayed levels that were comparable to progenitors (Figure S1C–D). These data indicated that progenitors contained a population of cytoplasmic granules harboring P-body proteins that were cleared during EC and EE differentiation.
Figure 1. Intestinal progenitor cells contain P-bodies.
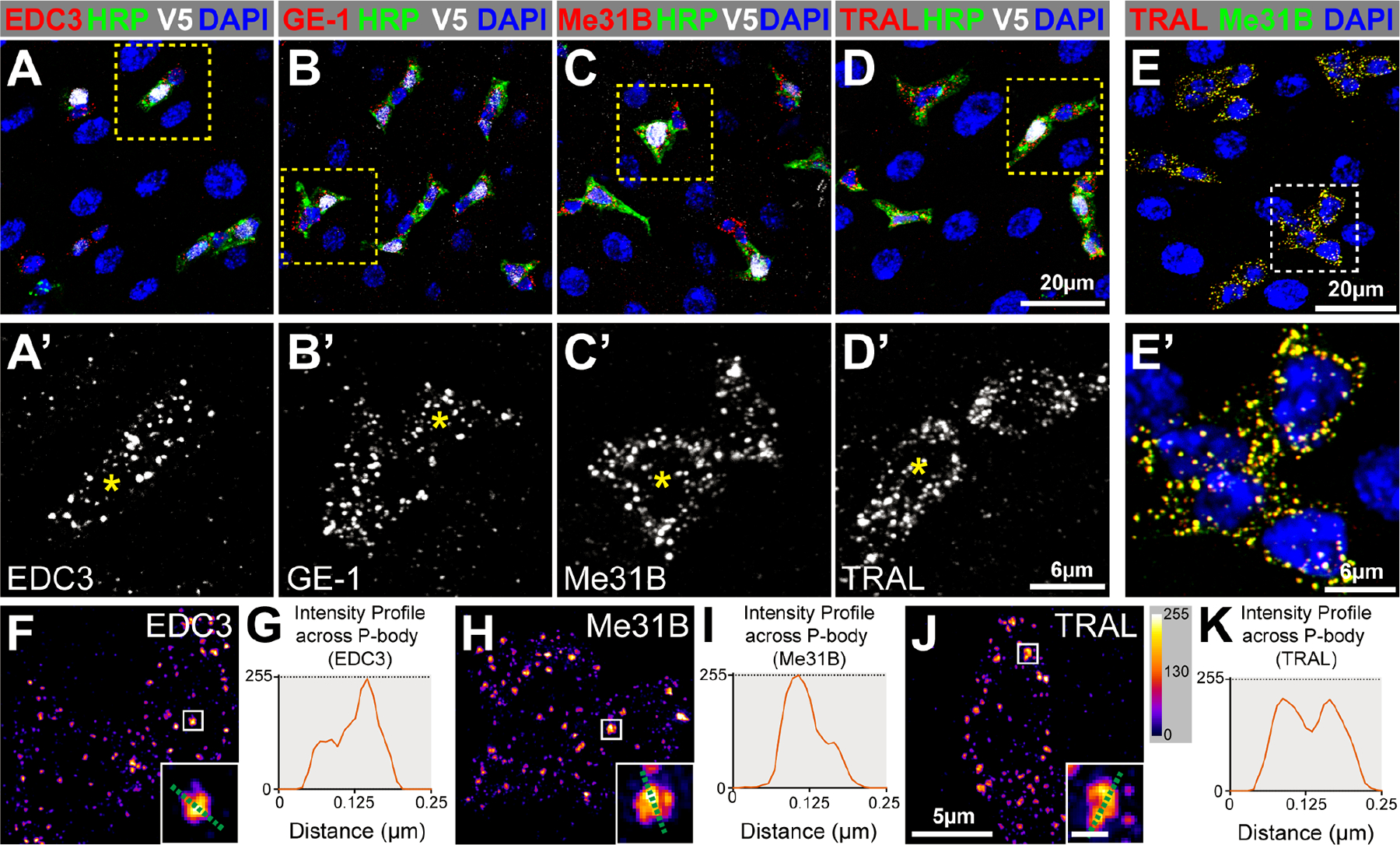
(A-D) Adult posterior midguts (PMGs) stained for EBs (V5, white), progenitor cells (HRP, green), all cells (DAPI, blue) and either EDC3, GE-1, Me31B, or TRAL (red). A’-D’ are enlargements with EBs labeled (asterisks). (E) PMG stained for TRAL (red), Me31B (green), and DAPI (blue). (F, H, J) Super-resolution micrographs of progenitor cells stained for EDC3, Me31B, or TRAL pseudocolored using intensity scale in J; insets are from boxed regions (scale 0.125μm). (G, I, K) Pixel-by-pixel intensity profiles along the green dotted line shown in insets. Full genotypes listed in Data S1F. See also Figure S1.
Intestinal P-bodies are ultrastructurally organized
Because the structural organization of P-bodies is not well understood,14 we assessed the ultrastructural organization of P-body protein complexes in progenitors. We stained dissected intestines with EDC3, Me31B, or TRAL, imaged the cytoplasmic distribution of these proteins using structured illumination super-resolution microscopy (SIM), and used the FIJI package of ImageJ to generate intensity maps and profiles of each protein.37 These revealed that the distribution of EDC3, Me31B, and TRAL was not uniform within a granule, with greater protein accumulation in central relative to peripheral domains of the granule (Figure 1F–K) and some granules containing multiple foci of staining (Figure 1J–K). The similarity of this non-uniform protein organization to the core-shell organization of stress granules suggested that P-bodies and stress granules share similar structural organizations and can contain multiple cores.38 Taken together, these data identified the existence of a structurally organized population of steady-state P-bodies in intestinal progenitors of adult Drosophila.
P-bodies and stress granules are distinct mRNPs in progenitor cells
The above analysis suggested that P-body complexes were distinct from intestinal progenitor stress granules (IPSGs) because, unliked IPSGs, P-bodies were present in unstressed progenitor cells.18 We therefore compared the subcellular locations and sizes of two P-body proteins, Me31B and TRAL, with two IPSG proteins, Fragile Mental Retardation Protein (FMRP) and Rasputin (RIN), in unstressed and stressed conditions using SIM. Unstressed samples were incubated ex-vivo in Krebs Ringer Bicarbonate Buffer (KRB) for 60 minutes. Stress was induced by incubation with 1mM Rapamycin in KRB for the same length of time. Rapamycin is known to induce stress granules via the inhibition of the mTORC1 complex.39 In the absence of stress, P-body proteins almost completely colocalized with each other but only partially colocalized with IPSG proteins (Figure S1E–J). Rapamycin caused P-body and IPSG protein staining to overlap substantially (Figure S1K–P). To quantify these complexes, we measured the sizes of TRAL and FMRP punctae. Under unstressed conditions, TRAL punctae (0.047 μm2; n=804 puncta in 24 cells) were bigger than FMRP punctae (0.026 μm2; n=629 puncta in 8 cells) (Figure S1Q–R, U). Stress caused TRAL punctae (0.097 μm2; n=657 puncta in 25 cells) and FMRP punctae (0.098 μm2; n=1215 puncta in 42 cells) to grow to the same size (Figure S1S–U). We concluded that P-body proteins localized to persistent mRNP complexes that were present in unstressed cells, and that stress led to the coalescence and enlargement of these mRNPs into IPSGs.
Identification of genetic modifiers of progenitor P-bodies using a targeted screen
To identify genes that altered P-bodies, we performed a RNAi screen targeting genes implicated in RNA-related processes. We generated a list of 600 candidates using the GO term analysis function at Flybase and screened the 485 with available RNAi lines (Data S1A); 186 genes were targeted with two or more RNAi strains, the remainder by just one RNAi line. RNAi was induced at the adult stage using the conditional progenitor cell driver escargot-GAL4TS (esgTS)30 and dissected seven days later. P-body morphology was scored using the P-body marker TRAL. This analysis identified 103 genes grouped into five phenotypic classes: big TRAL puncta, small TRAL puncta, diffuse TRAL puncta, both diffuse and big TRAL puncta, and no TRAL puncta (Figure 2A–B, Data S1B). While 39 (38%) of the identified genes were orthologs of proteins known to affect P-bodies in human cells (Data S1B),14 the remaining 64 (62%) genes were not previously implicated in P-body organization or assembly. Unexpectedly, regulators of nuclear processes (e.g. RNA splicing and nuclear export) as well as components of nuclear assemblies (e.g. the exon-exon junction complex) impacted the morphology of cytoplasmic TRAL granules. Knockdown of 24% of the positive genes (23/103 genes) caused loss of progenitor cells, raising the possibility that P-bodies were physiologically linked to the stemness of this progenitor cell population. Two percent of the total screened (11/485 genes) caused progenitor cell loss without noticeable changes to P-bodies, indicating that these genes were required for progenitor survival independent of P-body formation. To assess the specificity of the screen, a representative set of 25 positives were co-stained with IPSG-makers FMRP and ROX8 (Figure S2A–F, Data S1B). Most had no discernable effect on IPSGs, indicating that identified genes had specific effects on P-bodies; however, two genes, fne and CG13928, altered the expression profile of IPSG proteins and led to big or diffuse and big FMRP/ROX8 foci, respectively (Figure S2E, Data S1B). Altogether, this screen identified a large number of in vivo regulators of P-bodies, many of which have not been previously identified.
Figure 2. RNAi screen identifies PATR-1 as an essential component of progenitor cell P-bodies.
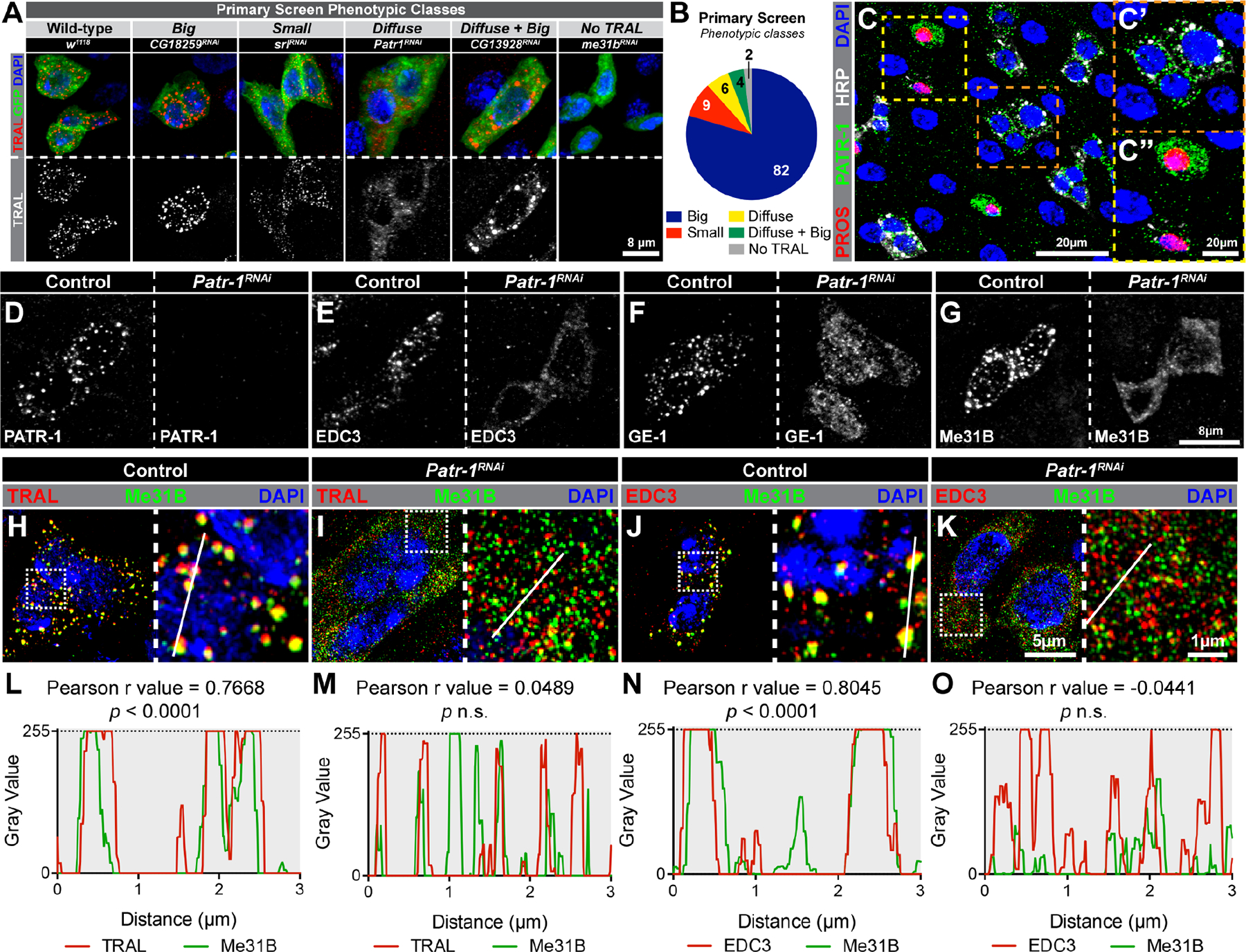
(A) Progenitor cells stained for TRAL (red), progenitor cells (green), and DAPI (blue) after knockdown of selected genes. (B) Numbers per phenotypic class. (C) PMG stained for PROS (red), PATR-1 (green), HRP (white) and DAPI (blue). C’ and C” are enlargements of indicated regions. (D-G) Progenitor cells from control and Patr-1 RNAi intestines stained for PATR-1, EDC3, GE-1, or Me31B. (H-K) Super-resolution micrographs of control and Patr-1 RNAi intestinal progenitors stained for Me31B (green), DAPI (blue) and either TRAL or EDC3 (red). (L-O) Normalized pixel-by-pixel fluorescent intensity profiles of indicated protein (TRAL, Me31B or EDC3) along white lines in H-K. Full genotypes listed in Data S1F. See also Figure S2.
PATR-1 is a P-body resident protein that is necessary for their formation
To investigate the function of P-bodies in progenitor cells, we focused on Patr-1, a member of the diffuse class and known nucleator of P-bodies in other systems.40–45 Using a verified antibody,45 we found PATR-1, like other mRNP proteins, was enriched in the cytoplasm with a distinctive granular organization (Figure 2C, 2C’). PATR-1 also showed strong but less punctate expression in ~45% of EEs (compare Figure S1C–D to Figure 2C”). PATR-1 colocalized with TRAL and Me31B (Figure S2G–H, J–K) but not with IPSG protein FMRP (Figure S2I, L). We therefore used the PATR-1 antibody as an independent P-body marker to verify the 103 genes identified above and found analogous effects in all cases except for the “No TRAL” category (Figure S2M–R). Smaller than normal PATR-1 granules were observed in this class (Figure S2R), suggesting that overall P-body morphology rather than just TRAL staining was affected. Collectively, these data identified PATR-1 as a P-body component in intestinal progenitors of adult Drosophila.
To test whether PATR-1 recruited other P-body proteins, we verified that Patr-1 RNAi eliminated PATR-1 protein expression (Figure 2D) and then analyzed its effect on EDC3, GE-1 and Me31B distribution. The prominent punctate pattern and colocalization of these proteins was lost (Figure 2E–O), indicating a failure in P-body assembly as well as P-body protein interaction. To verify these phenotypes, we generated a series of Patr-1 null alleles that were homozygous lethal, died as third instar larvae, displayed the same lethal phase when in trans to a deficiency, and lacked PATR-1 protein by Western blot (Figure S3A–B). Using two representative alleles, Patr-1P105Δ and Patr-1R107FS1, we generated Patr-1 mutant adult progenitors with the Mosaic Analysis with a Repressible Cell Marker (MARCM) technique.46 Seven days after clone induction, intestines were dissected and either stained for PATR-1 or scored for P-body morphology based on Me31B and EDC3 distribution. PATR-1 staining was not detected in either Patr-1P105Δ or Patr-1R107FS1 mutant cells (Figure S3C–D) but was restored by the P21M20 rescue transgene45 (Figure S3E). Like Patr-1 RNAi, P-body localization of Me31B and EDC3 was completely disrupted in Patr-1P105Δ and Patr-1R107FS1 clones (Figure S3F–G and I–J); these defects were rescued by P21M20 (Figure S3H and K). Taken together, these data indicated that PATR-1 was required for P-body assembly in intestinal progenitors.
Loss of PATR-1 leads to progenitor cell loss
We next used Patr-1 RNAi to analyze the effect of P-body loss in progenitor cells on the numbers of various intestinal cell types (Figure 3A). Intestinal progenitor cell number gradually declined over time and was almost completely absent after 15 days of Patr-1 RNAi (Figure 3B, F–I), consistent with other members of the diffuse class, all six of which displayed moderate to severe progenitor cell loss (Data S1B). It was also consistent with the absence of Delta-positive (Dl+) ISCs after 15 days of Patr-1 RNAi (Figure S3L–M). In contrast, EE and EC cell percentages remained unchanged or increased over 15 days (Figure 3C–D). The total number of cells per field, however, was significantly reduced at 15 days (Figure 3E) and intestines appeared shrunken (Figure 3G and I), at least in part reflecting the loss of progenitor cells. Consistent with these results, Patr-1P105Δ and Patr-1R107FS1 mutant clones were smaller and contained fewer ISCs compared to control (Figure S3P–X); these defects were rescued entirely by the P21M20 transgene. To further validate the loss of mitotic cells in the absence of PATR-1, we fed control and Patr-1 RNAi flies for 14 days with bleomycin, a chemical that induces rapid ISC proliferation, and quantified the number of mitotic cells one day later by staining for phosphorylated-histone 3 (pH3).47 Consistent with progenitor loss, intestines expressing Patr-1 RNAi for 15 days displayed significantly reduced numbers of pH3+ cells (Figure S3Y). Taken together, these data suggested that progenitors were lost and intestinal epithelial integrity was compromised due to a failure in P-body assembly.
Figure 3. PATR-1 maintains intestinal progenitor cells.
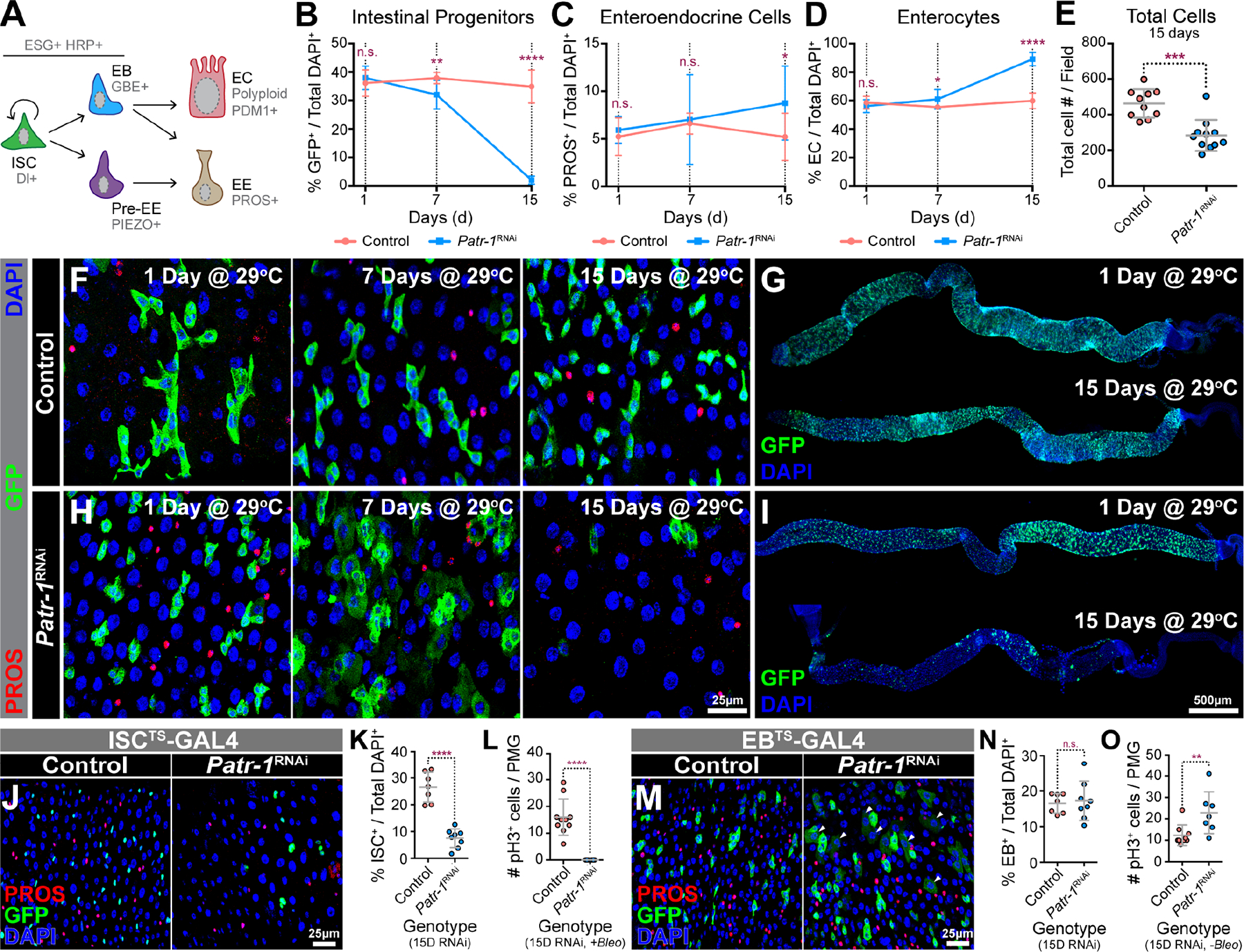
(A) Intestinal cell types and markers. (B-D) Normalized intestinal progenitor cell, EE cell, or EC percentages in esgTS (pink) and esgTS/Patr-1 RNAi (blue) PMGs (n=8-to-11) after 1, 7 and 15 days at 29°C. (E) Total cells per field in esgTS (n=10) and esgTS/Patr-1 RNAi (n=11) PMGs after 15 days at 29°C. (F, H) esgTS or esgTS/Patr-1 RNAi PMGs after 1, 7 and 15 days at 29°C stained for PROS (red), GFP (green) and DAPI (blue). (G, I) esgTS and esgTS/Patr-1 RNAi PMGs after 1 and 15 days at 29°C stained for GFP (green) and DAPI (blue). (J) ISC-KCKT-GAL4TS or ISC-KCKT-GAL4TS/Patr-1 RNAi PMGs after 15 days at 29°C stained for PROS (red), GFP (green) and DAPI (blue). (K-L) Normalized ISC percentage (n=7 or 8) or number of pH3+ cells per PMG after bleomycin feeding (n=10 or 7) of genotypes shown in J. (M) gbeTS or gbeTS/Patr-1 RNAi 15 days after shifting to 29°C and stained for PROS (red), GFP (green) and DAPI (blue). (N-O) Normalized EB percentage (n=7 or 8) or number of pH3+ mitotic cells per PMG (n=9 or 7) without bleomycin feeding of genotypes shown in M. Error bars on plots show mean±s.d. and asterisks denote statistical significance from Unpaired t-test (B-D, K, N O) or Mann-Whitney test (E, L). See also Figure S3.
Given that P-bodies were present in both ISCs and EBs, we assessed whether P-body function was necessary in each cell type by driving Patr-1 RNAi with either ISCTS- or EBTS-GAL4.48,49 Like esgTS-based knockdown, RNAi of Patr-1 in ISCs for 15 days led to a marked reduction in ISCs, as labeled by GFP (Figure 3J–K), Dl (Figure S3N–O), or pH3 after bleomycin feeding (Figure 3L). In contrast, RNAi using EBTS-GAL4 had no effect on EB cell number after 15 days (Figure 3M–N), although it did result in a marked increase in ISC proliferation (Figure 3O) and the emergence of a weakly GFP+ EC-like polyploid cell population (Figure 3M). These observations suggested that Patr-1 loss caused both an increase in the rate of EB differentiation and a commensurate increase in the rate of ISC proliferation that maintained EB number at normal levels, reminiscent of the recent finding that EB activation and loss of cell adhesion induces ISC proliferation.50 Taken together, these results indicated that P-bodies maintained ISCs but not EBs.
PATR-1 represses pro-differentiation genes in intestinal progenitors
To identify P-body mRNAs whose misregulation caused this ISC loss, we performed transcriptomic profiling of control and Patr-1-deficient intestinal progenitors collected via FACS (fluorescence activated cell sorting) (Figure 4A, S4A). Patr-1 knockdown upregulated 703 genes and downregulated 523 genes, most significantly Patr-1 mRNA, in progenitor cells (Figure 4B–D, Data S1C). Upregulated mRNAs were significantly longer and had a lower GC content than unchanged genes (3336.1 ± 143.9 bp vs 2698.4 ± 27.7 bp, and 44.1 ± 0.2 %GC vs 46.1 ± 0.1 %GC, respectively) (Figure S4B), consistent with recent studies.51 Gene Ontology (GO) analysis indicated cell differentiation genes were significantly upregulated, in agreement with the identities of mRNAs found in HEK293 P-bodies (Figure S4C).19 In contrast, key pro-apoptotic and -EE fate mRNAs were unchanged (Data S1C), suggesting that Patr-1 mutant progenitor cells were lost due to inappropriate differentiation caused by EC gene expression. Similar effects are associated with other progenitor proteins, including the Snail family transcription factor Esg and its co-factor Verthandi (Vtd, also known as Rad21).52,53 Notably, 22% (92/410 genes) and 18% (80/454 genes) of the significantly upregulated genes in esg-RNAi and vtd-RNAi overlapped with Patr-1 RNAi upregulated genes (Figure S4D–E, Data S1C–E). These included known EC-marker genes such as nubbin (nub, also known as POU-domain protein 1 or Pdm1), Myosin61F (Myo61F), and big bang (bbg). Taken together, these results suggested that esg, vtd, and Patr-1 act in progenitors to repress an overlapping cohort of pro-EC genes.
Figure 4. PATR-1 limits pro-differentiation gene expression in intestinal progenitor cells.
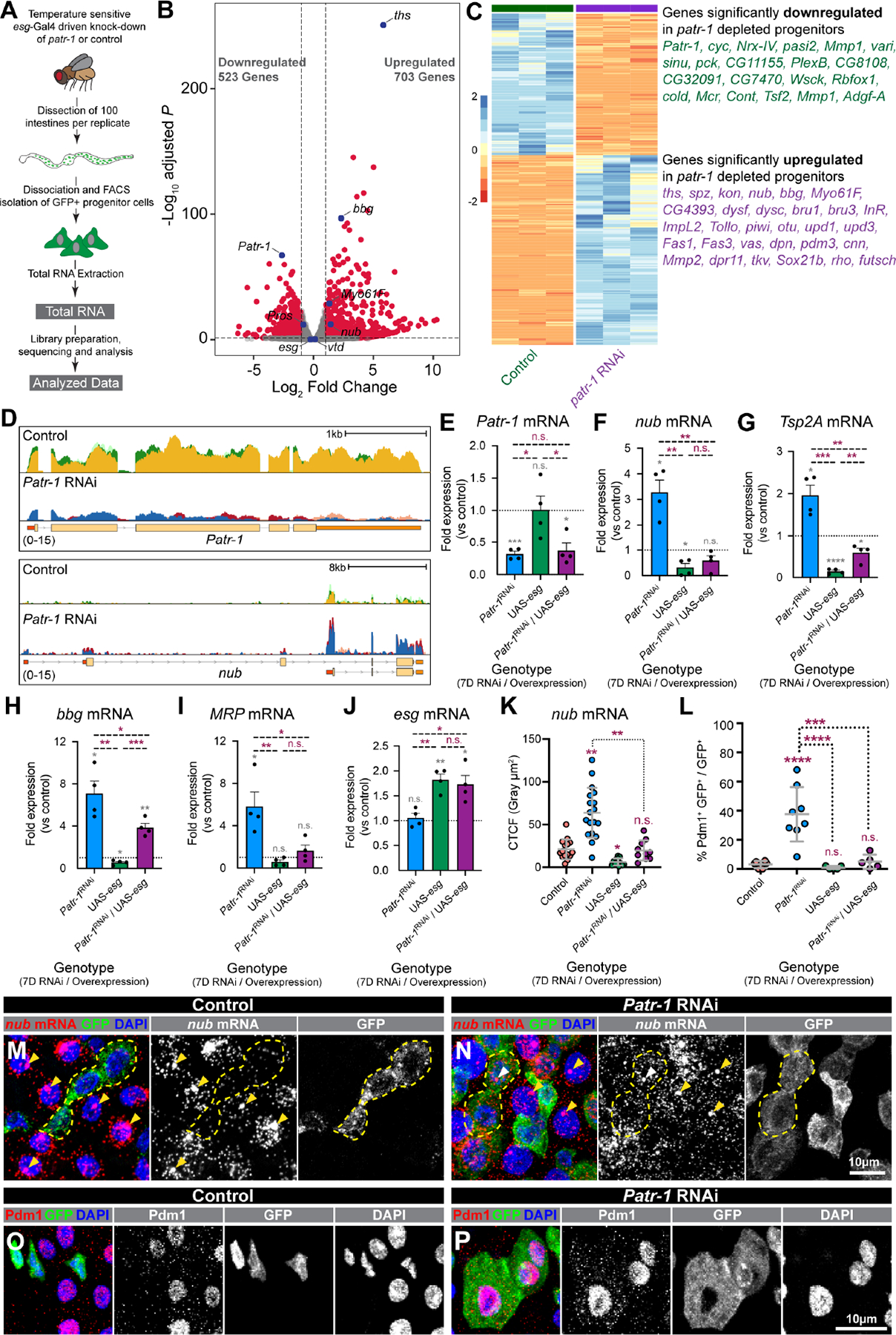
(A) Schematic of RNA-seq. (B-C) Volcano plot, heatmap, and selected list of differentially expressed genes in Patr-1 RNAi versus control. (D) CPM-normalized read counts at Patr-1 and nub loci from control and Patr-1 RNAi; replicates are overlayed. (E-J) qPCR levels of Patr-1, nub, Tsp2A, bbg, MRP, and esg mRNA in intestinal progenitors from indicated genotypes compared to control. (K) Normalized nub mRNA fluorescence intensity in progenitor cells of indicated genotypes (n=15, 19, 12, or 10 cells) (L) Percentage of intestinal progenitors with nuclear Pdm1 staining from indicated genotypes (n=6, 8, 5, or 5) after 7 days at 29°C. (M-P) Intestinal progenitors (outlined in yellow) from esgTS or esgTS/Patr-1 RNAi stained for GFP (green), DAPI (blue), and either nub mRNA (red) or Pdm1 (red). Yellow/white arrowheads indicate putative sites of active transcription in ECs/progenitors. Error bars show mean±s.d. and asterisks denote statistical significance from one sample t (E-J, grey), Unpaired t-test (E-J, purple), Kruskal-Wallis test (K) or ordinary one-way ANOVA with Turkey’s multiple comparison test (L). Full genotypes listed in Data S1F. See also Figure S4.
PATR-1 limits cytoplasmic nub mRNA levels in progenitor cells
This transcript profiling also indicated that EC genes were weakly expressed in wildtype progenitors despite the presence of transcriptional repressors like Esg known to target them.53 For example, low levels of nubbin as well as Tetraspanin 2A (Tsp2A), bbg, and Multidrug-Resistance like Protein 1 (MRP) were detected by RNA-sequencing (Figure 4D, S4F) and subsequently confirmed by quantitative PCR (qPCR), which also confirmed their upregulation in FACS-isolated Patr-1 mutant cells (Figure 4F–I). To directly confirm these observations, we analyzed the subcellular distribution and levels of nub transcript in wildtype intestines using RNAscope in situ hybridization probes verified for specificity after nub knockdown (Figure S4G–H). As expected, nub transcript was readily detected in ECs in both the cytoplasm as well as in bright nuclear punctae that were interpreted as sites of active transcription (Figure 4M, yellow arrowheads). In addition, nub transcript was more weakly detected in the cytoplasm of ~33% (32/98) wildtype progenitors, with low-to-no signal in the nuclei of these cells (Figure 4M, S4I). In comparison, the nub protein product Pdm1 was more rarely detected in just ~3% (9/272) of wildtype progenitors (Figure 4L, 4O). These results indicated that the transcriptional repression of pro-differentiation genes was incomplete and a cytoplasmic gene regulatory mechanism ensured the maintenance of progenitor identity in wildtype progenitors.
To analyze the role of P-bodies in this process, we analyzed nub mRNA and Pdm1 protein patterns and levels in progenitors after Patr-1 knockdown. In comparison to wildtype progenitors, cytoplasmic nub mRNA staining was noticeably higher in ~63% (59/94) Patr-1 deficient intestinal progenitors (Figure 4N, S4I) and, consistently, cellular nub mRNA staining levels were 3-fold higher in Patr-1-deficient vs. wildtype progenitors (Figure 4K). In addition, some Patr-1 intestinal progenitors also displayed nuclear punctae, which we had detected in wildtype ECs but rarely in wildtype progenitors. However, only a 40% (23/59) subset of the Patr-1 mutant progenitor cells with elevated cytoplasmic staining also displayed bright nuclear punctae (Figure 4N, white arrowhead and Figure S4I), indicating that elevated cytoplasmic expression did not require nuclear expression. In addition, the number of progenitors that expressed Pdm1 protein was significantly higher after Patr-1 RNAi in comparison to control (Figure 4L, 4P, S4J–M). RNA-seq and qPCR analysis indicated that neither esg nor vtd expression levels were affected by Patr-1 loss (Figure S4N–O and Figure 4J), suggesting that the increases in nub transcript and Pdm1 protein observed after Patr-1 RNAi were not likely due to dampened transcriptional repression. Collectively, these observations indicated that PATR-1-mediated P-body assembly in progenitors was required to limit the expression of pro-differentiation genes such as nub.
Loss of P-body assembly promotes progenitor-EC differentiation
The elevated Pdm1 protein and transcript levels detected in Patr-1 mutant progenitors suggested that P-body loss caused premature progenitor-to-EC differentiation. Consistently, forced expression of Pdm1 in progenitors also led to progenitor loss (Figure 5A). In addition, prior to their loss, both the cell and nuclear areas of Patr-1 mutant progenitors were significantly increased (Figure 5B–C), suggesting the acquisition of polyploid EC characteristics. EC differentiation was also supported by the absence of detectable increases in either GFP+ PROS+ or apoptotic cells (Figure S5A–C), ruling out EE differentiation or cell death as causes of progenitor loss. Further supporting the inverse correlation between EC cell fate and the presence of P-bodies, we noted that bleomycin ingestion, which leads ISCs to rapidly proliferate and differentiate,54 led to P-body dissolution (Figure 5D–G, S5D–E). These data showed that P-bodies were required for progenitor maintenance and were associated with the absence of EC features (e.g. large cell size, nuclear size, Pdm1 expression).
Figure 5. Loss of PATR-1 promotes progenitor-to-EC-differentiation.
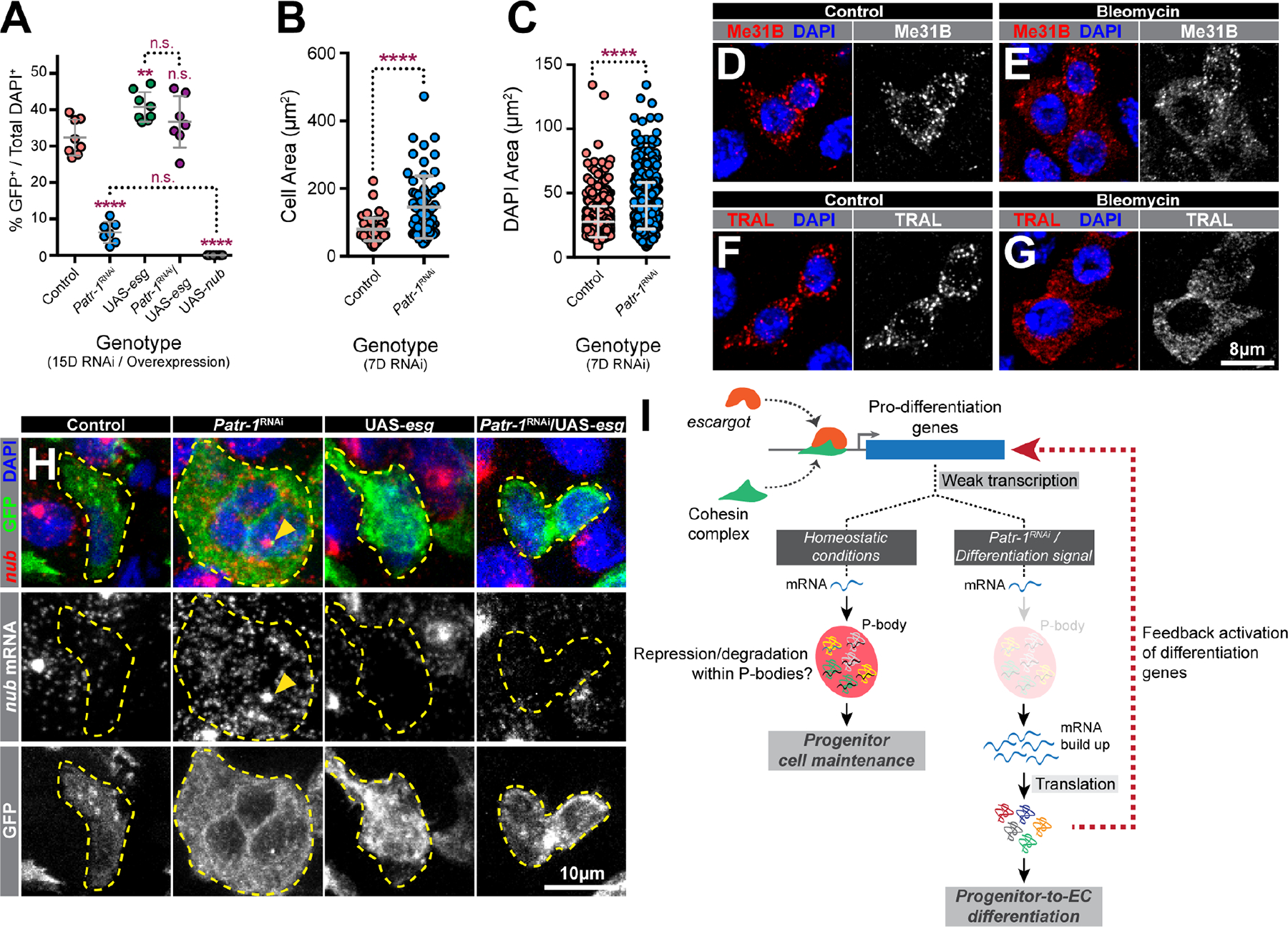
(A) Percentages of progenitor cells in PMGs of indicated genotypes (n=8, 7, 8, 7, or 8). (B-C) Cell or nuclear area of intestinal progenitor cells from esgTS or esgTS / Patr-1 RNAi intestines. (D-G) Progenitor cells from flies fed 5% sucrose (control) or 5% sucrose plus 25 μg/ml bleomycin (bleomycin) stained for DAPI (blue) and Me31B (red) or TRAL (red). (H) Intestinal progenitors (outlined) from indicated genotypes stained for nub mRNA (red), GFP (green) and DAPI (blue). Yellow arrowheads indicate putative sites of active transcription. (I) Model. Error bars on plots show mean±s.d. and asterisks denote statistical significance from Ordinary one-way ANOVA with Turkey’s multiple comparison test (A) or Mann-Whitney test (B-C). Full genotypes listed in Data S1F. See also Figure S5.
Overexpression of esg rescues Patr-1 RNAi mediated progenitor cell loss
The presence of nub transcript in wildtype progenitors suggested that P-bodies limited the expression of weakly transcribed pro-differentiation genes to maintain progenitor identity. To test whether the overexpression of the transcriptional repressor esg, which we hypothesized would block this weak transcription, eliminated the requirement of mature P-body formation to prevent differentiation, we analyzed the effects of progenitor expression of Patr-1 RNAi and esg transgenes, both individually and in combination, on progenitor cell numbers. As previously shown, knocking down Patr-1 reduced while esg overexpression increased the number of progenitors (Figure 5A, S5F). However, elevated nub/Pdm1 and the ensuing progenitor loss associated with Patr-1-RNAi expression was eliminated when esg was also co-expressed (Figure 4K, 4L, 5A, S5F–G). As expected, esg but not Patr-1 transcript abundance was elevated in progenitors expressing esg (Figure 4E, J). Consistent with our model, this ectopic esg expression significantly reduced nub mRNA level in both wildtype and Patr-1 mutant progenitors, detected by both qPCR and RNAscope (Figure 4F, 4K, 5H). Moreover, esg co-overexpression decreased the upregulation of Tsp2A, bbg, and MRP in the absence of Patr-1 (Figure 4G–I), indicating that this regulation is prevalent among pro-differentiation targets of Esg. To verify that the rescued progenitor cells in the Patr-1 RNAi/esg background included bona fide ISCs, we assessed the mitotic potential of intestines from these genetic backgrounds following bleomycin feeding. As expected, overexpression of esg in Patr-1 deficient intestinal progenitors elevated the total number of mitotic ISCs (Figure S5I). However, mature P-bodies were not restored, as indicated by diffuse TRAL staining in these progenitors (Figure S5H). Taken together, enhanced transcriptional repression of pro-differentiation genes associated with esg overexpression can prevent premature progenitor differentiation independent of mature P-body assembly.
Discussion
This study molecularly and functionally characterized stem cell mRNPs that we concluded are P-bodies based on three observations: (i) they contained colocalized protein complexes that include fly orthologs of proteins known to localize to P-bodies in mammalian and yeast cells, (ii) these mRNP granules were significantly larger than and show no colocalization with IPSG protein foci under controlled conditions, and (iii) acute stresses increased the size of these mRNPs and promoted colocalization with IPSGs. A targeted genetic screen identified 39 previously known and 64 new genes that influenced P-body morphology, including six required for P-body formation. To examine stem cell P-body function, we characterized one of this latter class, PATR-1, an evolutionary conserved protein with both translational repression and mRNA decay functions that is necessary for proper P-body assembly in Saccharomyces cerevisiae.15,40,43,44,55–57 Depletion of P-bodies in progenitor cells upregulated the expression of pro-differentiation genes, including nubbin. Loss of stem cell P-bodies, either by genetic depletion or differentiation, led to the increased translation as well as the cytoplasmic, but not nuclear, abundance of such transcripts. We therefore propose that mature P-bodies are necessary for stem cell maintenance by post-transcriptionally enforcing the repression of transcriptional programs that promote differentiation (Figure 5I).
We also used quantitative super-resolution microscopy to visualize substructures present within mature P-bodies. Consistent with the proposed “core-shell” structure of stress granules,14,38,58,59 P-bodies exhibit “cores” with high protein concentrations and “shells” with low protein concentration. Notably, Drosophila intestinal progenitor P-bodies have a ~125 nm diameter, as compared to larger P-bodies in HEK293 cells, which have a ~500 nm diameter.19 In addition, intestinal progenitors contain ~35–45 mature P-bodies while HEK293 cells contain only ~4–7 granules per cell, indicating that the size and number of mature P-bodies depends on cell type and species and may scale with overall cell size.
A recent study documented the presence of P-bodies in cultured human pluripotent stem cells and suggested their presence in adult stem cells.26 This analysis of DDX6, the ortholog of Drosophila Me31B, showed that DDX6-dependent P-bodies could both promote and repress stem cell identity, depending on context. For example, loss of DDX6 expanded endodermally derived Lgr5+ ISCs or ectodermally derived neural progenitor cell populations, but promoted the differentiation of other progenitor cell populations, including mesodermally derived progenitors. We confirm the presence of mature P-bodies in adult progenitor populations but show that they repress differentiation rather than increasing their proliferation, as in Lgr5+ ISCs. A few possible explanations could reconcile these results. Most simply, Drosophila intestinal progenitors behave more like mesodermally derived mammalian progenitors rather than endo- or ectodermally derived mammalian progenitors. Alternatively, the stem cell function of DDX6 might be affected by its roles in surrounding cells, since DDX6 was targeted in cells throughout mouse intestinal organoids, whereas PATR-1 was specifically targeted in progenitor cells in our study. Finally, DDX6-mediated P-body function might be modulated by signaling that is not fully recapitulated in in vitro-derived stem cell models.
The exact molecular function of P-bodies is a matter of current debate.44 Consistent with other recent studies,1,2,20 our analysis suggests intestinal progenitor P-bodies have both translational repressive and mRNA degratory functions. We found that Pdm protein was absent in progenitors despite the weak expression of nub mRNA, suggesting P-body dependent translational repression. In addition, RNAscope analysis showed that cytoplasmic nub transcript abundance was increased in Patr-1 mutant progenitors without an indication of nuclear transcription, suggesting the stabilization of transcripts that are targeted for degradation via P-bodies. Dual P-body roles in mRNA repression and degradation are also suggested in human cultured stem cells. For instance, P-bodies influence the translation of transcripts encoding fate-instructive transcription and chromatin factors in cultured embryonic and in vitro-derived adult stem cells.26 In addition, P-body proteins DDX6 and EDC3 are known to destabilize differentiation-inducing mRNAs such as KLF4 in human epidermal progenitor cells, although it is important to note that P-bodies have not been reported in these cells.61 Notably, the mammalian homolog of nub, OCT1/POU2F1, is one of the top 30 most enriched mRNAs of P-bodies in HEK293 cells, indicating evolutionary conservation of P-body targets.19
We propose that weak transcription of pro-differentiation genes likely maintains progenitors in a state primed for differentiation. The transcriptional repression of differentiation genes by the transcription factor esg is a key regulatory step of intestinal stem cell maintenance.52,53,62–65 The loss of esg gene expression or inability to localize the Esg protein to target genes promote progenitor loss via premature ISC-to-EC differentiation.52,53 Similar to Patr-1 RNAi, knocking down esg itself as well as either vtd, Nipped-B or polo, all of which are necessary for recruiting Esg to target promoters, markedly upregulated the expression of Pdm1 in intestinal progenitors.52,53 Notably, our transcriptomic profiling showed that the transcript level of neither esg nor any of the Esg-targeting proteins, vtd, Nipped-B or polo, was changed by the absence of mature P-bodies. These observations suggest that Esg protein level, its proper promoter targeting, and its transcriptional repression of EC-genes are all unlikely to be affected by the loss of PATR-1.
In addition to identifying 64 new genes affecting P-body morphology, we expect that the tissue-based stem cell P-body system identified and described here will prove critically useful in screening for chemicals, diet conditions and stress conditions that alter P-body assembly as well as performing larger, genome-wide screens to comprehensively characterize the molecular pathways that control P-body assembly. Moreover, similar approaches can be used to identify systemic signals that promotes P-body disassembly during the onset of differentiation as well as to identify molecular players of P-body disassembly.
STAR Methods
RESOURCE AVAILABILITY
Lead contact
Further information and requests for resources and reagents should be directed to and will be fulfilled by the lead contact, Nicholas S. Sokol (nssokol@gmail.com).
Materials availability
All fly strains and other materials used in these studies are available upon request.
Data and code availability
RNA-seq data have been deposited at GEO and are publicly available as of the date of publication. The accession number is listed in the key resources table.
This paper does not report original code.
Any additional information required to reanalyze the data reported in this paper is available from the lead contact upon request.
EXPERIMENTAL MODEL AND SUBJECT DETAILS
Drosophila strains and husbandry
Age matched female flies were used in all experiments. Fly strains were cultured on standard Bloomington Drosophila stock center media (https://bdsc.indiana.edu/information/recipes/bloomfood.html) and reared in 18°C, 25°C and 29°C incubators set for a 12hr light/dark schedule and 65% humidity. Flies were cultured in groups of 15–20 (typically 5 males and up to 15 females). All strains used in individual panels are listed in Data S1F. For temporal and regional gene expression-targeting (TARGET) experiments, flies were grown at 18°C, collected over 2 days, and reared in 29°C for up to 15 days before being dissected. For clonal analysis using mosaic analysis with repressible cell marker (MARCM) method, animals were reared at 25°C until eclosion, collected over 2 days and heat-shocked immediately at 37°C for 45 min in a Lauda circulating water bath. Subsequently, flies were reared at 25°C for 7–15 days.
METHOD DETAILS
Construction of new strains
CRISPR/cas9 mediated generation of Patr-1 alleles
New Patr-1 alleles were generated by co-injection of two guide RNA (gRNA) plasmids. The gRNA plasmids were generated by subcloning annealed oligos encoding gRNA 5’-CATGTTGAACATGTTATACATGG-3’ as well as annealed encoding gRNA 5’-TGTGACGAGACTGTCGGAAGGGG-3’ into the BbsI site of pU6-BbsI-chiRNA (Gratz et al 2013). The two resulting plasmids were sequence-verified, amplified, mixed, and co-injected into strain y1 sc* v1 sev21 ; P{nos-Cas9}attP2 (BL78782) by Rainbow Genetics (Camarillo CA). Stocks of F1 progeny were generated, and strains containing new Patr-1 alleles were selected based on non-complementation of the lethality associated with P{EPgy2}Patr-1EY10289 (BL19805). New Patr-1 alleles were molecularly defined by sequencing PCR products generated from genomic DNA obtained from homozygous mutant larvae with oligos flanking the gRNA locations. Two new Patr-1 alleles as well as P{EPgy2}Patr-1EY10289 were recombined onto P{FRT}82B-containing chromosomes using standard meiotic methods.
Antibody generation
Anti-TRAL antibodies were generated in rats (Cocalico Biologicals, Reamstown PA) against a 6XHIS-tagged version of an N-terminal portion of TRAL that was expressed and purified according to standard methods. The TRAL-encoding plasmid was generated by PCR amplifying a 477bp fragment that encodes the first 160 amino acids of TRAL from cDNA GH08269 (DGRC stock 4838) with high-fidelity Q5 polymerase (NEB), subcloning the resulting PCR product into the NcoI and EcoRI sites of pHIS.parallel using HiFi DNA Assembly Master Mix (NEB), and sequence-verifying the resulting plasmid to confirm the absence of any PCR-induced errors.
Anti-EDC3 antibodies were generated in rabbits and rats (MBL, Japan) against a 6XHIS-tagged version of an N-terminal portion of EDC3. The edc3 coding sequence (corresponding to amino acid 1–440) was PCR-amplified from an ovarian cDNA library using Phusion DNA polymerase (Finnzymes), with primers 5’-GGGAATTCATGGGTCCGACGGATCAAGA-3’ and 5’-GGGGCGGCCGCTCACTTATCGGCACTTATCTCGA-3’. The fragment was digested by EcoRI and NotI, and cloned into the pProEX HTa vector (Life Technologies). The 6×His-tagged Edc3 (1–440) protein was expressed in E. coli BL21 cells by IPTG induction, and purified using Ni-NTA agarose resin (Qiagen) under denaturing conditions. Protein was eluted from the resin and, based on Coomassie stain, the fractions with the most protein were further purified by disc preparative SDS-PAGE using the NA-1800 apparatus (Nihon Eido, Japan). Purified protein fractions were concentrated with Vivaspan-2 (10,000 MWCO PES; Sartorius), dialyzed against PBS containing 4M urea followed by PBS/2M urea. Polyclonal antibodies were affinity-purified with the same antigen immobilized on HiTrap NHS columns (GE Healthcare).
Dissections and immunostaining
Gastrointestinal (GI) tracts of adult female flies were dissected in ice-cold 1×PBS and fixed in 4% w/v paraformaldehyde (Electron Microscopy Sciences, Cat. No. 15714) in PBS for 45 min. These samples were washed with 1×PBT (1×PBS, 0.1% v/v Triton X-100) and then blocked (1×PBT, 0.5% w/v Bovine Serum Albumin) for at least 45 min. Subsequently, samples were incubated at 4°C overnight with primary antibodies, including rabbit anti-GFP (A11122, Life Technologies, 1:1000), mouse anti-V5 (MCA1360GA, Bio-Rad, 1:250), mouse anti-FLAG (F3165, Sigma, 1:1000), rabbit anti-HA (3724S, Cell Signaling Technology, 1:1000), mouse anti-FMR1 (5A11, Developmental Studies Hybridoma Bank, 1:100), mouse anti-Prospero (MR1A, Developmental Studies Hybridoma Bank, 1:100), mouse anti-Delta (C594.9B, Developmental Studies Hybridoma Bank, 1:500), rat anti-TRAL (this study, 1:1500), rabbit anti-Me31B32 (1:2000), mouse anti-Me31B34 (1:1500), rabbit anti-Ge-133 (1:500), rat anti-EDC3 (this study, 1:1500), rabbit anti-EDC3 (this study, 1:2000) and rabbit anti-Pdm1 (a gift from Xiaohang Yang) (1:1500). Samples were washed and incubated for 2–3 hours with secondary antibodies, including AlexaFluor-488 and −568-conjugated goat anti-rabbit, -mouse and -rat antibodies (Life Technologies, 1:1000). AlexaFluor-647 conjugated goat-HRP antibodies were used in the secondary antibody solution whenever required. Samples were washed and treated with DAPI (1:10000) and mounted in Vectashield (Vecta Laboratories). An alternative staining protocol was used for Delta staining as described in Buddika et al.,202048. First, intestines were dissected in ice-cold Grace’s insect medium (Lonza Bioscience) and fixed in a 1:1 (v/v) mixture of heptane (Sigma) and 4% w/v paraformaldehyde (Electron Microscopy Sciences) in water for 15 min. Subsequently, the bottom aqueous paraformaldehyde layer was removed, 500 μl of ice-cold methanol added, the mixture was shaken vigorously for 30 seconds, the methanol-heptane mixture was removed and intestines were incubated with 1 ml ice-cold methanol for 5 min. Next, samples were gradually rehydrated with a series of 0.3% v/v PBT (1× PBS, 0.3% v/v Triton X-100):methanol (3:7, 1:1, 7:3) washes, washed with 0.3% v/v PBT alone for another 5 min, blocked (0.3% v/v PBT, 0.5% w/v BSA) for at least 45 min and then the primary and secondary antibody staining were done as described above. Samples were mounted in ProLong Diamond mounting medium (Invitrogen). Cell death analysis was performed using the ApopTag® Fluorescein In-Situ Apoptosis Detection Kit (Sigma-Aldrich, Cat. No. S7110) following manufacturer’s instructions.
RNAscope in situ hybridization
Adult female flies were dissected in ice cold 1× PBS and fixed in 4% w/v paraformaldehyde (Electron Microscopy Sciences, Cat. No. 15714) in PBS for 45 min. Tissue was then washed in 1×PBT (1×PBS, 0.3% v/v Triton X-100) 3 times 5 min each. Next, samples were gradually dehydrated with a series of 0.3% v/v PBT (1×PBS, 0.3% v/v Triton X-100): Methanol (7:3, 1:1, 3:7) washes and incubated in Methanol for 10 min. Then tissue was rehydrated with a series of 0.3% v/v PBT: Methanol (3:7, 1:1, 7:3) washes and washed with 0.3% v/v PBT alone for another 5 min. For following steps, reagents from RNAscope Multiplex Fluorescent Reagent Kit v2 assay were used. Fixed tissue was transferred into 0.2 ml PCR tubes and incubated in RNAscope Protease III reagent for 5 min at 40°C (PCR thermal cycler was used for all the incubations at 40°C). Samples were immediately washed with 1× PBS twice and 20μl of RNAscope probes for nub (pre warmed to 40°C) (ACD Bio., Cat. No. 523981) was added. Samples were incubated at 40°C overnight. After overnight incubation, samples were washed twice with 1X RNAscope wash buffer. Next, RNAscope Multiplex FL v2 AMP 1, RNAscope Multiplex FL v2 AMP 2, RNAscope Multiplex FL v2 AMP 3 and RNAscope Multiplex FL v2 HRP-C1 steps were done as described in Chapter 4 of Fluorescent v2 assay manual, ACD Bio. Finally, samples were incubated for 30 min at 40°C with Opal 620 (AKOYA Biosciences, Cat. No. FP1495001KT, 1:1500 in TSA buffer) and washed with 1X RNAscope wash buffer and counterstained with DAPI. Samples were mounted in ProLong Diamond mounting medium (Invitrogen).
Microscopy and image processing
Images were collected on either a Leica SP8 Scanning Confocal microscope (Leica DMi8 inverted microscope platform; equipped with WLL 470–670 nm, 405 nm and 440 nm lasers, Huygens deconvolution software; controlled by Leica LAS-X software; image acquisition using Leica HC PL APO CS2 63x/1.40 or Leica HC PL APO CS2 40x/1.40 lens with Leica Type F Immersion Liquid (N = 1.518)) or an OMX 3D-SIM Super-Resolution microscope (DeltaVision OMX system; equipped with 405, 488, 561, 642 nm lasers; controlled by AquireSR software; image processing by SoftWorx imaging software; image acquisition using an Olympus PL APO N 60x/1.42 lens with Applied Precision Immersion Oil (N = 1.518)) available at the Light Microscopy Imaging Center, Indiana University, Bloomington. Whenever possible, samples to be compared were collected under identical settings on the same day, image files were adjusted simultaneously using Adobe Photoshop CC, and figures were assembled using Adobe Illustrator CC. Note that all the intestinal progenitor cells that we have imaged were verified to be progenitors by using HRP as a marker.
Ex vivo treatments
All ex-vivo treatments were performed as described in Buddika et al. 202018. Briefly, intestines from females aged 8–10 days on normal diet were incubated in Krebs-Ringer media/KRB (Alfa Aesar, Cat. No. J67591) or KRB supplemented with 1mM rapamycin (LC Laboratories, Cat. No. r-5000) for 60 min and then fixed.
Bleomycin feeding assay
Female flies were maintained on standard Bloomington Drosophila stock center media for an appropriate time and then separated into two cohorts. Cohort 1 and 2 were transferred to a vial with a chromatography paper soaked in either 5% w/v sucrose in water (control) or 5% w/v sucrose and 25 μg/ml bleomycin in water, respectively. Based on experiments, flies were dissected 24-to-36 hours after feeding.
Protein isolation and western blot analysis
Wandering L3 larvae or adult female flies were used for protein isolation. Collected larvae or adult flies were lysed in I-RIPA protein lysis buffer (150mM NaCl, 50mM Tris-HCl pH 7.5, 1mM EDTA, 1% v/v Triton X-100, 1% w/v Na Deoxycholic Acid, 1xprotease inhibitor cocktail), protein extracts were resolved on a 4–20% w/v gradient polyacrylamide gel (Bio-Rad, Cat. No. 456–1093), transferred to Immobilon®-P membrane (Millipore, Cat. No. IPVH00010) and probed with rat anti-TRAL (1:2000, this study), rat anti-PATR145 (1:2000) or mouse anti-a-tubulin (12G10, Developmental Studies Hybridoma Bank, 1:1000) antibodies. Subsequently, blots were washed extensively with 1×TBST (1×TBS, 0.1% v/v Tween-20) and incubated with anti-rat or -mouse conjugated HRP secondary antibodies. After extensive secondary washes with 1×TBST, blots were treated with ECL-detection reagent 1 and 2 (Thermo Scientific, Cat. No. 1859701 and 1859698) and finally exposed to chemiluminescence films (GE Healthcare, Cat. No. 28906839) and developed the signal.
FACS isolation of progenitor cells, RNA-seq library preparation and qPCR
Gastrointestinal (GI) tracts of 100 adult female flies (per replicate, reared at 29°C for 6 days) were dissected in ice-cold 1×PBS. Then cells were dissociated by treating intestines with 1mg/ml elastase at 27°C for 1 hour with agitation. Subsequently, ~30,000–50,000 GFP+ intestinal progenitor cells were sorted using a BD FACSAria™ II flow cytometer equipped with a 100μm nozzle at the IUB Flow Cytometry Core Facility. Total RNA was prepared using the TRIzol® LS reagent (Ambion). The rRNA-depleted libraries were prepared using the Ovation® SoLo RNA-seq system (Part No. 0502 including Parts 0407 and S02240) following manufacturer’s instructions. The quality and quantity of final libraries were assessed using Agilent 2200 TapeStation and KAPA Library Quantification Kit, respectively. For qPCR, resulting RNA was first treated with Turbo DNase (ThermoFisher, AM2239) and gDNA-free RNA used for cDNA synthesis with Superscript III (ThermoFisher, 56575). qPCR was performed using the PowerUp SYBR Green Master Mix (ThermoFisher, A25742) on a StepOnePlus machine (ThermoFisher). Primers for all targets detected are listed in the key resource table. Transcript levels were quantified in triplicates and normalized to Gapdh1. Fold enrichment was calculated as the ratio of transcript in genetic manipulation versus control.
RNA-seq data analysis
Transcriptomic data analysis was performed as described in Buddika et al., 202048 using a python based in-house pipeline (https://github.com/jkkbuddika/RNA-Seq-Data-Analyzer). Briefly, the quality of raw sequencing files was assessed using FastQC66 version 0.11.9, low quality reads were eliminated using Cutadapt67 version 2.9, and reads mapping to rRNAs were removed using TagDust268 version 2.2. Next, the remaining reads were mapped to the Berkeley Drosophila Genome Project (BDGP) assembly release 6.28 (Ensembl release 100) reference genome using STAR genome aligner69 version 2.7.3a and duplicated reads were removed using SAMtools70 version 1.10. Subsequently, the Subread71 version 2.0.0 function featureCounts was used to count the number of aligned reads to the nearest overlapping feature. Finally, bigWig files representing RNA-seq coverage were generated using deepTools72 version 3.4.2 with the settings --normalizeUsing CPM --binSize 1. Differential gene expression analysis was performed with the Bioconductor package DESeq2 (https://bioconductor.org/packages/release/bioc/html/DESeq2.html)73 version 1.26.0. Unless otherwise noted, significantly upregulated and downregulated genes were defined as FDR < 0.05; Log2 fold change > 1 and FDR < 0.05; Log2 fold change < −1, respectively and were used to identify enriched Gene Ontology (GO) terms using PANTHER overrepresentation analysis on GO Consortium (http://geneontology.org/). A selected significantly enriched GO categories were plotted. All data visualization steps were performed using custom scripts written using R.
QUANTIFICATION AND STATISTICAL ANALYSIS
Statistical analysis
GraphPad Prism, Version 9.0 was used for all statistical analyses. First, the normality of datasets was tested using D’Agostino-Pearson test. For comparisons involved in two datasets, if datasets follow [1] a parametric distribution, an Unpaired t-test or [2] a non-parametric distribution, a Mann-Whitney test was performed. Three or more datasets following a parametric distribution were analyzed using an ordinary one-way ANOVA test. Multiple comparisons of three or more datasets following a non-parametric distribution were analyzed using Kruskal-Wallis test. Unless otherwise noted, significance is indicated as follows: n.s., not significant; *p < 0.05; **p < 0.01; ***p < 0.001; ****p < 0.0001. ImageJ FIJI (https://fiji.sc/) Analyze Particles plugin was used to quantify the average area of granules using 0.01μm2 as the lower cut off of puncta size. The ImageJ FIJI Plot Profile plugin was used to generate line plots. Briefly, a line was drawn along the axis where fluorescent intensity values were needed to be quantified. This generates a matrix of distance along the line and corresponding fluorescent intensity value at each distance measurement for each channel. Line plots were generated by plotting distance values against fluorescent intensity values for each channel in a single plot. Moreover, these quantified fluorescent intensity values were used to calculate the Pearson’s correlation coefficients between different channels whenever needed.
Supplementary Material
Data S1. Supplementary data files. Related to Figure 5. (A) List of Bloomington Drosophila Stock Center (BDSC) RNAi strains screened and observed phenotypes. (B) List of positive genes and observed P-body phenotypes. (C) Differential gene expression results of control vs Patr-1 RNAi. (D) Differential gene expression results of control vs esg RNAi. (E) Differential gene expression results of control vs vtd RNAi. (F) List of complete genotypes of fly strains analyzed in each figure panel.
KEY RESOURCES TABLE
| REAGENT or RESOURCE | SOURCE | IDENTIFIER |
|---|---|---|
| Antibodies | ||
| Mouse anti-V5 | Bio-Rad | Cat# MCA1360GA; RRID:AB_567249 |
| Mouse anti-FLAG | Sigma-Aldrich | Cat# F3165; RRID:AB_259529 |
| Mouse anti-FMRP | DSHB | Cat# 5A11; RRID:AB_528252 |
| Mouse anti-Prospero | DSHB | Cat# MR1A; RRID:AB_528440 |
| Mouse anti-Delta | DSHB | Cat# C594.9B; RRID:AB_528194 |
| Mouse anti-Me31B | 34 | N/A |
| Mouse anti-Tubulin | DSHB | Cat# 12G10; RRID:AB_1157911 |
| Rabbit anti-EDC3 | This study | N/A |
| Rabbit anti-GFP | Thermo Fisher Scientific | Cat# A11122; RRID:AB_221569 |
| Rabbit anti-HA | Cell Signaling Technology | Cat# 3724S; RRID:AB_1549585 |
| Rabbit anti-Me31B | 32 | N/A |
| Rabbit anti-GE1 | 33 | N/A |
| Rabbit anti-Pdm1 | A gift from Xiaohang Yang | N/A |
| Rabbit anti-PATR-1 | 45 | N/A |
| Rat anti-TRAL | This study | N/A |
| Rat anti-EDC3 | This study | N/A |
| Chemicals, peptides, and recombinant proteins | ||
| Paraformaldehyde solution | Electron Microscopy Sciences | Cat# 15714 |
| Bleomycin | Calbiochem | Cat# 203408 |
| TRIzol® LS reagent | Ambion | Cat# 10296028 |
| Elastase | Sigma-Aldrich | Cat# E0258 |
| 10X PBS pH 7.4, RNase-free | Invitrogen | Cat# AM9624 |
| Heptane | Sigma-Aldrich | Cat# 246654 |
| Triton X-100 | Sigma-Aldrich | Cat# 11332481001 |
| Vectashield | Vector Laboratories | Cat# H-1000 |
| ProLong Diamond Antifade | Invitrogen | Cat# P36965 |
| Krebs-Ringer media | Alfa Aesar | Cat# J67591 |
| Rapamycin | LC Laboratories | Cat# r-5000 |
| ECL-detection reagent 1 and 2 | Thermo Fisher Scientific | Cat# 1859701 |
| SUPERasIn RNase Inhibitor | Invitrogen | Cat# AM2696 |
| cOmplete Protease Inhibitor | Roche | Cat# 1183617001 |
| Turbo DNase | Thermo Fisher Scientific | Cat# AM2239 |
| Superscript III | Thermo Fisher Scientific | Cat# 56575 |
| PowerUp SYBR Green Master Mix | Thermo Fisher Scientific | Cat# A25742 |
| Critical commercial assays | ||
| RNAscope Multiplex Fluorescent Reagent Kit v2 | ACD | Cat# 323100 |
| Ovation® SoLo RNA-seq system | Tecan | Cat# 0502 including 0407 |
| ApopTag® Fluorescein In situ Apoptosis Detection Kit | Sigma-Aldrich | Cat# S7110 |
| Deposited data | ||
| Patr-1 RNA-seq data: GSE183996 | This paper | GSE183996 |
| esg RNA-seq data: ArrayExpress E-MTAB-2915 | 53 | N/A |
| vtd RNA-seq data: Provided by H. Jasper | 52 | N/A |
| Experimental models: Organisms/strains | ||
| D. melanogaster: Patr-1P105FS1 / TM3, Sb[1] | This study | N/A |
| D. melanogaster: Patr-1P105FS2 / TM3, Sb[1] | This study | N/A |
| D. melanogaster: Patr-1P106FS1 / TM3, Sb[1] | This study | N/A |
| D. melanogaster: Patr-1P107FS1 / TM3, Sb[1] | This study | N/A |
| D. melanogaster: Patr-1P104Δ / TM3, Sb[1] | This study | N/A |
| D. melanogaster: Patr-1P105Δ / TM3, Sb[1] | This study | N/A |
| D. melanogaster: P21M20 | 45 | N/A |
| D. melanogaster: esg-Gal4 UAS-GFP tubGal80 ts | 30 | N/A |
| D. melanogaster: gbe-smGFP::V5::nls | 48 | N/A |
| D. melanogaster: hsFLP12 , tubGAL4, UAS-GFP / + ; P{lin-28 13kb::mCherry}attP40 / Cyo ; FRT82B tubP-GAL80LL3 / TM6, Tb[1] | This study | N/A |
| D. melanogaster: {3Xgbe-GAL80}ZH-2A ; {mira-KDRT>-dSTOP-KDRT>-GAL41438}attP40, P{tubP-GAL80[ts]}20; {CG10116-KD.PEST1107}attP2 | 48 | N/A |
| D. melanogaster: w; Su(h)Gbe-Gal4, UAS-mCD8GFP/ CyO; tubGal80ts / TM6B | B. Edgar | N/A |
| D. melanogaster: rin::HA | 18 | N/A |
| D. melanogaster: P{w[+mC]=UAS-rpr.C}27 | BDSC | RRID:BDSC_5823 |
| D. melanogaster: y[1] w[67c23]; P{y[+mDint2] w[+mC]=EPgy2}Patr-1[EY10289] / TM3, Sb[1] Ser[1] | BDSC | RRID:BDSC_19805 |
| D. melanogaster: P{y[+t7.7] v[+t1.8]=TRiP.HM05052}attP2 | BDSC | RRID:BDSC_28566 |
| D. melanogaster: P{y[+t7.7] v[+t1.8]=TRiP.HM05072}attP2 | BDSC | RRID:BDSC_28584 |
| D. melanogaster: P{y[+t7.7] v[+t1.8]=TRiP.JF03061}attP2 | BDSC | RRID:BDSC_28646 |
| D. melanogaster: P{y[+t7.7] v[+t1.8]=TRiP.HMS00857}attP2 | BDSC | RRID:BDSC_33914 |
| D. melanogaster: P{y[+t7.7] v[+t1.8]=TRiP.HMS01144}attP2 / TM3, Sb[1] | BDSC | RRID:BDSC_34667 |
| D. melanogaster: P{y[+t7.7] v[+t1.8]=TRiP.GL00680}attP2 | BDSC | RRID:BDSC_38908 |
| D. melanogaster: P{y[+t7.7] v[+t1.8]=TRiP.HMJ22374}attP40 | BDSC | RRID:BDSC_58288 |
| D. melanogaster: P{w+mC=UAS-Stinger}2 | BDSC | RRID:BDSC_65402 |
| D. melanogaster: P{UAS-nub.ORF.3xHA.GW}attP2 | BDSC | RRID:BDSC_83303 |
| D. melanogaster: y1 sc* v1 sev21; P{nos-Cas9}attP2 | BDSC | RRID:BDSC_78782 |
| D. melanogaster: P{EPgy2}Patr-1 EY10289 | BDSC | RRID:BDSC_19805 |
| D. melanogaster: M{UAS-esg.ORF.3xHA.GW}ZH-86Fb | FlyORF | RRID:FlyBase_FBst0501340 |
| Oligonucleotides | ||
|
nub qPCR forward primer: 5'-TGGTGGGGCATTGAATTTAACC-3' |
This study | N/A |
|
nub qPCR reverse primer: 5'-CTGTGGCCGATTTCACCGAAT-3' |
This study | N/A |
|
Patr-1 qPCR forward primer: 5'-TCGTTTTTCGGCTTTGACACG-3' |
This study | N/A |
|
Patr-1 qPCR reverse primer: 5'-GTCATTGAGGGCATCGTATTCC-3' |
This study | N/A |
|
esg qPCR forward primer: 5'-CTGAGATCCCAGCCATGTT-3' |
This study | N/A |
|
esg qPCR reverse primer: 5'-CTGCTTGAAGTTGTGTGTCTG-3' |
This study | N/A |
|
Tsp2A qPCR forward primer: 5'-CGTCTGGTTGAGGGCTGAG-3' |
This study | N/A |
|
Tsp2A qPCR reverse primer: 5'-CACGTACACGCCGATGTAG-3' |
This study | N/A |
|
MRP qPCR forward primer: 5'-AATCGAAAGTATGGCGTGCAG-3' |
This study | N/A |
|
MRP qPCR reverse primer: 5'-GGGGAATCGACAGCACAGT-3' |
This study | N/A |
|
bbg qPCR forward primer: 5'-AGGATGAGTCCTCTGCACCA-3' |
This study | N/A |
|
bbg qPCR reverse primer: 5'-TCCTGGCGATATACTTTCCTGG-3' |
This study | N/A |
| gRNA #1: 5'-CATGTTGAACATGTTATACATGG-3' | This study | N/A |
| gRNA #2: 5'-TGTGACGAGACTGTCGGAAGGGG-3' | This study | N/A |
| EDC3 amplification forward primer: 5'-GGGAATTCATGGGTCCGACGGATCAAGA-3' | This study | N/A |
| EDC3 amplification reverse primer: 5'-GGGGCGGCCGCTCACTTATCGGCACTTATCTCGA-3' | This study | N/A |
| Software and algorithms | ||
| ImageJ/Fiji | 37 | https://imagej.net/; RRID:SCR_003070 |
| Leica LAS-X | Leica | https://www.leica-microsystems.com/products/microscope-software/p/leica-las-x-ls/; RRID:SCR_013673 |
| Prism Version 9.0 | GraphPad Software | http://www.graphpad.com/; RRID:SCR_002798 |
| RNA-seq analysis pipeline | 48 | https://github.com/jkkbuddika/RNA-Seq-Data-Analyzer |
| Other | ||
| nub in situ probes | ACD | Cat# 523981 |
| Opal 620 | AKOYA Biosciences | Cat# FP1495001KT |
| Chemiluminescence films | GE Healthcare | Cat# 28906839 |
| Immobilon®-P membrane | Millipore | Cat# IPVH00010 |
| 4–20% gradient polyacrylamide gel | Bio-Rad | Cat# 456–1093 |
| Leica SP8 Scanning Confocal microscope | Leica | N/A |
| DeltaVision OMX system | DeltaVision | N/A |
Highlights.
Intestinal progenitors contain constitutive, ultrastructurally organized P-bodies.
P-body regulator, Patr-1, is required for intestinal progenitor cell maintenance.
Enterocyte gene, nub, is weakly transcribed but not translated in progenitors.
P-bodies repress enterocyte gene expression to promote stem cell maintenance.
Acknowledgements
We thank Scott Barbee, Anne Ephrussi, Xiaohang Yang, the Bloomington Drosophila Stock Center (supported by grant NIHP4OOD018537), the Drosophila Genome Resource Center (supported by grant NIH2P40OD010949), and the Developmental Studies Hybridoma Bank (created by the NICHD of the NIH) for reagents; Robert Policastro for bioinformatics expertise; the Light Microscopy Imaging Center (supported by grant NIH1S10OD024988-01) for access to the SP8 confocal and OMX super-resolution microscopes; the Flow Cytometry Core Facility at Indiana University, Bloomington for access to the BD FACSAria™ II flow cytometer; and the National Institute of General Medical Sciences (Award R01GM124220) for financial support.
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
Declaration of Interests
The authors declare no competing interests.
References
- 1.Anderson P, and Kedersha N (2006). RNA granules. J. Cell Biol 172, 803–808. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Decker CJ, and Parker R (2012). P-Bodies and Stress Granules: Possible Roles in the Control of Translation and mRNA Degradation. Cold Spring Harb. Perspect. Biol 4, a012286. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Kedersha N, Stoecklin G, Ayodele M, Yacono P, Lykke-Andersen J, Fritzler MJ, Scheuner D, Kaufman RJ, Golan DE, and Anderson P (2005). Stress granules and processing bodies are dynamically linked sites of mRNP remodeling. J. Cell Biol 169, 871–884. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Kedersha N, Cho MR, Li W, Yacono PW, Chen S, Gilks N, Golan DE, and Anderson P (2000). Dynamic Shuttling of Tia-1 Accompanies the Recruitment of mRNA to Mammalian Stress Granules. J. Cell Biol 151, 1257–1268. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Buchan JR, and Parker R (2009). Eukaryotic Stress Granules: The Ins and Outs of Translation. Mol. Cell 36, 932–941. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Teixeira D, Sheth U, Valencia-Sanchez MA, Brengues M, and Parker R (2005). Processing bodies require RNA for assembly and contain nontranslating mRNAs. RNA 11, 371–382. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Chan LY, Mugler CF, Heinrich S, Vallotton P, and Weis K (2018). Non-invasive measurement of mRNA decay reveals translation initiation as the major determinant of mRNA stability. Elife 7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Franks TM, and Lykke-Andersen J (2008). The Control of mRNA Decapping and P-Body Formation. Mol. Cell 32, 605–615. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Anderson P, and Kedersha N (2009). Stress granules. Curr. Biol 19, R397–8. [DOI] [PubMed] [Google Scholar]
- 10.Mugler CF, Hondele M, Heinrich S, Sachdev R, Vallotton P, Koek AY, Chan LY, and Weis K (2016). ATPase activity of the DEAD-box protein Dhh1 controls processing body formation. Elife 5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Protter DSW, and Parker R (2016). Principles and Properties of Stress Granules. Trends Cell Biol. 26, 668–679. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Martínez JP, Pérez-Vilaró G, Muthukumar Y, Scheller N, Hirsch T, Diestel R, Steinmetz H, Jansen R, Frank R, Sasse F, et al. (2013). Screening of small molecules affecting mammalian P-body assembly uncovers links with diverse intracellular processes and organelle physiology. RNA Biol. 10, 1661–9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Luo Y, Na Z, and Slavoff SA (2018). P-Bodies: Composition, Properties, and Functions. Biochemistry 57, 2424–2431. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Youn J-Y, Dyakov BJA, Zhang J, Knight JDR, Vernon RM, Forman-Kay JD, and Gingras A-C (2019). Properties of Stress Granule and P-Body Proteomes. Mol. Cell 76, 286–294. [DOI] [PubMed] [Google Scholar]
- 15.Sheth U, and Parker R (2003). Decapping and Decay of Messenger RNA Occur in Cytoplasmic Processing Bodies. Science (80-. ). 300, 805–808. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Parker R, and Sheth U (2007). P Bodies and the Control of mRNA Translation and Degradation. Mol. Cell 25, 635–646. [DOI] [PubMed] [Google Scholar]
- 17.Eulalio A, Behm-Ansmant I, Schweizer D, and Izaurralde E (2007). P-body formation is a consequence, not the cause, of RNA-mediated gene silencing. Mol. Cell. Biol 27, 3970–81. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Buddika K, Ariyapala IS, Hazuga MA, Riffert D, and Sokol NS (2020). Canonical nucleators are dispensable for stress granule assembly in Drosophila intestinal progenitors. J. Cell Sci 133. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Hubstenberger A, Courel M, Bénard M, Souquere S, Ernoult-Lange M, Chouaib R, Yi Z, Morlot J-B, Munier A, Fradet M, et al. (2017). P-Body Purification Reveals the Condensation of Repressed mRNA Regulons. Mol. Cell 68, 144–157.e5. [DOI] [PubMed] [Google Scholar]
- 20.Standart N, and Weil D (2018). P-Bodies: Cytosolic Droplets for Coordinated mRNA Storage. Trends Genet. 34, 612–626. [DOI] [PubMed] [Google Scholar]
- 21.Brengues M, Teixeira D, and Parker R (2005). Decapping and Decay of Messenger RNA Occur in Cytoplasmic Processing Bodies. Science (80-. ). 300, 805–808. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Bhattacharyya SN, Habermacher R, Martine U, Closs EI, and Filipowicz W (2006). Relief of microRNA-Mediated Translational Repression in Human Cells Subjected to Stress. Cell 125, 1111–1124. [DOI] [PubMed] [Google Scholar]
- 23.Stoecklin G, Mayo T, and Anderson P (2006). ARE-mRNA degradation requires the 5’–3’ decay pathway. EMBO Rep. 7, 72–77. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Souquere S, Mollet S, Kress M, Dautry F, Pierron G, and Weil D (2009). Unravelling the ultrastructure of stress granules and associated P-bodies in human cells. J. Cell Sci 122, 3619–3626. [DOI] [PubMed] [Google Scholar]
- 25.Chen Q, and Hu G (2017). Post-transcriptional regulation of the pluripotent state. Curr. Opin. Genet. Dev 46, 15–23. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Di Stefano B, Luo E-C, Haggerty C, Aigner S, Charlton J, Brumbaugh J, Ji F, Rabano Jiménez I, Clowers KJ, Huebner AJ, et al. (2019). The RNA Helicase DDX6 Controls Cellular Plasticity by Modulating P-Body Homeostasis. Cell Stem Cell 25, 622–638.e13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Luhur A, Buddika K, Ariyapala IS, Chen S, and Sokol NS (2017). Opposing Post-transcriptional Control of InR by FMRP and LIN-28 Adjusts Stem Cell-Based Tissue Growth. Cell Rep. 21, 2671–2677. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Chen C-H, Luhur A, and Sokol N (2015). Lin-28 promotes symmetric stem cell division and drives adaptive growth in the adult Drosophila intestine. Development 142, 3478–87. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Ohlstein B, and Spradling A (2006). The adult Drosophila posterior midgut is maintained by pluripotent stem cells. Nature 439, 470–474. [DOI] [PubMed] [Google Scholar]
- 30.Micchelli CA, and Perrimon N (2006). Evidence that stem cells reside in the adult Drosophila midgut epithelium. Nature 439, 475–479. [DOI] [PubMed] [Google Scholar]
- 31.Ayache J, Bénard M, Ernoult-Lange M, Minshall N, Standart N, Kress M, and Weil D (2015). P-body assembly requires DDX6 repression complexes rather than decay or Ataxin2/2L complexes. Mol. Biol. Cell 26, 2579–95. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Nakamura A, Amikura R, Hanyu K, K.S. (2001). Me31B silences translation of oocyte-localizing RNAs through the formation of cytoplasmic RNP complex during Drosophila oogenesis. Development 128, 3233–3242. [DOI] [PubMed] [Google Scholar]
- 33.Fan S-J, Marchand V, and Ephrussi A (2011). Drosophila Ge-1 Promotes P Body Formation and oskar mRNA Localization. PLoS One 6, e20612. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Barbee SA, Estes PS, Cziko A-M, Hillebrand J, Luedeman RA, Coller JM, Johnson N, Howlett IC, Geng C, Ueda R, et al. (2006). Staufen- and FMRP-containing neuronal RNPs are structurally and functionally related to somatic P bodies. Neuron 52, 997–1009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.O’Brien LE, Soliman SS, Li X, and Bilder D (2011). Altered Modes of Stem Cell Division Drive Adaptive Intestinal Growth. Cell 147, 603–614. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Miller DE, Kahsai L, Buddika K, Dixon MJ, Kim BY, Calvi BR, Sokol NS, Scott Hawley R, and Cook KR (2020). Identification and characterization of breakpoints and mutations on drosophila melanogaster balancer chromosomes. G3 Genes, Genomes, Genet. 10, 4271–4285. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Schindelin J, Arganda-Carreras I, Frise E, Kaynig V, Longair M, Pietzsch T, Preibisch S, Rueden C, Saalfeld S, Schmid B, et al. (2012). Fiji: an open-source platform for biological-image analysis. Nat. Methods 2012 97 9, 676–682. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Wheeler JR, Matheny T, Jain S, Abrisch R, and Parker R (2016). Distinct stages in stress granule assembly and disassembly. Elife 5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Panas MD, Ivanov P, and Anderson P (2016). Mechanistic insights into mammalian stress granule dynamics. J. Cell Biol 215, 313–323. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Pilkington GR, and Parker R (2008). Pat1 contains distinct functional domains that promote P-body assembly and activation of decapping. Mol. Cell. Biol 28, 1298–312. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Nissan T, Rajyaguru P, She M, Song H, and Parker R (2010). Decapping activators in Saccharomyces cerevisiae act by multiple mechanisms. Mol. Cell 39, 773–83. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.V R, KH S, and PK H (2011). The cAMP-dependent Protein Kinase Signaling Pathway Is a Key Regulator of P Body Foci Formation. Mol. Cell 43. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Coller J, and Parker R (2005). General translational repression by activators of mRNA decapping. Cell 122, 875–86. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Sachdev R, Hondele M, Linsenmeier M, Vallotton P, Mugler CF, Arosio P, and Weis K (2019). Pat1 promotes processing body assembly by enhancing the phase separation of the DEAD-box ATPase Dhh1 and RNA. Elife 8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Pradhan SJ, Nesler KR, Rosen SF, Kato Y, Nakamura A, Ramaswami M, and Barbee SA (2012). The conserved P body component HPat/Pat1 negatively regulates synaptic terminal growth at the larval Drosophila neuromuscular junction. J. Cell Sci 125, 6105–16. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Lee T, and Luo L (1999). Mosaic Analysis with a Repressible Cell Marker for Studies of Gene Function in Neuronal Morphogenesis. Neuron 22, 451–461. [DOI] [PubMed] [Google Scholar]
- 47.Amcheslavsky A, Jiang J, and Ip YT (2009). Tissue damage-induced intestinal stem cell division in Drosophila. Cell Stem Cell 4, 49–61. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Buddika K, Xu J, Ariyapala IS, and Sokol NS (2020). I-KCKT allows dissection-free RNA profiling of adult Drosophila intestinal progenitor cells. Development 148, dev.196568. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Zeng X, Chauhan C, and Hou SX (2010). Characterization of midgut stem cell- and enteroblast-specific Gal4 lines in drosophila. genesis 48, 607–611. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Villa SER, Meng FW, and Biteau B (2019). zfh2 controls progenitor cell activation and differentiation in the adult Drosophila intestinal absorptive lineage. PLOS Genet. 15, e1008553. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Courel M, Clément Y, Bossevain C, Foretek D, Cruchez OV, Yi Z, Bénard M, Benassy MN, Kress M, Vindry C, et al. (2019). Gc content shapes mRNA storage and decay in human cells. Elife 8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Khaminets A, Ronnen-Oron T, Baldauf M, Meier E, and Jasper H (2020). Cohesin controls intestinal stem cell identity by maintaining association of Escargot with target promoters. Elife 9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Korzelius J, Naumann SK, Loza-Coll MA, Chan JS, Dutta D, Oberheim J, Gläßer C, Southall TD, Brand AH, Jones DL, et al. (2014). Escargot maintains stemness and suppresses differentiation in Drosophila intestinal stem cells. EMBO J. 33, 2967–2982. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Tian A, Wang B, and Jiang J (2017). Injury-stimulated and self-restrained BMP signaling dynamically regulates stem cell pool size during Drosophila midgut regeneration. Proc. Natl. Acad. Sci. U. S. A 114, E2699–E2708. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Teixeira D, and Parker R (2007). Analysis of P-Body Assembly in Saccharomyces cerevisiae. Mol. Biol. Cell 18, 2274–2287. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Eulalio A, Behm-Ansmant I, and Izaurralde E (2007). P bodies: at the crossroads of post-transcriptional pathways. Nat. Rev. Mol. Cell Biol 8, 9–22. [DOI] [PubMed] [Google Scholar]
- 57.Scheller N, Resa-Infante P, de la Luna S, Galao RP, Albrecht M, Kaestner L, Lipp P, Lengauer T, Meyerhans A, and Díez J (2007). Identification of PatL1, a human homolog to yeast P body component Pat1. Biochim. Biophys. Acta - Mol. Cell Res 1773, 1786–1792. [DOI] [PubMed] [Google Scholar]
- 58.Jain S, Wheeler JR, Walters RW, Agrawal A, Barsic A, and Parker R (2016). ATPase-Modulated Stress Granules Contain a Diverse Proteome and Substructure. Cell 164, 487–498. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Niewidok B, Igaev M, Pereira da Graca A, Strassner A, Lenzen C, Richter CP, Piehler J, Kurre R, and Brandt R (2018). Single-molecule imaging reveals dynamic biphasic partition of RNA-binding proteins in stress granules. J. Cell Biol 217, 1303–1318. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Van Nostrand EL, Pratt GA, Shishkin AA, Gelboin-Burkhart C, Fang MY, Sundararaman B, Blue SM, Nguyen TB, Surka C, Elkins K, et al. (2016). Robust transcriptome-wide discovery of RNA-binding protein binding sites with enhanced CLIP (eCLIP). Nat. Methods 13, 508–514. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Wang Y, Arribas-Layton M, Chen Y, Lykke-Andersen J, and Sen GL (2015). DDX6 Orchestrates Mammalian Progenitor Function through the mRNA Degradation and Translation Pathways. Mol. Cell 60, 118–130. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Antonello ZA, Reiff T, Ballesta-Illan E, and Dominguez M (2015). Robust intestinal homeostasis relies on cellular plasticity in enteroblasts mediated by miR-8–Escargot switch. EMBO J. 34, 2025–2041. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Loza-Coll MA, Southall TD, Sandall SL, Brand AH, and Jones DL (2014). Regulation of Drosophila intestinal stem cell maintenance and differentiation by the transcription factor Escargot. EMBO J. 33, 2983–96. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Li Y, Pang Z, Huang H, Wang C, Cai T, and Xi R (2017). Transcription Factor Antagonism Controls Enteroendocrine Cell Specification from Intestinal Stem Cells. Sci. Rep 7, 988. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Liu Q, and Jin LH (2017). Tissue-resident stem cell activity: a view from the adult Drosophila gastrointestinal tract. Cell Commun. Signal 15, 33. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Andrews S (2010). FastQC: A quality control tool for high throughput sequence data. https://www.bioinformatics.babraham.ac.uk/projects/fastqc/.
- 67.Martin M (2011). Cutadapt removes adapter sequences from high-throughput sequencing reads. EMBnet.journal 17, 10. [Google Scholar]
- 68.Lassmann T (2015). TagDust2: a generic method to extract reads from sequencing data. BMC Bioinformatics 16, 24. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Dobin A, Davis CA, Schlesinger F, Drenkow J, Zaleski C, Jha S, Batut P, Chaisson M, and Gingeras TR (2013). STAR: ultrafast universal RNA-seq aligner. Bioinformatics 29, 15–21. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Li H, Handsaker B, Wysoker A, Fennell T, Ruan J, Homer N, Marth G, Abecasis G, Durbin R, and 1000 Genome Project Data Processing Subgroup (2009). The Sequence Alignment/Map format and SAMtools. Bioinformatics 25, 2078–9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Liao Y, Smyth GK, and Shi W (2019). The R package Rsubread is easier, faster, cheaper and better for alignment and quantification of RNA sequencing reads. Nucleic Acids Res. 47, e47. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Ramírez F, Ryan DP, Grüning B, Bhardwaj V, Kilpert F, Richter AS, Heyne S, Dündar F, and Manke T (2016). deepTools2: a next generation web server for deep-sequencing data analysis. Nucleic Acids Res. 44, W160–W165. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Love MI, Huber W, and Anders S (2014). Moderated estimation of fold change and dispersion for RNA-seq data with DESeq2. Genome Biol. 15, 550. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data S1. Supplementary data files. Related to Figure 5. (A) List of Bloomington Drosophila Stock Center (BDSC) RNAi strains screened and observed phenotypes. (B) List of positive genes and observed P-body phenotypes. (C) Differential gene expression results of control vs Patr-1 RNAi. (D) Differential gene expression results of control vs esg RNAi. (E) Differential gene expression results of control vs vtd RNAi. (F) List of complete genotypes of fly strains analyzed in each figure panel.
Data Availability Statement
RNA-seq data have been deposited at GEO and are publicly available as of the date of publication. The accession number is listed in the key resources table.
This paper does not report original code.
Any additional information required to reanalyze the data reported in this paper is available from the lead contact upon request.


