Abstract
The availability of effective smallpox vaccines was a critical element of the successful eradication of smallpox in 1980. Antibody responses play a primary role in protective immunity and neutralizing antibody is an established correlate of protection against smallpox. In this study we used a poxvirus proteome array to assess the antibody response to individual viral proteins in a cohort of 1,037 smallpox vaccine recipients. Several statistically significant differences were observed in the antibody response to immunodominant proteins between men and women, including B5R—a major target of neutralizing antibody in vaccinia immune globulin, and the membrane proteins D8L and A27L, both of which have been used as vaccine antigens providing protection in animal models. We also noted differences across racial/ethnic groups. In this cohort, which consisted of both ACAM2000 and Dryvax recipients, we noted minute differences in the antibody responses to a restricted number of viral proteins, providing additional support for the use of ACAM2000 as a replacement smallpox vaccine. Furthermore, our data indicate that poxvirus proteome microarrays can be valuable for screening and monitoring smallpox vaccine-induced humoral immune responses in large-scale serologic surveillance studies and prove useful in the guidance of developing novel smallpox candidate vaccines.
Keywords: Smallpox, Smallpox Vaccine, Immunity, Humoral, Antibodies, Neutralizing, Microarray Analysis
Introduction
Smallpox is the only human disease to have been eradicated. The eradication was made possible through a concerted, worldwide effort in surveillance and immunization campaigns, the lack of an animal reservoir for disease, as well as the availability of a temperature-stable, easily administered, effective vaccine containing live vaccinia virus (VACV) that induces cross-protective immunity against a broad range of poxviruses. The vaccine elicits strong humoral and cellular immunity in >97% of recipients; immune responses are detectable decades after immunization, and protection against disease lasts at least 5 years.1 Humoral immunity is believed to play a major role in protection and elimination of the virus from the host and neutralizing antibody (Ab) titer was identified as a correlate of protection during the eradication campaign.2, 3, 4 Data from more recent animal studies and clinical vaccine studies in humans also support the critical role of antibody responses in protection against poxvirus disease.5, 6, 7, 8 Poxviruses are dsDNA viruses with large genomes containing >200 open reading frames (ORFs). Infected cells produce two major forms of viral particles: 1) the intracellular mature virion (MV); and 2) the extracellular virion (EV), which contains one additional membrane and several unique proteins and is released from the surface of infected cells, mediating long-distance viral spreading within the infected host.9 MVs are the most abundant virion form produced, are environmentally stable, and are believed to be responsible for host-to-host transmission. The MV and EV virions differ antigenically and immune responses targeting both particles are believed to provide superiorprotection,10, 11, 12, 13, 14 although some EV proteins can be removed without compromising protection in animal models.15
Proteome microarrays containing the complete set of proteins for a given pathogen enable researchers to comprehensively screen the humoral response against that pathogen. An orthopox array was initially described in 2004.16 This array has been used to do the following: screen the diversity of humoral immune responses in Dryvax recipients;17, 18 evaluate vaccinia immune globulin (VIG) preparations;16 identify major targets of humoral immunity;16 compare modified vaccinia Ankara (MVA) with Dryvax in animal models and human vaccine recipients;19, 20 evaluate Dryvax, ACAM2000, and IMVAMUNE smallpox vaccines and monkeypox virus challenge;21 and assess antigenic differences in between MV and EV virions.22
In the current study we examined a cohort of 1,037 smallpox vaccine recipients in order to further characterize the breadth and depth of the humoral responses against vaccinia virus. We identified a number of viral proteins where the level of antibody (Ab) correlated with neutralizing antibody titer and/or cellular cytokine responses. Furthermore, we investigated race, ethnicity, and sex differences in antibody response following vaccination.
Methods
Subject Recruitment
The study cohort, which is a subset of a previously described cohort, consisted of 1,037 armed forces personnel who were recruited from 2011–2013.23 The individuals in this report were 18–40 years old and in good general health. 914 of the subjects had received one and only one dose of ACAM2000® 1 month–4 years prior to enrollment, while 123 subjects had received one and only one dose of Dryvax® in the same time period prior to enrollment. Subjects were recruited from Fort Bliss, Fort Campbell, Fort Lewis, and the Naval Health Research Center (NHRC) in San Diego, CA. The Institutional Review Boards for both the Mayo Clinic and NHRC approved all study procedures and written informed consent was obtained from each participant upon enrollment. Subjects underwent a single blood draw. Whole blood was collected in red-top collection tubes without anticoagulant. After incubation for at least 30 minutes at room temperature, serum was collected by centrifugation, aliquoted and stored at −80°C until use.
Neutralization assay
We used a vaccinia-specific neutralizing antibody assay developed by the Food and Drug Administration (FDA) and optimized for high throughput in our laboratory as previously described.24, 25 The vSC56 strain of vaccinia virus expressing β-galactosidase (obtained from Dr. Bernard Moss, National Institutes of Allergy and Infectious Diseases, NIAID) was grown, purified, and titered according to established protocols.26, 27Serum dilutions were incubated with a set quantity of VACV-vSC56 for 1 hour, the mixture was added to Hela cells overnight. Cells were lysed (Igepal CA630, Sigma-Aldrich, St. Louis, MS) and beta-galactosidase activity measured by the addition of a colorimetric substrate (CPRG, Calbiochem, San Diego, CA). The reaction was quenched with Na2CO3 and plates were read with a ThermoMax plate reader (Molecular Devices, Sunnyvale, CA). Sera were tested at least three times (Dryvax cohort) or twice (ACAM2000 cohort), and results were defined as the serum dilution that inhibits 50% of virus activity (ID50).24, 25
Orthopox pathogen array
Serum samples were used to probe Orthopox Full Proteome Microarrays in duplicate (Antigen Discovery; Irvine, CA) using the manufacturer’s established protocols.20 Briefly, serum samples were incubated on a protein microarray to allow VACV-specific antibodies to bind to the immobilized poxvirus proteins. Fluorochrome-labeled secondary reagents were used to detect the presence of bound serum antibody. Plates were scanned and fluorescent intensity values were used to determine the quantity of antibody specific for each protein on the array. Values are presented in fluorescence units: “Median Normalized Antibody Measurements”. The proteome microarray contained 246 proteins, 10 replicate spots coated with human IgG as internal positive controls, and negative controls - ‘noDNA’ spots comprising in vitro transcription/translation reaction products lacking template DNA.
Cytokine measurements
Cytokine responses to VACV were evaluated as previously described.23 Briefly, commercial ELISAs were used to measure cytokines present in cell culture supernatants following in vitro virus stimulation. Results are reported as pg/mL.
Statistical Analysis
For each sample, the median value of the 12 noDNA spots was calculated and the median of the noDNA spots was subtracted from each protein for the sample. After the control subtraction, a protein was considered a positive target of humoral immune responses if more than 50% of subjects in either vaccine group had detectable antibody response to that protein. The data were normalized using the Variance Stabilization and Calibration for Microarray Data method.28 This method introduces a transformation for the raw intensity values and an estimate of the variance that is constant across the intensity range. This allows one to calibrate the data, measure differences, and quantify measurement error. Penalized elastic-net logistic regression was used to identify predictors of high vs. low antibody response in the 194 ACAM2000 recipients who had the highest and lowest neutralizing antibody titers. Ten proteins were excluded from the model to help reduce noise, using Harrell’s redundancy analysis method.29 The model was chosen by selecting the penalty parameter “λ” utilizing 10-fold cross validation. This method selects a range of λ values, and then fits a model to 90% of the data, applies the model to the remaining 10% of the data and calculates the number that are not classified correctly, this is repeated 10 times, holding out a different 10% of the data. The λ value is selected as the one that minimizes the classification error.30 The predictive accuracy of the model is the area under the ROC for the elastic-net regression model. S pecific statistical tests for comparisons are documented in each table or figure. Time since vaccination was used as a controlling variable in the linear models, with vaccine as the predictor, i.e. log(neut) = ACAM2000 + log(Time.Since.Vacc).
Results
Cohort characteristics
We screened sera samples from a cohort of first-time smallpox vaccine recipients in order to characterize proteome-wide antibody responses following smallpox vaccination. Overall, the cohort had a median neutralizing antibody titer of 113.5 ID50 (serum dilution inhibiting 50% of viral activity), with a substantial amount of inter-individual variation as evidenced by the interquartile range of responses (ID50: 62.6 – 189.8). Comparisons between ACAM2000 recipients and Dryvax recipients did not identify any significant differences between cohorts in terms of sex, race, ethnicity, or neutralizing antibody titer (ID50). These comparisons are outlined in Table 1.
Table 1.
Cohort Demographics
| Variable | ACAM2000 (n=914) | Dryvax (n=123) | Total (n=1,037) | p-value |
|---|---|---|---|---|
| Sex | 0.302 | |||
| Female | 94 (10.3%) | 9 (7.3%) | 103 (9.9%) | |
| Male | 820 (89.7%) | 114 (92.7%) | 934 (90.1%) | |
| Race | 0.380 | |||
| American Indian/Alaska Native | 26 (2.8%) | 3 (2.4%) | 29 (2.8%) | |
| Asian | 16 (1.8%) | 2 (1.6%) | 18 (1.7%) | |
| Black or African American | 109 (11.9%) | 14 (11.4%) | 123 (11.9%) | |
| More Than One Race | 14 (1.5%) | 5 (4.1%) | 19 (1.8%) | |
| Native Hawaiian or Other Pacific | 10 (1.1%) | 3 (2.4%) | 13 (1.3%) | |
| Islander | ||||
| Unknown | 19 (2.1%) | 1 (0.8%) | 20 (1.9%) | |
| Caucasian | 720 (78.8%) | 95 (77.2%) | 815 (78.6%) | |
| Ethnicity | 0.362 | |||
| Don’t Know | 9 (1.0%) | 1 (0.8%) | 10 (1.0%) | |
| Hispanic | 160 (17.5%) | 28 (22.8%) | 188 (18.1%) | |
| Not Hispanic | 745 (81.5%) | 94 (76.4%) | 839 (80.9%) | |
| Neutralizing Ab titer (ID50) | 0.491 | |||
| Mean (SD) | 152.7 (189.8) | 140.3 (176.0) | 151.2 (188.2) | |
| Median | 117.0 | 98.6 | 113.5 | |
| Q1, Q3 | 63.4, 191.4 | 52.8, 178.4 | 62.6, 189.8 | |
| Range | 10.7 – 2979.8 | 15.9 – 1636.1 | 10.7 – 2979.8 |
Difference between vaccine types were calculated using Pearson’s chi-square test and one-way analysis of variance for continuous variables.
Comparison between ACAM2000 and Dryvax.
Our study cohort wass comprised of individuals vaccinated with two different smallpox vaccines, providing an opportunity to directly compare Dryvax recipients (n=123) with ACAM2000 recipients (n=914). The demographics for each sub-cohort are presented in Table 1 and demonstrate that neutralizing antibody titers are 8.8% higher in the ACAM2000 recipients, but the difference was not significant. However, linear modeling of neutralizing antibody titer as a function of vaccine received, after correcting for time since vaccination, indicated that this difference was not statistically significant (p=0.23). When we compared these vaccine groups by individual protein array response, we noted that antibody responses to five proteins exhibited significant differences between ACAM2000 and Dryvax vaccine recipients (Supplemental Table 1 and Supplemental Figure 1: Wilcoxon Rank-Sum test). Protein-specific antibody responses were higher in ACAM2000 recipients to the A13L (1.3% higher, p=0.000031), WR148 (0.5% higher, p=0.019), D13L (0.3% higher, 0.024), and B5R (0.3% higher, p=0.035) proteins and with slightly lower responses to D8L (6.4% lower, p=0.0018).
Correlation between neutralizing antibody titers and the humoral response to individual proteins
Within the cohort, only a portion of the viral proteome was targeted by humoral immune responses. We divided the cohort by vaccine received and calculated the percent of subjects with a positive protein antibody reading (i.e., above background) within each group (Table 2). A protein was considered a positive target of humoral immune responses if more than 50% of subjects in either vaccine group had detectable antibody response to that protein. These 28 positive proteins and an additional 11 proteins (i.e., A10L, A17L, A25L, A33R, A34R, B2R, B5R, E2L, F13L, L1R, L4R) found to be targets of neutralizing antibody in previously published reports were selected for further investigation. The relative magnitude of the immune response to each of these selected proteins is illustrated in Figure 1. A number of proteins were targeted at different rates in the two vaccine groups however, these were among the least immunogenic proteins where responses were barely above background and the differences are likely due to variation in the assay.
Table 2.
Percentage of Vaccinated Subjects with Antibodies Against Vaccinia Proteins
| > 75% of subjects | 50–75% of subjects | 25–50% of subjects | < 25% of subjects | |||
|---|---|---|---|---|---|---|
| A13L | A12L | A10L | A14.5L | B1R | F4L | WR002 |
| A14L | A19L | A11R | A15L | B2R | F5L | WR003 |
| A27L | A4L | A12L | A17L | B3R | F6L | WR004 |
| A40R | A9L | A16L | A18R | B4R | F8L | WR005 |
| A56R | B20R | A22R | A1L | B5R | F9L | WR006 |
| C5L | D8L | A36R | A20R | B6R | G1L | WR007 |
| D13L | E3L | A42R | A21L | B7R | G2R | WR008 |
| H5R | E4L | A43R | A23R | B8R | G3L | WR009 |
| I1L | F7L | A49R | A24R | C12L | G5R | WR010 |
| WR001 | H3L | A50R | A25L | C14L | G8R | WR011 |
| WR146 | H6R | A52R | A28L | C1L | G9R | WR012 |
| WR148 | J1R | B15R | A29L | C2L | H1L | WR013 |
| WR218 | N1L | B16R | A2L | C4L | H2R | WR014 |
| WR169 | B17L | A30L | C6L | H4L | WR015 | |
| WR214 | B19R | A31R | C7L | H7R | WR016 | |
| B9R | A32L | C8L | I2L | WR018 | ||
| C17L | A33R | D10R | I3L | WR019 | ||
| C3L | A34R | D11L | I4L | WR053.5 | ||
| E1L | A35R | D12L | I5L | WR083 | ||
| E5R | A37R | D1R | I7L | WR121 | ||
| E9L | A38L | D2L | I8R | WR149 | ||
| F11L | A3L | D3R | J2R | WR153.5 | ||
| F13L | A41L | D4R | J3R | WR161 | ||
| F14L | A44L | D5R | J4R | WR163 | ||
| F17R | A45R | D6R | J5L | WR164 | ||
| G4L | A46R | D7R | J6R | WR181.5 | ||
| G6R | A47L | D9R | K1L | WR199 | ||
| G7L | A48R | E10R | K3L | WR201 | ||
| I6L | A51R | E11L | K4L | WR204 | ||
| J1R | A53R | E2L | K5L | WR204.5 | ||
| K2L | A55R | E6R | K6L | WR204f | ||
| L4R | A57R | E7R | K7R | WR207 | ||
| L5R | A5R | E8R | L1R | WR209 | ||
| WR036 | A6L | F10L | L2R | WR210 | ||
| WR147 | A7L | F12L | L3L | WR211 | ||
| WR203 | A8R | F15L | M1L | WR212 | ||
| WR208 | B10R | F16L | M2L | WR213 | ||
| WR220 | B11R | F1L | N2L | WR215 | ||
| B12R | F2L | O1L | WR216 | |||
| B14R | F3L | O2L | WR217 | |||
Figure 1.
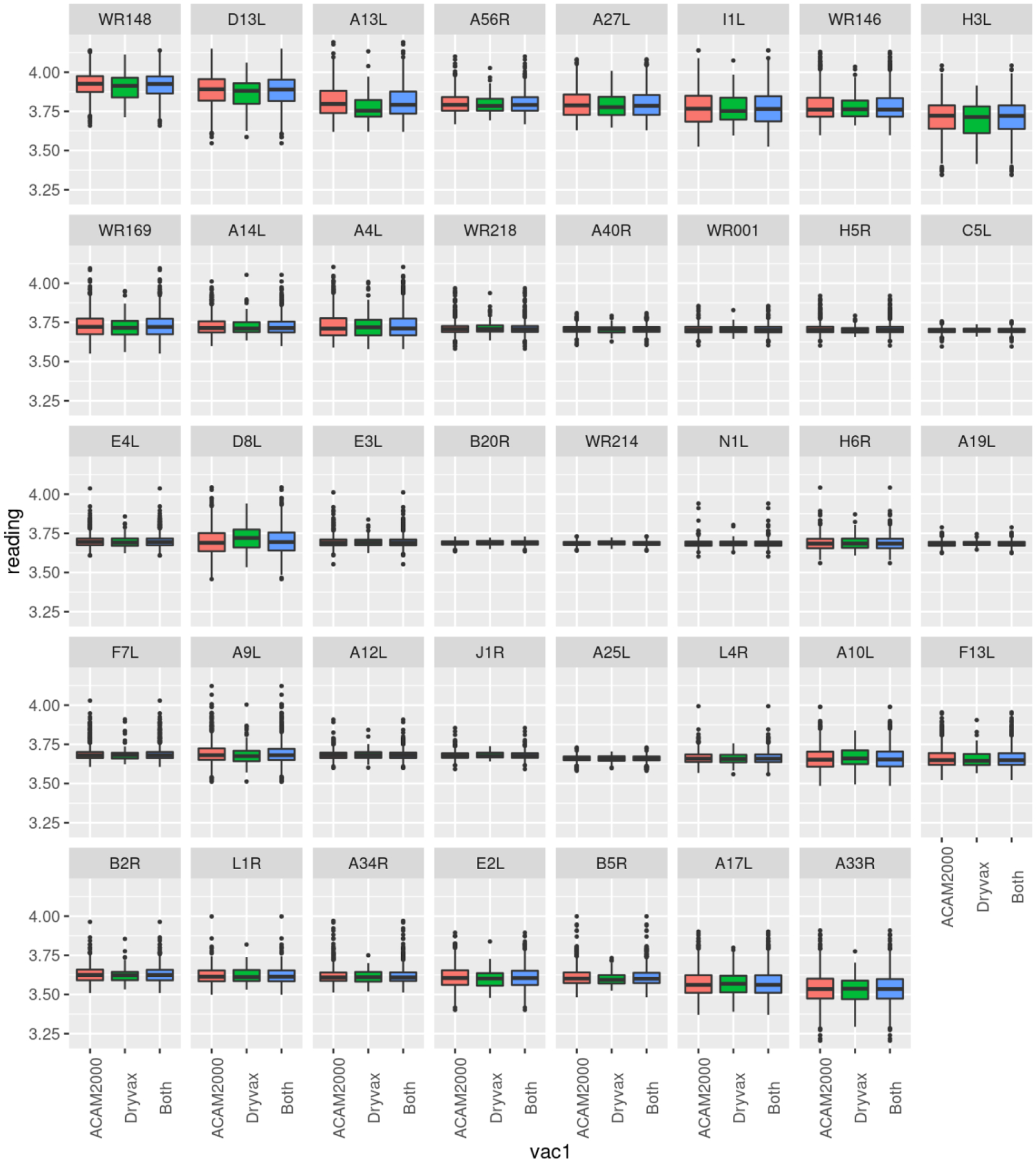
Median Antibody Responses for Selected VACV proteins. Each box and whisker plot provides the median normalized antibody measurements (log2 scale) for each selected protein in ACAM2000 recipients (red), Dryvax recipients (green), or the entire cohort (blue). The top and bottom of the box represent the 75th and 25th percentile respectively, with outliers above and below the 90th and 10th percentile shown as individual points.
Our data indicated that antibody levels to 22 viral proteins were significantly correlated with neutralizing antibody titer to MV (Table 3), suggesting that these proteins may be targets for neutralization.25 Note that four of the proteins in Table 3 (A33R, A34R, A56R, and B5R) are expressed on EV and not on MV. The strongest correlations were found with abundant membrane and structural proteins. From within the cohort, we selected the 194 ACAM2000 recipients with the highest 10% and lowest 10% of neutralizing antibody titers and performed regression modeling (elastic-net models) in order to determine whether or not antibody responses to a set of viral proteins could predict humoral immunity following vaccination. The modeling identified a set of viral proteins prominently on MV virions and expressed late in infection as targets of protein-specific antibody responses that collectively could predict individuals in the high and low neutralizing antibody titer groups a predictive ability of 84%. (Figure 2). Note that the antibody response to these proteins have variable correlations with one another (Supplemental Figure 2). These results largely overlap with the 22 proteins previously found to correlate with neutralizing antibody (Table 3). The exceptions include: A9L, WR218, F7L, WR146, A4L, C5L, and WR169, which are predominantly membrane and structural proteins.
Table 3.
Viral Proteins with Antibody Levels Significantly Correlated with Vaccinia-Specific Neutralizing Antibody Titer
| Protein ID | Correlationlation | p-value (raw) |
|---|---|---|
| Neutralizing Antibody (Median: 113.5, IQR: 62.6 – 189.8) | ||
| H3L | 0.356 | 2.31E-32 |
| A14L | 0.353 | 1.06E-31 |
| D8L | 0.321 | 2.44E-26 |
| A27L | 0.319 | 6.52E-26 |
| B5R | 0.277 | 1.10E-19 |
| I1L | 0.276 | 1.30E-19 |
| A10L | 0.268 | 1.69E-18 |
| A56R | 0.233 | 2.68E-14 |
| H6R | 0.232 | 3.88E-14 |
| A33R | 0.216 | 1.88E-12 |
| A17L | 0.214 | 3.02E-12 |
| D13L | 0.214 | 3.73E-12 |
| F13L | 0.199 | 9.64E-11 |
| WR148 | 0.196 | 1.84E-10 |
| E2L | 0.191 | 5.16E-10 |
| L4R | 0.187 | 1.23E-09 |
| H5R | 0.178 | 8.14E-09 |
| A13L | 0.158 | 3.43E-07 |
| E3L | 0.130 | 2.75E-05 |
| L1R | 0.112 | 0.00031 |
| B2R | 0.104 | 0.00077 |
| A34R | 0.104 | 0.001 |
| A40R | 0.067 | 0.032 |
| WR001 | 0.059 | 0.056 |
| E4L | 0.055 | 0.080 |
| A12L | 0.048 | 0.12 |
| F7L | 0.047 | 0.13 |
| C5L | 0.032 | 0.31 |
| N1L | 0.018 | 0.57 |
| WR218 | 0.006 | 0.86 |
| WR169 | 0.004 | 0.91 |
| A19L | −0.010 | 0.75 |
| J1R | −0.014 | 0.66 |
| WR214 | −0.017 | 0.58 |
| WR146 | −0.024 | 0.44 |
| A4L | −0.035 | 0.27 |
| A9L | −0.037 | 0.24 |
| A25L | −0.038 | 0.22 |
| B20R | −0.047 | 0.13 |
Spearman’s rank correlation
Correlations with p≤0.01 are in bold font.
Figure 2. Logistic Regression Predictive Model of Neutralizing Antibody Response .
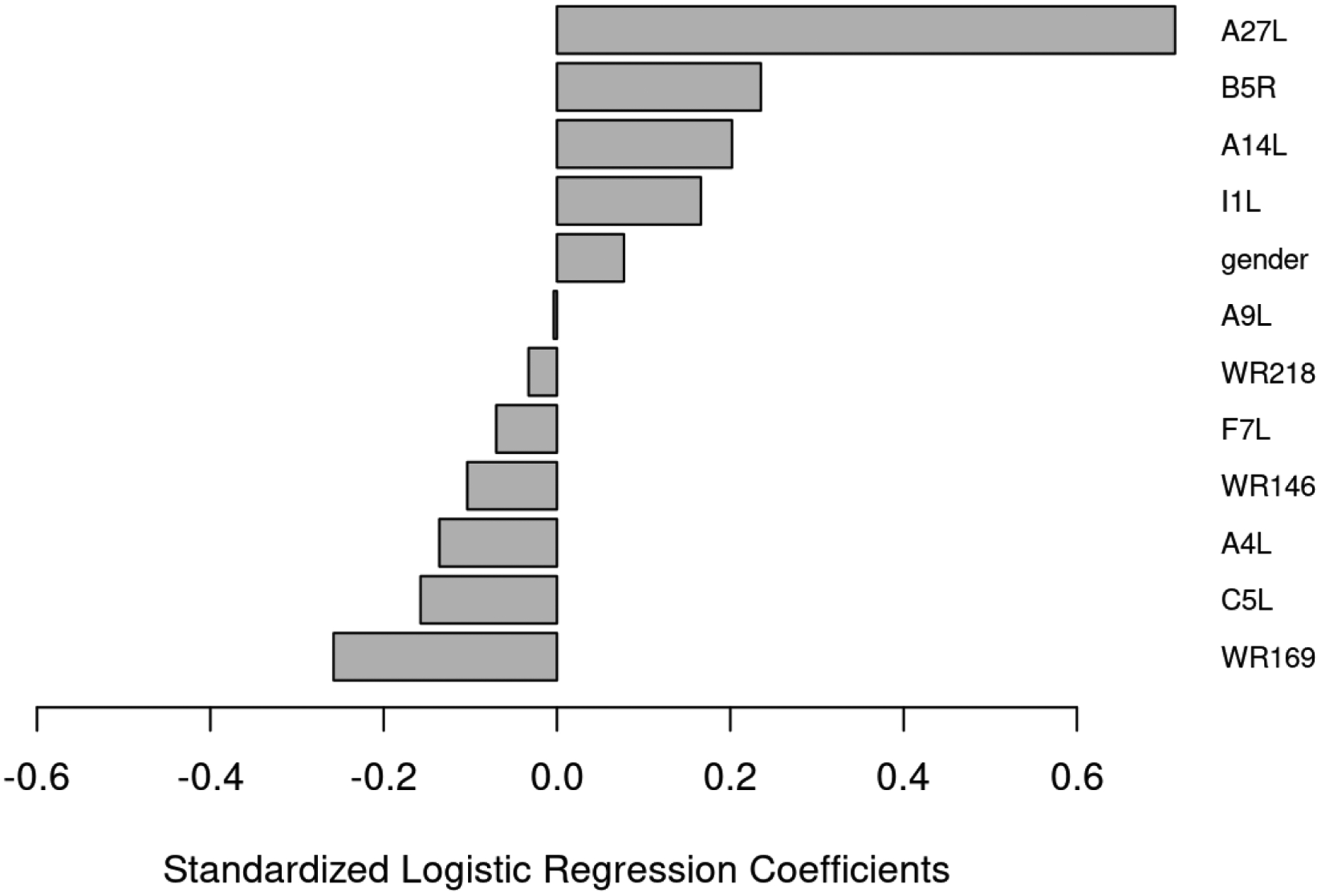
The bar graph depicts the regression coefficients for the variables present in the model with the lowest classification error. Providing a model to predict a vaccine recipient’s neutralizing antibody titer based on the observed antibody response to specific viral proteins.
Race and sex
The regression model also included sex, a result consistent with prior studies we and others have published on the known effect of sex on humoral responses to vaccines.31, 32, 33, 34, 35, 36, 37 When we compared antibody responses to individual proteins between men and women in the entire cohort, we identified five viral proteins in which antibody titers differed significantly by sex (Figure 3). We also examined potential differences in antibody responses to each protein due to race and/or ethnicity (Table 4). The proteins differentially targeted by race/ethnicity comprise both EV and MV proteins, membrane proteins, structural proteins, and secreted proteins. Compared to responses in Caucasian subjects, the consistent finding was that African-American subjects exhibited a trend to lower responses. Compared to non-Hispanic subjects, Hispanic subjects exhibited higher responses to D13L, WR148, A17L, and WR169.
Figure 3. Sex Specific Differences in Antibody Response.
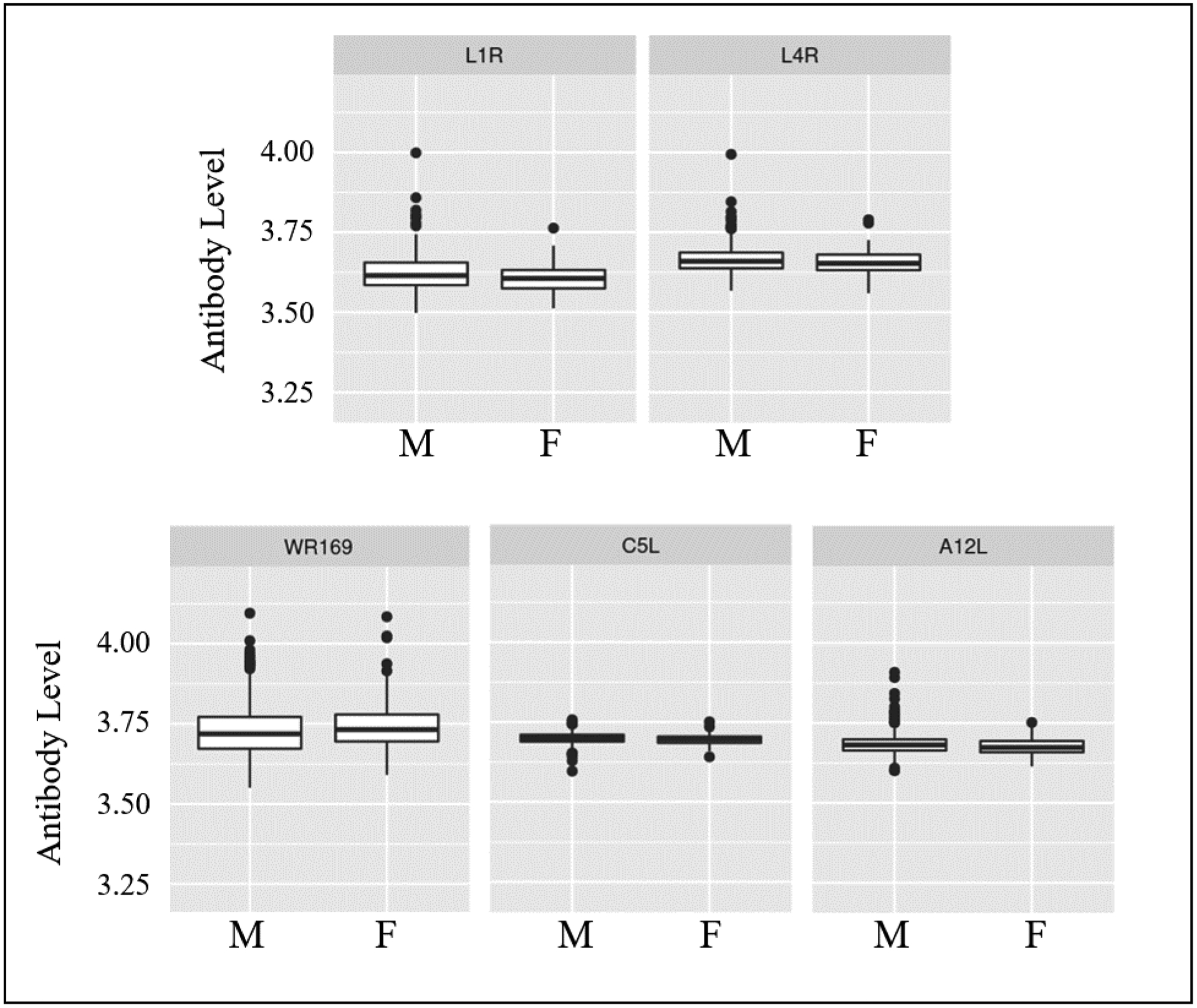
Median normalized antibody measurements for each protein (log2 scale) are provided in the box and whisker plots. M = male, F = female. p-values calculated using the Welch Two Sample t-test.
Table 4.
Race- and Ethnicity-Specific Differences in Antibody Responses
| Race | |||
|---|---|---|---|
| Protein | Race comparison | % Difference in Ab response | p-value* |
| D13L | Caucasian - Black or African American | −1.8% | 2.35E-11 |
| WR148 | Caucasian - Black or African American | −1.5% | 4.20E-11 |
| A33R | Caucasian - Black or African American | −1.6% | 5.98E-08 |
| H3L | Caucasian - Black or African American | −1.7% | 1.36E-07 |
| A27L | Caucasian - Black or African American | −1.2% | 1.65E-06 |
| E2L | Caucasian - Black or African American | −1.0% | 3.77E-06 |
| A14L | Caucasian - Black or African American | −0.7% | 0.00011 |
| F13L | Caucasian - Black or African American | −0.7% | 0.00019 |
| D8L | Caucasian - Black or African American | −1.0% | 0.00023 |
| B5R | Caucasian - Black or African American | −0.7% | 0.00079 |
| A17L | Caucasian - Black or African American | −0.9% | 0.0021 |
| A56R | Caucasian - Black or African American | −0.6% | 0.0021 |
| H5R | Caucasian - Black or African American | −0.3% | 0.0028 |
| A10L | Caucasian - Black or African American | −0.7% | 0.0047 |
| A13L | Caucasian - Black or African American | −0.9% | 0.0072 |
| E3L | Caucasian - Black or African American | −0.3% | 0.026 |
| C5L | Caucasian - Black or African American | −0.1% | 0.028 |
| L4R | Caucasian - Black or African American | −0.3% | 0.029 |
| Ethnicity | |||
| Protein | Race comparison | Difference | p-value |
| D13L | Non-Hispanic - Hispanic | −0.7% | 0.0025 |
| WR148 | Non-Hispanic - Hispanic | −0.6% | 0.0038 |
| WR169 | Non-Hispanic - Hispanic | −0.5% | 0.019 |
| A27L | Non-Hispanic - Hispanic | 1.8% | 0.043 |
| A17L | Non-Hispanic - Hispanic | −0.5% | 0.044 |
% Difference = difference in antibody response in African Americans compared to Caucasians or difference in antibody response in Hispanics compared to Non-Hispanic participants.
p-values calculated using one-way analysis of variance. Differences with p≤0.0001 are in bold font.
Correlation with cellular cytokine responses
We compared the humoral response to individual viral proteins with the cytokine response upon in vitro stimulation of each subject’s PBMCs with VACV.(ref 17) A number of significant associations were identified (Table 5). Each cytokine was associated with an overlapping but not identical set of protein-specific antibody responses. Antibody responses to A27L, A33R, and D8L were associated with both IL-2 and IFNγ secretion, while the antibody response to the L1R protein (an envelope protein required for cellular entry)38 was negatively associated with TNFα, IL-1β, and IL-12p40 production. Of note, a large number of protein-specific antibody response associations were observed with IFNγ and these matched the list of protein-specific antibody responses with the strongest associations with neutralizing antibody.
Table 5.
Correlations Between Antibody Responses to Individual Viral Proteins and Cellular Immune Responses
| Protein | Mean reading | R | p-value | protein | Mean reading | R | p-value |
|---|---|---|---|---|---|---|---|
| IFNα (Median: 184.3pg/ml, IQR: 102.4 – 284.4) | |||||||
| A40R | 3.71 | −0.09 | 0.003 | WR214 | 3.69 | −0.09 | 0.005 |
| IFNγ (Median: 274.1 pg/ml, IQR: 34.9 – 933.0) | |||||||
| H3L | 3.71 | 0.17 | 6.7E-08 | A10L | 3.66 | 0.12 | 7.7E-05 |
| D8L | 3.70 | 0.15 | 2.6E-06 | A13L | 3.81 | 0.12 | 0.00015 |
| A17L | 3.57 | 0.14 | 4.0E-06 | A14L | 3.73 | 0.12 | 0.00017 |
| B5R | 3.61 | 0.14 | 5.3E-06 | A56R | 3.80 | 0.11 | 0.0003 |
| A33R | 3.54 | 0.13 | 2.3E-05 | WR148 | 3.92 | 0.11 | 0.0003 |
| D13L | 3.88 | 0.13 | 5.5E-05 | E2L | 3.61 | 0.11 | 0.0007 |
| A27L | 3.80 | 0.12 | 7.5E-05 | F13L | 3.66 | 0.10 | 0.001 |
| IL-12p40 (Median: 94.4 pg/ml, IQR: 33.7 – 218.1) | |||||||
| L1R | 3.62 | −0.1 | 2.0E-5 | J1R | 3.68 | 0.09 | 0.003 |
| A25L | 3.66 | 0.1 | 0.0002 | ||||
| IL-1β (Median: 64.1 pg/ml, IQR: 25.2 – 160.9) | |||||||
| L1R | 3.62 | −0.09 | 0.002 | J1R | 3.68 | 0.08 | 0.009 |
| L4R | 3.66 | −0.09 | 0.004 | ||||
| IL-2 (Median: 3.8 pg/ml, IQR: −11.2 – 33.8) | |||||||
| A27L | 3.80 | 0.10 | 0.002 | A33R | 3.54 | 0.09 | 0.003 |
| D8L | 3.70 | 0.10 | 0.002 | ||||
| IL-6 (Median: 1,661.2 pg/ml, IQR: 547.3 – 3,439.7) | |||||||
| F7L | 3.69 | −0.12 | 0.0002 | ||||
| TNFα (Median: 115.5 pg/ml, IQR: 50.8 – 259.6) | |||||||
| L1R | 3.62 | −0.17 | 7.9E-9 | H6R | 3.69 | −0.10 | 0.002 |
| B2R | 3.63 | −0.14 | 9.0E-6 | B20R | 3.69 | 0.09 | 0.004 |
| L4R | 3.66 | −0.13 | 1.7E-5 | ||||
R=spearman correlation
Discussion
Our study examined antibody responses targeting specific vaccinia virus proteins in a large cohort of smallpox vaccine recipients. In terms of viral proteins targeted by humoral immune responses to the vaccine, our data were largely similar to what has been previously reported.17, 20, 21 Protein-specific antibody responses in our cohort targeted core vaccinia virus proteins (A4L, H5R, I1L), proteins found in mature virions (A9L, A13L, A27L, D8L, H3L), extracellular enveloped virion proteins (A33R, A34R, A56R, B5R, and F13L), immunomodulatory proteins (WR001, WR218, E3L, N1L) and hypothetical proteins (B20R, C5L, WR169, WR214). These targeted proteins included proteins expressed both early and late in the viral life cycle. Interestingly, our cohort did not respond strongly to several proteins previously described17, 20, 21 as commonly targeted by neutralizing antibodies. These included B5R, the major target of the neutralizing activity of vaccinia immune globulin39 that is known to contain multiple neutralization epitopes.40 Interestingly, titers of B5R-specific antibody were low and only 11.7% of our cohort had detectable responses to B5R – potentially due to the lack of proper folding after in vitro transcription and translation on the microarray. The WR148 (VACV: A26L) protein is expressed on MV and binds to laminin,41 is another protein reported to be widely recognized,17, 20, 21 and was not a major target in our cohort. These differences may reflect the nature of the protein antigens used in the array (expressed using an in vitro E. coli-based transcription/translation system) with possible loss of conformational epitopes found in the native context of viral particles.
Neutralizing antibody titer has served as a correlate of protection against poxvirus infection and previous studies have shown that antibody titers to specific poxvirus proteins exhibit strong correlations with plaque reduction neutralization assay titers. Davies et. al. reported that the average antibody signal across all proteins correlated with neutralizing antibody titer as measured by plaque reduction neutralization tests (r2=0.54).17 When we examined the correlation between protein-specific antibody levels and neutralizing antibody titer, we identified a large number of individual viral proteins whose antibody levels were significantly correlated with neutralizing antibody titer (Table 3). These include the following: H3L, the MV heparin binding protein; D8L, the MV membrane protein that binds chondroitin sulfate; A14L, an MV membrane phosphoprotein; A27L the MV attachment and fusion protein;42 I1L, a telomere-binding protein required for production of mature virions;44 and A10L, a major core protein.;45 the viral hemagglutinin protein A56R that also functions as a virulence factor;46 H6R, a DNA topoisomerase required for both RNA transcription and DNA replication;47 A17L, a viral membrane protein found on the intracellular mature virus that participates in viral assembly and morphogenesis;50 and D13L, a scaffold protein important for the structural integrity of viral membranes.51 The majority of these proteins are structural or membrane proteins and four of the five with the strongest correlation were membrane components of MV (i.e., H3L, A14L, D8L, A27L) with relatively high expression in infected cells and known roles in viral adhesion, binding, and/or entry. H3L is recognized as an important target of neutralizing antibody response, associated with protection in animal models on poxvirus infection.52 These results are not surprising, given that both vaccines consist almost entirely of MV, and the majority of the virus produced following immunization is also MV. Interestingly, neutralizing antibody titers also correlated with protein-specific antibody responses to several expressed on EV, despite the fact that the overwhelming majority of the viral particles present in our neutralization assay were MV. These proteins included: A33R is also a major target of antibody responses and can serve as a protective immunogen;53 B5R, a glycosylated membrane protein involved in release of EV from the cell;43, 54, 55 and A33R, a membrane glycoprotein found on extracellular virions that forms a complex with other membrane proteins (A34R, A36R, and B5R) enabling viral budding from the plasma membrane and cell to cell spread;48, 49 It is possible that the correlations noted in Table 3 simply reflect the fact that these EV proteins are highly immunogenic structural proteins expressed at levels similar to that of highly immunogenic structural proteins on MV virions. Of note, the protein with the fifth-strongest correlation, the EV protein B5R, was recognized by relatively few members of our cohort, which may reflect the fact that neutralizing antibodies to this protein are targeting highly conformational epitopes and are under-represented with the expression system used in the poxvirus antigen array.
The results from our regression modeling identified a collection of viral proteins for which antibody responses were predictive of neutralizing antibody titer. The four proteins with positive regression coefficients (A27L, B5R, A14L, and I1L) are each highly correlated with neutralizing antibody titer (Table 3). It remains to be determined why antibody responses to the other 7 proteins (A9L, WR218, F7L, WR146, A4L, C5L, WR169) have a negative impact on neutralizing antibody titer. One potential application for these results would be the development of a diagnostic test utilizing a subset of the proteins in order to predict protective efficacy in vaccine recipients.
We also assessed correlations between antibody titers to individual proteins and cellular immune responses. Interestingly, the protein-specific antibody responses that correlated with IFNγ production were exactly the same as those that were correlated with neutralizing antibody titers, suggesting that the two arms of immunity (i.e., cellular and humoral) are tightly linked for poxviruses. Some of the same protein-specific Ab responses were also observed with IL-2 (A27L, D8L, and A33R) but the correlations were smaller and the strength of the associations was considerably weaker. Antibody responses to several viral proteins where similar between TNFa, IL-12b, and IL-2 (Table 5) and we have previously reported a strong correlation between pro-inflammatory cytokines in smallpox vaccine recipients.56 Regarding the linkage between humoral and cellular immune responses, Sette et al. have previously reported on a structural linkage between CD4+ T cell epitopes and antibody epitopes.57 In that report, T cell help was only available when the T cell and B cell epitopes came from the same protein. This represents one possible mechanism to explain our results, but confirmation of this would require additional investigations. It should be noted that CD8+ T cells preferentially recognize proteins expressed early in infection with less frequent targeting of structural proteins, while CD4+ T cells preferentially target late antigens as well as structural proteins,58, 59, 60, 61 and are therefore more likely to share epitopes with proteins targeted by antibody responses.
With regard to sex, we have previously reported that women in our study cohort had higher neutralizing antibody titers than men (median ID50 144 vs 110, p=0.024).23 When we examined the antibody response to individual proteins, we found that men had statistically significant higher responses to five proteins (Figure 3), however the differences were extremely small (L1R: 0.4% increase, L4R: 0.28% increase, A12L: 0.19% increase, C5L: 0.11% increase) and a 0.49% decrease in the WR169-specific antibody response. These findings suggest that the prior reported sex differences in the overall antibody response to smallpox vaccination reflect small differences in response across most proteins rather than large differences in antibody titers to a subset of proteins. Another possible explanation is that neutralizing antibodies (found to be more prevalent in women)32 are targeting mostly conformational epitopes that are not readily detected on the proteome array chip. Interestingly, studies with IMVAMUNE, an MVA-based non-replicating smallpox vaccine, have reported higher antibody titers (ELISA, not neutralizing Ab) in men.62
Our study cohort had sufficient racial and ethnic diversity to examine differential antibody responses to individual viral proteins between racial/ethnic groups. Our findings suggested that the Caucasian subjects had potentially stronger humoral responses than African-American subjects to 18 viral proteins (with a wide array of expression kinetics, and functional category [membrane, structural, secreted, etc.], while individuals of Hispanic ethnicity had lower antibody responses to four viral proteins: A17L and D13L, both MV membrane proteins; WR148, a lamin-binding protein found on the envelope of MV particles; and WR169, an uncharacterized protein with unknown function. We do not know if these differences, which are all relatively minor, are biologically or clinically significant; nor is it clear whether or not they would affect protective efficacy at the population level. In fact, the effectiveness of the smallpox vaccines on a global scale would argue against this. Our results are similar to what we have previously reported on race and sex-based differences in cellular immune responses to smallpox vaccines.31 In that study, we found that Caucasian subjects had higher IFNγ ELISPOT responses than African-American and Hispanic subjects. Caucasian subjects also secreted higher levels of IL-2 and IFNα in response to in vitro vaccinia stimulation of subject’s PBMCs. In contrast, a study examining humoral immunity, side effects, and adverse events following smallpox vaccine response in a civilian smallpox vaccination campaign did not identify any associations between racial groups and adverse events, take rates, or antibody titer.63 These disparate results may reflect differences in pre-existing immunity, differences in race/sex/ethnicity between the two cohorts, differences in baseline immunity (our cohorts were confirmed first time vaccinees), or the assays used to measure the various immune response outcomes after vaccination.
Our study does have several limitations: 1) the cohort is predominantly male; nevertheless, we were able to detect sex-specific differences in individual protein responses; 2) the Dryvax subcohort was much smaller than the ACAM2000 cohort, which limited the power to detect small effects; 3) the protein microarray uses an in vitro translation system that may not fully present or display conformation epitopes targeted by some neutralizing antibodies. We found five statistically significant differences among antibody responses to individual proteins (Supplemental Table 1: A13L, D8L, WR148, D13L, B5R) between ACAM2000 and Dryvax recipients; however, the differences in magnitude were small and unlikely to be of clinical significance. The remaining 39 protein-specific antibody responses did not differ between vaccine groups, and the data provided in Figure 1 demonstrate that protein-specific antibody responses were similar between the two vaccines. Other groups have reported that the antigenic targets following vaccination with MVA and Dryvax are comparable,19, 20 and Townsend et al. found similarities between the humoral response in animal models of Dryvax, ACAM2000, and Imvamune vaccination.21 This matches our findings that (with only a few exceptions) antibody responses to individual viral proteins were similar in magnitude between our ACAM2000 and Dryvax recipients.
In conclusion, pathogen proteome microarrays provide a powerful tool for dissecting humoral immunity, which is increasingly important when dealing with complex pathogens such as poxviruses. Tools such as these also provide insights into humoral immune responses at the individual protein level. This finer resolution information can be valuable for understanding critical targets of humoral immunity and has implications for the development of novel vaccines.
Supplementary Material
Supplemental Figure 1. Comparison of Antibody Responses to Specific Viral Proteins in ACAM2000 and Dryvax Recipients. Each box and whisker plot provides the median normalized antibody measurements (log2 scale) for each selected protein in ACAM2000 recipients (red), Dryvax recipients (green), or the entire cohort (blue). The top and bottom of the box represent the 75th and 25th percentile respectively, with outliers above and below the 90th and 10th percentile shown as individual points.
Supplemental Figure 2. Correlation Matrix of the Antibody Levels to VACV-Proteins in the Predictive Model. The size and color of the circle represents the strength of the correlation as noted in the scale to the right. For comparison purposes, the strongest correlation was found between A27L and A14L (0.55).
Supplemental Table 1. Comparison of Antibody Responses to Individual VACV Proteins Between ACAM2000 and Dryvax Recipients.
Acknowledgments
The authors appreciate the subjects’ willing participation in this study as well as the dedicated clinical and research staff at the NHRC and Mayo Clinic who made this study possible - particularly, Drs. Megan Ryan and Kevin Russell.
Conflict of Interest/Disclosures
Dr. Poland is a paid scientific advisor for Johnson & Johnson/Janssen Global Services LLC. Dr. Poland is the chair of a Safety Evaluation Committee for novel investigational vaccine trials being conducted by Merck Research Laboratories. Dr. Poland offers consultative advice on vaccine development to Merck & Co., Medicago, GlaxoSmithKline, Sanofi Pasteur, Emergent Biosolutions, Dynavax, Genentech, Eli Lilly and Company, Kentucky Bioprocessing, Bavarian Nordic, AstraZeneca, Exelixis, Regeneron, Janssen, Vyriad, Moderna, and Genevant Sciences, Inc. Drs. Poland and Ovsyannikova hold patents related to vaccinia and measles peptide vaccines. Dr. Kennedy holds a patent related to vaccinia peptide vaccines. Drs. Poland, Kennedy, and Ovsyannikova have received grant funding from ICW Ventures for preclinical studies on a peptide-based COVID-19 vaccine. Dr. Kennedy has received funding from Merck Research Laboratories to study waning immunity to mumps vaccine. These activities have been reviewed by the Mayo Clinic Conflict of Interest Review Board and are conducted in compliance with Mayo Clinic Conflict of Interest policies. this research have been reviewed by the Mayo Clinic Conflict of Interest Review Board and are conducted in compliance with Mayo Clinic Conflict of Interest policies.
Funding Statement
This work was supported by the NIH through the NIAID Population Genetics Analysis Program Contract No. HHSN266200400065C and Contract No. HHSN272201000025C, and by the National Center for Research Resources grant 1 UL1 RR024150-01. The funding agencies had no role in study design, data collection and analysis, decision to publish, or preparation of this manuscript.
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References
- 1.Fenner F, Arita DAH,I, Jezek Z, Ladnyi ID. Smallpox and its Eradication, vol. 6. W.H.O.: Geneva, 1988. [Google Scholar]
- 2.Mack TM, Noble J Jr. & Thomas DB A prospective study of serum antibody and protection against smallpox. Am J Trop Med Hyg 21, 214–218 (1972). [DOI] [PubMed] [Google Scholar]
- 3.Sarkar JK, Mitra AC & Chakravarty MS Relationship of clinical severity, antibody level, and previous vaccination state in smallpox. Transactions of the Royal Society of Tropical Medicine and Hygiene 66, 789–792 (1972). [DOI] [PubMed] [Google Scholar]
- 4.Sarkar JK, Mitra AC & Mukherjee MK The minimum protective level of antibodies in smallpox. Bull World Health Organ 52, 307–311 (1975). [PMC free article] [PubMed] [Google Scholar]
- 5.Chaudhri G, Panchanathan V, Bluethmann H & Karupiah G Obligatory requirement for antibody in recovery from a primary poxvirus infection. J Virol 80, 6339–6344 (2006). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Panchanathan V, Chaudhri G & Karupiah G Protective immunity against secondary poxvirus infection is dependent on antibody but not on CD4 or CD8 T-cell function. J Virol 80, 6333–6338 (2006). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Panchanathan V, Chaudhri G & Karupiah G Correlates of protective immunity in poxvirus infection: where does antibody stand? Immunol Cell Biol 86, 80–86 (2008). [DOI] [PubMed] [Google Scholar]
- 8.Ramirez JC, Tapia E & Esteban M Administration to mice of a monoclonal antibody that neutralizes the intracellular mature virus form of vaccinia virus limits virus replication efficiently under prophylactic and therapeutic conditions. J Gen Virol 83, 1059–1067 (2002). [DOI] [PubMed] [Google Scholar]
- 9.Moss B Poxvirus entry and membrane fusion. Virology 344, 48–54 (2006). [DOI] [PubMed] [Google Scholar]
- 10.McCausland MM et al. Combination therapy of vaccinia virus infection with human anti-H3 and anti-B5 monoclonal antibodies in a small animal model. Antiviral therapy 15, 661–675 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Pacchioni SM et al. L1R, A27L, A33R and B5R vaccinia virus genes expressed by fowlpox recombinants as putative novel orthopoxvirus vaccines. Journal of translational medicine 11, 95 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Hooper JW, Custer DM & Thompson E Four-gene-combination DNA vaccine protects mice against a lethal vaccinia virus challenge and elicits appropriate antibody responses in nonhuman primates. Virology 306, 181–195 (2003). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Fogg C et al. Protective immunity to vaccinia virus induced by vaccination with multiple recombinant outer membrane proteins of intracellular and extracellular virions. J Virol 78, 10230–10237 (2004). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Law M, Putz MM & Smith GL An investigation of the therapeutic value of vaccinia-immune IgG in a mouse pneumonia model. J Gen Virol 86, 991–1000 (2005). [DOI] [PubMed] [Google Scholar]
- 15.Meseda CA et al. Effect of the deletion of genes encoding proteins of the extracellular virion form of vaccinia virus on vaccine immunogenicity and protective effectiveness in the mouse model. PLoS One 8, e67984 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Davies DH et al. Profiling the humoral immune response to infection by using proteome microarrays: high-throughput vaccine and diagnostic antigen discovery. Proc Natl Acad Sci U S A 102, 547–552 (2005). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Davies DH et al. Proteome-wide analysis of the serological response to vaccinia and smallpox. Proteomics 7, 1678–1686 (2007). [DOI] [PubMed] [Google Scholar]
- 18.Benhnia MR et al. Redundancy and plasticity of neutralizing antibody responses are cornerstone attributes of the human immune response to the smallpox vaccine. J Virol 82, 3751–3768 (2008). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Davies DH et al. Antibody profiling by proteome microarray reveals the immunogenicity of the attenuated smallpox vaccine modified vaccinia virus ankara is comparable to that of Dryvax. J Virol 82, 652–663 (2008). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Hermanson G et al. Measurement of antibody responses to Modified Vaccinia virus Ankara (MVA) and Dryvax((R)) using proteome microarrays and development of recombinant protein ELISAs. Vaccine 30, 614–625 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Townsend MB et al. Humoral immunity to smallpox vaccines and monkeypox virus challenge: proteomic assessment and clinical correlations. J Virol 87, 900–911 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Benhnia MR et al. Unusual features of vaccinia virus extracellular virion form neutralization resistance revealed in human antibody responses to the smallpox vaccine. J Virol 87, 1569–1585 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Ovsyannikova IG, Pankratz VS, Salk HM, Kennedy RB & Poland GA HLA alleles associated with the adaptive immune response to smallpox vaccine: a replication study. Hum Genet 133, 1083–1092 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Manischewitz J et al. Development of a novel vaccinia-neutralization assay based on reporter-gene expression. J Infect Dis 188, 440–448 (2003). [DOI] [PubMed] [Google Scholar]
- 25.Kennedy R et al. Statistical approach to estimate vaccinia- specific neutralizing antibody titers using a high throughput assay. Clin Vaccine Immunol (2009). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Chakrabarti S, Sisler JR & Moss B Compact, synthetic, vaccinia virus early/late promoter for protein expression. Biotechniques 23, 1094–1097 (1997). [DOI] [PubMed] [Google Scholar]
- 27.Chakrabarti S, Brechling K & Moss B Vaccinia virus expression vector: coexpression of beta-galactosidase provides visual screening of recombinant virus plaques. Molecular and cellular biology 5, 3403–3409 (1985). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Huber W, von Heydebreck A, Sultmann H, Poustka A & Vingron M Variance stabilization applied to microarray data calibration and to the quantification of differential expression. Bioinformatics 18 Suppl 1, S96–104 (2002). [DOI] [PubMed] [Google Scholar]
- 29.Harrell JFE Regression Modeling Strategies : With Applications to Linear Models, Logistic and Ordinal Regression, and Survival Analysis. Springer Series in Statistics,. 2nd ed. Cham: Springer International Publishing : Imprint: Springer,; 2015. pp. 1 online resource (XXV, 582 pages 157 illustrations, 553 illustrations in color. [Google Scholar]
- 30.Friedman J, Hastie T & Tibshirani R Regularization Paths for Generalized Linear Models via Coordinate Descent. J Stat Softw 33, 1–22 (2010). [PMC free article] [PubMed] [Google Scholar]
- 31.Haralambieva IH et al. Race and sex-based differences in cytokine immune responses to smallpox vaccine in healthy individuals. Hum Immunol 74, 1263–1266 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Kennedy RB et al. Gender effects on humoral immune responses to smallpox vaccine. Vaccine (2009). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Voigt EA et al. Sex Differences in Older Adults’ Immune Responses to Seasonal Influenza Vaccination. Front Immunol 10, 180 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Voigt EA et al. Genetically defined race, but not sex, is associated with higher humoral and cellular immune responses to measles vaccination. Vaccine 34, 4913–4919 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Haralambieva IH et al. Associations between race, sex and immune response variations to rubella vaccination in two independent cohorts. Vaccine 32, 1946–1953 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Denly L The effect of sex on responses to influenza vaccines. Hum Vaccin Immunother, 1–7 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Aaby P et al. The non-specific and sex-differential effects of vaccines. Nat Rev Immunol 20, 464–470 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Foo CH et al. Vaccinia virus L1 binds to cell surfaces and blocks virus entry independently of glycosaminoglycans. Virology 385, 368–382 (2009). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Bell E et al. Antibodies against the extracellular enveloped virus B5R protein are mainly responsible for the EEV neutralizing capacity of vaccinia immune globulin. Virology 325, 425–431 (2004). [DOI] [PubMed] [Google Scholar]
- 40.Aldaz-Carroll L et al. Epitope-mapping studies define two major neutralization sites on the vaccinia virus extracellular enveloped virus glycoprotein B5R. J Virol 79, 6260–6271 (2005). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Chiu WL, Lin CL, Yang MH, Tzou DL & Chang W Vaccinia virus 4c (A26L) protein on intracellular mature virus binds to the extracellular cellular matrix laminin. J Virol 81, 2149–2157 (2007). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Chung CS, Hsiao JC, Chang YS & Chang W A27L protein mediates vaccinia virus interaction with cell surface heparan sulfate. J Virol 72, 1577–1585 (1998). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Engelstad M & Smith GL The vaccinia virus 42-kDa envelope protein is required for the envelopment and egress of extracellular virus and for virus virulence. Virology 194, 627–637 (1993). [DOI] [PubMed] [Google Scholar]
- 44.Goebel SJ et al. The complete DNA sequence of vaccinia virus. Virology 179, 247–266, 517–263 (1990). [DOI] [PubMed] [Google Scholar]
- 45.Heljasvaara R et al. The major core protein P4a (A10L gene) of vaccinia virus is essential for correct assembly of viral DNA into the nucleoprotein complex to form immature viral particles. J Virol 75, 5778–5795 (2001). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.DeHaven BC, Gupta K & Isaacs SN The vaccinia virus A56 protein: a multifunctional transmembrane glycoprotein that anchors two secreted viral proteins. J Gen Virol 92, 1971–1980 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Beaud G Vaccinia virus DNA replication: a short review. Biochimie 77, 774–779 (1995). [DOI] [PubMed] [Google Scholar]
- 48.Roper RL, Payne LG & Moss B Extracellular vaccinia virus envelope glycoprotein encoded by the A33R gene. J Virol 70, 3753–3762 (1996). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Wolffe EJ, Weisberg AS & Moss B The vaccinia virus A33R protein provides a chaperone function for viral membrane localization and tyrosine phosphorylation of the A36R protein. J Virol 75, 303–310 (2001). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Wallengren K, Risco C, Krijnse-Locker J, Esteban M & Rodriguez D The A17L gene product of vaccinia virus is exposed on the surface of IMV. Virology 290, 143–152 (2001). [DOI] [PubMed] [Google Scholar]
- 51.Charity JC, Katz E & Moss B Amino acid substitutions at multiple sites within the vaccinia virus D13 scaffold protein confer resistance to rifampicin. Virology 359, 227–232 (2007). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Davies DH et al. Vaccinia virus H3L envelope protein is a major target of neutralizing antibodies in humans and elicits protection against lethal challenge in mice. J Virol 79, 11724–11733 (2005). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Paran N et al. Active vaccination with vaccinia virus A33 protects mice against lethal vaccinia and ectromelia viruses but not against cowpoxvirus; elucidation of the specific adaptive immune response. Virology journal 10, 229 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Law M, Carter GC, Roberts KL, Hollinshead M & Smith GL Ligand-induced and nonfusogenic dissolution of a viral membrane. Proc Natl Acad Sci U S A 103, 5989–5994 (2006). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Doceul V, Hollinshead M, Breiman A, Laval K & Smith GL Protein B5 is required on extracellular enveloped vaccinia virus for repulsion of superinfecting virions. J Gen Virol 93, 1876–1886 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Umlauf BJ et al. Correlations between vaccinia-specific immune responses within a cohort of armed forces members. Viral Immunol 24, 415–420 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Sette A et al. Selective CD4+ T cell help for antibody responses to a large viral pathogen: deterministic linkage of specificities. Immunity 28, 847–858 (2008). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Moutaftsi M et al. Uncovering the interplay between CD8, CD4 and antibody responses to complex pathogens. Future microbiology 5, 221–239 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Moutaftsi M et al. A consensus epitope prediction approach identifies the breadth of murine T(CD8+)-cell responses to vaccinia virus. Nat Biotechnol 24, 817–819 (2006). [DOI] [PubMed] [Google Scholar]
- 60.Oseroff C et al. HLA class I-restricted responses to vaccinia recognize a broad array of proteins mainly involved in virulence and viral gene regulation. Proc Natl Acad Sci U S A 102, 13980–13985 (2005). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Moutaftsi M et al. Vaccinia virus-specific CD4+ T cell responses target a set of antigens largely distinct from those targeted by CD8+ T cell responses. J Immunol 178, 6814–6820 (2007). [DOI] [PubMed] [Google Scholar]
- 62.Troy JD, Hill HR, Ewell MG & Frey SE Sex difference in immune response to vaccination: A participant-level meta-analysis of randomized trials of IMVAMUNE smallpox vaccine. Vaccine 33, 5425–5431 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Haselow D Vaccination-Related Side Effects, Humoral Immunity, and Adverse Events during the Civilian Smallpox Vaccination Campaign, Arkansas, 2003. Public Health Nurs (2015). [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Supplemental Figure 1. Comparison of Antibody Responses to Specific Viral Proteins in ACAM2000 and Dryvax Recipients. Each box and whisker plot provides the median normalized antibody measurements (log2 scale) for each selected protein in ACAM2000 recipients (red), Dryvax recipients (green), or the entire cohort (blue). The top and bottom of the box represent the 75th and 25th percentile respectively, with outliers above and below the 90th and 10th percentile shown as individual points.
Supplemental Figure 2. Correlation Matrix of the Antibody Levels to VACV-Proteins in the Predictive Model. The size and color of the circle represents the strength of the correlation as noted in the scale to the right. For comparison purposes, the strongest correlation was found between A27L and A14L (0.55).
Supplemental Table 1. Comparison of Antibody Responses to Individual VACV Proteins Between ACAM2000 and Dryvax Recipients.


