SUMMARY
During female meiosis I (MI), spindle positioning must be tightly regulated to ensure the fidelity of the first asymmetric division and faithful chromosome segregation. Although the role of F-actin in regulating these critical processes has been studied extensively, little is known whether microtubules (MTs) participate in regulating these processes. Using mouse oocytes as a model system, we characterize a subset of MT organizing centers that does not contribute directly to spindle assembly, termed mcMTOCs. Using laser ablation, STED super-resolution microscopy and chemical manipulation, we show that mcMTOCs are required to regulate spindle positioning and faithful chromosome segregation during MI. We discuss how forces exerted by F-actin on the spindle are balanced by mcMTOC-nucleated MTs to anchor the spindle centrally and to regulate its timely migration. Our findings provide a model for asymmetric cell division, complementing the current F-actin-based models, and implicate mcMTOCs as a major player in regulating spindle positioning.
Keywords: MTOC, Microtubule, Oocyte, Meiosis, Spindle positioning
eTOC blurb
Acentriolar microtubule organizing centers (MTOCs) are known for their role in assembling the meiotic spindle. Londoño-Vásquez et al. discover that a subset of cytoplasmic MTOCs (termed mcMTOCs) regulates spindle positioning by anchoring the spindle to the oocyte cortex.
Graphical Abstract
INTRODUCTION
Mammalian oocytes enter meiosis during early fetal life. Soon after birth, meiotic oocytes undergo a lengthy arrest at the dictyate stage of the prophase I of the first meiotic division (MI) (Von Stetina and Orr-Weaver, 2011). At the age of puberty, gonadotropin cues allow prophase I-arrested oocytes to resume MI evident by breakdown of the nuclear envelope (NEBD) and formation of a central bipolar spindle (Hashimoto and Kishimoto, 1988, Bennabi et al., 2016, Kitajima et al., 2011). Our recent preprint suggests that the central positioning of the spindle is required to establish proper kinetochore-microtubule (MT) attachments and to protect against aneuploidy (Kincade and Balboula, 2020). The position of the spindle dictates the plane of cell division (McCarthy and Goldstein, 2006) and therefore, in contrast to mitotic cells where a centrally positioned spindle allows symmetrical cell division, the meiotic spindle must migrate towards the cortex for the highly asymmetric meiotic divisions (Longo and Chen, 1985, McCarthy and Goldstein, 2006). Such peripheral positioning of the spindle is critical to extrude the tiny polar body (PB) thereby retaining the great majority of the cytoplasm containing maternal RNAs and proteins for the egg to support early embryonic development (Menezo, 2006, Ma et al., 2006). Therefore, it is essential to understand the critical events of spindle positioning and migration, which are likely required for the fidelity of chromosome transmission to the next generation.
To date, F-actin and its regulatory molecules represent the only cytoskeletal components known to regulate spindle migration and positioning in the mammalian oocyte. It has been shown that perturbation of F-actin, but not of MTs, impairs spindle migration in oocytes and that the resulting increased symmetry of cell division results in infertility (Schuh and Ellenberg, 2008, Azoury et al., 2008, Li et al., 2008, Longo and Chen, 1985). This contrasts with mitotically dividing somatic cells, where positioning of the spindle at the center of the cell is primarily regulated by the interaction of the astral MTs originated from spindle poles with the cell cortex (McNally, 2013). Such astral MTs are nucleated by centrosomes, centriole pairs surrounded by peri-centriolar material (PCM). By contrast, mammalian oocytes lack classic centrosomes (Szollosi et al., 1972) because centrioles are lost during early oogenesis through an unknown mechanism. The numerous acentriolar MT organizing centers (MTOCs) of the oocyte (Szollosi et al., 1972, Schuh and Ellenberg, 2007) are still able to nucleate astral-like MTs but these are short and unable to extend to the cortex when the spindle is centrally positioned (Schuh and Ellenberg, 2007, Xie et al., 2018, Schatten et al., 1986). These observations have enforced the notion that MTs have no role in regulating central spindle positioning and migration. It is therefore surprising that when mouse oocytes are treated with nocodazole, a MT depolymerizing agent, chromosomes migrate towards the cortex at a higher speed and at an earlier time than in control oocytes (Li et al., 2008, Verlhac et al., 2000). These unexplained observations suggest that MTs have, yet unknown, role(s) in regulating spindle migration and positioning in mammalian oocytes.
Based on MTOC behavior, three different classes of MTOCs were characterized in meiotic oocytes. In prophase I-arrested oocytes, the MTOCs (class I) are initially found in the perinuclear region (Szollosi et al., 1972). Then, before NEBD, these perinuclear MTOCs undergo distinct processes of decondensation, stretching and redistribution into a large number of smaller MTOCs (Clift and Schuh, 2015, Luksza et al., 2013). The fragmented MTOCs are then clustered and sorted to form two poles necessary to assemble a bipolar spindle (Schuh and Ellenberg, 2007, Balboula et al., 2016, Breuer et al., 2010). Another pool of MTOCs is also present in the cytoplasm during NEBD. Some of these cytoplasmic MTOCs (class II) migrate from the periphery to the center of the oocyte, where they too participate in spindle pole formation (polar MTOCs, pMTOCs; classes I and II) (Schuh and Ellenberg, 2007, Clift and Schuh, 2015). Another subset of cytoplasmic MTOCs (class III), hereafter referred to as metaphase cytoplasmic MTOCs (mcMTOCs) does not contribute to spindle formation and has yet unknown biological significance for oocyte meiosis.
Here, we show that the pMTOCs and the mcMTOCs of MI oocytes represent two different functional sets. We find by 3D time-lapse imaging that mcMTOCs localize asymmetrically opposite the site of F-actin enrichment where the first PB is extruded. Super-resolution Stimulated Emission Depletion (STED) microscopy reveals that mcMTOCs are able to nucleate MTs that connect the spindle to the cortex. Importantly, we show that by increasing mcMTOC numbers following treatment with an inhibitor of autophagy or by depleting them by laser ablation, the meiotic spindle becomes abnormally positioned leading to aneuploidy. Our results suggest a model whereby the role of F-actin in mediating movement of the meiotic spindle to the cortex is balanced by forces exerted from mcMTOCs to ensure the timely migration and accurate positioning of the spindle in the oocyte.
RESULTS
MTOCs in the cytoplasm and at the spindle poles of meiotic oocytes
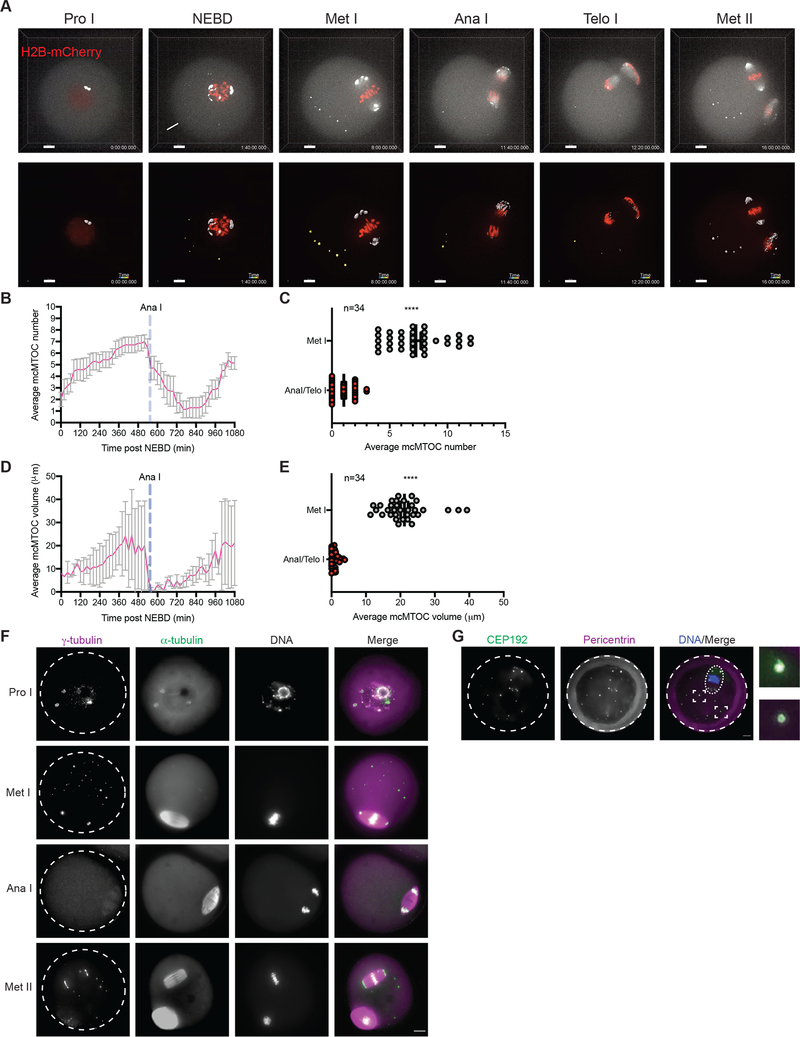
The multiple MTOCs of prophase I-arrested oocytes follow distinct patterns of behavior. Immediately before NEBD, perinuclear MTOCs become fragmented in three phases; decondensation, stretching and repositioning towards the spindle poles through the sequential actions of Polo-like kinase 1, BicD2-anchored dynein, and the KIF11 motor protein (Clift and Schuh, 2015, Luksza et al., 2013). Soon after NEBD, a second group of cytoplasmic MTOCs migrates from the periphery to the center of the oocyte, where it participates with perinuclear MTOCs in spindle assembly (Schuh and Ellenberg, 2007, Clift and Schuh, 2015). Finally, a third population of MTOCs were observed in the cytoplasm of oocytes at metaphase (here termed mcMTOCs) (Messinger and Albertini, 1991, Schatten et al., 1986, Houliston et al., 1987, Meng et al., 2004, Maro et al., 1988), whose role in meiosis is unknown. To follow the behavior of mcMTOCs during MI, we employed live imaging using 3D confocal microscopy to visualize prophase I-arrested oocytes (collected from CF-1 mice) expressing GFP-tagged Aurora A Kinase (AURKA-GFP, an integral component of MTOCs in mouse oocytes (Saskova et al., 2008, Solc et al., 2012, Bury et al., 2017) and H2B-mCherry (H2B-mCh) to label MTOCs and DNA, respectively. As previously reported, perinuclear MTOCs became fragmented into small multiple MTOCs at NEBD before they sorted and re-clustered at the two spindle poles as pMTOCs (Fig. 1A; Video S1). We also observed cytoplasmic MTOCs in the cytoplasm at NEBD, some of which migrated towards the oocyte center to contribute to spindle formation as pMTOCs, whereas others, mcMTOCs, remained free in the cytoplasm during metaphase I (Met I) after the bipolar spindle had formed (Fig. 1A; Video S1). We carried out 3D reconstruction of entire oocytes to examine the number (Fig. 1B,C) and volume (Fig. 1D, E) of the mcMTOCs (Video S2). In contrast to pMTOCs which undergo a time-dependent decrease in number and increase in volume (due to MTOC clustering) as the oocyte proceeds to Met I, the mcMTOCs displayed the opposite pattern and showed a time-dependent increase in number and volume during pro-Met I and Met I (Fig. 1A,B,D; Video S1). Each oocyte had a variable number of mcMTOCs at Met I (6.03±0.1 mcMTOCs per oocyte in average) located on different focal planes (Supplementary Fig. 1A; Video S2). The mcMTOCs became less distinct, appearing to be decreased in number and volume during anaphase I (Ana I) and telophase I (Telo I) before regaining their metaphase appearance as the oocytes arrested in Met II (Fig. 1A–E; Video S1). We confirmed that these AURKA-positive foci were indeed MTOCs by showing the colocalization of pericentrin, another integral component of PCM in meiotic oocytes (Baumann et al., 2017, Ma and Viveiros, 2014) (Supplementary Fig. 1B). Moreover, AURKA-GFP expression had no significant effect on mcMTOC numbers or volume (Supplementary. Fig. 1C–E). To confirm our observation, we fixed CF-1 oocytes at different developmental stages (Prophase I, Met I, Ana I/Telo I and Met II) and immunostained them to reveal the MTOC markers, γ-tubulin, pericentrin and CEP192 (Gueth-Hallonet et al., 1993, Clift and Schuh, 2015, So et al., 2019, Ma and Viveiros, 2014). This also revealed pMTOCs and mcMTOCs at Met I (Fig. 1F, G) that were associated with MT asters (α-tubulin staining, Fig. 1F). Similar findings were observed in Met I oocytes from C57BL/6 mice (Supplementary Fig. 1F). Just as we observed in time-lapse imaging of MTOCs in living oocytes, the mcMTOCs became less distinct in Ana I/Telo I of fixed oocytes (Fig. 1F).
Figure 1: Two different sets of MTOCs are present in acentriolar oocytes.
(A) Time-lapse confocal microscopy of MTOCs in live oocytes. Full-grown prophase-I oocytes were injected with cRNAs encoding H2b-mCherry (red) and Aurka-Gfp (pseudo white), incubated in milrinone-containing CZB medium for 3 h, followed by in vitro maturation. Shown are representative images (Z-projection of 16 sections every 3 μm) from a time course (see Video S1). Lower panels represent a 3D reconstruction of MTOCs from oocyte shown in upper panels (see Video S1). Fluorescence images were captured every 20 min. The scale bar represents 10 μm. The white arrowhead represents cytoplasmic MTOC that participate in spindle pole formation as a pMTOC. The white arrow represents cytoplasmic MTOC that remains in the cytoplasm during Met I (mcMTOC). (B,D) Quantification of average MTOC number and MTOC volume over time during meiosis I, respectively. Error bars show S.D. Dashed blue lines represent the time of Anaphase I onset. (C,E). Quantification of average MTOC number and MTOC volume, respectively, during metaphase I (Met I) and anaphase I/telophase I (Ana I/Telo I) stages. The data are expressed as mean ± SEM. Student t-test was used to analyze the data. Values with asterisks vary significantly, ****P < 0.0001. The total number of analyzed oocytes (from three independent replicates) is specified above each graph. (F) Fully grown prophase-I-arrested oocytes were in vitro matured for 0 h (prophase I, Pro I), 8 h (Met I), 9 h (Ana I/Telo I) or 16 h (Met II) prior to fixation and immunocytochemistry using γ-tubulin and α-tubulin antibodies to label MTOCs (magenta) and microtubules (green). DAPI was used to detect DNA (grey). (G) Fully grown prophase-I-arrested oocytes were in vitro matured for 8 h (Met I) followed by fixation and immunostaining using CEP192 (green) and pericentrin (magenta) antibodies to label MTOCs. DAPI was used to detect DNA (blue). Shown are representative confocal Z-projections. Arrowheads represent mcMTOCs. A total of 184 oocytes were examined. Scale bars represent 10 μm (zoomed panels, 2 μm).
mcMTOCs undergo three patterns of directional movement during MI
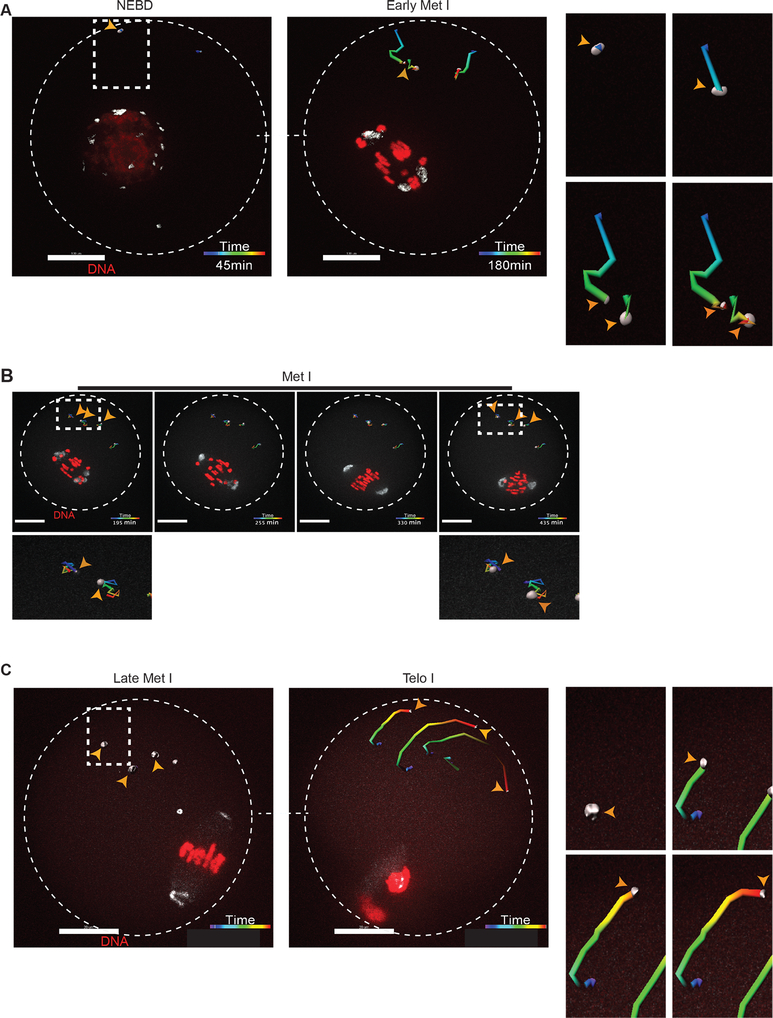
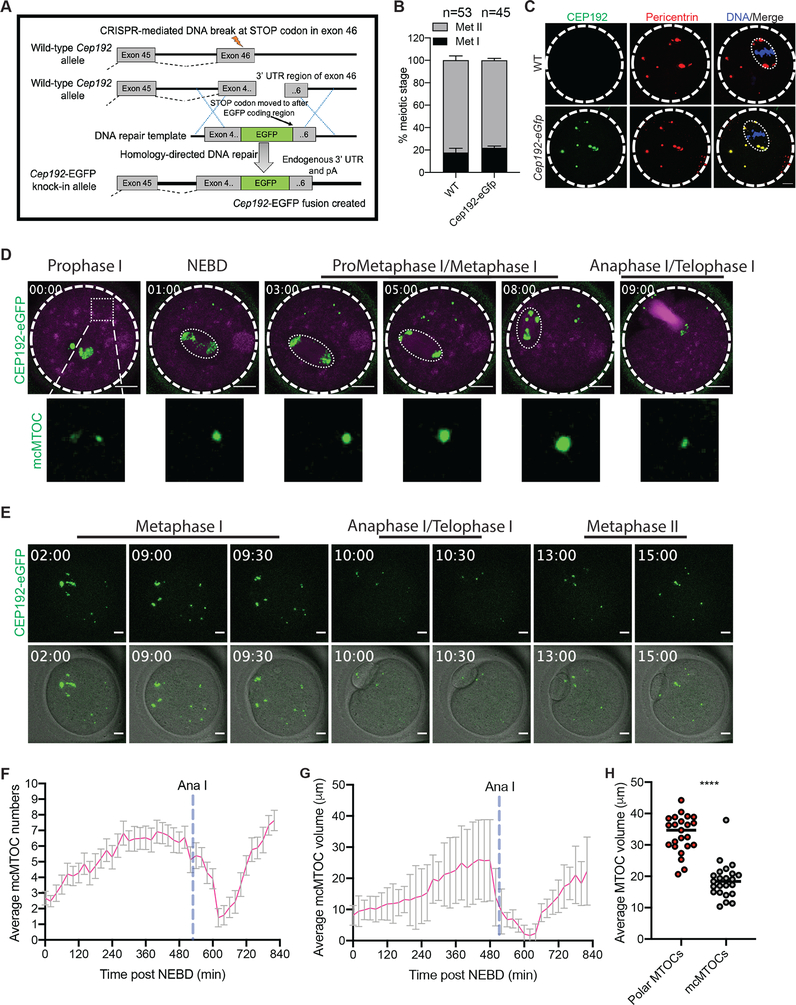
To dissect the directional motion and kinetics of the mcMTOCs, we tracked their movement using 3D reconstructions over time in oocytes expressing AURKA-GFP. In contrast to pMTOCs which primarily originate from perinuclear MTOCs at NEBD, the mcMTOCs formed at the periphery of the oocyte (Fig. 2A; Supplementary Movie 4 and 5) and then appeared to undergo three phases of directional movement (n = 42 examined oocytes). In the first phase, from NEBD to early Met I, the peripheral mcMTOCs moved towards the oocyte’s center with an average speed of 0.09 ± 0.007 μm min−1 (mean ± SEM) and a maximum speed of 0.18 ± 0.005 μm min−1. During this phase, the average volume of the mcMTOCs increased, in some cases due to mcMTOCs merging with each other (Fig. 2A; Video S3). The second was marked by the slowing of mcMTOC movement to an average speed of 0.06 ± 0.009 μm min−1 allowing them to remain in confined areas of the cytoplasm (Fig. 2B; Video S3). During this phase, the pMTOCs underwent active clustering whereas the mcMTOCs remained apart. The third phase occurred during Ana I and Telo I when the mcMTOCs showed a reversal of their behavior in phase I; they displayed a drastic reduction in volume and migrated towards the cortex with an average speed of 0.12 ± 0.01 μm min−1 and a maximum speed of 0.22 μm min−1 (Fig. 2C; Video S3). In as many as 50% of oocytes (14/28), we were not able to observe mcMTOCs during Ana I/Telo I but in all cases when they could be observed, the mcMTOCs migrated towards the region of cortex opposite direction the site of extrusion of the first PB. The mcMTOCs showed independent directional movement to cytoplasmic droplets and so do not reflect overall cytoplasmic movements (Supplementary Fig. 2). To exclude the possibility that the decrease in mcMTOC size during Ana I/Telo I is due to AURKA protein degradation, we generated a Cep192-eGfp reporter CF-1 mouse model (Fig. 3A) to allow for the precise tracking of mcMTOCs during MI. Briefly, a construct harboring the EGFP reporter gene was integrated into the mouse genome via CRISPR/Cas9-mediated homology-directed repair. The EGFP reporter was fused at the C-terminus of the endogenous mouse Cep192. Thus, in this model, CEP192 is just endogenously labeled, not perturbed or overexpressed. Accordingly, Cep192-eGfp oocytes expressed similar levels of CEP192 and are phenotypically normal to WT oocytes in terms of meiotic progression (Fig. 3B). Using immunocytochemistry, we confirmed that endogenous CEP192-eGFP colocalizes with all pericentrin-positive MTOCs (Fig. 3C). Importantly, CEP192-eGFP oocytes showed similar mcMTOC behavior during MI where variable number of mcMTOCs were scattered in the cytoplasm during Met I (Fig. 3D,E). Similar to AURKA-GFP-labeled mcMTOCs, endogenously labeled mcMTOCs (CEP192-eGFP) increase in number and volume towards Met I (Fig. 3D, F-H; Video S4). During Ana I/Telo I, endogenously labeled mcMTOCs underwent a decrease in volume and number while migrating towards the cortex (Fig. 3E, F and G; Video S4). Again, we were not able to observe mcMTOCs during Ana I/Telo I in ~ 46% of oocytes (35/76) prior to their reappearance again at Met II. Taken together, our observations confirm that meiotic oocytes have two different pools of MTOCs during Met I and raise the question whether mcMTOCs differ from pMTOCs in their function.
Figure 2: mcMTOCs exhibit three different directional behaviors during meiosis I.
Tracking of 3D reconstructed mcMTOCs from time-lapse confocal microscopy in live oocytes during three different phases: (A) nuclear envelope breakdown (NEBD)- early metaphase I (Met I), (B) Met I and (C) late Met I- telophase I (Telo I). Full-grown prophase-I oocytes were injected with cRNAs encoding H2b-mCherry (red) and Aurka-Gfp (pseudo white), incubated in milrinone-containing CZB medium for 3 h prior to in vitro maturation. Shown are representative images of 3D reconstructed Z-projection of 16 sections every 3 μm from a time course. Fluorescence images were captured every 15 min (time, h:min). The scale bar represents 20 μm (zoomed panels, 5 μm). Right panels show a zoomed area of mcMTOC tracking over time (See Video S3). The overall direction of displacement was calculated automatically by Bitplane Imaris software. Arrowheads represent mcMTOCs. The total number of analyzed oocytes is 60.
Figure 3: Determining the behavior of endogenously labeled mcMTOCs during meiosis I.
(A) Diagram for generating Cep192-eGfp reporter mice. (B) Full-grown prophase I-arrested oocytes were collected from wild-type (WT) and Cep192-eGfp reporter mice, in vitro matured for 16 h (Met II) and assessed for meiotic progression. (C) Full-grown prophase I-arrested oocytes were collected from Cep192-eGfp reporter mice (CEP192 MTOCs are labeled green) and in vitro matured for 7 h. Metaphase I oocytes were fixed and immunostained using pericentrin antibody to label MTOCs (red). DAPI was used to detect DNA (blue). Arrowheads represent colocalized CEP192 with pericentrin at mcMTOCs. Shown are representative Z-projection images. Scale bars represent 10 μm. (D) Representative images (Z-projection of 13 sections every 5 μm) of time-lapse confocal microscopy of a live Cep192-eGfp oocyte incubated with SiR-tubulin from a time course (see Video S4). Fluorescence images (lower panels) were captured every 30 min post NEBD (time, h:min). Scale bars represent 20 μm (zoomed panels, 2 μm). (E) Representative images (Z-projection of 13 sections every 5 μm) of time-lapse confocal microscopy of a live Cep192-eGfp oocyte from a time course (see Video S4). Fluorescence and bright-field images (lower panels) were captured every 30 min post NEBD (time, h:min). Scale bars represent 10 μm. (F,G) Quantification of average MTOC number and MTOC volume over time in Cep192-eGfp oocyte during meiosis I, respectively. Error bars show S.D. Dashed blue lines represent the time of Anaphase I onset. (H). Quantification of average mcMTOC and polar MTOC volume during metaphase I (Met I). The data are expressed as mean ± SEM. Student t-test was used to analyze the data. Values with asterisks vary significantly, ****P < 0.0001.
mcMTOCs localize asymmetrically to anchor the spindle to the cortex
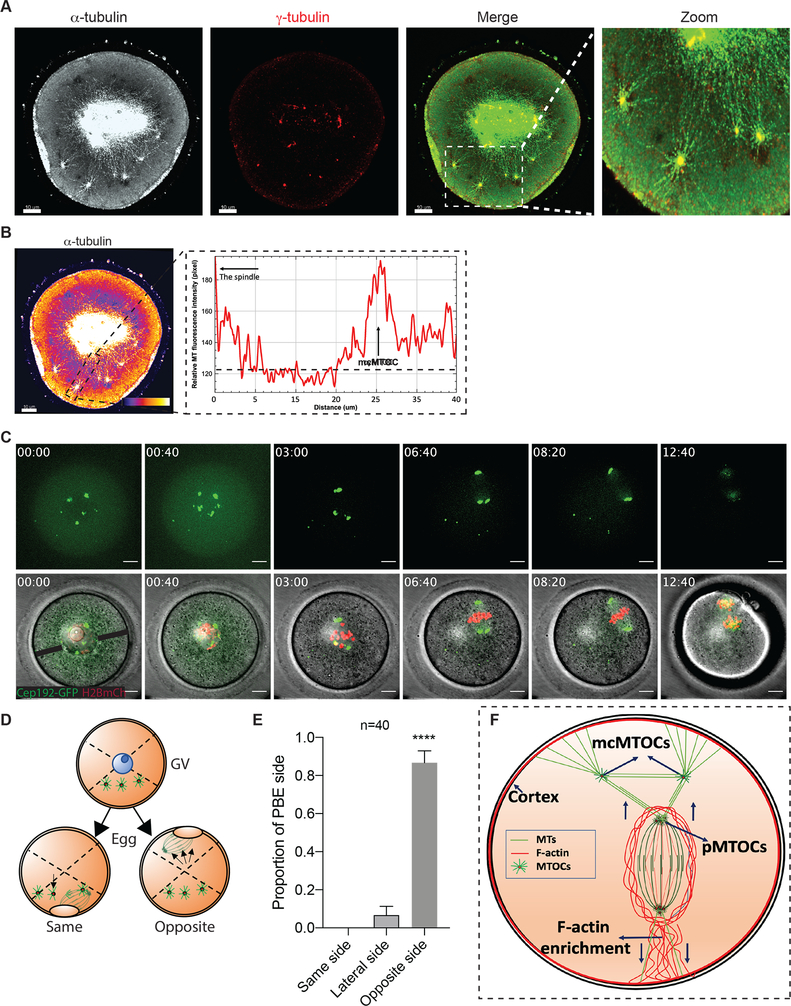
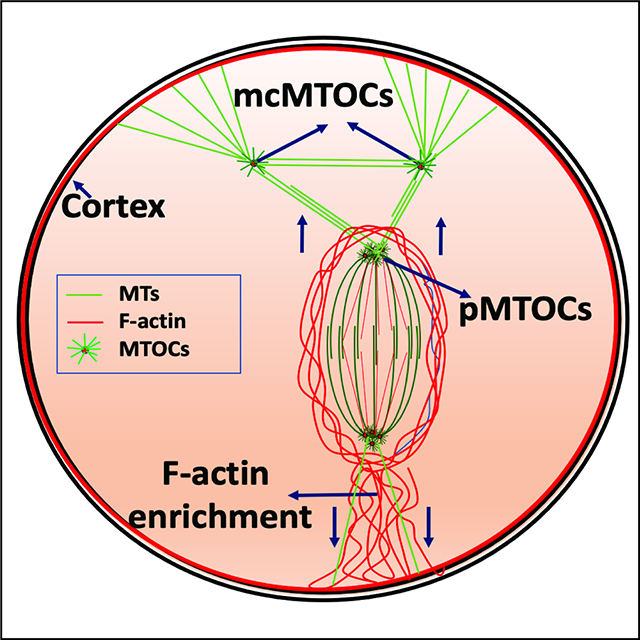
To determine whether the mcMTOCs were physically connected to pMTOCs, we employed immunocytochemistry and STED super-resolution microscopy to visualize the MTOCs in relation to MTs and F-actin. We confirmed that F-actin formed a cage around the spindle as previously reported (Azoury et al., 2008, Mogessie and Schuh, 2017) but could not detect any direct connection of F-actin between the pMTOCs, the mcMTOCs or the oocyte cortex (Supplementary Fig. 3A). In contrast, we detected MTs originating from the mcMTOCs and linking them to both the oocyte cortex and the pMTOCs and spindle (Fig. 4A,B; Supplementary Fig. 3B). In addition, we also observed MTs connecting mcMTOCs with each other. Relatively short astral-like MTs could not reach the oocyte cortex unless they bind mcMTOCs (Fig. 4A,B). Thus, mcMTOCs enable MTs to bridge the gap between the spindle and the cortex. It was also evident from our observations that the mcMTOCs were asymmetrically distributed in the oocyte cytoplasm leading us to consider their relationship in time and space to the positioning of the spindle itself. In prophase-arrested oocytes, the nucleus (Germinal vesicle, GV) is usually found in a centralized location (Brunet and Maro, 2007, Almonacid et al., 2015, Colin et al., 2020, Almonacid et al., 2019). Consequently, the spindle also forms around the chromatin at or near the center of the oocyte before it migrates towards the cortex during late Met I to allow asymmetrical cell division. In all examined oocytes, we found that the mcMTOCs were all asymmetrically positioned from the GV stage and throughout NEBD and early pro-Met I when the spindle is still localized centrally (Fig. 4C; Video S5). To date, F-actin is the sole cytoskeletal regulator for active spindle migration towards the PB cortical side (Li et al., 2008, Schuh and Ellenberg, 2008, Azoury et al., 2008). Strikingly, when the spindle began its actin-mediated migration, this took place towards the opposite side of the oocyte to that occupied by mcMTOCs in the majority of the oocytes (38 out of 40 examined, Fig. 4C–E; Video S5). These findings suggest a model whereby mcMTOCs localize asymmetrically to anchor the spindle to the cortex in such a way as to oppose the F-actin mediated force that builds to direct the spindle to the cortex on the opposite face of the oocyte for PB extrusion (Fig. 4F). This would account for the unexplained finding that nocodazole-induced MT depolymerization during MI results in the earlier migration of chromosomes towards the cortex at a relatively higher speed than in control oocytes (Supplementary Fig. 3C,D) and others (Li et al., 2008, Verlhac et al., 2000). Thus, the mcMTOCs appear to enable opposing MT forces that counter spindle/chromosome migration that are necessary to regulate its timing and the final position of the spindle.
Figure 4: mcMTOCs anchor the spindle to the cortex.
(A) Fully grown prophase-I-arrested oocytes were in vitro matured for 6 h (metaphase I) prior to fixation and immunocytochemistry using γ-tubulin and α-tubulin antibodies to label MTOCs (red) and microtubules (pseudo grey). Hoechst was used to detect DNA (blue). Fluorescence signals were detected under a 63X objective using STED super-resolution system. Shown is a representative image (Z-projection of 65 sections every 0.5 μm). (B) Example of fluorescence intensity of microtubules connecting pMTOCs, mcMTOCs and the cortex, using the ‘plot profiles’ function in ImageJ. The scale bar represents 10 μm. (C) Representative images (Z-projection of 16 sections every 3 μm) of time-lapse confocal microscopy of a live oocyte expressing AURKA-GFP (MTOCs) and H2B-mCherry (chromosomes) from a time course (see Video S5). Fluorescence and bright-field images (lower panels) were captured every 20 min (time, h:min). Arrowheads represent mcMTOCs. Scale bars represent 10 μm. (D) Schematic diagram shows how the proportion of polar body extrusion side in relation to mcMTOC position was assessed. (E) Quantification of the proportion of polar body extrusion side from “C” according to “D”. (F) Schematic model for spindle positioning in the meiotic oocyte. The data are expressed as mean ± SEM. Student t-test was used to analyze the data. Values with asterisks vary significantly, ****P < 0.0001. The total number of analyzed oocytes (from three independent replicates) is specified above the graph.
mcMTOCs regulate spindle positioning in acentriolar oocytes
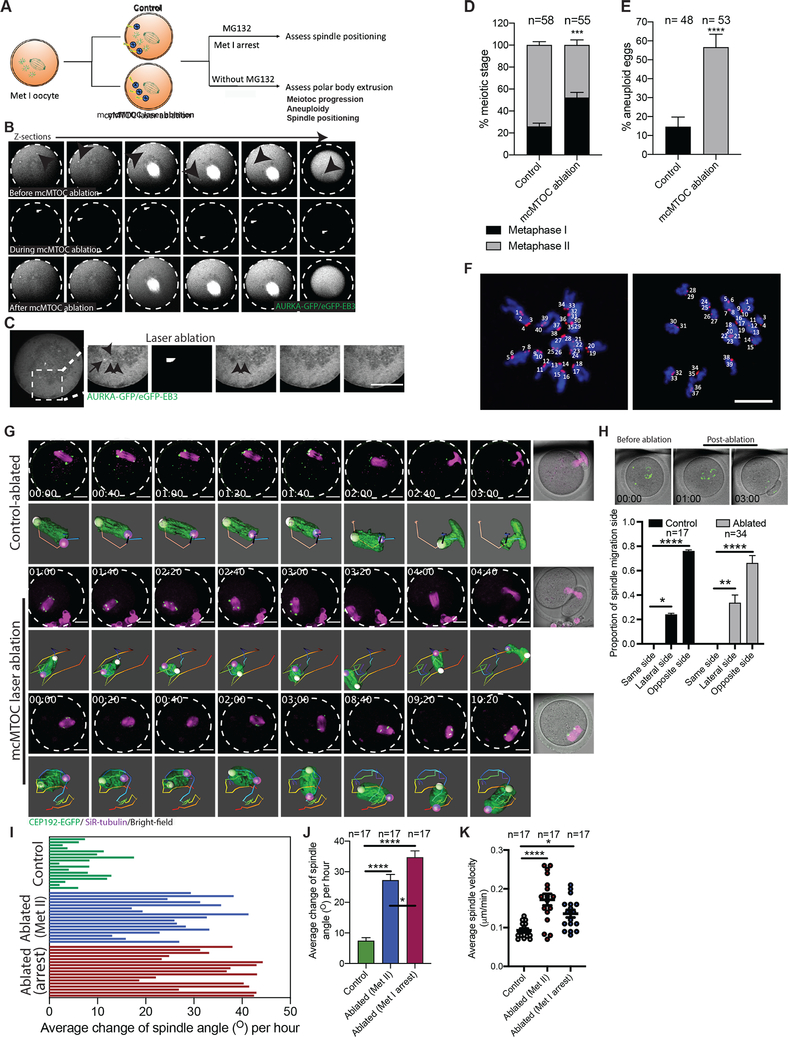
To determine the function(s) of mcMTOCs during MI, we selectively depleted mcMTOCs by two-photon laser ablation (Fig. 5A). The two-photon laser microscope has the advantage of offering deeper tissue penetration enabling efficient ablation and minimizing off-target effects (Denk et al., 1990). We first microinjected prophase I-arrested oocytes with cRNAs encoding Aurka-Gfp and eGFP-Eb3 to label MTOCs and MTs, respectively. We then marked small cuboidal regions surrounding each mcMTOC, which we then exposed to a 925 nm wavelength laser to ablate the mcMTOCs, (Fig. 5B; Video S6). We ensure reduction of fluorescence of each mcMTOC to background levels (compare images before ablation, Fig. 5B, upper panels, to after ablation, Fig. 5B, lower panels) before ablating the next. Importantly, laser ablation not only depleted the mcMTOCs but also disrupted their associated nucleation of MTs (Fig. 5C) without altering cytoplasmic F-actin (Supplementary Fig. 5F,G). We also exposed control oocytes to the same protocol by ablating random areas of the cytoplasm (same size and numbers of mcMTOCs) adjacent to but not overlapping with the mcMTOCs. We confirmed the efficiency of mcMTOC depletion by immunostaining a subset of oocytes to reveal γ-tubulin and were only able to detect γ-tubulin foci at spindle poles but not in the cytoplasm (Supplementary Fig. 4A). To assess spindle positioning, we arrested meiotic progression in Met I by incubating oocytes in meiotic maturation medium containing a proteasome inhibitor (MG-132). In this way, we could compare spindle positioning in such arrested oocytes for a longer time (Supplementary Fig. 4B). This revealed that whereas in control Met I-arrested oocytes, the spindle maintained its position over a period of 9 h (Supplementary Fig. 4B; Video S7), the position of the spindle in mcMTOC-depleted oocytes was not stable and displayed abnormal movement. Strikingly, the spindle in mcMTOC-depleted oocytes showed multiple migrations towards different cortical sides (Supplementary Fig. 4B; Video S7), likely due to the attempts of pMTOC-nucleated MTs to establish connections with the nearest cortical side. Consequently, the total distance traveled by the spindle and average spindle velocity (during 9 h time period) were significantly greater in mcMTOC-depleted oocytes than those in control oocytes (Supplementary Fig. 4C,D). Interestingly, after the spindle had lost its positioning, new mcMTOCs reappeared from the cortex, albeit smaller, in mcMTOC-depleted oocytes (Video S7). However, the newly emerged mcMTOCs were not able to rescue spindle positioning defects, likely due to the attempts of the spindle to establish new connections with the cortex. Following the loss of spindle positioning in mcMTOC-depleted oocytes, the spindle poles lost their integrity (Video S7). This secondary phenotype might be due to the imbalance of external (mcMTOC-mediated MTs) and internal (intra-spindle) forces on spindle poles.
Figure 5: mcMTOCs are required to regulate spindle positioning in meiotic oocytes.
(A) Schematic diagram shows the experimental design following mcMTOC depletion. (B,C) Oocytes expressing AURKA-GFP and eGFP-EB3 were in vitro maturated for 6 h (metaphase I, Met I), transferred to CZB medium with MG-132, followed by mcMTOC depletion using two-photon laser ablation. Small square area(s) surrounding mcMTOCs were marked and then exposed to a laser with 925 nm wavelength. Control oocytes were exposed to the same parameters except ablating random areas of the cytoplasm equal to the same size and number of mcMTOCs. (B) Z-sections of 3D time-lapse imaging of a live oocyte during mcMTOC ablation (see Video S6). Black arrowheads represent mcMTOCs before ablation (upper panels). White arrowheads represent laser beam targets (middle panels). (C) Z-sections of 3D time-lapse imaging of live oocyte to track microtubules (eGFP-EB3) following mcMTOC ablation. Black arrow represents an mcMTOC before ablation (−3 S). Black arrowheads track microtubules over time. The scale bar represents 20 μm. (D-J) Control and mcMTOC-depleted Cep192-eGfp oocytes were in vitro matured in MG-132-free medium until Met II (16 h) and assessed for meiotic progression (D) and aneuploidy (E). (F) Control-ablated and mcMTOC-ablated Met II oocytes were treated with monastrol followed by immunocytochemistry to label kinetochores with CREST anti-sera (red). DNA was stained with DAPI (blue). Kinetochore numbers were counted in each oocyte in serial confocal sections, and an aberration of 40 was scored as aneuploid. Shown are representative Z-projections. Scale bars represent 10 μm. (G) Representative Z-projection of the spindle from time-lapse confocal imaging (14 sections every 5 μm) of mcMTOC-ablated and control-ablated Cep192-eGfp oocytes (MTOCs are labeled green) in vitro matured in MG-132-free medium with SiR-tubulin (to label microtubules, magenta). Time-lapse imaging started 1 h post laser ablation. Scale bars represent 20 μm. (H) Quantification of the proportion of spindle migration side following laser ablation. (I) Quantification of the average change of spindle orientation angle per oocyte. Each bar represents an oocyte. (J) Quantification of the average change of spindle orientation angle per experimental group. (K) Quantification of average spindle velocity during migration until reaching the nearest point to the cortex. The data are expressed as mean ± SEM. Chi-square contingency test (D,E) and One-way ANOVA (H,J,K) was used to analyze the data. Values with asterisks vary significantly, *P < 0.05, ***P < 0.001, ****P < 0.0001. The total number of analyzed oocytes (from three independent replicates) is specified above each graph.
To assess the effect of mcMTOC depletion on meiotic progression and to further confirm the role of mcMTOCs in regulating spindle positioning, control and mcMTOC-depleted Met I oocytes were in vitro matured in MG-132-free medium. Depletion of mcMTOCs resulted in a significant increase in oocytes arrested at Met I compared to control oocytes (Fig. 5D). We previously implicated abnormal spindle positioning as a likely risk factor for aneuploidy (Kincade and Balboula, 2020). Using an in situ chromosome counting technique (Duncan et al., 2009, Balboula and Schindler, 2014), we examined the karyotype of mcMTOC-depleted oocytes that reached Met II (eggs). A relatively higher proportion of mcMTOC-depleted eggs were aneuploid compared to control-ablated oocytes (Fig. 5E,F). To assess spindle positioning, we tracked the spindle live in control-ablated and mcMTOC-ablated Cep192-eGfp oocytes using time-lapse confocal microscopy (Fig. 5G). Incubating the oocytes in MG-132-free medium entails mcMTOC depletion within a narrow time window (~3–4 h post-NEBD) to ensure that mcMTOC numbers reached their peak before mcMTOC ablation and that the spindle is still at the oocyte center before its migration. Oocytes with peripheral spindle positioning were excluded from spindle positioning assessment. In both control-ablated and mcMTOC-ablated oocytes, the spindle moved towards the cortex opposite the mcMTOC side (Fig. 5H). In contrast to control-ablated oocytes, in which the spindle migrated towards the cortex with a minimal change in spindle orientation, mcMTOC ablation was associated with spindle positioning and orientation defects. 3D reconstruction of the spindle and automatic tracking of the two spindle poles confirmed the increased incidence of abnormal spindle positioning/orientation in mcMTOC-depleted oocytes (both Met I-arrested oocytes or those extruded the PB), compared to control-ablated oocytes (Fig. 5G; Video S8). Accordingly, the average change of spindle orientation angle was significantly higher in mcMTOC-ablated oocytes when compared to control-ablated oocytes (Fig. 5I, J). Consistent with increased spindle orientation angle change, mcMTOC-depleted oocytes showed a significant increase in average spindle velocity during MI when compared to that in control-ablated oocytes (Fig. 5K). As a result of spindle erratic movement in mcMTOC-depleted oocytes, we observed cases in which the oocyte extruded the PB before the spindle had reached the cortex (Fig. 5G; Video S8), resulting in enlarged PBs (29.71±4.16% in mcMTOC-depleted oocytes vs. 0 % in control-ablated oocytes; P < 0.05). Again, the loss of spindle positioning in mcMTOC-depleted oocytes was associated with the loss of pMTOC integrity (Video S8) which likely contributes to increased Met I arrest and aneuploidy in mcMTOC-ablated oocytes. Together these findings suggest that the mcMTOCs are required to position the Met I spindle and play a critical role in regulating precise chromosome segregation.
Increasing mcMTOC numbers perturbs spindle positioning in meiotic oocytes
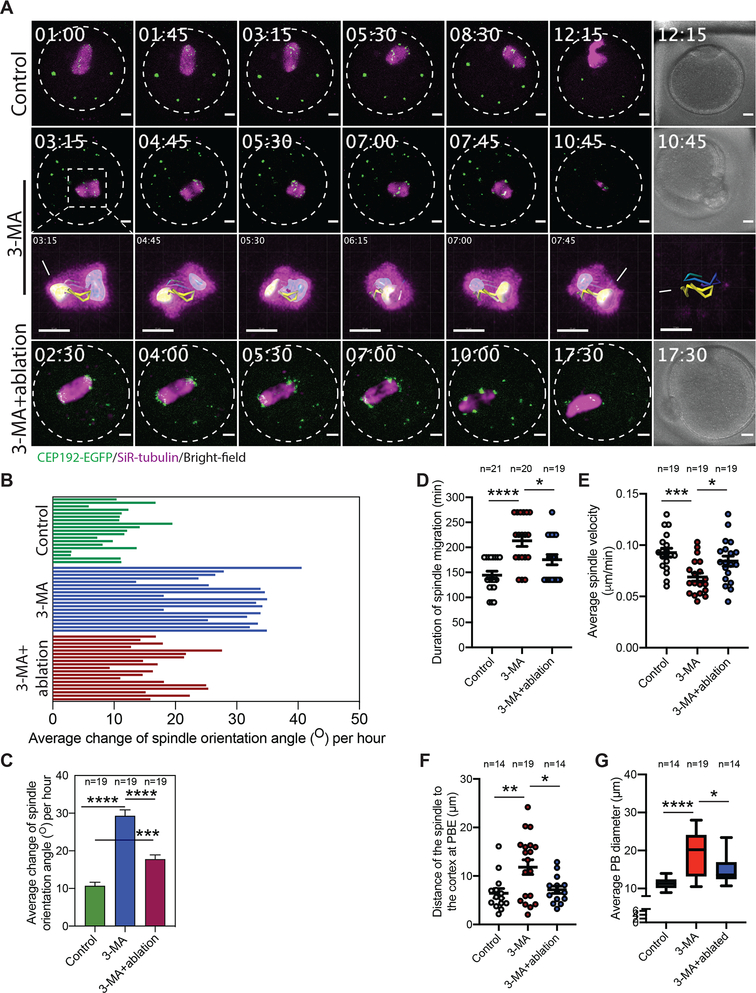
In mitotic cells, autophagy plays an important role in regulating and maintaining the proper number of centrosomes where autophagy-deficient cells contained multiple centrosomes (Watanabe et al., 2016, Honda and Shimizu, 2017). This led us to investigate the effects of inhibiting autophagy on mcMTOC numbers and the consequences for meiosis. To this end, we chose to treat oocytes with 3-Methyladenine (3-MA), which inhibits autophagy by blocking autophagosome formation via the inhibition of type III Phosphatidylinositol 3-kinases (PI-3K). We treated oocytes with 3-MA immediately after NEBD. 3-MA concentration was selected based on its efficiency to inhibit autophagy in mouse oocytes (Bang et al., 2014, Lin et al., 2018). We allowed control and 3-MA treated oocytes to mature for 5 h post-NEBD prior to fixation and immunostaining at Met I using anti-CEP192 and anti-α-tubulin antibodies to label MTOCs and the spindle, respectively. 3-MA treatment significantly increased (~ 2-fold increase) the number of mcMTOCs, but not pMTOCs, compared to control oocytes (Supplementary Fig. 5A–C). These mcMTOCs in 3-MA-treated oocytes are proficient to emanate astral-like MTs (Supplementary Fig. 5D). Interestingly, mcMTOC-nucleated MTs were significantly higher in 3-MA-treated oocytes, compared to control oocytes (Supplementary Fig. 5D,E). On the other hand, we did not observe any significant differences in cytoplasmic F-actin between control and 3-MA-treated oocytes (Supplementary Fig. 5F,G).
To determine the effect of increasing mcMTOC numbers by 3-MA treatment on spindle positioning, we employed time-lapse confocal microscopy to track spindle position over time. In contrast to control oocytes, increasing MTOC numbers following 3-MA treatment was associated with spindle positioning and orientation defects (Fig. 6A; Video S9). Indeed, 3D reconstruction of the spindle and automatic tracking of the two spindle poles showed abnormal spindle orientation and frequent spindle flipping (the phenotype that could not be observed in controls) during Met I in around 58% of 3-MA-treated oocytes (Fig. 6A; Video S9 and S10). During MI, the spindle migrates along its longitudinal axis towards the cortex with minimal change in spindle orientation. Consistent with abnormal spindle positioning/orientation phenotype, and in contrast to DMSO-treated controls (Fig. 6B,C; Video S9), 3-MA-treated oocytes exhibited a significant increase in the rate of spindle orientation change during Met I (Fig. 6B,C; Video S9). Moreover, in line with our model that mcMTOC-mediated MTs anchor the spindle to the cortex opposite the PB extrusion side to position the spindle centrally, we found that increasing mcMTOC numbers by 3-MA treatment resulted in a significant increase in the duration of spindle migration towards the cortex (Fig. 6D), resulting from their significantly (p < 0.001) reduced velocity (Fig. 6E) in comparison to control oocytes. Indeed, we frequently observed cases in which the chromosomes underwent segregation before the spindle had reached the cortex (Fig. 6F), resulting in enlarged PBs in around 21.05% of oocytes (Fig. 6G; Video S9).
Figure 6: mcMTOC numbers are critical for spindle positioning and timely spindle migration in meiotic oocytes.
(A) Z-projection (13 sections every 5 μm) of time-lapse confocal imaging of Cep192-eGfp oocytes (MTOCs are labeled green) in vitro matured with SiR-tubulin and DMSO (control), 3-MA or 3-MA+laser ablation (6 mcMTOCs were depleted, see Supplementary Fig. 6). Time-lapse imaging started 1 h post laser ablation. White arrows and asterisks indicate the two spindle poles. Scale bars represent 10 μm. (B) Quantification of the average change of spindle orientation angle per oocyte. Each bar represents an oocyte. (C) Quantification of the average change of spindle orientation angle per experimental group. (D) Quantification of the time spent by the spindle during its migration until reaching the nearest point to the cortex. (E) Quantification of average spindle velocity during migration until reaching the nearest point to the cortex. (F) Quantification of distance of the spindle to the nearest cortex at anaphase I. (G) Quantification of average polar body diameter. Scale bars represent 10 μm. The data are expressed as mean ± SEM. One-way ANOVA was used to analyze the data. Values with asterisks vary significantly, *P < 0.05, **P < 0.01, ***P < 0.001, ****P < 0.0001. The total number of analyzed oocytes (from three independent replicates) is specified above each graph.
To confirm that spindle positioning defects in 3-MA-treated oocytes are due to increased mcMTOC numbers, we asked whether reducing mcMTOC numbers in 3-MA-treated oocytes can rescue, at least in part, spindle positioning defects. Using laser ablation, we depleted the “extra” mcMTOCs to achieve physiological mcMTOC numbers (6 to 8) in 3-MA-treated oocytes (Supplementary Fig. 6). Strikingly, ablating the “extra” mcMTOCs in 3-MA-treated oocytes partially rescued spindle positioning defects as evidenced by the decrease of oocytes exhibiting spindle flipping phenotype (4 out of 19 examined oocytes) when compared to 3-MA-treated oocytes (11 out of 20 examined oocytes, Fig. 6A; Video S9). Indeed, the average change of spindle orientation angle during Met I was greatly reduced when some of these extra mcMTOCs were ablated in 3-MA-treated oocytes (Fig. 6B,C). Consistent with our model that mcMTOC-nucleated MTs anchor the spindle to the cortex opposite the PB extrusion side to position the spindle centrally, decreasing mcMTOC numbers in 3-MA-treated oocytes accelerated spindle migration towards the cortex and rescued the decrease in spindle velocity during migration (Fig. 6D,E). Importantly, decreasing mcMTOC numbers in 3-MA-treated oocytes by laser ablation greatly reduced the percentage of oocytes with enlarged PBs (5.26%), compared to non-ablated-3-MA oocytes (21.05%), resulting from the significant decrease in the distance between the spindle and the cortex at anaphase I (just before PB extrusion, Fig. 6F,G). Together, these data show that mcMTOC numbers must be regulated tightly during MI, and provide further evidence that mcMTOCs play an important role in regulating spindle positioning and its timely migration in mouse oocytes.
DISCUSSION
To date, the only known function of acentriolar MTOCs in mouse oocytes is to assemble the spindle. Using 3D time-lapse confocal microscopy, we characterize a subset of MTOCs (mcMTOCs) that remain free in the cytoplasm during Met I of meiosis and do not contribute to bipolar spindle assembly per se. The function of mcMTOCs in living mammalian oocytes is unknown. We demonstrated that mcMTOCs differ in several ways from the pMTOCs. First, the majority of pMTOCs, for example, originate from the perinuclear MTOCs, which never contribute to the mcMTOCs. Whereas the pMTOCs undergo a clustering-associated decrease in number and increase volume during pro-Met I/Met I, the mcMTOCs undergo a steady increase in both number and volume and rarely self-aggregate (~1.5% of all examined mcMTOCs). Second, inhibition of autophagy with 3-MA increased the number of mcMTOCs, but not pMTOCs. Finally, in contrast to pMTOCs, mcMTOCs participated in spindle positioning but never contributed directly to bipolar spindle assembly. Together, these observations suggest that mammalian oocytes have two different functional sets of MTOCs and raise the future important challenge to determine whether differences in their biochemical compositions underlie their differences in function.
The primary function of the spindle is to provide the machinery for faithful chromosome segregation. This is achieved in a series of critical, non-overlapping steps. First, during pro-Met I and early Met I, the spindle is assembled and positioned at or near the oocyte’s center. Second, during the late Met I, the spindle migrates towards a sub-cortical location to allow asymmetrical cell division. Third, the spindle rotates from a parallel to a perpendicular position in relation to the cortex to allow PB extrusion. Many studies have emphasized the roles of F-actin and its motor proteins in regulating spindle positioning and migration (Li et al., 2008, Azoury et al., 2008, Schuh and Ellenberg, 2008) and two models have been proposed to explain how F-actin regulates spindle positioning and migration. In the first, F-actin enrichment at the cortex provides a spindle pulling force (Schuh and Ellenberg, 2008, Azoury et al., 2008). In the second, a spindle pushing force is mediated by the cytoplasmic F-actin meshwork (Li et al., 2008). Both models enforce the notion that an F-actin-mediated force on the spindle, whether pushing or bulling, acts towards the nearest cortical side through which the PB is extruded. On the other hand, spindle orientation at the oocyte cortex seems to be dependent on both F-actin and MTs (Xie et al., 2018). Astral-like MTs are only able to reach the cortex when the spindle is in very close proximity to the cortex (Xie et al., 2018). Because astral-like MTs are relatively short and cannot easily reach the cortex, they can only establish contacts with MTs nucleated by mcMTOCs which, in turn, could act as amplifying sites that anchor the spindle to the cortex. This model, which we here propose, thus depends on the presence of two opposing forces: mcMTOC-mediated MTs at one side and F-actin at the other side of the spindle. These opposing forces would be essential to position the spindle centrally during pro-Met I and to prevent premature spindle migration, a possible risk factor for aneuploidy (Kincade and Balboula, 2020). Our model is consistent with three sets of observations: 1) mcMTOCs are exclusively localized asymmetrically, opposite the site of PB extrusion (the side of F-actin enrichment); 2) mcMTOCs undergo a significant decrease in number and volume during late Met I and Ana I/Telo I, allowing the F-actin mediated force to drive spindle migration and to extrude the PB; and 3) nocodazole-mediated MT depolymerization advances the timing of chromosome migration to the cortex, which takes place at a relatively higher speed (Supplementary Fig. 3B,C and others (Li et al., 2008, Verlhac et al., 2000) whereas increasing mcMTOC numbers delays chromosome migration to the cortex, which occurs at a relatively reduced speed compared to controls.
Depletion of mcMTOCs, disruption of mcMTOC numbers and MT depolymerization were each associated with abnormal spindle positioning and/or perturbed spindle migration. For example, increasing mcMTOC numbers by 3-MA treatment significantly decreased chromosome speed and delayed chromosome migration towards the cortex. This phenotype accords with increased mcMTOC-mediated MT forces that oppose F-actin; thereby preventing proper spindle migration. Such phenotype was rescued, at least in part, by reducing mcMTOC numbers to the physiological range (6 to 8). Conversely, depleting mcMTOCs using laser ablation or MT depolymerization has the reciprocal effect (Li et al., 2008, Verlhac et al., 2000). These findings indicate that the numbers of mcMTOCs must be regulated tightly to regulate spindle positioning and timely spindle migration.
In almost all mammals, including humans (George et al., 1996, Roeles and Tsiavaliaris, 2019), meiotic oocytes contain numerous acentriolar MTOCs. This is in contrast to somatic mitotic cells, which contain only a pair of centrosomes that are sufficient to assemble and position the spindle centrally. Positioning the spindle at the center of mitotic cells depends on nucleating symmetrical astral MTs that anchor the spindle to the cell cortex. However, the mechanism appears different during MI. Mammalian eggs are large and the astral MTs from acentriolar MTOCs are relatively short. Yet, the cell must divide asymmetrically, something that would likely be difficult for a pair of symmetrical centrosomes to achieve. Our proposed model may, therefore, account for why meiotic oocytes rely upon two different functional sets of numerous MTOCs rather than a pair of typical centrosomes.
Limitations of the study
Our data suggest that mcMTOCs differ from pMTOCs in several ways. For example, polar MTOCs originate from perinuclear MTOCs, cluster together, and contribute to spindle assembly, whereas mcMTOCs initially appear near the cortex, rarely self-aggregate and, never contribute directly to spindle assembly but regulate spindle positioning. This finding would be strengthened by determining whether biochemical composition differences between mcMTOCs and pMTOCs underpin such spatiotemporally and functionally divergent behaviors.
STAR METHODS
Resource availability
Lead contact
Further information and requests for resources and reagents should be directed to and will be fulfilled by the lead contact, Ahmed Z. Balboula (abalboula@missouri.edu).
Materials availability
All new materials generated in this study will be available upon request from Ahmed Z. Balboula (abalboula@missouri.edu).
Data and code availability
Data reported in this paper will be shared by the lead contact upon request.
This paper does not report original code.
Any additional information required to reanalyze the data reported in this paper is available from the lead contact upon request.
Experimental model and subject details
Mouse
All animals were kept and experiments were conducted in accordance with the University of Missouri (Animal Care Quality Assurance Ref. Number, 9695) and UK Home Office regulations. C57BL/6 and CF-1 mice were purchased from Envigo, Indianapolis, IN, USA. Cep192-eGfp reporter mice were generated by integrating a construct harboring the EGFP reporter gene into CF-1 mouse genome via CRISPR/Cas9-mediated homology-directed repair. The EGFP reporter was fused at the C-terminus of the endogenous mouse Cep192 (Fig. 3A). Homozygous breeding pairs of Cep192-eGfp reporter mice were used to maintain the colony. All animals were maintained in cages (up to 4 animals/cage) at 21 °C and 55% humidity in 12 h light/dark cycle with lights on from 7:00 am-7:00 pm and ad libitum access to food and water.
Method details
Cloning and in vitro cRNA synthesis
Generation of Aurka-Gfp, H2b-mCherry and eGfp-Eb3 were described previously (Shuda et al., 2009, Bury et al., 2017). DNA linearization of Aurka-Gfp and H2b-mCherry constructs was carried out using Nde I (New England BioLabs), whereas DNA linearization of eGfp-Eb3 construct was carried out using SfiI (New England BioLabs). Purification of linearized DNA was carried out according to the manufacturer’s protocol (Qiagen, QIAquick PCR Purification). Purified DNA was in vitro transcribed using an mMessage mMachine T7 kit (Ambion) to generate Aurka-Gfp and H2b-mCherry cRNAs or mMessage mMachine T3 kit (Ambion) to generate eGfp-Eb3 cRNA according to the manufacturer’s instructions. cRNA purification was performed using an RNAEasy kit (Qiagen) and stored at −80°C.
Oocyte collection, microinjection and culture
Full-grown GV-arrested oocytes were isolated from CF-1, C57BL/6 or Cep192-eGfp reporter CF-1 female mice (6–8-week-old) previously primed (~44 h before collection), with pregnant mare serum gonadotropin (Lee BioSolutions #493–10-10) according to (Schultz et al., 1983, Stein and Schindler, 2011). Unless otherwise specified, CF-1 mice were used to conduct the experiments. Cumulus oocyte complexes (COCs) were collected and denuded using mechanical pipetting in bicarbonate-free minimal essential medium (MEM) containing 3 mg/ml polyvinylpyrolidone (PVP) and 25 mM Hepes (pH 7.3) supplemented with 2.5 μM milrinone (MilliporeSigma, St. Louis, MO, USA # M4659), a phospho diesterase inhibitor to arrest the oocytes at prophase I (Tsafriri et al., 1996). Prophase I-arrested oocytes were microinjected with 10–15 pl of cRNAs encoding fluorescently labeled proteins while cultured in milrinone-containing MEM medium. Microinjected oocytes were then cultured in Chatot, Ziomek, and Bavister (CZB) medium (Chatot et al., 1989) supplemented with milrinone in a humidified incubator with 5% CO2 in air at 37°C for ~3 h to allow protein expression before releasing into milrinone-free CZB medium and initiating in vitro maturation. Met I, Ana I/Telo I or Met II oocytes were collected at 4, 7 or 14 h after NEBD.
Nocodazole (MilliporeSigma #M1404), MG-132 (MilliporeSigma #474790), and 3-Methyladenine (3-MA, Cayman Chemical, Ann Arbor, MI, USA #13242) were dissolved in dimethyl sulfoxide (DMSO) and used at a final concentration of 5 μM, 7.5 μM and 10 mM, respectively. In vitro maturation was carried out in organ culture dishes under humidified conditions (Becton Dickinson #353037).
Immunocytochemistry and fluorescence microscopy
Meiotic oocytes were fixed for 20 min at room temperature in freshly prepared 2 % paraformaldehyde solution (MilliporeSigma #P6148) dissolved in phosphate buffer saline (PBS). To preserve MTs, 0.05 % glutaraldehyde was added to the fixative solution (Akera et al., 2017, Dujardin et al., 1998). Fixed oocytes were permeabilized in 0.1% Triton X-100 in PBS for 20 min prior to incubation for an additional 20 min in PBS containing 0.3% BSA and 0.01% Tween-20 (blocking solution). Primary antibody incubation was performed at room temperature for 1 h. Oocytes were then washed three times (8–9 min each) prior to incubation with secondary antibodies for 1 h. Oocytes were washed again three times in blocking solutions for 8–9 min. To detect F-actin, phalloidin (Texas Red X Phalloidin, ThermoFisher Scientific #T7471; 1:50) was added to secondary antibody solutions. Oocytes were mounted on slides using Vectashieled with 4’,6-Diamidino-2-Phenylindole, Dihydrochloride (DAPI; Vector Laboratories, Burlingame, CA, USA) to stain DNA. To label DNA for STED super-resolution imaging, 5 mg/ml Hoechst 33342 was used (Molecular Probes H3570). The following primary antibodies were used in immunofluorescence: α-tubulin-Alexa Fluor 488 conjugate (Life Technologies #322 588; 1:100), CEP192 (AbFrontier #AR07-PA0001; 1:100), γ-tubulin (Millipore-Sigma #T6557; 1:75), Pericentrin (BD Biosciences #611814; 1:100), CREST autoimmune serum (Antibodies Incorporated #15–234; 1:25). Omitting the primary antibody served as a negative control. Fluorescence signals were detected under a 63X objective using Leica TCS SP8 confocal microscope equipped with 3-color, 3-D STED super-resolution 3X system. Images were captured to span the entire oocyte at 3 or 5 μm Z-intervals (confocal microscopy) or 0.5 μm Z-intervals (STED super-resolution microscopy). All images were acquired using the same laser power when the intensity of fluorescence is quantified.
Time-lapse confocal microscopy
Oocytes expressing fluorescently labeled proteins were transferred to milrinone-free CZB medium and imaged over time under a 63X objective using Leica TCS SP8 confocal microscope equipped with microenvironmental chamber to maintain the oocytes at controlled CO2 (5%) and temperature (37 °C) in a humidified air. DIC, GFP and mCherry image acquisitions were started at prophase I stage or as indicated and images were captured every certain time according to each experimental design (as indicated in corresponding figure legends). Images were captured to span the entire oocyte at 3 or 5 μm Z-intervals.
Depletion of mcMTOCs using laser ablation
Depletion of mcMTOCs was carried out using two-photon laser ablation which has the advantage of offering deeper tissue penetration, efficient ablation and minimizing off-target effects (Denk et al., 1990). Two different microscopes were used. Oocytes expressing fluorescently labeled proteins were transferred to milrinone-free MEM medium and mcMTOCs were ablated using a laser with 925 nm wavelength of upright LaVision BioTec TriM Scope II (with controlled temperature at 37 °C) or to milrinone-free CZB medium and mcMTOCs were ablated using a laser with 820 nm wavelength if Leica TCP SP8 two-photon inverted microscope (equipped with microenvironmantal chamber to control CO2 and temperature) was used. In both cases, a small square area(s) surrounding mcMTOCs were marked and then exposed to a laser beam. We compared the first image (before ablation) and the second image (after ablation) in the time series cycle to ensure that after ablation, the fluorescence in the targeted mcMTOCs decreased to that observed at the background levels. Next, we moved the focal plane and ablated the remaining mcMTOCs. McMTOC-depleted oocytes underwent live imaging using the same parameters. Control oocytes were exposed to the same protocol except ablating random areas of the cytoplasm, just adjacent and equal to the same size and number of mcMTOCs.
In situ chromosome counting
Oocytes at Met II stage (12 h post-NEBD) were treated with 100 μM monastrol (MilliporeSigma #M8515), an Eg5-kinesin inhibitor to induce monopolar spindle formation with subsequent chromosome dispersion (Duncan et al., 2009, Balboula and Schindler, 2014). Oocytes were fixed and immunostained as previously mentioned with CREST autoimmune serum to detect kinetochores. Oocytes were then mounted onto a glass slide using Vectashield with DAPI (Vector Laboratories) to label DNA. Confocal microscopy was used to image the entire region of the chromosomes at 0.7-μm Z-intervals to capture all kinetochores. Serial confocal sections were analyzed and the total number of kinetochores were counted using NIH image J software.
Quantification and statistical analysis
Image processing and analysis
Images acquired using 3-D STED super-resolution microscopy were deconvolved using Huygens Professional software before image analysis. NIH image J software (National Institute of Health, Bethesda, MD, USA) was used to process and analyze the images of fixed oocytes. The speed and average distance of chromosome/spindle/MTOCs at their final position over time were analyzed using the manual tracking function of NIH image J software. The point of intersection between the line connecting the two dominant spindle poles and spindle midzone was used to determine the position of the spindle. The point of intersection between the two lines representing the minor axis length and the major axis length of chromosomes was used to determine the position of all chromosomes (Bennabi et al., 2020). The speed and distance for each MTOC were analyzed separately before calculating the average for all MTOCs within each oocyte. 3D reconstruction of MTOCs, MTOC number and volume were processed and analyzed using isosurface spot analysis feature of Imaris software (Bitplane, Zürich, Switzerland) according to (Clift and Schuh, 2015). Briefly, based on MTOC signal to noise, the threshold value was adjusted on an oocyte-to-oocyte basis followed by MTOC surface segmentation. MTOC number was calculated using the spot analysis feature, whereas MTOC volume was analyzed using surface analysis feature. mcMTOCs were quantified after excluding the pMTOCs manually, and vice versa. Same processing parameters were applied for each experimental analysis. Spindle orientation was analyzed using Imaris measurement points. The two dominant spindle poles were determined automatically based on increased MT fluorescent intensity at spindle poles. A line connecting the two spindle poles defined the spindle axis at each time point. The angle between two successive spindle axes was computed to determine the spindle orientation angle.
Statistical analysis
One-way ANOVA, Student t-test and chi-square contingency test were used to evaluate the differences between groups using GraphPad Prism. ANOVA test was followed by the Tukey post hoc test to allow the comparison among groups. The differences of P < 0.05 were considered significant. The data were expressed as means ± SEM.
Key resources table
Supplementary Material
Video S1: Time-lapse confocal microscopy of MTOCs in live oocyte. Related to Figure 1. The full-grown prophase-I oocyte was injected with cRNAs encoding H2b-mCherry (red) and Aurka-Gfp (pseudo grey), followed by in vitro maturation. Fluorescence images (Z-projection of 16 sections every 3 μm) were captured every 20 min (time, h:min). Same oocyte as shown in Fig. 1A. The scale bar represents 10 μm.
Video S2: 3D reconstruction of MTOCs in metaphase I oocyte. Related to Figure 1.
Video S3: Tracking of mcMTOCs during oocyte meiosis I. Related to Figure 2. The full-grown prophase-I oocyte was injected with cRNAs encoding H2b-mCherry (red) and Aurka-Gfp (pseudo grey), followed by time-lapse confocal microscopy to track mcMTOCs during meiosis I. Fluorescence images (Z-projection of 16 sections every 3 μm) were captured every 15 min (time, h:min). The scale bar represents 20 μm.
Video S4: Tracking of mcMTOCs in Cep192-eGfp reporter oocyte during meiosis I. Related to Figure 3. Full-grown prophase I-arrested oocytes were collected from Cep192-eGfp reporter mice (MTOCs are labeled green) and in vitro matured in CZB medium. Images (Z-projection of 13 sections every 5 μm) were captured every 30 min (time post NEBD, h:min). The scale bar represents 20 μm.
Video S5: mcMTOCs localize asymmetrically opposite the direction of spindle migration. Related to Figure 4. The full-grown prophase-I oocyte was injected with cRNAs encoding H2b-mCherry (red) and Aurka-Gfp (pseudo grey), followed by in vitro maturation. Fluorescence and bright-field images (Z-projection of 16 sections every 3 μm) were captured every 20 min (time, h:min). Same oocyte as shown in Fig. 4C (lower panels). The scale bar represents 10 μm.
Video S6: Time-lapse imaging of live oocyte expressing AURKA-GFP (pseudo grey) and eGFP-EB3 (pseudo grey) during mcMTOC ablation. Related to Figure 5. The white squares represent the actual laser beam targets. Same oocyte as shown in Fig. 5B.
Video S7: Tracking the spindle over time in control-ablated and mcMTOC-ablated oocytes cultured in MG-132-containing medium. Related to Figure 5. Oocytes expressing AURKA-GFP (pseudo grey) and eGFP-EB3 (pseudo grey) were cultured for 9 h and imaged using 3D time-lapse microscopy. Fluorescence images were captured every 3 min (time post laser ablation, h:min). The scale bar represents 10 μm. Same oocyte as shown in Supplementary Fig. 4B.
Video S8: Tracking the spindle over time in control-ablated and mcMTOC-ablated oocytes cultured in MG-132-free medium. Related to Figure 5. Full-grown prophase I-arrested oocytes were collected from Cep192-eGfp reporter mice (MTOCs are labeled green) and in vitro matured in CZB medium containing SiR-tubulin (to label microtubules, magenta) until Met I. mcMTOCs were depleted using laser ablation prior to time-lapse confocal microscopy. Control oocytes were exposed to ablating multiple areas of the cytoplasm adjacent but not overlapping with mcMTOCs. Fluorescence and bright-field images (Z-projection of 18 sections every 3 μm) were captured every 30 min (time post laser ablation, h:min). Same oocytes as shown in Fig. 5G. The scale bar represents 25 μm.
Video S9: Tracking the spindle over time in DMSO-treated, 3-MA-treated and 3-MA-treated oocytes exposed to partial mcMTOC laser ablation during meiosis I. Related to Figure 6. Full-grown prophase I-arrested oocytes were collected from Cep192-eGfp reporter mice (MTOCs are labeled green) and in vitro matured in CZB medium containing SiR-tubulin (to label microtubules, magenta). Time-lapse imaging started at prometaphase I (3 h post NEBD). Fluorescence images (Z-projection of 13 sections every 5 μm) were captured every 45 min (time, h:min). Same oocytes as shown in Fig. 6A. The scale bar represents 10 μm.
Video S10: Tracking of 3D reconstructed spindle poles in 3-MA-treated oocyte. Related to Figure 6. Same oocyte as shown in Fig. 6A (middle panels).
Key resources table.
| REAGENT or RESOURCE | SOURCE | IDENTIFIER |
|---|---|---|
| Antibodies | ||
| α-tubulin-Alexa Fluor 488 conjugate | Life Technologies | Cat# 322588; RRID: AB_1502303 |
| Pericentrin | BD Biosciences | Cat# 611814; RRID: AB_399294 |
| CREST autoimmune serum | Antibodies Incorporated | Cat# 15–234; RRID: AB_2687472 |
| γ-tubulin | Millipore-Sigma | Cat# T6557; RRID: AB_477584 |
| CEP192 | AbFrontier | Cat# AR07-PA0001 |
| Chemicals, peptides, and recombinant proteins | ||
| Nocodazole | MilliporeSigma | Cat# M1404 |
| MG-132 | MilliporeSigma | Cat# 474790 |
| 3-Methyladenine | Cayman Chemical | Cat# 13242 |
| Paraformaldehyde | MilliporeSigma | Cat# P6148 |
| Monastrol | MilliporeSigma | Cat# M8515 |
| Pregnant mare serum gonadotropin | Lee BioSolutions | Cat# 493–10–10 |
| Milrinone | MilliporeSigma | Cat# M4659 |
| Phalloidin | ThermoFisher Scientific | Cat# T7471 |
| Hoechst 33342 | Molecular Probes | Cat# H3570 |
| SiR-tubulin | Cytoskeleton | NC0958386 |
| Critical commercial assays | ||
| mMESSAGE mMACHINE kit | Invitrogen (Ambion) | Cat#AM1340, AM1344&AM1348 |
| RNeasy mini kit | Qiagen | Cat# 74104 |
| Experimental models: Organisms/strains | ||
| Mouse: C57BL/6 | Envigo | NHsd |
| Mouse: CF-1 | Envigo | Hsd:NSA |
| Mouse: Cep192-eGfp reporter CF-1 | This paper | N/A |
| Recombinant DNA | ||
| pIVT Aurka-Gfp | Shuda et al., 2009 | N/A |
| pIVT H2b-mCherry | Shuda et al., 2009 | N/A |
| RN3P eGfp-Eb3 | Bury et al., 2017 | N/A |
| Software and algorithms | ||
| Image J | NIH | N/A |
| Graphpad Prism | Graphpad Software | N/A |
| Imaris | Oxford instruments | N/A |
| Huygens Software | Scientific Volume Imaging | N/A |
Highlights.
Two different sets of MTOCs are present in acentriolar mouse oocytes
mcMTOCs exhibit three different directional behaviors during meiosis I
mcMTOC-nucleated microtubules anchor the spindle to the oocyte cortex
mcMTOCs are required to regulate spindle positioning and timely spindle migration
ACKNOWLEDGMENTS
The authors would like to thank Dr. David Glover, California Institute of Technology, USA for critical reading and editing the manuscript, and for providing resources. The authors would like to thank all members of the Glover lab and the Balboula lab for valuable help and discussions. This study was supported by laboratory start-up funding from the University of Missouri, Marie Sklodowska-Curie Fellowship 706170 (Horizon 2020, European Commission) and the R35 GM142537 (NIGMS, NIH) to A.Z.B.
Footnotes
DECLARATION OF INTEREST
The authors declare no competing interests.
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
REFERENCES
- AKERA T, CHMATAL L, TRIMM E, YANG K, AONBANGKHEN C, CHENOWETH DM, JANKE C, SCHULTZ RM & LAMPSON MA 2017. Spindle asymmetry drives non-Mendelian chromosome segregation. Science, 358, 668–672. [DOI] [PMC free article] [PubMed] [Google Scholar]
- ALMONACID M, AHMED WW, BUSSONNIER M, MAILLY P, BETZ T, VOITURIEZ R, GOV NS & VERLHAC MH 2015. Active diffusion positions the nucleus in mouse oocytes. Nat Cell Biol, 17, 470–9. [DOI] [PubMed] [Google Scholar]
- ALMONACID M, AL JORD A, EL-HAYEK S, OTHMANI A, COULPIER F, LEMOINE S, MIYAMOTO K, GROSSE R, KLEIN C, PIOLOT T, MAILLY P, VOITURIEZ R, GENOVESIO A & VERLHAC MH 2019. Active Fluctuations of the Nuclear Envelope Shape the Transcriptional Dynamics in Oocytes. Dev Cell, 51, 145–157 e10. [DOI] [PubMed] [Google Scholar]
- AZOURY J, LEE KW, GEORGET V, RASSINIER P, LEADER B & VERLHAC MH 2008. Spindle positioning in mouse oocytes relies on a dynamic meshwork of actin filaments. Curr Biol, 18, 1514–9. [DOI] [PubMed] [Google Scholar]
- BALBOULA AZ, NGUYEN AL, GENTILELLO AS, QUARTUCCIO SM, DRUTOVIC D, SOLC P & SCHINDLER K 2016. Haspin kinase regulates microtubule-organizing center clustering and stability through Aurora kinase C in mouse oocytes. J Cell Sci, 129, 3648–3660. [DOI] [PMC free article] [PubMed] [Google Scholar]
- BALBOULA AZ & SCHINDLER K 2014. Selective disruption of aurora C kinase reveals distinct functions from aurora B kinase during meiosis in mouse oocytes. PLoS Genet, 10, e1004194. [DOI] [PMC free article] [PubMed] [Google Scholar]
- BANG S, SHIN H, SONG H, SUH CS & LIM HJ 2014. Autophagic activation in vitrified-warmed mouse oocytes. Reproduction, 148, 11–9. [DOI] [PubMed] [Google Scholar]
- BAUMANN C, WANG X, YANG L & VIVEIROS MM 2017. Error-prone meiotic division and subfertility in mice with oocyte-conditional knockdown of pericentrin. J Cell Sci, 130, 1251–1262. [DOI] [PMC free article] [PubMed] [Google Scholar]
- BENNABI I, CROZET F, NIKALAYEVICH E, CHAIGNE A, LETORT G, MANIL-SEGALEN M, CAMPILLO C, CADART C, OTHMANI A, ATTIA R, GENOVESIO A, VERLHAC MH & TERRET ME 2020. Artificially decreasing cortical tension generates aneuploidy in mouse oocytes. Nat Commun, 11, 1649. [DOI] [PMC free article] [PubMed] [Google Scholar]
- BENNABI I, TERRET ME & VERLHAC MH 2016. Meiotic spindle assembly and chromosome segregation in oocytes. J Cell Biol, 215, 611–619. [DOI] [PMC free article] [PubMed] [Google Scholar]
- BREUER M, KOLANO A, KWON M, LI CC, TSAI TF, PELLMAN D, BRUNET S & VERLHAC MH 2010. HURP permits MTOC sorting for robust meiotic spindle bipolarity, similar to extra centrosome clustering in cancer cells. J Cell Biol, 191, 1251–60. [DOI] [PMC free article] [PubMed] [Google Scholar]
- BRUNET S & MARO B 2007. Germinal vesicle position and meiotic maturation in mouse oocyte. Reproduction, 133, 1069–72. [DOI] [PubMed] [Google Scholar]
- BURY L, COELHO PA, SIMEONE A, FERRIES S, EYERS CE, EYERS PA, ZERNICKA-GOETZ M & GLOVER DM 2017. Plk4 and Aurora A cooperate in the initiation of acentriolar spindle assembly in mammalian oocytes. J Cell Biol, 216, 3571–3590. [DOI] [PMC free article] [PubMed] [Google Scholar]
- CHATOT CL, ZIOMEK CA, BAVISTER BD, LEWIS JL & TORRES I 1989. An improved culture medium supports development of random-bred 1-cell mouse embryos in vitro. J Reprod Fertil, 86, 679–88. [DOI] [PubMed] [Google Scholar]
- CLIFT D & SCHUH M 2015. A three-step MTOC fragmentation mechanism facilitates bipolar spindle assembly in mouse oocytes. Nat Commun, 6, 7217. [DOI] [PMC free article] [PubMed] [Google Scholar]
- COLIN A, LETORT G, RAZIN N, ALMONACID M, AHMED W, BETZ T, TERRET ME, GOV NS, VOITURIEZ R, GUEROUI Z & VERLHAC MH 2020. Active diffusion in oocytes nonspecifically centers large objects during prophase I and meiosis I. J Cell Biol, 219. [DOI] [PMC free article] [PubMed] [Google Scholar]
- DENK W, STRICKLER JH & WEBB WW 1990. Two-photon laser scanning fluorescence microscopy. Science, 248, 73–6. [DOI] [PubMed] [Google Scholar]
- DUJARDIN D, WACKER UI, MOREAU A, SCHROER TA, RICKARD JE & DE MEY JR 1998. Evidence for a role of CLIP-170 in the establishment of metaphase chromosome alignment. J Cell Biol, 141, 849–62. [DOI] [PMC free article] [PubMed] [Google Scholar]
- DUNCAN FE, CHIANG T, SCHULTZ RM & LAMPSON MA 2009. Evidence that a defective spindle assembly checkpoint is not the primary cause of maternal age-associated aneuploidy in mouse eggs. Biol Reprod, 81, 768–76. [DOI] [PMC free article] [PubMed] [Google Scholar]
- GEORGE MA, PICKERING SJ, BRAUDE PR & JOHNSON MH 1996. The distribution of alpha- and gamma-tubulin in fresh and aged human and mouse oocytes exposed to cryoprotectant. Mol Hum Reprod, 2, 445–56. [DOI] [PubMed] [Google Scholar]
- GUETH-HALLONET C, ANTONY C, AGHION J, SANTA-MARIA A, LAJOIE-MAZENC I, WRIGHT M & MARO B 1993. gamma-Tubulin is present in acentriolar MTOCs during early mouse development. J Cell Sci, 105 ( Pt 1), 157–66. [DOI] [PubMed] [Google Scholar]
- HASHIMOTO N & KISHIMOTO T 1988. Regulation of meiotic metaphase by a cytoplasmic maturation-promoting factor during mouse oocyte maturation. Dev Biol, 126, 242–52. [DOI] [PubMed] [Google Scholar]
- HONDA S & SHIMIZU S 2017. Autophagy controls centrosome number. Oncotarget, 8, 14277–14278. [DOI] [PMC free article] [PubMed] [Google Scholar]
- HOULISTON E, PICKERING SJ & MARO B 1987. Redistribution of microtubules and pericentriolar material during the development of polarity in mouse blastomeres. J Cell Biol, 104, 1299–308. [DOI] [PMC free article] [PubMed] [Google Scholar]
- KINCADE JN & BALBOULA AZ 2020. Initial spindle formation at the oocyte center protects against incorrect kinetochore-microtubule attachment and aneuploidy in mice. bioRxiv, 2020.06.28.176594. [DOI] [PMC free article] [PubMed] [Google Scholar]
- KITAJIMA TS, OHSUGI M & ELLENBERG J 2011. Complete kinetochore tracking reveals error-prone homologous chromosome biorientation in mammalian oocytes. Cell, 146, 568–81. [DOI] [PubMed] [Google Scholar]
- LI H, GUO F, RUBINSTEIN B & LI R 2008. Actin-driven chromosomal motility leads to symmetry breaking in mammalian meiotic oocytes. Nat Cell Biol, 10, 1301–8. [DOI] [PubMed] [Google Scholar]
- LIN FH, ZHANG WL, LI H, TIAN XD, ZHANG J, LI X, LI CY & TAN JH 2018. Role of autophagy in modulating post-maturation aging of mouse oocytes. Cell Death Dis, 9, 308. [DOI] [PMC free article] [PubMed] [Google Scholar]
- LONGO FJ & CHEN DY 1985. Development of cortical polarity in mouse eggs: involvement of the meiotic apparatus. Dev Biol, 107, 382–94. [DOI] [PubMed] [Google Scholar]
- LUKSZA M, QUEGUIGNER I, VERLHAC MH & BRUNET S 2013. Rebuilding MTOCs upon centriole loss during mouse oogenesis. Dev Biol, 382, 48–56. [DOI] [PubMed] [Google Scholar]
- MA J, ZENG F, SCHULTZ RM & TSENG H 2006. Basonuclin: a novel mammalian maternal-effect gene. Development, 133, 2053–62. [DOI] [PubMed] [Google Scholar]
- MA W & VIVEIROS MM 2014. Depletion of pericentrin in mouse oocytes disrupts microtubule organizing center function and meiotic spindle organization. Mol Reprod Dev, 81, 1019–29. [DOI] [PMC free article] [PubMed] [Google Scholar]
- MARO B, HOULISTON E & PAINTRAND M 1988. Purification of meiotic spindles and cytoplasmic asters from mouse oocytes. Dev Biol, 129, 275–82. [DOI] [PubMed] [Google Scholar]
- MCCARTHY EK & GOLDSTEIN B 2006. Asymmetric spindle positioning. Curr Opin Cell Biol, 18, 79–85. [DOI] [PMC free article] [PubMed] [Google Scholar]
- MCNALLY FJ 2013. Mechanisms of spindle positioning. J Cell Biol, 200, 131–40. [DOI] [PMC free article] [PubMed] [Google Scholar]
- MENEZO YJ 2006. Paternal and maternal factors in preimplantation embryogenesis: interaction with the biochemical environment. Reprod Biomed Online, 12, 616–21. [DOI] [PubMed] [Google Scholar]
- MENG XQ, FAN HY, ZHONG ZS, ZHANG G, LI YL, CHEN DY & SUN QY 2004. Localization of gamma-tubulin in mouse eggs during meiotic maturation, fertilization, and early embryonic development. J Reprod Dev, 50, 97–105. [DOI] [PubMed] [Google Scholar]
- MESSINGER SM & ALBERTINI DF 1991. Centrosome and microtubule dynamics during meiotic progression in the mouse oocyte. J Cell Sci, 100 ( Pt 2), 289–98. [DOI] [PubMed] [Google Scholar]
- MOGESSIE B & SCHUH M 2017. Actin protects mammalian eggs against chromosome segregation errors. Science, 357. [DOI] [PubMed] [Google Scholar]
- ROELES J & TSIAVALIARIS G 2019. Actin-microtubule interplay coordinates spindle assembly in human oocytes. Nat Commun, 10, 4651. [DOI] [PMC free article] [PubMed] [Google Scholar]
- SASKOVA A, SOLC P, BARAN V, KUBELKA M, SCHULTZ RM & MOTLIK J 2008. Aurora kinase A controls meiosis I progression in mouse oocytes. Cell Cycle, 7, 2368–76. [DOI] [PMC free article] [PubMed] [Google Scholar]
- SCHATTEN H, SCHATTEN G, MAZIA D, BALCZON R & SIMERLY C 1986. Behavior of centrosomes during fertilization and cell division in mouse oocytes and in sea urchin eggs. Proc Natl Acad Sci U S A, 83, 105–9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- SCHUH M & ELLENBERG J 2007. Self-organization of MTOCs replaces centrosome function during acentrosomal spindle assembly in live mouse oocytes. Cell, 130, 484–98. [DOI] [PubMed] [Google Scholar]
- SCHUH M & ELLENBERG J 2008. A new model for asymmetric spindle positioning in mouse oocytes. Curr Biol, 18, 1986–92. [DOI] [PubMed] [Google Scholar]
- SCHULTZ RM, MONTGOMERY RR & BELANOFF JR 1983. Regulation of mouse oocyte meiotic maturation: implication of a decrease in oocyte cAMP and protein dephosphorylation in commitment to resume meiosis. Dev Biol, 97, 264–73. [DOI] [PubMed] [Google Scholar]
- SHUDA K, SCHINDLER K, MA J, SCHULTZ RM & DONOVAN PJ 2009. Aurora kinase B modulates chromosome alignment in mouse oocytes. Mol Reprod Dev, 76, 1094–105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- SO C, SERES KB, STEYER AM, MONNICH E, CLIFT D, PEJKOVSKA A, MOBIUS W & SCHUH M 2019. A liquid-like spindle domain promotes acentrosomal spindle assembly in mammalian oocytes. Science, 364. [DOI] [PMC free article] [PubMed] [Google Scholar]
- SOLC P, BARAN V, MAYER A, BOHMOVA T, PANENKOVA-HAVLOVA G, SASKOVA A, SCHULTZ RM & MOTLIK J 2012. Aurora kinase A drives MTOC biogenesis but does not trigger resumption of meiosis in mouse oocytes matured in vivo. Biol Reprod, 87, 85. [DOI] [PMC free article] [PubMed] [Google Scholar]
- STEIN P & SCHINDLER K 2011. Mouse oocyte microinjection, maturation and ploidy assessment. J Vis Exp. [DOI] [PMC free article] [PubMed] [Google Scholar]
- SZOLLOSI D, CALARCO P & DONAHUE RP 1972. Absence of centrioles in the first and second meiotic spindles of mouse oocytes. J Cell Sci, 11, 521–41. [DOI] [PubMed] [Google Scholar]
- TSAFRIRI A, CHUN SY, ZHANG R, HSUEH AJ & CONTI M 1996. Oocyte maturation involves compartmentalization and opposing changes of cAMP levels in follicular somatic and germ cells: studies using selective phosphodiesterase inhibitors. Dev Biol, 178, 393–402. [DOI] [PubMed] [Google Scholar]
- VERLHAC MH, LEFEBVRE C, GUILLAUD P, RASSINIER P & MARO B 2000. Asymmetric division in mouse oocytes: with or without Mos. Curr Biol, 10, 1303–6. [DOI] [PubMed] [Google Scholar]
- VON STETINA JR & ORR-WEAVER TL 2011. Developmental control of oocyte maturation and egg activation in metazoan models. Cold Spring Harb Perspect Biol, 3, a005553. [DOI] [PMC free article] [PubMed] [Google Scholar]
- WATANABE Y, HONDA S, KONISHI A, ARAKAWA S, MUROHASHI M, YAMAGUCHI H, TORII S, TANABE M, TANAKA S, WARABI E & SHIMIZU S 2016. Autophagy controls centrosome number by degrading Cep63. Nat Commun, 7, 13508. [DOI] [PMC free article] [PubMed] [Google Scholar]
- XIE B, ZHANG L, ZHAO H, BAI Q, FAN Y, ZHU X, YU Y, LI R, LIANG X, SUN QY, LI M & QIAO J 2018. Poly(ADP-ribose) mediates asymmetric division of mouse oocyte. Cell Res, 28, 462–475. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Video S1: Time-lapse confocal microscopy of MTOCs in live oocyte. Related to Figure 1. The full-grown prophase-I oocyte was injected with cRNAs encoding H2b-mCherry (red) and Aurka-Gfp (pseudo grey), followed by in vitro maturation. Fluorescence images (Z-projection of 16 sections every 3 μm) were captured every 20 min (time, h:min). Same oocyte as shown in Fig. 1A. The scale bar represents 10 μm.
Video S2: 3D reconstruction of MTOCs in metaphase I oocyte. Related to Figure 1.
Video S3: Tracking of mcMTOCs during oocyte meiosis I. Related to Figure 2. The full-grown prophase-I oocyte was injected with cRNAs encoding H2b-mCherry (red) and Aurka-Gfp (pseudo grey), followed by time-lapse confocal microscopy to track mcMTOCs during meiosis I. Fluorescence images (Z-projection of 16 sections every 3 μm) were captured every 15 min (time, h:min). The scale bar represents 20 μm.
Video S4: Tracking of mcMTOCs in Cep192-eGfp reporter oocyte during meiosis I. Related to Figure 3. Full-grown prophase I-arrested oocytes were collected from Cep192-eGfp reporter mice (MTOCs are labeled green) and in vitro matured in CZB medium. Images (Z-projection of 13 sections every 5 μm) were captured every 30 min (time post NEBD, h:min). The scale bar represents 20 μm.
Video S5: mcMTOCs localize asymmetrically opposite the direction of spindle migration. Related to Figure 4. The full-grown prophase-I oocyte was injected with cRNAs encoding H2b-mCherry (red) and Aurka-Gfp (pseudo grey), followed by in vitro maturation. Fluorescence and bright-field images (Z-projection of 16 sections every 3 μm) were captured every 20 min (time, h:min). Same oocyte as shown in Fig. 4C (lower panels). The scale bar represents 10 μm.
Video S6: Time-lapse imaging of live oocyte expressing AURKA-GFP (pseudo grey) and eGFP-EB3 (pseudo grey) during mcMTOC ablation. Related to Figure 5. The white squares represent the actual laser beam targets. Same oocyte as shown in Fig. 5B.
Video S7: Tracking the spindle over time in control-ablated and mcMTOC-ablated oocytes cultured in MG-132-containing medium. Related to Figure 5. Oocytes expressing AURKA-GFP (pseudo grey) and eGFP-EB3 (pseudo grey) were cultured for 9 h and imaged using 3D time-lapse microscopy. Fluorescence images were captured every 3 min (time post laser ablation, h:min). The scale bar represents 10 μm. Same oocyte as shown in Supplementary Fig. 4B.
Video S8: Tracking the spindle over time in control-ablated and mcMTOC-ablated oocytes cultured in MG-132-free medium. Related to Figure 5. Full-grown prophase I-arrested oocytes were collected from Cep192-eGfp reporter mice (MTOCs are labeled green) and in vitro matured in CZB medium containing SiR-tubulin (to label microtubules, magenta) until Met I. mcMTOCs were depleted using laser ablation prior to time-lapse confocal microscopy. Control oocytes were exposed to ablating multiple areas of the cytoplasm adjacent but not overlapping with mcMTOCs. Fluorescence and bright-field images (Z-projection of 18 sections every 3 μm) were captured every 30 min (time post laser ablation, h:min). Same oocytes as shown in Fig. 5G. The scale bar represents 25 μm.
Video S9: Tracking the spindle over time in DMSO-treated, 3-MA-treated and 3-MA-treated oocytes exposed to partial mcMTOC laser ablation during meiosis I. Related to Figure 6. Full-grown prophase I-arrested oocytes were collected from Cep192-eGfp reporter mice (MTOCs are labeled green) and in vitro matured in CZB medium containing SiR-tubulin (to label microtubules, magenta). Time-lapse imaging started at prometaphase I (3 h post NEBD). Fluorescence images (Z-projection of 13 sections every 5 μm) were captured every 45 min (time, h:min). Same oocytes as shown in Fig. 6A. The scale bar represents 10 μm.
Video S10: Tracking of 3D reconstructed spindle poles in 3-MA-treated oocyte. Related to Figure 6. Same oocyte as shown in Fig. 6A (middle panels).
Data Availability Statement
Data reported in this paper will be shared by the lead contact upon request.
This paper does not report original code.
Any additional information required to reanalyze the data reported in this paper is available from the lead contact upon request.