Summary
Ferroptosis is an emerging cancer suppression strategy. However, how to select cancer patients for treating with ferroptosis-inducers remains challenging. Here, we develop photochemical activation of membrane lipid peroxidation (PALP) that uses targeted lasers to induce localized polyunsaturated fatty acyl (PUFA)-lipid peroxidation for reporting ferroptosis sensitivity in cells and tissues. PALP captured by BODIPY-C11 can be suppressed by lipophilic antioxidants and iron chelation, and is dependent on PUFA-lipid levels. Moreover, we develop PALPv2, for studying lipid peroxidation on selected membranes along the z-axis in live cells using two-photon microscopes. Using PALPv1, we detect PUFA-lipids in multiple tissues, and validate a PUFA-phospholipid reduction during muscle aging as previously reported. Patterns of PALPv1 signals across multiple cancer cell types in vitro and in vivo are concordant with their ferroptosis susceptibility and PUFA-phospholipid levels. We envision that PALP will enable rapid stratification of ferroptosis sensitivity in cancer patients and facilitate PUFA-lipid research.
eTOC Blurb
Selecting the right cancer patients for ferroptosis–inducing therapies remains difficult. Wang et al. have developed an imaging technique, photochemical activation of membrane lipid peroxidation (PALP) that utilizes the propensity of polyunsatura ted phospholipids to undergo laser–induced peroxidation to estimate ferroptosis susceptibility in live cells and tissues in situ.
Graphical Abstract
Introduction
Ferroptosis, triggered by aberrant polyunsaturated phospholipid peroxidation, contributes to the development of various diseases including neurodegeneration, ischemia/reperfusion-induced damages in the liver, kidney, heart and brain, and hemolysis (Zou and Schreiber, 2020). Vulnerability to ferroptosis induction is also a common feature of various cancers including clear-cell carcinomas from the kidney and ovary, pancreatic cancer, diffuse large B-cell lymphoma, hepatocellular carcinoma, colorectal cancer, and therapy resistant cancer cells (Badgley et al., 2020; Hangauer et al., 2017; Singhal et al., 2021; Sun et al., 2016; Viswanathan et al., 2017; Yang et al., 2014; Zou et al., 2019). Hence, inducing polyunsaturated phospholipid peroxidation and ferroptosis in human tumor cells has emerged as a promising strategy for cancer therapy (Jiang et al., 2021) This concept is supported by the tumor suppressive effects of ferroptosis-inducing agents including imidazole ketone erastin (IKE) and its combination with other targeted therapies (Yi et al., 2020; Zhang et al., 2019), recombinant cysteinase (Badgley et al., 2020), and direct administration of polyunsaturated fatty acids including arachidonic acid, conjugated linolenic acid (Beatty et al., 2021) and dihomo-γ-linolenic acid (Perez et al., 2020). More potent and more specific ferroptosis inducers for in vivo use will likely emerge from extensive ongoing investigations (Eaton et al., 2020). However, rapidly stratifying cancer patients for their likelihood to respond to ferroptosis-inducing therapies remains a major challenge in developing ferroptosis-targeted anti-cancer treatments. This issue is further highlighted by the facts that no specific primary cancer lineage or genotype is predictive of ferroptosis responsiveness, and biomarkers that are indicative of ferroptosis sensitivity in vivo remain largely unavailable.
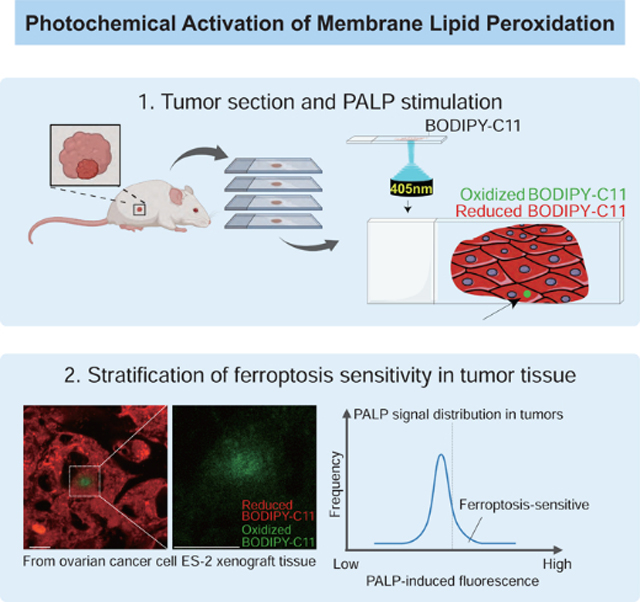
Herein, we establish a technique termed photochemical activation of membrane lipid peroxidation (PALP) to detect polyunsaturated phospholipids as well as report ferroptosis sensitivity in live cells and tissues in situ. Specifically, we use high-power laser pulses to induce localized lipid peroxidation in membranes, which subsequently results in oxidation of a fluorescent BODIPY-C11 probe (Drummen et al., 2002). We demonstrate that PALPv1-induced fluorescence is dependent on membrane polyunsaturated phospholipid levels. Moreover, replacing the light source with the two-photon laser microscope system (PALPv2) further improves the spatial resolution on the z-axis. In addition to live cells, the PALP technique series is applicable for rapid polyunsaturated phospholipid detection and ferroptosis sensitivity prediction in various normal and tumor tissues in situ. We envision that the PALP techniques can have broad utilities in basic and clinical research involving polyunsaturated lipid peroxidation and metabolism as well as facilitating the development of ferroptosis-targeted therapies.
Results
Targeted laser pulses induce localized lipid peroxidation in live cells
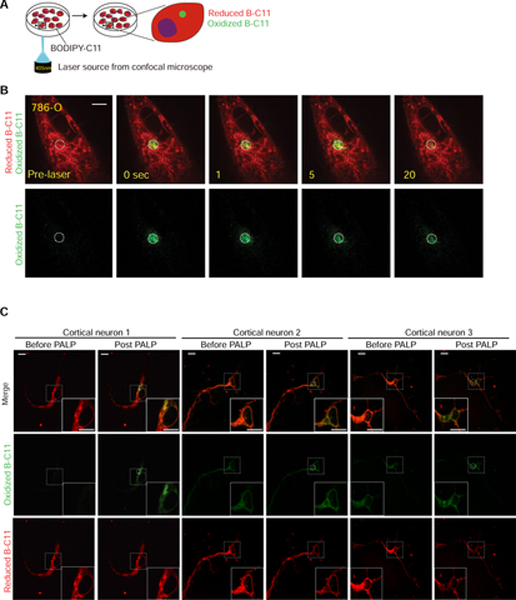
To assess the dynamics of lipid peroxidation, we pre-treated the ferroptosis-susceptible 786-O clear-cell renal cell carcinoma (ccRCC) cells (Zou et al., 2019) with BODIPY-C11 (B-C11). B-C11 can be oxidized by membrane-associated lipophilic reactive oxygen species (ROS) and changes from orange (591 nm) to green (503 nm) fluorescence after oxidation (Pap et al., 1999). We applied 5 pulses of 405 nm light from a high-power laser source on a confocal microscope (Andor) to a randomly selected, localized region in the cell over a 5-second period (Figure 1A). The target cells were imaged for oxidized B-C11 (oxB-C11) fluorescence for 25 seconds at 1-second intervals (Figure 1A). We observed strong oxB-C11 fluorescence signals in 786-O cells immediately following the laser pulses, and the fluorescence gradually decayed after reaching the peak intensity (normalized maximum intensity, PALP I-max) (Figure 1B Supplementary Figure 1A, Supplementary Video 1). In addition to the Andor revolution spinning disk confocal microscope system we initially used, PALP-induced fluorescence in 786-O cells can also be stimulated and detected with similar efficiency on two other imaging systems, including the Nikon A1R HD25 confocal microscope and the Nikon spinning disk CSU-W1 confocal system (Supplementary Figures 1B,C, Supplementary Videos 2,3). Herein, the PALP technique performed using laser sources from these regular confocal microscopes are designated as PALPv1.
Figure 1. Targeted laser pulses induce localized lipid peroxidation in live cells.
A. Schematic diagram describing the procedures of the photochemical activation of membrane lipid peroxidation (PALPv1) technique. BODIPY-C11 at a concentration of 5 μM was added to the cells approximately 30 min prior to imaging. Then a region in the cell was targeted by 5 pulses of the 405 nm laser. Each pulse lasts for 1 second. The 488 nm channel intensity was then recorded for 25 seconds at 1-second intervals to monitor the oxidized BODIPY-C11 signal.
B. Representative fluorescent images showing the time course of the reduced and oxidized BODIPY-C11 signals before and after the application of 405 nm laser pulses in wildtype 786-O cells. Scale bar indicates 10 μm. Five independent experiments were performed.
C. Representative fluorescent images showing the reduced and oxidized BODIPY-C11 signal before and after the application of 405 nm laser pulses in primary cortical neurons after 2-day culture in vitro. Scale bar indicates 10 μm. This experiment was performed once and 9 neurons were analyzed.
To evaluate the generality of high-power laser-induced lipid peroxidation further, we applied the PALPv1 technique to a clear-cell ovarian carcinoma cell line ES-2, and two non-human originated cell lines – the normal rat kidney (NRK) epithelial cell line and L929 mouse C3H/An connective tissue cell line. These experiments were performed on a Nikon A1R Confocal imaging system. Similar to the 786-O ccRCC cells, 5 seconds of laser stimulation induced strong oxB-C11 fluorescence in all three cell line models (Supplementary Figure 1D–F, Supplementary Videos 4,5).
One of the key cellular functions of polyunsaturated lipids is to support the neuronal myelin sheath structure and synapse activity (Breckenridge et al., 1972; Innis, 2007; Salem et al., 2001). We next evaluated whether PALPv1 can be applied to detect polyunsaturated phospholipids and induce lipid peroxidation in complex cellular structures such as neurons. In a primary culture of mouse cortical neurons, PALPv1 induced significant activation of lipid peroxidation at the axon hillock structures (Figure 1C, Supplementary Figure 2A). Comparing the PALP-induced fluorescence intensities from multiple analyzed neurons suggested a nearly normal I-max distribution (Supplementary Figure 2B). Fluorescent images showed the colocalization of cytochrome P450 oxidoreductase (POR), used as an endoplasmic reticulum tracker (Yan et al., 2021), and alkynated-polyunsaturated fatty acids in neurons (Supplementary Figure 2C). Together, these results suggest that PALPv1 can be used to induce lipid peroxidation non-invasively in a broad array of live cell cultures.
PALPv1-induced fluorescence is suppressed by lipid radical scavengers and iron chelation
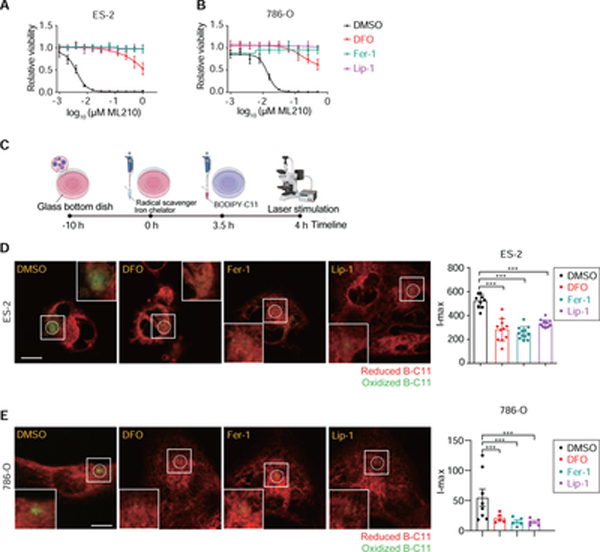
We then assessed whether the oxB-C11 fluorescence is caused by membrane lipid peroxidation. Key features of lipid peroxidation include its iron-dependency and ability to be quenched by lipophilic radical-trapping antioxidants (RTAs) (Conrad and Pratt, 2019; Zilka et al., 2017). Treatment with iron chelator deferoxamine (DFO), or lipophilic RTAs ferrostatin-1 (Fer-1) and liproxstatin-1 (Lip-1) (Dixon et al., 2012; Friedmann Angeli et al., 2014; Zilka et al., 2017) effectively decreased the sensitivity to ML210, a ferroptosis inducer inhibiting glutathione peroxidase 4 (GPX4), in both ES-2 and 786-O cells (Figure 2A,B). Treatment with each of the three chemicals 4 hours prior to PALP stimulation could significantly reduce the I-max signal of PALPv1-induced fluorescence in ES-2 and 786-O cells (Figure 2C–E), suggesting that PALPv1 induces oxidative lipid damage in an iron-dependent and lipid radical-dependent manner. These features resemble that of lipid peroxidation induced by chemical or genetic inhibition of GPX4 (Yang et al., 2014; Zou et al., 2019).
Figure 2. PALPv1-induced fluorescence is suppressed by lipid radical scavengers and iron chelation.
A. Viability curves of ES-2 cells treated with DMSO, ferrostatin-1 (Fer-1), liproxstatin-1 (Lip-1) or deferoxamine (DFO). n = 6; error bar, mean±s.d. This plot is representative data from experiments performed in duplicate.
B. Viability curves of 786-O cells treated with DMSO, ferrostatin-1 (Fer-1), liproxstatin-1 (Lip-1) or deferoxamine (DFO). n = 6; error bar, mean±s.d. This plot is representative data from experiments performed in duplicate.
C. Graphic scheme summarizing the experimental procedure in Figures D,E. Briefly, DMSO, Fer-1, Lip-1 or DFO were added 4 hours prior to laser stimulation, whereas B-C11 was added 30min prior to stimulation. The scheme was created using Biorender (https://biorender.com/).
D,E. Representative fluorescent images showing the I-max (0 sec) post the application of 405 nm laser pulses to ES-2 (D) and 786-O (E) cells treated with DMSO, Fer-1, Lip-1 or DFO. Green, oxidized BODIPY-C11 signal; red, reduced BODIPY-C11 signal. On the right, quantifications of the I-max fluorescence intensities. The number of cells analyzed in each experiment are indicated by the plots. Lines and error bars indicate mean±s.d. Statistical analysis was performed using two-tailed, unpaired student’s T-test. ***, p<0.001. Representative results from three independent experiments.
Lipid peroxidation can spread non-enzymatically via autoxidation or enzymatically via POR and its associated redox partners in multiple neoplastic cell types including 786-O ccRCC cells (Yan et al., 2021; Zou et al., 2020a), and through activities of arachidonate lipoxygenases (ALOXs) in certain other cellular contexts (Wenzel et al., 2017; Yang et al., 2016). To determine whether laser-induced lipid peroxidation is dependent on POR activity, we applied laser pulses to 786-O POR−/− single-cell clones (SCP) generated previously (Supplementary Figure 3A) (Zou et al., 2020a). We observed that POR-depletion did not significantly alter the I-max signal of PALPv1-induced fluorescence (Supplementary Figure 3B,C). This result suggests that POR activity is not required for PALPv1 induction. This likely reflects that high-power laser is bypassing POR to initiate lipid peroxidation photochemically. Consistent with recent characterizations in in vitro reconstituted liposomes (Yan et al., 2021), our data supports that POR is involved in the early initiation, rather than the propagation of cellular lipid peroxidation reactions in ferroptosis. Together, these results suggest that PALPv1 is largely a photochemical process that is independent of enzymatic activities to induce lipid peroxidation. We hence postulated that the local polyunsaturated phospholipid concentration at the laser-targeted membrane region might be one of the key contributing factors determining the I-max signal of PALPv1-induced fluorescence.
PALPv1-induced fluorescence is dependent on local polyunsaturated phospholipid levels
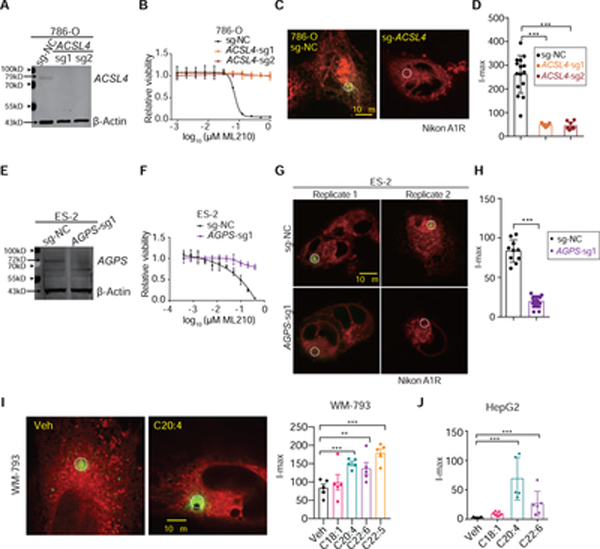
To investigate the relationship between cellular polyunsaturated phospholipid content and the I-max signal of PALPv1-induced fluorescence, we first depleted acyl-CoA synthetase long chain family member 4 (ACSL4) in 786-O cells using CRISPR/Cas9 (Figure 3A). ACSL4 catalyzes the conversion of long-chain polyunsaturated fatty acids (PUFAs) into fatty acyl-CoA, and is a key requirement for polyunsaturated phospholipid synthesis as well as a common mediator of ferroptosis susceptibility in multiple cellular contexts (Dixon et al., 2015; Doll et al., 2017; Kagan et al., 2017; Zou et al., 2019). Lipidomic analysis showed that ACSL4-depletion selectively reduced the levels of cellular PUFA-lipid levels, including PUFA-triacylglycerides (TG) and PUFA-phospholipids (PL) (Supplementary Figure 4A, Supplementary Table 1). As a result, ACSL4-depleted cells exhibited significantly lower susceptibility to GPX4 inhibition-induced ferroptosis (Figure 3B, Supplementary Figure 4B). Importantly, the I-max signal of PALPv1-induced fluorescence was also significantly suppressed in ACSL4-depleted cells (Figure 3C,D, Supplementary Figures 4C–E), suggesting that high levels of PUFA-phospholipids are necessary to potentiate strong PALPv1-induced fluorescence.
Figure 3. PALPv1-induced fluorescence is dependent on membrane polyunsaturated phospholipid levels.
A. Immunoblot analysis of ACSL4 protein levels in 786-O cells expressing non-targeting negative control sgRNA (sg-NC) or ACSL4-targeting sgRNAs. β-Actin was used as a loading control. This experiment was repeated twice.
B. Viability curves of 786-O cells expressing sg-NC or sgRNAs targeting ACSL4 treated with indicated concentrations of GPX4 inhibitor ML210 for 48h. n=6; dots and error bars, mean±s.d. This experiment was repeated three times.
C. Representative fluorescent images showing the I-max (0 sec) post the application of 405 nm laser pulses to 786-O cells expressing sg-NC or ACSL4-targeting sgRNA. Green, oxidized BODIPY-C11 signal; red, reduced BODIPY-C11 signal. Representative images from three independent experiments.
D. Quantifications of the I-max fluorescence intensities in sgNC or ACSL4-sgRNA expressing 786-O cells. Dots and error bars, mean±s.d. Statistical analysis was performed using two-tailed unpaired student’s T-test. ***, p<0.001.
E. Immunoblot analysis of AGPS protein levels in ES-2 cells expressing sg-NC or AGPS-targeting sgRNAs. β-Actin was used as a loading control. This experiment was performed once, and the efficacy of the sgRNA was previously validated (Zou et al., 2020b).
F. Viability curves of ES-2 cells expressing sg-NC or sgRNAs targeting AGPS treated with indicated concentrations of GPX4 inhibitor ML210 for 48h. n=6; error bar, mean±s.d. This experiment was repeated twice.
G. Representative fluorescent images showing the I-max (0 sec) post the application of laser pulses to ES-2 cells expressing sg-NC or AGPS-targeting sgRNA. Green, oxidized BODIPY-C11 signal; red, reduced BODIPY-C11 signal. This experiment was performed once and 5 different cells were targeted and analyzed for each group.
H. Box-scatter plots showing the I-max of PALP signals in WM-793 melanoma cells treated with vehicle (veh) or indicated free fatty acids at 20 μM concentration for 3 days. Dots and error bars, mean±s.d. Statistical analysis was performed using two-tailed unpaired T-test, ***, p<0.001.
I. Representative fluorescent images showing the I-max (0 sec) post the application of laser pulses to WM-793 cells treated with vehicle (Veh, 5% BSA) or BSA conjugated arachidonic acid (C20:4). Scale bar indicates 10 μm. Green, oxidized BODIPY-C11 signal; red, reduced BODIPY-C11 signal. Dots and error bars, mean±s.d. Statistical analysis was performed using two-tailed unpaired T-test, **, p<0.01, ***, p<0.001. Representative results from three independent experiments.
J. Box-scatter plots showing the I-max of PALP signals in HepG2 hepatoma cells pretreated with vehicle (veh) or indicated free fatty acids at 20 μM concentration for 3 days. Dots and error bars, mean±s.d. Statistical analysis was performed using two-tailed unpaired T-test, ***, p<0.001.
We and others recently reported that depletion of peroxisomal enzymes such as alkylglycerone phosphate synthetase (AGPS) blocks the synthesis of polyunsaturated ether-phospholipids in multiple cancer cell types and suppresses cellular sensitivity to ferroptosis induction (Cui et al., 2021; Zou et al., 2020b) (Figure 3E,F, Supplementary Figure 4F). To test whether PALPv1 can detect the peroxidation of polyunsaturated ether-phospholipids, we applied PALP to AGPS-depleted ES-2 cells and found that these cells exhibited reduced I-max signals of PALPv1-induced fluorescence (Figure 3G,H). These results confirm that polyunsaturated ether-phospholipids contribute to lipid peroxidation, and suggest that PALPv1 can also be used to detect this subtype of polyunsaturated phospholipids.
To demonstrate the correlation between cellular PUFA-lipid level and the intensity of PALPv1-induced fluorescence further, we used chemical approaches to complement the genetic modulation of PUFA-phospholipid levels. We pre-treated the human melanoma cell line WM-793 with various synthetic fatty acids for three days prior to PALPv1 application. These fatty acids include monounsaturated fatty acid (MUFA) oleic acid (OA, C18:1), and PUFAs including arachidonic acid (AA, C20:4, ω6), docosapentaenoic acid (DPA, C22:5, ω3), and docosahexaenoic acid (DHA, C22:6, ω3). Treatment with AA, DPA or DHA, but not OA, significantly enhanced the I-max signal of PALPv1-induced fluorescence; the extent of enhancement is largely correlated with the ferroptosis sensitization activities of these fatty acids characterized here and previously (Zou et al., 2020a) (Figures 3I, Supplementary Figure 4G). Similar effects were observed in human hepatoma HepG2 cells (Figure 3J). In another cell line BFTC-909, a urothelial carcinoma model, OA treatment even inhibited the I-max signal of PALPv1-induced fluorescence (Supplementary Figure 4H,I). The PALPv1-suppressive effect of MUFA is consistent with the recent identification of exogenous MUFAs as ferroptosis inhibitory agents (Magtanong et al., 2019). Taken together, these results support the positive correlations among cellular susceptibility to ferroptosis, the polyunsaturated phospholipid levels, and the I-max signals of PALPv1-induced fluorescence, and further demonstrate that PALPv1 can be used to detect polyunsaturated phospholipids and report ferroptosis sensitivity in live cells (Supplementary Figure 4J).
PALPv2: lipid peroxidation induced by two-photon excitation microscopes enables targeting specific membranes along the z-axis
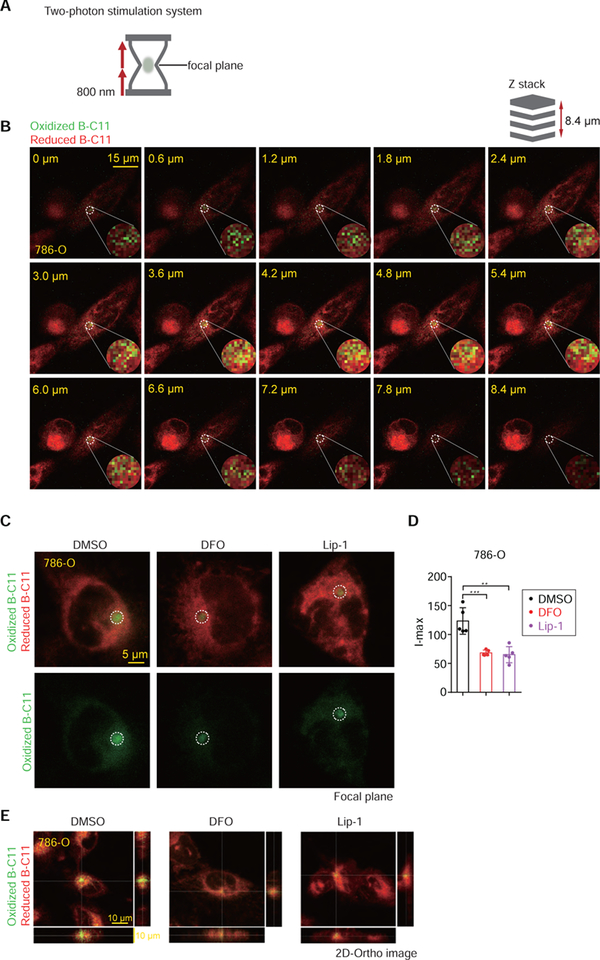
One caveat of PALPv1 using regular confocal laser scanning microscope is that the high-power laser penetrates all membranes distributed along the selected z-axis, precluding its usage for mechanistic studies involving the oxidative damage of specific membranes at high spatial resolution in live cells. To enable the photochemical induction of lipid peroxidation on selected membranes along the z-axis of cells, we developed PALPv2 using the double 800 nm photon sources from a Zeiss 880 two-photon confocal microscope (Figure 4A). In PALPv2, multiple 800 nm low-energy photons excite the same target molecule on a single focal plane and induce lipid peroxidation, whereas membrane structures outside of the focal plane won’t be oxidized. In a proof-of-principle study, we targeted the photon sources towards one layer of intracellular membrane that contains extensive reduced B-C11 distribution, presumably an endoplasmic reticulum membrane structure, and scanned through the z-axis for oxidized B-C11 signals at 0.6 μm internals over the 8.4 μm thickness of the cell (Figure 4A). This analysis showed that only the membranes close to the targeted focal plane exhibited strong oxB-C11 signals (Figure 4B–E, Supplementary video 6) and the I-max signal was significantly quenched by treatment with iron chelator DFO, or lipophilic RTA Lip-1 (Figure 4C–E), suggesting that PALPv2 using the two-photon excitation microscope system can potentially be used for assessing polyunsaturated phospholipid levels and track lipid peroxidation dynamics on selected membranes along the z-axis in live cells.
Figure 4. Laser sources from two-photon confocal microscopes improve the z-axis resolution in PALPv2.
A. Schematic diagram of two-photon excitation confocal microscope system. The focal plane was stimulated by two 800 nm infrared photon beams, while the non-focal-plane could not be excited.
B. Representative fluorescent images showing the Z-stack series of the reduced and oxidized BODIPY-C11 signal before and after the application of 100% 800 nm two-photon excitation in wildtype 786-O cells. Scale bar indicates 20 μm. Every 0.6 μm thickness of the 786-O cell was captured along the z-stack. This experiment was performed once and 3 different cells were targeted and analyzed.
C. Representative fluorescent images showing the I-max signals on the focal plane in 786-O cells pretreated with DMSO, DFO or Lip-1 for 4 hours and then stimulated by PALPv2. Scale bar indicates 5 μm.
D. Box-scatter plots showing the I-max of PALP signals in Figure C. Statistical analysis was performed using two-tailed unpaired T-test, **, p<0.01, ***, p<0.001. Dots and error bars, mean±s.d. This experiment was performed once.
E. 2-D orthogonal image presentation showing the representative fluorescent images of 786-O cells pretreated with DMSO, DFO or Lip-1 for 4 hours and then stimulated by PALPv2.
PALPv1 enables detection of polyunsaturated phospholipids in various tissues in situ
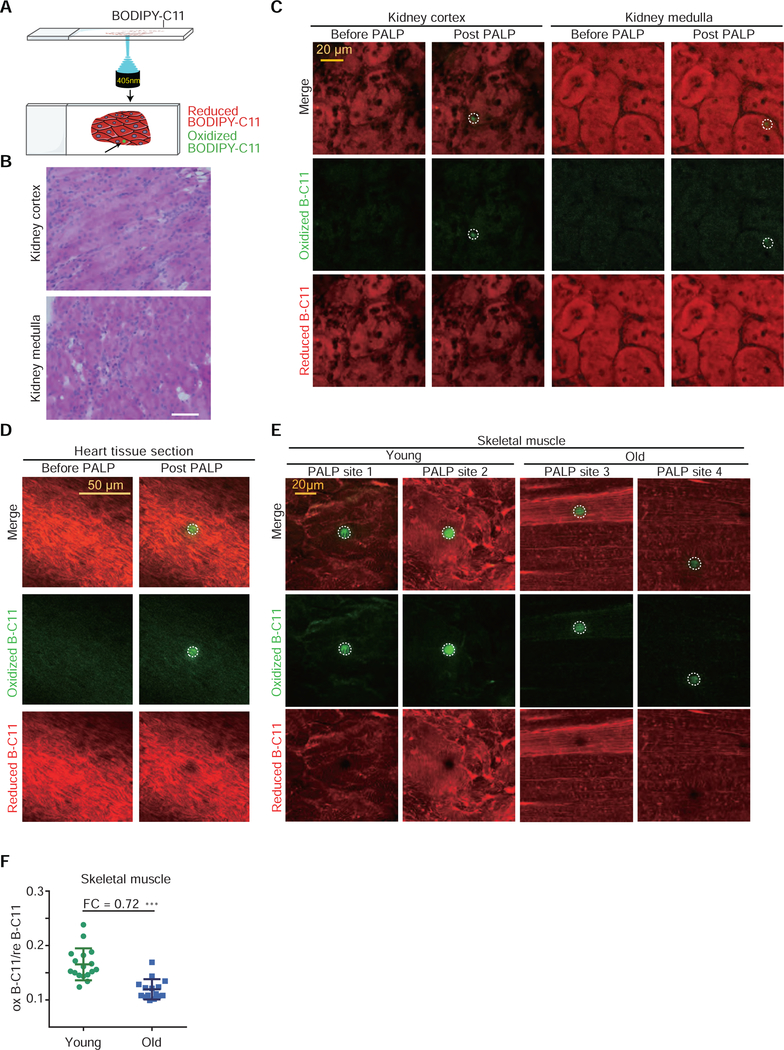
The above results indicated that high-power laser pulses can bypass enzymatic requirements and induce targeted lipid peroxidation in live cells. To explore the broader utility of the PALP technique series, we next assessed whether PALP could induce lipid peroxidation in tissue sections (Figure 5A). We reasoned that paraformaldehyde fixation might disrupt the membrane lipid organization, and chose to apply PALPv1, the more accessible and quantitative approach, on non-fixed, cryo-sectioned tissue samples. By applying PALPv1 to cryo-sectioned mouse kidney slides, we found that PALPv1 under standard laser settings induces strong oxB-C11 fluorescence in both kidney cortex and medulla regions (Figure 5B,C, Supplementary Figure 5A). Similar results were obtained in randomly selected regions of mouse heart tissue sections (Figure 5D, Supplementary Figure 5B,C). These results highlight the applicability of PALPv1 technique in detecting polyunsaturated phospholipids and potentially stratifying ferroptosis sensitivity in cryo-sectioned tissue samples.
Figure 5. PALPv1 enables detection of polyunsaturated phospholipids in multiple tissues in situ.
A. Schematic diagram describing the procedures of PALPv1 technique for tissue sections. BODIPY-C11 at a concentration of 5 μM was added to the cells approximately 30 min prior to imaging. Each 405 nm confocal laser was set at zero HV mode and standard gain with 4 loops of 120 ms exposure laser. Scan speed was set at 8 Fps and pixel dwell was 11.6. The 488 nm channel intensity was then recorded for the oxidized BODIPY-C11 signal.
B. Histologic examination of transverse section of mouse kidney cortex and medulla using hematoxylin and eosin (H&E) staining. Original magnification X 400.
C. Representative fluorescent images showing the reduced and oxidized BODIPY-C11 signal before and after the application of 405 nm laser pulses in cryo-sectioned mouse kidney tissue sections. Green, oxidized BODIPY-C11 signal; red, reduced BODIPY-C11 signal. This experiment was performed once and 8 random regions were targeted and analyzed for either the kidney cortex or medulla.
D. Representative fluorescent images showing the reduced and oxidized BODIPY-C11 signal before and after the application of 405 nm laser pulses in cryo-sectioned mouse heart tissue sections. This experiment was performed once and 5 random regions were targeted and analyzed.
E. Representative fluorescent images showing the reduced and oxidized BODIPY-C11 signal before and after the application of 405 nm laser pulses in cryo-sectioned skeletal muscle tissue sections from young and old mice. This experiment was performed once.
F. Scatter plot showing the quantitation of the oxidized/reduced BODIPY-C11 fluorescence in laser stimulated region in the skeletal muscle tissue samples from young and old mice. PALP signal was normalized by the reduced BODIPY-C11 fluorescence prior to laser stimulation. Dots and error bars, mean±s.d. Two-tailed, unpaired student’s T-test. ***, p<0.001. FC, fold-change.
To further validate the utility of PALP in detecting and measuring polyunsaturated phospholipid levels in situ, we applied the PALPv1 technique to aging mouse skeletal muscle tissues based on a recent report showing that polyunsaturated phosphatidylcholine (PC) and phosphatidylethanolamine (PE) levels decrease in mouse skeletal muscle tissues during aging (Lee et al., 2020). We dissected and cryo-sectioned the skeletal muscle tissues from one 39-week-old and one 116-week-old mouse from the same genetic background (Figure 5E, Supplementary Figure 5D), and performed PALPv1 analysis in 12 random regions from each section. The results revealed that the oxB-C11 signals of PALPv1-induced fluorescence obtained from the longitudinal muscle section of the old mouse are significantly weaker (fold change = 0.72) than those from the young mouse (Figure 5F). This experiment supports that PALPv1 can discern the differences in polyunsaturated phospholipid levels and likely changes in ferroptosis sensitivity in animal tissues in situ.
PALPv1 technique enables the stratification of ferroptosis sensitivity in vitro and in vivo
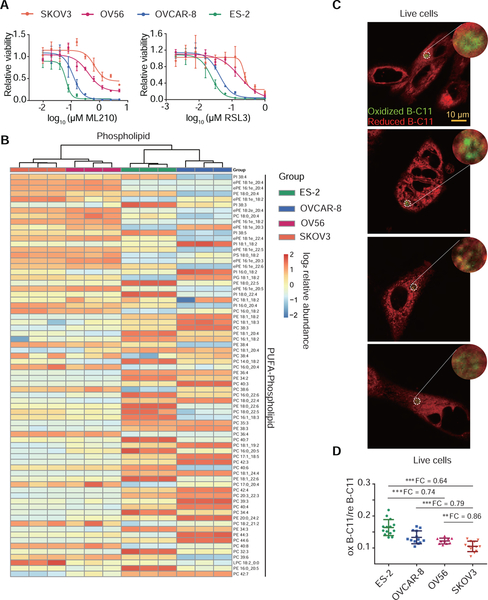
In addition to its applicability in live cells and in situ, we evaluated whether the PALP techniques could assist in predicting and stratifying ferroptosis sensitivity in minute tumor samples to guide patient selection for ferroptosis-targeted therapies. Our prior studies showed that some but not all ovarian cancer cells are sensitive to ferroptosis induction (Zou et al., 2019, 2020b). To determine whether PALPv1 can distinguish ferroptosis-sensitive from -insensitive cells and tumors, we selected four ovarian cancer cell lines that exhibit a wide range of sensitivity to GPX4 inhibitors ML210 and RSL3 in vitro, with the sensitivity rank of ES2 > OVCAR-8 > OV56 > SKOV3 (Figure 6A). Among these four cell lines, untargeted lipidomic analysis showed that ES-2 and OVCAR-8 cells exhibited the highest PUFA-phospholipid and lowest triacylglyceride levels compared with that of OV56 and SKOV3 cells (Figure 6B, Supplementary Figure 6A,B, Supplementary Table 1). Importantly, the randomly induced PUFA-lipid peroxidation, represented by the oxB-C11 signals of PALPv1-induced fluorescence of ES-2 and OVCAR-8 cells are the strongest, followed by those of OV56 and SKOV3 cells (Figure 6C,D). Hence, PALPv1-induced fluorescence signal is concordant with the PUFA-phospholipid levels and sensitivities to ferroptosis induction in tumor cell lines in vitro.
Figure 6. PALPv1 differentiates the differences in ferroptosis sensitivity across a collection of ovarian cancer cell lines in culture.
A. Viability curves of SKOV3, OV56, OVCAR-8, and ES-2 cells treated with indicated concentrations of GPX4 inhibitors ML210 or RSL3 for 48h. n=4; dots and error bars, mean±s.d. Representative results from experiments performed twice.
B. Heatmap presentation of lipidomic analysis results showing the polyunsaturated phospholipid distribution among ES-2, OVCAR-8, OV56, and SKOV3 ovarian cancer cells. Red, high intensity; blue, low intensity. Three biological replicates were analyzed by lipidomics for each cell line. This experiment was performed once. SFA, MUFA, PUFA, saturated, monounsaturated, or polyunsaturated fatty acids.
C. Representative fluorescent images showing the I-max (0 sec) post the application of laser pulses to ES-2, OVCAR-8, OV56, and SKOV3 cells. Green, oxidized BODIPY-C11 signal; red, reduced BODIPY-C11 signal. Scale bar indicates 10 μm.
D. Scatter plots showing the oxidized/reduced BODIPY-C11 fluorescence of PALP signals in ES-2, OVCAR-8, OV56, and SKOV3 ovarian cancer cells. PALP signal was normalized by the reduced BODIPY-C11 fluorescence prior to laser stimulation. Dots and error bars, mean±s.d. Two-tailed unpaired T-test, **, p<0.01, ***, p<0.001. n=15. FC, fold-change.
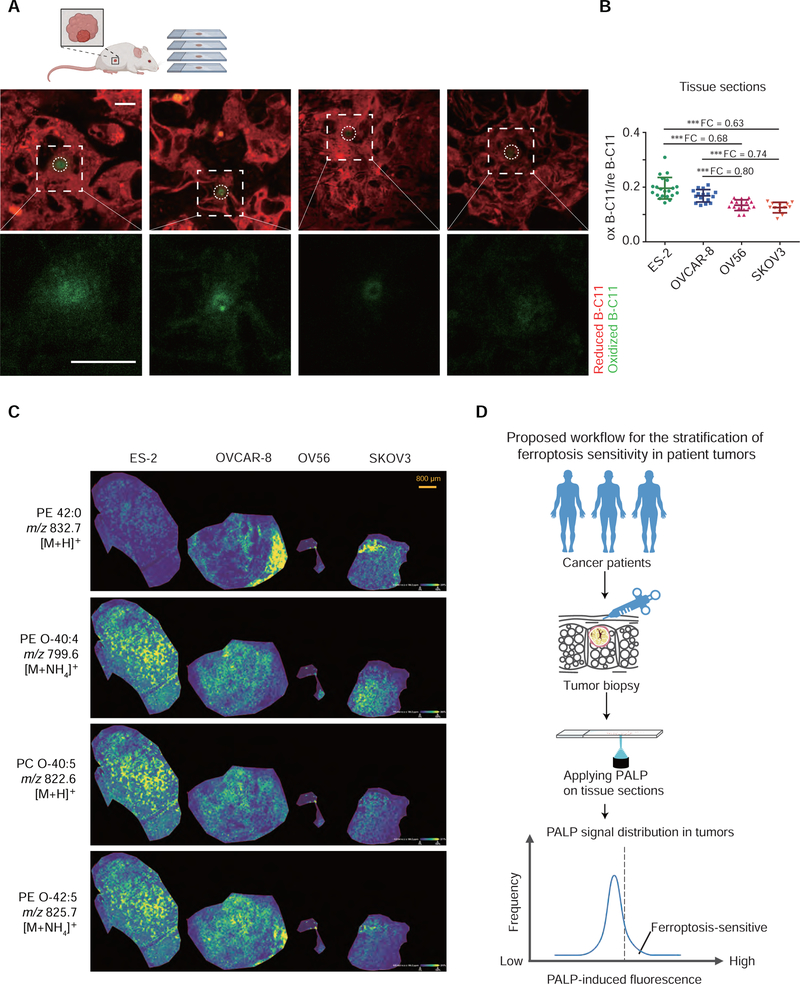
To assess whether PALPv1 can also be employed to stratify ferroptosis sensitivity of tumors in situ, we subcutaneously implanted the four ovarian cancer cells to immunocompromised mice. The high-power laser pulses were applied to thaw-mounted xenograft tumor sections for at least 15 random regions per tumor type (Figure 7A, Supplementary Figure 7A). The rank order of PALPv1 signals of ovarian xenograft tumor sections was ES-2 > OVCAR-8 > OV56 > SKOV3 (Figure 7A,B). To confirm that the PALPv1 signal in situ is reflective of the polyunsaturated phospholipid levels of the tumor samples, we utilized the matrix assisted laser desorption/ionization-time of flight (MALDI-TOF) mass spectrometry imaging (MSI) technique to determine the spatial distribution of various lipid components. Using 2,5-dihydroxybenzoic acid (DHB) as the ionization matrix, we successfully detected a major fraction of the most abundant phospholipid species in these samples though the precise chemical structures of these lipid molecules remain to be validated with additional approaches (Supplementary Figure 7B). Among the detected lipids, polyunsaturated phospholipids including PE O-40:4, PC O-40:5, PE O-42:5, PC O-38:4, PC 36:5, PE 44:5, and PC O-34:2 are highly enriched in ES-2 and OVCAR-8 tumors compared with those in OV56 and SKOV3 tumors (Figure 7C, Supplementary Figure 7C). Collectively, these results are highly consistent with the patterns of in vitro PALPv1 signals, untargeted lipidomic characterizations, as well as ferroptosis sensitivities (Figure 7B), supporting that PALPv1 is likely useful for stratifying ferroptosis sensitivity in tumors in situ in clinically relevant settings.
Figure 7. PALPv1-induced fluorescence is correlated with polyunsaturated phospholipid levels and ferroptosis in human ovarian xenograft tumors in vivo.
A. Representative fluorescent images showing the I-max (0 sec) post the application of laser pulses to xenograft tumor sections including human ovarian ES-2, OVCAR-8, OV56, and SKOV3 cells. Green, oxidized BODIPY-C11 signal; red, reduced BODIPY-C11 signal. Scale bar indicates 10 μm. The xenograft experiment was performed once. The scheme was created using Biorender (https://biorender.com/).
B. Scatter plots showing the oxidized/reduced BODIPY-C11 fluorescenceI of PALP signals in xenograft tumor sections including human ovarian ES-2, OVCAR-8, OV56, and SKOV3 cells. PALP signal was normalized by the reduced BODIPY-C11 fluorescence prior to laser stimulation. n=15. Dots and error bars, mean±s.d. Two-tailed unpaired T-test, ***, p<0.001. FC, fold-change. This experiment was performed once and x random regions were targeted and analyzed.
C. Mass spectrometry images showing the spatial distribution of representative phospholipid on xenograft tumor sections including human ovarian ES-2, OVCAR-8, OV56, and SKOV3 cells. Yellow, high intensity; Blue, low intensity. Scale bar indicates 800 μm.
D. Schematic diagram showing the proposed workflow for using PALP to stratify the ferroptosis sensitivity of patient tumors. We suggest collecting the tumor biopsy samples from cancer patients using fine needle aspiration, and then apply the PALP technique to cryo-sectioned tumor tissue samples. Relative sensitivity to ferroptosis of the tested tumor may be speculated based on the distribution of PALP signals relative to a validated reference dataset.
Discussion
Polyunsaturated phospholipids are the major substrates of lipid peroxidation and determinants of cellular sensitivity to ferroptosis. Ferroptosis-targeted therapies have broad implications in suppressing tumor progression and metastasis (Jiang et al., 2021; Ubellacker et al., 2020; Zou and Schreiber, 2020). However, how to stratify the tumor ferroptosis sensitivity and predict drug efficacy in specific patients remains unsolved. Here, we present PALP, a rather simple and cost-effective imaging technique that enables quick detection of polyunsaturated phospholipids and assessment of sensitivity to ferroptosis in live cells and tissues in situ. Based on our characterizations in animal models, we propose a tentative workflow for rapidly stratifying ferroptosis sensitivity in human cancer patients using biopsy samples integrated with the PALP techniques (Figure 7D).
Key innovations in the PALP technique include: 1) leveraging the unique chemical propensity of polyunsaturated fatty acids to undergo lipid peroxidation and using the abundances of peroxidation products to detect polyunsaturated phospholipids in cellular membranes and estimate the relative cellular sensitivity to ferroptosis induction at high spatial resolution; 2) using an existing fluorescent probe, BODIPY-C11 that can easily penetrate live cells and tissue sections, to report and quantify laser-induced lipid peroxidation levels; 3) employing high-power lasers – a readily available resource on confocal microscopes from many research labs – to photochemically induce localized lipid peroxidation. Restricting the laser to a small area in the cell both ensures efficient induction of lipid peroxidation and prevents over-activation of lipid peroxidation that may trigger ferroptotic cell death. In contrast to cellular lipid peroxidation, which is dependent on activities of oxidoreductases such as POR (Yan et al., 2021; Zou et al., 2020a), PALP bypasses enzymatic lipid peroxidation. Hence, PALP-induced fluorescence is not sensitive to the variable expression levels of lipid peroxidation enzymes, but is largely determined by the local abundances of polyunsaturated phospholipids in the laser-targeted membrane region. With these features, PALP can be practiced in most labs with access to a confocal microscope.
PALP can possibly induce lipid peroxidation via two mechanisms: photosensitized or photochemical oxidations. In the former model, photons from UV irradiation or high-power laser pulses can be absorbed by intracellular chromophores including porphyrins, riboflavin, and heme-containing proteins. These chromophores in the excited state can then react with molecular oxygen (O2) to generate ROS including singlet oxygens and superoxide anion radicals. Singlet oxygens may directly react with PUFA-phospholipids via the 1,4-pentadiene functional group. Superoxide anion radicals can be converted to hydrogen peroxides (H2O2), which react with intracellular ferrous ions to produce hydroxyl radicals. Excess ROS fuels lipid peroxidation, which disrupts membrane homeostasis and induces DNA damage (Girotti, 2001; McMillan et al., 2008; Yin et al., 2011). Consistent with this mechanism, a recent study showed that pervasive photodynamic therapy was sufficient to induce non-enzymatic lipid peroxidation and ferroptosis-like cell death (Shui et al., 2021). On the other hand, in photochemical oxidation, UV irradiation and high-power laser pulses can directly elicit lipid peroxidation by targeting the highly oxidable 1,4-pentadiene functional group in PUFA-phospholipids. The precise mechanism by which laser sources on fluorescent microscopes induce membrane lipid peroxidation warrants further characterization.
In addition to contributing to lipid peroxidative damage and susceptibility to ferroptosis, polyunsaturated lipids also play diverse roles in cellular physiology, including reducing membrane bending rigidity to assist membrane deformation, supporting transmembrane receptor activities and signal transduction, and serving as precursors of signaling molecules involved in immunity (Calder, 2005; Gimple et al., 2019; Pinot et al., 2014). Dysregulation of polyunsaturated lipid metabolism contributes to the progression of cancer, obesity, cardiovascular diseases, rheumatoid arthritis, and neurodegeneration (D’Angelo et al., 2020; Maki et al., 2018; Shahidi and Ambigaipalan, 2018; Veselinovic et al., 2017), and loss of polyunsaturated lipids is a hallmark of aging in certain tissues (Lee et al., 2020). Currently, lipid profiling in cells and tissue samples are largely achieved by untargeted lipidomic analysis, which requires a substantial amount of materials and loses the spatial information about lipid distribution (Han, 2016); mass spectrometry imaging, a complementary approach, exhibits low sensitivity and resolution, requires highly-specialized equipment, and can only report limited information about the total level of polyunsaturated lipids as a chemically distinct group (Neumann et al., 2020). The PALP techniques we developed here will enable the rapid detection of polyunsaturated phospholipids in cells and tissues for better dissecting their cellular and physiological functions.
Given the relative simplicity of the technique, we envision that PALP may additionally be applicable to the following research settings: 1) to study the mechanisms of ferroptosis at high spatio-temporal resolution and to detect polyunsaturated phospholipids in heterogeneous cell populations; 2) to evaluate how novel chemicals and genetic perturbations affect the dynamics of lipid peroxidation in cells and tissues. We expect that compounds that exhibit lipid radical scavenging or iron chelating activity will be able to suppress PALP signals; 3) to assess the functions and dynamics of polyunsaturated phospholipids in aging-related diseases such as Alzheimer’s Disease and organ degeneration. Overall, we believe the PALP technique series will be widely useful for studying ferroptosis, lipid metabolism, membrane lipid damage and repair, and human diseases associated with aberrant lipid metabolic states.
Significance
Ferroptosis induction has been implicated as a new therapeutic strategy for treating various primary and therapy resistant cancers. However, the sensitivity to ferroptosis varies widely among cancers sharing the same lineage-of-origin or genotype, and how to select the right patients for treatment with bioavailable ferroptosis-inducing agents remains unsolved. We present the photochemical activation of membrane lipid peroxidation (PALP) technique that can potentially address this challenge at low-cost, and propose a potential clinical workflow for using PALP to stratify ferroptosis sensitivity in patient tumors. PALP uses high-power laser pulses to induce localized membrane lipid peroxidation – a chemical property unique to the 1,4-pentadiene structures in polyunsaturated lipids – that can be captured by the oxidation of a fluorescent probe, and the sensitivity to ferroptosis can be predicted by the fluorescent intensity both in cultured cells and tissues in situ. Moreover, we developed PALP version2, which enables studying lipid peroxidation dynamics on specific membranes across the vertical axis of live cells. The PALP technique series may be useful for predicting ferroptosis susceptibility in tissue samples, and for characterizing polyunsaturated lipid metabolism during normal development, aging, tumorigenesis, and other relevant disease settings.
STAR Methods
Resource Availability
Lead Contact
Further information and requests for resources and reagents should be directed to and will be fulfilled by the Lead Contact, Yilong Zou (zouyilong@westlake.edu.cn).
Material Availability
This study did not generate new unique reagents.
Data and Code Availability
Raw videos showing the dynamics of BODIPY-C11 fluorescences in live cells are provided as multiple Supplementary Videos. Lipidomics datasets are presented in the Supplementary Tables. These data are publicly available as of the date of publication and are listed in the key resources table.
This paper does not report original code.
Any additional information required to reanalyze the data reported in this paper is available from the Lead Contact upon request.
KEY RESOURCES TABLE.
| REAGENT or RESOURCE | SOURCE | IDENTIFIER |
|---|---|---|
| Antibodies | ||
| Anti-FACL4 antibody | Abcam | Cat# ab155282, RRID: AB_2714020 |
| Anti-AGPS | Abcam | Cat# ab236621 |
| β Actin Mouse mAb | Cell Signaling Technologies | Cat# MA5–11116, RRID: AB_10985704 |
| IRDye 800CW goat-anti-Rabbit secondary antibodies | LiCor | Cat# 925–32211, RRID: AB_2651127 |
| IRDye 680RD donkey-anti-Mouse secondary antibodies | LiCor | Cat# 925–68072, RRID: AB_2814912 |
| Anti-Cytochrome P450 Reductase antibody | Abcam | Cat# ab180597 |
| Alexa Fluor 647 anti-rabbit secondary antibodies | Beyotime | Cat# P0180 |
| Chemicals, peptides, and recombinant proteins | ||
| penicillin/streptomycin | Gibco | Cat# 15140–122 |
| GlutMax | Gibco | Cat# 35050061 |
| B27 | Gibco | Cat# 17504001 |
| ML210 | Cayman | Item No. 23282 |
| 1S,3R-RSL3 | Cayman | Item No. 19288 |
| ferrostatin-1 | Cayman | Item No. 17729 |
| liproxstatin-1 | Cayman | Item No. 17730 |
| deferoxamine | Cayman | Item No. 14595 |
| BODIPY-C11 | Life Technologies | Cat# D3861 |
| DMSO | Sigma-Aldrich | Cat# D2650 |
| Oleic acid | Cayman | Item No. 90260 |
| arachidonic acid | Cayman | Item No. 10007268 |
| docosapentaenoic acid | Cayman | Item No. 90165 |
| docosahexaenoic acid | Cayman | Item No. 17950 |
| BSA | Sigma-Aldrich | Item No. 10010566 |
| puromycin | Gibco | Cat# A11138–03 |
| Lipofectamine™ 2000 Transfection Reagent | Thermo Fisher | Cat# 11668019 |
| polybrene | Sigma-Aldrich | Cat# TR-1003-G |
| lithium dodecyl sulfate sample buffer | Novus | Cat# NP0008 |
| isopropanol, HPLC Grade | Thermo Fisher | CAS: 67–63-0 |
| ammonium acetate | Sigma-Aldrich | Cat# 73594 |
| methanol | Sigma-Aldrich | Cat# 34885 |
| formic acid, HPLC LiiChropur | Supelco | Cat# 5.33002 |
| 2,5-dihydroxybenzoic acid | Sigma-Aldrich | Cat# 39319 |
| trifluoroacetic acid | Sigma-Aldrich | Cat# 80457 |
| arachidonic acid-alkyne | Cayman | Item No. 10538 |
| Critical commercial assays | ||
| Lenti-X Go-Stix Plus | TakaraBio | Cat# 631280 |
| CellTiter-Glo Luminescence Assay | Promega | Cat# G7571 |
| BCA Protein Assay kit | Pierce | Cat# 23225 |
| Click-iT® Lipid Peroxidation Detection with Linoleamide Alkyne | Invitrogen | Cat# C10446 |
| Deposited data | ||
| Lipidomics data of ACSL4-depleted 786-O cell | This paper | Supplementary Table1 |
| Lipidomics data of ES-2, OVCAR8, SKOV3 and OV56 cell lines | This paper | Supplementary Table2 |
| Experimental models: Cell lines | ||
| Human clear-cell renal cell carcinoma cell: 786-O | ATCC | Cat# CRL-1932, RRID: CVCL_1051 |
| Human clear-cell ovarian carcinoma cell: ES-2 | ATCC | Cat# CRL-1978, RRID: CVCL_3509 |
| Human melanoma cell: WM-793 | ATCC | Cat# CRL-2806, RRID: CVCL_8787 |
| Human urothelial carcinoma cell: BFTC-909 | DSMZ | Cat# ACC-367, RRID: CVCL_1084 |
| Human ovarian cancer cell: OVCAR-8 | NCI-DTP | Cat# OVCAR-8, RRID: CVCL_1629 |
| Human ovarian cancer cell: OV56 | ECACC | Cat# 96020759, RRID: CVCL_2673 |
| Human ovarian cancer cell: SKOV3 | NICR | 4201PAT-CCTCC01017 |
| Human hepatoma cell: HepG2 | NICR | 1101HUM-PUMC000035 |
| Normal rat kidney (NRK) epithelial cell line | NICR | 1101RAT-PUMC000198 |
| L929 mouse C3H/An connective tissue cell line | NICR | 1101MOU-PUMC000403 |
| HEK-293T | NICR | 1101HUM-PUMC000010 |
| Experimental models: Organisms/strains | ||
| Mouse: C57BL/6J | The Jackson Laboratory | N/A |
| Mouse: athymic nude mice | The Jackson Laboratory | N/A |
| Oligonucleotides | ||
| ACSL4-sg1, GTGTGTCTGAGGAGATAGCG | This paper | N/A |
| ACSL4-sg2, GCATCATCACTCCCTTAGGT | This paper | N/A |
| AGPS-sg1, GTACCAATGAGTGCAAAGCG | This paper | N/A |
| Recombinant DNA | ||
| LentiCRISPR V2 plasmid | (Sanjana et al., 2014) | Addgene, Cat# 52961 |
| psPAX2 plasmid | Trono Lab Packaging and Envelope Plasmids (unpublished) | Addgene, Cat# 12260 |
| pMD2.G plasmid | Trono Lab Packaging and Envelope Plasmids (unpublished) | Addgene, Cat# 12259 |
| Software and algorithms | ||
| ImageStudio software | LiCor | https://www.licor.com/bio/image-studio/ |
| Metamorph software | Molecular Devices, Andor | |
| ZEN 3.3 blue edition | Zeiss Microscope | https://www.zeiss.com/microscopy/int/products/microscope-software/zen.html |
| ImageJ 1.52P | Fiji | https://imagej.net/software/fiji/ |
| GraphPad Prism | GraphPad software Inc | https://www.graphpad.com/scientific-software/prism/ |
| Nikon NIS Elements software | Nikon NIS Microscope | https://www.microscope.healthcare.nikon.com/products/software/nis-elements/viewer |
| Profinder 10.0 | Agilent | N/A |
| Lipid Annotator | Agilent | N/A |
| LIPID MAPS | (Sud et al., 2006) | https://www.lipidmaps.org |
| Human Metabolome Database | (Wishart et al., 2018) | http://www.hmdb.ca/ |
| Adobe Illustrator | Adobe | www.adobe.com |
| Spearman correlation and cluster analyses | R-3.3.3 | |
| Biorender | https://biorender.com/ | |
| SciLS Lab v.2021c Pro software | Bruker Daltonik | www.bruker.com |
| Flex control software | Bruker Daltonik | www.bruker.com |
| Flex imaging software | Bruker Daltonik | www.bruker.com |
| Other | ||
| Accucore C8 column (100 × 2.1 mm, 2.6 μm) | Thermo Fisher Scientific | Cat# 17226–102130 |
| RPMI 1640 | Gibco | Cat# C11875500BT |
| DMEM | Gibco | Cat# C11995500BT |
Experimental Model and Subject Details
All mice were acquired from Shanghai SLAC Laboratory Animal Co.,Ltd (Shanghai, China) and were maintained in approved pathogen- and viral-free institutional housing facilities. Animal experiments were carried out in accordance with the animal protocol approved by the Institutional Animal Care and Use Committee (IACUC) of Westlake University.
Normal mouse tissue collection:
Male C57BL/6J mice were 39-week-old for the young group, while the aged mouse was 116-week-old. Mice weighing 21–23 g were used in this study. Mice were sacrificed by cervical dislocation. The kidney, heart, and skeletal muscle tissues were rapidly dissected, snap-frozen in liquid nitrogen, and stored at −80 °C until use.
Mouse xenografts:
Female athymic nude mice (5 weeks old, 18–20 g) were housed and adapted to the breeding environment for one week before the experiment. One group of mice (n=3) were subcutaneously injected with 2×106 human OVCAR-8 cells on the left flank and with 1×106 ES-2 cells on the right flank. Another group of mice (n=3) were subcutaneously injected with 1×106 OV56 cells on the left flank and with 1×106 SKOV3 cells on the right flank. The mice were sacrificed when the tumor volume reached about 60 mm3. The dissected tumors were snap-frozen in liquid nitrogen and kept in −80 °C until use.
Primary neuron isolation:
Primary cortical neurons were prepared from neonatal (p0) C57BL/6J mice. The mouse was sacrificed by cutting the head before neuron isolation following the above referenced IACUC protocol.
Cell lines and culture conditions:
786-O, ES-2, WM-793, OVCAR8, OV56 and SKOV3 cells were cultured with RPMI 1640 (Gibco) medium. HepG2, BFTC-909 cells and adherent cells (normal rat kidney, NRK; L929) were cultured with DMEM (Gibco) medium. 786-O, ES-2, OVCAR-8, OV56, BFTC-909 and WM-793 cells were obtained from the Cancer Cell Line Encyclopedia (CCLE) distributed and authenticated by the Broad Institute Biological Samples Platform and Genetic Perturbation Platform. HepG2, BFTC-909, L929 and SKOV3 cells were purchased from Chinese National Infrastructure of Cell Line Resource (NICR). All culture media were supplemented with 10% fetal bovine serum (Gibco) and 1% penicillin/streptomycin. 786-O cells expressing ACSL4-sgRNA (Zou et al., 2018), ES-2 expressing AGPS-sgRNA, and POR−/− single cell clones were generated and validated in prior studies (Zou et al., 2020a). All cells were tested as mycoplasma-negative prior to experiments.
Neuronal cell culture:
Primary cortical neurons were prepared from neonatal (p0) C57BL/6J mice. Dissected cortices were digested with papain (2 mg/ml) for 30 min at 37 °C and digestion was stopped using 10% FBS. Cortical neurons (500 cells/mm2) were plated on poly-D-lysine pre-coated glass dishes in DMEM (Sigma-Aldrich) supplemented with 10% FBS (Gibco) and 1% penicillin-streptomycin (Gibco) for 4 hours. Afterwards, the medium was replaced with neurobasal medium A (Gibco) containing 1% penicillin-streptomycin, 1% GlutMax (Gibco) and 2% B27 (Gibco). The PALP imaging experiment was performed with primary neurons cultured for 2 days in vitro (DIV 2).
Method Details
Sources and usage concentrations of chemicals
ML210, 1S,3R-RSL3 (RSL3), ferrostatin-1, liproxstatin-1 and deferoxamine (DFO) were obtained from commercial sources (Cayman, 23282, 19288, 17729, 17730, and 14595, respectively). The final concentration of DFO, ferrostatin-1, and liproxstatin-1 in the assays was 50 μM,5 μM, and1 μM, respectively. Cells were treated with DFO, ferrostatin-1, and liproxstatin-1 for 4 hours prior to imaging analysis, and for 2 days prior to viability assay. BODIPY-C11 (Life Technologies) was reconstituted in DMSO to 5 mM stock concentration. Thirty minutes prior to live-cell imaging, the medium was replaced with a complete medium containing 5 μM BODIPY-C11 final concentration.
Free fatty acid treatment
Oleic acid (OA, C18:1), arachidonic acid (AA, C20:4), docosapentaenoic acid (DPA, C22:5), and docosahexaenoic acid (DHA, C22:6) were purchased from Cayman Chemicals and conjugated with fatty-acid free BSA (Sigma-Aldrich) using previously described protocols(Hannah et al., 2001). Briefly, a 0.1M stock solution of each fatty acid was prepared by diluting the free fatty acid in ethanol and neutralizing it with the excessive NaOH at 0.25 M final concentration. The precipitated sodium salt was then evaporated under nitrogen gas, reconstituted with 10% defatted BSA in 0.15 M NaCl. Each solution was adjusted to pH 7.4 with NaOH and stored in multiple aliquots at −20 °C protected from light and oxygen. The cells were treated with 20 μM indicated free fatty acid for 3 days prior to viability assays or imaging analysis.
Viability assays
For cellular viability assays, cells were seeded in 384-well opaque white tissue culture and assay plates (Corning) at 500 cells/well. 12–18 hours after seeding, cells were treated with compounds at the indicated concentrations for 48 hours. Cellular ATP levels were quantified using CellTiter-Glo Luminescence Assay (Promega) on an Envision multi-plate reader (PerkinElmer). Relative viability was normalized to the respective untreated condition of each cell line using RStudio and plotted in PRISM 9 (GraphPad software). For data presentation, the mean and standard deviation (s.d.) for the four or six biological replicates of each data point in a representative experiment is presented. Sigmoidal non-linear regression models were used to compute the regression fit curves.
CRISPR/Cas9-mediated genome editing
Cells were engineered for simultaneously Cas9 and ACSL4-sgRNA expression using the lentiCRISPR v2 vector (Addgene 52961), which contains the puromycin-resistance gene and Cas9 gene driven by an EF1α promoter. For AGPS depletion, the U6-sgRNA was inserted into pCDH-CD513B-CMV-Cas9 plasmid with puromycin-resistance gene. Lentiviruses were generated from lentiviral Cas9 and sgRNA constructs in HEK-293T packaging cells. Lipofectamine 2000 (Life Technologies) was used as transfection reagents to deliver plasmids to cells following manufacturer’s instructions. Second generation V2 packaging plasmids, including pMD2.G and pPAX2, were used for lentiviral production. CD513B lentiviral packaging plasmids included pLP1, pLP2, and pLP/VSVG. Lentivirus titer was briefly assessed with Lenti-X Go-Stix Plus (TakaraBio). Target cells were infected with lentiviruses in the presence of 5 μg/mL of polybrene (Millipore). Infected cells were selected with 2 μg/mL of puromycin and propagated for further analysis. Sequences of sgRNAs used in CRISPR experiments are: ACSL4-sg1, GTGTGTCTGAGGAGATAGCG; ACSL4-sg2, GCATCATCACTCCCTTAGGT; AGPS-sg1, GTACCAATGAGTGCAAAGCG.
Immunoblotting
Adherent cells were briefly washed twice with ice-cold PBS and lysed with 1% SDS lysis buffer containing 10 mM EDTA and 50 mM Tris-HCl, pH 8.0. Lysates were collected, briefly sonicated, then incubated at 95 °C for 10 min and the protein concentrations were determined using the BCA Protein Assay kit (Pierce) following manufacturer’s instructions. Calibrated samples were diluted with 4x lithium dodecyl sulfate (LDS) sample buffer (Novus), separated by SDS–PAGE using SurePAGE 4–20% Bis-Tris protein gels (GenScript), and transferred to 0.45 μm PVDF membranes by an eBlot L1 protein-transfer system (Genscript). Membranes were blocked with 5% skim milk (Solarbio) diluted with 0.1% Tween-20-containing Tris buffered saline (TBST) and immunoblotted with antibodies against ACSL4 (Abcam, ab155282, produced in rabbit, used at 1:1000 dilution), AGPS (Abcam, ab236621, produced in rabbit, used at 1:1000 dilution), cytochrome P450 oxidoreductase (POR) (Abcam, ab180597, produced in rabbit, used at 1: 1000 dilution) and β-Actin (8H10D10, no. 3700, Cell Signaling Technologies, produced in mouse, used at 1:5,000 dilution). Membranes were then washed with TBST and incubated with IRDye 800CW goat-anti-Rabbit or 680RD donkey-anti-Mouse secondary antibodies (LiCor). Immunoblotting images were acquired on an Odyssey equipment (LiCor) according to the manufacturer’s instructions, and analyzed in the ImageStudio software (LiCor). β-Actin was used as a loading control.
Confocal microscope imaging and data analysis
For PALPv1 experiments performed on an Andor Revolution Spinning Disk Confocal, FRAPPA and TIRF microscope:
Prior to high-power laser application, steady state images were acquired to visualize the distributions of reduced and oxidized BODIPY-C11 in cells respectively. Each confocal laser was set at 3 mHz and standard gain with a 200 ms exposure. FRAPPA bleaching was done with a 405 nm laser for 5 pulses and the 488 nm confocal channel was used to collect images for another 25 seconds following the laser pulses. Images are acquired using the Metamorph software (Molecular Devices) associated with the equipment.
For PALP performed on a Nikon A1R HD25 Confocal microscope: two loops of 120 ms pulse stimulation at 100% 405 nm laser were used to stimulate a certain region of the cell. Scan speed was set at 8 Fps and pixel dwell was 11.6. The synchronized laser was in Zero HV mode. The 488 nm and 561 nm confocal channel was used to collect images before and after the laser stimulation. The grey intensity of the stimulated region was analyzed using Nikon NIS Elements software.
For PALP performed on a Nikon Spinning Disk Confocal CSU-W1 microscope: the 100% 405 nm stimulation laser was used to scan a certain region. Pulse time: 100 ms; stimulation time: 22 ms; dwell time: 20 μs; scanning time: 39 ms; disk speed: 4000 rpm. The 488 nm and 561 nm confocal channel was used to collect images before and after the laser stimulation. The grey intensity of the stimulated region was analyzed using Nikon NIS Elements software.
For PALP performed on Zeiss LSM 880 NLO multiphoton confocal microscope:
Differ from laser scanning confocal microscopy, two-photon excitation microscopy utilizes two photons with longer wavelengths to excite target molecules simultaneously. Specific z-axis was stimulated by a 800 nm two-photon laser at 100% power output on a Zeiss 880 multi-photon confocal microscope. Every 0.6 μm thickness of the 786-O cell was captured along the z-stack during the imaging acquisition stage. The 488 nm and 561 nm confocal channel was used to collect images for BODIPY-C11 fluorescence before and after the laser stimulation. The grey intensity of the stimulated region was analyzed using ZEN 3.3 blue edition.
Imaging data analysis:
Downstream image analysis was performed using Fiji ImageJ (1.52P). ROIs were imported from Metamorph (Molecular Devices), Nikon NIS Elements and ZEN blue edition software to locate laser bleaching regions. Each series measurement was analyzed using ImageJ ROI manager multi-measurement tool at a 10 px radius around the region of interest. I-max was normalized to the time point before bleaching occurred and the resulting plot was fitted with a non-linear regression to find I-max (Y0), T50, Plateau and T20 in Prism 9 (GraphPad). We recommend using the ratio of oxidized/reduced B-C11, i.e. the oxidized BODIPY-C11 intensity post-stimulation divided by the reduced BODIPY-C11 fluorescence pre-stimulation, to normalize and present the rate of BODIPY-C11 oxidation across different sample conditions.
Lipidomic analysis
Lipidomic analysis was conducted using an LC-MS system comprising a Agilent 1290 Infinity II UHPLC system tandem with Agilent 6545 Q-TOF/MS (Agilent). Lipids were extracted from cells with 0.8 mL isopropanol per sample condition (HPLC Grade; Fisher). Three replicates were analyzed for each cell line condition. Cell extracts were centrifuged at 10,000 g for 10 minutes to remove residual cellular debris prior to injecting 10 μL onto an Accucore C8 column (100 × 2.1 mm, 2.6 μm; Thermo Fisher Scientific). The column was eluted with 80% mobile phase A (95:5:0.1 vol/vol/vol 10 mM ammonium acetate/methanol/formic acid) for 1 minute followed by a linear gradient to 80% mobile-phase B (99.9:0.1 vol/vol methanol/formic acid) over 2 min, a linear gradient to 100% mobile phase B over 7 min, then 5 min at 100% mobile-phase B. MS data were acquired using electrospray ionization in both positive and negative ion mode over 200–1100 m/z. Other MS settings include: sheath gas temperature 300 °C, sheath gas flow 11 l/min, VCap 3500 V, Capillary 0.09 μA, Nozzle voltage 0 V, gas temperature 275°C, fragmentor 150 V, skimmer 65 V. Raw data were processed using Profinder 10.0 (Agilent) for peak detection, alignment and integration. Lipids were identified based on comparison to reference standards and Lipid Annotator (Agilent) and were denoted by the total number of carbons in the lipid acyl chain(s) and total number of double bonds in the lipid acyl chain(s).
For downstream lipidomics data analysis, total peak area normalization was performed among samples from the same experiment. Differential analysis was performed among annotated lipids using two-tailed Student’s T-test. For fold-change analysis, each group dataset was normalized to the mean of the wildtype cell condition for each lipid species, and the ratio between Test/WT groups was log2 transformed and presented as volcano plots. P values are false discovery rates (FDR)-adjusted for multiple-test correction using Benjamini-Hochberg correction method and presented as −log10 p adj. Heatmaps were obtained based on spearman correlation and cluster analyses using R-3.3.3.
Cryo-tissue sectioning and PALPv1 analysis of mouse tissues
For PALPv1 imaging, tissue sections of 10 μm thickness were prepared at −20 °C using a cryostat (CM1950, Leica, Germany), and thaw-mounted onto microscope slides (CITOTEST). 30 minutes prior to PALP imaging, BODIPY-C11 (Life Technologies) prepared in PBS at 5 μM was added to the surface of the tissue section and mounted with coverslips. PALPv1 imaging was performed on a Nikon A1R HD25 Confocal microscope. Each confocal laser was set at zero HV mode and standard gain with 4 loops of 120 ms exposure laser. Scan speed was set at 8 Fps and pixel dwell was 11.6. The 488 nm and 561 nm confocal channels were used for image acquisition before and after the laser stimulation.
MALDI imaging analysis on mouse tumors
10 μm-thick cryo-sectioned tumor tissue slices were thaw-mounted onto indium tin oxide coated glass slides. The slices were dried under vacuum for 30 min. After that, the sections were covered with 15 mg/ml 2,5-dihydroxybenzoic acid (DHB) in 70% methanol/ 30% H2O/ 0.003% trifluoroacetic acid (TFA) at a 0.1 ml/min speed using an HTX TM-Sprayer (HTX technologies, LLC) equipment. Mass spectrometry imaging was performed using a Bruker RapifleX MALDI-TOF system (Bruker Daltonik, Bremen, Germany) in positive mode. The tissue specimen was then raster-scanned from left to right at a spatial resolution of 20 μm, generating a mass spectrum for each measuring pixel. Ions were detected over a mass range of m/z 200–1400.
For MALDI data analysis, the signal of each pixel was normalized against the total ion count to remove variations in pixel-to-pixel intensity, along with the baseline subtraction, smoothing and alignment using SciLS Lab v.2021c Pro software. The mass signals of each m/z species detected were then presented as color intensity maps which allows the visualization of molecule distribution patterns. To identify structural features of the metabolites, the m/z value was matched with accurate theoretical mass in curated public databases including LIPID MAPS (https://www.lipidmaps.org) and Human Metabolome Database (http://www.hmdb.ca/) with 10 ppm variation tolerance.
Alkynated-polyunsaturated fatty acid treatment and imaging
Primary cortical neurons were incubated with 5 μM arachidonic acid-alkyne (AAA, Cayman 10538) for 18 hours so that AAA were incorporated into cellular membranes. Alkyne-containing lipids can be subsequently detected using Click-iT chemistry (Invitrogen C10446). Briefly, AAA-incorporated neurons were immediately fixed using 4% formaldehyde in PBS, permeabilized using 0.5% Triton X-100 in PBS and blocked by 1% BSA. Neurons were treated with Click-iT reaction cocktail and then immunostained with antibodies against cytochrome P450 oxidoreductase (POR) (Abcam, ab180597, used at 1: 300 dilution) followed by incubation with fluorescently labeled secondary antibodies (Beyotime P0180). Immunofluorescent images were acquired on a Nikon A1R confocal microscopy.
QUANTIFICATION AND STATISTICAL ANALYSIS
Data are generally expressed as mean±s.d. unless otherwise indicated. No statistical methods were used to predetermine sample sizes. Statistical significance was determined using a two-tailed, unpaired student’s T-test using Prism 9 software (GraphPad) unless otherwise indicated. Cut-off for statistical significance was set at p <= 0.05 unless otherwise indicated. Detailed statistical methods were indicated in the figure legend.
Supplementary Material
Supplementary Table 1. Lipidomics analysis of ACSL4-depleted 786-O cells, related to STAR METHOD and Supplementary Figure 4A, and lipidomics analysis of ovarian cancer cells including ES-2, OVCAR-8, OV56 and SKOV3, related to STAR METHOD, Figure 6B, and Supplementary Figure 6.
Supplementary Video 1. Time-lapse imaging of oxidized BODIPY-C11 signal in 786-O cells stimulated with PALPv1 on an Andor Revolution Spinning Disk confocal microscope, related to Figure 1B. White signal, oxidized BODIPY-C11 fluorescence. The time scale is accelerated at 3x speed.
Supplementary Video 2. Time-lapse imaging of BODIPY-C11 fluorescent signals in 786-O cells stimulated with PALPv1 on a Nikon A1R confocal microscope, related to Supplementary Figure 1B.
Supplementary Video 3. Time-lapse imaging of BODIPY-C11 fluorescent signals in 786-O cells stimulated with PALPv1 on a Nikon spinning disk microscope, related to Supplementary Figure 1C.
Supplementary Video 4. Time-lapse imaging of BODIPY-C11 fluorescent signals in ES-2 cells stimulated with PALPv1 on a Nikon A1R microscope, related to Supplementary Figure 1D.
Supplementary Video 5. Time-lapse imaging of BODIPY-C11 fluorescent signals in rat NRK cells stimulated with PALPv1 on a Nikon A1 confocal microscope, related to Supplementary Figure 1E.
Supplementary Video 6. Z-stack imaging of oxidized BODIPY-C11 signal in 786-O cells stimulated with PALPv2 on Zeiss 880 multi-photon confocal microscope, related to Figure 4B.
Highlights.
Targeted high-power laser induces lipid peroxidation on cellular membranes.
PALP signals are dependent on polyunsaturated phospholipid levels on membranes.
PALPv1 can assist estimating sensitivity to ferroptosis in cells and tissue sections.
PALPv2 enables studying lipid peroxidation on selected cellular membranes.
Acknowledgements
We thank members of the Zou lab for insightful discussions, the Westlake University Microscopic Imaging Platform for assisting the microscope training and usage, and the Mass Spectrometry and Metabolomics Platform for assisting the lipidomics analyses and MALDI-TOF mass spectrometry imaging training. This work was supported by the National Cancer Institute’s Office of Cancer Genomics Cancer Target Discovery and Development (CTD^2) initiative (grant number U01CA217848, awarded to S.L.S.), and by the Westlake Education Foundation, the Westlake Laboratory of Life Sciences and Biomedicine, the Zhejiang Provincial Natural Science Foundation of China (LQ19H090009, awarded to J-M.J.), as well as the National Institute of General Medical Sciences (grant number R35GM127045, awarded to S.L.S.).
Declaration of interests
S.L.S. serves on the Board of Directors of the Genomics Institute of the Novartis Research Foundation (“GNF”); is a shareholder and serves on the Board of Directors of Jnana Therapeutics; is a shareholder of Forma Therapeutics; is a shareholder and advises Kojin Therapeutics, Kisbee Therapeutics, Decibel Therapeutics and Eikonizo Therapeutics; serves on the Scientific Advisory Boards of Eisai Co., Ltd., Ono Pharma Foundation, Exo Therapeutics, and F-Prime Capital Partners; and is a Novartis Faculty Scholar. Kojin Therapeutics in particular explores the medical potential of cell plasticity related to ferroptosis. Y. Z. is a consultant of Keen Therapeutics. The remaining authors declare no competing interests. A patent describing the utility of PALP in clinical samples was filed by Y.Z., F.W., Z.S., N.N., Y.W., G.X. and X.W.
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References
- Badgley MA, Kremer DM, Maurer HC, DelGiorno KE, Lee H-J, Purohit V, Sagalovskiy IR, Ma A, Kapilian J, Firl CEM, et al. (2020). Cysteine depletion induces pancreatic tumor ferroptosis in mice. Science 368, 85–89. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Beatty A, Singh T, Tyurina YY, Tyurin VA, Samovich S, Nicolas E, Maslar K, Zhou Y, Cai KQ, Tan Y, et al. (2021). Ferroptotic cell death triggered by conjugated linolenic acids is mediated by ACSL1. Nat. Commun 12, 2244. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Breckenridge WC, Gombos G, and Morgan IG (1972). The lipid composition of adult rat brain synaptosomal plasma membranes. Biochim. Biophys. Acta 266, 695–707. [DOI] [PubMed] [Google Scholar]
- Calder PC (2005). Polyunsaturated fatty acids and inflammation. Biochem. Soc. Trans 33, 423–427. [DOI] [PubMed] [Google Scholar]
- Conrad M, and Pratt DA (2019). The chemical basis of ferroptosis. Nat. Chem. Biol 15, 1137–1147. [DOI] [PubMed] [Google Scholar]
- Cui W, Liu D, Gu W, and Chu B (2021). Peroxisome-driven ether-linked phospholipids biosynthesis is essential for ferroptosis. Cell Death Differ. 28, 2536–2551. [DOI] [PMC free article] [PubMed] [Google Scholar]
- D’Angelo S, Motti ML, and Meccariello R (2020). ω−3 and ω−6 Polyunsaturated Fatty Acids, Obesity and Cancer. Nutrients 12, 2751. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dixon SJ, Lemberg KM, Lamprecht MR, Skouta R, Zaitsev EM, Gleason CE, Patel DN, Bauer AJ, Cantley AM, Yang WS, et al. (2012). Ferroptosis: An Iron-Dependent Form of Nonapoptotic Cell Death. Cell 149, 1060–1072. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dixon SJ, Winter GE, Musavi LS, Lee ED, Snijder B, Rebsamen M, Superti-Furga G, and Stockwell BR (2015). Human Haploid Cell Genetics Reveals Roles for Lipid Metabolism Genes in Nonapoptotic Cell Death. ACS Chem. Biol 10, 1604–1609. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Doll S, Proneth B, Tyurina YY, Panzilius E, Kobayashi S, Ingold I, Irmler M, Beckers J, Aichler M, Walch A, et al. (2017). ACSL4 dictates ferroptosis sensitivity by shaping cellular lipid composition. Nat. Chem. Biol 13, 91–98. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Drummen GPC, van Liebergen LCM, den Kamp JAFO, and Post JA (2002). C11-BODIPY581/591, an oxidation-sensitive fluorescent lipid peroxidation probe: (micro)spectroscopic characterization and validation of methodology. Free Radic. Biol. Med 33, 473–490. [DOI] [PubMed] [Google Scholar]
- Eaton JK, Furst L, Ruberto RA, Moosmayer D, Hilpmann A, Ryan MJ, Zimmermann K, Cai LL, Niehues M, Badock V, et al. (2020). Selective covalent targeting of GPX4 using masked nitrile-oxide electrophiles. Nat. Chem. Biol 16, 497–506. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Friedmann Angeli JP, Schneider M, Proneth B, Tyurina YY, Tyurin VA, Hammond VJ, Herbach N, Aichler M, Walch A, Eggenhofer E, et al. (2014). Inactivation of the ferroptosis regulator Gpx4 triggers acute renal failure in mice. Nat. Cell Biol 16, 1180–1191. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gimple RC, Kidwell RL, Kim LJY, Sun T, Gromovsky AD, Wu Q, Wolf M, Lv D, Bhargava S, Jiang L, et al. (2019). Glioma Stem Cell–Specific Superenhancer Promotes Polyunsaturated Fatty-Acid Synthesis to Support EGFR Signaling. Cancer Discovery 9, 1248–1267. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Girotti AW (2001). Photosensitized oxidation of membrane lipids: reaction pathways, cytotoxic effects, and cytoprotective mechanisms. J. Photochem. Photobiol. B 63, 103–113. [DOI] [PubMed] [Google Scholar]
- Han X (2016). Lipidomics for studying metabolism. Nat. Rev. Endocrinol 12, 668–679. [DOI] [PubMed] [Google Scholar]
- Hangauer MJ, Viswanathan VS, Ryan MJ, Bole D, Eaton JK, Matov A, Galeas J, Dhruv HD, Berens ME, Schreiber SL, et al. (2017). Drug-tolerant persister cancer cells are vulnerable to GPX4 inhibition. Nature 551, 247–250. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hannah VC, Ou J, Luong A, Goldstein JL, and Brown MS (2001). Unsaturated fatty acids down-regulate srebp isoforms 1a and 1c by two mechanisms in HEK-293 cells. J. Biol. Chem 276, 4365–4372. [DOI] [PubMed] [Google Scholar]
- Innis SM (2007). Dietary (n-3) Fatty Acids and Brain Development. J. Nutr 137, 855–859. [DOI] [PubMed] [Google Scholar]
- Jiang X, Stockwell BR, and Conrad M (2021). Ferroptosis: mechanisms, biology and role in disease. Nat. Rev. Mol. Cell Biol 22, 266–282. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kagan VE, Mao G, Qu F, Angeli JPF, Doll S, Croix CS, Dar HH, Liu B, Tyurin VA, Ritov VB, et al. (2017). Oxidized arachidonic and adrenic PEs navigate cells to ferroptosis. Nat. Chem. Biol. 13, 81–90. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lee S-M, Lee SH, Jung Y, Lee Y, Yoon JH, Choi JY, Hwang CY, Son YH, Park SS, Hwang G-S, et al. (2020). FABP3-mediated membrane lipid saturation alters fluidity and induces ER stress in skeletal muscle with aging. Nat. Commun 11, 5661. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Magtanong L, Ko P-J, To M, Cao JY, Forcina GC, Tarangelo A, Ward CC, Cho K, Patti GJ, Nomura DK, et al. (2019). Exogenous Monounsaturated Fatty Acids Promote a Ferroptosis-Resistant Cell State. Cell Chem Biol 26, 420–432.e9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Maki KC, Eren F, Cassens ME, Dicklin MR, and Davidson MH (2018). ω−6 Polyunsaturated Fatty Acids and Cardiometabolic Health: Current Evidence, Controversies, and Research Gaps. Adv. Nutr 9, 688–700. [DOI] [PMC free article] [PubMed] [Google Scholar]
- McMillan TJ, Leatherman E, Ridley A, Shorrocks J, Tobi SE, and Whiteside JR (2008). Cellular effects of long wavelength UV light (UVA) in mammalian cells. J. Pharm. Pharmacol 60, 969–976. [DOI] [PubMed] [Google Scholar]
- Neumann EK, Djambazova KV, Caprioli RM, and Spraggins JM (2020). Multimodal Imaging Mass Spectrometry: Next Generation Molecular Mapping in Biology and Medicine. J. Am. Soc. Mass Spectrom 31, 2401–2415. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pap EH, Drummen GP, Winter VJ, Kooij TW, Rijken P, Wirtz KW, Op den Kamp JA, Hage WJ, and Post JA (1999). Ratio-fluorescence microscopy of lipid oxidation in living cells using C11-BODIPY(581/591). FEBS Lett. 453, 278–282. [DOI] [PubMed] [Google Scholar]
- Perez MA, Magtanong L, Dixon SJ, and Watts JL (2020). Dietary Lipids Induce Ferroptosis in Caenorhabditiselegans and Human Cancer Cells. Dev. Cell 54, 447–454.e4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pinot M, Vanni S, Pagnotta S, Lacas-Gervais S, Payet L-A, Ferreira T, Gautier R, Goud B, Antonny B, and Barelli H (2014). Lipid cell biology. Polyunsaturated phospholipids facilitate membrane deformation and fission by endocytic proteins. Science 345, 693–697. [DOI] [PubMed] [Google Scholar]
- Salem N, Litman B, Kim H-Y, and Gawrisch K (2001). Mechanisms of action of docosahexaenoic acid in the nervous system. Lipids 36, 945–959. [DOI] [PubMed] [Google Scholar]
- Sanjana NE, Shalem O, and Zhang F (2014) Improved vectors and genome-wide libraries for CRISPR screening. Nat Methods 11(8):783–4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shahidi F, and Ambigaipalan P (2018). Omega-3 Polyunsaturated Fatty Acids and Their Health Benefits. Annu. Rev. Food Sci. Technol 9, 345–381. [DOI] [PubMed] [Google Scholar]
- Shui S, Zhao Z, Wang H, Conrad M, and Liu G (2021). Non-enzymatic lipid peroxidation initiated by photodynamic therapy drives a distinct ferroptosis-like cell death pathway. Redox Biol 45, 102056. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Singhal R, Mitta SR, Das NK, Kerk SA, Sajjakulnukit P, Solanki S, Andren A, Kumar R, Olive KP, Banerjee R, et al. (2021). HIF-2α activation potentiates oxidative cell death in colorectal cancers by increasing cellular iron. J. Clin. Invest 131, e143691. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sud M, Fahy E, Cotter D, Brown A, Dennis E, Glass C, Murphy R, Raetz C, Russell D, and Subramaniam S (2006) LMSD: LIPID MAPS® structure database. Nucleic Acids Research 35, D527–32. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sun X, Ou Z, Chen R, Niu X, Chen D, Kang R, and Tang D (2016). Activation of the p62-Keap1-NRF2 pathway protects against ferroptosis in hepatocellular carcinoma cells. Hepatology 63, 173–184. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ubellacker JM, Tasdogan A, Ramesh V, Shen B, Mitchell EC, Martin-Sandoval MS, Gu Z, McCormick ML, Durham AB, Spitz DR, et al. (2020). Lymph protects metastasizing melanoma cells from ferroptosis. Nature 585, 113–118. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Veselinovic M, Vasiljevic D, Vucic V, Arsic A, Petrovic S, Tomic-Lucic A, Savic M, Zivanovic S, Stojic V, and Jakovljevic V (2017). Clinical Benefits of n-3 PUFA and ɤ-Linolenic Acid in Patients with Rheumatoid Arthritis. Nutrients 9, 325. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Viswanathan VS, Ryan MJ, Dhruv HD, Gill S, Eichhoff OM, Seashore-Ludlow B, Kaffenberger SD, Eaton JK, Shimada K, Aguirre AJ, et al. (2017). Dependency of a therapy-resistant state of cancer cells on a lipid peroxidase pathway. Nature 547, 453–457. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wenzel SE, Tyurina YY, Zhao J, St Croix CM, Dar HH, Mao G, Tyurin VA, Anthonymuthu TS, Kapralov AA, Amoscato AA, et al. (2017). PEBP1 Wardens Ferroptosis by Enabling Lipoxygenase Generation of Lipid Death Signals. Cell 171, 628–641.e26. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wishart DS, Feunang YD, Marcu A, Guo AC, Liang K, et al. , (2018). HMDB 4.0 — The Human Metabolome Database for 2018. Nucleic Acids Res 46(D1):D608–17. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yan B, Ai Y, Sun Q, Ma Y, Cao Y, Wang J, Zhang Z, and Wang X (2021). Membrane Damage during Ferroptosis Is Caused by Oxidation of Phospholipids Catalyzed by the Oxidoreductases POR and CYB5R1. Mol. Cell 81, 355–369.e10. [DOI] [PubMed] [Google Scholar]
- Yang WS, SriRamaratnam R, Welsch ME, Shimada K, Skouta R, Viswanathan VS, Cheah JH, Clemons PA, Shamji AF, Clish CB, et al. (2014). Regulation of ferroptotic cancer cell death by GPX4. Cell 156, 317–331. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yang WS, Kim KJ, Gaschler MM, Patel M, Shchepinov MS, and Stockwell BR (2016). Peroxidation of polyunsaturated fatty acids by lipoxygenases drives ferroptosis. Proc. Natl. Acad. Sci. U. S. A 113, E4966–E4975. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yi J, Zhu J, Wu J, Thompson CB, and Jiang X (2020). Oncogenic activation of PI3K-AKT-mTOR signaling suppresses ferroptosis via SREBP-mediated lipogenesis. Proc. Natl. Acad. Sci. U. S. A 117, 31189–31197. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yin H, Xu L, and Porter NA (2011). Free Radical Lipid Peroxidation: Mechanisms and Analysis. Chemical Reviews 111, 5944–5972. [DOI] [PubMed] [Google Scholar]
- Zhang Y, Tan H, Daniels JD, Zandkarimi F, Liu H, Brown LM, Uchida K, O’Connor OA, and Stockwell BR (2019). Imidazole Ketone Erastin Induces Ferroptosis and Slows Tumor Growth in a Mouse Lymphoma Model. Cell Chem Biol 26, 623–633.e9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zilka O, Shah R, Li B, Friedmann Angeli JP, Griesser M, Conrad M, and Pratt DA (2017). On the Mechanism of Cytoprotection by Ferrostatin-1 and Liproxstatin-1 and the Role of Lipid Peroxidation in Ferroptotic Cell Death. ACS Cent Sci 3, 232–243. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zou Y, and Schreiber SL (2020). Progress in Understanding Ferroptosis and Challenges in Its Targeting for Therapeutic Benefit. Cell Chem Biol 27, 463–471. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zou Y, Palte MJ, Deik AA, Li H, Eaton JK, Wang W, Tseng Y-Y, Deasy R, Alimova M, Dančík V, et al. (2018). HIF-2α drives an intrinsic vulnerability to ferroptosis in clear cell renal cell carcinoma. bioRxiv. [Google Scholar]
- Zou Y, Palte MJ, Deik AA, Li H, Eaton JK, Wang W, Tseng Y-Y, Deasy R, Kost-Alimova M, Dančík V, et al. (2019). A GPX4-dependent cancer cell state underlies the clear-cell morphology and confers sensitivity to ferroptosis. Nat. Commun 10, 1617. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zou Y, Li H, Graham ET, Deik AA, Eaton JK, Wang W, Sandoval-Gomez G, Clish CB, Doench JG, and Schreiber SL (2020a). Cytochrome P450 oxidoreductase contributes to phospholipid peroxidation in ferroptosis. Nat. Chem. Biol 16, 302–309. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zou Y, Henry WS, Ricq EL, Graham ET, Phadnis VV, Maretich P, Paradkar S, Boehnke N, Deik AA, Reinhardt F, et al. (2020b). Plasticity of ether lipids promotes ferroptosis susceptibility and evasion. Nature 585, 603–608. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Supplementary Table 1. Lipidomics analysis of ACSL4-depleted 786-O cells, related to STAR METHOD and Supplementary Figure 4A, and lipidomics analysis of ovarian cancer cells including ES-2, OVCAR-8, OV56 and SKOV3, related to STAR METHOD, Figure 6B, and Supplementary Figure 6.
Supplementary Video 1. Time-lapse imaging of oxidized BODIPY-C11 signal in 786-O cells stimulated with PALPv1 on an Andor Revolution Spinning Disk confocal microscope, related to Figure 1B. White signal, oxidized BODIPY-C11 fluorescence. The time scale is accelerated at 3x speed.
Supplementary Video 2. Time-lapse imaging of BODIPY-C11 fluorescent signals in 786-O cells stimulated with PALPv1 on a Nikon A1R confocal microscope, related to Supplementary Figure 1B.
Supplementary Video 3. Time-lapse imaging of BODIPY-C11 fluorescent signals in 786-O cells stimulated with PALPv1 on a Nikon spinning disk microscope, related to Supplementary Figure 1C.
Supplementary Video 4. Time-lapse imaging of BODIPY-C11 fluorescent signals in ES-2 cells stimulated with PALPv1 on a Nikon A1R microscope, related to Supplementary Figure 1D.
Supplementary Video 5. Time-lapse imaging of BODIPY-C11 fluorescent signals in rat NRK cells stimulated with PALPv1 on a Nikon A1 confocal microscope, related to Supplementary Figure 1E.
Supplementary Video 6. Z-stack imaging of oxidized BODIPY-C11 signal in 786-O cells stimulated with PALPv2 on Zeiss 880 multi-photon confocal microscope, related to Figure 4B.
Data Availability Statement
Raw videos showing the dynamics of BODIPY-C11 fluorescences in live cells are provided as multiple Supplementary Videos. Lipidomics datasets are presented in the Supplementary Tables. These data are publicly available as of the date of publication and are listed in the key resources table.
This paper does not report original code.
Any additional information required to reanalyze the data reported in this paper is available from the Lead Contact upon request.
KEY RESOURCES TABLE.
| REAGENT or RESOURCE | SOURCE | IDENTIFIER |
|---|---|---|
| Antibodies | ||
| Anti-FACL4 antibody | Abcam | Cat# ab155282, RRID: AB_2714020 |
| Anti-AGPS | Abcam | Cat# ab236621 |
| β Actin Mouse mAb | Cell Signaling Technologies | Cat# MA5–11116, RRID: AB_10985704 |
| IRDye 800CW goat-anti-Rabbit secondary antibodies | LiCor | Cat# 925–32211, RRID: AB_2651127 |
| IRDye 680RD donkey-anti-Mouse secondary antibodies | LiCor | Cat# 925–68072, RRID: AB_2814912 |
| Anti-Cytochrome P450 Reductase antibody | Abcam | Cat# ab180597 |
| Alexa Fluor 647 anti-rabbit secondary antibodies | Beyotime | Cat# P0180 |
| Chemicals, peptides, and recombinant proteins | ||
| penicillin/streptomycin | Gibco | Cat# 15140–122 |
| GlutMax | Gibco | Cat# 35050061 |
| B27 | Gibco | Cat# 17504001 |
| ML210 | Cayman | Item No. 23282 |
| 1S,3R-RSL3 | Cayman | Item No. 19288 |
| ferrostatin-1 | Cayman | Item No. 17729 |
| liproxstatin-1 | Cayman | Item No. 17730 |
| deferoxamine | Cayman | Item No. 14595 |
| BODIPY-C11 | Life Technologies | Cat# D3861 |
| DMSO | Sigma-Aldrich | Cat# D2650 |
| Oleic acid | Cayman | Item No. 90260 |
| arachidonic acid | Cayman | Item No. 10007268 |
| docosapentaenoic acid | Cayman | Item No. 90165 |
| docosahexaenoic acid | Cayman | Item No. 17950 |
| BSA | Sigma-Aldrich | Item No. 10010566 |
| puromycin | Gibco | Cat# A11138–03 |
| Lipofectamine™ 2000 Transfection Reagent | Thermo Fisher | Cat# 11668019 |
| polybrene | Sigma-Aldrich | Cat# TR-1003-G |
| lithium dodecyl sulfate sample buffer | Novus | Cat# NP0008 |
| isopropanol, HPLC Grade | Thermo Fisher | CAS: 67–63-0 |
| ammonium acetate | Sigma-Aldrich | Cat# 73594 |
| methanol | Sigma-Aldrich | Cat# 34885 |
| formic acid, HPLC LiiChropur | Supelco | Cat# 5.33002 |
| 2,5-dihydroxybenzoic acid | Sigma-Aldrich | Cat# 39319 |
| trifluoroacetic acid | Sigma-Aldrich | Cat# 80457 |
| arachidonic acid-alkyne | Cayman | Item No. 10538 |
| Critical commercial assays | ||
| Lenti-X Go-Stix Plus | TakaraBio | Cat# 631280 |
| CellTiter-Glo Luminescence Assay | Promega | Cat# G7571 |
| BCA Protein Assay kit | Pierce | Cat# 23225 |
| Click-iT® Lipid Peroxidation Detection with Linoleamide Alkyne | Invitrogen | Cat# C10446 |
| Deposited data | ||
| Lipidomics data of ACSL4-depleted 786-O cell | This paper | Supplementary Table1 |
| Lipidomics data of ES-2, OVCAR8, SKOV3 and OV56 cell lines | This paper | Supplementary Table2 |
| Experimental models: Cell lines | ||
| Human clear-cell renal cell carcinoma cell: 786-O | ATCC | Cat# CRL-1932, RRID: CVCL_1051 |
| Human clear-cell ovarian carcinoma cell: ES-2 | ATCC | Cat# CRL-1978, RRID: CVCL_3509 |
| Human melanoma cell: WM-793 | ATCC | Cat# CRL-2806, RRID: CVCL_8787 |
| Human urothelial carcinoma cell: BFTC-909 | DSMZ | Cat# ACC-367, RRID: CVCL_1084 |
| Human ovarian cancer cell: OVCAR-8 | NCI-DTP | Cat# OVCAR-8, RRID: CVCL_1629 |
| Human ovarian cancer cell: OV56 | ECACC | Cat# 96020759, RRID: CVCL_2673 |
| Human ovarian cancer cell: SKOV3 | NICR | 4201PAT-CCTCC01017 |
| Human hepatoma cell: HepG2 | NICR | 1101HUM-PUMC000035 |
| Normal rat kidney (NRK) epithelial cell line | NICR | 1101RAT-PUMC000198 |
| L929 mouse C3H/An connective tissue cell line | NICR | 1101MOU-PUMC000403 |
| HEK-293T | NICR | 1101HUM-PUMC000010 |
| Experimental models: Organisms/strains | ||
| Mouse: C57BL/6J | The Jackson Laboratory | N/A |
| Mouse: athymic nude mice | The Jackson Laboratory | N/A |
| Oligonucleotides | ||
| ACSL4-sg1, GTGTGTCTGAGGAGATAGCG | This paper | N/A |
| ACSL4-sg2, GCATCATCACTCCCTTAGGT | This paper | N/A |
| AGPS-sg1, GTACCAATGAGTGCAAAGCG | This paper | N/A |
| Recombinant DNA | ||
| LentiCRISPR V2 plasmid | (Sanjana et al., 2014) | Addgene, Cat# 52961 |
| psPAX2 plasmid | Trono Lab Packaging and Envelope Plasmids (unpublished) | Addgene, Cat# 12260 |
| pMD2.G plasmid | Trono Lab Packaging and Envelope Plasmids (unpublished) | Addgene, Cat# 12259 |
| Software and algorithms | ||
| ImageStudio software | LiCor | https://www.licor.com/bio/image-studio/ |
| Metamorph software | Molecular Devices, Andor | |
| ZEN 3.3 blue edition | Zeiss Microscope | https://www.zeiss.com/microscopy/int/products/microscope-software/zen.html |
| ImageJ 1.52P | Fiji | https://imagej.net/software/fiji/ |
| GraphPad Prism | GraphPad software Inc | https://www.graphpad.com/scientific-software/prism/ |
| Nikon NIS Elements software | Nikon NIS Microscope | https://www.microscope.healthcare.nikon.com/products/software/nis-elements/viewer |
| Profinder 10.0 | Agilent | N/A |
| Lipid Annotator | Agilent | N/A |
| LIPID MAPS | (Sud et al., 2006) | https://www.lipidmaps.org |
| Human Metabolome Database | (Wishart et al., 2018) | http://www.hmdb.ca/ |
| Adobe Illustrator | Adobe | www.adobe.com |
| Spearman correlation and cluster analyses | R-3.3.3 | |
| Biorender | https://biorender.com/ | |
| SciLS Lab v.2021c Pro software | Bruker Daltonik | www.bruker.com |
| Flex control software | Bruker Daltonik | www.bruker.com |
| Flex imaging software | Bruker Daltonik | www.bruker.com |
| Other | ||
| Accucore C8 column (100 × 2.1 mm, 2.6 μm) | Thermo Fisher Scientific | Cat# 17226–102130 |
| RPMI 1640 | Gibco | Cat# C11875500BT |
| DMEM | Gibco | Cat# C11995500BT |