Abstract
Epitranscriptomic reader, writer, and eraser (RWE) proteins recognize, install, and remove modified nucleosides in RNA, which are known to play crucial roles in RNA processing, splicing, and stability. Here, we established a liquid chromatography-parallel-reaction monitoring (LC–PRM) method for high-throughput profiling of a total of 152 epitranscriptomic RWE proteins. We also applied the LC–PRM method, in conjunction with stable isotope labeling by amino acids in cell culture (SILAC), to quantify these proteins in two pairs of matched parental/radioresistant breast cancer cells (i.e., MDA-MB-231 and MCF-7 cells and their corresponding radioresistant C5 and C6 clones), with the goal of assessing the roles of these proteins in radioresistance. We found that eight epitranscriptomic RWE proteins were commonly altered by over 1.5-fold in the two pairs of breast cancer cells. Among them, TRMT1 (an m2,2G writer) may play a role in promoting breast cancer radioresistance due to its clinical relevance and its correlation with DNA repair gene sets. To our knowledge, this is the first report of a targeted proteomic method for comprehensive quantifications of epitranscriptomic RWE proteins. We envision that the LC–PRM method is applicable for studying the roles of these proteins in the metastatic transformation of cancer and therapeutic resistance of other types of cancer in the future.
Graphical Abstract
Unlike the extensively studied DNA methylation and histone post-translational modifications, the investigations about RNA modifications did not gain wide attention in the scientific community until the availability of a high-throughput sequencing method rendered transcriptome-wide profiling of N6-methyladenosine (m6A) in 2012.1 RNA is known to contain more than 170 types of modifications, among which m6A is the most abundant internal modification in mRNA.2 m6A-modifying enzymes (“writers” and “erasers”) install or remove m6A, whereas m6A-binding proteins (“readers”) recognize m6A to confer downstream effects. m6A is involved in regulating various cellular processes, including mRNA stability, splicing, translation, and decay.3–6 Aside from m6A, other RNA modifications also regulate biological processes through their reader, writer, and eraser (RWE) proteins. For instance, ALYREF and YTHDF2, which are 5-methylcytidine (m5C) reader proteins, modulate mRNA export and rRNA maturation, respectively.7,8 In addition, NSUN2 (m5C writer) and YBX1 (m5C reader) drive the pathogenesis of human bladder urothelial carcinoma by targeting the m5C site in the mRNA of the HDGF gene.9
Breast cancer represents the second most common cancer among women in the United States. Radiation therapy harnesses ionizing radiation to eliminate local malignant cells and prevent cancer recurrence. It delivers high-energy X-rays to target tissues and elicits DNA damage in rapidly dividing cancer cells. Although more than 83% of breast cancer patients benefit from radiation therapy,10 some patients suffer from tumor recurrence due to the development of resistance to radiation therapy.11 Many genes involved in DNA damage repair and cell cycle checkpoints have been documented to modulate radioresistance, including AKT, HER2, BRCA2, CDK1, and CHK1.12–15
Several studies also unveiled the functions of m6A RWE proteins in modulating radioresistance ofcancer cells. METTL3, the catalytic subunit of the major m6A writer complex, promotes radioresistance in glioblastoma by regulating m6A modification of SOX2 mRNA and enhancing its stability.16 m6A eraser, ALKBH5 augments radioresistance by modulating homologous recombination in glioblastoma.17 m6A reader YTHDC2 promotes radioresistance of nasopharyngeal carcinoma via enhancing IGF1R mRNA and activating the IGF1R-AKT/S6 i signaling pathway.18 Little, however, is known about the roles of other epitranscriptomic RWE proteins, such as those for N1-methyladenosine (m1A), m5C, and pseudouridine (Ψ) in RNA, in modulating the sensitivity of cancer cells to radiation therapy.
Parallel-reaction monitoring (PRM)-based targeted proteomics, which can be performed on hybrid quadrupole-Orbitrap or quadrupole time-of-flight (TOF) mass spectrometers, can be used to quantify hundreds of peptides in complex sample matrixes in a single LC–MS/MS run.19 Since the MS/MS are acquired on a high-resolution mass analyzer, PRM offers highly selective and reliable identification and quantification of target peptides. Moreover, the mass spectrometer can be programed to collect MS/MS of precursor ions in predefined retention time windows with the use of normalized retention time (iRT), which provides improved throughput of the LC–PRM method.20
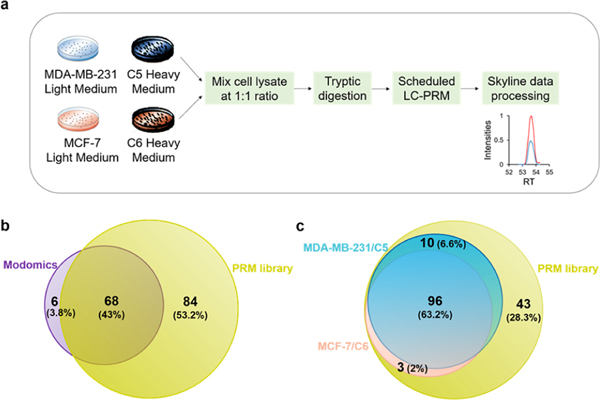
To investigate systematically the roles of epitranscriptomic RWE proteins in modulating radioresistance in breast cancer, we established an LC–PRM method, coupled with stable isotope labeling by amino acids in cell culture (SILAC), to examine the differences in expression levels of the proteins in MDA-MB-231 and MCF-7 breast cancer cells relative to their corresponding radioresistant C5 and C6 clones (Figure 1a). We first developed a Skyline21 PRM library, which includes all the 68 human epitranscriptomic RWE proteins deposited in the Modomics database2 and another 84 RWE proteins retrieved from several recent review articles (Figure 1b, Table S1).22–27 Each RWE protein is represented by two or three unique peptides, whose MS/MS were acquired from previously published shotgun proteomic analyses and imported into the Skyline library.28
Figure 1.
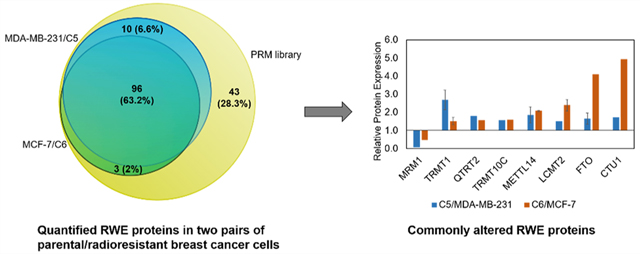
LC–PRM method for uncovering alterations in expression of epitranscriptomic RWE proteins associated with the development of radioresistance. (a) A SILAC-based LC–PRM workflow. The parental cells (i.e., MDA-MB-231 and MCF-7) and their radioresistant counterparts (i.e., C5 and C6) were labeled in light- or heavy- amino acid-containing media for over six cell doubling times. In the forward SILAC labeling experiments, light-isotope-labeled C5 and C6 cell lysates were mixed at a 1:1 ratio (by mass) with heavy-isotope-labeled MDA-MB-231 and MCF-7 cell lysates, respectively. In the reverse SILAC labeling experiments, light-isotope-labeled MDA-MB-231 and MCF-7 cell lysates were mixed at a 1:1 ratio (by mass) with heavy-isotope-labeled C5 and C6 cell lysates, respectively. The mixed cell lysate was tryptic digested and subjected to LC–PRM analysis. Data were processed using Skyline. (b,c) Venn diagrams showing the number and percentage of human epitranscriptomic RWE proteins deposited in the Modomics database (purple) compared with those included in the PRM library of this study (yellow) (b) and illustrating the number and percentage of quantified epitranscriptomic RWE proteins in MDA-MB-231/C5 and MCF-7/C6 pairs of breast cancer cells from LC–PRM analyses, compared with those deposited in the PRM library (c). Blue and pink circles in parts b and c designate the numbers of quantified epitranscriptomic RWE proteins in MDA-MB-231/C5 and MCF-7/C6 pairs of breast cancer cells, respectively.
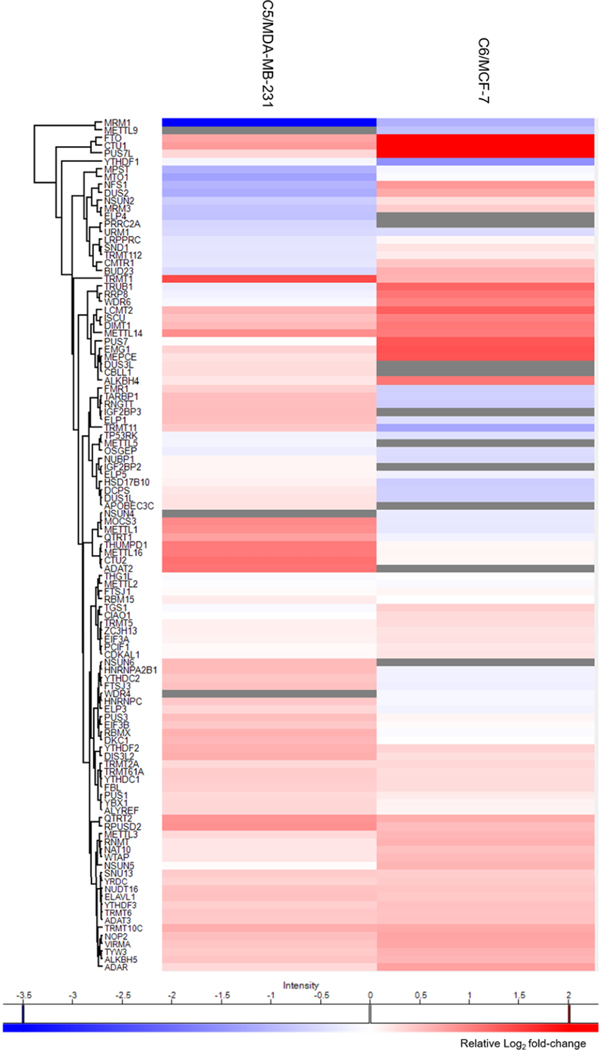
To achieve high-throughput analysis of these proteins, we employed scheduled LC–PRM with a 7 min retention time window and a maximum of 40 concurrent precursor ions. In this vein, iRT of each peptide was derived from the linear regression of RT with iRT by analyzing a tryptic digestion mixture of bovine serum albumin (BSA) under the same chromatographic conditions. With this method, the 152 epitranscriptomic RWE proteins (i.e., 444 unique peptides, and 888 precursor ions for SILAC) could be monitored in three LC–MS/MS runs with a 125 min gradient. The LC–PRM analysis enabled the quantifications of 106 and 99 epitranscriptomic RWE proteins from two forward and two reverse SILAC experiments in the MDA-MB-231/C5 and the MCF-7/C6 pairs, respectively, accounting for approximately 70% and 65% of proteins in the PRM library (Figure 1c). The quantification result of each RWE protein was calculated from the average ratios of all detected tryptic peptides of the protein, where the ratio of each peptide was calculated in Skyline based on LC–PRM results from the four replicates of SILAC experiments. A total of 96 epitranscriptomic RWE proteins were commonly quantified in the two pairs of cell lines. We also performed hierarchical clustering analysis to illustrate the differential expression of the quantified epitranscriptomic RWE proteins in the radioresistant C5 and C6 lines relative to the corresponding parental MDA-MB-231 and MCF-7 lines (Figure 2). Such analysis revealed similarities and differences in alterations in expression of epitranscriptomic RWE proteins accompanied by the development of radioresistance in the two breast cancer cell lines 1 (Figure 2).
Figure 2.
Hierarchical clustering displaying the Log2 transformed expression fold differences of epitranscriptomic RWE proteins in radioresistant C5 cells relative to parental MDA-MB-231 cells and radioresistant C6 cells relative to parental MCF-7 cells. The expression fold differences were averaged from two forward and two reverse SILAC experiments. Hierarchical clustering was generated using Perseus, where red and blue boxes designate proteins up- and down-regulated in radioresistant breast cancer cells compared with the corresponding parental lines, respectively; gray boxes represent missed data. Genes were clustered using the Euclidean distance.
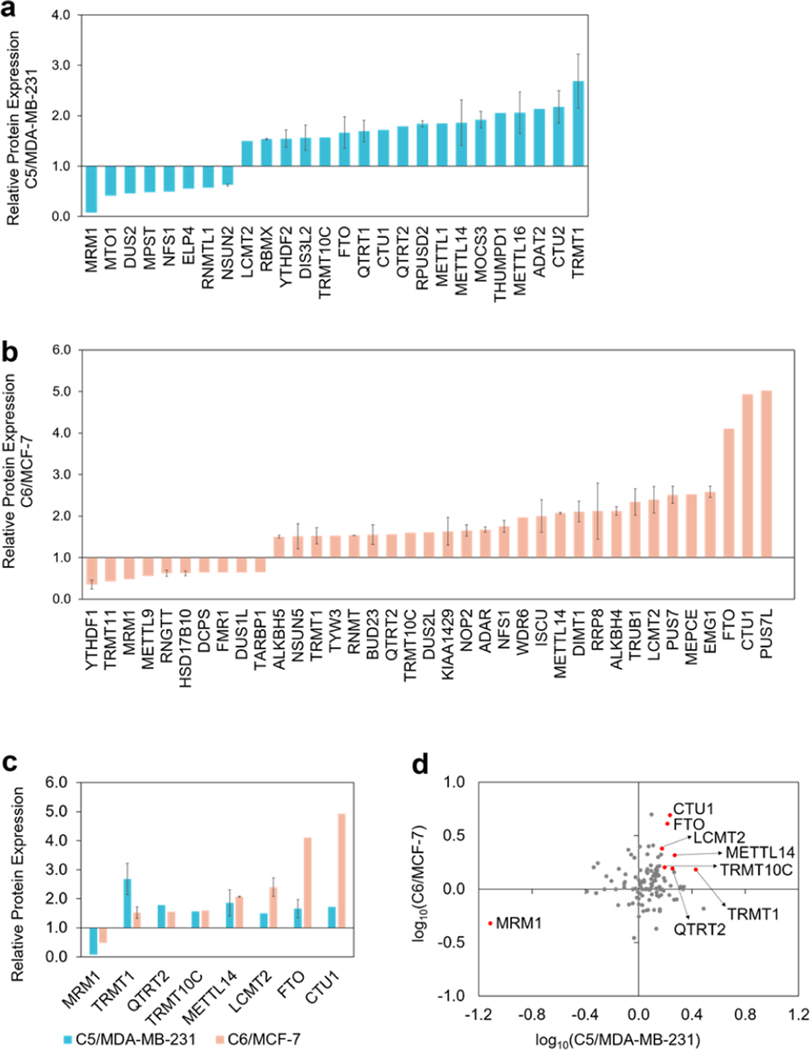
Our LC–PRM data revealed that 8 and 11 epitranscriptomic RWE proteins were down-regulated by more than 1.5-fold, and 18 and 27 epitranscriptomic RWE proteins were up-regulated by over 1.5-fold in the radioresistant C5 and C6 lines relative to their corresponding parental lines, respectively (Figure 3a,b). Gene ontology (GO) analysis of these differentially expressed proteins showed that the up-regulated epitranscriptomic RWE proteins are mainly involved in tRNA modification, tRNA processing, and rRNA base methylation (Figure S1a). The down-regulated epitranscriptomic RWE proteins play roles in tRNA methylation, the oxidation-reduction process, and tRNA 1 dihydrouridine synthesis (Figure S1a). In this context, it is worth noting that over 100 types of modifications have been detected in tRNA,2 including Ψ, m1A, N1-methylguanosine (m1G), and N6-threonyl-carbamoyl-adenosine (t6A), where many tRNA modifications regulate the stabilities of tRNA.29,30
Figure 3.
(a,b) Bar graphs depicting the LC–PRM results for those epitranscriptomic RWE proteins with expression differences of over 1.5-fold or less than 0.67-fold in radioresistant cells relative to the corresponding parental cells. (c) Bar graphs illustrating epitranscriptomic RWE proteins that were commonly altered by over 1.5-fold in the two pairs of matched breast cancer cells. (d) Scatter plot displaying log10 transformed expression ratios of the quantified epitranscriptomic RWE proteins in the two pairs of matched breast cancer cells. Eight commonly altered RWE proteins from both pairs by over 1.5-fold were labeled in red dots. The data in parts a-c display the means and standard deviations of the quantified ratios of different peptides representing a specific epitranscriptomic RWE protein, where the ratio of each peptide was averaged from the quantification results of two forward and two reverse SILAC experiments. Error bars were displayed for those epitranscriptomic RWE proteins with more than one peptide being quantified.
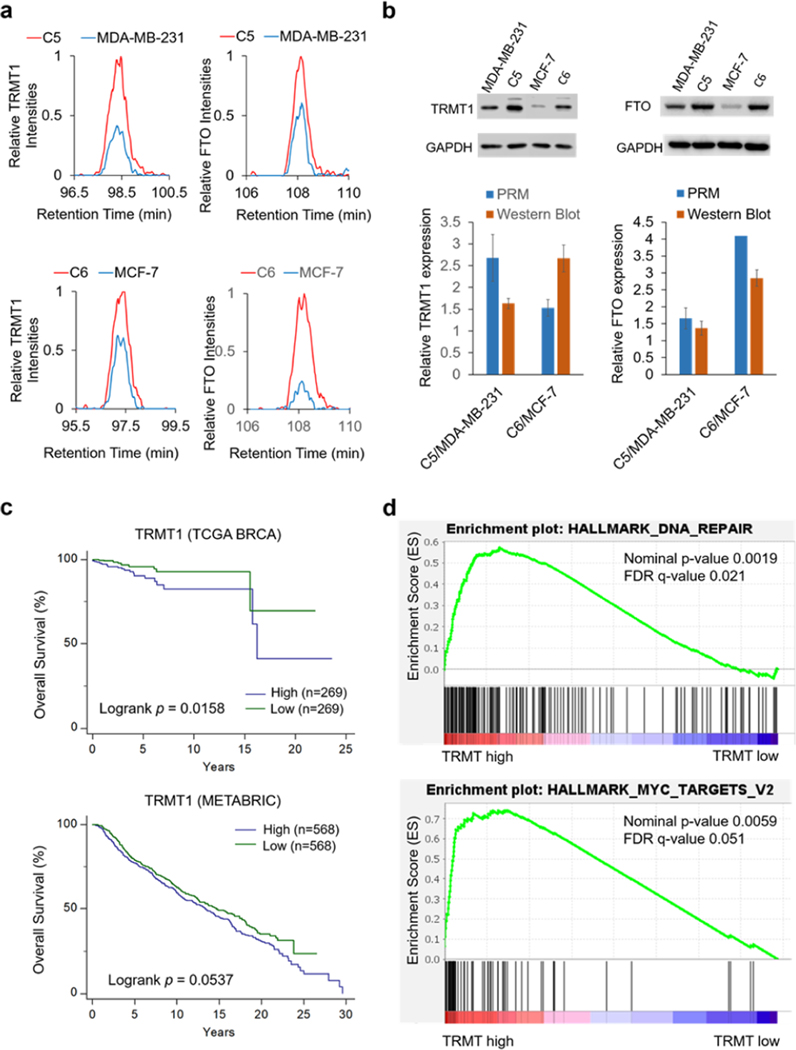
Among the epitranscriptomic RWE proteins that are up- or down-regulated by at least 1.5-fold, eight were commonly altered in both pairs of breast cancer cell lines (Figure 3c,d). For instance, MRM1 was pronouncedly down-regulated, whereas FTO and CTU1 were markedly up-regulated in radioresistant lines compared to parental lines in both pairs of breast cancer cells (Figure 2). Figure 4a illustrates the PRM traces of representative peptides from TRMT1 and FTO, two ofthe eight commonly altered proteins, in two pairs of matched radioresistant/parental breast cancer cells. The up-regulations of TRMT1 and FTO in the radioresistant cells were further validated by Western blot analysis (Figure 4b).
Figure 4.
(a) PRM traces of representative peptides, FALEVPGLR from TRMT1 and FTVPWPVK from FTO, in C5/MDA-MB-231 and C6/MCF-7 pairs of breast cancer cells. (b) Western blots of TRMT1 and FTO proteins in MDA-MB-231 and MCF-7 pairs of radioresistant/parental breast cancer cells. Relative quantification results of TRMT1 and FTO obtained from PRM and Western blot analysis were shown. The PRM results represent the mean and standard deviation of quantification results of different peptides from a given epitranscriptomic RWE protein, where the ratio of each peptide in the radioresistant over parental cells was averaged from the quantification data of two forward and two reverse SILAC experiments. Western blot data represent the mean and standard deviation ofresults obtained from three separate experiments. (c) Kaplan-Meier survival analysis of TRMT1 gene in the TCGA-BRCA and METABRIC cohort of patients who received radiation therapy. Breast cancer patients were stratified by the mRNA expression of TRMT1 using its median value as a cutoff. The survival plots and log-rank p-values were generated and calculated by using MedCalc software. (d) GSEA enrichment plots were generated using GSEA 4.1.0, where the number of permutations was set at 1000.
Our proteomic results showed that the established LC–PRM method coupled with SILAC affords a highly efficient, selective, sensitive, and reproducible peptide quantification. The efficiency of the method is manifested by its high throughput, where 888 precursor ions of 444 tryptic peptides derived from the 152 epitranscriptomic RWE proteins could be monitored in three LC–MS/MS runs. Additionally, the high consistency of the quantification results of TRMT1 and FTO obtained from PRM and Western blot analyses underscores the high accuracy of the method. Moreover, the relatively high coverage (i.e., 70% and 65%) of the epitranscriptomic RWE proteins in the library indicates the high sensitivity of the PRM method. The PRM method is also highly reproducible, as reflected by the small mean relative standard deviations of the quantification results obtained from two forward and two reverse SILAC experiments, i.e., 11.7% and 9.1% in the MDA-MB-231/C5 and MCF-7/C6 and pairs of breast cancer cells, respectively (Table S2). In this context, it is worth noting that our PRM method does not take into account post-translational modifications (PTMs) in the peptides employed for the quantifications of the epitranscriptomic RWE proteins. Hence, differences in PTMs between the radioresistant and parental breast cancer cells may contribute, in part, to variations in quantification results obtained from different peptides of the same protein (Table S2).
Considering that the above-mentioned proteomic results were acquired from breast cancer cell lines derived from two patients, we next asked if the findings could be extended to breast cancer patients in general. To this end, we performed Kaplan-Meier survival analyses in two breast cancer patient cohorts, i.e., The Cancer Genome Atlas-Breast Invasive Carcinoma (TCGA-BRCA) and Molecular Taxonomy of Breast Cancer International Consortium (METABRIC). We placed our emphasis on patients who received radiation therapy and explored the correlation between the mRNA expression level of each commonly altered epitranscriptomic RWE protein and patient survival. Our results showed that a higher level of mRNA expression of TRMT1 is significantly correlated with poorer survival of breast cancer patients who received radiation therapy in both TCGA-BRCA and METABRIC cohorts (Figure 4c). This result is in keeping with our proteomic data showing that TRMT1 is up-regulated in C5 and C6 cells compared with parental MDA-MB-231 and MCF-7 cells. For the other commonly altered epitranscriptomic RWE proteins in both pairs, only the Kaplan-Meier survival analysis of CTU1 gene in the METABRIC cohorts who received radiation therapy corroborates with proteomics results (Figure S1b). The lack of correlation for other proteins may be due to the differences in the mRNA and protein expression levels of epitranscriptomic RWE proteins and/or the heterogeneity of breast cancer.31
To explore the potential mechanism of TRMT1 in radioresistant breast cancer, we carried out gene set enrichment analysis (GSEA). TCGA-BRCA data set was stratified by the high and low mRNA expression of TRMT1 using its median value as a cutoff. Upon performing GSEA between the stratified TCGA data set and the hallmark gene sets downloaded from the GSEA Molecular Signatures Database,32 we observed that, among 23 gene sets, 4 are significantly (at FDR < 25%) up-regulated in the high-TRMT1-expression group. The DNA repair gene set is the most significantly enriched (Figure 4d). Since radioresistance is known to be associated with the enhanced ability to repair radiation-induced DNA damage,11 this finding again suggests a role of TRMT1 in promoting radioresistance. Additionally, two other hallmark gene sets, i.e., Myc_target_V2 (Figure 4d) and Myc_target_V1 (Figure S1c), were also enriched significantly with the high-TRMT1-expression group; hence, TRMT1 may be associated with Myc target genes. Moreover, the hallmark gene set UV_response_up, i.e., up-regulated in response to ultraviolet (UV) radiation, was also associated with high expression of TRMT1 (Figure S1d).
Radiation therapy is known to enhance cancer metastasis through activating epithelial-mesenchymal transition (EMT) transcription factors, including Snail, Slug, ZEB1, and ZEB.33 Additionally, radioresistant breast cancer cells exhibit increased metastatic potential,34 and breast cancer distant metastasis was shown to promote resistance to radiation therapy.35 Based on the observed co-occurrence between metastasis and radioresistance, several reports interrogated their cross-regulation and revealed several common pathways, including PI3K/AKT/mTOR, MAPK, Wnt/β-catenin, NF-kB, EMT, and reactive oxygen species scavenging.36–40
TRMT1 dimethylates the N2 position of guanosine 26 in most tRNAs to give m2,2G. It was documented that the urinary level of m2,2G was elevated in 35.1% or 57% in two cohorts of metastatic breast cancer patients.41,42 TRMT1 is the only known writer of m2,2G in humans;43 thus, the augmented levels of m2,2G in metastatic breast cancer patients also suggest a role of TRMT1 in the metastatic transformation of breast cancer.
In summary, we established, for the first time, a high-throughput scheduled LC–PRM method for profiling simultaneously a total of 152 epitranscriptomic RWE proteins. We also employed this method to explore the roles of these proteins in radioresistance in breast cancer cells, and we found that eight epitranscriptomic RWE proteins were commonly altered by over 1.5-fold in the MDA-MB-231/C5 and MCF-7/C6 pairs of breast cancer cells. Among them, TRMT1 may play a role in promoting radioresistance in breast cancer and be involved in breast cancer metastatic transformation. Thus, TRMT1 could be a target for overcoming radioresistance in breast cancer therapy. In addition, other differentially expressed epitranscriptomic RWE proteins in matched radioresistant/parental breast cancer cell lines revealed from this study may provide a comprehensive understanding of epitranscriptomic RWE proteins in modulating radiation sensitivity in breast cancer. Moreover, we envision that the LC–PRM method developed in this study can also be employed to examine, in the future, the roles of epitranscriptomic RWE proteins in the metastatic transformation of cancer and therapeutic resistance of other types of cancer.
Supplementary Material
ACKNOWLEDGMENTS
This work was supported by the National Institutes of Health (R35 Grant ES031707). The authors also would like to thank Prof. Jian-Jian Li of University of California, Davis, for providing the radioresistant cell lines used in the current study.
Footnotes
The authors declare no competing financial interest.
All the LC–MS/MS raw files and Skyline PRM library were deposited to the ProteomeXchange Consortium via the PRIDE44 partner repository with the data set identifier PXD030387.
ASSOCIATED CONTENT
Supporting Information
The Supporting Information is available free of charge at https://pubs.acs.org/doi/10.1021/acs.analchem.1c05441.
Detailed experimental procedures; Figure S1, Gene ontology (GO) biological pathway (BP) analysis and Kaplan–Meier survival analysis; and Table S1, list of epitranscriptomic RWE proteins included in the PRM library (PDF)
Table S2, expression ratios of epitranscriptomic RWE proteins in MDA-MB-231/C5 and MCF-7/C6 cells (XLSX)
Complete contact information is available at: https://pubs.acs.org/10.1021/acs.analchem.1c05441
Contributor Information
Tianyu F. Qi, Environmental Toxicology Graduate Program, University of California Riverside, Riverside, California 92521-0403, United States
Weili Miao, Department of Chemistry, University of California Riverside, Riverside, California 92521-0403, United States.
Yinsheng Wang, Environmental Toxicology Graduate Program and Department of Chemistry, University of California Riverside, Riverside, California 92521-0403, United States.
REFERENCES
- (1).Dominissini D; Moshitch-Moshkovitz S; Schwartz S; Salmon-Divon M; Ungar L; Osenberg S; Cesarkas K; Jacob-Hirsch J; Amariglio N; Kupiec M; Sorek R; Rechavi G. Nature 2012, 485, 201–206. [DOI] [PubMed] [Google Scholar]
- (2).Boccaletto P; Machnicka MA; Purta E; Piatkowski P; Baginski B; Wirecki TK; de Crécy-Lagard V; Ross R; Limbach PA; Kotter A; Helm M; Bujnicki JM Nucleic Acids Res. 2018, 46, D303–d307. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (3).Wang X; Lu Z; Gomez A; Hon GC; Yue Y; Han D; Fu Y; Parisien M; Dai Q; Jia G; Ren B; Pan T; He C. Nature 2014, 505, 117–120. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (4).Xiao W; Adhikari S; Dahal U; Chen YS; Hao YJ; Sun BF; Sun HY; Li A; Ping XL; Lai WY; Wang X; Ma HL; Huang CM; Yang Y; Huang N; Jiang GB; Wang HL; Zhou Q; Wang XJ; Zhao YL; et al. Mol. Cell 2016, 61, 507–519. [DOI] [PubMed] [Google Scholar]
- (5).Liu T; Wei Q; Jin J; Luo Q; Liu Y; Yang Y; Cheng C; Li L; Pi J; Si Y; Xiao H; Li L; Rao S; Wang F; Yu J; Yu J; Zou D; Yi P. Nucleic Acids Res. 2020, 48, 3816–3831. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (6).Shi H; Wang X; Lu Z; Zhao BS; Ma H; Hsu PJ; Liu C; He C. Cell Res. 2017, 27, 315–328. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (7).Yang X; Yang Y; Sun BF; Chen YS; Xu JW; Lai WY; Li A; Wang X; Bhattarai DP; Xiao W; Sun HY; Zhu Q; Ma HL; Adhikari S; Sun M; Hao YJ; Zhang B; Huang CM; Huang N; Jiang GB; et al. Cell Res. 2017, 27, 606–625. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (8).Dai X; Gonzalez G; Li L; Li J; You C; Miao W; Hu J; Fu L; Zhao Y; Li R; Li L; Chen X; Xu Y; Gu W; Wang Y. Anal. Chem. 2020, 92, 1346–1354. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (9).Chen X; Li A; Sun BF; Yang Y; Han YN; Yuan X; Chen RX; Wei WS; Liu Y; Gao CC; Chen YS; Zhang M; Ma XD; Liu ZW; Luo JH; Lyu C; Wang HL; Ma J; Zhao YL; Zhou FJ; et al. Nat. Cell Biol. 2019, 21, 978–990. [DOI] [PubMed] [Google Scholar]
- (10).Delaney G; Jacob S; Featherstone C; Barton M. Cancer 2005, 104, 1129–1137. [DOI] [PubMed] [Google Scholar]
- (11).Boreham DR; Mitchel RE J. Radiat. Res. 1993, 135, 365–371. [PubMed] [Google Scholar]
- (12).Guo L; Xiao Y; Fan M; Li JJ; Wang YJ Proteome Res. 2015, 14, 193–201. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (13).Xia F; Taghian DG; DeFrank JS; Zeng ZC; Willers H; Iliakis G; Powell SN Proc. Natl Acad. Sci. U. S. A. 2001, 98, 8644–8649. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (14).Zhou H; Kim YS; Peletier A; McCall W; Earp HS; Sartor CI Int. J. Radiat. Oncol Biol. Phys. 2004, 58, 344–352. [DOI] [PubMed] [Google Scholar]
- (15).Liang K; Jin W; Knuefermann C; Schmidt M; Mills GB; Ang KK; Milas L; Fan Z. Mol. Cancer Ther. 2003, 2, 353–360. [PubMed] [Google Scholar]
- (16).Visvanathan A; Patil V; Arora A; Hegde AS; Arivazhagan A; Santosh V; Somasundaram K. Oncogene 2018, 37, 522–533. [DOI] [PubMed] [Google Scholar]
- (17).Kowalski-Chauvel A; Lacore MG; Arnauduc F; Delmas C; Toulas C; Cohen-Jonathan-Moyal E; Seva C. Cancers 2021, 13, 40. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (18).He JJ; Li Z; Rong ZX; Gao J; Mu Y; Guan YD; Ren XX; Zi YY; Liu LY; Fan Q; Zhou M; Duan YM; Zhou Q; Deng YZ; Sun LQ Front. Oncol. 2020, 10, 1166. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (19).Ronsein GE; Pamir N; von Haller PD; Kim DS; Oda MN; Jarvik GP; Vaisar T; Heinecke JW Journal of proteomics 2015, 113, 388–399. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (20).Escher C; Reiter L; MacLean B; Ossola R; Herzog F; Chilton J; MacCoss MJ; Rinner O. Proteomics 2012, 12, 1111–1121. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (21).MacLean B; Tomazela DM; Shulman N; Chambers M; Finney GL; Frewen B; Kern R; Tabb DL; Liebler DC; MacCoss MJ Bioinformatics 2010, 26, 966–968. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (22).de Crécy-Lagard V; Boccaletto P; Mangleburg CG; Sharma P; Lowe TM; Leidel SA; Bujnicki JM Nucleic Acids Res. 2019, 47, 2143–2159. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (23).Mathlin J; Le Pera L; Colombo T. Int. J. Mol. Sci. 2020, 21, 4684. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (24).Kadumuri RV; Janga SC Trends Mol. Med. 2018, 24, 886–903. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (25).Suzuki T; Suzuki T. Nucleic Acids Res. 2014, 42, 7346–7357. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (26).Boriack-Sjodin PA; Ribich S; Copeland RA Nature reviews. Drug discovery 2018, 17, 435–453. [DOI] [PubMed] [Google Scholar]
- (27).Begik O; Lucas MC; Liu H; Ramirez JM; Mattick JS; Novoa EM Genome Biol. 2020, 21, 97. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (28).Miao WL; Li L; Wang YS Anal. Chem. 2018, 90, 6835–6842. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (29).Liu F; Clark W; Luo G; Wang X; Fu Y; Wei J; Wang X; Hao Z; Dai Q; Zheng G; Ma H; Han D; Evans M; Klungland A; Pan T; He C. Cell 2016, 167, 1897. [DOI] [PubMed] [Google Scholar]
- (30).Yarian CS; Basti MM; Cain RJ; Ansari G; Guenther RH; Sochacka E; Czerwinska G; Malkiewicz A; Agris PF Nucleic Acids Res. 1999, 27, 3543–3549. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (31).Holm J; Eriksson L; Ploner A; Eriksson M; Rantalainen M; Li J; Hall P; Czene K. Cancer Res. 2017, 77, 3708–3717. [DOI] [PubMed] [Google Scholar]
- (32).Subramanian A; Tamayo P; Mootha VK; Mukherjee S; Ebert BL; Gillette MA; Paulovich A; Pomeroy SL; Golub TR; Lander ES; Mesirov JP Proc. Natl. Acad. Sci. U.S.A. 2005, 102, 15545–15550. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (33).Zhang P; Sun Y; Ma L. Cell cycle (Georgetown, Tex.) 2015, 14, 481–487. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (34).Gray M; Turnbull AK; Ward C; Meehan J; Martfnez-Pérez C; Bonello M; Pang LY; Langdon SP; Kunkler IH; Murray A; Argyle D. Radiat. Oncol. 2019, 14, 64. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (35).Hara T; Iwadate M; Tachibana K; Waguri S; Takenoshita S; Hamada N. Strahlenther. Onkol 2017, 193, 848–855. [DOI] [PubMed] [Google Scholar]
- (36).Che Y; Li Y; Zheng F; Zou K; Li Z; Chen M; Hu S; Tian C; Yu W; Guo W; Luo M; Deng W; Zou L. Cancer Lett. 2019, 452, 1–13. [DOI] [PubMed] [Google Scholar]
- (37).You G-R; Chang JT; Li Y-L; Chen Y-J; Huang Y-C; Fan K-H; Chen Y-C; Kang C-J; Cheng A-J Frontiers in oncology 2021, 11, 681717–681717. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (38).Xu T; Zeng Y; Shi L; Yang Q; Chen Y; Wu G; Li G; Xu SJ Exp. Clin. Cancer Res. 2020, 39, 183. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (39).Hou Y; Liang H; Rao E; Zheng W; Huang X; Deng L; Zhang Y; Yu X; Xu M; Mauceri H; Arina A; Weichselbaum RR; Fu YX Immunity 2018, 49, 490–503. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (40).Tsao T; Beretov J; Ni J; Bai X; Bucci J; Graham P; Li Y. Cancer Lett. 2019, 465, 94–104. [DOI] [PubMed] [Google Scholar]
- (41).Tormey DC; Waalkes TP; Gehrke CW Journal of surgical oncology 1980, 14, 267–273. [DOI] [PubMed] [Google Scholar]
- (42).Tormey DC; Waalkes TP; Ahmann D; Gehrke CW; Zumwatt RW; Snyder J; Hansen H. Cancer 1975, 35, 1095–1100. [DOI] [PubMed] [Google Scholar]
- (43).Boccaletto P; Machnicka MA; Purta E; Piatkowski P; Bagmski B; Wirecki TK; de Crécy-Lagard V; Ross R; Limbach PA; Kotter A; Helm M; Bujnicki JM Nucleic Acids Res. 2018, 46, D303–D307. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (44).Perez-Riverol Y; Csordas A; Bai J; Bernal-Llinares M; Hewapathirana S; Kundu DJ; Inuganti A; Griss J; Mayer G; Eisenacher M; Perez E; Uszkoreit J; Pfeuffer J; Sachsenberg T; Yilmaz S; Tiwary S; Cox J; Audain E; Walzer M; Jarnuczak AF; et al. Nucleic Acids Res. 2019, 47, 442–450. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.