Abstract
Objective:
To develop a machine learning-based prediction model for incident radiographic osteoarthritis (OA) of the knee over 8 years using MRI-based cartilage biochemical composition and knee joint structure, demographics, and clinical predictors including muscle strength and symptoms.
Design:
Individuals (n=1044) with baseline Kellgren Lawrence (KL) grade 0–1 in the right knee from the Osteoarthritis Initiative database were analyzed. 3T MRI at baseline was used to quantify knee cartilage T2, and Whole-Organ Magnetic Resonance Imaging Scores (WORMS) were obtained for cartilage, meniscus, and bone marrow. The outcome was set as true if a subject developed KL grade 2–4 OA in the right knee over 8 years (n=183) and false if the subject remained at KL 0–1 over 8 years (n=861). We developed and compared three models: Model 1: 112 predictors based on OA risk factors; Model 2: top ten predictors based on feature importance score from Model 1 and clinical relevance; Model 3: Model 2 without the imaging predictors. We compared the models using the area under the R OC curve derived from holdout data.
Results:
The 10-predictor model (Model 2, that includes cartilage and meniscus WORMS scores and cartilage T2) had a slightly lower AUC (0.772) compared to the model with 112 predictors (Model 1: AUC=0.792, p=0.739); and had a significantly higher AUC compared to the model without MR imaging predictors (Model 3, AUC=0.669, p=0.011).
Conclusions:
A 10-predictor model including MRI parameters coupled with demographics, symptoms, muscle, and physical activity scores provides good prediction of incident radiographic OA over 8 years.
Keywords: Osteoarthritis, Cartilage imaging, MRI, XGboost, Machine Learning
INTRODUCTION
Osteoarthritis (OA) is a heterogeneous joint disease that affects approximately 250 million people globally1 and causes severe disability2. Total knee arthroplasty (TKA) is the primary treatment for severe OA, but it is invasive and often requires secondary revision surgeries3. The ability to predict at an early stage which patients will develop knee OA using multivariate modeling would enable preemptive measures (such as weight loss or lifestyle changes4) prior to disease development, with prospects of preventing the disability and pain associated with OA progression. Such a prediction model would include established risk factors for OA such as obesity, genetic predisposition, and joint injury5, 6; we hypothesize that integrating imaging features will significantly improve risk prediction for OA.
In addition to demographic risk factors, magnetic resonance imaging (MRI) features have been associated with knee OA. Such features include meniscus tears7 and cartilage lesions8 seen on MRI, and cartilage T2 values9, which are markers of cartilage matrix abnormalities, specifically collagen architecture and changes in hydration10. We have previously developed a Tool for Osteoarthritis Risk Prediction (TOARP)11 demographic and MR imaging predictors, followed by best subsets variable selection and by cross-validation, yielding an AUC of 0.72. However, we have not used ensemble machine learning approaches for prediction and, we have not included clinical risk factors such as muscle strength, physical activity, and symptoms in our previous models.
Knowing which combination of variables best predict OA, and which patients are at risk for OA development may benefit clinical practice by providing a model that is clinically viable and easy to implement. Another application of such a model would be to define inclusion criteria for clinical trials by identifying subjects with a high probability of OA progression; often clinical trials include patients that may not show signs of OA progression, and thus the effect of the tested treatment cannot be observed12.
The purpose of this study was to develop a clinically feasible machine learning-based prediction model for incident radiographic OA over 8 years using MR imaging-based cartilage biochemical composition and knee joint structure, demographics, and clinical features including muscle strength and symptoms. We hypothesized that a XGBoost ensemble learning algorithm could be used to develop a prediction model for the development of radiographic OA, and that by adding MRI-based T2 and WORMS scoring features the prediction compared to a model with only symptoms and demographics will be improved. Such a model would be designed with a goal of feasibility, having readily obtainable inputs for widespread clinical implementation.
METHOD
Patient Data
This study utilizes public use limited datasets from the Osteoarthritis Initiative (OAI; https://www.niams.nih.gov/grants-funding/funded-research/osteoarthritis-initiative)13, a multi-center (n=4), longitudinal study of persons with increased risk for knee OA aged 45–79 years at enrollment, aimed at assessing biomarkers in knee OA including those derived from MR imaging. The study protocol, amendments, and informed consent documentation were reviewed and approved by the local institutional review boards of all participating centers.
For the present study, we retrospectively analyzed a sample of OAI subjects by selecting all subjects that had a Kellgren Lawrence score (KL) = 0 or 1 in the right knee from which we had previously obtained both T2 relaxation time and semi-quantitative joint morphology measures performed by our research group; these readings have been recorded in our database and the results have been previously published14–18. Knees with KL 0 or 1 on the baseline weight-bearing, semi-flexed PA radiograph were included to study subjects without definite radiographic OA of the tibiofemoral joint. The OAI exclusion criteria were: (i) inflammatory arthropathies (including rheumatoid arthritis and seronegative spondylarthropathies), (ii) 3T MRI contraindications, (iii) use of ambulatory aids and co-morbid conditions that may affect the ability to participate in the study. For this analysis we excluded knees that had at baseline (i) a history of knee injury with post-traumatic deformity of the knee joint, (ii) total joint replacements at the lower extremities, (iii) MRI evidence of fractures or abnormalities, that did not fit into the spectrum of OA such as tumor or inflammation at baseline. A total of 1044 individuals were included in the analysis. Incident radiographic OA was defined as development of KL grade 2–4 OA in the right knee during up to 8 years (n=183) and negative if a subject remained grade KL 0 or 1 over 8 years in the right knee (n=861) as shown in Figure 1. In our model we included questionnaire, upper leg strength, physical activity, and imaging data.
Figure 1:
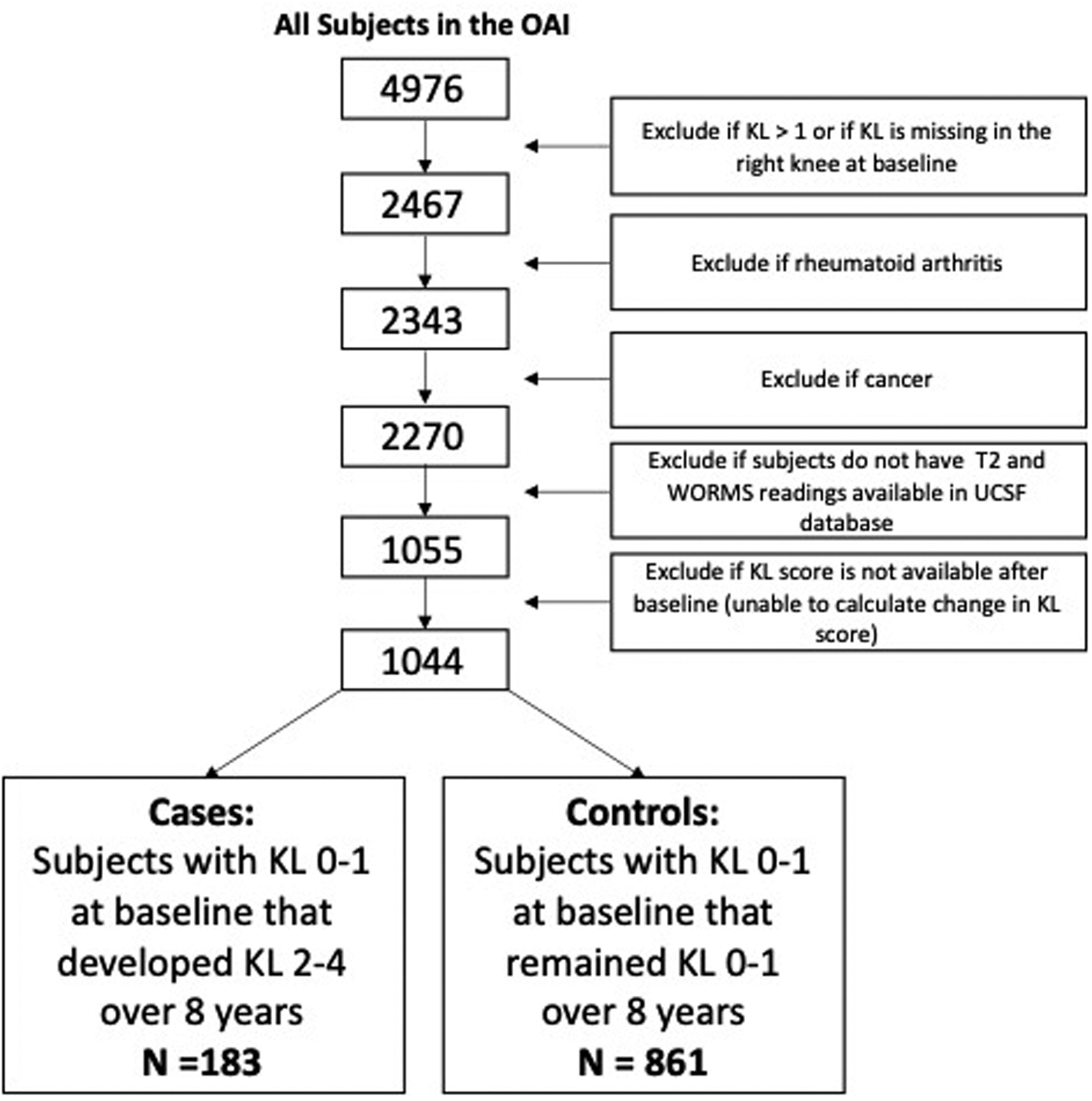
Subject Selection Diagram.
Questionnaires:
(a). WOMAC (Western Ontario McMaster Universities Osteoarthritis) Index:
The WOMAC (Western Ontario McMaster Universities Osteoarthritis) Index19 is a well-established questionnaire used to obtain a complete assessment of potential symptoms related to knee OA including function, pain and stiffness.
(b). Knee Injury and Osteoarthritis Outcome Score (KOOS):
The Knee Injury and Osteoarthritis Outcome Score (KOOS) provides complimentary information to the WOMAC index concerning knee symptoms and function with an additional focus on sport and recreation as well as quality of life during the last 7 days. This score was designed to extend the target population of the WOMAC index to younger and middle aged subjects with knee injuries and post-traumatic arthritis20, 21.
Upper Leg Strength
Bilateral isometric knee extensor and flexor strength were measured using the Good Strength isometric strength chair (Metitur, Jyvaskyla, Finland)22. The maximal force produced during isometric contraction were measured during isometric contractions of the right quadriceps and hamstring muscles at a knee angle of 60 degrees from full extension. The coefficient of variation between two consecutive measurements performed two weeks apart was 6.3% (SD 5.7) for knee extension and flexion strength22.
Imaging of the knee
Radiographs:
Fixed flexion knee radiographs were obtained at baseline, and radiographic KL grades23 were provided in the OAI dataset. Subjects with baseline KL grades of 0–1 were selected.
MR Imaging:
MR images were obtained using four identical 3.0 Tesla (Siemens Magnetom Trio, Erlangen, Germany) scanners in Columbus, Ohio; Baltimore, Maryland; Pittsburgh, Pennsylvania; Pawtucket, Rhode Island. The following four sequences were obtained for the morphological analysis: (i) 2D intermediate-weighted fast spin echo (FSE) sequences with fat suppression in the sagittal plane (3200/30 milliseconds (ms), repetition time (TR)/ echo time (TE)); (ii) 2D proton density-weighted FSE sequences in the sagittal plane (2700/20 ms, TR/TE); (iii) 3D T1-weighted fast low-angle shot (FLASH) gradient-echo sequences (20/7.6 ms/12°, TR/TE/flip angle), 512×512 matrix and (iv) 3D dual echo steady-state gradient-echo (DESS) obtained in the sagittal plane (16.3/4.7 ms/25°, TR/TE/flip angle), 307×384 matrix. Further details about the image acquisition are available in the OAI MR protocol24. A sagittal 2D multi-slice multi-echo sequence (MSME, TR=2700ms, TE1-TE7=10–70ms, spatial resolution=0.313mmx0.446mm, slice thickness=3.0mm, and 0.5mm gap) was used for cartilage T2 measurements25.
MR Image Analysis
WORMS Scoring
WORMS scoring was performed at baseline. MR images of the right knee obtained at the baseline visit were reviewed on picture archiving communication system (PACS) workstations (Agfa, Ridgefield Park, NJ, USA). Three radiologists with 8, 6- and 6-years of experience graded all knee abnormalities. In equivocal cases, a consensus reading was performed with a musculoskeletal radiologist with 25-years of experience.
Baseline cartilage, meniscus, and bone marrow morphology were assessed using a modified semi-quantitative whole-organ magnetic resonance imaging score (WORMS) as previously described26, 27. Compartment specific scores as well as maximum and sum scores were obtained. The maximum (MAX) cartilage, meniscus or bone marrow edema pattern (BMEP) scores were defined as the maximum score in any compartment. MRI was evaluated for the presence or absence of a knee effusion.
The reproducibility results for WORMS reading have been previously published27: The ICCs for intraobserver agreement were 0.85 (95% CI: 0.79, 0.93) for meniscus WORMS, 0.87 (95% CI: 0.81, 0.92) for cartilage WORMS, and 0.89 (95% CI: 0.85, 0.91) for BMEP. The ICCs for interobserver agreement were 0.83 (95% CI: 0.76, 0.91) for meniscus WORMS, 0.80 (95% CI: 0.74, 0.87) for cartilage WORMS, and 0.88 (95% CI: 0.81, 0.94) for BMEP.
T2 measurements
Cartilage T2 measurements were performed at baseline. Semi-automatic cartilage segmentation of lateral/medial femur, lateral/medial tibia, and patella regions was performed as previously described, using an in-house, spline-based software based on MATLAB (MathWorks, Natick, Massachusetts)28. T2 maps were computed from the MSME images on a pixel-by-pixel basis using 6 echoes (TE=20–70ms) and 3 parameter fittings accounting for noise29, 30, and averaged over all of the slices in each cartilage compartment. The first echo (TE=10ms) was not included in the T2 fitting procedure in order to reduce potential errors resulting from stimulated echoes. The average cartilage T2 value in the knee was defined as the average T2 in all the regions described above. The average values in each region and in the knee were predictors in the ML model. Trained investigators segmented the entire cartilage but used rigorous criteria to exclude sections with compromised image quality.
Validated methods for obtaining a T2 map of the cartilage have been previously published by our group9, 28. The cartilage T2 reproducibility results have been described previously9, 18, 28. Inter-reader reproducibility was assessed between the two readers in 10 patients and was 1.66% over all compartments. CVs for single compartments were as follows: 1.28% for the lateral femur, 1.11% for the lateral tibia, 1.29% for the medial femur, 2.01% for the medial tibia, and 2.42% for the patella18. For intra-reader reproducibility analysis, the same reader performed repeated T2 measurements in 10 randomly selected patients with readings separated by at least 14 days. Intra-reader CVs were calculated for each compartment using these repeated measurements and compartment specific and overall CVs were as follows: 0.92% for the lateral femur, 1.14% for the lateral tibia, 1.07% for the medial femur, 1.63% for the medial tibia, 2.33% for the patella, and 1.42% over all compartments18.
Machine Learning Model Development
Ground Truth:
The outcome was set as true if the subject developed KL grade 2–4 OA in the right knee over 8 years (n=183) and false if the patient remained at KL 0–1 over 8 years (n=861). All patients had KL grade 0–1 at baseline. The KL grade was used for the ground truth label generation, described in detail in by Lawrence et al23, as it has been validated and extensively used for OA classification31. A study of image assessment in the OAI found good reliability for KL grading between baseline and the 36-month follow-up visit, with κ values of about 0.70 to 0.80. However, the 8-year reliability readings have not been reported.
Exploratory Data Analysis and Preprocessing
A total of 112 predictors based on existing literature5, 6, 32–34 and clinical relevance including (Model 1) baseline participant demographics, family history of OA, symptoms, muscle strength, cartilage T2, physical activity, WORMS scores, physical exam, medication, and knee alignment were used to predict the development of OA (KL=2–4) over 8 years using XGboost (Python version 3.7, Python Software Foundation, Wilmington, DE, Figure 2). Supplementary Table 1 lists the resulting 112 features, by category. No rescaling was performed to the predictor variables included in the ML model.
Figure 2:
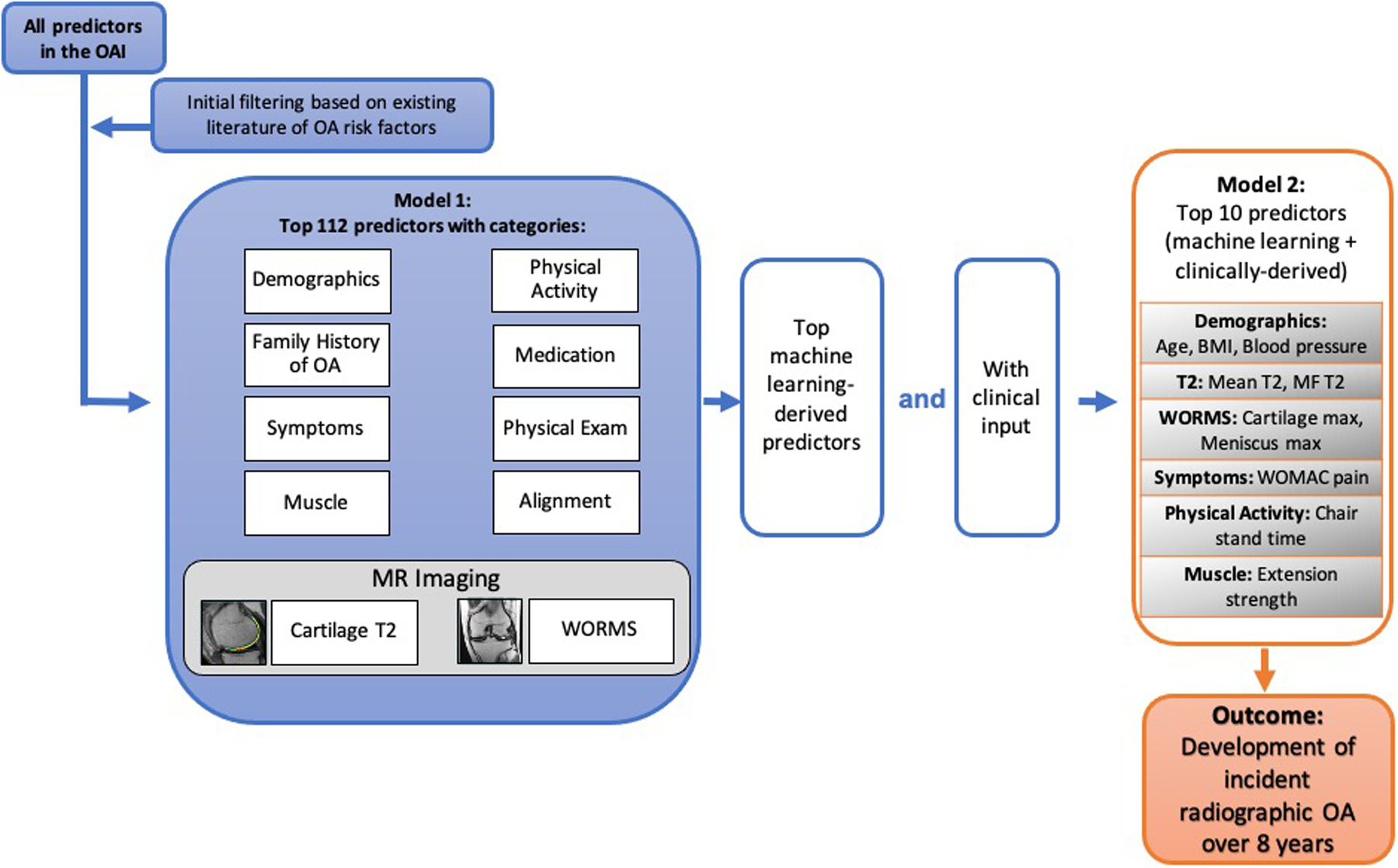
Machine learning model development schema. For the initial model with 112 predictors (Model 1), predictors are shown in categories for graphical representation; in the predictor selection process (Model 2: 10 predictors), we aimed to integrate predictors that were most important for model performance and relatively easy to obtain in the clinical setting. XGBOOST was used for the machine learning algorithm. Abbreviations: Medial Femur (MF); Whole-Organ Magnetic Resonance Imaging Scores (WORMS); Western Ontario and McMaster Universities Arthritis Index (WOMAC), Body Mass Index (BMI), Osteoarthritis (OA).
Single imputation was used to address missing data using STATA version 16 software (StataCorp LP, College Station, TX). Supplementary Table 2 reports the percentages of missing data for each predictor in the final hybrid model. The imputed dataset had a similar distribution of predictor values compared to the non-imputed dataset for all imputed predictors.
A 15% holdout test set (on which the final three models were evaluated) was randomly selected and was not used for the model training and validation process. The remaining 85% of the data was used for training and validation in a 5-fold cross-validation schema (randomly split training set of 68% and validation set of 17%). 5-fold cross validation was used to confirm the reliability of the final chosen hyperparameters.
Model Development and Hyperparameter Optimization
The study used a supervised XGBoost machine learning model35. XGBoost is a high-performance decision tree-based algorithm with advantages of high accuracy due to its ensemble learning and high interpretability due to its feature importance calculation. The feature importance, which is determined by the level at which the decision tree split occurs, identifies which features contribute most to the optimized prediction algorithm. It was calculated for a single decision tree based on the magnitude that each attribute split point improved the performance measure (weighted by the number of observations the node was responsible for). Then, each feature importance measure was averaged across all of the decision trees within the model to obtain an overall “importance” score for each predictor36. The relative importance (F score) of each variable is scaled so that the sum of all F scores adds to 100, with higher numbers indicating stronger influence on the response37.
To tune the hyperparameters for the tree booster, we exhaustively searched the combinations of the following parameters: number of estimators, maximum depth, minimum child weight, L1 regularization alpha, gamma, step size shrinkage, lambda, maximum delta step allowed, subsample ratio of the training instances, and subsample ratio of columns in constructing a tree on the imputed dataset. The form of the XGboost algorithm was set as multiple logistic.
In order to target the clinical utility of the ML model, we also report a model with only 10 predictors (Model 2) optimized on variables chosen based on the XGBoost feature importance score and clinically relevant parameters (i.e. disease risk factors such physical activity and BMI) described in the recent literature5, 6, 32–34. The predictors in Model 2 included demographics, cartilage T2, WORMS scores, symptoms, physical activity, and muscle strength. We also report the performance of a clinically relevant model excluding MR imaging predictors/variables, for comparison purposes as well as for potential usage in patients without imaging (Model 3). We then obtained an area under the ROC curve (AUC) for these models using the testing dataset. In summary, we developed and compared three models: Model 1: 112 predictors based on OA risk factors; Model 2: top ten predictors based the F score from Model 1 and clinical relevance based on high yield parameters from previous literature; Model 3: Model 2 without the imaging predictors.
Model Evaluation
Diagnostic performance for predicting radiographic incidence of OA using the three machine learning models was determined using area under the receiver operator characteristic (ROC) analysis on the 15% holdout test set. The ROC AUCs of the three models were compared in a combined and pairwise fashion using the DeLong test and confidence intervals were also generated similarly38.
Statistical Analysis:
Statistical analysis was performed using SAS Studio version 3.8 (SAS Institute Inc., Cary, NC, USA). Descriptive statistics were performed using a SAS macro program called “Tablen”39. Differences in continuous parameters between groups (i.e. age, BMI) were assessed using Kruskal Wallis tests, and differences in categorical parameters between groups (i.e. sex and race) were assessed using Chi-squared tests. Differences in AUCs were compared between the models pairwise using the Delong test38.
RESULTS
Subject Characteristics
The full dataset (including training/validation and holdout test set) contained 1044 participants; of those 183 were cases that developed KL 2, 3 or 4 over 8 years, and 861 were controls with KL grades 0 or 1 at baseline and up to 8 years. The subject characteristics are listed in Table 1.
Table 1:
Subject Characteristics (descriptive).
| Total (N=1044) |
|
|---|---|
| Age (years) | |
| N | 1044 |
| Mean (SD) | 56.9 (8.32) |
| Median | 55.0 |
| Range | 45.0 – 79.0 |
| BMI (kg/m2) | |
| N | 1044 |
| Mean (SD) | 27.8 (4.38) |
| Median | 27.4 |
| Range | 16.9 – 42.4 |
| Sex, n (%) | |
| Male | 457 (43.8%) |
| Female | 587 (56.2%) |
| Race, n (%) | |
| Other Non-white | 18 (1.7%) |
| White or Caucasian | 823 (78.9%) |
| Black or African American | 195 (18.7%) |
| Asian | 7 (0.7%) |
| Missing | 1 |
| KL grade right knee, n (%) | |
| 0 | 704 (67.4%) |
| 1 | 340 (32.6%) |
| WOMAC* pain score (right knee) | |
| N | 1044 |
| Mean (SD) | 1.8 (2.86) |
| Median | 0.0 |
| Range | 0.0 – 16.0 |
| PASE score | |
| N | 1039 |
| Mean (SD) | 175.1 (87.05) |
| Median | 167.0 |
| Range | 5.0 – 526.0 |
WOMAC: The Western Ontario and McMaster Universities Arthritis Index;
PASE: physical activity scale for the elderly
Model 1 – initial model with 112 predictors:
The model with 112 predictors had an AUC of 0.792 (Figure 3), and the 10 top features for prediction (listed in order of importance) were radial pulse, systolic blood pressure, Western Ontario and McMaster Universities Arthritis Index (WOMAC) total score, medial femur cartilage T2, maximum cartilage WORMS score, abdominal circumference, knee muscle extension strength, patella cartilage T2, chair stand time, and BMI (Figure 4). The final optimized hyperparameters for the model with 112 predictors were: number of estimators [100], maximum depth [8], minimum child weight [3], L1 regularization alpha [0], gamma [1.5], step size shrinkage [0.2], lambda [20], maximum delta step allowed [100], subsample ratio of the training instances [0.5], and subsample ratio of columns [0.6].
Figure 3:
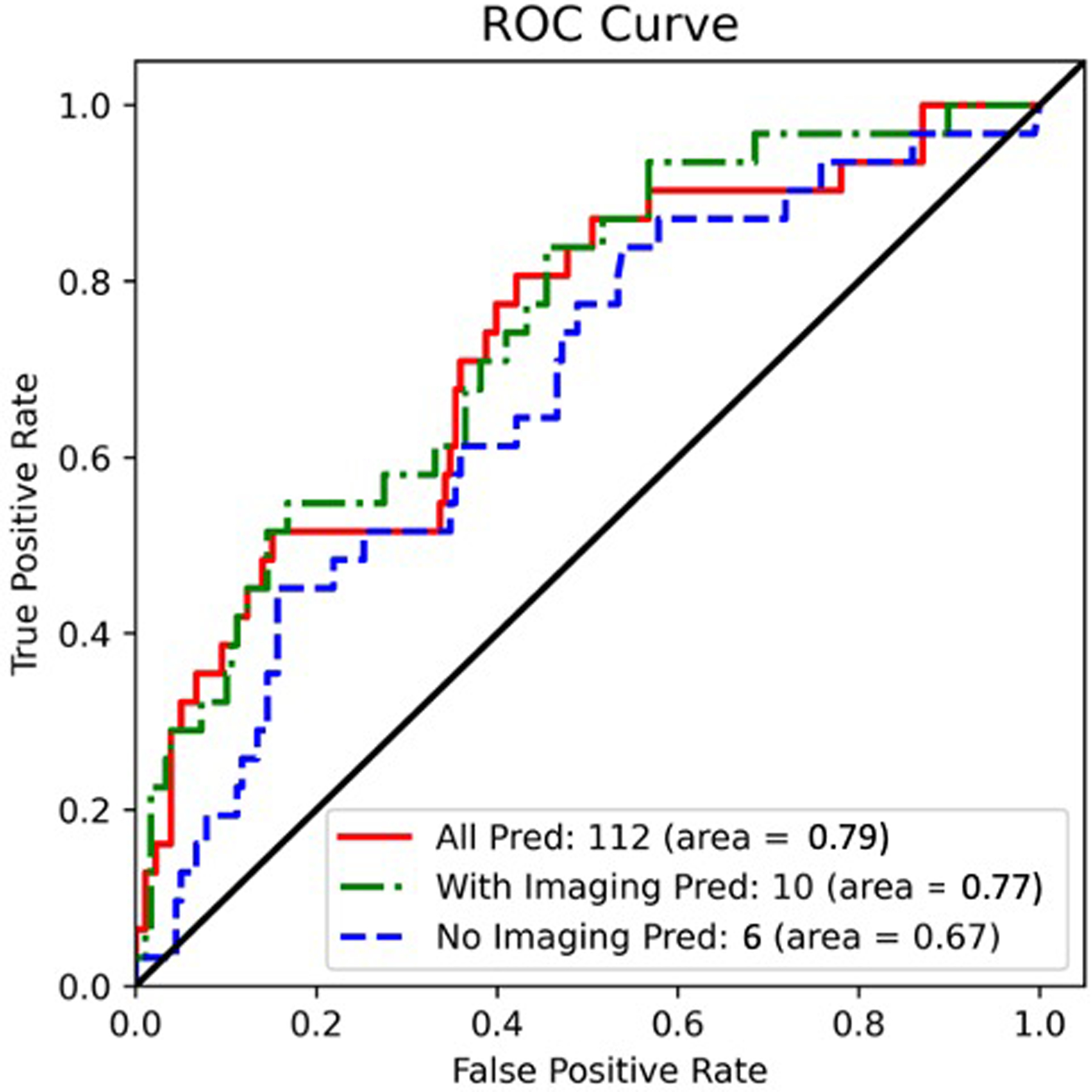
ROC curves for a) Model 1: full model with 112 predictors, b) Model 2: 10 predictors (final proposed model), and c) Model 3: Model 2 without MR imaging features (for comparison; 6 predictors). The 10-predictor model (Model 2, that includes cartilage and meniscus WORMS scores and cartilage T2) had a slightly lower AUC (0.772) compared to the model with 112 predictors (Model 1: AUC=0.792, p=0.739); and had a significantly higher AUC compared to the model without MR imaging predictors (Model 3, AUC=0.669, p=0.011).
Figure 4:
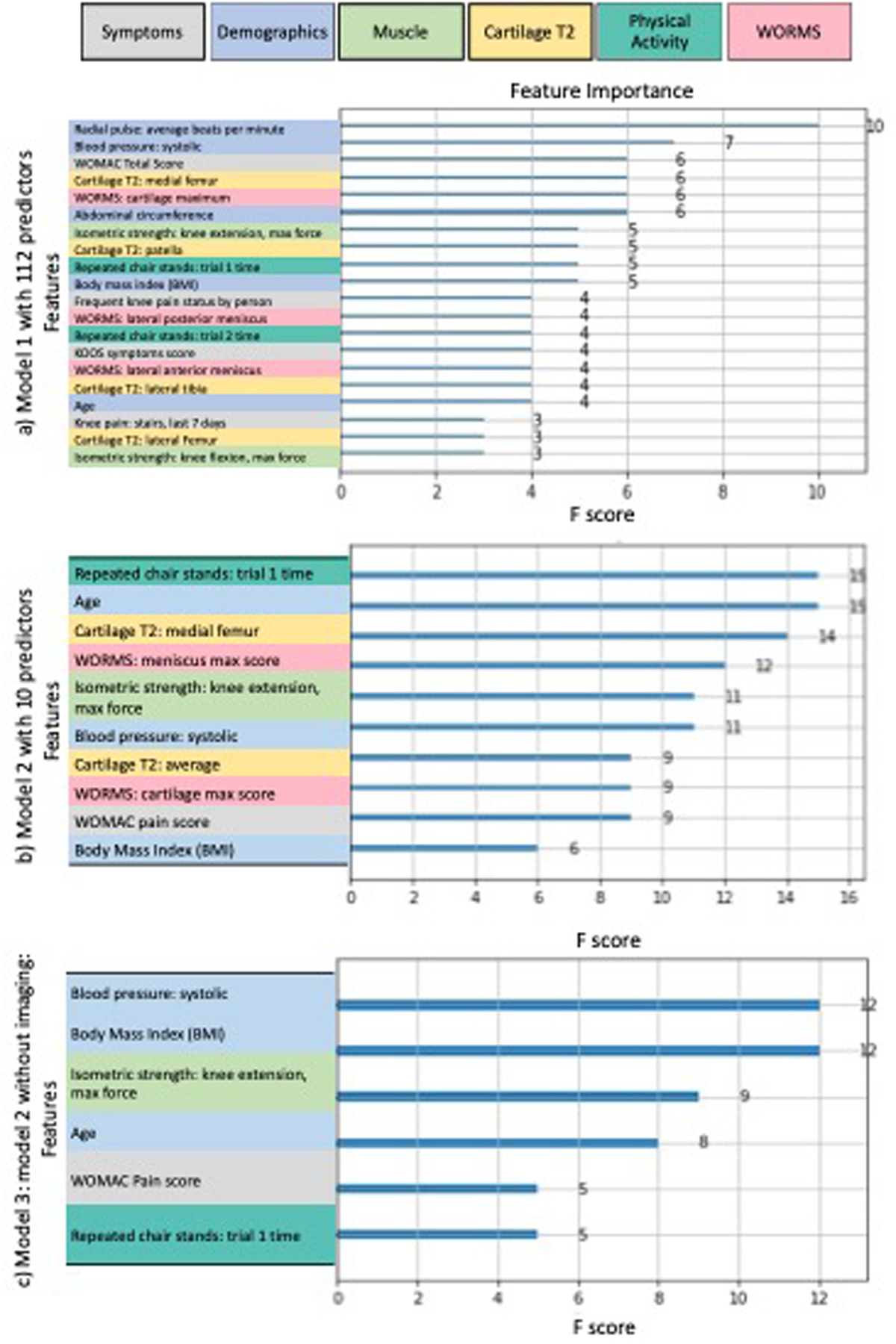
Feature importance charts, color coded by predictor category for a) model with 112 predictors (Model 1), b) model using 10 predictors (Model 2, final proposed model), and c) Model 2 without MR imaging features (Model 3, for comparison; 6 predictors). The relative importance (F score) of each variable is scaled so that the sum of all variable importance adds to 100, with higher numbers indicating stronger influence on the response37. Note that predictor categories: physical exam, medication, alignment, and family history of OA were not chosen as the top predictors in any of the models, and were therefore excluded from the Figure. Also, note that the magnitude of the feature importance scores indicate the relative importance of covariates within a model, and are not comparable between different models.
Model 2: 10 predictors (final proposed model):
The final hybrid model with 10 predictors had an AUC of 0.772 (Figure 3), and the predictors (listed in order of importance) were: chair stand time, age, medial femur cartilage T2, maximum meniscus WORMS score, knee muscle extension strength, systolic blood pressure, mean cartilage T2 (in all regions), maximum cartilage WORMS score, WOMAC pain score, and BMI (Figure 4). The final optimized hyperparameters for the hybrid model were: number of estimators [100], maximum depth [10], minimum child weight [1], L1 regularization alpha [2], gamma [0], step size shrinkage [0.3], lambda [20], maximum delta step allowed [100], subsample ratio of the training instances [0.6], and subsample ratio of columns [0.6], which can be implemented by other researchers on independent datasets.
Model 3: Model 2 without imaging predictors (for comparison):
To assess the value of MR imaging predictors on model performance, we ran an additional model including only non-imaging predictors (chair stand time, age, knee muscle extension strength, systolic blood pressure, WOMAC pain score, and BMI), which yielded an AUC of 0.669 (Figure 3). The feature importance chart is illustrated in Figure 4. The final optimized hyperparameters for the hybrid model without imaging predictors were: number of estimators [100], maximum depth [10], minimum child weight [5], L1 regularization alpha [2], gamma [0], step size shrinkage [0.4], lambda [20], maximum delta step allowed [100], subsample ratio of the training instances [0.4], and subsample ratio of columns [0.4].
Model Comparison:
The 10-predictor model (Model 2, that includes cartilage and meniscus WORMS scores and cartilage T2) had a slightly lower AUC = 0.772 (95% CI = 0.680 to 0.863) compared to the model with 112 predictors (Model 1: AUC=0.792, 95% CI = 0.694 to 0.890), p=0.739; and had a significantly higher AUC compared to the model without MR imaging predictors (Model 3, AUC=0.669, 95% CI = 0.567 to 0.770, p=0.011). The specificities of Models 1, 2, and 3 respectively were: 90.05% [95%CI = 85.6%−94.5%], 90.05% [95%CI = 85.6%−94.5%], and 87.35% [95%CI = 82.4%−92.3%]. The sensitivities of Models 1, 2, and 3 respectively were: 36.84% [95%CI = 21.5%−52.1%], 36.84% [95%CI = 21.5%−52.1%], and 25.71% [95%CI = 11.2%−40.2%]. The threshold was determined by maximizing the product of sensitivity and specificity; however, a more clinically relevant, highly sensitive model with a low threshold is also described in the discussion section. The model performance results on the validation set are provided in Supplementary Figure 1, and the confusion matrices are provided in Supplementary Figure 2. The full code can be found on https://bit.ly/38xVu5w.
Missing Data Analysis
The full 112 predictor dataset (Model 1) had missing data of 3.8% on average ranging from 0.0% to 26.0%). Of the 10 chosen predictors in the final proposed model (Model 2), missing data average of 2.0% (range: 0.0% to 17.6%). Supplementary Table 2 reports the detailed percentages of missing data for each predictor in the final proposed model (Model 2).
DISCUSSION
We have developed machine learning models that can predict the future development of radiographic knee OA over 8 years in subjects without radiographic OA at baseline. The model consisting of 10 predictors (our final proposed model) had only a slightly lower AUC (0.772) compared to the full model with 112 predictors (0.792), but was substantially easier to use due to fewer number of predictors, most of which could be easily obtained in the clinical setting. The 10-predictor model had a significantly higher AUC compared to the model without imaging predictors (0.669, p=0.011). A unique feature of this study is assessment of both MRI cartilage T2 and WORMS scores in the machine learning models; these imaging parameters improved diagnostic performance when comparing AUCs from the model with and without imaging predictors. The results suggest a valuable impact of imaging biomarkers in the ML model, which may also eventually translate into improved and more socioeconomically favorable outcomes. Another unique aspect of this study is specifically selecting subjects without knee OA at baseline to target and predict the development of incident radiographic OA over 8 years. This study suggests that using a model with 10 predictors that includes MR imaging features may be clinically viable for prediction of radiographic OA development over 8 years.
While the model with 112 predictors had a slightly higher prediction accuracy compared to the model with 10 predictors, implementing a model with over 100 predictors in a clinical setting is not feasible. Utilizing a model with only 10 predictors is a viable alternative for a clinical setting given its comparable diagnostic accuracy to the larger model and a substantially lower number of predictors. The model with 10 predictors incorporates features that are easily obtainable during clinical intake including demographic information, pain, blood pressure, and chair stand time. While MR imaging features may add intricacy for clinical implementation, they are essential for model diagnostic performance as removing imaging features from the model significantly decreases the AUC (AUC=0.669 for the model without MR imaging features vs. 0.772 for the hybrid model with MR imaging features, p=0.011). To simplify the implementation of cartilage T2 quantification in clinical practice, novel techniques for automatic segmentation have been recently developed having high accuracy and reproducibility40. Semi-quantitative evaluation of WORMS scores is also clinically feasible as MRI sequences for WORMS grading are routinely acquired and a radiologist can perform the gradings relatively quickly.
Prognosis of knee OA is challenging due to the heterogeneity and multifactorial nature of the disease, and because its pathophysiology is still poorly understood. Machine learning models for prediction of OA have been developed using varied algorithms, clinical and structural outcomes, and a range of predictors41. Outcome measures have included pain, radiographic joint space12, KL grade42, OARSI grade, TKA42, 43; predictors have included demographics, biomechanics, biomarkers, and MR images and extracted features41. Some machine learning studies have utilized raw images42 as an inputs using deep learning (data-driven approach), while others have derived features from an image (i.e. KL score) and have used those as model inputs in order to reduce dimensionality. We have previously developed a Tool for Osteoarthritis Risk Prediction (TOARP)11 demographic and MR imaging predictors, using logistic regression, best subsets variable selection, and by cross-validation, yielding an AUC of 0.72. A machine learning study by Pedoia et al. utilized MRI cartilage T2 features and demographics for the prediction of knee OA, reporting that T2 values and demographic features alone had AUCs of 0.56 and 0.67, respectively; however, a joint model using principal components of T2 relaxometry patterns and demographics had an AUC of 0.7740. Our AUC were similar to that of other reported studies as described in a recent review and demonstrated the importance of MR features for model prediction, similar to Jamshidi et al44. With clinical feasibility in mind, our study used a unique hybrid approach bridging data-driven ensemble machine learning to identify key features with clinical input thereby creating a model geared toward high performance and ease of use in the clinic.
XGboost was our machine learning approach of choice given the known strengths of ensemble learning in classification tasks that involve standard tabular data. XGboost is a type of tree-based boosting algorithm with more regularized model formalization to control over-fitting, which ultimately improves model performance. This ensemble tree-based model is currently one of the most popular if not the most popular one in Kaggle machine learning competition community45, has significant capabilities for parameter tuning including tree parameters, regularization and cross-validation, and can be robust to the curse of dimensionality46. XGboost also allows the identification of features that were important for final model classification, enhancing interpretability that ultimately allows for convenient feature selection.
While previous studies have reported that hypertension and OA are associated47, 48, we are not aware of any studies that have reported a relationship between radial pulse and OA. Since these associations are largely unexplored in the literature, we did not include radial pulse in Model 2. Blood pressure had high feature importance in Models 1 and 3, and may be considered a proxy for general health; however, the direct mechanisms responsible for the associations between this parameter and OA are not known. Studies have reported a relationship between hypertension and OA, possibly due to microvascular remodeling, leading to reduced blood flow to the subchondral bone and compromised nutrient and oxygen exchange to the articular cartilage47,48. Additional research may be needed to study the mechanisms by which pulse is related to OA.
Given only minimal risk involved with early lifestyle modification and close clinical follow up, we would suggest setting a low threshold for the XGboost output value (for example, 0.0879) to achieve a highly sensitive model with sensitivity of 94 percent at specificity of 43 percent on the ROC curve. Such a sensitive model could serve as a useful screening tool to identify those who may progress to knee OA and may warrant further workup / follow up.
Several limitations were pertinent to this study including lack of external testing in independent cohorts though the dataset was acquired from four institutions, and the challenges and costs to obtain standardized MR imaging and quantify T2/WORMS scoring. While other quantitative cartilage compositional measurements would be ideal to implement in the machine learning models, only cartilage T2 relaxation time was included as the OAI only provided images for T2 quantification. In addition, we only analyzed the right knee as MR images for T2 quantification were only acquired in the right knee (not left). Recently, algorithms for automatic T2 quantification have been developed49 and there is ongoing work to standardize T2 mapping among acquisition methods, vendors, coils and post-processing techniques through the Quantitative Imaging Biomarker Alliance (QIBA), thus enabling future implementation of T2 quantification use in a clinical setting. We were unable to perform external validation as, to the best of our knowledge, no large longitudinal databases with cartilage T2 and knee MRIs performed at 3T exist. While ideally cases and controls would have been age- and BMI-matched, this study did not have a matched design due to the subject inclusion/exclusion criteria, availability of readings, and sample size. We instead included age and BMI in the final model and algorithmically adjusted for these predictors. WORMS and T2 quantification have inherent subjectivity due inter-reader variation; however, the ICCs in this study were deemed as having “good reliability”50. The effects of inter-reader variability on the ML model output were mitigated by including other predictors in the ML model, and by training with standardized objective data points. Thus, we do not expect the inter-reader variation to have a large effect on the accuracy of the model output. We acknowledge that the dataset is imbalanced, have explored balanced options, and concluded the unbalanced dataset performed the best, potentially due to higher subject numbers (greater power). Despite these limitations, we believe this study is significant as it is the first to study using both cartilage compositional measurements and knee morphologic grading combined with machine learning for prediction of radiographic knee OA.
In conclusion, we have developed an ensemble machine learning model that uses 10 demographic, clinical, and MR imaging variables to predict the development of radiographic OA over 8 years with an AUC of 0.772. The 10-predictor model (had a slightly lower AUC compared to the model with 112 predictors (AUC=0.792), and had a significantly higher AUC compared to the model without MR imaging predictors (AUC=0.669, p=0.011). The 10 predictor model may be used to identify people at risk for radiographic OA, thus providing guidance on inclusion criteria for clinical trials and for patient management prior to irreversible joint degeneration.
Supplementary Material
Supplemental Figure 1: Model performance results on the validation set. Although we show these results for completeness, the final performance is based on the holdout test set as it represents the most unbiased dataset that the model had not seen or optimized on.
Supplemental Figure 2: Confusion matrices for models 1, 2, and 3.
Supplemental Table 1: List of 112 predictors included in data exploration
Supplemental Table 2: The number of and percentage of missing data for each predictor in the model with 10 predictors (final proposed model).
ROLE OF FUNDING SOURCE
The analyses performed in this study were funded by NIH R01-AR064771 and NIH R01-AR078917. The OAI is a public-private partnership comprised of five contracts (N01-AR-2-2258; N01-AR-2-2259; N01-AR-2-2260; N01-AR-2-2261; N01-AR-2-2262) funded by the National Institutes of Health, a branch of the Department of Health and Human Services, and conducted by the OAI Study Investigators. Private funding partners include Merck Research Laboratories; Novartis Pharmaceuticals Corporation, GlaxoSmithKline; and Pfizer, Inc. Private sector funding for the OAI is managed by the Foundation for the National Institutes of Health.
Footnotes
CONFLICT OF INTEREST:
Competing interest statement: There are no conflicts of interest.
REFERENCES:
- 1.Murray CJ, Vos T, Lozano R, Naghavi M, Flaxman AD, Michaud C, et al. Disability-adjusted life years (DALYs) for 291 diseases and injuries in 21 regions, 1990–2010: a systematic analysis for the Global Burden of Disease Study 2010. Lancet 2012; 380: 2197–2223. [DOI] [PubMed] [Google Scholar]
- 2.Murphy L, Helmick CG. The impact of osteoarthritis in the United States: a population-health perspective. Am J Nurs 2012; 112: S13–19. [DOI] [PubMed] [Google Scholar]
- 3.Center ME-bP. Total Knee Replacement. In: Services UDoHaH Ed. Evidence Report/Technology Assessment, vol. 86. Minneapolis: Agency for Healthcare and Research Quality; 2003. [Google Scholar]
- 4.Roddy E, Doherty M. Changing life-styles and osteoarthritis: what is the evidence? Best Pract Res Clin Rheumatol 2006; 20: 81–97. [DOI] [PubMed] [Google Scholar]
- 5.Rogers MW, Wilder FV. The association of BMI and knee pain among persons with radiographic knee osteoarthritis: a cross-sectional study. BMC musculoskeletal disorders 2008; 9: 163. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Felson DT, Lawrence RC, Dieppe PA, Hirsch R, Helmick CG, Jordan JM, et al. Osteoarthritis: new insights. Part 1: the disease and its risk factors. Ann Intern Med 2000; 133: 635–646. [DOI] [PubMed] [Google Scholar]
- 7.Sharma L, Nevitt M, Hochberg M, Guermazi A, Roemer FW, Crema M, et al. Clinical significance of worsening versus stable preradiographic MRI lesions in a cohort study of persons at higher risk for knee osteoarthritis. Ann Rheum Dis 2016; 75: 1630–1636. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Baum T, Joseph GB, Arulanandan A, Nardo L, Virayavanich W, Carballido-Gamio J, et al. Association of magnetic resonance imaging-based knee cartilage T2 measurements and focal knee lesions with knee pain: data from the Osteoarthritis Initiative. Arthritis care & research 2012; 64: 248–255. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Joseph GB, Baum T, Alizai H, Carballido-Gamio J, Nardo L, Virayavanich W, et al. Baseline mean and heterogeneity of MR cartilage T2 are associated with morphologic degeneration of cartilage, meniscus, and bone marrow over 3 years--data from the Osteoarthritis Initiative. Osteoarthritis and cartilage 2012; 20: 727–735. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Xia Y Magic-angle effect in magnetic resonance imaging of articular cartilage: a review. Invest Radiol 2000; 35: 602–621. [DOI] [PubMed] [Google Scholar]
- 11.Joseph GB, McCulloch CE, Nevitt MC, Neumann J, Gersing AS, Kretzschmar M, et al. Tool for osteoarthritis risk prediction (TOARP) over 8 years using baseline clinical data, X-ray, and MRI: Data from the osteoarthritis initiative. J Magn Reson Imaging 2018; 47: 1517–1526. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Widera P, Welsing PMJ, Ladel C, Loughlin J, Lafeber F, Petit Dop F, et al. Multi-classifier prediction of knee osteoarthritis progression from incomplete imbalanced longitudinal data. Sci Rep 2020; 10: 8427. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Peterfy C, Schneider E, Nevitt M. The osteoarthritis initiative: report on the design rationale for the magnetic resonance imaging protocol for the knee. Osteoarthritis and Cartilage 2008; 16: 1433–1441. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Baum T, Stehling C, Joseph GB, Carballido-Gamio J, Schwaiger BJ, Muller-Hocker C, et al. Changes in knee cartilage T2 values over 24 months in subjects with and without risk factors for knee osteoarthritis and their association with focal knee lesions at baseline: data from the osteoarthritis initiative. Journal of magnetic resonance imaging : JMRI 2012; 35: 370–378. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Joseph GB, Baum T, Carballido-Gamio J, Nardo L, Virayavanich W, Alizai H, et al. Texture analysis of cartilage T2 maps: individuals with risk factors for OA have higher and more heterogeneous knee cartilage MR T2 compared to normal controls--data from the osteoarthritis initiative. Arthritis research & therapy 2011; 13: R153. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Stehling C, Lane NE, Nevitt MC, Lynch J, McCulloch CE, Link TM. Subjects with higher physical activity levels have more severe focal knee lesions diagnosed with 3T MRI: analysis of a non-symptomatic cohort of the osteoarthritis initiative. Osteoarthritis Cartilage 2010; 18: 776–786. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Kretzschmar M, Lin W, Nardo L, Joseph GB, Dunlop DD, Heilmeier U, et al. Association of Physical Activity Measured by Accelerometer, Knee Joint Abnormalities, and Cartilage T2 Measurements Obtained From 3T Magnetic Resonance Imaging: Data From the Osteoarthritis Initiative. Arthritis Care Res (Hoboken) 2015; 67: 1272–1280. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Gersing AS, Solka M, Joseph GB, Schwaiger BJ, Heilmeier U, Feuerriegel G, et al. Progression of cartilage degeneration and clinical symptoms in obese and overweight individuals is dependent on the amount of weight loss: 48-month data from the Osteoarthritis Initiative. Osteoarthritis Cartilage 2016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Bellamy N, Buchanan W, Goldsmith C, Campbell J, Stitt L. Validation study of WOMAC: a health status instrument for measuring clinically important patient relevant outcomes to antirheumatic drug therapy in patients with osteoarthritis of the hip or knee. J Rheumatol 1988; 15: 1833–1840. [PubMed] [Google Scholar]
- 20.Roos EM, Lohmander LS. The Knee injury and Osteoarthritis Outcome Score (KOOS): from joint injury to osteoarthritis. Health Qual Life Outcomes 2003; 1: 64. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Roos EM, Roos HP, Lohmander LS, Ekdahl C, Beynnon BD. Knee Injury and Osteoarthritis Outcome Score (KOOS)--development of a self-administered outcome measure. J Orthop Sports Phys Ther 1998; 28: 88–96. [DOI] [PubMed] [Google Scholar]
- 22.Nevitt MC, Felson DT, Lester G. THE OSTEOARTHRITIS INITIATIVE: protocol for the cohort study. UC San Francisco; Boston University; National Institude of Arthritis, Musculoskeletal and Skin Diseases; 2006. [Google Scholar]
- 23.Kellgren J, Lawrence J. Radiologic assessment of osteoarthritis. Ann Rheum Dis 1957; 16: 494–502. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Peterfy CG, Schneider E, Nevitt M. The osteoarthritis initiative: report on the design rationale for the magnetic resonance imaging protocol for the knee. Osteoarthritis Cartilage 2008; 16: 1433–1441. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Peterfy C, Schneider E, Nevitt M. The osteoarthritis initiative: report on the design rationale for the magnetic resonance imaging protocol for the knee. Osteoarthritis and cartilage/OARS, Osteoarthritis Research Society 2008; 16: 1433. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Peterfy CG, Guermazi A, Zaim S, Tirman PF, Miaux Y, White D, et al. Whole-Organ Magnetic Resonance Imaging Score (WORMS) of the knee in osteoarthritis. Osteoarthritis Cartilage 2004; 12: 177–190. [DOI] [PubMed] [Google Scholar]
- 27.Gersing AS, Schwaiger BJ, Nevitt MC, Joseph GB, Chanchek N, Guimaraes JB, et al. Is Weight Loss Associated with Less Progression of Changes in Knee Articular Cartilage among Obese and Overweight Patients as Assessed with MR Imaging over 48 Months? Data from the Osteoarthritis Initiative. Radiology 2017; 284: 508–520. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Stehling C, Baum T, Mueller-Hoecker C, Liebl H, Carballido-Gamio J, Joseph GB, et al. A novel fast knee cartilage segmentation technique for T2 measurements at MR imaging--data from the Osteoarthritis Initiative. Osteoarthritis Cartilage 2011; 19: 984–989. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Miller AJ, Joseph PM. The use of power images to perform quantitative analysis on low SNR MR images. Magn Reson Imaging 1993; 11: 1051–1056. [DOI] [PubMed] [Google Scholar]
- 30.Raya J, Dietrich O, Horng A, Weber J, Reiser M, Glaser C. T2 measurement in articular cartilage: Impact of the fitting method on accuracy and precision at low SNR. Magnetic Resonance in Medicine 2010; 63: 181–193. [DOI] [PubMed] [Google Scholar]
- 31.Kohn MD, Sassoon AA, Fernando ND. Classifications in Brief: Kellgren-Lawrence Classification of Osteoarthritis. Clin Orthop Relat Res 2016; 474: 1886–1893. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Alexos A, Moustakidis S, Kokkotis C, Tsaopoulos D. Physical activity as a risk factor in the progression of osteoarthritis: a machine learning perspective. International Conference on Learning and Intelligent Optimization: Springer 2020:16–26. [Google Scholar]
- 33.Alghadir AH, Anwer S, Sarkar B, Paul AK, Anwar D. Effect of 6-week retro or forward walking program on pain, functional disability, quadriceps muscle strength, and performance in individuals with knee osteoarthritis: a randomized controlled trial (retro-walking trial). BMC Musculoskelet Disord 2019; 20: 159. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Joseph GB, Baum T, Alizai H, Carballido-Gamio J, Nardo L, Virayavanich W, et al. Baseline mean and heterogeneity of MR cartilage T2 are associated with morphologic degeneration of cartilage, meniscus, and bone marrow over 3 years--data from the Osteoarthritis Initiative. Osteoarthritis Cartilage 2012; 20: 727–735. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Chen T, Guestrin C. XGBoost: A Scalable Tree Boosting System. Proceedings of the 22nd ACM SIGKDD International Conference on Knowledge Discovery and Data Mining. San Francisco, California, USA: Association for Computing Machinery; 2016:785–794. [Google Scholar]
- 36.Friedman JH, Meulman JJ. Multiple additive regression trees with application in epidemiology. Stat Med 2003; 22: 1365–1381. [DOI] [PubMed] [Google Scholar]
- 37.Elith J, Leathwick JR, Hastie T. A working guide to boosted regression trees. J Anim Ecol 2008; 77: 802–813. [DOI] [PubMed] [Google Scholar]
- 38.DeLong ER, DeLong DM, Clarke-Pearson DL. Comparing the areas under two or more correlated receiver operating characteristic curves: a nonparametric approach. Biometrics 1988; 44: 837–845. [PubMed] [Google Scholar]
- 39.Meyers J Paper AD-088: Demographic Table and Subgroup Summary Macro %TABLEN. Pharmaceuticals SAS Users Group conference. San Francisco, CA2020. [Google Scholar]
- 40.Pedoia V, Lee J, Norman B, Link TM, Majumdar S. Diagnosing osteoarthritis from T2 maps using deep learning: an analysis of the entire Osteoarthritis Initiative baseline cohort. Osteoarthritis Cartilage 2019; 27: 1002–1010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Jamshidi A, Pelletier JP, Martel-Pelletier J. Machine-learning-based patient-specific prediction models for knee osteoarthritis. Nat Rev Rheumatol 2019; 15: 49–60. [DOI] [PubMed] [Google Scholar]
- 42.Tiulpin A, Klein S, Bierma-Zeinstra SMA, Thevenot J, Rahtu E, Meurs JV, et al. Multimodal Machine Learning-based Knee Osteoarthritis Progression Prediction from Plain Radiographs and Clinical Data. Sci Rep 2019; 9: 20038. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Leung K, Zhang B, Tan J, Shen Y, Geras KJ, Babb JS, et al. Prediction of Total Knee Replacement and Diagnosis of Osteoarthritis by Using Deep Learning on Knee Radiographs: Data from the Osteoarthritis Initiative. Radiology 2020; 296: 584–593. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Jamshidi A, Leclercq M, Labbe A, Pelletier JP, Abram F, Droit A, et al. Identification of the most important features of knee osteoarthritis structural progressors using machine learning methods. Ther Adv Musculoskelet Dis 2020; 12: 1759720X20933468. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Becker D XGBoost. Learn Machine Learning 2018. [Google Scholar]
- 46.Nielsen D Tree boosting with xgboost-why does xgboost win” every” machine learning competition? : NTNU; 2016. [Google Scholar]
- 47.Ashmeik W, Joseph GB, Nevitt MC, Lane NE, McCulloch CE, Link TM. Association of blood pressure with knee cartilage composition and structural knee abnormalities: data from the osteoarthritis initiative. Skeletal Radiol 2020; 49: 1359–1368. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Zhang YM, Wang J, Liu XG. Association between hypertension and risk of knee osteoarthritis: A meta-analysis of observational studies. Medicine (Baltimore) 2017; 96: e7584. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Razmjoo A, Caliva F, Lee J, Liu F, Joseph GB, Link TM, et al. T2 analysis of the entire osteoarthritis initiative dataset. J Orthop Res 2021; 39: 74–85. [DOI] [PubMed] [Google Scholar]
- 50.Koo TK, Li MY. A Guideline of Selecting and Reporting Intraclass Correlation Coefficients for Reliability Research. J Chiropr Med 2016; 15: 155–163. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Supplemental Figure 1: Model performance results on the validation set. Although we show these results for completeness, the final performance is based on the holdout test set as it represents the most unbiased dataset that the model had not seen or optimized on.
Supplemental Figure 2: Confusion matrices for models 1, 2, and 3.
Supplemental Table 1: List of 112 predictors included in data exploration
Supplemental Table 2: The number of and percentage of missing data for each predictor in the model with 10 predictors (final proposed model).


