Abstract
Genome-wide association studies (GWAS) provide an unbiased first look at genetic loci involved in aging and noise-induced sensorineural hearing loss and tinnitus. The hearing phenotype, whether audiogram-based or self-report, is regressed against genotyped information at representative single nucleotide polymorphisms (SNPs) across the genome. Findings include the fact that both hearing loss and tinnitus are polygenic disorders, with up to thousands of genes, each of effect size of < 0.02. Smaller human GWAS’ were able to use objective measures and identified a few loci, however, hundreds of thousands of participants have been required for the statistical power to identify significant variants, and GWAS is unable to assess rare variants with mean allele frequency < 1%. Animal studies are required as well because of inability to access the human cochlea. Mouse GWAS builds on linkage techniques and the known phenotypic differences in auditory function between inbred strains. With the advantage that the laboratory environment can be controlled for noise and aging, the Hybrid Mouse Diversity Panel (HDMP) combines 100 strains sequenced at high resolution. Lift-over regions between mice and humans have identified over 17,000 homologous genes. Since most significant SNPs are either intergenic or in introns, and binding sites between species are poorly preserved between species, expression quantitative trait locus information is required to bring humans and mice into agreement. Transcriptome-wide analysis studies (TWAS) can prioritize putative causal genes and tissues. Diverse species, each making a distinct contribution, carry a synergistic advantage in the quest for treatment and ultimate cure of sensorineural hearing difficulties.
Keywords: Hearing loss, tinnitus, genome-wide association studies, Hybrid mouse diversity panel
The World Health Organization estimates that 1.4 billion (18.7% of the global population) live with hearing loss, and 90% of those with moderate to profound hearing loss reside in low- and middle-income countries (Bolajoko et al. 2019). The economic impact is estimated at over 750 billion US dollars yearly, including health care costs, loss of productivity due to unemployment, and societal costs of disability-adjusted life years (DALYs). Although a portion of this burden is secondary to a controllable environmental exposure such as noise, heritability studies of sensorineural hearing loss (SNHL) indicate a genetic contribution of 53%-65% (Karlsson et al. 1997; Kvestad et al. 2012; Bogo et al. 2017), while tinnitus appears to be 27-56% heritable (Bogo et al. 2017; Maas et al. 2017). Yet, despite its pervasiveness, there is currently no known cure for either non-congenital sensorineural hearing loss or tinnitus.
In order to focus on genetic intervention and treatment, the identification of genes, variants, and pathways associated with human hearing dysfunction is necessary. Genome-wide association studies (GWAS) have become a core initial step, particularly in polygenic disorders such as these. The burgeoning of statistical techniques in the analysis of large genomic and phenotypic data sets has allowed a “first look” at human and animal DNA analysis, tentatively narrowing the search for relevant genes in diseases to specific haplotype blocks of DNA prior to fine-mapping of relevant variants and genes responsible for population-wide auditory disorders. This review will focus on hearing loss etiologies responsible for the majority of permanent sensorineural hearing loss (SNHL) in the world, specifically age-related and chronic noise-induced hearing loss, and tinnitus, rather than rarer syndromic and non-syndromic congenital hearing loss. We will discuss GWAS in animal and human studies, the polygenic nature of these disorders dictating large datasets, disparate phenotypes, and the challenging task of correlating between species.
A. Genotype considerations in GWAS of human hearing difficulties.
Genome-wide association studies offer an impartial examination of the human genome and consist of either linear regression on each SNP against audiogram data in the case of continuous variables, or logistic regression for case/control studies of those with hearing difficulties versus normal hearing controls. Briefly, the phenotype, i.e., the definition of hearing difficulty, is regressed against participants’ allele findings at representative loci along the genome (Hoffmann et al. 2016; Wells et al. 2019). Current microarrays contain over 800,000 single nucleotide polymorphisms (SNPs) and can be augmented with imputation (Howie et al. 2009; Herzig et al. 2018) of other variants with their linkage disequilibrium values (LD) to narrow findings to smaller loci. While a general discussion of GWAS is beyond the scope of this paper, several thorough manuscripts regarding its scope and requirements can be found (Visscher, PM et al. 2012; Visscher et al. 2017).
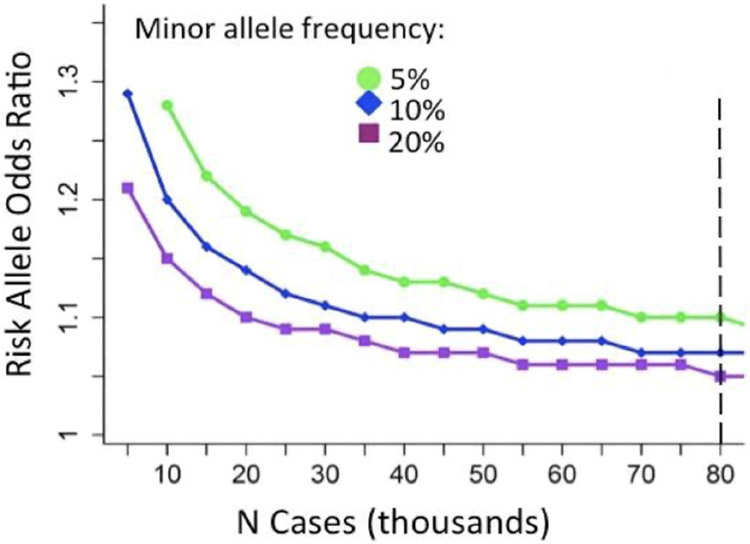
Both hearing loss and tinnitus have been shown to be highly polygenic – their heritable portion consists of multiple variants, each individual SNP having a low effect size, or “beta” (ß) (Gilles et al. 2017). Polygenicity is estimated at 92-95% for hearing difficulties and 96% for tinnitus (Wells et al. 2019; Clifford et al. 2020), indicating that thousands of variants and genes are involved in these disorders, each with small but additive impact. Because of the small effect size of individual SNPs, in order to achieve significance, a study ideally requires at least tens of thousands of cases to arrive at the accepted significant p-value of 5 x 10−8 (See Figure 1), depending on its minor allele frequency (MAF) in the studied population. The overwhelming majority of significant GWAS SNPs in tinnitus and hearing loss have each demonstrated an effect ß of less than 0.02 (Wells et al. 2019; Kalra et al. 2020; Clifford et al. 2020), requiring 100’s of thousands of participants for statistical significance. Inherent in GWAS, the power to detect significance in minor allele frequencies (MAF) of less than 1% is low. At least partly because of this limitation, the largest hearing GWAS studies have explained only up to 19% of the heritability associated with the trait, far short of the broad-sense heritability of 40-75% noted in twin studies (Kvestad et al. 2012; Wells et al. 2019; Kalra et al. 2020).
Fig. 1.
Power calculations showing relationship between increasing sample size and ability to detect loci of different population mean allele frequencies for case-control analysis.
Nevertheless, GWAS in large cohorts, such as the UK Biobank (UKB) or Million Veteran Program (MVP) that have aggregated individual genotyping associated with phenotype information, either medical records or self-report, have allowed a first look at the complexities of these disorders and elicited multiple SNPs, genes, and pathways for further analysis (Gaziano et al. 2016; Bycroft et al. 2018).
B. Human phenotype considerations.
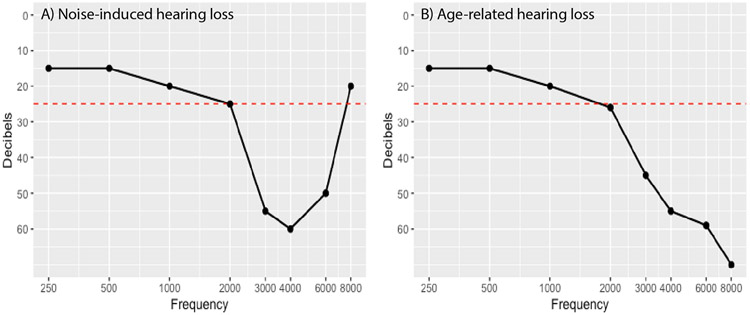
In previous years, when numbers of cases were limited to thousands, studies were able to use audiogram measures. The “shape” of the audiogram can indicate the etiology of hearing loss, i.e., classically, age-related hearing loss shows a slope in the higher frequencies, while noise-induced loss audiograms have a “notch” from 3-6 kHz. Studies have shown highly significant heritability for the concavity, slope, and individual threshold values (Figure 2) (Rabinowitz et al. 2006; Wingfield et al. 2007; Demeester et al. 2010). In order to include both threshold levels and “shapes” in the phenotype, researchers employed audiogram values and principal components (PCs) of the audiogram, where an “eigenvalue” is the total variability explained for that PC. Studies indicate that PC1 measures the overall amount of loss, PC2 indicates high frequency loss, and PC3 accounts for the “shape” of the audiogram (Smith 2002; Huyghe et al. 2008; Demeester et al. 2010; van Laer et al. 2010; Girotto et al. 2011; Anwar and Oakes 2012; Nagtegaal et al. 2019).
Fig. 2.
Examples of hearing loss of different etiologies. A) Noise-induced hearing loss classically shows a “notch” between the frequencies of 3-6 kHz with an upslope in the higher frequencies. B) Age-related hearing loss demonstrates a down-sloping shape in the higher frequencies. Normal hearing can be defined as a hearing threshold at 25 dB or above in all frequencies (red dotted line). The first three principal components capture approximately 87% of the variability in the audiogram and have been shown to constitute 1. Overall amount of hearing loss, 2. High frequency loss, and 3. Overall ‘shape of the audiogram, relevant to the “noise notch” or the age-related “slope” (Huyghe et al. 2008; Demeester et al.).
Because of budget constraints, larger data sets have used self-report or International Classification of Diagnosis (ICD) from the medical record for analysis (Hoffmann et al. 2016; Wells et al. 2019; Kalra et al. 2020; Clifford et al. 2020). Self-report increases the number of participants, as audiograms are time-consuming and expensive to administer, however correlation of self-report with objective measures such as audiograms range from 0.30 to 0.84 (Cherny et al. 2020). McCullagh, et al., noted that although 42% of workers at a factory had documented hearing loss (a threshold of > 25 dB at the frequencies of 2, 3, or 4 k), 76% of them noted their hearing to be excellent or good (McCullagh et al. 2011). In addition, use of self-report makes hearing loss a threshold model, where those who have an aggregate number or certain combination of variants above a threshold “value” are susceptible, and an “exposure” such as age, traumatic brain injury, or chronic noise is required to elicit the disorder (Cederroth et al. 2019).
The UK Biobank self-report includes a separate question on speech intelligibility in background noise which has been absorbed into other hearing phenotypes (Wells et al. 2019; Kalra et al. 2020); speech-in-noise is 25% heritable after age-adjustment (95% CI, 16%, 33%) (Momi et al. 2015). Difficulties in understanding speech may be related to a “hidden hearing loss.” This “hidden hearing loss”, secondary to loss of synapses between inner hair cells and cochlear nerve fibers, occurs prior to loss of outer or inner hair cells in the cochlea, and is associated with no visible pure-tone audiogram loss (Liberman and Kujawa 2017). This loss of comprehension in the face of normal audiograms has been noted in tinnitus as well (Gabriela et al. 2010; Gilles et al. 2016; Liberman et al. 2016). It will be important going forward to have objective measurements of both threshold values as well as a measure of comprehension of speech in noise, as different phenotypes may elicit different variants in GWAS analysis.
Tinnitus remains a subjective disorder that shares 0.489 (SE 0.072) of its genetic architecture with hearing loss (Clifford et al. 2020). In some analyses it has been subsumed into a general “hearing difficulties” phenotype (Kalra et al. 2020); variants identified in larger studies may be within genes and pathways shared between hearing loss and tinnitus. Anatomically, while hearing loss is thought to relate directly to cochlear sensory damage, current models associate tinnitus with central neural changes, dictating different gene isoforms in different anatomic locations (Elgoyhen et al. 2015). In addition to genes related directly to the perception of ringing in the ears, self-reports of annoyance from the disorder may measure variants in portions of the brain and brainstem other than the auditory pathway. Imaging studies in tinnitus demonstrate multiple connections between intracranial areas including prefrontal, orbitofrontal, anterior cingulate, parahippocampus, in both gray and white matter (Demopoulos et al. 2020; Lin et al. 2020). These intracranial areas outside the auditory pathway are coupled with emotions, cognition, and memory, all of which have been associated both epidemiologically and genetically with tinnitus (Leaver et al. 2012; Tegg-Quinn et al. 2016; Lin et al. 2020; Clifford et al. 2020; Prewitt et al. 2021).
C. Human hearing and tinnitus GWAS.
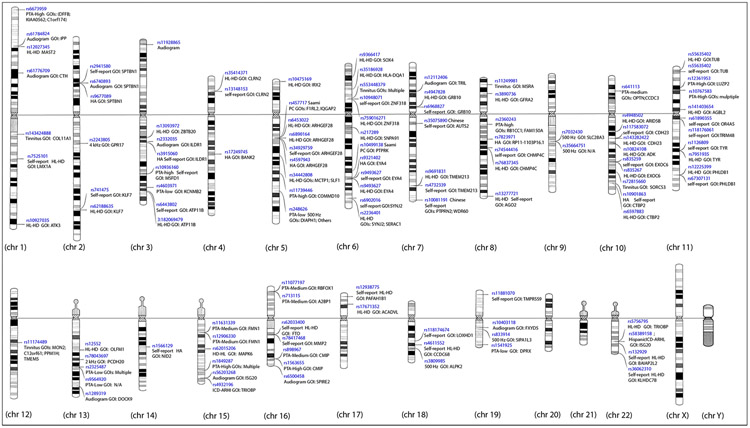
The first GWAs on SNHL were family-based studies to control for heterogeneity and population stratification. Early reports using twins or multiple kinships regressed audiogram data to identify large chromosomal loci (DeStefano et al. 2003; Garringer et al. 2006; Huyghe et al. 2008). When it became clear that presbycusis and noise-induced hearing loss were polygenic disorders, larger isolated populations were identified in order to increase population homozygosity (van Laer et al. 2010; Girotto et al. 2011). Friedman, et al., pooled study results from eight European centers and used an Affymetrix 500K GeneChip to identify GRM7 (glutamate receptor, metabotropic 7), with replication in different communities and relevant expression of the gene in cochlear cells (Friedman et al. 2009). See Figure 3 and S1 for human chromosome map of loci identified through 2021. Although these studies set the standard by using the first gene-chip arrays, utilizing objective measures of hearing, and reporting suggestive SNPs, they individually did not achieve statistical significance (Fransen et al. 2003). The largest study thus far with objective data from audiograms was a meta-analysis which identified three significant and seven suggestive variants, five of which were ultimately replicated in UKB (Nagtegaal et al. 2019; Wells et al. 2019). SNP heritability was measured at 11-15%, and SNPs demonstrated differential effect based on low versus high frequencies. In an effort to narrow the phenotype to age-related hearing loss, a study from Kaiser Hospitals used diagnosis codes from the medical record chart to identify 6,527 cases/45,882 controls, finding two significant SNPs (Hoffmann et al. 2016) in TRIOBP and ISG20, which were later replicated in the larger studies.
Fig. 3.
Chromosome map of significant loci identified in humans GWAS with phenotypes and genes of interest. Except as noted, loci were identified in those of Caucasian ancestry. Abbr. rs – reference sequence for single nucleotide polymorphism; HL-HD - hearing loss or hearing difficulty; HA – self-report of hearing aid use; GOI – gene of interest; PTA-Low – pure tone average of lower frequencies; PTA-High – pure tone average of higher frequencies; kHz – kilohertz (DeStefano et al. 2003; Garringer et al. 2006; Huyghe et al. 2008; Newman et al. 2008; van Laer et al. 2010; Girotto et al. 2011; Vuckovic et al. 2015; Hoffmann et al. 2016; Nagtegaal et al. 2019; Wells et al. 2019; Kalra et al. 2020; Clifford et al. 2020; Niu et al. 2021).
Case/control studies that have used hundreds of thousands of participants and an aggregate of self-reported answers have been the most productive in identifying significant variants. Wells, et al. identified 44 independent loci and Kalra, et. al., then duplicated 23 of these and found eight in addition, using a different combination of self-reported hearing loss and tinnitus questions from the same UKB cohort (Wells et al. 2019; Kalra et al. 2020). For tinnitus, using a broad phenotype of regularly having tinnitus for greater than 5 minutes at a time, past or present, allowed identification of six significant SNPs and 27 genes. Three out of six SNPs and 8/27 genes were replicated in the Million Veteran Program cohort (Clifford et al. 2020). Eight of these 27 tinnitus genes were identified in hearing GWAS in the same UKB cohort, namely COL11A1, ZNF318, AGO2, SLC22A7, CRIP3, MSRA, MON2, and C12orf61, suggesting that some of these may be in shared pathways involved in reaction to damage in the auditory system. Using a different “extreme” phenotype of ringing in the ears “almost all of the time, RCOR1 was reported in tinnitus GWAS using UKB data (Wells et al. 2020).
Thus, success in identifying variants related to population hearing difficulties so far has been with large homogeneous populations and wide-ranging phenotypes, including self-reported hearing difficulty, wearing hearing aids, tinnitus, and speech comprehension.
D. Limitations of Human GWAS and future directions.
By design, GWAS is an unbiased “sampling” of the genome based on common variants. On the other hand, rare variants are defined as those with minor allele frequencies less than 1% and because these rare variants mask bias secondary to population structure, they are usually excluded from analysis. Nevertheless, they may be rare for a reason – they can carry a higher combined annotation dependent depletion score (CADD), indicating deleterious causal variants (Rentzsch et al. 2019). In order to complete the picture that begins with GWAS, recently researchers have utilized whole genome sequencing (WGS) and whole exome sequencing (WES) to quantify the percentage of rare variants in cases compared to controls without hearing loss.
In looking at rare variants, it is critical to use an appropriate control group. Using WES, the same percentage of variants associated with deafness-associated genes was found in cases versus 1000 Genomes Project controls (Lewis et al. 2018). In contrast, when controls having normal hearing were used instead of the general population, a difference between the numbers of rare deleterious variants between cases and controls was identified. Genes in the pathway of “sensory perception of sound” were the most enriched in a European population study (Vuckovic et al. 2018).
Although the analysis of WES and WGS requires a different approach and is beyond the scope of this paper, recent studies using deep sequencing has uncovered a burden of rare variants in significant GWAS genes. GWAS has identified SNPs within EYA4, a known syndromic hearing loss gene identified as significant in GWAS for hearing loss, was studied within the context of adult-onset hearing loss. Those with hearing loss were found to have a significant burden of deleterious rare variants within EYA4 (Ahmadmehrabi et al. 2021). In other studies, analysis by selecting extreme phenotypes, i.e., those with early onset of disease, or severe hearing loss, has recently been utilized to examine complex diseases (Amanat et al. 2020). Using the standard Tinnitus Handicap Inventory questionnaire, subjects scoring ≥ 76 as extreme and between 56 and 76 as “almost extreme” and focusing on synaptic genes, Amanat, et al identified a burden of rare missense variants in ANK2, AKAP9, and TSC2 associated with severe tinnitus (Amanat et al. 2021). Fine mapping following unbiased GWAS rare variants will be required to pinpoint genetic variations associated with common, complex auditory difficulties.
Since heritability has been noted to be frequency-specific both in humans and in animal models (Wingfield et al. 2007; Demeester et al. 2010), in order to pinpoint genes and pathways directly related to hearing and/or comprehension it will be important to utilize an objective measure of hearing loss. This may involve a combination of principal components of audiogram thresholds and/or a measure of speech comprehension. Large genotype datasets integrated with health records including audiograms and speech audiometry will enable researchers to identify genes and pathways specific to narrower phenotypes, i.e., speech in noise, frequency-specific genomic influences, and constant versus intermittent tinnitus, for example.
E. Pre-GWAS attempts to identify non-congenital hearing loss genes in mice.
Phenotypic differences in auditory function between inbred strains were well-documented before the high-throughput molecular technologies era (Henry 1982). The C57BL6 strain shows a mild progressive hearing loss in high frequencies beginning as early as 9 months of age that spreads to mid-frequencies by 12 months. A/J and DBA-related strains have an earlier onset with more severe hearing loss. In contrast, CAST/Ei and CBA-related strains maintain good hearing through 15 months of age (Zheng et al. 1999). This phenotypic variation among inbred strains reflects the idea that age-related hearing loss (AHL) consists of quantitative trait loci (QTLs) appropriate for genetic mapping. Linkage analysis techniques were applied to those strains and other genome-tagged mice to identify ahl loci: Ahl1 (Johnson et al. 1997), Ahl2 (Johnson and Zheng 2002), Ahl3 (Nemoto et al. 2004), Ahl4 (Zheng et al. 2009), Ahl5 and Ahl6 (Drayton and Noben-Trauth 2006), and Ahl8 (Johnson et al. 2006); progressive hearing loss (Phl) −1 and −2 (Mashimo et al. 2006); and sensorineural hearing loss loci (Snhl) −1, −2, −3, and −4 (Noben-Trauth et al. 2010; Latoche et al. 2011). QTLs associated with noise damage susceptibility or recovery ability were identified either by mapping ABR thresholds (White et al. 2009)or endocochlear potential variation (Ohlemiller et al. 2016).
While linkage analysis in inbred and genome-wide tagged strains were successful in mapping the firsts ahl loci, lower resolution of the mapped QTLs hindered identification of specific genes and loci. Only two genes within these loci have been pinpointed: a mutation in the cadherin 23 (Cdh23) gene was found to be responsible for the effect of the Ahl1 locus (Noben-Trauth et al. 2003), while a nonsynonymous substitution in the mouse fascin-2 gene (Fscn2) was associated with the Ahl8 locus (Shin et al. 2010).
F. The advent of mouse genome-wide association studies.
The usefulness of human GWAS in hearing investigations has been limited by the inability to control or measure environmental noise exposure throughout a lifetime. In contrast, GWAS in multiple inbred strains permits careful control of environmental factors where measurements can be replicated in genetically identical animals, increasing the proportion of the variability explained solely by genetic variation. The classic advantages of mice are their close evolutionary relationship to human genome where up to 99% of genes in mice have a human orthologue, early onset of sexual maturity, and relatively short life span. The structural and physiological similarities in auditory functions between mice and humans and the extensive overlap of genes and proteins critical for these functions make the mouse a crucial model system for the study of the functional genomics of the auditory system. Complex traits have been shown to have higher heritability in mice, and genetic loci often have stronger effects in mice compared to humans (Lindblad-Toh et al. 2000; Wiltshire et al. 2003; Yalcin et al. 2004).
The Hybrid Mouse Diversity Panel (HMDP) provides much higher resolution for associated loci than traditional approaches with QTL mapping. The HMDP is a collection of 30 classical inbred strains (CI) and 70 recombinant inbred strains (RI) that together provide genetic resolution and allelic diversity. Power calculations have demonstrated that this panel is superior to traditional linkage analysis and is capable of detecting loci responsible for 5% of the overall phenotypic variance (Bennett et al. 2010; Ghazalpour et al. 2012; Flint and Eskin 2012).
GWAS analysis of complex traits using this panel with an efficient mixed-model algorithm (EMMA) (Kang et al. 2008) has revealed several candidate genes for age-related hearing loss (Lusis et al. 2016). The first AHL GWAS on the HMDP panel utilized 99 strains mapped for their auditory brainstem response (ABR) across six frequencies (4, 8, 12, 16, 24, and 32 kHz). Wave I of ABR is a signal generated from the distal portion of cranial nerve VIII where it exits the cochlea. Results indicated a two- to five-fold variation in each audiogram frequency between strains and identification of nine frequency-specific significant loci (Crow et al. 2015). The data sets from this cohort along with four other cohorts (Zheng et al. 1999; Johnson et al. 2008) were combined in a meta-analysis for a genome-wide association study (GWAS) including 937 samples that identified five genome-wide significant loci. One of these loci confirmed Ahl8 and narrowed its candidate region on chromosome 11 to a SNP 450 kb upstream from the responsible gene (Ohmen et al. 2014). Another locus identified a SNP mapped to chromosome 8 in an interval of approximately 2 Mb containing Nrp1, a transmembrane receptor type I protein known to bind both vascular endothelial growth factor beta (Vegfb) and semaphorin classes. Salehi, et al., noted that Nrp1 mRNA expression coincided with early postnatal stages of cochlear hair cell innervation. Nrp1−/− mice demonstrated an elevated hearing threshold from 4 to 32kHz at 2-months of age that progressed with age. In addition, young Nrp1 conditional knockout mice showed disorganized outer spiral bundles, enlarged microvessels of the stria vascularis, and a reduction of presynaptic ribbons in outer hair cells (Salehi et al. 2017).
Besides age-related studies, HMDP strains have also been utilized for noise-induced hearing loss GWAS. ABR data collected from mice prior and after 2-hr exposure to 10-kHz octave band noise at 108 dB sound pressure level showed large frequency-specific strain variation in susceptibility to NIHL, suggesting a gene-by-environment interaction. GWAS analysis with correction for population structure yielded multiple genome-wide significant loci, with a peak for susceptibility to NIHL on chromosome 17 within a haplotype block containing NADPH oxidase-3 (Nox3). Nox3 mutants and heterozygotes showed a greater susceptibility to NIHL specifically at 8 kHz on ABR with normal distortion-product otoacoustic emissions, a measure of outer hair cell function. Synaptic ribbons of inner hair cells in the mutant animals were specifically diminished at 8 kHz (Lavinsky et al. 2015, 2016).
In order to validate an objective phenotype for hearing in the HDMP, we recently measured variation in Wave 1 amplitude in 102 strains pre- and post-noise exposure. Wave 1 amplitude correlated with the density of the auditory neurons and the complement of synaptic ribbons within the inner hair cells. Amplitude change pre- and post-noise exposure varied up to 7.5-fold through frequencies tested (4 kHz, 8 kHz, 12 kHz, 16 kHz, 24 kHz, and 32 kHz). Immunolabeling of paired synaptic ribbons and glutamate receptors of strains with the highest and lowest Wave 1 values revealed significant correlation with synaptic ribbon counts pre- and post-exposure. Via microarray profiling for cis-eQTLs (expression quantitative trait loci) in whole cochleae, 17 candidate genes were prioritized. GWAS analysis defined two genomic regions associated with Wave 1 amplitude prior to noise exposure and an additional two associated after noise exposure (Boussaty et al. 2020).
Although GWAS has identified multiple loci for age-related and noise induced hearing loss, limitations of using the HMDP include a finite number of inherited alleles in these inbred strains. Commercial outbred strains which are produced to foster genetic diversity both in terms of homozygosity/heterozygosity and haplotypes are more advantageous in complex trait mapping (Chia et al. 2005). The decreasing cost of sequencing makes it possible to genotype large numbers of animals required to capture the full diversity in these outbred strains. The introduction of a new generation RNA techniques such as single-cell RNAseq will enable comparison of the function of genes within cell types in the development and homeostasis of the inner ear.
To our knowledge, no HMDP GWAS has been performed on a phenotype of tinnitus. Animal models of tinnitus consist of either pharmaceutical or noise induction, followed by gap pre-pulse inhibition coordinated with normal ABR Wave I, fear conditioning, reward learning, and other combinations of behavioral testing (Heffner and Heffner 2012) Important considerations in tinnitus GWAS in animals will be the existence of concomitant hearing loss, different response to the inducing method by different strains, permanence of behavioral training for testing, and ease of testing hundreds of animals of different strains.
G. Combining mouse with human results – beyond GWAS.
Lift-over regions between mice and humans have been well documented, and it is possible to identify homologous genomic loci (http://genome.ucsc.edu/goldenPath/help.hgTracksHelp.html) in over 17,000 mouse genes (Maynard and Ackert-Bicknell 2019). Nevertheless, despite anatomic and pathophysiological similarities, to date, genes identified in mice GWAS have not correlated with human association studies. In part, this may be due to disparate phenotypic definitions of hearing. Mouse GWAS has measured auditory brainstem response (ABR) Wave I as an objective trait, with identification of frequency-specific loci. Larger human studies with more statistical power to identify variants have used broad, self-reported definitions of hearing, which measure a perception rather than a threshold value. In addition, as noted before, murine environmental noise exposure can be controlled, thus unlike human studies, age-related hearing loss can be separated from noise-induced loss, with specific parameters for noise and age. The largest hearing cohort available to date, the Million Veteran Program, consists of over 850,000 subjects who have been exposed to ships, aircraft, artillery, guns, tanks, and other military hardware at a young age. These two mechanisms of injury, i.e., age-related, and noise-induced, may have different pathways of injury and repair as evidenced by different audiograms for the two disorders (Rabinowitz et al. 2006).
Translation from mice to humans has been problematic. Uniformly, no significant HDML genes or loci have translated to humans and vice versa. Part of the problem of disparate GWAS’ is that causative variants are located predominantly in intronic and intragenic areas, more in line with regulatory sequences. Comparison of transcription factor binding between species indicates that no more than 20% of binding sites are conserved (Breschi et al. 2017), however pathways and functions are preserved. Recently, computational models have attempted to bridge the species gap. Semi-supervised learning in publicly available datasets has been shown to be more effective in inter-species translation than empiric comparison of genes (Brubaker et al. 2019). Other data-driven statistical models have been able to more successfully predict relevant human genes (Normand, 2018) which greatly prioritizes genes for translational research. The ENCODE program has expanded their database of cis- and trans- regulatory elements to coordinate 926,535 human with 339,815 mouse genes (The ENCODE Project Consortium, 2020).
Coordination of results from mouse to human GWAS is crucial because ultimately, validation is only possible in the laboratory, i.e., modification of mouse genomes for proof of function. Transcription-wide association studies (TWAS) have been used with success (Pastina et al. 2019) to identify homologous functions. TWAS uses eQTL data to identify gene/phenotype associations from GWAS in order to prioritize putative causal genes and tissues. A TWAS pathway method, JEPEGMIX2-P, tests for association between pathway expression and phenotype (Chatzinakos et al. 2020).
Because of constraints of access to the live human cochlea, the mouse can be studied in regard to genomic expression data in the unique cell types of the inner ear. In addition, the murine noise exposure environment can be controlled and objectively measured where the human is subject to a lifetime of varied environmental sound.
Diverse species, each making a distinct contribution, carry a synergistic advantage in the quest for treatment and ultimate cure of sensorineural hearing difficulties.
Acknowledgments:
This study was supported by NIDCD Grant 1R01DC018566-01A1. The authors are grateful to the Mills Auditory Foundation. We thank all the participants in the United Kingdom Biobank and the Veterans Administration Million Veteran Program.
Declarations
This study was supported by Grant 1R01DC018566-01A1 of National Institutes on Deafness and Other Communications Disorders (NIDCD)
Footnotes
Conflicts of Interest/Competing Interests – On behalf of all authors, the corresponding author states that there is no conflict of interest.
Ethics approval – not applicable.
Consent – not applicable.
Data – not applicable.
Material availability – not applicable.
Publisher's Disclaimer: This AM is a PDF file of the manuscript accepted for publication after peer review, when applicable, but does not reflect post-acceptance improvements, or any corrections. Use of this AM is subject to the publisher's embargo period and AM terms of use. Under no circumstances may this AM be shared or distributed under a Creative Commons or other form of open access license, nor may it be reformatted or enhanced, whether by the Author or third parties. See here for Springer Nature's terms of use for AM versions of subscription articles: https://www.springernature.com/gp/open-research/policies/accepted-manuscript-terms
References
- Ahmadmehrabi S, Li B, Park J, et al. (2021) Genome-first approach to rare EYA4 variants and cardio-auditory phenotypes in adults. Human Genetics 140:957–967. 10.1007/s00439-021-02263-6 [DOI] [PubMed] [Google Scholar]
- Amanat S, Gallego-Martinez A, Sollini J, et al. (2021) Burden of rare variants in synaptic genes in patients with severe tinnitus: An exome based extreme phenotype study. EBioMedicine 66: 10.1016/j.ebiom.2021.103309 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Amanat S, Requena T, Lopez-Escamez JA (2020) A Systematic Review of Extreme Phenotype Strategies to Search for Rare Variants in Genetic Studies of Complex Disorders. Genes 11: 10.3390/genes11090987 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Anwar MN, Oakes MP (2012) Data mining of audiology patient records: Factors influencing the choice of hearing aid type. BMC Medical Informatics and Decision Making 12:1–8. 10.1186/1472-6947-12-S1-S6 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bennett BJ, Farber CR, Orozco L, et al. (2010) A high-resolution association mapping panel for the dissection of complex traits in mice. Genome Research 20: 10.1101/gr.099234.109 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bogo R, Farah A, Karlsson KK, et al. (2017) Prevalence, Incidence Proportion, and Heritability for Tinnitus. Ear Hear 38:292–300. 10.1097/AUD.0000000000000397 [DOI] [PubMed] [Google Scholar]
- Bolajoko O, Davis AC, Hoffman HJ (2019) Hearing loss: Rising prevalence and impact. Bulletin of the World Health Organization 97:646–646A. 10.2471/BLT.19.224683 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Boussaty EC, Gillard D, Lavinsky J, et al. (2020) The Genetics of Variation of the Wave 1 Amplitude of the Mouse Auditory Brainstem Response. Journal of the Association for Research in Otolaryngology 21: 10.1007/s10162-020-00762-3 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Breschi A, Gingeras TR, Guigo R (2017) Comparative transcriptomics in human and mouse. Nature Reviews Genetics 18:425–440. 10.1038/nrg.2017.19 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brubaker DK, Proctor EA, Haigis KM, Lauffenburger DA (2019) Computational translation of genomic responses from experimental model systems to humans. PLoS computational biology 15:e1006286. 10.1371/journal.pcbi.1006286 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bycroft C, Freeman C, Petkova D, et al. (2018) Genome-wide genetic data on 500,000 UK Biobank participants. Nature 562:203–209. Epub 2018 Oct 10.30305743 [Google Scholar]
- Cederroth CR, Pirouzifard M, Trpchevska N, et al. (2019) Association of Genetic vs Environmental Factors in Swedish Adoptees with Clinically Significant Tinnitus. JAMA Otolaryngology - Head and Neck Surgery 145:222–229. 10.1001/jamaoto.2018.3852 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chatzinakos C, Georgiadis F, Lee D, et al. (2020) TWAS pathway method greatly enhances the number of leads for uncovering the molecular underpinnings of psychiatric disorders. American Journal of Medical Genetics Part B: Neuropsychiatric Genetics 183: 10.1002/ajmg.b.32823 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cherny SS, Livshits G, Wells HRR, et al. (2020) Self-reported hearing loss questions provide a good measure for genetic studies: a polygenic risk score analysis from UK Biobank. European Journal of Human Genetics 28:1056–1065. 10.1038/s41431-020-0603-2 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chia R, Achilli F, Festing MFW, Fisher EMC (2005) The origins and uses of mouse outbred stocks. Nature Genetics 37: 10.1038/ng1665 [DOI] [PubMed] [Google Scholar]
- Clifford RE, Maihofer AX, Stein MB, et al. (2020) Novel Risk Loci in Tinnitus and Causal Inference with Neuropsychiatric Disorders Among Adults of European Ancestry. JAMA otolaryngology-- head & neck surgery 146:1015–25. 10.1001/jamaoto.2020.2920 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Crow AL, Ohmen J, Wang J, et al. (2015) The Genetic Architecture of Hearing Impairment in Mice: Evidence for Frequency-Specific Genetic Determinants. G3 Genes | Genomes | Genetics 5: 10.1534/g3.115.021592 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Demeester K, van Wieringen A, Hendrickx J jaap, et al. (2010) Heritability of audiometric shape parameters and familial aggregation of presbycusis in an elderly Flemish population. Hearing Research 265:1–10. 10.1016/j.heares.2010.03.007 [DOI] [PubMed] [Google Scholar]
- Demopoulos C, Duong X, Hinkley LB, et al. (2020) Global resting-state functional connectivity of neural oscillations in tinnitus with and without hearing loss. Human Brain Mapping 41:2846–2861. 10.1002/hbm.24981 [DOI] [PMC free article] [PubMed] [Google Scholar]
- DeStefano AL, Gates GA, Heard-Costa N, et al. (2003) Genome-wide linkage analysis to presbycusis in the Framingham Heart Study. Archives of Otolaryngology - Head and Neck Surgery 129:285–289. 10.1001/archotol.129.3.285 [DOI] [PubMed] [Google Scholar]
- Drayton M, Noben-Trauth K (2006) Mapping quantitative trait loci for hearing loss in Black Swiss mice. Hearing Research 212: 10.1016/j.heares.2005.11.006 [DOI] [PubMed] [Google Scholar]
- Elgoyhen AB, Langguth B, De Ridder D, Vanneste S (2015) Tinnitus: perspectives from human neuroimaging. Nature Reviews Neuroscience 16:632–642. 10.1038/nrn4003 [DOI] [PubMed] [Google Scholar]
- Flint J, Eskin E (2012) Genome-wide association studies in mice. Nature Reviews Genetics 13:807–817. 10.1038/nrg3335 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Friedman RA, van Laer L, Huentelman MJ, et al. (2009) GRM7 variants confer susceptibility to age-related hearing impairment. Human Molecular Genetics 18:785–796. 10.1093/hmg/ddn402 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gabriela S, Sanches G, Samelli AG, Nishiyama AK (2010) GIN Test (Gaps-in-Noise) in normal listeners with and without tinnitus. SciElo Brasil 22:257–262. https://doi.org/S0104-56872010000300017 [DOI] [PubMed] [Google Scholar]
- Garringer HJ, Pankratz ND, Nichols WC, Reed T (2006) Hearing impairment susceptibility in elderly men and the DFNA18 locus. Archives of Otolaryngology - Head and Neck Surgery 132:506–510. 10.1001/archotol.132.5.506 [DOI] [PubMed] [Google Scholar]
- Gaziano JM, Concato J, Brophy M, et al. (2016) Million Veteran Program: A mega-biobank to study genetic influences on health and disease. Journal of Clinical Epidemiology 70:214–223. 10.1016/j.jclinepi.2015.09.016 [DOI] [PubMed] [Google Scholar]
- Ghazalpour A, Rau CD, Farber CR, et al. (2012) Hybrid mouse diversity panel: a panel of inbred mouse strains suitable for analysis of complex genetic traits. Mammalian Genome 23: 10.1007/s00335-012-9411-5 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gilles A, van Camp G, van De Heyning P, Fransen E (2017) A Pilot Genome-Wide Association Study Identifies Potential Metabolic Pathways Involved in Tinnitus. Frontiers in Neuroscience 11:1–10. 10.3389/fnins.2017.00071 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gilles A, Schlee W, Rabau S, et al. (2016) Decreased speech-in-noise understanding in young adults with tinnitus. Frontiers in Neuroscience 10:1–14. 10.3389/fnins.2016.00288 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Girotto G, Pirastu N, Sorice R, et al. (2011) Hearing function and thresholds: A genome-wide association study in European isolated populations identifies new loci and pathways. Journal of Medical Genetics 48:369–374. 10.1136/jmg.2010.088310 [DOI] [PubMed] [Google Scholar]
- Heffner HE, Heffner RS (2012) Behavioral Tests for Tinnitus in Animals. In: 1st ed. Springer Science and Business Media, New York, pp 21–58 [Google Scholar]
- Henry KR (1982) Influence of genotype and age on noise-induced auditory losses. Behavior Genetics 12: 10.1007/BF01070410 [DOI] [PubMed] [Google Scholar]
- Herzig AF, Nutile T, Babron MC, et al. (2018) Strategies for phasing and imputation in a population isolate. Genetic Epidemiology. 10.1002/gepi.22109 [DOI] [PubMed] [Google Scholar]
- Hoffmann TJ, Keats BJ, Yoshikawa N, et al. (2016) A Large Genome-Wide Association Study of Age-Related Hearing Impairment Using Electronic Health Records. PLoS Genetics 12:1–20. 10.1371/journal.pgen.1006371 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Howie BN, Donnelly P, Marchini J (2009) A flexible and accurate genotype imputation method for the next generation of genome-wide association studies. PLoS Genetics 5: 10.1371/journal.pgen.1000529 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Huyghe JR, van Laer L, Hendrickx JJ, et al. (2008) Genome-wide SNP-Based Linkage Scan Identifies a Locus on 8q24 for an Age-Related Hearing Impairment Trait. American Journal of Human Genetics 83:401–407. 10.1016/j.ajhg.2008.08.002 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Johnson K, Zheng Q (2002) Ahl2, a second locus affecting age-related hearing loss in mice. Genomics 80:461–464 [PMC free article] [PubMed] [Google Scholar]
- Johnson KR, Erway LC, Cook SA, et al. (1997) A major gene affecting age-related hearing loss in C57BL/6J mice. Hearing Research 114: 10.1016/S0378-5955(97)00155-X [DOI] [PubMed] [Google Scholar]
- Johnson KR, Longo-Guess C, Gagnon LH, et al. (2008) A locus on distal chromosome 11 (ahl8) and its interaction with Cdh23ahl underlie the early onset, age-related hearing loss of DBA/2J mice. Genomics 92: 10.1016/j.ygeno.2008.06.007 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Johnson KR, Zheng QY, Noben-Trauth K (2006) Strain background effects and genetic modifiers of hearing in mice. Brain Research 1091: 10.1016/j.brainres.2006.02.021 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kalra G, Milon B, Casella AM, et al. (2020) Biological insights from multi-omic analysis of 31 genomic risk loci for adult hearing difficulty. PloS Genetics . 16:1–32. 10.1371/journal.pgen.1009025 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kang HM, Zaitlen NA, Wade CM, et al. (2008) Efficient Control of Population Structure in Model Organism Association Mapping. Genetics 178: 10.1534/genetics.107.080101 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Karlsson KK, Harris JR, Svartengren M (1997) Description and Primary Results from an Audiometric Study of Male Twins. Ear and Hearing 18(2). https://journals.lww.com/ear-hearing/Fulltext/1997/04000/Description_and_Primary_Results_from_an.3.aspx [DOI] [PubMed] [Google Scholar]
- Kvestad E, Czajkowski N, Krog NH, et al. (2012) Heritability of hearing loss. Epidemiology 23:328–331. 10.1097/EDE.0b013e318245996e [DOI] [PubMed] [Google Scholar]
- Latoche JR, Neely HR, Noben-Trauth K (2011) Polygenic inheritance of sensorineural hearing loss (Snhl2, −3, and −4) and organ of Corti patterning defect in the ALR/LtJ mouse strain. Hearing Research 275: 10.1016/j.heares.2010.12.017 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lavinsky J, Crow AL, Pan C, et al. (2015) Genome-wide association study identifies nox3 as a critical gene for susceptibility to noise-induced hearing loss. PLoS genetics 11:e1005094. 10.1371/journal.pgen.1005094 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lavinsky J, Ge M, Crow AL, et al. (2016) The Genetic Architecture of Noise-Induced Hearing Loss: Evidence for a Gene-by-Environment Interaction. G3 Genes | Genomes | Genetics 6: 10.1534/g3.116.032516 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Leaver AM, Seydell-Greenwald A, Turesky TK, et al. (2012) Cortico-limbic morphology separates tinnitus from tinnitus distress. Frontiers in systems neuroscience 6:21. 10.3389/fnsys.2012.00021 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lewis MA, Nolan LS, Cadge BA, et al. (2018) Whole exome sequencing in adult-onset hearing loss reveals a high load of predicted pathogenic variants in known deafness-associated genes and identifies new candidate genes. BMC Medical Genomics 11:1–12. 10.1186/s12920-018-0395-1 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liberman MC, Epstein MJ, Cleveland SS, et al. (2016) Toward a differential diagnosis of hidden hearing loss in humans. PLoS ONE 11:1–15. 10.1371/journal.pone.0162726 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liberman MC, Kujawa SG (2017) Cochlear synaptopathy in acquired sensorineural hearing loss: Manifestations and mechanisms. Hearing Research 349:138–147. 10.1016/j.heares.2017.01.003 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lin X, Chen Y, Wang M, et al. (2020) Altered Topological Patterns of Gray Matter Networks in Tinnitus: A Graph-Theoretical-Based Study. Frontiers in Neuroscience 14:1–12. 10.3389/fnins.2020.00541 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lindblad-Toh K, Winchester E, Daly MJ, et al. (2000) Large-scale discovery and genotyping of single-nucleotide polymorphisms in the mouse. Nature Genetics 24: 10.1038/74215 [DOI] [PubMed] [Google Scholar]
- Lusis AJ, Seldin MM, Allayee H, et al. (2016) The Hybrid Mouse Diversity Panel: a resource for systems genetics analyses of metabolic and cardiovascular traits. Journal of Lipid Research 57: 10.1194/jlr.R066944 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Maas IL, Brüggemann P, Requena T, et al. (2017) Genetic susceptibility to bilateral tinnitus in a Swedish twin cohort. Genetics in Medicine 1–6. 10.1038/gim.2017.4 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mashimo T, Erven AE, Spiden SL, et al. (2006) Two quantitative trait loci affecting progressive hearing loss in 101/H mice. Mammalian Genome 17: 10.1007/s00335-004-2438-5 [DOI] [PubMed] [Google Scholar]
- Maynard RD, Ackert-Bicknell CL (2019) Mouse Models and Online Resources for Functional Analysis of Osteoporosis Genome-Wide Association Studies. Frontiers in Endocrinology 10: 10.3389/fendo.2019.00277 [DOI] [PMC free article] [PubMed] [Google Scholar]
- McCullagh M, Raymond D, Kerr M, Lusk S (2011) Prevalence of hearing loss and accuracy of self-report among factory workers. Noise Health 13:340–7. [DOI] [PubMed] [Google Scholar]
- Momi SK, Wolber LE, Fabiane SM, et al. (2015) Genetic and Environmental Factors in Age-Related Hearing Impairment. Twin Research and Human Genetics 18:383–392. 10.1017/thg.2015.35 [DOI] [PubMed] [Google Scholar]
- Nagtegaal AP, Broer L, Zilhao NR, et al. (2019) Genome-wide association meta-analysis identifies five novel loci for age-related hearing impairment. Scientific Reports 9:1–10. 10.1038/s41598-019-51630-x [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nemoto M, Morita Y, Mishima Y, et al. (2004) Ahl3, a third locus on mouse chromosome 17 affecting age-related hearing loss. Biochemical and Biophysical Research Communications 324: 10.1016/j.bbrc.2004.09.186 [DOI] [PubMed] [Google Scholar]
- Newman D, Fisher L, Ohmen J, et al. (2008) GRM7 variants associated with age-related hearing loss based on auditory perception. Hear Res 64:2391–2404. 10.1038/jid.2014.371 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Niu Y, Xie C, Du Z, et al. (2021) Genome-wide association study identifies 7q11.22 and 7q36.3 associated with noise-induced hearing loss among Chinese population. Journal of Cellular and Molecular Medicine 25:411–420. 10.1111/jcmm.16094 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Noben-Trauth K, Latoche JR, Neely HR, Bennett B (2010) Phenotype and Genetics of Progressive Sensorineural Hearing Loss (Snhl1) in the LXS Set of Recombinant Inbred Strains of Mice. PLoS ONE 5: 10.1371/journal.pone.0011459 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Noben-Trauth K, Zheng QY, Johnson KR (2003) Association of cadherin 23 with polygenic inheritance and genetic modification of sensorineural hearing loss. Nature Genetics 35: 10.1038/ng1226 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ohlemiller KK, Kiener AL, Gagnon PM (2016) QTL Mapping of Endocochlear Potential Differences between C57BL/6J and BALB/cJ mice. Journal of the Association for Research in Otolaryngology 17: 10.1007/s10162-016-0558-8 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ohmen J, Kang EY, Li X, et al. (2014) Genome-wide association study for age-related hearing loss (AHL) in the mouse: a meta-analysis. Journal of the Association for Research in Otolaryngology: JARO 15:335–352. 10.1007/s10162-014-0443-2 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pastina MM, Pinto LR, Oliveira KM, et al. (2019) Opportunities and chanllenges of TWAS. Nature Genetics 51:117–148. 10.1038/s41588-019-0385-z [DOI] [PMC free article] [PubMed] [Google Scholar]
- Prewitt A, Harker G, Gilbert TA, et al. (2021) Mental Health Symptoms Among Veteran VA Users by Tinnitus Severity:A Population-based Survey. Military medicine 186:167–175. 10.1093/milmed/usaa288 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rabinowitz PM, Galusha D, Slade MD, et al. (2006) Audiogram notches in noise-exposed workers. Ear and hearing 27:742–50. 10.1097/01.aud.0000240544.79254.bc [DOI] [PubMed] [Google Scholar]
- Rentzsch P, Witten D, Cooper GM, et al. (2019) CADD: predicting the deleteriousness of variants throughout the human genome. Nucleic Acids Research 47: 10.1093/nar/gky1016 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Salehi P, Ge MX, Gundimeda U, et al. (2017) Role of Neuropilin-1/Semaphorin-3A signaling in the functional and morphological integrity of the cochlea. PLOS Genetics 13: 10.1371/journal.pgen.1007048 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shin J-B, Longo-Guess CM, Gagnon LH, et al. (2010) The R109H Variant of Fascin-2, a Developmentally Regulated Actin Crosslinker in Hair-Cell Stereocilia, Underlies Early-Onset Hearing Loss of DBA/2J Mice. Journal of Neuroscience 30: 10.1523/JNEUROSCI.1541-10.2010 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Smith LI (2002) A tutorial on Principal Components Analysis Introduction. http://www.cs.otago.ac.nz/cosc453/student_tutorials/principal_components.pdf [Google Scholar]
- Tegg-Quinn S, Bennett RJ, Eikelboom RH, Baguley DM (2016) The impact of tinnitus upon cognition in adults: A systematic review. International Journal of Audiology 55:533–540. 10.1080/14992027.2016.1185168 [DOI] [PubMed] [Google Scholar]
- van Laer L, Huyghe J, Hannula S, et al. (2010) A genome-wide association study for age-related hearing impairment in the Saami. Eur J Hum Genet 18:685–693 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Visscher PM, McCarthy M, Yang J (2012) Five years of GWAS discovery. Am J Hum Genet 90:7–24. https://doi.org/0.1016/j.ajhg.2011.11.029 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Visscher PM, Wray NR, Zhang Q, et al. (2017) 10 Years of GWAS Discovery: Biology, Function, and Translation. American Journal of Human Genetics 101:5–22. 10.1016/j.ajhg.2017.06.005 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vuckovic D, Dawson S, Scheffer DI, et al. (2015) Genome-wide association analysis on normal hearing function identifies PCDH20 and SLC28A3 as candidates for hearing function and loss. Human Molecular Genetics 24:5655–5664. 10.1093/hmg/ddv279 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vuckovic D, Mezzavilla M, Cocca M, et al. (2018) Whole-genome sequencing reveals new insights into age-related hearing loss: cumulative effects, pleiotropy and the role of selection. European Journal of Human Genetics 26:1167–1179. 10.1038/s41431-018-0126-2 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wells H, Abidin F, Freidin M, et al. (2020) Genetic variation in RCOR1 is associated with tinnitus in UK Biobank. Bioreview Sept 24:1–20 [Google Scholar]
- Wells HRR, Freidin MB, Abidin FNZ, et al. (2019) Genome-wide association study identifies 44 independent genomic loci for self-reported adult hearing difficulty in the UK Biobank cohort. Am J Hum Genet 1–15. 10.1101/549071 [DOI] [PMC free article] [PubMed] [Google Scholar]
- White CH, Ohmen JD, Sheth S, et al. (2009) Genome-wide screening for genetic loci associated with noise- induced hearing loss. Mamm Genome 20:207–213. 10.1007/s00335-009-9178-5 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wiltshire T, Pletcher MT, Batalov S, et al. (2003) Genome-wide single-nucleotide polymorphism analysis defines haplotype patterns in mouse. Proceedings of the National Academy of Sciences 100: 10.1073/pnas.0130101100 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wingfield A, Panizzon M, Grant MDM, et al. (2007) A twin-study of genetic contributions to hearing acuity in late middle age. J Gerontol A Biol Sci Med Sci 62:1294–1299. 10.1111/j.1365-2958.2011.07804.x [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yalcin B, Fullerton J, Miller S, et al. (2004) Unexpected complexity in the haplotypes of commonly used inbred strains of laboratory mice. Proceedings of the National Academy of Sciences 101: 10.1073/pnas.0401189101 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zheng QY, Ding D, Yu H, et al. (2009) A locus on distal chromosome 10 (ahl4) affecting age-related hearing loss in A/J mice. Neurobiology of Aging 30. 10.1016/j.neurobiolaging.2007.12.011 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zheng QY, Johnson KR, Erway LC (1999) Assessment of hearing in 80 inbred strains of mice by ABR threshold analyses. Hearing Research 130: 10.1016/S0378-5955(99)00003-9 [DOI] [PMC free article] [PubMed] [Google Scholar]