Abstract
The ability to monitor DNA replication fork directionality at the genome-wide scale is paramount for a greater understanding of how genetic and environmental perturbations can impact replication dynamics in human cells. Here, we describe a detailed protocol for isolating and sequencing Okazaki fragments from asynchronously-growing mammalian cells, termed Okazaki fragment sequencing (Ok-seq), for the purpose of quantitatively determining replication initiation and termination frequencies around specific genomic loci by meta-analyses. Briefly, cells are pulsed with 5-ethynyl-2′-deoxyuridine (EdU) to label newly synthesized DNA, and collected for DNA extraction. After size-fractionation on a sucrose gradient, Okazaki fragments are concentrated and purified before click chemistry is used to tag the EdU label with a biotin conjugate that is cleavable under mild conditions. Biotinylated Okazaki fragments are then captured on streptavidin beads and ligated to Illumina adapters before library preparation for Illumina sequencing. The use of Ok-seq to interrogate genome-wide replication fork initiation and termination efficiencies can be applied to all unperturbed, asynchronously-growing mammalian cells or under conditions of replication stress, and the assay can be performed in less than 2 weeks.
Introduction
DNA replication in human cells is a highly regulated process and any errors could potentially lead to genomic instability, a hallmark of cancer1, 2. In eukaryotes, replication origins are first licensed by pre-replication complexes (pre-RC) in the G1 phase of the cell cycle, before they are activated or ‘fired’ in S phase2. Interestingly, there are usually ~20-fold more licensed origins than required for completing full genome duplication, implying a flexibility of usage of these origins and varying origin firing efficiencies23. During DNA replication however, individual replication forks may be slowed down or stalled due to the presence of endogenous or exogenous sources of replication stress. How do cells overcome perturbed replication forks to finish genome duplication in a timely manner? A critical response is to fire additional licensed origins to complete replication within the intervening regions of the stalled forks; these backup replication origins are commonly referred to as “dormant origins”4. Under conditions of replication stress, the importance of dormant origins has been demonstrated to ensure complete replication and prevent genomic instability5, 6. While replication origin locations have been widely studied and several techniques developed to map them, origin firing efficiencies can be difficult to quantify, and mapping sites of replication termination, which is especially pertinent during conditions of replication stress, has been a long-standing challenge7.
Synthesis of DNA after the firing of origins is asymmetric: the leading strand is synthesized continuously while the lagging strand is synthesized discontinuously; short fragments known as Okazaki fragments are synthesized in a direction that opposes fork progression8. From each activated replication origin, two replication forks move bi-directionally away from each other, with the replisome at each fork, until they encounter a converging fork. This bi-directional replisome movement protects DNA from under-replication in the event of fork stalling or arrest as there is no need to wait for the fork to restart once the inhibition on replication is lifted. Okazaki fragments mapping to the Watson and Crick strands are generated by leftward- and rightward-moving forks respectively.
Okazaki fragment sequencing (Ok-seq) was first developed in Saccharomyces cerevisiae as a method to quantitatively analyze replication initiation, progression, and termination9–11. This method was also used in Caenorhabditis elegans and required the repression or knockdown of DNA ligase to increase the amount of Okazaki fragments synthesized12. No mammalian system had been interrogated with DNA ligase repression until the recently published GLOE-seq method13, but instead, a new method, first described by the Hyrien group, has since been developed to isolate and sequence Okazaki fragments in asynchronously-growing human cell lines without the need for ligase repression. The method uaes pulse-labelling of newly synthesized DNA with 5-Ethynyl-deoxyuridine (EdU) or 5-Ethynyl-2’-deoxycytidine (EdC) instead, before size fractionation to isolate the Okazaki Fragments. The use of Ok-seq has been applied to several recent studies which looked at lagging strand synthesis and processing10, as well as the replication dynamics involved in replication initiation, elongation, and termination in unperturbed yeast and mammalian cells, and under conditions of replication stress10, 12, 14–18. In addition, using Ok-seq over other developed methods for mapping origins, allowed for not only the identification of replication origin locations but also in-depth meta-analyses of specific genomic classes; these other methods are compared to Ok-seq below (see ‘Comparison with other Methods’).
Overview of the protocol
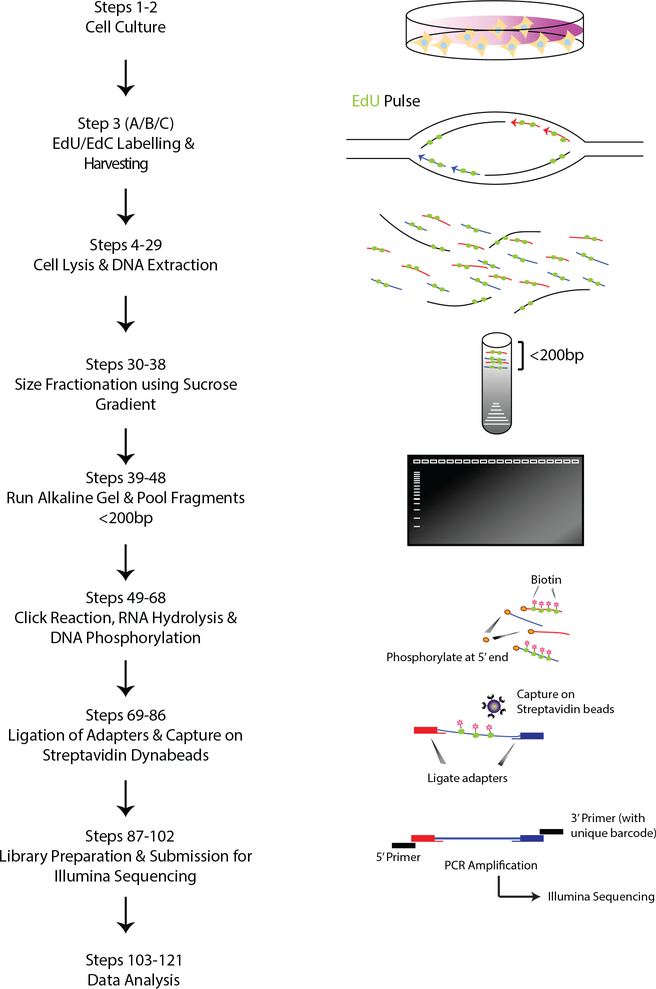
The Ok-seq methodology relies on the isolation and purification of newly synthesized Okazaki fragments that are made during the S-phase of replicating cells. In order to capture Okazaki fragments, EdU/EdC incorporation into the newly synthesized DNA of replicating cells is utilized. EdU, a nucleoside analog for thymidine, contains an alkyne group that allows a covalent linkage to biotin azide by click chemistry (copper-catalyzed cycloaddition reaction)19, 20. The procedure starts by culturing a sufficient number of asynchronously-growing mammalian cells and pulse-labelling them with EdU for a very short duration of 2 min, which is sufficient for its incorporation into the Okazaki fragments, before cells are harvested (Fig. 1, Steps 1–3). The harvested cells can be frozen as pellets before they are lysed and their DNA extracted (Steps 4–29). To separate the Okazaki fragments from the larger-sized DNA fragments, a 5–30% linear sucrose gradient is used (Steps 30–38). Upon size-fractionation, fractions of the gradient can be collected and a portion from each is run on an alkaline gel to determine which fractions contain the desired size of DNA fragments (DNA smear corresponding to <200bp) (Steps 39–42). These fractions are then pooled together and further concentrated before subjecting the sample to the click reaction to conjugate a cleavable (disulfide bond) biotin-azide to the incorporated EdU (Steps 43–54). After subsequent RNA hydrolysis and phosphorylation of the 5’ ends, adapters containing Illumina adapter index sequences are ligated to the fragments in a two-step procedure, punctuated by an incubation with Streptavidin beads to capture the EdU-labelled and biotin-conjugated Okazaki fragments (Steps 55–86). Finally, after stringent washes, the library of Okazaki fragments can be prepared via PCR amplification using Phusion enzyme and PCR primers which contain unique index barcodes (Steps 87–97). The final library is then submitted for paired-end 50 bp Illumina sequencing (NovaSeq) after completing quality controls using Agilent Tapestation or qPCR to ensure sufficient yield and purity (Steps 98–102). Post-sequencing, the data is analyzed and visualized using our bioinformatics pipeline (Steps 103–121).
Fig. 1|.
Schematic overview of the Ok-seq protocol. After cells are scaled to a sufficient amount, they are pulsed with EdU before harvesting and lysis. Extracted DNA is subjected to a size fractionation by ultracentrifugation on a 5–30% linear sucrose gradient, and the recovered fractions can then be visualized on an alkaline agarose gel. Fragments of the desired size (<200bp) are pooled and concentrated prior to biotinylation using a Click reaction. Subsequently, any existing RNA is hydrolyzed and the 5’ ends of DNA fragments are phosphorylated before a two-step adapter ligation procedure that is punctuated by an incubation with Streptavidin beads to capture biotin-conjugated Okazaki fragments. Following stringent washes, the library of Okazaki fragments can be prepared via PCR amplification and submitted for paired-end sequencing. Post-sequencing, the data would be analyzed using our bioinformatics pipeline.
Applications of the method
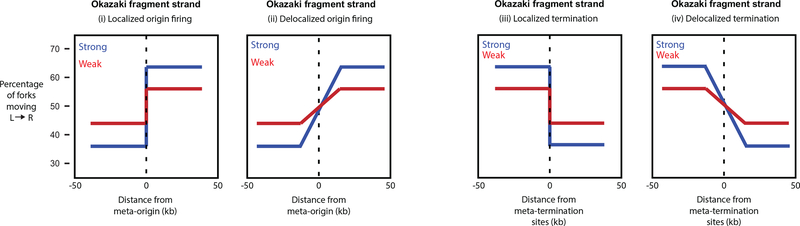
The principle of the mammalian Ok-seq method is to sequence isolated Okazaki fragments from an asynchronous population of dividing cells and detect proportions of replication forks moving either rightward (Crick strand) or leftward (Watson strand). Thus far, the method has two main applications14. Firstly, it enables the user to detect replication initiation and termination sites based on the proportions of forks moving in each direction14. One downstream analysis approach looks at the replication fork directionality (RFD) profile where the RFD is computed as the difference between the proportions of rightward- and leftward-moving forks within each 1kb window. Upward slopes in the RFD profile mark initiation sites while the downward slopes mark the termination sites. Another approach that is preferred by our group is to perform a meta-analysis on the percentage of forks moving from left to right at specific classes of genomic locus, namely transcription start sites (TSS), transcription termination sites (TTS) and enhancers, to report on Okazaki fragment strand bias transitions (Fig. 2)17. A derivative of this percentage can directly report on initiation or termination frequency along the 50-kb window that is analyzed. In order to capture both early- and late-firing origins, cells are maintained asynchronously but at a 60–70% confluency to ensure most, if not all, cells are replicating in S phase.
Fig. 2|.
Schematic representation of expected Okazaki fragment distributions for replication initiation and termination sites. A strong localized origin firing or termination will have a larger amplitude (i, iii), while the gradient of the Okazaki fragment strand bias transition can provide information on the spatial localization of origins or termination sites upstream or downstream of the meta-element (ii, iv).
A second application is the measurement of replication origin firing efficiencies. Looking at the Okazaki fragment strand bias transitions, we can compute the firing efficiency of an origin from the amplitude of shift and the spatial localization from the gradient (Fig. 2)14. A more efficient and localized origin would result in a larger amplitude of shift with a gradient equivalent to 0, as shown in Fig. 2. In a recent report by our group, we analyzed the efficiency of origin firing and replication termination in transcribed regions based on previous genome-wide studies17. The ability to measure origin firing efficiencies and replication termination provides a platform to identify determinants that regulate replication dynamics under different genetic or environmental perturbations. For example, this protocol provides the option for treatment with hydroxyurea (HU), a ribonucleotide reductase inhibitor, as a way to introduce replication stress and slow the replication forks, thus activating adjacent flexible or dormant origins21. Applying different replication stress inducers (for example, chemotherapeutic drugs), we can also compare the efficiency of how flexible or dormant origins are activated in different cell types or in diseased states, such as in cancer cells. Ok-seq is applicable to both adherent human cells and cells in suspension, but a difference lies in the way the cells are labelled and harvested, which is a critical step of the protocol.
Comparison with other methods
The quest to determine the precise locations of mammalian origins and understand origin firing has led to the development of several methods of mapping genome-wide replication origins based on certain hallmarks of an initiation event22. These include short nascent strand sequencing (SNS-Seq), Bubble-Seq, chromatin immunoprecipitation (ChIP) sequencing of DNA fragments bound to origin recognition complex (ORC) proteins, initiation site sequencing (Ini-Seq), and EdUseq-HU23–27. More recently, a new method involving genome-wide ligation of 3’-hydroxy (3’-OH) ends followed by sequencing (GLOE-seq) and a high-resolution Repli-seq method have also been published13, 28.
SNS-Seq enriches for size-selected nascent strands that are purified by treatment with excess lambda-exonuclease before they are subjected to deep sequencing29, 30. This method identifies discrete start sites for replication, but the enrichment is only for strong initiation events and is unable to differentiate between abortive and productive initiation27. Inefficient digestion by lambda-exonuclease of the GC-rich sequences can also result in higher number of false-positive sites detected31. Bubble-Seq takes advantage of the 2N “bubble” replication intermediate that can be trapped in agarose plugs during gel electrophoresis and it provides a more sensitive detection of low-efficiency initiation events over broad regions23. However, it lacks the precision of SNS-seq, generating more false positives, and it can be biased against asymmetrically located origins or those on small restriction fragments. ChIP-Seq depends on ORC binding to DNA sequences but these sequences are not limited to origin-sites; non-specific signals may appear in the results using this method. EdUseq-HU and Ini-Seq are newer methods which map early S-phase replication origins. Both require cell synchronization, with Ini-Seq allowing for synthesis of small stretches of nascent DNA from activated origins under cell-free, controlled biochemical conditions24, 25. However, synchronization subjects cells to a large number of perturbations and manipulations which may not reflect the natural biological replication processes. While high resolution Repli-seq enriches for cells in the S-phase by sorting and isolating cells containing nascent strands labelled with BrdU instead of using cell synchronization, the limitation with these methods (EdUseq-HU, Ini-Seq, and Repli-seq) is the underrepresentation of origins that fire later in S-phase28.
The Ok-seq method is derived from techniques that use labelling of nascent DNA with a tagged analog of the thymidine nucleoside, uridine, and sucrose gradients for isolating the desired DNA fragments 26. Its advantage over other methods is its ability to quantify proportions of rightward and leftward moving forks throughout the genome, thus providing a direct and fully quantitative view about replication fork directionality14. This enables users to gain important information such as the sites and frequencies of replication-transcription conflicts. In addition to directly reporting origin firing efficiencies, Ok-seq can report on fork progression and termination efficiencies as well. The recently published GLOE-seq method is comparable to Ok-seq in that its sequenced data shows a similar RFD profile albeit with lower peaks in human cells13. GLOE-seq detects the 3’-hydroxy (3’-OH) ends of single-strand break (SSB) sites, which allows for the mapping of naturally-occurring SSBs that arise spontaneously and as intermediates of many DNA transactions, such as from DNA damage, DNA repair, and DNA replication13. Although its advantages of fewer starting cells (~700, 000 cells) and the absence of fragment size selection and enrichment of replicated DNA via EdU pulse labeling are potentially appealing, GLOE-seq still requires manipulation of cells to deplete nuclear DNA ligase 1, which is not reflective of the natural biological replication processes. Importantly, GLOE-seq also captures spontaneously-occurring SSBs that are independent of DNA synthesis13.
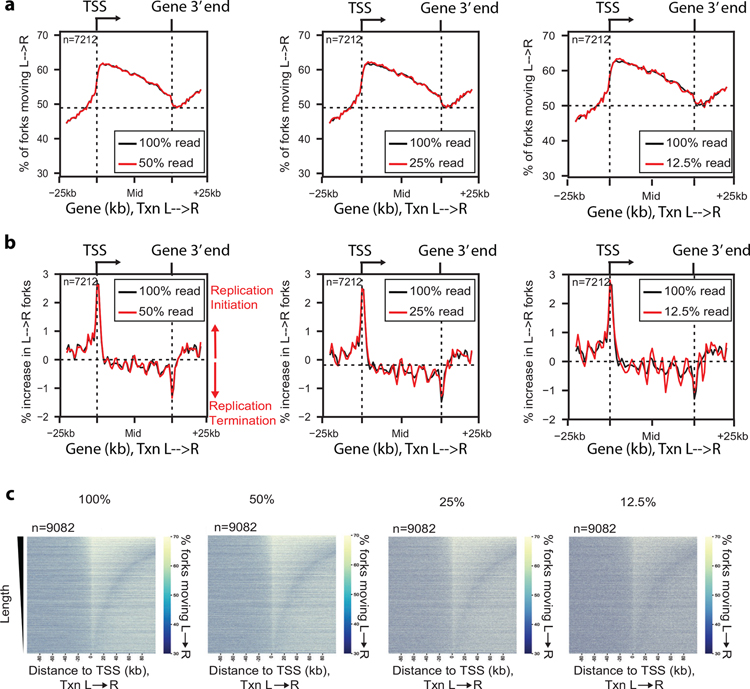
Another strength of Ok-seq lies in the meta-analyses performed at specific genomic loci which provides a quantitative view of replication origin firing around those elements. In addition, the method can capture both early and late firing origins, and even dormant origins by including a low-dose HU treatment of the cells to slow the replication forks21. Additionally, an advantage of performing such meta-analyses is that a low number of reads can still produce a sufficient amount of usable data. In Petryk et al.’s study, a large number of reads was required to predict origins from the data14. However, by looking for predictable origin firing distributions around genomic elements, fewer reads will suffice, down to 12.5% of the reads compared to a full read count (Fig. 3). This can substantially improve cost-efficiency of sequencing the fragments as more samples with unique barcodes can be sequenced in a single lane in NovaSeq, which is cheaper and faster than the other Illumina sequencing platforms.
Fig. 3.
Anticipated results from RPE-1 cells. a, Percentage of replication forks moving left to right across gene body, for actively transcribed genes (FPKM>median) normalized by gene length. Comparison is of data computed from 100% sequenced reads with 3 downsampling levels: (left) 50%, (middle) 25%, (right) 12.5%. (n, total number of genes analyzed) b, Replication initiation frequency, calculated as the first derivative of Okazaki fragment strand bias as in a. c, Heat map representation of the change in replication direction around TSS for actively transcribed genes (FPKM>median), sorted by transcriptional length. (n, total number of genes analyzed).
Experimental Design
Several parameters can be changed within the Ok-seq protocol depending on the cell type and the desired experimental outcome. First, adherent cells and suspension cells require different cell culturing techniques and therefore, the way they are labelled with EdU and subsequently harvested would differ. It is also important to note that different cell lines have different growth rates and cell sizes that could affect the number of dishes one would require. We recommend that at least 500 million cells are used as starting material and that the cells are no more than 70% confluent to obtain the best data from sequencing. However, given that down-sampling of the number of reads can still provide sufficient usable data, it may be possible to decrease the starting number of cells to 100 million but we cannot guarantee that this would produce sufficient Okazaki fragment libraries for sequencing.
Secondly, we provide an option for users to look at dormant origin firing efficiencies in cells that undergo replication stress. Additional steps are included in the protocol to treat cells with low-dose HU (200 μM) for 4 hours prior to EdU labelling and harvesting of the cells. This specific dose and duration was chosen as it serves to slow replication forks and reduce inter-origin distances without influence from checkpoint kinases5. However, given the large number of cells to be treated, it is recommended to stagger the HU treatment and EdU labelling and harvesting in order to ensure the accuracy of each component. EdU labelling time should also be doubled in order to ensure sufficient labelling of the newly replicated fragments32.
To achieve a robust analysis of the sequencing data, at least two biological replicates (e.g. samples prepared under the same conditions from separate biological samples) should be performed in order to ensure reproducibility in the results generated. Both replicates should demonstrate agreement in strand bias transitions in order for the results to be significant. This protocol would also require users to have sufficient knowledge of cell culturing techniques and molecular biology techniques such as running agarose gel electrophoresis and polymerase chain reactions. For data analysis, a moderate level of expertise in bioinformatics is needed, including a basic understanding of Python.
Limitations
The most important limitation of Ok-seq in comparison to SNS-seq is the inability to identify origins de novo unless they are very efficient and well-localized. Similarly, identifying terminators de novo, except for a small subset of strong, localized terminators, is difficult using this procedure. Broad initiation zones were observed instead, where Okazaki fragment strand transitions occurred over large domains between 6–150kb14. Counteracting this limitation, if users limit Ok-seq to analyzing changes in replication fork direction around rationally selected genomic elements, a lot of useful information can be obtained.
Possible background caused by the inevitable contamination of small genomic fragments, that are not Okazaki fragments, generated during DNA purification is another limitation of this procedure. The greater the proportion of these fragments, the more globally depressed the calculated origin firing efficiencies will be. Comparisons between biological samples would therefore require several replicates to ensure a higher rate of precision and accuracy. Due to the complexity of the mammalian genome, another limitation lies in the difficulty of distinguishing two identical sequences that could be replicated in opposite directions. Any non-unique sequence would have to be excluded from the analysis.
While the Ok-seq procedure is easily adaptable for use in a wide range of cell lines from different metazoan organisms, one final limitation is in the large numbers of cells required in the starting material. As presented here, the protocol requires an ideal initial number of 5 × 108 cells that are between 60–70% confluency to be labelled with EdU and harvested. This is to ensure sufficient amounts of labelled Okazaki fragments by the end of the procedure after they have been purified and are ready for sequencing. The protocol does not require any cell synchronization; this is strongly discouraged since many techniques used to synchronize cells require drug treatments that directly or indirectly alter the transcriptome and replication dynamics. Instead, cell types that have more replicating cells during the log growth phase (higher percentage of S-phase cells) may be more advantageous for the Ok-seq procedure to reduce the total number of cells needed at collection.
Materials
Biological materials
Cell line of interest. To date, the protocol has been successfully applied to RPE-1 (ATCC Cat# CRL-4000https://scicrunch.org/resolver/CVCL_4388) and HCT116 cell lines (ATCC Cat# CCL-247, https://scicrunch.org/resolver/CVCL_0291) from our lab, and numerous other non-cancerous and patient-derived cancer cells from other groups33. The data shown in this protocol are for RPE-1 and HCT116 cells. !Caution The cell lines used in your research should be regularly checked to ensure authenticity and that they are free from mycoplasma contamination.
Reagents
-
Cell Culture Medium (For RPE-1 cells: Dulbecco’s modified Eagle’s medium: Nutrient Mixture F-12 (DMEM/F-12) Media, Thermo Fisher Scientific, cat. no. 11320–033; 10% (vol/vol) Fetal Bovine Serum, Atlantic Biologicals, cat. No. S11150H; 3% (wt/vol) Sodium Bicarbonate, Thermo Fisher Scientific, cat. no. 25080–094; 1% (vol/vol) Penicillin-Streptomycin (pen-strep), Thermo Fisher Scientific, cat. no. 35050–061; For HCT116 cells: McCoy’s 5A Medium (1X), Thermo Fisher Scientific, cat. no. 25030–081; 10% (vol/vol) Fetal Bovine Serum, Atlantic Biologicals, cat. No. S11150H; 1x L-Glutamine 200 mM, Thermo Fisher Scientific, cat. no. 25030–081); 1% (vol/vol) Penicillin-Streptomycin (pen-strep), Thermo Fisher Scientific, cat. no. 35050–061)
*Critical* The medium may be adapted to the cell line of interest.
5-Ethynyl-deoxyuridine (5-EdU) 50mg (Thermo Fisher Scientific, cat. no. A10044)
5-Ethynyl-2’-deoxycytidine (5-EdC), 10mg pack (Jena Bioscience, cat. no. CLK-N003–10)
Hydroxyurea (HU; Sigma-Aldrich, cat. no. H8627–25G)
Dimethyl Sulfoxide (DMSO; Fisher Scientific, cat. no. BP231–100) !Caution Dimethyl Sulfoxide is harmful upon contact with skin and is a combustible liquid. Wear gloves.
Copper (II) Sulphate (CuSO4; Jena Bioscience, cat. no. CLK-MI004–50) !Caution It is toxic if swallowed. It can cause skin or eye irritation. Wear gloves.
UltraPure™ Phenol:Chloroform:Isoamyl Alcohol (25:24:1, v/v) (Thermo Fisher Scientific, cat. no. 15593049) !Caution Phenol:Chloroform:Isoamyl Alcohol is flammable and toxic when swallowed or inhaled, or upon contact with the skin. Wear gloves and work under a fume hood.
SYBR® Gold nucleic acid gel stain (Thermo Fisher Scientific, cat. no. S-11494)
Dynabeads MyOne streptavidin T1 (Thermo Fisher Scientific, cat. no. 65602)
Biotin-TEG azide 25mg (Berry & Associates, cat. no. BT 1085) *Critical This may be substituted with other Biotin Azide, not necessarily Biotin-TEG azide.
100bp DNA Ladder (Thermo Fisher Scientific, cat. no. 15628019)
T4 DNA ligase (Enzymatics, cat. #L6030-HC-L) *Critical It is important that this particular reagent is obtained from this supplier in order for an optimal ligation of adapters.
T4 Polynucleotide Kinase (10 U/μl) (New England Biolabs Inc., cat. no. M0201L)
Proteinase K (Roche, cat. no. 3115828001)
Trypsin-EDTA (0.5%), no phenol red (Thermo Fisher Scientific, cat. no. 15400–054)
Phosphate-Buffered Saline (PBS) (Corning, cat. no. 21–040-CV)
Ethanol, Absolute (200 Proof), Molecular Grade (Fisher Scientific, cat. no. BP2818–500) !Caution Ethanol is flammable and causes severe eye irritation.
Vacuum Grease (Dow Corning, cat. no. 1597418)
10% (wt/vol) SDS solution (Thermo Fisher Scientific, cat. no. 15553027) !Caution SDS can cause irritation to the eyes and upon contact with skin. Wear gloves.
Tween™ 20 (Fisher Scientific, cat. no. BP337–500) !Caution Tween™ 20 may cause eye, skin and respiratory tract irritation. Wear gloves and avoid inhalation.
Sucrose (Fisher Scientific, cat. no. BP220)
Tris (1M, pH 8, RNase-free; Thermo Fisher Scientific, cat. no. AM9855G)
EDTA (0.5M, pH 8, RNase-free; Thermo Fisher Scientific, cat. no. AM9260G)
TE Buffer (Thermo Fisher Scientific, cat. no. AM9849)
Sodium Chloride (NaCl; 5M; Millipore Sigma, cat. no. SX0420–5)
Hydrochloric Acid (HCl; Fisher Scientific, cat. no. A144–500)
Tris-boric-EDTA Buffer (TBE; Fisher scientific, cat. no. FERB52)
Tris-acetate-EDTA Buffer (TAE; Fisher scientific, cat. no. FERB49)
Sodium Acetate (NaOAc; 3M, pH 5.2; Sigma-Aldrich, cat. no. S7899–100ML)
Acrylamide/bis-Acrylamide 30% solution (Sigma-Aldrich, cat. no. A3574)
TEMED (Sigma, cat. no. T9281) !Caution TEMED is flammable and toxic if swallowed or inhaled. It can cause severe skin burns and eye damage upon contact. Wear gloves.
Agarose (GeneMate, cat. no. E-3120–500)
Acetic Acid (Fisher Scientific, cat. no. 38S-212) !Caution Acetic Acid is flammable and corrosive, able to cause severe skin burns and eye damage. Wear gloves and work under fume hood.
Sodium Hydroxide (NaOH; Fisher Scientific, cat. no. 15696780) !Caution Sodium Hydroxide can cause eye and skin irritation. Wear gloves.
L-Ascorbic Acid (Sigma-Aldrich, cat. no. A92902)
Ammonium acetate (Sigma-Aldrich, cat. no. A1542)
Bromophenol Blue (Fisher Scientific, cat. no. BP115–25)
Bromocresol Green (Fisher Scientific, cat. no. B383–5)
Xylene Cyanole (Fisher Scientific, cat. no. BP565–10)
Gel Loading Dye, Purple (6X) (New England Biolabs Inc, cat. no. B7024S)
Ficoll PM400 (Sigma-Aldrich, cat. no. F4375)
dNTPs Mix (10 mM each) (Thermo Fisher scientific, cat. no. R0192)
Phusion® HF Buffer (New England Biolabs, cat. no. B0518S)
Phusion® High-Fidelity DNA Polymerase (New England Biolabs, cat. no. M0530)
SPRIselect beads (Beckman Coulter cat. no. B23317)
Oligonucleotides from IDT technologies (see info on sequences)
Adapters (Illumina; see Table 1 for sequences)
PCR primers for library amplification. The libraries are amplified using the PE PCR Primer 1.0 (5’-AATGATACGGCGACCACCGAGATCTACACTCTTTCCCTACACGACGCTCTTCCGATCT-3’) and TruSeq PCR Primers, Index 1–24 (see Table 2 for sequences)
Qubit® dsDNA HS Assay Kit, for use with the Qubit® 2.0 Fluorometer (100 assays) (Thermo Fisher Scientific, cat. no. Q32851)
KAPA Library Quantification Kit (Roche, cat. no. KK4824)
Dry Ice
Milli-Q water
Table 1:
Illumina Adapter Sequences
| Adapterstrand | Sequence |
|---|---|
| 5’ Adaptertop | 5’-ACACTCTTTCCCTACACGACGCTCTTCCGATCT-3’ |
| 5’ Adapterbottom | 5’-NNNNNNAGATCGGAAGAGCGTCGTGTAGGGAAAGAGTGT-3’ |
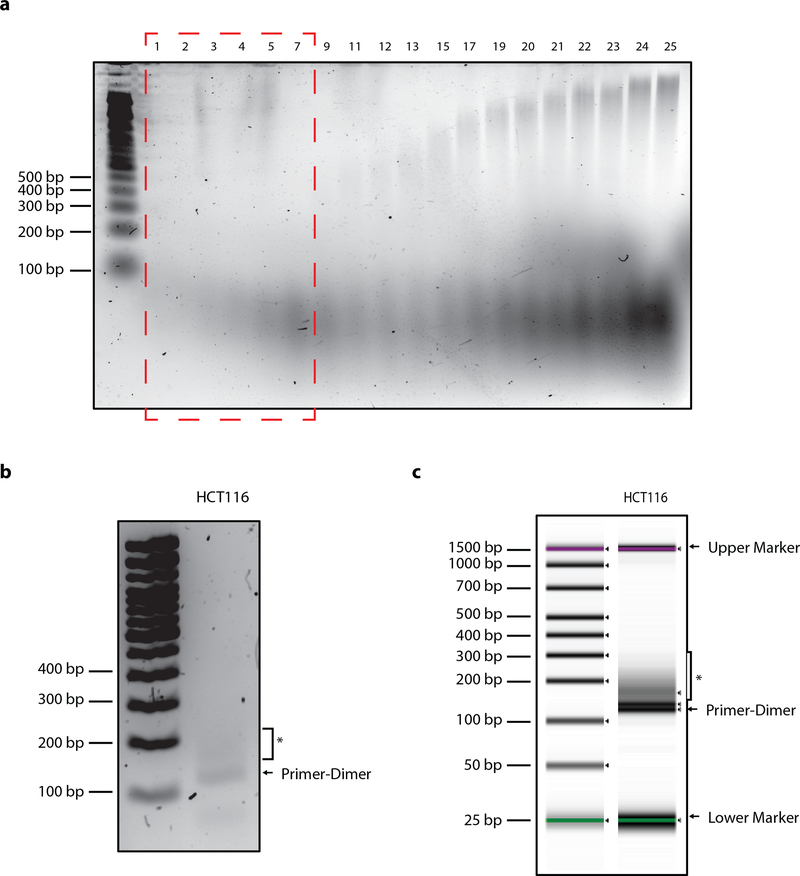
| 3’ Adaptertop | 5’-[Phos]- AGATCGGAAGAGCACACGTCTGAACTCCAGTCAC-3’ |
| 3’ Adapterbottom | 5’-GTGACTGGAGTTCAGACGTGTGCTCTTCCGATCTNNNNNN-3’ |
Table 2:
TruSeq PCR Primers Used for PCR Amplification of Okazaki Fragments Library Preparation
| Index # | Index Seq | rev comp index | Generic PCR R primer | specific primer |
|---|---|---|---|---|
| generic | nnnnnn | nnnnnn | CAAGCAGAAGACGGCATACGAGATnnnnnnGTGACTGGAGTTCAGACGTGTGCTCTTCCGATCt | CAAGCAGAAGACGGCATACGAGATnnnnnnGTGACTGGAGTTCAGACGTGTGCTCTTCCGATCt |
| 1 | ATCACG | cgtgat | illumina_PCR-R_barc_1 | CAAGCAGAAGACGGCATACGAGATcgtgatGTGACTGGAGTTCAGACGTGTGCTCTTCCGATCt |
| 2 | CGATGT | acatcg | illumina_PCR-R_barc_2 | CAAGCAGAAGACGGCATACGAGATacatcgGTGACTGGAGTTCAGACGTGTGCTCTTCCGATCt |
| 3 | TTAGGC | gcctaa | illumina_PCR-R_barc_3 | CAAGCAGAAGACGGCATACGAGATgcctaaGTGACTGGAGTTCAGACGTGTGCTCTTCCGATCt |
| 4 | TGACCA | tggtca | illumina_PCR-R_barc_4 | CAAGCAGAAGACGGCATACGAGATtggtcaGTGACTGGAGTTCAGACGTGTGCTCTTCCGATCt |
| 5 | ACAGTG | cactgt | illumina_PCR-R_barc_5 | CAAGCAGAAGACGGCATACGAGATcactgtGTGACTGGAGTTCAGACGTGTGCTCTTCCGATCt |
| 6 | GCCAAT | attggc | illumina_PCR-R_barc_6 | CAAGCAGAAGACGGCATACGAGATattggcGTGACTGGAGTTCAGACGTGTGCTCTTCCGATCt |
| 7 | CAGATC | gatctg | illumina_PCR-R_barc_7 | CAAGCAGAAGACGGCATACGAGATgatctgGTGACTGGAGTTCAGACGTGTGCTCTTCCGATCt |
| 8 | ACTTGA | tcaagt | illumina_PCR-R_barc_8 | CAAGCAGAAGACGGCATACGAGATtcaagtGTGACTGGAGTTCAGACGTGTGCTCTTCCGATCt |
| 9 | GATCAG | ctgatc | illumina_PCR-R_barc_9 | CAAGCAGAAGACGGCATACGAGATctgatcGTGACTGGAGTTCAGACGTGTGCTCTTCCGATCt |
| 10 | TAGCTT | aagcta | illumina_PCR-R_barc_10 | CAAGCAGAAGACGGCATACGAGATaagctaGTGACTGGAGTTCAGACGTGTGCTCTTCCGATCt |
| 11 | GGCTAC | gtagcc | illumina_PCR-R_barc_11 | CAAGCAGAAGACGGCATACGAGATgtagccGTGACTGGAGTTCAGACGTGTGCTCTTCCGATCt |
| 12 | CTTGTA | tacaag | illumina_PCR-R_barc_12 | CAAGCAGAAGACGGCATACGAGATtacaagGTGACTGGAGTTCAGACGTGTGCTCTTCCGATCt |
| 13 | AGTCAA | ttgact | illumina_PCR-R_barc_13 | CAAGCAGAAGACGGCATACGAGATttgactGTGACTGGAGTTCAGACGTGTGCTCTTCCGATCt |
| 14 | AGTTCC | ggaact | illumina_PCR-R_barc_14 | CAAGCAGAAGACGGCATACGAGATggaactGTGACTGGAGTTCAGACGTGTGCTCTTCCGATCt |
| 15 | ATGTCA | tgacat | illumina_PCR-R_barc_15 | CAAGCAGAAGACGGCATACGAGATtgacatGTGACTGGAGTTCAGACGTGTGCTCTTCCGATCt |
| 16 | CCGTCC | ggacgg | illumina_PCR-R_barc_16 | CAAGCAGAAGACGGCATACGAGATggacggGTGACTGGAGTTCAGACGTGTGCTCTTCCGATCt |
| 17 | GTAGAG | ctctac | illumina_PCR-R_barc_17 | CAAGCAGAAGACGGCATACGAGATctctacGTGACTGGAGTTCAGACGTGTGCTCTTCCGATCt |
| 18 | GTCCGC | gcggac | illumina_PCR-R_barc_18 | CAAGCAGAAGACGGCATACGAGATgcggacGTGACTGGAGTTCAGACGTGTGCTCTTCCGATCt |
| 19 | GTGAAA | tttcac | illumina_PCR-R_barc_19 | CAAGCAGAAGACGGCATACGAGATtttcacGTGACTGGAGTTCAGACGTGTGCTCTTCCGATCt |
| 20 | GTGGCC | ggccac | illumina_PCR-R_barc_20 | CAAGCAGAAGACGGCATACGAGATggccacGTGACTGGAGTTCAGACGTGTGCTCTTCCGATCt |
| 21 | GTTTCG | cgaaac | illumina_PCR-R_barc_21 | CAAGCAGAAGACGGCATACGAGATcgaaacGTGACTGGAGTTCAGACGTGTGCTCTTCCGATCt |
| 22 | CGTACG | cgtacg | illumina_PCR-R_barc_22 | CAAGCAGAAGACGGCATACGAGATcgtacgGTGACTGGAGTTCAGACGTGTGCTCTTCCGATCt |
| 23 | GAGTGG | ccactc | illumina_PCR-R_barc_23 | CAAGCAGAAGACGGCATACGAGATccactcGTGACTGGAGTTCAGACGTGTGCTCTTCCGATCt |
| 24 | GGTAGC | gctacc | illumina_PCR-R_barc_24 | CAAGCAGAAGACGGCATACGAGATgctaccGTGACTGGAGTTCAGACGTGTGCTCTTCCGATCt |
All sequences are listed in the 5’–3’ direction. Primers are resuspended in TE buffer at a 100-μM concentration and then further diluted in molecular biology–grade water at a working concentration of 10 μM
Equipment
Cell Culture Incubator (37°C, 5% CO2)
Biological Safety Cabinet
Centrifuge (Thermo Scientific, Sorvall Legend X1R)
Microcentrifuge (Fisher Scientific SPROUT™ HSE 49480)
Benchtop centrifuge (Eppendorf 5415R or 5415D)
Ultracentrifuge (Beckman Coulter, Optima XPN-100 Ultracentrifuge)
Water Bath
Vortex
Digital Dry Bath / Heatblock (Fisherband™ Isobrand; Fisher Scientific, cat. no. 11–715-125DQ
Thermomixer (Eppendorf™; Fisher Scientific, cat. no. 05–400-205)
Thermal Cycler (T100™; Bio-Rad, cat. no. 1861096EDU)
Falcon® tissue culture dishes150 mm (VWR, cat. no. 25383–103)
Falcon conical tubes 50ml Cellstar® (Greiner bio-one, cat. no. 227–261)
Falcon conical tubes 15ml Cellstar® (Greiner bio-one, cat. no. 188–271)
1.5-mL microcentrifuge tubes (Seal-Rite™; USA Scientific, cat. no. 1615–5500)
0.2-mL PCR tubes (TempAssure; USA Scientific, cat. no. 1402–4300)
Vacuum Filter/Storage bottle System, 0.22-μm pore (Corning, cat. no. 431154)
Cell lifter (Biologix group cat. no. 70–2180)
Pasteur pipettes (glass; VWR, cat. no. 14672–380)
10-mL syringe (Fisher Scientific, cat. no. 14–829-22A)
20-mL syringe (Fisher Scientific, cat. no. 22–124-967)
Laboratory pipetting needle with 90° blunt ends (Sigma-Aldrich, cat. no. CAD7941–12EA)
38.5-mL Open-top Thinwall, Ultra-clear Tube, 25 × 89 mm (Beckman coulter, cat. no. 344058)
Analytical Balance (Mettler Toledo™ MS104TS; Fisher Scientific, cat. no. 01–913-930)
Precision Balance (Mettler Toledo™ MS1602TS; Fisher Scientific, cat. no. 01–913-937)
Micro Bio-spin 30 columns (Bio-Rad, cat. no. 732–6250)
Centrifugal filter Amicon Ultra-15 Ultracel-10 membrane (Millipore, cat. no. UFC901024)
Centrifugal unit Amicon Ultra-0,5 Ultracel-3 membrane (Millipore, cat. no. UFC500396)
Gradient Maker (Biocomp Instruments Ltd., cat. no. 108)
1” (25mm) Magnabase™ holder and marker block for Sw28/Sw32 rotors long and short caps (Biocomp Instruments Ltd., cat. 105–925-IR)
Magnet Dynamag™−2 (Life technologies, cat. no. 123.21D)
SW28/SW32Ti Rotor (Beckman Coulter)
Sub-Cell GT Horizontal Electrophoresis System, 15 × 10cm tray (Bio-Rad, cat no. 1704401)
20-Well Comb (Bio-Rad, cat. no. 170448)
Sub-Cell GT Gel Caster (Bio-Rad, cat. no. 1704412)
RunOne™ Electrophoresis System with Timer, 100–120V (EmbiTec®, cat. no. EP-2100)
Qubit® 2.0 Fluorometer (Thermo Fisher Scientific)
Illumina NovaSeq next-generation sequencer (Illumina)
Tapestation 2200 parts and accessories (Agilent Technologies)
Hardware: High Performance Computing (HPC) cluster, and/or standard desktop/laptop computer running MacOS or Linux.
Software: Python v3.5 or higher; the following bioinformatics tools (freely available for download): FastQC and Trim Galore (Babraham Bioinformatics), bowtie234, samtools35, bedtools36, Picard and deepTools37; additional custom python scripts at https://github.com/FenyoLab/Ok-Seq_Processing (further described in the Data Analysis section.)
Example data: An example data file is included in the software link. Additional example data files are available here: GSE114017.
Reagent Setup
DNA lysis buffer
Prepare 100 mL of DNA lysis buffer by mixing 1 mL of 1 M Tris (pH 8.0), 5 mL of 0.5M EDTA (pH 8.0) and 2 mL of 5 M NaCl, then adjust the volume to 100 mL with Milli-Q water (10 mM tris, 25 mM EDTA and 100 mM NaCl final). Store the buffer at room temperature (20–25 °C) for up to 6 months. Resuspend the cells, and then add 0.5% (vol/vol) SDS final and 0.1 mg/ml Proteinase K final.
TEN 10X
Prepare 100 mL of TEN 10X by mixing 10 mL of 1 M Tris (pH 8.0), 2 ml of 0.5 M EDTA (pH 8.0) and 20 mL of 5 M NaCl, then adjust the volume to 100 mL with Milli-Q water (100 mM tris, 10 mM EDTA and 1 M NaCl final). Store the buffer at room temperature for up to 6 months. *Critical Autoclave before use.
5% and 30% sucrose in TEN buffer
Prepare 400 mL each of 5% (wt/vol) and 30% (wt/vol) sucrose solution by weighing 20 g and 120 g of sucrose, respectively. Adjust the volume to 300 mL with Milli-Q water to dilute the sucrose. Then add 40 mL of 10X TEN and adjust to a final volume of 400 mL for each solution. The solution can be stored one month at room temperature or up to six months at 4 °C.
B&W buffer + Tween 20, 2X
Prepare 100 mL of 2X B&W buffer by mixing 1 mL of 1 M Tris (pH7.5), 200 μL of 0.5M EDTA (pH 8.0), 40 mL of 5 M NaCl and 0.1% (vol/vol) Tween 20, then adjust the volume to 100 mL with Milli-Q water (10 mM tris, 1 mM EDTA and 2 M NaCl final). Store the buffer at room temperature for up to 6 months. *Critical Filter the solution with a 0.22 μm vacuum filter/storage bottle system before use.
Alkaline gel running buffer, 1X
Prepare 500 mL of 1X alkaline gel running buffer by mixing 25 mL of 1 M NaOH and 1 mL of 0.5 M EDTA (pH 8.0), then adjust the volume to 100 mL with Milli-Q water (50 mM NaOH, 1 mM EDTA final). Prepare buffer fresh.
Alkaline loading dye buffer 6X
Prepare 6 mL stock of 6X buffer by mixing 1.8 mL of 1 M NaOH, 72 μL of 0.5 M EDTA (pH 8.0), 1.08 g of Ficoll, 0.009 g of Bromocresol Green and 0.015 g of Xylene Cyanol FF, then adjusting the volume to 6 mL with Milli-Q water (300 mM NaOH, 6 mM EDTA, 18% Ficoll (wt/vol), 0.15% (wt/vol) Bromocresol Green, 0.25% (wt/vol) Xylene Cyanol FF final). Store the dye in 4 °C for up to 6 months.
Procedure
Cell culture preparation – Timing 1–7 d
-
1
Determine number of 150 mm dishes or 75cm2 flasks of cells for EdU labelling. Number of dishes/flasks may vary depending on cell line, anywhere from 15 – 60 dishes/flasks per sample. Ideally, 5 × 108 normal cells are required while a smaller number of cells would suffice for cancer cells.
-
2
Expand cell cultures 1 – 2 days prior to EdU labelling to ensure optimal exponential growth conditions, using 20 mL final media per dish or 50 mL final media per flask. Ensure uniform cell distribution with careful splitting technique and balanced incubator placement.
*Critical Step* Cells must be in log phase growth for proper EdU incorporation. For RPE-1 cells, aim for 50 – 60% confluency at time of labelling, attained by splitting a 60–70% confluent 150 mm dish into 10 × 150 mm dishes 48 h prior to labelling. A total of 15 × 150 mm dishes of RPE-1 cells are used per condition. For HCT116 cells, aim for 60–70% confluency at the time of labelling, attained by splitting a 70% confluent 150 mm dish (~1.6 × 107 cells) into 2×150 mm dishes 20 h prior to labelling. A total of 30 × 150 mm dishes of HCT116 cells are used per condition.
EdU/EdC Labelling & Harvesting of Cells – Timing 2 −5 h
-
3
Follow the desired protocol: (A) Adherent cells, (B) Cells in Suspension (C) Cells with HU treatment
(A). Adherent cells
Place on ice: 1× 500 mL PBS and labelled 50 mL conical tubes (1 tube for every 5–8 150 mm dishes, to be pooled later).
-
Pre-warm cell culture media as well as an empty glass bottle in a 37 °C water bath. Thaw a 50 mM stock of EdU or EdC at room temperature.
!Caution Keep EdU or EdC protected from light. *Critical Step* EdU would be preferred as the level of incorporation tends to be higher than EdC but EdC can still serve as an alternative38, 39.
Aspirate 10 mL of conditioned media from each dish into the pre-warmed glass bottle, noting total volume. Place dishes back into the incubator.
Add 20–30 mL prewarmed media to the glass bottle to ensure there is an excess of media. Note new final volume. Add 50 mM EdU or EdC stock to the glass bottle to a final concentration of 50 μM.
Take 7–8 dishes out of the incubator and place inside a biological safety cabinet.
Add 10 mL of 50 μM EdU or EdC conditioned media to each dish (final concentration of 25 μM EdU). Start a 2-minute timer after adding media to the first dish. Gently swirl each dish after addition of media to ensure proper mixing. Take care to note the order of the dishes. Depending on technique, by the time media is added to the 7–8th dish, the 2-minute timer should be complete.
Rapidly aspirate all the media from each dish in the same order, starting with the dish to which EdU or EdC was added first.
-
Add 5 mL cold PBS to each dish. Eject volume onto plates so that PBS readily covers surface of plate.
*Critical Step* Steps 3A(v)-(viii) should be performed as quickly as possible to ensure that the 2-minute EdU or EdC labelling occurs as close to 37 °C as possible. Ice cold PBS is necessary to terminate DNA replication and prevent over-representation of EdU or EdC in leading strand synthesis.
Scrape and collect the cells from all dishes and pool into the same 50 mL conical tube. Keep this tube on ice.
Rinse all the empty plates with a total of 5 mL of cold PBS to ensure complete collection of cells. Note final volume should be 40–45 mL per tube, and color should be clear to light pink. If color is darker pink to red, ensure proper media aspiration before cold PBS addition.
Repeat steps (v) through (x) until all dishes from same sample are harvested. It’s possible to label more than one sample at a time, but only if the number of dishes per sample is low. If number of dishes per sample is high, stagger the labelling of the extra samples.
Centrifuge cells for 5 min at 300g at 4 °C.
Aspirate PBS.
Wash each cell pellet in 5 mL ice cold PBS and combine multiple pellets from the same sample into one or two tubes depending on sizes of cell pellets.
Centrifuge for 5 min at 500g at 4 °C. (xvi) Aspirate PBS.
#Pause Point # Cell pellets can be frozen and stored at −80 °C for weeks to months. Snap freeze pellets using dry ice and 70% (vol/vol) ethanol.
(B). Suspension Cells
Grow suspension cells in 20–25 mL of media in flasks.
Pre-warm equal amounts of media and prepare 20 mM EdU in warmed glass bottle, and mix gently.
Add equal volume of EdU/EdC conditioned media back to each flask for final concentration of 10 μM and incubate flasks for 2 min at room temperature.
Rapidly chill flasks in an ice-cold water bath and add 250 μL of 0.5 M EDTA.
Collect cells from all flasks by spinning down at 300g for 10 min at 4 °C.
Wash the pellet once with cold PBS and centrifuge 300g, 10 min, 4 °C. (vii) Aspirate PBS.
#Pause Point # Cell pellet can be frozen and stored at −80 °C for weeks to months. Snap freeze pellets using dry ice and 70% (vol/vol) ethanol.
(C). Cells with HU treatment
CRITICAL: For 0.2 mM HU treatment, doubling the EdU labelling time was sufficient to achieve a similar amount of replicated DNA to the untreated 2 minute labelling (compensating for the reduced replication fork speed32).
Treat cells with 0.2 mM HU by adding it to the cells for 4 hours prior to EdU labelling and harvesting. Stagger the dishes so that the cells are all equally treated for the same duration.
Aspirate 10 mL (adherent cells) or 40 mL (suspension cells) of conditioned HU media from each dish or flask into the pre-warmed glass bottle as in A(ii), noting total volume. Place dishes back into incubator.
Add 20–30 mL prewarmed media to the glass bottle to ensure there is an excess. Add enough HU to the bottle to compensate for the 20–30 mL of untreated media (final concentration of 0.2 mM HU). Note the new final volume. Add 50 mM EdU or EdC stock to the glass bottle to a final concentration of 50 μM.
Add EdU or EdC conditioned media into each dish as per Step 3A(vi) but start the timer for 4 min instead. Due to the increased labelling time, place dishes back in incubator after adding media to all plates. Take care to note the order of the dishes when transferring from incubator to hood.
Perform steps 3A(v) - (x) for adherent cells until cells from all dishes of the same sample are harvested. Perform steps 2A(iv) – (vii) for suspension cells until cells from all flasks of the same sample are harvested.
#Pause Point # Cell pellet can be frozen and stored at −80 °C for weeks to months. Snap freeze pellets using dry ice and 70% (vol/vol) ethanol.
Cell Lysis – Timing 30 min to O/N
-
4
Resuspend the cell pellet from Step 3A(xvi), 3B(vi), or 3C(v) in 9.5 mL DNA lysis buffer. The volume of lysis buffer needed may vary depending on cell pellet size - maximum volume is 15 mL. If two tubes of cell pellets were collected for one sample, resuspend in 9.5 mL DNA lysis buffer for each pellet.
-
5
Add Proteinase K to a final concentration of 0.1 mg/mL.
-
6
Add enough 10% (wt/vol) SDS solution for a final concentration of 0.5% (wt/vol) SDS. For 10 mL lysis volume, that is 0.5 mL 10% SDS into 9.5 mL DNA lysis buffer. Mix carefully by swirling and inverting tube several times.
*Critical Step* Do not vortex.
-
7
Incubate at 50 °C overnight.
DNA Extraction – Timing 3 h to O/N
-
8
Take the tube containing the cell lysate out of the 50 °C incubator and bring to room temperature. Ensure phenol chloroform, 7.5 M ammonium acetate, and centrifuge are at room temperature.
-
9
Add approximately 5 mL of Corning vacuum grease to one or two 50 mL conical tubes per sample depending on the final number of cell lysate tubes. The easiest way to do this is by squeezing grease into 10 mL syringes and using the syringes to expel grease to the bottom of tube. The purpose of the grease functions like a phase-lock gel that will help to separate the aqueous layer from the phenol:chloroform:isoamyl layer.
-
10
Centrifuge these tubes for 5 min at 1,500g at room temperature. Grease should pellet nicely on the bottom.
-
11
Carefully pour full amount of cell lysate from each tube into the labelled conical tube(s) containing pelleted vacuum grease.
-
12
Add an equal volume phenol:chloroform:isoamyl alcohol. For 10 mL lysate, add 10 mL phenol:chloroform:isoamyl alcohol.
!Caution Work under a fume hood when manipulating phenol:chloroform:isoamyl alcohol. The chemicals and contaminated tubes must be disposed as hazardous waste.
-
13
Carefully mix cell lysate and phenol chloroform well by slowly inverting tubes back and forth. Do not vortex.
*Critical Step* Care must be taken to avoid shearing of any genomic DNA, as that will create smaller DNA fragments that are not Okazaki fragments, resulting in higher background and leading to poor sequencing results. Make sure tubes are capped tightly, and mix until when the tube held upright, there is no longer a readily developed clear layer at the bottom of the tube. This may take at least 5 min.
-
14
Centrifuge for 5 min at 1,500g at room temperature.
-
15
Ensure three distinct layers are seen: aqueous (top layer), vacuum grease (middle layer), and phenol:chloroform:isoamyl alcohol phase (bottom layer). If the layers are not distinct, repeat Step 14 again.
-
16
Transfer the top soluble layer into a new labelled 50 mL conical tube containing 5 mL vacuum grease. Repeat steps 12–15 for a second phenol chloroform extraction.
*Critical Step* After the second extraction, the top soluble layer should be clear of any white wispy contamination. If you still see any, simply perform a third extraction by repeating steps 1115.
-
17
Transfer the top soluble layer into a clean labelled 50 mL conical tube by carefully pouring, being sure not to disturb the grease barrier trapping the chloroform at the bottom.
-
18
Slowly add 0.5 volumes of 7.5 M ammonium acetate while gently swirling the tube. Continue to swirl gently to adequately mix. Do not vortex.
-
19
Add 2 volumes 100% (vol/vol) ethanol. Mix by successive careful inversions until DNA precipitate is formed.
-
20
Incubate the mixture for 15 min at room temperature with occasional inversions to ensure complete DNA precipitation.
-
21
Using a glass Pasteur pipette with a finger covering the open wide end, carefully swirl the precipitate to capture it on the pointy end of the pipette. Transfer the precipitate into a new conical tube containing 35 mL 70% (vol/vol) Ethanol at room temperature.
*Critical Step* Be careful when transferring precipitate. This method is preferred over centrifugation followed by ethanol wash and aspiration to reduce potential loss of material due to repeated aspirations.
-
22
Gently mix the precipitate by careful inversions, then incubate for 5 minutes at room temperature (Wash 1).
-
23
Using same technique as in step 21, transfer the precipitate into new tube containing 35 mL 70% (vol/vol) Ethanol for a second wash.
-
24
Incubate the second wash for 1 h at room temperature. Protect from light.
-
25
Using same technique as in step 21, carefully transfer the precipitate into a new empty 50 mL conical tube, pushing towards bottom. During transfer, allow residual Ethanol on precipitate to drip off the walls of the tube in step 23 before transferring DNA into the new tube.
-
26
Using a P200 pipette and pipette tip, carefully press the pellet to gently squeeze excess Ethanol and gently move the pellet around conical tube. To further remove excess Ethanol, carefully use the tip to fold the pellet over itself with gentle squeezing.
?Troubleshooting
-
27
Dry briefly for 5 min at room temperature.
-
28
Add 2–4 mL of TE pH 8, ensuring the volume covers the pellet at bottom of the tube (1 mL of TE pH 8.0 per 1–1.5 × 108 starting cells is ideal).
-
29
Let the pellet dissolve overnight at 4 °C, protected from light.
#Pause Point# Resuspended DNA may be kept at 4 °C protected from light for a week.
Size fractionation using 5%−30% linear sucrose gradient – Timing 22–24 h
-
30
Allow resuspended DNA to come to room temperature. If the pellet is still not completely resuspended, incubate at 50 °C for 2–3 hours prior to ultracentrifugation.
-
31
Generate a 5–30% linear gradient. First, add 18 mL of 5% (wt/vol) sucrose to a 38.5 mL clear ultracentrifuge tube. Next, using a 20 mL syringe fitted with a needle, add 18 mL of 30% (wt/vol) sucrose to the bottom of the tube. (See Reagent Setup for sucrose solution preparation). Finally, cap the tubes with the long caps and use the gradient maker to generate the 5–30% linear gradient. Select the desired rotor (SW28 or SW32Ti) on the gradient maker and the settings are: Long_Sucr_05–30%_wv_1St.
-
32
Warm dissolved genomic DNA (gDNA) at 50 °C for ~1 h to lower the viscosity of the gDNA before preparing 1mL aliquots into 1.5 mL Eppendorf tubes using cut tips, which will make it easier to pipette the gDNA.
-
33
Denature gDNA for 10 min at 95 °C.
-
34
Immediately place on ice for 10 min.
-
35
Carefully layer the DNA solution on the top of each gradient. For gradients prepared in the 38.5 mL clear ultracentrifuge tubes, one sample can be loaded onto 2–4 gradients (maximum ~1.5 × 108 cells/gradient), 1 mL onto each. The gDNA should be less viscous at this point than when initial placement into Eppendorf tubes due to denaturation.
*Critical Step* Add the gDNA very slowly onto the top of the gradients, being sure to avoid any sharp jets that will disturb the gradient and lead to poorer resolution of DNA size.
-
36
Balance the samples by weighing them against each other on a scale, using 5% sucrose as excess if necessary.
*Critical Step* The tubes containing the gradients and gDNA need to be balanced in order for the rotor to run smoothly and safely in the ultracentrifuge at its operating speed.
-
37
Centrifuge in an ultracentrifuge in a SW28/SW32Ti rotor at 122,000g, 20 °C for 17h with maximum acceleration and minimum deceleration.
*Critical Step* Minimize the time between gradient formation in step 31 and beginning the ultracentrifuge spin in step 37 to prevent diffusion of the gDNA and maintain the linear gradient. Be very careful when transporting gradients to and from the ultracentrifuge, minimizing any sudden disruptions that may again affect linear nature of the gradient.
-
38
Collect 1 mL fractions from the top of the gradient using cut tips. Ensure tip is just below meniscus layer so that collection is coming from the top.
#Pause Point# Fractions can be stored at −20 °C for up to 3 days before concentrating the desired fractions.
Alkaline gel electrophoresis – Timing 1 h to O/N
-
39
Prepare a 1.5% (wt/vol) alkaline agarose gel (50 mM NaOH, 1 mM EDTA) in Milli-Q water. *Critical Step* Add NaOH and EDTA after agarose has cooled slightly to avoid hydrolyzing agarose.
-
40
Mix at least 30 μL of each fraction with 6X alkaline loading dye and run gel either overnight at 20 V or 4 h at 60 V at 4 °C in alkaline running buffer.
-
41
Neutralise gel in 100 mL 1X TBE for 15 min before staining gel with SybrGold diluted 1/10,000 in 200 mL of 1X TBE for 15 min on the rocker.
-
42
Image gel, determine the number of fractions that contain DNA fragments shorter or equal to 200 bp. For HCT116, these are typically fractions 1–8 (Fig. 3a).
Pool fractions and concentrate – Timing 1–2 h
-
43
Pool all fractions containing DNA shorter than or equal to 200 bp as determined by alkaline gel into one or more Millipore Amicon 10k tube(s) (holds up to 15 mL per tube).
-
44
Spin Millipore Amicon 10k tube at 4,000g for 15 min at room temperature.
-
45
Discard eluate in the bottom of the tube, add 5 mL Milli-Q water to the column, and spin at 4,000g at room temperature for 15 min.
-
46
Perform a second wash by repeating step 45.
-
47
Perform a third wash by repeating step 45. If using more than 1 Millipore Amicon 10k tube, combine the concentrated DNA into one of the concentrator tubes using a P200 pipette before performing the third wash.
-
48
Using a P200, transfer the concentrated DNA (approximately 320–380 μL) into an Eppendorf tube.
#Pause Point# Concentrated fractions can be stored at −20 °C for 5 days.
Biotinylation in Click reaction – Timing 2 h
-
49Set up the click reaction by mixing the following components with the concentrated fractions in the order listed
Reagent Volume (μl) Final concentration DNA sample (Step 48) 320–380 Click-it reaction buffer 10× or Tris-HCl pH 8 100mM 50 10 mM CuSO4 100mM 10 2 mM Biotin TEG Azide 100mM 10 2 mM Ascorbate Na 100mM 50 10 mM Milli-Q Water to 500 Total 500 *Critical Step* Water may be added first to the DNA to bring up the volume to 380 μL before adding the rest in the order listed. Flick to mix and do a short spin after adding each component.
-
50
Incubate at room temperature for 45 min protected from light.
-
51
Purify using a Millipore Amicon 3K column. Transfer the sample (500 μL) to a column and centrifuge at 14,000g, at room temperature, for 15 min.
-
52
Discard the eluate in the bottom of the tube, add 400 μL Milli-Q water to the column, and spin 14,000g at 15 min room temperature.
-
53
Perform two more washes (for a total of three) by repeating Step 52 twice.
-
54
Centrifuge column upside down in a new collection tube at 1000g, at room temperature, for 2 min to collect final purified sample. Transfer this sample into a 1.5 mL microcentrifuge tube and note the volume.
RNA Hydrolysis – Timing 1 h
-
55
Add NaOH 1 M at a final concentration of 250 mM to the sample and incubate 30 min at 37 °C.
-
56
Add 2.5 M Acetic Acid at a final concentration of 250 mM to neutralize the pH. !Caution Work under a fume hood when manipulating Acetic Acid.
-
57
Purify sample on 2 Bio-rad Micro bio-spin 30 columns (Steps 57–59). Firstly, snap the end tip and place each column in the collection tube for 5 min to allow gravity to drain the buffer.
-
58
Centrifuge the empty columns at room temperature, 1,000g, for 2 min.
-
59
Place each column in a collection tube and add half of the sample from step 56 to one of the columns. Centrifuge at room temperature, 1,000g, for 4 min to collect the sample (the two eluates are to be combined).
Phosphorylation of DNA Fragments – Timing 3–18 h
-
60Mix the following components:
Volume DNA sample (Step 59) 70–100 μL 10X T4 Ligase Buffer 20 μL T4 PNK Enzyme 10 μL Milli-Q Water Final 200 μL *CRITICAL STEP*10X T4 Ligase Buffer is used here as the buffer already contains 10 mM ATP required for the reaction. Alternatively, you may use the 10X PNK Buffer with 20 μL of 10 mM ATP.
-
61
Incubate the reaction for 30 min at 37 °C.
-
62
Inactivate the enzyme for 20 min at 65 °C.
-
63
Precipitate DNA by adding 0.1 vol of 3 M NaAcetate, pH 5.2 and 2.5 vol of 100% (vol/vol) ethanol. Incubate overnight at −20 °C. Alternatively, incubate 1 h at −80 °C, then 1 h at −20 °C.
#Pause Point# The precipitate can be stored overnight at −20 °C.
DNA Recovery – Timing 1 h
-
64
Centrifuge samples at max speed (or 20,800g) at 4 °C for 30 min.
-
65
Discard the supernatant and wash the pellet with 200 μL of 70% (vol/vol) ethanol (enough to cover pellet).
-
66
Centrifuge again for 10 min at max speed, 4 °C.
-
67
Remove all ethanol and dry briefly at room temperature to evaporate remaining ethanol.
-
68
Resuspend the pellet well in 29 μL of Milli-Q water by flicking to mix. Incubate at 37 °C for 15 min to completely dissolve DNA. This dissolved DNA would be the inserts for the subsequent ligation reaction.
Ligation of Adapters, Part I – Timing 3–4 h
*Critical* Adapters must be annealed and then gel purified on a native 10% (wt/vol) polyacrylamide gel before use. Once gel purified, they must be aliquoted and kept at −20 °C for a maximum of 6 months. They can be freeze-thawed for a maximum of 2 times. See Box 1 for details on annealing and gel-purifying your own adapters. Alternatively, they may also be ordered from IDT.
Box 1|. Annealing of Adapters – Timing 24 h.
Take 20 μl of each adapter (each at 1 mM stock concentration) and mix together in a PCR tube. Adapter sequences are listed in Table 2.
Add 0.4 μL of 5 M NaCl (50 mM final).
Denature and reanneal in a PCR machine set as follows: 2 min 90 °C, then 0.1 °C/s decrease until 16 °C.
Add 6X neutral PAGE gel buffer.
- Prepare a 10% (wt/vol) gel by mixing:
Reagent Amount 5X TBE 6 mL 40% (wt/vol) Acrylamide 7.5 mL H2O 16.5 mL (Make up to a total of 30 mL) Total 30 mL Add 30 μL of TEMED and 300 μL of 10% (wt/vol) APS right before pouring gel (polymerization starts immediately after APS addition).
Pour in a 25 × 20.5 cm glass plate. Let the gel polymerize for about 15 minutes, load the 40 μl of annealed adapters in two consecutives wells at the center of the gel and migrate for at least 2 h at 250 V in 1X TBE buffer.
Use saran wrap to remove the gel from the glass plate.
Use a UV lamp to locate the annealed adapters on the gel.
Cut adaptor band with a clean razorblade and put in a microcentrifuge tube.
Once in the tube, cut the band in very small pieces in order to facilitate the subsequent elution.
Add 50 μl of STE buffer (TE +150 mM NaCl final).
Leave to elute overnight RT under max agitation in a thermomixer.
Proceed to a short spin and pipet the maximum volume of eluate avoiding pipetting small gel pieces.
Purify on a G50 column (to remove all acrylamide traces) following the manufacturer’s instructions. Recover the eluate obtained after the last centrifugation of the column (STE).
Add 2.5 volumes of absolute ethanol in order to precipitate the adapters. Leave it 1 hour at −80°C, then 1 hour at 4°C and centrifuge 30 min at max speed. Wash with 500 μl of 75% ethanol and centrifuge 10min at max speed. Let the pellet dry for 5 or 10 min.
-
Resuspend in 20 μl of STE.
Measure the concentration using a Nanodrop.
Dilute to a 2 μg/μL concentration and aliquot (the number of aliquots will vary upon the total amount of adapters recovered).
Adapter are now ready to use and can be stored aliquots at −20 °C
-
69
Denature DNA on a heat block for 5 min and quench on ice for 5 min.
-
70Mix the following components:
Reagent Volume (μL) Inserts (resuspended DNA in water; Step 68) 29 Purified 5’ adapter pair [2 μg/μl] 1.5 Purified 3’ adapter pair [2 μg/μl] 1.5 T4 Ligase Buffer 4 T4 DNA Ligase 4 Total 40 *CRITICAL STEP* Incubate inserts and adapters for a couple of minutes before adding ligase buffer and enzyme.
-
71
Incubate reaction at room temperature for at least 2 h, protected from light.
Capture on Streptavidin Dynabeads – Timing 1–2 h
-
72
Dispense the required amount of MyOne T1 Streptavidin dynabeads into an Eppendorf tube. Use 10 μL dynabeads per sample.
-
73
Place the tube on a magnet and let beads settle for 2 min and remove supernatant.
-
74
Prewash beads twice with 200 μL 1X B&W Buffer + 0.05% (vol/vol) Tween 20.
-
75
Dilute beads in 40 μL of 2X B&W Buffer + 0.1% (vol/vol) Tween 20.
-
76
Add 40 μL of beads to 40 μL of ligation mix from Step 71. Top up with 70 μL 1X B&W Buffer + 0.05% (vol/vol) Tween 20 to ensure the mixture can tumble up and down well.
-
77
Mix well and rotate for at least 20 min at room temperature, protected from light.
*Critical Step* Ensure beads remain in suspension all the time.
-
78
Place the tube on a magnetic rack and let the beads settle. Remove the supernatant and store it at −20 °C in an Eppendorf tube as a back-up in case the beads did not bind well. Wash the beads 5 times with 200 μL 1X B&W Buffer + 0.05% (vol/vol) Tween 20. Flick gently or invert tube gently to mix the beads in the wash buffer each time.
-
79
Perform 2 washes with 200 μL 10 mM Tris pH 8.0 + 0.05% (vol/vol) Tween 20. Flick gently or invert tube gently to mix the beads in the wash buffer each time.
-
80
Perform 1 wash with 200 μL Milli-Q water. Flick gently or invert tube gently to mix the beads in the wash buffer each time.
-
81
Resuspend beads in 28 μL Milli-Q water (the total volume will be larger as beads take up volume too).
Ligation of adapters, Part II – Timing 18–20 h
-
82Mix the following components (Total 40 μL reaction):
Reagent Volume (μL) Inserts (resuspended DNA in water; Step 80) 29 Purified 5’ adapter pair [2 μg/μl] 1.5 Purified 3’ adapter pair [2 μg/μl] 1.5 T4 Ligase Buffer 4 T4 DNA Ligase 4 Total 40 -
83
Mix by pipetting/flicking, then incubate at 16 °C in thermoblock overnight.
-
84
Place the tube on a magnetic rack and remove the ligation supernatant. Store this supernatant in an Eppendorf tube at −20 °C as a back-up in case the reaction was insufficient or incomplete. Perform 5 washes of the beads with 200 μL 1X B&W Buffer + 0.05% (vol/vol) Tween 20. Flick gently to mix or invert tube gently.
-
85
Perform 2 washes with 200 μL 10 mM Tris pH 8.0 + 0.05% (vol/vol) Tween 20. Flick gently to mix or invert tube gently.
-
86
Perform 1 wash with 200 μL Milli-Q water. Flick gently to mix or invert tube gently.
-
87
Resuspend beads in 9 μL of Milli-Q water.
#Pause Point# The beads can be kept at 4 °C for several weeks or at −20 °C for several months.
Library Construction by PCR Amplification – Timing 1 h
-
88Mix on ice the following components:
Reagent Volume (μL) Beads (step 80) 10 PE PCR Primer 1 10 μM (Reagents) 1 TruSeq PCR Primer (Index #1–24) 10 μM (Table 2) 1 dNTPs 10 mM 1 5X Phusion Buffer 10 Milli-Q water 26 Phusion polymerase 1 Total 50 *CRITICAL STEP*A control sample could be run without the beads alongside this PCR reaction to ensure that the library is constructed from the samples that are bound to the beads.
*Critical Step* Select TruSeq PCR Primers with a different index number for different samples so they can be pooled and sequenced in the same lane on the NovaSeq 6000.
-
89Samples should be placed in thermocycler right away after assembling the reactions as the beads settle quickly. Run the following program:
Cycle Denature Extend Anneal Hold 1 95 °C 2 min 2–14 98 °C 30 s 62.5 °C 30 s 72 °C 30 s 15 72 °C 5 min 16 4 °C -
90
After PCR, transfer the sample into a 1.5 mL Eppendorf tubes and place on magnetic rack in order to capture the beads and recover the PCR product (supernatant).
-
91
Wash the beads twice in 200 μL Tris 10 mM, pH 8 + 0.05% (vol/vol) Tween 20, and resuspend them in 10 μL Tris 10 mM pH 8 + 0.05% (vol/vol) Tween 20 for further re-amplification if necessary.
#Pause Point# Beads can stored in −20 °C for several months. The PCR product (supernatant from step 89) can be stored in −20 °C for a few days.
PCR Test of Okazaki Fragments Library on Agarose gel – Timing 1 h
-
92
Prepare 2% (wt/vol) agarose gel in 1X TAE Buffer with SYBR Safe stain (1/10000 dilution).
-
93
Run 1/5th of the PCR product (supernatant from Step 89) i.e. 10 μL from 50 μL final volume of supernatant, with 100 bp DNA ladder on gel @ 75 V for 35–45 min or until the bands are about 3/4 down the length of the gel.
-
94
Visualize the gel to check the size distribution and purity of the library. The Okazaki fragments should form a smear above 130 bp. A primer dimer band is almost always present but separated by a gap from the smear at a discrete position around 120 bp (Fig. 3).
?Troubleshooting
Purification of Okazaki Fragments Library – Timing 30 min
-
95
Purify the PCR products and eliminate primer dimers (Steps 93–96). Firstly, add an equal volume of SPRIselect beads (vortex before use) to the PCR product from Step 89 and incubate on the bench for at least 5 min.
-
96
Remove and discard the supernatant and wash the beads twice with 200 μL 80% (vol/vol) ethanol (prepared fresh), keeping beads for 30 s on the magnetic rack for each wash.
-
97
After the 2nd wash, wait 3–5 min to allow any excess ethanol to evaporate (beads will become less shiny).
-
98
Elute DNA by resuspending beads in 20–25 μL TE Buffer and incubate for at least 2 min on the bench before transferring the supernatant (containing the DNA) to a new tube.
*CRITICAL STEP* Always add 1 μL more than the final volume that you want to avoid pipetting the beads as well when transferring the supernatent. E.g. Add 21 μL if you want at least 20 μL of the supernatant.
*CRITICAL STEP* The SPRI beads will help remove the primer dimer band so in the Tapestation result, it should appear to be reduced compared to the agarose gel. However, SPRIselect beads may remove less primer dimers if the library contains a high concentration of products that are similar in size to the primer dimers.
-
99
Check the size distribution and quality of the library on Tapestation. Similarly to the agarose gel, a smear above 130 bp that is not too faint would indicate good distribution and quality.
#Pause Point# DNA can be kept at 4 °C overnight or −20 °C for a few days.
?Troubleshooting
Library QC and Sequencing – Timing variable, typically 5–24 h
-
100
Quantify the libraries with a Qubit fluorometer, by using the Qubit® dsDNA HS Kit (typical yield is 0.5–2 ng/μL).
-
101
Check the proper amplification of libraries by performing a qPCR reaction using the KAPA Library Quantification Kit following standard manufacturer’s protocol.
-
102
Prepare the final library (or pool) at the required Illumina sequencer concentration. 10 μL of 2 nM DNA is required for the flow cell.
-
103
Submit the libraries for Illumina sequencing using NovaSeq SP100 (paired-end 50 bp sequencing).
*CRITICAL STEP* More than 1 library (up to 8 libraries) can be run on one single lane if they have unique barcodes (on the 3’ Primers listed in Table 2). A good coverage would be around 150–200 million reads per library but our down-sampling results show that even 12.5% of reads (19–25 million reads) produced sufficient usable data for analysis (Fig. 4).
Fig. 4|.
Agarose gel and tapestation results. a, Agarose gels visualized after sucrose gradient fractionation of genomic DNA from human cell lines (as depicted here by HCT116 cells). Only the fractions containing fragments that are not longer than 200 bp are pooled together and concentrated. The first 8 fractions (shown within the red box) typically contain the fragments of the desired sizes. b, Library preparation by PCR amplification as indicated by the smear (*) above the primer-dimer band that is present at around 120 bp. c, Tapestation analysis of size-distribution and quality of library. Library size distribution is indicated by the smear (*) above the primer-dimer band. The green and pink bands mark the upper and lower markers respectively.
?Troubleshooting
Data Analysis – Timing 1–2 d
CRITICAL Script files for performing both the initial processing and further downstream analysis are included in our public GitHub repository: https://github.com/FenyoLab/Ok-Seq_Processing. Also included are an example input file for the downstream analysis and the plots produced as output. A full set of example data, including raw read files and Watson/Crick read density tables for Ok-seq of RPE-1 cells can be found at GSE114017. Below is a detailed description of the analysis workflow.
Initial processing and QC
CRITICAL The initial processing is run on a High Performance Computing (HPC) cluster and can be executed by invoking the Python script process_okseq_SE_PE.py. If no HPC cluster is available, the steps can be performed on a standard laptop or desktop computer running a Linux or Mac operating system. In this case, the user is advised to check the documentation of the bioinformatics tools listed below for processing and memory requirements. All other custom Python code provided will run on Linux, Mac or Windows operating systems, provided Python v3.5 or higher has been installed on the computer. The output of this script is a text file of read counts on the Watson and Crick strands in 1kb bins. We use 1kb bins to allow for the analysis of relatively sparse sequencing data without a high level of noise. Moreover, we do not feel that our analysis can provide greater spatial resolution than 1 kb regardless of bin size.
Detailed below are the steps performed by the Python script. The following bioinformatics tools are used: FastQC and Trim Galore (Babraham Bioinformatics), bowtie234, samtools35, bedtools36, Picard and deepTools37.
-
104Starting from FASTQ files for paired-end reads, run FastQC for each read file. Open resulting html file and check that “Per base sequence quality” is high (green check mark). We have not encountered problems with sequence quality and typically sequence quality for Illumina should be high. If this is not the case, it could indicate a problem with the sequencing. The quality can also be re-evaluated after step 104 (Trim Galore), where quality trimming is performed. The library will most likely contain “Overrepresented sequences” and/or “Adaptor content” at this stage.
fastqc R1.fastq.gz fastqc R2.fastq.gz
-
105Run Trim Galore for the read files. This will perform quality and adaptor trimming. With the fastqc option it will also run FastQC on the resulting trimmed fastq files. The FastQC html file can be viewed to check that adaptors have been trimmed from the sequences and if needed, for an improvement in sequence quality. Any reads where one or both read pairs do not meet the minimum length after adaptor trimming are removed. Default minimum length of 20bp is used here.
trim_galore -–paired -–fastqc -–output_dir mydir R1.fastq R2.fastq
-
106Align reads to human genome with bowtie2. The index (hg19) can be downloaded from the bowtie2 manual page (http://bowtie-bio.sourceforge.net/bowtie2/manual.shtml).
bowtie2 -p 8 -x hg19 −1 R1_val_1.fq.gz −2 R2_val_2.fq.gz > aligned.sam
-
107
Filter for alignments with both reads aligning and MAPQ score > 30.
samtools view -q 30 -f 0×2 -b aligned.sam > aligned.bam
-
108
Remove duplicates with Picard. Picard requires a sorted bam file as input.
samtools sort aligned.bam > sorted.bamjava -jar picard.jar MarkDuplicates INPUT=sorted.bam OUTPUT=dupl_rem.bam METRICS_FILE=metrics.txt REMOVE_DUPLICATES=true ASSUME_SORT_ORDER=coordinate
-
109Convert to bedpe file. This requires the bam file to be sorted by query name. In bedpe format, the two ends of a paired-end alignment are reported on a single line. -mate1 is specified to always report mate1 first.
java -jar picard.jar SortSam I=dupl_rem.bam O=sorted_by_name.bam sort_order=queryname bedtools bamtobed -I sorted_by_name.bam -bedpe -mate1 > bedpefile.bedpe
-
110Combine read pairs from the bedpe file to a single fragment and split fragments mapped to the forward and reverse strands to separate files. Custom python code was written for this step. See the function convert_bedpe_to_bed in the script process_okseq_SE_PE.py. The output of this function is fwd.bed and rev.bed, bed files containing the fragments mapped to the forward and reverse strands.
convert_bedpe_to_bed(bedpefile.bedpe, fwd.bed, rev.bed)
CRITICAL STEP Steps 110 and 111 below should be executed for both fwd.bed and rev.bed
-
111Convert back to bam format. Conversion to bam format requires a sorted bed file. bedToBam requires the genome file “human.hg19.genome”, which contains the chromosome sizes (https://genome.ucsc.edu/goldenpath/help/hg19.chrom.size).
sort -k 1,1 fwd.bed > fwd_sorted.bed bedToBam -i fwd_sorted.bed -g human.hg19.genome > fwd.bam
-
112Run bamCoverage (deepTools) to obtain bedgraph files with binsize 1000. BamCoverage requires a sorted bam file and an index that can be created with samtools.
samtools sort fwd.bam > fwd.sorted.bam samtools index fwd_sorted.bam bamCoverage -b fwd_sorted.bam -o fwd.bedGraph -binsize 1000 -outFileFormat bedgraph
-
113Create final text output file with columns for Watson and Crick strand coverage density for strict 1000bp bins. The bamCoverage tool outputs a bedgraph file (step 111) that combines consecutive bins with matching read counts. Therefore some bins are larger than 1000bp. The function convert_bedGraph_to_txt in the script process_okseq_SE_PE.py will split these combined bins into 1000bp each, and also combine the read counts for both Watson and Crick strands to a single tab-delimited text file.
convert_bedGraph_to_txt(fwd.bedGraph, rev.bedGraph, outfile.txt)
Visualization and Quantification of origin firing at genomic loci
CRITICAL Further processing and visualization can now be performed to assess origin firing localization and efficiency (Fig. 2) around genomic loci. Python scripts for producing plots of strand bias around gene TSS and TTS and the plots shown in Figure 4 (strand bias across length normalized genes, derivative plots and heatmaps) are included in the GitHub repository and described below. This code can be run on any operating system where Python v3.5 or higher has been installed. The tables referenced in step 113 are also included in our GitHub repository. The RNA-Seq data included is from GSE89413.
-
114
Obtain additional data from online resources. (i) Download the gene table for all human genes (GRCh37/hg19) from the UCSC table browser (https://genome.ucsc.edu/cgi-bin/hgTables). This will include locations of gene TSS and TTS sites. (ii) Download pre-processed RNA-Seq data for the relevant cell type. This data should provide a table of gene names and normalized read counts, e.g. FPKM or TPM. (iii) Download a gene name table from HGNC (https://www.genenames.org). Specify for the table to contain gene aliases and previous names. This table can be useful for matching the gene symbols from the RNA-Seq data to the gene symbols listed in the table from UCSC.
-
115Pre-process the gene table file: (i) Filter the gene table file for only protein coding genes. Many protein coding genes have more than one associated TSS/TTS. These can be filtered by only taking the first set of positions listed, or by taking the positions listed as “canonical” (this column must be selected when downloading the gene table file from UCSC). Both choices resulted in similar output in our analysis. (ii) Add the RNA-Seq information to the gene table file, so that there is a column with RNA-Seq read counts for each gene in the table. Genes without RNA-Seq read count data are filtered prior to producing the plots described below.
pre_process_gene_table_file.py data_dir gene_table_file rna_seq_file data_col gene_alias_file data_dir: the directory where the files are located gene_table_file: the genes table downloaded from UCSC (step 113) rna_seq_file: the data table containing the RNA Seq data (step 113) data_col: column from rna_seq_file containing e.g. FPKM values gene_alias_file: HGNC gene names table (step 113)
Pre-processed gene table file with Rna-Seq data is saved in data_dir.
-
116Add Watson and Crick read density data to the gene table file. Extract the W Watson and Crick read counts from 100kb upstream to 100kb downstream of the TSS and TTS sites, in 1kb bins. This data is added to the gene table as columns, i.e. 201 columns are added for TSS and 201 for TTS.
add_okazaki_data_to_site_list.py data_dir okseq_file gene_table_file data_col results_dir data_dir: the directory where files are located okseq_file: the text file containing the okseq read counts (step 112) gene_table_file: processed genes table file (step 114) data_col: same as in step 114 results_dir: directory where results file is saved
Gene table file with read count data is saved in results_dir.
CRITICAL The python script make_okseq_plots.py creates all plots and statistical comparisons (steps 116–121). The code is annotated with descriptions of the type of plots being produced. Below are additional descriptions of each of these plots.make_okseq_plots.py data_dir loci input_file data_dir: the directory where the files are located loci: genomic loci of interest, e.g. TSS or TTS file_name: output file from step 115 (gene table file with read count data)
All plots are saved in data_dir.
-
117
Plot average strand bias curves for all genes. Strand bias curves are useful for assessing origin localization and efficiency (Fig. 2). The curve is plotted as the ratio of Crick reads to total read count (Watson+Crick) and centered around genomic loci of interest. The code has been provided for making these plots for gene TSS and TTS sites. In order to visualize replication fork initiation and termination as it relates to positions upstream/downstream of the TSS or TTS, the strand bias curve should be normalized for transcriptional direction by reversing and calculating the ratio of Watson strand reads for genes on the Crick strand. Strand bias curves for both Watson and Crick genes can then be averaged together to produce an accurate picture of replication dynamics around gene TSS and TTS sites17.
-
118
Plot strand bias curves at gene TSS/TTS by transcript levels. Strand bias curves are plotted e.g. by FPKM level divided into 4 quartiles to examine the relation between transcription and replication. Transcriptional volume (FPKM x gene length) is another useful measure for plotting17.
-
119
Normalize by gene length to visualize strand bias over the entire gene region (Fig. 4a). This plot summarizes replication dynamics across actively transcribed genes, from upstream of the TSS to downstream of the TTS. In order to average data over this region for all actively transcribed genes, a length normalization is performed. The strand bias values for each gene were transformed to a standard length of 50kb by up-sampling values for genes shorter and down-sampling values for genes longer than the standard length. Very short (<5kb) or long (>100kb) genes were removed from this analysis.
-
120
Plot the derivative of the strand bias curve (Fig. 4b). The derivative of the strand bias curve indicates an increase in replication initiation frequency (positive values) or termination (negative values). For example, the positive increase in the derivative curves in Fig. 4b indicates localized replication initiation at the TSS sites of actively transcribed genes. Immediately downstream of the TSS, the derivative curve becomes negative indicating an absence of replication initiation17.
-
121
Plot strand bias curves as a heatmap (Fig. 4c). Heatmaps can serve as an alternative and powerful visualization of strand bias. In the heatmaps, each row represents a gene and the genes are sorted by length.
-
122
Statistics. In order to test for a significant difference between strand bias curves, the difference between ranges downstream and upstream of the site being analyzed can be calculated for each gene. Then, a non-parametric test such as the Kruskal-Wallace H-test can be used to compare strand bias for multiple groups of genes or results from different experimental conditions.17 The function calculate_pvalue in the plotting script will produce p-values for a comparison of strand bias curves between data sets. The plotting script will print these p-values as output for the example data set. Please refer to the comments in the script file for additional details.
Timing
Steps 1 – 2, Cell Culture Preparation: 1–7 d
Step 3, EdU/EdC Labelling & Termination of Cells: 2–4 h
Steps 4 – 7, Cell Lysis: 30 min to O/N
Steps 8 – 29, DNA Extraction: 3 h to O/N
Steps 30 – 38, Size Fractionation using 5–30% Linear Sucrose Gradient: 22–24 h
Steps 39 – 42, Alkaline Gel Electrophoresis: 1–18 h
Steps 43 – 48, Pool and Concentrate Fractions: 1–2 h
Steps 49 – 54, Biotinylation in Click Reaction: 2 h
Steps 55 – 59, RNA Hydrolysis: 1 h
Steps 60 – 63, Phosphorylation of DNA Fragments: 1–18 h
Steps 64 – 68, DNA Recovery: 1 h
Steps 69 – 71, Ligation of Adapters Part I: 3–4 h
Steps 72 – 80, Capture on Streptavidin Dynabeads: 1–2 h
Steps 81 – 86, Ligation of Adapters Part 2: 18–20 h
Steps 87 – 90, Library Construction by PCR Amplification: 1 h
Steps 91 – 93, PCR Test of Okazaki Fragments Library on Agarose gel: 1 h
Steps 94 – 98, Purification of Okazaki Fragments Library: 30 min
Steps 99 – 102, Library QC and Sequencing: variable, typically 5–24 h
Steps 103 – 121, Data Analysis: 1–2 d
Troubleshooting
Troubleshooting advice can be found in Table 3.
Table 3:
Troubleshooting Advice
| Step | Problem | Possible reason | Solution |
|---|---|---|---|
| 26 | DNA pellet is gooey | DNA is partially dissolved during the washes | Add small amounts of 7.5M Ammonium Acetate (drop by drop) to precipitate the DnA fully again. |
| 93 | Excess of dimers | Not enough starting material | Increase the number of starting cells. Cut the agarose gel above the end of the smear and perform a gel purification when preparing libraries. |
| 93 | Absence of smear containing Okazaki fragments | Insufficient yield | Perform a second amplification on the same batch of beads. The yield is significantly higher with this second PCR. |
| 93 | Absence of smear containing Okazaki fragments | dNTPs are degraded | Use a fresh aliquot of dNTPs and perform a second amplification on the same batch of beads. |
| 93 | Heavy banding instead of a smear | Adapters may have degraded | Use fresh aliquot or prepare new adapter on a new sample. |
| 98 | Smear on Tapestation is very faint with an excess of signals from the primer dimers | Not enough material | Increase the number of starting cells. |
| 102 | High number of reads containing primer-dimer sequences or decreased library complexity | Excess of primers, low amount of produced library | Increase input material. Optimize the number of PCR cycles to prevent excessive PCR duplicates. Test first the libraries on a low throughput machine (e.g. MiSeq) to assess library quality. |
Anticipated Results
Typically, Okazaki fragments of <200 bp are found in the first 8 fractions of the sucrose gradient (Fig. 4a). After ligation of the adapters, the fragments are amplified to prepare a library for paired-end sequencing (2 × 50 bp). Visualization of the prepared library on a 1% agarose gel should show a smear above the primer-dimer bands at 120 bp, with a discrete gap in between (Fig. 4b). The size distribution and quality of the library as determined on Tapestation after removal of the adapter-dimers would show a similar smear (Fig. 4c).
Bioinformatics analysis of the sequenced library is performed, focusing on a specific genomic locus and a typical expected output of the data of a meta-analysis surrounding transcription start and termination sites is shown in Fig. 3a, with comparisons between the original data, published previously by Chen et al., and 3 downsampling levels: 50%, 25% and 12.5% respectively17. Fig. 3a represents information on the Okazaki fragment strand bias while Fig. 3b reports on the replication initiation frequencies calculated as the first derivative of Okazaki fragment strand bias around TSS as in Fig. 3a. Heat map representations of the change in replication direction around TSS of the data are shown in Fig. 3c. The results of the various downsampling levels demonstrate that usable data can be obtained even downsampling to 12.5% of total reads from the sequenced data.
Acknowledgements
We thank Adriana Heguy and Paul Zappile from the NYU Genome Technology Center for assistance with TapeStation and sequencing. Work in T.T.H. laboratory is supported by grants from the NIH (ES025166), V Foundation BRCA Research and Basser Innovation Award. Work in D.J.S. laboratory is supported by grant (R35 GM134918) from the NIH.
Footnotes
Competing Interests
The authors declare no competing interests.
Additional Information
Code Availability
All code is publicly available under the GNU General Public License v2.0 in our GitHub repository: https://github.com/FenyoLab/Ok-Seq_Processing. Included in the repository is example input data for creating plots as shown in Figure 4, as well as additional plots as described in the Data Analysis section. The expected output of all scripts is also included as pdf/jpg files. Documentation is provided as a readme file and specific instructions on parameters to functions are embedded as inline comments in the code. The current version of the software is v1.0, tagged as a release in GitHub: https://github.com/FenyoLab/Ok-Seq_Processing/releases/tag/v1.0.
EDITORIAL SUMMARY This protocol describes an approach to quantify DNA replication dynamics (initiation and termination frequencies and origin firing efficiencies) at defined genomic loci in asynchronously growing cells.
TWEET New protocol for measuring DNA replication dynamics at selected genomic loci in asynchronously growing cells.
COVER TEASER Quantifying DNA replication dynamics
Related links
Key reference(s) using this protocol
Chen, Y. et al. Nat Struct Mol Biol 26, 67–77 (2019): https://doi.org/10.1038/s41594-018-01710
Key data used in this protocol
Orbán-Németh, Z. et al. Nat. Protoc. 13, 478–494 (2018) [URL]
Data Availability
Data used for the non-down-sampled (100%) curves in Figure 4 is provided at GSE114017 The down-sampled curves can be produced by repeatedly removing one-half of the reads from the fastq files and then re-running the processing pipeline on the reduced data set.
References
- 1.Macheret M & Halazonetis TD DNA replication stress as a hallmark of cancer. Annu Rev Pathol 10, 425–448 (2015). [DOI] [PubMed] [Google Scholar]
- 2.Fragkos M, Ganier O, Coulombe P & Mechali M DNA replication origin activation in space and time. Nat Rev Mol Cell Biol 16, 360–374 (2015). [DOI] [PubMed] [Google Scholar]
- 3.Ibarra A, Schwob E & Méndez J Excess MCM proteins protect human cells from replicative stress by licensing backup origins of replication. Proc Natl Acad Sci U S A 105, 8956–8961 (2008). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.McIntosh D & Blow JJ Dormant origins, the licensing checkpoint, and the response to replicative stresses. Cold Spring Harb Perspect Biol 4 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Ge XQ, Jackson DA & Blow JJ Dormant origins licensed by excess Mcm2–7 are required for human cells to survive replicative stress. Genes Dev 21, 3331–3341 (2007). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Kawabata T et al. Stalled fork rescue via dormant replication origins in unchallenged S phase promotes proper chromosome segregation and tumor suppression. Mol Cell 41, 543–553 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Hyrien O Peaks cloaked in the mist: The landscape of mammalian replication origins. Journal of Cell Biology 208, 147–160 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Okazaki R, Okazaki T, Sakabe K, Sugimoto K & Sugino A Mechanism of DNA chain growth, I. Possible discontinuity and unusual secondary structure of newly synthesized chains. Proc Natl Acad Sci U S A 59, 598–605 (1968). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Smith DJ, Yadav T & Whitehouse I Detection and Sequencing of Okazaki Fragments in S. cerevisiae, in DNA Replication 141–153 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Smith DJ & Whitehouse I Intrinsic coupling of lagging-strand synthesis to chromatin assembly. Nature 483, 434–438 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Bielinsky A-KG, Susan A Chromosomal ARS1 Has a SingleLeading Strand Start Site. Molecular Cell 3, 477–486 (1999). [DOI] [PubMed] [Google Scholar]
- 12.Pourkarimi E, Bellush JM & Whitehouse I Spatiotemporal coupling and decoupling of gene transcription with DNA replication origins during embryogenesis in C. elegans. Elife 5 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Sriramachandran AM et al. Genome-wide Nucleotide-Resolution Mapping of DNA Replication Patterns, Single-Strand Breaks, and Lesions by GLOE-Seq. Mol Cell 78, 975–985 e977 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Petryk N et al. Replication landscape of the human genome. Nat Commun 7, 10208 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Osmundson JS, Kumar J, Yeung R & Smith DJ Pif1-family helicases cooperatively suppress widespread replication-fork arrest at tRNA genes. Nat Struct Mol Biol 24, 162–170 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Tubbs A et al. Dual Roles of Poly(dA:dT) Tracts in Replication Initiation and Fork Collapse. Cell 174, 1127–1142.e1119 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Chen Y-H et al. Transcription shapes DNA replication initiation and termination in human cells. Nature Structural & Molecular Biology 26, 67–77 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Kahli M, Osmundson JS, Yeung R & Smith DJ Processing of eukaryotic Okazaki fragments by redundant nucleases can be uncoupled from ongoing DNA replication in vivo. Nucleic Acids Res 47, 1814–1822 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Meldal M & Tornøe CW Cu-catalyzed azide-alkyne cycloaddition. Chemical Review 108, 2952–3015 (2008). [DOI] [PubMed] [Google Scholar]
- 20.Salic A & Mitchison TJ A chemical method for fast and sensitive detection of DNA synthesis in vivo. Proceedings of the National Academy of Sciences of the United States of America 105, 2415–2420 (2008). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Koç A, Wheeler LJ, Mathews CK & Merrill GF Hydroxyurea Arrests DNA Replication by a Mechanism That Preserves Basal dNTP Pools. Journal of Biological Chemistry 279, 223–230 (2004). [DOI] [PubMed] [Google Scholar]
- 22.Gilbert DM Evaluating genome-scale approaches to eukaryotic DNA replication. Nat Rev Genet 11, 673–684 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Mesner LD et al. Bubble-seq analysis of the human genome reveals distinct chromatin-mediated mechanisms for regulating early- and late-firing origins. Genome Research 23, 1774–1788 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Langley AR, Gräf S, Smith JC & Krude T Genome-wide identification and characterisation of human DNA replication origins by initiation site sequencing (ini-seq). Nucleic Acids Research (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Macheret M & Halazonetis TD Monitoring early S-phase origin firing and replication fork movement by sequencing nascent DNA from synchronized cells. Nature Protocols 14, 51–67 (2018). [DOI] [PubMed] [Google Scholar]
- 26.Giacca M, Pelizon C & Falaschi A Mapping Replication Origins by Quantifying Relative Abundance of Nascent DNA Strands Using Competitive Polymerase Chain Reaction. Methods: A Companion to Methods in Enzymology 13, 301–312 (1997). [DOI] [PubMed] [Google Scholar]
- 27.Besnard E et al. Best practices for mapping replication origins in eukaryotic chromosomes. Curr Protoc Cell Biol 64, 22 18 21–13 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Zhao PA, Sasaki T & Gilbert DM High-resolution Repli-Seq defines the temporal choreography of initiation, elongation and termination of replication in mammalian cells. Genome Biol 21, 76 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Vassilev L & Johnson EM Mapping initiation sites of DNA replication in vivo using polymerase chain reaction amplification of nascent strand segments. Nucleic Acids Res 17, 7693–7705 (1989). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Bielinsky A-K & Gerbi SA Discrete start sites for DNA synthesis in the yeast ARS1 origin. Science 279, 95–98 (1998). [DOI] [PubMed] [Google Scholar]
- 31.Foulk MS, Urban JM, Casella C & Gerbi SA Characterizing and controlling intrinsic biases of lambda exonuclease in nascent strand sequencing reveals phasing between nucleosomes and G-quadruplex motifs around a subset of human replication origins. Genome Res 25, 725–735 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Chen YH et al. ATR-mediated phosphorylation of FANCI regulates dormant origin firing in response to replication stress. Mol Cell 58, 323–338 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Wu X et al. Developmental and cancer-associated plasticity of DNA replication preferentially targets GC-poor, lowly expressed and late-replicating regions. Nucleic Acids Res 46, 10157–10172 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Langmead B & Salzberg SL Fast gapped-read alignment with Bowtie 2. Nature Methods 9, 357–359 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Li H et al. The Sequence Alignment/Map format and SAMtools. Bioinformatics 25, 2078–2079 (2009). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Quinlan AR & Hall IM BEDTools: a flexible suite of utilities for comparing genomic features. Bioinformatics 26, 841–842 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Ramírez F et al. deepTools2: a next generation web server for deep-sequencing data analysis. Nucleic Acids Research 44, W160–W165 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Ligasova A et al. Dr Jekyll and Mr Hyde: a strange case of 5-ethynyl-2’-deoxyuridine and 5-ethynyl-2’-deoxycytidine. Open Biol 6, 150172 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Manska S, Octaviano R & Rossetto CC 5-Ethynyl-2’-deoxycytidine and 5-ethynyl-2’-deoxyuridine are differentially incorporated in cells infected with HSV-1, HCMV, and KSHV viruses. J Biol Chem 295, 5871–5890 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Data Availability Statement
Data used for the non-down-sampled (100%) curves in Figure 4 is provided at GSE114017 The down-sampled curves can be produced by repeatedly removing one-half of the reads from the fastq files and then re-running the processing pipeline on the reduced data set.