Abstract
Background: Bufavirus (BuV), Human Cosavirus (HCoSV), and Saffold (SAFV) virus are three newly discovered viruses and have been suggested as possible causes of gastroenteritis (GE) in some studies. The aim of the present study was to estimate the overall prevalence of viruses and their association with GE.
Methods: A comprehensive systematic search was conducted in Scopus, Web of Science, PubMed, and Google scholar between 2007 and 2021 to find studies on the prevalence of BuV, HCoSV, and SAFV viruses.
Result: Meta-analysis of the 46 included studies showed the low prevalence of BuV (1.%, 95% CI 0.6–1.5%), HCoSV (0.8%, 95% CI 0.4–1.5%), and SAFV (1.9%, 95% CI 1.1–3.1%) worldwide. Also, no significant association between these viruses and GE was observed. BuV was isolated from patients with GE in Africa, while SAFV was more common in Europe. BuV1 and BuV2 have the same prevalence between the three identified genotypes of BuV. HCoSV-C was the most prevalent genotype of HCoSV, and SAFV2 was the commonest genotype of SAFV. All of these viruses were more prevalent in children older than 5 years of age.
Conclusion: This was the first meta-analysis on the prevalence and association of BuV, HCoSV, and SAFV with GE. While no significant association was found between infection with these viruses and GE, we suggest more studies, especially with case-control design and from different geographical regions in order to enhance our knowledge of these viruses.
Keywords: Bufavirus, Saffold virus, Cosavirus, gastroenteritis, meta-analysis
Introduction
Gastroenteritis (GE) is one of the most common illnesses in both children and adults worldwide. The high importance of GE is due to both high morbidity and mortality and also the financial burdens of the disease. Children, the elderly, and immunocompromised individuals are at higher risk of severe GE (1). Infectious agents, particularly viruses are the main cause of GE worldwide (2). Before the implication of Rotavirus vaccination, Rotavirus was the leading cause of viral GE, while other enteric viruses, such as Noroviruses, Astroviruses, and Human adenoviruses, are now the most prevalent viruses causing GE (3). Besides the aforementioned enteric viruses, the list of enteric viruses is continuously growing due to the discovery of emerging viruses (4, 5). Since still 40% of cases of GE are of unknown etiology (6), these newly discovered viruses may likely be involved in causing the GE (7).
The Parvoviridae family consists of small, non-enveloped, icosahedral-shaped viruses, which have a single-stranded DNA genome. Members of this family can infect both vertebrates and invertebrates (8). For about 3 decades, Parvovirus B19 was taught to be the only human pathogen in this family (9). In 2005, Human bocavirus 1 was isolated from the nasopharyngeal swab of children with respiratory symptoms. Since 2009, three other types of the virus, named Human bocavirus 2–4, have been isolated from a stool specimen of children with or without GE (10). In 2012, the metagenomic survey of stool samples of children with acute diarrhea in Burkina Faso resulted in the discovery of a new member of this family, which was named Bufavirus (BuV) (9). Human BuVs belong to the genus Protoparvovirus, and, so far, three genotypes of Human BuV have been identified (11).
The Picornaviridae family contains non-enveloped, icosahedral-shaped viruses with a positive-sense single-stranded RNA genome (12). Unlike the Parvoviridae, viruses in the Picornaviridae family are not able to infect invertebrates (13). This family contains a growing number of viruses, which cause a variety of diseases that can affect different organs of the body. In 2007, a new member of this family was isolated from a child with a fever of an unknown origin in the United States. This virus was later named Saffold virus (SAFV); this name was derived from the lead author of the research, Morris Saffold Jones. Phylogenetic analysis showed that this virus is closely related to theilovirus species in the Cardiovirus genus of this family (14). Since then, eight genotypes of SAFV have been identified (15). The other virus in this family is the Cosavirus (CoSV), which was discovered in 2008 in pediatric patients with acute flaccid paralysis and later found in patients with GE (7). These three novel viruses were isolated from patients with different clinical and epidemiologic patterns (4). They were isolated from patients with GE (6, 16) and neurological disorders (17–19). While GE is a threat to global health, the causative agents of many cases still remained unclear (4). Therefore, we conducted this systematic review and meta-analysis to (1) elucidate the possible role of these viruses in development of GE and (2) understand the current epidemiologic pattern of these viruses in different parts of the world.
Methods
Search Strategy
This systematic and meta-analysis review was performed using the recommendations of the PRISMA (Preferred Reporting Items for Systematic Reviews and Meta-Analyses) (20). We comprehensively searched from multiple electronic databases, including Web of Science, PubMed, Google scholar, and Scopus. English-language-related articles published from January 2007 to April 2021 were searched by two investigators independently (AK and MZ) using the following keywords: “Bufavirus” OR “BuV” OR “novel human picornavirus” OR “Saffold virus” OR “SAFV” OR “HCosV” OR “Human Cosavirus” AND “prevalence” OR “epidemiology” OR “molecular prevalence” AND “acute gastroenteritis” OR “diarrhea” OR “gastroenteritis” OR “gastrointestinal complications. In addition, the reference list of all relevant articles and narrative reviews were retrieved in full to search for additional eligible studies. All selected studies were imported to the EndNote software versionX8 (Thomson Reuters, California) for criteria analysis.
Inclusion and Exclusion Criteria
The inclusion criteria for the studies were as follows: (1) All observational studies (case-control, cohort, and cross-sectional studies); (2) Published: 2007 to 2021 for SAFV, between 2012 and 2021 for BuV, and between 2008 and 2021 for HCosV; and (3) Studies reporting the molecular techniques of Bufavirus, Saffoldvirus, and Cosavirus among patients with GE across the world. Papers were excluded from this review if (1) Samples were selected entirely from patients with Bufavirus, Saffold virus, and Cosavirus; (2) Research provides incomplete data; and (3) Review articles, congress abstracts, conference papers, meta-analysis, or systematic reviews, and articles in languages other than English.
Data Extraction
The data were extracted from 46 selected studies by two researchers separately and independently, including the first author's name, location, year of publication, continent, number of investigated patients, number of isolated viruses, target gene, molecular technique, and genotypes. If necessary, any issue related to the selection of studies was resolved by the first and corresponding authors.
Data Synthesis and Statistical Analysis
We used a random-effect model to estimate the overall prevalence of the BuV, SAFV, and HCosV, and results are shown in the forest plot with a 95% confidence interval. Furthermore, evaluation of the prevalence of the viruses was performed on continental, country, diagnostic method, and age as well as gender subgroups. Also, the prevalence of the viruses and their association with GE were estimated and reported by odds ratio (OR). The Egger's test and I2 statistic/Cochran's Q statistic were used to determining publication bias and heterogeneity assessments, respectively, and p < 0.05 was considered statistically significant. All analyses of the present study were performed with comprehensive meta-analysis (V2.2, Bio stat) software.
Results
Search Results and Studies Characteristics
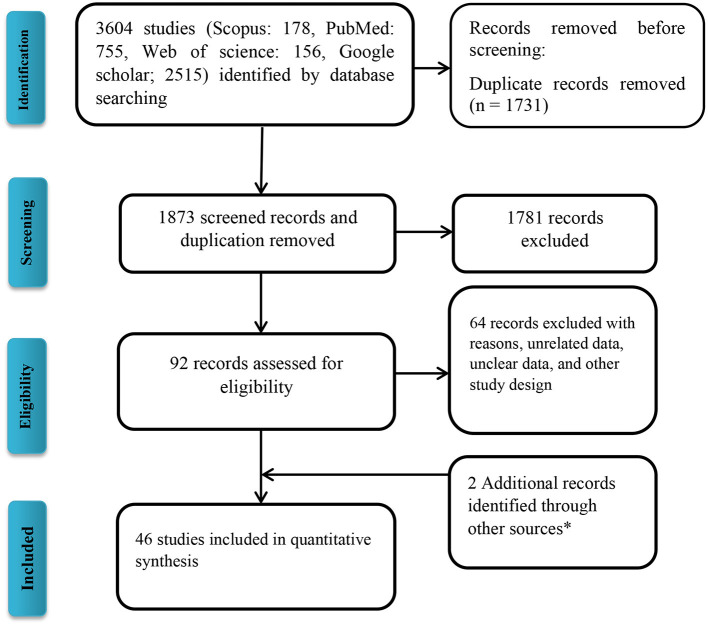
Following the initial search strategy in the aforementioned databases, 3,604 original related articles were identified (PubMed: 755, Scopus: 178, Web of Science: 156, Google scholar: 2,515). A total of 46, observational articles, which included 30 cross-sectional (BuV: 6, SAFV: 12, and HCosV: 12), seven case-control (BuV: 1, SAFV: 3, and HCosV: 3), and nine cohort (BuV: 5, SAFV: 3, and HCosV: 1) studies were included based on our inclusion criteria. A summary of the research selection process and the reasons for exclusion is shown in Figure 1. In the case of Bufavirus, five articles were conducted in Europe, four in Asia, and three in Africa. About the Cosavirus, nine in Asia, four in Europe, one in Africa, and two articles were done in America. In the case of Saffold virus, 15 and three were performed in Asia Europe, respectively. Characteristics of the included 46 articles are shown in Tables 1–3.
Figure 1.
Flow diagram of the literature search for studies included in the meta-analysis. *Including manual search and library records.
Table 1.
The general characterization of Bufavirus studies.
| References | Study type | Country | Continent | Publishing year | Cases | Positive | Target | Method | Not distinguished Genotype | BuV1 | BuV2 | BuV3 |
|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Phan et al. (21) | Cross-sectional | Burkina Faso | Africa | 2012 | 98 | 4 | NS1 | Nested RT-PCR | 3 | 1 | ||
| Phan et al. (21) | Cross-sectional | Tunisia | Africa | 2012 | 100 | 0 | NS1 | Nested RT-PCR | ||||
| Smits et al. (22) | Cross-sectional | Netherlands | Europe | 2014 | 27 | 1 | NS1 | Real-time RT-PCR | 1 | |||
| Vaisanen et al. (9) | Cross-sectional | Finland | Europe | 2014 | 629 | 7 | VP2 | Real-time RT-PCR | 7 | |||
| Yahiro et al. (23) | Cross-sectional | Bhutan | Asia | 2014 | 393 | 3 | NS1 | Nested RT-PCR | 3 | |||
| Huang et al. (16) | Cross-sectional | China | Asia | 2015 | 1877 | 9 | NS1 | Real-time RT-PCR | 4 | 5 | ||
| Altay et al. (24) | Case-control | Turkey | Europe | 2015 | 583 | 8 | RT-PCR | 8 | ||||
| Chieochansin et al. (25) | Cohort | Thailand | Asia | 2015 | 1414 | 1 | NS1 | Nested RT-PCR | 1 | |||
| Chieochansin et al. (25) | Cohort | Thailand | Asia | 2015 | 81 | 3 | NS1 | Nested RT-PCR | 3 | |||
| Ayouni et al. (7) | Cohort | Tunisia | Africa | 2016 | 203 | 2 | NS1 | Nested RT-PCR | 2 | |||
| Vaisanen et al. (11) | Cohort | Finland | Europe | 2016 | 410 | 3 | NS1 | Real-time RT-PCR | 3 | |||
| Mohammad et al. (26) | Cross-sectional | Kuwait | Asia | 2020 | 84 | 1 | Multiplex RT-PCR | 1 | ||||
| Dapra et al. (5) | Cohort | Italy | Europe | 2021 | 160 | 0 | Real-time RT-PCR | |||||
| Mohanraj et al. (27) | Cohort | Finland | Europe | 2021 | 243 | 4 | NS1 | Multiplex real-time qPCR | 4 | |||
| Mohanraj et al. (27) | Cohort | Finland | Europe | 2021 | 386 | 3 | NS1 | Multiplex real-time qPCR | 3 | |||
| Mohanraj et al. (27) | Cohort | Finland | Europe | 2021 | 955 | 3 | NS1 | Multiplex real-time qPCR | 3 | |||
| Mohanraj et al. (27) | Cohort | Latvia | Europe | 2021 | 115 | 0 | NS1 | Multiplex real-time qPCR | 0 | |||
| Mohanraj et al. (27) | Cohort | Malawi | Africa | 2021 | 164 | 1 | NS1 | Multiplex real-time qPCR | 1 |
Table 3.
The general characterization of Cosavirus studies.
| References | Study | Publishing year | Country | Continent | Cases | Positive |
|---|---|---|---|---|---|---|
| Nielsen et al. (33) | Cohort | 2013 | Denmark | Europe | 386 | 0 |
| Stocker et al. (44) | Case-control | 2012 | Brazil | America | 359 | 13 |
| Vizzi et al. (45) | Case-control | 2021 | Venezuela | America | 82 | 5 |
| Yu et al. (46) | Case-control | 2017 | China | Asia | 461 | 8 |
| Ayouni et al. (7) | Cross-sectional | 2016 | Tunisia | Africa | 203 | 2 |
| Dapra et al. (38) | Cross-sectional | 2018 | Italy | Europe | 164 | 0 |
| Dapra et al. (5) | Cross-sectional | 2021 | Italy | Europe | 160 | 0 |
| Khamrin et al. (47) | Cross-sectional | 2012 | Thailand | Asia | 300 | 1 |
| Khamrin et al. (48) | Cross-sectional | 2014 | Thailand | Asia | 411 | 1 |
| Kim et al. (40) | Cross-sectional | 2020 | South Korea | Asia | 801 | 0 |
| Menage et al. (6) | Cross-sectional | 2017 | Thailand | Asia | 1,093 | 16 |
| Mohammad et al. (26) | Cross-sectional | 2020 | Kuwait | Asia | 84 | 1 |
| Okitsu et al. (49) | Cross-sectional | 2014 | Japan | Asia | 630 | 1 |
| Rovida et al. (50) | Cross-sectional | 2013 | Italy | Europe | 689 | 1 |
| Thongprachum et al. (35) | Cross-sectional | 2017 | Japan | Asia | 751 | 1 |
| Kochjan et al. (51) | Cross-sectional | 2016 | Thailand | Asia | 21 | 1 |
Table 2.
The general characterization of Saffold virus studies.
| References | Study | Country | Continent | Publishing year | Cases | Positive | Target | Method | SAFV-1 | SAFV-2 | SAFV-3 | SAFV-4 | SAFV-6 |
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Ren et al. (28) | Cross-sectional | China | Asia | 2009 | 373 | 12 | 5′ UTR | Nested RT-PCR | 12 | ||||
| Khamrin et al. (29) | Cross-sectional | Thailand | Asia | 2011 | 150 | 4 | 5′ UTR | Nested RT-PCR | 4 | ||||
| Dai et al. (30) | Case-control | China | Asia | 2011 | 577 | 6 | 5′ UTR | Nested RT-PCR | 3 | ||||
| Zhang et al. (31) | Cohort | China | Asia | 2012 | 2,013 | 12 | 5′ UTR | Real-time RT-PCR | 4 | 5 | |||
| Khamrin et al. (32) | Cross-sectional | Japan | Asia | 2013 | 454 | 7 | 5′ UTR | Nested RT-PCR | 5 | 2 | |||
| Nielsen et al. (33) | Cohort | Denmark | Europe | 2013 | 386 | 10 | VP1 | Real-time RT-PCR | 10 | ||||
| Yodmeeklin et al. (34) | Cross-sectional | Thailand | Asia | 2015 | 608 | 9 | 5′ UTR | Nested RT-PCR | 1 | 5 | 2 | 1 | |
| Thongprachum et al. (35) | Cross-sectional | Japan | Asia | 2017 | 751 | 4 | 5′ UTR | Multiplex RT-PCR | |||||
| Kumthip et al. (36) | Cross-sectional | Thailand | Asia | 2017 | 73 | 1 | 5′ UTR | Nested RT-PCR | |||||
| Menage et al. (6) | Cross-sectional | Thailand | Asia | 2017 | 1,093 | 18 | 5′ UTR | Nested RT-PCR | 3 | 9 | 6 | ||
| Li et al. (37) | Case-control | China | Asia | 2017 | 461 | 7 | VP1 | Nested RT-PCR | 3 | 4 | |||
| Dapra et al. (38) | Cross-sectional | Italy | Europe | 2018 | 164 | 1 | NR* | ||||||
| Malasao et al. (39) | Cross-sectional | Thailand | Asia | 2019 | 2,002 | 30 | NR | ||||||
| Kim et al. (40) | Cross-sectional | South Korea | Asia | 2020 | 801 | 0 | Multiplex RT-PCR | ||||||
| Mohammad et al. (26) | Cross-sectional | Kuwait | Asia | 2020 | 84 | 1 | Metagenomics sequencing | ||||||
| Vandesande et al. (41) | Cohort | Sweden | Europe | 2021 | 209 | 11 | 5′ UTR | Semi-nested RT-PCR | 1 | ||||
| Yaghobi et al. (42) | Cross-sectional | Iran | Asia | 2020 | 160 | 26 | 5′ UTR | RT-PCR | |||||
| Taghinejad et al. (43) | Cross-sectional | Iran | Asia | 2020 | 160 | 11 | RT-PCR |
NR, Not reported.
Pooled Prevalence of Bufavirus in the Patients With Gastroenteritis
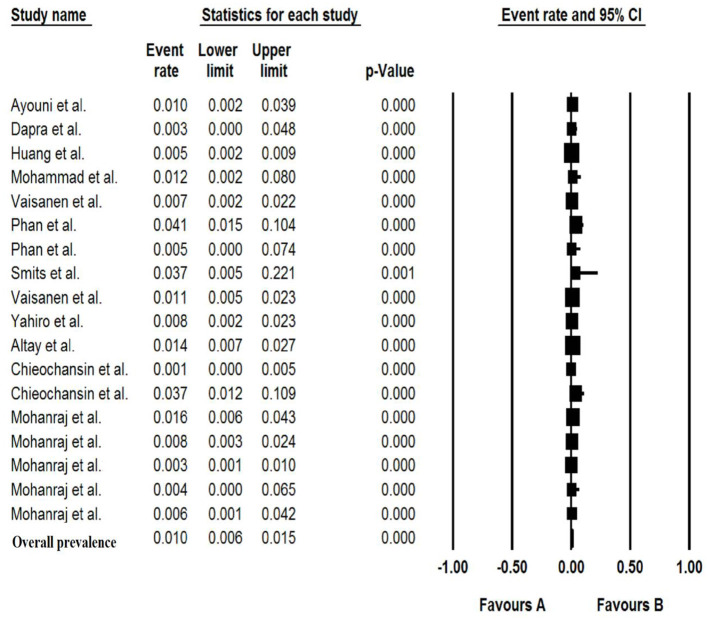
The total number of patients with GE included in this meta-analysis was 7,922 from children and adults based on 11 articles. The pooled prevalence of Bufavirus infection among patients with GE was 1.% (95% CI, 0.6–1.5%) based on a random-effects meta-analysis (Figure 2). In subgroup analysis by continent, the highest prevalence of Bufavirus was seen in Africa (1.4%, 95% CI, 0.5–4.1%) while the lowest prevalence was observed in Asia (0.7%, 95% CI, 0.2–2.1%) (Table 4). Highest prevalence of virus belongs to older than 5 years old subgroups (3.7%, 95% CI: 1.4–9.5%). As well, in three genotypes of BuV, BuV1 (1.%, 95% CI: 0.3–3.4%), and BuV2 (1.%, 95% CI: 0.1–6.9%) were of the same prevalence, while BuV3 (0.7%, 95% CI: 0.3–1.7%) was less prevalent.
Figure 2.
Forest plot of the pooled prevalence for BuV.
Table 4.
The Bufavirus prevalence based on subgroups and studies heterogeneity.
| Characteristics | Categories | Data sets |
Pooled prevalence (%) (95% CI) |
Heterogeneity | ||
|---|---|---|---|---|---|---|
| Q value | P-value | I2% | ||||
| Overall | – | 18 | 1.0 (0.6–1.5) | 35.005 | 0.006 | 51.435 |
| Continent | Africa | 4 | 1.4 (0.5–4.1) | 5.486 | 0.139 | 45.319 |
| Asia | 5 | 0.7 (0.2–2.1) | 15.201 | 0.004 | 73.685 | |
| Europe | 9 | 1.0 (0.7–1.4) | 9.203 | 0.325 | 13.071 | |
| Method | Nested RT-PCR | 5 | 1.1 (0.4–3.1) | 18.311 | 0.003 | 72.694 |
| Real-time RT-PCR | 5 | 0.8 (0.4–1.4) | 5.853 | 0.210 | 31.660 | |
| multiplex real-time qPCR | 5 | 0.7 (0.4–1.4) | 4.975 | 0.290 | 19.599 | |
| Genotype | BuV1 | 6 | 1.0 (0.3–3.4) | 27.351 | 0.000 | 81.719 |
| BuV2 | 1 | 1.0 (0.1–6.9) | 0.000 | 1.000 | 0.000 | |
| BuV3 | 4 | 0.7 (0.3–1.7) | 8.548 | 0.036 | 0.501 | |
| Co–infection | NoV | 6 | 0.3 (0.1–0.5) | 4.103 | 0.535 | 0.000 |
| HBoV | 2 | 0.3 (0.1–0.9) | 0.078 | 0.780 | 0.000 | |
| RoV | 2 | 0.6 (0.2–2.2) | 1.307 | 0.253 | 23.480 | |
| AdV | 1 | 1.0 (0.2–3.9) | 0.000 | 1000 | 0.000 | |
| Age | Under 5 | 5 | 1.4 (0.6–2.9) | 7.381 | 0.117 | 45.804 |
| Over 5 | 2 | 3.7 (1.4–9.5) | 0.000 | 1.000 | 0.000 | |
| Sex | Male | 4 | 0.9 (0.2–4.4) | 12.447 | 0.006 | 75.898 |
| Female | 4 | 0.6 (0.2–1.8) | 4.279 | 0.233 | 29.883 | |
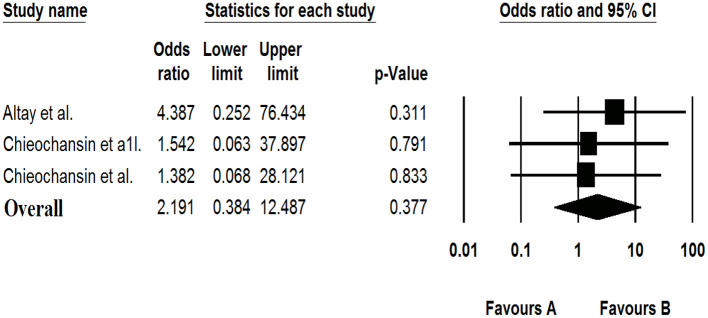
The Association of Bufavirus With Gastroenteritis
In three data sets, the meta-analysis showed that Bufavirus was not associated with GE [OR: 2.191 (95% CI; 0.384–12.487), I2: 0%] (Figure 3).
Figure 3.
Forest plot of odds ratios for the BuV based on case-control studies.
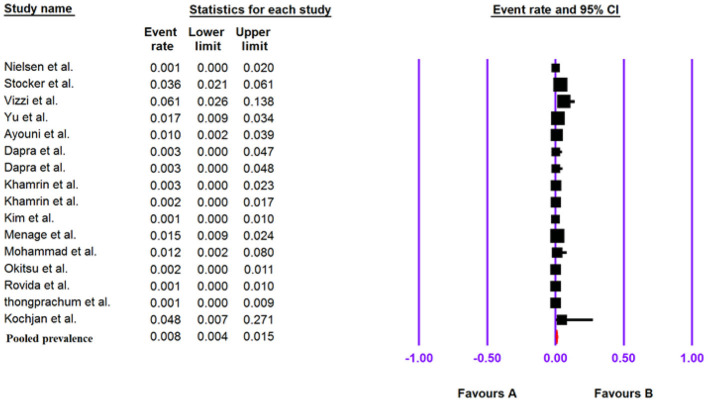
Pooled Prevalence of Saffold Virus in the Patients With Gastroenteritis
The results of analysis of Saffold virus based on random-effects meta-analysis are summarized in Table 4. Using random-effects meta-analysis, the pooled prevalence of Saffold virus in the studied patients was 1.9% (95% CI, 1.1–3.1%) (Figure 4). Among included studies, the maximum and minimum pooled prevalence of Saffold virus among patients with GE was found in Europe and Asia, respectively (2.9, 95% CI: 1.2–6.5% vs. 1.7, 95% CI: 0.9–3.1%) (Table 5). The highest prevalence of the virus was detected in children younger than 5 years of old (2.4%, 95% CI: 0.6–0.9). Among the eight genotypes of SAFV, SAFV-2 was the most prevalent genotype (1.%, 95% CI: 0.5–1.9%), and SAFV-4 was the least prevalent (0.2%, 95% CI: 0–1.2%) in patients with GE.
Figure 4.
Forest plot of the pooled prevalence for SAFV.
Table 5.
The Saffold virus prevalence based on subgroups and studies heterogeneity.
| Characteristics | Categories |
No. of Datasets |
Pooled prevalence (%) (95% CI) |
Heterogeneity | ||
|---|---|---|---|---|---|---|
| Q value | P-value | I2% | ||||
| Overall | – | 18 | 1.9 (1.1–3.1) | 174.465 | 0.000 | 90.256 |
| Continent | Asia | 15 | 1.7 (0.9–3.1) | 165.693 | 0.000 | 91.553 |
| Europe | 3 | 2.9 (1.2–6.5) | 5.965 | 0.051 | 66.471 | |
| Genotype | SAFV-1 | 5 | 0.9 (0.3–2.6) | 25.159 | 0.000 | 84.101 |
| SAFV-2 | 7 | 1.0 (0.5–1.9) | 23.800 | 0.001 | 74.790 | |
| SAFV-3 | 6 | 0.6 (0.2–1.5) | 23.853 | 0.000 | 79.038 | |
| SAFV-4 | 1 | 0.2 (0.0–1.2) | 0.000 | 1.000 | 0.000 | |
| SAFV-6 | 1 | 0.5 (0.2–1.2) | 0.000 | 1.000 | 0.000 | |
| Co-infection | NoV | 6 | 0.6 (0.3–1.0) | 8.635 | 0.125 | 42.097 |
| HBoV | 2 | 0.4 (0.1–1.5) | 1.457 | 0.227 | 31.352 | |
| RoV | 8 | 0.4 (0.2–0.9) | 19.395 | 0.007 | 63.909 | |
| AdV | 4 | 0.2 (0.1–0.5) | 2.624 | 0.453 | 0.000 | |
| Method | Multiplex RT-PCR | 2 | 0.3 (0.0–1.9) | 2.052 | 0.152 | 51.263 |
| Nested RT-PCR | 7 | 2.3 (1.5–3.5) | 14.417 | 0.025 | 58.383 | |
| RT-PCR | 2 | 10.9 (4.6–24.) | 6.505 | 0.011 | 84.627 | |
| Age | Under 5 | 8 | 1.6 (0.5–4.5) | 70.138 | 0.000 | 90.020 |
| Over 5 | 3 | 2.4 (0.6–0.9) | 4.183 | 0.124 | 52.184 | |
| Sex | Male | 2 | 0.3 (0.0–2.2) | 0.984 | 0.321 | 0.000 |
| Female | 2 | 0.9 (0.0–19.7) | 3.846 | 0.050 | 73.999 | |
The Association of Saffold Virus With Gastroenteritis
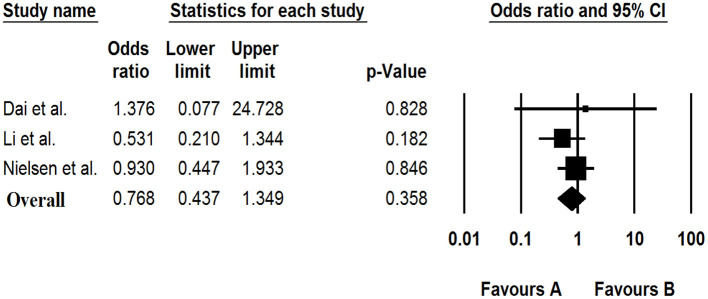
Based on the meta-analysis of three case-control studies, there was no significant association between the Saffold virus and GE [OR: 0.768 (95% CI: 0.437–1.349), I2: 0%] (Figure 5).
Figure 5.
Forest plot of odds ratios for the SAFV based on case-control studies.
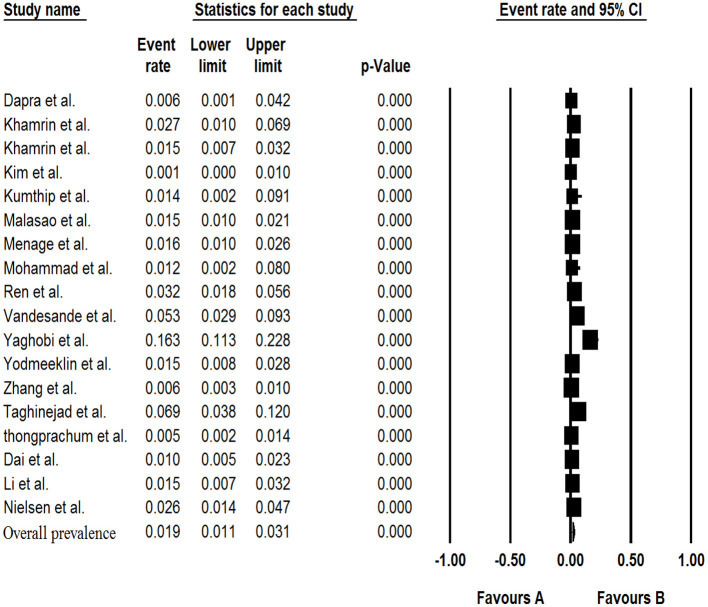
Pooled Prevalence of Human Cosavirus in the Patients With Gastroenteritis
The total number of patients with GE included in this meta-analysis was 6,595 based on 16 included articles. Based on a random-effect meta-analysis, the pooled prevalence of the human Cosavirus infection among patients with GEs was 0.8% (95% CI, 0.4–1.5%) (Figure 6). In subgroup analysis by continent, the highest prevalence of Cosavirus was seen in America (4.2%, 95% CI, 2.6–6.6%), whereas Europe (0.2%, 95% CI, 0.1–0.7%) observed the lowest prevalence (Table 6).
Figure 6.
Forest plot of the pooled prevalence for HCosV.
Table 6.
The Cosavirus prevalence based on subgroups and studies heterogeneity.
| Characteristics | Categories |
No. of Data sets |
Pooled prevalence (%) (95% CI) |
Heterogeneity | ||
|---|---|---|---|---|---|---|
| Q value | P-value | I2% | ||||
| Overall | – | 16 | 0.8 (0.4–1.5) | 28.29 | 0.000 | 92.932 |
| WHO regions | Africa | 1 | 1.0 (0.2–3.9) | 0.000 | 1.000 | 0.000 |
| America | 2 | 4.2 (2.6–6.6) | 1.022 | 0.312 | 2.185 | |
| Asia | 9 | 0.7 (0.3–1.4) | 21.240 | 0.007 | 62.335 | |
| Europe | 4 | 0.2 (0.1–0.7) | 0.377 | 0.945 | 0.000 | |
| Genotype | HCoSV-A | 3 | 0.5 (0.1–2.1) | 6.292 | 0.043 | 68.213 |
| HCoSV-C | 1 | 0.1 (0.0–0.6) | 0.000 | 1.000 | 0.000 | |
| HCoSV-D | 2 | 0.2 (0.0–0.7) | 0.837 | 0.360 | 0.000 | |
| Co-infection | NoV | 2 | 0.2 (0.0–1.1) | 1.420 | 0.233 | 29.561 |
| EV | 3 | 0.7 (0.1–3.3) | 5.932 | 0.052 | 66.286 | |
| RoV | 3 | 0.4 (0.2–0.8) | 1.384 | 0.500 | 0.000 | |
| AdV | 5 | 0.6 (0.1–2.1) | 9.329 | 0.053 | 57.122 | |
| Age | <5 | 10 | 0.5 (0.2–1.1) | 21.031 | 0.013 | 57.207 |
| <15 | 7 | 1.2 (0.5–2.9) | 18.564 | 0.005 | 67.680 | |
| >15 | 2 | 0.4 (0.1–1.8) | 0.319 | 0.517 | 0.000 | |
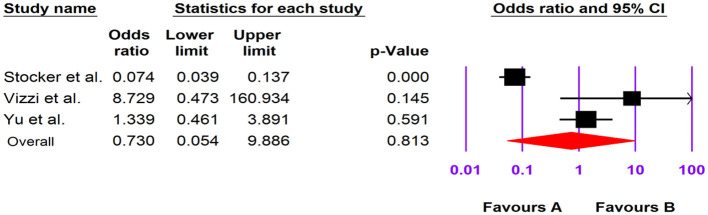
The Association of Human Cosavirus With Gastroenteritis
Of the four included case-control studies, one study could not be analyzed due to zero values for cases and controls (33), and, according to the three analyzed studies, human Cosavirus was not associated with GE [OR: 0.730 (95% CI; 0.054–9.886), I2: 0%] (Figures 3, 7).
Figure 7.
Forest plot of odds ratios for the HCosV based on case-control studies.
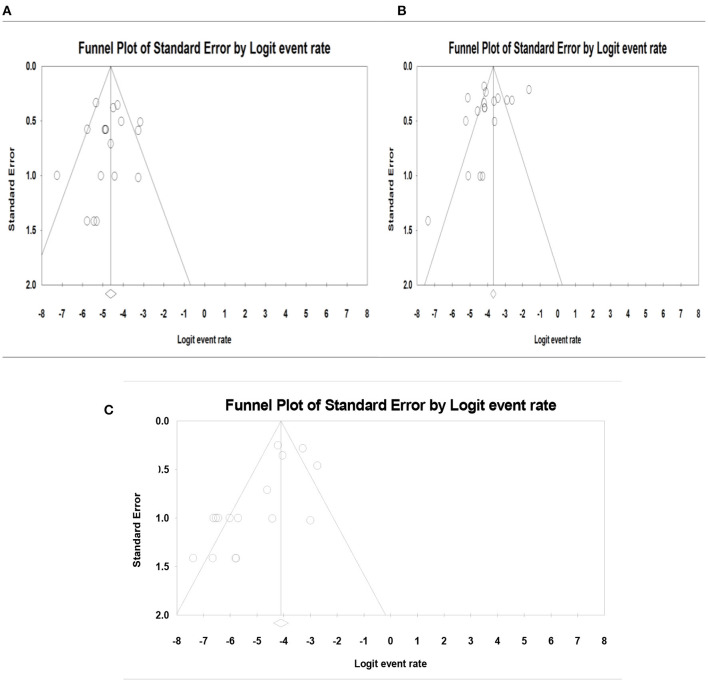
Publication Bias and Heterogeneity Assessment
The publication bias results were not significant for two viruses (SAFV and BuV) and significant for Cosavirus prevalence reports by applying Egger's regression test (P = 0.1912 for SAFV, P = 0.5667 for BuV, vs. P = 0.0031 for Cosavirus) (as shown in Figure 8). Also, the heterogeneity results of the studies according to the I2 statistics and Cochran's Q statistics were statistically significant for BuV (Q = 35.005, p < 0.006, I2 = 51.435%), SAFV (Q = 174.465, p < 0, I2 = 90.256%), and Cosavirus (Q = 28.29, P = 0, I2 = 92.932) (Tables 4–6).
Figure 8.
Funnel plot for publication bias assessment in BuV (A), SAFV (B), and CosV (C).
Discussion
Rapid progressions in sequencing technologies, bioinformatics, and metagenomic have led to the discovery of new viruses in recent years. However, while some studies stated the isolation of new viruses from fecal samples of patients with GE, there is still no solid evidence of the association of these viruses with GE (4, 52, 53). They are often neglected in epidemiological studies as they cause milder or asymptomatic infection, and researchers have a higher tendency to detect common enteric viruses and other infectious agents in patients with GE (54–56). In the present meta-analysis, we investigated the role of three emerging discovered viruses in the development of GE. Our results show no association between infection with Bufavirus (OR; 2.91, 95% CI: 0.384–12.487), Cosavirus (OR; 0.73, 95% CI: 0.054–9.886), and Saffold virus (OR; 0.77, 95% CI: 0.44–1.35) with GE. Also, a low prevalence of BuV (1.%, 95% CI: 0.6–1.5%), HCoSV (0.8%, 95% CI: 0.4–1.5%), and SAFV (1.9%, 95% CI: 1.1–3.1%) was observed. In general, the prevalence of SAFV was higher than BuV, and the least prevalence was observed in the case of HCoSV. The highest prevalence of BuV was in Africa (1.4%, 95% CI: 0.5–4.1%), where it was discovered (21), and the least prevalence was in Asia (0.7%, 95% CI: 0.2–2.1%). This might be due to poor hygiene and lack of access to safe water in African countries. Given the fact that these viruses were detected in environmental and sewage samples from various parts of the world (57–62), they possibly transmit through the oral-fecal route.
About the three genotypes of BuV, BuV1, and BuV2 were of the same prevalence, while BuV3 was less common in patients with GE; this lower prevalence of BuV3 might be due to the later discovery of this genotype in 2014 (23). SAFV consists of eight genotypes, of which five (SAFV1-4 and 6) were found in the included studies. SAFV-2 was the most prevalent genotype, and SAFV-4 was the least prevalent in patients with GE. It should be pointed out that, although SAFV genotypes 5, 7, and 8 were not detected in the included studies, Blinkova et al. isolated them along with other genotypes in children with non-polio acute flaccid paralysis (63). Also, some of the included studies did not investigate the genotypes of isolated SAFVs. Therefore, we cannot conclude that they are not present in fecal samples of patients with GE. The genotype A of HCoSV was more frequently (0.5%, 95% CI: 0.1–2.1%) isolated from patients with GE. Other founded genotypes were Genotype D (0.2%, 95% CI: 0–0.7%) and C (0.1%, 95% CI: 0–0.6%).
The presence of common enteric viruses, such as Rotavirus (RoV), human bocavirus (HBoV), Adenovirus (AdV), and Norovirus (NoV), was observed in patients that are BuV and SAFV infected. According to the Tables 4–6, co-infection with NoV was more common in patients infected with SAFV than BuV. There was a similar situation in the case of HBoV in which more prevalence of this virus was seen in SAFV than patients who are BuV infected. Contrastingly, RoV infection was more frequent in patients infected with BuV than SAFV. Similarly, AdV infection was more common in patients with BuV than SAFV infection. EVs have the highest proportion of co-infection with HCoSV followed by AdVs, RoVs, and NoVs. The high rate of co-infection with classic enteric viruses may indicate the role of these viruses in causing symptoms in patients infected with these newly discovered viruses (6, 46). The other possible point that is against the pathologic role of these viruses in the development of GE is the low viral load in patients with GE, which might be due to transient infection and the lack of replication in the gastrointestinal tract (44). Also, the high presence of these viruses in healthy individuals raises the likelihood that they are a part of the human virome (6).
Three studied viruses can infect people of all age groups (16, 41). Our analysis showed that BuV and SAFV are more common in individuals older than 5 years of age. In contrast, HCoSV was more common in the children younger than 15 years old. While GE is known as a prevalent disease in children younger than 5 years of age and common enteric viruses such as RoV and NoV are mostly found in this age group (64, 65), interestingly, our analysis showed that these viruses are more prevalent in older patients. These results might be due to reason that outdoor activities further expose people to viral agents (52).
BuV and SAFV are differently distributed among males and females, while BuV is more prevalent in males than females; SAFV is more common in females (42). However, these slight differences do not implicate that these viruses have a higher tendency to infect people of a specific gender.
All included studies had a molecularly based diagnosis with relatively close sensitivity and specificity. However, in the case of SAFV, RT-PCR had the highest detection, while nested-PCR showed the highest detection rate for BuV. It is noteworthy to mention that it requires more studies on the sensitivity and specificity of these methods to conclude which one is more suitable.
The present study faced some limitations. There were a few studies on adults, and details of participants (gender, clinical signs, and age groups) were insufficient in some studies. The genotypes of the viruses were not reported from some studies, and also some of research conducted without a healthy control group. The prevalence of these viruses had not been reported in many countries and geographical areas. In addition, some of the included studies did not evaluate the co-infection of the novel viruses with common enteric viruses. In addition, the language limitations of many studies and lack of association assessments of genotypes and clinical signs were the other main limitations of the present study. Hence, we suggest further studies, especially in case-control design, and more comprehensive studies from different geographical areas to overcome these limitations.
Conclusion
Progression in the development of molecular and metagenomics methods has facilitated discovering and studying emerging viruses. In the present meta-analysis, we investigated the prevalence and role of three recently discovered viruses in the development of GE. The pooled prevalence of three viruses was low, and neither was associated with GE. These results might be due to the few numbers of studies conducted. Therefore, we suggest more comprehensive studies with large cohorts of symptomatic and healthy patients in order to enhance our knowledge about these newly identified viruses. Also, we recommend in vitro studies to investigate the possible effects of these viruses on the gastrointestinal cell lines. In addition, the possible role of these emerging viruses in the etiology of other complications, such as respiratory symptoms, neurological diseases, and fever of an unknown origin, should not be neglected.
Data Availability Statement
The original contributions presented in the study are included in the article/supplementary materials, further inquiries can be directed to the corresponding author/s.
Author Contributions
MR and AK designed the study and collaborated in the manuscript writing. MR and MZ collaborated in the studies search, data extraction, and double checking. MZ helped in revision. All authors commented on the drafts of the manuscript and approved the final version of the article.
Conflict of Interest
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Publisher's Note
All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article, or claim that may be made by its manufacturer, is not guaranteed or endorsed by the publisher.
References
- 1.Eckardt AJ, Baumgart DC. Viral gastroenteritis in adults. Recent Patents Anti Infect Drug Discov. (2011) 6:54–63. 10.2174/157489111794407877 [DOI] [PubMed] [Google Scholar]
- 2.Kesheh MM, Khatami A, Saadati H, Jabbari M, Razizadeh MH, Fatemipour M, et al. Salivirus Infection: Systematic Review and Meta-Analysis of AssociationWith Gastrointestinal Symptoms in Children. New York, NY: Wiley Online Library. [DOI] [PubMed] [Google Scholar]
- 3.Arashkia A, Bahrami F, Farsi M, Nejati B, Jalilvand S, Nateghian A, et al. Molecular analysis of human adenoviruses in hospitalized children < 5 years old with acute gastroenteritis in Tehran, Iran. J Med Virol. (2019) 91:1930–6. 10.1002/jmv.25539 [DOI] [PubMed] [Google Scholar]
- 4.Smits SL, Osterhaus AD, Koopmans MP. Newly identified viruses in human gastroenteritis: pathogens or not? Pediatr Infect Dis J. (2016) 35:104–7. 10.1097/INF.0000000000000950 [DOI] [PubMed] [Google Scholar]
- 5.Daprà V, Galliano I, Montanari P, Zaniol E, Calvi C, Alliaudi C, et al. Bufavirus, Cosavirus, and Salivirus in diarrheal Italian infants. Intervirology. (2021) 64:165–8. 10.1159/000514384 [DOI] [PubMed] [Google Scholar]
- 6.Menage L, Yodmeeklin A, Khamrin P, Kumthip K, Maneekarn N. Prevalence of human cosavirus and saffold virus with an emergence of saffold virus genotype 6 in patients hospitalized with acute gastroenteritis in Chiang Mai, Thailand, 2014–2016. Infect Genet Evol. (2017) 53:1–6. 10.1016/j.meegid.2017.05.005 [DOI] [PubMed] [Google Scholar]
- 7.Ayouni S, Estienney M, Hammami S, Neji Guediche M, Pothier P, Aouni M, et al. Cosavirus, salivirus and bufavirus in diarrheal Tunisian infants. PLoS ONE. (2016) 11:e0162255. 10.1371/journal.pone.0162255 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Cotmore SF, Agbandje-McKenna M, Canuti M, Chiorini JA, Eis-Hubinger A-M, Hughes J, et al. ICTV virus taxonomy profile: parvoviridae. J Gen Virol. (2019) 100:367–8. 10.1099/jgv.0.001212 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Väisänen E, Kuisma I, Phan TG, Delwart E, Lappalainen M, Tarkka E, et al. Bufavirus in feces of patients with gastroenteritis, Finland. Emerg Infect Dis. (2014) 20:1077–9. 10.3201/eid2006.131674 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Christensen A, Kesti O, Elenius V, Eskola AL, Døllner H, Altunbulakli C, et al. Human bocaviruses and paediatric infections. Lancet Child AdolescHealth. (2019) 3:418–26. 10.1016/S2352-4642(19)30057-4 [DOI] [PubMed] [Google Scholar]
- 11.Väisänen E, Paloniemi M, Kuisma I, Lithovius V, Kumar A, Franssila R, et al. Epidemiology of two human protoparvoviruses, bufavirus and tusavirus. Sci Rep. (2016) 6:39267. 10.1038/srep39267 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Zell R, Delwart E, Gorbalenya AE, Hovi T, King AMQ, Knowles NJ, et al. ICTV virus taxonomy profile: picornaviridae. J Gen Virol. (2017) 98:2421–2. 10.1099/jgv.0.000911 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Zell R. Picornaviridae—the ever-growing virus family. Arch Virol. (2018) 163:299–317. 10.1007/s00705-017-3614-8 [DOI] [PubMed] [Google Scholar]
- 14.Tan SZK, Tan MZY, Prabakaran M. Saffold virus, an emerging human cardiovirus. Rev Med Virol. (2017) 27:e1908. 10.1002/rmv.1908 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Himeda T, Ohara Y. Saffold virus, a novel human cardiovirus with unknown pathogenicity. J Virol. (2012) 86:1292–6. 10.1128/JVI.06087-11 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Huang D-D, Wang W, Lu Q-B, Zhao J, Guo C-T, Wang H-Y, et al. Identification of bufavirus-1 and bufavirus-3 in feces of patients with acute diarrhea, China. Sci Rep. (2015) 5:1–4. 10.1038/srep13272 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Rezig D, Farhat EB, Touzi H, Meddeb Z, Salah AB, Triki H. Prevalence of human cosaviruses in Tunisia, North Africa. J Med Virol. (2015) 87:940–3. 10.1002/jmv.24076 [DOI] [PubMed] [Google Scholar]
- 18.Altay Koçak A, Öcal M, Polat M, Kanik Yüksek S, Aktaş Tapisiz A, Tezer H, et al. [Multicenter investigation of bufavirus in the etiology of viral central nervous system infections of adults and children]. Mikrobiyol Bul. (2017) 51:191–4. 10.5578/mb.54035 [DOI] [PubMed] [Google Scholar]
- 19.Naeem A, Hosomi T, Nishimura Y, Alam MM, Oka T, Zaidi SSZ, et al. Genetic diversity of circulating Saffold viruses in Pakistan and Afghanistan. J Gen Virol. (2014) 95:1945–57. 10.1099/vir.0.066498-0 [DOI] [PubMed] [Google Scholar]
- 20.Moher D, Liberati A, Tetzlaff J, Altman DG, Group P. Preferred reporting items for systematic reviews and meta-analyses: the PRISMA statement. PLoS Med. (2009) 6:e1000097. 10.1371/journal.pmed.1000097 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Phan TG, Vo NP, Bonkoungou IJ, Kapoor A, Barro N, O'Ryan M, et al. Acute diarrhea in West African children: diverse enteric viruses and a novel parvovirus genus. J Virol. (2012) 86:11024–30. 10.1128/JVI.01427-12 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Smits SL, Schapendonk CM, van Beek J, Vennema H, Schürch AC, Schipper D, et al. New viruses in idiopathic human diarrhea cases, the Netherlands. Emerg Infect Dis. (2014) 20:1218. 10.3201/eid2007.140190 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Yahiro T, Wangchuk S, Tshering K, Bandhari P, Zangmo S, Dorji T, et al. Novel human bufavirus genotype 3 in children with severe diarrhea, Bhutan. Emerg Infect Dis. (2014) 20:1037. 10.3201/eid2006.131430 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Altay A, Yahiro T, Bozdayi G, Matsumoto T, Sahin F, Ozkan S, et al. Bufavirus genotype 3 in Turkish children with severe diarrhoea. Clin Microbiol Infect. (2015) 21:965.e1-4. 10.1016/j.cmi.2015.06.006 [DOI] [PubMed] [Google Scholar]
- 25.Chieochansin T, Vutithanachot V, Theamboonlers A, Poovorawan Y. Bufavirus in fecal specimens of patients with and without diarrhea in Thailand. Arch Virol. (2015) 160:1781–4. 10.1007/s00705-015-2441-z [DOI] [PubMed] [Google Scholar]
- 26.Mohammad HA, Madi NM, Al-Nakib W. Analysis of viral diversity in stool samples from infants and children with acute gastroenteritis in Kuwait using Metagenomics approach. Virol J. (2020) 17:10. 10.1186/s12985-020-1287-5 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Mohanraj U, Jokinen M, Thapa RR, Paloniemi M, Vesikari T, Lappalainen M, et al. Human Protoparvovirus DNA and IgG in Children and Adults with and without Respiratory or Gastrointestinal Infections. Viruses. (2021) 13:483. 10.3390/v13030483 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Ren L, Gonzalez R, Xiao Y, Xu X, Chen L, Vernet G, et al. Saffold cardiovirus in children with acute gastroenteritis, Beijing, China. Emerg Infect Dis. (2009) 15:1509. 10.3201/eid1509.081531 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Khamrin P, Chaimongkol N, Nantachit N, Okitsu S, Ushijima H, Maneekarn N. Saffold cardioviruses in children with diarrhea, Thailand. Emerg Infect Dis. (2011) 17:1150. 10.3201/eid1706.101983 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Dai X, Yuan C, Yu Y, Zhao W, Yang Z. Molecular detection of Saffold virus in children in Shanghai, China. J Clin Virol. (2011) 50:186–7. 10.1016/j.jcv.2010.11.004 [DOI] [PubMed] [Google Scholar]
- 31.Zhang X-A, Lu Q-B, Wo Y, Zhao J, Huang D-D, Guo C-T, et al. Prevalence and genetic characteristics of Saffold cardiovirus in China from 2009 to 2012. Sci Rep. (2015) 5:1–7. 10.1038/srep07704 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Khamrin P, Thongprachum A, Kikuta H, Yamamoto A, Nishimura S, Sugita K, et al. Three clusters of Saffold viruses circulating in children with diarrhea in Japan. Infect Genet Evol. (2013) 13:339–43. 10.1016/j.meegid.2012.11.004 [DOI] [PubMed] [Google Scholar]
- 33.Nielsen ACY, Gyhrs ML, Nielsen LP, Pedersen C, Böttiger Gastroenteritis B, and the novel picornaviruses aichi virus, cosavirus, saffold virus, and salivirus in young children . J Clin Virol. (2013) 57:239–42. 10.1016/j.jcv.2013.03.015 [DOI] [PubMed] [Google Scholar]
- 34.Yodmeeklin A, Khamrin P, Chuchaona W, Saikruang W, Malasao R, Chaimongkol N, et al. Saffold viruses in pediatric patients with diarrhea in Thailand. J Med Virol. (2015) 87:702–7. 10.1002/jmv.24114 [DOI] [PubMed] [Google Scholar]
- 35.Thongprachum A, Khamrin P, Pham NTK, Takanashi S, Okitsu S, Shimizu H, et al. Multiplex RT-PCR for rapid detection of viruses commonly causing diarrhea in pediatric patients. J Med Virol. (2017) 89:818–24. 10.1002/jmv.24711 [DOI] [PubMed] [Google Scholar]
- 36.Kumthip K, Khamrin P, Ushijima H, Maneekarn N. Multiple enterovirus genotypes circulating in children hospitalized with acute gastroenteritis in Thailand. Infect Genet Evol. (2017) 55:324–31. 10.1016/j.meegid.2017.10.002 [DOI] [PubMed] [Google Scholar]
- 37.Li L-l, Liu N, Yu J-m, Ao Y-y, Li S, Stine OC, et al. Analysis of Aichi virus and Saffold virus association with pediatric acute gastroenteritis. J Clin Virol. (2017) 87:37–42. 10.1016/j.jcv.2016.12.003 [DOI] [PubMed] [Google Scholar]
- 38.Daprà V, Montanari P, Rassu M, Calvi C, Galliano I, Bergallo M. Prevalence of human cosavirus and saffold virus in young children with gastroenteritis, Northern Italy. Minerva Pediatr. (2018). 10.23736/s0026-4946.18.05219-2 [DOI] [PubMed] [Google Scholar]
- 39.Malasao R, Khamrin P, Kumthip K, Ushijima H, Maneekarn N. Molecular epidemiology and genetic diversity of human parechoviruses in children hospitalized with acute diarrhea in Thailand during 2011–2016. Arch Virol. (2019) 164:1743–52. 10.1007/s00705-019-04249-2 [DOI] [PubMed] [Google Scholar]
- 40.Kim GR, Kim SH, Jeon GW, Shin JH. Prevalence of eleven infectious viruses causing diarrhea in Korea. Jpn J Infect Dis. (2020) 73:427–30. 10.7883/yoken.JJID.2020.069 [DOI] [PubMed] [Google Scholar]
- 41.Vandesande H, Edman K, Rondahl E, Falkeborn T, Serrander L, Lindberg AM. Saffold virus infection in elderly people with acute gastroenteritis in Sweden. J Med Virol. (2021) 93:3980–4. 10.1002/jmv.26452 [DOI] [PubMed] [Google Scholar]
- 42.Yaghobi TS, Bahrami H, Harzandi N, Asadi A, Shareghi M, Firouzjani MH, et al. First molecular detection of saffold virus in children with acute gastroenteritis in iran. Novelty in Biomedicine. 9:11–6.31729639 [Google Scholar]
- 43.Taghinejad M, Ghaderi M, Mousavi-Nasab SD. First molecular detection of aichivirus in pediatric patients with acute gastroenteritis in Iran. Novelty Biomed. (2020) 8:20–5. 10.22037/nbm.v7i4.26729 [DOI] [PubMed] [Google Scholar]
- 44.Stöcker A, Souza BFdCD, Ribeiro TCM, Netto EM, Araujo LO, Corrêa JI, et al. Cosavirus infection in persons with and without gastroenteritis, Brazil. Emerg Infect Dis. (2012) 18:656. 10.3201/eid1804.111415 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Vizzi E, Fernández R, Angulo LA, Blanco R, Pérez C. Human cosavirus infection in HIV subjects with diarrhoea: persistent detection associated with fatal outcome. J Clin Virol. (2021) 139:104825. 10.1016/j.jcv.2021.104825 [DOI] [PubMed] [Google Scholar]
- 46.Yu J-M, Ao Y-Y, Li L-L, Duan Z-J. Identification of a novel cosavirus species in faeces of children and its relationship with acute gastroenteritis in China. Clin Microbiol Infect. (2017) 23:550–4. 10.1016/j.cmi.2017.02.018 [DOI] [PubMed] [Google Scholar]
- 47.Khamrin P, Chaimongkol N, Malasao R, Suantai B, Saikhruang W, Kongsricharoern T, et al. Detection and molecular characterization of cosavirus in adults with diarrhea, Thailand. Virus Genes. (2012) 44:244–6. 10.1007/s11262-011-0700-y [DOI] [PubMed] [Google Scholar]
- 48.Khamrin P, Maneekarn N. Detection and genetic characterization of cosavirus in a pediatric patient with diarrhea. Arch Virol. (2014) 159:2485–9. 10.1007/s00705-014-2091-6 [DOI] [PubMed] [Google Scholar]
- 49.Okitsu S, Khamrin P, Thongprachum A, Nishimura S, Kalesaran AF, Takanashi S, et al. Detection and molecular characterization of human cosavirus in a pediatric patient with acute gastroenteritis, Japan. Infect Genet Evol. (2014) 28:125–9. 10.1016/j.meegid.2014.09.019 [DOI] [PubMed] [Google Scholar]
- 50.Rovida F, Campanini G, Piralla A, Adzasehoun KMG, Sarasini A, Baldanti F. Molecular detection of gastrointestinal viral infections in hospitalized patients. Diagnostic microbiology and infectious disease. (2013) 77:231–5. 10.1016/j.diagmicrobio.2013.07.020 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Kochjan P, Khamrin P, Kumthip K, Maneekarn N. Detection and Characterization of Human Bocavirus in Pediatric Patients with Acute Gastroenteritis in Chiang Mai, Thailand. [DOI] [PubMed] [Google Scholar]
- 52.Razizadeh MH, Khatami A, Zarei M. Global molecular prevalence and genotype distribution of Sapovirus in children with gastrointestinal complications: a systematic review and meta-analysis. Rev Med Virol. (2021) e2302. 10.1002/rmv.2302 [DOI] [PubMed] [Google Scholar]
- 53.Kesheh MM, Khatami A, Saadati H, Jabbari M, Razizadeh MH, Fatemipour M, et al. Salivirus infection: systematic review and meta-analysis of association with gastrointestinal symptoms in children. Rev Med Virol. (2021) e2238. 10.1002/rmv.2238 [DOI] [PubMed] [Google Scholar]
- 54.Taghipour A, Bahadory S, Badri M, Yadegar A, Mirsamadi ES, Mirjalali H, et al. A systematic review and meta-analysis on the co-infection of Helicobacter pylori with intestinal parasites: public health issue or neglected correlation? Int J Environ Health Res. (2020) 1–11. 10.1080/09603123.2020.1798890 [DOI] [PubMed] [Google Scholar]
- 55.Taghipour A, Olfatifar M, Foroutan M, Bahadory S, Malih N, Norouzi M. Global prevalence of Cryptosporidium infection in rodents: a systematic review and meta-analysis. Prev Vet Medi. (2020) 105119. 10.1016/j.prevetmed.2020.105119 [DOI] [PubMed] [Google Scholar]
- 56.Khatami A, Bahadory S, Ghorbani S, Saadati H, Zarei M, Soleimani A, et al. Two rivals or colleagues in the liver? Hepatit B virus and Schistosoma mansoni co-infections: a systematic review and meta-analysis. Microbial Pathogen. (2021) 104828. 10.1016/j.micpath.2021.104828 [DOI] [PubMed] [Google Scholar]
- 57.López GR, Martinez LM, Freyre L, Freire MC, Vladimirsky S, Rabossi A, et al. Persistent detection of Cosavirus and Saffold cardiovirus in Riachuelo River, Argentina. Food Environ Virol. (2021) 13:64–73. 10.1007/s12560-020-09451-z [DOI] [PubMed] [Google Scholar]
- 58.Guerrero-Latorre L, Romero B, Bonifaz E, Timoneda N, Rusiñol M, Girones R, et al. Quito's virome: metagenomic analysis of viral diversity in urban streams of Ecuador's capital city. Sci Total Environ. (2018) 645:1334–43. 10.1016/j.scitotenv.2018.07.213 [DOI] [PubMed] [Google Scholar]
- 59.Nantachit N, Khamrin P, Kumthip K, Malasao R, Maneekarn N. Molecular surveillance and genetic analyses of bufavirus in environmental water in Thailand. Infect Genet Evol. (2019) 75:104013. 10.1016/j.meegid.2019.104013 [DOI] [PubMed] [Google Scholar]
- 60.Thongprachum A, Fujimoto T, Takanashi S, Saito H, Okitsu S, Shimizu H, et al. Detection of nineteen enteric viruses in raw sewage in Japan. Infect Genet Evol. (2018) 63:17–23. 10.1016/j.meegid.2018.05.006 [DOI] [PubMed] [Google Scholar]
- 61.Bonanno Ferraro G, Mancini P, Veneri C, Iaconelli M, Suffredini E, Brandtner D, et al. Evidence of Saffold virus circulation in Italy provided through environmental surveillance. Lett Appl Microbiol. (2020) 70:102–8. 10.1111/lam.13249 [DOI] [PubMed] [Google Scholar]
- 62.Aminipour M, Ghaderi M, Harzandi N. First occurrence of Saffold virus in sewage and river water samples in Karaj, Iran. Food Environ Virol. (2020) 12:75–80. 10.1007/s12560-019-09415-y [DOI] [PubMed] [Google Scholar]
- 63.Blinkova O, Kapoor A, Victoria J, Jones M, Wolfe N, Naeem A, et al. cardioviruses are genetically diverse and cause common enteric infections in South Asian children. J Virol. (2009) 83:4631–41. 10.1128/JVI.02085-08 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Bányai K, Estes MK, Martella V, Parashar UD. Viral gastroenteritis. Lancet. (2018) 392:175–86. 10.1016/S0140-6736(18)31128-0 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Nguyen GT, Phan K, Teng I, Pu J, Watanabe T. A systematic review and meta-analysis of the prevalence of norovirus in cases of gastroenteritis in developing countries. Medicine. (2017) 96:e8139. 10.1097/MD.0000000000008139 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Ghaderi M. First molecular detection of Saffold virus in children with acute gastroenteritis in Iran. Novelty Biomed. (2021) 9:11–6. 10.22037/nbm.vi.3166231729639 [DOI] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Data Availability Statement
The original contributions presented in the study are included in the article/supplementary materials, further inquiries can be directed to the corresponding author/s.