Abstract
Motivation
N 6-methyladenosine (m6A) is the most prevalent modification in eukaryotic messenger RNAs. MicroRNAs (miRNAs) are abundant post-transcriptional regulators of gene expression. Correlation between m6A and miRNA-targeting sites has been reported to suggest possible involvement of m6A in miRNA-mediated gene regulation. However, it is unknown what the regulatory effects might be. In this study, we performed comprehensive analyses of high-throughput data on m6A and miRNA target binding and regulation.
Results
We found that the level of miRNA-mediated target suppression is significantly enhanced when m6A is present on target mRNAs. The evolutionary conservation for miRNA-binding sites with m6A modification is significantly higher than that for miRNA-binding sites without modification. These findings suggest functional significance of m6A modification in post-transcriptional gene regulation by miRNAs. We also found that methylated targets have more stable structure than non-methylated targets, as indicated by significantly higher GC content. Furthermore, miRNA-binding sites that can be potentially methylated are significantly less accessible without methylation than those that do not possess potential methylation sites. Since either RNA-binding proteins or m6A modification by itself can destabilize RNA structure, we propose a model in which m6A alters local target secondary structure to increase accessibility for efficient binding by Argonaute proteins, leading to enhanced miRNA-mediated regulation.
Availability and implementation
N/A.
1 Introduction
Methylation of the N6 position of adenosine (m6A) is the most prevalent modification in eukaryotic mRNAs (Liu and Pan, 2016; Meyer and Jaffrey, 2017; Roundtree et al., 2017). m6A methylation is mediated by methyltransferases (m6A writers) that include methyltransferase-like 3 (METTL3) and METTL14. The modification can be erased by RNA demethylases (m6A erasers), such as ALKBH5 (Zheng et al., 2013). Recognition of m6A by m6A-binding proteins (m6A readers) is a major mechanism for effects of the modification (Meyer and Jaffrey, 2017). Transcriptome-wide single-nucleotide resolution mapping has revealed that m6A sites are enriched in the 3′-untranslated regions (3′-UTRs) and near stop codon, and that there is an association or correlation between m6A and predicted microRNA (miRNAs)-binding sites on mRNAs (Das Mandal and Ray, 2021; Ke et al., 2015; Meyer et al., 2012). m6A modification plays important regulatory roles in a variety of fundamental cellular processes. Examples are regulation of pre-mRNA splicing (Louloupi et al., 2018), control of cell fate transition (Batista et al., 2014), regulation of mRNA stability (Wang et al., 2014), processing of primary miRNAs (Alarcon et al., 2015) and cell reprogramming (Chen et al., 2015). Emerging evidence also suggests an association between m6A modification and cancer progression (Chen et al., 2019). N6-adenosine methylation in some miRNAs has been observed (Berulava et al., 2015; Ke et al., 2015). It was recently reported that miRNA-mediated loss of m6A increases nascent translation in glioblastoma (Zepecki et al., 2021).
miRNAs are an abundant class of small non-coding RNAs of about ∼22 nt that have been found in plants, animals and some viruses (Ambros, 2004; Bartel, 2004) A mature miRNA contained in an RNA-induced silencing complex hybridizes to partially complementary sequences typically in the 3'-UTRs of the target mRNAs, leading to translational repression and/or mRNA degradation of the target mRNA. miRNAs play important roles in development, differentiation, apoptosis and proliferation (Bartel, 2004; Harfe, 2005). Moreover, mis-regulation in miRNA activity has been found to be associated with cancer and other human diseases (Erson and Petty, 2008; Esau and Monia, 2007).
Computational identification of target-binding sites has been primarily based on the seed, a key sequence feature for miRNA targeting (Lewis et al., 2005). In addition, the importance of target structural accessibility for miRNA targeting has been established (Long et al., 2007). With the development of the cross-linking immunoprecipitation (CLIP) technique (Chi et al., 2009; Hafner et al., 2010), it has become possible to identify short Argonaute (AGO)-crosslinked sequences containing miRNA-binding sites. By utilizing such data and a comprehensive list of sequence, thermodynamic and target structure features, models and software were developed for statistical prediction of both seed and seedless-binding sites (Liu et al., 2013; Rennie et al., 2014). Through the addition of a ligation step in the CLIP framework, the CLASH method allows direct observation of miRNA: target interactions (Helwak et al., 2013). Therefore, the CLASH study presented a high-quality dataset of high-confidence miRNA-binding sites for analysis.
Since m6A methylation is enriched in the 3'-UTRs, the primary target regions of miRNAs, the question arises whether there is any regulatory impact of m6A modification on post-transcriptional regulation by miRNAs beyond the reported correlation between the methylation and miRNA-targeting sites (Das Mandal and Ray, 2021). To investigate this, we utilized multiple high-throughput data on m6A in human transcriptome (Sun et al., 2016), and miRNA target binding and regulation (Baek et al., 2008; Helwak et al., 2013). We also performed conservation analysis as well as structural accessibility analysis for interpretation of our findings.
2 Methods
2.1 m6A modification data
We downloaded the data of m6A modification sites in mRNAs from the RMBase database (Sun et al., 2016). A total of 140 574 single-nucleotide modification sites for human mRNAs and 84 539 for mouse mRNAs were available from the database and were all included in our analyses. The m6A modifications in RMBase database were all assembled from various studies involving high-throughput m6A-seq experiments. This method integrates immunoprecipitation of methylated, randomly fragmented RNA using a highly specific anti-m6A antibody to obtain an enriched population of modified fragments and massively parallel sequencing, resulting in mapping of this modification throughout the transcriptome.
2.2 Classification of miRNA targets and binding sites
In this study, a transcript is termed as m6A+ if it possesses at least one m6A modification site, and m6A− otherwise. Similarly, a miRNA-binding site is m6A+ if it overlaps with at least one m6A methylation site, and m6A− otherwise.
2.3 miRNA targeting data
For regulatory effects of mRNA targeting, we used high-throughput mRNA expression data from overexpression of human miR-1, miR-181 and miR-124, and knockout of mouse miR-223 (Baek et al., 2008). Our dataset selection criteria were: (i) data reported the gene expression consequences after perturbing the expression of a single miRNA (overexpression or KO) in either human or mouse cells; (ii) microarray or RNAseq studies were performed before and after miRNA manipulation to allow for estimates of mRNA fold changes; and (iii) accurate assembly of seed targets among all mRNAs in the experimental system was provided in the publication. For human miRNA overexpression, there were 19 864 expression measurements from microarray. For miR-223 knockout, there were 20 334 expression measurements. Protein expression data were also available from this study. However, the size of this protein dataset is only about 15–20% of the mRNA dataset. For example, for miR-124, only 13 m6A− transcripts with either an 8mer or a 7mer seed site have ≥6 independent protein measurements, a consideration in the previous data analysis (Baek et al., 2008). For our analyses, the proteomic data were too limited in size to be included. On the other hand, it has been shown that, to a great extent, changes in mRNA levels reflect the impact of mammalian miRNAs on gene expression (Guo et al., 2010). For these reasons, our study focused on the analyses of the large mRNA data.
The second dataset for miRNA binding was based on the CLASH method to reveal high-confidence miRNA: target interactions (Helwak et al., 2013), which allows for accurate identification of miRNA-binding sites on target mRNAs, albeit with unknown regulatory impact. This study identified ∼18 500 miRNA: target interactions for 7390 transcripts and 399 miRNAs. A majority of the interactions did not have canonical seed base pairing (i.e. binding sites were seedless).
2.4 Identification of miRNA-binding sites
For CLASH data, the STarMir program (Rennie et al., 2014) was used to identify both seed and seedless binding sites in the target regions within the CLASH chimeras. STarMir incorporates the RNAhybrid program in model-based predictions (Liu et al., 2013; Rehmsmeier et al., 2004). For mRNAs with expression measurements from overexpression of human miR-1, miR-181 and miR-124, and knockout of mouse miR-223, the STarMir program was used for prediction of miRNA-binding sites.
2.5 Computation of site conservation, structural accessibility and statistical significance
For each miRNA-binding site, we computed a site conservation score as the average of individual nucleotide conservation scores available from the UCSC genome browser (Kent et al., 2002; Siepel et al., 2005). For analysis on target site accessibility, we computed ΔGtotal, a miRNA-target-hybridization-model-based quantitative measure of local structural accessibility (Long et al., 2007). Because the calculation was based on RNA thermodynamics for unmodified nucleotides (Mathews et al., 1999), the analysis only revealed the degree of accessibility before any potential effects by m6A methylation. To assess the statistical significance of pairwise distributional comparisons, the P-values from Kolmogorov–Smirnov tests were reported.
3 Results
3.1 Higher levels of regulation for targets with m6A methylation
A previous study (Baek et al., 2008) presented data of mRNA fold changes (in log2 scale) for target mRNAs with an 8mer or a 7mer (A1 or m8) binding site, showing that such sites are more effective for miRNA targeting. Thus, to examine whether m6A methylation on mRNAs has any effects on regulation by miRNAs, we focused our analysis on these targets. Among 2879 targets for human miR-1, 1519 were m6A+ and 1360 were m6A−. For targets of human miR-124, 1841 were m6A+ and 1361 were m6A−. For human miR-181, 1920 targets were m6A+ and 1597 were m6A−. For targets of mouse miR-223, 1276 were m6A+ and 922 were m6A−. For miR-1 overexpression, the levels of down-regulation for m6A+ targets were significantly higher than m6A− targets (Fig. 1A; P-value of 5.70e-06). Similarly, for overexpression of miR-124 (Fig. 1B; P-value of 3.20e-08) or miR-181 (Fig. 1C; P-value of 1.23e-07), m6A+ targets were down-regulated to a greater extent than m6A− targets. For miR-223 knockout, for up-regulated targets (i.e. log2-fold change >0), the levels of up-regulation for m6A+ targets were significantly higher than m6A− targets (Fig. 1D; P-value of 0.002). All together, these results indicate that m6A modification on target transcripts significantly enhances effects of regulation by miRNAs.
Fig. 1.
Impact of m6A methylation on miRNA regulation. Comparison of cumulative distributions of mRNA fold changes (in log2 scale) for m6A+ targets (red), m6A− targets (green) and ‘Others’ (blue), in response to (A) miR-1 overexpression (number of targets is 2879), with P-values of 5.70e-06 for red versus green, <2.2e-16 for red versus blue and 0.011 for green versus blue; (B) miR-124 overexpression (number of targets is 3202), with P-values of 3.20e-08 for red versus green, <2.2e-16 for red versus blue and 4.71e-09 for green versus blue; (C) miR-181 overexpression (number of targets is 3517), with P-values of 1.23e-07 for red versus green, <2.2e-16 for red versus blue and 2.78e-06 for green versus blue; and (D) miR-223 knockdown (number of targets is 2268), with P-values of 0.002 for red versus green (among the up-regulated), 2.687e-14 for red versus blue and 6.22e-06 for green versus blue
Also plotted in Figure 1 is the empirical cumulative distribution for ‘Others’. This group was formed from the whole set of mRNAs by removing 7-mer, 8-mer targets and 6mer targets with a probability of 0.5 or higher as predicted by STarMir. The probability is a model-based measure of confidence that a predicted site is bound by AGO (Liu et al., 2013). Thus, the group does not include higher confidence seed targets; however, it could contain other targets harboring only seedless sites. The levels of regulation for this group were significantly less than either the m6A+ targets (P-value <2.2e-16 for miR-1, miR-124, miR-181 and 2.687e-14 for miR-223), or the m6A− targets (P-value of 0.011, 4.71e-09, 2.78e-06 and 6.22e-06, for miR-1, miR-124, miR-181 and miR-223, respectively).
3.2 Higher conservation for m6A+ miRNA sites
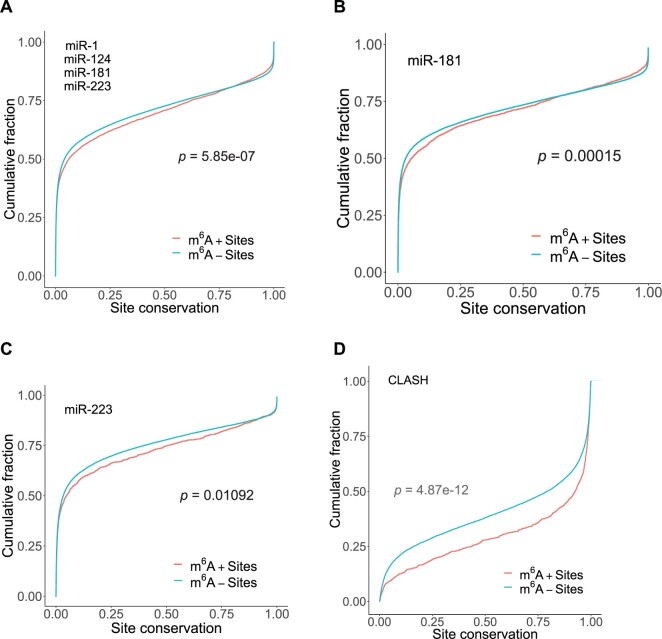
We investigated whether the presence of m6A has any effects on the conservation of miRNA-binding sites. For miR-1, miR-124, miR-181 and miR-223, we predicted miRNA-binding sites using the STarMir program (Rennie et al., 2014) to identify the nucleotide positions of both seed and seedless sites in all mRNAs. Position information for seed sites was not provided in the previous study (Baek et al., 2008). Pooling the four miRNAs to achieve greater statistical power, we found that the m6A+ binding sites have significantly higher conservation than m6A− binding sites (Fig. 2A, P-value of 5.85e-07). For the individual miRNAs, significance was observed for miR-1 (P-value of 0.005688), miR-181 (P-value of 0.0001492, Fig. 2B) and miR-223 (P-value of 0.01092, Fig. 2C). There was a higher conservation for miR-124, but it was insignificant at 0.05 level (P-value of 0.1387).
Fig. 2.
m6A+ miRNA sites are more conserved. Comparison of cumulative distributions of conservation scores between m6A+ binding site (red) and m6A− binding sites (blue), for (A) pooled miRNA-binding sites for miR-1 (928 m6A+ sites, 28 876 m6A− sites), miR-124 (1715 m6A+ sites, 51 467 m6A− sites), miR-181 (2460 m6A+ sites, 52 900 m6A− sites) and miR-223 (1283 m6A+ sites, 48 989 m6A− sites), with a P-value of 5.85e-07; (B) miR-181, with a P-value of 0.0001492; (C) miR-223, with a P-value of 0.01092; and (D) miRNA sites identified from CLASH (number of miRNAs is 399; total number of sites is 18 500), with a P-value of 4.87e-12
For CLASH data on 399 miRNAs and 7390 transcripts, the conservation for m6A+ miRNA-binding sites was also significantly higher than for m6A− miRNA-binding sites (Fig. 2D; P-value of 4.87e-12). Higher conservation suggests functional relevance of m6A in gene regulation by miRNAs.
3.3 Analyses of potential confounding factors
3.3.1. Distance from m6A site to miRNA-binding site
To investigate whether the distance from the site of m6A modification to the site of miRNA binding has any effect on miRNA-mediated regulation, we divided m6A+ targets into two subsets: one with distances under 100 nt (‘shorter distance subset’), and the other with distances of 100 nt or greater (‘longer distance subset’). In cases having multiple m6A sites on the same target 3′-UTR, the one with the shortest distance was used. The nucleotide positions of the 7mer or 8mer binding sites were identified using the RNAhybrid program (Rehmsmeier et al., 2004). In cases with both a 7mer and an 8mer site present on the same 3′-UTR, the 8mer site was used due to its generally stronger regulation (Bartel, 2004); in cases of multiple 7mer sites or multiple 8mer sites, the one with the stronger hybridization energy was used to represent the corresponding site type.
For miR-1 and miR-124, the shorter distance target subset had a significantly higher level of regulation than the longer distance target subset (P-value of 0.045 for miR-1 and 0.049 for miR-124). However, this effect was not observed for miR-181 (P-value of 0.138) or miR-223 (P-value of 0.664). These observations suggest that for some miRNAs, a shorter distance from the m6A site to the miRNA-binding site exerts a greater level of regulatory enhancement.
3.3.2. Target 3′-UTR length
We compared the lengths of 3′-UTRs between m6A+ and m6A− targets. For all four miRNAs, we found that m6A+ targets had significantly longer 3′-UTRs as compared to m6A− targets (P-values are 8.128e-10 for miR-1, 5.917e-13 for miR-124 and <2.2e-16 for miR-181 and miR-223).
To control for 3′-UTR length in a reanalysis, we set up length bin of 500 nt, i.e. (0 nt, 500 nt), (500 nt, 1000 nt)… (4500 nt, 5000 nt) and (5000 nt, +∞). For each bin, with a preset common sampling size, we randomly sampled UTRs for m6A+ targets and m6A− targets. For example, in the human data, for bin (500 nt, 1000 nt), there were 5505 m6A+ targets and 3232 m6A− targets. We set the sampling size at 3000. Because each bin was equally represented by m6A+ targets and m6A− targets, the cumulative distributions of the 3′-UTR length for the sampled m6A+ targets and the sampled m6A− targets were nearly identical, with insignificant P-values (data not shown). For the sampled targets, we repeated the previous analysis. We found that, for miR-181 and miR-124, the level of regulation was higher for the sampled m6A+ targets as compared to the sampled m6A− targets (P-value of 3.169e-06 for miR-181 and 4.203e-07 for miR-124). For miR-1 and miR-223, however, there was no significant enhancement by m6A (P-value of 0.539 for miR-1 and 0.3548 for miR-223). These findings suggest that, for some miRNAs, longer 3′-UTR has a positive effect on the enhancement of regulation.
3.3.3. miRNA-binding location within 3′-UTR
We next examined whether there was a difference in the location of miRNA-binding sites between the m6A+ targets and the m6A− targets. For the kth nucleotide in a 3′-UTR of n nucleotides, the relative location within the 3′-UTR was (k/n) ×100%. For miR-1 and miR-124, miRNA-binding sites for the m6A+ targets were located significantly farther away from the start of 3′-UTR, in comparison to the m6A− targets (P-value of 0.003285 for miR-1 and 0.02981 for miR-124). For miR-181 and miR-223, the difference in location was not significant (P-value of 0.6407 for miR-181 and 0.88 for miR-223).
3.3.4. Target basal expression
Using expression data for wild-type cells (i.e. control sample for mRNA overexpression or KO), we found there was no significant difference in basal expression levels between m6A+ targets and m6A− targets for all three human datasets (P-value of 0.5618 for miR-1 control sample; 0.8651 for miR-124 control sample; and 0.06 for miR-181 control sample). For the mouse miR-223 control sample, however, there was a significantly higher basal expression for the m6A+ targets as compared to the m6A− targets (P-value of 8.164e-10).
3.3.5. Proportion of 7mer or 8mer binding sites
For each miRNA, we compared the proportion of 7mer or 8mer binding sites between the m6A+ targets and the m6A− targets. We did not observe an appreciable difference (data not shown).
3.3.6. Proximal m6A sites within 200 nt of miRNA-binding sites
In this study, a m6A+ site required an overlap between the m6A site and a miRNA-binding site. However, it is of interest to consider proximal m6A sites that are outside the miRNA-binding site. It has been reported that 60% of m6A sites are within 200 nt of miRNA-binding sites (Ke et al., 2015). For the m6A+ targets used in the analysis for Figure 1, we removed those with m6A sites overlapping with miRNA-binding sites as well as those with m6A sites located outside the 200 nt window. The remaining targets were compared with the m6A− targets. We found that there was significant regulatory enhancement for miR-1 and miR-124 (P-value of 0.0001369 and 0.001936, respectively). This suggests that in some cases, proximal m6A sites can have a positive effect on gene regulation. The effect may depend on other factors, such as structural accessibility.
In summary, of the factors examined above, we could not identify one that could potentially explain the findings in Figure 1 for all four miRNAs. A remaining factor to be examined was the GC content, which is reported in the next subsection, as it has implications for RNA structural stability.
3.4 Higher GC content for methylated target and lower structural accessibility for m6A+ binding sites
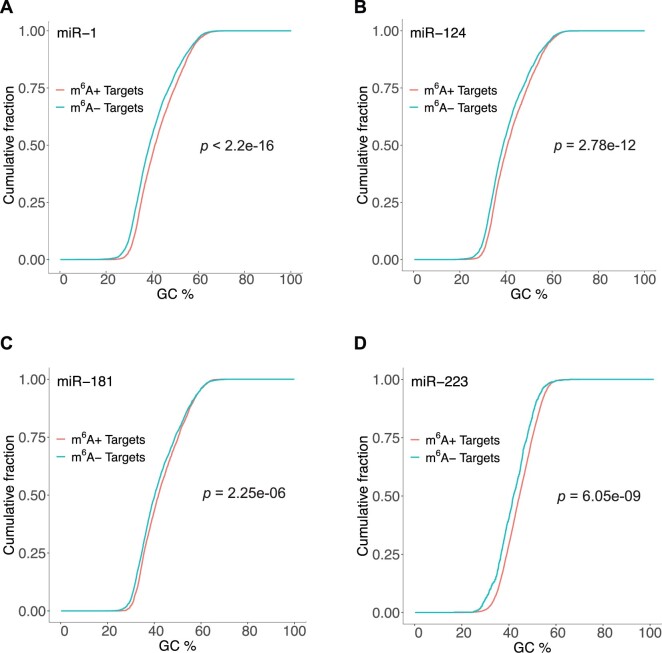
We compared the GC content of the 3′-UTR between the m6A+ targets and the m6A− targets. We found that for each of the four miRNAs, the GC content for the m6A+ targets was significantly higher than that for the m6A− targets (miR-1: Fig. 3A, P-value <2.2e-16; miR-124: Fig. 3B, P-value of 2.78e-12; miR-181: Fig. 3C, P-value of 2.25e-06; miR-223: Fig. 3D, P-vale of 6.05e-09). Because GC pairing is thermodynamically more stable than AU pairing, the finding indicates that m6A+ targets are structurally more stable than m6A− targets.
Fig. 3.
Methylated target 3′-UTRs have higher GC content. Comparison of cumulative distributions of GC% between m6A+ targets (red) and m6A− targets (green). A significant higher GC% is observed for (A) miR-1 targets with a P-value <2.2e-16; (B) miR-124 targets with a P-value of 2.78e-12; (C) miR-181 targets with a P-value of 2.25e-06; and (D) miR-223 targets with a P-value of 6.05e-09
To further investigate potential interplay between m6A and the target secondary structure, we performed a target accessibility analysis for all predicted binding sites for miR-1, miR-124, miR-181 and miR-223. We computed ΔGtotal, the total energy change of miRNA: target hybridization, which is an energetic measure of target structural accessibility (Long et al., 2007). For the binding sites pooled together for all four miRNAs, ΔGtotal was significantly higher for m6A+ sites than m6A− binding sites (Fig. 4A; P-value of 4.55e-15). For each individual miRNA, a higher ΔGtotal was observed for m6A+ sites in comparison to m6A− sites. Two examples are shown for miR-1 (Fig. 4B, P-value of 7.78e-07) and miR-124 (Fig. 4C; P-value of 2.2e-16). A higher ΔGtotal indicates lower accessibility. For target structure predictions and free energy calculation, the RNA thermodynamics in the computation are for unmodified nucleotides. Thus, the observation on accessibility is for unmodified targets. The finding indicates that a miRNA-binding site that can be m6A modified is structurally less accessible in the absence of methylation. We did not observe such an effect with statistical significance for the CLASH data. This could be due to differences in cellular environments for different experimental systems. For RNA/RNA interaction, e.g. when both molecules are present at high concentrations, the equilibrium would favor hybridization, regardless of local structural accessibility. In contrast, difference in conservation is an evolutionary signal that does not depend on experimental conditions.
Fig. 4.
MiRNA-binding sites that can be potentially m6A modified are structurally less accessible based on free energies of unmodified nucleotides. Comparison of cumulative distributions of ΔGtotal for m6A+ binding sites (red) and m6A− binding sites (blue), for (A) miR-1, miR-124, miR-181 and miR-223 pooled together, with a P-value of 4.55e-15; (B) miR-1, with a P-value of 7.78e-07; and (C) miR-124, with a P-value of 2.2e-16
3.5 A secondary structure-based model
Two previous studies have reported enhancement or suppression of miRNA targeting through alteration of a local target secondary structure upon binding by RNA-binding proteins (RBPs) (Kedde et al., 2010; Xue et al., 2013). In our study context, enhanced target site accessibility and miRNA targeting could possibly be facilitated through the binding of a m6A reader protein.
The effects of m6A on RNA structure have been studied by RNA structure probing using RNases V1 and nuclease S1 (Liu et al., 2015), and by high-throughput structure probing using icSHAPE (Spitale et al., 2015). Both studies reported that m6A can alter the base-pairing status of neighboring nucleotides from paired to unpaired, thereby increasing the local structural accessibility. It has been reported that modified adenosines including m6A reduce stability of RNA duplexes (Kierzek and Kierzek, 2003). In a biochemical study, it was reported that N6 methylation within a helical region can have destabilizing effect, whereas methylation of an unpaired base can increase the stability of single base stacking (Roost et al., 2015). These findings suggest that m6A alone could be sufficient for modulation of regulation.
To answer the question of why regulation is enhanced for modified targets, given that miRNA-binding sites that can be modified are less accessible before methylation than unmodified sites, we propose two specific secondary structure-based mechanistic models (Fig. 5). In the first model, m6A within the miRNA-binding site is recognized and bound by an m6A reader protein(s), leading to opening of the local target secondary structure, which facilitates binding by AGO for enhanced regulation. In the second m6A reader-independent model, the presence of m6A alone is sufficient for alteration of the local structure, leading to increased site accessibility for Ago binding.
Fig. 5.
A proposed model of m6A function in miRNA targeting. m6A alters the local secondary structure of the target RNA to increase accessibility for binding by AGO. (A) In the absence of m6A, the local target structure at the miRNA-binding site is structurally inaccessible; (B) m6A is recognized and bound by an m6A reader, and this interaction opens the local structure (top); m6A alone is sufficient to significantly weaken the local target secondary structure (bottom), resulting in a structurally accessible miRNA-binding site facilitating AGO binding and miRNA-mediated regulation
4 Discussions
In this study to explore the potential interaction between m6A modification and gene regulation by miRNAs, we performed comprehensive analyses of high-throughput data on m6A in human and mouse transcriptome, miRNA target binding and regulation. There are two possible outcomes of miRNA-mediated gene regulation: mRNA degradation or translational inhibition. The levels of mRNAs in an experimental system can be easily measured by microarrays or RNAseq. However, measurements of levels of large number of proteins are much more difficult and can be very costly. Our analyses are thus limited to available high throughout mRNA data. On the other hand, it has been reported that miRNAs predominantly act to decrease target mRNA levels, and destabilization of target mRNAs is the predominant reason for reduced protein output (Guo et al., 2010).
We found that the level of regulation is significantly higher when m6A is present on target mRNAs. The evolutionary conservation for miRNA-binding sites with m6A modification is significantly higher than that for miRNA-binding sites without modification. These findings strongly indicate the functional significance of m6A modification in miRNA-mediated gene regulation.
The RNA modification database used in this study compiles experimental data from different experiments (Sun et al., 2016). Differences in cell lines and techniques in these experiments can present different m6A profiles, as m6A modification is cell-type specific. For these reasons, some of the m6A+ targets may not be methylated in the specific cell line for the microarray study (Baek et al., 2008). It is impossible to determine which of the m6A+ targets are methylated and which are not. The observed effects of the enhanced miRNA regulation by methylation could be diluted by the inclusion of targets that are not methylated. In other words, the true regulatory effects by m6A could be greater than what were observed here.
To explore a model-based explanation of our findings, we performed a target site accessibility analysis based on a target secondary structure prediction and miRNA-target hybridization modeling with RNA thermodynamic parameters for unmodified nucleotides. While modeling the secondary structure of RNAs with modified nucleotides would be ideal, complete thermodynamic parameters for chemical modifications including m6A are not available. We found that miRNA-binding sites that can be potentially m6A modified are significantly less accessible in the absence of methylation than those that do not have potential methylation sites. This is in sync with the finding that m6A+ targets are structurally more stable than m6A− targets, as indicated by their higher GC content.
Our findings and known role of m6A in destabilizing RNA structure led to a proposed model in which m6A can alter local target structure to increase accessibility for binding by AGO, leading to enhanced regulation. Specifically, we propose an m6A reader-dependent model and an m6A reader-independent model (Fig. 5). Our model is limited to modification within miRNA-binding sites. Because long-range base-pairing interaction is possible in mRNAs, we speculate that in some cases modification outside miRNA-binding sites can also enhance regulation if the modification unpairs base-pairs involving nucleotides within a miRNA-binding site.
In one recent computational study, spatial correlation among m6A, AGO binding and binding of RBPs led to a proposed model for a three way interplay mediated through alteration of the target RNA secondary structure (Das Mandal and Ray, 2021). For two of the four modes in the model, methylation facilitated target binding by miRNAs. In another recent study with a focus on RBPs and miRNA targeting, most RBPs were found to enhance miRNA targeting by increasing target site accessibility (Kim et al., 2021). For miRNA targeting, the importance of target structure and binding site accessibility was established over 14 years ago (Long et al., 2007). The findings from this study and the two recent studies signal the emergence of target structure as a common theme in our expanding understanding of miRNA-mediated gene regulation.
We hope our key finding of enhanced miRNA targeting by m6A and the two proposed models can be directly tested through properly designed experiments. To this end, m6A writer knockouts, nucleotide mutagenesis at the modification site that preserves base-pairing status, and an assay for miRNA regulatory activity can be useful tools. Local base-pairing status can be assessed either by experimental structure probing or by computational prediction (Ding et al., 2004; Ding and Lawrence, 2003). For a measurement of local structural accessibility, ΔGtotal is available from STarMir (Rennie et al., 2014).
Acknowledgements
The Computational Molecular Biology and Statistics Core at the Wadsworth Center is acknowledged for supporting computing resources for this work. The authors thank Maneesh Kumar for helpful suggestions.
Funding
This work was supported by the National Institutes of Health (GM099811, GM138856 to Y.D. and J.L.).
Conflict of Interest: none declared.
References
- Alarcon C.R. et al. (2015) N6-methyladenosine marks primary microRNAs for processing. Nature, 519, 482–485. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ambros V. (2004) The functions of animal microRNAs. Nature, 431, 350–355. [DOI] [PubMed] [Google Scholar]
- Baek D. et al. (2008) The impact of microRNAs on protein output. Nature, 455, 64–71. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bartel D.P. (2004) MicroRNAs: genomics, biogenesis, mechanism, and function. Cell, 116, 281–297. [DOI] [PubMed] [Google Scholar]
- Batista P.J. et al. (2014) m(6)A RNA modification controls cell fate transition in mammalian embryonic stem cells. Cell Stem Cell, 15, 707–719. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Berulava T. et al. (2015) N6-adenosine methylation in MiRNAs. PLoS One, 10, e0118438. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen T. et al. (2015) m(6)A RNA methylation is regulated by microRNAs and promotes reprogramming to pluripotency. Cell Stem Cell, 16, 289–301. [DOI] [PubMed] [Google Scholar]
- Chen X.Y. et al. (2019) The role of m(6)A RNA methylation in human cancer. Mol. Cancer, 18, 103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chi S.W. et al. (2009) Argonaute HITS-CLIP decodes microRNA-mRNA interaction maps. Nature, 460, 479–486. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Das Mandal S., Ray P.S. (2021) Transcriptome-wide analysis reveals spatial correlation between N6-methyladenosine and binding sites of microRNAs and RNA-binding proteins. Genomics, 113, 205–216. [DOI] [PubMed] [Google Scholar]
- Ding Y., Lawrence C.E. (2003) A statistical sampling algorithm for RNA secondary structure prediction. Nucleic Acids Res., 31, 7280–7301. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ding Y. et al. (2004) Sfold web server for statistical folding and rational design of nucleic acids. Nucleic Acids Res., 32, W135–W141. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Erson A.E., Petty E.M. (2008) MicroRNAs in development and disease. Clin. Genet., 74, 296–306. [DOI] [PubMed] [Google Scholar]
- Esau C.C., Monia B.P. (2007) Therapeutic potential for microRNAs. Adv. Drug. Deliv. Rev., 59, 101–114. [DOI] [PubMed] [Google Scholar]
- Guo H. et al. (2010) Mammalian microRNAs predominantly act to decrease target mRNA levels. Nature, 466, 835–840. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hafner M. et al. (2010) Transcriptome-wide identification of RNA-binding protein and microRNA target sites by PAR-CLIP. Cell, 141, 129–141. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Harfe B.D. (2005) MicroRNAs in vertebrate development. Curr. Opin. Genet. Dev., 15, 410–415. [DOI] [PubMed] [Google Scholar]
- Helwak A. et al. (2013) Mapping the human miRNA interactome by CLASH reveals frequent noncanonical binding. Cell, 153, 654–665. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ke S. et al. (2015) A majority of m6A residues are in the last exons, allowing the potential for 3' UTR regulation. Genes Dev., 29, 2037–2053. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kedde M. et al. (2010) A Pumilio-induced RNA structure switch in p27-3' UTR controls miR-221 and miR-222 accessibility. Nat. Cell Biol., 12, 1014–1020. [DOI] [PubMed] [Google Scholar]
- Kent W.J. et al. (2002) The human genome browser at UCSC. Genome Res., 12, 996–1006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kierzek E., Kierzek R. (2003) The thermodynamic stability of RNA duplexes and hairpins containing N6-alkyladenosines and 2-methylthio-N6-alkyladenosines. Nucleic Acids Res., 31, 4472–4480. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kim S. et al. (2021) The regulatory impact of RNA-binding proteins on microRNA targeting. Nat. Commun., 12, 5057. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lewis B.P. et al. (2005) Conserved seed pairing, often flanked by adenosines, indicates that thousands of human genes are microRNA targets. Cell, 120, 15–20. [DOI] [PubMed] [Google Scholar]
- Liu C. et al. (2013) CLIP-based prediction of mammalian microRNA binding sites. Nucleic Acids Res., 41, e138. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu N., Pan T. (2016) N6-methyladenosine-encoded epitranscriptomics. Nat. Struct. Mol. Biol., 23, 98–102. [DOI] [PubMed] [Google Scholar]
- Liu N. et al. (2015) N(6)-methyladenosine-dependent RNA structural switches regulate RNA-protein interactions. Nature, 518, 560–564. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Long D. et al. (2007) Potent effect of target structure on microRNA function. Nat. Struct. Mol. Biol., 14, 287–294. [DOI] [PubMed] [Google Scholar]
- Louloupi A. et al. (2018) Transient N-6-methyladenosine transcriptome sequencing reveals a regulatory role of m6A in splicing efficiency. Cell Rep., 23, 3429–3437. [DOI] [PubMed] [Google Scholar]
- Mathews D.H. et al. (1999) Expanded sequence dependence of thermodynamic parameters improves prediction of RNA secondary structure. J. Mol. Biol., 288, 911–940. [DOI] [PubMed] [Google Scholar]
- Meyer K.D., Jaffrey S.R. (2017) Rethinking m(6)A readers, writers, and erasers. Annu. Rev. Cell Dev. Biol., 33, 319–342. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Meyer K.D. et al. (2012) Comprehensive analysis of mRNA methylation reveals enrichment in 3' UTRs and near stop codons. Cell, 149, 1635–1646. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rehmsmeier M. et al. (2004) Fast and effective prediction of microRNA/target duplexes. RNA, 10, 1507–1517. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rennie W. et al. (2014) STarMir: a web server for prediction of microRNA binding sites. Nucleic Acids Res., 42, W114–W118. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Roost C. et al. (2015) Structure and thermodynamics of N6-methyladenosine in RNA: a spring-loaded base modification. J. Am. Chem. Soc., 137, 2107–2115. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Roundtree I.A. et al. (2017) Dynamic RNA modifications in gene expression regulation. Cell, 169, 1187–1200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Siepel A. et al. (2005) Evolutionarily conserved elements in vertebrate, insect, worm, and yeast genomes. Genome Res., 15, 1034–1050. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Spitale R.C. et al. (2015) Structural imprints in vivo decode RNA regulatory mechanisms. Nature, 519, 486–490. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sun W.J. et al. (2016) RMBase: a resource for decoding the landscape of RNA modifications from high-throughput sequencing data. Nucleic Acids Res., 44, D259–D265. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang X. et al. (2014) N6-methyladenosine-dependent regulation of messenger RNA stability. Nature, 505, 117–120. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xue Y. et al. (2013) Direct conversion of fibroblasts to neurons by reprogramming PTB-regulated microRNA circuits. Cell, 152, 82–96. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zepecki J.P. et al. (2021) miRNA-mediated loss of m6A increases nascent translation in glioblastoma. PLoS Genet., 17, e1009086. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zheng G. et al. (2013) ALKBH5 is a mammalian RNA demethylase that impacts RNA metabolism and mouse fertility. Mol. Cell, 49, 18–29. [DOI] [PMC free article] [PubMed] [Google Scholar]