Abstract
Acinetobacter baumannii has emerged as one of the most problematic bacterial pathogens responsible for hospital-acquired and community infections worldwide. Besides its high capacity to acquire antibiotic resistance mechanisms, it also presents high adhesion abilities on inert and living surfaces leading to biofilm development. This lifestyle confers additional protection against various treatments and allows it to persist for long periods in various hospital niches. Due to their remarkable antimicrobial tolerance, A. baumannii biofilms are difficult to control and ultimately eradicate. Further insights into the mechanism of biofilm development will help to overcome this challenge and to develop novel antibiofilm strategies. To unravel critical determinants of this sessile lifestyle, the proteomic profiles of two A. baumannii strains (ATTC17978 and SDF) grown in planktonic stationary phase or in mature solid–liquid (S-L) biofilm were compared using a semiquantitative proteomic study. Of interest, among the 69 common proteins determinants accumulated in the two strains at the S-L interface, we sorted out the MacAB-TolC system. This tripartite efflux pump played a role in A. baumannii biofilm formation as demonstrated by using ΔmacAB-tolC deletion mutant. Complementary approaches allowed us to get an overview of the impact of macAB-tolC deletion in A. baumannii physiology. Indeed, this efflux pump appeared to be involved in the envelope stress response occurring in mature biofilm. It contributes to maintain wild type (WT) membrane rigidity and provides tolerance to high osmolarity conditions. In addition, this system is probably involved in the maintenance of iron and sulfur homeostasis. MacAB-TolC might help this pathogen face and adapt to deleterious conditions occurring in mature biofilms. Increasing our knowledge of A. baumannii biofilm formation will undoubtedly help us develop new therapeutic strategies to tackle this emerging threat to human health.
Keywords: solid–liquid interface, biofilm, efflux pump, eDNA, envelop stress response
1. Introduction
Over the last decades, Acinetobacter baumannii has emerged as one of the most problematic opportunistic pathogens involved in hospital-acquired infections and community infections worldwide (Lin and Lan, 2014). The pathogenicity of this member of the ESKAPE group of bacterial pathogens (Enterococcus faecium, Staphylococcus aureus, Klebsiella pneumoniae, A. baumannii, Pseudomonas aeruginosa, and Enterobacter spp.) (Boucher et al., 2009) and its success as an infective agent appear to be related to multiple factors, and especially its ability to form biofilms. Indeed, its high capacity to acquire antibiotic resistance mechanisms has led to the increasing occurrence of outbreaks of infection involving multi- or pan-drug-resistant A. baumannii (Lee et al., 2017; Nasr, 2020). Furthermore, it also presents remarkable adhesion abilities on inert and leaving surfaces, leading to biofilm development that allows it to survive desiccation (Gayoso et al., 2014), oxidative stress (Soares et al., 2010), or disinfectants (Peleg et al., 2008; Harding et al., 2018) and hence to persist for long periods in various hospital environments. This concerning public health threat was therefore ranked on the global priority pathogens list established by the World Health Organization (WHO) for which there is an urgent need for new antibiotic development.
Biofilms are structural communities of interface-associated bacteria organized as microcolonies embedded within a complex hydrated polymeric matrix composed of extracellular polymeric substances (EPSs), such as exopolysaccharides, proteins, nucleic acids, and other compounds (Monds and O’Toole, 2009). The regulatory process of biofilm formation is highly dynamic and influenced by environmental factors that allow the transition between free-floating cells and biofilm lifestyles. The sessile growth mode provides clear ecological and physiological advantages to microorganisms that inherently benefit of protection against adverse environments, host immune system clearance, antibiotics, and other antimicrobial agents and protection from starvation through carbon storage (Yan and Bassler, 2019; Zhang et al., 2020). In addition, bacterial biofilm appears also to be an ideal environment for the horizontal exchange of genetic material between microorganisms through genetic mutations and rearrangements and also integration of determinants carried by mobile genetic elements, thus reinforcing bacterial genetic plasticity (Soucy et al., 2015). It is now well established that bacterial biofilm is involved in lots of infectious diseases and in a variety of medical device-related infections (Zhang et al., 2020). Indeed, the pathogenic potential of sessile microorganisms is much higher than the one of planktonic cells.
The ability of A. baumannii to form a biofilm is one of the leading mechanisms that has largely contributed to its success as a human pathogen. This Gram-negative bacterium may cause severe nosocomial infections including hospital-acquired and ventilator-associated pneumonia, bacteremia, endocarditis, skin and soft tissue infections, urinary tract infections, or meningitis (Peleg et al., 2008; Nasr, 2020). Biofilms are commonly referred to as solid-attached structures, but they can develop on a wide variety of interfaces including solid–liquid (S-L), air–liquid (A-L), liquid–liquid, or air–solid interfaces. A. baumannii biofilms grow at S-L interfaces, e.g., between a biological or an abiotic surface and an aqueous medium, but this organism has also been characterized for its ability to develop A-L interface biofilms, also known as pellicles, which constitute more complex structures than classical surface-attached biofilms in terms of development, level of organization, and mechanics (Marti et al., 2011b). Some of our investigations have revealed that Acinetobacter species forming pellicles, such as A. baumannii and A. nosocomialis, are those mainly involved in nosocomial infections, suggesting a correlation between this sedentary lifestyle and bacterial pathogenicity (Marti et al., 2011a; Kentache et al., 2017).
Despite the abundant literature on biofilm lifestyles and their widespread distribution in diseases, some issues remain unclear. Owing to their increasing resilience to antimicrobial treatments, A. baumannii biofilms are difficult to control and ultimately eradicate. Our understanding of this pathogen is that biofilm formation rather facilitates and/or prolongs its survival in harsh conditions likely by adopting a “persist and resist” strategy as previously proposed (Harding et al., 2018). Therefore, it is urgently needed to decipher mechanisms involved in A. baumannii biofilm formation and thus to identify key determinants that can be potential targets, aiming at developing novel anti-biofilm strategies. In this context, efflux pumps constitute critical determinants of this sessile lifestyle and have emerged as promising targets, as their inhibition may allow to fight pathogens at various levels, antibiotic resistance, but also biofilm formation. Indeed, in some bacterial species, such as Escherichia coli, tripartite efflux pumps have been previously reported to be involved in biofilm formation (Alav et al., 2018). In A. baumannii, it was envisaged that the Pmt [putative major facilitator superfamily (MFS) transporter-like] protein could be associated with the release of eDNA and adhesion on biotic and abiotic surfaces (Sahu et al., 2012). Another MFS transporter, AbeF, involved in fosfomycin efflux, was also proposed to participate in the secretion of biofilm matrix (Sharma et al., 2017). The contribution of resistance-nodulation-division (RND)-efflux pumps, like AdeABC, to this growth mode was also demonstrated but especially in terms of adhesion (Richmond et al., 2016). Finally, deletion of the efflux pump genes emrA/emrB resulted in a decrease of biofilm formation in A. baumannii, even though their precise roles remained to be clarified (Lin et al., 2020).
In the current study, we have compared the biofilm-forming ability of two strains of A. baumannii harboring specific features. We used the SDF strain that interestingly produces an abundant biofilm, but not pellicle, without presenting the main classical determinants associated with virulence of biofilm (such as Csu pili, PgaABCD, and type IV pili) (Antunes et al., 2011; Eijkelkamp et al., 2014), and the A. baumannii ATCC 17978 strain as a reference strain. The proteome profiles of bacteria grown in planktonic stationary phase with those of bacteria grown in mature S-L biofilm were compared using a proteomic semiquantitative study. This analysis highlights adhesins that could contribute to initiation and development of the SDF biofilm. Of interest, among the 69 common protein determinants accumulated by the two A. baumannii strains at the S-L interface, we sorted out the MacAB-TolC system. This pump has been reported to actively extrude various substrates, including macrolide antibiotics and virulence factors in E. coli and other Gram-negative bacteria. It was also involved in the transport of outer membrane glycolipids, lipopeptides, and protoporphyrin (reviewed in Fitzpatrick et al., 2017). Interestingly, this tripartite efflux pump appears to be a noteworthy determinant of A. baumannii mature biofilms as demonstrated by using macAB-tolC deletion mutant. Complementary approaches allowed us to suggest its contribution to iron and sulfur homeostasis and to demonstrate its involvement in cell wall rigidity and osmotic protection.
2. Materials and Methods
2.1 Bacterial Strains and Growth Conditions
Strains and plasmids used in this study are listed in Supplementary Table 1. A. baumannii SDF strain was selected based on its failure to form pellicle and the lack of the main classically defined determinants of biofilm (Fournier et al., 2006). The ATCC 17978 strain, lacking the pAB3 plasmid (pAB3-) as checked by PCR amplification and sulfamethoxazole/trimethoprim (SXT) susceptibility testing (Weber et al., 2015), was chosen because of its high capacity to form biofilms compared to ATCC 17978 pAB3+ strain. The SDF strain was grown in Luria Bertani medium (LB, Difco; Antunes et al., 2011). ATCC 17978 and its derivative strains were grown in Mueller–Hinton broth (MHB, Difco). The mutant strains complemented with pWH1266 (ΔMac_e) or pWH1266::macAB-tolC (ΔMac_c) were selected on MHB supplemented with 10 μg/ml ticarcillin. E. coli DH5α (pCR-Blunt) and CC118λpir (pKNG101) were selected on MHB containing 50 μg/ml kanamycin and 50 μg/ml streptomycin, respectively.
2.2. Mutant and Complemented Strain Construction
Deletion mutant was constructed from A. baumannii ATCC 17978 using overlapping PCRs and recombination events according to the protocol of Richardot et al. (2016). Briefly, the 5′ region of tolC gene (ABYAL0571, 703 bp) and the 3′ region of macA gene (ABYAL0574, 816 bp) were amplified by PCR with specific primers (Supplementary Table 2). The resulting PCR products were used as templates for overlapping PCRs to generate the mutagenic DNA insert ΔmacAB-tolC. The insert was cloned into pCR-Blunt plasmid, then digested with BamHI/ApaI. The generated fragment was subcloned into pKNG101, then the resulting plasmid pKNG101::ΔmacAB-tolC was transferred into E. coli CC118λpir. The suicide vector was introduced into A. baumannii strain by triparental mattings using E. coli HB101 (pRK2013) helper strain. A. baumannii with pKNG101::ΔmacAB-tolC was selected on MH agar supplemented with 800 μg/ml streptomycin, and E. coli was counterselected with 30 μg/ml chloramphenicol. Suicide vector pKNG101 with the macAB-tolC genes was excised by selection on M9 medium agar plates supplemented with 5% sucrose. The plasmid loss was confirmed by negative selection on MH agar with 800 μg/ml streptomycin, and the deletion of 4,524 bp was checked by PCR and sequencing (Supplementary Table 2). The entire operon macAB-tolC from A. baumannii ATCC 17978 was amplified using specific primers containing a complemented sequence from the expression vector pWH1266 (Supplementary Table 2). The plasmid pWH1266 was linearized with HindIII enzyme and then reassembled with the macAB-tolC PCR product using NEBuilder DNA Hifi Assembly kit (New England Biolabs). The resulting plasmid pWH1266::macAB-tolC was transferred into E. coli DH5a by transformation and then into ΔMac strain by electroporation.
2.3. Proteomic Analyses of Planktonic and Sessile Bacterial Cultures
Biofilms were grown on 30 g of glass wool in 800 ml of rich medium using 107 [Colony Forming Unit (CFU)/ml] as an inoculum (Crouzet et al., 2017). They were incubated at 37°C for 4 days with slight shaking (90 rpm) to avoid pellicle formation. Then, glass wool was washed three times with phosphate buffer saline (PBS) to remove unattached cells. Biofilm bacteria were recovered from glass wool by vigorous shaking with 30 g of glass beads and a subsequent centrifugation (6,000 × g, 15 min, 4°C). One-day-old planktonic cultures were performed similarly but with shaking at 140 rpm and without glass wool. Total protein extraction from planktonic and biofilm cells was performed as already described (Kentache et al., 2017). Protein samples were prepared at least in biological triplicate for each condition. Then, enzymatic digestion of protein extracts and quantitative analysis by mass spectrometry analyses were performed according to Kentache et al. (2017). Protein abundances in the wild type (WT) and ΔMac were compared using Progenesis LC-MS software for protein quantification. False discovery rates (FDRs) were calculated using a decoy-fusion approach in Mascot (version 2.6.0.0). Identified peptide spectrum matches with −10logP value higher than 14 were kept at an FDR threshold of 1%, and proteins identified with less than two peptides were discarded. The mass spectrometry proteomics data have been deposited to the ProteomeXchange Consortium via the PRIDE partner repository with the dataset identifier PXD028619.
2.4. Biofilm Assays
To compare A. baumannii ATCC 17978 and derivatives strains, biofilm formation and metabolic activity were measured using 2,3-bis(2-methoxy-4-nitro-5-sulfophenyl)-5-[(phenylamino)carbonyl]-2H-tetrazolium hydroxide (XTT) assays as previously described (Orsinger-Jacobsen et al., 2013) with some modifications. Briefly, MHB in a 96-well flat-bottomed polystyrene plate was inoculated with 150 μl per well at 5.107 CFU/ml of a fresh overnight culture. The plate was incubated at 37°C without shaking in darkness. After 24 h, the plate was read at 595 nm, and the medium and the pellicle were discarded. Biofilm was washed twice with 200 μl of ultrapure water. XTT solution was added, and samples were incubated for 3 h at 37°C. The optical density (OD) at 490 nm was then measured. Biomass quantification between the different strains was performed by crystal violet (CV) method using a 24-well flat-bottomed polystyrene plate inoculated with 1 ml per well at 5.107 CFU/ml. The plate was incubated at 37°C without shaking in darkness for 48 h. Then, OD at 600 nm of cultures was read. Biofilm was washed once and stained with 1 ml of 0.1% CV for 15 min. After CV removal, wells were washed twice with 1 ml of ultrapure water. CV attached to biomass was solubilized by 1 ml of acetic acid at 30%. Wells were homogenized to measure OD at 580 nm. All assays were performed at least in triplicate in a minimum of three independent experiments.
2.5. Confocal Laser Scanning Microscopy
Biofilm formation at the S-L interface was achieved in glass coverslips as described (Le et al., 2021). Briefly, aliquots of 1 ml of bacteria in MHB (inoculum 5.107 CFU/ml) were transferred into each well (24-well flat-bottomed plate) containing a glass coverslip ø12 mm (Supplementary Figure 1). The plate was incubated at 37°C without shaking in darkness for 48 h. The medium was discarded, and biofilms were washed twice with PBS. Biofilms were finally stained with Syto9 (Thermo Fisher Scientific) for 30 min following the manufacturer’s protocol prior to microscopy. Biofilm formation at the solid–liquid–air interface was prepared using a previously described protocol (Fulaz et al., 2019) with some modifications. A 10-ml volume of bacteria in MHB (inoculum 5.107 CFU/ml) was added to a sterile 50-ml Falcon centrifuge tube containing a glass coverslip (24 mm × 50 mm) (Supplementary Figure 1). Biofilm formation in the presence of DNase I from bovine pancreas (Sigma-Aldrich) was performed by supplementing medium with DNase I at 100 μg/ml (Tahrioui et al., 2019). After 24 h of incubation at 37°C without shaking, the coverslip was washed with PBS, and biofilms were stained with Syto9 (Filmtracer LIVE/DEAD Biofilm Viability Kit, Invitrogen). The coverslip was then assembled onto a glass microscope slide using Mowiol 4–88 mounting medium. Image acquisitions were performed using Leica TCS SP8 CFS confocal microscope with fixed stature (Leica Microsystems), equipped with diode laser (Coherent) at 488 nm for Syto9. Fluorescence emission was detected sequentially by a hybrid detector (Leica Microsystems) in photon counting mode with a specific band from 500 to 540 nm for Syto9. Image processing was performed with Imaris software.
2.6. Drug Susceptibility Assays
The minimum inhibitory concentrations (MICs) of antibiotics (azithromycin, erythromycin, spiramycin, ticarcillin, erythromycin, colistin, gentamicin, tobramycin, novobiocin, tetracycline, tigecycline, imipenem, and ciprofloxacin; Sigma-Aldrich) and antiseptic (chlorhexidine gluconate, Sigma-Aldrich) on ATCC 17978 WT and derivative strains were determined by the standard microdilution method in MH or MH-cation-adjusted broth using an initial inoculum of 5.105 CFU/ml, as recommended by the Clinical and Laboratory Standards Institute (CLSI) (2015). The minimum biofilm eradication concentration (MBEC), defined as the lowest concentration of an antibiotic that prevents visible growth in the recovery medium used to collect biofilm cells (Macia et al., 2014), was determined using Calgari Biofilm Device (Innovotech, Canada) as previously described (Ceri et al., 1999). Briefly, MH or MH-cation-adjusted broth was inoculated with 107 CFU/ml from an overnight culture in a 96-well plate and incubated at 37°C for 24 h with shaking. Biofilms grew around the plastic pegs on the lid of the plate. Pegs were washed with PBS at 10 mM and challenged with increasing concentrations of antimicrobial agents for an additional 24 h at 37°C. Then, biofilms were washed and removed from pegs by sonication (ultrasonic bath) for 20 min in fresh sterile MHB (recovery plate). The recovery plate was incubated for 24 h at 37°C. OD650 of each well was measured to determine MBEC values. MIC and MBEC experiments were performed in three independent assays.
2.7. Growth Assays
MHB was inoculated at 107 CFU/ml with fresh overnight cultures of A. baumannii ATCC 17978 WT or derivative strains. Strains were grown to mid-log phase and harvested by centrifugation (2,000 × g for 5 min). Spotting assay method on supplemented M9 agar plate (Harding et al., 2017) was then used to quantify the impact of 10 mM L-phenylalanine, 256 μg/ml phenylacetic acid (PAA), and 2,048 μg/ml gallic acid (GA) and tannic acid (TA) (Cerqueira et al., 2014; Lin et al., 2015). High-osmolarity adaptation was achieved by measuring for 24 h A. baumannii growth in MHB supplemented with 500 mM sucrose (Fluka). Conventional dilution series and plating techniques were carried out to evaluate bacterial survival. Three independent experiments were performed.
2.8. Fluorescence Anisotropy Assay
Planktonic or biofilm cultures of A. baumannii ATCC 17978 WT, ΔMac, and ΔMac_c were grown in MHB at 37°C for 24 or 48 h, respectively, and cell membrane fluidity was investigated as previously described (Tahrioui et al., 2020). Briefly, cell pellets were washed twice in 10 mM MgSO4 and resuspended to reach 0.1 OD600. Then, 1 ml of the resuspended cultures was incubated with 4 μM 1,6-diphenyl-1,3,5-hexatriene (DPH; Sigma-Aldrich) in the dark for 30 min at 37°C. Measurement of the fluorescence anisotropy was performed using the Spark 20 M multimode Microplate Reader (Tecan Group Ltd.). Excitation and emission wavelengths were set to 365 and 425 nm, respectively. The anisotropy was calculated according to Lakowicz (2006). The relationship between anisotropy and membrane fluidity is an inverse one, where decreasing anisotropy values correspond to a more fluid lipid membrane and vice versa. All values are reported as means of at least triplicate analyses for each experimental variable.
2.9. Chrome Azurol S Assay
Quantification of secreted siderophores was performed as previously described (Penwell and Actis, 2019). Briefly, 250 ml Erlenmeyer for preculture and 24-well plate for culture were both conditioned with 0.5 M HCl and then rinsed three times with MiliQ water before sterilization (autoclaving or 30-min UV treatment). For the preculture, 50 ml of the succinate medium (Penwell and Actis, 2019) was inoculated with three colonies and then incubated at 37°C during 48 h under 140 rpm agitation. For the culture, 2 ml of succinate medium were inoculated from the preculture at 0.01 OD600 and incubated for 48 h at 37°C without agitation. Then, 1 ml of the culture was centrifuged during 20 min at 10,000 × g, and 150 μl of the supernatant were transferred to a 96-well plate, at least in triplicate. Finally, 30 μl of the chrome azurol S (CAS) reagent were added, and kinetic absorbance at 630 nm was performed for 60 min. All assays were repeated three times in triplicate.
2.10. Bacterial Adhesion to A549 Human Alveolar Epithelial Cells
A549 human lung adenocarcinoma cells from ATCC were grown as monolayer cultures in Dulbecco’s modified Eagle’s medium (DMEM) or in Ham’s F-12 Nutrient Mixture for at least 20 days to allow differentiation to an alveolar type II (ATII)-like phenotype, as indicated, supplemented with 10% heat-inactivated fetal bovine serum and antibiotics (100 U/ml of penicillin G and 100 μg/ml of streptomycin) (Cooper et al., 2016). Cells were maintained at 37°C in a humidified atmosphere of 5% CO2. All cell culture media and supplements were purchased from Thermo Fisher Scientific. Then, cells were trypsinized and transferred to 24-well plates to get a monolayer of 105 cells per well. After 24 h of incubation under the same conditions, A549 cells were washed twice with PBS and fresh medium without antibiotic was added. A. baumannii ATCC 17978 WT, ΔMac, and ΔMac_c strains were added to the cells at a ratio of bacteria to host cells of 20:1 [multiplicity of infection (MOI) of 20]. The cells infected with bacteria were incubated at 37°C under an atmosphere of 5% CO2 for 5 or 24 h. To determine bacterial adhesion, they were washed five times with PBS, fixed with ice-cold methanol for 20 min, and stained with Giemsa solution. Routinely, 10 microscopic fields were examined along the length of the coverslip. In each field, 10 epithelial cells were examined. The adhesion index was calculated as the total bacterial count divided by 100. The cells were examined using a Nikon Eclipse Ci-S microscope. All assays were repeated three times in triplicate. Student’s t-test was performed to evaluate the statistical significance of the observed differences.
2.11 Statistical Analysis
Proteomic data were statistically analyzed using Progenesis LC-MS software with ANOVA. Except for the proteomic data, the statistical analyses were carried out with the GraphPad Prism8 software. We used the non-parametric t-test, which is a Mann–Whitney test. Mean and standard deviation (SD) calculated from at least three independent experiments were presented.
3. Results and Discussion
Biofilms are key microbial ecosystems. To highlight critical determinants of A. baumannii sessile lifestyle that can be potential targets against biofilms, we compared protein profiles of bacteria grown in mature S-L biofilms with those of their planktonic stationary phase counterparts. For this comparison, we used the SDF strain, since it does not present classical biofilm determinants (such as Csu pili, PgaABCD, and type IV pili) (Antunes et al., 2011; Eijkelkamp et al., 2014) and may therefore express less characterized and interesting protein systems involved in biofilm development. We compared its proteome with those of the well-studied ATCC 17978 strain that was cultivated in similar conditions.
3.1 Proteomic Study of Solid–Liquid Biofilms Formed by SDF and ATCC 17978 Strains
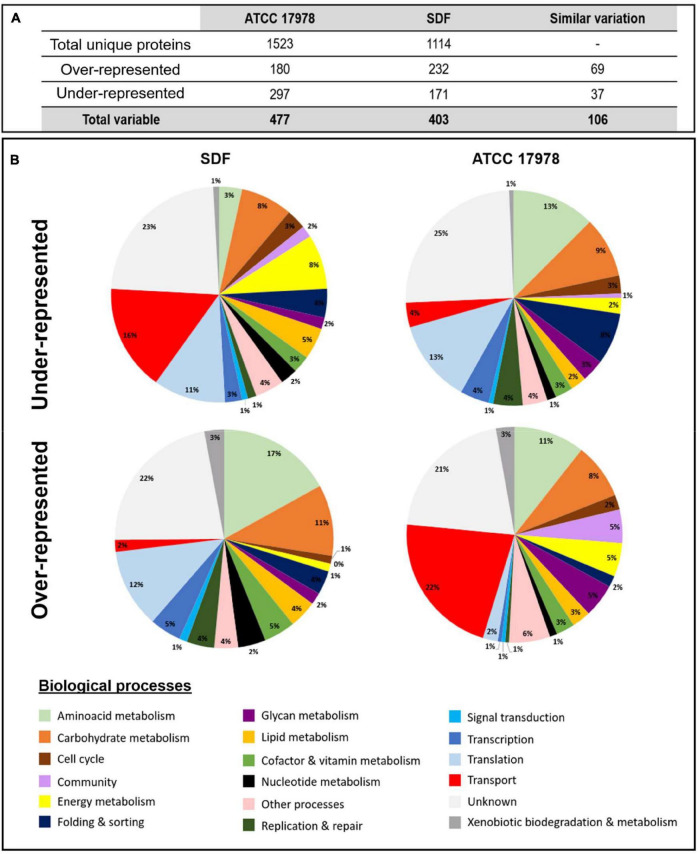
The S-L interface is not the favored interface to grow as biofilm for some A. baumannii strains (Marti et al., 2011b), and thus, for an optimized biomass–surface ratio, S-L biofilms were grown on glass wool (Crouzet et al., 2017). A slight shaking was performed to prevent the pellicle development that could happen with the ATCC 17978 strain (Kentache et al., 2017). The proteomic quantitative study revealed that among the 1,523 and 1,114 unique proteins identified in the overall samples (planktonic and biofilm) from ATCC 17978 and SDF, respectively (Figure 1A), 477 and 403 proteins showed a significant variation of abundance (fold ≥ 2) according to the mode of growth [Supplementary Table 3 (ATCC) and Supplementary Table 4 (SDF)]. Indeed, two protein populations were distinguished in S-L biofilms: (i) underrepresented and (ii) overrepresented proteins (Figure 1B). Among all the differentially represented proteins, 106 out of 143 proteins common to both analyses presented the same dynamics of variation. Taken together, A. baumannii S-L biofilms were characterized by decreased accumulation of proteins involved in bacterial fitness including metabolic proteins and in important surface remodeling such as membrane proteins belonging to transport systems. Indeed, underrepresented proteins were mainly distributed in four functional groups according to the kyoto encyclopedia of genes and genomes (KEGG) pathway: (1) amino acid metabolism (37/297 in ATCC 17978 and 29/171 in SDF), (2) carbohydrate metabolism (27/297 and 18/171, respectively), (3) genetic information processes corresponding to replication and repair, transcription, translation, folding, and sorting (87/297 of underrepresented population in ATCC 17978 and 41/171 in SDF), and (4) unknown functions (74/297 and 38/171 proteins, respectively). Even though in both strains overrepresented proteins were more heterogeneously distributed (Figure 1B), we were able to distinguish two main groups: (1) proteins involved in transport systems (39/180 in ATCC 17978 and 36/232 in SDF) and (2) proteins with unknown functions (37/180 of overrepresented proteins in ATCC 17978 and 54/232 overrepresented proteins in SDF).
FIGURE 1.
Proteomic analyses of solid–liquid (S-L) biofilms formed by A. baumannii ATCC 17978 and SDF strains compared to their planktonic counterparts. (A) Number of proteins identified by proteomic analyses of A. baumannii ATCC 17978 and SDF strains and number of proteins with modified abundance in S-L biofilms compared to their planktonic counterparts. (B) Classification of these differentially represented proteins from A. baumannii SDF (left) and ATCC 17978 (right) according to their biological processes using the kyoto encyclopedia of genes and genomes (KEGG) pathway.
3.2 Specific Determinants of SDF Solid-Liquid Biofilm
We looked for specific determinants of SDF S-L biofilm to understand how this strain could produce and maintain a biofilm as dense as the reference strain ATCC 17978, while lacking the main critical biofilm determinants. Of interest, SDF S-L biofilm was characterized by the accumulation of proteins involved in translation (Figure 1B). Indeed, the SDF strain accumulated 22 ribosomal subunits in S-L biofilm (with maximum fold changes ranging from 3.3 to 48; Supplementary Table 4). These proteins are usually characteristics of the physiological growing state (Bosdriesz et al., 2015) like in ATCC 17978 planktonic growth mode (Cabral et al., 2011; Kentache et al., 2017). However, analysis of P. aeruginosa PAO1 biofilms showed that the ribosomic mRNA expression level was stably maintained between dividing and slowly growing cells (Williamson et al., 2012). The importance of these ribosomal proteins was also reported in Bacillus subtilis where the deletion of rpsU and rpsK genes highly decreased biofilm formation (Takada et al., 2014). Even though S-L biofilms were 4 days old, we cannot exclude that the SDF strain continues to divide, with a substantial part of ribosomes translating community determinants.
In addition, our analysis also revealed the accumulation of 20 proteins being part of the energy metabolism and that belong to the complex I (NuoABCDFGHI, 7.7- to 37-fold changes), the complex II (SdhCAB, 17- to 252-fold changes), the cytochrome b0 oxidase (CyoAB, 9- and 57-fold changes, respectively) or the F1F0 ATP synthase (3 proteins) and three ubiquinone biosynthesis proteins (UbiG, 10-fold; UbiB, 18-fold; and UbiE, fivefold), and the flavoprotein-ubiquinone oxidoreductase EftD (13-fold), indicating that the function of the energetic respiratory chain was strongly exacerbated here. This suggests that a high ATP requirement and a potential redox regulation may increase the membrane potential to promote biofilm development (Qin et al., 2019).
Finally, we observed the overexpression of 11 proteins involved in the lipid metabolism, several of them being enzymes implicated in complex lipid biosyntheses. For example, PlsB (13-fold change) synthesizes phosphatidic acid precursors (Lehner and Kuksis, 1996), which are essential for the adaptation to environmental stresses through the modification of membrane composition and fluidity (Dubois-Brissonnet et al., 2016; Tao et al., 2021). As already reported, it may also contribute to modify the SDF adhesiveness character (Benamara et al., 2011; Lattif et al., 2011; Dubois-Brissonnet et al., 2016). Other enzymes, like Acr1 (54-fold change) and Wax-dgaT (fivefold change) are characteristics of environmental bacteria. The bifunctional acyltransferase Wax-dgaT is involved in the synthesis of triacylglycerols (TAGs) from diacylglycerols as well as in the synthesis of wax-ester (WE) (Lehner and Kuksis, 1996; Ishige et al., 2002). Accumulation of Wax-dgaT might so promote lipid storage as a carbon source to survive in nutrient deprivation. TAG storage could also contribute to bacterial desiccation tolerance (Alvarez et al., 2004; Alvarez, 2016).
This S-L biofilm proteomic quantitative analysis showed that SDF did not synthesize adhesion/community determinants known to be expressed in A. baumannii, like the Acinetin locus, the Csu pili or the P pilus or the PNAG polymer transporter PgaA, determinants detected in ATCC 17978 S-L biofilm. Interestingly, two systems may participate to the maintenance and the cohesion of the SDF S-L biofilm: (i) the type III pilus adhesion factors, FilC (71-fold change) and FilF (12-fold change), already described in ATCC 17978 pellicle communities (Marti et al., 2011b; Kentache et al., 2017), and (ii) a two-partner secretion system (TPS) FhaB/C. Indeed, we identified the protein ABSDF3544 (fourfold change), which is a hemagglutinin/hemolysin type protein. It may correspond to the secreted protein of a TPS system, with FhaB being the passenger domain and FhaC, the translocator (identified here with a 17.8-fold change). ABSDF3544 is not conserved between A. baumannii species, but it had two homologous in the SDF genome (not identified here). In A. baumannii, FhaB/C systems are involved in the adhesion to human epithelial and bronchial cells (Astaneh et al., 2014, 2017; Pérez et al., 2017). In AbH12O-A2 strain, the exoprotein AbFhaB (also called TpsA, 31.8% id. to ABSDF3544) contributes to the tridimensional A. baumannii aggregation (Pérez et al., 2017). Here, ABSDF3544 may participate in cell–cell interactions but also in SDF biofilm formation, as it was already shown for FhaB/C system in Bordetella pertussis (Serra et al., 2011).
3.3 Common Determinants of Solid–Liquid Biofilm Formation
Despite the small genome of SDF (3.2 Mb and 3,050 open reading frames) (Fournier et al., 2006), we highlighted 69 commonly overexpressed proteins in both ATCC 17978 and SDF S-L biofilm cells. Among them, we identified proteins belonging to already characterized metabolic pathways and several systems necessary for environmental exchanges in sessile bacteria such as: (i) arginine catabolism (Cabral et al., 2011; Kentache et al., 2017), (ii) some adhesion factors like OmpA (Gaddy et al., 2009), (iii) the polysaccharide export system (Wza-Wzc-Wzi) (Kenyon and Hall, 2013), (iv) the T6SS secretion system, and (v) transport systems for surface modulation (Bam, Tam) or environmental exchanges (OmpW, Omp25, OprD, CarO, and ABYAL0223 porins) and also the AdeIJK efflux pump (Kentache et al., 2017). Interestingly our comparative analysis revealed that both SDF and ATCC 17978 S-L biofilm cells overexpressed also two proteins of a tripartite efflux pump ABSDF2985 and ABSDF2983 (40- and 22-fold changes, respectively) and ABYAL0573-74 and ABYAL0571 (four and sixfold changes, respectively) that were annotated MacAB-TolC. The overexpression of the A1S_0538 gene from this system was also highlighted in a transcriptomic approach of A. baumannii ATCC1978 24-h S-L biofilms (Rumbo-Feal et al., 2013).
In A. baumannii, the MacAB-TolC system is a tripartite efflux pump where MacB is an atypical ABC family transporter with a recently determined atomic structure (Okada et al., 2017), MacA is a membrane fusion protein, and TolC is an outer membrane protein. This well-conserved system was first identified in E. coli (Kobayashi et al., 2001). It handles the efflux of substrates either from the periplasm and/or from the cytoplasm to the extracellular environment of the bacterial cell (Crow et al., 2017; Fitzpatrick et al., 2017). For many species like E. coli, Stenotrophomonas maltophilia, or K. pneumoniae, it is involved in the resistance to macrolides, aminoglycosides, polymyxins, and cyclines (Lin Y. T. et al., 2014; Fitzpatrick et al., 2017; Zheng et al., 2018). In addition, MacAB was shown to extrude various compounds such as toxins (enterotoxin STII in E. coli; Yamanaka et al., 2008), protoporphyrin IX (Turlin et al., 2014), or lipopeptides and siderophores in Pseudomonas species (Imperi et al., 2009; Greene et al., 2018) and can be involved in virulence (Nishino et al., 2006). MacAB was recently described to protect Salmonella enterica serovar typhimurium from oxidative stress through linearized siderophore product secretion (Bogomolnaya et al., 2020). Thus, the MacAB system appears to fulfill numerous transport functions for a wide range of substrates. In addition, its contribution to biofilm formation has been described for S. maltophilia (Lin Y. T. et al., 2014). In A. baumannii, MacAB-TolC shares 83% of amino acid sequence similarity with its E. coli counterpart and, therefore, may be involved in the efflux of macrolides and novobiocin (Okada et al., 2017; Pérez-Varela et al., 2019). So far, to our knowledge, the function of the MacAB-TolC pump in A. baumannii biofilm cells has never been considered.
3.4 Involvement of MacAB-TolC in Biofilm Formation
To unravel the function of MacAB-TolC tripartite efflux system of A. baumannii in biofilm formation, we have attempted unsuccessfully to generate a ΔmacAB-tolC deletion mutant in the SDF strain. We, however, succeeded to make this deletion mutant (ΔMac) in the ATCC 17978 strain and also generated a complemented strain (ΔMac_c) harboring pWH1266::macAB-tolC. We checked that deletion of macAB-tolC did not affect growth of ΔMac mutant neither in planktonic nor in biofilm cultures (Supplementary Figure 2). For ΔMac_c strain, the cell metabolic activity was slightly decreased in biofilm probably due to the presence of the complementation plasmid, since the same phenotype was observed in the strain carrying the empty plasmid (Supplementary Figure 2).
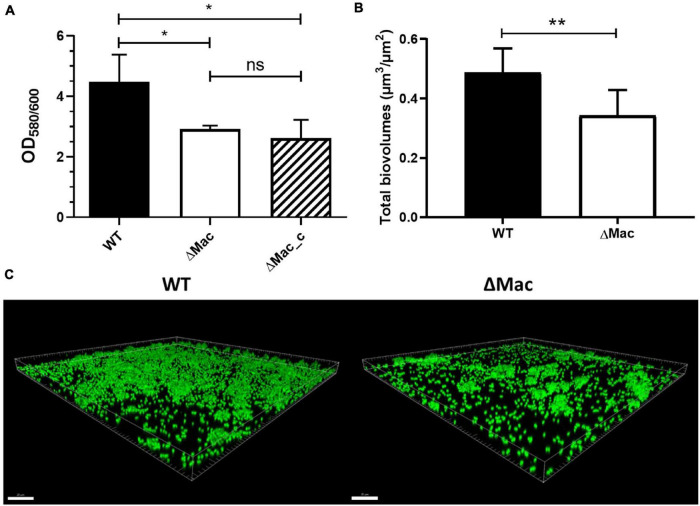
Interestingly, our results showed that deletion of the Mac system negatively affected the biomass amount that was decreased by 33% (p < 0.05) after 48 h of biofilm growth (Figure 2A). Complementation did not, however, restore the phenotype to a level comparable to the level reached by the WT strain. Since the antibiotic selection pressure was not applied in our experiment owing to its influence on biofilm formation (Peleg et al., 2008; Penesyan et al., 2019), the complementation plasmid may have been lost within the time frame of the experiment. We, however, failed to demonstrate by numeration a difference between the ΔMac and the ΔMac_c strains (data not shown). In parallel, confocal laser scanning microscopy (CLSM) analysis of biofilms was performed using Syto9 staining. Again, the total biovolume subsequent to the ΔMac deletion was reduced by 23% after 48 h biofilm formation (Figures 2B,C). Our results are consistent with the study performed in S. maltophilia, where the deletion of the MacAB-TolC system induces a 48% decrease of biofilm formation (Lin Y. T. et al., 2014). Taken together, inactivation of genes encoding the MacAB-TolC tripartite efflux system of A. baumannii results in impaired biofilm formation.
FIGURE 2.
Impact of the macAB-tolC deletion on A. baumannii biofilm formation at the solid–liquid interface. (A) Biomass quantification by crystal violet staining of A. baumannii ATCC 17978 wild type (WT), ΔMac, and ΔMac_c (ΔMac complemented strain). (B) Quantification of biovolume (μm3/μm2) of A. baumannii ATCC 17978 WT and ΔMac biofilms based on the confocal fluorescence images. (C) Representative confocal laser scanning microscopy (CLSM) images of A. baumannii ATCC 17978 WT (left) and ΔMac biofilms (right) labeled with Syto9. Data shown represent mean values (±SD) from at least three independent biological experiments (ns, not significant; *p < 0.05; **p < 0.01). Scale bar: 20 μm. OD, optical density.
3.5 Involvement of MacAB-TolC in Antibiotic Resistance
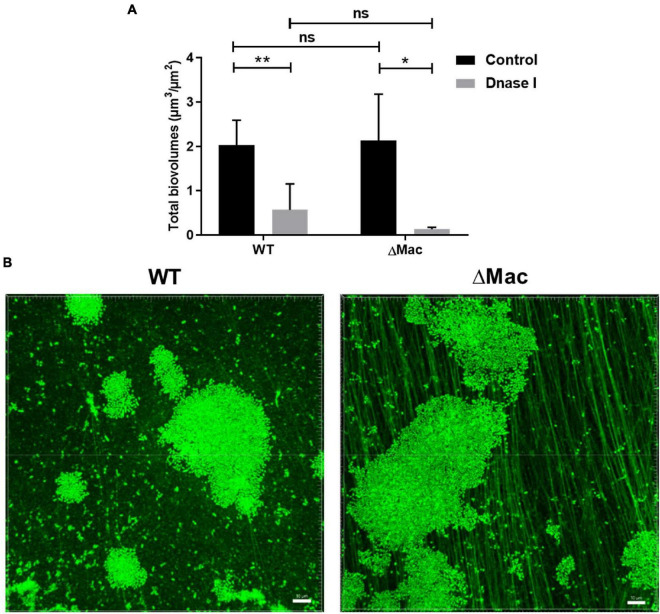
As the MacAB-TolC system was already shown to contribute to the antibiotic resistance in A. baumannii and various bacterial species (Greene et al., 2018; Pérez-Varela et al., 2019), we compared MICs of WT and ΔMac strains. In the present study, we did not observe any difference between both strains (Supplementary Table 5). The deletion of the MacB transporter was, however, shown to be associated with a slight decrease in erythromycin (from 4 to 2 μg/ml) and novobiocin MICs (from 8 to 2 μg/ml; Pérez-Varela et al., 2019). This discrepancy might be linked to the deletion of the entire system instead of macB alone. Here, MacAB-TolC does not seem to participate in the antibiotic efflux in A. baumannii in a planktonic growth mode or the expression of another efflux pump may counteract the deletion of the Mac system in the ΔMac strain. Regarding the antibiotic tolerance in biofilms, MBEC assays revealed that ΔMac was surprisingly more tolerant to aminoglycosides, such as gentamicin (128 μg/ml) and tobramycin (64 μg/ml), than the WT strain (32 μg/ml). ΔMac_c strain showed a restored phenotype with MBEC at 16 μg/ml for gentamicin and 32 μg/ml for tobramycin. Hence, to investigate this difference, a biofilm model at the solid–liquid–air interface (three-phase interface), similar to the one present in the Calgary biofilm device, was performed for CLSM imaging (Supplementary Figure 1). After 24 h of incubation, all bacteria (live and dead cells) and biofilm matrices were stained with Syto9. There was no difference in the biovolume at the three-phase interface (Figure 3A) contrary to the one observed at the S-L interface (Figure 2B). This is consistent with our proteomic data pointing out that the overexpression of MacAB-TolC happens essentially when A. baumannii grows at the S-L interface and not in pellicle (Kentache et al., 2017). However, ΔMac biofilm images at the three-phase interface showed a well-developed fiber-like network within the EPS matrix. These fibers are extracellular DNA (eDNA), since this component was labeled with Syto9 and was also degraded by DNAse I (Figure 3). Indeed, DNase I treatment led to a significant reduction of biofilm formation for both WT and ΔMac (72 and 95%, respectively) after 24-h incubation (Figure 3A). In A. baumannii, eDNA was shown to be released either by an active mode, in a free form or encapsulated in membrane vesicles during early biofilm growth phase, or by cell lysis contributing to the regrowth of freshly dispersed cells (Sahu et al., 2012). A kinetic profile of biofilm formation at the three-phase interface was also performed in our study. As these eDNA-containing fibers were mainly observed from 24 h of growth (data not shown), these eDNA fibers may mainly originate from cell lysis.
FIGURE 3.
Impact of macAB-tolC deletion on biofilm formation at the solid-liquid-air interface. (A) Total biovolume (μm3/μm2) of 24-h-old A. baumannii ATCC 17978 WT and ΔMac biofilms after exposure to DNase I (100 μg/ml) compared to untreated control biofilms. Quantification of biovolume was based on the confocal fluorescence images of Syto9 stained biofilms. Data shown represent mean values (±SD) from at least three independent biological experiments (ns, not significant; *p < 0.05; **p < 0.01). (B) Representative confocal fluorescence images of A. baumannii ATCC 17978 WT (left) and ΔMac biofilms (right) labeled with Syto9 at the solid–liquid–air interface (three-phase interface). Image construction was carried out using Imaris software. Scale bar: 10 μm.
It has been largely reported that aminoglycosides could have an impaired penetration in P. aeruginosa biofilms due to their interaction with negative components of the matrix (alginate or eDNA; Alipour et al., 2009). In line with this observation, exogenous DNA addition to biofilm growth medium was shown to provide a shield effect against aminoglycosides (Chiang et al., 2013). Here, in the ΔMac mutant, it is likely that an excess of eDNA could promote divalent cation sequestration and block aminoglycoside antibiotic diffusion within the matrix, thus explaining the observed tobramycin and gentamicin tolerance increase (Tseng et al., 2013). Although colistin can interfere with the electrostatic network of the EPS matrix (Klinger-Strobel et al., 2017), eDNA does not seem here to induce a colistin biofilm tolerance.
3.6 MacAB-TolC Contributes to the Envelope Stress Response
Gram-negative bacteria possess a complex envelope to adapt their physiology to environmental conditions. This adaptation is highly controlled by two-component systems (TCSs) (De Silva and Kumar, 2019). In A. baumannii, MacAB-TolC is regulated by BaeSR, a TCS that detects environmental stresses, like specific envelope-damaging agents and high osmolarity conditions. Moreover, BaeSR modulates the expression of other efflux pumps such as AdeIJK and AdeABC, which are involved in cell detoxification and maintenance (Lin M. F. et al., 2014; Lin et al., 2015).
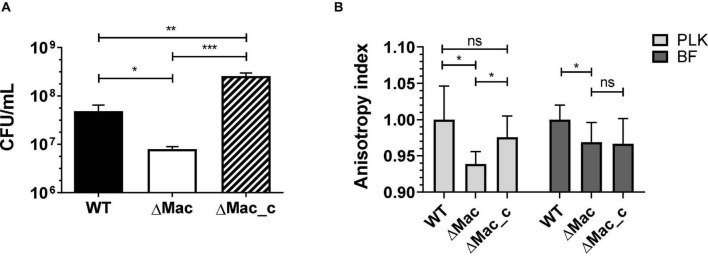
Lin et al. (2015) have demonstrated by phenotype microarray experiment the ΔbaeR mutant susceptibility and an upregulation of macB (6.2-fold) in response to a tannic acid (TA) treatment. Herein, we determined TA MICs on WT and ΔMac deletion mutant and found accordingly that the WT strain was more resistant to TA (512 μg/ml) than the ΔMac strain (128 μg/ml). Complementation partially restored the resistance to this tannin (256 μg/ml in ΔMac_c). The ΔMac susceptibility to TA was confirmed using spot assay (Supplementary Figure 3). At least 108 CFU/ml of ΔMac were inhibited by 2,048 μg/ml of TA, whereas the same concentration of tannin inhibited only 105 CFU/ml of WT and 106 CFU/ml of the complemented strain. The same experiments were performed with GA, a degradation product of TA (Tahmourespour et al., 2016), but no significant difference between the WT and the ΔMac strains was observed (Supplementary Figure 3). Thus, MacAB-TolC may be required to allow A. baumannii survival in the presence of high doses of TA through either a TA efflux or a degradation process. Since TA is also an iron-chelating and antioxidant agent (Lin et al., 2015, 2020), the incapacity of the ΔMac strain to survive at high doses of TA could also be due to its impairment in maintaining iron homeostasis.
Moreover, we compared the tolerance to envelope stress of the WT, ΔMac, and ΔMac_c strains in high-osmolarity conditions. Interestingly, the ΔMac mutant was less tolerant to 500 mM sucrose after 24 h of growth as compared to the WT strain (Figure 4A). Susceptibility to this high-osmolarity condition was restored in ΔMac_c complemented mutant. These results are consistent with the study demonstrating that the expression of baeR was increased by twofold in response to 20% sucrose (Lin M. F. et al., 2014). To counteract high-osmolarity conditions and to survive this environmental stress, microorganisms may change their membrane composition, thus impacting membrane fluidity (Beney and Gervais, 2001). Accordingly, we measured the membrane fluidity of WT, ΔMac, and ΔMac_c strains by fluorescence anisotropy method (Figure 4B). The anisotropy index indicated that the ΔMac mutant presented a higher membrane fluidity than that in the WT strain, either in planktonic suspension or in biofilm. The membrane fluidity of ΔMac_c was partially and significantly restored as compared to the WT one, in planktonic but not in sessile lifestyle, probably due to the lack of antibiotic selection pressure.
FIGURE 4.
Tolerance to sucrose and membrane fluidity analyses of WT vs. ΔMac and ΔMac_c derivative strains of A. baumannii. (A) The 24 h growth measurements in high-osmolarity condition (500 mM sucrose). (B) Fluorescence anisotropy measurements of planktonic (PLK) growth culture (24 h; clear gray) or biofilm (BF) growth culture (48 h; dark gray). Data shown represent mean values (±SD) from at least three independent experiments. The data were statistically analyzed using unpaired t-test to calculate p-values (ns, not significant; *p < 0.05; **p < 0.01; ***p < 0.001). CFU/mL, colony-forming unit per ml.
These results demonstrated that MacAB-TolC contributes to maintain WT membrane rigidity and allow the tolerance to high-osmolarity conditions. This is in agreement with the study of Henry et al. (2012), highlighting that the rebuilding and rigidity maintenance of the membrane of a lipopolysaccharide (LPS)-deficient mutant is concomitant with the overexpression (28- to 39-fold) of the macAB-tolC system under the regulation of BaeS/R. In a similar manner, colistin treatment causing major membrane damages, i.e., an important envelope stress, induced the overexpression of macAB-tolC as a cell wall maintenance response (Henry et al., 2014). Of note, the emrAB efflux pump contributes in a similar manner to osmotic stress and colistin resistance in A. baumannii (Lin et al., 2017). Recently, it was also reported to be involved in biofilm formation (Lin et al., 2020). As mentioned above, BaeS/R positively regulates AdeIJK and AdeABC together with MacAB-TolC (Lin M. F. et al., 2014; Lin et al., 2015). In our proteomic data (Supplementary Tables 3, 4), these efflux pumps were overrepresented in biofilm and could therefore contribute to antibiotic tolerance in this mode of growth.
When the deletion of AdeB led to a significant decrease of biofilm formation similarly to what the deletion of MacAB-TolC did, the deletion of BaeR moderately impacted biofilm development (Lin et al., 2020). It is tempting to speculate that an effective antibiofilm strategy would be directed toward the design of a broad efflux pump inhibitor rather than to prevent a TCS activity.
3.7 Biofilm Proteome Reveals Disrupted Iron Homeostasis in ΔMac
To further understand the contribution of MacAB-TolC to A. baumannii biofilm formation, we proceeded to a large-scale proteomic analysis of WT and ΔMac strains in the conditions used to highlight MacAB-TolC overexpression (see Proteomic Analyses of Planktonic and Sessile Bacterial Cultures section). We analyzed intracellular and membrane compartments and identified 48 proteins with varying expression levels. Among them, 37 proteins were underexpressed (Table 1), whereas 11 were overexpressed in the ΔMac strain (Table 2).
TABLE 1.
Proteins under-represented in solid–liquid biofilm of ΔMac.
| Label ABYAL | Label A1S_ | Fold change | Gene | Description | Fraction | Peptides | Confidence | Anova (p) |
| MacAB-TolC system | ||||||||
| ABYAL0571 | A1S_0535 | 18.0 | tolC | Outer membrane protein | M | 2 | 89.64 | 1.1E-03 |
| ABYAL0574 | A1S_0538 | 12.9 | macA | ABC tripartite efflux pump membrane fusion protein | M | 5 | 216.38 | 1.5E-10 |
| Siderophore | ||||||||
| ABYAL1976 | A1S_1657 | 2.6 | bfnL | Baumannoferrin biosynthesis protein | I | 3 | 137.34 | 5.1E-03 |
| ABYAL2846 | A1S_2375 | 3.1 | barB | Siderophore ABC transporter | M | 13 | 716.65 | 1.6E-03 |
| ABYAL2852 | A1S_2380 | 3.2 | basF | Isochorismatase | I+M | 3 | 109.43 | 3.8E-03 |
| ABYAL2853 | A1S_2381 | 2.8 | basE | 2,3-Dihydroxybenzoate-AMP ligase/S-dihydroxybenzoyltransferase | I | 2 | 168.79 | 5.1E-03 |
| ABYAL2858 | A1S_2386 | 2.3 | bauB | Acinetobactin periplasmic binding protein | M | 7 | 389.76 | 7.5E-03 |
| ABYAL2863 | A1S_2390 | 2.3 | basB | Acinetobactin biosynthesis protein | M | 3 | 161.11 | 7.6E-03 |
| Sulfur | ||||||||
| ABYAL0038 | A1S_0028 | 2.2 | ssuD | FMNH(2)-dependent alkanesulfonate monooxygenase | I+M | 4 | 202.00 | 3.4E-03 |
| ABYAL0039 | A1S_0029 | 2.5 | ssuA | Aliphatic sulfonate ABC transporter periplasmic | M | 2 | 103.65 | 4.2E-04 |
| ABYAL0040 | A1S_0030 | 2.5 | ssuA | Aliphatic sulfonate ABC transporter periplasmic | M | 2 | 88.15 | 1.3E-03 |
| ABYAL3025 | A1S_2537 | 2.3 | ssuR | DNA-binding transcriptional activator (LysR-family) | M | 3 | 147.98 | 2.1E-04 |
| ABYAL3888 | A1S_3305 | 2.1 | msuE | NADH-dependent FMN reductase | M | 2 | 96.85 | 3.7E-03 |
| ABYAL3889 | A1S_3306 | 3.1 | msuD | FMNH(2)-dependent dimethylsulfone monooxygenase | I+M | 4 | 259.99 | 3.8E-04 |
| ABYAL1751 | A1S_1485 | 2.4 | metQ | Methionine ABC transporter permease | M | 8 | 392.49 | 4.3E-04 |
| PAA degradation | ||||||||
| ABYAL1576 | A1S_1335 | 15.2 | paaZ | Oxepin-CoA hydrolase/dehydrosuberyl-CoA semialdehyde dehydrogenase | I+M | 5 | 221.52 | 4.9E-03 |
| ABYAL1577 | A1S_1336 | 9.6 | paaA | 1,2-phenylacetyl-CoA epoxidase subunit A | I | 3 | 113.1 | 7.7E-04 |
| ABYAL1583 | A1S_1342 | 9.2 | paaG | 2-(1,2-epoxy-1.2-dihydrophenyl)acetyl-CoA isomerase | I+M | 8 | 394.61 | 3.1E-03 |
| ABYAL1584 | A1S_1343 | 9.6 | paaH | 3-hydroxybutyryl-CoA dehydrogenase | I | 3 | 178.95 | 2.6E-03 |
| ABYAL1585 | A1S_1344 | 12.4 | paaJ | Beta-ketoadipyl-CoA thiolase | I | 2 | 123.36 | 6.4E-03 |
| Secretion system | ||||||||
| ABYAL1534 | A1S_1296 | 2.5 | hcp1 | Type VI secretion system effector | M | 14 | 1017.28 | 2.6E-07 |
| ABYAL3106 | A1S_2602 | 4.1 | rbtA | Rhombotarget A | M | 3 | 102.26 | 3.2E-04 |
| Others | ||||||||
| ABYAL0608 | A1S_0569 | 2.0 | Short-chain dehydrogenase/reductase | M | 2 | 86.33 | 1.1E-03 | |
| ABYAL1300 | A1S_1126 | 3.6 | Baeyer–Villiger monooxygenase | M | 3 | 149.25 | 1.5E-04 | |
| ABYAL1493 | A1S_1264 | 2.2 | Class A β-lactamase-related serine hydrolase | M | 2 | 105.9 | 1.4E-03 | |
| ABYAL1530 | A1S_1292 | 2.4 | Conserved hypothetical protein | M | 2 | 48.99 | 3.4E-05 | |
| ABYAL1698 | A1S_1439 | 2.3 | Luciferase-like monooxygenase | I | 2 | 129.35 | 9.8E-05 | |
| ABYAL1742 | A1S_1478 | 2.4 | Conserved hypothetical protein | I | 3 | 128.33 | 9.3E-05 | |
| ABYAL1831 | A1S_1551 | 2.1 | parA | ATPase chromosome partitioning protein | M | 2 | 127.04 | 8.0E-05 |
| ABYAL2029 | A1S_1700 | 2.0 | acoB | Acetoin:2,6-dichlorophenolindophenol oxidoreductase beta subunit | M | 2 | 127.54 | 1.5E-03 |
| ABYAL2289 | A1S_1922 | 2.5 | Ribokinase | M | 2 | 96.2 | 3.3E-04 | |
| ABYAL2361 | 2.2 | Conserved hypothetical protein | I | 3 | 127.91 | 2.2E-03 | ||
| ABYAL2518 | A1S_2084 | 3.4 | pheA | Secreted chorismate mutase | M | 3 | 180.53 | 1.8E-05 |
| ABYAL2931 | A1S_2452 | 3.6 | styD | Phenylacetaldehyde dehydrogenase | I+M | 8 | 448.54 | 1.3E-04 |
| ABYAL3342 | A1S_2820 | 2.8 | Conserved hypothetical protein | I | 2 | 122.24 | 1.7E-03 | |
| ABYAL3515 | A1S_2957 | 2.2 | Zn-dependent hydrolase | M | 2 | 101.57 | 2.3E-05 | |
| ABYAL4020 | A1S_3418 | 2.7 | hpd | 4-hydroxyphenylpyruvate dioxygenase | I | 4 | 210.88 | 6.0E-03 |
“M” for membrane fraction and “I” for intracellular fraction.
TABLE 2.
Over-represented proteins in solid–liquid biofilm of ΔMac.
| Label ABYAL | Label A1S_ | Fold change | Gene | Description | Fraction | Peptides | Confidence | Anova (p) |
| Quorum sensing | ||||||||
| ABYAL0138 | A1S_0115 | 2.7 | Non-ribosomal peptide synthetase (NRPS) | I | 2 | 93.95 | 9.4E-04 | |
| ABYAL0139 | A1S_0116 | 2.3 | Resistance-nodulation-division (RND) transporter (Ac-505 secretion) | M | 5 | 259.28 | 2.0E-03 | |
| Others | ||||||||
| ABYAL1401 | A1S_1191 | 2.1 | pyrX | Aspartate carbamoyltransferase | M | 4 | 214.67 | 1.6E-03 |
| ABYAL1402 | A1S_1192 | 2.3 | pyrX | Aspartate carbamoyltransferase | M | 2 | 169.65 | 4.2E-05 |
| ABYAL1640 | A1S_1387 | 2.0 | yhdF | Short-chain dehydrogenase reductase | M | 2 | 56.02 | 9.3E-03 |
| ABYAL2806 | A1S_2338 | 2.2 | maeB | Malate dehydrogenase | M | 20 | 1084.58 | 2.4E-05 |
| ABYAL2984 | A1S_2501 | 2.2 | gap | Glyceraldehyde-3-phosphate dehydrogenase | I | 2 | 97.82 | 1.3E-05 |
| ABYAL3089 | A1S_2586 | 2.0 | dgt2 | Deoxyguanosinetriphosphate triphosphohydrolase-like protein | M | 2 | 92.13 | 6.3E-04 |
| ABYAL3806 | A1S_3231 | 2.7 | cat | Succinyl-CoA coenzyme A transferase | I | 2 | 97.74 | 4.1E-06 |
| ABYAL3914 | A1S_3327 | 2.2 | aceF | Dihydrolipoamide acyltransferase (E2) component | M | 10 | 587.81 | 2.8E-05 |
| ABYAL4005 | A1S_3403 | 2.0 | hutI | Imidazolonepropionase | I | 3 | 147.31 | 1.0E-04 |
“M” for membrane fraction and “I” for intracellular fraction.
All bacterial cells need iron and have thus developed iron-uptake pathways to scavenge iron from their host during infection. A. baumannii ATCC 17978 produces up to 10 siderophores that chelate iron with high affinity from three different loci, named acinetobactin, baumannoferrin (A and B), and the fimsbactins (A–F) (Sheldon and Skaar, 2020). Acinetobactin and the fimsbactins are mixed catechol-hydroxamate-type siderophores, whereas baumannoferrin has solely a hydroxamate-type structure (Yamamoto et al., 1994; Proschak et al., 2013; Penwell et al., 2015). It was already reported that these siderophores and iron requirement were critical for the development of A. baumannii communities (Nait Chabane et al., 2014; Kentache et al., 2017). Accordingly, the ATCC 17978 S-L biofilm proteomic analysis pointed out the underexpression of the negative regulator Fur (Ferric uptake regulator). It inhibits the expression of siderophore synthesis and promoted iron storage (Cornelis et al., 2011). Consistently, BauA (10-fold) was involved in acinetobactin import, and proteins involved in baumannoferrin transport (BfnH, threefold; TonB-dependent receptor, 26-fold) were accumulated (Supplementary Table 3).
Interestingly, in the ΔMac proteome analysis, the BfnL protein, involved in the biosynthesis of the baumannoferrin, and five other proteins related to the acinetobactin locus (BasB, BasE, and BasF for biosynthesis; BauB for import; and BarB for export) were underexpressed compared to those in the WT strain (Table 1). However, a potential decrease of baumannoferrin biosynthesis in ΔMac should be considered with caution, since only the BfnL amount decreased (Sheldon and Skaar, 2020). Nevertheless, there was no doubt regarding the decrease of acinetobactin production. Likewise, in P. aeruginosa, a deletion mutant of PvdRT-OpmQ, an efflux pump sharing a high structural similarity with MacAB-TolC, presented a downregulation of pyoverdin biosynthesis (Imperi et al., 2009). This PvdRT-OpmQ system was proposed to be responsible for pyoverdin recycling and/or required for its secretion when newly synthesized (Imperi et al., 2009; Hannauer et al., 2010). It is thus tempting to propose that the MacAB-TolC system could participate in the secretion and/or recycling of acinetobactin from the periplasm to the extracellular medium. We performed CAS assays to examine this hypothesis, but as already mentioned (Sheldon and Skaar, 2020), we did not detect any variation of siderophore activity in ΔMac compared to WT in disrupting only the acinetobactin pathway (Supplementary Figure 4A).
In ΔMac, the sulfonate-sulfur utilization step of cysteine biosynthesis pathway was notably affected with the downregulation of five proteins (Table 1) with: (i) and (ii) SsuA involved in aliphatic sulfonate import and SsuD involved in desulfonation of aliphatic sulfonates, both are members of the SsuEADCB system; (iii) SsuR the regulator of this system; (iv) and (v) MsuE and MsuD also involved in desulfonation of other aliphatic sulfonate (Kertesz et al., 1999). Moreover, MetQ responsible for the methionine import, an organosulfur source other than sulfonates (Kertesz et al., 1999), was also underexpressed (Table 1). Altogether, these results showed a potential decrease of the cysteine biosynthesis. This particular amino acid is crucial for [Fe-S] cluster biosynthesis. Indeed, iron–sulfur cluster (ISC) and sulfur formation (SUF) pathways, directly regulated by cellular iron status, use free cysteine to liberate sulfur atoms for [Fe-S] cluster assembly (Guédon and Martin-Verstraete, 2006; Ayala-Castro et al., 2008). In ΔMac, iron homeostasis deregulation might compromise [Fe-S] cluster status. Of interest, IscR regulator of [Fe-S] cluster synthesis (Ayala-Castro et al., 2008) and two chaperones involved in this pathway, HscA (3-fold) and HscB (4-fold) were downregulated in ATCC 17978 S-L biofilm (Supplementary Table 3) consistently with an iron limitation in these growth conditions (Vickery and Cupp-Vickery, 2007).
Finally, the highest protein fold changes (up to 15-fold) in ΔMac were obtained for five proteins: PaaA, PaaG, PaaZ, PaaJ, and PaaH, which were highly underrepresented (Table 1). These proteins belong to the phenylacetate (PAA) catabolic pathway that allows the degradation of aromatic compounds to produce acetyl-coA and succinyl-coA for the trichloroacetic acid (TCA) cycle (Teufel et al., 2010; Cerqueira et al., 2014). Considering the function of this pathway, we investigated the capacity of ΔMac to grow on M9 agar plate with phenylacetic acid or phenylalanine. We did not observe a difference between the ΔMac and the WT strains (Supplementary Figure 4B). However, iron availability and/or the unbalanced cellular iron status of the ΔMac mutant may influence the synthesis of this operon as described (Nwugo et al., 2011). Here, the unbalanced cellular iron status on the ΔMac mutant may also influence the PAA expression. It is known that the PAA catabolic pathway, under the control of the global virulence regulator GacA, is involved in A. baumannii virulence (Cerqueira et al., 2014). Virulence assays conducted on the ΔMac strain in the model organism Caenorhabditis elegans model did not, however, allow us to observe attenuated virulence compared to WT (data not shown, Pérez-Varela et al., 2019). Similarly, in the ATCC 17978 S-L biofilm, five proteins of the PAA pathway and GacA were downregulated (Supplementary Table 3). This is remarkably different from our proteomic analysis of A. baumannii pellicle (Kentache et al., 2017) and suggests that A. baumannii virulence mediated by paa locus and GacA is strictly associated with pellicle formation (Marti et al., 2011a).
3.8 MacAB-TolC Limits Adhesion to Human Alveolar Epithelial Cells
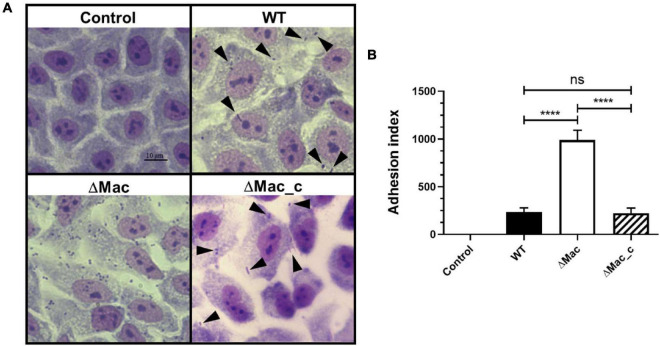
Bacterial adherence to target cells is the first step of the infectious process. Interestingly, in our proteomic study, we identified two proteins, ABYAL0138 (A1S_0115, 2.7-fold) and ABYAL0139 (A1S_0116, 2.3-fold), overrepresented in the ΔMac mutant that are involved in the synthesis and transport of acinetin. They are part of the A1S_0112-A1S_0119 operon already described to contribute to biofilm formation and potential acinetin secretion but also to the interaction with eukaryotic cells and in virulence (Rumbo-Feal et al., 2017). The biological effect of the macAB-tolC deletion was therefore tested using A549 human alveolar epithelial cells as a model, since they represent a host cell that could be targeted by A. baumannii during respiratory infections. The ΔMac strain shows a remarkable ability (fivefold more) to attach to ATII-like phenotype A549 differentiated cells compared to the A. baumannii 17978 parental strain (Figure 5). The results obtained with the ATII-like phenotype A549 cells cultured in Ham’s F-12 Nutrient Mixture for at least 20 days were also confirmed by infecting A549 cells cultured in DMEM with the A. baumannii WT or the ΔMac derivative strains (data not shown). When the mutant strain was complemented, the phenotype was restored. The observed increase of bacterial adherence to epithelial cells when macAB-tolC was deleted is presumably due to the fact that this strain produced more eDNA that is a cell–cell interconnecting compound (Whitchurch et al., 2002) and overexpressed the A1S_0112-A1S_0119 operon contributing to the interaction with eukaryotic cells (Rumbo-Feal et al., 2017).
FIGURE 5.
Adhesion of A. baumannii to A549 human alveolar epithelial cells. (A) ATII-like phenotype A549 differentiated cells were infected with A. baumannii ATCC 17978 (WT), ΔMac, and ΔMac_c (ΔMac complemented) at a multiplicity of infection (MOI) of 20. Negative control corresponds to cells with no bacteria added. Black arrows indicate bacteria attached to A549 cells (magnification, ×400). (B) Quantification of the adherence of A. baumannii ATCC 17978 WT, ΔMac, and ΔMac_c strains to A549 cells. Index of attached bacteria after 24 h of infection. Data shown represent mean values (±SD) from at least three independent experiments performed at least in triplicate. Student’s t-test was used to validate the experimental data (ns, not significant; ****p < 0.0001).
4. Conclusion
Even though a positive correlation between biofilm formation and antimicrobial resistance is still debated, clinical A. baumannii strains presenting concomitantly a biofilm-forming capacity and a multidrug resistance are currently isolated (Badave and Kulkarni, 2015; Qi et al., 2016; Wang et al., 2018). Indeed, increasing evidence demonstrated that efflux pumps are key actors of antibiotic resistance and also play a role in biofilm formation. They could efflux QS or quorum quenching molecules, as well as EPSs, but also harmful accumulated molecules and can thus promote or regulate biofilm formation (Alav et al., 2018). In A. baumannii, MFS and RND-efflux pumps may participate in eDNA release, transport of autoinducer molecules, or adhesion process, but this involvement was suggested to be strain-dependent (Sahu et al., 2012; He et al., 2015; Yoon et al., 2015; Richmond et al., 2016; Lin et al., 2020). Here, we demonstrated that the MacAB-TolC pump is commonly overexpressed in mature S-L biofilms of SDF and ATCC17978 strains. This system, being involved in osmotic protection and probably maintenance of iron homeostasis, may help A. baumannii not only to face deleterious conditions present in mature biofilms, where severe ionic gradients can develop. It could help to detoxify cell to persist and to fit in harsh environments, even though the precise substrates of this pump remain to be characterized. Its role in cell wall maintenance demonstrated here is in agreement with its overexpression when bacteria are facing membrane-targeted antibiotics such as colistin (Henry et al., 2014). Shedding some light on the roles of efflux pumps in the biofilm formation may help to develop therapeutic strategies and to improve treatments of biofilm-related infections as well. Design of broad-spectrum and safe efflux pump inhibitors would be valuable tools to both decrease the bacterial biofilm development and restore the activity of antimicrobials.
Data Availability Statement
The datasets presented in this study can be found in online repositories. The names of the repositories and accession number(s) can be found in the “Materials and Methods” section.
Author Contributions
AP, VP, and ED contributed to conception and design of the study. MN, BR, HL, AT, AS, J-BV, DV, OL, and VP performed the experiments. MN, BR, HL, AT, TJ, AP, VP, and ED wrote the article. All authors approved the submitted version.
Conflict of Interest
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Publisher’s Note
All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article, or claim that may be made by its manufacturer, is not guaranteed or endorsed by the publisher.
Acknowledgments
Images were obtained on PRIMACEN (https://primacen.crihan.fr), the Cell Imaging Platform of Normandy, IRIB, Faculty of Sciences, University of Rouen, 76821 Mont-Saint-Aignan.
Funding
This work was supported by the Normandie Region (SéSAD Research Network, France) and European Union. Europe gets involved in Normandie with European Regional Development Fund. BR was a recipient of a French fellowship of the Ministère de l’Enseignement Supérieur et de la Recherche.
Supplementary Material
The Supplementary Material for this article can be found online at: https://www.frontiersin.org/articles/10.3389/fmicb.2021.785161/full#supplementary-material
References
- Alav I., Sutton J. M., Rahman K. M. (2018). Role of bacterial efflux pumps in biofilm formation. J. Antimicrob. Chemother. 73 2003–2020. 10.1093/jac/dky042 [DOI] [PubMed] [Google Scholar]
- Alipour M., Suntres Z. E., Omri A. (2009). Importance of DNase and alginate lyase for enhancing free and liposome encapsulated aminoglycoside activity against Pseudomonas aeruginosa. J. Antimicrob. Chemother. 64 317–325. 10.1093/JAC/DKP165 [DOI] [PubMed] [Google Scholar]
- Alvarez H. M. (2016). Triacylglycerol and wax ester-accumulating machinery in prokaryotes. Biochimie 120 28–39. 10.1016/J.BIOCHI.2015.08.016 [DOI] [PubMed] [Google Scholar]
- Alvarez H. M., Silva R. A., Cesari A. C., Zamit A. L., Peressutti S. R., Reichelt R., et al. (2004). Physiological and morphological responses of the soil bacterium Rhodococcus opacus strain PD630 to water stress. FEMS Microbiol. Ecol. 50 75–86. 10.1016/j.femsec.2004.06.002 [DOI] [PubMed] [Google Scholar]
- Antunes L. C. S., Imperi F., Carattoli A., Visca P. (2011). Deciphering the multifactorial nature of Acinetobacter baumannii pathogenicity. PLoS One 6:e22674. 10.1371/JOURNAL.PONE.0022674 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Astaneh S. D. A., Rasooli I., Gargari S. L. M. (2017). Filamentous hemagglutinin adhesin FhaB limits A baumannii biofilm formation. Front. Biosci. Elit. 9:266–275. 10.2741/e801 [DOI] [PubMed] [Google Scholar]
- Astaneh S. D. A., Rasooli I., Mousavi Gargari S. L. (2014). The role of filamentous hemagglutinin adhesin in adherence and biofilm formation in Acinetobacter baumannii ATCC19606T. Microb. Pathog. 74 42–49. 10.1016/j.micpath.2014.07.007 [DOI] [PubMed] [Google Scholar]
- Ayala-Castro C., Saini A., Outten F. W. (2008). Fe-S cluster assembly pathways in bacteria. Microbiol. Mol. Biol. Rev. 72 110–125. 10.1128/mmbr.00034-07 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Badave G. K., Kulkarni D. (2015). Biofilm producing multidrug resistant Acinetobacter baumannii: an emerging challenge. J. Clin. Diagnostic Res. 9 DC08–DC10. 10.7860/JCDR/2015/11014.5398 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Benamara H., Rihouey C., Jouenne T., Alexandre S. (2011). Impact of the biofilm mode of growth on the inner membrane phospholipid composition and lipid domains in Pseudomonas aeruginosa. Biochim. Biophys. Acta Biomembr. 1808 98–105. 10.1016/J.BBAMEM.2010.09.004 [DOI] [PubMed] [Google Scholar]
- Beney L., Gervais P. (2001). Influence of the fluidity of the membrane on the response of microorganisms to environmental stresses. Appl. Microbiol. Biotechnol. 57 34–42. 10.1007/s002530100754 [DOI] [PubMed] [Google Scholar]
- Bogomolnaya L. M., Tilvawala R., Elfenbein J. R., Cirillo J. D., Andrews-Polymenis H. L. (2020). Linearized siderophore products secreted via MacAB efflux pump protect Salmonella enterica serovar Typhimurium from oxidative stress. MBio 11:e00528-20. 10.1128/mBio.00528-20 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bosdriesz E., Molenaar D., Teusink B., Bruggeman F. J. (2015). How fast-growing bacteria robustly tune their ribosome concentration to approximate growth-rate maximization. FEBS J. 282 2029–2044. 10.1111/FEBS.13258 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Boucher H. W., Talbot G. H., Bradley J. S., Edwards J. E., Gilbert D., Rice L. B., et al. (2009). Bad bugs, no drugs: no ESKAPE! An update from the infectious diseases society of America. Clin. Infect. Dis. 48 1–12. 10.1086/595011 [DOI] [PubMed] [Google Scholar]
- Cabral M. P., Soares N. C., Aranda J., Parreira J. R., Rumbo C., Poza M., et al. (2011). Proteomic and functional analyses reveal a unique lifestyle for Acinetobacter baumannii biofilms and a key role for histidine metabolism. J. Proteome Res. 10 3399–3417. 10.1021/pr101299j [DOI] [PubMed] [Google Scholar]
- Ceri H., Olson M. E., Stremick C., Read R. R., Morck D., Buret A. (1999). The calgary biofilm device: new technology for rapid determination of antibiotic susceptibilities of bacterial biofilms. J. Clin. Microbiol. 37 1771–1776. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cerqueira G. M., Kostoulias X., Khoo C., Aibinu I., Qu Y., Traven A., et al. (2014). A global virulence regulator in Acinetobacter baumannii and its control of the phenylacetic acid catabolic pathway. J. Infect. Dis. 210 46–55. 10.1093/infdis/jiu024 [DOI] [PubMed] [Google Scholar]
- Chiang W. C., Nilsson M., Jensen P. Ø, Høiby N., Nielsen T. E., Givskov M., et al. (2013). Extracellular DNA shields against aminoglycosides in Pseudomonas aeruginosa biofilms. Antimicrob. Agents Chemother. 57 2352–2361. 10.1128/AAC.00001-13 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Clinical and Laboratory Standards Institute (CLSI) (2015). M07-A10: Methods for Dilution Antimicrobial Susceptibility Tests for Bacteria That Grow Aerobically. Wayne, PA: CLSI. [Google Scholar]
- Cooper J. R., Abdullatif M. B., Burnett E. C., Kempsell K. E., Conforti F., Tolley H., et al. (2016). Long term culture of the a549 cancer cell line promotes multilamellar body formation and differentiation towards an alveolar type II pneumocyte phenotype. PLoS One 11:e0164438. 10.1371/JOURNAL.PONE.0164438 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cornelis P., Wei Q., Andrews S. C., Vinckx T. (2011). Iron homeostasis and management of oxidative stress response in bacteria. Metallomics 3 540–549. 10.1039/C1MT00022E [DOI] [PubMed] [Google Scholar]
- Crouzet M., Claverol S., Lomenech A.-M., Le Sénéchal C., Costaglioli P., Barthe C., et al. (2017). Pseudomonas aeruginosa cells attached to a surface display a typical proteome early as 20 minutes of incubation. PLoS One 12:e0180341. 10.1371/journal.pone.0180341 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Crow A., Greene N. P., Kaplan E., Koronakis V. (2017). Structure and mechanotransmission mechanism of the MacB ABC transporter superfamily. Proc. Natl. Acad. Sci. U.S.A. 114 12572–12577. 10.1073/pnas.1712153114 [DOI] [PMC free article] [PubMed] [Google Scholar]
- De Silva P. M., Kumar A. (2019). Signal transduction proteins in Acinetobacter baumannii: role in antibiotic resistance, virulence, and potential as drug targets. Front. Microbiol. 10, 1–12. 10.3389/fmicb.2019.00049 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dubois-Brissonnet F., Trotier E., Briandet R. (2016). The biofilm lifestyle involves an increase in bacterial membrane saturated fatty acids. Front. Microbiol. 7:1673. 10.3389/fmicb.2016.01673 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Eijkelkamp B. A., Stroeher U. H., Hassan K. A., Paulsen I. T., Brown M. H. (2014). Comparative analysis of surface-exposed virulence factors of Acinetobacter baumannii. BMC Genomics 15:1020. 10.1186/1471-2164-15-1020 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fitzpatrick A. W. P., Llabrés S., Neuberger A., Blaza J. N., Bai X.-C., Okada U., et al. (2017). Structure of the MacAB–TolC ABC-type tripartite multidrug efflux pump. Nat. Microbiol. 2:17070. 10.1038/nmicrobiol.2017.70 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fournier P. E., Vallenet D., Barbe V., Audic S., Ogata H., Poirel L., et al. (2006). Comparative genomics of multidrug resistance in Acinetobacter baumannii. PLoS Genet. 2:e0020007. 10.1371/journal.pgen.0020007 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fulaz S., Hiebner D., Barros C. H. N., Devlin H., Vitale S., Quinn L., et al. (2019). Ratiometric imaging of the in Situ pH distribution of biofilms by use of fluorescent mesoporous silica nanosensors. ACS Appl. Mater. Interfaces 11 32679–32688. 10.1021/ACSAMI.9B09978 [DOI] [PubMed] [Google Scholar]
- Gaddy J. A., Tomaras A. P., Actis L. A. (2009). The Acinetobacter baumannii 19606 OmpA protein plays a role in biofilm formation on abiotic surfaces and in the interaction of this pathogen with eukaryotic cells. Infect. Immun. 77 3150–3160. 10.1128/IAI.00096-09 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gayoso C. M., Mateos J., Méndez J. A., Fernández-Puente P., Rumbo C., Tomás M., et al. (2014). Molecular mechanisms involved in the response to desiccation stress and persistence in Acinetobacter baumannii. J. Proteome Res. 13 460–476. 10.1021/pr400603f [DOI] [PubMed] [Google Scholar]
- Greene N. P., Kaplan E., Crow A., Koronakis V. (2018). Antibiotic resistance mediated by the MacB ABC transporter family: a structural and functional perspective. Front. Microbiol. 9:950. 10.3389/fmicb.2018.00950 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Guédon E., Martin-Verstraete I. (2006). “Cysteine metabolism and its regulation in bacteria,” in Amino Acid Biosynthesis ~ Pathways, Regulation and Metabolic Engineering, ed. Wendisch V. F. (Berlin: Springer; ), 195–218. 10.1007/7171_2006_060 [DOI] [Google Scholar]
- Hannauer M., Yeterian E., Martin L. W., Lamont I. L., Schalk I. J. (2010). An efflux pump is involved in secretion of newly synthesized siderophore by Pseudomonas aeruginosa. FEBS Lett. 584 4751–4755. 10.1016/j.febslet.2010.10.051 [DOI] [PubMed] [Google Scholar]
- Harding C. M., Hennon S. W., Feldman M. F. (2018). Uncovering the mechanisms of Acinetobacter baumannii virulence. Nat. Rev. Microbiol. 16 91–102. 10.1038/nrmicro.2017.148 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Harding C. M., Pulido M. R., Di Venanzio G., Kinsella R. L., Webb A. I., Scott N. E., et al. (2017). Pathogenic Acinetobacter species have a functional type I secretion system and contact-dependent inhibition systems. J. Biol. Chem. 292:90759087. 10.1074/jbc.M117.781575 [DOI] [PMC free article] [PubMed] [Google Scholar]
- He X., Lu F., Yuan F., Jiang D., Zhao P., Zhu J., et al. (2015). Biofilm formation caused by clinical Acinetobacter baumannii isolates is associated with overexpression of the adefgh efflux pump. Antimicrob. Agents Chemother. 59 4817–4825. 10.1128/AAC.00877-15 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Henry R., Crane B., Powell D., Lucas D. D., Li Z., Aranda J., et al. (2014). The transcriptomic response of Acinetobacter baumannii to colistin and doripenem alone and in combination in an in vitro pharmacokinetics/pharmacodynamics model. J. Antimicrob. Chemother. 70 1303–1313. 10.1093/jac/dku536 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Henry R., Vithanage N., Harrison P., Seemann T., Coutts S., Moffatt J. H., et al. (2012). Colistin-resistant, lipopolysaccharide-deficient Acinetobacter baumannii responds to lipopolysaccharide loss through increased expression of genes involved in the synthesis and transport of lipoproteins, phospholipids, and poly-β-1,6-N-acetylglucosamine. Antimicrob. Agents Chemother. 56 59–69. 10.1128/AAC.05191-11 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Imperi F., Tiburzi F., Visca P. (2009). Molecular basis of pyoverdine siderophore recycling in Pseudomonas aeruginosa. Proc. Natl. Acad. Sci.U.S.A. 106 20440–20445. 10.1073/pnas.0908760106 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ishige T., Tani A., Takabe K., Kawasaki K., Sakai Y., Kato N. (2002). Wax ester production from n-alkanes by Acinetobacter sp. strain M-1: ultrastructure of cellular inclusions and role of acyl coenzyme a reductase. Appl. Environ. Microbiol. 68 1192–1195. 10.1128/AEM.68.3.1192-1195.2002 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kentache T., Ben Abdelkrim A., Jouenne T., Dé E., Hardouin J. (2017). Global dynamic proteome study of a pellicle-forming Acinetobacter baumannii strain. Mol. Cell. Proteomics 16 100–112. 10.1074/mcp.M116.061044 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kenyon J. J., Hall R. M. (2013). Variation in the complex carbohydrate biosynthesis loci of Acinetobacter baumannii Genomes. PLoS One 8:e62160. 10.1371/JOURNAL.PONE.0062160 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kertesz M. A., Schmidt-Larbig K., Wüest T. (1999). A novel reduced flavin mononucleotide-dependent methanesulfonate sulfonatase encoded by the sulfur-regulated msu operon of Pseudomonas aeruginosa. J. Bacteriol. 181 1464–1473. 10.1128/jb.181.5.1464-1473.1999 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Klinger-Strobel M., Stein C., Forstner C., Makarewicz O., Pletz M. W. (2017). Effects of colistin on biofilm matrices of Escherichia coli and Staphylococcus aureus. Int. J. Antimicrob. Agents 49 472–479. 10.1016/j.ijantimicag.2017.01.005 [DOI] [PubMed] [Google Scholar]
- Kobayashi N., Nishino K., Yamaguchi A. (2001). Novel macrolide-specific ABC-type efflux transporter in Escherichia coli. J. Bacteriol. 183 5639–5644. 10.1128/JB.183.19.5639-5644.2001 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lakowicz J. R (ed) (2006). “Fluorescence anisotropy,” in Principles of Fluorescence Spectroscopy, (Boston, MA: Springer; ), 353–382. 10.1007/978-0-387-46312-4_10 [DOI] [Google Scholar]
- Lattif A. A., Mukherjee P. K., Chandra J., Roth M. R., Welti R., Rouabhia M., et al. (2011). Lipidomics of Candida albicans biofilms reveals phase-dependent production of phospholipid molecular classes and role for lipid rafts in biofilm formation. Microbiology 157:3232. 10.1099/MIC.0.051086-0 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Le H., Arnoult C., Dé E., Schapman D., Galas L., Le Cerf D., et al. (2021). Antibody-conjugated nanocarriers for targeted antibiotic delivery: application in the treatment of bacterial biofilms. Biomacromolecules 22 1639–1653. 10.1021/ACS.BIOMAC.1C00082 [DOI] [PubMed] [Google Scholar]
- Lee C. R., Lee J. H., Park M., Park K. S., Bae I. K., Kim Y. B., et al. (2017). Biology of Acinetobacter baumannii: pathogenesis, antibiotic resistance mechanisms, and prospective treatment options. Front. Cell. Infect. Microbiol. 7:55. 10.3389/fcimb.2017.00055 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lehner R., Kuksis A. (1996). Biosynthesis of triacylglycerols. Prog. Lipid Res. 35 169–201. 10.1016/0163-7827(96)00005-7 [DOI] [PubMed] [Google Scholar]
- Lin M.-F., Lan C.-Y. (2014). Antimicrobial resistance in Acinetobacter baumannii: from bench to bedside. World J. Clin. Cases WJCC 2:787. 10.12998/WJCC.V2.I12.787 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lin M. F., Lin Y. Y., Lan C. Y. (2015). The role of the two-component system BaeSR in disposing chemicals through regulating transporter systems in Acinetobacter baumannii. PLoS One 10:e0132843. 10.1371/journal.pone.0132843 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lin M.-F., Lin Y.-Y., Lan C.-Y. (2017). Contribution of EmrAB efflux pumps to colistin resistance in Acinetobacter baumannii. J. Microbiol. 55 130–136. 10.1007/s12275-017-6408-5 [DOI] [PubMed] [Google Scholar]
- Lin M.-F., Lin Y.-Y., Lan C.-Y. (2020). Characterization of biofilm production in different strains of Acinetobacter baumannii and the effects of chemical compounds on biofilm formation. PeerJ 8:e9020. 10.7717/peerj.9020 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lin M.-F., Lin Y.-Y., Yeh H.-W., Lan C.-Y. (2014). Role of the BaeSR two-component system in the regulation of Acinetobacter baumannii adeAB genes and its correlation with tigecycline susceptibility. BMC Microbiol. 14:119. 10.1186/1471-2180-14-119 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lin Y.-T., Huang Y.-W., Liou R.-S., Chang Y.-C., Yang T.-C. (2014). MacABCsm, an ABC-type tripartite efflux pump of Stenotrophomonas maltophilia involved in drug resistance, oxidative and envelope stress tolerances and biofilm formation. J. Antimicrob. Chemother. 69 3221–3226. 10.1093/jac/dku317 [DOI] [PubMed] [Google Scholar]
- Macia M. D., Rojo-Molinero E., Oliver A. (2014). Antimicrobial susceptibility testing in biofilm-growing bacteria. Clin. Microbiol. Infect. 20 981–990. 10.1111/1469-0691.12651 [DOI] [PubMed] [Google Scholar]
- Marti S., Rodríguez-Bão J., Catel-Ferreira M., Jouenne T., Vila J., Seifert H., et al. (2011b). Biofilm formation at the solid-liquid and air-liquid interfaces by Acinetobacter species. BMC Res. Notes 4:5. 10.1186/1756-0500-4-5 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Marti S., Chabane Y. N., Alexandre S., Coquet L., Vila J., Jouenne T., et al. (2011a). Growth of Acinetobacter baumannii in pellicle enhanced the expression of potential virulence factors. PLoS One 6:e0026030. 10.1371/journal.pone.0026030 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Monds R. D., O’Toole G. A. (2009). The developmental model of microbial biofilms: ten years of a paradigm up for review. Trends Microbiol. 17 73–87. 10.1016/j.tim.2008.11.001 [DOI] [PubMed] [Google Scholar]
- Nait Chabane Y., Marti S., Rihouey C., Alexandre S., Hardouin J., Lesouhaitier O., et al. (2014). Characterisation of pellicles formed by Acinetobacter baumannii at the air-liquid interface. PLoS One 9:e111660. 10.1371/journal.pone.0111660 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nasr P. (2020). Genetics, epidemiology, and clinical manifestations of multidrug-resistant Acinetobacter baumannii. J. Hosp. Infect. 104 4–11. 10.1016/j.jhin.2019.09.021 [DOI] [PubMed] [Google Scholar]
- Nishino K., Latifi T., Groisman E. A. (2006). Virulence and drug resistance roles of multidrug efflux systems of Salmonella enterica serovar Typhimurium. Mol. Microbiol. 59 126–141. [DOI] [PubMed] [Google Scholar]
- Nwugo C. C., Gaddy J. A., Zimbler D. L., Actis L. A. (2011). Deciphering the iron response in Acinetobacter baumannii: a proteomics approach. J. Proteomics 74 44–58. 10.1016/j.jprot.2010.07.010 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Okada U., Yamashita E., Neuberger A., Morimoto M., Van Veen H. W., Murakami S. (2017). Crystal structure of tripartite-type ABC transporter MacB from Acinetobacter baumannii. Nat. Commun. 8 1–11. 10.1038/s41467-017-01399-2 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Orsinger-Jacobsen S. J., Patel S. S., Vellozzi E. M., Gialanella P., Nimrichter L., Miranda K., et al. (2013). Use of a stainless steel washer platform to study Acinetobacter baumannii adhesion and biofilm formation on abiotic surfaces. Microbiol. (United Kingdom) 159 2594–2604. 10.1099/mic.0.068825-0 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Peleg A. Y., Seifert H., Paterson D. L. (2008). Acinetobacter baumannii: emergence of a successful pathogen. Clin. Microbiol. Rev. 21 538–582. 10.1128/CMR.00058-07 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Penesyan A., Nagy S. S., Kjelleberg S., Gillings M. R., Paulsen I. T. (2019). Rapid microevolution of biofilm cells in response to antibiotics. NPJ Biofilms Microbiomes 51 1–14. 10.1038/s41522-019-0108-3 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Penwell W. F., Actis L. A. (2019). “Isolation and Characterization of the acinetobactin and baumannoferrin siderophores produced by Acinetobacter baumannii,” in Acinetobacter baumannii Methods and Protocols, eds Biswas I., Rather P. N. (New York, NY: Humana Press; ), 259–270. 10.1007/978-1-4939-9118-1_24 [DOI] [PubMed] [Google Scholar]
- Penwell W. F., DeGrace N., Tentarelli S., Gauthier L., Gilbert C. M., Arivett B. A., et al. (2015). Discovery and characterization of new hydroxamate siderophores, baumannoferrin A and B, produced by Acinetobacter baumannii. ChemBioChem 16 1896–1904. 10.1002/cbic.201500147 [DOI] [PubMed] [Google Scholar]
- Pérez A., Merino M., Rumbo-Feal S., Álvarez-Fraga L., Vallejo J. A., Beceiro A., et al. (2017). The FhaB/FhaC two-partner secretion system is involved in adhesion of Acinetobacter baumannii AbH12O-A2 strain. Virulence 8 959–974. 10.1080/21505594.2016.1262313 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pérez-Varela M., Corral J., Aranda J., Barbé J. (2019). Roles of efflux pumps from different superfamilies in the surface-associated motility and virulence of Acinetobacter baumannii ATCC 17978. Antimicrob. Agents Chemother. 63 1–11. 10.1128/AAC.02190-18 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Proschak A., Lubuta P., Grün P., Löhr F., Wilharm G., De Berardinis V., et al. (2013). Structure and biosynthesis of fimsbactins a-f, Siderophores from Acinetobacter baumannii and Acinetobacter baylyi. ChemBioChem 14 633–638. 10.1002/cbic.201200764 [DOI] [PubMed] [Google Scholar]
- Qi L., Li H., Zhang C., Liang B., Li J., Wang L., et al. (2016). Relationship between antibiotic resistance, biofilm formation, and biofilm-specific resistance in Acinetobacter baumannii. Front. Microbiol. 7:483. 10.3389/fmicb.2016.00483 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Qin Y., He Y., She Q., Larese-Casanova P., Li P., Chai Y. (2019). Heterogeneity in respiratory electron transfer and adaptive iron utilization in a bacterial biofilm. Nat. Commun. 2019 101 1–12. 10.1038/s41467-019-11681-0 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Richardot C., Juarez P., Jeannot K., Patry I., Plésiat P., Llanes C. (2016). Amino acid substitutions account for most mexS alterations in clinical nfxC mutants of Pseudomonas aeruginosa. Antimicrob. Agents Chemother. 60 2302–2310. 10.1128/AAC.02622-15 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Richmond G. E., Evans L. P., Anderson M. J., Wand M. E., Bonney L. C., Ivens A., et al. (2016). The Acinetobacter baumannii two-component system aders regulates genes required for multidrug efflux, biofilm formation, and virulence in a strain-specific manner. MBio 7:e00430-16. 10.1128/mBio.00430-16 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rumbo-Feal S., Gómez M. J., Gayoso C., Álvarez-Fraga L., Cabral M. P., Aransay A. M., et al. (2013). Whole transcriptome analysis of Acinetobacter baumannii assessed by RNA-sequencing reveals different mrna expression profiles in biofilm compared to planktonic cells. PLoS One 8:e72968. 10.1371/journal.pone.0072968 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rumbo-Feal S., Pérez A., Ramelot T. A., Álvarez-Fraga L., Vallejo J. A., Beceiro A., et al. (2017). Contribution of the A. baumannii A1S_0114 Gene to the Interaction with eukaryotic cells and virulence. Front. Cell. Infect. Microbiol. 7:108. 10.3389/fcimb.2017.00108 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sahu P. K., Iyer P. S., Oak A. M., Pardesi K. R., Chopade B. A. (2012). Characterization of eDNA from the clinical strain Acinetobacter baumannii AIIMS 7 and Its role in biofilm formation. Sci. World J. 2012 1–10. 10.1100/2012/973436 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Serra D. O., Conover M. S., Arnal L., Sloan G. P., Rodriguez M. E., Yantorno O. M., et al. (2011). FHA-mediated cell-substrate and cell-cell adhesions are critical for bordetella pertussis biofilm formation on abiotic surfaces and in the mouse nose and the trachea. PLoS One 6:e28811. 10.1371/journal.pone.0028811 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sharma A., Sharma R., Bhattacharyya T., Bhando T., Pathania R. (2017). Fosfomycin resistance in Acinetobacter baumannii is mediated by efflux through a major facilitator superfamily (MFS) transporter—AbaF. J. Antimicrob. Chemother. 72 68–74. 10.1093/jac/dkw382 [DOI] [PubMed] [Google Scholar]
- Sheldon J. R., Skaar E. P. (2020). Acinetobacter baumannii can use multiple siderophores for iron acquisition, but only acinetobactin is required for virulence. PLoS Pathog. 16:e1008995. 10.1371/journal.ppat.1008995 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Soares N. C., Cabral M. P., Gayoso C., Mallo S., Rodriguez-Velo P., Fernández-Moreira E., et al. (2010). Associating growth-phase-related changes in the proteome of Acinetobacter baumannii with increased resistance to oxidative stress. J. Proteome Res. 9 1951–1964. 10.1021/pr901116r [DOI] [PubMed] [Google Scholar]
- Soucy S. M., Huang J., Gogarten J. P. (2015). Horizontal gene transfer: building the web of life. Nat. Rev. Genet. 168 472–482. 10.1038/nrg3962 [DOI] [PubMed] [Google Scholar]
- Tahmourespour A., Tabatabaee N., Khalkhali H., Amini I. (2016). Tannic acid degradation by Klebsiella strains isolated from goat feces. Iran. J. Microbiol. 8 14–20. [PMC free article] [PubMed] [Google Scholar]
- Tahrioui A., Duchesne R., Bouffartigues E., Rodrigues S., Maillot O., Tortuel D., et al. (2019). Extracellular DNA release, quorum sensing, and PrrF1/F2 small RNAs are key players in Pseudomonas aeruginosa tobramycin-enhanced biofilm formation. NPJ Biofilms Microbiomes 5 1–11. 10.1038/s41522-019-0088-3 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tahrioui A., Ortiz S., Azuama O. C., Bouffartigues E., Benalia N., Tortuel D., et al. (2020). Membrane-interactive compounds from Pistacia lentiscus L. Thwart Pseudomonas aeruginosa virulence. Front. Microbiol. 11:1068. 10.3389/FMICB.2020.01068 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Takada H., Morita M., Shiwa Y., Sugimoto R., Suzuki S., Kawamura F., et al. (2014). Cell motility and biofilm formation in Bacillus subtilis are affected by the ribosomal proteins, S11 and S21. Biosci. Biotechnol. Biochem. 78 898–907. 10.1080/09168451.2014.915729 [DOI] [PubMed] [Google Scholar]
- Tao Y., Acket S., Beaumont E., Galez H., Duma L., Rossez Y. (2021). Colistin treatment affects lipid composition of Acinetobacter baumannii. Antibiotics 10:528. 10.3390/antibiotics10050528 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Teufel R., Mascaraque V., Ismail W., Voss M., Perera J., Eisenreich W., et al. (2010). Bacterial phenylalanine and phenylacetate catabolic pathway revealed. Proc. Natl. Acad. Sci. U.S.A. 107 14390–14395. 10.1073/pnas.1005399107 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tseng B. S., Zhang W., Harrison J. J., Quach T. P., Song J. L., Penterman J., et al. (2013). The extracellular matrix protects Pseudomonas aeruginosa biofilms by limiting the penetration of tobramycin. Environ. Microbiol. 15 2865–2878. 10.1111/1462-2920.12155 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Turlin E., Heuck G., Simões Brandão M. I., Szili N., Mellin J. R., Lange N., et al. (2014). Protoporphyrin (PPIX) efflux by the MacAB-TolC pump in Escherichia coli. Microbiologyopen 3 849–859. 10.1002/mbo3.203 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vickery L. E., Cupp-Vickery J. R. (2007). Molecular chaperones HscA/Ssq1 and HscB/Jac1 and their roles in iron-sulfur protein maturation. Crit. Rev. Biochem. Mol. Biol. 42 95–111. 10.1080/10409230701322298 [DOI] [PubMed] [Google Scholar]
- Wang Y.-C., Huang T.-W., Yang Y.-S., Kuo S.-C., Chen C.-T., Liu C.-P., et al. (2018). Biofilm formation is not associated with worse outcome in Acinetobacter baumannii bacteraemic pneumonia. Sci. Rep. 8:7289. 10.1038/s41598-018-25661-9 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Weber B. S., Ly P. M., Irwin J. N., Pukatzki S., Feldman M. F. (2015). A multidrug resistance plasmid contains the molecular switch for type VI secretion in Acinetobacter baumannii. Proc. Natl. Acad. Sci. U.S.A. 112 9442–9447. 10.1073/pnas.1502966112 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Whitchurch C. B., Tolker-Nielsen T., Ragas P. C., Mattick J. S. (2002). Extracellular DNA required for bacterial biofilm formation. Science 295:1487. 10.1126/science.295.5559.1487 [DOI] [PubMed] [Google Scholar]
- Williamson K. S., Richards L. A., Perez-Osorio A. C., Pitts B., McInnerney K., Stewart P. S., et al. (2012). Heterogeneity in Pseudomonas aeruginosa biofilms includes expression of ribosome hibernation factors in the antibiotic-tolerant subpopulation and hypoxia-induced stress response in the metabolically active population. J. Bacteriol. 194 2062–2073. 10.1128/JB.00022-12 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yamamoto S., Okujo N., Sakakibara Y. (1994). Isolation and structure elucidation of acinetobactin., a novel siderophore from Acinetobacter baumannii. Arch. Microbiol. 162 249–254. 10.1007/BF00301846 [DOI] [PubMed] [Google Scholar]
- Yamanaka H., Kobayashi H., Takahashi E., Okamoto K. (2008). MacAB Is involved in the secretion of escherichia coli heat-stable enterotoxin II. J. Bacteriol. 190 7693–7698. 10.1128/JB.00853-08 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yan J., Bassler B. L. (2019). Surviving as a community: antibiotic tolerance and persistence in bacterial biofilms. Cell Host Microbe 26 15–21. 10.1016/J.CHOM.2019.06.002 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yoon E.-J., Nait Chabane Y., Goussard S., Snesrud E., Courvalin P., Dé E., et al. (2015). Contribution of resistance-nodulation-cell division efflux systems to antibiotic resistance and biofilm formation in Acinetobacter baumannii. MBio 6:e00309-15. 10.1128/mBio.00309-15 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang K., Li X., Yu C., Wang Y. (2020). Promising therapeutic strategies against microbial biofilm challenges. Front. Cell. Infect. Microbiol. 10:359. 10.3389/FCIMB.2020.00359 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zheng J., Lin Z., Sun X., Lin W., Chen Z., Wu Y., et al. (2018). Overexpression of OqxAB and MacAB efflux pumps contributes to eravacycline resistance and heteroresistance in clinical isolates of Klebsiella pneumoniae. Emerg. Microbes Infect. 7 1–11. 10.1038/s41426-018-0141-y [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
The datasets presented in this study can be found in online repositories. The names of the repositories and accession number(s) can be found in the “Materials and Methods” section.