Abstract
Laboratory differentiation of erythromycin resistance phenotypes is poorly standardized for pneumococci. In this study, 85 clinical isolates of erythromycin-resistant (MIC ≥ 1 μg/ml) Streptococcus pneumoniae were tested for the resistance phenotype by the erythromycin-clindamycin double-disk test (previously used to determine the macrolide resistance phenotype in Streptococcus pyogenes strains) and by MIC induction tests, i.e., by determining the MICs of macrolide antibiotics without and with pre-exposure to 0.05 μg of erythromycin per ml. By the double-disk test, 65 strains, all carrying the erm(AM) determinant, were assigned to the constitutive macrolide, lincosamide, and streptogramin B resistance (cMLS) phenotype, and the remaining 20, all carrying the mef(E) gene, were assigned to the recently described M phenotype; an inducible MLS resistance (iMLS) phenotype was not found. The lack of inducible resistance to clindamycin was confirmed by determining clindamycin MICs without and with pre-exposure to subinhibitory concentrations of erythromycin. In macrolide MIC and MIC-induction tests, whereas homogeneous susceptibility patterns were observed among the 20 strains assigned to the M phenotype by the double-disk test, two distinct patterns were recognized among the 65 strains assigned to the cMLS phenotype by the same test; one pattern (n = 10; probably that of the true cMLS isolates) was characterized by resistance to rokitamycin also without induction, and the other pattern (n = 55; designated the iMcLS phenotype) was characterized by full or intermediate susceptibility to rokitamycin without induction turning to resistance after induction, with an MIC increase by more than three dilutions. A triple-disk test, set up by adding a rokitamycin disk to the erythromycin and clindamycin disks of the double-disk test, allowed the easy differentiation not only of pneumococci with the M phenotype from those with MLS resistance but also, among the latter, of those of the true cMLS phenotype from those of the iMcLS phenotype. While distinguishing MLS from M resistance in pneumococci is easily and reliably achieved, the differentiation of constitutive from inducible MLS resistance is far more uncertain and is strongly affected by the antibiotic used to test inducibility.
Ribosomal target site modification due to methylases encoded by erm class genes is the most common and extensively investigated mechanism of erythromycin resistance in streptococci (28). It has long been known that ribosome methylation causes reduced binding of and coresistance to macrolide, lincosamide, and streptogramin B (MLS) antibiotics and that in streptococci MLS resistance can be expressed either constitutively (cMLS phenotype) or inducibly (iMLS phenotype) (9, 12, 27). Only recently has a macrolide efflux mechanism been described for streptococci (24), in which it is associated with a new resistance pattern (M phenotype) characterized by resistance to 14- and 15-membered macrolides and susceptibility to 16-membered macrolides, lincosamides, and streptogramin B (21, 24).
While M resistance is similar in Streptococcus pyogenes and Streptococcus pneumoniae, being mediated in both species by similar determinants—mef(A) (4) and mef(E) (26), respectively, recently recommended to be considered a single gene, mef(A) (18)—encoding similar membrane proteins responsible for the macrolide efflux, MLS resistance appears to be more varied. From a genotypic point of view, in S. pyogenes MLS resistance is mediated by two classes of methylase genes, i.e., the conventional erm(AM) determinant (12), belonging to gene class erm(B) (18), and the recently described erm(TR) determinant (22), belonging to gene class erm(A) (18). In S. pneumoniae, only the former methylase gene has been extensively documented (12, 24), even though the presence of erm(TR) has been recently demonstrated in particular isolates of erythromycin-resistant pneumococci (3, 25). From a phenotypic point of view, erythromycin-resistant strains of S. pyogenes can be differentiated into three phenotypes (cMLS, iMLS, and M) by a simple double-disk (erythromycin plus clindamycin) test (21) or—as easily but more accurately—into five phenotypes by a triple-disk (erythromycin plus clindamycin and josamycin) test, which allows further differentiation of inducibly resistant strains into three distinct types (iMLS-A, iMLS-B, and iMLS-C) (8). Furthermore, in S. pyogenes the erm(AM) determinant can be associated with both constitutive (cMLS phenotype) and inducible (iMLS-A phenotype) resistance, whereas the erm(TR) determinant is usually associated with inducible resistance (iMLS-B and iMLS-C phenotypes) (8). By contrast, the discrimination between constitutive and inducible MLS resistance in S. pneumoniae strains is uncertain, and laboratory differentiation of macrolide resistance phenotypes is poorly standardized.
In this study, several clinical isolates of erythromycin-resistant S. pneumoniae were tested for the resistance phenotype by comparing results of erythromycin-clindamycin double-disk (ECDD) tests used as in erythromycin-resistant S. pyogenes strains and MIC induction tests, i.e., by determining the MICs of some MLS antibiotics without and with pre-exposure to a subinhibitory concentration of erythromycin.
(Part of these data were presented at the 40th Interscience Conference on Antimicrobial Agents and Chemotherapy, Toronto, Canada, 17 to 20 September 2000.)
MATERIALS AND METHODS
Bacterial strains.
A total of 85 clinical isolates of erythromycin-resistant S. pneumoniae were collected from several Italian laboratories between September 1998 and June 2000. Multiple isolates from the same patient were avoided. Strain identification was confirmed in our laboratory by conventional laboratory tests such as susceptibility to optochin and solubility in bile (20) and by using the API system (bioMérieux, Marcy-I'Etoile, France). Erythromycin resistance (MIC ≥ 1 μg/ml) was also confirmed in our laboratory by the broth microdilution method (see below).
ECDD test.
The ECDD test was carried out by a modification (i.e., using commercial disks) of the assay described by Seppälä et al. (21) for S. pyogenes strains. Disks (Oxoid Ltd., Basingstoke, United Kingdom) of erythromycin (15 μg) and clindamycin (2 μg) were placed 15 to 20 mm apart on Mueller-Hinton agar (BBL Microbiology Systems, Cockeysville, Md.) supplemented with 5% sheep blood, which had been inoculated with a swab dipped into a bacterial suspension with a turbidity equivalent to that of a 0.5 McFarland standard. After 18 h of incubation at 37°C, the absence of a zone of inhibition around the two disks indicated constitutive resistance (cMLS phenotype): blunting of the clindamycin zone of inhibition proximal to the erythromycin disk indicated inducible resistance (iMLS phenotype); and susceptibility to clindamycin with no blunting of the zone of inhibition around the clindamycin disk indicated the M phenotype.
Antibiotics.
Erythromycin and clindamycin were purchased from Sigma Chemical Co. (St. Louis, Mo.). The other antibiotics were obtained from the following sources: clarithromycin, Abbott Laboratories (Abbott Park, Ill.); azithromycin, Pfizer Inc. (New York, N.Y.); josamycin, ICN Biomedicals (Costa Mesa, Calif.); and rokitamycin, Prodotti Formenti (Milan, Italy).
Susceptibility tests.
MICs were determined by the broth microdilution method according to the procedure recommended by the National Committee for Clinical Laboratory Standards (NCCLS) (14). Mueller-Hinton II broth (BBL) supplemented with 3% lysed horse blood was used as the test medium. The antibiotics were tested at final concentrations (prepared from twofold dilutions) that ranged from 0.015 to 128 μg/ml. S. pneumoniae ATCC 49619 was used for quality control. The MIC breakpoints suggested by the NCCLS (14) were used for erythromycin, clindamycin, and clarithromycin (susceptible, ≤0.25 μg/ml; intermediate, 0.5 μg/ml; resistant, ≥1 μg/ml) and for azythromycin (susceptible, ≤0.5 μg/ml; intermediate, 1 μg/ml; resistant, ≥2 μg/ml), and those suggested by the French Society for Microbiology (5) for 16-membered macrolides were used for josamycin and rokitamycin (susceptible, ≤1 μg/ml; intermediate, 2 μg/ml; resistant, ≥4 μg/ml).
Induction of MLS resistance (MIC induction tests).
Induction of MLS resistance was evaluated by pregrowth (3 h at 37°C) in erythromycin at a subinhibitory concentration (0.05 μg/ml). As described previously (8), the culture was then washed, and the cells were used to prepare the inoculum for MIC testing by the usual broth microdilution method.
Detection of erythromycin resistance genes.
The presence of erythromycin resistance genes was investigated by PCR. Primer pairs specific for the detection of erm(AM) and mef(E) (expected PCR product sizes, 639 and 348 bp, respectively) were as reported by Sutcliffe et al. (23). The primers designated III8 and III10 by Seppälä et al. (22) were used to detect the erm(TR) gene (expected PCR product size, 208 bp). DNA preparation and amplification and electrophoresis of PCR products were carried out by established procedures (10, 22, 23).
RESULTS
ECDD test.
All 85 erythromycin-resistant S. pneumoniae strains studied were tested using the ECDD assay; 65 (76.5%) were assigned to the cMLS phenotype and 20 (23.5%) to the M phenotype (Table 1). Inducibly resistant isolates (iMLS phenotype) were not found by this method.
TABLE 1.
Macrolide resistance phenotype in 85 clinical isolates of erythromycin-resistant S. pneumoniae by the ECDD test and correlations with clindamycin susceptibility and erythromycin resistance genes
| Phenotype of macrolide resistance | No. (%) of strains | Clindamycin susceptibility (MIC [μg/ml])a of strains
|
No. of strains with the following gene:
|
||||||
|---|---|---|---|---|---|---|---|---|---|
| Without induction
|
After inductionb
|
||||||||
| Range | 50% | 90% | Range | 50% | 90% | erm(AM) | mef(E) | ||
| cMLS | 65 (76.5) | 8–>128 | 64 | >128 | 16–>128 | >128 | >128 | 65 | |
| M | 20 (23.5) | 0.03–0.12 | 0.03 | 0.12 | 0.03–0.12 | 0.03 | 0.12 | 20 | |
50% and 90%, MICs at which 50 and 90% of isolates were inhibited, respectively.
Induction was performed by pregrowth in 0.05 μg of erythromycin per ml.
Clindamycin MIC induction tests.
The lack of inducible resistance to clindamycin was confirmed by MIC induction tests, by determining clindamycin MICs without and with pre-exposure to 0.05 μg of erythromycin per ml. All the isolates assigned to the cMLS phenotype by the ECDD test were found to be clindamycin resistant both without (MICs, 8 to >128 μg/ml) and with (MICs, 16 to >128 μg/ml) induction, whereas those assigned to the M phenotype remained equally susceptible under both conditions (MIC range, 0.03 to 0.12 μg/ml in both instances) (Table 1).
Erythromycin resistance genes.
All strains identified as having the cMLS phenotype by the ECDD test had the erm(AM) gene, whereas all those identified as having the M phenotype had the mef(E) gene; no strain showed both erm(AM) and mef(E) (Table 1). No strain had the erm(TR) gene.
Macrolide MICs and MIC-induction tests.
The MICs of two 14-membered (erythromycin and clarithromycin), one 15-membered (azithromycin), and two 16-membered (josamycin and rokitamycin) macrolides were determined and compared (Table 2). Homogeneous susceptibility patterns were observed in the 20 strains assigned to the M phenotype by the ECDD test; all these isolates were resistant to the 14- and 15-membered macrolides (with MICs not exceeding 16 μg/ml for erythromycin and clarithromycin and 32 μg/ml for azithromycin) and susceptible to the 16-membered macrolides. By contrast, heterogeneous susceptibility patterns were observed among the 65 strains assigned to the cMLS phenotype by the ECDD test; all these isolates were resistant (with widely variable MIC levels) to the 14- and 15-membered macrolides, whereas the MICs of josamycin and rokitamycin ranged from susceptibility (0.5 and 0.06 μg/ml, respectively) to high-level resistance (>128 μg/ml).
TABLE 2.
Susceptibility to macrolides of 85 clinical isolates of erythromycin-resistant S. pneumoniae, subdivided into macrolide resistance phenotypes by the ECDD test
| Phenotype (no. of strains) | Antibiotica | MIC (μg/ml)b
|
||
|---|---|---|---|---|
| Range | 50% | 90% | ||
| cMLS (65) | Erythromycin | 4–>128 | 64 | >128 |
| Clarithromycin | 2–>128 | 128 | >128 | |
| Azithromycin | 8–>128 | >128 | >128 | |
| Josamycin | 0.5–>128 | 16 | 128 | |
| Rokitamycin | 0.06–>128 | 0.25 | 16 | |
| Josamycin (ind.) | 16–>128 | >128 | >128 | |
| Rokitamycin (ind.) | 4–>128 | 128 | >128 | |
| M (20) | Erythromycin | 2–16 | 4 | 16 |
| Clarithromycin | 1–16 | 4 | 8 | |
| Azithromycin | 2–32 | 16 | 16 | |
| Josamycin | 0.03–0.25 | 0.06 | 0.12 | |
| Rokitamycin | 0.03–0.06 | 0.06 | 0.06 | |
| Josamycin (ind.) | 0.06–0.25 | 0.06 | 0.25 | |
| Rokitamycin (ind.) | 0.03–0.12 | 0.06 | 0.06 | |
ind., after induction by pregrowth in 0.05 μg of erythromycin per ml.
50% and 90%, MICs at which 50 and 90% of isolates were inhibited, respectively.
Josamycin and rokitamycin MICs were also determined after induction with erythromycin by the pregrowth procedure used for clindamycin (Table 2). While the susceptibilities of the 20 M phenotype isolates were substantially unaffected by induction, all 65 cMLS strains were found to be resistant, although at variable levels, to both josamycin (MIC range, 16 to >128 μg/ml) and rokitamycin (MIC range, 4 to >128 μg/ml) after induction.
Altogether, two distinct patterns of macrolide resistance were recognized among the 65 isolates assigned to the cMLS phenotype by the ECDD test (Table 3); one, observed in 10 isolates (15.4%), was characterized by resistance to rokitamycin also without induction (MICs, ≥4 μg/ml), and the other, observed in 55 isolates (84.6%), by full or intermediate susceptibility to rokitamycin without induction (MICs, ≤2 μg/ml) turning to resistance after induction (MICs, 4 to >128 μg/ml), with an MIC increase of more than three dilutions. The first pattern was also characterized by high-level resistance to the 14- and 15-membered macrolides (MICs, ≥128 μg/ml) and to josamycin (MIC range, 32 to >128 μg/ml) also without induction; the second pattern was also characterized by variable-level resistance to the 14- and 15-membered macrolides and by susceptibility to moderate resistance to josamycin without induction (MIC range, 0.5 to 32 μg/ml) turning to uniform, mostly high-level resistance after induction.
TABLE 3.
Susceptibility to MLS antibiotics of 65 clinical isolates of erythromycin-resistant S. pneumoniae, all identified as cMLS phenotype by the ECDD test and differentiated into cMLS and iMcLS types on the basis of macrolide MICs and MIC-induction tests
| Phenotype in MIC-induction tests (no. of strains) | Antibiotica | MIC (μg/ml)b
|
||
|---|---|---|---|---|
| Range | 50% | 90% | ||
| cMLS (10) | Erythromycin | 128–>128 | >128 | >128 |
| Clarithromycin | 128–>128 | >128 | >128 | |
| Azithromycin | 128–>128 | >128 | >128 | |
| Josamycin | 32–>128 | 128 | >128 | |
| Rokitamycin | 4–>128 | 64 | >128 | |
| Clindamycin | 16–>128 | 128 | >128 | |
| Josamycin (ind.) | 128–>128 | >128 | >128 | |
| Rokitamycin (ind.) | 128–>128 | >128 | >128 | |
| Clindamycin (ind.) | 32–>128 | >128 | >128 | |
| iMcLS (55) | Erythromycin | 2–>128 | 64 | >128 |
| Clarithromycin | 4–>128 | 64 | >128 | |
| Azithromycin | 8–>128 | 128 | >128 | |
| Josamycin | 0.5–32 | 8 | 32 | |
| Rokitamycin | 0.06–2 | 0.25 | 1 | |
| Clindamycin | 8–>128 | 32 | >128 | |
| Josamycin (ind.) | 16–>128 | >128 | >128 | |
| Rokitamycin (ind.) | 4–>128 | 64 | >128 | |
| Clindamycin (ind.) | 16–>128 | 128 | >128 | |
ind., after induction by pregrowth in 0.05 μg of erythromycin per ml.
50% and 90%, MICs at which 50 and 90% of isolates were inhibited, respectively.
Triple-disk test.
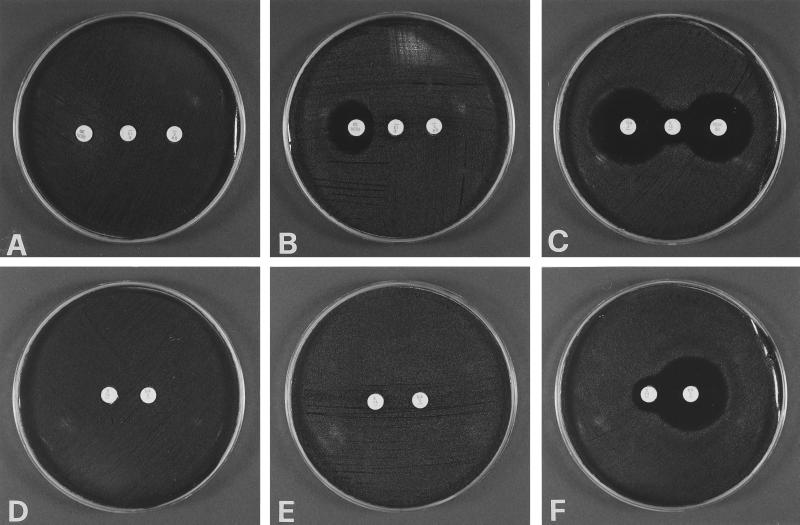
In order to easily differentiate, within erythromycin-resistant pneumococci, not only the isolates of the M phenotype from those with MLS resistance but also, among the latter, cMLS from iMcLS isolates, a triple-disk test was set up by adding a rokitamycin disk (30 μg; BBL) to the erythromycin and clindamycin disks of the conventional ECDD test. The erythromycin disk was placed at the center of the agar plate with the clindamycin and rokitamycin disks placed 15 to 20 mm apart on either side. All strains were tested by this triple-disk (ECRTD) assay. The iMcLS strains (Fig. 1B) were characterized by no significant zone of inhibition around either the erythromycin or the clindamycin disk, in line with their resistance to both drugs, but presented a zone of inhibition around rokitamycin that was blunted on the side proximal to the erythromycin disk, in line with the inducibility of their rokitamycin resistance. By the ECDD test (Fig. 1E) these strains would be identified as cMLS, no clindamycin zone of inhibition being appreciable. The true cMLS phenotype (Fig. 1A and D), characterized by the absence of a significant zone of inhibition around the three disks, and the M phenotype (Fig. 1C and F), characterized by susceptibility to clindamycin and rokitamycin with no blunting of the relevant zones of inhibition, were identified by the ECRTD test as easily as by the ECDD test.
FIG. 1.
Phenotypes of erythromycin-resistant pneumococci as determined in representative strains by the ECRTD and the ECDD tests. In the ECRTD test (A to C), the erythromycin disk (15 μg each) is at the center in each panel, with the clindamycin disk (2 μg each) on the right and the rokitamycin disk (30 μg each) on the left. In the ECDD test (D to F), the erythromycin disk is on the left and the clindamycin disk on the right in each panel. A and D, cMLS phenotype; B and E, iMcLS phenotype; C and F, M phenotype.
DISCUSSION
The predominance among erythromycin-resistant pneumococci of the isolates carrying the erm(AM) gene over those carrying the mef(E) gene observed in this study (76.5 versus 23.5%) is consistent with the results of other recent studies from European countries (2, 3, 13, 16, 25), South Africa (11), and Japan (15). It is worth noting that in some European reports the rate of isolates carrying the mef(E) gene was particularly low (<10%) (2, 3, 13, 16). The newly described erm(TR) gene (22), so far investigated more extensively in S. pyogenes than in S. pneumoniae isolates, was not detected in any of our erythromycin-resistant pneumococci. Very recently, a similar negative finding has been documented in France (2), whereas one pneumococcus carrying both erm(TR) and erm(B) has been reported in Spain (3), and erm(TR) was the only resistance determinant detected in 2.9% of the erythromycin-resistant pneumococci surveyed in a Greek study (25).
On the other hand and of more interest, the results of this study indicated that, while distinguishing MLS from M resistance in pneumococci is easily and reliably achieved, the differentiation between constitutive and inducible MLS resistance is far more uncertain and is strongly affected by the antibiotic used to test inducibility.
The ECDD test, conventionally used to identify three resistance phenotypes (cMLS, iMLS, and M) among erythromycin-resistant strains of S. pyogenes, appears to be less applicable to erythromycin-resistant pneumococci because in this test the constitutive or inducible character of MLS resistance is inferred from the response to clindamycin. Among S. pyogenes strains, the cMLS phenotype is associated with resistance to clindamycin without induction, whereas the iMLS phenotype is associated with susceptibility to clindamycin without induction, turning to high-level resistance after induction; of course, susceptibility to clindamycin without induction is shared by M phenotype isolates, which also however remain clindamycin susceptible after induction (8, 21). Among pneumococci, on the other hand, susceptibility to clindamycin is characteristic of the strains of the M phenotype, carrying the mef(E) gene but not of those with MLS resistance, carrying the erm(AM) gene, which, as a rule, also are clindamycin resistant without induction. As reported previously (6), possible false susceptibilities produced by the NCCLS microdilution method in the detection of pneumococcal resistance to clindamycin can be corrected by the extension of incubation time to 48 h, incubation in CO2, or the use of a disk diffusion method. Therefore, pneumococci with MLS resistance when tested by the ECDD assay are almost invariably, as in the present study, assigned to the cMLS phenotype (3, 7, 11, 13, 17, 24).
The fact that inducible resistance to clindamycin is not usually encountered in pneumococci is unlikely to mean that MLS resistance is only constitutively expressed in these organisms. Indeed, 16-membered macrolides, which are particularly effective in vitro against streptococci, including some erythromycin-resistant isolates (12, 21), appear to be better suited than lincosamides to the detection of inducible MLS resistance in pneumococci. This applies especially to rokitamycin, which has more powerful antipneumococcal activity in vitro than other 16-membered macrolides (15) and has actually been used in the past to tentatively distinguish inducible from constitutive MLS resistance in pneumococci (1, 15, 19).
Among the S. pneumoniae isolates with MLS resistance [genotypically characterized by the erm(AM) gene and usually assigned to the cMLS phenotype based on the ECDD test], those also resistant to rokitamycin without induction are likely to represent the veritable cMLS phenotype, whereas those becoming rokitamycin resistant only after induction are in fact likely to represent an iMLS phenotype. We designated the latter type as iMcLS to underline that inducibility regards macrolides (particularly 16-membered ones, with emphasis on rokitamycin) but not lincosamides, to which these strains are resistant also without induction (we have as yet no data about group B streptogramins). A triple-disk ECRTD test, set up by adding a rokitamycin disk to the erythromycin and clindamycin disks of the ECDD test, allowed easy differentiation not only of the pneumococci of the M phenotype from those with MLS resistance, but also, among the latter, of those of a narrower but probably truer cMLS phenotype from those of the iMcLS phenotype.
In any case, the meaning of inducible MLS resistance appears to be different in S. pneumoniae from that in S. pyogenes. Further investigations are warranted to better understand the underlying mechanisms.
REFERENCES
- 1.Agouridas C, Bonnefoy A, Chantot J F. Antibacterial activity of RU 64004 (HMR 3004), a novel ketolide derivative active against respiratory pathogens. Antimicrob Agents Chemother. 1997;41:2149–2158. doi: 10.1128/aac.41.10.2149. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Angot P, Vergnaud M, Auzou M, Leclercq R Observatoire de Normandie du Pneumocoque. Macrolide resistance phenotypes and genotypes in French clinical isolates of Streptococcus pneumoniae. Eur J Clin Microbiol Infect Dis. 2000;19:755–758. doi: 10.1007/pl00011229. [DOI] [PubMed] [Google Scholar]
- 3.Betriu C, Redondo M, Palau M L, Sánchez A, Gómez M, Culebras E, Boloix A, Picazo J J. Comparative in vitro activities of linezolid, quinupristin-dalfopristin, moxifloxacin, and trovafloxacin against erythromycin-susceptible and -resistant streptococci. Antimicrob Agents Chemother. 2000;44:1838–1841. doi: 10.1128/aac.44.7.1838-1841.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Clancy J, Petitpas J, Dib-Hajj F, Yuan W, Cronan M, Kamath A V, Bergeron J, Retsema J A. Molecular cloning and functional analysis of a novel macrolide resistance determinant, mefA, from Streptococcus pyogenes. Mol Microbiol. 1996;22:867–879. doi: 10.1046/j.1365-2958.1996.01521.x. [DOI] [PubMed] [Google Scholar]
- 5.Comité de I'Antibiogramme de la Société Française de Microbiologie. Statement 1996 CA-SFM: zone sizes and MIC breakpoints for non-fastidious organisms. Clin Microbiol Infect. 1996;2(Suppl. 1):46–49. [PubMed] [Google Scholar]
- 6.Fasola E, Bajaksouzian S, Appelbaum P C, Jacobs M R. Variation in erythromycin and clindamycin susceptibilities of Streptococcus pneumoniae by four test methods. Antimicrob Agents Chemother. 1997;41:129–134. doi: 10.1128/aac.41.1.129. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Giovanetti E, Montanari M P, Marchetti F, Varaldo P E. In vitro activity of ketolides telithromycin and HMR 3004 against Italian isolates of Streptococcus pyogenes and Streptococcus pneumoniae with different erythromycin susceptibility. J Antimicrob Chemother. 2000;46:905–908. doi: 10.1093/jac/46.6.905. [DOI] [PubMed] [Google Scholar]
- 8.Giovanetti E, Montanari M P, Mingoia M, Varaldo P E. Phenotypes and genotypes of erythromycin-resistant Streptococcus pyogenes strains in Italy and heterogeneity of inducibly resistant strains. Antimicrob Agents Chemother. 1999;43:1935–1940. doi: 10.1128/aac.43.8.1935. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Hyder S L, Streitfeld M M. Inducible and constitutive resistance to macrolide antibiotics and lincomycin in clinically isolated strains of Streptococcus pyogenes. Antimicrob Agents Chemother. 1973;4:327–331. doi: 10.1128/aac.4.3.327. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Hynes W L, Ferretti J J, Gilmore M S, Segarra R A. PCR amplification of streptococcal DNA using crude cell lysates. FEMS Microbiol Lett. 1992;94:139–142. doi: 10.1016/0378-1097(92)90597-h. [DOI] [PubMed] [Google Scholar]
- 11.Klugman K P, Capper T, Widdowson C A, Koornhof H J, Moser W. Increased activity of 16-membered lactone ring macrolides against erythromycin-resistant Streptococcus pyogenes and Streptococcus pneumoniae: characterization of South African isolates. J Antimicrob Chemother. 1998;42:729–734. doi: 10.1093/jac/42.6.729. [DOI] [PubMed] [Google Scholar]
- 12.Leclercq R, Courvalin P. Bacterial resistance to macrolide, lincosamide, and streptogramin antibiotics by target modification. Antimicrob Agents Chemother. 1991;35:1267–1272. doi: 10.1128/aac.35.7.1267. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Marchese A, Tonoli E, Debbia E A, Schito G C. Macrolide resistance mechanisms and expression of phenotypes among Streptococcus pneumoniae circulating in Italy. J Antimicrob Chemother. 1999;44:461–464. doi: 10.1093/jac/44.4.461. [DOI] [PubMed] [Google Scholar]
- 14.National Committee for Clinical Laboratory Standards. Methods for dilution antimicrobial susceptibility tests for bacteria that grow aerobically. 5th ed. 2000. Approved standard M7–A5. National Committee for Clinical Laboratory Standards, Wayne, Pa. [Google Scholar]
- 15.Nishijima T, Saito Y, Aoki A, Toriya M, Toyonaga Y, Fujii R. Distribution of mefE and ermB genes in macrolide-resistant strains of Streptococcus pneumoniae and their variable susceptibility to various antibiotics. J Antimicrob Chemother. 1999;43:637–643. doi: 10.1093/jac/43.5.637. [DOI] [PubMed] [Google Scholar]
- 16.Oster P, Zanchi A, Cresti S, Lattanzi M, Montagnani F, Cellesi C, Rossolini G M. Patterns of macrolide resistance determinants among community-acquired Streptococcus pneumoniae isolates over a 5-year period of decreased macrolide susceptibility. Antimicrob Agents Chemother. 1999;43:2510–2512. doi: 10.1128/aac.43.10.2510. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Reinert R R, Bryskier A, Lütticken R. In vitro activities of the new ketolide antibiotics HMR 3004 and HMR 3647 against Streptococcus pneumoniae in Germany. Antimicrob Agents Chemother. 1998;42:1509–1511. doi: 10.1128/aac.42.6.1509. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Roberts M C, Sutcliffe J, Courvalin P, Jensen L B, Rood J, Seppälä H. Nomenclature for macrolide and macrolide-lincosamide-streptogramin B resistance determinants. Antimicrob Agents Chemother. 1999;43:2823–2830. doi: 10.1128/aac.43.12.2823. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Rosato A, Vicarini H, Leclercq R. Inducible or constitutive expression of resistance in clinical isolates of streptococci and enterococci cross-resistant to erythromycin and lincomycin. J Antimicrob Chemother. 1999;43:559–562. doi: 10.1093/jac/43.4.559. [DOI] [PubMed] [Google Scholar]
- 20.Ruoff K L, Whiley R A, Beighton D. Streptococcus. In: Murray P R, Baron E J, Pfaller M A, Tenover F, Yolken R H, editors. Manual of clinical microbiology. 7th ed. Washington, D.C.: American Society for Microbiology; 1999. pp. 283–296. [Google Scholar]
- 21.Seppälä H, Nissinen A, Yu Q, Huovinen P. Three different phenotypes of erythromycin-resistant Streptococcus pyogenes in Finland. J Antimicrob Chemother. 1993;32:885–891. doi: 10.1093/jac/32.6.885. [DOI] [PubMed] [Google Scholar]
- 22.Seppälä H, Skurnik M, Soini H, Roberts M C, Huovinen P. A novel erythromycin resistance methylase gene (ermTR) in Streptococcus pyogenes. Antimicrob Agents Chemother. 1998;42:257–262. doi: 10.1128/aac.42.2.257. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Sutcliffe J, Grebe T, Tait-Kamradt A, Wondrack L. Detection of erythromycin-resistant determinants by PCR. Antimicrob Agents Chemother. 1996;40:2562–2566. doi: 10.1128/aac.40.11.2562. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Sutcliffe J, Tait-Kamradt A, Wondrack L. Streptococcus pneumoniae and Streptococcus pyogenes resistant to macrolides but sensitive to clindamycin: a common resistance pattern mediated by an efflux system. Antimicrob Agents Chemother. 1996;40:1817–1824. doi: 10.1128/aac.40.8.1817. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Syrogiannopoulos G A, Grivea I N, Tait-Kamradt A, Katopodis G D, Beratis N G, Sutcliffe J, Appelbaum P C, Davies T A. Identification of an erm(A) erythromycin resistance methylase gene in Streptococcus pneumoniae isolated in Greece. Antimicrob Agents Chemother. 2001;45:342–344. doi: 10.1128/AAC.45.1.342-344.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Tait-Kamradt A, Clancy J, Cronan M, Dib-Hajj F, Wondrack L, Yuan W, Sutcliffe J. mefE is necessary for erythromycin-resistant M phenotype in Streptococcus pneumoniae. Antimicrob Agents Chemother. 1997;41:2251–2255. doi: 10.1128/aac.41.10.2251. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Weisblum B. Inducible resistance to macrolides, lincosamides and streptogramin type B antibiotics: the resistance phenotype, its biological diversity, and structural elements that regulate expression—a review. J Antimicrob Chemother. 1985;16(Suppl. A):63–90. doi: 10.1093/jac/16.suppl_a.63. [DOI] [PubMed] [Google Scholar]
- 28.Weisblum B. Erythromycin resistance by ribosome modification. Antimicrob Agents Chemother. 1995;39:577–585. doi: 10.1128/AAC.39.3.577. [DOI] [PMC free article] [PubMed] [Google Scholar]