Abstract
The dengue NS2B-NS3 protease existing in equilibrium between the active and inactive forms is essential for virus replication, thus representing a key drug target. Here, myricetin, a plant flavonoid, was characterized to noncompetitively inhibit the dengue protease. Further NMR study identified the protease residues perturbed by binding to myricetin, which were utilized to construct the myricetin–protease complexes. Strikingly, in the active form, myricetin binds to a new allosteric site (AS2) far away from the active site pocket and the allosteric site (AS1) for binding curcumin, while in the inactive form, it binds to both AS1 and AS2. To decipher the mechanism for the allosteric inhibition by myricetin, we conducted molecular dynamics simulations on different forms of dengue NS2B-NS3 proteases. Unexpectedly, the binding of myricetin to AS2 is sufficient to disrupt the active conformation by displacing the characteristic NS2B C-terminal β-hairpin from the active site pocket. By contrast, the binding of myricetin to AS1 and AS2 results in locking the inactive conformation. Therefore, myricetin represents the first small molecule, which allosterically inhibits the dengue protease by both disrupting the active conformation and locking the inactive conformation. The results enforce the notion that a global allosteric network exists in the dengue NS2B-NS3 protease, which is susceptible to allosteric inhibition by small molecules such as myricetin and curcumin. As myricetin has been extensively used as a food additive, it might be directly utilized to fight the dengue infections and as a promising starting material for further design of potent allosteric inhibitors.
1. Introduction
Dengue virus (DENV) belongs to the Flaviviridae family, which also includes Zika (ZIKV), West Nile (WNV), Japanese encephalitis, and yellow fever viruses. DENV is the most prevalent human pathogen transmitted by Aedes mosquitoes with ∼3.6 billion people at risk in >120 countries, particularly in tropical and subtropical regions such as Singapore.1−3 It still remains a huge challenge to develop an effective vaccine, and no marketed antiviral drug exists so far to treat dengue infection despite exhaustive studies.4,5
Common to all Flaviviridae members, DENV has an ∼11 kb single-stranded positive sense RNA genome, which is translated into a large polyprotein by the host cell machinery. Intriguingly, the large polyprotein is subsequently processed into 10 proteins, including three structural (capsid, membrane, and envelope) and seven nonstructural (NS1, NS2A/B, NS3, NS4A/B, and NS5) proteins. The cleavage of the polyprotein is conducted by host cell proteases such as furin and signalaseas, as well as a virus-encoded protease, which has been established to be a key target for drug design to treat DENV and other flavivirus infections.4−26
The dengue protease domain is over the N-terminal region of the NS3 protein, which folds into a chymotrypsin-like fold consisting of two β-barrels with each composed of six β-strands and the catalytic triad (His51-Asp75-Ser135) located at the cleft between the two β-barrels. Intriguingly, unlike other chymotrypsin-like proteases such as 3C-like proteases of coronaviruses,23 the flavivirus proteases including the dengue protease require an additional segment of ∼40 amino acids within the cytosolic region of the NS2B protein for its correct folding and catalysis, thus called a two-component NS2B-NS3 protease.7−24 In crystal structures of flaviviral NS2B-NS3 proteases determined so far, the NS3 protease domains adopt highly similar structures, while the NS2B cofactor was shown to assume two distinctive conformations, namely, the active or closed form and the inactive or open form (Figure S1). Noticeably, in the active form, the NS2B cofactor becomes wrapped around the NS3 protease domain with its C-terminal residues Ser75-Ser79 and Gly82-Ile86 forming a short β-hairpin to serve as part of the active site pocket.7−10 Furthermore, nuclear magnetic resonance (NMR) studies revealed that in solution the two forms undergo significant exchanges even in the absence of any substrates, thus leading to a high conformational dynamics on a microsecond–millisecond time scale.12−15 Strikingly, it appears that a global allosteric network exists in the dengue NS2B-NS3 protease and the perturbation of key residues/sites of the network is sufficient to modulate the conformational equilibrium, thus manifesting as the allosteric effect. For example, the chemical modification of residues around Ala125 led to the conformational equilibrium being shifted to and then locked in the inactive form (Figure S1), which has both the NS2B cofactor and the NS3 protease domain well defined in the crystal structure.17 Furthermore, as we recently showed by NMR and molecular dynamics (MD) simulations, the binding of a natural product curcumin to a pocket close to Ala125 without any overlap with the active site resulted in the allosteric inhibition of the dengue protease by destabilizing the active conformation.26
To date, tremendous efforts have been dedicated to drug design by targeting flaviviral NS2B-NS3 proteases, but the results revealed the major challenge in developing inhibitors for their active sites, which appear to be relatively flat.8−26 Therefore, one promising strategy to overcome this challenge is to discover/design their allosteric inhibitors. Previously, to combat ZIKV and DENV in Singapore, we have carried out intense efforts to identify inhibitors for Zika and dengue NS2B-NS3 proteases from various natural products isolated from edible plants. As a result, we have successfully identified curcumin and a group of flavonoids with significant inhibitory effects on both proteases, which include myricetin, quercetin, luteolin, isorhamnetin, and apigenin, out of which myricetin inhibits the Zika NS2B-NS3 protease with the highest activity (IC50 of 1.3 μM and Ki of 0.8 μM) in a noncompetitive mode. In fact, plant flavonoids have been previously shown to inhibit the dengue protease,24,25 but so far, their binding sites and inhibitory mechanisms have not been experimentally characterized such as by X-ray crystallography or NMR spectroscopy, most likely due to the challenge in studying the dengue NS2B-NS3 protease with largely provoked dynamics upon binding to these flavonoids.
Recently, with a protocol we previously developed for selectively isotope-labeling the NS2B cofactor or the NS3 protease domain,15 we have successfully utilized NMR spectroscopy to identify the binding-involved residues by curcumin, although the binding by curcumin did dramatically enhance the conformational changes on the microsecond–millisecond time scale.26 Consequently, we were able to construct the structures of the curcumin–protease complexes with NMR-derived constraints and conducted further MD simulations. The results revealed that despite binding to a pocket without any overlap with the active site, curcumin imposes the allosteric inhibition by disrupting the active conformation of the dengue protease.26
Here, we aimed to decode the mechanism by which myricetin inhibits the dengue protease by enzymatic assay, NMR characterization, molecular docking, and MD simulations. Briefly, the enzymatic assay showed that myricetin inhibits the dengue protease also in a noncompetitive manner. Further NMR studies identified the binding-perturbed residues by myricetin, which are dramatically different from those binding to curcumin.26 With NMR-derived constraints, the structures of the myricetin–protease complexes have been successfully constructed for both active and inactive forms. Very unexpectedly, in the active form, myricetin binds to a new allosteric site (AS2) far away from the allosteric site (AS1), which was previously identified for binding curcumin (Figure S1B).26 By contrast, in the inactive form, myricetin binds to both AS1 and AS2. Subsequent MD simulations decode that the myricetin binding to the active form at AS2 far away from the active site pocket is sufficient to allosterically destabilize the active conformation. By contrast, the myricetin binding to the inactive form at both AS1 and AS2 locks the inactive conformation. The results together indicate that the dengue protease has more than one allosteric sites for natural products and is susceptible to allosteric inhibition. Therefore, allosteric inhibitors could be designed to target dengue NS2B-NS3 protease not only by disrupting the active conformation but also by locking the inactive conformation.
2. Results
2.1. Myricetin Inhibits the Dengue NS2B-NS3 Protease in a Noncompetitive Manner
Previously to obtain the active recombinant dengue NS2B-NS3 protease in E. coli cells, the enzyme was extensively constructed by joining the NS2B cofactor and the NS3 protease domain covalently with an engineered linker. However, this artificial form does not exist in vivo and in particular has NMR spectra of poor quality.12−15 Therefore, to solve this problem, we have previously developed a protocol to recombinantly generate the dengue protease with the NS2B cofactor and the NS3 protease domain unlinked,15 which also allowed us to selectively isotope-label either NS2B or NS3 in the dengue NS2B-NS3 protease complex for detailed NMR studies, as demonstrated in our recent delineation of the mechanism for the allosteric inhibition by curcumin.26
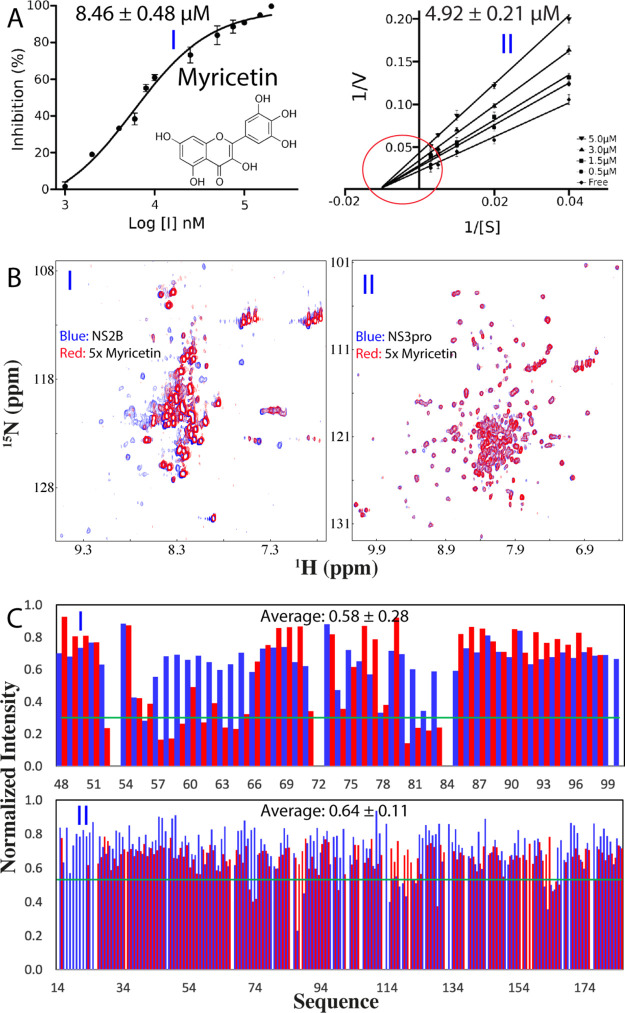
Here, we utilized the same unlinked dengue protease for all enzymatic and NMR experiments. We have determined its Km to be 89.39 ± 6.62 μM and Kcat to be 0.12 ± 0.01 s–1, which are almost identical to our previous results with a Km of 92.39 ± 9.94 μM and a Kcat of 0.15 ± 0.01 s–1.15,26 In our preliminary screening,20 we found that myricetin showed significant inhibition on both Zika and dengue NS2B-NS3 proteases. Here, we further determined its IC50 to be 8.46 ± 0.48 μM and inhibitory constant Ki to be 4.92 ± 0.21 μM on the dengue protease (Figure 1A), which are very similar to those of curcumin on the dengue protease (IC50 of 7.18 and Ki of 4.35 μM).26 Intriguingly, the inhibitory activity of myricetin on the dengue protease appears to be lower than those on the Zika protease (IC50 of 1.30 μM and Ki of 0.80 μM),20 which might be due to distinctive physicochemical properties, conformations, or/and dynamics of the two proteases. However, similar to what we previously observed on the Zika protease,20 myricetin also inhibited the dengue protease by changing Vmax but not Km (Figure 1A), thus suggesting that myricetin acts as a noncompetitive inhibitor for the dengue protease.
Figure 1.
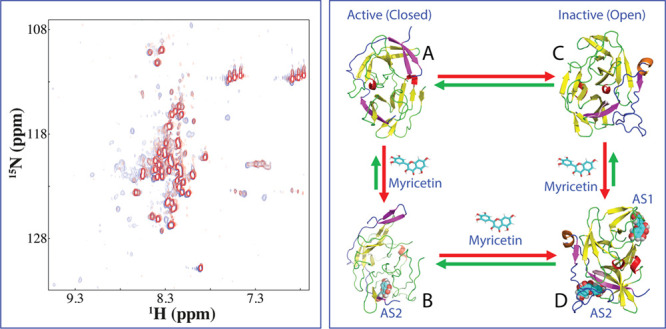
Enzymatic and NMR characterization of the inhibition of the dengue NS2B-NS3 protease by myricetin. (A) Chemical structure of myricetin and the inhibitory data used for fitting IC50 for myricetin. The Lineweaver–Burk plot for determining the inhibitory constant (Ki) of myricetin. [S] is the substrate concentration; v is the initial reaction rate. The red circle is used to indicate that the inhibition is noncompetitive characteristic of the same Km but varying Vmax values. (B) (I) NMR 1H-15N HSQC spectra of the dengue NS2B-NS3 complex with NS2B selectively 15N-labeled at a protein concentration of 100 μM in the absence (blue) and in the presence of myricetin (red) at 1:5 (protease:myricetin). (II) HSQC spectra of the protease complex with NS3 selectively 15N-labeled at a protein concentration of 100 μM in the absence (blue) and in the presence of myricetin (red) at 1:5. (C) (I) Normalized HSQC peak intensity of the 15N-labeled NS2B complexed with the unlabeled NS3 in the presence of myricetin at 1:5. (II) Normalized HSQC peak intensity of the 15N-labeled NS3 complexed with the unlabeled NS2B in the presence of myricetin at 1:5. Significantly perturbed residues are defined to be those with the normalized intensity <0.53 for NS2B and <0.59 for NS3 (average value – 1 standard deviation).
2.2. NMR Characterization of the Binding of Myricetin to the Dengue NS2B-NS3 Protease
To determine the binding modes of myricetin, here, we prepared the dengue protease samples with either the NS2B cofactor or the NS3 protease domain selectively 15N-labeled. As shown in Figure 1B, in the unlinked dengue protease complex, both 15N-labeled NS2B (I of Figure 1B) and NS3 (II of Figure 1B) have well-dispersed HSQC spectra, indicating that both of them are well folded in the protease complex. In particular, the chemical shifts of their HSQC peaks are very similar to what were previously reported.13,15,26
We then titrated the dengue protease samples with myricetin at molar ratios of 1:0.5, 1:1; 1:2.5, 1:5, and 1:10 (protease:myricetin). Intriguingly, no significant shifts were observed for most HSQC peaks of NS2B (I of Figure 1C) and NS3 (II of Figure 1C), indicating no major structural change upon binding, which is very similar to what was observed on the binding of curcumin to the dengue protease.26 On the other hand, their HSQC peaks became stepwise broadened and consequently the intensity of the peaks gradually reduced. At 1:10, many well-dispersed HSQC peaks became too weak to be detectable. Usually, the line-broadening of HSQC peaks upon binding results from the micromolar dissociation constants and/or binding-induced increase of conformational exchanges particularly on the microsecond–millisecond time scale.11−15,26−28 Indeed, for a folded but dynamic protein, a slight unfolding/destabilization of the native structure is sufficient to induce the dramatic increase of conformational exchange on a microsecond–millisecond time scale and, consequently, resulting in the broadening/disappearance of many well-dispersed HSQC peaks.28−30 Here, the myricetin binding-provoked increase of conformational exchanges as detected by NMR might rationalize the results of our ITC measurements on the binding of both myricetin and curcumin26 to the dengue protease, all generated the data with unstable baselines.
The current results indicate that as we recently observed on curcumin,26 the binding of myricetin also led to a significant increase of conformational dynamics on the microsecond–millisecond time scale, in a contrast to a recent NMR report on an active site inhibitor of the dengue protease in which the inhibitor binding resulted in a dramatically reduced dynamics, thus showing much better quality of NMR spectra.14 As such, despite intense attempts, the weakening of the intensity of HSQC peaks prevented us from further acquiring high-quality NMR relaxation data to derive their backbone dynamics in complex with curcumin or myricetin as we previously conducted on other proteins to gain insights into the picosecond–nanosecond and microsecond–millisecond dynamics.27,28
2.3. Distinctive Binding Modes of Myricetin for the Active and Inactive Forms
Due to the significant increase of the protein dynamics upon binding to myricetin or curcumin,26 we were also not able to determine the structures of the dengue protease in complex with myricetin or curcumin by NMR. Furthermore, we also intensely attempted to crystalize their complex samples but all failed. Nevertheless, the ability to selectively label the NS2B cofactor and the NS3 protease domain offered us to follow the intensity changes of HSQC peaks upon adding myricetin at different molar ratios.
Figure 1C presents the normalized peak intensities of NS2B (I of Figure 1C) and NS3 (II of Figure 1C) in the presence of myricetin at 1:5 (red bars) as well as curcumin at 1:5 (blue bars). The NS2B residues have an average intensity of 0.58 and 0.63 for their HSQC peaks, while the NS3 residues have an average intensity of 0.64 and 0.76 in the presence of myricetin and curcumin, respectively, implying that both myricetin and curcumin similarly triggered slightly higher dynamics for the NS2B cofactor than for the NS3 protease domain. Very unexpectedly, however, at residue-specific resolution, myricetin and curcumin induced the distinctive patterns of the intensity changes for both NS2B and NS3 residues. For example, for the NS2B cofactor, myricetin additionally triggered the significant reduction (<average – 1 SD) of the HSQC peak intensity of the residues over 56–64 and 80–83 (I of Figure 1C), while for the NS3 protease domain, myricetin induced the complete disappearance of HSQC peaks of the residue over 18–23 (II of Figure 1C). Very interestingly, the majorities of the perturbed residues of both NS2B and NS3 are conserved in the proteases of four subtypes of DENV (Figure S2). Therefore, the present NMR result also rationalizes the previous enzymatic results that flavonoids including myricetin were able to inhibit the proteases of different DENV subtypes.
We thus mapped these significantly perturbed residues back to the dengue protease structures in either active (Figure 2A) or inactive (Figure 3A) forms. With these NMR-derived constraints, we were able to construct the structures of the myricetin–protease complexes for the active (Figure 2B) or inactive (Figure 3B) form by the well-established molecular docking program HADDOCK,31 which we extensively utilized to build up the protease–curcumin complex26 and complexes of other proteins with small molecules including ATP.32,33
Figure 2.
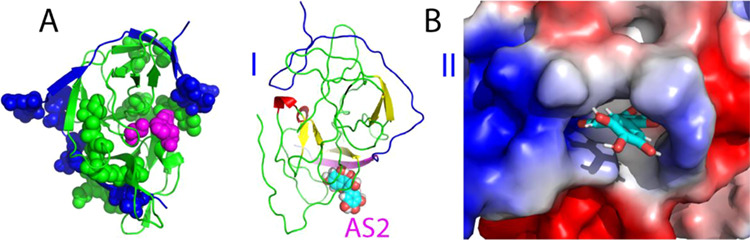
Active form of the dengue NS2B-NS3 protease complexed with myricetin. (A) Structure of the active form of the dengue protease with the significantly perturbed residues displayed in spheres. NS2B is colored in blue while NS3 in green. The catalytic triad residues His51-Asp75-Ser135 are displayed in purple spheres. (B) Lowest energy docking structures in the ribbon (I) and the electrostatic potential surface (II) of the active form of the protease complexed with myricetin at a new allosteric site (AS2). The β-strand is colored in purple and the loop in blue for NS2B, while the β-strand is in yellow, the helix in red, and the loop in green for NS3. Myricetin is displayed in spheres (I) and sticks (II), respectively.
Figure 3.
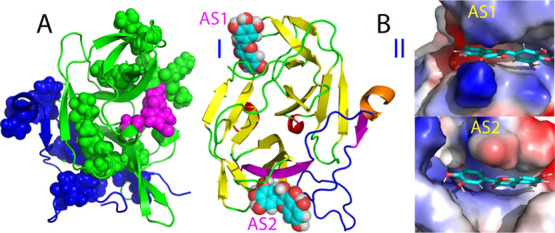
Inactive form of the dengue NS2B-NS3 protease complexed with myricetin. (A) Structure of the inactive form of the dengue NS2B-NS3 protease with the significantly perturbed residues displayed in spheres. NS2B is colored in blue while NS3 in green. The catalytic triad residues His51-Asp75-Ser135 are displayed in purple spheres. (B) Lowest energy docking structures in the ribbon (I) and the electrostatic potential surface (II) of the inactive form of the protease complexed with myricetin at two allosteric sites (AS1 and AS2). The β-strand is colored in purple, the helix in brown, and the loop in blue for NS2B, while the β-strand is in yellow, the helix in red, and the loop in green for NS3. Myricetin is displayed, respectively, as spheres (I) and sticks (II).
Figure 2B presents the lowest energy structure of the myricetin–protease complex for the active form. Very unexpectedly, completely different from the curcumin–protease complex for the active form we previously obtained (Figure S1B) in which curcumin binds to the dengue protease at an allosteric site (designated as AS1) close to Ala125,17,26 here, myricetin binds to the dengue protease at a new allosteric site (designated as AS2), which is far away from both the active site pocket and AS1. It is very interesting to note that even in the complex, the short β-hairpin characteristic of the active form over the NS2B residues Ser75-Ser79 and Gly82-Ile86 is distorted to some degree, although it is still located at the original position, implying that the binding of myricetin to AS2 could perturb the conformation of this β-hairpin despite being located far away. A close examination revealed that AS2 is constituted mostly by the NS3 residues Lys15-Asp20 in the loop and Gly21-Arg24 in the β-stand, as well as the NS2B residues Leu54-Val59 (I of Figure 2B). Consistent with the complex structure, in the presence of myricetin at 1:5, HSQC peaks of many of these involved NS3 residues became too broad to be detectable, while those of the NS2B residues also have significantly reduced intensities (Figure 1C), both of which are unique for the myricetin binding and were not observed on the curcumin binding (Figure 1C). Moreover, the AS2 is relatively positively charged as shown in II of Figure 2B.
On the other hand, for the inactive form of the dengue protease,17 myricetin was identified to bind not only to AS2 but also to AS1 as we previously found for binding curcumin. For AS2 of the inactive form, in addition to the NS3 and NS2B residues observed in the active form (Figure 2B), the NS2B residues Thr77-Gly82 further fold back to have direct contact with myricetin (Figure 3B). For AS1 of the inactive form, only NS3 residues are involved in contacting myricetin, including Val72-Asp75 and T118-T122 as well as G151-V154, which form a β-sheet with A160-A166. Again, consistent with the complex structure, many of these involved NS3 residues have significantly reduced intensities of their HSQC peaks upon binding to myricetin but not to curcumin (II of Figure 1C). Interestingly, both AS1 and AS2 of the inactive form have similar electrostatic properties: relatively positively charged (II of Figure 3B) very similar to AS2 of the active form (II of Figure 2B).
2.4. Myricetin Destabilizes the Active Conformation of the Dengue NS2B-NS3 Protease
In the present study, myricetin has been identified to bind one allosteric site (AS2) in the active form but to two allosteric sites (AS1 and AS2) in the inactive form of the dengue NS2B-NS3 protease. Intriguingly, both AS1 and AS2 have no overlap with the active site. Therefore, an immediate question to be answered is how the binding of myricetin to these sites can lead to the allosteric inhibition of the catalytic activity of the dengue NS2B-NS3 protease. To address this question, we subsequently conducted MD simulations on two myricetin–protease complexes up to 50 ns. The MD simulation represents a powerful method to obtain the dynamic properties of proteins underlying their functions including the enzymatic catalysis.34−38 Particularly, it can pinpoint the dynamically driven allosteric mechanisms for the enzymatic catalysis and inhibition. For example, previously with MD simulations, we have successfully revealed the existence of a global allosteric network in the SARS 3C-like protease, the mutation perturbation of which could lead to either inactivation37 or enhancement of the catalytic activity,38 although all these mutations have no significant effect on the crystal structures. Very recently, with MD simulations, we delineated the dynamically driven mechanism for the allosteric inhibition of the dengue NS2B-NS3 protease by curcumin.26
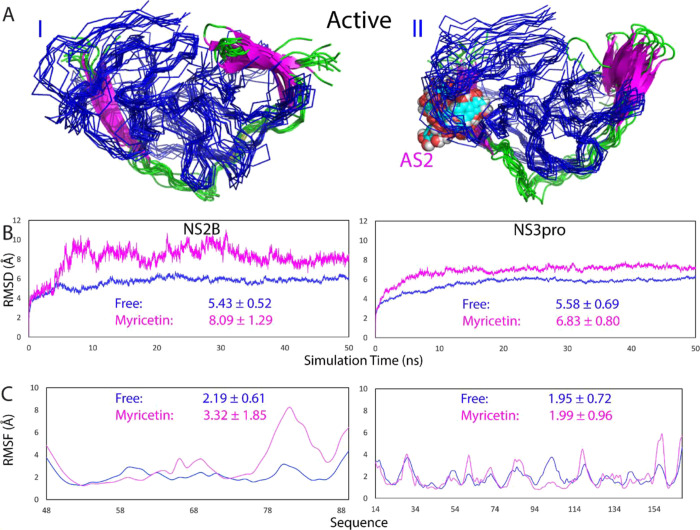
Therefore, here, to explore the mechanism by which myricetin allosterically inhibits the dengue NS2B-NS3 protease, we first set up three independent MD simulations up to 50 ns on the myricetin–protease complex in the active form. Figure 4A presents the structure snapshots in the first set of MD simulations for the active form of the dengue protease in the unbound state (I of Figure 4A) and in complex with myricetin at AS2 (II of Figure 4A). Figure 4B presents the trajectories of RMSD of Cα atoms averaged over three independent simulations of the NS2B cofactor and the NS3 protease domain of the dengue protease in the unbound state (blue) we previously performed for understanding the allosteric inhibition of curcumin26 as well as in complex with myricetin we currently conducted (purple). For NS2B, the averaged RMSD values are 5.43 ± 0.52 and 8.09 ± 1.29 Å, respectively, for the unbound and myricetin-bound states. For the NS3 protease domain, the averaged RMSD values are 5.58 ± 0.69 and 6.83 ± 0.80 Å, respectively, for the unbound and myricetin-bound states. This set of the results clearly indicates that myricetin binding led to a large increase of conformational dynamics in NS2B but only a slight change in NS3. This observation is in a general agreement with the NMR results that upon binding to myricetin, the NS2B factor becomes more dynamic than the NS3 protease domain (Figure 1C).
Figure 4.
Overall dynamic behaviors of the active form complexed with myricetin. (A) Superimposition of 11 structures (one structure for a 5 ns interval) of the first set of MD simulations for the active dengue NS2B-NS3 protease in the free state (I) and in complex with myricetin (II). For clarity, NS3 is displayed in the blue ribbon and NS2B in cartoon with the β-strand colored in purple and the loop in green, as well as myricetin in spheres. (B) Root-mean-square deviations (RMSD) of the Cα atoms (from their positions in the initial structures used for MD simulations after the energy minimization) averaged over three independent MD simulations of NS2B and NS3, respectively. (C) Root-mean-square fluctuations (RMSF) of the Cα atoms averaged over three independent MD simulations of NS2B and NS3, respectively.
Similar dynamic behaviors are also reflected by the RMSF of the Cα atoms in the MD simulations. Figure 4C presents the averaged RMSF of three independent simulations in which for the NS2B residues, the averaged RMSF values are 2.19 ± 0.61 and 3.32 ± 1.85 Å, respectively, for the unbound and myricetin-bound states, while for the NS3 residues, the averaged RMSF values are 1.95 ± 0.72 and 1.99 ± 0.96 Å, respectively, for the unbound and myricetin-bound states. The RMSF values also indicate that during simulations, no significant difference was observed on the overall conformational fluctuations of the NS3 residues in the unbound and myricetin-bound states. On the other hand, upon binding to myricetin, the conformational fluctuations of the NS2B residues increased. Indeed, the C-terminal residues Ile76-Glu89 become highly dynamic, while residues 48–52 and 65–69 also showed high dynamics. On the other hand, it is important to point out that the current 50 ns simulations are insufficient to capture the large conformational transition from the closed (active) to the open (inactive) conformations, which is expected to occur over the microsecond–second time scale.
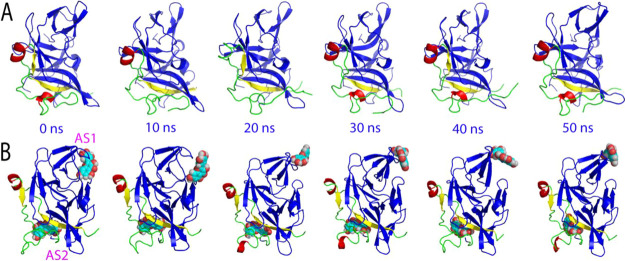
The structural changes in simulations can be better visualized in the structure snapshot at different simulation time points. As well established, in the active conformation, the short antiparallel β-hairpin formed over the NS2B residues Ser75-Ser79 and Gly82-Ile86 wraps the NS3 protease domain to serve as part of the active site pocket, and indeed in the previous study its tip was found to have direct contact with the substrate peptide, which therefore, represents a characteristic feature of the active form (Figure S1A). As we have previously shown,26 although the active form in the unbound state is dynamic, this NS2B β-hairpin retains and only has small fluctuations over its original position in the 50 ns simulation. By contrast, for the myricetin-bound state, after 10 ns of the simulation, this short β-hairpin was significantly displaced from its original position to become highly exposed to bulk solvent. Another significant change in the myricetin-bound state is that the long NS2B β-strand over residues Asp50-Ala57, which has direct contact with myricetin, became much shorter after 20 ns, only over Leu51-Glu54 (Figure 5), while this β-strand showed no significant changes over the 50 ns simulation for the active form in the unbound state.26 Together with our present simulation results on the active form in the unbound dengue protease,26 the MD simulation results reveal that the binding of myricetin at AS2 not only disrupts the NS2B β-strand over Asp50-Ala57, which has direct contact with myricetin, but unexpectedly displaced the characteristic β-hairpin from the active site pocket to become highly exposed to bulk solvent. Therefore, the binding of myricetin to AS2 of the active form is sufficient to disrupt the active conformation and, consequently, achieves the allosteric inhibition of the dengue NS2B-NS3 protease.
Figure 5.
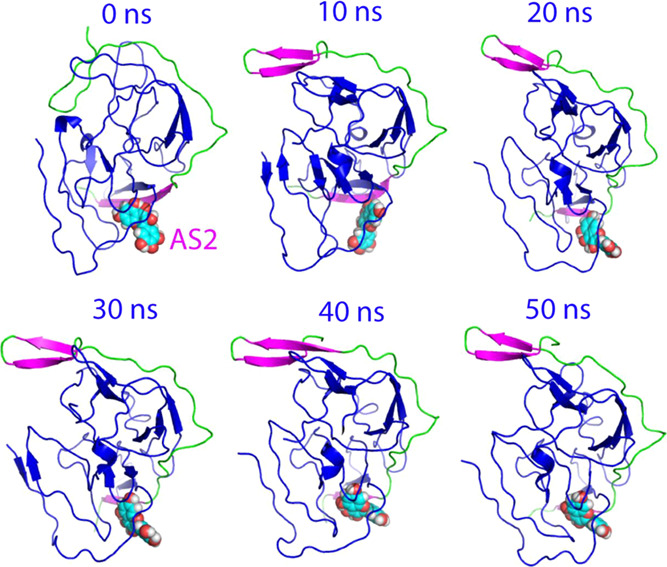
Structure snapshots of MD simulations of the active form. Six individual structures of the first set of MD simulations for the active form of the dengue NS2B-NS3 protease complexed with myricetin at different simulation time points. For clarity, NS3 is displayed in the blue cartoon and NS2B with the β-strand colored in purple and the loop in green, as well as myricetin in spheres.
2.5. Myricetin Binds and Locks the Inactive Conformation of the Dengue NS2B-NS3 Protease
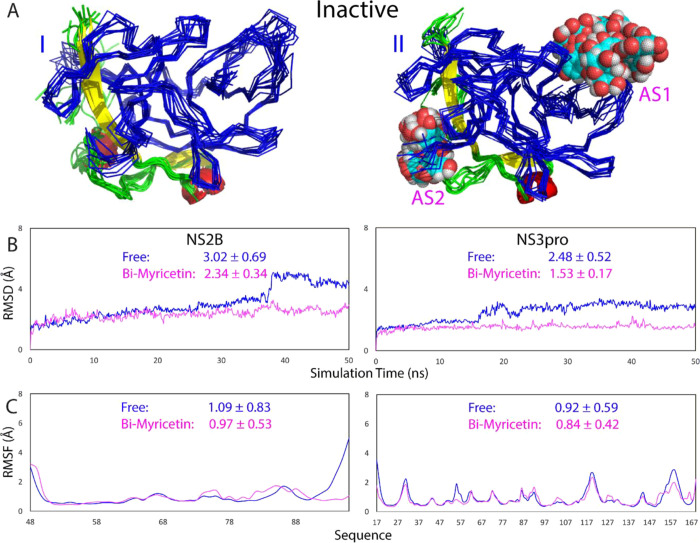
We further performed three independent MD simulations up to 50 ns on the inactive form of the dengue NS2B-NS3 protease in the unbound and myricetin-bound states. Figure 6A presents the structure snapshots in the first sets of MD simulations in the inactive form in the unbound state (I of Figure 6A) and in complex with two myricetin molecules (II of Figure 6A). Strikingly, overall, the inactive form in the unbound state appears to be much less dynamic than that of the active form (Figure 4A), while the inactive form in the myricetin-bound state became even less dynamic than the inactive form in the unbound state (Figure 6A).
Figure 6.
Overall dynamic behaviors of the inactive form complexed with myricetin. (A) Superimposition of 11 structures (one structure for the 5 ns interval) of the first set of MD simulations for the inactive form of the dengue NS2B-NS3 protease in the free state (I) and in complex with myricetin at two allosteric sites (II). For clarity, NS3 is displayed in the blue ribbon and NS2B with the β-strand colored in yellow, the helix in red, and the loop in green, as well as myricetin in spheres. (B) RMSD of the Cα atoms (from their positions in the initial structures used for MD simulations after energy minimization) averaged over three independent MD simulations of NS2B and NS3, respectively. (C) RMSF of the Cα atoms averaged over three independent MD simulations of NS2B and NS3, respectively.
Figure 6B presents the trajectories of RMSD of Cα atoms averaged over three independent simulations of the NS2B cofactor and the NS3 protease domain of the inactive form of the dengue protease in the unbound state (blue) and in complex with two myricetin molecules (purple). For NS2B, the averaged RMSD values are 3.02 ± 0.69 and 2.34 ± 0.34 Å, respectively, for the unbound and myricetin-bound proteases. For the NS3 protease domain, the averaged RMSD values are 2.48 ± 0.52 and 1.53 ± 0.17 Å, respectively, for the unbound and myricetin-bound proteases. The results clearly indicate that: (1) even in the unbound state, the inactive form is much less dynamic than the active form for both the NS2B cofactor and the NS3 protease domain. (2) Most strikingly, unlike the active form for which the myricetin binding provoked the increase in conformational dynamics particularly over the NS2B cofactor, the binding of two myricetin molecules to AS1 and AS2 of the inactive form by contrast led to the reduction of conformational dynamics for both NS2B and NS3.
Similar dynamic behaviors are also reflected by the RMSF of the Cα atoms in the MD simulations. Figure 6C presents the averaged RMSF of three independent simulations in which for NS2B, the averaged RMSF values are 1.09 ± 0.83 and 0.97 ± 0.53 Å, respectively, for the unbound and myricetin-bound proteases, while for the NS3 protease domain, the averaged RMSF values are 0.92 ± 0.59 and 0.84 ± 0.42 Å, respectively, for the unbound and myricetin-bound proteases. The RMSF values indicate that during simulations, no significant difference was observed on the overall conformational fluctuations of the NS3 residues in the unbound and myricetin-bound states. On the other hand, upon binding to two myricetin molecules, the conformational fluctuations of the NS2B residues decreased. Particularly, the RMSF of residues Glu92-Arg100 become significantly reduced (I of Figure 6C). Moreover, as shown in the structure snapshots (Figure 7), the inactive forms in both the unbound and bound with two myricetin molecules are much less dynamic than those of the active forms (Figure 5).
Figure 7.
Structure snapshots of MD simulations of the inactive form. (A) Six individual structures of the first set of MD simulations for the inactive form of the dengue NS2B-NS3 protease in the free state. (B) Six individual structures of the first set of MD simulations for the inactive form of the dengue NS2B-NS3 protease complexed with myricetin at two allosteric sites at different simulation time points. For clarity, NS3 is displayed in the blue cartoon and NS2B with the β-strand colored in yellow, the helix in red, and the loop in green, as well as myricetin in spheres.
Therefore, the MD simulation results indicate that the inactive form is less dynamic than the active form, and most unexpectedly, the binding of two myricetin molecules to the inactive form further reduced the conformational dynamics of both the NS2B cofactor and the NS3 protease domain. Therefore, myricetin appears to allosterically inhibit the dengue NS2B-NS3 protease also by binding to both AS1 and AS2 to lock the inactive conformation.
3. Discussion and Conclusions
The flaviviral NS2B-NS3 proteases are highly conserved and therefore have been established to be key targets for discovery/design of inhibitors for treating flavivirus infections.4−24,39−42 Currently, most studies have focused on the active sites for discovery/design of the competitive inhibitors.21−23 However, due to the intrinsic features, the design of the active site inhibitors has been demonstrated to be largely challenging. In this regard, one solution is to discover/design their allosteric inhibitors capable of binding to the cavities other than the active sites. Nevertheless, because of the general challenge in understanding the mechanisms of the allosteric processes, only limited attempts have been directed to developing allosteric inhibitors of flaviviral proteases.17−24,26,39−42 Very recently, we have delineated the mechanism for the allosteric inhibition of the dengue NS2B-NS3 protease by curcumin, which binds an allosteric site (AS1) close to but having no direct overlap with the active site pocket.26
In the present study, we further explored the mechanism for the inhibition of the dengue protease by myricetin, a flavonoid extensively existing in edible plants, with the well-recognized nutraceuticals value.43 Very unexpectedly, although myricetin and curcumin both allosterically inhibit the dengue NS2B-NS3 protease, NMR characterization revealed that the pattern of the perturbed NS2B and NS3 residues upon binding to myricetin is fundamentally different from that upon binding to curcumin.26 With NMR-derived constraints, the myricetin–protease complexes were successfully constructed for both active and inactive forms of the dengue NS2B-NS3 protease. Indeed, consistent with NMR results, for the active form, myricetin binds to a new allosteric site (AS2), which is far away from both the active site pocket and AS1 previously identified for binding curcumin. By contrast, for the inactive form, myricetin has been shown to bind to both AS1 and AS2.
Therefore, a key question to be addressed is how myricetin is able to allosterically inhibit dengue NS2B-NS3 proteases if its two binding sites have no overlap with the active site pocket. To answer this question, we subsequently conducted MD simulations on both active and inactive forms in the free state and in complex with myricetin. Strikingly, for the active form, the binding of myricetin to AS2 far away from the active site pocket as well as the NS2B β-hairpin characteristic of the active form is sufficient to displace this β-hairpin to become highly exposed to bulk solvent, thus disrupting the active conformation. By contrast, for the inactive form, the binding of myricetin to AS1 and AS2 acts to reduce the conformational dynamics of both NS2B and NS3, thus locking the dengue protease in the inactive conformation. In this context, here, we propose a speculative mechanism for myricetin to allosterically inhibit the dengue NS2B-NS3 protease (Figure 8). Briefly, in solution, the active and inactive forms of the protease coexist to reach equilibrium of conformational exchanges on the microsecond–millisecond time scale, which, however, could be modulated by many factors. For the active form (Figure 8A), only AS2 is highly accessible and suitable for binding myricetin, and this binding results in the disruption of the active conformation as reflected by the displacement of the characteristic β-hairpin (Figure 8B). On the other hand, for the inactive form (Figure 8C), both AS1 and AS2 become accessible and suitable for binding myricetin and the binding leads to the locking of the dengue NS2B-NS3 protease in the inactive conformation (Figure 8D). Moreover, it might also be possible that for the active form, upon displacing the β-hairpin by binding of one myricetin molecule at AS2, AS1 then becomes accessible to binding another myricetin molecule, which will transform the active form to the inactive form. Unfortunately, as this large structural transition is anticipated to occur on the microsecond–millisecond time scale, it still remains challenging to be assessed by MD simulations. Nevertheless, the present study reveals that unlike curcumin, which allosterically inhibits the dengue NS2B-NS3 protease only by binding to AS1 to destabilize the active conformation, myricetin allosterically inhibits the enzyme by binding to AS2 of the active form to destabilize the active conformation as well as by binding to both AS1 and AS2 of the inactive form to lock the enzyme in the inactive conformation.
Figure 8.
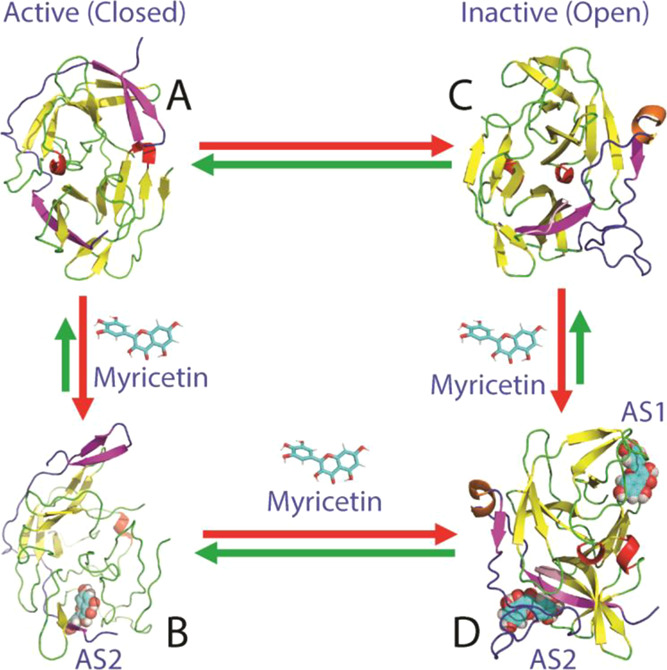
Speculative model for myricetin to allosterically inhibit the dengue protease. In solution, the dengue NS2B-NS3 protease exists in conformational equilibrium between the active (A) and inactive (C) forms. Myricetin achieves the allosteric inhibition of the dengue NS2B-NS3 protease by binding to the active form at AS2 to destabilize the active conformation (B) or/and by binding to the inactive form at both AS1 and AS2 to stabilize/lock the inactive conformation (D), both of which lead to the inhibition of the catalytic activity although AS1 and AS2 have no overlap with the active site.
In conclusion, here, we have first successfully established the binding modes of myricetin in both active and inactive forms of the dengue NS2B-NS3 protease. Subsequent MD simulations decrypted that myricetin is able to allosterically inhibit the protease not only by disrupting the active conformation as curcumin does but additionally by locking the protease in the inactive conformation. The current results also enforce the notion that the dengue NS2B-NS3 protease does own a global allosteric network and perturbation of this network at key sites is sufficient to achieve allosteric inhibition. The high susceptibility of the dengue NS2B-NS3 protease to the allosteric regulation highlights the promising potential to discover/design its allosteric small molecules for therapeutic applications. Finally, as flavonoids from edible plants have been extensively used for beneficial ingredients of various foods and beverages, myricetin might be directly used to inhibit the dengue infections or/and serve as a starting lead for the design of potent allosteric inhibitors for the dengue NS2B-NS3 protease.
4. Experimental Section
4.1. Plasmid Construction, Protein Expression, and Purification
Here, we used the expression plasmids we previously obtained,15,26 which encodes DENV2 NS3 (14–185) and NS2B (48–100).13 The NS2B and NS3 were expressed in Escherichia coli BL21 (DE3) Star cells as we performed before.15,26 The isotope-labeled proteins were expressed in M9 medium with the addition of (15NH4)2SO4 for 15N labeling.15,26 The protein concentration was determined in the presence of 8 M urea.44
4.2. Enzymatic Kinetic Assay
Enzymatic kinetic assays followed the protocol we previously established for dengue and Zika NS2B-NS3 proteases,15,20,26 which were performed in triplicate. Bz-Nle-Lys-Arg-Arg-AMC (Bz-nKRR-AMC) was purchased from GenScript (Piscataway, NJ), while HPLC-purified myricetin was from Sigma-Aldrich with the purity >95%. The protease is dissolved in 50 mM Tris–HCl (pH 7.5), 0.001% Triton X-100, and 0.5 mM EGTA. To determine IC50 for myricetin, 50 nM protease was incubated with various concentrations of myricetin at 37 °C for 30 min and then Bz-nKRR-AMC was added to 250 μM to initiate enzymatic reactions. To determine the Ki for myricetin, the kinetic assays were performed with different final concentrations of Bz-nKRR-AMC and myricetin. The enzymatic reaction was monitored with fluorescence at a λex of 380 nm and a λem of 450 nm. Relative fluorescence (units/s) were fitted to the noncompetitive inhibition model in GraphPad Prism 7.0. Ki was obtained with fitting to equation: Vmaxinh = Vmax/(1 + I/Ki), while I is the concentration of the inhibitor.15,20,26
4.3. NMR Characterization of the Binding
2D 1H-15N HSQC NMR experiments were acquired on an 800 MHz Bruker Avance spectrometer as described previously.15,20,26 NMR samples of 100 μM protease were prepared in 10 mM phosphate buffer, pH 7.5, 5% DMSO, and 10% D2O for NMR spin-lock. All NMR experiments were carried out at 25 °C.
4.4. Molecular Docking
In this study, the crystal structures of the dengue protease in the closed form (PDB code: 3U1I)9 and in the open form (PDB ID of 4M9T)17 were used for docking. The structural geometry of myricetin was generated and optimized with Avogadro.45 NMR-derived constraints were used to guide the docking by HADDOCK software31 and CNS.46 CNS topology and force field parameters of myricetin is converted from the PRODRG server.47 The docking of the myricetin–protease complexes was conducted as we extensively performed.32,33 The myricetin–protease structures with the lowest energy scores were selected for the detailed analysis and display by Pymol (PyMOL Molecular Graphics System, Version 0.99rc6 Schrödinger, LLC).
4.5. MD Simulations
The crystal structures of the dengue NS2B-NS3 protease in the active form (PDB code of 3U1I) and in the inactive form (PDB ID of 4M9T) were used as the free states, while the docking structures of the myricetin–protease complexes in the active and inactive forms were used as the bound form for MD simulations with three independent simulations. The electrostatic potential was first calculated with the 6-31G(d,p) basis set using the Gaussian 16 program and then converted into partial charge of individual atoms using the restrained electrostatic potential (RESP) procedure in the Antechamber program.48 The topology parameters of myricetin were obtained using GAFF.49 All MD simulations reaching 50 ns were performed by the use of GROMACS50 with AMBER99SB-IDLN all-atom force field51 parameters.
The simulation system is a periodic cubic box with about 13,000 water molecules (TIP3P model). Na+ ions were randomly added to neutralize the charge of myricetin–protease complexes. The long-range electrostatic interactions were treated using the fast particle-mesh Ewald summation method,52 while bond lengths including hydrogen atoms were constrained by the LINCS algorithm.53 The time step was set as 2 fs. The initial structures were relaxed by 500 steps of energy minimization, followed by 100 ps equilibration with a harmonic restraint potential before MD simulations.
Acknowledgments
This study is supported by the Ministry of Education of Singapore (MOE) Tier 1 Grant R-154-000-B45-114 and R-154-000-B92-114 to J. S.
Supporting Information Available
The Supporting Information is available free of charge at https://pubs.acs.org/doi/10.1021/acsomega.1c05569.
The equilibrium of the conformational exchange of the dengue NS2B-NS3 protease in the active and inactive forms (Figure S1), as well as sequence alignment of four subtypes of the DENV proteases (Figure S2) (PDF)
Author Contributions
† M.D. and L.L. contribute equally.
The authors declare no competing financial interest.
Supplementary Material
References
- Beatty M. E.; Stone A.; Fitzsimons D. W.; Hanna J. N.; Lam S. K.; Vong S.; Guzman M. G.; Mendez-Galvan J. F.; Halstead S. B.; Letson G. W.; Kuritsky J.; Mahoney R.; Margolis H. S.; Best practices in dengue surveillance: a report from the Asia-Pacific and Americas Dengue Prevention Boards. PLoS Neglected Trop. Dis. 2010, 4, e8902 10.1371/journal.pntd.0000890. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bhatt S.; Gething P. W.; Brady O. J.; Messina J. P.; Farlow A. W.; Moyes C. L.; Drake J. M.; Brownstein J. S.; Hoen A. G.; Sankoh O.; Myers M. F.; George D. B.; Jaenisch T.; Wint G. R.; Simmons C. P.; Scott T. W.; Farrar J. J.; Hay S. I. The global distribution and burden of dengue. Nature 2013, 496, 504–507. 10.1038/nature12060. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Guzman M. G.; Halstead S. B.; Artsob H.; Buchy P.; Farrar J.; Gubler D. J.; Hunsperger E.; Kroeger A.; Margolis H. S.; Martínez E.; Nathan M. B.; Pelegrino J. L.; Simmons C.; Yoksan S.; Peeling R. W. Dengue: a continuing global threat. Nat. Rev. Microbiol. 2010, 8, S7–S16. 10.1038/nrmicro2460. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Behnam M. A. M.; Nitsche C.; Boldescu V.; Klein C. D. The Medicinal Chemistry of Dengue Virus. J. Med. Chem. 2016, 59, 5622–5649. 10.1021/acs.jmedchem.5b01653. [DOI] [PubMed] [Google Scholar]
- Timiri A. K.; Sinha B. N.; Jayaprakash V. Progress and prospects on DENV protease inhibitors. Eur. J. Med. Chem. 2016, 117, 125–143. 10.1016/j.ejmech.2016.04.008. [DOI] [PubMed] [Google Scholar]
- Perera R.; Kuhn R. J. Structural proteomics of dengue virus. Curr. Opin. Microbiol. 2008, 11, 369–377. 10.1016/j.mib.2008.06.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Noble C. G.; Shi P. Y. Structural biology of dengue virus enzymes: towards rational design of therapeutics. Antiviral Res. 2012, 96, 115–126. 10.1016/j.antiviral.2012.09.007. [DOI] [PubMed] [Google Scholar]
- Erbel P.; Schiering N.; D’Arcy A.; Renatus M.; Kroemer M.; Lim S. P.; Yin Z.; Keller T. H.; Vasudevan S. G.; Hommel U. Structural basis for the activation of flaviviral NS3 proteases from dengue and West Nile virus. Nat. Struct. Mol. Biol. 2006, 13, 372–373. 10.1038/nsmb1073. [DOI] [PubMed] [Google Scholar]
- Noble C. G.; She C. C.; Chao A. T.; Shi P. Y. Ligand-bound structures of the dengue virus protease reveal the active conformation. J. Virol. 2012, 86, 438–446. 10.1128/JVI.06225-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Luo D.; Vasudevan S. G.; Lescar J. The flavivirus NS2B-NS3 protease helicase as a target for antiviral drug development. Antiviral Res. 2015, 118, 148–158. 10.1016/j.antiviral.2015.03.014. [DOI] [PubMed] [Google Scholar]
- de la Cruz L.; Nguyen T. H.; Ozawa K.; Shin J.; Graham B.; Huber T.; Otting G. Binding of low molecular weight inhibitors promotes large conformational changes in the dengue virus NS2B-NS3 protease: fold analysis by pseudocontact shifts. J. Am. Chem. Soc. 2011, 133, 19205–19215. 10.1021/ja208435s. [DOI] [PubMed] [Google Scholar]
- de la Cruz L.; Chen W. N.; Graham B.; Otting G. Binding mode of the activity-modulating C-terminal segment of NS2B to NS3 in the dengue virus NS2B-NS3 protease. FEBS J. 2014, 281, 1517–1533. 10.1111/febs.12729. [DOI] [PubMed] [Google Scholar]
- Kim Y. M.; Gayen S.; Kang C.; Joy J.; Huang Q.; Chen A. S.; Wee J. L.; Ang M. J.; Lim H. A.; Hung A. W.; Li R.; Noble C. G.; Lee L. T.; Yip A.; Wang Q. Y.; Chia C. S.; Hill J.; Shi P. Y.; Keller T. H. NMR analysis of a novel enzymatically active unlinked dengue NS2B-NS3 protease complex. J. Biol. Chem. 2013, 288, 12891–12900. 10.1074/jbc.M112.442723. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gibbs A. C.; Steele R.; Liu G.; Tounge B. A.; Montelione G. T. Inhibitor Bound Dengue NS2B-NS3pro Reveals Multiple Dynamic Binding Modes. Biochemistry 2018, 57, 1591–1602. 10.1021/acs.biochem.7b01127. [DOI] [PubMed] [Google Scholar]
- Gupta G.; Lim L.; Song J. NMR and MD studies reveal that the isolated dengue NS3 protease is an intrinsically disordered chymotrypsin fold which absolutely requests NS2B for correct folding and functional dynamics. PLoS One 2015, 10, e0134823 10.1371/journal.pone.0134823. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li J.; Lim S. P.; Beer D.; Patel V.; Wen D.; Tumanut C.; Tully D. C.; Williams J. A.; Jiricek J.; Priestle J. P.; Harris J. L.; Vasudevan S. G. Functional profiling of recombinant NS3 proteases from all four serotypes of dengue virus using tetra-peptide and octapeptide substrate libraries. J. Biol. Chem. 2005, 280, 28766–28774. 10.1074/jbc.M500588200. [DOI] [PubMed] [Google Scholar]
- Yildiz M.; Ghosh S.; Bell J. A.; Sherman W.; Hardy J. A. Allosteric inhibition of the NS2B-NS3 protease from dengue virus. ACS Chem. Biol. 2013, 8, 2744–2752. 10.1021/cb400612h. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yao Y.; Huo T.; Lin Y. L.; Nie S.; Wu F.; Hua Y.; Wu J.; Kneubehl A. R.; Vogt M. B.; Rico-Hesse R.; Song Y. Discovery, X-ray Crystallography and Antiviral Activity of Allosteric Inhibitors of Flavivirus NS2B-NS3 Protease. J. Am. Chem. Soc. 2019, 141, 6832–6836. 10.1021/jacs.9b02505. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lei J.; Hansen G.; Nitsche C.; Klein C. D.; Zhang L.; Hilgenfeld R. Crystal structure of Zika virus NS2B-NS3 protease in complex with a boronate inhibitor. Science 2016, 353, 503–505. 10.1126/science.aag2419. [DOI] [PubMed] [Google Scholar]
- Roy A.; Lim L.; Srivastava S.; Lu Y.; Song J. Solution conformations of Zika NS2B-NS3pro and its inhibition by natural products from edible plants. PLoS One 2017, 12, e0180632 10.1371/journal.pone.0180632. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nitsche C. Proteases from dengue, West Nile and Zika viruses as drug targets. Biophys. Rev. 2019, 11, 157–165. 10.1007/s12551-019-00508-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Majerová T.; Novotnỳ P.; Krỳsová E.; Konvalinka J. Exploiting the unique features of Zika and Dengue proteases for inhibitor design. Biochimie 2019, 166, 132–141. 10.1016/j.biochi.2019.05.004. [DOI] [PubMed] [Google Scholar]
- Lim L.; Gupta G.; Roy A.; Kang J.; Srivastava S.; Shi J.; Song J. Structurally- and dynamically-driven allostery of the chymotrypsin-like proteases of SARS, Dengue and Zika viruses. Prog. Biophys. Mol. Biol. 2019, 143, 52–66. 10.1016/j.pbiomolbio.2018.08.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- de Sousa L. R.; Wu H.; Nebo L.; Fernandes J. B.; da Silva M. F.; Kiefer W.; Kanitz M.; Bodem J.; Diederich W. E.; Schirmeister T.; Vieira P. C. Flavonoids as noncompetitive inhibitors of Dengue virus NS2B-NS3 protease: inhibition kinetics and docking studies. Bioorg. Med. Chem. 2015, 23, 466–470. 10.1016/j.bmc.2014.12.015. [DOI] [PubMed] [Google Scholar]
- Sarwar M. W.; Riaz A.; Dilshad S. M. R.; Al-Qahtani A.; Nawaz-Ul-Rehman M. S.; Mubin M. Structure activity relationship (SAR) and quantitative structure activity relationship (QSAR) studies showed plant flavonoids as potential inhibitors of dengue NS2B-NS3 protease. BMC Struct. Biol. 2018, 18, 6. 10.1186/s12900-018-0084-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lim L.; Dang M.; Roy A.; Kang J.; Song J. Curcumin Allosterically Inhibits the Dengue NS2B-NS3 Protease by Disrupting Its Active Conformation. ACS Omega 2020, 5, 25677–25686. 10.1021/acsomega.0c00039. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Qin H.; Lim L. Z.; Song J. Dynamic principle for designing antagonistic/agonistic molecules for EphA4 receptor, the only known ALS modifier. ACS Chem. Biol. 2015, 10, 372–378. 10.1021/cb500413n. [DOI] [PubMed] [Google Scholar]
- Qin H.; Lim L.; Wei Y.; Song J. TDP-43 N terminus encodes a novel ubiquitin-like fold and its unfolded form in equilibrium that can be shifted by binding to ssDNA. Proc. Natl. Acad. Sci. U. S. A. 2014, 111, 18619–18624. 10.1073/pnas.1413994112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Song J.; Jamin N.; Gilquin B.; Vita C.; Menez A. A gradual disruption of tight side-chain packing: 2D 1H- NMR characterization of acid-induced unfolding of CHABII. Nat. Struct. Biol. 1999, 6, 129–134. 10.1038/5815. [DOI] [PubMed] [Google Scholar]
- Wei Z.; Song J. Molecular mechanism underlying the thermal stability and pH-induced unfolding of CHABII. J. Mol. Biol. 2005, 348, 205–218. 10.1016/j.jmb.2005.02.028. [DOI] [PubMed] [Google Scholar]
- Dominguez C.; Boelens R.; Bonvin A. M. HADDOCK: a protein- protein docking approach based on biochemical or biophysical information. J. Am. Chem. Soc. 2003, 125, 1731–1737. 10.1021/ja026939x. [DOI] [PubMed] [Google Scholar]
- Qin H.; Shi J.; Noberini R.; Pasquale E. B.; Song J. Crystal structure and NMR binding reveal that two small molecule antagonists target the high affinity ephrin-binding channel of the EphA4 receptor. J Biol Chem. 2008, 283, 29473–29484. 10.1074/jbc.M804114200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kang J.; Lim L.; Song J. ATP binds and inhibits the neurodegeneration-associated fibrillization of the FUS RRM domain. Commun. Biol. 2019, 2, 223. 10.1038/s42003-019-0463-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Karplus M.; McCammon J. A. Dynamics of proteins: elements and function. Annu. Rev. Biochem. 1983, 52, 263–300. 10.1146/annurev.bi.52.070183.001403. [DOI] [PubMed] [Google Scholar]
- Ma B.; Nussinov R. Enzyme dynamics point to stepwise conformational selection in catalysis. Curr. Opin. Chem. Biol. 2010, 14, 652–659. 10.1016/j.cbpa.2010.08.012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pang Y. P. Three-dimensional model of a substrate-bound SARS chymotrypsin-like cysteine proteinase predicted by multiple molecular dynamics simulations: catalytic efficiency regulated by substrate binding. Proteins 2004, 57, 747–757. 10.1002/prot.20249. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shi J.; Han N.; Lim L.; Lua S.; Sivaraman J.; Wang L.; Mu Y.; Song J. Dynamically-driven inactivation of the catalytic machinery of the SARS 3C-like protease by the N214A mutation on the extra domain. PLoS Comput. Biol. 2011, 7, e1001084 10.1371/journal.pcbi.1001084. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lim L.; Shi J.; Mu Y.; Song J. Dynamically-driven enhancement of the catalytic machinery of the SARS 3C-like protease by the S284-T285-I286/A mutations on the extra domain. PLoS One 2014, 9, e101941 10.1371/journal.pone.0101941. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wu H.; Bock S.; Snitko M.; Berger T.; Weidner T.; Holloway S.; Kanitz M.; Diederich W. E.; Steuber H.; Walter C.; Hofmann D.; Weissbrich B.; Spannaus R.; Acosta E. G.; Bartenschlager R.; Engels B.; Schirmeister T.; Bodem J. Novel dengue virus NS2B/NS3 protease inhibitors. Antimicrob. Agents Chemother. 2015, 59, 1100–1109. 10.1128/AAC.03543-14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brecher M.; Li Z.; Liu B.; Zhang J.; Koetzner C. A.; Alifarag A.; Jones S. A.; Lin Q.; Kramer L. D.; Li H. A conformational switch highthroughput screening assay and allosteric inhibition of the flaviviru NS2B-NS3 protease. PLoS Pathog. 2017, 13, e1006411 10.1371/journal.ppat.1006411. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shiryaev S. A.; Farhy C.; Pinto A.; Huang C. T.; Simonetti N.; Elong Ngono A.; Dewing A.; Shresta S.; Pinkerton A. B.; Cieplak P.; Strongin A. Y.; Terskikh A. V. Characterization of the Zika virus two- component NS2B-NS3 protease and structure-assisted identification of allosteric small-molecule antagonists. Antiviral Res. 2017, 143, 218–229. 10.1016/j.antiviral.2017.04.015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nitsche C.; Passioura T.; Varava P.; Mahawaththa M. C.; Leuthold M. M.; Klein C. D.; Suga H.; Otting G. De novo discovery of nonstandard macrocyclic peptides as noncompetitive inhibitors of the Zika virus NS2B-NS3 protease. ACS Med. Chem. Lett. 2019, 10, 168–174. 10.1021/acsmedchemlett.8b00535. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Semwal D. K.; Semwal R. B.; Combrinck S.; Viljoen A. myricetin: A Dietary Molecule with Diverse Biological Activities. Nutrients 2016, 8, 90. 10.3390/nu8020090. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pace C. N.; Vajdos F.; Fee L.; Grimsley G.; Gray T. How to measure and predict the molar absorption coefficient of a protein. Protein Sci. 1995, 4, 2411–2423. 10.1002/pro.5560041120. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hanwell M. D.; Curtis D. E.; Lonie D. C.; Vandermeersch T.; Zurek E.; Hutchison G. R. Avogadro: an advanced semantic chemical editor, visualization, and analysis platform. Aust. J. Chem. 2012, 4, 17. 10.1186/1758-2946-4-17. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brünger A. T.; Adams P. D.; Clore G. M.; DeLano W. L.; Gros P.; Grosse-Kunstleve R. W.; Jiang J. S.; Kuszewski J.; Nilges M.; Pannu N. S.; Read R. J.; Rice L. M.; Simonson T.; Warren G. L. Crystallography & NMR system: A new software suite for macromolecular structure determination. Acta Crystallogr., Sect. D: Biol. Crystallogr. 1998, 54, 905–921. 10.1107/S0907444998003254. [DOI] [PubMed] [Google Scholar]
- Schüttelkopf A. W.; Van Aalten D. M. PRODRG: a tool for high-throughput crystallography of protein–ligand complexes. Acta Crystallogr., Sect. D: Biol. Crystallogr. 2004, 60, 1355–1363. 10.1107/S0907444904011679. [DOI] [PubMed] [Google Scholar]
- Bayly C. I.; Cieplak P.; Cornell W.; Kollman P. A. A well-behaved electrostatic potential based method using charge restraints for deriving atomic charges: the RESP model. J. Phys. Chem. 1993, 97, 10269–10280. 10.1021/j100142a004. [DOI] [Google Scholar]
- Wang J.; Wolf R. M.; Caldwell J. W.; Kollman P. A.; Case D. A. Development and testing of a general amber force field. J. Comput. Chem. 2004, 25, 1157–1174. 10.1002/jcc.20035. [DOI] [PubMed] [Google Scholar]
- Hess B.; Kutzner C.; Van Der Spoel D.; Lindahl E. GROMACS 4: algorithms for highly efficient, load-balanced, and scalable molecular simulation. J. Chem. Theory Comput. 2008, 4, 435–447. 10.1021/ct700301q. [DOI] [PubMed] [Google Scholar]
- Lindorff-Larsen K.; Piana S.; Palmo K.; Maragakis P.; Klepeis J. L.; Dror R. O.; Shaw D. E. Improved side-chain torsion potentials for the Amber ff99SB protein force field. Proteins: Struct., Funct., Bioinf. 2010, 78, 1950–1958. 10.1002/prot.22711. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Essmann U.; Perera L.; Berkowitz M. L.; Darden T.; Lee H.; Pedersen L. G. A smooth particle mesh Ewald method. J. Chem. Phys. 1995, 103, 8577–8593. 10.1063/1.470117. [DOI] [Google Scholar]
- Hess B.; Bekker H.; Berendsen H. J.; Fraaije J. G. LINCS: a linear constraint solver for molecular simulations. J. Comput. Chem. 1997, 18, 1463–1472. . [DOI] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.