Abstract

Small molecules that modulate biological functions are targets of modern-day drug discovery efforts. A new series of novel 1H-benzo[d]imidazoles (BBZs) were designed and synthesized with different functional groups at the phenyl ring and variable lengths of the alkyl chain at the piperazine end as anticancer agents. We identified human topoisomerase I (Hu Topo I) as a probable target of these molecules through a computational study and DNA relaxation assay, a functional assay of the Hu Topo I enzyme. UV absorption, fluorescence, and circular dichroism spectroscopy were used to study interactions between BBZ and DNA. Out of 16 compounds, 11a, 12a, and 12b showed strong binding affinity and thermal stabilization of AT sequence-specific DNA. BBZs were screened against a panel of 60 human cancer cell lines at National Cancer Institute, USA. Most potent molecules 11a, 12a, and 12b showed 50% growth inhibition (GI50) in a concentration range from 0.16 to 3.6 μM cancer cells. Moreover, 12b showed 50% inhibition of the relaxation of DNA by Hu Topo I at 16 μM. Furthermore, flow cytometry revealed that 11a, 12a, and 12b cause prominent G2M arrest of cancer cells. In view of the above, we propose that 12b deserves to be further evaluated for its therapeutic use as an anticancer agent.
Introduction
Bisbenzimidazole derivatives have been found to have a variety of biological activities. Bisbenzimidazoles are DNA minor groove-binding ligands (MGBLs) that form non-covalent interactions with the minor groove of DNA and continue to be promising drug candidates in the discovery of potential new anticancer agents.1−4 Based on the specific mode of binding of MGBL, that is, via hydrogen bonding, electrostatic, and van der Waal’s interactions,5−7 these molecules are used for the treatment of infectious diseases and as chemotherapeutic agents against cancer.8,9 MGBL’s in the majority bind to AT-rich sequences of DNA. These agents are classified according to their interaction through a direct or indirect irreversible interaction with nucleotides causing permanent DNA damage, for example, mitomycin C, anthramycin, CC-1065,10,11 bleomycins,12,13 and others that interact only physically with DNA and cause reversible inhibition of DNA-dependent functions such as distamycin A, diarylamidines, DAPI, berenil, pentamidine, and 1H-benzo[d]imidazoles (BBZs) such as Hoechst-33258, 33342.14−20 The diverse substitutions at the benzimidazole core gave rise to its antihelmintic,21,22 antifungal,23−25 antitumor,26,27 and antiviral28 activities. Different substituted benzimidazoles were also evaluated as H2 receptor blockers and proton pump inhibitors.29 Due to the interference in DNA-mediated enzymatic processes, these BBZ derivatives act through various modes to inhibit the proliferation of cancer cells. The inhibitory activity of DNA topoisomerases advances BBZ to one of the most interesting classes of the therapeutic agent.30−38 Previously, our group has reported two BBZ analogues,39 DMA and TBZ (Figure 1), containing disubstituted phenyl ring carrying electron-donating groups such as 3,4-dimethoxy, 3-methoxy 4-hydroxy, and N-methyl piperazine at another end. These substitutions resulted in increased lipophilicity and improved sequence specificity that prevent cells from radiation-induced DNA damage. Our previous studies have encouraged us to apply different approaches to correlate the position and identify the terminal substituent of BBZ at the phenyl end and piperazine end, to synthesize new derivatives with enhanced anticancer potency. Different BBZ derivatives were synthesized by substituting electron-withdrawing and electron-donating groups at the phenyl end and insertion of different lengths of the carbon chain at the N1 position of piperazine with propyl, butyl, and N,N-dimethylaminoethyl chains. Previous studies on MGBL by spectroscopic techniques identified sequence-specific binding of the molecules to the minor groove of DNA.40−44
Figure 1.
Previously synthesized bisbenzimidazole derivatives.
In addition, we have performed a molecular docking study to correlate our findings with a theoretical approach. We have also investigated the binding affinity of the BBZ derivatives with B-DNA using UV–vis, thermal denaturation, fluorescence, and circular dichroism (CD) spectroscopic techniques. The synthesized BBZ derivatives were evaluated for the anticancer potential at the National Cancer Institute (NCI), USA, against a panel of NCI 60 cell lines.45−50 Out of all molecules, 11a, 12a, and 12b were observed to be most potent molecules with the lowest 50% growth inhibition (GI50) values against 60 different cancer cell lines. Compound 12b inhibited human topoisomerase I (Hu Topo I) enzyme at 16 μM (IC50) concentration, which was comparable to IC50 of camptothecin (CPT) in our assay. Furthermore, flow cytometric studies provided evidence that 11a, 12a, and 12b cause the G2/M phase of the cell cycle arrest. Cell cycle arrest at the G2/M phase indicates that the DNA damage is challenging to be repaired.
Results and Discussion
Chemistry
The previously reported methodology for the synthesis of benzimidazole includes the condensation of o-phenylenediamine with carboxylic acid under harsh dehydrating reaction conditions such as hydrochloric acid, p-toluenesulfonic acid, boric acid, and polyphosphoric acid.51−53 Similarly, another methodology involving condensation of o-phenylenediamine with aldehydes in the presence of oxidizing agents such as benzoquinone, copper(II) acetate, indium perfluorooctanesulfonates, mercuric oxide, nitrobenzene, lead tetra-acetate, iodine, and even air has been reported in the literature.54−58 We have used sodium metabisulfite (Na2S2O5) as an oxidizing agent, and condensation was carried out in ethanol to synthesize novel bis-benzimidazoles.59 The electronic effects of the variably substituted aldehydes, both with electron-withdrawing groups (trifluoromethyl) and electron-donating groups (dimethoxy, diethoxy, trimethoxy), have been investigated for the anticancer property.
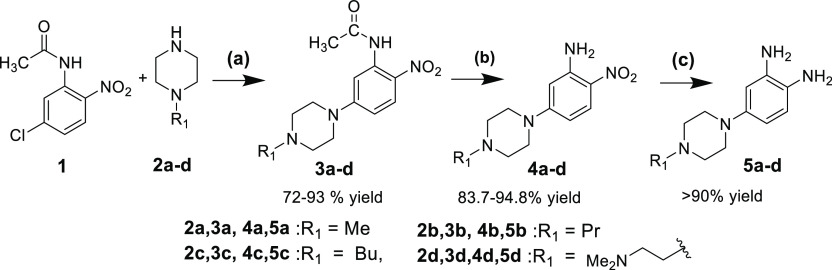
The synthesis procedure for the preparation of target bisbenzimidazole compounds and their intermediates is shown in Schemes 1–3. BBZ derivatives, 10a–c, 11a–d, 12a–e, and 13a–d, were synthesized, as per the reported method59 (Scheme 3). The scheme for the first diamine intermediates is a multi-step process involving reactions like acetylation, deacetylation, hydrogenation, and so forth (Scheme 1). The diamine intermediates, 5a–d, were prepared by a nucleophilic substitution reaction of 5-chloro-2-nitroacetanilide 1 (1.46 mmol) with the N-substituted piperazines (1.5 equiv), 2a–d, in the presence of triethylamine (TEA, 3 equiv) in DMSO with stirring at 120 °C to produce 2-nitroacetanilide derivatives 3a–d with the 71.7–98% yield. Deacetylation of 3a–d was carried out using 10% sulfuric acid to obtain 4a–d with excellent yields. The deacetylated intermediates, 4a–d, were reduced by catalytic hydrogenation using 10% Pd/C at 40 psi at rt to obtain diamine derivatives, 5a–d, in >90% yield (Scheme 1).
Scheme 1. Synthesis of N-Substituted Piperazin-1-yl Benzene-1,2-diamine Derivatives (5a–d).
Reaction conditions: (a) 1 (6.98 mmol, l equiv), 2a–d (8.3 mmol, 1.2 equiv), (Et)3N, (20.94 mmol, 3 equiv), DMSO, 120 °C for 6 h, 72–93%; (b) 3a–d (3.59 mmol) 10% H2SO4, 80 °C for 1.5 h., 83.7–94.8%; and (c) 4a–d 10% Pd/C, H2 at 40 psi, EtOAc:MeOH (4:1) 100 mL for 5 h.
Scheme 3. Synthesis of BBZ Compounds (10a–c, 11a–d, 12a–e, and 13a–d).

Reaction conditions: (a) 5a–d (1.46 mmol, 1 equiv), 9a–e (2.2 mmol, 1.5 equiv), Na2S2O5 (0.73 mmol, 0.5 equiv) in 1 mL of water, ethanol, reflux for 24 h, 60–72% isolated yield. The structures of all the synthesized final molecules are mentioned here.
Second intermediates, 2-aryl-5-cyano-1H-benzimidazoles, 8a–e, were synthesized by reacting 4-cyano-1,2-phenylenediamine 6 (3.75 mmol) with the appropriately substituted benzaldehydes, 7a–e, using the reported method (Scheme 2).59 Catalytic reduction of 8a–e by Ni–Al alloy in the presence of 75% formic acid in the water provides intermediates 2-aryl-5-formyl-1H-benzimidazole 9a–e, with the 67–91% quantitative yield (Scheme 2).
Scheme 2. Synthesis of 2-Aryl-5-formylbenzimidazoles (9a–e).
Reaction conditions: (a) 6 (3.75 mmol), 7a–e (1.5 equiv, 5.6 mmol), Na2S2O5 (0.5 equiv, 1.9 mmol) in 2.5 mL of water, ethanol, reflux for 4–6 h, 64–78% isolated yield; (b) 8a–e (2.6 mmol), Ni–Al alloy (0.036 mmol 1.2 eqiuv), 75% HCOOH, 95 °C, 3 h, 67–91% isolated yield.
Condensation of two intermediates, 4-piperazinyl-phenylenediamine derivatives, 5a–d, (1.46 mmol 1 equiv), and aldehydes require 9a–e (2.2 mmol, 1.2 equiv) in Na2S2O5 in the ethanol solvent. The oxidizing agent Na2S2O5 forms sodium bisulfite (NaHSO3) in water which then forms an adduct with aldehyde and reacts with the diamine derivatives to give the final compounds viz; 10a–c, 11a–d, 12a–e, and 13a–d in 62–72% isolated yield (Scheme 3). All the final compounds and intermediates were confirmed through 1H NMR, 13C NMR (Figures S1–S52), and MS/HRMS (Figures S53–S69). To increase the water solubility of all the newly synthesized benzimidazole derivatives, the compounds were further converted to their respective hydrochloride salts by passing dry HCl gas into a methanolic solution.
Molecular Docking Study of Ligands with Human Topoisomerase I (Hu-TopoI) DNA Complex
Molecular docking studies helped us in better understanding the binding interactions of synthesized active compounds within the Hu Topo I DNA complex (Figure 2a,b). The molecular docking of the compounds 11a, 12a, and 12b was performed with Hu Topo I DNA complex (PDB entry 1EJ9)60 (https://www.rcsb.org) and compared with CPT, a standard Hu Topo I inhibitor. Molecular docking was carried out using Glide.61 The analysis of the X-ray crystal structure of Hu Topo I covalently joined to double-stranded DNA and bound to the anticancer agent topotecan (a synthetic analogue of CPT) suggests that topotecan binds at the site of DNA cleavage by intercalating between the upstream (−1) and downstream (+1) DNA base pairs. Intercalation displaces the downstream DNA, thus preventing relegation of the cleaved strand. Topotecan acts as an uncompetitive inhibitor by specifically binding to the enzyme–substrate complex.63 The 2D and 3D models of the ternary complex BBZs:DNA:Hu Topo I were constructed through Ligplots+64 and BIOVIA discovery studio65 (Table 1). The CPT displayed the strong hydrogen bonding with Gly490, Asn491, and Lys493 at a distance of 2.95, 3.25, and 312 Å, respectively, and van der Waals interactions with His367, Arg364, Arg488, Thr501, Val502, Gly503, and Asp533 (Figure 2c). The binding affinities of the compounds were as 11a (−5.453 kcal/mol), 12a (−5.429 kcal/mol), and 12b (−5.512 kcal/mol). The 11a displayed H-bonds with Asp533 at a distance of 3.01 Å along with the hydrophobic and van der Waals interactions with residues like Arg362, Gly363, Arg364, Arg488, Ala489, Gly490, Asn491, Lys493, Val502, and Gly503 (Figure 2d). Besides, 12a interacts with Hu Topo I with H-bonds involving Arg362 and Lys374 along with hydrophobic and van der Waal interactions with Met263, Leu360, Phe361, Gln421, Lys425, Kys493, Thr498, Thr501, Val502, Gly503, Lys532, and Asp533, displaying the strong affinity (Figure 2e). 12b interacts with Hu Topo I through hydrogen bonds with Gly490 and Lys493 along with hydrophobic and other van der Waal interactions mediated by Pro357, Phe361, Atg364, Pro357, Gly359, Lys374, Thr501, Lys532, Leu360, Arg362, and Asp533 (Figure 2, 11b). The compounds 11a, 12a, and 12b showed considerably good binding affinity to Hu Topo I and interacts through H bonds, hydrophobic interactions, and van der Waals interactions (Figure 2d–f). Since approximately equal number of poses and similar energies were obtained for compounds 11a, 12a, and 12b, the conformations were kept for further analysis.
Figure 2.
Molecular docking interactions and orientations of CPT, 11a, 12a, and 12b and with Hu Topo I enzyme: DNA complex using Glide and construction of the 2D and 3D model through Ligplots and BIOVIA discovery studio. (a) Structure of the Hu Topo I DNA complex without CPT. (b) Structure of the Hu Topo I DNA complex with CPT. The binary complex is diagrammed with protein (green) and DNA (red). The 2D docking interactions of Hu Topo I with compound (c) CPT, (d) 11a, (e) 12a, and (f) 12b. The right panel shows a schematic representation of interactions made by these BBZs with Hu Topo I. H bond and hydrophobic analysis of docking poses were carried out using Ligplots (blue line—ligand bonds; red line non-ligand bonds; dotted lines—hydrogen bonds and its length; half red circle non-ligand residues involved in the hydrophobic contacts; and black dots—corresponding atoms involved in the hydrophobic contacts).
Table 1. Binding Affinities of the Given Compounds with Human Topoisomerase I: DNA Complex.
| ligands | Glide score (kcal/mol) | hydrogen bonding | hydrophobic interaction |
|---|---|---|---|
| camptothecin | –6.001 | Gly490, Asn491, Lys493 | His367, Arg364, Arg488, Thr501, Val502, Gly503, Asp533 |
| 11a | –5.453 | Asp533 | Arg362, Gly363, Arg364, Arg488, Ala489, Gly490, Asn491, Lys493, Val502, Gly503 |
| 12a | –5.429 | Arg362, Lys374 | Met263, Leu360, Phe361, Gln421, Lys425 Kys493, Thr498, Thr501, Val502, Gly503, Lys532, Asp533, |
| 12b | –5.512 | Gly490, Asn491, Lys493 | Pro357, Phe361, Atg364, Pro357, Gly359, Lys374, Thr501, Lys532, Leu360, Arg362, Asp533 |
Rationale for Choice of Oligonucleotides
The interaction of all BBZ analogs (Scheme 2) with DNA has been studied by biophysical techniques. The X-ray crystal structure study of the complex of Hoechst 33258 and the DNA dodecamer CGCG-AATTCGCG suggests that Hoechst 33258 is located in the narrow central AATT region of the minor groove.66,67 Earlier studies suggested that at 25 and 100 °C, the Hoechst 33258/DNA binding ratio (r) is 1:1, whereas this ratio changed to 2:1 at 0 °C. The dihedral angles between the rings of Hoechst 33258 appear to change in a temperature-dependent manner. Thus, we have selected two oligonucleotides, oligo-1: d(CGCGAATTCGCG)2 and oligo-2: d(CGATGTACATCG)2 for our study.68−70
Determination of the Ligand-DNA UV Melting Profile
We performed thermal denaturation experiments of oligo-1 {d(CGCGAATTCGCG)2} and oligo-2 {d(CGATGTACATCG)2} (2.5 μM) with and without BBZs. All 16 BBZ derivatives (Figure 3) showed binding with both the oligos (Tables 2 and S1). BBZs had a higher affinity of binding with oligo-1 as compared to oligo-2. The 10a–b, 11a–d, 12a–e, and 13a–13c showed ΔTm in the range 1 to 5 °C with oligo-1 at a BBZ/oligomer ratio, r = 0.5 (Table 2). When we increased the BBZ/oligomer ratio, r = 2, ΔTm value also increased and was observed in the range of 5 to 9 °C. The 10c and 13d displayed no significant improvement in ΔTm at r = 1.5 or 2 with both the oligonucleotides (Table 2). Throughout duplex formation, the sigmoid curves were observed as a new distinct peak as the first derivative of all BBZs and proved to be minor groove binders without any deformation. These results suggest that the longer alkylated chain at the piperazine end along with the mono- and disubstituted phenyl ring of the BBZ moiety increases its binding in the minor groove of oligo-1. However, N,N-di-methyl aminoethyl alkyl chain inserted at the piperazine end and trisubstitution on the phenyl ring was observed to be an unfavorable modification in the compounds (11d, 12d, 13a, 13c, and 13d) for binding affinity with oligo-1. With oligo-2, 10a–b, 11a–d, 12a–b, 12d–e, and 13b showed maximum change in melting temperatures, ΔTm = 3 and 4 °C at r = 0.5 to 2. However, the 10c, 12c, and 13b–d showed no such gradual increase in melting temperature with an increase in the ratio from r = 0.5 to 2. The variations between the two sequences in melting temperature are visible with distinct melting patterns (Figures 3 and S69–S72 and Table S1), which suggest that how bisbenzimidazoles recognize the different sequences of DNA differentially. This proves that BBZs can differentiate between the different conformations of DNA.
Figure 3.

Distinct UV melting profiles of oligomer-1 and 2, i.e., d(CGCGAATTCGCG)2 and d(CGATGTACATCG)2 alone and in the presence of ligands 11a, 12a, and 12b at a ligand/oligomer ratio, r = 0–2. Samples of DNA (2.5 μM) were mixed with ligand (1.25–5 μM) in buffer [20 mM sodium cacodylate, 100 mM NaCl (pH 7.2)] before being heated at 95 °C for 5 min and slowly annealed to 4 °C before UV analysis at 260 nm from 15 to 95 °C at a heating rate of 0.2 °C/min. Tm values were determined by first-derivative analysis.
Table 2. Tm (°C) of Synthetic Duplex Oligo-1: d(CGCGAATTCGCG)2 and Oligo-2: d(CGATGTACATCG)2 with All Ligands (r = Ligand/DNA)a.
| r = 0 |
r = 0.5 |
r = 1 |
r = 1.5 |
r = 2.0 |
|||||
|---|---|---|---|---|---|---|---|---|---|
| study system | Tm | Tm | ΔTm | Tm | ΔTm | Tm | ΔTm | Tm | ΔTm |
| oligo-1 + 11a | 54.7 | 57.7 | 3 | 58.7 | 4 | 60.7 | 6 | 62.7 | 8 |
| oligo-2 + 11a | 56.9 | 57.9 | 1 | 58.9 | 2 | 58.9 | 2 | 5.9 | 2 |
| oligo-1 + 12a | 54.7 | 56.7 | 2 | 57.7 | 3 | 58.7 | 4 | 61.7 | 7 |
| oligo-2 + 12a | 56.9 | 57.9 | 1 | 58.9 | 2 | 58.9 | 2 | 59.9 | 3 |
| oligo-1 + 12b | 54.7 | 57.7 | 3 | 58.7 | 4 | 59.7 | 5 | 61.7 | 7 |
| oligo-2 + 12b | 56.9 | 57.9 | 1 | 58.9 | 2 | 58.9 | 2 | 59.9 | 3 |
UV/vis absorption studies.
UV/Vis Absorption Studies
All the BBZs exhibit absorption maxima between 347 and 355 nm. To assess the sequence specificity, the binding of the BBZs with specific AT sequence and GC mixed oligomer was studied by calorimetric methods at a fixed ligand/oligomer ratio, r = 1. There was a progressive increase in absorption intensity, and a continued red shift was observed in absorption spectra with decreasing BBZ/DNA ratio. A significant red shift of 18 nm was observed with 12b after binding to oligo-1. Hoechst-33342 exhibits an 18 nm of redshift with oligo-1, which was the same as that to 12b (Table 3), suggesting that the 12b has higher specificity for AT-rich DNA. The absorption maxima for 10a, 11a, 11b, 12a, and 12e showed a redshift of 14, 15, 16, 16, 16 nm, respectively, at BBZ/DNA ratio, r = 1.0. With GC mix oligo-2, 10a, 11b, 12a, 12e, 13a, and 13b showed the redshift in the range of 9–12 nm (Table 3), and Hoechst-33342 showed a redshift of 9 nm, which was comparatively similar to the synthesized BBZ. A higher red shift observed with these ligands can be correlated to random binding with oligo-2 (Figures S73 and S74). The hypochromicity/hyperchromicity in the absorption spectra is mostly attributed to the interaction between the electronic states of the ligand chromophore and those of the DNA bases. On the other hand, the red shift is associated with a decrease in the energy gap between the highest and lowest occupied molecular orbits when the ligand binds to DNA. The extent of the red shift follows the order d(CGATGTACATCG)2 < d(CGCGAATTCGCG)2 in the case of all synthesized BBZs and suggests that BBZ binds in a non-cooperative manner with no differential recognition of mixed DNA due to the substitution of bulky ethoxy and methoxy groups at ortho, meta, and para positions, respectively.
Table 3. Binding Affinity Obtained from Fluorescence Titration Experiments and Red-Shift Observed from UV–Visible Titration of Oligo–Ligand Complexes.
|
Ka |
Ka |
|||
|---|---|---|---|---|
| BBZs | binding affinity with oligo-1 (M–1) | UV titration redshift (nm) | binding affinity with oligo-2 (M1) | UV titration redshift (nm) |
| 10a | 7 × 106 | 14 | 2 × 106 | 10 |
| 10b | 7 × 106 | 10 | 3 × 106 | 8 |
| 10c | 8 × 106 | 11 | 2 × 106 | 8 |
| 11a | 3 × 107 | 15 | 5 × 106 | 8 |
| 11b | 7 × 106 | 16 | 2 × 106 | 10 |
| 11c | 7 × 106 | 8 | 3 × 106 | 2 |
| 11d | 8 × 106 | 12 | 1 × 106 | 5 |
| 12a | 1 × 107 | 16 | 4 × 106 | 10 |
| 12b | 1 × 107 | 18 | 2 × 106 | 9 |
| 12c | 6 × 106 | 9 | 2 × 106 | 3 |
| 12d | 9 × 106 | 13 | 4 × 106 | 5 |
| 12e | 1 × 107 | 16 | 3 × 106 | 6 |
| 13a | 9 × 106 | 12 | 2 × 106 | 12 |
| 13b | 7 × 106 | 12 | 2 × 106 | 9 |
| 13c | 5 × 106 | 7 | 1 × 106 | 3 |
| 13d | 5 × 106 | 11 | 1 × 106 | 4 |
| H 33342 | 2 × 107 | 18 | 5 × 106 | 12 |
Fluorescence Spectroscopy Studies
The fluorescence spectrum of free ligands from 10a–10c, 11a–11d, 12a–12e, and 13b–13d contains a single broad emission band from 410 to 560 nm. BBZ analogs, the 2, 5 di-methoxy and CF3-substituted BBZs viz. 10c, 11c, 12c, 13c, 10b, 11b, 12b, 13b, and 13c, have comparatively weak fluorescence intensity. 11d, 12d, 11a, 12a, 12e, and 13a displayed moderate and strong fluorescence intensity. In fluorescence titration, most BBZs showed a blue shift of 20–35 nm, while Hoechst 33342 showed a blue shift of 20 nm with oligo-1. For oligo-2, the minimum value of the blueshift change was 2–3 nm with the synthesized BBZ, while Hoechst 33342 showed a blueshift of 15 nm with oligo-2. The fluorescence of 11a, 12a, and 12b upon binding with oligo-1 has increased significantly compared to oligo-2 at a BBZ/oligomer ratio, r = 2.
Further evaluation of their binding strength with oligomeric duplex demonstrated the strong binding affinity for 11a (K = 3 × 107), 12a (K = 1 × 107), 12b (K = 1 × 107), and Hoechst 33342 (K = 2 × 107) with the oligo-1. The obtained binding affinity values confirm the increased sequence selective binding by these BBZs (Table 3). However with oligo-2, relative fluorescence enhancement after BBZ complexation was comparatively less than the oligo-1 (Figure 4). The results indicate that the BBZ analogs 11a, 12a, and 12b, including Hoechst 33342, have a strong affinity with the oligo-1 sequence, where more AT bases are located in the minor groove. It was also shown that the ligand binding to DNA was AT sequence-specific rather than random mix sequences (Figure 4). The fluorescence spectra of other molecules are given in the Supporting Information. The binding affinity was calculated from the Scatchard plots (r/Cf vs r) and fitted to data (Figures S75 and S76). Based on the obtained results, it can be suggested that selectivity was established for minor groove-oriented BBZs due to the following reason: (a) B-DNA’s electrostatic potential is most negative in the AT-rich region of the minor groove and thus facilitates the binding of cationic drugs and (b) the small groove width is narrower in the AT region, thereby stabilizing the desired bound molecule through the desirable van der Waals interactions. The results indicate that BBZ analogs with 3,4-diethoxy, 4-ethoxy (11a, 12a, and 12b) substituted at the phenyl end act as strong binders relative to the multiple substitutions at the phenyl ring. Apart from this, it is also reported that the NH group of the imidazole ring has strong H-bond interaction with the nucleic acid base in DNA. Hence, an increase in electron density at N-atom upon substitution with an electron-donating group at the phenyl end increases the feasibility and strength of H-bond formation between NH-imidazole and electronegative “O”-atoms of nucleic acid bases. However, compound 12b, having a strong electron-withdrawing group, CF3, at the phenyl ring end, also showed strong binding to oligo-1. Piperazine itself and three to four carbon long side chains on piperazine favor the binding of BBZs in the minor groove with a specific AT sequence. From overall observations, we found that bi/mono substitution at the phenyl end and chain lengthening at the piperazine end of BBZ analogs (10a, 11a, 11b, 12a, 12b, and 12e) enable them to interact more firmly with both DNA sequences as compared to other bulkier analogs like tri-methoxy substitutions at the phenyl ring and N,N-dimethylaminoethyl alkylating chain substituted at the piperazine end of BBZ.
Figure 4.
Fluorescence titration binding study of oligo-1: d(CGCGAATTCGCG)2 and oligo-2: d(CGATGTACATCG)2 with ligands (11a, 12a, and 12b). 1–20 μL of oligos (stock 0.1 mM) were added to a solution of ligand (1 mL of a 1 mM solution), and emission scanning was carried out from 360 to 680 at 2 nm/s speed; 5 nm slit width; T = 20 °C in a buffer containing 20 mM sodium cacodylate (pH 7.2) and 100 mM Na.
Circular Dichroism Studies
CD titration is an effective method of evaluating the binding mode and the saturation limit for BBZ binding with oligos. The conformation of DNA is influenced by the binding of the compounds to DNA. The changes in CD signals were observed upon the interaction of DNA with compounds which may often be assigned to the corresponding changes in the DNA structure. Neither oligo-1 and oligo 2 nor the ligands (11a, 12a, 12b, and Hoechst 33342) alone exhibited CD signals, but the asymmetry in the optical properties of the ligands due to the effect of the DNA field results in differentially induced CD spectra of the complexes at 340–360 nm. The CD spectroscopic studies were carried out with three molecules, 11a, 12a, 12b, and Hoechst 33342 (as control) complexes with each of the oligo 1 and oligo 2 as these molecules exhibit potent anticancer activity in in vitro studies. These BBZ showed an induced cotton effect due to the complexation with oligo-1 at different wavelengths, but peak intensity was centered at around 340–360 nm (Figure 5) with varying shapes of the induced CD (ICD) bands obtained for different BBZs. All the spectra were characterized by isoelliptic points that are observed at around 235 and 300 nm for the BBZ: oligo complexes. An upbeat ICD band was observed at 347 to 351 nm on continuous addition of 11a, 12a, 12b, and Hoechst 33342 to oligos, with intensity kept augmenting with increasing BBZ/oligo ratios. With oligo-1, upon addition of BBZ, we observed saturation in the ellipticity value at r = 2 that suggests binding of ligands to DNA was 1:1 per binding site. We observed a shoulder at around 272–290 nm in the CD spectra of complexes of 11a, 12a, and 12b with oligo-1, which is similar to Hoechst 33342. These positive ICD signals indicate a minor groove-binding mode of these ligands. The interaction of the BBZ does not alter the conserved CD spectrum of the oligo-1. In the case of oligo-2, all the BBZs, including Hoechst 33342, gave ICD signals on the interaction with oligo-2 in the 348–352 nm region (Figure 5). In general, the strength of the induced band due to complex formation between BBZ and oligo-2 is weak compared with oligo-1 complexes. With oligo-2, the intensity of CD signals increased with increasing the r-value and saturation observed at a higher ratio of 4–4.5 (Figure S77). Thus, the constant increase in the CD signal upon increasing the ligand concentration suggests non-specific interactions with oligo 2.
Figure 5.
CD scans of oligomer-1 and oligomer-2, i.e., d(CGCGAATTCGCG)2 and d(CGATGTACATCG)2 with increasing concentrations of ligands (11a, 12a, 12b, and Hoechst 33342) at 20 °C. Samples of the oligomer (10 μM) were scanned from 450 to 190 nm after serial additions of the concentrated ligand with stirring. Peaks around 347–352 nm correspond to the ligand–oligomer complex. Buffer: 20 mM sodium cacodylate, 100 mM NaCl (pH 7.2); r stands for the ligand/oligomer ratio.
Overall, we established that the newly synthesized ligands have strong and stable binding with AT-rich oligo-1: d(CGCGAATTCGCG)2 than GC mix oligo-2, and the BBZ/oligo-1 complex is stabilized by hydrogen bonds between the two N–H hydrogen-bond donors in the imidazole rings and adenine N3 and thymine O2 acceptors in the minor groove of the oligonucleotide. A general preference for AT regions is conferred by electrostatic potential and by narrowing the walls of the groove. We hypothesize that local point-by-point AT specificity follows from close van der Waals contacts between ring hydrogen atoms in BBZ and the C2 hydrogens of adenines. It was reported in the literature that replacement of one benzimidazole ring by purine can make that particular molecule GC tolerant, which was also observed by the substitution of the imidazole ring in place of pyrrole in lexitropsins. This supports our observation that BBZs preferentially bind to AT-rich DNA.
Cell Viability Assay of BBZs on the Panel of 60 Cancer Cell Lines
The preliminary screening by cytotoxicity assay on cancerous cell lines was performed to identify the anticancer potential of BBZs.71−73 Only 13 BBZs, out of 16 were submitted to the NCI, USA, for evaluation of their single-dose (10 μM) in vitro anticancer activity against complete NCI60 cell lines (Supporting File II), representing mammalian leukemia, melanoma, lung, colon, brain, breast, ovary, kidney, and prostate cancer cell line panels. The selected compounds were further tested for five dose screening (0.01, 0.1, 1, 10 and 100 μM) (Supporting File III). The HCl salt of 13a, 13d, and 13c was precipitating in cell culture media; hence, these were not used for screening (Table 4).
Table 4. Calculated Values of GI50, TGI, and LC50 (in μM) of the Selected BBZs against NCI Cell Line Panels.
|
12b |
11a |
12a |
|||||||
|---|---|---|---|---|---|---|---|---|---|
| 60 cancer cell lines | GI50 | TGI | LC50 | GI50 | TGI | LC50 | GI50 | TGI | LC50 |
| Leukemia | |||||||||
| CCRF-CEM | 0.3 | 1.03 | 5.04 | 0.4 | 3.6 | >100 | 0.53 | 3.06 | >100 |
| HL-60(TB) | 1.2 | 2.8 | 6.6 | 0.37 | 2.7 | >100 | 0.70 | 3.18 | 67.9 |
| K-562 | 0.22 | >100 | 0.31 | 1.5 | 0.52 | 2.27 | 7.53 | ||
| MOLT-4 | 1.07 | 2.5 | 6.3 | 0.59 | 4.4 | >100 | 1.22 | 3.86 | 31.6 |
| RPMI-8226 | 0.25 | 0.62 | 0.47 | 2.5 | >100 | 0.43 | 2.06 | >100 | |
| SR | 0.26 | 0.65 | 0.29 | >100 | 0.31 | 1.19 | >100 | ||
| Non-Small Cell Lung | |||||||||
| A549/ATCC | 0.35 | 1.19 | 1.17 | 3.02 | 1.18 | 2.93 | |||
| EKVX | 1.5 | 2.9 | 5.7 | 1.73 | 13.3 | >100 | 1.38 | 3.18 | |
| HOP-62 | 1.3 | 3.05 | 1.98 | 5.2 | 23.4 | 1.81 | 3.40 | ||
| HOP-92 | 1.18 | 2.78 | 6.5 | 1.2 | 4.5 | 26.3 | 1.23 | 3.17 | 8.15 |
| NCI–H226 | 1.57 | 3.59 | 0.8 | 3.5 | 37.5 | 1.29 | 3.95 | >100 | |
| NCI–H23 | 1.65 | 3.36 | 6.8 | 0.25 | 0.7 | 4.24 | 0.78 | 2.35 | |
| NCI–H23M | 1.39 | 2.74 | 5.39 | 1.1 | 3.2 | 9.18 | 1.27 | 2.67 | 5.65 |
| NCI–H460 | 0.23 | 0.5 | 1.62 | 0.43 | 1.51 | 4.38 | 0.53 | 1.86 | 5.31 |
| NCI–H522 | 1.6 | 3.6 | 0.45 | 2.11 | 1.20 | 3.97 | >100 | ||
| Colon Cancer | |||||||||
| COLO 205 | 1.3 | 3.1 | 0.16 | 1.29 | 5.4 | 0.23 | 1.52 | 6.18 | |
| HCC-2998 | 1.8 | 3.5 | 6.6 | 1.24 | 2.62 | 5.5 | 1.71 | 3.22 | |
| HCT-116 | 0.2 | 0.42 | 0.24 | 1.23 | 4.7 | 0.43 | 1.66 | 4.30 | |
| HCT-15 | 0.27 | 0.74 | 2.42 | 18.9 | >100 | 1.56 | 3.32 | 7.05 | |
| HT29 | 0.34 | 1.42 | 0.45 | 2.3 | >100 | 0.80 | 2.97 | 100 | |
| KM12 | 0.67 | 2.04 | 4.9 | 1.26 | 2.8 | 6.4 | 1.42 | 2.99 | 6.28 |
| SW620 | 0.21 | 0.45 | 0.45 | 0.30 | 1.2 | 4.3 | 0.47 | 1.79 | 5.15 |
| CNS | |||||||||
| SF-268 | 1.7 | 3.3 | 6.4 | 1.9 | 11.5 | 68.8 | 2.66 | 22.0 | >100 |
| SF-265 | 1.34 | 2.7 | 5.4 | 0.5 | 2.3 | 7.8 | 0.70 | 2.26 | |
| SF-539 | 1.6 | 3.05 | 5.8 | 0.79 | 2.1 | 4.8 | 1.26 | 2.56 | 5.20 |
| SNB-19 | 2.18 | 5.6 | 1.22 | 2.61 | 5.58 | ||||
| SNB-75 | 1.05 | 2.25 | 4.8 | 0.71 | 1.9 | 4.3 | 0.65 | 2.01 | 4.54 |
| U251 | 0.24 | 0.56 | 0.42 | 1.8 | 4.8 | 1.28 | 2.69 | 5.65 | |
| Melanoma | |||||||||
| LOX IMVI | 0.38 | 1.1 | 0.2 | 0.44 | 0.96 | 0.42 | 1.62 | ||
| MALME-3M | 1.4 | 3.0 | 0.08 | 1.2 | 3.9 | 0.19 | 1.18 | ||
| M14 | 0.37 | 2.4 | 0.25 | 1.5 | 5 | 0.31 | 1.55 | 4.68 | |
| MDA-MB-435 | 1.4 | 2.8 | 5.5 | 0.67 | 1.9 | 4.5 | 1.09 | 2.39 | 5.24 |
| SK-MEL-2 | 1.8 | 3.3 | 6.3 | 14 | 3.1 | 1.68 | 3.66 | ||
| SK-MEL-28 | 1.7 | 3.1 | 5.9 | 0.91 | 2.2 | 5.2 | 1.01 | 2.22 | 4.86 |
| SK-MEL-5 | 1.6 | 3.2 | 6.2 | 0.63 | 2.01 | 4.6 | 1.09 | 3.16 | 9.22 |
| UACC-257 | 1.7 | 3.5 | 7.2 | 0.79 | 2.8 | 9.4 | 1.21 | 10.5 | >100 |
| UACC-62 | 1.5 | 2.9 | 5.8 | 0.4 | 1.9 | 5.3 | 0.68 | 2.18 | 5.42 |
| Ovarian | |||||||||
| IGROV1 | 1.7 | 3.4 | 0.76 | 4.1 | 37.8 | 1.15 | 2.60 | . | |
| OVCAR-3 | 0.21 | 0.4 | 0.73 | 0.26 | 0.58 | 1.8 | 0.35 | 0.85 | 2.92 |
| OVCAR-4 | 0.6 | 1.8 | 5.2 | 0.69 | 2.8 | 9.5 | 1.48 | 3.21 | 6.95 |
| OVCAR-5 | 1.8 | 3.5 | 5.9 | 2.09 | 5.01 | 15.9 | 1.90 | 3.40 | 6.08 |
| OVCAR-8 | 1.1 | 2.8 | 0.45 | 2.16 | >100 | 0.89 | 3.06 | ||
| NCI/ADR-RES | 2.1 | 57.7 | >100 | >100 | >100 | >100 | 100 | 100 | >100 |
| SK-OV-3 | 2.1 | 10.2 | 9.9 | 1.33 | 3.27 | 8.01 | |||
| Renal | |||||||||
| 786–0 | 1.47 | 3.04 | 1.13 | 2.50 | 5.54 | 1.41 | 2.84 | 5.72 | |
| A498 | 1.69 | 3.31 | 6.49 | 0.47 | 1.79 | 4.38 | 1.18 | 2.47 | 5.16 |
| ACHN | 1.32 | 2.6 | 5.14 | 0.82 | 2.1 | 4.77 | 0.92 | 2.12 | 4.65 |
| CAKI-1 | 1.29 | 2.58 | 5.15 | 1.04 | 3.03 | 8.8 | 1.04 | 2.52 | 6.12 |
| RXF393 | 0.47 | 1.5 | 0.26 | 0.9 | 3.6 | 1.01 | 2.35 | . | |
| SN12c | 1.54 | 3.02 | 0.45 | 2.06 | 6.75 | 1.03 | 2.61 | . | |
| TK-10 | 1.22 | 4.9 | 2.47 | 1.66 | 5.17 | >100 | |||
| UO-31 | 1.77 | 3.24 | 5.9 | 1.67 | 3.82 | 8.76 | 2.00 | 5.80 | 43.8 |
| Prostate | |||||||||
| PC-3 | 0.86 | 2.31 | 5.5 | 0.38 | 1.92 | 6.12 | 1.08 | 2.44 | 5.49 |
| DU-145 | 0.45 | 1.68 | 4.3 | 0.44 | 2.02 | 12.4 | 1.00 | 2.21 | 4.88 |
| Breast | |||||||||
| MCF7 | 0.17 | 0.57 | 0.35 | 1.53 | 0.19 | 1.42 | |||
| MDA-MB-231 | 1.46 | 2.92 | 5.84 | 1.38 | 3.06 | 6.78 | 1.69 | 3.28 | 6.38 |
| HS578T | 1.26 | 3.05 - | 1.09 | 3.68 | >100 | 1.54 | 3.90 | ||
| BT-549 | 1.62 | 3.15 | 1.22 | 2.72 | 6.07 | 1.55 | 3.07 | ||
| T-47D | 1.24 | 3.23 | 0.91 | 9.11 | >100 | 1.36 | 7.88 | >100 | |
| MDA-MB-468 | 1.66 | 3.34 | 0.43 | 1.82 | 7.07 | 1.27 | 3.09 | ||
The 12b showed the highest toxicity against K562 hematopoietic malignant cells and human lymphoma cells (SR) among leukemia cells. While in melanoma cell line panel LOX IMVI, human epithelial melanoma cells, MALME-3M, metastatic malignant melanoma, and M14, human melanoma cells were more sensitive to 11a, 12a, and 12b out of 13 molecules. On the other hand, lesser activity of 11a and 12a was found against non-small cell lung, renal, and ovarian carcinoma cells with mean GI50 ≥ 5 μM. These molecules were ineffective against NCI/ADR-RES of the ovarian cancer cell line panel. The mean GI50 value suggests the potent anticancer activity of 12b against leukemia and colon carcinoma cell lines. However, 12b has been found to be highly sensitive to a few of the most frequently diagnosed cancer types worldwide, such as lung cancer (A549 and NCI460), breast cancer (MCF-7), and prostate cancer (PC-3 and DU-145). The LC50 value indicates the toxicity profile of the molecules, that is, the higher the LC50, the lower the toxicity. Noticeably, 11a was found to be the least toxic for all the cell lines of the leukemia panel (LC50 > 100). However, both the molecules, 11a and 12b are toxic in renal cancer, CNS cancer, and prostate cancer cell line panels (LC50 < 10). Interestingly, all the three screened BBZs, that is, 11a, 12a, and 12b have shown toxicity in melanoma cell line panels (LC50 < 10). The cytotoxicity study was also performed in normal transformed cell lines (HEK and NIH3T3) using 3-(4,5-dimethylthiazole-2-yl)2,5-diphenyltetrazolium (MTT), and 50% inhibitory concentration (IC50) was calculated, as shown in Table 5. The molecules were found to be less toxic to normal cells as compared to cancerous cell lines.
Table 5. Cytotoxicity Study on the Normal Cell Line by MTT Assay.
| IC50 (μM) 48 h |
|||
|---|---|---|---|
| s. no. | compounds | HEK | NIH3T3 |
| 1 | 11a | 14.4 | 2.3 |
| 2 | 12a | 2.7 | 8.2 |
| 3 | 12b | 2.7 | 3.2 |
Inhibition of DNA Relaxation Activity HuTopoI Enzyme by BBZ Derivatives
Furthermore, these molecules were screened against Hu Topo I to check whether they have any effect on HuTopoI activity as parent analogues Hoechst 33342 and 33258 are known as HuTopoI inhibitors. DNA relaxation inhibition assays74,75 were performed on 13 molecules, but only three compounds, 11a, 12a, and 12b, showed inhibition of Hu Topo I (Figure 6a–c). We have included the results of only those three here. The compound 12b showed 50% inhibition of topoI activity at 16 μm, but the other two compounds, 11a and 12a, did show IC90 at 140 and 152 μM concentration (Figure 6d). On the other hand, CPT showed 85% inhibition at 25 μM concentration.
Figure 6.
Plasmid DNA relaxation mediated by Hu TopoI in the presence of an increasing concentration of compound CPT, 11a, 12a, and 12b by Hu Topo I.FR (fully relaxed); PR (partially relaxed); and SC (supercoiled). (a–c) Ethidium bromide-stained agarose gel showing: lane 1, pHOT1 plasmid DNA (C); lane 2, relaxation of plasmid DNA by HuTopoI (CT); lane 3, plasmid DNA in the presence of compound (CL); and lanes 4–12, inhibition of relaxation of plasmid DNA by HuTopoI in the presence of 15, 25, 50, 75, 100, 125, 150, 175, and 200 μM of compounds, respectively. The CPT was used at a fixed concentration of 25 μM in this experiment (d); densitometry of relaxation gel was carried out, values were normalized against enzyme control and control, and the final values were plotted in Graph Pad Prism 9. The 50% inhibitory concentrations (IC50) of compounds were calculated by Origin 2019. All experiments were carried out in both biological and technical triplicates.
Effect of BBZ Derivatives on Cell Cycle Progression in MDA-MB-231 Cells
The triple-negative breast cancers are highly aggressive, with low survival rates and recurrence in patients worldwide. The treatment of triple-negative cancer is still a challenge.76 To further evaluate the BBZ-mediated inhibition of cell proliferation and its correlation with cell cycle progression, MDA-MB-231 was treated with 11a, 12a, and 12b at their respective GI50 concentration (Table 4). After the treatment of cells with the molecules, the cell cycle distribution at different time points was assessed by flow cytometry after propidium iodide (PI) staining (Figure 7a). A significant G2/M arrest was observed through cell cycle analysis with 11a and 12b (Figure 7b,c). These results indicate that 11a and 12b treated cells increase the G2/M cell population from 2 h onward.
Figure 7.
(a) Representative cell cycle analysis at 6 and 12 h post-treatment with three molecules. (b) After treatment with 11a and 12b, the % of cells at 6 h in the G2/M phase rises to 38.8 and 44%, respectively, from 27% in untreated cells (average values of three biological repeats). (c) Marked increase in the G2/M phase was prominently maintained till 12 h with 37.8 and 39% in 11a and 12b, respectively. 11a and 12b showed marked G2/M arrest and increase in sub-G1 population as compared to control at 6 and 12 h, respectively. Data shown are expressed as ±SD repeated in triplicates for each ligand; *p < 0.05 and **p < 0.01, respectively. There were no significant changes observed in the cell cycle distribution of 12a treated cells as compared to untreated cells.
The 12b treated MDA-MB-231 cells showed an enhanced sub-G1 region, that is, apoptotic cells. At 6 and 12 h, 7.4% of cells was observed in the sub-G1 phase, which is much higher than control cells (1–2%). Thus, an increase in the sub-G1 and G2/M phase reflects the irreparable double-strand DNA damage, causing apoptotic cell death. Our most potent molecule emerged to be 12a, causing maximum apoptotic cell death.
Conclusions
The results obtained from this work identify various areas of valuable information relating to ligand-DNA interactions which could be useful for rational drug design and helpful in the development of their potential pharmaceutical and biological effect. In our study, by substituting BBZ with different groups, we synthesized compounds with strong binding affinity with AT sequence of DNA and potent anticancer activity. The docking study of these compounds showed considerable binding affinity and probable interactions with the binary complex of the DNA + Hu Topo I enzyme. The binding affinity was observed to be the highest for three molecules, 11a, 12a, and 12b. Electrophoretic methods showed that the 11a, 12a, and 12b compounds influence the activity of Hu TopoI as a minor groove binder. Topoisomerase inhibitors inhibit DNA synthesis, which can lead to the arrest of the cell cycle in the G2 phase. However, only 11a and 12b showed a prominent G2/M arrest of the cell cycle, leading to the cell death. Interestingly, we found that 11a and 12a having electron-donating groups at the phenyl ring, viz, 3,4-diethoxy and increased carbon chain length at the piperazine ring showed better cytotoxicity to cancerous cells. However, 12b with the electron-withdrawing group, CF3, at the phenyl ring showed strong cytotoxicity against cancerous cell lines. The other synthesized BBZ molecules with tri-methoxy and 2,5-dimethoxy substitution at the phenyl end and N,N-dimethyl aminoethyl alkyl chain at the piperazine end demonstrated less binding affinity due to the steric hindrance. This suggests that longer propyl and butyl carbon chains at the piperazine end are responsible for increased binding affinity and efficacy. Also, it was evident by our spectroscopic studies that BBZ molecules bind in a non-cooperative manner to DNA while increasing the ligand-DNA ratio to 2. Thus, experimental studies proved that a combination of the −CF3 group at the phenyl ring and the longer alkyl chain at the piperazine ring is one of the probable combinations leading to toxicity to cancer cells, and therefore, it can be exploited further for the development of new selective small molecules as a drug candidate.
Materials and Methods
All melting points were determined using BUCHI B540 appliances. The 1H (400/500 MHz) and 13C NMR (400 MHz) spectra were observed with the solvents CDCl3 and DMSO-d6 on an ECX-400P. Jeol chemical shifts are identified in ppm by assigning CDCl3 and DMSO-d6 with resonance as 7.26 and 2.49 ppm in the 1H NMR spectrum and 77.23 and 39.50 ppm in the 13C NMR spectrum. The coupling constant (J) in hertz (Hz) is reported. On the Agilent 6520 Accurate Mass Q-TOF LC/MS mass spectrometer, HRMS spectra were measured, and IR spectra were taken on a PerkinElmer, FT-IR system. The development of all reactions was recorded by thin-layer chromatography (TLC), which was completed with silica gel 60 (F-254, Merck) pre-coated aluminum sheets with a thickness of 0.25 mm on 20 cm. The TLC plates developed were seen under ultraviolet light (254–366 nm) and treated with iodine vapor. For the purification of intermediate and final compounds, chromatography of the silica gel column was performed. The purity of compounds was found to be greater than 90%. Commercially available reagents were used without further purification unless otherwise stated. The anhydrous organic solvents (e.g., ether, ethyl acetate, chloroform, methanol, ethanol, ACN, DMF, hexane, toluene, etc.) were purchased from commercial sources and used as stated. Chemical reagents, synthesized oligos, d(CGCGAATTCGCG)2, d(CGATGTACATCG)2, QI-Aquick nucleotide removal kit (Qiagen catalogue 28304), and Dulbecco’s modified Eagle’s medium (DMEM) were purchased from Sigma. The MDA-MB-231 and A549 cell lines were procured from National Center for Cell Sciences, Pune, India.
General Procedure for the Synthesis of N-Substituted ((Piperazin-1-yl)-2-nitrophenyl) Acetamide (3a–d)
Synthesis of 2-nitroacetanilide derivatives was carried out using the reported procedure.39 The mixture of N-(5-chloro-2-nitrophenyl)acetamide 1 (6.98 mmol) with the respective N-substituted piperazine 2a–d (1.2 equiv, 8.37 mmol) and TEA (3.0 equiv, 20.94 mmol) in 10 mL of DMSO was taken in a 250 mL round-bottom flask. The resulting reaction mixture was stirred at 120 °C for 6 h. The reaction was followed by TLC till the completion of the reaction; the reaction mixture was poured on ice-cold water, resulting in a solid precipitate, followed by filtration and drying overnight will give the crude product mixture. Then, the crude mixture was purified by column chromatography over silica gel (60–120 mesh size) using 10–15% MeOH in DCM as the mobile phase, and the product was obtained as a yellow solid with a quantitative yield 72–98% of the (piperazin-1-yl)-2-nitrophenyl) acetamide compounds. The spectra of 3a, 3b, and 3d are not included in the Supporting Information because these compounds are synthesized using the reported procedure.59
N-(5-(4-Methylpiperazin-1-yl)-2-nitrophenyl)acetamide (3a)
This compound was synthesized using the reported procedure:39 yellow solid (92.7% yield, 1.8 g); mp 152.5–153 °C. HRMS (ESI) m/z: calcd for C13H18N4O3 [M + H]+, 279.1478; obsd, 279.1475.
N-(5-(4-Allylpiperazin-1-yl)-2-nitrophenyl)acetamide (3b)
This compound was synthesized using the reported procedure:59 yellow solid (93% yield, 2.1 g); mp 143.5–144.5 °C. HRMS (ESI) m/z: calcd for C15H22N4O3 [M + H]+, 305.1567; obsd, 305.1563.
N-(5-(4-Butylpiperazin-1-yl)-2-nitrophenyl)acetamide (3c)
The product was obtained as a yellow solid (73.8% yield, 1.65 g); mp 95.6–98.4 °C. 1H NMR (400 MHz, DMSO-d6): δ ppm 0.85–0.89 (t, J = 7.64 Hz, 3H), 1.31–1.27 (m, 2H), 1.41–1.34 (m, 2H), 2.13 (s, 3H), 2.28–2.26 (m, 2H), 2.49–2.43 (m, 4H), 3.39–3.32 (m, 4H), 6.19 (s, 1H), 6.67 (dd, J = 9.9, 3.0 Hz, 1H), 7.67 (d, 1H), 7.96 (d, J = 9.9, 1H), 10.4 (s 1H). 13C NMR (100 MHz, DMSO-d6): δ ppm 14.0, 20.6, 25.2, 28.8, 46.7, 52.7, 57.8, 104.8, 109.2, 128.2, 128.4, 136.6, 155.1, 169.5. HRMS (ESI) m/z: calcd for C16H24N4O3 [M + H]+, 321.1878; obsd, 321.1875.
5-[4-(2-(N,N-Dimethyaminoethyl))-1-piperazinyl]-2 Nitroacetanilide (3d)
This compound was synthesized using the reported procedure:59 yellow solid (81.2% yield, 1.9 g); mp 166.2–168.7 °C. HRMS (ESI) m/z: calcd for C16H25N5O3 [M + H]+, 336.2035; obsd, 336.2032.
General Procedure for the Synthesis of N-Substituted (Piperazin-1-yl)-2-nitroaniline (4a–d)
Synthesis of 2-nitroaniline derivatives was carried out using the reported procedure.59 Deacetylation of the compound 3a–d (3.59 mmol 1 equiv) was carried out with 10% H2SO4 in a round-bottom flask, and H2SO4 solution was added to it (10 mL/g). The resulting reaction mixture was heated at 80 °C till the reaction completion. After the reaction completion, the reaction mixture was brought to room temperature and poured onto the crushed ice. This resulted in a yellow-colored solid deacetylated compound. For complete precipitation of the compound, the pH was maintained neutral with 30% ammonia solution. The resulting solid was then filtered, washed with water, and dried to yield the title compound in good yield. The deacetylated products 4a, 4b, and 4d are synthesized using the reported procedure; therefore, the spectra of these compounds are not added in the Supporting Information.59
5-(4-Methylpiperazin-1-yl)-2-nitroaniline (4a)
This compound was synthesized using the reported procedure:38 yellow solid (83.7% yield, 0.710 g); mp 150.2–151.5 °C. HRMS (ESI) m/z: calcd for C11H16N4O2 [M + H]+, 236.1367; obsd, 237.1362.
(4-Allyl-1-piperazinyl)-2-nitroaniline (4b)
This compound was synthesized using the reported procedure:59 yellow solid (94.8% yield, 0.900 g); mp 83.6–85.4 °C. HRMS (ESI) m/z: calcd for C13H18N4O2 [M + H]+, 263.1508; obsd, 263.1508.
5-(4-Butyllpiperazin-1-yl)-2-nitroaniline (4c)
The product was obtained as a yellow solid (92% yield, 0.919 g); mp 83.6–85.4 °C. 1H NMR (400 MHz, CdCl3): δ ppm 0.956 (t, J = 6.0 Hz, 3H), 1.39–1.35 (m, 2H), 2.41–2.38 (m, J = 6.4 Hz, 2H), 2.57 (s 4H), 3.39 (s, 4H), 5.96 (s, 1H) 6.16 (s, 2H), 6.30 (d, J = 7.6 Hz, 1H), 8.03 (d, J = 7.6 Hz, 1H). 13C NMR (100 MHz, DMSO-d6): δ ppm 14.4, 20.6, 28.6, 46.4, 52.7, 58.0, 97.6, 106.1, 123.3, 127.8, 148.8, 155.6. HRMS (ESI) m/z: calcd for C14H22N4O3 [M + H]+, 279.1816; obsd, 279.1811.
5-[4-(2-(N,N-Dimethyl amino ethyl))-1-piperazinyl]-2-nitroaniline (4d)
This compound was synthesized using the reported procedure:59 yellow solid (91.2% yield, 0.960 g); mp 142.2–143.8 °C. HRMS (ESI) m/z: calcd for C14H23N5O2 [M + H]+, 294.1930; obsd, 294.2047.
General Procedure for the Synthesis of N-4-(4-Substituted piperazinyl)-1-phenylenediamine (5a–d)
Compounds 4a–d dissolved in 100 mL, with a ratio of EtOAc/MeOH (4:1), was hydrogenated at room temperature under 40 psi H2 pressure with a catalytic amount of 10% Pd/C until TLC showed the disappearance of starting material, and the reaction mixture becomes colorless. The reaction mixture was filtered through a Celite bed, and the filtrate was used for the next step without further purification and delay.
General Procedure for Preparation of 2-Aryl-5-cyanobenzimidazoles (8a–e)
Synthesis of 2-aryl-5-cyanobenzimidazoles derivatives was carried out using the reported procedure.59 To freshly prepared ethanolic solution of 4-cyano-1,2-phenylenediamine 6 (3.75 mmol, 1.0 equiv), reacting with the mixture of respective aldehyde 7a–e (1.5 equiv, 5.6 mmol) and solution of Na2S2O5 (0.5 equiv, 1.9 mmol) in water 2.5 mL (1 mL/150 mg) were added. The resulting solution was stirred at reflux for 5–6 h, then cooled to room temperature, and filtered through a Celite bed. The solvents were evaporated under reduced pressure. The crude residue was purified by chromatography on silica gel (60–120 mesh size) in EtOAc/hexane as a solvent mixture to achieve a solid in 64–78% isolated yield. The spectra of 8e are not included in the Supporting Information because these compounds are synthesized using the reported procedure.59
2-(3,4-Diethoxyphenyl)-1H-benzo[d]imidazole-6-carbonitrile (8a)
This compound was purified by column chromatography using a gradient of 35% of EtOAc in hexane, and the product was obtained as an off-white solid (71.5% yield, 0.826 g); mp 198 °C. 1H NMR (400 MHz, DMSO-d6): δ ppm 1.3–1.4 (m, 6H), 4.03 (m, 4H), 7.11 (dd, J = 18.32, 8.4 Hz, 1H), 7.55 (d, J = 7.64 Hz, 1H), 7.68 (d, J = 9.92 Hz, 1H). 7.72 (s, 1H), 7.76 (s, 1H), 8.13 (s, 1H), 13.2 (s, 1H). 13C NMR (100 MHz, DMSO-d6); 14.7, 63.9, 96.8, 103.6, 111.2, 112.9, 113.2, 115.6, 120.0, 121.4, 122.4, 125.5, 134.6, 140.2, 148.2, 150.3. HRMS (ESI) m/z: calcd for C18H17N3O2 [M + H]+, 308.1399; obsd, 308.1387.
2-(4-(Trifluoromethyl)phenyl)-1H-benzo[d]imidazole-6-carbonitrile (8b)
This compound was purified by column chromatography using a gradient of 30% of EtOAc in hexane, and the product was obtained as an off-white solid (69.5% yield, 0.750 g); mp 215–217 °C. 1H NMR (400 MHz, DMSO-d6): δ ppm 7.61 (d, J = 7.64 Hz, 1H), 7.77 (d, J = 7.64 Hz 1H), 7.93 (d, J = 8.4 Hz, 2H), 8.10 (s, 1H), 8.30 (d, J = 8.4 Hz, 2H). 13C NMR (100 MHz, DMSO-d6): δ ppm 104.5, 119.8, 12.6, 125.3, 126.0 (q, J = 3.86 Hz, 1C), 126.76 (d J = 3.86 Hz, 1C), 128.0, 129.8, 130.2, 130.4, 130.8, 132.9, 152.8. HRMS (ESI) m/z: calcd for C15H8N3F3 [M + H]+, 288.0739; obsd, 288.0745.
2-(2,5-Dimethoxyphenyl)-1H-benzo[d]imidazole-5-carbonitrile (8c)
This compound was purified by column chromatography using a gradient of 35% of EtOAC in hexane, and the product was obtained as an off-white solid (66.7% yield, 0.700 g); mp 235–237 °C. 1H NMR (400 MHz, DMSO-d6): δ ppm 3.81 (s, 1H), 3.83 (s, 1H), 7.10 (d, J = 8.4 Hz, 1H), 7.52 (d, J = 8.0 Hz, 1H), 7.74 (s, 2H), 7.67 (s, 1H), 8.07 (s, 1H), 13.26 (s, 1H). 13C NMR (100 MHz, DMSO-d6): δ ppm 56.13, 104.13, 110.42, 112.30, 120.50, 120.69, 122.10, 125.98, 149.45, 151.48. HRMS (ESI) m/z: calcd for C16 H14N2O2 [M + H]+, 280.1086; obsd, 280.1060.
2-(3,4,5-Trimethoxyphenyl)-1H-benzo[d]imidazole-6-carbonitrile (8d)
This compound was synthesized using the reported procedure39 and purified by column chromatography using a gradient of 35% of EtOAc in hexane, and the product was eluted as an off-white solid (64.5% yield, 0.750 g); mp 240–238 °C. 1H NMR (400 MHz, DMSO-d6): δ ppm 3.73 (s, 3H), 3.89 (s, 6H), 7.54 (d, J = 11.44 Hz, 2H), 7.58 (d, J = 10.68 Hz, 1H), 7.70 (d, J = 9.16 Hz, 1H), 8.12 (d, J = 9.16 Hz, 1H), 13.3 (s, 1H). 13C NMR (100 MHz, DMSO-d6): δ ppm 56.11, 60.2, 104.2, 112.6, 116.3, 119.7, 119.9, 123.3, 124.3, 125.3, 125.9, 138.1, 139.5, 143.2, 153.3, 154.0, 154.8. HRMS (ESI) m/z: calcd for C17H16N2O4 [M + H]+, 310.1180; obsd, 310.1187.
2-(4-Ethoxyphenyl)-1H-benzo[d]imidazole-6-carbonitrile (8e)
This compound was synthesized using the known procedure59 and purified by silica gel column chromatography using a gradient of 25% of EtOAc in hexane, and the product was eluted as a white solid (72.8% yield, 0.720 g); mp 223.1–224.8 °C. HRMS (ESI) m/z: calcd for C16H13N3O [M + H]+, 264.1137; obsd, 264.1129.
General Procedure for the Preparation of 2-Aryl-5-formylbenzimidazoles (9a–e)
Ni–Al alloy (1.2 equiv, 0.036 mmol) was added to a solution of 8a–8f (2.6 mmol 1 equiv) in 75% aqueous formic acid. The reaction mixture was heated at 95 °C for 3 h under an inert atmosphere. The hot mixture was filtered through a Celite bed, and the reaction flask was rinsed with water (2 × 10 mL). The aqueous solution was concentrated to dryness. After the addition of water to this residue, a precipitate was formed. The pH of this suspension was adjusted to 9.0 by the dropwise addition of 2 N NaOH, and the product was then extracted into ethyl acetate. The organic layers were dried over anhydrous Na2SO4 and filtered. The solvents were evaporated under reduced pressure. The residue was purified by chromatography on silica gel (60–120 mesh size) using the gradient MeOH/EtOAc in the range of 3:97 to 6:94 to obtain the product as a solid in 67–93% yield. The spectra of 9e, 9f are not added in the Supporting Information because these compounds are synthesized using the reported procedure.59
2-(3,4-Diethoxyphenyl)-1H-benzo[d]imidazole-6-carbaldehyde (9a)
The product was obtained as an off-white solid (80% yield, 0.650 g); mp 110 °C. 1H NMR (400 MHz, DMSO-d6): δ ppm 1.40–1.30 (m, 6H), 4.13–4.02 (m, 4H), 5.73 (br, s, 1H), 7.08 (d, J = 9.16, 1H), 7.69 (d, J = 8.4 Hz, 1H), 7.73 (d, J = 8.4 Hz, 1H), 7.70 (s, 1H), 8.10 (s, 1H), 8.20 (d, J = 8.4 Hz, 1H), 10.01 (s, 1H). 13C NMR (100 MHz, DMSO-d6): δ ppm 14.7, 63.9, 111.2, 112.9, 115.1, 117.8, 120.0, 121.7, 122.4, 122.9, 131.0, 148.2, 150.3, 154.7, 163.6, 192.5. HRMS (ESI) m/z: calcd for C18H19N2O3 [M + H]+, 311.1396; obsd, 311.1398.
2-(4-(Trifluoromethyl)phenyl)-1H-benzo[d]imidazole-6-carbaldehyde (9b)
The product was obtained as an off-white solid (93.5% yield, 0.713 g); mp 160 °C. 1H NMR (400 MHz, DMSO-d6): δ ppm 5.80 (s, 1H), 7.87 (d, J = 8.4 Hz, 1H), 7.92 (d, J = 8.4 Hz, 2H), 8.11 (d, J = 18.2 Hz, 1H), 8.25 (s, 1H), 8.37 (d, J = 16.8 Hz, 2H), 10.04 (s, 1H). 13C NMR (100 MHz, DMSO-d6): δ ppm 112.4, 120.1, 122.6, 125.39, 126.05 (q, J = 3.83 Hz), 125.86 (d, J = 3.83 Hz, 1C), 126.9, 127.4, 128.18, 130.0, 131.5, 133.2, 152.2, 192.5. HRMS (ESI) m/z: calcd C15H10N2F3O [M + H]+, 291.0745; obsd, 291.0748.
2-(2,5-Dimethoxyphenyl)-1H-benzo[d]imidazole-5-carbaldehyde (9c)
The product was obtained as an off-white solid (91.7% yield, 0.680 g); mp 230–232 °C. 1H NMR (400 MHz, CDCl3): δ ppm 3.81 (s, 3H), 3.92 (s, 3H), 6.92 (d, J = 6.4 Hz, 1H), 7.67 (d, J = 6.8 Hz, 2H), 7.75 (s, 1H), 7.8 (d, J = 6.8 Hz, 1H), 8.10 (s, 1H), 10.04 (s 1H). 13C NMR (100 MHz, DMSO-d6): δ ppm 56.10, 110.31, 112.31, 114.20, 119.27, 120.28, 122.37, 123.53, 131.56, 135.53, 140.25, 144.24, 149.45, 151.29, 154.51, 193.1. HRMS (ESI) m/z: calcd C16H15N2O3 [M + H]+, 283.1083; obsd, 283.1060.
2-(3,4,5-Trimethoxyphenyl)-1H-benzo[d]imidazole-6-carbaldehyde (9d)
The compound was synthesized using the reported procedure:39 off-white solid (84% yield, 0.690 g); mp 198 °C. 1H NMR (400 MHz, DMSO-d6): δ ppm 3.69 (s, 3H), 3.84 (s, 6H), 7.54 (s, 2H), 7.75 (s, 2H), 8.13 (s 1H), 10.03 (s, 1H). 13C NMR (100 MHz, DMSO-d6): δ ppm 56.09, 60.2, 104.2, 123.0, 124.6, 131.2, 139.3, 153.3, 163.6, 192.5. HRMS (ESI) m/z: calcd for C17H17N2O4 [M + H]+, 313.119; obsd, 313.1190.
2-(4-Ethoxyphenyl)-1H-benzo[d]imidazole-6-carbaldehyde (9e)
This compound was synthesized using the reported procedure:59 off-white solid (70.8% yield, 0.499 g); mp 266.8–268.6 °C. HRMS (ESI) m/z: calcd for C16H14N2O2 [M + H]+, 267.1133; obsd, 267.1125.
General Procedure for Preparation of BBZs (10a–c, 11a–d, and 12a–e)
To the freshly prepared ethanolic solution of N-4-(4-substituted piperazinyl)-1phenylenediamine (1.46 mmol, 1 equiv) 5a–d, a mixture of respective aldehyde 9a–e (2.2 mmol, 1.5 equiv) and solution of Na2S2O5 (0.73 mmol, 0.5 equiv) in water (1 mL/150 mg) were added in 20 mL of ethanol. The resulting solution was stirred at reflux for 24 h, then cooled to room temperature, and filtered through a bed of Celite. The solvents were evaporated under reduced pressure. The crude residue was purified by chromatography on silica gel (100–200 mesh size) in the MeOH/DCM solvent mixture to afford the solid title compounds in 60–72% yield.
2-(2-(3,4-Diethoxyphenyl)-1H-benzo[d]imidazol-5-yl)-5-(4-methylpiperazin-1-yl)-1H-benzo[d]imidazole (10a)
This compound was purified by column chromatography using a gradient of 12% of MeOH in DCM, and the product was obtained as a light brown crystalline solid (63% yield, 0.450 g); mp 185–186.1 °C. 1H NMR (400 MHz, DMSO-d6): δ ppm 1.35–1.25 (m, 6H), 2.35 (s, 3H), 2.70 (s, 4H), 3.14 (s, 4H), 4.12–3.99 (m, 4H), 6.88 (d, J = 7.2 Hz, 1H), 7.00 (s, 1H), 7.05 (d, J = 8.4 Hz, 1H), 7.41 (d, J = 8.8 Hz, 1H), 7.63 (d, J = 8.4 Hz, 1H), 7.78–7.72 (m, 1H), 7.98 (d, J = 8.4 Hz, 1H), 8.20 (s, 1H), 8.29 (s, 1H). 13C NMR (100 MHz, DMSO-d6): δ ppm 15.1, 43.6, 48.6, 53.6, 64.5, 102.0, 111.6, 113.7, 114.9, 120.1, 121.4, 122.3, 124.6, 135.6, 5, 147.2, 148.6, 150.5, 152.5, 153.9, 167.2. HRMS (ESI) m/z: calcd for C29H32N6O2 [M + H]+, 497.2610; obsd, 497.2631.
5-(4-Methylpiperazin-1-yl)-2′-(4-(trifluoromethyl)phenyl)-1H,1′H-2,5′-Bibenzo[d]imidazole (10b)
This compound was purified by column chromatography using a gradient of 12% of MeOH in DCM, and the product was obtained as a brown crystalline solid (67.7% yield, 0.470 g); mp 284–285.2 °C. 1H NMR (400 MHz, DMSO-d6): δ ppm 2.28–2.26 (s, 3H), 2.58–2.52 (s, 4H), 3.15–3.12 (s, 4H), 6.97–6.93 (d, J = 6.8, 2.28 Hz, 1H), 7.04–7.01 (s, 1H), 7.47–7.44 (d, J = 9.16 Hz, 1H), 7.73–7.78 (d, J = 8.4 Hz, 1H), 7.95–7.99 (d, J = 8.4 Hz, 2H), 8.1–8.08 (d, J = 9.16 Hz, 1H), 8.24–8.27 (s, 1H), 8.38 (s, 1H), 8.48–8.44 (d, J = 8.4 Hz, 1H). 13C NMR (100 MHz, DMSO-d6): δ ppm 45.27, 49.6, 54.6, 113.8, 121.5, 122.8, 125.24, 126.03 (q, J = 3.83 Hz, 4C), 127.0, 127.3, 129.7, 130.0, 133.7, 147.6, 151.1. HRMS (ESI) m/z: calcd for C26H23F3N6 [M + H]+, 477.2003; obsd, 477.2004.
2′-(2,5-Dimethoxyphenyl)-6-(4-methylpiperazin-1-yl)-1H,3′H-2,5′-bibenzo[d]imidazole (10c)
Purification of this compound by column chromatography using a gradient of 12% of MeOH in DCM, and the product was eluted as a brown solid (70.4% yield, 0.480 g); mp 232–235 °C. 1H NMR (400 MHz, DMSO-d6): δ ppm 2.40 (s, 3H), 2.75 (s, 4H), 3.18 (s, 4H), 3.78 (s, 3H), 3.96 (s, 3H), 6.90 (d, J = 8.3 Hz, 1H), 7.05–7.01 (m, 2H), 7.16 (d, J = 9.0 Hz, 1H), 7.43 (d, J = 8.5 Hz, 1H), 7.73 (d, J = 8.0 Hz, 1H), 7.86 (s, 1H), 8.01 (s, 1H), 8.40 (s, 1H), 12.34 (s, 1H). 13C NMR (100 MHz, DMSO-d6): δ 153.72, 152.30, 151.71, 150.71, 147.62, 125.01, 118.74, 117.92, 114.27, 114.00, 56.71, 56.08, 54.56, 49.59, 44.99. HRMS (ESI) m/z: calcd for C27H29N6O2 [M + H]+, 469.2352; obsd, 469.2374.
2-(2-(3,4-Diethoxyphenyl)-1H-benzo[d]imidazol-5-yl)-5-(4-propylpiperazin-1-yl)-1H-benzo[d]imidazole (11a)
Purification of this compound by column chromatography using a gradient of 10% of MeOH in DCM, and the product was obtained as a brown crystalline solid (60% yield, 0.460 g); mp 230–231.7 °C. 1H NMR (400 MHz, DMSO-d6): δ ppm 0.79 (t, J = 6.4 Hz, 3H), 1.34–1.27 (m, 6H), 1.43–1.40 (m, 2H), 2.32 (m, 2H), 2.58 (s, 4H), 3.07 (s, 4H), 4.09–4.01 (m, 4H), 6.84 (d, J = 8.4 Hz, 1H), 6.95 (s, 1H), 7.05 (d, J = 8.4 Hz, 1H), 7.37 (d, J = 8.4 Hz, 1H), 7.61 (d, J = 8.4 Hz, 1H), 7.73 (d, J = 5.2 Hz, 2H), 7.95 (d, J = 8.0 Hz, 1H), 8.14 (s, 1H), 8.24 (s, 1H). 13C NMR (100 MHz, DMSO-d6): δ ppm 11.6, 14.7, 19.03, 49.6, 52.6, 59.7, 111.1, 113.0, 114.0, 119.4, 120.8, 122.3, 124.4, 147.1, 147.1, 150.0, 151.5, 153.3, 163.8. HRMS (ESI) m/z: calcd for C31H36N6O2 [M + H]+, 525.2963; obsd, 525.2970.
2-(4-(Trifluoromethyl)phenyl)-5-(5-(4-propylpiperazin-1-yl)-1H-benzo[d]imidazol-2-yl)-1H-benzo[d]imidazole (11b)
The compound was purified by column chromatography using a gradient of 10% of MeOH in DCM, and the product was eluted as a light yellow crystalline solid (63.8% yield, 0.470 g); mp 268–270.2 °C. 1H NMR (400 MHz, DMSO-d6): δ ppm 0.82–0.9 (t, J = 6.84 Hz, 3H), 1.42–1.5 (m, J = 6.84 Hz, 2H), 2.26–2.34 (t, J = 6.8 Hz, 2H), 2.52–2.6 (m, 4H), 3.12–3.06 (m, 4H), 6.92 (d, J = 8.4 Hz, 1H), 7.44 (s, 1H), 7.69 (d, J = 6.12 Hz, 1H), 7.82 (d, J = 6.12 Hz, 1H), 7.95 (d, J = 8.4 Hz, 2H), 8.08 (dd, J = 16.8 Hz, 16.8 Hz, 1H), 8.3 (s, 1H), 8.42 (d, J = 8.4 Hz, 2H), 12.6 (s, 1H), 13.3 (s, 1H). 13C NMR (100 MHz, DMSO-d6): δ ppm 11.83, 19.4, 49.89, 52.89, 59.7, 109.1, 120.9, 122.8, 125.4, 126.04 (q, J = 3.83 Hz, CF3), 127.28, 129.33, 129.69, 131.10, 130.31, 133.62, 147.7, 151.1. HRMS (ESI) m/z: calcd for C28H27F3N6 [M + H]+, 505.2312; obsd, 505.2319.
2′-(2,5-Dimethoxyphenyl)-6-(4-propylpiperazin-1-yl)-1H,3′H-2,5′-bibenzo[d]imidazole (11c)
Purification of this compound by column chromatography using a gradient of 10% of MeOH in DCM, and the product was eluted as a brown solid (71.7% yield, 0.520 g); mp 238–241 °C. 1H NMR (400 MHz, DMSO-d6): δ ppm 0.91 (t, J = 7.3 Hz, 3H), 1.60 (s, 2H), 2.82 (s, 2H), 3.06 (s, 4H), 3.31 (s, 4H), 3.82 (s, 3H), 4.00 (s, 3H), 6.97 (d, J = 6.6 Hz, 1H), 7.11–7.01 (m, 2H), 7.21 (d, J = 9.1 Hz, 1H), 7.49 (d, J = 7.0 Hz, 1H), 7.77 (d, J = 7.5 Hz, 1H), 7.90 (s, 1H), 8.03 (s, 1H), 8.04 (s, 1H). 13C NMR (100 MHz, DMSO-d6): δ ppm 164.3, 153.4, 152.1, 151.0, 149.4, 147.8, 124.8, 122.9, 121.2, 120.15, 114.2, 112.2, 110.43, 57.5, 56.17, 53.0, 49.9, 28.2, 20.5, 14.3. HRMS (ESI) m/z: calcd for C29H33N6O2 [M + H]+, 497.2665; obsd, 497.2684.
5-(4-Propylpiperazin-1-yl)-2′-(3,4,5-trimethoxyphenyl)-1H,1′H-2,5′-bibenzo[d]imidazole (11d)
Purification of this compound by column chromatography using a gradient of 10% of MeOH in DCM, and the product was eluted as a brown crystalline solid (70.2% yield, 0.540 g); mp 250–252.6 °C. 1H NMR (400 MHz, DMSO-d6): δ ppm 0.82 (t, J = 9.2 Hz, 3H), 1.47–141 (m, 2H), 2.35–2.33 (m, 2H), 2.59 (s, 4H), 3.09 (s, 4H), 3.70 (s, 3H), 3.87 (s, 6H), 6.90–6.84 (dd, J = 9.2, 1H), 6.97 (s, 1H), 7.40 (d, J = 7.2 Hz, 1H), 7.53 (s, 1H), 7.68 (d, J = 8.0 Hz, 1H), 8.00 (d, J = 8.4 Hz, 1H), 8.17 (s, 1H), 8.29 (s 1H). 13C NMR (100 MHz, DMSO-d6): δ ppm 11.7, 19.0, 49.5, 52.5, 56.1, 59.3, 60.2, 104.1, 114.2, 121.3, 124.4, 125.7, 139.2, 147.1, 151.9, 153.0, 153.3, 164.1. HRMS (ESI) m/z: calcd for C30H34N6O3 [M + H]+, 527.2749; obsd, 527.2755.
5-(4-Butylpiperazin-1-yl)-2-(2-(3,4-diethoxyphenyl)-1H-benzo[d]imidazol-5-yl)-1H-benzo[d]imidazole (12a)
Purification of this compound by column chromatography using a gradient of 15% of MeOH in DCM, and the product was eluted as a brown crystalline solid (60.2% yield, 0.465 g); mp 255–256.2 °C. 1H NMR (400 MHz, DMSO-d6): δ ppm 0.82 (d, J = 6.4 Hz, 3H), 1.29–121 (m, 6H), 1.32 (m, 2H), 1.42 (m, 2H), 2.42 (s, 2H), 2.69 (s, 4H) 3.11 (s, 4H), 4.04 (m, 4H), 6.85 (d, J = 8.4 Hz, 1H), 6.96 (s, 1H), 7.04 (d, J = 8.4 Hz, 1H), 7.37 (d, J = 8.0 Hz, 1H), 7.59 (d, J = 8.0 Hz, 1H), 7.74–7.68 (m, 1H), 7.93 (d, J = 8.0 Hz, 1H), 8.10 (s, 1H), 8.23 (s, 1H). 13C NMR (100 MHz, DMSO-d6): δ ppm 13.8, 14.7, 19.9, 27.6, 49.2, 52.3, 55.1, 56.7, 64.0, 111.0, 112.5, 114.1, 119.3, 120.3, 122.7, 123.9, 146.9, 148.3, 150.4, 152.09, 153.2, 163.5. HRMS (ESI) m/z: calcd for C32H38N6O2 [M + H]+, 539.3122; obsd, 539.3117.
5-(5-(4-Butylpiperazin-1-yl)-1H-benzo[d]imidazol-2-yl)2(4(trifluoromethyl)phenyl)-1H-benzo[d]imidazole (12b)
The product was obtained as a light brown crystalline solid (66% yield, 0.500 g); mp 265–266.5 °C. 1H NMR (400 MHz, DMSO-d6): δ ppm, 0.82–0.78 (t, J = 7.2 Hz, 3H), 1.25–1.15 (m, 2H), 1.39–1.29 (m, J = 2H), 2.28–2.24 (t, J = 7.6 Hz, 2H), 2.49 (m, 4H), 3.047 (m, 4H), 6.86 (dd, J = 8.8, J = 1.6 Hz, 1H), 6.96 (s, 1H), 7.39 (d, J = 8.4, Hz, 1H), 7.71 (d, J = 8.0 Hz, 1H), 7.87 (d, J = 8.8 Hz, 2H), 8.00 (d, J = 8.4 Hz, 1H), 8.29 (d, J = 8.0 Hz, 1H), 8.33 (d, J = 8.0 Hz, 2H). 3C NMR (100 MHz, DMSO-d6): δ ppm 13.9, 20.1, 28.3, 49.9, 52.9, 57.4, 113.6, 120.0, 122.7, 125.5, 126.0 (q, J = 3.83 Hz, CF3), 127.7, 128.2, 129.2, 129.7, 130.0, 130.4, 133.6, 147.7, 151.1. HRMS (ESI) m/z: calcd for C29H29F3N6 [M + H]+, 519.2462; obsd, 519.2467.
6-(4-Butylpiperazin-1-yl)-2′-(2,5-dimethoxyphenyl)-1H,3′H-2,5′-bibenzo[d]imidazole (12c)
Purification of this compound by column chromatography using a gradient of 15% of MeOH in DCM, and the product was obtained as a pale yellow crystalline solid (61.7% yield, 0.460 g); mp 272.5–275 °C. 1H NMR (400 MHz, DMSO-d6): δ ppm, 0.84–0.80 (t, J = 7.02 Hz, 3H), 1.26–1.18 (m, 2H), 1.42–1.37 (m, 2H), 2.45–2.39 (m, 2H), 2.62 (s, 4H), 3.10 (s, 4H), 3.75 (s, 3H), 3.93 (s, 3H), 6.85 (d, J = 8.6 Hz, 1H), 6.96 (s, 1H), 7.02 (dd, J = 8.8, 2.8 Hz, 1H), 7.13 (d, J = 8.8 Hz, 1H), 7.38 (d, J = 8.4 Hz, 1H), 7.69 (d, J = 8.4 Hz, 1H), 7.84 (d, J = 3.2 Hz, 1H), 8.16 (s, 1H), 8.36 (s, 1H). 13C NMR (100 MHz, DMSO-d6): δ ppm 164.4, 153.6, 152.2, 151.6, 150.6, 147.8, 124.9, 121.3, 118.7, 117.8, 114.2, 113.9, 57.4, 56.6, 56.0, 52.9, 49.9, 28.1, 20.4, 14.3. HRMS (ESI) m/z: calcd for C30H34N6O2 [M + H]+, 511.2810; obsd, 511.2815.
5-(4-Butylpiperazin-1-yl)-2-(2-(3,4,5-trimethoxyphenyl)-1H-benzo[d]imidazol-5-yl)-1H-benzo[d]imidazole (12d)
Purification of this compound by column chromatography using a gradient of 15% of MeOH in DCM, and the product was eluted as a brown crystalline solid (64.4% yield, 0.509 g); mp 235–237.1 °C. 1H NMR (400 MHz, DMSO-d6): δ ppm 0.90–0.87 (t, J = 7.6 Hz, 3H), 1.36–1.27 (m 2H), 1.52–1.42 (m 2H), 2.4–2.39 (t, J = 7.6 Hz, 2H), 2.62 (m, 4H), 3.14 (s, 4H), 3.74 (s, 3H), 3.91 (s, 6H), 6.92 (d, J = 9.16 Hz, 1H), 7.01 (s, 1H), 7.44 (d, J = 8.4 Hz, 1H), 7.56 (s, 2H), 7.71 (d, J = 8.4 Hz, 1H), 8.03 (d, J = 8.4 Hz, 1H), 8.18 (s, 1H), 8.31 (s, 1H). 13C NMR (100 MHz, DMSO-d6): δ ppm 13.9, 20.0, 28.4, 49.6, 52.7, 56.1, 57.3, 60.2, 104.0, 114.4, 120.7, 125.0, 125.6, 139.3, 147.0, 150.6, 152.4, 153.2, 163.8. HRMS (ESI) m/z: calcd for C31H36N6O3 [M + H]+, 541.2907; obsd, 541.2917.
5-(4-Butylpiperazin-1-yl)-2′-(4-ethoxyphenyl)-1H,1′H-2,5′-bibenzo[d]imidazole (12e)
Purification of this compound by column chromatography using a gradient of 20% of MeOH in DCM, and the product was obtained as a brown crystalline solid (63.7% yield, 0.460 g); 284–285.2 °C. 1H NMR (400 MHz, DMSO-d6): δ ppm 0.86 (t, J = 7.6 Hz, 3H), 1.31–1.26 (m, 2H), 1.39–1.31 (m, 3H), 1.46–1.38 (m, 2H), 2.47–2.51 (m, 2H), 2.71 (s, 4H), 3.16 (s, 4H), 4.12–4.06 (m 2H), 6.90 (dd, J = 8.4 Hz, 1H), 7.02 (m, 1H), 7.09 (d, J = 9.16 Hz, 2H), 7.44 (d, J = 9.16 Hz, 1H) 7.65 (d, J = 8.4 Hz, 1H), 8.02 (d, J = 8.4 Hz 1H), 8.15 (d, J = 9.12 Hz, 1H), 8.2 (s, 1H), 8.3 (s, 1H). 13C NMR (100 MHz, DMSO-d6): δ ppm 13.8, 20.0, 27.6, 49.4, 52.4, 57.2, 63.2, 113.2, 114.3, 120.3, 122.3, 124.3, 128.3, 147.2, 152.1, 153.0, 159.3, 164.0, 172.4. HRMS (ESI) m/z: calcd for C30H34N6O2 [M + H]+, 495.2856; obsd, 495.2855.
2-(4-(2′-(3,4-Diethoxyphenyl)-1H,3′H-[2,5′-bibenzo[d]imidazol]-6-yl)piperazin-1-yl)N,N-dimethylethan-1-amine (13a)
Purification of this compound by column chromatography using a gradient of 20% of MeOH in DCM, and the product was eluted as a brown crystalline solid (61.8% yield, 0.500 g); mp 262–2565 °C. 1H NMR (400 MHz, DMSO-d6): δ ppm 1.37–1.30 (m 6H). 2.43 (s, 6H), 2.55 (m, 2H), 2.56 (s, 4H), 2.77 (m, 2H), 3.09 (s, 4H), 4.14–4.04 (m, 4H), 6.88 (d, J = 8.8 Hz, 1H), 6.98 (s 1H), 7.08 (d, J = 8.8 Hz, 1H), 7.40 (d, J = 8.4 Hz, 1H), 7.63 (d, J = 7.2 Hz, 1H), 7.77 (d, J = 8.4 Hz, 1H), 7.81 (s 1H), 7.99 (d, J = 8.4 Hz, 1H) 8.30 (s, 1H), 13.2 (br, 2H). 13C NMR (100 MHz, DMSO-d6): δ 153.4, 152.0, 150.9, 149.4, 148.1, 124.9, 122.9, 121.1, 120.1, 114.1, 112.2, 110.4, 109.8, 56.1, 55.4, 54.7, 53.5, 50.4, 44.7. HRMS (ESI) m/z: calcd for C32H39N7O3 [M + H]+, 553.3238; obsd, 553.3235.
2-(4-(2′-(4-(Tifluoromethyl)phenyl)-1H,3′H-[2,5′-bibenzo[d]imidazol]-6-yl)piperazin-1-yl) N,N-Dimethylethan-1-amine (13b)
Purification of this compound by column chromatography using a gradient of 18% of MeOH in DCM, and the product was eluted as a light brown crystalline solid (63.5% yield, 0.495 g); mp 250–254 °C. 1H NMR (400 MHz, DMSO-d6): δ ppm 2 (s, 6H), 2.37 (s, 4H), 2.50 (s, 4H), 3.04 (s, 4H), 6.86 (d, J = 8.8 Hz, 1H), 6.95 (s, 1H), 7.38 (d, J = 8.4 Hz, 1H), 7.71 (d, J = 8.4 Hz, 1H), 7.89 (d, J = 8.4 Hz, 2H), 8.04 (d, J = 8.4 Hz, 1H), 8.35 (s, 1H), 8.40 (d, J = 8.0 Hz, 2H). 13C NMR (100 MHz, DMSO-d6): δ ppm 151.57, 148.25, 134.21, 130.45, 130.15, 127, 126, 125.97, 123.45, 122.01, 114.20, 60.78, 56.52, 52.72, 53.76, 50.53, 45.80. HRMS (ESI) m/z: calcd for C29H31F3N7 [M + H]+, 534.2593; obsd, 534.2610.
2-(4-(2′-(2,5-Dimethoxyphenyl)-1H,3′H-[2,5′-bibenzo[d]imidazol]-6-yl)piperazin-1-yl)-N,N-dimethylethan-1-amine (13c)
Purification of this compound by column chromatography using a gradient of 18% of MeOH in DCM, and the product was obtained as a crystalline solid (61.2% yield, 0.470 g); mp 238–240 °C. 1H NMR (400 MHz, DMSO-d6): δ ppm 2.36 (s, 6H), 2.47 (m, 2H), 2.55 (m, 4H), 2.67 (m, 2H), 3.07 (s, 4H), 3.81 (s, 3H), 3.87 (s, 3H), 6.88 (d, J = 6.4 Hz, 1H), 6.98 (s, 1H), 7.10 (d, J = 6.0 Hz, 1H), 7.40 (s, 1H), 7.64 (d, J = 6.4 Hz, 2H), 7.83 (s, 2H), 8.00 (d, J = 5.6 Hz, 1H), 8.31 (s, 1H). 13C NMR (100 MHz, DMSO-d6): δ ppm 153.38, 152.08, 151.04, 149.40, 148.33, 148.13, 124.9, 122.9, 121.1, 120.0, 114.0, 112.2, 110.4, 56.13, 55.48, 54.71, 53.56, 50.49, 44.77. HRMS (ESI) m/z: calcd for C30H34N7O2 [M + H]+, calcd 526.2916; obsd, 525.2914.
2-(4-(2′-(3,4,5-Trimethoxyphenyl)-1H,3′H-[2,5′-bibenzo[d]imidazol]-6-yl)piperazin-1-yl) N,N-Dimethylethan-1-amine (13d)
Purification of this compound by column chromatography using a gradient of 18% of MeOH in DCM, and the product was eluted as a light brown crystalline solid (60.9% yield, 0.495 g). 1H NMR (400 MHz, DMSO-d6): δ ppm 2.50 (m, 6H), 2.56 (m, 4H), 2.88 (m, 2H), 3.03 (s, 2H), 3.06 (s, 4H), 3.66 (s, 3H), 3.83 (s, 6H), 6.85 (d, J = 8.4 Hz, 1H), 7.37 (d, J = 8.4 Hz, 1H), 7.50 (d, J = 16.8 Hz, 1H), 7.57 (s, 1H), 7.62 (s, 2H), 8.00 (d, J = 7.2 Hz, 1H), 8.33 (s, 1H). 13C NMR (100 MHz, DMSO-d6): δ 153.65, 147.20, 139.56, 124.87, 115.40, 104.43, 60.78, 56.52, 52.72, 48.95, 47.92, 43.31 (d, J = 17.8 Hz). HRMS (ESI) m/z: calcd for C29H31F3N7 [M + H]+, 556.3018; obsd,. 555.3024.
Molecular Docking Study
The 3D X-crystallographic structure of the crystal structure of Hu Topo I DNA complex (PDB ID: 1EJ9) was retrieved from the RCSB Protein Data Bank (https://www.rcsb.org) and used as a model for molecular docking. The PDB used for this study had 22 bp duplex DNA oligonucleotide along with Hu Topo I protein. We used the glide module60−62 of the Schrödinger suite for docking of molecules in Hu Topo I: DNA complex. All peptides were prepared using the LigPrep module of the Schrodinger suite and docked in the binding site of protein using the SP-peptide mode of Glide. The receptor grid was generated using the receptor grid generation in the Glide application by specifying the binding (active) site residues, which was identified using the SiteMap tool.63 The docked conformers were evaluated using Glide (G) Score, and the best-docked pose with the lowest Glide score value was recorded. The most favorable ligand orientation with the lowest free energy (binding affinity) was selected finally for further structural analysis of the protein–ligand complex, which included calculations of hydrogen bonds, hydrophobic interactions, bond lengths, and so on, using Discovery studio visualizer,64 Maestro of glide module60−62 of Schrödinger suite, and Ligplot+tools.65
UV–Visible Spectroscopy
UV–vis absorbance measurements were performed using a Varian, Cary 300 spectrophotometer instrument with a Peltier temperature control unit. Spectra were acquired using a 1 cm path length cuvette. The 100 μm stock solution of oligo 1 and 2; oligo 1 d(CGCGAATTCGCG)2 and oligo 2 d(CGATGTACATCG)2, were used for the study. Duplex DNA of d(GCGCAATTGCGC)2 and d(CGATGTACATCG)2 were made by annealing the complimentary oligonucleotide strands (10 μM each strand) in 20 mM sodium cacodylate buffer containing 100 mM NaCl at pH 7.2. The stock solutions of the ligands (BBZs) of 10 mM concentration were prepared in water and stored at −20 °C. The buffer used for all the studies was 20 mM sodium cacodylate, 100 mM NaCl (pH 7.2). Milli-Q water was used for making buffers. The 1 μm concentration of oligo was titrated 15 times into 5 μM solution of the ligand in cacodylate buffer. DNA binding was marked by hypochromicity or hyperchromicity and red shift of the absorption maxima.69,70
UV-Thermal Denaturation
A Cary Varian 300 spectrophotometer equipped with a Peltier thermal programmer was used for the thermal denaturation experiments using stoppered quartz cuvettes of 1 cm optical path length and 1 mL volume. The temperature of the cell holder was increased from 15 to 95 °C and vice versa at a rate of 0.2 °C/min. Tm experiments with oligos 1 and 2 (2.5 μM) in the absence and presence of ligands were carried out at a wavelength of 260 nm as a function of temperature. For Tm experiments, the ligand-oligo ratio, r = 0.5–2 per synthetic oligomers was used. The Tm was determined by plotting the first derivative of the absorbance with respect to temperature (dA260/dT), and Tm is the temperature corresponding to the maximum in the plot. The ΔTm can be obtained by subtracting the melting temperature of free oligo from that of complex (ligand-oligo).70 The accuracy of the Tm value is ±1 °C.
Fluorescence Measurement
A Varian, Cary Eclipse instrument was used for equilibrium-binding experiments using a solution of ligand (serially diluted to working concentrations) being excited at its respective λmax (slits of 5 nm), and the resulting emission curves (from 360 to 680 nm) were recorded after serial additions of a concentrated DNA solution (stock 0.1 mM) at 20 °C. After each addition, the solution was mixed by pipetting up and down. The sample equilibrium was monitored by continually exciting and scanning the sample at different times and was usually reached within 3 min. A Scatchard plot of r versus r/c was obtained from the titration data, where r is the ratio of bound ligands (Hoechst 33342, other synthetic analogues) concentration to the total oligo concentrations in base pairs, and c is the concentration of free ligands. The total fluorescence intensity (It) was measured by the sum of the contributions from both free and bound ligands as It = Io(Ct – Cb) + IbCb, where Io and Ib are fluorescence intensities of the free and fully bound ligand, and Ct and Cb correspond to the total and bound concentrations. From the slope of the plot, the binding constant k is determined for the ligand with oligonucleotides.69,70
Circular Dichroism Titrations
CD experiments were carried out at 20 °C using a Jasco J-810 spectropolarimeter. The spectra were recorded at a scan rate of 1 nm/min from 450 to 190 nm wavelength, and an average of three scans was taken. The ICD of the ligands was monitored in the 340–360 nm range. Measurements were carried out in quartz cuvettes of 1 cm optical path length. Duplex DNA of d(GCGCAATTGCGC)2 and d(CGATGTACATCG)2 were made by annealing the complementary oligonucleotide strands (10 μM each strand) in 20 mM sodium cacodylate buffer containing 100 mM NaCl at pH 7.2. The ligand was added to DNA at an increasing ligand/DNA ratio (r = 0.2). Between each ligand addition, an interval of 5 min was maintained, and the samples were mixed by pipetting up and down with a Pasteur pipette.69,70
Anticancer Screening Assay
The mammalian cancerous cell lines from the NCI screening panel are grown in RPMI 1640 medium with glutamine, bicarbonate, and 5% fetal calf serum. The cells were inoculated into 96-well plates in 0.2 mL of growth medium at the inoculum densities that depended on the growth characteristics or doubling time of particular cell lines. The compounds to be tested were evaluated at a single dose of 10 μM and then at five different doses prepared by 10-fold dilution starting from as high as 100 μM. The 96-well plates after the addition of respective test compounds were incubated for 48 h at 37 °C in an incubator with 5% CO2 atmosphere. The cells were fixed in 50 mL of cold 50% (w/v) trichloroacetic acid (TCA), bringing the final concentration of TCA to 10%, and incubated for 60 min at 4 °C. The supernatant was discarded, and the plate was washed five times with tap water to remove TCA and then air-dried. The staining was carried out by adding 100 mL of sulforhodamine B solution at 0.4% (w/v) in 1% acetic acid being added to each well, and the plates were incubated for 30 min at rt. The unbound dye was removed by washing four times with 1% acetic acid, and the plates were air-dried. The bound dyes were then solubilized in a 10 mM Tris base (pH 10.5), and the absorbance was recorded on an automated plate reader at a wavelength of 564 nm. The percentage growth is calculated using the seven absorbance measurements at time zero (T0), control growth (C), and five different concentrations of test agents or compounds (TD). Percentage growth inhibition is calculated as 100 × [(T – T0)/(C – T0)].71,72 Three parameters were calculated for each test agent. The 50% growth inhibition (GI50) is the concentration of the test agent, where 100 × [(T – T0)/(C – T0)] = 50. The ligand concentration resulting in total growth inhibition (TGI) is calculated when: 100 × [(T – T0)/(C – T0)] = 0. The lethal concentration, LC50, is calculated as where 100 × [(T – T0)/T0] = 50.
Cytotoxicity Assay
Cytotoxicity was determined using the MTT [3-(4,5-dimethylthiazol-2-yl)-2,5-diphenyl-2H-tetrazoliumbromide] assay using a 96-well microtiter plate. 3000 cells per well were plated in 200 μL of the complete medium, and treatment with these ligands was performed 48 h after plating. For IC50 determination, cells were exposed continuously with varying concentrations of BBZ, and MTT assays were performed at the end of the fourth day. At the end of the treatment, control and treated cells were incubated with MTT at a final concentration of 0.5 mg/mL for 2 h at 37 °C, and then, the medium was removed. The cells were lysed, and the formazan crystals were dissolved using 150 μL of DMSO. The absorbance was read at 570 nm using 630 nm as the reference wavelength using the TECAN Pro200 reader.39
Relaxation Assay of Hu Topo I
Relaxation of negatively supercoiled plasmid DNA by Hu Topo I was assayed in 20 μL of reaction buffer (10 mM Tris–HCl, pH 7.9, 150 mM NaCl, 0.1% BSA, 0.1 mM spermidine, and 5% glycerol) containing 500 ng of supercoiled pHOT1 plasmid DNA and 1 unit of the Hu Topo I enzyme. After incubation at 37 °C for 30 min, the reactions were terminated and analyzed by 1% agarose gel electrophoresis at voltage, as described for Hu Topo I relaxation assay.74
Cell Cycle Analysis
Breast cancer cell line MDA-MB 231 was seeded in 35 mm culture plates at a density of 3 × 105 cells/million and cultured for 24 h in DMEM with 1% antibiotic antimycotic solution (Invitrogen) and 10% fetal bovine serum in a 5% humidified CO2 atmosphere at 37 °C. The cells were then treated with the respective GI50 value observed in the MDA-MB-231 cell line of compounds 11a, 12b, and 12a, respectively, for 2 h. The cell was trypsinized, collected, and washed twice with PBS solution at 4 °C. The supernatant was discarded, and the cells were fixed with 70% ethanol. The fixed cells were rinsed with PBS, then stained with the DNA fluorochrome PI (25 μg/mL) and RNase (50 μL of 10 mg/mL), and incubated for 30 min at 37 °C. The samples were then analyzed with a Flow-Activated Cell Sorting equipment (LSR Fortessa, BD Biosciences Pvt. Ltd.) using an FL2 filter (585/40 nm bandpass filter). A minimum of 10,000 events were analyzed for each sample.
Statistical Analysis
Statistical analysis data from three independent experiments are presented as mean ± SD. GI50 and LC50 values and SD values were calculated by SPSS version 10.0 (SPSS, Inc., Chicago, IL, USA).
Acknowledgments
We are thankful to USIC, the University of Delhi and JNU for the instrumentation facility. S.P. is thankful to DU Non-Net & Council of Scientific and Industrial Research (CSIR) for fellowship. We are grateful to the Department of Biotechnology (DBT) (project no. BT/PR20794/MED/29/1057/2016, dated 27.06.2017), Department of Science and Technology SERB(DST-SERB) (project no. EMR/2017/000454, dated 21.03.2018), DST PURSE (Phase II), UPOE-II Jawaharlal Nehru University, and UGC Mid Term Award grant for funding.
Supporting Information Available
The Supporting Information is available free of charge at https://pubs.acs.org/doi/10.1021/acsomega.1c05743.
Spectra of 1H NMR, 13C NMR, and HRMS of the synthesized compounds, Tm of the binary complex, UV–visible spectral changes, fluorescence titration spectra, and CD of all BBZ analogues with oligo-1 and oligo-2 (PDF)
Single dose screening of BBZs (PDF)
Five dose screening of selected BBZs against NCI panel of 60 cancer cell lines (PDF)
The authors declare no competing financial interest.
Supplementary Material
References
- Mann J.; Baron A.; Opoku-Boahen Y.; Johansson E.; Parkinson G.; Kelland L. R.; Neidle S. A new class of symmetric bisbenzimidazole-based DNA minor groove-binding agents showing antitumor activity. J. Med. Chem. 2001, 44, 138–144. 10.1021/jm000297b. [DOI] [PubMed] [Google Scholar]
- Baraldi P. G.; Bovero A.; Fruttarolo F.; Preti D.; Tabrizi M. A.; Pavani M. G.; Romagnoli R. DNA minor groove binders as potential antitumor and antimicrobial agents. Med. Res. Rev. 2004, 24, 475–528. 10.1002/med.20000. [DOI] [PubMed] [Google Scholar]
- Matsuba Y.; Edatsugi H.; Mita I.; Matsunaga A.; Nakanishi O. A novel synthetic DNA minor groove binder, MS-247: Antitumor activity and cytotoxic mechanism. Cancer Chemother. Pharmacol. 2000, 46, 1–9. 10.1007/s002800000120. [DOI] [PubMed] [Google Scholar]
- Khalaf A. I.; Bourdin C.; Breen D.; Donoghue G.; Scott F. J.; Suckling C. J.; Macmillan D.; Clements C.; Fox K.; Sekibo D. A. T. Design, synthesis and antibacterial activity of minor groove binders: The role of non-cationic tail groups. Eur. J. Med. Chem. 2012, 56, 39–47. 10.1016/j.ejmech.2012.08.013. [DOI] [PubMed] [Google Scholar]
- Bhaduri S.; Ranjan N.; Arya D. P. An overview of recent advances in duplex DNA recognition by small molecules. Beilstein J. Org. Chem. 2018, 14, 1051–1086. 10.3762/bjoc.14.93. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang L.; Bailly C.; Kumar A.; Ding D.; Bajic M.; Boykin D. W.; Wilson W. D. Specific molecular recognition of mixed nucleic acid sequences: An aromatic dication that binds in the DNA minor groove as a dimer. Proc. Natl. Acad. Sci. U.S.A. 2000, 97, 12–16. 10.1073/pnas.97.1.12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cai X.; Gray P. J.; Von Hoff D. D. DNA minor groove binders: Back in the groove. Cancer Treat. Rev. 2009, 35, 437–450. 10.1016/j.ctrv.2009.02.004. [DOI] [PubMed] [Google Scholar]
- Palchaudhuri R.; Hergenrother P. J. DNA as a target for anticancer compounds: methods to determine the mode of binding and the mechanism of action. Curr. Opin. Biotechnol. 2007, 18, 497–503. 10.1016/j.copbio.2007.09.006. [DOI] [PubMed] [Google Scholar]
- Seaton A.; Higgins C.; Mann J.; Baron A.; Bailly C.; Neidle S.; van den Berg H. Mechanistic and anti-proliferative studies of two novel, biologically active bis-benzimidazoles. Eur. J. Cancer 2003, 39, 2548–2555. 10.1016/s0959-8049(03)00621-x. [DOI] [PubMed] [Google Scholar]
- Lenglet G.; David-Cordonnier M.-H. DNA-destabilizing agents as an alternative approach for targeting DNA: Mechanisms of action and cellular consequences. J. Nucleic Acids 2010, 2010, 1–17. 10.4061/2010/290935. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tse W. C.; Boger D. L. Sequence-selective DNA recognition: Natural products and nature’s lessons. J. Chem. Biol. 2004, 11, 1607–1617. 10.1016/j.chembiol.2003.08.012. [DOI] [PubMed] [Google Scholar]
- Weber G. F.DNA Damaging Drugs. Molecular Therapies of Cancer; Springer: Cham, 2015; pp 9–112. [Google Scholar]
- Bales B. C.; Kodama T.; Weledji Y. N.; Pitié M.; Meunier B.; Greenberg M. M. Mechanistic studies on DNA damage by minor groove binding copper-phenanthroline conjugates. Nucleic Acids Res. 2005, 33, 5371–5379. 10.1093/nar/gki856. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gupta S.; Tiwari N.; Munde M. A Comprehensive Biophysical Analysis of the Effect of DNA Binding Drugs on Protamine-induced DNA Condensation. Sci. Rep. 2019, 9, 5891. 10.1038/s41598-019-41975-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lam K. S.; Veitch J. A.; Golik J.; Krishnan B.; Klohr S. E.; Volk K. J.; Forenza S.; Doyle T. W. Biosynthesis of Esperamicin A1, an Enediyne Antitumor Antibiotic. J. Am. Chem. Soc. 1993, 115, 12340–12345. 10.1021/ja00079a014. [DOI] [Google Scholar]
- Banerjee D.; Pal S. K. Dynamics in the DNA recognition by DAPI: Exploration of the various binding modes. J. Phys. Chem. B 2008, 112, 1016–1021. 10.1021/jp077090f. [DOI] [PubMed] [Google Scholar]
- Paul A.; Guo P.; Boykin D.; Wilson W. A new generation of minor-groove-binding—heterocyclic diamidines that recognize G·C base pairs in an at sequence context. Molecules 2019, 24, 946. 10.3390/molecules24050946. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pjura P. E.; Grzeskowiak K.; Dickerson R. E. Binding of Hoechst 33258 to the minor groove of B-DNA. J. Mol. Biol. 1987, 197, 257–271. 10.1016/0022-2836(87)90123-9. [DOI] [PubMed] [Google Scholar]
- Drozdowska D.; Bruzgo I.; Midura-Nowaczek K. Carbocyclic potential DNA minor groove binders and their biological evaluation. J. Enzyme Inhib. Med. Chem. 2010, 25, 629–634. 10.3109/14756360903389872. [DOI] [PubMed] [Google Scholar]
- Fairley T. A.; Tidwell R. R.; Donkor I.; Naiman N. A.; Ohemeng K. A.; Lombardy R. J.; Bentley J. A.; Cory M. Structure, DNA minor groove binding, and base pair specificity of alkyl- and aryl-Linked Bis(amidinobenzimidazoles) and Bis(amidinoindoles). J. Med. Chem. 1993, 36, 1746–1753. 10.1021/jm00064a008. [DOI] [PubMed] [Google Scholar]
- Townsend L. B.; Wise D. S. The synthesis and chemistry of certain anthelmintic benzimidazoles. Parasitol. Today 1990, 6, 107–112. 10.1016/0169-4758(90)90226-t. [DOI] [PubMed] [Google Scholar]
- Merino G.; Jonker J. W.; Wagenaar E.; Pulido M. M.; Molina A. J.; Alvarez A. I.; Schinkel A. H. Transport of anthelmintic benzimidazole drugs by breast cancer resistance protein (BCRP/ABCG2). Drug Metab. Dispos. 2005, 33, 614–618. 10.1124/dmd.104.003319. [DOI] [PubMed] [Google Scholar]
- Agh-Atabay N.; Dulger B.; Gucin F. Synthesis, and investigation of antimicrobial activity of some bisbenzimidazole-derived chelating agents. Eur. J. Med. Chem. 2003, 38, 875–881. 10.1016/s0223-5234(03)00146-6. [DOI] [PubMed] [Google Scholar]
- Arjmand F.; Mohani B.; Ahmad S. Synthesis, antibacterial, antifungal activity and interaction of CT-DNA with a new benzimidazole derived Cu(II) complex. Eur. J. Med. Chem. 2005, 40, 1103–1110. 10.1016/j.ejmech.2005.05.005. [DOI] [PubMed] [Google Scholar]
- Ansari K. F.; Lal C. Synthesis, physicochemical properties and antimicrobial activity of some new benzimidazole derivatives. Eur. J. Med. Chem. 2009, 44, 4028–4033. 10.1016/j.ejmech.2009.04.037. [DOI] [PubMed] [Google Scholar]
- Kim J. S.; Sun Q.; Yu C.; Liu A.; Liu L. F.; Lavoie E. J. Quantitative structure-activity relationships on 5-substituted terbnzimidazoles as topoisomerase I poison and antitumor agents. Bioorg. Med. Chem. 1998, 6, 163–172. 10.1016/s0968-0896(97)10021-9. [DOI] [PubMed] [Google Scholar]
- Wang X.-J.; Chu N.-Y.; Wang Q.-H.; Liu C.; Jiang C.-g.; Wang X.-Y.; Ikejima T.; Cheng M.-S. Newly synthesized bis-benzimidazole derivatives exerting antitumor activity through induction of apoptosis and autophagy. Bioorg. Med. Chem. Lett. 2012, 22, 6297–6300. 10.1016/j.bmcl.2012.06.102. [DOI] [PubMed] [Google Scholar]
- Fonseca T.; Gigante B.; Marques M. M.; Gilchrist T. L.; De Clercq E. Synthesis and antiviral evaluation of benzimidazoles, quinoxalines, and indoles from dehydroabietic acid. Bioorg. Med. Chem. 2004, 12, 103–112. 10.1016/j.bmc.2003.10.013. [DOI] [PubMed] [Google Scholar]
- Malaty H.; El-Zimaity H. M. T.; Genta R. M.; Cole R. A.; Graham D. Y. High-dose proton pump inhibitor plus Amoxycillin for the treatment or retreatment of helicobacter pylori infection. Aliment. Pharmacol. Ther. 1996, 10, 1001–1004. 10.1046/j.1365-2036.1996.77248000.x. [DOI] [PubMed] [Google Scholar]
- Zhang X.; Kiechle F. 33342-induced apoptosis is associated with decreased immunoreactive topoisomerase I and topoisomerase I-DNA complex formation. Ann. Clin. Lab. Sci. 2001, 31, 187–198. [PubMed] [Google Scholar]
- Thomas A.; Pommier Y. Targeting Topoisomerase I in the Era of Precision Medicine. Clin. Cancer Res. 2019, 25, 6581–6589. 10.1158/1078-0432.ccr-19-1089. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang J. C. DNA topoisomerases. Annu. Rev. Biochem. 1996, 65, 635–692. 10.1146/annurev.bi.65.070196.003223. [DOI] [PubMed] [Google Scholar]
- Coban G.; Zencir S.; Zupkó I.; Réthy B.; Gunes H. S.; Topcu Z. Synthesis and biological activity evaluation of 1H-benzimidazoles via mammalian DNA topoisomerase I and cytostaticity assays. Eur. J. Med. Chem. 2009, 44, 2280–2285. 10.1016/j.ejmech.2008.06.018. [DOI] [PubMed] [Google Scholar]
- Singh R.; Pandey S.; Sur S.; Tandon V. PPEF: A bisbenzimdazole potent antimicrobial agent interacts at acidic triad of catalytic domain of E. coli topoisomerase IA. Biochim. Biophys. Acta, Gen. Subj. 2019, 1863, 1524–1535. 10.1016/j.bbagen.2019.05.015. [DOI] [PubMed] [Google Scholar]
- Singh M.; Tandon V. Synthesis and biological activity of novel inhibitors of topoisomerase I: 2-Aryl-substituted 2-bis-1H-benzimidazoles. Eur. J. Med. Chem. 2011, 46, 659–669. 10.1016/j.ejmech.2010.11.046. [DOI] [PubMed] [Google Scholar]
- Bansal S.; Bajaj P.; Pandey S.; Tandon V. Topoisomerases: Resistance versus Sensitivity, How Far We Can Go?. Med. Res. Rev. 2017, 37, 404–438. 10.1002/med.21417. [DOI] [PubMed] [Google Scholar]
- Nitiss J. L. DNA topoisomerase II and its growing repertoire of biological functions. Nat. Rev. Cancer 2009, 9, 327–337. 10.1038/nrc2608. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pourqier P.; Lansiaux A. Molecular determinants of response to topoisomerase I inhibitors. Bull. Cancer 2011, 98, 1287–1298. 10.1684/bdc.2011.1474. [DOI] [PubMed] [Google Scholar]
- Tawar U.; Jain A. K.; Dwarakanath B. S.; Chandra R.; Singh Y.; Chaudhury N. K.; Khaitan D.; Tandon V. Influence of phenyl ring disubstitution on bisbenzimidazole and terbenzimidazole cytotoxicity: Synthesis and biological evaluation as radioprotectors. J. Med. Chem. 2003, 46, 3785–3792. 10.1021/jm030114w. [DOI] [PubMed] [Google Scholar]
- Fudickar W.; Linker T. Structural motives controlling the binding affinity of 9,10-bis(methylpyridinium) anthracenes towards DNA. Bioorg. Med. Chem. 2020, 28, 115432. 10.1016/j.bmc.2020.115432. [DOI] [PubMed] [Google Scholar]
- Rosu F.; Gabelica V.; Houssier C.; Edwin De. P. Determination of affinity, stoichiometry, and sequence selectivity of minor groove binder complexes with double-stranded oligodeoxynucleotides by electrospray ionization mass spectrometry. Nucleic Acids Res. 2002, 30, e82 10.1093/nar/gnf081. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bostock-Smith C. E.; Harris S. A.; Laughton C. A.; Searle M. S. Induced fit DNA recognition by a minor groove binding analogue of Hoechst 33258: Fluctuations in DNA A tract structure investigated by NMR and molecular dynamics simulations. Nucleic Acids Res. 2001, 29, 693–702. 10.1093/nar/29.3.693. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ranjan N.; Kellish P.; King A.; Arya D. P. Impact of Linker Length and Composition on Fragment Binding and Cell Permeation: Story of a Bisbenzimidazole Dye Fragment. Biochemistry 2017, 56, 6434–6447. 10.1021/acs.biochem.7b00929. [DOI] [PubMed] [Google Scholar]
- Amirbekyan K.; Duchemin N.; Benedetti E.; Joseph R.; Colon A.; Markarian S. A.; Bethge L.; Vonhoff S.; Klussmann S.; Cossy J.; et al. Design, Synthesis, and Binding Affinity Evaluation of Hoechst 33258 Derivatives for the Development of Sequence-Specific DNA-Based Asymmetric Catalysts. ACS Catal. 2016, 6, 3096–3105. 10.1021/acscatal.6b00495. [DOI] [Google Scholar]
- Shoemaker R. H. The NCI60 human tumour cell line anticancer drug screen. Nat. Rev. Cancer 2006, 6, 813–823. 10.1038/nrc1951. [DOI] [PubMed] [Google Scholar]
- Selwood D. Anticancer Drug Development: Preclinical Screening, Clinical Trials, and Approval. Br. J. Cancer 2004, 91, 1000. 10.1038/sj.bjc.6602070. [DOI] [Google Scholar]
- Shaharyar M.; Abdullah M. M.; Bakht M. A.; Majeed J. Pyrazoline bearing benzimidazoles: Search for anticancer agent. Eur. J. Med. Chem. 2010, 45, 114–119. 10.1016/j.ejmech.2009.09.032. [DOI] [PubMed] [Google Scholar]
- Rashid M.; Husain A.; Mishra R. Synthesis of benzimidazoles bearing oxadiazole nucleus as anticancer agents. Eur. J. Med. Chem. 2012, 54, 855–866. 10.1016/j.ejmech.2012.04.027. [DOI] [PubMed] [Google Scholar]
- Paul K.; Sharma A.; Luxami V. Synthesis and in vitro antitumor evaluation of primary amine substituted quinazoline linked benzimidazole. Bioorg. Med. Chem. Lett. 2014, 24, 624–629. 10.1016/j.bmcl.2013.12.005. [DOI] [PubMed] [Google Scholar]
- Hearn J. M.; Romero-Canelón I.; Qamar B.; Liu Z.; Hands-Portman I.; Sadler P. J. Organometallic Iridium(III) anticancer complexes with new mechanisms of action: NCI-60 screening, mitochondrial targeting, and apoptosis. ACS Chem. Biol. 2013, 8, 1335–1343. 10.1021/cb400070a. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Phillips M. A. CCCXVII. The formation of 2-substituted benziminazoles. J. Chem. Soc. 1928, 2393–2399. 10.1039/jr9280002393. [DOI] [Google Scholar]
- Singh M. P.; Sasmal S.; Lu W.; Chatterjee M. N. Synthetic utility of catalytic Fe(III)/Fe(II) redox cycling towards fused heterocycles: A facile access to substituted benzimidazole, bis-benzimidazole, and imidazopyridine derivatives. Synthesis 2000, 1380–1390. 10.1055/s-2000-7111. [DOI] [Google Scholar]
- Wright J. B. The Chemistry of the Benzimidazoles. Chem. Rev. 1951, 48, 397–541. 10.1021/cr60151a002. [DOI] [PubMed] [Google Scholar]
- Bahrami K.; Khodaei M.; Kavianinia I. A simple and efficient one-pot synthesis of 2-substituted benzimidazoles. Synthesis 2007, 547–550. 10.1055/s-2007-965878. [DOI] [Google Scholar]
- Shen M.; Driver T. G. Iron (II) bromide-catalyzed synthesis of benzimidazoles from aryl azides. Org. Lett. 2008, 10, 3367–3370. 10.1021/ol801227f. [DOI] [PubMed] [Google Scholar]
- Hegedüs A.; Hell Z.; Potor A. Zeolite-catalyzed environmentally friendly synthesis of benzimidazole derivatives. Synth. Commun. 2006, 36, 3625–3630. 10.1080/00397910600943865. [DOI] [Google Scholar]
- Curini M.; Epifano F.; Montanari F.; Rosati O.; Taccone S. Ytterbium triflate promoted synthesis of benzimidazole derivatives. Synlett 2004, 1832–1834. 10.1055/s-2004-829555. [DOI] [Google Scholar]
- Deng X.; McAllister H.; Mani N. S. CuI-catalyzed amination of arylhalides with guanidines or amidines: A facile synthesis of 1-H-2-substituted benzimidazoles. J. Org. Chem. 2009, 74, 5742–5745. 10.1021/jo900912h. [DOI] [PubMed] [Google Scholar]
- Nimesh H.; Sur S.; Sinha D.; Yadav P.; Anand P.; Bajaj P.; Virdi J. S.; Tandon V. Synthesis and Biological Evaluation of Novel Bisbenzimidazoles as Escherichia coli Topoisomerase IA Inhibitors and Potential Antibacterial Agents. J. Med. Chem. 2014, 57, 5238–5257. 10.1021/jm5003028. [DOI] [PubMed] [Google Scholar]
- Redinbo M. R.; Champoux J. J.; Hol W. G. J. Novel Insights into Catalytic Mechanism from a Crystal Structure of Human Topoisomerase I in Complex with DNA. Biochemistry 2000, 39, 6832–6840. 10.1021/bi992690t. [DOI] [PubMed] [Google Scholar]
- Friesner R. A.; Murphy R. B.; Repasky M. P.; Frye L. L.; Greenwood J. R.; Halgren T. A.; Sanschagrin P. C.; Mainz D. T. Xtra Precision Glide:Docking and Scoring incorporating a model of hydrophobic enclosure for Protein–Ligand Complexes. J. Med. Chem. 2006, 49, 6177–6196. 10.1021/jm051256o. [DOI] [PubMed] [Google Scholar]
- Halgren T. A.; Murphy R. B.; Friesner R. A.; Beard H. S.; Frye L. L.; Pollard W. T.; Banks J. L. Glide: a new approach for rapid, accurate docking and scoring. 2. Enrichment factors in database screening. J. Med. Chem. 2004, 47, 1750–1759. 10.1021/jm030644s. [DOI] [PubMed] [Google Scholar]
- Bart L.; Hjerrild K.; Feese M. D.; Behnke C. A.; Burgin A. B.; Stewart L. The mechanism of topoisomerase I poisoning by a camptothecin analog. Proc. Natl. Acad. Sci. U.S.A. 2002, 99, 15387–15392. 10.1073/pnas.242259599. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Laskowski R. A.; Swindells M. B. LigPlot+: multiple ligand-protein interaction diagrams for drug discovery. J. Chem. Inf. Model. 2011, 51, 2778–2786. 10.1021/ci200227u. [DOI] [PubMed] [Google Scholar]
- Biovia . Dassault Systèmes. Discovery Studio Visualizer: San Diego, CA, USA, 936, 2017.
- Carrondo M. A. A. F. d. C. T.; Coll M.; Aymami J.; Wang A. H. J.; Van der Marel G. A.; Van Boom J. H.; Rich A. Binding of a Hoechst Dye to d(CGCGATATCGCG) and Its Influence on the Conformation of the DNA Fragment. Biochemistry 1989, 28, 7849–7859. 10.1021/bi00445a047. [DOI] [PubMed] [Google Scholar]
- Quintana J. R.; Lipanov A. A.; Dickerson R. E. Low-Temperature Crystallographic Analyses of the Binding of Hoechst 33258 to the Double-Helical DNA Dodecamer C-G-C-G-A-A-T-T-C-G-C-G. Biochemistry 1991, 30, 10294–10306. 10.1021/bi00106a030. [DOI] [PubMed] [Google Scholar]
- Guo P.; Farahat A. A.; Paul A.; Harika N. K.; Boykin D. W.; Wilson W. D. Compound Shape Effects in Minor Groove Binding Affinity and Specificity for Mixed Sequence DNA. J. Am. Chem. Soc. 2018, 140, 14761–14769. 10.1021/jacs.8b08152. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Han F.; Taulier N.; Chalikian T. V. Association of the minor groove binding drug Hoechst 33258 with d(CGCGAATTCGCG)2: Volumetric, calorimetric, and spectroscopic characterizations. Biochemistry 2005, 44, 9785–9794. 10.1021/bi047374f. [DOI] [PubMed] [Google Scholar]
- Singh M.; Sur S.; Rastogi G. K.; Jayaram B.; Tandon V. Bi and tri-substituted phenyl rings containing bisbenzimidazoles bind differentially with DNA duplexes: A biophysical and molecular simulation study. Mol. BioSyst. 2013, 9, 2541–2553. 10.1039/c3mb70169g. [DOI] [PubMed] [Google Scholar]
- Boyd M. R.; Paull K. D. Some Practical Considerations and Applications of the National Cancer Institute In Vitro Anticancer Drug Discovery Screen. Drug Dev. Res. 1995, 34, 91–109. 10.1002/ddr.430340203. [DOI] [Google Scholar]
- Skehan P.; Storeng R.; Scudiero D.; Monks A.; McMahon J.; Vistica D.; Warren J. T.; Bokesch H.; Kenney S.; Boyd M. R. New colorimetric cytotoxicity assay for anticancer-drug screening. J. Natl. Cancer Inst. 1990, 82, 1107–1112. 10.1093/jnci/82.13.1107. [DOI] [PubMed] [Google Scholar]
- Chabner B. A. NCI-60 Cell Line Screening: A Radical Departure in Its Time. J. Natl. Cancer Inst. 2016, 108, djv388. 10.1093/jnci/djv388. [DOI] [PubMed] [Google Scholar]
- Staker B. L.; Hjerrild K.; Feese M. D.; Behnke C. A.; Burgin A. B.; Stewart L. The mechanism of topoisomerase I poisoning by a camptothecin analog. Proc. Natl. Acad. Sci. U.S.A. 2002, 99, 15387–15392. 10.1073/pnas.242259599. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pommier Y. Topoisomerase I inhibitors: camptothecins and beyond. Nat. Rev. Cancer 2006, 6, 789–802. 10.1038/nrc1977. [DOI] [PubMed] [Google Scholar]
- Jian-Song J.; Gao Q.-L.; Wu B.-W.; Li D.; Shi L.; Zhu T.; Lou J.-F.; Jin C.-Y.; Zhang Y.-B.; Zhang S.-Y.; Liu H.-M. Novel tertiary sulfonamide derivatives containing benzimidazole moiety as potent anti-gastric cancer agents: Design, synthesis, and SAR. Eur. J. Med. Chem. 2019, 183, 111731. 10.1016/j.ejmech.2019.111731. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.