Abstract
Liver cancer has high rates of morbidity and mortality, and its treatment is a global health challenge. Hepatocellular carcinoma (HCC) accounts for 90% of all primary liver cancer cases. B-lymphoma Mo-MLV insertion region 1 (BMI1) has been identified as a proto-oncogene, which contributes to the initiation and progression of many malignant tumors. BMI1 expression is upregulated in HCC, and it influences the occurrence and development of HCC by various mechanisms, such as the INK4a/ARF locus, NF-κB signaling pathway, and PTEN/PI3K/AKT signaling pathway. In addition, the expression of BMI1 is related to prognosis and recurrence of HCC. Hence, there is clear evidence that BMI1 is a novel and valid therapeutic target for HCC. Accordingly, the development of therapeutic strategies targeting BMI1 has been a focus of recent research, providing new directions for HCC treatment. This review summarizes the role of BMI1 in the occurrence and treatment of HCC, which will provide a basis for using BMI1 as a potential target for the development of therapeutic strategies for HCC.
Keywords: hepatocellular carcinoma, BMI1, miR-218, miR-203, INK4a/ARF locus
Introduction
Liver cancer poses a global health challenge, with 905 677 new cases detected in 2020 and an estimated incidence of >1 million cases by 2025. 1 Liver cancer is the sixth most common cancer worldwide and the fourth leading cause of cancer deaths globally. Hepatocellular carcinoma (HCC) accounts for more than 90% of all primary liver cancer cases and is the most common type of liver cancer. Current therapeutic methods for liver cancer mainly include surgical resection, liver transplantation, local ablation, external radiation, trans-arterial therapies, chemotherapy, targeted therapy, and immunotherapy. 2 However, the efficacies of these strategies remain insufficient, and the mortality rate of liver cancer is still high. Therefore, to improve survival and the quality of life, new and effective treatment methods for HCC are required.
Similar to the processes involved in other cancers, the occurrence and invasion of HCC involve complicated processes, involving multiple factors and processes. A series of physiological processes involving the polycomb group (PcG) gene family, including cellular differentiation, stem cell self-renewal, and correct gene silencing, are critical for normal tissue functioning. B-lymphoma Mo-MLV insertion region 1 (BMI1) encodes a key PcG protein that ubiquitylates histone H2A by forming a stable heterodimer with RING1B. 3 BMI1 has been identified as a proto-oncogene. It encodes a protein with 324 amino acids, which is principally localized in the nucleus and contributes to the generation of mouse pre-B cell lymphomas. 4 BMI1 has been found in various normal cells and is highly conserved evolutionarily; moreover, it contains a type of zinc finger motif. 5 Accumulating evidence indicates that BMI1 is expressed at significantly high levels and is closely related to the occurrence, development, and outcomes of various human malignancies.6–8 In endometrial adenocarcinoma, the overexpression of BMI1 is a poor prognostic factor correlated with the invasion of the myometrium and lymph nodes. 9 In addition, in gastric cancer, the BMI1 gene regulates tumor growth, metastasis, and chemoresistance by downregulating Raf kinase inhibitor protein (RKIP) expression. 10 Therefore, a clear understanding of the role of BMI1 in HCC is essential for the development of new therapeutic strategies. This review describes the roles of BMI1 in the occurrence, progression, and prognosis of HCC as well as the implications of recent research for treatment strategies for HCC.
BMI1 Expression Is Upregulated in HCC
Neo et al identified 218 differentially expressed genes between HCC and normal liver tissues; these genes included BMI1. 11 BMI1 expression is markedly higher in HCC tissues than in normal and adjacent non-cancerous liver tissues,12–14 consistent with the results for other cancers.8,15,16 However, changes in the expression of BMI1 in HCC at different stages are still controversial. Sasaki et al established that the percentage of BMI1-positive cells is higher in poorly differentiated HCC (91.22 ± 5.64%) than in moderately differentiated HCC (56.94 ± 28.55%) and well-differentiated HCC (8.48 ± 11.97%). 17 In contrast, Effendi et al concluded that BMI1 expression level is the highest in well-differentiated HCC (including early HCC samples), followed by the expression in moderately differentiated and poorly differentiated HCC. 18 These conflicting results could be because of differences in analysis methods, evaluation criteria, and population differences. Both were semi-quantitative analyses; however, Sasaki et al evaluated the proportion of BMI1-positive cells in 27 HCC tissue samples (among >100 cells) under a microscope, 17 while Effendi et al evaluated tissues resected from patients with HCC by immunohistochemical staining based on a 3-point scale (0 for no staining, 1 + for focal and weak distribution, and 2 + for diffuse and clear distribution). 18 To further explore the relationship between the expression of BMI1 and the degree of HCC differentiation, Effendi et al determined BMI1 mRNA expression levels in HCC samples with different levels of differentiation and obtained similar results. Both studies proved that BMI1 is highly expressed in the nuclei in HCC. Nevertheless, correlations between high BMI1 expression levels and clinical characteristics of patients with HCC should be investigated further. Based on data for 62 patients with HCC, Li et al determined that high BMI1 expression is not statistically correlated with age, gender, satellite foci, tumor location and number, and the level of alpha-fetoprotein (AFP) but was highly correlated with neoplasm size, distant metastasis, vascular infiltration, and AJCC TNM stage. 19 Sasaki et al also detected correlations between high BMI1 expression levels and venous invasion and cancer cell proliferation, thus revealing that BMI1 accelerates disease progression and is a poor prognostic factor in HCC. 17 Surprisingly, Wang et al obtained contradictory results. They found that the expression of BMI1 was not related to the basic clinical characteristics of patients with HCC or to metastasis and postoperative recurrence, although they reported that high BMI1 levels promoted the development and progression of HCC. 20 These inconsistent findings may be related to the differences in disease stage and indicate that more comprehensive and objective analyses are urgently required.
Mechanism by Which BMI1 Contributes to the Initiation of HCC
Excessive proliferation of cells plays a pivotal role in the early formation of cancers. 21 Several studies have demonstrated that BMI1 not only participates in the self-renewal of normal hematopoietic stem cells but also contributes greatly to the continuous proliferation of acute myelocytic leukemia stem cells without senescence and apoptosis.22,23 It has been reported that BMI1 is required for hepatic stem cell expansion. 24 Interestingly, BMI1 promotes the proliferation, colony formation, and cell cycle progression of hepatic progenitor cells in vitro, and high BMI1 expression in hepatic progenitor cells results in the occurrence of poorly differentiated HCC in nude mice. 25
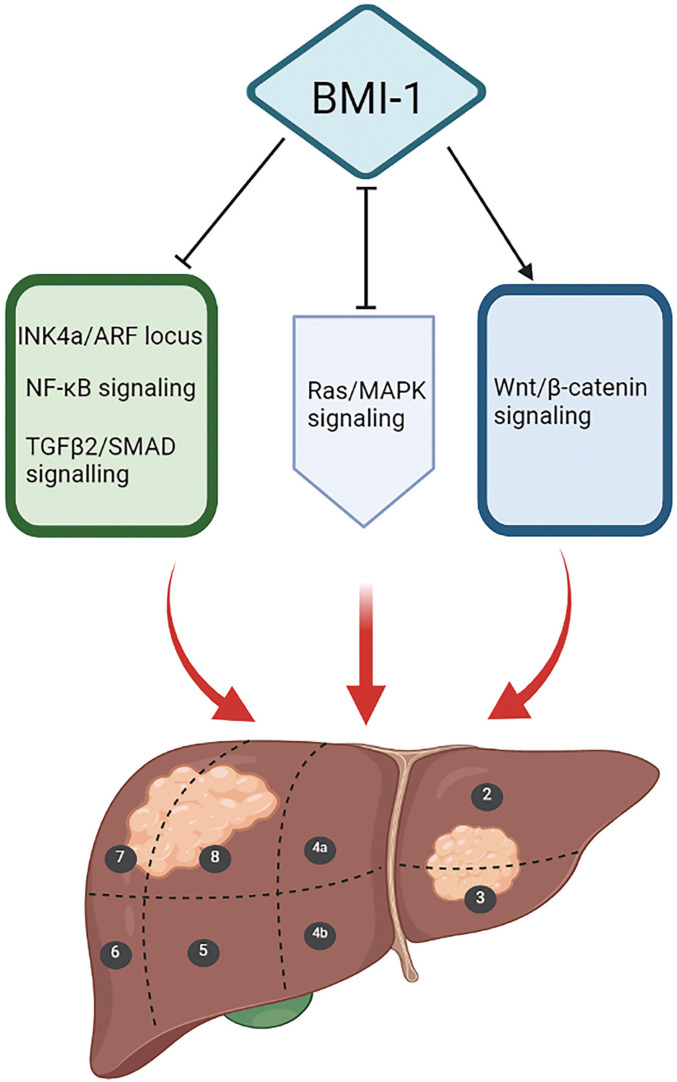
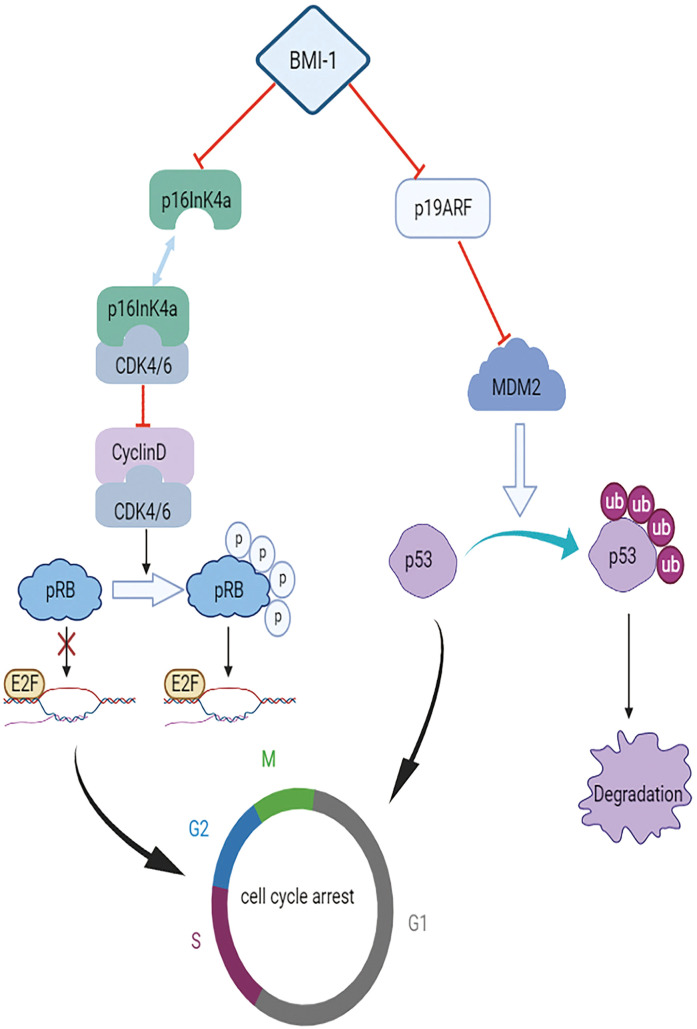
The initiation of HCC by BMI1 is mediated by many mechanisms (Figure 1). Chiba et al revealed that BMI1 determines the self-renewal capability of side population cells purified from HCC tissues and demonstrated the high proliferative potential and anti-apoptotic properties of these cells, 26 which directly contribute to its tumorigenic potential. 27 Although BMI1 can promote the self-renewal of hepatic stem cells, it has no influence on the overall cell cycle, and there is no evidence that BMI1 promotes the growth or self-renewal of differentiated cells. 28 However, Ma et al reported that the downregulation of BMI1 expression could induce CD133+ Huh7 cell cycle arrest in the G0/G1 and S phases by blocking the NF-κB signaling pathway, thereby promoting cell apoptosis. 29 It has previously been reported that BMI1 regulates the cell cycle, apoptosis, and senescence by repressing INK4a/ARF, encoding p16ink4a, which inhibits the activity of cyclin-dependent kinase (CDK), as well as p19ARF, a well-known tumor suppressor (Figure 2).30–32 Direct binding of p16INK4a to CDK4 and CDK6 maintains Rb in a hypophosphorylated state. Hypophosphorylated Rb represses E2F-dependent transcription, causing cell cycle arrest and senescence. 33 p19ARF suppresses MDM2, which mediates the ubiquitin-dependent degradation of p53, and subsequently activates p53 target genes involved in cell cycle arrest and apoptosis.34,35 The increase in cell self-renewal ability induced by BMI1 in neural stem cells and glioma cells is not completely INK4a/ARF-dependent.36,37 In a study by Chiba et al the expression levels of INK4a and ARF were increased in PLC/PRF/5 cells upon BMI1 knockdown, with no remarkable changes in Huh7 cells compared with levels in the control group. 27 Intriguingly, BMI1 increased the expression of p16INK4a and inhibited the expression of p19ARF in mouse hepatocytes. 38 Hence, Chiba et al further explored the role of the INK4a/ARF tumor suppressor gene locus in hepatic stem cell expansion and hepatocarcinogenesis. They ascertained that transfection with BMI1 alone could enhance the renewal ability of Dlk+ cells, without INK4a/ARF expression, demonstrating that the INK4a/ARF locus does not directly contribute to early hepatoma formation. 39 Additionally, they identified five candidate genes downstream of BMI1 in hepatic stem/progenitor cells, representing a major step forward. Among the five candidate genes, Prom1 (CD133) and EpCAM are highly expressed in hepatic stem cells, whereas carbamoyl phosphate synthase I (Cps1), Mat1a, and Gjb2 show low expression in hepatocyte-differentiated cells. 39 Knockout of the Mat1a gene in mice enhanced the sensitivity of hepatocytes to oxidative stress and accelerated the formation of HCC, providing powerful evidence that Mat1a is a hepatoma marker. 40 Furthermore, Cps1 may be a key component in the progression of hepatocytes to malignant HCC cells by metabolic reprograming. 41 Nevertheless, further research is necessary to determine the relationships between these five candidate downstream targets and carcinogenesis. Chiba et al ultimately determined that BMI1 promotes hepatic stem cell expansion and tumorigenicity in both INK4a/ARF-dependent and INK4a/ARF-independent manners in mice. 39 This conclusion is supported by the results of Fu et al who found that BMI1 knockdown induces G1-phase arrest and activates the p14ARF and p16INK4a signaling pathways in HepG2 cells, 42 as well as the results of Xu et al who established that the silencing of BMI1 expression does not lead to the upregulation of p14ARF and p16INK4a in Huh7 and Hep3B cells. 38
Figure 1.
Mechanism underlying the initiation of hepatocellular carcinoma (HCC) by BMI1. Previous studies have shown that BMI1 promotes the occurrence of HCC by blocking the INK4a/ARF locus, NF-κB signaling pathway, and TGFβ2/SMAD signaling axis and by stimulating the Wnt/β-catenin signaling axis. Besides, BMI1 and Ras/MAPK signaling work together to activate the initiation of HCC. Black and red arrows represent promotion, T-shaped symbols represent inhibition, and H-shaped symbols represent interactive effects.
Figure 2.
Mechanism by which INK4a/ARF mediates the effects of BMI1. The BMI1 gene downregulates the expression of p16INK4a and p19ARF. Direct binding of p16INK4a to CDK4 and CDK6 blocks the binding of CDK4/6 to cyclin D, resulting in the dephosphorylated state of Rb. Dephosphorylated Rb does not dissociate the repressive complexes of E2F, whose protein products dictate regular and correct DNA transcription, and thus prevents the release of E2F, causing cell cycle arrest and senescence. p19ARF suppresses MDM2, which mediates the ubiquitination and degradation of p53, and can initiate related processes to promote abnormal cell cycle arrest and apoptosis during uncontrolled proliferation or irreparable damage.
Furthermore, some additional INK4a/ARF-independent mechanisms have been reported. It has been proposed that BMI1 functions synergistically with activated Ras/MAPK signaling and inhibits the TGFβ2/SMAD signaling axis to induce the occurrence of HCC.38,43 BMI1 stimulates the Wnt/β-catenin signaling axis to increase the expression levels of cyclin D1, c-myc, and E-cadherin, thereby activating HCC cell proliferation. 44 These findings unequivocally demonstrate that BMI1 mediates the occurrence of HCC by complex and diverse mechanisms. In the future, these mechanisms should be explored further to provide more ideas for the treatment of HCC.
Relationship Between BMI1 and the Progression of HCC
Metastasis and invasion are crucial hallmarks of cancer progression. BMI1 is important for the invasion and metastasis of various cancers,45,46 including HCC. 19 Cancer invasion and metastasis are complex and multi-factorial processes that involve tumor angiogenesis, extracellular matrix degradation, and the epithelial-to-mesenchymal transition (EMT). 47 Li et al have revealed that BMI1 can inhibit the PTEN/PI3K/AKT signaling pathway in HCC, ultimately stimulating metastasis by increasing the expression and activity of vascular endothelial growth factor (VEGF), matrix metalloproteinase (MMP)-9, and MMP-2. 19 PTEN that encodes the protein phosphatase phosphatase and tensin homolog is mutated or lost in various human cancers, including HCC. 48 PTEN acts as a negative regulator of the PI3K/AKT signaling pathway in cells. 49 Tumor growth and migration are inseparable from the nutritional support provided by the blood; VEGF has important roles in tumor angiogenesis and metastasis. Matrix metalloproteinases are the common extracellular matrix-degrading enzymes. The expression levels of MMP-2 and MMP-9 are markedly upregulated in later TNM stages of HCC, and there is a significant correlation between MMP-9 (but not MMP-2) and PTEN in HCC. 50 MMP-9 is upregulated via the PI3K/AKT/NF-κB signaling pathway, which promotes HCC metastasis and invasion. 51
BMI1 overexpression accelerates the progression of HCC by inducing EMT. Vimentin is beneficial for EMT, whereas E-cadherin inhibits this process. 52 Zhang et al demonstrated that high BMI1 expression decreased E-cadherin levels and increased vimentin levels in CD133+ HepG2 cells, which are regarded as representative HCC stem cells. 53 These results are consistent with those of studies on breast and colon cancer.6,45 The NF-κB signaling pathway has been identified as a key regulator of EMT in a variety of cancers.54,55 Intriguingly, Ma et al obtained similar results and found that BMI1 knockdown inhibits the metastasis of CD133+ Huh7 cells by inhibiting the NF-κB pathway. 29 In addition, Twist2, a primary helix-loop-helix transcription factor, regulates the progression of EMT in many cancers.56–58 A previous study has indicated that Twist2-expressing HepG2 cells exhibit an increase in the expression of the stem cell marker BMI1. 59 Therefore, further research is necessary to identify the links between the BMI1 proto-oncogene and EMT in HCC.
Recently, new mechanisms by which BMI1 affects the invasion and metastasis of cancers have been reported. BMI1 negatively regulates RKIP expression via miR-27a and miR-155, thus promoting tumor metastasis and chemoresistance. 10 These findings provide new directions for research on the mechanisms mediating the effect of BMI1 on HCC progression.
High BMI1 Expression Effectively Indicates Prognosis in HCC
Consistent with the association between BMI1 and prognosis in other cancers,9,60 high BMI1 levels are related to relatively poor outcomes in HCC, indicating that BMI1 is a reliable prognostic factor. 19 It is striking that BMI1 knockdown can enhance the sensitivity of HCC cells to sorafenib and 5-fluorouracil, which are promising and frequently used drugs for HCC.61,62 Wang et al found that the 5-year survival rate of patients with HCC from China with low BMI1 expression is 46%, compared with only 15% for the patients with high BMI1 expression after routine surgery without prior radiotherapy or chemotherapy. 20 There was an approximately three-fold gap in the survival rate between the two groups, which was quite substantial. Of note, there was no significant difference in the survival rate of patients with high BMI1 levels and those with low BMI1 levels in 2 years, as determined by a Kaplan–Meier curve analysis. 20 These findings suggest that BMI1 is an important biomarker for predicting the long-term survival of patients with HCC. However, Yonemitsu et al concluded that the expression level of BMI1 had no apparent effect on the survival of patients with HCC after resection in Japan, although high BMI1 protein levels were correlated with the recurrence rate after hepatectomy. 63 These inconsistent results imply that the prognostic value of BMI1 expression in HCC patients differs among regions/territories. A meta-analysis suggested that high BMI1 expression predicts an adverse overall survival rate for cancer patients in Asian populations and as a favorable predictor in Caucasian populations. 64 Another meta-analysis also suggested that high BMI1 expression is associated with lower overall survival rates in patients with colorectal cancer in Asia and is associated with higher overall survival rates in European populations. 65
Overview of HCC Therapy Targeting BMI1
The link between BMI1 and the initiation and progression of HCC is well-established. Therefore, several researchers have investigated the potential of BMI1 as a target for the treatment of HCC (Table 1).
Table 1.
The Overview of Potential Therapeutic Strategies Targeting BMI1 in HCC Treatment.
| Therapeutic strategies targeting BMI1 | Category | Reference |
|---|---|---|
| Compounds inhibiting BMI1 expression | PTC-209 | 66 |
| PTC-028 | 67 | |
| PTC-596 | 68 | |
| QW24 | 69 | |
| RU-A1 | 70 | |
| Increasing the expression of MicroRNAs downregulating BMI1 level | 1,6,7-trihydroxyxanthone(THA) upregulating miR-218 | 76 |
| Luteolin and sevoflurane upregulating miR-203 | 81 to 83 | |
| Short-chain fatty acid sodium butyrate (NaB) upregulating miR-200c | 87 | |
| Natural extracts suppressing BMI1 expression | Jiedu Xiaozheng Yin (JXY) | 88 |
| Wallichoside | 89 | |
| Combinations of chemotherapeutical agent and BMI1- siRNA | Nanoplatin and siRNA co-loaded CaP nanoparticles (NPSC) delivering cisplatin and BMI1- siRNA | 92 |
| Nanocapsules delivering BMI1- siRNA and cisplatin cationic | 93 | |
| Genome editing | Clustered regularly interspaced short palindromic repeats and CRISPR associated protein-9(CRISPR-Cas9) | 97 to 99 |
Previous studies have identified BMI1 inhibitors, including PTC-209, PTC-028, PTC-596, and QW24. PTC-209 was the first reported BMI1 inhibitor. It is a low-molecular-weight compound identified by high-throughput screening using the GEMS (Gene Expression Modulation by Small Molecules) platform. 66 Kreso et al demonstrated that PTC-209 decreases BMI1 expression at the protein level and causes the loss of tumor-initiating cells in primary colorectal cancer as well as irreversible intra-tumor injury. 66 PTC-028 decreases BMI1 function via hyper-phosphorylation and induces apoptosis in epithelial ovarian cancer cells. 67 Likewise, PTC-596, identified as a novel BMI1 inhibitor, can promote the apoptosis of acute myeloid leukemia progenitor cells. 68 Wang et al demonstrated that QW24, a small-molecule inhibitor against BMI1, could reduce the stability of the BMI1 protein and downregulate BMI1 expression via the autophagy-lysosome pathway, without affecting BMI1 mRNA levels in colorectal cancer cells. 69 Bartucci et al identified BMI1 inhibitors, named RU-A1 compounds, by medicinal chemistry and chemical engineering, which can inhibit HCC cell growth, chemosensitivity, and tumor-initiating capacity, with more effective tumor-suppressive effects than those of PTC-209. 70 These BMI1 inhibitors may be effective therapeutic agents for cancer in the future. However, BMI1 inhibitors have not been used in HCC clinical treatment to date and the efficacy of these inhibitors against HCC is not clear.
MicroRNAs are a group of endogenous noncoding single-stranded RNAs that negatively regulate the expression of target genes. 71 MiR-218 has suppressive effects in many cancers.72–74 Furthermore, accumulating evidence indicates that miR-218 negatively regulates BMI1 expression in HCC14,75,76 and is an independent prognostic factor for HCC. 77 Hence, reducing the expression of BMI1 by increasing the expression of miR-218 may be an effective treatment strategy for HCC. Fu et al found that 1,6,7-trihydroxyxanthone (THA), an active ingredient derived from Goodyera oblongifolia, could increase miR-218 expression and decrease BMI1 expression, thereby strongly inhibiting cancer cell growth and inducing abnormal cell apoptosis in HCC. 76 Similarly, some studies have demonstrated that miR-203 has suppressive effects in numerous tumors. 78 A study has shown that miR-203 levels are lower in HCC tissue samples than in normal tissues and the overexpression of miR-203 could suppress the proliferation and invasion of HCC cells. 79 Interestingly, miR-203 can enhance the radiosensitivity of HCC cells by directly targeting BMI1. 80 Therefore, miR-203 may be a valid therapeutic target for HCC, as it inhibits BMI1 expression. Recently, some progress has been made in the elucidation of miR-203-based regulation. For example, luteolin, a natural flavonoid, possesses anti-breast cancer properties, as it upregulates miR-203 expression. 81 Strikingly, sevoflurane, an inhalational anesthetic, suppresses cell proliferation in breast cancer and migration in colorectal cancer by upregulating miR-203 expression.82,83 In addition, BMI1 is a well-known target gene of the miR-200 family, which plays an essential role in cancer progression and metastasis.84,85 A recent study has proven that miR-200c suppresses the initiation of HCC, at least in part by inhibiting BMI1 expression. 86 Interestingly, the short-chain fatty acid sodium butyrate (NaB) produced by bacterial fermentation of dietary fiber in the colon limits colorectal cancer liver metastasis by downregulating BMI1 expression by enhanced miR-200c expression. 87 These results provide potential therapeutic agents for HCC.
Many studies have inspired new ideas for the development of HCC treatments targeting BMI1. Jiedu Xiaozheng Yin (JXY), a Chinese herbal decoction, inhibits HCC cell growth by suppressing BMI1 expression. 88 Kaneta et al identified that wallichoside, a methanol-soluble extract from Beaumontia murtonii and Eugenia operculate, suppressed BMI1 promoter activity and inhibited the growth of Huh7 human hepatocellular carcinoma cells. 89 Small interfering RNAs (siRNAs) can silence specific cancer genes; thus, siRNA therapeutics are very promising and are a focus of cancer research. 90 Cisplatin (cis-dichlorodiamminoplatinum [II]; CDDP) is an anticancer agent used widely in HCC treatment. 91 Li et al designed Nanoplatin and siRNA co-loaded CaP nanoparticles to deliver the chemotherapeutical agent CDDP and BMI1 siRNA to effectively and safely treat HCC; the upregulation of BMI1 expression under CDDP-exposure contributed to drug resistance in HCC cell lines. 92 Yang et al found that BMI1 siRNA and cisplatin cationic nanocapsules delivered to HCC-bearing mice had higher anti-tumor activity than that of mice treated with nanocapsules alone. 93
Genome editing is an emerging area of research because of its ability to precisely alter genomes of target cells and regulate functional genes using targeted nucleases. Clustered regularly interspaced short palindromic repeats and CRISPR associated protein-9 (CRISPR-Cas9) acts as “molecular scissors” and is quicker, more precise, and more effective than alternative genome-editing strategies. 94 CRISPR-Cas9 has also shown great potential in the treatment of cancer, including HCC.95,96 Wang et al reported that the knockout of CXC chemokine receptor 4 (CXCR4), associated with a poor prognosis in HCC, by CRISPR/Cas9 not only suppresses proliferation and invasion but also increases sensitivity to the anticancer drug cisplatin in HCC. 97 Another study has shown that the CRISPR/Cas9-mediated knockout of nuclear receptor binding SET domain-containing protein 1 inhibits the ability of HCC cells to proliferate and migrate. 98 In addition, Zhao et al applied the CRISPR/Cas9 technique to knock out BMI1 in epithelial ovarian cancer cells and found that the knockout of BMI1 could promote cell apoptosis and reduce cell growth and metastasis. 99 These results suggest that CRISPR/Cas9-mediated BMI1 knockout is feasible for the treatment of HCC, and relevant research is urgently needed.
Conclusion
BMI1 is a key gene in tumorigenesis and contributes to the development of many cancers, making it a well-established therapeutic target. In this review, we introduced the characteristics of BMI1 expression in the occurrence and development of HCC and briefly summarized the mechanisms by which BMI1 influences disease progression. Accumulating evidence shows that in some cancers, suppressing the expression and activity of BMI1 by direct inhibition or by suppressing upstream factors is a highly efficient treatment strategy. In addition, considering the biological availability of BMI1 inhibitors in clinical application, innovative drug combinations and agents have also shown promising results. Although such inhibitors are not widely used in clinical practice, there is sufficient evidence that BMI1 is an effective therapeutic target for HCC. Therefore, further studies to elucidate the basic mechanism of action of BMI1 in HCC and its clinical correlations are needed.
Abbreviations
- AFP
Alpha-fetoprotein
- BMI1
B-lymphoma Mo-MLV insertion region 1
- CDK
cyclin-dependent kinase
- CDDP
cis-dichlorodiamminoplatinum (II)
- Cps1
carbamoyl phosphate synthase I
- CRISPR-Cas9
clustered regularly interspaced short palindromic repeats and CRISPR associated protein-9
- CXCR4
CXC chemokine receptor 4
- ECM
the extracellular matrix
- EMT
epithelial-to-mesenchymal transition
- GEMS
Gene Expression Modulation by Small Molecules
- HCC
Hepatocellular carcinoma
- HPCs
hepatic progenitor cells
- JXY
Jiedu Xiaozheng Yin
- MMP
matrix metalloproteinase
- NPSC
Nanoplatin and siRNA co-loaded CaP nanoparticles
- PcG
polycomb group
- RKIP
Raf kinase inhibitor protein
- siRNAs
small interfering RNAs
- THA
1,6,7-trihydroxyxanthone
- VEGF
vascular endothelial growth factor.
Footnotes
Declaration of Conflicting Interests: The authors declared no potential conflicts of interest with respect to the research, authorship, and/or publication of this article.
Funding: The authors disclosed receipt of the following financial support for the research, authorship, and/or publication of this article: This study was sponsored by the National Natural Science Foundation of China (grant number: 82071964), Shanghai Leading Talent Program funded by Shanghai Human Resources and Social Security Bureau (grant number: 03.05.19005), Shanghai Shenkang Three-year Action Project (grant number: SHDC2020CR2054B), Shanghai Tenth People's Hospital (grant number: 2021SYPDRC065 and 04.01.20065), and Shanghai Municipal Health Commission (grant number: 202040043; GWV-10.1-XK09 and 20214Y0159).
ORCID iDs: Ru Wang https://orcid.org/0000-0002-7939-7359
Hengwei Fan https://orcid.org/0000-0002-4761-8217
References
- 1.Llovet JM, Kelley RK, Villanueva A, et al. Hepatocellular carcinoma. Nat Rev Dis Primers. 2021;7(1):6. doi: 10.1038/s41572-020-00240-3 [DOI] [PubMed] [Google Scholar]
- 2.European Association for the Study of the Liver. EASL Clinical practice guidelines: management of hepatocellular carcinoma. J Hepatol. 2018;69(1):182‐236. doi: 10.1016/j.jhep.2018.03.019 [DOI] [PubMed] [Google Scholar]
- 3.Simon JA, Kingston RE. Mechanisms of polycomb gene silencing: knowns and unknowns. Nat Rev Mol Cell Biol. 2009;10(10):697‐708. doi: 10.1038/nrm2763 [DOI] [PubMed] [Google Scholar]
- 4.van Lohuizen M, Verbeek S, Scheijen B, et al. Identification of cooperating oncogenes in E mu-myc transgenic mice by provirus tagging. Cell. 1991;65(5):737‐752. doi: 10.1016/0092-8674(91)90382-9 [DOI] [PubMed] [Google Scholar]
- 5.Haupt Y, Alexander WS, Barri G, et al. Novel zinc finger gene implicated as myc collaborator by retrovirally accelerated lymphomagenesis in E mu-myc transgenic mice. Cell. 1991;65(5):753‐763. doi: 10.1016/0092-8674(91)90383-a [DOI] [PubMed] [Google Scholar]
- 6.Guo BH, Feng Y, Zhang R, et al. Bmi-1 promotes invasion and metastasis, and its elevated expression is correlated with an advanced stage of breast cancer. Mol Cancer. 2011;10(1):10. doi: 10.1186/1476-4598-10-10 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Li DW, Tang HM, Fan JW, et al. Expression level of Bmi-1 oncoprotein is associated with progression and prognosis in colon cancer. J Cancer Res Clin Oncol. 2010;136(7):997‐1006. doi: 10.1007/s00432-009-0745-7 [DOI] [PubMed] [Google Scholar]
- 8.Kang MK, Kim RH, Kim SJ, et al. Elevated Bmi-1 expression is associated with dysplastic cell transformation during oral carcinogenesis and is required for cancer cell replication and survival. Br J Cancer. 2007;96(1):126‐133. doi: 10.1038/sj.bjc.6603529 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Yu J, Chen L, Bao Z, et al. BMI-1 promotes invasion and metastasis in endometrial adenocarcinoma and is a poor prognostic factor. Oncol Rep. 2020;43(5):1630‐1640. doi: 10.3892/or.2020.7539 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Li Y, Tian Z, Tan Y, et al. Bmi-1-induced miR-27a and miR-155 promote tumor metastasis and chemoresistance by targeting RKIP in gastric cancer. Mol Cancer. 2020;19(1):109. doi: 10.1186/s12943-020-01229-y [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Neo SY, Leow CK, Vega VB, et al. Identification of discriminators of hepatoma by gene expression profiling using a minimal dataset approach. Hepatology. 2004;39(4):944‐953. doi: 10.1002/hep.20105 [DOI] [PubMed] [Google Scholar]
- 12.Yu J, Hu M, Chen W, et al. Expression of B cell-specific moloney murine leukemia virus integration site 1 (BMI-1) and WW domain-containing oxidoreductase (WWOX) in liver cancer tissue and normal liver tissue. Med Sci Monit. 2018;9(24):6673‐6679. doi: 10.12659/msm.909675 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Ma S, Chan KW, Hu L, et al. Identification and characterization of tumorigenic liver cancer stem/progenitor cells. Gastroenterology. 2007;132(7):2542‐2556. doi: 10.1053/j.gastro.2007.04.025 [DOI] [PubMed] [Google Scholar]
- 14.Tu K, Li C, Zheng X, et al. Prognostic significance of miR-218 in human hepatocellular carcinoma and its role in cell growth. Oncol Rep. 2014;32(4):1571‐1577. doi: 10.3892/or.2014.3386 [DOI] [PubMed] [Google Scholar]
- 15.Breuer RH, Snijders PJ, Smit EF, et al. Increased expression of the EZH2 polycomb group gene in BMI-1-positive neoplastic cells during bronchial carcinogenesis. Neoplasia. 2004;6(6):736‐743. doi: 10.1593/neo.04160 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Tateishi K, Ohta M, Kanai F, et al. Dysregulated expression of stem cell factor Bmi1 in precancerous lesions of the gastrointestinal tract. Clin Cancer Res. 2006;12(23):6960‐6966. doi: 10.1158/1078-0432.Ccr-06-0449 [DOI] [PubMed] [Google Scholar]
- 17.Sasaki M, Ikeda H, Itatsu K, et al. The overexpression of polycomb group proteins Bmi1 and EZH2 is associated with the progression and aggressive biological behavior of hepatocellular carcinoma. Lab Invest. 2008;88(8):873‐882. doi: 10.1038/labinvest.2008.52 [DOI] [PubMed] [Google Scholar]
- 18.Effendi K, Mori T, Komuta M, et al. Bmi-1 gene is upregulated in early-stage hepatocellular carcinoma and correlates with ATP-binding cassette transporter B1 expression. Cancer Sci. 2010;101(3):666‐672. doi: 10.1111/j.1349-7006.2009.01431.x [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Li X, Yang Z, Song W, et al. Overexpression of Bmi-1 contributes to the invasion and metastasis of hepatocellular carcinoma by increasing the expression of matrix metalloproteinase (MMP)-2, MMP-9 and vascular endothelial growth factor via the PTEN/PI3 K/Akt pathway. Int J Oncol. 2013;43(3):793‐802. doi: 10.3892/ijo.2013.1992 [DOI] [PubMed] [Google Scholar]
- 20.Wang H, Pan K, Zhang HK, et al. Increased polycomb-group oncogene Bmi-1 expression correlates with poor prognosis in hepatocellular carcinoma. J Cancer Res Clin Oncol. 2008;134(5):535‐541. doi: 10.1007/s00432-007-0316-8 [DOI] [PubMed] [Google Scholar]
- 21.Wicha MS, Liu S, Dontu G. Cancer stem cells: an old idea--a paradigm shift. Cancer Res. 2006;66(4):1883‐1890; discussion 1895-6. doi: 10.1158/0008-5472.Can-05-3153 [DOI] [PubMed] [Google Scholar]
- 22.Park IK, Qian D, Kiel M, et al. Bmi-1 is required for maintenance of adult self-renewing haematopoietic stem cells. Nature. 2003;423(6937):302‐305. doi: 10.1038/nature01587 [DOI] [PubMed] [Google Scholar]
- 23.Lessard J, Sauvageau G. Bmi-1 determines the proliferative capacity of normal and leukaemic stem cells. Nature. 2003;423(6937):255‐260. doi: 10.1038/nature01572 [DOI] [PubMed] [Google Scholar]
- 24.Fan L, Xu C, Wang C, et al. Bmi1 is required for hepatic progenitor cell expansion and liver tumor development. PLoS One. 2012;7(9):e46472. doi: 10.1371/journal.pone.0046472 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Zhang R, Wu WR, Shi XD, et al. Dysregulation of Bmi1 promotes malignant transformation of hepatic progenitor cells. Oncogenesis. 2016;5(2):e203. doi: 10.1038/oncsis.2016.6 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Chiba T, Kita K, Zheng YW, et al. Side population purified from hepatocellular carcinoma cells harbors cancer stem cell-like properties. Hepatology. 2006;44(1):240‐251. doi: 10.1002/hep.21227 [DOI] [PubMed] [Google Scholar]
- 27.Chiba T, Miyagi S, Saraya A, et al. The polycomb gene product BMI1 contributes to the maintenance of tumor-initiating side population cells in hepatocellular carcinoma. Cancer Res. 2008;68(19):7742‐7749. doi: 10.1158/0008-5472.Can-07-5882 [DOI] [PubMed] [Google Scholar]
- 28.Chiba T, Zheng YW, Kita K, et al. Enhanced self-ren-ewal capability in hepatic stem/progenitor cells drives cancer initiation. Gastroenterology. 2007;133(3):937‐950. doi: 10.1053/j.gastro.2007.06.016 [DOI] [PubMed] [Google Scholar]
- 29.Ma DQ, Zhang YH, Ding DP, et al. Effect of Bmi-1-mediated NF-κB signaling pathway on the stem-like properties of CD133+ human liver cancer cells. Cancer Biomark. 2018;22(3):575‐585. doi: 10.3233/cbm-181329 [DOI] [PubMed] [Google Scholar]
- 30.Sparmann A, van Lohuizen M. Polycomb silencers control cell fate, development and cancer. Nat Rev Cancer. 2006;6(11):846‐856. doi: 10.1038/nrc1991 [DOI] [PubMed] [Google Scholar]
- 31.Park IK, Morrison SJ, Clarke MF. Bmi1, stem cells, and senescence regulation. J Clin Invest. 2004;113(2):175‐179. doi: 10.1172/jci20800 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Jacobs JJ, Kieboom K, Marino S, et al. The oncogene and polycomb-group gene bmi-1 regulates cell proliferation and senescence through the ink4a locus. Nature. 1999;397(6715):164‐168. doi: 10.1038/16476 [DOI] [PubMed] [Google Scholar]
- 33.Kent LN, Leone G. The broken cycle: e2F dysfunction in cancer. Nat Rev Cancer. 2019;19(6):326‐338. doi: 10.1038/s41568-019-0143-7 [DOI] [PubMed] [Google Scholar]
- 34.Gil J, Peters G. Regulation of the INK4b-ARF-INK4a tumour suppressor locus: all for one or one for all. Nat Rev Mol Cell Biol. 2006;7(9):667‐677. doi: 10.1038/nrm1987 [DOI] [PubMed] [Google Scholar]
- 35.Kanapathipillai M. Treating p53 mutant aggregation-associated cancer. Cancers (Basel). 2018;10(6):154. doi: 10.3390/cancers10060154 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Fasano CA, Dimos JT, Ivanova NB, et al. shRNA knockdown of Bmi-1 reveals a critical role for p21-Rb pathway in NSC self-renewal during development. Cell Stem Cell. 2007;1(1):87‐99. doi: 10.1016/j.stem.2007.04.001 [DOI] [PubMed] [Google Scholar]
- 37.Bruggeman SW, Hulsman D, Tanger E, et al. Bmi1 co-ntrols tumor development in an Ink4a/Arf-independent manner in a mouse model for gliom-a. Cancer Cell. 2007;12(4):328‐341. doi: 10.1016/j.ccr.2007.08.032 [DOI] [PubMed] [Google Scholar]
- 38.Xu CR, Lee S, Ho C, et al. Bmi1 functions as an oncogene independent of Ink4A/Arf repression in hepatic carcinogenesis. Mol Cancer Res. 2009;7(12):1937‐1945. doi: 10.1158/1541-7786.Mcr-09-0333 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Chiba T, Seki A, Aoki R, et al. Bmi1 promotes hepatic stem cell expansion and tumorigenicity in both Ink4a/Arf-dependent and -independent manners in mice. Hepatology. 2010;52(3):1111‐1123. doi: 10.1002/hep.23793 [DOI] [PubMed] [Google Scholar]
- 40.Lu SC, Alvarez L, Huang ZZ, et al. Methionine adenosyltransferase 1A knockout mice are predisposed to liver injury and exhibit increased expression of genes involved in proliferation. Proc Natl Acad Sci U S A. 2001;98(10):5560‐5565. doi: 10.1073/pnas.091016398 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Cancer Genome Atlas Research Network. Comprehensive and integrative genomic characterization of hepatocellular carcinoma. Cell. 2017;169(7):1327‐41.e23. doi: 10.1016/j.cell.2017.05.046 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Fu WM, Zhu X, Wang WM, et al. Hotair mediates hepatocarcinogenesis through suppressing miRNA-218 expression and activating P14 and P16 signaling. J Hepatol. 2015;63(4):886‐895. doi: 10.1016/j.jhep.2015.05.016 [DOI] [PubMed] [Google Scholar]
- 43.Li B, Chen Y, Wang F, et al. Bmi1 drives hepatocarcinogenesis by repressing the TGFβ2/SMAD signalling axis. Oncogene. 2020;39(5):1063‐1079. doi: 10.1038/s41388-019-1043-8 [DOI] [PubMed] [Google Scholar]
- 44.Chen MH, Fu LS, Zhang F, et al. LncAY controls BMI1 expression and activates BMI1/Wnt/β-catenin signaling axis in hepatocellular carcinoma. Life Sci. 2021;9(280):119748. doi: 10.1016/j.lfs.2021.119748 [DOI] [PubMed] [Google Scholar]
- 45.Zhang Z, Bu X, Chen H, et al. Bmi-1 promotes the invasion and migration of colon cancer stem cells through the downregulation of E-cadherin. Int J Mol Med. 2016;38(4):1199‐1207. doi: 10.3892/ijmm.2016.2730 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Gao FL, Li WS, Liu CL, et al. Silencing Bmi-1 enhances the senescence and decreases the metastasis of human gastric cancer cells. World J Gastroenterol. 2013;19(46):8764‐8769. doi: 10.3748/wjg.v19.i46.8764 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Su Z, Yang Z, Xu Y, et al. Apoptosis, autophagy, necroptosis, and cancer metastasis. Mol Cancer. 2015;2(14):48. doi: 10.1186/s12943-015-0321-5 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Chalhoub N, Baker SJ. PTEN And the PI3-kinase pathway in cancer. Annu Rev Pathol. 2009;7(4):127‐150. doi: 10.1146/annurev.pathol.4.110807.092311 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Stambolic V, Suzuki A, de la Pompa JL, et al. Negative regulation of PKB/Akt-dependent cell survival by the tumor suppressor PTEN. Cell. 1998;95(1):29‐39. doi: 10.1016/s0092-8674(00)81780-8 [DOI] [PubMed] [Google Scholar]
- 50.Chen JS, Wang Q, Fu XH, et al. Involvement of PI3 K/PTEN/AKT/mTOR pathway in invasion and metastasis in hepatocellular carcinoma: association with MMP-9. Hepatol Res. 2009;39(2):177‐186. doi: 10.1111/j.1872-034X.2008.00449.x [DOI] [PubMed] [Google Scholar]
- 51.Cheng JC, Chou CH, Kuo ML, et al. Radiation-enhanced hepatocellular carcinoma cell invasion with MMP-9 expression through PI3 K/Akt/NF-kappaB signal transduction pathway. Oncogene. 2006;25(53):7009‐7018. doi: 10.1038/sj.onc.1209706 [DOI] [PubMed] [Google Scholar]
- 52.Onder TT, Gupta PB, Mani SA, et al. Loss of E-cadherin promotes metastasis via multiple downstream transcriptional pathways. Cancer Res. 2008;68(10):3645‐3654. doi: 10.1158/0008-5472.Can-07-2938 [DOI] [PubMed] [Google Scholar]
- 53.Zhang Z, Wang Q, Bu X, et al. Overexpression of Bmi-1 promotes epithelial-mesenchymal transition in CD133 + Hep G2 cells. Mol Med Rep. 2017;16(5):6156‐6161. doi: 10.3892/mmr.2017.7347 [DOI] [PubMed] [Google Scholar]
- 54.Lin C, Zong J, Lin W, et al. EBV-miR-BART8-3p induces epithelial-mesenchymal transition and promotes metastasis of nasopharyngeal carcinoma cells through activating NF-κB and Erk1/2 pathways. J Exp Clin Cancer Res. 2018;37(1):283. doi: 10.1186/s13046-018-0953-6 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Hseu YC, Lin YC, Rajendran P, et al. Antrodia salmonea suppresses invasion and metastasis in triple-negative breast cancer cells by reversing EMT through the NF-κB and Wnt/β-catenin signaling pathway. Food Chem Toxicol. 2019;2(124):219‐230. doi: 10.1016/j.fct.2018.12.009 [DOI] [PubMed] [Google Scholar]
- 56.Fang X, Cai Y, Liu J, et al. Twist2 contributes to breast cancer progression by promoting an epithelial-mesenchymal transition and cancer stem-like cell self-renewal. Oncogene. 2011;30(47):4707‐4720. doi: 10.1038/onc.2011.181 [DOI] [PubMed] [Google Scholar]
- 57.Li Y, Wang W, Wang W, et al. Correlation of TWIST2 up-regulation and epithelial-mesenchymal transition during tumorigenesis and progression of cervical carcinoma. Gynecol Oncol. 2012;124(1):112‐118. doi: 10.1016/j.ygyno.2011.09.003 [DOI] [PubMed] [Google Scholar]
- 58.Yang J, Zhang X, Zhang Y, et al. HIF-2α promotes epithelial-mesenchymal transition through regulating Twist2 binding to the promoter of E-cadherin in pancreatic cancer. J Exp Clin Cancer Res. 2016;2(35):26. doi: 10.1186/s13046-016-0298-y [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Liu AY, Cai Y, Mao Y, et al. Twist2 promotes self-renewal of liver cancer stem-like cells by regulating CD24. Carcinogenesis. 2014;35(3):537‐545. doi: 10.1093/carcin/bgt364 [DOI] [PubMed] [Google Scholar]
- 60.Peng HX, Liu XD, Luo ZY, et al. Upregulation of the proto-oncogene Bmi-1 predicts a poor prognosis in pediatric acute lymphoblastic leukemia. BMC Cancer. 2017;17(1):76. doi: 10.1186/s12885-017-3049-3 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Ruan ZP, Xu R, Lv Y, et al. Bmi1 knockdown inhibits hepatocarcinogenesis. Int J Oncol. 2013;42(1):261‐268. doi: 10.3892/ijo.2012.1693 [DOI] [PubMed] [Google Scholar]
- 62.Zhang R, Xu LB, Yue XJ, et al. Bmi1 gene silencing inhibits the proliferation and invasiveness of human hepatocellular carcinoma cells and increases their sensitivity to 5-fluorouracil. Oncol Rep. 2013;29(3):967‐974. doi: 10.3892/or.2012.2189 [DOI] [PubMed] [Google Scholar]
- 63.Yonemitsu Y, Imazeki F, Chiba T, et al. Distinct expression of polycomb group proteins EZH2 and BMI1 in hepatocellular carcinoma. Hum Pathol. 2009;40(9):1304‐1311. doi: 10.1016/j.humpath.2009.01.017 [DOI] [PubMed] [Google Scholar]
- 64.Shao Y, Geng Y, Gu W, et al. Prognostic role of high Bmi-1 expression in asian and caucasian patients with solid tumors: a meta-analysis. Biomed Pharmacother. 2014;68(8):969‐977. doi: 10.1016/j.biopha.2014.10.017 [DOI] [PubMed] [Google Scholar]
- 65.Pourjafar M, Samadi P, Karami M, et al. Assessment of clinicopathological and progn-ostic relevance of BMI-1 in patients with colorectal cancer: a meta-analysis. Biotechnol Appl Biochem. 2021,68(6):1313-1322. doi: 10.1002/bab.2053 [DOI] [PubMed] [Google Scholar]
- 66.Kreso A, van Galen P, Pedley NM, et al. Self-renewal as a therapeutic target in human colorectal cancer. Nat Med. 2014;20(1):29‐36. doi: 10.1038/nm.3418 [DOI] [PubMed] [Google Scholar]
- 67.Dey A, Xiong X, Crim A, et al. Evaluating the mechanism and therapeutic potential of PTC-028, a novel inhibitor of BMI-1 function in ovarian cancer. Mol Cancer Ther. 2018;17(1):39‐49. doi: 10.1158/1535-7163.Mct-17-0574 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Nishida Y, Maeda A, Kim MJ, et al. The novel BMI-1 inhibitor PTC596 downregulates MCL-1 and induces p53-independent mitochondrial apoptosis in acute myeloid leukemia progenitor cells. Blood Cancer J. 2017;7(2):e527. doi: 10.1038/bcj.2017.8 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Wang J, Xing Y, Wang Y, et al. A novel BMI-1 inhibitor QW24 for the treatment of stem-like colorectal cancer. J Exp Clin Cancer Res. 2019;38(1):422. doi: 10.1186/s13046-019-1392-8 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Bartucci M, Hussein MS, Huselid E, et al. Synthesis and characterization of novel BMI1 inhibitors targeting cellular self-renewal in hepatocellular carcinoma. Target Oncol. 2017;12(4):449‐462. doi: 10.1007/s11523-017-0501-x [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Ambros V. The functions of animal microRNAs. Nature. 2004;431(7006):350‐355. doi: 10.1038/nature02871 [DOI] [PubMed] [Google Scholar]
- 72.Yang Y, Ding L, Hu Q, et al. MicroRNA-218 functions as a tumor suppressor in lung cancer by targeting IL-6/STAT3 and negatively correlates with poor prognosis. Mol Cancer. 2017;16(1):141. doi: 10.1186/s12943-017-0710-z [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Liu Z, Mao L, Wang L, et al. Mir-218 functions as a tumor suppressor gene in cervical cancer. Mol Med Rep. 2020;21(1):209‐219. doi: 10.3892/mmr.2019.10809 [DOI] [PMC free article] [PubMed] [Google Scholar] [Retracted]
- 74.Wang T, Xu L, Jia R, et al. MiR-218 suppresses the metastasis and EMT of HCC cells via targeting SERBP1. Acta Biochim Biophys Sin (Shanghai). 2017;49(5):383‐391. doi: 10.1093/abbs/gmx017 [DOI] [PubMed] [Google Scholar]
- 75.Wu J, Jiang ZM, Xie Y, et al. miR-218 suppresses the growth of hepatocellular carcinoma by inhibiting the expression of proto-oncogene Bmi-1. J Buon. 2018;23(3):604‐610. [PubMed] [Google Scholar]
- 76.Fu WM, Tang LP, Zhu X, et al. MiR-218–targeting-Bmi-1 mediates the suppressive effect of 1,6,7-trihydroxyxanthone on liver cancer cells. Apoptosis. 2015;20(1):75‐82. doi: 10.1007/s10495-014-1047-3 [DOI] [PubMed] [Google Scholar]
- 77.Tu K, Li C, Zheng X, et al. Prognostic significance of miR-218 in human hepatocellular carcinoma and its role in cell growth. Oncol Rep. 2014;32(4):1571‐1577. doi: 10.3892/or.2014.3386 [DOI] [PubMed] [Google Scholar]
- 78.Du SL, Xu LY, Gao P, et al. MiR-203 regulates DJ-1 expression and affects proliferation, apoptosis and DDP resistance of pancreatic cancer cells. Eur Rev Med Pharmacol Sci. 2019;23(20):8833‐8840. doi: 10.26355/eurrev_201910_19278 [DOI] [PubMed] [Google Scholar]
- 79.Yang F, Lv LZ, Cai QC, et al. Potential roles of EZH2, Bmi-1 and miR-203 in cell proliferation and invasion in hepatocellular carcinoma cell line Hep3B. World J Gastroenterol. 2015;21(47):13268‐13276. doi: 10.3748/wjg.v21.i47.13268 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80.Shao Y, Zhang D, Li X, et al. MicroRNA-203 increases cell radiosensitivity via directly targeting Bmi-1 in hepatocellular carcinoma. Mol Pharm. 2018;15(8):3205‐3215. doi: 10.1021/acs.molpharmaceut.8b00302 [DOI] [PubMed] [Google Scholar]
- 81.Gao G, Ge R, Li Y, et al. Luteolin exhibits anti-breast cancer property through up-regulating miR-203. Artif Cells Nanomed Biotechnol. 2019;47(1):3265‐3271. doi: 10.1080/21691401.2019.1646749 [DOI] [PubMed] [Google Scholar]
- 82.Liu J, Yang L, Guo X, et al. Sevoflurane suppresses proliferation by upregulating microRNA-203 in breast cancer cells. Mol Med Rep. 2018;18(1):455‐460. doi: 10.3892/mmr.2018.8949 [DOI] [PubMed] [Google Scholar]
- 83.Fan L, Wu Y, Wang J, et al. Sevoflurane inhibits the migration and invasion of colorectal cancer cells through regulating ERK/MMP-9 pathway by up-regulating miR-203. Eur J Pharmacol. 2019;5(850):43‐52. doi: 10.1016/j.ejphar.2019.01.025 [DOI] [PubMed] [Google Scholar]
- 84.Liu Y, Sánchez-Tilló E, Lu X, et al. The ZEB1 transcription factor acts in a negative feedback loop with miR200 downstream of Ras and Rb1 to regulate Bmi1 expression. J Biol Chem. 2014;289(7):4116‐4125. doi: 10.1074/jbc.M113.533505 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 85.Xu Z, Tao J, Chen P, et al. Sodium butyrate inhibits colorectal cancer cell migration by downregulating Bmi-1 through enhanced miR-200c expression. Mol Nutr Food Res. 2018;62(6):e1700844. doi: 10.1002/mnfr.201700844 [DOI] [PubMed] [Google Scholar]
- 86.Xu L, Lin J, Deng W, et al. EZH2 Facilitates BMI1-dependent hepatocarcinogenesis through epigenetically silencing microRNA-200c. Oncogenesis. 2020;9(11):101. doi: 10.1038/s41389-020-00284-w [DOI] [PMC free article] [PubMed] [Google Scholar]
- 87.Xu Z, Zhou Z, Zhang J, et al. Targeting BMI-1-mediated epithelial-mesenchymal transition to inhibit colorectal cancer liver metastasis. Acta Pharm Sin B. 2021;11(5):1274‐1285. doi: 10.1016/j.apsb.2020.11.018 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 88.Chen XZ, Cao ZY, Li JN, et al. Ethyl acetate extract from jiedu Xiaozheng Yin inhibits the proliferation of human hepatocellular carcinoma cells by suppressing polycomb gene product Bmi1 and Wnt/β-catenin signaling. Oncol Rep. 2014;32(6):2710‐2718. doi: 10.3892/or.2014.3541 [DOI] [PubMed] [Google Scholar]
- 89.Kaneta Y, Arai MA, Ishikawa N, et al. Identification of BMI1 promoter inhibitors from Beaumontia murtonii and Eugenia operculata. J Nat Prod. 2017;80(6):1853‐1859. doi: 10.1021/acs.jnatprod.7b00138 [DOI] [PubMed] [Google Scholar]
- 90.Subhan MA, Torchilin VP. Efficient nanocarriers of siRNA therapeutics for cancer treatment. Transl Res. 2019;12(214):62‐91. doi: 10.1016/j.trsl.2019.07.006 [DOI] [PubMed] [Google Scholar]
- 91.Lee JE, Bae SH, Choi JY, et al. Epirubicin, cisplatin, 5-FU comb-ination chemotherapy in sorafenib-refractory metastatic hepatocellular carcinoma. World J Gastroenterol. 2014;20(1):235‐241. doi: 10.3748/wjg.v20.i1.235 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 92.Li M, Zhao P, Fan T, et al. Biocompatible co-loading vehicles for delivering both nanoplatin cores and siRNA to treat hepatocellular carcinoma. Int J Pharm. 2019;12(572):118769. doi: 10.1016/j.ijpharm.2019.118769 [DOI] [PubMed] [Google Scholar]
- 93.Yang T, Chen Y, Zhao P, et al. Enhancing the therapeutic effect via elimination of hepatocellular carcinoma stem cells using Bmi1 siRNA delivered by cationic cisplatin nanocapsules. Nanomedicine. 2018;14(7):2009‐2021. doi: 10.1016/j.nano.2018.05.012 [DOI] [PubMed] [Google Scholar]
- 94.Lino CA, Harper JC, Carney JP, et al. Delivering CRISPR: a review of the challenges and approaches. Drug Deliv. 2018;25(1):1234‐1257. doi: 10.1080/10717544.2018.1474964 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 95.Daisy PS, Shreyas KS, Anitha TS. Will CRISPR-Cas9 have cards to play against cancer? An update on its applications. Mol Biotechnol. 2021;63(2):93‐108. doi: 10.1007/s12033-020-00289-1 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 96.Wu X, Ma W, Mei C, et al. Description of CRISPR/Cas9 development and its prospect in hepatocellular carcinoma treatment. J Exp Clin Cancer Res. 2020;39(1):97. doi: 10.1186/s13046-020-01603-0 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 97.Wang X, Zhang W, Ding Y, et al. CRISPR/Cas9-mediated genome engineering of CXCR4 decreases the malignancy of hepatocellular carcinoma cells in vitro and in vivo. Oncol Rep. 2017;37(6):3565‐3571. doi: 10.3892/or.2017.5601 [DOI] [PubMed] [Google Scholar]
- 98.Zhang S, Zhang F, Chen Q, et al. CRISPR/Cas9-mediated knockout of NSD1 suppresses the hepatocellular carcinoma development via the NSD1/H3/Wnt10b signaling pathway. J Exp Clin Cancer Res. 2019;38(1):467. doi: 10.1186/s13046-019-1462-y [DOI] [PMC free article] [PubMed] [Google Scholar]
- 99.Zhao Q, Qian Q, Cao D, et al. Role of BMI1 in epithelial ovarian cancer: investigated via the CRISPR/Cas9 system and RNA sequencing. J Ovarian Res. 2018;11(1):31. doi: 10.1186/s13048-018-0406-z [DOI] [PMC free article] [PubMed] [Google Scholar]