Abstract
Background
Natural phenolic compounds and Phenolics-rich medicinal plants are also of great interest in the management of diabetes. The current study was aimed to analyze phenolics in P. hydropiepr L extracts via HPLC-DAD analysis and assess their anti-diabetic potentials using in-vitro and in-silico approaches.
Methods
Plant crude methanolic extract (Ph.Cme) was evaluated for the presence of phenolic compounds using HPLC-DAD analysis. Subsequently, samples including crude (Ph.Cr), hexane (Ph.Hex), chloroform (Ph.Chf), ethyl acetate (Ph.EtAc), butanol (Ph.Bt), aqueous (Ph.Aq) and saponins (Ph.Sp) were tested for α-glucsidase and α-amylase inhibitory potentials and identified compounds were docked against these target enzymes using Molecular Operating Environment (MOE) software. Fractions were also analyzed for the nutritional contents and acute toxicity was performed in animals.
Results
In HPLC-DAD analysis of Ph.Cme, 24 compounds were indentfied and quantified. Among these, Kaemferol-3-(p-coumaroyl-diglucoside)-7-glucoside (275.4 mg g− 1), p-Coumaroylhexose-4-hexoside (96.5 mg g− 1), Quercetin-3-glucoronide (76.0 mg g− 1), 4-Caffeoylquinic acid (58.1 mg g− 1), Quercetin (57.9 mg g− 1), 5,7,3′-Trihydroxy-3,6,4′,5′-tetramethoxyflavone (55.5 mg g− 1), 5-Feruloylquinic acid (45.8 mg g− 1), Cyanidin-3-glucoside (26.8 mg g− 1), Delphinidin-3-glucoside (24 mg g− 1), Quercetin-3-hexoside (20.7 mg g− 1) were highly abundant compounds. In α-glucosidase inhibition assay, Ph.Sp were most effective with IC50 value of 100 μg mL-1. Likewise in α-amylase inhibition assay, Ph.Chf, Ph.Sp and Ph.Cme were most potent fractions displayed IC50 values of 90, 100 and 200 μg mL-1 respectively. Docking with the α-glucosidase enzyme revealed top ranked conformations for majority of the compounds with Kaemferol-3-(p-coumaroyl-diglucoside)-7-glucoside as the most active compound with docking score of − 19.80899, forming 14 hydrogen bonds, two pi-H and two pi-pi linkages with the Tyr 71, Phe 158, Phe 177, Gln 181, Arg 212, Asp 214, Glu 276, Phe 300, Val 303, Tyr 344, Asp 349, Gln 350, Arg 439, and Asp 408 residues of the enzyme. Likewise, docking with α-amylase revealed that most of the compounds are well accommodated in the active site residues (Trp 59, Tyr 62, Thr 163, Leu 165, Arg 195, Asp 197, Glu 240, Asp 300, His 305, Asp 356) of the enzyme and Cyanidin-3-rutinoside displayed most active compound with docking score of − 15.03757.
Conclusions
Phytochemical studies revealed the presence of highly valuable phenolic compounds, which might be responsible for the anti-diabetic potentials of the plant samples.
Keywords: HPLC-DAD analysis, Phenolics, P. hydropiper, Diabetes, Saponins, Molecular docking
Background
Diabetes mellitus (DM) is a chronic metabolic disorder of glucose processing and characterized by hyperglycemia. DM occur as a result of some abnormalities in insulin production, secretion or its action, dysfunction in carbohydrate, protein and fat metabolism and other complications [1, 2]. This state of hyperglycemia produces classical symptoms of polyuria, polydipsia and polyphagia [3]. Globally, it has been estimated that the occurrence of diabetes has increased, from 4% in 1995 to 5.4% by the year 2025 [4]. About 450 million peoples have been effected by DM worldwide and its prevalence is expected to increase 690 million by 2044 [5]. Diabetes is one of the most challenging serious metabolic disorder and is the leading cause of death worldwide. Long term high level of glucose can result in number of acute or chronic complications [6], and failure of various organs such as eyes, kidneys, liver, nerves, heart, and blood vessels [7]. Type 1 and Type 2 are two prominent types of DM [8]. Type-1 diabetes is associated with auto-immune destruction of pancreatic β-cells and characterized by absolute deficiency of insulin secretion [9]. Whereas, Type-2 diabetes accounts for 90% of cases and is caused by resistance of tissues to insulin action and decrease insulin secretion [10]. Type 2 diabetes can be prevented by managing obesity, diet control and with anti-diabetic drugs [11]. Regarding drug development against type-2 diabetes, one of the most important strategy is inhibition of enzymes implicated in glucose absorption from gastrointestinal tract. For instance, α-amylase and α-glucosidase enzymes are responsible for the breakdown of starch and oligosaccharides to glucose and their inhibition play a significant role to decrease the absorption of glucose in the intestine [12]. Consequently, inhibitors of these enzymes are the potential targets in the development of anti-diabetic drugs.
Since ancient times, medicinal plants and natural products have been employed as sources of medicine for the treatment of diabetes and alleviating human suffering mostly in developing countries [13–15]. More than 400 traditional plants have been reported for DM treatment, but only few of these have received scientific and medical evaluation to assess their efficacy [16]. Natural products such as galegine, andrographolide, and acarbose are used for type-2 diabetes treatment. Plant containing polyphenols have been reported to inhibit α–amylase and α–glucosidase enzymes associated with type 2 diabetes and to exhibit insulin like activities in the utilization of glucose [17]. Phenolic phytochemicals are secondary metabolites of plant origin, possess preventive management of various chronic diseases linked with oxidation such as diabetes and cardiovascular disease [18]. Large number of α-amylase and α-glucosidase inhibitors are produced by different microorganisms and plants to regulate the activities of these enzymes [19]. α-amylase inhibitors decrease the hyperglycemia that usually occur after eating meal by reducing the speed of starch conversion into glucose. Hence low alpha amylase level is needed in diabetic patients for keeping their sugar level under control.
Family Polygonaceae also known as knotwood or smartweed family, consist of 59 genera and 1300 species which are distributed worldwide [20]. Polygonum, Persicaria, Coccoloba, Calligonum, Rumex and Rheum are the largest genera of Polygonaceae family. Traditionally numerous species of this family are used in folk medicine and as vegetables [21]. The Persicaria genus having 100 species, is found throughout the world, plays a vital role as alternative medicines. Persicaria hydropiper L. also known as water-pepper, belonging to Polygonaceae family, that can be search out in South East Asia. The medicinal uses of P. hydropiper has been reported in epilepsy, inflammation, edema, rheumatoid arthritis, joint pain, headache, colic pain, fever and other infectious diseases. It can also be used as diuretic, central nervous system (CNS) stimulant, anthelmintic and in the treatment of hypertension, hemorrhoids, kidney diseases, diarrhea, bleeding, parasitic worms, piles and angina [22]. We reported the plant and some bioactive metabolites for neuroprotective [23–26], gastroprotective [27], antimicrobial [28] and cytotoxic potentials [29–31]. P. hydropiper contains flavonoids, chalcone derivatives, phenylpropanoid derivatives, phenolic compounds, antraquinon, isocumarine, terpenoids and steroids [20]. Among the phenolic compounds in the ethanolic extract of P. hydropiper, rutin has been reported for its anti-diabetic, antioxidant and anti-inflammation activity [32]. Apart from this, the anti-diabetic potential of the ethanolic extract of P. hydropiper leaves has also been reported in mice during oral glucose tolerance tests [33]. The current project was aimed to investigate the plant for detailed phenolic composition via HPLC-DAD analysis, evaluate its in-vitro and in-silico anti-diabetic as well as nutritional potentials.
Materials and methods
Plant material, extraction and fractionation
Several species being reported for efficacy in diabetes, the current plant Persicaria hydropiper (L.) Delarbre, F. Polygonaceae was selected for the study and was collected in consultation with botanical taxonomist (Dr. Gul Rahim) from a marshy area of Talash Dir Pakistan during the month of July, 2013. Whole study protocol on the selected plant complies with institutional, national and international guidelines for the use of plants. After identification by the taxonomist, a dried sample was deposited at the herbarium of University of Malakand, Chakdara (Dir), Pakistan with voucher (H.UOM.BG.107). After collection, plant was properly cleansed with distilled water and subjected to shade drying for about 30 days. Subsequently, the dried pant material was coarsely crushed with a cutter machine and resulted powder (4.5 kg) was transferred to stainless steel container and 22 L of 80% methanol was added for crude extraction purpose. Powder material was kept for about 15 days in the solvent with occasional shaking to fully remove any soluble constituents. Thereafter, solvent was removed, filtered and evaporated via a rotary evaporator (Heidolph Laborota 4000, Schwabach, Germany) [34]. Finally, we got about 290 g (6.44%) of crude methanolic extact (Ph.Cme). To get further sub-fractions, 250 g Ph.Cme was suspended in 500 ml of distilled water in a separating funnel and gradually washed with solvents (polarity directed) including Ph.Hex (3 × 500 ml), Ph.Chf (3 × 500 ml), Ph.EtAc (3 × 500 ml) Ph.Bt (3 × 500 ml) and H2O (3 × 500 ml). Lastly, we got 68 g (27.2%) of Ph.Hex, 27 g (10.8%) of Ph.Chf, 13 g (5.2%) of Ph.EtAc, 11 g (4.4%) of Ph.Bt and 37 g (14.8%) of Ph.Aq [35, 36]. These were stored in tight containers and kept at refrigerator temperature till further use.
Extraction of saponins
For the isolation of crude saponins, about 60 g powder material was added to 100 ml of ethanol (20%) using a conical flask. The mixture was heated for 4 h via water bath (55 °C) with appropriated gradual shaking. Thereafter, the solvent was filtered and the powder material was again extracted with 200 ml of ethanol. The ethanol was combined and was placed in water bath until its volume was reduced to 40 ml. The fluid was transferred to separating funnel and with the subsequent addition of 20 ml diethyl ether. The mixture was shaked vigorously. Within the funnel two layers were formed, the diethyl ether and water. The diethyl ether layer was discarded and 60 mL of n-butanol was added to the aqueous layer. The resultant mixture was twice washed with 5% NaCl and finally the solvents were evaporated via water bath and 9 g of saponins residue was obtained [37, 38].
HPLC -DAD analysis of Ph.Cme
For sample preparation, 100 mg extract was dissolved in 10 mL methanol (100%) and shaken for 1 h. Samples were filtered by syringe filter (PFTE filter, 0.45 μ, Agilent Technologies, Germany) in to HPLC vials (2 mL). Injection volume was 50 μL. Chromatographic analysis was performed following our previously reported standard procedure [39–41]. In brief, Agilent 1260 infinity HPLC system equipped with quaternary pump, degasser, auto-sampler and coupled with diode array detector was used for phenolics quantification of the test sample. Compounds separation was done via an Agilent rapid resolution Zorbax Eclipse plus C18 column with dimensions of 4.6 X 100 mm and 3.5 μm, and maintained at temperature of 25 °C and a flow rate of 1 ml min− 1. Chromatogram was obtained at 320 nm while absorption spectra was scanned at wide range of 200-600 nm and only higher purity peaks (95%) were quantified [39]. Phenolic compounds were identified by comparison of the retention time as well as absorption spectra with standards available analyzed simultaneously. Other compounds were identified via comparison of absorption spectra with published literature [42–44]. For unknown compounds, calibration curves of standards with same chromatographic response factor were used.
In-vitro anti-diabetic studies
α-glucosidase inhibition assay
The inhibitory activity of our samples against α-glucosidase enzyme was evaluated using the established method of McCue et al. (2005) [45]. In brief, solutions of the α-glucosidase enzyme was prepared by dissolving 0.5 unit mL− 1 in a 0.1 M phosphate buffer (pH 6.9). The final enzyme solution contain 20 μl α-glucosidase (0.5 unit mL− 1) and 120 μl 0.1 M phosphate buffer. Substrate solution consisting of p-Nitrophenyl-α-D-glucopyranoside (5 mM) was prepared in the same buffer (pH 6.9). Test samples at concentration range of 31.25-1000 μg mL− 1 were prepared and were mixed with enzyme solution followed by incubation for 15 min at 37 °C. Finally, 20 μl substrate solution was added to the enzyme mixture and was again incubated for 15 min at 37 °C. The reaction was completed by the addition of 80 μl of 0.2 M sodium carbonate solution. Absorbance were measured at 405 nm using UV visible spectrophotometer (Thermo electron corporation USA). The system without α-glucosidase act as blank, and acarbose was used as positive control. Each experiment was conducted in triplicate and percent inhibition were calculated using formula;
α-amylase inhibition assay
In-vitro amylase inhibition of our samples were performed according to the previously reported protocol [46]. Briefly, 100 μL of test samples were added to 200 μL of enzyme solution and 100 μL (2 mM) of phosphate buffer (pH -6.9). Thereafter, the mixture was incubated for 20 min and subsequently, 100 μL of 1% starch solution will be added to it. The same was repeated for the controls where 200 μL of the enzyme will replaced by buffer. After incubation for 5 min, 500 μL of dinitrosalicylic acid reagent was added to both control and test groups. Both samples were incubated for 10 min and absorbance’s were recorded at 580 nm via spectrophotometer. Percent inhibition were calculated using the formula;
Where A = absorbance of test and B = absorbance of enzyme control.
Molecular docking with HPLC-DAD identified compounds
In-silico docking is an important tool to assess the mode of molecular interactions of new compounds within the target molecule as a potential inhibitor or activating agent [47]. The binding interactions of identified compounds in the active sites of our target enzymes α-glucosidase and α-amylase were elucidated via MOE-Dock software. The crystal structure of α-glucosidase is not available yet, so, we used homology model as described by Ming Liu et al [48] while the 3D crystal structure of the α-amylase (4 W93) was retrieved from the Protein Databank (PDB). Prior to molecular docking, all water molecules and ions were removed from the retrieved crystal structure using the Molecular Operating Environment software (www.chemcomp.com). The hydrogen atoms were added to the protein structures by 3D protonation and then energy minimization were carried out by using the default parameters of the MOE (gradient: 0.05, Force Field: Amber99).
The structures of the compounds were built in MOE and energy minimized using the default parameters of the MOE [49]. Both α-glucosidase and α-amylase were allowed to dock to the compounds using MOE by the default parameters i.e., Placement: Triangle Matcher, Refinement: Induced Fit, Rescoring: London dG. For each ligand ten conformations were generated. The top-ranked conformation of each compound was used for further analysis. After the molecular docking, the best poses having polar, arene-arene, H-pi and pi-H interactions were analyzed by Pymol software.
Nutritional contents
Assessment of moisture content
Loss on drying (LOD) method was followed for the determination of moisture content of the plant sample. A weighed quantity of powdered plant sample was taken in a suitable container and allowed to dry at 105 °C in oven till the achievement of constant weight. Thus the amount of moisture present in the powdered plant sample was figured out from the difference of dried weight of sample and the total weight of the sample.
Assessment of ash content
Incineration procedure was followed for determination of ash content of powdered plant sample. A weighed amount of sample was put in a crucible and transferred into the muffle furnace and allowed to incinerate at 550 °C for 24 h. Similarly total ash content was figured out after conversion of dried mass of powdered plant sample into ashes.
Assessment of crude fat
Soxhlet method was followed for the determination of total fats in the sample. Briefly, 2 g of dried powdered plant sample was transferred into a soxhlet extractor and petroleum ether was added to the flask of the extractor. The extraction was carried out for 6 h till the exhaustion of sample from fat content. The obtained petroleum ether was filtered and the filtrate obtained was allowed to be evaporated in a weighed beaker. Similarly, the total fats were calculated as the total increase in weight of the beaker.
Assessment of crude protein
For determination of crude protein the method of microkjeldahl nitrogen method was followed. This method involved the digestion of plant sample with concentrated sulphuric acid and catalyst for the conversion of organic nitrogen into ammonium sulfate in the solution. After which the decomposition of ammonium sulfate was carried out via NaOH. The liberated ammonia was distilled into 5% boric acid. After this the titration of trapped ammonia was carried out with 0.05 N HCl for the deduction of nitrogen from ammonia. The indicators used were methylene red and blue both. The percent proteins were calculated from the value of nitrogen obtained multiplied by 6.25.
Toxicity evaluations
Animals and ethical committee approval
BALB/c albino mice (18-35 g) mixed breed were used in the acute toxicity study. Animals were provided appropriate food and water ad libitum. Our study was evaluated and approved by Departmental Research Ethics Committee (DREC) via reference no DREC/2016052/01. Animals studies were performed following rules of Institute of Laboratory Animal Resources Commission on life sciences, National research council 1996 [50].
Acute toxicity study
Test samples were evaluated for acute toxicity in mice after oral administration of increasing doses up to 2000 mg kg− 1. Animals were observed for lethality and aberrant behavioral changes [51].
Haemagglutination study
Haemagglutination activity was performed pursuing the procedure followed by Naqvi et al. [52]. Blood taken from healthy individuals consisting of different groups was centrifuged and 2% suspension of RBCs of each blood group was prepared in phosphate buffer (pH 7). Serial dilutions of each plant sample were prepared and 1 ml of each dilution was combined with 1 ml of each RBCs suspension. The solutions were kept for a while in test tubes at 25 °C. Negative haemagglutination activity was shown by the formation of smooth button at the bottom of test tube while positive activity was indicated by the formation of rough granular deposition. The intensity of activity was measured by the extent of smooth button formation or deposition.
Statistical analysis
All tests were performed in triplicate and results were presented as Mean ± SEM. Results were expressed as % inhibition (mean ± SEM of n = 3) and IC50. IC50 were calculated from dose-response curve along the doses tested in the inhibition studies. Values significantly different as compare to standard drug One way ANOVA followed by multiple comparison DUNNETT test was applied to the data for comparison with the standard group. *: p < 0.05, **: p < 0.01, ***: p < 0.001. ns: Results not significantly different in comparison to standard drug.
Results and discussion
HPLC-DAD phenolic-profiling
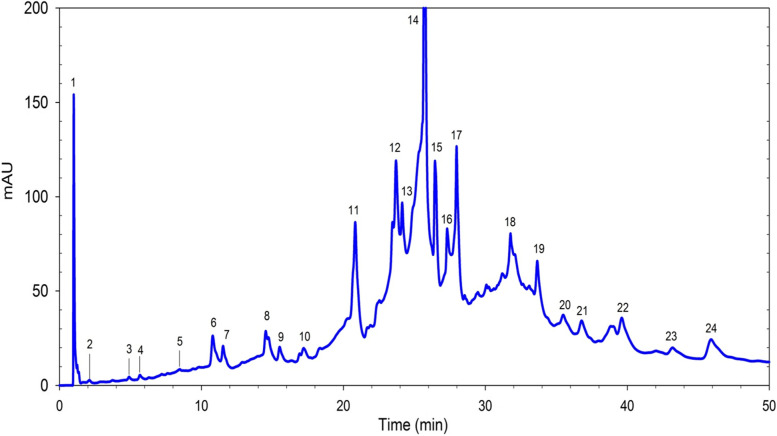
HPLC-DAD analysis of Ph.Cme is summarized in Table 1 and Fig. 1. Chromatogram exhibit identification of 24 phhenolic compounds. The most abundant identified compounds were Kaemferol-3-(p-coumaroyl-diglucoside)-7-glucoside (275.4 mg/g peak 14), p-Coumaroylhexose-4-hexoside (96.5 mg/g peak 11) and Quercetin-3-glucoronide (76.0 mg/g peak 17). Other abundant compounds were 4-Caffeoylquinic acid (58.1 mg/g), Quercetin (57.9 mg/g), 5,7,3′-Trihydroxy-3,6,4′,5′-tetramethoxyflavone (55.5 mg/g) Ellagic acid (50.4 mg/g), 5-Feruloylquinic acid (45.8 mg/g), Cyanidin-3-glucoside (26.8 mg/g), Delphinidin-3-glucoside (24 mg/g), Quercetin-3-hexoside (20.7 mg/g), 5,7-dihydroxy-4′-methoxyflavone (15.2 mg/g) of the sample. Among the other compounds were Hydroxybenzoic acid (2.8 mg/g), Gallic acid (0.2 mg/g), Hydroxybenzoylhexose (0.2 mg/g), Caffeic acid (0.2 mg/g), Syringic Acid (0.2 mg/g), p-Coumaric acid (8.8 mg/g), 5-Coumaroylquinic acid (5.2 mg/g), 3-Caffeoylquinic acid (6.2 mg/g), 3-Coumaroylquinic Acid (3.8 mg/g), p-Coumaroylhexose (5.1 mg/g), Malvidin-3-glucoside (3.8 mg/g), Cyanidin-3-rutinoside (12.0 mg/g) respectively (Table 1, Fig. 1).
Table 1.
Phenolic profile of Ph.Cme extract (mg/g)
| Peak | Rt (min) | Identity | Mean Composition (mg/g) of sample | STD |
|---|---|---|---|---|
| 1 | 1 | Hydroxybenzoic acid | 2.8 | 0.02 |
| 2 | 2.1 | Gallic acid | 0.2 | 0.01 |
| 3 | 4.9 | Hydroxybenzoylhexose | 0.2 | 0.01 |
| 4 | 5.6 | Caffeic acid | 0.2 | 0.01 |
| 5 | 8.4 | Syringic Acid | 0.2 | 0.01 |
| 6 | 10.7 | p-Coumaric acid | 8.8 | 0.2 |
| 7 | 11.5 | 5-Coumaroylquinic acid | 5.2 | 0.2 |
| 8 | 14.5 | 3-Caffeoylquinic acid | 6.2 | 0.3 |
| 9 | 15.5 | 3-Coumaroylquinic Acid | 3.8 | 0.1 |
| 10 | 17.2 | p-Coumaroylhexose | 5.1 | 0.1 |
| 11 | 20.8 | p-Coumaroylhexose-4-hexoside | 96.5 | 2.4 |
| 12 | 23.7 | 4-Caffeoylquinic acid | 58.1 | 1.0 |
| 13 | 24.1 | 5-Feruloylquinic acid | 45.8 | 1.0 |
| 14 | 25.7 | Kaemferol-3-(p-coumaroyl-diglucoside)-7-glucoside | 275.4 | 6.5 |
| 15 | 26.4 | Ellagic acid | 50.4 | 0.5 |
| 16 | 27.3 | Quercetin | 57.9 | 1.3 |
| 17 | 27.9 | Quercetin-3-glucoronide | 76.0 | 1.2 |
| 18 | 31.6 | 5,7-dihydroxy-4′-methoxyflavone | 15.2 | 0.2 |
| 19 | 33.6 | 5,7,3′-Trihydroxy-3,6,4′,5′-tetramethoxyflavone | 55.5 | 1.2 |
| 20 | 35.4 | Cyanidin-3-glucoside | 26.8 | 0.3 |
| 21 | 36.7 | Delphinidin-3-glucoside | 24.0 | 0.4 |
| 22 | 39.6 | Quercetin-3-hexoside | 20.7 | 0.8 |
| 23 | 43.1 | Malvidin-3-glucoside | 3.8 | 0.1 |
| 24 | 45.9 | Cyanidin-3-rutinoside | 12.0 | 0.2 |
Standard compounds used were; Hydroxybenzoic acid, Gallic acid, Caffeic acid, Syringic acid, p-coumaric acid, 3-Caffeoylquinic acid, Quercetin, Ellagic acid and Cyanidin-3-glucoside
Fig. 1.
Chromatogram of the HPLC-DAD analysis of Ph.Cme. Peak numbers represent individual compounds and their details are provided in Table 1
Enzymes inhibition studies
Natural phenolics are widely known and scientifically validated for efficacy in DM. For instance, mulberry polyphenolic compounds such as syringic acid and galloylcyanidin-glycoside are reported to inhibit α-glucosidase activity while quercetin and cyanidin-glycosides are essential for cellular antioxidant activity [53]. Quercetin is reported to control glucose homeostasis of whole-body by interacting with various molecular targets in small intestine, pancreas, skeletal muscle, liver and adipose tissue. Quercetin mechanisms of action include intestinal glucose absorption inhibition, insulin-sensitizing and secreting activities and increased utilization of glucose in peripheral tissues [54]. Ellagic acid seems to play an anti-diabetic activity. The anti-diabetic effect of ellagic acid through the action on pancreas β-cells, decreasing glucose intolerance and stimulation of insulin secretion has been reported by Fatima et al., [55]. Likewise, fruit extract of Emblica officinalis exhibit anti-diabetic potentials via increased insulin sensitization preimirilymediated by the presence of gallic acid [56]. The antioxidant and anti-diabetic potential of caffeic acid in a streptozotocin-induced diabetic rat model has been evaluated which showed a significant increase in serum insulin level, and decrease glucose level in the blood of diabetic rat models [57]. It has also been demonstrated that cyanidin-3-O-glucoside inhibit glucosidase enzyme which result in decrease glucose absorption in intestine [58]. Anti-diabetic and antioxidant activity of sweet cherries [59] and Prunus avium [60] has been reported which may be due to the identified phenolic contents, including hydroxybenzoic acid. The hydroxybenzoic acid and p-coumaric acid are probably responsible for the anti-diabetic activity investigated in edible mushrooms by D.Stojkovic et al., 2019 [61]. Among the phenolic compounds in the ethanolic extract of P. hydropiper, rutin has been reported for its anti-diabetic, antioxidant and anti-inflammation activity [32]. The identified phenolics might contribute to the overall anti-diabetic potentials of our test samples.
In the present study, Ph.Sp was found highly active against α-glucosidase enzyme as shown in Table 2. Overall a concentration dependent inhibition was observed against the enzyme. Ph.Sp exhibited 71.50 ± 0.28% inhibitory activity at the high tested dose (1000 μg mL− 1). Acarbose inhibitory activity at the same dose was 77.30 ± 0.61%. The inhibitory activity of Ph.Sp was comparable to the standard drug acarbose at the same concentrations. The IC50 for Ph.Sp and acrabose were 100 and 18 μg/ml respectively. Among the other fractions, Ph.Cr, Ph.Hex, Ph.Chf, Ph.EtAc, Ph.Bt and Ph.Aq have displayed concentration dependent inhibitions with IC50 of 400, 1800, 320, 680, 1000 and 700 μg mL− 1 respectively. Ph.Cr, Ph.Chf and Ph.Sp are most active samples and need further in-vivo studies for potential effectiveness against type 2 DM. The Ph.Cr, Ph.Chf can be subjected to column chromatography for isolation of bioactive compounds.
Table 2.
Results of α-glucosidase and α-amylase inhibitory potentials of Persicaria hydropiper
| Samples | α-glucosidase assay | α-amylase assay | |||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Conc. | % Inhibition | IC50 μg/mL | Sample | Conc. | % Inhibition | IC50 μg/mL | Sample | % Inhibition | IC50 μg/mL | Samples | % Inhibition | IC50 μg/mL | |
| Ph.Cr |
1000 500 250 125 62.5 31.25 |
67.33 ± 1.52* 51.86 ± 3.09*** 42.50 ± 1.32*** 35.20 ± 2.52*** 29.66 ± 2.56*** 25.50 ± 1.32*** |
400 | Ph.Bt |
1000 500 250 125 62.5 31.25 |
49.67 ± 1.52*** 43.00 ± 1.73*** 38.67 ± 0.57*** 32.00 ± 0.00*** 24.00 ± 0.00*** 19.20 ± 2.25*** |
1000 | Ph.Cr |
70.89 ± 0.55** 59.93 ± 1.45** 51.02 ± 0.78* 43.83 ± 0.38 ns 32.56 ± 2.12* 19.55 ± 1.56** |
200 | Ph.Bt |
57.58 ± 0.15ns 51.68 ± 2.33* 33.90 ± 1.22*** 20.17 ± 0.88*** 11.55 ± 2.50*** 05.40 ± 1.90*** |
550 |
| Ph.Hex |
1000 500 250 125 62.5 31.25 |
32.66 ± 2.52*** 26.16 ± 2.75*** 21.50 ± 1.32*** 17.73 ± 0.51*** 15.69 ± 1.04*** 11.93 ± 1.61*** |
1800 | Ph.Aq |
1000 500 250 125 62.5 31.25 |
53.83 ± 1.04*** 45.00 ± 1.00*** 41.86 ± 3.09*** 34.33 ± 2.02*** 30.36 ± 0.57** 22.00 ± 0.00** |
700 | Ph.Hex |
55.78 ± 0.55*** 42.33 ± 1.10*** 29.00 ± 2.41*** 12.88 ± 0.91*** 07.96 ± 2.44*** 03.63 ± 1.91*** |
750 | Ph.Aq |
41.99 ± 2.40** 35.71 ± 3.20*** 26.21 ± 1.50*** 15.00 ± 0.62*** 10.94 ± 2.86** 4.99 ± 1.00*** |
920 |
| Ph.Chf |
1000 500 250 125 62.5 31.25 |
66.44 ± 1.50* 54.73 ± 0.51*** 47.23 ± 1.05*** 40.16 ± 1.02* 34.50 ± 1.32 ns 29.00 ± 0.00 |
320 | Ph.Sp |
1000 500 250 125 62.5 31.25 |
71.50 ± 0.28 ns 65.00 ± 0.86* 58.00 ± 0.28* 51.00 ± 1.15 ns 39.00 ± 0.00 ns 30.36 ± 0.57* |
100 | Ph.Chf |
87.32 ± 2.45* 77.54 ± 0.87ns 68.49 ± 1.33 ns 60.95 ± 2.40 ns 48.97 ± 0.51 ns 27.88 ± 1.20** |
90 | Ph.Sp |
90.06 ± 0.45 ns 80.13 ± 1.77 ns 68.83 ± 0.64 ns 58.56 ± 0.84 ns 47.26 ± 2.35 ns 29.33 ± 1.68** |
100 |
| Ph.EtAc |
1000 500 250 125 62.5 31.25 |
55.00 ± 1.00*** 43.60 ± 1.76*** 34.00 ± 1.00*** 28.20 ± 1.04*** 22.00 ± 0.00*** 17.00 ± 1.00*** |
680 | P.C |
1000 500 250 125 62.5 31.25 |
77.30 ± 0.61 73.00 ± 0.00 69.00 ± 0.00 55.50 ± 1.04 49.83 ± 0.44 41.00 ± 0.00 |
18 | Ph.EtAc |
68.00 ± 0.51ns 49.60 ± 2.23* 37.95 ± 0.77** 21.40 ± 2.25** 15.86 ± 1.33*** 09.71 ± 2.55*** |
480 | P.C |
77.30 ± 0.61 73.00 ± 0.00 69.00 ± 0.00 55.50 ± 1.04 49.83 ± 0.44 41.00 ± 0.00 |
18 |
Results were expressed as % inhibition (mean ± SEM of n = 3) and IC50. Values significantly different as compare to standard drug (Acarbose), *: p < 0.05, **: p < 0.01, ***: p < 0.001. ns: Results not significantly different in comparison to standard drug. P.C = Acarbose
In amylase inhibition studies, all fractions displayed a concentration dependent inhibition of α-amylase enzyme with Ph.Sp and Ph.Chf with highest percent inhibitions. Ph.Sp and Ph.Chf exhibited 90.06 ± 0.45% and 87.32 ± 2.45 inhibitions at highest tested concentration (1000 μg mL− 1) respectively (Table 2). The IC50 for Ph.Sp and Ph.Chf were 100 and 90 μg mL− 1 respectively. Percent inhibitions of these fractions were very comparable with standard inhibitions. Among the other fractions Ph.Cr, Ph.Bt and Ph.EtAc showed moderate inhibitory activity with IC50 of 200, 550 and 480 μg mL− 1 respectively.
Natural products of enormous structural miscellany are still major source for the development of new drugs including inhibitors of glucose metabolizing enzymes [62, 63]. α-glucosidase inhibitors (AGI’s), like acarbose, voglibose in microorganisms and nojirimycin, 1-deoxynojirimycin has been reported from plants [64–66]. Commercially accessible AGI’s for instance acarbose, miglitol and voglibose are widely employed for the treatment of type 2 DM. These AGI’s are shown to diminish the insulin requirements for type 1 diabetes as well as improves reactive hypoglycemia [67]. As the AGI’s show therapeutic effect by restraining carbohydrate absorption, the undigested carbohydrate dislocate to the colon go through fermentation by colonic flora to result in adverse effects such as flatulence, abdominal discomfort and diarrhea [68]. But the undesirable effects are dose dependent and diminishes with the duration of therapy [69]. Recently, numerous efforts have been made to find out more effective drugs against type 2 diabetes from natural sources to develop physiologic functional food or isolate new and more effective compounds [70]. Several AGI present as phyto-constituents including alkaloids, glycosides, flavonoids, terpenoids and phenolic compounds have been reported from plant origin [71]. Thus, there is an urgent need to search for novel drugs from several sources, including natural products, with increased potency and lesser adverse effects than the existing drugs to fight global health problems posed by DM.
Docking analysis of α-glucosidase
The docking results of the compounds with the alpha glucosidase enzyme have given good information about the nature of the binding mode. Our current docking findings revealed that majority of the compounds exhibited good confirmations in alpha glucosidase enzyme and were involved in various type of interactions with the active site residues of the target enzymes. The detail of docking scores and interactions for all compounds are listed in Table 3. From the docking conformation of the compounds, it was revealed that the top most active compound was compound 14 (docking score = − 19.80899) formed 14 hydrogen bonds, two pi-H and two pi-pi linkages with the Tyr 71, Phe 158, Phe 177, Gln 181, Arg 212, Asp 214, Glu 276, Phe 300, Val 303, Tyr 344, Asp 349, Gln 350, Arg 439, and Asp 408 residues of the binding pocket of the α-glucosidase as shown in Fig. 2. The high potency of the ligand might be due to the presence of the electron donating group (−OH) as well as the electron cloud system of the compound.
Table 3.
Results of molecular docking studies with the identified compounds against α-glucosidase
| S. No | Ligand | Receptor | Interaction | Distance | E (kcal/mol) | Docking score | |||
|---|---|---|---|---|---|---|---|---|---|
| 1 | O | 14 | ND2 | ASN | 347 | H-acceptor | 3.29 | −2.9 | −7.43843651 |
| O | 15 | ND2 | ASN | 347 | H-acceptor | 3.02 | −2.4 | ||
| 2 | O | 9 | OE2 | GLU | 276 | H-donor | 2.58 | −1.2 | −11.3185654 |
| O | 13 | O | ASP | 349 | H-donor | 2.77 | −1.9 | ||
| O | 18 | NH1 | ARG | 439 | H-acceptor | 2.74 | −3 | ||
| 3 | O | 33 | OE1 | GLN | 350 | H-donor | 3.04 | −0.6 | −9.3436832 |
| O | 24 | ND2 | ASN | 347 | H-acceptor | 3.24 | −1.3 | ||
| O | 24 | 6-ring | PHE | 300 | H-pi | 3.36 | −0.5 | ||
| 4 | O | 12 | OD2 | ASP | 408 | H-donor | 2.99 | −2 | −10.572752 |
| O | 19 | NE2 | HIS | 111 | H-acceptor | 3.27 | −3.5 | ||
| O | 20 | NH1 | ARG | 212 | H-acceptor | 3.29 | − 2.9 | ||
| O | 20 | NH2 | ARG | 212 | H-acceptor | 3.15 | −2.1 | ||
| O | 20 | NH1 | ARG | 212 | ionic | 3.29 | −2.8 | ||
| O | 20 | NH2 | ARG | 212 | ionic | 3.15 | −3.6 | ||
| 5 | O | 14 | ND2 | ASN | 347 | H-acceptor | 2.93 | −3.4 | −8.25566006 |
| C | 16 | 6-ring | PHE | 300 | H-pi | 3.99 | −0.5 | ||
| 6 | O | 18 | NE2 | HIS | 348 | H-acceptor | 2.99 | −0.8 | −7.69557381 |
| 7 | O | 18 | OE1 | GLN | 350 | H-donor | 3.36 | −0.6 | −9.5489674 |
| O | 22 | OE1 | GLN | 350 | H-donor | 3.06 | −2.5 | ||
| O | 41 | 5-ring | HIS | 279 | H-pi | 3.6 | − 2.6 | ||
| 8 | C | 5 | O | ASP | 349 | H-donor | 3.21 | −0.1 | −18.5305271 |
| O | 17 | OE1 | GLN | 350 | H-donor | 2.77 | −2 | ||
| C | 43 | OD2 | ASP | 349 | H-donor | 3.78 | −0.1 | ||
| C | 45 | OE1 | GLU | 276 | H-donor | 3.86 | −0.1 | ||
| C | 52 | OD2 | ASP | 68 | H-donor | 3.21 | −0.2 | ||
| O | 56 | OD2 | ASP | 68 | H-donor | 2.79 | −6.8 | ||
| O | 19 | CD2 | PHE | 300 | H-acceptor | 3.17 | −0.1 | ||
| O | 41 | ND2 | ASN | 347 | H-acceptor | 3.35 | −0.1 | ||
| O | 16 | NH2 | ARG | 312 | ionic | 2.76 | −6.3 | ||
| −6 | ring | CZ | PHE | 177 | pi-H | 4.24 | −0.1 | ||
| −6 | ring | NH1 | ARG | 439 | pi-cation | 4.27 | −0.4 | ||
| 9 | C | 26 | OD1 | ASP | 214 | H-donor | 3.52 | −0.2 | −13.9830008 |
| C | 30 | OD1 | ASP | 214 | H-donor | 3.7 | −0.1 | ||
| O | 35 | OD2 | ASP | 349 | H-donor | 2.89 | −3.7 | ||
| O | 40 | OE1 | GLN | 181 | H-donor | 3.57 | −0.1 | ||
| O | 35 | NH1 | ARG | 439 | H-acceptor | 2.86 | −0.3 | ||
| O | 38 | NE2 | HIS | 348 | H-acceptor | 2.78 | −5.9 | ||
| O | 39 | NE2 | HIS | 348 | H-acceptor | 2.89 | −2.1 | ||
| O | 40 | NE2 | HIS | 111 | H-acceptor | 3.1 | −1.6 | ||
| O | 38 | NH1 | ARG | 212 | ionic | 3.25 | −3 | ||
| O | 38 | NH2 | ARG | 212 | ionic | 2.91 | −5.1 | ||
| O | 39 | NH2 | ARG | 212 | ionic | 3.31 | − 2.7 | ||
| C | 26 | 6-ring | TYR | 71 | H-pi | 4.79 | −0.1 | ||
| O | 33 | 6-ring | PHE | 177 | H-pi | 3.06 | −0.2 | ||
| 10 | C | 5 | O | ASP | 349 | H-donor | 3.34 | −0.4 | −13.088253 |
| O | 22 | OE1 | GLN | 350 | H-donor | 3.06 | −0.2 | ||
| C | 32 | OD2 | ASP | 68 | H-donor | 3.16 | −0.2 | ||
| O | 40 | OE1 | GLN | 181 | H-donor | 2.72 | −1.6 | ||
| O | 22 | CG2 | VAL | 303 | H-acceptor | 3.26 | −0.1 | ||
| C | 35 | 6-ring | PHE | 177 | H-pi | 3.49 | −0.1 | ||
| −6 | ring | NE2 | HIS | 111 | pi-H | 3.91 | −0.1 | ||
| 11 | C | 26 | O | PHE | 157 | H-donor | 3.55 | −0.1 | −13.8564711 |
| C | 41 | OD2 | ASP | 408 | H-donor | 3.37 | −0.3 | ||
| O | 21 | CE1 | PHE | 177 | H-acceptor | 3.78 | −0.1 | ||
| O | 35 | N | ARG | 312 | H-acceptor | 3.4 | −0.3 | ||
| O | 45 | CB | ARG | 312 | H-acceptor | 3.64 | −0.1 | ||
| O | 45 | NE | ARG | 312 | H-acceptor | 3.12 | −1.9 | ||
| O | 64 | CD1 | PHE | 158 | H-acceptor | 3.78 | −0.1 | ||
| O | 64 | CE1 | PHE | 158 | H-acceptor | 3.8 | −0.1 | ||
| O | 64 | CD1 | PHE | 177 | H-acceptor | 3.26 | −0.1 | ||
| C | 1 | 6-ring | PHE | 157 | H-pi | 4.44 | −0.1 | ||
| C | 3 | 6-ring | PHE | 157 | H-pi | 4.44 | −0.1 | ||
| −6 | ring | NH1 | ARG | 439 | pi-cation | 3.14 | −0.1 | ||
| 12 | C | 4 | OD1 | ASP | 214 | H-donor | 3.49 | −0.1 | −15.2827396 |
| O | 10 | OE1 | GLN | 181 | H-donor | 2.78 | −2.4 | ||
| C | 23 | O | ASP | 349 | H-donor | 3.61 | −0.1 | ||
| O | 20 | NH2 | ARG | 212 | H-acceptor | 3.15 | −0.8 | ||
| O | 20 | CZ | PHE | 300 | H-acceptor | 3.89 | −0.1 | ||
| O | 39 | NE | ARG | 312 | H-acceptor | 2.92 | −0.9 | ||
| O | 39 | NH2 | ARG | 312 | H-acceptor | 2.94 | −0.5 | ||
| O | 40 | CD | ARG | 312 | H-acceptor | 3.22 | −0.1 | ||
| O | 39 | NE | ARG | 312 | ionic | 2.92 | −5.1 | ||
| O | 39 | NH2 | ARG | 312 | ionic | 2.94 | −4.9 | ||
| O | 40 | NE | ARG | 312 | ionic | 2.97 | −4.7 | ||
| −6 | ring | NE2 | HIS | 111 | pi-H | 4.84 | −0.1 | ||
| 13 | C | 3 | OD1 | ASP | 214 | H-donor | 3.16 | − 0.3 | −12.4345493 |
| O | 16 | OD1 | ASP | 214 | H-donor | 2.8 | −5.3 | ||
| O | 42 | OE2 | GLU | 304 | H-donor | 2.75 | −3.7 | ||
| O | 42 | CD2 | PHE | 300 | H-acceptor | 3.68 | −0.1 | ||
| O | 42 | CD | ARG | 312 | H-acceptor | 3.1 | −0.1 | ||
| O | 44 | NE2 | HIS | 245 | H-acceptor | 2.91 | −7.1 | ||
| O | 44 | CD2 | HIS | 279 | H-acceptor | 3.23 | −0.3 | ||
| O | 45 | CD2 | LEU | 218 | H-acceptor | 4.03 | −0.1 | ||
| O | 45 | NE2 | HIS | 245 | H-acceptor | 3.38 | −1.1 | ||
| O | 39 | 6-ring | PHE | 157 | H-pi | 4.32 | −0.4 | ||
| 14 | O | 42 | OD1 | ASP | 214 | H-donor | 3.41 | −0.1 | − 19.8089981 |
| C | 22 | OE1 | GLU | 276 | H-donor | 3.45 | −0.5 | ||
| O | 27 | OD2 | ASP | 408 | H-donor | 2.98 | −1 | ||
| C | 31 | OE1 | GLN | 181 | H-donor | 3.43 | −0.1 | ||
| O | 40 | O | TYR | 71 | H-donor | 2.59 | −1 | ||
| O | 44 | OE2 | GLU | 276 | H-donor | 2.96 | −0.6 | ||
| O | 53 | OD2 | ASP | 349 | H-donor | 2.62 | −3.6 | ||
| C | 64 | O | VAL | 303 | H-donor | 3.36 | −0.1 | ||
| O | 21 | CE2 | PHE | 300 | H-acceptor | 3.68 | −0.1 | ||
| O | 42 | NH2 | ARG | 212 | H-acceptor | 2.98 | −1.6 | ||
| O | 44 | CZ | PHE | 300 | H-acceptor | 3.7 | −0.1 | ||
| O | 53 | NH1 | ARG | 439 | H-acceptor | 2.72 | −0.7 | ||
| O | 55 | CD | ARG | 439 | H-acceptor | 3.14 | −0.2 | ||
| O | 68 | OH | TYR | 344 | H-acceptor | 2.96 | −0.5 | ||
| −6 | ring | CE1 | PHE | 158 | pi-H | 3.35 | −0.2 | ||
| −6 | ring | CG | GLN | 350 | pi-H | 3.54 | −0.2 | ||
| −6 | ring | 6-ring | PHE | 177 | pi-pi | 3.13 | 0 | ||
| −6 | ring | 6-ring | PHE | 300 | pi-pi | 3.97 | 0 | ||
| 15 | O | 23 | OD2 | ASP | 68 | H-donor | 2.63 | −7 | −15.9700079 |
| O | 25 | O | ASP | 349 | H-donor | 2.66 | −4.3 | ||
| O | 14 | NE2 | HIS | 348 | H-acceptor | 3.25 | −0.1 | ||
| O | 19 | ND2 | ASN | 347 | H-acceptor | 2.86 | −3.7 | ||
| O | 23 | NE2 | HIS | 111 | H-acceptor | 3.44 | −0.2 | ||
| −6 | ring | CD | ARG | 439 | pi-H | 4.04 | −0.2 | ||
| 16 | O | 24 | OE1 | GLN | 181 | H-donor | 2.88 | −2.5 | −12.9178295 |
| O | 23 | NE2 | HIS | 111 | H-acceptor | 3.03 | −0.7 | ||
| O | 23 | CG2 | THR | 215 | H-acceptor | 3.53 | −0.1 | ||
| O | 26 | CE2 | PHE | 300 | H-acceptor | 4.05 | −0.1 | ||
| −6 | ring | 6-ring | PHE | 177 | pi-pi | 3.8 | 0 | ||
| 17 | O | 29 | OD2 | ASP | 408 | H-donor | 2.75 | −2.2 | −17.0351429 |
| O | 29 | OD1 | ASP | 408 | H-donor | 3.44 | −0.1 | ||
| O | 44 | O | ASP | 349 | H-donor | 2.97 | −1.4 | ||
| O | 23 | NE2 | HIS | 245 | H-acceptor | 3.22 | −2.2 | ||
| O | 26 | ND2 | ASN | 241 | H-acceptor | 2.93 | −1 | ||
| O | 48 | ND2 | ASN | 347 | H-acceptor | 2.93 | −2.6 | ||
| O | 26 | ND1 | HIS | 279 | ionic | 3.88 | −0.8 | ||
| O | 26 | NE2 | HIS | 279 | ionic | 3.7 | −1.2 | ||
| O | 50 | NH1 | ARG | 439 | ionic | 3.36 | −2.5 | ||
| O | 48 | 6-ring | PHE | 300 | H-pi | 3.32 | −0.2 | ||
| −6 | ring | 5-ring | HIS | 279 | pi-pi | 3.91 | 0 | ||
| 18 | C | 10 | OD2 | ASP | 349 | H-donor | 3.45 | −0.8 | −13.8796387 |
| C | 19 | OD1 | ASN | 347 | H-donor | 3.6 | −0.1 | ||
| O | 29 | OE1 | GLN | 181 | H-donor | 2.79 | −3.8 | ||
| O | 26 | NH1 | ARG | 212 | H-acceptor | 3.2 | −0.3 | ||
| O | 26 | NH2 | ARG | 212 | H-acceptor | 2.87 | −0.8 | ||
| O | 26 | NE2 | HIS | 348 | H-acceptor | 3.34 | −1.6 | ||
| O | 27 | CG2 | VAL | 108 | H-acceptor | 3.54 | −0.1 | ||
| C | 17 | 5-ring | HIS | 348 | H-pi | 4.44 | −1.4 | ||
| −6 | ring | NE2 | HIS | 111 | pi-H | 4.45 | −0.1 | ||
| 19 | O | 43 | OD1 | ASP | 408 | H-donor | 3.32 | −0.8 | −16.4973335 |
| O | 25 | NE | ARG | 312 | ionic | 3.65 | −1.4 | ||
| O | 25 | NH1 | ARG | 312 | ionic | 2.9 | −5.1 | ||
| C | 21 | 6-ring | PHE | 300 | H-pi | 3.73 | −0.3 | ||
| C | 29 | 5-ring | HIS | 279 | H-pi | 4.66 | −0.2 | ||
| −6 | ring | CD | ARG | 312 | pi-H | 3.45 | −0.1 | ||
| 20 | C | 29 | OD2 | ASP | 408 | H-donor | 3.35 | −0.8 | − 18.9063892 |
| O | 43 | OD1 | ASP | 408 | H-donor | 3.63 | −0.1 | ||
| O | 45 | OD2 | ASP | 408 | H-donor | 3.02 | −1.2 | ||
| O | 47 | O | PHE | 157 | H-donor | 3.13 | −1.3 | ||
| O | 45 | CE1 | TYR | 313 | H-acceptor | 3.37 | −0.2 | ||
| O | 45 | CD | ARG | 439 | H-acceptor | 3.51 | − 0.2 | ||
| C | 22 | 6-ring | PHE | 157 | H-pi | 3.81 | −0.8 | ||
| −6 | ring | NH1 | ARG | 439 | pi-cation | 4.19 | −0.1 | ||
| −6 | ring | 6-ring | PHE | 177 | pi-pi | 3.94 | 0 | ||
| 21 | O | 44 | OE1 | GLU | 276 | H-donor | 2.98 | −1.5 | −17.2185078 |
| O | 50 | OE2 | GLU | 304 | H-donor | 2.7 | −3.3 | ||
| O | 52 | OD2 | ASP | 408 | H-donor | 2.83 | −2.9 | ||
| O | 44 | CG2 | THR | 215 | H-acceptor | 3.28 | −0.1 | ||
| O | 44 | CE2 | PHE | 300 | H-acceptor | 3.55 | −0.1 | ||
| −6 | ring | N | ARG | 312 | pi-H | 4.7 | −0.2 | ||
| −6 | ring | CB | ARG | 312 | pi-H | 3.76 | −0.1 | ||
| −6 | ring | CD | ARG | 312 | pi-H | 4.37 | − 0.5 | ||
| 22 | C | 20 | OD2 | ASP | 408 | H-donor | 3.25 | −0.6 | −18.1408939 |
| C | 22 | OD1 | ASN | 347 | H-donor | 3.34 | −0.3 | ||
| O | 32 | OD2 | ASP | 408 | H-donor | 2.97 | −4.7 | ||
| O | 46 | OE2 | GLU | 276 | H-donor | 3.03 | −0.6 | ||
| O | 38 | NH2 | ARG | 212 | H-acceptor | 2.98 | −0.4 | ||
| O | 38 | NE2 | HIS | 348 | H-acceptor | 3.15 | −5.8 | ||
| O | 44 | ND2 | ASN | 347 | H-acceptor | 2.95 | −2.2 | ||
| O | 38 | NH1 | ARG | 212 | ionic | 3.6 | −1.5 | ||
| O | 38 | NH2 | ARG | 212 | ionic | 2.98 | −4.6 | ||
| C | 6 | 6-ring | PHE | 177 | H-pi | 3.96 | −0.6 | ||
| C | 24 | 6-ring | PHE | 300 | H-pi | 3.6 | −0.1 | ||
| C | 39 | 5-ring | HIS | 348 | H-pi | 3.98 | −0.3 | ||
| −6 | ring | CB | PHE | 157 | pi-H | 4.63 | −0.4 | ||
| −6 | ring | CE1 | PHE | 177 | pi-H | 3.41 | −0.4 | ||
| −6 | ring | NH2 | ARG | 212 | pi-cation | 4.71 | −0.1 | ||
| 23 | O | 46 | OD2 | ASP | 68 | H-donor | 2.8 | −1.8 | −16.2608795 |
| O | 48 | OD2 | ASP | 68 | H-donor | 3.1 | −2.6 | ||
| O | 52 | OD1 | ASP | 214 | H-donor | 2.65 | −2.4 | ||
| C | 55 | OE2 | GLU | 276 | H-donor | 3.31 | −0.3 | ||
| O | 14 | CE2 | PHE | 300 | H-acceptor | 3.64 | −0.1 | ||
| O | 46 | CZ | PHE | 158 | H-acceptor | 3.67 | −0.1 | ||
| O | 46 | NH1 | ARG | 439 | H-acceptor | 3.02 | −2.6 | ||
| O | 46 | NH2 | ARG | 439 | H-acceptor | 2.92 | −2.8 | ||
| O | 52 | CG2 | THR | 215 | H-acceptor | 3.78 | −0.1 | ||
| C | 35 | 6-ring | PHE | 177 | H-pi | 3.6 | −0.3 | ||
| C | 55 | 6-ring | PHE | 300 | H-pi | 4.77 | − 0.1 | ||
| 24 | C | 2 | OE1 | GLN | 350 | H-donor | 3.49 | −0.3 | −18.0597305 |
| O | 24 | OD2 | ASP | 68 | H-donor | 3.06 | −1.2 | ||
| C | 38 | O | PHE | 157 | H-donor | 3.67 | −0.1 | ||
| C | 42 | OD2 | ASP | 408 | H-donor | 3.11 | −0.1 | ||
| C | 65 | O | PRO | 309 | H-donor | 3.57 | −0.1 | ||
| O | 71 | OD1 | ASN | 241 | H-donor | 3.05 | −0.2 | ||
| O | 35 | CE1 | PHE | 177 | H-acceptor | 3.79 | −0.1 | ||
| O | 69 | ND2 | ASN | 241 | H-acceptor | 3.04 | −1.5 | ||
| O | 71 | ND2 | ASN | 241 | H-acceptor | 2.93 | −0.6 | ||
| C | 18 | 6-ring | PHE | 177 | H-pi | 3.95 | − 0.1 | ||
| O | 26 | 6-ring | PHE | 177 | H-pi | 3.14 | −0.1 | ||
| C | 44 | 6-ring | PHE | 157 | H-pi | 4.48 | −0.7 | ||
| C | 54 | 6-ring | PHE | 157 | H-pi | 3.99 | −0.2 | ||
| −6 | ring | NH1 | ARG | 439 | pi-cation | 3.66 | −0.1 | ||
Fig. 2.
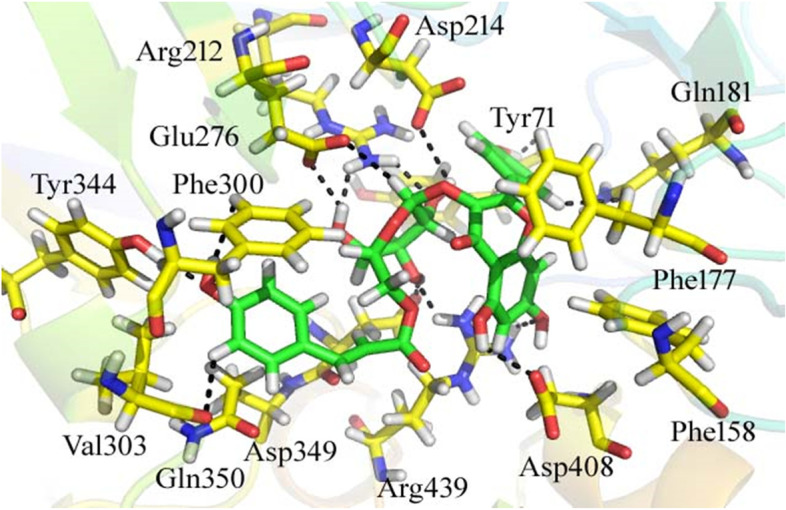
Molecular docking conformations of compound 14 against α-glucosidase
Docking analysis of α-amylase
Docking against revealed that the identified compounds were well accommodated in the active site residues (Trp 59, Tyr 62, Thr 163, Leu 165, Arg 195, Asp 197, Glu 240, Asp 300, His 305, Asp 356) of the target enzyme α-Amylase. From the docking conformation of the compounds, it was observed that compound 24 was the top active compound (docking score = − 15.03757). This compound formed 11 hydrogen bonds, three H-pi and one pi-H contacts with the active site residues of α-amylase (Fig. 3). The interactions detail of the compound is mentioned in Table 4. The inhibition of this compound might be due to the availability of the electron donating group (−OH) and electronic cloud system may be the reason of the excellent in-silico activity of the compound.
Fig. 3.
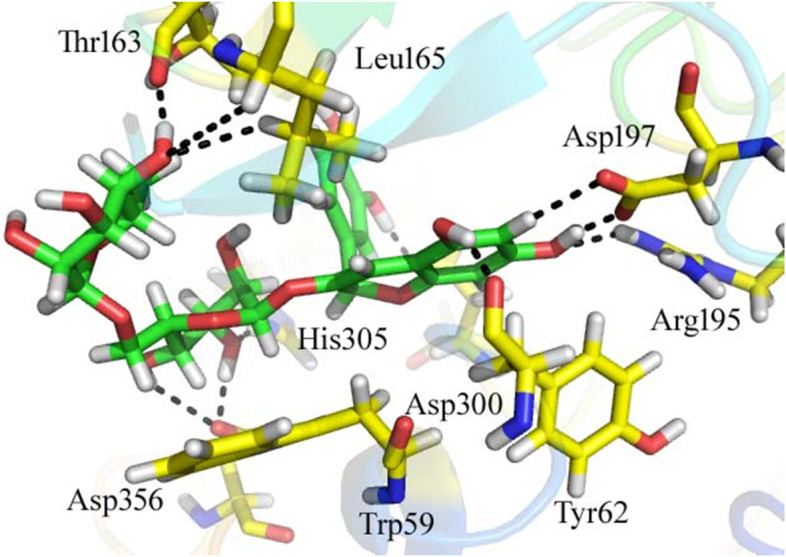
Docking conformation of compound 24 in the active site of α-amylase
Table 4.
Results of molecular docking studies with the identified compounds against α-amylase
| S. No | Ligand | Receptor | Interaction | Distance | E (kcal/mol) | Docking score | ||||
|---|---|---|---|---|---|---|---|---|---|---|
| 1 | O | 16 | NE2 | HIS | 299 | (A) | H-acceptor | 2.88 | −3.7 | −7.69861555 |
| 2 | O | 18 | NH2 | ARG | 195 | (A) | H-acceptor | 2.92 | −1.9 | −8.24910164 |
| O | 18 | NE2 | HIS | 299 | (A) | H-acceptor | 3.45 | −0.8 | ||
| O | 9 | 6-ring | TRP | 58 | (A) | H-pi | 4.64 | −0.7 | ||
| 3 | O | 24 | OH | TYR | 151 | (A) | H-acceptor | 3.06 | −0.3 | −9.24960327 |
| O | 24 | NE2 | HIS | 201 | (A) | H-acceptor | 3.15 | −1.1 | ||
| 4 | O | 20 | NE2 | GLN | 63 | (A) | H-acceptor | 3.1 | −1.8 | −7.81459236 |
| −6 | ring | CZ3 | TRP | 58 | (A) | pi-H | 4.05 | −0.1 | ||
| −6 | ring | CH2 | TRP | 58 | (A) | pi-H | 4.17 | −0.1 | ||
| 5 | O | 12 | NE2 | HIS | 299 | (A) | H-acceptor | 3.11 | −1.1 | −8.02968025 |
| O | 13 | CZ3 | TRP | 58 | (A) | H-acceptor | 3.83 | −0.1 | ||
| O | 13 | NE2 | HIS | 299 | (A) | H-acceptor | 2.97 | −6.9 | ||
| O | 12 | NH2 | ARG | 195 | (A) | ionic | 3.23 | −3.1 | ||
| 6 | O | 18 | NE2 | HIS | 101 | (A) | H-acceptor | 3.23 | −3.3 | −6.82410812 |
| O | 19 | NH2 | ARG | 195 | (A) | H-acceptor | 3.4 | −1.1 | ||
| O | 19 | NH2 | ARG | 195 | (A) | ionic | 3.4 | −2.3 | ||
| 7 | O | 18 | O | TYR | 62 | (A) | H-donor | 3.25 | −0.5 | −10.6273155 |
| O | 22 | O | TYR | 62 | (A) | H-donor | 2.95 | −0.9 | ||
| O | 16 | CH2 | TRP | 58 | (A) | H-acceptor | 3.73 | −0.1 | ||
| −6 | ring | CG2 | THR | 163 | (A) | pi-H | 3.86 | −0.3 | ||
| 8 | O | 17 | OD1 | ASP | 197 | (A) | H-donor | 3.02 | −3 | −11.8703232 |
| O | 39 | OD1 | ASP | 356 | (A) | H-donor | 3.32 | −0.3 | ||
| O | 16 | NH2 | ARG | 195 | (A) | H-acceptor | 2.96 | −1.7 | ||
| O | 16 | N | ALA | 198 | (A) | H-acceptor | 3.38 | −0.1 | ||
| O | 19 | CD1 | LEU | 165 | (A) | H-acceptor | 3.46 | −0.1 | ||
| O | 36 | NE2 | GLN | 63 | (A) | H-acceptor | 3.02 | −1.3 | ||
| O | 41 | CH2 | TRP | 58 | (A) | H-acceptor | 3.68 | −0.1 | ||
| O | 16 | NH2 | ARG | 195 | (A) | ionic | 2.96 | −4.7 | ||
| −6 | ring | CG2 | ILE | 235 | (A) | pi-H | 4.07 | −0.1 | ||
| −6 | ring | CD2 | HIS | 305 | (A) | pi-H | 3.44 | −0.1 | ||
| −6 | ring | N | ALA | 307 | (A) | pi-H | 4.28 | −0.4 | ||
| −6 | ring | CB | ALA | 307 | (A) | pi-H | 4.29 | −0.3 | ||
| 9 | O | 33 | OD1 | ASP | 197 | (A) | H-donor | 3.05 | −2.2 | −9.00002575 |
| O | 33 | OD2 | ASP | 197 | (A) | H-donor | 3.11 | −1.4 | ||
| O | 40 | OD1 | ASP | 197 | (A) | H-donor | 2.83 | −2.6 | ||
| O | 19 | CB | TRP | 59 | (A) | H-acceptor | 3.24 | −0.1 | ||
| O | 33 | NE2 | HIS | 101 | (A) | H-acceptor | 3.23 | −0.7 | ||
| O | 40 | NH2 | ARG | 195 | (A) | H-acceptor | 3.07 | −0.2 | ||
| O | 40 | CB | ALA | 198 | (A) | H-acceptor | 3.5 | −0.1 | ||
| C | 9 | 5-ring | TRP | 59 | (A) | H-pi | 4.35 | −0.1 | ||
| 10 | O | 22 | OE1 | GLU | 240 | (A) | H-donor | 3.38 | −0.4 | − 10.8587704 |
| O | 22 | OE2 | GLU | 240 | (A) | H-donor | 3.17 | −0.2 | ||
| O | 40 | OD1 | ASP | 197 | (A) | H-donor | 3.13 | −2.5 | ||
| O | 22 | CG | LEU | 237 | (A) | H-acceptor | 3.48 | −0.1 | ||
| O | 22 | CD1 | LEU | 237 | (A) | H-acceptor | 3.47 | −0.1 | ||
| O | 39 | CG2 | ILE | 235 | (A) | H-acceptor | 3.78 | −0.1 | ||
| O | 39 | N | ALA | 307 | (A) | H-acceptor | 2.97 | −4.6 | ||
| −6 | ring | CB | ALA | 198 | (A) | pi-H | 4.15 | −0.5 | ||
| 11 | O | 62 | OD2 | ASP | 197 | (A) | H-donor | 3.12 | −1.3 | −10.0222139 |
| O | 19 | CG2 | ILE | 235 | (A) | H-acceptor | 3.47 | −0.1 | ||
| C | 55 | 5-ring | HIS | 101 | (A) | H-pi | 4.64 | −0.5 | ||
| C | 60 | 6-ring | TRP | 58 | (A) | H-pi | 4.77 | −0.1 | ||
| O | 62 | 5-ring | HIS | 101 | (A) | H-pi | 4.84 | −0.1 | ||
| −6 | ring | CB | TYR | 62 | (A) | pi-H | 3.74 | −0.3 | ||
| 12 | O | 41 | O | THR | 163 | (A) | H-donor | 2.98 | −1.1 | −7.80944586 |
| 13 | O | 23 | NE2 | GLN | 63 | (A) | H-acceptor | 3 | −2.5 | −9.71675777 |
| O | 45 | NE2 | HIS | 299 | (A) | H-acceptor | 2.97 | −6.2 | ||
| O | 44 | NH2 | ARG | 195 | (A) | ionic | 3.99 | −0.5 | ||
| O | 45 | NH1 | ARG | 195 | (A) | ionic | 3.92 | −0.7 | ||
| O | 45 | NH2 | ARG | 195 | (A) | ionic | 2.95 | −4.8 | ||
| 14 | O | 25 | OE1 | GLU | 233 | (A) | H-donor | 2.89 | −2.8 | −11.215291 |
| O | 68 | OD1 | ASP | 356 | (A) | H-donor | 3.05 | −4 | ||
| O | 53 | NE2 | GLN | 63 | (A) | H-acceptor | 2.98 | −2.3 | ||
| 15 | O | 22 | OD1 | ASP | 197 | (A) | H-donor | 2.93 | −2.3 | −14.5967274 |
| O | 20 | CG | LEU | 162 | (A) | H-acceptor | 3.69 | −0.1 | ||
| O | 21 | NH2 | ARG | 195 | (A) | H-acceptor | 3.11 | −1.5 | ||
| O | 21 | NE2 | HIS | 299 | (A) | H-acceptor | 3.61 | −1 | ||
| O | 21 | NH2 | ARG | 195 | (A) | ionic | 3.11 | −3.8 | ||
| −6 | ring | CB | ALA | 198 | (A) | pi-H | 4.48 | −0.2 | ||
| −6 | ring | CG2 | ILE | 235 | (A) | pi-H | 4.56 | −0.3 | ||
| −6 | ring | CD1 | ILE | 235 | (A) | pi-H | 3.76 | −0.1 | ||
| 16 | C | 7 | OD1 | ASP | 300 | (A) | H-donor | 3.59 | −0.1 | −11.8797693 |
| C | 7 | OD2 | ASP | 300 | (A) | H-donor | 3.53 | −0.1 | ||
| O | 24 | OE1 | GLU | 233 | (A) | H-donor | 3.73 | −0.1 | ||
| O | 23 | NH2 | ARG | 195 | (A) | H-acceptor | 2.94 | −1.7 | ||
| O | 23 | NE2 | HIS | 299 | (A) | H-acceptor | 3.15 | −2.3 | ||
| O | 31 | CB | TYR | 62 | (A) | H-acceptor | 3.45 | −0.1 | ||
| O | 23 | NH1 | ARG | 195 | (A) | ionic | 3.79 | −1 | ||
| O | 23 | NH2 | ARG | 195 | (A) | ionic | 2.94 | −4.9 | ||
| C | 11 | 6-ring | TRP | 58 | (A) | H-pi | 4.83 | −0.4 | ||
| C | 14 | 5-ring | TRP | 59 | (A) | H-pi | 4.38 | −0.2 | ||
| O | 27 | 5-ring | TRP | 59 | (A) | H-pi | 3.66 | −2.4 | ||
| −6 | ring | NE2 | GLN | 63 | (A) | pi-H | 3.67 | −0.1 | ||
| −6 | ring | CD1 | LEU | 165 | (A) | pi-H | 4.89 | −0.3 | ||
| 17 | O | 24 | O | TYR | 62 | (A) | H-donor | 3.05 | −2.1 | −10.7860909 |
| C | 33 | O | HIS | 305 | (A) | H-donor | 3.51 | −0.2 | ||
| C | 39 | O | HIS | 305 | (A) | H-donor | 3.49 | −0.2 | ||
| O | 26 | NE2 | GLN | 63 | (A) | H-acceptor | 2.98 | −5.6 | ||
| O | 26 | CD1 | LEU | 165 | (A) | H-acceptor | 3.97 | −0.1 | ||
| −6 | ring | CB | TYR | 62 | (A) | pi-H | 4.83 | −0.1 | ||
| −6 | ring | CD1 | LEU | 165 | (A) | pi-H | 4 | −0.2 | ||
| −6 | ring | CG2 | ILE | 235 | (A) | pi-H | 4.31 | −0.1 | ||
| 18 | O | 27 | O | TYR | 62 | (A) | H-donor | 3.06 | −1 | −7.62582397 |
| 19 | C | 18 | O | HIS | 305 | (A) | H-donor | 3.65 | −0.2 | −11.0418396 |
| O | 25 | NE2 | HIS | 305 | (A) | H-acceptor | 3.04 | −2.1 | ||
| C | 29 | 5-ring | HIS | 305 | (A) | H-pi | 4.89 | −0.1 | ||
| − 6 | ring | CH2 | TRP | 58 | (A) | pi-H | 4.87 | −0.1 | ||
| −6 | ring | CA | GLY | 306 | (A) | pi-H | 3.75 | −0.1 | ||
| −6 | ring | CB | ALA | 307 | (A) | pi-H | 3.65 | −0.4 | ||
| 20 | O | 53 | OD2 | ASP | 356 | (A) | H-donor | 3.19 | −1.3 | −12.1673326 |
| O | 24 | NE2 | HIS | 305 | (A) | H-acceptor | 3.04 | −1.9 | ||
| O | 43 | CD1 | LEU | 165 | (A) | H-acceptor | 3.94 | −0.1 | ||
| O | 45 | NE2 | GLN | 63 | (A) | H-acceptor | 3.07 | −0.5 | ||
| C | 40 | 5-ring | TRP | 59 | (A) | H-pi | 4.1 | −0.1 | ||
| O | 45 | 5-ring | TRP | 59 | (A) | H-pi | 3.98 | −0.5 | ||
| O | 45 | 6-ring | TRP | 59 | (A) | H-pi | 4.8 | −0.2 | ||
| 21 | C | 1 | OD1 | ASP | 197 | (A) | H-donor | 3.25 | −0.1 | −12.8208132 |
| O | 34 | OD1 | ASP | 197 | (A) | H-donor | 2.92 | −3.6 | ||
| O | 42 | OH | TYR | 151 | (A) | H-acceptor | 2.9 | −1.1 | ||
| O | 42 | CD1 | LEU | 162 | (A) | H-acceptor | 3.77 | −0.1 | ||
| O | 44 | CA | GLY | 306 | (A) | H-acceptor | 3.39 | −0.1 | ||
| O | 44 | N | ALA | 307 | (A) | H-acceptor | 2.96 | −2.2 | ||
| −6 | ring | CB | ALA | 198 | (A) | pi-H | 4.1 | −0.8 | ||
| 22 | C | 28 | OD1 | ASP | 300 | (A) | H-donor | 3.13 | −0.5 | − 10.3951654 |
| O | 36 | O | TYR | 62 | (A) | H-donor | 2.83 | −1.5 | ||
| O | 52 | OD1 | ASP | 300 | (A) | H-donor | 3.36 | −0.1 | ||
| O | 31 | CZ3 | TRP | 58 | (A) | H-acceptor | 3.55 | −0.1 | ||
| 23 | O | 23 | OD1 | ASP | 197 | (A) | H-donor | 2.9 | −0.5 | −13.9205046 |
| C | 60 | OD2 | ASP | 300 | (A) | H-donor | 3.45 | −0.2 | ||
| O | 46 | NE2 | HIS | 305 | (A) | H-acceptor | 3.44 | −0.6 | ||
| O | 50 | NE2 | HIS | 305 | (A) | H-acceptor | 3.2 | −0.5 | ||
| O | 52 | CD2 | LEU | 165 | (A) | H-acceptor | 3.7 | −0.1 | ||
| −6 | ring | CB | ALA | 198 | (A) | pi-H | 4.43 | −0.2 | ||
| −6 | ring | CB | ALA | 307 | (A) | pi-H | 4.85 | −0.1 | ||
| 24 | C | 2 | OD2 | ASP | 197 | (A) | H-donor | 3.47 | −0.5 | −15.0375738 |
| O | 26 | OD1 | ASP | 300 | (A) | H-donor | 2.55 | −3 | ||
| O | 28 | OD1 | ASP | 197 | (A) | H-donor | 2.5 | −3.6 | ||
| O | 30 | O | TYR | 62 | (A) | H-donor | 2.64 | −2.2 | ||
| O | 47 | OD1 | ASP | 356 | (A) | H-donor | 2.64 | −1.8 | ||
| O | 49 | OD1 | ASP | 356 | (A) | H-donor | 3.36 | −0.3 | ||
| O | 73 | O | THR | 163 | (A) | H-donor | 2.72 | −2.2 | ||
| O | 28 | NH2 | ARG | 195 | (A) | H-acceptor | 3.08 | −0.1 | ||
| O | 47 | NE2 | HIS | 305 | (A) | H-acceptor | 3 | −2.9 | ||
| O | 73 | CG | LEU | 165 | (A) | H-acceptor | 3.68 | −0.1 | ||
| O | 73 | CD2 | LEU | 165 | (A) | H-acceptor | 3.5 | −0.1 | ||
| C | 33 | 5-ring | TRP | 59 | (A) | H-pi | 3.59 | −1 | ||
| C | 38 | 5-ring | TRP | 59 | (A) | H-pi | 4.09 | −0.2 | ||
| C | 44 | 6-ring | TRP | 59 | (A) | H-pi | 3.86 | −0.2 | ||
| −6 | ring | CB | TYR | 62 | (A) | pi-H | 3.68 | −0.1 | ||
Nutritional studies
In preliminary nutritional analysis of crude powder, 17.38% proteins contents, 15.70% moisture content, 10.73% ash content and 4.18% fat content as summarized in Table 5. These presence of these food contents signify the nutritional potentials of the plant. The plant is used as tea decoction in some countries and is used as salad. Nutritional finding suggests that the plant might be a useful source for the dietary management of proteins and fats. Further, due to the moisture contents the powder materials may need proper storage to avoid fungi growth and deterioration [72].
Table 5.
Nutritional contents of P. hydropiper crude powder
| S. No | Proteins % Contents | |||||
| Weight of sample | Vol. of titer | Bulk | Titer - bulk | N% | Protein % | |
| 1 | 0.552 | 23.8 | 3.3 | 20.5 | 2.859601 | 17.87251 |
| 2 | 0.6028 | 25 | 3.3 | 21.7 | 2.771898 | 17.32436 |
| 3 | 0.6042 | 24.6 | 3.3 | 21.3 | 2.714499 | 16.96562 |
| % Moisture Contents | ||||||
| S. No | Empty Dish weight | Sample + Dish | Sample weight | After heating | Moisture weight | % Moisture |
| 1 | 16.3058 | 18.24 | 1.9342 | 17.9325 | 0.3075 | 15.89805 |
| 2 | 16.3003 | 18.4246 | 2.1243 | 18.0682 | 0.3564 | 16.77729 |
| 3 | 14.6234 | 15.841 | 1.2176 | 15.6652 | 0.1758 | 14.43824 |
| % Ash Contents | ||||||
| S. No | Empty dish wt | Sample + dish wt. | Sample wt. | Wt. after heating | Ash wt. | % ash |
| 1 | 23.1155 | 24.1033 | 0.9878 | 23.2244 | 0.1089 | 11.0245 |
| 2 | 20.9005 | 22.1234 | 1.2229 | 21.0294 | 0.1289 | 10.54052 |
| 3 | 29.3161 | 31.2188 | 1.9027 | 29.5186 | 0.2025 | 10.64277 |
| % Fat Contents | ||||||
| S. No | Sample weight | Empty bk. wt | BK + Oil Wt. | Oil Wt. | % Fat | |
| 1 | 2.2468 | 29.6871 | 29.7832 | 0.0961 | 4.277194 | – |
| 2 | 2.6881 | 22.654 | 22.7482 | 0.0942 | 3.504334 | – |
| 3 | 1.9867 | 28.3641 | 28.4588 | 0.0947 | 4.766699 | – |
Toxicological assessments
Acute toxicity studies reveled no lethality in animal groups as well as no abnormal behavioral changes in animals up to 24 h of samples administration. In this study Ph.Cr and Ph.Hex were found most effective against different blood groups. Haemagglutination activity of Ph.Cr was most prominent (+++) against AB+, AB−, O+ and O− blood groups. Ph.Hex was highly effective against A+, A− and B+ blood groups at 1:1 concentration (Table 6).
Table 6.
Result of hemagglutination effect of P. hydropiper extracts and saponins on different blood groups at different concentrations
| Blood groups | Ph.Cr | Ph.Hex | Ph.Chf | Ph.EtAc | Ph.Bt | Ph.Aq | ||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| 1:1 | 1:2 | 1:4 | 1:1 | 1:2 | 1:4 | 1:1 | 1:2 | 1:4 | 1:1 | 1:2 | 1:4 | 1:1 | 1:2 | 1:4 | 1:1 | 1:2 | 1:4 | |
| A+ | – | – | – | +++ | ++ | + | – | – | – | – | – | – | – | – | – | – | – | – |
| A- | ++ | + | + | +++ | ++ | ++ | – | – | – | + | + | – | – | – | – | + | + | + |
| B+ | – | – | – | +++ | +++ | +++ | – | – | – | – | – | – | – | – | – | + | + | + |
| B- | – | – | – | – | – | – | – | – | – | – | – | – | – | – | – | – | – | – |
| AB+ | +++ | ++ | ++ | +++ | ++ | ++ | – | – | – | ++ | + | – | +++ | ++ | ++ | – | – | – |
| AB- | +++ | ++ | ++ | +++ | ++ | ++ | ++ | ++ | + | ++ | + | + | +++ | ++ | ++ | – | – | – |
| O+ | +++ | ++ | ++ | +++ | ++ | ++ | – | – | – | – | – | – | +++ | ++ | ++ | – | + | + |
| O- | +++ | ++ | + | – | – | – | – | – | – | – | – | – | – | – | – | – | – | – |
+++: High hemagglutination activity, ++: Intermediate activity, +: Low activity and - = No activity
Plant agglutinins, also called phytohemagglutinins, cause haemagglutination of human and animal erythrocytes (RBCs). These phyto-hemagglutinins/phytolectins have wide range of applications as research tools in diverse biological activities like mitogenic action, cancer chemotherapy and cell membrane structure analysis [73]. These are also utilized as a drug targets, separation and characterization of glycoconjugates, glycopeptides, in histochemistry and cell differentiations techniques [74, 75]. Traditionally P. hydropiper is used in bleeding disorders and to repair ruptured blood vessels [76].
Conclusions
This study revealed that P. hydropiper, exhibit considerable amount of important secondary metabolites which might contribute to the α-glucosidase and α-amylase inhibition potentials of the plant. The same was confirmed by molecular simulation studies performed on identified compounds against these enzymes. Plant has significant proteins, fat contents, could be a good source of important valuable plant lectins which justify its ethnomedicinal uses in bleeding disorders and is safe at the test concentrations in animals. Further in-vivo anti-diabetic studies are required for potential uses of the plant in type-2 diabetes.
Acknowledgments
The authors would like to express their gratitude to the Deanship of Scientific Research, Najran University for their financial and technical support under code number [NU/-/MRC/10/357].
Authors’ contributions
MA, AS, FU, AN performed lab work analyzed data and collected relevant literature AZ performed HPLC-DAD analysis. MG performed Molecular docking studies. MHM, YSA, BAA, AOA helped in study design, drafted and refined the manuscript for publication. SAA helped in revising the manuscript, improved scientific presentation and quality of revised paper. All authors read and approved the final manuscript for publication.
Funding
Not available.
Availability of data and materials
Data related to the current paper can be provided upon request to the corresponding author.
Declarations
Ethics approval and consent to participate
Our study was evaluated and approved by Departmental Research Ethics Committee, Department of Pharmacy, University of Malakand (DREC-Pharmacy) via reference no DREC/2016052/01. Animals studies were performed in accordance with the ARRIVE guideline and following rules of Institute of Laboratory Animal Resources Commission on life sciences, National research council 1996. For the haemagglutination study, blood samples were collected from participants subsequent to “Written informed consent” obtained from them. And all procedures related to the use of human blood was in accordance with the relevant guidelines.
Consent for publication
Not Applicable.
Competing interests
Authors declare to have no conflict of interest in relation to the current paper.
Footnotes
Publisher’s Note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Contributor Information
Mater H. Mahnashi, Email: matermaha@gmail.com
Yahya S. Alqahtani, Email: yahyasalqahtani0@gmail.com
Bandar A. Alyami, Email: alyamibandar1@gmail.com
Ali O. Alqarni, Email: aoqarni@gmail.com
Sultan A. Alqahl, Email: sagohal@gmail.com
Farhat Ullah, Email: farhataziz80@hotmail.com.
Abdul Sadiq, Email: sadiquom@yahoo.com.
Alam Zeb, Email: azebuom@gmail.com.
Mehreen Ghufran, Email: mehreen@awkum.edu.pk.
Alexey Kuraev, Email: stat.stat2016@yandex.ru.
Asif Nawaz, Email: asifnawaz2446@gmail.com.
Muhammad Ayaz, Email: ayazuop@gmail.com.
References
- 1.Arky R. Clinical correlates of metabolic derangements of diabetes mellitus. Philadelphia: Complications of diabetes mellitus WB Saunders; 1982. pp. 16–20. [Google Scholar]
- 2.Booth G, Lipscombe L, Butalia S, Dasgupta K, Eurich D, Goldenberg R, Khan N, MacCallum L, Shah B, Simpson S. Pharmacologic management of type 2 diabetes: 2016 interim update. Can J Diabetes. 2016;40(6):484–486. doi: 10.1016/j.jcjd.2016.09.003. [DOI] [PubMed] [Google Scholar]
- 3.Ahmed F, Urooj A. Antihyperglycemic activity of Ficus glomerata stem bark in streptozotocin-induced diabetic rats. Glob J Pharmacol. 2008;2(3):41–45. [Google Scholar]
- 4.Kumar A, Ilavarasan R, Jayach T, Deecaraman M, Aravindan P, Padmanabhan N, Krishan M. Anti-diabetic activity of Syzygium cumini and its isolated compound against streptozotocin-induced diabetic rats. J Med Plant Res. 2013;2(9):246–249. [Google Scholar]
- 5.Cho N, Shaw J, Karuranga S, Huang Y, da Rocha Fernandes J, Ohlrogge A, Malanda B. IDF diabetes atlas: global estimates of diabetes prevalence for 2017 and projections for 2045. Diabetes Res Clin Pract. 2018;138:271–281. doi: 10.1016/j.diabres.2018.02.023. [DOI] [PubMed] [Google Scholar]
- 6.Löe H. Periodontal disease: the sixth complication of diabetes mellitus. Diabetes Care. 1993;16(1):329–334. [PubMed] [Google Scholar]
- 7.Ramachandran S, Asokkumar K, Uma Maheswari M, Ravi T, Sivashanmugam A, Saravanan S, Rajasekaran A, Dharman J. Investigation of antidiabetic, antihyperlipidemic, and in vivo antioxidant properties of Sphaeranthus indicus Linn. in type 1 diabetic rats: an identification of possible biomarkers. Evid Based Complement Alternat Med. 2010;2011:571721. doi: 10.1155/2011/571721. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Del Prete GF, Betterle C, Padovan D, Erle G, Toffolo A, Bersahi G. Incidence and significance of islet-cell autoantibodies in different types of diabetes mellitus. Diabetes. 1977;26(10):909–915. doi: 10.2337/diab.26.10.909. [DOI] [PubMed] [Google Scholar]
- 9.Bottini N, Vang T, Cucca F, Mustelin T. Role of PTPN22 in type 1 diabetes and other autoimmune diseases. Semin Immunol. 2006;18:207–213. doi: 10.1016/j.smim.2006.03.008. [DOI] [PubMed] [Google Scholar]
- 10.Manam DVK, Murugesan S. Biogenic silver nanoparticles by Halymenia poryphyroides and its in vitro anti-diabetic efficacy. J Chem Pharm Res. 2013;5(12):1001–1008. [Google Scholar]
- 11.Ohlson L-O, Larsson B, Svärdsudd K, Welin L, Eriksson H, Wilhelmsen L, Björntorp P, Tibblin G. The influence of body fat distribution on the incidence of diabetes mellitus: 13.5 years of follow-up of the participants in the study of men born in 1913. Diabetes. 1985;34(10):1055–1058. doi: 10.2337/diab.34.10.1055. [DOI] [PubMed] [Google Scholar]
- 12.Gin H, Rigalleau V. Post-prandial hyperglycemia. post-prandial hyperglycemia and diabetes. Diabetes Metab. 2000;26(4):265–272. [PubMed] [Google Scholar]
- 13.Petrovska BB. Historical review of medicinal plants’ usage. Pharmacogn Rev. 2012;6(11):1. doi: 10.4103/0973-7847.95849. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Elkhawas YA, Elissawy AM, Elnaggar MS, Mostafa NM, Al-Sayed E, Bishr MM, Singab ANB, Salama OM. Chemical diversity in species belonging to soft coral genus Sacrophyton and its impact on biological activity: A review. Mar Drugs. 2020;18(1):41. doi: 10.3390/md18010041. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Todirascu-Ciornea E, El-Nashar HA, Mostafa NM, Eldahshan OA, Boiangiu RS, Dumitru G, Hritcu L, Singab ANB. Schinus terebinthifolius essential oil attenuates scopolamine-induced memory deficits via cholinergic modulation and antioxidant properties in a zebrafish model. Evid Based Complement Alternat Med. 2019;2019:5256781. doi: 10.1155/2019/5256781. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Bailey CJ, Day C. Traditional plant medicines as treatments for diabetes. Diabetes Care. 1989;12(8):553–564. doi: 10.2337/diacare.12.8.553. [DOI] [PubMed] [Google Scholar]
- 17.Reddy N, Anarthe SJ, Raghavendra N. In vitro antioxidant and antidiabetic activity of Asystasia gangetica (Chinese Violet) Linn.(Acanthaceae) Int J Res Pharmaceut Biomed Sci. 2010;1(2):72–75. [Google Scholar]
- 18.Shetty K. Phytochemicals: Biotechnology of phenolic phytochemicals for food preservatives and functional food applications. Wiley Encyclopedia Food Sci Technol. 1999;2:1901–1909. [Google Scholar]
- 19.Choudhury A, Maeda K, Murayama R, DiMagno EP. Character of a wheat amylase inhibitor preparation and effects on fasting human pancreaticobiliary secretions and hormones. Gastroenterology. 1996;111(5):1313–1320. doi: 10.1053/gast.1996.v111.pm8898646. [DOI] [PubMed] [Google Scholar]
- 20.Ayaz M, Ahmad I, Sadiq A, Ullah F, Ovais M, Khalil AT, Devkota HP. Persicaria hydropiper (L.) Delarbre: A review on traditional uses, bioactive chemical constituents and pharmacological and toxicological activities. J Ethnopharmacol. 2020;251:112516. doi: 10.1016/j.jep.2019.112516. [DOI] [PubMed] [Google Scholar]
- 21.Orbán-Gyapai O, Lajter I, Hohmann J, Jakab G, Vasas A. Xanthine oxidase inhibitory activity of extracts prepared from Polygonaceae species. Phytother Res. 2015;29(3):459–465. doi: 10.1002/ptr.5275. [DOI] [PubMed] [Google Scholar]
- 22.Ayaz M, Junaid M, Ahmed J, Ullah F, Sadiq A, Ahmad S, Imran M. Phenolic contents, antioxidant and anticholinesterase potentials of crude extract, subsequent fractions and crude saponins from Polygonum hydropiper L. BMC Complement Altern Med. 2014;14(1):1–9. doi: 10.1186/1472-6882-14-145. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Ayaz M, Junaid M, Ullah F, Subhan F, Sadiq A, Ali G, Ovais M, Shahid M, Ahmad A, Wadood A. Anti-Alzheimer’s Studies on β-Sitosterol Isolated from Polygonum hydropiper L. Front Pharmacol. 2017;8:697. doi: 10.3389/fphar.2017.00697. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Ayaz M, Ullah F, Sadiq A, Kim MO, Ali T. Natural products-based drugs: potential therapeutics against Alzheimer’s disease and other neurological disorders. Front Pharmacol. 2019;10:1417. doi: 10.3389/fphar.2019.01417. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Tong X, Li X, Ayaz M, Ullah F, Sadiq A, Ovais M, Shahid M, Khayrullin M, Hazrat A. Neuroprotective studies on Polygonum hydropiper L. essential oils using transgenic animal models. Front Pharmacol. 2020;11:580069. doi: 10.3389/fphar.2020.580069. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Ayaz M, Junaid M, Ullah F, Sadiq A, Khan MA, Ahmad W, Shah MR, Imran M, Ahmad S. Comparative chemical profiling, cholinesterase inhibitions and anti-radicals properties of essential oils from Polygonum hydropiper L: a preliminary anti-Alzheimer’s study. Lipids Health Dis. 2015;14(1):141. doi: 10.1186/s12944-015-0145-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Ayaz M, Junaid M, Ullah F, Sadiq A, Shahid M, Ahmad W, Ullah I, Ahmad A, Syed N-i-H. GC-MS analysis and gastroprotective evaluations of crude extracts, isolated saponins, and essential oil from Polygonum hydropiper L. Front Chem. 2017;5:58. doi: 10.3389/fchem.2017.00058. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Ayaz M, Junaid M, Ullah F, Sadiq A, Ovais M, Ahmad W, Zeb A. Chemical profiling, antimicrobial and insecticidal evaluations of Polygonum hydropiper L. BMC Complement Altern Med. 2016;16(1):1–14. doi: 10.1186/s12906-016-1491-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Ayaz M, Sadiq A, Wadood A, Junaid M, Ullah F, Khan NZ. Cytotoxicity and molecular docking studies on phytosterols isolated from Polygonum hydropiper L. Steroids. 2019;141:30–35. doi: 10.1016/j.steroids.2018.11.005. [DOI] [PubMed] [Google Scholar]
- 30.Ayaz M, Junaid M, Ullah F, Sadiq A, Subhan F, Khan MA, Ahmad W, Ali G, Imran M, Ahmad S. Molecularly characterized solvent extracts and saponins from Polygonum hydropiper L. show high anti-angiogenic, anti-tumor, brine shrimp, and fibroblast NIH/3T3 cell line cytotoxicity. Front Pharmacol. 2016;7:74. doi: 10.3389/fphar.2016.00074. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Mahnashi MH, Alqahtani YS, Alyami BA, Alqarni AO, Ullah F, Wadood A, Sadiq A, Shareef A, Ayaz M. Cytotoxicity, anti-angiogenic, anti-tumor and molecular docking studies on phytochemicals isolated from Polygonum hydropiper L. BMC Complement Med Ther. 2021;21(1):1–14. doi: 10.1186/s12906-021-03411-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Chua LS. A review on plant-based rutin extraction methods and its pharmacological activities. J Ethnopharmacol. 2013;150(3):805–817. doi: 10.1016/j.jep.2013.10.036. [DOI] [PubMed] [Google Scholar]
- 33.Oany AR, Hossain MU, Islam R, Emran A-A. A preliminary evaluation of cytotoxicity, antihyperglycemic and antinociceptive activity of Polygonum hydropiper L. ethanolic leaf extract. Clin Phytosci. 2017;2(1):1–6. [Google Scholar]
- 34.Konan AB, Datte JY, Yapo P. Nitric oxide pathway-mediated relaxant effect of aqueous sesame leaves extract (Sesamum radiatum Schum. & Thonn.) in the guinea-pig isolated aorta smooth muscle. BMC Complement Altern Med. 2008;8:23. doi: 10.1186/1472-6882-8-23. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Zeb A, Sadiq A, Ullah F, Ahmad S, Ayaz M. Investigations of anticholinesterase and antioxidant potentials of methanolic extract, subsequent fractions, crude saponins and flavonoids isolated from Isodon rugosus. Biol Res. 2014;47:76. doi: 10.1186/0717-6287-47-76. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Kamal Z, Ullah F, Ayaz M, Sadiq A, Ahmad S, Zeb A, Hussain A, Imran M. Anticholinesterse and antioxidant investigations of crude extracts, subsequent fractions, saponins and flavonoids of Atriplex laciniata L.: potential effectiveness in Alzheimers and other neurological disorders. Biol Res. 2015;48:1–11. doi: 10.1186/s40659-015-0011-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Ahmad S, Ullah F, Ayaz M, Sadiq A, Imran M. Antioxidant and anticholinesterase investigations of Rumex hastatus D. Don: potential effectiveness in oxidative stress and neurological disorders. Biol Res. 2015;48:1–8. doi: 10.1186/s40659-015-0010-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Zeb A, Sadiq A, Ullah F, Ahmad S, Ayaz M. Phytochemical and toxicological investigations of crude methanolic extracts, subsequent fractions and crude saponins of Isodon rugosus. Biol Res. 2014;47:57. doi: 10.1186/0717-6287-47-57. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Zeb A. A reversed phase HPLC-DAD method for the determination of phenolic compounds in plant leaves. Anal Methods. 2015;7(18):7753–7757. [Google Scholar]
- 40.Saleem U, Khalid S, Zaib S, Anwar F, Ahmad B, Ullah I, Zeb A, Ayaz M. Phytochemical analysis and wound healing studies on ethnomedicinally important plant Malva neglecta Wallr. J Ethnopharmacol. 2020;249:112401. doi: 10.1016/j.jep.2019.112401. [DOI] [PubMed] [Google Scholar]
- 41.Zohra T, Ovais M, Khalil AT, Qasim M, Ayaz M, Shinwari ZK, Ahmad S, Zahoor M. Bio-guided profiling and HPLC-DAD finger printing of Atriplex lasiantha Boiss. BMC Complement Altern Med. 2019;19(1):4. doi: 10.1186/s12906-018-2416-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Papetti A, Maietta M, Corana F, Marrubini G, Gazzani G. Polyphenolic profile of green/red spotted Italian Cichorium intybus salads by RP-HPLC-PDA-ESI-MSn. J Food Compos Anal. 2017;63:189–197. [Google Scholar]
- 43.Santos J, Oliveira M, Ibáñez E, Herrero M. Phenolic profile evolution of different ready-to-eat baby-leaf vegetables during storage. J Chromatogr A. 2014;1327:118–131. doi: 10.1016/j.chroma.2013.12.085. [DOI] [PubMed] [Google Scholar]
- 44.Weisz GM, Kammerer DR, Carle R. Identification and quantification of phenolic compounds from sunflower (Helianthus annuus L.) kernels and shells by HPLC-DAD/ESI-MSn. Food Chem. 2009;115(2):758–765. [Google Scholar]
- 45.McCue P, Kwon YI, Shetty K. Anti-amylase, anti-glucosidase and anti-angiotensin −1 converting enzyme potential of selected foods. J Food Biochem. 2005;29(3):278–294. [Google Scholar]
- 46.Bernfeld P, Colowick S. Methods in enzymology. New York: by SP Colowick and NO Kaplan, Academic Press Inc; 1955. p. 149. [Google Scholar]
- 47.Leach AR, Shoichet BK, Peishoff CE. Prediction of protein− ligand interactions. Docking and scoring: successes and gaps. J Med Chem. 2006;49(20):5851–5855. doi: 10.1021/jm060999m. [DOI] [PubMed] [Google Scholar]
- 48.Liu M, Zhang W, Wei J, Lin X. Synthesis and α-glucosidase inhibitory mechanisms of bis (2, 3-dibromo-4, 5-dihydroxybenzyl) ether, a potential marine bromophenol α-glucosidase inhibitor. Mar Drugs. 2011;9(9):1554–1565. doi: 10.3390/md9091554. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Ayaz M, Wadood A, Sadiq A, Ullah F, Anichkina O, Ghufran M. In-silico evaluations of the isolated phytosterols from polygonum hydropiper L against BACE1 and MAO drug targets. J Biomol Struct Dyn. 2021:1–9. [DOI] [PubMed]
- 50.Council NR . Institute of laboratory animal resources, commission on life sciences. 1996. p. 27. [Google Scholar]
- 51.Nascimento DK, Souza IA, Oliveira AFD, Barbosa MO, Santana MA, Pereira DF, Lira EC, Vieira JR. Phytochemical screening and acute toxicity of aqueous extract of leaves of Conocarpus erectus Linnaeus in swiss albino mice. An Acad Bras Cienc. 2016;88:1431–1437. doi: 10.1590/0001-3765201620150391. [DOI] [PubMed] [Google Scholar]
- 52.Naqvi SBS, Sheikh D, Usmanghani K, Shameel M, Sheikh R. Screening of marine algae of Karachi for haemagglutinin activity. Pak J Pharm Sci. 1992;5(2):129–138. [PubMed] [Google Scholar]
- 53.Li F, Zhang B, Chen G, Fu X. The novel contributors of anti-diabetic potential in mulberry polyphenols revealed by UHPLC-HR-ESI-TOF-MS/MS. Food Res Int. 2017;100:873–884. doi: 10.1016/j.foodres.2017.06.052. [DOI] [PubMed] [Google Scholar]
- 54.Eid MH, Haddad SP. The antidiabetic potential of quercetin: underlying mechanisms. Curr Med Chem. 2017;24(4):355–364. doi: 10.2174/0929867323666160909153707. [DOI] [PubMed] [Google Scholar]
- 55.Derosa G, Maffioli P, Sahebkar A. Ellagic acid and its role in chronic diseases. Adv Exp Med Biol. 2016;928:473–479. doi: 10.1007/978-3-319-41334-1_20. [DOI] [PubMed] [Google Scholar]
- 56.Variya BC, Bakrania AK, Patel SS. Antidiabetic potential of gallic acid from Emblica officinalis: improved glucose transporters and insulin sensitivity through PPAR-γ and Akt signaling. Phytomedicine. 2020;73:152906. doi: 10.1016/j.phymed.2019.152906. [DOI] [PubMed] [Google Scholar]
- 57.Xu W, Luo Q, Wen X, Xiao M, Mei Q. Antioxidant and anti-diabetic effects of caffeic acid in a rat model of diabetes. Trop J Pharm Res. 2020;19(6):1227–1232. [Google Scholar]
- 58.Cásedas G, Les F, González-Burgos E, Gómez-Serranillos MP, Smith C, López V. Cyanidin-3-O-glucoside inhibits different enzymes involved in central nervous system pathologies and type-2 diabetes. S Afr J Bot. 2019;120:241–246. [Google Scholar]
- 59.Gonçalves AC, Bento C, Silva BM, Silva LR. Sweet cherries from Fundão possess antidiabetic potential and protect human erythrocytes against oxidative damage. Food Res Int. 2017;95:91–100. doi: 10.1016/j.foodres.2017.02.023. [DOI] [PubMed] [Google Scholar]
- 60.Jesus F, Gonçalves AC, Alves G, Silva LR. Exploring the phenolic profile, antioxidant, antidiabetic and anti-hemolytic potential of Prunus avium vegetal parts. Food Res Int. 2019;116:600–610. doi: 10.1016/j.foodres.2018.08.079. [DOI] [PubMed] [Google Scholar]
- 61.Stojkovic D, Smiljkovic M, Ciric A, Glamoclija J, Van Griensven L, Ferreira IC, Sokovic M. An insight into antidiabetic properties of six medicinal and edible mushrooms: Inhibition of α-amylase and α-glucosidase linked to type-2 diabetes. S Afr J Bot. 2019;120:100–103. [Google Scholar]
- 62.Ullah I, Subhan F, Alam J, Shahid M, Ayaz M. Suppression of cisplatin-induced vomiting by Cannabis sativa in pigeons: neurochemical evidences. Front Pharmacol. 2018;9:231. doi: 10.3389/fphar.2018.00231. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Sadiq A, Rashid U, Ahmad S, Zahoor M, AlAjmi MF, Ullah R, Noman OM, Ullah F, Ayaz M, Khan I. Treating hyperglycemia from Eryngium caeruleum M. Bieb: in-vitro α-glucosidase, antioxidant, in-vivo antidiabetic and molecular docking-based approaches. Front Chem. 2020;8:1064. doi: 10.3389/fchem.2020.558641. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Truscheit E, Frommer W, Junge B, Müller L, Schmidt DD, Wingender W. Chemistry and biochemistry of microbial α-glucosidase inhibitors. Angew Chem Int Ed Eng. 1981;20(9):744–761. [Google Scholar]
- 65.Wehmeier UF, Piepersberg W. Biotechnology and molecular biology of the α-glucosidase inhibitor acarbose. Appl Microbiol Biotechnol. 2004;63(6):613–625. doi: 10.1007/s00253-003-1477-2. [DOI] [PubMed] [Google Scholar]
- 66.Inouye S, Tsuruoka T, Ito T, Niida T. Structure and synthesis of nojirimycin. Tetrahedron. 1968;24(5):2125–2144. doi: 10.1016/0040-4020(68)88115-3. [DOI] [PubMed] [Google Scholar]
- 67.van de Laar FA. Alpha-glucosidase inhibitors in the early treatment of type 2 diabetes. Vasc Health Risk Manag. 2008;4(6):1189. doi: 10.2147/vhrm.s3119. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Suzuki Y, Sano M, Hayashida K, Ohsawa I, Ohta S, Fukuda K. Are the effects of α-glucosidase inhibitors on cardiovascular events related to elevated levels of hydrogen gas in the gastrointestinal tract? FEBS Lett. 2009;583(13):2157–2159. doi: 10.1016/j.febslet.2009.05.052. [DOI] [PubMed] [Google Scholar]
- 69.Coniff RF, Shapiro JA, Robbins D, Kleinfield R, Seaton TB, Beisswenger P, McGill JB. Reduction of glycosylated hemoglobin and postprandial hyperglycemia by acarbose in patients with NIDDM: a placebo-controlled dose-comparison study. Diabetes Care. 1995;18(6):817–824. doi: 10.2337/diacare.18.6.817. [DOI] [PubMed] [Google Scholar]
- 70.Matsuda H, Nishida N, Yoshikawa M. Antidiabetic principles of natural medicines. V. Aldose reductase inhibitors from Myrcia multiflora DC.(2): structures of myrciacitrins III, IV, and V. Chem Pharm Bull. 2002;50(3):429–431. doi: 10.1248/cpb.50.429. [DOI] [PubMed] [Google Scholar]
- 71.Kumar S, Narwal S, Kumar V, Prakash O. α-Glucosidase inhibitors from plants: a natural approach to treat diabetes. Pharmacogn Rev. 2011;5(9):19. doi: 10.4103/0973-7847.79096. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Ahmad S, Ullah F, Ayaz M, Ahmad A, Sadiq A, Mohani SN-U-H. Nutritional and medicinal aspects of Rumex hastatus D. Don along with in vitro anti-diabetic activity. Int J Food Prop. 2019;22(1):1733–1748. [Google Scholar]
- 73.Lis H, Sharon N. The biochemistry of plant lectins (phytohemagglutinins) Annu Rev Biochem. 1973;42(1):541–574. doi: 10.1146/annurev.bi.42.070173.002545. [DOI] [PubMed] [Google Scholar]
- 74.Rüdiger H, Gabius H-J. Plant lectins: occurrence, biochemistry, functions and applications. Glycoconj J. 2001;18(8):589–613. doi: 10.1023/a:1020687518999. [DOI] [PubMed] [Google Scholar]
- 75.Liener I. The lectins: properties, functions, and applications in biology and medicine. Orlando: Elsevier; ACADEMI C PRESS, INC.; 1986.
- 76.Ayaz M, Junaid M, Ahmed J, Ullah F, Sadiq A, Ahmad S, et al. Phenolic contents, antioxidant and anticholinesterase potentials of crude extract, subsequent fractions and crude saponins from Polygonum hydropiper L. BMC Complement Altern Med. 2014. 10.1186/1472-6882-14-145. [DOI] [PMC free article] [PubMed]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Data Availability Statement
Data related to the current paper can be provided upon request to the corresponding author.