Abstract
Modifications to the constituents of the gut microbiome influence bone density and tissue-level strength, but the specific microbial components that influence tissue-level strength in bone are not known. Here we selectively modify constituents of the gut microbiota using narrow spectrum antibiotics to identify components of the microbiome associated with changes in bone mechanical and material properties. Male C57BL/6J mice (4 weeks) were divided into seven groups (n=7–10/group) and had taxa within the gut microbiome removed through dosing with: 1) ampicillin; 2) neomycin; 3) vancomycin; 4) metronidazole; 5) a cocktail of all four antibiotics together (with zero-calorie sweetener to ensure intake); 6) zero-calorie sweetener only or; 7) no additive (untreated) for 12 weeks. Individual antibiotics remove only some taxa from the gut while the cocktail of all four removes almost all microbes. After accounting for differences in geometry, whole bone strength was reduced in animals with gut microbiome modified by neomycin (−28%, p=0.002) and was increased in the group in which the gut microbiome was altered by sweetener alone (+ 39%, p < 0.001). Analysis of the fecal microbiota detected seven lower ranked taxa differentially abundant in animals with impaired tissue-level strength and 14 differentially abundant taxa associated with increased tissue-level strength. Histological and serum markers of bone turnover and trabecular BV/TV did not differ among groups. These findings demonstrate that modifications to the taxonomic components of the gut microbiome have the potential to decrease or increase tissue-level strength of bone independent of bone quantity and without noticeable changes in bone turnover.
Introduction
Whole bone strength and risk of fragility fracture are influenced by bone quantity and bone quality. Bone quantity is assessed clinically through measures of bone mineral density. Most pharmaceutical interventions primarily address bone mineral density (1), but have an indirect and/or small effect on bone quality at the tissue level(2). To move the field forward and reduce fracture risk beyond what is currently possible, there is a need for new approaches to address bone tissue quality.
The microbiome has recently been identified as a factor that can influence bone quantity and bone quality(3–6). The gut microbiome is composed of the microbial organisms that inhabit the intestines and their genetic components. The gastrointestinal tract of an individual includes hundreds of distinct microbial species(7). The gut microbiota can influence the host by regulating nutrient and energy absorption, producing vitamins and other useful metabolic byproducts, and by stimulating the host immune system at the gut lining(5,8). The gut microbiome is an enticing target for therapies because microbiome-based therapies have the potential to be inexpensive, have few side effects, and the potential to provide long-term benefits from a single intervention (such as a fecal microbiota transplantation), thereby avoiding daily dosing/supplements(9).
Recent investigations in animal models have shown that the constituents of the gut microbiome influence bone mass and bone remodeling(10–16). In mice, the complete absence of the gut microbiota and/or decimation of the gut microbial population with broad-spectrum antibiotics alters bone mineral density and/or bone volume fraction, but the effect depends on animal age, sex, genotype, and duration of dysbiosis-inducing stimuli(3,10,11,13–16). Recently, we found that changing the constituents of the gut microbiota using chronic oral antibiotics (ampicillin+neomycin) in mice leads to impaired whole bone strength that could not be explained by alterations in bone geometry, indicating bone impaired strength of the bone matrix itself (3). The strength of the bone matrix is referred to here as “tissue strength” to differentiate it from “whole bone strength” (whole bone strength is determined by both bone geometry and tissue strength). The finding demonstrates that the gut microbiota can influence bone tissue quality. Subsequent metagenomic analysis of the gut microbiota-associated reductions in bone tissue strength with reductions in: a) the capacity of the gut microbiota to produce vitamin K; b) microbe-derived vitamin K in the gut, kidney and liver; and c) concentrations of the vitamin K dependent protein osteocalcin in bone matrix(4).
Our past studies showed that modifications to the microbiota can alter bone tissue quality, but there is limited information about which of the hundreds of distinct microbes within the gut microbiota influence bone tissue quality. While analysis of microbial sequence data can help to identify taxonomic and/or functional components of the gut microbiota associated with a phenotype(4), such studies are enhanced by experimental strategies that generate distinct gut microbial communities. One experimental approach is to selectively deplete (i.e. “knock out”) components of the gut microbiome using different narrow spectrum antibiotics to remove only some components of the gut microbiota and determine which of the modifications to the gut microbiome result in a phenotype of interest. Although selective removal of some components of the gut microbiota with narrow spectrum antibiotics has been used to isolate gut microbial communities that influence resistance to cancer cell growth(17), the approach has not yet been applied to identify pathways linking the microbiome to bone.
The long-term goal of the current research is to identify microbiome-based interventions that can enhance bone tissue quality and reduce fracture risk. In the present study, we selectively deplete components of the gut microbiota as a means of identifying microbial components associated with impaired bone tissue strength.
Methods
Study Design
Animal procedures were approved by the local Institutional Animal Care and Use Committee. C57BL/6J mice were acquired (Jackson Laboratory, Bar Harbor, ME, USA) and bred via homozygous mating in a conventional animal facility. Breeder animals were housed in plastic cages filled with ¼-inch corn cob bedding (The Andersons’ Lab Bedding, Maumee, OH, USA), provided standard laboratory chow (Teklad LM-485 Mouse/Rat Sterilizable Diet including 80 mg/kg of menadione as a source of vitamin K activity, Envigo Diets, Madison WI, USA) and water ad libitum, and provided a cardboard refuge environmental enrichment hut (Ketchum Manufacturing; Brockville, ON, Canada). Mice were housed with their dam until weaning at 3 weeks of age. At 4 weeks of age, male mice (n=64) were divided into seven groups (n=7–10/group). A sample size of n=7 was expected to detect differences in whole bone strength relative to section modulus observed in our prior work (3), although additional animals were available in some litters and were included in the study. Animals were housed in cages consisting of 3–5 animals taken from 2–3 different breeding cages. Bedding from breeding cages was mixed and placed in each new experimental cage. A distinct gut microbiota was induced in each of the groups using the following supplements in drinking water: ampicillin (Amp, n=8, 1g/L, MilliporeSigma, Burlington, MA, USA), neomycin (Neo, n=9, 1g/L, MilliporeSigma, Burlington, MA, USA), metronidazole (Metro, n=7, 1g/L, Covetrus, Portland, ME, USA), vancomycin (Vanco, n=7, 0.5 g/L, PhyloTech Labs, Lenexa, KS, USA), a cocktail of the four antibiotics in drinking water with zero-calorie sweetener (18)(n=7, ampicillin 1g/L, neomycin 1g/L, metronidazole 1g/L, vancomycin 0.5 g/L, sweetener 10g/L), zero-calorie sweetener only control (n=9, 10g/L) or untreated control (n=10, Supplementary Table S1). The cocktail of all four antibiotics is commonly used in microbiome research and depletes the gut microbial population (99.9% removal(19)) thereby mimicking the complete absence of a gut microbiome seen in germ-free mice(18). Under veterinary advice, artificial zero-calorie sweetener was included with the cocktail of antibiotics to ensure sufficient water intake by the animals and mask the bitterness of the combined antibiotics. The zero-calorie sweetener consisted of aspartame, dextrose with maltodextrin, and acesulfame potassium (Equal, Merisant Company, Chicago, IL, USA, 10 g/L). The individual antibiotics each primarily target distinct sub-populations of the bacterial community (Gram +, Gram -, anaerobes, etc., Supplementary Table S1). Hence, these interventions were selected to generate seven distinct gut microbiota and were not intended to be a comparison of different antibiotics and/or artificial sweetener. Treatment throughout growth simulates the effect of alterations of the microbiome on the development of the bone phenotype at skeletal maturity(20). At 16 weeks of age animals were fasted for four hours, and blood serum and fecal pellets were collected immediately before euthanasia. Epididymal fat pad, cecal contents, and colons were collected immediately after euthanasia.
Taxonomic Composition of the Fecal Microbiota
Fecal samples were collected at 16 weeks of age from two mice per cage (n=6/group, gut microbiota shows little variation within a cage). DNA was isolated from fecal samples using the DNeasy PowerFood Microbial Kit (QIAGEN, Hilden, Germany) according to the manufacturer’s instructions. DNA isolation and next-generation sequencing were conducted according to previously published methods(3,21,22). The core-metrics-phylogenetic method in QIIME2 was used to compute alpha diversity (Shannon Diversity, within sample) and beta diversity (UniFrac distance, between samples). Taxonomies were assigned using QIIME’s machine learning classifier trained on Greengenes sequences (version 13_8). Samples from the cocktail group were excluded from microbiota analysis because the cocktail dosing depletes the microbial population (95% reduction in population as measured by qPCR reads, Fig. S1). Sequences are publicly available through the Sequence Read Archive under the accession number PRJNA625754 (http://www.ncbi.nlm.nih.gov/Traces/sra, National Center for Biotechnology Information, Bethesda, Maryland, USA)(23).
Femoral Geometry of Mid-diaphyseal Cortical Bone
The right femora were harvested, wrapped in saline soaked gauze and stored in an airtight container at −20°C. Images of the femoral diaphyseal cross-section were obtained by micro-computed X-ray tomography with a voxel size of 25 μm (eXplore CT 120, GE, Fairfield, CT, USA; 80 kVp, 32 μA, 100 ms integration time). A Gaussian filter was used to remove noise and a global threshold (determined as the average of those selected by a user across all samples, 1220 HU) was used to segment mineralized tissue from surrounding nonmineralized tissue. Cross-sectional geometry of the mid-diaphyseal cortical bone was determined using a volume of interest extending 2.5% of total bone length(3) and centered midway between the greater trochanter and lateral condyle (BoneJ, version 1.3.3). The total area, cortical cross-sectional area, cortical thickness, marrow area, moment of inertia (I) about the medial-lateral axis (the direction of loading), distance from the neutral axis to the edge of the bone surface (c), and section modulus (I/c) were determined. Geometric measures are reported as original, unadjusted values (values adjusted for body weight are provided in the Supplementary Material(20)).
Mechanical Testing
The right femora were thawed to room temperature in saline and, while hydrated, tested in the anterior-to-posterior direction to failure in three-point bending at a rate of 0.1 mm/s using a span length of 6.25 mm between outer loading pins (858 Mini Bionix; MTS, Eden Prairie, MN, USA). Force and displacement measurements were gathered using a 45 kg (100 lbs.) load cell (SSM-100; Transducer Techniques, Temecula, CA, USA, accuracy confirmed through manual calibration each week) and a linear variable differential transducer at a 100-Hz sampling rate. Peak bending moment was determined using standard techniques(20). Additionally, the relationship between whole bone strength and section modulus was examined as an indicator of modifications in tissue-level strength, referred to here as tissue strength (such an approach avoids assumptions used when calculating ultimate stress using beam theory (24)).
Bone Histomorphometry, Blood Serum Analysis, and Metaphyseal Cancellous Bone Morphology
Intraperitoneal injections of the bone formation marker calcein (20mg calcein/ Kg body weight) were given to the mice at 3 and 11 days before euthanasia. The left femur was embedded undecalcified in methyl methacrylate, and sectioned using a diamond wafering saw (Buehler, Lake Bluff, Illinois). Images were collected using confocal microscopy with a 488nm argon laser line and a 0.6 pixels/micron resolution (LSM880 Confocal/Multiphoton Inverted Microscope (i880), Zeiss, Oberkochen, Germany). Mineralizing surface and mineral apposition rate of cortical bone on the mid-diaphysis of the femur were traced using ImageJ (version 1.52a, National Institute of Health, Bethesda, Maryland, USA) and calculated using standard approaches(25).
Whole blood was collected at euthanasia via cardiac puncture. Serum was isolated and sent to the Duke Molecular Physiology Institute Biomarkers Shared Resource for an ELISA analysis (RatLaps™ (CTX-I) EIA and Rat/Mouse PINP EIA, immunodiagnostic systems, East Boldon, UK). Serum was analyzed for cross linked c-telopeptide of type I collagen (CTX-I, a marker of bone resorption) and procollagen I N-terminal propeptide (PINP, a marker of bone formation).
The distal ends of the left femurs that were embedded in methyl methacrylate and imaged (n=6 per group) using microcomputed tomography (μCT35; Scanco Medical AG, Switzerland; 55 kVp, 145 mA, 600 ms integration time, 10 μm voxel size). Images were collected at the distal metaphysis of the femur and the trabecular bone segmented from the cortical shell manually. An average global threshold for all samples (3090 HU) was used to identify mineralized trabecular bone regions (26). Measurements of trabecular microarchitecture were made using BoneJ to determine bone volume fraction (BV/ TV), trabecular thickness (Tb.Th), trabecular number (Tb.N), and trabecular separation (Tb.Sp).
Fourier-transform Infrared (FTIR) Spectroscopy Analysis
FTIR spectroscopy was used to evaluate the compositional properties of the femoral mid-diaphysis of tissue from experimental groups that showed significantly altered tissue-level strength. After mechanical testing, the broken femora were homogenized into pellets for FTIR analysis as described previously(27). Briefly, bone tissue specimens were powdered by cryomilling, combined with a KBr in a 1:100 bone powder:KBr ratio, and pressed into pellets with a hydraulic press. Absorbance spectra were gathered via FTIR over the range 800–2000 cm−1. Spectra were analyzed using chemical imaging software (ISys, Malvern Panalytical Ltd, Malvern, UK), in which mineral and organic peak areas and intensities were calculated to quantify bone tissue composition. Five compositional outcomes were calculated 1) the mineral:matrix ratio (ν1- ν3 PO4 [916–1180 cm−1]: Amide I [1596–1712 cm−1]) a measure of the extent of mineralization of the collagen matrix(28), 2) the carbonate:phosphate ratio (ν2 CO3 [852–890 cm−1]: ν1- ν3 PO4 [916–1180 cm−1]) a measure of the extent of carbonate substitution into the mineral crystal lattice, 3) mineral crystallinity(XST) (peak intensity ratio 1030 cm−1: 1020 cm−1) a measure of bone mineral crystal size and stoichiometric perfection(29), 4) collagen maturity (XLR) (intensity ratio 1660 cm−1:1690 cm−1) a measure of the extent of enzymatic crosslinking in the secondary structure of collagen(30), and 5) mineral acid phosphate content (intensity ratio 1126 cm−1: 1096 cm−1) a measure of the extent of protonation of phosphate molecules in the mineral crystal lattice(31).
Associations between Gut Microbiota and Bone Phenotypes
The functional capacity of the microbial community was estimated from the 16S rRNA sequence results with the PICRUSt software (version 1.1.4, https://picrust.github.io/picrust/index.html)(32) using reference genomes to predict functional pathways for experimental groups that showed significantly altered tissue strength. In our previous study, vitamin K was linked to decreased tissue quality(4), and therefore PICRUSt results were analyzed using a biased pathway-focused approach focused on vitamin K biosynthesis pathways.
Cecal vitamin K content (phylloquinone and menaquinones 4–13 (MK4-13)) was measured and characterized (n=6/group, the same animals used for fecal microbiota analysis) using high-performance liquid chromatography-mass spectrometry with atmospheric pressure chemical ionization (LC-APCI-MS), previously described(33). The characterization system consisted of an Agilent 6130 Quadrupole MSD with an APCI source connected to an Agilent series 1260 HPLC instrument (Agilent Technologies, Santa Clara, CA, USA). The limit of detection for phylloquinone and each type of menaquinone were as followed: phylloquinone and MK4, 30 pmol/g; MK6, 10 pmol/g; MK5, MK7-MK9, MK11-13, 5 pmol/g; MK10, 1 pmol/g. For findings below the detection limit a value of one half the detection limit was used when making statistical comparisons.
Statistical Analysis
Homogenous variance was tested using the Brown-Forsythe test. If parametric assumptions were met, differences in the gut microbiome associated with bone, fat pad mass, body mass, bone geometry adjusted/unadjusted for body mass and serum concentration measurements was determined using a one-way ANOVA with group as the factor followed by Tukey post hoc for multiple comparisons (JMP Pro, v.9, 2013; SAS Institute Inc., Cary, NC, USA). If parametric assumptions were not met, a log transformation was applied to attain homogenous variance and normality. ANCOVA using the Tukey post hoc was applied to determine differences in the relationship between whole bone strength and section modulus (the covariate) among groups. ANCOVA of whole bone strength with section modulus as a covariate is more sensitive to differences between groups than ANOVA of ultimate stress(34). For Unless otherwise noted, statistical tests were performed with alpha= 0.05.
The linear discriminant analysis effect size (LEfSe) method was applied to taxonomic data to determine the features (taxonomic assignments ranging from phylum to genus) that differed in relative abundance between experimental groups with altered tissue strength and those that did not have an altered tissue strength(35). The LEfSe method is ideal for this approach because it allows for comparisons of the gut microbiota from animals displaying phenotypes (bone tissue strength) and in the event that multiple study groups show the same phenotype ensures that the detected differences are not caused by a single study group (referred to as classes in the LEfSe software). The software package STAMP (version 2.1.3, 2015,GNU General Public License, Halifax, Nova Scotia) was used to identify differences among groups in terms of microbial function determined from PICRUSt(36).
Results
Taxonomic Composition of the Fecal Microbiota
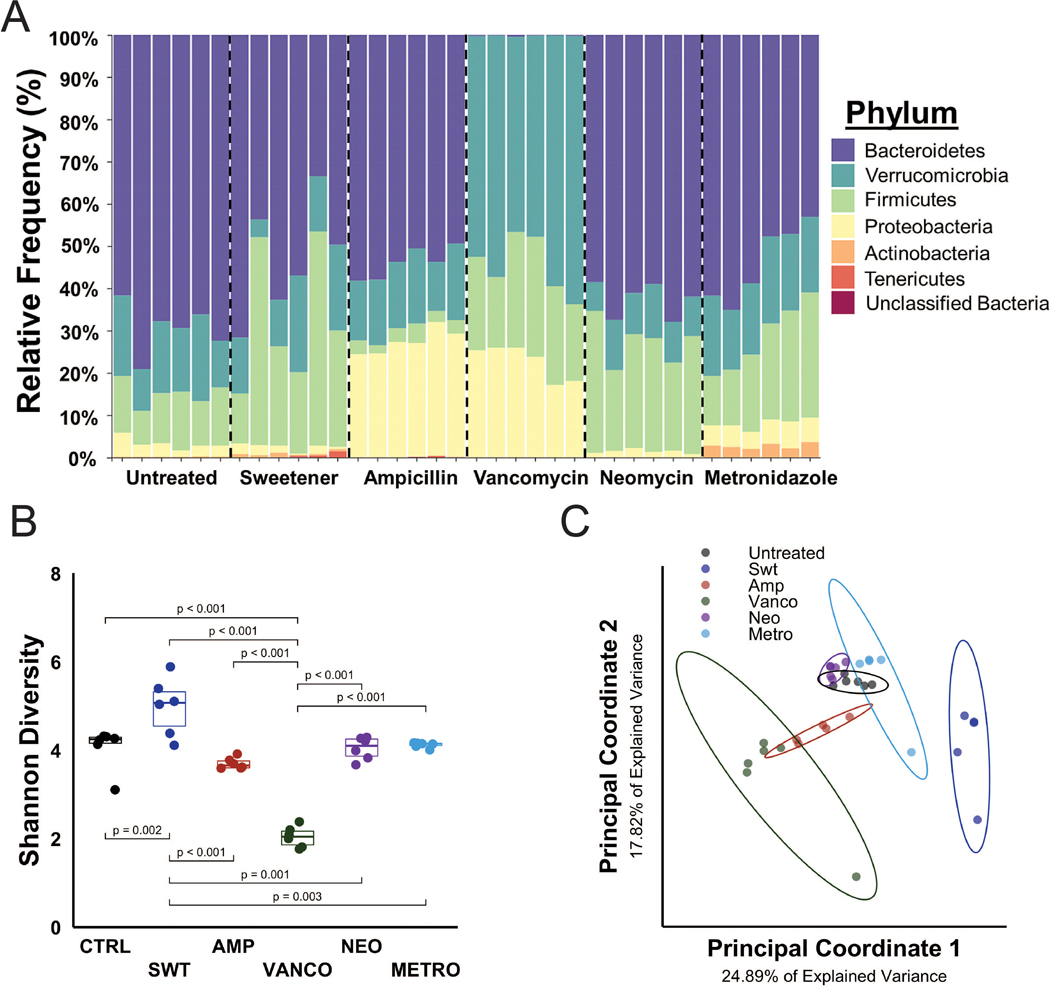
The composition of the gut microbiota differed substantially among groups, including in the animals given sweetener only (Fig. 1A). Shannon diversity, a quantitative measure of alpha diversity and community richness, was lower in animals with a microbiota altered by vancomycin (p<0.001) and greater in animals with microbiota altered by sweetener (p<0.001) (Fig. 1B). Unweighted UniFrac, a qualitative measure of beta diversity, was distinct in animals receiving vancomycin and sweetener, as determined by the overlap of the data clusters (Fig. 1C).
Figure 1:
The study included seven different gut microbiota (six different microbial communities and one cocktail group with a depleted gut microbiota). A) The relative abundance at the phylum level is shown. Each column represents the gut microbiota composition of a single mouse. B) Alpha diversity (Shannon diversity) was decreased in the vancomycin group and increased in the sweetener group. C) Beta diversity (unweighted UniFrac distance), represented in a principal coordinates plot, shows distinct beta diversity among groups. The ellipses represent 95% confidence.
Bone Geometry and Mechanical Properties
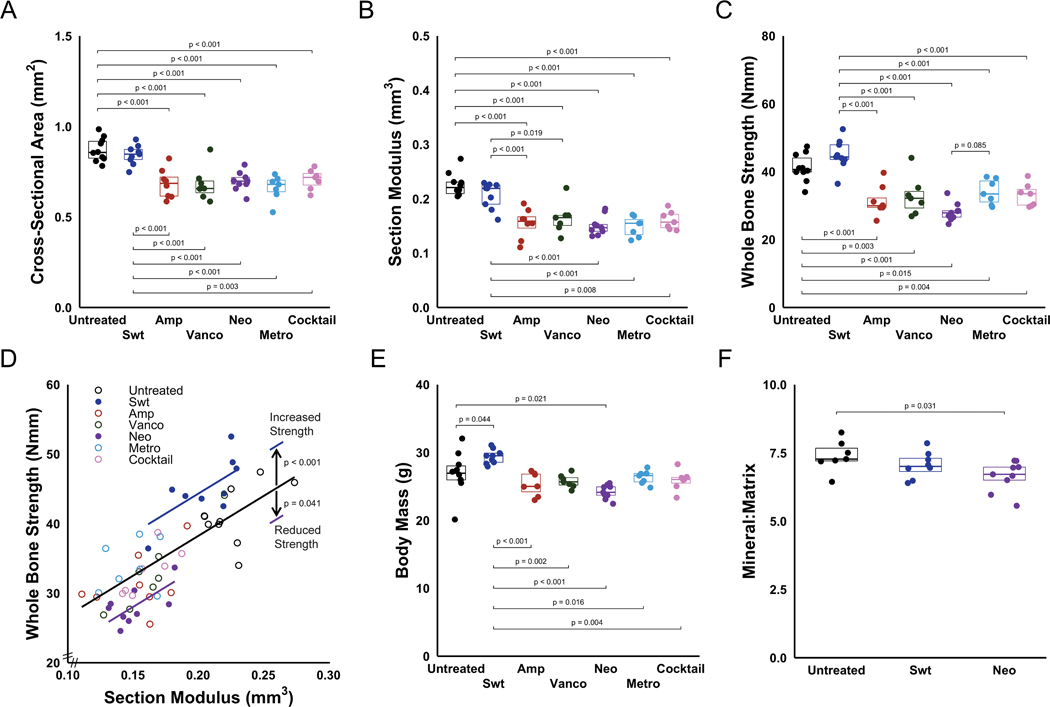
Removal of components of the gut microbiota decreased cross-sectional area (Fig. 2A), moment of inertia (Supplemental Fig. S2A), and section modulus (Fig. 2B) of the mid-diaphyseal cortical bone compared to untreated animals (p<0.001). Animals receiving sweetener alone did not show observable differences in cross sectional area, moment of inertia and section modulus from untreated animals.
Figure 2:
Removal of components of the gut microbiota using narrow spectrum antibiotics decreased (A) cross-sectional area and (B) section modulus. Mechanical and material properties of bone were influenced by neomycin dosing. (C) Removal of components of the gut microbiota using narrow spectrum antibiotics decreased whole bone strength. (D) A graphical representation of the data and ANCOVA analysis (lines) is shown. Alterations to the gut microbiota caused by dosing with neomycin led to bone strength lower than would be expected from geometry (ANCOVA, p < 0.001), indicating impaired tissue-level strength. The whole bone strength in animals receiving only sweetener was greater than would be expected from geometry (ANCOVA, p < 0.001), indicating increased tissue-level strength. Lines represent ANCOVA coefficients determined with generalized linear models. (E) Compared to untreated animals, body mass was decreased in animals receiving neomycin and increased in animals receiving sweetener. Bone material properties were analyzed in groups with altered tissue-level strength (sweetener and neomycin groups). F) Mineral-to-matrix ratio was decreased in animals dosed with only neomycin. p values are indicated for all comparisons in which p < 0.10.
In addition to reducing bone cross-sectional geometry, selective depletion of some components of the gut microbiota decreased whole bone strength as compared to untreated animals with an unaltered gut microbiome (p<0.001, Fig. 2C). Animals in the sweetener group had similar whole bone strength to the animals with an unaltered gut microbiome (untreated). However, differences in whole bone strength were not completely explained by differences in bone geometry, suggesting differences in tissue strength among groups. The relationship between whole bone strength and section modulus was altered in the neomycin group, on average, 29.28% less than other groups with similar section modulus (p=0.002, as determined by ANCOVA coefficients and indicated graphically by the difference in lines in Fig. 2D, Supplementary Table S2), indicating reduced tissue strength. After accounting for geometry, whole bone strength was increased in animals receiving sweetener by 39.64% (p < 0.001, as determined by ANCOVA coefficients and indicated by differences in lines in Fig. 2D, Supplementary Table S2). Additional whole-bone mechanical properties derived from 3-point bending, such as maximum load, ultimate stress, stiffness, and Young’s Modulus, can be found in Supplementary Table S3.
Compared to untreated animals, body mass was decreased by 10% in animals receiving neomycin (p=0.021), and increased by 9% in animals treated with sweetener alone (p=0.044) (Fig. 2E. Epididymal fat pad mass was decreased by 44% in animals receiving an antibiotic cocktail (p<0.001) and increased by 32% in animals receiving only sweetener (p=0.005) when compared to untreated animals (Fig. S2B). Compared to untreated animals, femur length was increased in animals receiving sweetener, and decreased in animals receiving ampicillin, vancomycin, neomycin or cocktail (Fig. S2C). After adjusting for body weight, similar trends in cross-sectional geometry and femur length were observed, although the increased body mass in the sweetener group altered some comparisons (Supplemental Fig. S2D–F).
Fourier-transform Infrared (FTIR) Spectroscopy Analysis
Compositional properties of the femoral mid-diaphysis was analyzed in tissue from experimental groups that showed significantly altered tissue-level strength. The amount of mineral per unit of collagen in the matrix (Mineral-to-matrix ratio) at the femoral mid-diaphysis was decreased in animals from the neomycin group (p=0.038, Fig. 2F), while bone from the animals in the sweetener group showed an intermediate mineral-to-matrix ratio. Crystallinity, collagen maturity and carbonate to phosphate ratio did not noticeably differ among the groups (Supplemental Fig. S3A–C).
Bone Histomorphometry, Blood Serum Analysis, and Metaphyseal Cancellous Bone Morphology
Mineralizing surface of the endosteum and periosteum at the femoral mid-diaphysis did not differ among groups (Table 1). The mineral apposition rate at the endosteum and periosteum of the mid-diaphysis of the femur did not differ among groups (Table 1). No differences in serum procollagen I N-terminal propeptide (a marker of bone formation, p=0.095, Table 1) or C-telopeptide of type I collagen (a marker of bone resorption, p=0.545, Table 1) were observed among groups. Animals in the sweetener group had increased bone volume fraction (BV/TV) when compared to all other groups (p<0.001, Table 1) as well as increased trabecular thickness (Tb.Th.) (p<0.001, Table 1). Trabecular separation (Tb.Sp) and trabecular number (Tb.N) did not differ among groups (Table 1).
Table 1:
Cortical bone histomorphometry, serum bone turnover markers, and metaphyseal cancellous bone microcomputed tomography results for all groups (mean ± standard deviation, range in parentheses). Selective removal of components of the gut microbiota via antibiotics did not alter cortical bone histomorphometry, serum bone turnover markers, and metaphyseal Tb.N, Th.Sp or TBMD.
| Untreated | Sweetener | Ampicillin | Vancomycin | Neomycin | Metronidazole | Cocktail | ||
|---|---|---|---|---|---|---|---|---|
| Cortical Bone Histomorphometry (n=6/group) | Periosteum Mineralizing Surface | 0.42 ± 0.12 (0.23, 0.58) | 0.68 ± 0.14 (0.51, 0.87) | 0.46 ± 0.26 (0.00, 0.73) | 0.34 ± 0.19 (0.13, 0.57) | 0.54 ± 0.26 (0.23, 0.83) | 0.46 ± 0.45 (0.01, 1.25) | 0.35 ± 0.22 (0.01, 0.58) |
| Periosteum MAR (μm/day) | 2.59 ± 1.09 (1.39, 4.54) | 3.05 ± 0.60 (2.17, 3.60) | 2.04 ± 1.15 (0.00, 3.11) | 1.17 ± 1.40 (0.00, 3.31) | 3.23 ± 1.10 (1.85, 4.50) | 2.10 ± 1.85 (0.00, 4.46) | 1.68 ± 1.34 (0.00, 3.03) | |
| Endosteum Mineralizing Surface | 0.71 ± 0.28 (0.32, 1.04) | 0.51 ± 0.12 (0.34, 0.63) | 0.65 ± 0.34 (0.00, 0.91) | 0.65 ± 0.21 (0.40, 0.99) | 0.85 ± 0.27 (0.46, 1.28) | 0.69 ± 0.30 (0.32, 1.08) | 0.74 ± 0.31 (0.20, 1.13) | |
| Endosteum MAR (μm/day) | 2.62 ± 1.01 (1.33, 4.19) | 2.60 ± 0.85 (1.65, 3.74) | 2.07 ± 1.19 (0.00, 3.42) | 1.82 ± 1.10 (0.00, 3.02) | 2.23 ± 0.71 (1.67, 3.54) | 2.23 ± 0.44 (1.71, 2.79) | 2.08 ± 1.10 (0.00, 2.89) | |
| Serum Bone Turnover Markers (n=7–10/group) | Serum PINP (ng/mL) | 22.38 ± 3.71 (18.16, 29.23) | 22.45 ± 7.38 (9.71, 32.91) | 22.27 ± 5.36 (16.92, 31.80) | 19.70 ± 9.86 (4.41, 32.39) | 24.01 ± 5.35 (17.52, 34.68) | 21.95 ± 11.51 (3.17, 38.73) | 26.97 ± 4.96 (17.81, 31.91) |
| Serum CTX-I (ng/mL) | 25.04 ± 16.45 (11.96, 58.05) | 19.94 ± 3.24 (17.00, 27.26) | 22.96 ± 20.66 (10.61, 69.27) | 21.51 ± 6.16 (14.66, 31.27) | 17.63 ± 2.93 (13.07, 21.84) | 17.07 ± 4.69 (11.81, 24.57) | 35.54 ± 18.81 (19.34, 63.31) | |
| Metaphyseal Cancellous Bone (n=6/group) | BV/TV | 0.15 ± 0.05 (0.10, 0.22) | 0.21 ± 0.02 $ (0.19, 0.23) | 0.13 ± 0.04 (0.08, 0.18) | 0.13 ± 0.03 (0.09, 0.17) | 0.11 ± 0.02 (0.08, 0.14) | 0.13 ± 0.04 (0.09, 0.20) | 0.13 ± 0.02 (0.10, 0.16) |
| Tb.Th (mm) | 0.05 ± 0.005 (0.04, 0.06) | 0.05 ± 0.006 # (0.05, 0.06) | 0.04 ± 0.004 (0.04, 0.05) | 0.04 ± 0.004 (0.04, 0.05) | 0.04 ± 0.001 (0.040, 0.043) | 0.04 ± 0.005 (0.04, 0.05) | 0.04 ± 0.004 (0.04, 0.05) | |
| Tb.N (1/mm) | 4.69 ± 0.48 (4.20, 5.60) | 5.20 ± 0.24 (4.90, 5.63) | 4.92 ± 0.38 (4.30, 5.24) | 5.13 ± 0.18 (4.82, 5.32) | 4.83 ± 0.48 (4.25, 5.38) | 4.89 ± 0.32 (4.32, 5.19) | 5.01 ± 0.32 (4.63, 5.53) | |
| Tb.Sp (mm) | 0.21 ± 0.02 (0.17, 0.23) | 0.18 ± 0.01 (0.17, 0.19) | 0.20 ± 0.02 (0.19, 0.23) | 0.19 ± 0.01 (0.18, 0.20) | 0.20 ± 0.02 (0.18, 0.23) | 0.20 ± 0.02 (0.19, 0.23) | 0.19 ± 0.01 (0.17, 0.21) |
Sweetener v. Untreated (p=0.036), Ampicillin (p = 0.005), Vancomycin (p=0.002), Neomycin (p < 0.001), Metronidazole (p=0.003), Cocktail (p =0.003)
Sweetener v. Ampicillin (p = 0.009), Vancomycin (p < 0.001), Neomycin (p = 0.001), Metronidazole (p = 0.022), Cocktail (p =0.001)
Associations between Gut Microbiota and Bone Phenotypes
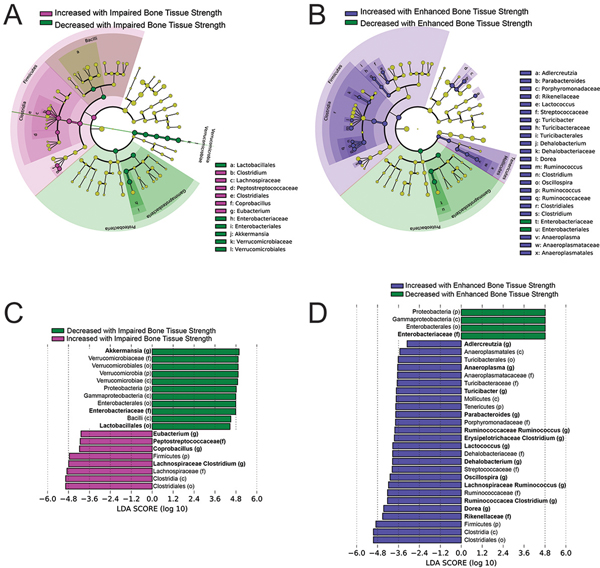
Linear discriminant analysis comparing animals with reduced tissue-level strength (neomycin group) and the groups with normal bone tissue strength (untreated, ampicillin, vancomycin and metronidazole subgroups) identified 19 features at all taxonomic levels that differed by an effect size greater than |2| (Fig. 3A). Of these 19 features, many are detected as a result of differences in abundance at lower ranks (for example, the significant differential abundance of the phyla Verrucomicrobia is dominated by the differential abundance of the genus Akkermansia). Focusing on these influential lower-ranked taxa, seven show differential abundance among groups including greater abundance of the genera Lachnospiraceae Clostridium, Coprobacillus and Eubacterium and the taxa from the family Peptostreptococcacea in mice with impaired bone tissue-level strength (Fig. 3A, B and Supplemental Fig. S4). Additionally, mice with impaired bone tissue strength showed reduced abundance of the genera Akkermanisa, the family Enterobacteriaceae and the order Lactobacillales.
Figure 3:
Results from linear discriminant analysis (LDA) are shown including cladograms (A,B) and LDA scores (C,D). The cladogram shows different taxonomic ranks represented by rings with phyla in the innermost ring and genera in the outermost ring. Colored circles represent individual taxa and are sized in proportion to relative abundance. Features with an effect size greater than |2| are highlighted. The taxa in each level are colored indicating increased or decreased abundance in (A) impaired bone tissue-level strength (neomycin) or (B) enhanced bone tissue-level strength (sweetener) from that in animals with normal bone strength (untreated, ampicillin, vancomycin, metronidazole, LEfSe analysis confirms that the differences between groups are valid for all subgroups). The corresponding LDA scores are illustrated for p <0.05 (C,D) with taxonomic rank is shown in parentheses: (p) phylum, (c) class, (o) order, (f) family, (g) genus. Taxa in bold are lower rank taxa mentioned in the text that appear to dictate the differences at the higher ranks.
Linear discriminant analysis detected 30 taxonomic features with an effect size greater than |2| that differ between animals with increased tissue-level strength (sweetener group) and animals with normal bone tissue strength (untreated, ampicillin, vancomycin and metronidazole) (Fig. 3C,D). Focusing on the 14 lower ranked taxonomic features reveals greater abundance of the genera Dorea, Ruminococcaceae Clostridium, Lachnospiraceae Ruminococcus, Oscillospira, Dehalobacterium, Lactoccocus, Erysipelotrichaceae Clostridium, Ruminococcaeceae Ruminococus, Parabacteroides, Turicibacter, Anaeroplasma, Adlercruetzia, and the family Rikencellaceae in mice with increased bone tissue strength (Fig. 3C, D and Supplemental Fig. S4). Additionally, taxa from the family Enterobacteriaceae were reduced in abundance in animals with increased bone tissue strength.
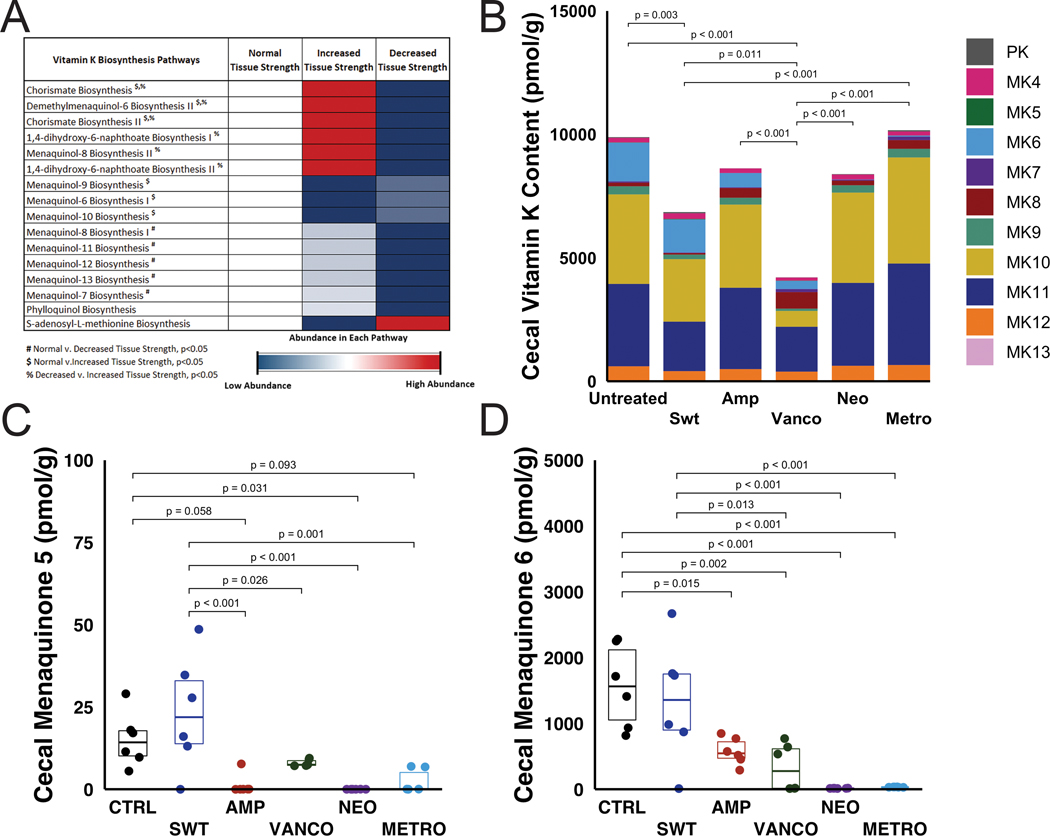
A comparison of the functional capacity of the gut microbiota estimated from the taxonomic information using PICRUSt between animals with normal (untreated), decreased (neomycin) and increased (sweetener) tissue-level strength resulted in 285 differential pathways out of 489 detected MetaCyc pathways (the twenty most differential pathways are listed in Supplemental Fig. S5). Fourteen vitamin K biosynthesis pathways were among the differential pathways identified. A comparison of vitamin K biosynthesis pathways between animals with normal (untreated), decreased (neomycin) and increased (sweetener) tissue-level strength showed reduced functional capacity to synthesize vitamin K in the neomycin group (on average, 37 ± 2% reduction) compared to untreated animals (Fig. 4A). A comparison of vitamin K biosynthesis pathways between all groups showed that animals with decreased tissue-level strength (neomycin) had the most vitamin K pathways that were decreased in abundance (Supplemental Fig. S6).
Figure 4.
A) Predicted functional capacity from PICRUSt results show a decreased abundance in vitamin K biosynthesis in animals with decreased tissue-level strength (dosed with neomycin). Pathways are listed and ordered by ascending p-value. B) Cecal vitamin K content varied among groups. Each color in a column represents the average concentration of phylloquinone (PK) or menaquinone (MK). C) Cecal menaquinone 5 (detection limit 5 pmol/g) and D) cecal menaquinone 6 (detection limit 10 pmol/g) was reduced in the neomycin group when compared to the untreated group. p values for statistical comparisons are indicated for comparisons where p < 0.10.
There were differences in the concentrations of cecal vitamin K (phylloquinone and menaquinones) among groups. On average, microbe-derived menaquinones (MK5-MK13) accounted for 97.3 ± 0.8% of the total vitamin K content (Fig. 4B). Animals with reduced bone tissue strength (the neomycin group) had reduced amounts of MK5 (Fig. 4C) and MK6 (Fig. 4D) when compared to untreated and sweetener groups. The total vitamin K content was composed primarily of MK10 (35.6 ± 10.4%, on average) and MK11 (37.5 ± 5.1%, on average). The average total cecal vitamin K content, including phylloquinone and menaquinones, was greater in untreated animals and animals dosed with metronidazole (Fig. 4B). Animals with an intestinal microbiome altered by intake of vancomycin and sweetener had reduced levels of cecal vitamin K (Fig. 4B). Alterations in the gut microbiota associated with neomycin and ampicillin groups led to total cecal vitamin K content in between vancomycin/sweetener and untreated groups.
Discussion
The gut microbiome consists of a complex network of interacting microorganisms that can modulate bone quantity and quality(5). Here we generated seven different gut microbial populations in mice. One microbial population was associated with impaired bone tissue-level strength and another was associated with to increased bone tissue-level strength. In contrast, no noticeable changes in trabecular bone volume fraction or bone formation were observed. Analysis of the fecal microbiota revealed differential abundance of 7 lower ranked taxonomic features associated with impaired tissue strength and 14 with increased tissue strength (as compared to groups with unaltered tissue strength). These findings indicate that modifications in the gut microbiome can alter cortical bone tissue-level strength independent of bone quantity, bone formation and bone remodeling.
Of the distinct gut microbiota generated in the study, one (the neomycin group) resulted in reductions in bone tissue-level strength. This finding is consistent with our prior work that found alterations to the gut microbiota caused by a combination of ampicillin and neomycin led to reductions in whole bone strength and tissue-level strength (3). Neomycin is not absorbed in the gut, hence impairment of bone tissue strength is not due to the effect of the antibiotic, but rather to the changes in the gut microbial community caused by the antibiotic. The reductions in bone tissue-level strength may be a result of: 1) the elimination of microbial taxa (i.e. organisms that have reduced abundance, indicated in green in Fig. 3A,B for example Akkermansia); and/or 2) increases in abundance of microbial taxa (indicated in pink Fig. 3A, for example Lachnospiraceae Clostridium). The relationship between taxonomic abundance and the bone tissue-level strength is complicated; a differentially abundant taxonomic feature may contribute to the bone tissue-level phenotype or may simply be differentially abundant due to interdependencies with other taxa that more directly influence the bone phenotype. For example, the genus Akkermansia is reduced in abundance in mice with impaired tissue strength. Akkermansia muciniphila, the only species of this genera, is used as a probiotic and has been negatively associated with diabetes and obesity in humans (37). However, it is unclear in the current study if Akkermansia helps regulate bone tissue strength or is simply an indicator of the health of the gut mictobiota. Further studies are required to determine if the correlation between the abundance of A. muciniphila or other taxa and bone tissue strength is causative.
Of the seven gut microbiota considered in this experiment, one resulted in an increased bone tissue strength (the microbiota generated by zero-calorie sweetener). This finding was unexpected. Sweetener is often used in experimental manipulations of the gut microbiota with antibiotic cocktails in drinking water as a means of masking the taste of the antibiotics to ensure adequate water intake. How the sweetener by itself led to increased bone tissue strength is not clear and difficult to interpret since the study was not designed to evaluate the effects of sweetener (sweetener was only used in two study groups). The aspartame-based sweetener used in our study has negligible caloric content yet led to increased body mass and fat pad mass (consistent with the literature(38)). Sucrose and aspartame have dissimilar chemical compositions, thus it unlikely that artificial sweeteners have the same effect as sugar/natural sweetener on bone. Sweetener substantially changed the constituents of the gut microbiota as indicated by both alpha diversity and linear discriminant analysis. Our findings correlate increases in bone tissue strength with differential abundance of 14 lower rank taxonomic features (Fig. 3C,D). The increase in bone tissue strength may be associated with reductions in taxa from the family Enterobacteriaceae or may be a result of increased abundance of the other 13 lower rank taxonomic features. For example, the genera Oscillospira and Lachnospiraceae Ruminococcus are increased in abundance in the group with enhanced bone tissue strength. Oscillospira sp. And Lachnospiraceae Ruminococcus sp. are recognized as producers of the short chain fatty acid butyrate which has many beneficial effects on the host (although mechanisms relating butyrate to bone tissue mechanical properties have not been proposed). As with the features associated with impaired bone tissue strength above, additional experiments are required to determine a causative effect and mechanisms that lead to increased bone tissue strength. However, we can conclude that aspartame-based sweeteners– a common additive in microbiome studies in mice – should be used with caution as the additives could potentially influence bone phenotypes.
Our observation that alterations in the gut microbiome caused by neomycin alone led to impaired bone tissue strength while the cocktail (which included neomycin and sweetener) did not noticeably alter bone tissue strength may at first seem odd. There are two possible explanations: 1) the microbes that lead to altered bone tissue strength were depleted in the cocktail group; the cocktail removes 95% of the microbes from the gut, hence other changes caused by neomycin or sweetener could not occur; and 2) it is possible that sweetener in the cocktail group counteracted the effects of the changes caused by neomycin. Further study of the relationships between the gut microbiota and bone tissue strength are required to differentiate between these two possibilities and whether or not the changes in the microbiota that promote reduced bone tissue strength are the same as those that promote increased bone tissue strength. It is likely that there are distinct mechanisms leading to impaired bone tissue strength and enhanced bone tissue strength. For example, the reduced tissue strength in the neomycin group is consistent with the reduced mineral-to-matrix ratio (an assay of tissue composition positively correlated with bone tissue strength (39)) yet the increased tissue strength in the sweetener group was not associated with increased mineral-to-matrix ratio as compared to mice with normal bone tissue strength.
Our prior work associated microbiome-induced reductions in bone tissue strength (from oral neomycin+ampicillin) with reduced functional capacity to synthesize vitamin K (determined directly through shotgun metagenomic sequencing), cecal vitamin K concentrations and osteocalcin concentrations in bone (4). Vitamin K has long been associated with bone health. Although diet is a source of vitamin K (predominately phylloquinone) the gut microbiome is an important source of forms of vitamin K known as menaquinones. There are a number of vitamin K-dependent proteins in bone matrix including osteocalcin, the most abundant non-collagenous protein in bone (40,41). In the presence of vitamin K, osteocalcin synthesized by osteoblasts is carboxylated and can then bind to bone mineral. In the absence of osteocalcin, bone tissue is altered in ways that may reduce measures of bone tissue strength or brittleness (42,43). Alterations to the microbiome caused by oral antibiotics have been linked to decreased abundance of menaquinone producers and decreased abundances of menaquinone biosynthesis genes(5). The current findings, however, are mixed with regard to vitamin K. The only group to show impaired bone tissue strength (the neomycin group) also displayed large reductions in many of the genes required to synthesize vitamin K, consistent with the idea that the impaired bone tissue strength is secondary to reduced vitamin K. Consistent with this finding, the concentration of the MK5 and MK6 forms of vitamin K were reduced in the cecum of animals in the neomycin group. However, while total cecal vitamin K concentrations in the neomycin group were lower than untreated controls (consistent with our prior work), some groups in the current study with similar or greater bone tissue strength showed similar or even lower concentrations of total cecal vitamin K. One possible explanation is that different forms of menaquinone either have different effects on bone or differences in the bioavailability and/or transport (45). Additional experiments that directly modulate dietary vitamin K and measure matrix-bound vitamin K-dependent proteins are required to further evaluate the vitamin K-based mechanism linking the gut microbiota to bone.
The current study has a number of strengths. First, the experimental design is novel among studies of the bone and microbiome in studying seven distinct gut microbial communities. An additional strength is that the different gut microbiota were generated using oral antibiotics which cause more immediate, consistent and sustained changes in the gut microbial community than manipulations such as pre- or probiotics(46). Second, the analysis includes biomechanical, chemical and structural analysis of the bone as well as measures of systemic factors. Lastly the current study focuses on the bone phenotype of skeletally-mature animals. Many studies of the gut microbiota and bone have evaluated the bone phenotype in young animals (12 weeks of age or younger) in which the rapidly growing skeleton can make it difficult to identify differences in bone phenotype (47).
Some limitations must be considered when interpreting our results. First, although most antibiotics chosen in this study are poorly absorbed in the gut, we cannot ignore the possibility that some antibiotics could have a small direct effect on bone. Ampicillin and metronidazole have recognized oral bioavailability and are distributed systemically; however, dosing with either of these antibiotics alone did not lead to alterations in bone tissue strength. In contrast, neomycin, the antibiotic we have associated with impaired bone tissue strength, has a zero oral bioavailability, strongly suggesting that observed reductions in tissue strength are the result of the alterations to the gut microbiome. Second, the gut microbiome may regulate bone by causing inflammation at the gut lining which can cause a “leaky gut” phenomenon that enhances translocation of gut microbial products into the systemic circulation and/or initiates inflammatory processes and altering the function of local and circulating T cell and cytokines(5,48). While we did not directly measure these alternate mechanisms in the current study, prior work suggests that neither of these mechanisms are likely explanations for changes in bone tissue strength. Increases in gut permeability lead to increased translocation of microbial proteins into systemic circulation(49), however we have previously shown that animals receiving ampicillin and neomycin over a similar time period did not have noticeably altered serum concentrations of the bacterial protein lipopolysaccharide(3), which would be expected if gut permeability had increased. Inflammatory processes associated with the gut microbiome regulate trabecular bone remodeling and trabecular bone volume fraction by altering the function of T cells that migrate to the bone marrow (50). However, we did not observe differences in trabecular bone volume fraction in the neomycin group. Furthermore, while T cells are known to regulate bone remodeling, no mechanisms through which T cell activity would lead to altered bone tissue strength have been proposed/identified. Third, the study did not include measurement of food and water intake during the experiment so there may have been differences in nutrient intake. The gut microbiota may influence both appetite (51) and nutrient absorption (5) in complex feedback loops. However, it is unlikely that alterations in nutrient absorption are responsible for the observed differences in bone tissue strength because there were only minor differences in whole bone length and undetectable differences in serum markers of bone remodeling and periosteal bone formation. Hence if there were differences in nutrient absorption they were unlikely to have a major effect on bone tissue strength. Another limitation is that sweetener was only used in two groups and the study therefore does not provide a detailed effect of sweetener as a treatment. However, the current study does provide a comparison of the bone phenotypes resulting from seven distinct gut microbiota. Lastly, we must emphasize that the assessment of the functional capacity of the gut microbiome using the PICRUSt software is not direct and is predicted from taxonomy(32). PICRUSt uses reference genomes to predict functions or pathways from taxonomic information, therefore, any gaps or inaccuracies in the pathway databases will be reflected in the results. Although PICRUSt has limitations, it provides functional predictions that are useful to generate hypotheses. Complete metagenomic sequencing of the gut microbiota is required to confirm predictions made by PICRUSt and to directly determine the functional components of the gut microbiome associated with impaired bone tissue quality.
Many pharmaceuticals and mechanistic pathways are known to influence bone quantity by regulating bone remodeling. In contrast, there are few treatments and interventions that act by modulating bone tissue quality. Our findings show that the gut microbiome can modulate bone tissue quality and potentially provide a new therapeutic target to address bone fragility.
Supplementary Material
Acknowledgments
The research was supported by the Office of the Assistant Secretary of Defense for Health Affairs through the office of the Congressionally Directed Medical Research Programs under award number W81XWH-15-1-0239, the National Institutes of Health under award numbers R56AG067997, R21AR068061, R21AR073454, and R21AR071534, the National Science Foundation Graduate Research Fellowships Program under the award number DGE-1650441, the USDA Agricultural Research Service under Cooperative Agreement No. 58-1950-7-707, and the Alfred P. Sloan Foundation. The content of the work is solely the responsibility of the authors and does not necessarily represent the official views of the funding sources.
The authors thank the Cornell University Biotechnology Resource Center and their funding for shared Zeiss LSM880 confocal/multiphoton microscope from the National Institute of Health under the award number S10OD018516. The authors also thank Hayat Benlarbi of the Hospital for Special Surgery microCT facility, Marjory Xavier Rodrigues for her assistance with gut microbiome sequencing, and the Cornell CARE staff for their assistance with animal care.
Footnotes
Disclosures:
M. Luna has nothing to disclose. J.D. Guss has nothing to disclose.
L.S. Vasquez-Bolanos has nothing to disclose. M. Castaneda has nothing to disclose.
M. Vargas Rojas has nothing to disclose. J.M. Strong has nothing to disclose.
D.A. Alabi has nothing to disclose. S.D. Dornevil has nothing to disclose.
J.C. Nixon has nothing to disclose. E.A. Taylor has nothing to disclose.
E. Donnelly has nothing to disclose. X. Fu has nothing to disclose.
M.K. Shea has nothing to disclose. S.L. Booth has nothing to disclose.
R. Bicahlo has nothing to disclose. C.J. Hernandez has nothing to disclose.
Data Availability Statement:
Supporting data are publicly available through the Sequence Read Archive under the accession number PRJNA625754 (http://www.ncbi.nlm.nih.gov/Traces/sra, National Center for Biotechnology Information, Bethesda, Maryland, USA).
References
- 1.Tu KN, Lie JD, Wan CKV, Cameron M, Austel AG, Nguyen JK, Van K, Hyun D. Osteoporosis: A Review of Treatment Options. Pharm. Ther. 2018. Feb;43(2):92–104. [PMC free article] [PubMed] [Google Scholar]
- 2.Alliston T. Biological Regulation of Bone Quality. Curr. Osteoporos. Rep. 2014. Sep;12(3):366–75. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Guss JD, Horsfield MW, Fontenele FF, Sandoval TN, Luna M, Apoorva F, Lima SF, Bicalho RC, Singh A, Ley RE, Meulen van der MC, Goldring SR, Hernandez CJ Alterations to the Gut Microbiome Impair Bone Strength and Tissue Material Properties. J. Bone Miner. Res. 2017;32(6):1343–53. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Guss JD, Taylor E, Rouse Z, Roubert S, Higgins CH, Thomas CJ, Baker SP, Vashishth D, Donnelly E, Shea MK, Booth SL, Bicalho RC, Hernandez CJ. The microbial metagenome and bone tissue composition in mice with microbiome-induced reductions in bone strength. Bone. 2019. Oct 1;127:146–54. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Hernandez CJ, Guss JD, Luna M, Goldring SR. Links Between the Microbiome and Bone. J. Bone Miner. Res. Off. J. Am. Soc. Bone Miner. Res. 2016;31(9):1638–46. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Behera J, Ison J, Tyagi SC, Tyagi N. The role of gut microbiota in bone homeostasis. Bone. 2020. Jun 1;135:115317. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Sears CL. A dynamic partnership: celebrating our gut flora. Anaerobe. 2005. Oct;11(5):247–51. [DOI] [PubMed] [Google Scholar]
- 8.Kho ZY, Lal SK. The Human Gut Microbiome - A Potential Controller of Wellness and Disease. Front. Microbiol. 2018;9:1835. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Gupta S, Allen-Vercoe E, Petrof EO. Fecal microbiota transplantation: in perspective. Ther. Adv. Gastroenterol. 2016. Mar;9(2):229–39. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Sjögren K, Engdahl C, Henning P, Lerner UH, Tremaroli V, Lagerquist MK, Bäckhed F, Ohlsson C. The gut microbiota regulates bone mass in mice. J. Bone Miner. Res. 2012. Jun;27(6):1357–67. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Schwarzer M, Makki K, Storelli G, Machuca-Gayet I, Srutkova D, Hermanova P, Martino ME, Balmand S, Hudcovic T, Heddi A, Rieusset J, Kozakova H, Vidal H, Leulier F. Lactobacillus plantarum strain maintains growth of infant mice during chronic undernutrition. Science. 2016. Feb 19;351(6275):854–7. [DOI] [PubMed] [Google Scholar]
- 12.Yan J, Herzog JW, Tsang K, Brennan CA, Bower MA, Garrett WS, Sartor BR, Aliprantis AO, Charles JF. Gut microbiota induce IGF-1 and promote bone formation and growth. Proc. Natl. Acad. Sci. U. S. A. 2016. 22;113(47):E7554–63. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Cho I, Yamanishi S, Cox L, Methé BA, Zavadil J, Li K, Gao Z, Mahana D, Raju K, Teitler I, Li H, Alekseyenko AV, Blaser MJ. Antibiotics in early life alter the murine colonic microbiome and adiposity. Nature. 2012. Aug 30;488(7413):621–6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Cox LM, Yamanishi S, Sohn J, Alekseyenko AV, Leung JM, Cho I, Kim SG, Li H, Gao Z, Mahana D, Zárate Rodriguez JG, Rogers AB, Robine N, Loke P, Blaser MJ Altering the Intestinal Microbiota during a Critical Developmental Window Has Lasting Metabolic Consequences. Cell. 2014. Aug 14;158(4):705–21. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Nobel YR, Cox LM, Kirigin FF, Bokulich NA, Yamanishi S, Teitler I, Chung J, Sohn J, Barber CM, Goldfarb DS, Raju K, Abubucker S, Zhou Y, Ruiz VE, Li H, Mitreva M, Alekseyenko AV, Weinstock GM, Sodergren E, Blaser MJ. Metabolic and metagenomic outcomes from early-life pulsed antibiotic treatment. Nat. Commun. 2015. Jun 30;6:7486. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Hathaway-Schrader JD, Steinkamp HM, Chavez MB, Poulides NA, Kirkpatrick JE, Chew ME, Huang E, Alekseyenko AV, Aguirre JI, Novince CM. Antibiotic Perturbation of Gut Microbiota Dysregulates Osteoimmune Cross Talk in Postpubertal Skeletal Development. Am. J. Pathol. 2019. Feb 1;189(2):370–90. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Tanoue T, Morita S, Plichta DR, Skelly AN, Suda W, Sugiura Y, Narushima S, Vlamakis H, Motoo I, Sugita K, Shiota A, Takeshita K, Yasuma-Mitobe K, Riethmacher D, Kaisho T, Norman JM, Mucida D, Suematsu M, Yaguchi T, Bucci V, Inoue T, Kawakami Y, Olle B, Roberts B, Hattori M, Xavier RJ, Atarashi K, Honda K. A defined commensal consortium elicits CD8 T cells and anti-cancer immunity. Nature. 2019. Jan;565(7741):600–5. [DOI] [PubMed] [Google Scholar]
- 18.Kennedy EA, King KY, Baldridge MT. Mouse Microbiota Models: Comparing Germ-Free Mice and Antibiotics Treatment as Tools for Modifying Gut Bacteria. Front. Physiol. 2018;9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Reikvam DH, Erofeev A, Sandvik A, Grcic V, Jahnsen FL, Gaustad P, McCoy KD, Macpherson AJ, Meza-Zepeda LA, Johansen F-E. Depletion of murine intestinal microbiota: effects on gut mucosa and epithelial gene expression. PloS One. 2011. Mar 21;6(3):e17996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Jepsen KJ, Silva MJ, Vashishth D, Guo XE, van der Meulen MC. Establishing biomechanical mechanisms in mouse models: practical guidelines for systematically evaluating phenotypic changes in the diaphyses of long bones. J Bone Min. Res. 2015. Jun;30(6):951–66. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Lima SF, Teixeira AGV, Higgins CH, Lima FS, Bicalho RC. The upper respiratory tract microbiome and its potential role in bovine respiratory disease and otitis media. Sci. Rep. 2016. Jul 1;6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Caporaso JG, Lauber CL, Walters WA, Berg-Lyons D, Huntley J, Fierer N, Owens SM, Betley J, Fraser L, Bauer M, Gormley N, Gilbert JA, Smith G, Knight R. Ultra-high-throughput microbial community analysis on the Illumina HiSeq and MiSeq platforms. ISME J. 2012. Aug;6(8):1621–4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Leinonen R, Sugawara H, Shumway M. The Sequence Read Archive. Nucleic Acids Res. 2011. Jan;39:D19–21. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Nyman JS, Vashishth D. Assessment of Bone Mass, Structure, and Quality in Rodents. Primer Metab. Bone Dis. Disord. Miner. Metab. John Wiley & Sons, Ltd; 2018. p. 93–100. [Google Scholar]
- 25.Dempster DW, Compston JE, Drezner MK, Glorieux FH, Kanis JA, Malluche H, Meunier PJ, Ott SM, Recker RR, Parfitt AM. Standardized nomenclature, symbols, and units for bone histomorphometry: A 2012 update of the report of the ASBMR Histomorphometry Nomenclature Committee. J Bone Min. Res. 2013. Jan;28(1):2–17. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Bouxsein ML, Boyd SK, Christiansen BA, Guldberg RE, Jepsen KJ, Muller R. Guidelines for assessment of bone microstructure in rodents using micro-computed tomography. J Bone Min. Res. 2010. Jul;25(7):1468–86. [DOI] [PubMed] [Google Scholar]
- 27.Hunt HB, Torres AM, Palomino PM, Marty E, Saiyed R, Cohn M, Jo J, Warner S, Sroga GE, King KB, Lane JM, Vashishth D, Hernandez CJ, Donnelly E. Altered Tissue Composition, Microarchitecture, and Mechanical Performance in Cancellous Bone From Men With Type 2 Diabetes Mellitus. J. Bone Miner. Res. 2019;34(7):1191–206. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Taylor EA, Lloyd AA, Salazar-Lara C, Donnelly E. Raman and Fourier Transform Infrared (FT-IR) Mineral to Matrix Ratios Correlate with Physical Chemical Properties of Model Compounds and Native Bone Tissue. Appl. Spectrosc. SAGE Publications Ltd STM; 2017. Oct 1;71(10):2404–10. [DOI] [PubMed] [Google Scholar]
- 29.Gadaleta SJ, Paschalis EP, Betts F, Mendelsohn R, Boskey AL. Fourier transform infrared spectroscopy of the solution-mediated conversion of amorphous calcium phosphate to hydroxyapatite: New correlations between X-ray diffraction and infrared data. Calcif. Tissue Int. 1996. Jan 1;58(1):9–16. [DOI] [PubMed] [Google Scholar]
- 30.Paschalis EP, Verdelis K, Doty SB, Boskey AL, Mendelsohn R, Yamauchi M. Spectroscopic characterization of collagen cross-links in bone. J Bone Min. Res. 2001. Oct;16(10):1821–8. [DOI] [PubMed] [Google Scholar]
- 31.Spevak L, Flach CR, Hunter T, Mendelsohn R, Boskey A. Fourier Transform Infrared Spectroscopic Imaging Parameters Describing Acid Phosphate Substitution in Biologic Hydroxyapatite. Calcif. Tissue Int. 2013. May 1;92(5):418–28. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Langille MGI, Zaneveld J, Caporaso JG, McDonald D, Knights D, Reyes JA, Clemente JC, Burkepile DE, Vega Thurber RL, Knight R, Beiko RG, Huttenhower C. Predictive functional profiling of microbial communities using 16S rRNA marker gene sequences. Nat. Biotechnol. 2013. Sep;31(9):814–21. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Karl JP, Fu X, Dolnikowski GG, Saltzman E, Booth SL. Quantification of phylloquinone and menaquinones in feces, serum, and food by high-performance liquid chromatography–mass spectrometry. J. Chromatogr. B. 2014. Jul 15;963:128–33. [DOI] [PubMed] [Google Scholar]
- 34.Philippas D. Analysis of Covariance (ANCOVA). In: Michalos AC, editor. Encycl. Qual. Life Well- Res. Dordrecht: Springer Netherlands; 2014. p. 157–61. [Google Scholar]
- 35.Segata N, Izard J, Waldron L, Gevers D, Miropolsky L, Garrett WS, Huttenhower C. Metagenomic biomarker discovery and explanation. Genome Biol. 2011. Jun 24;12(6):R60. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Parks DH, Tyson GW, Hugenholtz P, Beiko RG. STAMP: statistical analysis of taxonomic and functional profiles. Bioinformatics. 2014. Nov 1;30(21):3123–4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Jayachandran M, Chung SSM, Xu B. A critical review of the relationship between dietary components, the gut microbe Akkermansia muciniphila, and human health. Crit Rev Food Sci Nutr. 2020;60(13):2265–76. [DOI] [PubMed] [Google Scholar]
- 38.Palmnäs MSA, Cowan TE, Bomhof MR, Su J, Reimer RA, Vogel HJ, Hittel DS, Shearer J. Low-dose aspartame consumption differentially affects gut microbiota-host metabolic interactions in the diet-induced obese rat. PloS One. 2014;9(10):e109841. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Bi X, Patil CA, Lynch CC, Pharr GM, Mahadevan-Jansen A, Nyman JS. Raman and mechanical properties correlate at whole bone- and tissue-levels in a genetic mouse model. J Biomech. 2011. Jan 11;44(2):297–303. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Palermo A, Tuccinardi D, D’Onofrio L, Watanabe M, Maggi D, Maurizi AR, Greto V, Buzzetti R, Napoli N, Pozzilli P, Manfrini S. Vitamin K and osteoporosis: Myth or reality? Metabolism. 2017;70:57–71. [DOI] [PubMed] [Google Scholar]
- 41.Wen L, Chen J, Duan L, Li S. Vitamin K-dependent proteins involved in bone and cardiovascular health. Mol. Med. Rep. 2018. Jul;18(1):3–15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Hernandez CJ, van der Meulen MC. Understanding Bone Strength Is Not Enough. J Bone Min. Res. 2017. Jun;32(6):1157–62. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Poundarik AA, Diab T, Sroga GE, Ural A, Boskey AL, Gundberg CM, Vashishth D. Dilatational band formation in bone. Proc. Natl. Acad. Sci. U. S. A. 2012. Nov 20;109(47):19178–83. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Quinn L, Sheh A, Ellis JL, Smith DE, Booth SL, Fu X, Muthupalani S, Ge Z, Puglisi DA, Wang TC, Gonda TA, Holcombe H, Fox JG. Helicobacter pylori antibiotic eradication coupled with a chemically defined diet in INS-GAS mice triggers dysbiosis and vitamin K deficiency resulting in gastric hemorrhage. Gut Microbes. 2020. Jul 3;11(4):820–41. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Thijssen HH, Drittij-Reijnders MJ. Vitamin K status in human tissues: tissue-specific accumulation of phylloquinone and menaquinone-4. Br. J. Nutr. 1996. Jan;75(1):121–7. [DOI] [PubMed] [Google Scholar]
- 46.Antonopoulos DA, Huse SM, Morrison HG, Schmidt TM, Sogin ML, Young VB. Reproducible community dynamics of the gastrointestinal microbiota following antibiotic perturbation. Infect. Immun. 2009. Jun;77(6):2367–75. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Jepsen KJ, Hu B, Tommasini SM, Courtland HW, Price C, Cordova M, Nadeau JH. Phenotypic integration of skeletal traits during growth buffers genetic variants affecting the slenderness of femora in inbred mouse strains. Mamm Genome. 2009. Jan;20(1):21–33. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Mu Q, Kirby J, Reilly CM, Luo XM. Leaky Gut As a Danger Signal for Autoimmune Diseases. Front. Immunol. 2017. May 23;8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Chakaroun RM, Massier L, Kovacs P. Gut Microbiome, Intestinal Permeability, and Tissue Bacteria in Metabolic Disease: Perpetrators or Bystanders? Nutrients. 2020. Apr 14;12(4). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Pacifici R. Bone Remodeling and the Microbiome. Cold Spring Harb Perspect Med. 2018. Apr 2;8(4). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Vijay-Kumar M, Aitken JD, Carvalho FA, Cullender TC, Mwangi S, Srinivasan S, Sitaraman SV, Knight R, Ley RE, Gewirtz AT. Metabolic syndrome and altered gut microbiota in mice lacking Toll-like receptor 5. Science. 2010. Apr 9;328(5975):228–31. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.