Abstract
The coronavirus disease 2019 (COVID-19) is a respiratory disease caused by a virus called the severe acute respiratory syndrome coronavirus 2 (SARS-CoV-2). In this paper, we analyze a within-host SARS-CoV-2/HIV coinfection model. The model is made up of eight ordinary differential equations. These equations describe the interactions between healthy epithelial cells, latently infected epithelial cells, productively infected epithelial cells, SARS-CoV-2 particles, healthy CD T cells, latently infected CD T cells, productively infected CD T cells, and HIV particles. We confirm that the solutions of the developed model are bounded and nonnegative. We calculate the different steady states of the model and derive their existence conditions. We choose appropriate Lyapunov functions to show the global stability of all steady states. We execute some numerical simulations to assist the theoretical contributions. Based on our results, weak CD T cell immunity in SARS-CoV-2/HIV coinfected patients causes an increase in the concentrations of productively infected epithelial cells and SARS-CoV-2 particles. This may lead to severe SARS-CoV-2 infection in HIV patients. This result agrees with many studies that discussed the high risk of severe infection and death in HIV patients when they get SARS-CoV-2 infection. On the other hand, increasing the death rate of infected epithelial cells during the latency period can reduce the severity of SARS-CoV-2 infection in HIV patients. More studies are needed to understand the dynamics of SARS-CoV-2/HIV coinfection and find better ways to treat this vulnerable group of patients.
Introduction
The coronavirus disease 2019 (COVID-19) is a new epidemic that emerged in China in late 2019. It is a respiratory disease ascribed to a virus called the severe acute respiratory syndrome coronavirus 2 (SARS-CoV-2). According to COVID-19 weekly epidemiological update of October 13, 2021, by the World Health Organization (WHO) [1], the cumulative number of confirmed cases reported globally exceeded 237 million and the total number of deaths reached over 4.8 million [1]. The number of new weekly COVID-19 cases has showed a decline since late August 2021 in most countries of the world [1]. SARS-CoV-2/HIV coinfection has become a concern especially in HIV patients who are not receiving antiretroviral therapy (ART) or have low CD T cell counts [2, 3]. As there were approximately 37.7 million people living with HIV at the end of 2020 [4], understanding SARS-CoV-2/HIV coinfection should take a special attention.
SARS-CoV-2 is an RNA virus and it is a member of the Coronaviridae family [5]. It binds to the angiotensin-converting enzyme 2 (ACE2) receptor of epithelial cells [5, 6]. The principal target of SARS-CoV-2 is the alveolar epithelial type 2 cells of the lungs [7]. ACE2 is also expressed in many organs like the kidney, liver, and heart [8]. SARS-CoV-2 is mainly transmitted through respiratory droplets which carry virus particles [9]. Many COVID-19 therapies are being clinically tested to evaluate their effectiveness and safety [10]. The U.S. Food and Drug Administration (FDA) has approved the antiviral drug Veklury to treat COVID-19 in adults and some pediatric patients who need hospitalization [10]. There are seven vaccines approved for use by WHO: Pfizer/BioNTech, Moderna, Janssen(Johnson & Johnson), Oxford/AstraZeneca, Serum Institute of India, Sinpharm (Beijing), and Sinovac [11].
On the other hand, HIV is a member of RNA lentiviruses [9]. The principal receptor of HIV is CD4 receptor [6, 9]. CD4 is expressed in different immune cells like CD T cells, macrophages, and dendritic cells [6]. Nevertheless, CD T cells are the primary target of HIV. CD T cells help other immune cells like CD T cells and B cells in fighting against viral infections [12]. Targeting CD T cells by HIV causes a reduction in the number of these cells. Therefore, the body of HIV patient becomes susceptible to other viral infections [13]. HIV is transmitted through blood or sexual contact [6]. Antiretroviral therapy (ART) is used to treat HIV infection, which reduces the viral load and prevents the development to the acquired immunodeficiency syndrome (AIDS) [14]. Notably, no HIV vaccines have been approved yet [6].
The first case of SARS-CoV-2/HIV coinfection was reported for a 61-year-old man from China [9]. Other coinfection cases were reported in Spain, Italy, and the USA [9]. The most typical symptoms of coinfection were fever, cough, and shortness of breath [9]. It has been found that HIV patients are more likely to experience severe COVID-19 when infected [3]. The risk of severe infection increases further in HIV patients who do not receive antiretroviral therapy or have low CD T cell counts [3, 15]. Furthermore, the severity risk increases in the presence of other comorbidities like hypertension, diabetes, respiratory disease, cardiovascular disease, and chronic kidney disease [3, 16, 17]. Based on WHO recommendations [18], many COVID-19 vaccines are safe for people living with HIV.
Mathematical modeling has been considered a significant tool for studying and investigating viral infections. HIV within-host models have received great attention and lead to the significant results. These models were formulated using ordinary differential equations (ODEs) [19–23], delay differential equations (DDEs) [24–27], partial differential equations (PDEs) [28–31], delay partial differential equations (DPDEs) [13, 32–34], and fractional differential equations (FDEs) [35, 36]. These models exhibit mainly the interactions between HIV, uninfected CD T cells, different types of infected CD T cells, and the immune system.
However, very few models have studied so far to investigate the dynamics of SARS-CoV-2 within the human body. For example, Li et al. [37] formulated a within-host ODE model to characterize the interactions between uninfected epithelial cells, infected epithelial cells, and SARS-CoV-2 particles. Du and Yuan [38] analyzed a similar model with taking into consideration the effect of antiviral drugs which prevent either infection or the production of SARS-CoV-2 particles. Al Agha et al. [7] used a within-host DDE to depict the effect of SARS-CoV-2 infection on cancer patients and the impact of infection on immune responses. Pinky and Dobrovolny [39] established a within-host model to study SARS-CoV-2 coinfection with other viruses like influenza A virus and human rhinovirus. Fadai et al. [40] proposed an ODE model with the assumption that uninfected epithelial cells follow logistic growth.
To the best of our knowledge, no within-host SARS-CoV-2/HIV models have been studied so far. However, it is worth mentioning that Bellomo et al. [41, 42] studied the within-host dynamics of SARS-CoV-2 within a multiscale approach and the mathematical theory of active particles. The multiscale approach accounts for the interaction of different spatial scales where the dynamics at the high scale of individuals depends on the dynamics at the microscopic scale. Microscopic scale is determined by the competition between virus particles and the immune cells. Thus, the multiscale approach can be used to predict the time evolution of the number of healthy, infected, recovered, and dead individuals. Nevertheless, in this paper we concentrate on the interactions at the microscopic scale of coinfection between SARS-CoV-2 and HIV. Coinfection models are needed to help understand the dynamics of SARS-CoV-2 infection in HIV patients and the role of the immune system, to support medical research, and to find better ways to treat this vulnerable group of patients. In this paper, we establish a within-host model of SARS-CoV-2/HIV coinfection. For this model, we (i) demonstrate that all solutions are bounded and nonnegative, (ii) calculate all steady-state solutions and the corresponding conditions of their existence, (iii) show the global stability of the steady states, (iv) execute some numerical simulations to enhance the results of computations, (v) discuss the effect of low concentration of CD T cells on coinfected patients, (vi) test the impact of latency on the number of SARS-CoV-2 particles and HIV particles, and (vii) suggest some possible future works.
The paper is organized as follows. Section 2 describes the model under consideration. Section 3 shows that all solutions are bounded and greater than or equal zero. In addition, it lists all steady states with the positivity conditions of their components. Section 4 proves the global stability of all steady states computed in Sect. 3. Section 5 displays some numerical simulations. Finally, Sect. 6 discusses the results with some suggestions for future works.
SARS-CoV-2/HIV coinfection model with latency
This section describes the model intended to be studied in this paper. The proposed model takes the form
| 1 |
where denote the concentrations of uninfected epithelial cells, latently infected epithelial cells, actively infected epithelial cells, free SARS-CoV-2 particles, uninfected CD T cells, latently infected CD T cells, actively infected CD T cells, and free HIV particles at time t. Epithelial cells are produced from a source at a constant rate , die at rate , and get infected by SARS-CoV-2 at rate . Latently infected epithelial cells proliferate at rate , turn into active infected cells at rate kN, and die at rate . Actively infected epithelial cells die at rate and are indirectly eliminated by CD T cells at rate . SARS-CoV-2 particles are produced from infected cells at rate aY and die at rate . Uninfected CD T cells are produced at a constant rate , stimulated by infected epithelial cells at rate uYS, die at rate , and get infected by HIV at rate . A fraction of new infected CD T cells will be active and the rest will be latent. Latently infected CD T cells are transmitted into active cells at rate and die at a natural death rate . Actively infected CD T cells die at a natural death rate . HIV particles are produced by infected cells at rate and die at rate . The descriptions of the different parameters are summarized in Table 1.
Table 1.
Values of parameters of model (1)
| Par. | Description | Value | References |
|---|---|---|---|
| Recruitment rate of uninfected epithelial cells | 0.02241 | [43] | |
| Death rate constant of uninfected epithelial cells | [43] | ||
| Infection rate constant of epithelial cells | Varied | – | |
| k | Transmission rate constant of latently infected epithelial cells into active cells | 4.08 | [39] |
| Death rate constant of latently infected epithelial cells | [39] | ||
| Death rate constant of actively infected epithelial cells | 0.11 | [43] | |
| Indirect killing rate constant of CD T cells | Varied | – | |
| a | Production rate constant of SARS-CoV-2 by actively infected epithelial cells | 0.24 | [43] |
| Death rate constant of free SARS-CoV-2 particles | Varied | – | |
| Recruitment rate of uninfected CD T cells | 10 | [21] | |
| u | Stimulation rate constant of CD T cells | 0.1 | [44] |
| Death rate constant of uninfected CD T cells | 0.01 | [45] | |
| Infection rate constant of CD T cells | Varied | – | |
| Transmission rate constant of latently infected CD T cells into active cells | 0.2 | [46] | |
| Death rate constant of latently infected CD T cells | 0.02 | [46] | |
| b | A fraction of newly infected CD T cells that become active | 0.7 | [46] |
| Death rate constant of actively infected CD T cells | 0.5 | [47] | |
| Production rate constant of HIV by actively infected cells | 5 | [48] | |
| Death rate constant of free HIV particles | 2 | [48] |
Basic properties
This section verifies the nonnegativity and boundedness of solutions of model (1). Moreover, it calculates all possible steady states with the associated threshold conditions.
Nonnegativity and boundedness
We define a compact set
where , .
Proposition 1
The set is positively invariant for model (1).
Proof
From model (1), we have
Thus, we get for all when .
To prove the boundedness of all state variables, we define
Then, we get
where . It follows that
Hence, if for , where . As X, N, Y, V, S, T, W and H are nonnegative, we have , , if , where , , and . This shows that the set is positively invariant.
Steady states
In this subsection, we calculate all possible steady states of model (1) and conclude the threshold conditions that cover the existence of these steady states.
To compute the steady states of the model we solve the following system of algebraic equations:
We find that model (1) has four steady states:
-
(i)
The uninfected steady state , where and .
-
(ii)The single HIV-infection steady state , where
where . Here, is the basic reproduction number of HIV infection. It determines the establishment of HIV infection in the body. We see that and are always positive, while , and are positive if . Therefore, exists when . -
(iii)The single SARS-CoV-2–infection steady state , where
and satisfies the following equation:2
where3
Now we show that there exists a positive root for Eq. (3). We define a function G(V) as
We have
where . This implies that when . Moreover, we find that
It follows that there exists such that . From Eq. (2) we get , and . As a result, exists when . The parameter is the basic reproduction number of SARS-CoV-2 infection. It determines the establishment of SARS-CoV-2 infection in the body. -
(iv)The SARS-CoV-2/HIV coinfection steady state , where
It follows that , and only when . On the other hand, , and only when . Thus, we can rewrite the components of as
where
Therefore, exists when and . At this point, and are threshold numbers that determine the occurrence of SARS-CoV-2/HIV coinfection.
All steady states of model (1) and their existence conditions are summarized in Table 2.
Table 2.
Steady states of model (1) and their existence conditions
| Steady state | Definition | Existence conditions |
|---|---|---|
| Uninfected steady state | None | |
| Single HIV–infection steady state | ||
| Single SARS-CoV-2–infection steady state | ||
| SARS-CoV-2/HIV coinfection steady state | and |
The four threshold parameters are given as follows:
Global stability of steady states
In this section, we prove the global asymptotic stability of all steady states by constructing Lyapunov functions following the method presented in [49]. We define . We will use the arithmetic–geometric mean inequality
which yields
| 4 |
| 5 |
| 6 |
Theorem 1
If and , then is globally asymptotically stable (G.A.S).
Proof
Construct a Lyapunov function as:
Clearly, for all , and . By calculating along the solutions of model (1), we get
Using and , we obtain
As and , we get for all and when , and . Define and let be the largest invariant subset of . The solutions of model (1) converge to . The set includes elements with , and , and hence . The third and last equations of model (1) yield
Thus, for all t. The second and seventh equations give
Thus, for all t. Therefore, and by applying Lyapunov–LaSalle asymptotic stability theorem [50–52] we get that is G.A.S.
Theorem 2
If and , then is globally asymptotically stable (G.A.S).
Proof
Define a Lyapunov function as
By differentiating , we obtain
Using steady-state conditions for , we get
Then, we obtain
As and according to inequalities (4) and (5), we get for all . Moreover, when , , , , and . The solutions of model (1) converge to the largest invariant subset of . The set includes , and then . The third equation of model (1) implies that
which yields for all t. We get from the second equation that
which yields for all t. Hence, and is G.A.S using Lyapunov–LaSalle asymptotic stability theorem.
Theorem 3
If and , then is globally asymptotically stable (G.A.S).
Proof
Define a Lyapunov function as:
By differentiating , we get
By using the steady-state conditions for
we obtain
Hence, if , then does not exist since , and . This implies that
It follows that for all . Thus, and by using inequality (6), we get for all with equality holding when , , , , and . The solutions of model (1) converge to , the largest invariant subset of . contains elements with , and then . Using the last equation of model (1), we obtain
which gives for all t. Using the seventh equation of model (1), we obtain
which gives for all t. Therefore, and by applying Lyapunov–LaSalle asymptotic stability theorem we get that is G.A.S.
Theorem 4
If and , then is globally asymptotically stable (G.A.S).
Proof
Define a Lyapunov function as:
Differentiating with respect to t gives
By using the steady-state conditions of
we get
Since and using inequalities (4), (5) and (6) we get for all . Moreover, when , , , , , , and . The solutions of model (1) converge to the largest invariant subset of . Hence, and is G.A.S using Lyapunov–LaSalle asymptotic stability theorem.
The global stability conditions of all steady states are summarized in Table 3.
Table 3.
Global stability conditions of the steady states of model (1)
| Steady state | Global stability conditions |
|---|---|
| and | |
| and | |
| and | |
| and |
Numerical simulations
This section presents some numerical simulations to assist the results obtained in the previous parts. In addition, it shows the impact of low number of CD T cells on SARS-CoV-2/HIV coinfection. Furthermore, it illustrates the effect of death rates during the latency periods on viral loads. To achieve these goals, we consider three sets of initial conditions as follows:
Set 1 , , , , , , , .
Set 2 , , , , , , , .
Set 3 , , , , , , , .
The selection of these values is optional. Furthermore, it is divided into three sets to ensure that the global stability of the steady states is not affected by the choice of initial conditions. We use the MATLAB solver ode45 to solve system (1). According to the global stability of the steady states , , , and in Theorems 1–4, we split the simulations into four cases. In these cases, we vary the values of , , , and of model (1). The values of all other parameters are fixed and listed in Table 1. The four cases are given as follows:
-
(i)
We take , , , and . The thresholds in this case are given by and . In harmony with Theorem 1, the steady state is G.A.S (see Fig. 1). This is an optimal situation when the person does not have neither SARS-CoV-2 infection nor HIV infection.
-
(ii)
We choose , , , and . This provides us with and . According to Theorem 2, the steady state is G.A.S (see Fig. 2). This point represents the case when a person has HIV infection with low CD T cell counts, while SARS-CoV-2 infection is not detected.
-
(iii)
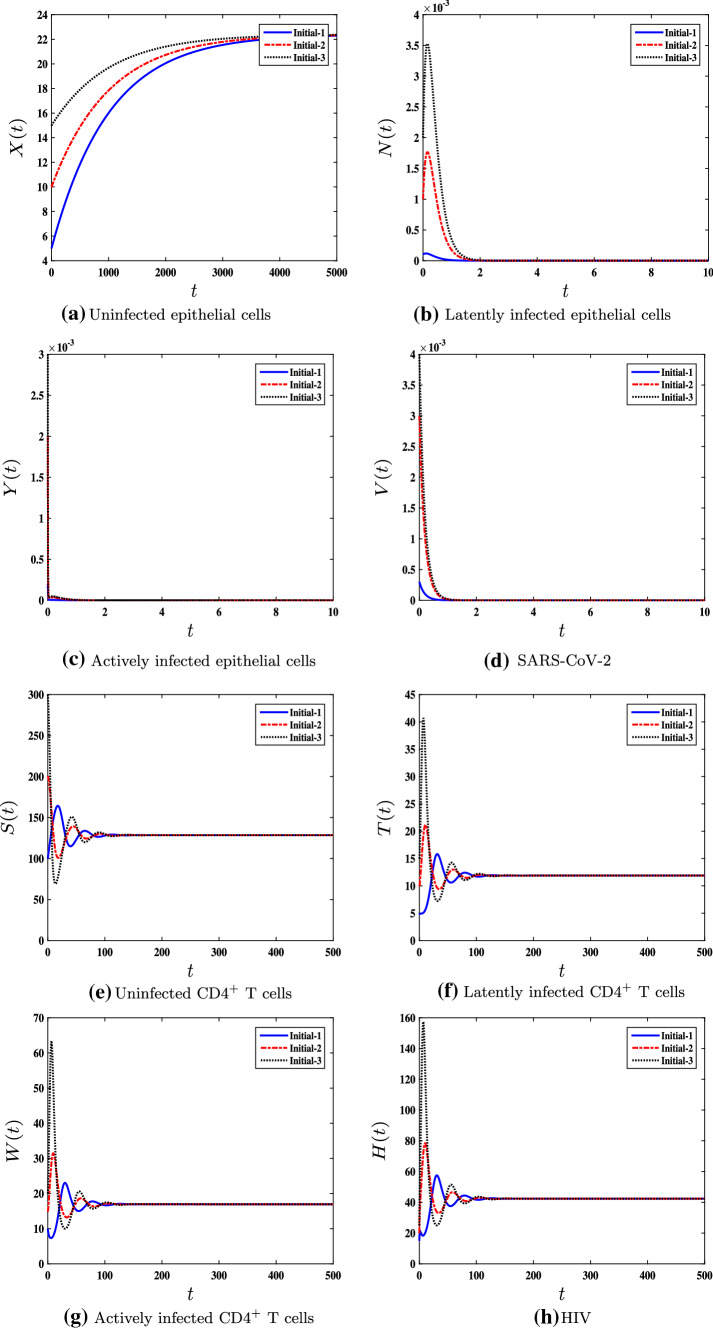
We select , , , and . This gives and . In this case, the solutions globally converge to the steady state . This result agrees with Theorem 3 and is displayed in Fig. 3. This case represents a person with SARS-CoV-2 infection, but he does not have HIV disease.
-
(iv)
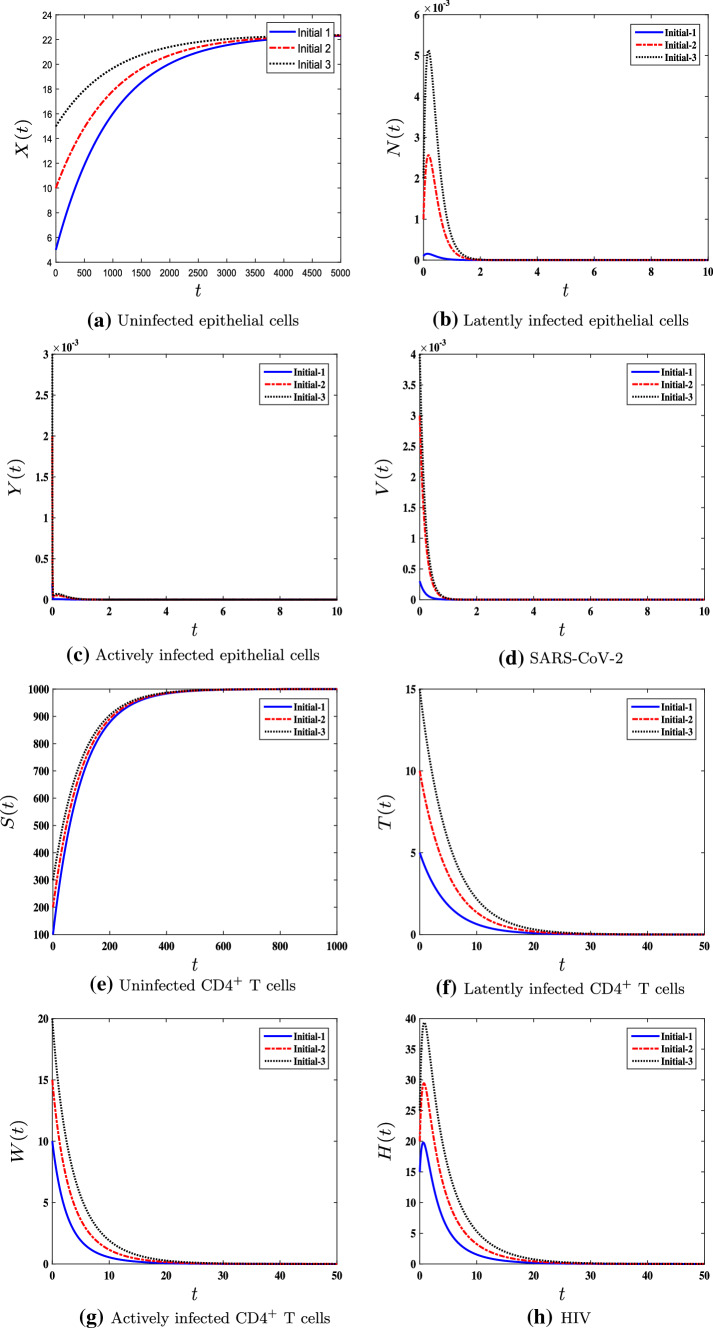
We consider , , , and . This implies that , , and . In harmony with Theorem 4, the steady state is G.A.S (see Fig. 4). In this situation, SARS-CoV-2/HIV coinfection occurs, where an HIV patient gets infected with COVID-19. CD T cells are stimulated to eliminate SARS-CoV-2 infection from the body. Nevertheless, if the patient has low CD T cell counts, the clearance of SARS-CoV-2 may not be achieved. This can lead to severe infection and death.
For further verification of the asymptotic stability of , we calculate the Jacobian matrix of model (1) at the steady state as
Next, we calculate the eigenvalues () of the Jacobian matrix at all possible steady states. For asymptotic stability of , we need to prove that
and all other steady states have eigenvalues with positive real parts. The computations are organized in Table 4.
Fig. 1.
The numerical simulations of model (1) for , , , and with three different sets of initial conditions. The uninfected steady state is G.A.S
Fig. 2.
The numerical simulations of model (1) for , , , and with three different sets of initial conditions. The single HIV–infection steady state is G.A.S
Fig. 3.
The numerical simulations of model (1) for , , , and with three different sets of initial conditions. The single SARS-CoV-2–infection steady state is G.A.S
Fig. 4.
The numerical simulations of model (1) for , , , and with three different sets of initial conditions. The SARS-CoV-2/HIV coinfection steady state is G.A.S
Table 4.
Local stability of the steady state
| Case | The steady sates | Re(), | Stability |
|---|---|---|---|
| (iv) | (− 19.907, − 4.94688, − 3.70454, 1.29544, 0.562873, − 0.310899, − 0.01, − 0.001) | Unstable | |
| (− 4.46776, − 4.46776, − 2.37868, 2.07443 , − 0.340978, − 0.0390795, − 0.0390795, − 0.001) | Unstable | ||
| (− 20.2784, − 4.20655, − 3.71538, 1.30623, − 0.310848, − 0.00380659, − 0.00380659, − 0.00987671) | Unstable | ||
| (− 3.81897, − 3.04464, − 2.3788, − 0.340867, − 0.0395697, − 0.0395697, − 0.0273365, − 0.0273365) | Stable |
The effect of the CD T cells killing rate
To check the impact of changing the value of on the stability of model (1), we take the same values used in case (iv) (, , and ) with increasing the value of from 0.02 to 1.5. It follows that and . Thus, the steady state is G.A.S. Mathematically, increasing the value of switches the value of from to . This means that there is a bifurcation at . Accordingly, the steady state becomes unstable and becomes G.A.S.
In addition, to characterize the effect of increasing or decreasing the value of on the number of actively infected epithelial cells and SARS-CoV-2 particles, we examine case (iv) with different values of (Fig. 5). We find out that decreasing the value of increases the concentration of actively infected epithelial cells and, accordingly, the concentration of SARS-CoV-2 particles is increased. On the other hand, increasing the value of decreases SARS-CoV-2 viral load.
Fig. 5.

The effect of decreasing on the concentrations of SARS-CoV-2 particles V(t). The parameters considered are , , and with initial conditions
Biologically, these results imply that high killing rate of CD T cells is needed to remove SARS-CoV-2 from the body of HIV patient. Conversely, low killing rates can cause severe SARS-CoV-2 infection for HIV patient.
The effect of latency
To see the effect of the eclipse phase on the production of SARS-CoV-2 particles, we take the same values considered in case (iv) with increasing the value of . We observe from Fig. 6a that increasing the death rate of latently infected epithelial cells decreases the concentration of SARS-CoV-2 particles in coinfected patients. Similarly, increasing the death rate () of latently infected CD T cells decreases the density of HIV particles (See Fig. 6b). Thus, the death rates during the latency periods can have a strong impact on the viral loads.
Fig. 6.
The effect of increasing the death rates during the latency periods on SARS-CoV-2 and HIV particles. The parameters considered are , , and with initial conditions
Discussion
In this paper, we developed a within-host SARS-CoV-2/HIV coinfection model that investigates the interactions between eight components: uninfected epithelial cells, latently infected epithelial cells, productively infected epithelial cells, SARS-CoV-2 particles, uninfected CD T cells, latently infected CD T cells, productively infected CD T cells, and HIV particles. The model has four steady states as the following:
The uninfected steady state always exists. It is G.A.S when and . This represents the healthy state when the person does not suffer from neither SARS-CoV-2 infection nor HIV infection.
The HIV infection steady state is defined if , and it is G.A.S if . This represents the case of a patient carrying only HIV infection.
The SARS-CoV-2–infection steady state is defined if , and it is G.A.S if . This represents the case of a patient carrying only SARS-CoV-2 infection.
The SARS-CoV-2/HIV coinfection steady state is defined and G.A.S if and . This case simulates the occurrence of SARS-CoV-2 infection in HIV patients.
The numerical results are totally compatible with the theoretical results. We found that decreasing the killing rate () of CD T cells increases the concentrations of both productively infected epithelial cells and SARS-CoV-2 particles. This implies that low CD T cell counts can increase the severity of SARS-CoV-2 infection in HIV patients. This result comes in agreement with many results that discussed that HIV patients with low CD T cell counts or who do not receive ART are at higher risk of death when they get infected by SARS-CoV-2. In addition, we observed that increasing the death rate () of infected epithelial cells during the latency period decreases SARS-CoV-2 viral load in the body. Increasing means that more cells will die in the eclipse phase before converting into productively infected cells. This can have a positive effect on reducing the severity of SARS-CoV-2 infection in HIV patients. Comparing with previous studies, the model considered in this work is the first model that takes into consideration the coinfection of HIV with SAR-CoV-2. The results obtained in this paper can be examined and used to (i) understand SARS-CoV-2/HIV coinfection, (ii) estimate the values of the parameters that are needed to clear SARS-CoV-infection from the body of HIV patient, (iii) test the effect of increasing the killing rate () on SARS-CoV-2 viral load, and (iv) examine the effect of death rates during the latency periods on the concentrations of viral particles. The main limitation of this work is that we did not use real data due to its unavailability. Therefore, these results can be examined when more data become available.
The model studied in this paper can be improved by considering the effect of time delays that are associated with many biological processes. Furthermore, adding the effect of treatments may lead to the important results that can help to find treatments for this group of patients. In addition, the coinfection dynamics of SARS-CoV-2 and HIV can be studied within a multiscale approach [41, 42] which can provide a deeper understanding and help develop vaccines and antiviral therapies. Finally, the results can be developed by using real data to find an accurate estimation of the parameters of model (1).
Acknowledgements
This work was supported by the Deanship of Scientific Research (DSR), King Abdulaziz University, Jeddah, under grant No. (D-74-130-1443). The authors, therefore, gratefully acknowledge the DSR technical and financial support.
Declarations
Conflict of interest
The authors declare that they have no conflict of interest.
References
- 1.Coronavirus disease (COVID-19), weekly epidemiological update (13 October 2021), World Health Organization (WHO) (2021). Available online: https://www.who.int/docs/default-source/coronaviruse/situation-reports/20211013_weekly_epi_update_61.pdf?sfvrsn=5092f979_3&download=true
- 2.Coronavirus disease (COVID-19): HIV and antiretrovirals, World Health Organization (WHO) (2020). Available online: https://www.who.int/news-room/q-a-detail/coronavirus-disease-covid-19-hiv-and-antiretrovirals
- 3.Ssentongo P, Heilbrunn ES, Ssentongo AE, Advani S, et al. Epidemiology and outcomes of COVID-19 in HIV-infected individuals: a systematic review and meta-analysis. Sci. Rep. 2021;11(1):1–12. doi: 10.1038/s41598-021-85359-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.HIV/AIDs: Fact sheets, World Health Organization (WHO) (2021). Available online: https://www.who.int/news-room/fact-sheets/detail/hiv-aids
- 5.Bakouny Z, Hawley JE, Choueiri TK, Peters S, et al. COVID-19 and cancer: current challenges and perspectives. Cancer Cell. 2020;38:629–646. doi: 10.1016/j.ccell.2020.09.018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Gatechompol S, Avihingsanon A, Putcharoen O, Ruxrungtham K, Kuritzkes DR, Res AIDS. Therapy. 2021 doi: 10.1186/s12981-021-00335-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.A. Al Agha, S. Alshehaiween, A. Elaiw, M. Alshaikh, Global analysis of delayed SARS-CoV-2/cancer model with immune response. Mathematics 9(11), 1–27 (2021)
- 8.Derosa L, Melenotte C, Griscelli F, Gachot B, et al. The immuno-oncological challenge of COVID-19. Nat. Cancer. 2020;1(10):946–964. doi: 10.1038/s43018-020-00122-3. [DOI] [PubMed] [Google Scholar]
- 9.Evans N, Martinez E, Petrosillo N, Nichols J. SARS-CoV-2 and human immunodeficiency virus: pathogen pincer attack. HIV/AIDS Res. Palliat. Care. 2021;13:361–375. doi: 10.2147/HIV.S300055. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.The U.S. Food and Drug Administration, Know your treatment options for COVID-19 (2021). Available online: https://www.fda.gov/consumers/consumer-updates/know-your-treatment-options-covid-19
- 11.Coronavirus disease (COVID-19), Vaccine tracker, World Health Organization (WHO) (2021). Available online: https://covid19.trackvaccines.org/agency/who/
- 12.Sette A, Crotty S. Adaptive immunity to SARS-CoV-2 and COVID-19. Cell. 2021;184(4):861–880. doi: 10.1016/j.cell.2021.01.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Elaiw AM, AlAgha AD. Stability of a general HIV-1 reaction-diffusion model with multiple delays and immune response. Physica A Stat. Mech. Appl. 2019;536:1–20. [Google Scholar]
- 14.Peng X, Ouyang J, Isnard S, Lin J, et al. Sharing CD4+ T cell loss: when COVID-19 and HIV collide on immune system. Front. Immunol. 2020;11:1–11. doi: 10.3389/fimmu.2020.596631. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Sharov KS. HIV/SARS-CoV-2 co-infection: T cell profile, cytokine dynamics and role of exhausted lymphocytes. Int. J. Infect. Dis. 2021;102:163–169. doi: 10.1016/j.ijid.2020.10.049. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Ambrosioni J, Blanco J, Reyes-Urueña JM, Davies M, et al. Overview of SARS-CoV-2 infection in adults living with HIV. Lancet HIV. 2021;8(5):E294–E305. doi: 10.1016/S2352-3018(21)00070-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Kanwugu ON, Adadi P. HIV/SARS-CoV-2 coinfection: a global perspective. J. Med. Virol. 2020;93(2):726–732. doi: 10.1002/jmv.26321. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.COVID-19 vaccines, World Health Organization (WHO) (2021). Available online: https://www.who.int/news-room/q-a-detail/coronavirus-disease-(covid-19)-covid-19-vaccines-and-people-living-with-hiv
- 19.Nowak MA, Bangham CRM. Population dynamics of immune responses to persistent viruses. Science. 1996;272:74–79. doi: 10.1126/science.272.5258.74. [DOI] [PubMed] [Google Scholar]
- 20.Elaiw AM. Global properties of a class of HIV models. Nonlinear Anal. Real World Appl. 2010;11(4):2253–2263. [Google Scholar]
- 21.Perelson A, Kirschner D, De Boer R. Dynamics of HIV infection of CD4+ T cells. Math. Biosci. 1993;114(1):81–125. doi: 10.1016/0025-5564(93)90043-a. [DOI] [PubMed] [Google Scholar]
- 22.Perelson A, Essunger P, Cao Y, Vesanen M, Hurley A, Saksela K, Markowitz M, Ho D. Decay characteristics of HIV-1-infected compartments during combination therapy. Nature. 1997;387:188–191. doi: 10.1038/387188a0. [DOI] [PubMed] [Google Scholar]
- 23.Culshaw R, Ruan S, Spiteri R. Optimal HIV treatment by maximising immune response. J. Math. Biol. 2004;48(5):545–562. doi: 10.1007/s00285-003-0245-3. [DOI] [PubMed] [Google Scholar]
- 24.Pawelek K, Liu S, Pahlevani F, Rong L. A model of HIV-1 infection with two time delays: mathematical analysis and comparison with patient data. Math. Biosci. 2012;235(1):98–109. doi: 10.1016/j.mbs.2011.11.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Shu H, Wang L, Watmough J. Global stability of a nonlinear viral infection model with infinitely distributed intracellular delays and CTL immune responses. SIAM J. Appl. Math. 2013;73(3):1280–1302. [Google Scholar]
- 26.Li Y, Xu R, Mao S. Global dynamics of a delayed HIV-1 infection model with CTL immune response. Discrete Dyn. Nat. Soc. 2011;2011:1–13. [Google Scholar]
- 27.Xu R. Global stability of an HIV-1 infection model with saturation infection and intracellular delay. J. Math. Anal. Appl. 2011;375:75–81. [Google Scholar]
- 28.AlAgha AD, Elaiw AM. Stability of a general reaction-diffusion HIV-1 dynamics model with humoral immunity. Eur. Phys. J. Plus. 2019;134(8):1–18. [Google Scholar]
- 29.Ren X, Tian Y, Liu L, Liu X. A reaction-diffusion within-host HIV model with cell-to-cell transmission. J. Math. Biol. 2018;76:1831–1872. doi: 10.1007/s00285-017-1202-x. [DOI] [PubMed] [Google Scholar]
- 30.Lai X, Zou X. A reaction diffusion system modeling virus dynamics and CTL response with chemotaxis. Discrete Contin. Dyn. Syst. 2016;21(8):2567–2585. [Google Scholar]
- 31.Zhuang K. Dynamics of a diffusive viral model with Beddington-DeAngelis incidence rate and CTL immune response. J. Nonlinear Sci. Appl. 2017;10(11):5753–5762. [Google Scholar]
- 32.Polyanin AD, Sorokin VG, Vyazmin AV. Reaction-diffusion models with delay: some properties, equations, problems, and solutions. Theor. Found. Chem. Eng. 2018;52(3):334–348. [Google Scholar]
- 33.Kang C, Miao H, Chen X, Xu J, Huang D. Global stability of a diffusive and delayed virus dynamics model with Crowley-Martin incidence function and CTL immune response. Adv. Differ. Equ. 2017;2017(324):1–16. [Google Scholar]
- 34.Miao H, Teng Z, Abdurahman X, Li Z. Global stability of a diffusive and delayed virus infection model with general incidence function and adaptive immune response. Comput. Appl. Math. 2018;37(3):3780–3805. [Google Scholar]
- 35.Bachraoui M, Hattaf K, Yousfi N. Global stability of a fractional order HIV infection model with cure of infected cells in eclipse stage. Revue Africaine de la Recherche en Informatique et Mathématiques Appliquéaes. 2019 doi: 10.46298/arima.4359. [DOI] [Google Scholar]
- 36.Carvalho ARM, Pinto CMA, Baleanu D. HIV/HCV coinfection model: a fractional-order perspective for the effect of the HIV viral load. Adv. Differ. Equ. 2018;2018(2):1–22. [Google Scholar]
- 37.Li C, Xu J, Liu J, Zhou Y. The within-host viral kinetics of SARS-CoV-2. Math. Biosci. Eng. 2020;17(4):2853–2861. doi: 10.3934/mbe.2020159. [DOI] [PubMed] [Google Scholar]
- 38.Du SQ, Yuan W. Mathematical modeling of interaction between innate and adaptive immune responses in COVID-19 and implications for viral pathogenesis. J. Med. Virol. 2020;92(9):1615–1628. doi: 10.1002/jmv.25866. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Pinky L, Dobrovolny HM. SARS-CoV-2 coinfections: Could influenza and the common cold be beneficial? J. Med. Virol. 2020;92:1–8. doi: 10.1002/jmv.26098. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Fadai NT, Sachak-Patwa R, Byrne HM, Maini PK, Bafadhel M, Nicolau DV. Infection, inflammation and intervention: mechanistic modelling of epithelial cells in COVID-19. J. R. Soc. Interface. 2021 doi: 10.1098/rsif.2020.0950. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Bellomo N, Bingham R, Chaplain MAJ, Dosi G, et al. A multiscale model of virus pandemic: heterogeneous interactive entities in a globally connected world. Math. Models Methods Appl. Sci. 2020;30(8):1591–1651. doi: 10.1142/s0218202520500323. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Bellomo N, Burini D, Dosi G, Gibelli L, et al. What is life? A perspective of the mathematical kinetic theory of active particles. Math. Models Methods Appl. Sci. 2021;31(9):1821–1866. [Google Scholar]
- 43.Nath BJ, Dehingia K, Mishra VN, Chu Y, Sarmah HK. Mathematical analysis of a within-host model of SARS-CoV-2. Adv. Differ. Equ. 2021;2021(1):1–11. doi: 10.1186/s13662-021-03276-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Prakash M, Rakkiyappan R, Manivannan A, Cao J. Dynamical analysis of antigen-driven T-cell infection model with multiple delays. Appl. Math. Comput. 2019;354:266–281. [Google Scholar]
- 45.Callaway DS, Perelson AS. HIV-1 infection and low steady state viral loads. Bull. Math. Biol. 2002;64:29–64. doi: 10.1006/bulm.2001.0266. [DOI] [PubMed] [Google Scholar]
- 46.Elaiw AM, AlShamrani NH, Hobiny AD. Mathematical modeling of HIV/HTLV co-infection with CTL-mediated immunity. AIMS Math. 2021;6(2):1634–1676. [Google Scholar]
- 47.Perelson AS, Nelson PW. Mathematical analysis of HIV-1 dynamics in vivo. SIAM Rev. 1999;41(1):3–44. [Google Scholar]
- 48.Adak D, Bairagi N. Analysis and computation of multi-pathways and multi-delays HIV-1 infection model. Appl. Math. Model. 2018;54:517–536. [Google Scholar]
- 49.Korobeinikov A. Global properties of basic virus dynamics models. Bull. Math. Biol. 2004;66:879–883. doi: 10.1016/j.bulm.2004.02.001. [DOI] [PubMed] [Google Scholar]
- 50.E.A. Barbashin, Introduction to the Theory of Stability (Wolters-Noordhoff, 1970)
- 51.J.P. La Salle, The Stability of Dynamical Systems (Society for Industrial and Applied Mathematics, 1976)
- 52.Lyapunov AM. The general problem of the stability of motion. Int. J. Control. 1992;55(3):531–534. [Google Scholar]