Summary.
More than 50% of the world’s population lives in urban centers. As collection basins for landscape activity, urban waters are an interface between human activity and the natural environment. The microbiome of urban waters could provide insight into the impacts of pollution, the presence of human health risks, or the potential for long-term consequences for these ecosystems and the people who depend upon them. An integral part of the urban water cycle is sewer infrastructure. Thousands of miles of pipes line cities as part of wastewater and stormwater systems. As stormwater and sewage are released into natural waterways, traces of human and animal microbiomes reflect the sources and magnitude of fecal pollution and indicate the presence of pollutants, such as nutrients, pathogens, and chemicals. Non-fecal organisms are also released as part of these systems. Runoff from impervious surfaces delivers microbes from soils, plants and the built environment to stormwater systems. Further, urban sewer infrastructure contains its own unique microbial community seemingly adapted to this relatively new artificial habitat. High microbial densities are conveyed via pipes to waterways, and these organisms can be found as an urban microbial signature imprinted on the natural community of rivers and urban coastal waters. The potential consequences of mass releases of non-indigenous microorganisms into natural waters include creation of reservoirs for emerging human pathogens, altered nutrient flows into aquatic food webs, and increased genetic exchange between two distinct gene pools. This review highlights the recent characterization of the microbiome of urban sewer and stormwater infrastructure and its connection to and potential impact upon freshwater systems.
Keywords: urban freshwaters, infrastructure and sanitation, next generation sequencing, human health, aquatic food webs
Introduction
Microbes underpin the integrity of clean water. In past centuries, removing harmful microorganisms and remediating wastewater in urban areas proved very difficult and frequent disease outbreaks occurred. As urban areas grew and urban infrastructure advanced, humans recognized the need for technologies capable of capturing and later, also treating wastewater to maintain high water quality in surrounding surface waters. Although these technologies have resulted in vastly improved water quality, human waste is still found in urban waterways. Further, large-scale urban infrastructure created in the past 100 to 150 years has created a number of relatively new ecological niches for colonization by microorganisms (e.g., sewer conveyance pipes, secondary wastewater treatment, drinking water infrastructure). Our understanding is limited with regards to the microbial communities now inhabiting these systems, how these communities interface with natural environments, and how new human pathogens may evolve or emerge.
Sources and transport of urban microbes
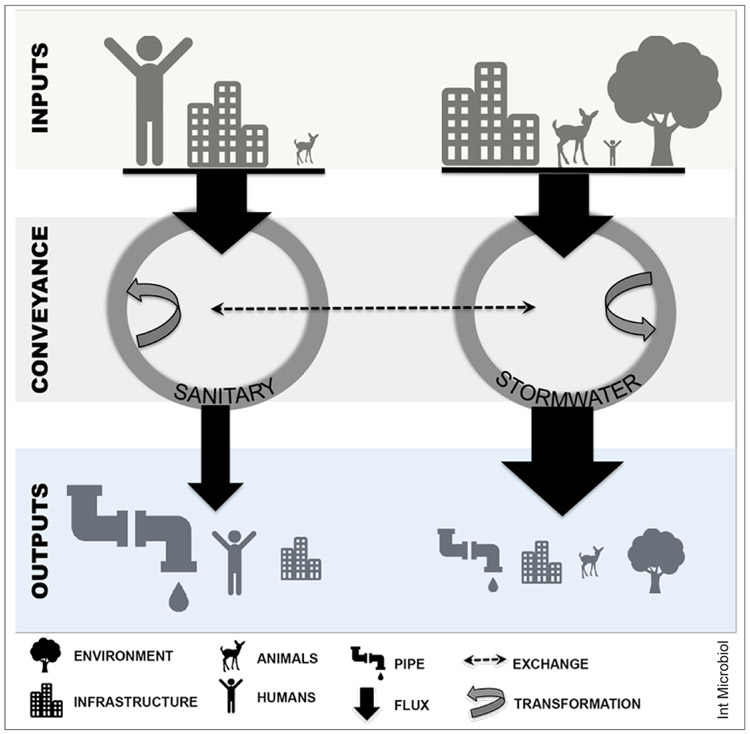
The urban water cycle is a clear example of how human activity interfaces with aquatic environments (Fig. 1), where pipes and impervious surfaces serve as pathways for urban- and human-derived microbes to enter waterways. Within a large metropolitan area, sewer infrastructure can consist of thousands of miles of pipes that transport human waste and/or stormwater away from homes and city buildings to surface waters (rivers, estuaries, lakes). During the Industrial Revolution in Europe and North America, pipes were built to carry water from streets to prevent flooding and were later connected to houses and businesses to carry sanitary sewage waste directly to waterways [54]. These early systems were eventually connected to wastewater treatment plants, forming what are known as combined sewers. Older cities in Europe and the USA still have combined sewer systems, which often overflow, releasing untreated sewage mixed with stormwater into local rivers. In some combined sewer systems, only a few mm of rain can cause overflows, but the capacity varies greatly among cities [35,42]. In a single combined sewer overflow (CSO), millions to billions of gallons of stormwater runoff and untreated sewage can be released [57]. Bacterial densities in stormwater and sanitary sewage are much higher than the receiving waters [33]; therefore these overflow events could leave a significant imprint on the natural bacterial community.
Fig. 1.
Sanitary sewers act as collectors of organisms from “indoor” microbiomes, including bacteria associated with the human body and waste, food, and pipes. Stormwater sewers collect organisms from “outdoor” microbiomes, such as soil, impervious surfaces, plants, and animal feces. Sewers serve as transporters that deliver bacteria to aquatic environments; but microbial communities are also transformed within the sewers, including death of some organisms and growth of others within the pipe.
Newer sewer infrastructure (post 1920s) generally consists of separated sewers, where sanitary sewage is conveyed to wastewater treatment plants and stormwater is collected in a separate set of pipes and discharged directly to waterways. Separated sewer systems can also be a source of human microbial waste to area waterways, typically from sanitary sewage overflows during heavy rain or following pipe deterioration and sewage exfiltration. Fecal bacteria are not the only microbial inputs into sanitary sewage systems. These systems also collect and aggregate the microorganisms associated with grey water waste, such as those on human skin and in the oral cavity, food waste, industrial waste, pet waste, and miscellaneous waste items flushed from homes [21]. The majority of sanitary waste is treated at wastewater treatment plants; however, the resulting effluent contains residual influent microbes and newly introduced microbes from the treatment plant system. Although treated effluent has much lower cell densities than untreated sewage [69], it is a continuous source of urban derived microbes to receiving waters and has been shown to alter the makeup of communities in the natural environment [9,63].
Within separated sewer systems, stormwater systems collect runoff from impervious surfaces to prevent flooding. Rain events are essentially a citywide “cleansing”, thereby washing microbes from exposed surfaces within the urban built (e.g., buildings, roads) and natural (e.g. plants, soils, animals) environments into the pipe conveyance system and ultimately into area waterways via stormwater outfalls. While urban wildlife and domestic pet waste are the primary sources of fecal microbes in stormwater, human sewage also may migrate into these systems from leaking or failing sanitary sewer pipes [45,46] and through illicit pipe connections.
Human and animal microbiomes as tracers of fecal pollution in the environment
The introduction of fecal pollution from urban discharge to surface waters is the most recognized and studied connection between urban water systems. Throughout history, self-perpetuating cycles of waterborne disease occurred with greater frequency in densely populated areas that lacked proper sanitation. Cholera outbreaks plagued major European cities throughout the mid-1800s, including multiple outbreaks in London that eventually led to an understanding of disease transmission [48]. Humans carry pathogenic bacteria, viruses, and protozoa; and even today, fecal pollution of drinking water sources and recreational waters creates a risk for waterborne disease transmission [12,19,24]. Cultivation of Escherichia coli or enterococci, two organisms found in fecal waste, has been used conventionally to assess fecal pollution in waters. However, the use of E. coli and enterococci as fecal pollution indicators does not identify fecal sources, since humans and the majority of animals carry these organisms. In urban areas, fecal pollution from humans is more likely to carry human pathogens than other fecal sources such as urban wildlife, like birds, squirrels, rabbits, and raccoons. Information on the source of fecal pollution is necessary to determine human health risk and provide direction for remediation efforts [12].
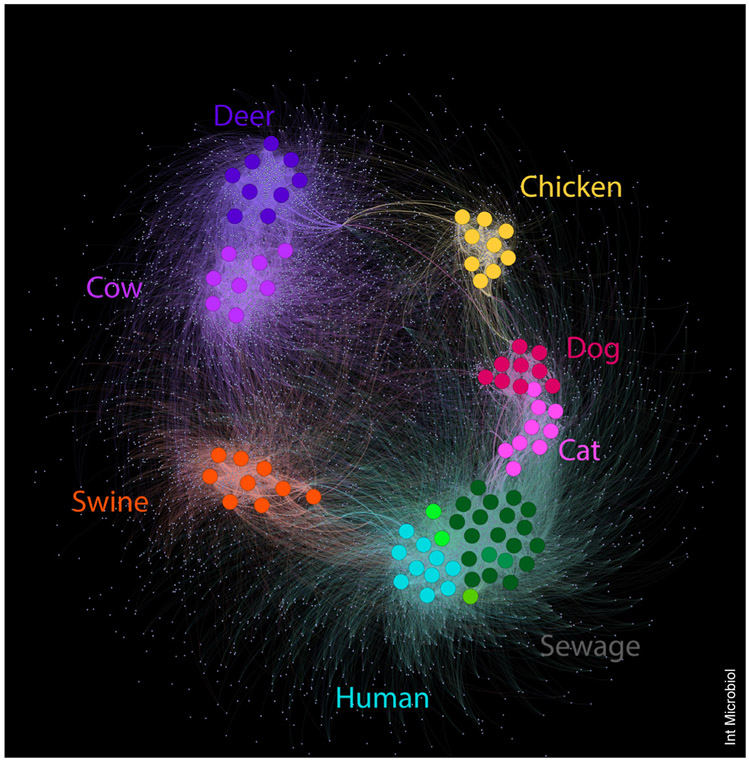
Recent discoveries in microbiome research have shed light on the complex communities associated with humans and animal fecal waste. Using the unique microbial assemblages of a host as a signature or profile is quickly becoming a feasible approach for characterizing pollution sources in surface waters [4,32]. Distinct sequence patterns (Fig. 2) within the microbiome create host signatures that include both unique organisms and organisms with differential relative abundance [13]. Animals of a given species as well as those with closely related physiology (e.g., ruminants) tend to have more similar fecal bacterial communities [26]. Recent studies also showed that domestic animals (e.g., pets) tended to share a higher similarity with cohabiting humans [49]. Bacterial signatures associated with humans and specific animals can be used to identify the relative contributions of each group to urban waters [56]. Upstream of urban areas, agricultural inputs are often the major source of fecal pollution; but this signal transitions to a mixed signal of urban wildlife, domestic pet, and human fecal signatures within cities.
Fig. 2.
Network analysis of the family Lachnospiraceae in fecal communities of animals, humans, and sewage. Large dots represent individual samples, small dots represent operational taxonomic units (OTUs). Lines connect samples and OTUs to show connections among different individuals from the same and from different host species. Sewage samples from Spain, Brazil, Malawi, and the USA are indicated by distinct shades of green. Clear trends within host species are present, as well as OTUs that are shared among hosts or associated with an individual sample.
There is high variability within individual human microbiomes [20,29,55,67]; therefore defining a “typical” human microbiome signature is challenging. Recent work by Newton et al. [37] demonstrated that sewage systems provide an integrative sample of individuals within a city and influent reflects the composite or population-level collection of a city’s human fecal microbial community. Sanitary sewage from 72 cities in the USA had highly similar fecal microbiomes, but also exhibited subtle differences that distinguished the cities. These differences included composition changes that reflected population demographics known to associate with the gut microbiome, in this case obesity levels [37].
Global differences in the gut microbiome have also been demonstrated in studies of humans in different geographic locations [1,66,67], sewage from different countries [11,23], and in source tracking studies of impacted waters [23,43]. While diet can affect the gut microbiome at an individual level [66], several studies showed that significant differences in the human microbiome were observed among geographically and culturally distinct groups as whole [1,7,66,67]. Certain groups of bacteria are more common and more abundant in different groups of humans, and may be more applicable for assessing human fecal pollution. While organisms from the order Bacteroidales have been the primary target for alternative fecal pollution indicators in the USA and Europe, they lack effectiveness in regions where these bacteria are in low abundance in the human gut due to diet or other factors [23,43]. Lachnospiraceae, although thus far less thoroughly explored, may be a preferable target, as these organisms are abundant and widely distributed in diverse human populations [11,23,34].
Urban infrastructure as a new niche for microbial communities
The earliest known rudimentary stormwater and sanitary sewers were initially used for flood control (stormwater); they were later utilized to move human waste out of dense population centers. Despite their significance in early human settlements, this urban infrastructure is a relatively novel environment for microorganisms compared to natural ecosystems like soils, oceans, animals, or even human hosts.
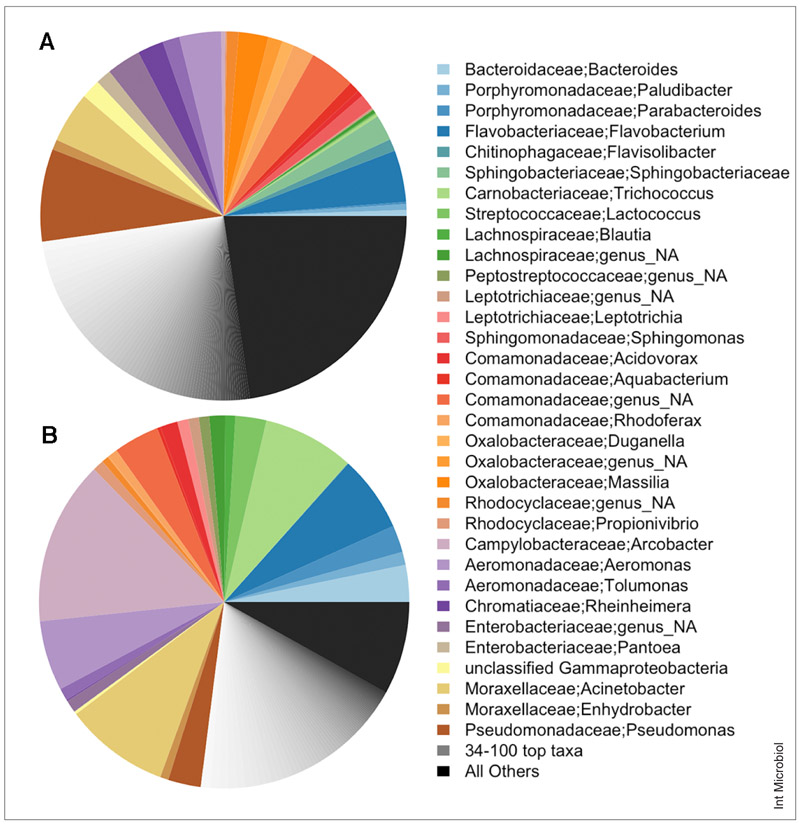
Sewer pipe-derived communities have been investigated primarily to study concrete-corroding biofilms [25,65]. However, recent analysis of bacterial sequences from untreated sewage influent samples from around the USA revealed that the majority of organisms did not match sequences from these biofilms or with human fecal bacteria. On average, nearly 35% of sewage communities were comprised of only five genera: Acinetobacter, Aeromonas, Arcobacter, Pseudomonas, and Trichococcus [12,34,38,50] (Fig. 3). Stormwater collected from pipes or directly from outfalls also contained relatively high proportions of Acinetobacter, Aeromonas, and Pseudomonas, with Arcobacter and Trichococcus nearly absent unless the stormwater had sewage contamination [15]. These organisms appeared to be resident in both sewer systems, as they were consistently present and not readily found in uncontaminated surface water or human or animal microbiomes [13,14,60,67]. Furthermore, the organisms known to cause concrete corrosion are rarely found in influent sewage communities, suggesting that there is a pipe-associated community within loose sediments that are more easily mobilized by turbulent water flow in pipes. This phenomenon has been observed in both sewer and drinking water systems: biofilm communities release very few organisms, while loose sediments contribute the bulk of organisms to flowing water [25,27].
Fig. 3.
Dominant bacterial taxa found in (A) stormwater (n = 30) and (B) untreated sewage influent (n = 6). Several abundant taxa were shared between the two environments and were mainly of non-fecal origin. Both communities were very diverse, with a total of 1709 and 1491 designated taxa in stormwater and untreated sewage, respectively. (Figure adapted from Fisher et al. [15].)
The idea of niche growth in pipe infrastructure is further supported by observations of ecological shifts in populations. Recent work has demonstrated a shift in the distribution of non-fecal organisms in response to geography and season that appears to be driven by temperature differences. In one study of sewage across the USA, fecal communities were largely stable in a given city from season to season, however, the non-fecal organisms changed significantly [37]. A long-term study of sewage from two WWTPs in Milwaukee, WI, USA, showed seasonal variation in two Acinetobacter V6 sequences—sequence was more abundant in summer and fall, while sequence increased in abundance during winter and spring. These two sequences corresponded to different clades of Acinetobacter, confirming that they are different organisms that appear to have different growth optima [60]. Similarly, Arcobacter sequences, which were abundant in sewage from multiple cities in the USA and Reus, Spain, demonstrated temperature dynamics, where two distinct strains of A. cryaerophilus, showed reciprocal abundance trends at temperatures above or below 20°C. Cities with a moderate, consistent climate showed little seasonal variation in the distribution of Arcobacter sequences, while cities that experience more extreme temperature variation showed markedly different communities [14].
The ecology of these very closely related organisms (i.e. populations within a genus) appears to be tied to fine scale factors such as temperature, while the pipe environment itself is the larger driver for selection of these genera as a whole. It is interesting but not fully understood how such similar organisms (at the genus or species level) maintain these highly abundant and ubiquitous populations but have variants that are driven by the same factors that are often major determinants of assemblages in natural environments. One primary concern is how these organisms may survive and function outside of the pipe environment, as these organisms become part of the natural environment along with the other organisms conveyed during storm runoff and sewage overflows. With the exception of Trichococcus, the pipe-associated genera all contain species with some degree of pathogenicity to humans. In the previous Arcobacter example, the two strains of. A. cryaerophilus represent a known clinical strain and an environmental strain [14]; thus the relative human health risk associated with a sewage release may be greater when a pathogenic species is in higher abundance. Depending on the particular strains of the genera present, the pipe environment may represent a new source of pathogens that contribute to waterborne illness.
Urban signature in surface water communities
Organisms from the urban microbiome are consistently transported to natural waters via sewer systems and urban runoff. Stormwater runoff and sanitary sewage (untreated waste in overflows and treated wastewater effluent) are two major sources of urban-associated bacteria to environmental waters. Little crossover exists between bacteria in natural aquatic communities and those in urban effluent; therefore, the imprint of the urban signature can be seen in contaminated waterways. This signature includes both fecal and infrastructure-associated organisms, and their presence increases in magnitude proportionally with storm intensity and duration. Short-term observations reveal a small but persistent community of urban organisms present in chronically impacted aquatic resources, but the long-term consequences in terms of fate and function of the community are unknown.
Evidence of urban impacts on the natural microbial community composition can be observed by both imprints of organisms constituting an urban infrastructure signal [15,48], or a human and animal fecal signal [21,42-45], or by changes in the composition [26,46] and/or functional output [47,48] of naturally occurring aquatic microbes. Annual fluxes of urban microbes from runoff, stormwater, and CSO/SSO depend on the number and intensity of storm events. In heavily urbanized cities with high impervious surface cover, rainfall with intensity of 10 mm h−1 produces >2.2 × 105 m3 day−1 of runoff for every 1 km2, and can deliver trillions of bacteria to surface waters. Stormwater alone is the cause of 32% of impaired estuaries in the USA [58], and CSOs introduce >107 m3 of combined sewage and stormwater in both North America and Europe every year [16]. Table 1 highlights bacterial taxa commonly associated with urban sources.
Table 1.
Bacterial taxa associated with urban sources
| Source | Dominant urban-associated organisms | Citation |
|---|---|---|
| Treated effluent | Vibrio, Mycobacterium, Serratia, TM7, Clostridium XI, Arcobacter, Rhodobacter, Pseudomonas, Legionella, Acinetobacter, Aeromonas, Dechloromonas, Thiothrix, Zooglea | [2,68, Unpublished data] |
| Stormwater | Oxalobacteraceae, Acinetobacter, Pseudomonas, Aeromonas, Tolumonas, Enterobacteriaceae, Pantoea | [13,15.36] |
| Combined sewer overflow | Pseudomonas, Enterobacteriaceae, Acinetobacter, Arcobacter, Trichococcus, Bacteroidaceae, Lachnospiraceae, Porphyromonadaceae, Clostridiaceae, Ruminococcaceae | [36] |
Wastewater effluent is often discharged to surface waters surrounding urban areas and this effluent is not free from microbes, particularly effluent from treatment plants that do not disinfect their treated product [31,59]. The effluent community depends on the treatment processes used [2,28,68], but effluent flows are a constant source of microbes to natural waterways. The effects of WWTP effluent on aquatic communities have focused mainly on impacts to benthic communities in rivers or the analysis of indicator organisms [6,9,17,31,63,64]. Changes in surface water communities include increased densities of fecal coliforms, heterotrophic bacteria, and an altered species composition within the genus Acinetobacter [17]. An increased prevalence of culturable Arcobacter was observed at all urbanized sites downstream of a clean water reference cite in Catalonia, and both influent and treated wastewater effluent yielded isolates [6].
Our understanding of the alterations to natural aquatic microbial communities from the perspective of both acute environmental scenarios, such as following rain and heavy urban discharge, and the long-term influence of constant urban microbial input remain relatively obscure. Most analyses indicate persistent or widespread contamination of surface waters with microbes originating from fecal pollution sources [65,62,38], but fecal-derived organisms are typically a small portion (<20%) of the flow of microbes from pipes and urban run-off [37,47]. Two comprehensive analyses of urban-derived bacterial assemblages present in an urban estuary of Lake Michigan suggest these organisms make-up 1–10% of the bacterial community present, and this proportion is influenced heavily by recent rain intensity [15,36]. Therefore, by sheer mass effects, the flux of organisms coming from urban environments could have significant impacts on the microbial communities naturally present. Urban-derived microbes can compete for resources, and also could represent a large and supplemental food source for microbial predators, which could have implications for aquatic food webs and/or nutrient cycles.
The urban water interface could also create new pathways for gene flow among microbial communities. Antibiotic resistant bacteria are selected for in human populations, and untreated sewage as well as treated sanitary effluent have a high occurrence of antibiotic resistant bacteria [5,30]. Goñi-Urriza el al. reported an increase in the number of antibiotic resistant Aeromonas and Enterobacteriaceae isolates downstream of WWTP effluent release in the Arga River (Spain), as well as increased instance of acquired resistance in the Aeromonas spp. (64). Similar results were observed for Acinetobacter spp. in the Huron River (MI, USA) with a higher prevalence of antibiotic resistance in isolates from downstream compared to upstream of the WWTP. Additionally, although the total abundance of Acinetobacter was reduced from influent to effluent, the percentage of isolates displaying multiple drug resistance increased significantly [69].
Chemicals, nutrients, and solid waste from urban areas can also alter the microbial community. For example, landscape changes and chemicals corresponded to microbial community changes in the Mississippi River, USA [51-53]. Microplastics released from WWTPs, can act as both a growth substrate and as a vector for bacteria [31]. Notably, the organisms that tend to be enriched on the plastics are from the same families of organisms that are present in sewage as pipe-associated, namely Campylobacteraceae, Pseudomonadaceae, and Moraxellaceae [15,61]. Emerging contaminants have been noted for their effects on wastewater treatment plant sludge organisms [41,70], but their impact on natural aquatic communities has yet to be been determined.
Conclusions
While stormwater and sanitary sewage infrastructure are clearly important for both the growth and transport of urban bacteria to urban waters, they are only two facets of the extensive urban water system, which in turn is only a part of the larger urban microbiome. King [70] discussed the idea of distinct urban microbiomes associated with the atmosphere, internal and external building surfaces, impervious surfaces such as roads and sidewalks, and vegetation, in addition to water distribution systems, waste treatment, and mobile organisms (humans and animals), that also interact with one another. Just as humans are constantly shedding their bacteria onto surfaces as they move around, the urban environment as a whole is shedding its bacteria via water conveyance systems that collect and disperse these microorganisms – with surface waters being the primary recipient.
Urban water conveyance systems have a great potential to tell us about the microorganisms associated with cities, about health characteristics of human populations, and if or how these organisms influence the human interface with the natural environment. The microbial communities of urban surface waters (e.g., rivers, lakes, estuaries) also could provide significant insight into the degree of human impacts on these freshwater systems and provide clues as to the short and long term ecosystem alteration caused by human activity. The magnitude of stormwater and sanitary sewage fluxes make these systems important to study, but understanding the long-term fate of organisms derived from this infrastructure must be pursued. We are currently in a new area of exploration in which next generation sequencing can provide a wealth of information on the microorganisms inhabiting any environment of the world around us. Continued monitoring of urban aquatic microbiomes has the potential to benefit both human and ecological health and must be prioritized.
Acknowledgements.
We would like to thank our colleague Maria Josefa Figueras Salvat (University Rovira Virgili, Reus, Spain) for insightful discussion on infrastructure niche organisms and our colleague Mitchell Sogin (Marine Biological Laboratory, Woods Hole, MA, USA), who has worked with us over the years to gain invaluable insight into urban microbial communities using next generation sequencing.
Footnotes
Competing interests. None declared.
References
- 1.Arumugam M, Raes J, Pelletier E, Le Paslier D, Yamada T, Mende DR, Fernandes GR, Tap J, et al. , (2011) Enterotypes of the human gut microbiome. Nature 473:174–180 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Cai L, Ju F, Zhang T (2014) Tracking human sewage microbiome in a municipal wastewater treatment plant. Appl Microbiol Biot 98:3317–3326 [DOI] [PubMed] [Google Scholar]
- 3.Cao Y, Van De Werfhorst LC, Sercu B, Murray JL, Holden PA (2011) Application of an integrated community analysis approach for microbial source tracking in a coastal creek. Environ Sci Technol 45:7195–7201 [DOI] [PubMed] [Google Scholar]
- 4.Cao Y, Van De Werfhorst LC, Dubinsky EA, Badgley BD, Sadowsky MJ, Andersen GL, Griffith JF, Holden PA (2013) Evaluation of molecular community analysis methods for discerning fecal sources and human waste. Water Res 47:6862–6872 [DOI] [PubMed] [Google Scholar]
- 5.Caplin JL, Hanlon GW, Taylor HD (2008) Presence of vancomycin and ampicillin-resistant Enterococcus faecium of epidemic clonal complex-17 in wastewaters from the south coast of England. Environ Microbiol 10:885–892 [DOI] [PubMed] [Google Scholar]
- 6.Collado L, Kasimir G, Pérez U, Bosch A, Pinto R, Saucedo G, Huguet JM, Figueras MJ (2010) Occurrence and diversity of Arcobacter spp. along the Llobregat River catchment, at sewage effluents and in a drinking water treatment plant. Water Res 44:3696–3702 [DOI] [PubMed] [Google Scholar]
- 7.De Filippo C, Cavalieri D, Di Paola M, Ramazzotti M, Poullet JB, Massart S, Collini S, Pieraccini G, Lionetti P (2010) Impact of diet in shaping gut microbiota revealed by a comparative study in children from Europe and rural Africa. Proc Natl Acad Sci USA 107:14691–14696 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Doubek J, Carey C, Cardinale B (2015) Anthropogenic land use is associated with N-fixing cyanobacterial dominance in lakes across the continental United States. Aquat Sci 77:681–694 [Google Scholar]
- 9.Drury B, Rosi-Marshall E, Kelly JJ (2013) Wastewater treatment effluent reduces the abundance and diversity of benthic bacterial communities in urban and suburban rivers. Appl Environ Microbiol 79:1897–1905 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Dubinsky EA, Esmaili L, Hulls JR, Cao Y, Griffith JF, Andersen GL (2012) Application of phylogenetic microarray analysis to discriminate sources of fecal pollution. Environ Sci Technol 46:4340–4347 [DOI] [PubMed] [Google Scholar]
- 11.Eren AM, Sogin ML, Morrison HG, Vineis JH, Fisher JC, Newton RJ, McLellan SL (2015) A single genus in the gut microbiome reflects host preference and specificity. ISME J 9:90–100 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Field KG, Samadpour M (2007) Fecal source tracking, the indicator paradigm, and managing water quality. Water Res 41:3517–3538 [DOI] [PubMed] [Google Scholar]
- 13.Fisher JC, Eren AM, Green HC, Shanks OC, Morrison HG, Vineis JH, Sogin ML, McLellan SL (2015) Comparison of sewage and animal fecal microbiomes using oligotyping reveals potential human fecal indicators in multiple taxonomic groups. Appl Environ Microbiol 81:7023–7033 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Fisher JC, Levican A, Figueras MJ, McLellan SL(2014) Population dynamics and ecology of Arcobacter in sewage. Front Microbiol 5:525. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Fisher J, Newton RJ, Dila DK, McLellan SL (2015) Urban microbial ecology of a freshwater estuary of Lake Michigan. Elem Sci Anth 3:000064. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Gasperi J, Garnaud S, Rocher V, Moilleron R (2008) Priority pollutants in wastewater and combined sewer overflow. Sci Total Environ 407:263–272 [DOI] [PubMed] [Google Scholar]
- 17.Goñi-Urriza M, Capdepuy M, Raymond N, Quentin C, Caumette P (1999) Impact of an urban effluent on the bacterial community structure in the Arga River (Spain), with special reference to culturable Gramnegative rods. Can J Microbiol 45:826–832 [PubMed] [Google Scholar]
- 18.Goñi-Urriza M, Capdepuy M, Arpin C, Raymond N, Caumette P, Quentin C (2000) Impact of an urban effluent on antibiotic resistance of riverine Enterobacteriaceae and Aeromonas spp. Appl Environ Microbiol 66:125–132 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Harwood VJ, Staley C, Badgley BD, Borges K, Korajkic A (2014) Microbial source tracking markers for detection of fecal contamination in environmental waters: relationships between pathogens and human health outcomes. FEMS Microbiol Rev 38:1–40 [DOI] [PubMed] [Google Scholar]
- 20.Huttenhower C, Gevers D, Knight R, Abubucker S, Badger JH, Chinwalla AT, Creasy HH, Earl AM, et al. , (2012) Structure, function and diversity of the healthy human microbiome. Nature 486:207–214 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Keely SP, Brinkman NE, Zimmerman BD, Wendell D, Ekeren KM, De Long SK, Sharvelle S, Garland JL (2015) Characterization of the relative importance of human- and infrastructure-associated bacteria in grey water: a case study. J Appl Microbiol 119:289–301 [DOI] [PubMed] [Google Scholar]
- 22.King GM (2014) Urban microbiomes and urban ecology: how do microbes in the built environment affect human sustainability in cities? J Microbiol 52:721–728 [DOI] [PubMed] [Google Scholar]
- 23.Koskey AM, Fisher JC, Eren AM, Ponce-Terashima R, Reis MG, Blanton RE, McLellan SL (2014) Blautia and Prevotella sequences distinguish human and animal fecal pollution in Brazil surface waters. Environ Microbiol Rep 6:696–704 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Leclerc H, Schwartzbrod L, Dei-Cas E (2002) Microbial agents associated with waterborne diseases. Crit Rev Microbiol 28:371–409 [DOI] [PubMed] [Google Scholar]
- 25.Leung HD, Chen G, Sharma K (2005) Effect of detached/re-suspended solids from sewer sediment on the sewage phase bacterial activity. Water Sci Technol 52:147–152 [PubMed] [Google Scholar]
- 26.Ley RE, Hamady M, Lozupone C, Tumbaugh PJ, Ramey RR, Bircher JS, Schlegel ML, Tucker TA, Schrenzel MD, Knight R, Gordon JI (2008) Evolution of mammals and their gut microbes. Science 320:1647–1651 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Liu G, Bakker GL, Li S, Vreeburg JH, Verberk JQ, Medema GJ, Liu WT, Van Dijk JC (2014) Pyrosequencing reveals bacterial communities in unchlorinated drinking water distribution system: an integral study of bulk water, suspended solids, loose deposits, and pipe wall biofilm. Environ Sci Technol 48:5467–5476 [DOI] [PubMed] [Google Scholar]
- 28.Liu XC, Zhang Y, Yang M, Wang ZY, Lv WZ (2007) Analysis of bacterial community structures in two sewage treatment plants with different sludge properties and treatment performance by nested PCR-DGGE method. J Environ Sci (China) 19:60–66 [DOI] [PubMed] [Google Scholar]
- 29.Lozupone CA, Stombaugh JI, Gordon JI, Jansson JK, Knight R (2012) Diversity, stability and resilience of the human gut microbiota. Nature 489:220–230 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Mao D, Yu S, Rysz M, Luo Y, Yang F, Li F, Hou J, Mu Q, Alvarez PJ (2015) Prevalence and proliferation of antibiotic resistance genes in two municipal wastewater treatment plants. Water Res 85:458–466 [DOI] [PubMed] [Google Scholar]
- 31.McCormick A, Hoellein TJ, Mason SA, Schluep J, Kelly JJ (2014) Microplastic is an abundant and distinct microbial habitat in an urban river. Environ Sci Technol 48:11863–11871 [DOI] [PubMed] [Google Scholar]
- 32.McLellan SL, Eren AM (2014) Discovering new indicators of fecal pollution. Trends Microbiol 22:697–706 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.McLellan SL, Huse SM, Mueller-Spitz SR, Andreishcheva EN, Sogin ML (2010) Diversity and population structure of sewage-derived microorganisms in wastewater treatment plant influent. Environ Microbiol 12:378–392 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.McLellan SL, Newton RJ, Vandewalle JL, Shanks OC, Huse SM, Eren AM, Sogin ML (2013) Sewage reflects the distribution of human faecal Lachnospiraceae. Environ Microbiol 15:2213–2227 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Montserrat A, Bosch L, Kiser MA, Poch M, Corominas L (2015) Using data from monitoring combined sewer overflows to assess, improve, and maintain combined sewer systems. Sci Total Environ 505:1053–1061 [DOI] [PubMed] [Google Scholar]
- 36.Newton RJ, Bootsma MJ, Morrison HG, Sogin ML, McLellan SL(2013) A microbial signature approach to identify fecal pollution in the waters off an urbanized coast of Lake Michigan. Microb Ecol 65:1011–1023 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Newton RJ, McLellan SL, Dila DK, Vineis JH, Morrison HG, Eren AM, Sogin ML (2015) Sewage reflects the microbiomes of human populations. mBio 6:e02574. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Newton RJ, Vandewalle JL, Borchardt MA, Gorelick MH, McLellan SL (2011) Lachnospiraceae and Bacteroidales alternative fecal indicators reveal chronic human sewage contamination in an urban harbor. Appl Environ Microbiol 77:6972–6981 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Nogales B, Lanfranconi MP, Pina-Villalonga JM, Bosch R (2011) Anthropogenic perturbations in marine microbial communities. FEMS Microbiol Rev 35:275–298 [DOI] [PubMed] [Google Scholar]
- 40.Okabe S, Odagiri M, Ito T, Satoh H (2007) Succession of sulfur-oxidizing bacteria in the microbial community on corroding concrete in sewer systems. Appl Env Microbiol 73:971–980 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Ortiz de García SA, Pinto Pinto G, García-Encina PA, Irusta-Mata R (2014) Ecotoxicity and environmental risk assessment of pharmaceuticals and personal care products in aquatic environments and wastewater treatment plants. Ecotoxicology 23:1517–1533 [DOI] [PubMed] [Google Scholar]
- 42.Patz JA, Vavrus SJ, Uejio CK, McLellan SL (2008) Climate change and waterborne disease risk in the Great Lakes region of the U.S. Am J Prev Med 35:451–458 [DOI] [PubMed] [Google Scholar]
- 43.Reischer GH, Ebdon JE, Bauer JM, et al. (2013) Performance characteristics of qPCR assays targeting human- and ruminant-associated bacteroidetes for microbial source tracking across sixteen countries on six continents. Environ Sci Technol 47:8548–8556 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Satoh H, Odagiri M, Ito T, Okabe S (2009) Microbial community structures and in situ sulfate-reducing and sulfur-oxidizing activities in biofilms developed on mortar specimens in a corroded sewer system. Water Res 43:4729–4739 [DOI] [PubMed] [Google Scholar]
- 45.Sauer EP, VandeWalle JL, Bootsma MJ, McLellan SL (2011) Detection of the human specific Bacteroides genetic marker provides evidence of widespread sewage contamination of stormwater in the urban environment. Water Res 45:4081–4091 [DOI] [PubMed] [Google Scholar]
- 46.Sercu B, Van De Werfhorst LC, Murray J, Holden PA (2009) Storm drains are sources of human fecal pollution during dry weather in three urban southern California watersheds. Environ Sci Technol 43:293–298 [DOI] [PubMed] [Google Scholar]
- 47.Shanks OC, Newton RJ, Kelty CA, Huse SM, Sogin ML, McLellan SL (2013) Comparison of the microbial community structures of untreated wastewaters from different geographic locales. Appl Environ Microbiol 79:2906–2913 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Shi ode N, Shiode S, Rod-Thatcher E, Rana S, Vinten-Johansen P (2015) The mortality rates and the space-time patterns of John Snow’s cholera epidemic map. Int J Health Geogr 14:21. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Song SJ, Lauber C, Costello EK, et al. (2013) Cohabiting family members share microbiota with one another and with their dogs. eLife 2:e00458. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Staley C, Gould TJ, Wang P, Phillips J, Cotner JB, Sadowsky MJ (2014) Core functional traits of bacterial communities in the Upper Mississippi River show limited variation in response to land cover. Front Microbiol 5:414. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Staley C, Gould TJ, Wang P, Phillips J, Cotner JB, Sadowsky MJ (2014) Bacterial community structure is indicative of chemical inputs in the Upper Mississippi River. Front Microbiol 5:524. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Staley C, Sadowsky MJ (2015) Application of metagenomics to assess microbial communities in water and other environmental matrices. J Mar Biol Asso UK First View: 1–9 [Google Scholar]
- 53.Staley C, Unno T, Gould TJ, Jarvis B, Phillips J, Cotner JB, Sadowsky MJ (2013) Application of Illumina next-generation sequencing to characterize the bacterial community of the Upper Mississippi River. J Appl Microbiol 115:1147–1158 [DOI] [PubMed] [Google Scholar]
- 54.Thomas GB, Crawford D (2011) London Tideway Tunnels: tackling London’s Victorian legacy of combined sewer overflows. Water Sci Technol 63:80–87 [DOI] [PubMed] [Google Scholar]
- 55.Tumbaugh PJ, Hamady M, Yatsunenko T, Cantarel BL, Duncan A, Ley RE, Sogin ML, Jones WJ, Roe BA, Affourtit JP, Egholm M, Henrissat B, Heath AC, Knight R, Gordon JI (2009) A core gut microbiome in obese and lean twins. Nature 457:480–484 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Unno T, Jang J, Han D, Kim JH, Sadowsky MJ, Kim OS, Chun J, Hur HG (2010) Use of barcoded pyrosequencing and shared OTUs to determine sources of fecal bacteria in watersheds. Env Sci Technol 44:7777–7782 [DOI] [PubMed] [Google Scholar]
- 57.USEPA (2004) Report to Congress: Impacts and Control of CSOs and SSOs. EPA 833-R-04-001
- 58.USEPA (2004) The National Water Quality Inventory: Report to Congress, Reporting Cycle. EPA-841-R-08-00
- 59.USEPA (2007) Report to Congress: Combined Sewer Overflows to the Lake Michigan Basin. Env Prot Agency USA, Office of Water, Washington, DC [Google Scholar]
- 60.Vandewalle JL, Goetz GW, Huse SM, Morrison HG, Sogin ML, Hoffmann RG, Yan K, McLellan SL (2012). Acinetobacter, Aeromonas and Trichococcus populations dominate the microbial community within urban sewer infrastructure. Environ Microbiol 14:2538–2552 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.VandeWalle JL, Goetz GW, Huse SM, Morrison HG, Sogin ML, Hoffmann RG, Yan K, McLellan SL (2012) Acinetobacter, Aeromonas and Trichococcus populations dominate the microbial community within urban sewer infrastructure. Environ Microbiol 14:2538–2552 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Verhougstraete MP, Martin SL, Kendall AD, Hyndman DW, Rose JB (2015) Linking fecal bacteria in rivers to landscape, geochemical, and hydrologic factors and sources at the basin scale. Proc Natl Acad Sci USA 112:10419–10424 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Wakelin S A, Colloff MJ, Kookana RS (2008) Effect of wastewater treatment plant effluent on microbial function and community structure in the sediment of a freshwater stream with variable seasonal flow. Appl Environ Microbiol 74:2659–2668 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Wéry N, Monteil C, Pourcher AM, Godon JJ (2010) Human-specific fecal bacteria in wastewater treatment plant effluents. Water Res 44:1873–1883 [DOI] [PubMed] [Google Scholar]
- 65.Wu CH, Sercu B, Van de Werfhorst LC, Wong J, DeSantis TZ, Brodie EL, Hazen TC, Holden PA, Andersen GL (2010) Characterization of coastal urban watershed bacterial communities leads to alternative community-based indicators. PLoS One 5 :e11285. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Wu GD, Chen J, Hoffmann C, Bittinger K, Chen YY, Keilbaugh SA, Bewtra M, Knights D, Walters WA, Knight R, Sinha R, Gilroy E, Gupta K, Baldassano R, Nessel L, Li H, Bushman FD, Lewis JD (2011) Linking long-term dietary patterns with gut microbial enterotypes. Science 334:105–108 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Yatsunenko T, Rey FE, Manary MJ, Trehan I, Domínguez-Bello MG, Contreras M, Magris M, Hidalgo G, et al. , (2012) Human gut microbiome viewed across age and geography. Nature 486:222–227 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Ye L, Zhang T (2013) Bacterial communities in different sections of a municipal wastewater treatment plant revealed by 16S rDNA 454 pyrosequencing. Appl Microbiol Biot 97:2681–2690 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Zhang Y, Marrs CF, Simon C, Xi C (2009) Wastewater treatment contributes to selective increase of antibiotic resistance among Acinetobacter spp. Sci Total Environ 407:3702–3706 [DOI] [PubMed] [Google Scholar]
- 70.Zheng X, Huang H, Su Y, Wei Y, Chen Y (2015) Long-term effects of engineered nanoparticles on enzyme activity and functional bacteria in wastewater treatment plants. Water Sci Technol 72:99–105 [DOI] [PubMed] [Google Scholar]