ABSTRACT
Wastewater-based severe acute respiratory syndrome coronavirus 2 (SARS-CoV-2) surveillance of Frankfurt Airport by genome sequencing was used to detect SARS-CoV-2 variants entering the region. In November 2021, we found all characteristic mutations of Omicron in wastewater originating from Frankfurt Airport before the first confirmed clinical report from an arriving passenger on 26 November 2021.
ANNOUNCEMENT
Since the initial report from South Africa of the new severe acute respiratory syndrome coronavirus 2 (SARS-CoV-2) (genus Betacoronavirus, family Coronaviridae) variant that is known as the Omicron variant (WHO nomenclature) (1) or B.1.1.529 (PANGO lineage) (2), it has already been detected in 13 European Union and European Economic Area (EU/EEA) countries (3), most often from confirmed cases with a history of travel to southern African countries. The first positive report of Omicron from Frankfurt, Germany, was on 29 November 2021; the sample was collected on 26 November 2021 from a traveler who had arrived at Frankfurt Airport. In this work, we aimed to assess the earliest import of Omicron in Frankfurt by monitoring two wastewater streams, one partly originating from Frankfurt Airport and the other from the City of Frankfurt, using sequencing analysis. Using genomic sequencing could help overcome current limitations in detecting Omicron. Accordingly, the European Centre for Disease Prevention and Control (ECDC) suggested using genomic sequencing wastewater-based epidemiology (WBE) surveillance from incoming flights to track the import of Omicron (4).
For WBE surveillance of SARS-CoV-2 variants in wastewaters from Frankfurt Airport and Frankfurt City, we collected 24-h composite samples on 2 November and 23 November 2021. The samples were (i) sewage samples from a canal receiving wastewater from Frankfurt Airport and (ii) influent samples from the wastewater treatment plant (WWTP) of Frankfurt City. For sequencing, 500 mL of the untreated wastewater was concentrated by ultrafiltration with 100-kDa Centricon Plus-70 centrifugal ultrafilters (Merck), and RNA was extracted using the MagMAX Microbiome Ultra Nucleic Acid isolation kit (Thermo Fisher Scientific) according to the manufacturer’s protocol. Library preparation was performed according to a previous study (5). Briefly, the library was prepared using the Ion AmpliSeq SARS-CoV-2 research panel (Thermo Fisher Scientific) according to the manufacturer’s instructions. This panel consists of 237 primer pairs, resulting in an amplicon length range of 125 to 275 bp, which cover the nearly full genome of SARS-CoV-2. Libraries were multiplexed and sequenced using an Ion Torrent 540 chip on an Ion S5 sequencer (Thermo Fisher Scientific).
For data analysis, we used the directly installed software packages in Ion Torrent Suite v5.12.2 of our Ion S5 sequencer. We determined the genome coverage of our samples by mapping them to a SARS-CoV-2 reference genome (Wuhan-Hu-1 [GenBank accession numbers NC_045512 and MN908947.3]), using TMAP software included in the Ion Torrent Suite, after the unaligned reads were filtered using default parameters. The sequencing data for the samples are summarized in Table 1. For identification of mutations, all single-nucleotide variants (SNVs) were called using Variant Caller v5.16.0.5 with default parameters.
TABLE 1.
Summary of the sequencing data for the samples
| Sample location and collection date (day-mo-yr) | Total no. of reads | No. of mapped reads | Avg target base coverage depth (×) | Avg read identity vs target (%)a | GC content (%) | BioSample accession no. | SRA accession no. |
|---|---|---|---|---|---|---|---|
| Airport | |||||||
| 4-11-2021 | 1,169,001 | 345,149 | 319.5 | 95.61 | 62.9 | SAMN24156791 | SRR17258655 |
| 23-11-2021 | 8,299,055 | 3,245,669 | 9,693 | 98.36 | 55.8 | SAMN24156792 | SRR17258654 |
| City | |||||||
| 4-11-2021 | 1,972,935 | 620,663 | 753.4 | 98.72 | 62.2 | SAMN24156793 | SRR17258653 |
| 23-11-2021 | 9,908,030 | 3,825,450 | 11,834 | 98.02 | 56.8 | SAMN24156794 | SRR17258652 |
The target sequence was the SARS-CoV-2 reference genome (Wuhan-Hu-1 [GenBank accession numbers NC_045512 and MN908947.3]).
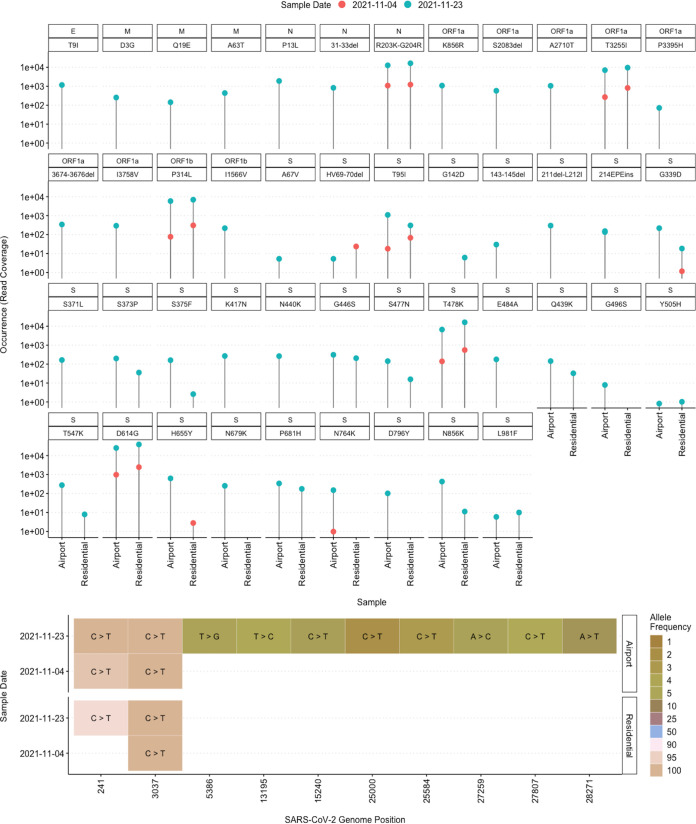
We determined the numbers of reads corresponding to each of the characteristic mutations of the Omicron variant to estimate its prevalence in the samples. The Omicron variant has several mutations, compared to the reference Wuhan SARS-CoV-2 genome, including 37 mutations in the spike (S) protein, 3 mutations in the nucleocapsid (N) protein, 1 mutation in the envelope (E) protein, 3 mutations in the membrane (M) protein, and 10 synonymous mutations (6). Overall, we found the following S protein mutations in Omicron: A67V, HV69–70del, T95I, G142D, 143–145del, 211del-L212I, 214EPEins, G339D, S371L, S373P, S375F, K417N, N440K, G446S, S477N, T478K, E484A, Q439K, G496S, Y505H, T547K, D614G, H655Y, N679K, P681H, N764K, D796Y, N856K, and L981F (Fig. 1). Also, N protein mutations, an E protein mutation, M protein mutations, and mutations in the open reading frame 1ab (ORF1ab) replication complex were found. However, all of the Omicron-specific amino acid changes were detected only in the airport wastewater sample from 23 November 2021. We show that the surveillance of wastewater from transportation hubs, such as airports, using sequencing can support tracking of the import of SARS-CoV-2 variants.
FIG 1.
(Top) Occurrence (i.e., the numbers of reads corresponding to each mutation) of characteristic S, E, M, and N protein mutations and ORF1ab mutations of the Omicron variant. (Bottom) Heatmap showing the allele frequency and the alternate nucleotide of the characteristic synonymous mutations of the Omicron variant. Airport, wastewater sample with a significant fraction from Frankfurt Airport; Residential, influent wastewater sample collected from the WWTP of Frankfurt City.
Data availability.
The raw metagenomic sequence data are available in the NCBI Sequence Read Archive (SRA) under BioProject number PRJNA789814.
ACKNOWLEDGMENTS
We thank the employees of the Stadtentwässerung Frankfurt (SEF) and members of our research group for their support with sample collection.
This work has been funded by the European Commission Joint Research Centre.
Contributor Information
Shelesh Agrawal, Email: s.agrawal@iwar.tu-darmstadt.de.
Simon Roux, DOE Joint Genome Institute.
REFERENCES
- 1.World Health Organization. 26 November 2021. Classification of Omicron (B.1.1.529): SARS-CoV-2 variant of concern. https://www.who.int/news/item/26-11-2021-classification-of-omicron-(b.1.1.529)-sars-cov-2-variant-of-concern.
- 2.Rambaut A, Holmes EC, O'Toole Á, Hill V, McCrone JT, Ruis C, Du Plessis L, Pybus OG. 2020. A dynamic nomenclature proposal for SARS-CoV-2 lineages to assist genomic epidemiology. Nat Microbiol 5:1403–1407. doi: 10.1038/s41564-020-0770-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.European Centre for Disease Prevention and Control. 2021. Risk related to the spread of new SARS-CoV-2 variants of concern in the EU/EEA: first update. European Centre for Disease Prevention and Control, Solna, Sweden. [Google Scholar]
- 4.European Centre for Disease Prevention and Control. 2021. Implications of the emergence and spread of the SARS-CoV-2 B.1.1.529 variant of concern (Omicron) for the EU/EEA. European Centre for Disease Prevention and Control, Solna, Sweden. [Google Scholar]
- 5.Agrawal S, Orschler L, Schubert S, Zachmann K, Heijnen L, Tavazzi S, Gawlik BM, de Graaf M, Medema G, Lackner S. 2021. A pan-European study of SARS-CoV-2 variants in wastewater under the EU Sewage Sentinel System. medRxiv 2021.06.11.21258756. doi: 10.1101/2021.06.11.21258756. [DOI] [Google Scholar]
- 6.Hodcroft EB. 2021. CoVariants: SARS-CoV-2 mutations and variants of interest. https://covariants.org. Accessed 17 December 2021.
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Data Availability Statement
The raw metagenomic sequence data are available in the NCBI Sequence Read Archive (SRA) under BioProject number PRJNA789814.