Dear Editor,
Most recently, a new SARS-CoV-2 variant of concern (VOC), Omicron (B.1.1.529), was first reported to the World Health Organization (WHO) from South Africa and then quickly spread to many countries,1,2 posing a serious threat to current vaccine prevention and antibody therapeutic strategies. Several studies have reported that the Omicron variant successfully escapes from neutralizing antibodies elicited by COVID-19 vaccines or from COVID-19 convalescent patients.3,4 Therefore, the development of potent anti-Omicron agents is urgently needed.
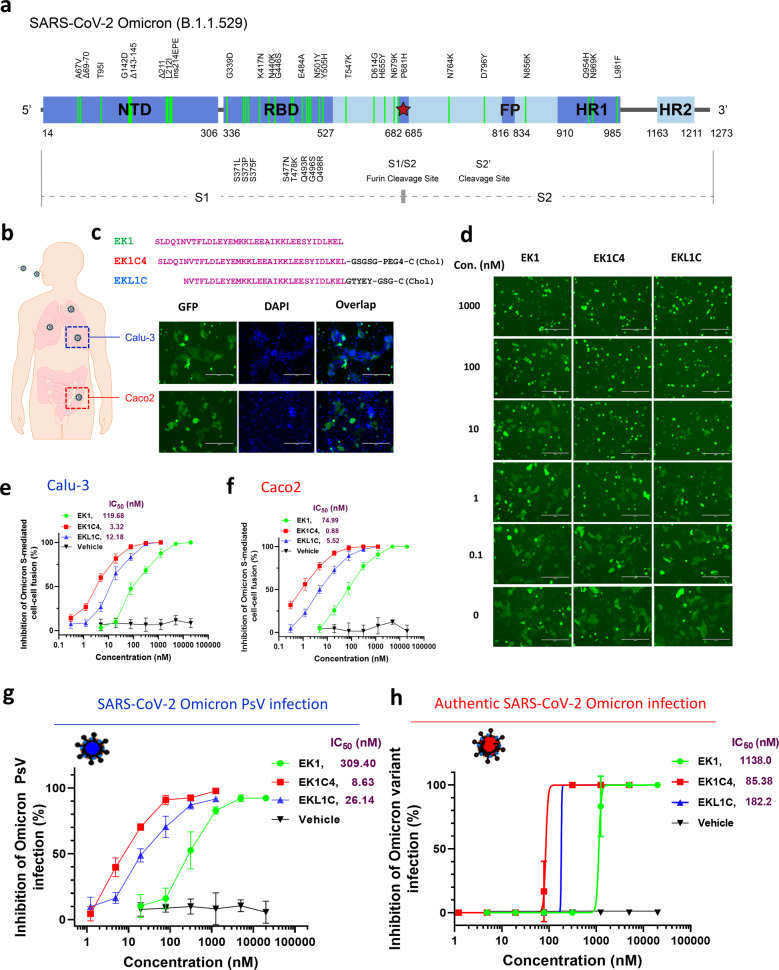
The Omicron variant is documented to have more than 30 mutations in its spike (S) protein, including A67V, del69-70, T95I, G142D, del143-145, del211, L212I, ins214EPE, G339D, S371L, S373P, S375F, K417N, N440K, G446S, S477N, T478K, E484A, Q493R, G496S, Q498R, N501Y, Y505H, T547K, D614G, H655Y, N679K, P681H, N764K, D796Y, N856K, Q954H, N969K, and L981F (Fig. 1a). These mutations seem to empower the virus to evade neutralizing antibodies against SARS-CoV-2 wild-type (WT) strain. To determine the functional basis for this mutant S, we developed an S-mediated cell–cell fusion assay using 293T cells co-expressing Omicron S protein and EGFP (293T/Omicron/EGFP), as the effector cells, and human ACE2 (hACE2)-expressing Calu-3 cells derived from human lung tissue, or Caco2 cells derived from human intestine tissue, as the target cells. As expected, after coculture of effector and target cells at 37 °C for 24 h, effector cells effectively fused with both Calu-3 cells and Caco2 cells. As shown in Fig. 1b, the fused cells showed larger size and darker fluorescent light than normal cells, and they contained multiple nuclei. Similarly, effector cells bearing Omicron S protein could also fuse with 293T/hACE2 cells, but not 293T cells (Supplementary information, Fig. S1). These results suggest that Omicron variant mutant S protein can interact with hACE2 to mediate viral fusion and infection in human lung and intestine tissues. Previous studies have reported that the fusogenicity found in S protein of SARS-CoV-2 variants favors viral transmissibility and pathogenicity.5,6 This calls for the development of effective fusion inhibitors against Omicron.
Fig. 1. Potent inhibitory activity of peptide-based pan-CoV fusion inhibitors against SARS-CoV-2 Omicron infection.
a Schematic representation of SARS-CoV-2 Omicron-S protein and its mutations. The S1 subunit consists of two main functional domains, including N-terminal domain (NTD) and receptor-binding domain (RBD). The S2 subunit consists of three main functional domains, including fusion peptide (FP), heptad repeat 1 (HR1) and heptad repeat 2 (HR2). b Representative images of S protein-mediated cell–cell fusion between 293 T/SARS-CoV-2(Omicron)/EGFP cells (effector cells) and Calu-3 or Caco2 cells (target cells) after their coculture for 24 h. Scale bars, 200 µm. c Amino-acid sequences of EK1, EK1C4 or EKL1C peptides. d Images of SARS-CoV-2 Omicron-S protein-mediated cell–cell fusion in the presence of EK1, EK1C4 or EKL1C at indicated concentrations after coculture of the effector (293T/SARS-CoV-2(Omicron)/EGFP) and target (Calu-3) cells for 4 h. Scale bars, 200 µm. e, f Inhibitory activities of EK1, EK1C4 or EKL1C peptides against Omicron S-mediated cell–cell fusion between 293T/SARS-CoV-2(Omicron)/EGFP and Calu-3 cells (e) or Caco2 cells (f). Samples were tested in triplicate, and the experiment was performed twice. Data from a representative experiment are presented as means ± SD. g Inhibitory activities of EK1, EK1C4, or EKL1C peptides against Omicron S-pseudovirus infection. Samples were tested in triplicate, and the experiment was performed twice. Data from a representative experiment are presented as means ± SD. h Inhibitory activities of EK1, EK1C4, or EKL1C peptides against infection by authentic SARS-CoV-2 Omicron variant. Samples were tested in triplicate, and the experiment was performed twice. Data from two experiments are presented as means ± SD.
We previously reported a series of peptide-based pan-coronavirus (CoV) fusion inhibitors, including EK1, EK1C4, and EKL1C (Fig. 1c), all of which have exhibited potent and broad-spectrum inhibitory activity against multiple human CoVs by targeting the conserved HR1 region in the S2 subunit of S protein.7–10 Given that some mutations in Omicron S protein locate in the HR1 region (e.g., Q954H, N969K, and L981F), we herein assessed the inhibitory activity of our HR1-targeting pan-CoV fusion inhibitors on Omicron-S-mediated cell-cell fusion. As shown in Fig. 1d, e, EK1, EK1C4, and EKL1C all potently inhibited Omicron S-mediated cell-cell fusion between 293T/Omicron/EGFP and Calu-3 cells in a dose-dependent manner with IC50 (half maximal inhibitory concentration) values of 119.68, 3.32 and 12.18 nM, respectively. EK1, EK1C4, and EKL1C could also block Omicron S-mediated cell–cell fusion between 293T/Omicron/EGFP and Caco2 cells with IC50s of 74.99, 0.88, and 5.52 nM, respectively (Fig. 1f). EK1, EK1C4, and EKL1C also showed high potency against the cell–cell fusion mediated by the S proteins of SARS-CoV-2 Delta variant (IC50: 131.8, 4.04 and 14.42 nM, respectively) and D614G strain (IC50: 314.6, 2.57, and 11.77 nM, respectively) (Supplementary information, Figs. S2 and S3). Overall, these pan-CoV fusion inhibitors showed equal, or even higher, potency to block Omicron-S- and Delta-S-mediated cell–cell fusion when compared with the inhibitory potency of these same peptides against SARS-CoV-2 D614G-S-mediated cell-cell fusion.
Consistent with our cell–cell fusion results, EK1, EK1C4, and EKL1C effectively inhibited Omicron pseudovirus (PsV) infection with IC50 values of 309.4, 8.63, and 26.14 nM, respectively (Fig. 1g). EK1, EK1C4, and EKL1C also exhibited high potency against infection by PsV of SARS-CoV-2 Delta variant (IC50: 427.55, 9.83 and 31.99 nM, respectively) and D614G variant (IC50: 414.85, 5.58 and 23.6 nM, respectively) (Supplementary information, Figs. S2 and S3).
More importantly, EK1, EK1C4, and EKL1C could effectively inhibit authentic SARS-CoV-2 Omicron (hCoV-19/Hong Kong/HKU-344/2021) infection in a dose-dependent manner with IC50 values of 1138, 85.38, and 182.2 nM, respectively (Fig. 1h), consistent with their inhibitory activities against infection by authentic SARS-CoV-2 WT strain (nCoV-SH01)9,10 reported previously. Collectively, these results confirm that the SARS-CoV-2 Omicron variant is as sensitive to these three pan-CoV fusion inhibitors as both D614G and Delta variants.
As noted above, the SARS-CoV-2 Omicron variant shows higher transmissibility than other VOCs, possibly because of its escape from neutralizing antibody responses elicited by COVID-19 vaccines and SARS-CoV-2 infection.3,4 In 2017, we began to develop peptide-based pan-CoV fusion inhibitors targeting the HR1 in S2 subunit of S proteins of hCoVs. These extensive efforts resulted in the development of EK1 and its cholesterol-modified lipopeptides EK1C4 and EKL1C. We have previously demonstrated the efficacy of these peptides in inhibiting infection by SARS-CoV, MERS-CoV, and SARS-CoV-2, as well as their variants, by blocking their fusion with and entry into the host cells, in in vitro systems and in vivo animal models.7–10 As shown in Supplementary information, Fig. S2, although Delta and Omicron variants contain many point mutations in the pre-fusion spike, only a few mutations are located in the post-fusion spike, such as D950 in Delta variant, or Q954 and N969 in Omicron variant, whereas none of them may affect the interaction between EK1 inhibitor and HR1 domain (Supplementary information, Fig. S4). Thus, it is not surprising that the data from this study show that these same peptide-based pan-CoV fusion inhibitors can also potently inhibit the SARS-CoV-2 Omicron variant and that they can be further developed as clinically applicable antiviral agents to meet the challenge of Omicron’s continuing worldwide spread.
Supplementary information
Acknowledgements
This work was supported by the National Key R&D Program of China (2021YFC2300703 to L.L.); the National Natural Science Foundation of China (82041025 and 92169112 to S.J.; 82002142 to S.X.); Program of Shanghai Academic/Technology Research Leader (20XD1420300 to L.L.); the Health and Medical Research Fund (COVID1903010—Project 7 to J.F-.W.C.), the Food and Health Bureau, The Government of the Hong Kong Special Administrative Region; Health@InnoHK, Innovation and Technology Commission, the Government of the Hong Kong Special Administrative Region (to K.-Y.Y.); the National Program on Key Research Project of China (2020YFA0707500 and 2020YFA0707504 to J.F.-W.C); and Sanming Project of Medicine in Shenzhen, China (SZSM201911014 to K.-Y.Y.). The funding sources had no role in the study design, data collection, analysis, interpretation, or writing of the report.
Author contributions
S.J., L.L., and K.-Y.Y. conceived, planned and supervised the experiments; S.X., J.F.-W. C., L.W., F.J., K.K.-H.C., H.C., Q.L., W.X., and Q.W. performed the experiments and analyzed the data; C.W. synthesized the peptides; S.X., and L.W. wrote the draft, while L.L., S.J., K.-Y.Y., and J.F.-W. C. revised the manuscript.
Competing interests
S.J., L.L., S.X., W.X. and Q.W. are the inventors in the patent or patent application covering the peptides EK1, EK1C4 and EKL1C. J.F.-W.C. has received travel grants from Pfizer Corporation Hong Kong and Astellas Pharma Hong Kong Corporation Limited and was an invited speaker for Gilead Sciences Hong Kong Limited and Luminex Corporation. Other authors declare that they have no competing interest.
Footnotes
These authors contributed equally: Shuai Xia, Jasper Fuk-Woo Chan, Lijue Wang.
Contributor Information
Kwok-Yung Yuen, Email: kyyuen@hku.hk.
Lu Lu, Email: lul@fudan.edu.cn.
Shibo Jiang, Email: shibojiang@fudan.edu.cn.
Supplementary information
The online version contains supplementary material available at 10.1038/s41422-022-00617-x.
References
- 1.Callaway E. Nature. 2021;600:21. doi: 10.1038/d41586-021-03552-w. [DOI] [PubMed] [Google Scholar]
- 2.He X, et al. MedComm. 2021;2:838–845. doi: 10.1002/mco2.110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Lu L, et al. Clin. Infect. Dis. 2021 doi: 10.1093/cid/ciab1041. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Zhang L, et al. Emerg. Microbes. Infect. 2021;11:1–5. doi: 10.1080/22221751.2021.2017757. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Saito A, et al. Nature. 2021 doi: 10.1038/s41586-021-04266-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Zhang J, et al. Science. 2021;374:1353–1360. doi: 10.1126/science.abl9463. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Xia S, et al. Sci. Adv. 2019;5:eaav4580. doi: 10.1126/sciadv.aav4580. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Xia S, et al. Cell Res. 2020;30:343–355. doi: 10.1038/s41422-020-0305-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Xia S, et al. Signal Transduct. Target. Ther. 2021;6:288. doi: 10.1038/s41392-021-00712-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Zhou J, et al. Acta Pharm. Sin. B. 2021 doi: 10.1016/j.apsb.2021.07.026. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.