Abstract
Campylobacter jejuni is one of the most common causes of bacterial diarrhea worldwide and is the primary bacterial cause of food-borne illness. Adherence to and invasion of epithelial cells are the most important pathogenic mechanisms of Campylobacter diarrhea. Molecular characterization of invasive and noninvasive Campylobacter isolates from children with diarrhea and symptom-free children was performed by random amplified polymorphic DNA techniques (RAPD). A distinct RAPD profile with a DNA band of 1.6 kb was observed significantly more frequently among invasive (63%) than among noninvasive (16%) Campylobacter isolates (P = 0.000005). The 1.6-kb band was named the invasion-associated marker (IAM). Using specifically designed primers, a fragment of 518 bp of the iam locus was amplified in 85% of invasive and 20% of noninvasive strains (P = 0.0000000). Molecular typing with a PCR-restriction fragment length polymorphism assay which amplified the entire iam locus showed a HindIII restriction fragment polymorphism pattern associated mainly with invasive strains. Although cluster analysis of the RAPD fingerprinting showed genetic diversity among strains, two main clusters were identified. Cluster I comprised significantly more pathogenic and invasive isolates, while cluster II grouped the majority of nonpathogenic, noninvasive isolates. These data indicate that most of the invasive Campylobacter strains could be differentiated from noninvasive isolates by RAPD analysis and PCR using specific primers that amplify a fragment of the iam locus.
Campylobacter jejuni is the most common cause of diarrhea in children of developing countries (4) and the primary cause of food-borne enteritis in industrialized regions (21). Variability in the clinical expression of Campylobacter infection has been observed for many years. In a study on the natural history of this infection in children, the clinical picture ranged from asymptomatic infections to secretory diarrhea and, less frequently, inflammatory diarrhea (4). Other clinical presentations of Campylobacter infection are meningitis (12), bacteremia (32), localized extraintestinal infections (5), and immunoreactive complications such as Guillain-Barré syndrome (17, 24) and reactive arthritis (2). This wide range of clinical manifestations cannot be explained as pertaining only to the host's response; characteristics of the bacterial pathogen may contribute. Recently, some phenotypic traits of infecting strains have been associated with the clinical presentation. In enteritis, three pathogenic mechanisms have been proposed: production of a cholera-like enterotoxin (28), production of a cytotoxin (35), and the ability to adhere to and invade epithelial cells, as demonstrated in vitro (6, 22, 30). The latter is considered essential for intestinal infection and production of disease (14). There is a good correlation between the clinical presentation of diarrhea and the isolation of Campylobacter strains that adhere to and invade HEp-2 cells. In a study in Mexico, we found that 70% of C. jejuni and Campylobacter coli isolates from children with diarrhea were invasive, as determined by the HEp-2 cell chamber-slide monolayer method, while 83% of isolates from asymptomatic children were nonadherent and noninvasive (29).
Variability in the clinical expression and in the phenotypic traits of isolates may be related to genetic diversity of Campylobacter strains. Several studies have focused on the characterization of C. jejuni adhesins and binding factors that enable some strains to adhere to and invade epithelial cells (10, 13, 31). Most of these genetic studies, however, have employed a single strain or reference strains, and to date no studies have examined genetic diversity in a population of Campylobacter isolated from symptomatic and symptom-free infections and its relation to adherence and invasion of epithelial cells.
Random amplified polymorphic DNA (RAPD) is a PCR-based molecular method that has been widely used for bacterial inter- and intraspecies discrimination (36, 37). The RAPD methodology does not require previous knowledge of the DNA template to be analyzed, and only a small quantity of DNA is needed to generate a fingerprint. Single, short, arbitrary nucleotide sequences can be amplified by PCR assay under low stringency conditions, thus generating polymorphic fingerprints that may be used for clustering pathogenic and nonpathogenic organisms and for demonstrating genetic diversity (25, 39).
The present study was designed to evaluate whether RAPD techniques could be applied to (i) identify genetic markers of pathogenicity in Campylobacter and (ii) generate fingerprints that distinguish invasive from noninvasive Campylobacter strains. Here we report the identification of a new, chromosomal 1.6-kb genetic marker of Campylobacter strains that was preferentially associated with adherence to and invasion of HEp-2 cells in vitro and was named the invasion-associated marker (IAM). Subsequently, using RAPD-generated fingerprints to construct dendrograms, we identified clusters of Campylobacter isolates which carried the IAM and were invasive and diarrhea associated.
MATERIALS AND METHODS
Bacterial strains and growth conditions.
A total of 119 Campylobacter strains from Mexico were studied, 60 from children with diarrhea (56 C. jejuni and 4 C. coli isolates) and 59 from asymptomatic children (53 C. jejuni, 4 C. coli, and 2 Campylobacter sp. isolates) (4, 29). Campylobacter strains were routinely subcultured at 42°C under microaerophilic conditions on brain heart infusion agar plates supplemented with 0.4% activated charcoal. C. jejuni 287ip (invasive and IAM positive) and 49sp (noninvasive and IAM negative) were used as prototype strains.
Adherence and invasion assays.
All Campylobacter strains were tested for adherence and invasion in HEp-2 cell monolayer chamber-slide assays, as previously described (18, 29). This technique allowed discrimination between strains with high and low indices of adherence to and invasion of HEp-2 cells; Campylobacter strains with ≥20% association were considered invasive. Using this method, 70 strains were invasive (66 C. jejuni and 4 C. coli) and 49 were noninvasive (45 C. jejuni and 4 C. coli).
DNA extraction and RAPD fingerprinting.
Genomic DNA was isolated using the guanidinium thiocyanate method (27). For initial screening, RAPD fingerprints were generated for 21 strains by using seven different arbitrary primers of 10-mers each, with different G+C contents: R2, 5′-AGTACAGGTC (11); 1290, 5′-GTGGATGCGA; 1283, 5′-CGATCCCCA; 1247, 5′-AAGAGCCCGT (1); HLWL74, 5′-ACGTATCTGC; HLWL85, 5′-ACAACTGCTC (20); and Wil2, 5′-TCACGATGCA (38). Approximately 10 ng of purified Campylobacter genomic DNA was used as a template for RAPD amplification in a volume of 20 μl containing 10 mM Tris-HCl (pH 8.3), 50 mM KCl, 2.5 mM MgCl2, 0.2 mM of each deoxynucleotide triphosphate, 1 U of Taq DNA polymerase (Boehringer, Mannheim, Germany), and 30 pmol of 10-mer primer (Operon Technologies, Alameda, Calif.). This reaction was overlaid with 1 drop of mineral oil and placed in a thermal cycler (model 9600; Perkin-Elmer, Norwalk, Conn.). The amplification program consisted of 35 cycles of 15 s at 92°C, 45 s at 36°C, and 1 min at 72°C. The PCR-amplified products were analyzed in a horizontal 1.4% agarose gel by electrophoresis in 0.5× Tris-borate–EDTA buffer and then visualized under UV light after ethidium bromide staining.
Amplification of a fragment of the iam locus.
PCR assay for amplification of a 518-bp DNA fragment (nucleotides 316 to 834) of the 1.6-kb band was standardized using a pair of primers selected from the iam locus sequence of the C. jejuni 287ip strain. A 19-nucleotide forward primer, 1.6F (GCG CAA AAT ATT ATC ACC C), corresponding to positions 316 to 334 of the iam locus, and an 18-nucleotide reverse primer, 1.6R (TTC ACG ACT ACT ATG CGG), corresponding to positions 817 to 834, were selected. PCR amplification was carried out with a DNA thermal cycler (model PTC 200 thermocycler; MJ Research, Cambridge, Mass.) using final volumes of 40 μl containing 30 pmol each of specific primers 1.6F and 1.6R. The amplification program consisted of 30 cycles of 30 s at 92°C, 1 min at 52°C, and 1 min at 72°C, with a final extension step at 72°C for 5 min. Five-microliter volumes of the products were analyzed in horizontal 1.2% agarose gels stained with ethidium bromide.
Degenerated PCR and PCR-RFLP analysis.
A degenerated PCR was developed to amplify the iam locus of all invasive and noninvasive strains. The DNA sequence of the iam locus was aligned with the DNA sequence of ABC transporter proteins from Helicobacter pylori (GenBank accession no. AE000646) (33), Haemophilus influenzae (U32744) (7), and Escherichia coli (D90705). After comparison of the DNA sequences of the ABC transporter proteins, a pair of degenerated primers, p77F [GG(A)CCT TTA GG(A)G AAG CTG] and p1415R [CTT TAA AT(A)T(G) GAA TC(G)A CG(T)GG], was designed and used to amplify an ∼1,360-bp fragment from genomic DNA of invasive and noninvasive Campylobacter strains. DNA amplification consisted of 30 cycles of 30 s at 92°C, annealing of primers at 49°C for 1 min, and extension at 72°C for 1 min. A final extension cycle at 72°C for 10 min was included. To determine whether there were any differences in DNA polymorphisms between invasive and noninvasive strains, a PCR-restriction fragment length polymorphism (PCR-RFLP) of the iam locus was done by digesting PCR-amplified products with HindIII endonuclease, and the profiles were checked in horizontal agarose gels.
Southern blot analysis of PCR-RFLP products.
PCR-RFLP products separated in agarose gels were transferred to nylon membranes by capillarity, using 6× SSC (1× SSC is 0.15 M NaCl plus 0.015 M sodium citrate). Membranes were probed with the entire DNA sequence of the IAM fragment labeled with digoxigenin-11–ddUTP (DIG-ddUTP) by random priming (DIG DNA labeling kit; Boehringer). Hybridization was carried out at 45 to 48°C. Prehybridization, hybridization, washes, solutions, and detection of the DIG-labeled probes were done according to the manufacturer's recommendations (Boehringer).
Computer-assisted analysis of RAPD fingerprints.
Numerical analysis of the RAPD fingerprinting data was done to examine associations between the genotypic heterogeneity of the iam locus, as detected by PCR and RAPD techniques, and the host's status, invasive or noninvasive phenotypes, and PCR-RFLP patterns. Primer 1290 RAPD amplification gel images of all 119 Campylobacter isolates were digitized with a Gel Doc 1000 system (Bio-Rad Laboratories, Hercules, Calif.) and stored as tagged image file format files; images were later converted, normalized, and analyzed with GelCompar software, version 4.0 (Applied Maths, Kortrijk, Belgium). The similarity matrix and clustering dendrograms were calculated using the Jaccard coefficient and Ward algorithm, respectively. Bands with faint intensity and high molecular weights were not reproducible and were excluded from the final analysis. Campylobacter strains with a level of similarity of ≥95% were considered to have the same RAPD type.
Statistical analysis.
A basic descriptive analysis was done using percentages. Associations were determined using the χ2 test. Results were considered significant when P was ≤0.05. All statistical analyses were done using Stata 7 (Stata Corporation, College Station, Tex.).
Nucleotide sequence accession number.
The iam locus, including the 518-bp DNA fragment described in this study, was registered by us in GenBank with accession number AF023133.
RESULTS
Identification of an RAPD marker associated with invasive Campylobacter strains.
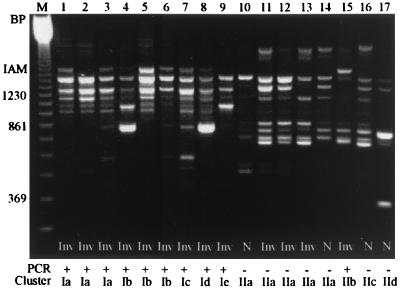
Seven primers were selected for initial screening by RAPD fingerprinting of 21 Campylobacter strains (15 invasive and 6 noninvasive). Four of these primers (Wil2, 1290, 1283, and 1247) produced distinct fingerprints. Primer 1290 produced the most distinct pattern, with up to 13 bands ranging in size from 0.2 to 3.0 kb, and was selected to test the remaining 98 invasive and noninvasive Campylobacter strains. All strains were typeable (Fig. 1), displaying patterns that allowed discrimination of strains with high adherence and invasion indices from those with low indices. A 1.6-kb band was predominantly found in invasive strains (44 of 70; 63%) but was present in very few of the noninvasive isolates (8 of 49; 16%; χ2 = 25.15; P = 0.00000053) (Table 1). This is the fragment that was named the invasion-associated marker.
FIG. 1.
RAPD amplification products of invasive and noninvasive Campylobacter strains using the arbitrary primer 1290. The IAM band size was estimated as 1.6 kb, marked on the left side. Results of the iam PCR amplification and their location in cluster analysis is shown. Lanes: M, 123-bp ladder marker; 1, C. jejuni 84sp; 2, C. jejuni 135ip; 3, C. jejuni 227sp; 4, C. jejuni 33K; 5, C. jejuni 287ip; 6, C. jejuni 151sp; 7, C. jejuni 268ip; 8, C. jejuni 401ip; 9, C. jejuni 188K; 10, C. jejuni 63sp; 11, C. jejuni 221sp; 12, C. jejuni 286sp; 13, C. jejuni 246sp; 14, C. jejuni 349K; 15, C. jejuni 180ip; 16, C. jejuni 128sp; 17, C. coli 49sp. Inv, invasive strains; N, noninvasive strains.
TABLE 1.
Detection of the 1.6-kb IAM by RAPD fingerprinting and by PCR assay for amplification of the iam locus in invasive and noninvasive Campylobacter isolates
| Strain type (n) | No. (%) of isolates with 1.6-kb IAM
|
|||
|---|---|---|---|---|
| RAPD (primer 1290)a
|
PCR of iam locus (primers 1.6F and 1.6R)b
|
|||
| Positive | Negative | Positive | Negative | |
| Invasive (70) | 44 (63) | 26 (37) | 60 (85) | 10 (14) |
| Noninvasive (49) | 8 (16) | 41 (84) | 10 (20) | 39 (79) |
| Total | 52 | 67 | 70 | 49 |
χ2 = 25.15; P = 0.00000053.
χ2 = 50.75; P = 0.00000000.
Specific amplification of the 518-bp fragment of the iam locus.
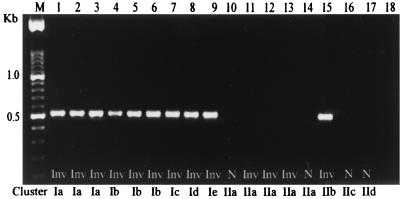
When the iam locus was amplified by PCR using an internal pair of primers, 1.6F and 1.6R, as expected a unique 518-bp PCR product (Fig. 2) was detected in 60 of 70 (85%) invasive strains, whereas only 10 of 49 (20%) of noninvasive isolates presented this band (χ2 = 45.12; P = 0.0000000) (Table 1). According to species, 64 of 109 (58%) C. jejuni, 5 of 8 (62%) C. coli, and 0 of 2 Campylobacter spp. were iam PCR positive. The sensitivity of the PCR was 85% and the specificity 74%, with a false-positive rate for true negative (noninvasive) of 25%, and a false-negative rate for true positive (invasive) of 14%. In addition, Campylobacter iam PCR positivity was a significant risk factor for intestinal infection associated with diarrhea (odds ratio, 18.53; 95% confidence interval, 2.21 to 6.38). This 518-bp iam locus fragment could not be amplified from the DNA of the following microorganisms: Candida spp., Enterobacter aerogenes, E. coli, Klebsiella pneumoniae, Lactobacillus reuterii, Pseudomonas aeruginosa, Proteus mirabilis, Proteus vulgaris, Salmonella enterica serovar Typhimurium, Shigella dysenteriae, Shigella sonnei, Staphylococcus aureus, Streptococcus faecalis, Vibrio cholerae, and Yersinia enterocolitica. To confirm that the 1.6-kb band was a homogeneous DNA fragment, a 518-bp iam locus DIG-labeled probe was hybridized in Southern blots with RAPD fingerprints from the 119 Campylobacter isolates. Only the 1.6-kb product from the RAPD amplification hybridized with the DIG-labeled probe (data not shown).
FIG. 2.
iam PCR amplification, using primers 1.6F and 1.6R, of the genomic DNA of noninvasive and invasive Campylobacter strains. The location in cluster analysis is shown. Lanes: M, 100-bp ladder; 1, C. jejuni 84sp; 2, C. jejuni 135ip; 3, C. jejuni 227sp; 4, C. jejuni 33K; 5, C. jejuni 287ip; 6, C. jejuni 151sp; 7, C. jejuni 268ip; 8, C. jejuni 401ip; 9, C. jejuni 188K; 10, C. jejuni 63sp; 11, C. jejuni 221sp; 12, C. jejuni 286sp; 13, C. jejuni 246sp; 14, C. jejuni 349K; 15, C. jejuni 180ip; 16, C. jejuni 128sp; 17, C. coli 49sp; 18, negative control.
PCR amplification using degenerate primers and PCR-RFLP analysis of the iam locus.
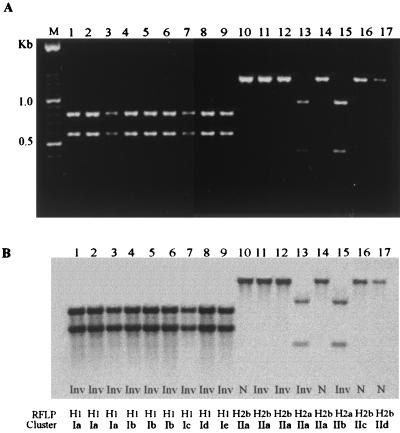
To determine the genetic polymorphism of the iam locus, we used degenerate primers (p77F and p1415R) to amplify a fragment of ∼1,360 bp from the 119 strains. In 115 (96.6%), we were able to obtain PCR products. A unique band of the expected size was amplified in 113 of 119 (95%); in addition, a PCR product of ∼0.8 kb was observed in 2 noninvasive strains. Three main HindIII RFLP genotypes, named H1, H2a, and H2b, were observed for the 1.3-kb degenerated PCR products (Fig. 3). The majority (52 of 60) of the invasive and iam PCR-positive strains were typed as H1 (Table 2), which showed a characteristic RFLP pattern by digestion of the 1,360-bp PCR of two fragments, 0.78 and 0.58 kb, cut at the site expected for HindIII (Fig. 3A, lanes 1 to 9). On the other hand, the majority (30 of 39) of the noninvasive, PCR-negative strains were typed as H2b. None of the PCR products of the H2b pattern were cut with HindIII (Fig. 3A, lanes 10 to 12, 14, 16 and 17). Ten strains, four of which were invasive, were typed as H2a. This PCR-RFLP pattern yielded 0.4- and 0.96-kb fragments (Fig. 3A, lanes 13 and 15). PCR-RFLP analysis and hybridization with the DIG-labeled 1.6-kb fragment showed a strong homologous signal with H1 patterns and showed a less strong signal for H2a and H2b patterns (Fig. 3B). These findings show that fragments amplified with the degenerate primers were homologous to the iam locus.
FIG. 3.
(A) Electrophoresis showing PCR-RFLP analysis of the iam locus. An ∼1,360-bp PCR product amplified with degenerated primers (p77F and p1415R) was digested with HindIII, and three patterns were identified: H1 (lanes 1 to 9), H2a (lanes 13 to 15), and H2b (lanes 10 to 12, 14, 16, and 17). (B) Southern blot of the same gel with the entire 1.6-kb probe labeled with DIG. Lanes: M, 100-bp ladder; 1, C. jejuni 84sp; 2, C. jejuni 135ip; 3, C. jejuni 227sp; 4, C. jejuni 33K; 5, C. jejuni 287ip; 6, C. jejuni 151sp; 7, C. jejuni 286ip; 8, C. jejuni 401ip; 9, C. jejuni 188K; 10, C. jejuni 63sp; 11, C. jejuni 221sp; 12, C. jejuni 286sp; 13, C. jejuni 246sp; 14, C. jejuni 349K; 15, C. jejuni 180ip; 16, C. jejuni 128sp; 17, C. coli 49sp.
TABLE 2.
Correlation between invasive phenotype, HindIII-RFLP genotypes, and iam PCR in 119 Campylobacter isolates
| Phenotype (n) | HindIII-RFLP genotypes | No. of isolates by iam PCR
|
|
|---|---|---|---|
| Positive | Negative | ||
| Invasive (70) | H1 | 52 | 1 |
| H2a | 2 | 2 | |
| H2b | 2 | 7 | |
| Nontypeable | 4 | 0 | |
| Subtotal | 60 | 10 | |
| Noninvasive (49) | H1 | 10 | 1 |
| H2a | 0 | 6 | |
| H2b | 0 | 30 | |
| Nontypeable | 0 | 2 | |
| Subtotal | 10 | 39 | |
Cluster analysis of RAPD fingerprint patterns and relationship between host status, invasive phenotype, and genetic markers.
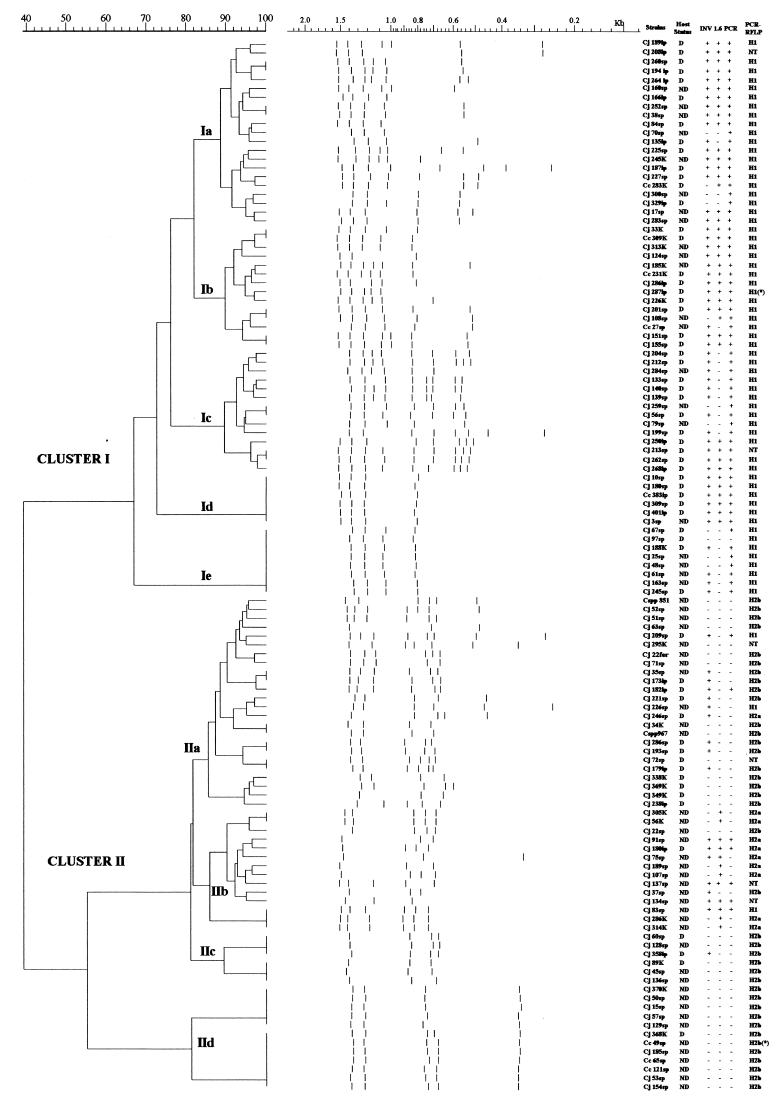
The genetic relationship between isolates based on their RAPD fingerprinting is represented in the dendrogram shown in Fig. 4. Using primer 1290, we found 42 RAPD types among the 119 isolates. Analysis using the Jaccard coefficient followed by Ward algorithm revealed two major clusters. Cluster I, at a similarity level of 67%, contained 63 strains in five subgroups (Ia to Ie), each of them with 6 to 21 isolates. Prototype invasive strain 287ip was clustered in Ib, with a similarity level of 89%. Subgroups Id and Ie were not distinguishable by RAPD fingerprinting. Cluster II, at a similarity level of 54.8%, comprised four subgroups (IIa to IId), each one containing 6 to 24 isolates. Subgroups IIc and IId showed distinctive fingerprints with clonal characteristics, including the prototype noninvasive strain 49sp.
FIG. 4.
Oligonucleotide 1290 RAPD fingerprinting dendrogram shows cluster analysis results of 119 invasive and noninvasive Campylobacter strains. Tracks show the band pattern after conversion, normalization, and GelCompar numerical analysis. Prototype strains 287ip and 49sp are marked with an asterisk (∗). On the right side of the figure are columns describing strain denomination, host's status, invasive phenotype, presence of IAM, PCR amplification of iam locus, and the PCR-RFLP pattern. D, diarrhea; ND, nondiarrhea; +, positive; −, negative.
The strains' denomination, host status, invasive phenotype, presence of the genetic marker, and PCR-RFLP patterns are also shown in Fig. 4. The number of strains isolated from patients with diarrhea was significantly larger in cluster I (42 of 63) than in cluster II (18 of 56) (χ2 = 14.02; P = 0.00018). More significantly, the majority of invasive strains were grouped in cluster I (52 of 63 versus 18 of 56; χ2 = 30.83; P = 0.00000000). PCR-RFLP genotypes were homogeneously distributed between the two main clusters. Of 63 isolates from cluster I, 61 were genotype H1; the remaining 2 isolates could not be typed. All 49 isolates belonging to genotype H2 were grouped in cluster II; 10 were genotype H2a, 9 of which belonged to cluster IIb. The remaining 39 were genotype H2b and were distributed in clusters IIa, IIc, and IId. The PCR fragment of iam was amplified in 62 of 63 isolates from cluster I and in only 8 of 56 from cluster II (χ2 = 85.90; P = 0.00000). The fragment was amplified in most of the invasive strains (52 of 56) of cluster I and in more than half of the invasive isolates of cluster II (10 of 18). None of the 38 noninvasive strains from cluster II carried the iam locus. The 1.6-kb band of RAPD fingerprinting was observed in 40 of 63 strains from cluster I and in 12 of 46 from cluster II (χ2 = 21.14; P = 0.0000043).
DISCUSSION
An undeniable benefit of studying well-characterized populations of Campylobacter isolates selected from symptomatic and symptom-free individuals and phenotypically defined as invasive or noninvasive is that when using methods that can screen complete bacterial genomic DNA, such as RAPD, it is possible to group strains according to their genotypic and phenotypic features and to identify genetic markers of virulence. Results of the present study demonstrate genetic differences between invasive and noninvasive Campylobacter strains. A strong association was found between the invasive phenotype, a particular RAPD fingerprint, and the presence of a specific DNA region of 1.6 kb. This DNA fragment was identified significantly more frequently among invasive strains than among noninvasive strains and was equally distributed among C. jejuni and C. coli isolates.
RAPD has been used to identify specific DNA regions associated with a given phenotype of different microorganisms (11, 25, 38). A novel DNA marker, with significant similarity to some negative transcriptional regulators, was identified in epidemic clinical strains of Burkholderia cepacia isolated from patients with cystic fibrosis (19); this DNA marker was absent in nonepidemic and environmental strains. Recently, using random amplification of different O serotypes of C. jejuni isolated from patients with Guillain-Barré syndrome, an association of a clonal population with virulence was observed (9). Moreover, an RAPD marker of 1.4 kb that differentiated O19-positive from O19-negative C. jejuni strains was cloned, sequenced, and characterized (23). In our study, RAPD techniques proved to be excellent molecular tools for typing Campylobacter strains, consistent with other reports (9, 20, 23). Although several investigators have successfully used RAPD in Campylobacter isolates as an epidemiological tool to identify the source of infection (20), we are unaware of studies that apply this method to discriminate between invasive and noninvasive clinical isolates.
RAPD techniques allowed us to compare polymorphisms of the entire bacterial genome in a population of invasive and noninvasive strains and to identify an invasion-associated DNA marker. However, we found that some of the invasive strains lacked this RAPD marker (Fig. 1, lanes 2 and 9). This could be explained by mismatching sequences at the binding site of random primer 1290 due to a greater polymorphism among invasive strains or by a low sensitivity of RAPD. To improve the sensitivity and more accurately differentiate invasive strains, we designed a PCR using specific primers that amplified a fragment of the iam locus. This PCR appears to be useful for the identification of invasive strains, since it accurately classified 82% of strains, with a sensitivity of 87%. However, this method misclassified as positive 10 of 49 noninvasive strains (specificity of 74%; false-positive rate for true negative of 25%). We do not have a clear explanation of why this DNA locus present in invasive strains also was amplified in some noninvasive strains, but we could speculate that there may be internal mutations in the IAM fragment of noninvasive strains. It will be interesting to sequence the amplicons from these PCR-positive noninvasive strains and to compare them with amplicons from PCR-positive invasive strains. The fact that some invasive strains also were not identified by PCR supports the existence of important polymorphism and high heterogeneity in the iam locus or suggests that there may be other genetic markers of invasion in different loci. In previous studies on the molecular characterization of genes associated with the phenotype of adherence to and invasion of epithelial cells, several Campylobacter strains have been studied and some genetic loci have been identified: peb1 (26), peb4A (3), cadF (15), and fla (13, 34), and more recently the gene that encodes antigen B (16) and the galE gene, involved in lipopolysaccharide synthesis and virulence (8).
When polymorphism of the iam locus was further explored in all Campylobacter strains by PCR-RFLP with HindIII endonuclease, it was possible to amplify this locus in most of the isolates by using degenerated primers. Invasive strains had a specific HindIII site (genotype H1), while most of the noninvasive strains lacked this site (genotype H2b), and a few invasive and noninvasive strains had this restriction site at a different position (genotype H2a). These findings confirm the polymorphism of the iam locus and the genetic diversity of strains. It will be important to sequence the iam locus from invasive and noninvasive strains that have a different polymorphism to determine whether there are changes in other sites of the locus which could have been overlooked by RAPD or PCR-RFLP.
Dendrograms constructed by numerical analysis of the RAPD fingerprints also confirmed the genetic diversity of isolates. Two main clusters were clearly defined. Cluster I grouped most invasive strains from diarrhea cases, which corresponded to the H1 genotype of RFLP and were iam PCR positive. By contrast, most strains from cluster II were noninvasive, were isolated from asymptomatic individuals, corresponded to genotype H2b of RFLP, and were iam PCR negative. In cluster II there was an interesting subcluster that grouped invasive isolates from asymptomatic individuals; these strains had an H2a genotype, which has a HindIII site in a different position from H1, and some were iam PCR positive. These genetic differences with H1 genotype strains could explain differences in virulence, since all but one of the strains were isolated from symptom-free individuals.
Another important finding was the presence of RAPD genotypes where strains had identical fingerprintings, i.e., clusters Id, Ie, and IId, which is a characteristic of clonal populations. Clonality has also been observed in some C. jejuni strains isolated from patients with Guillain-Barré syndrome (9, 23). It is then possible that some virulent Campylobacter strains are clonal populations. Finally, the presence of this molecular marker of invasion is not restricted to C. jejuni but is also present in C. coli. It would be interesting if this marker were carried by other less common Campylobacter species, such as C. lari or C. jejuni subsp. doylei.
We propose the use of RAPD or even more specific PCR assays as molecular tools for typing and studying different populations of invasive and noninvasive Campylobacter strains. Using a single PCR, we were able to detect most of the invasive isolates studied. These findings suggest that RAPD and PCR assays are effective molecular methods to discriminate invasive from noninvasive Campylobacter strains.
ACKNOWLEDGMENTS
This work was supported by U.S. Public Health Service grant PO HD 13021-22 from the National Institute of Child Health and Human Development and by a scholarship for A. C. T. Carvalho from Conselho Nacional de Desenvolvimiento Cientifico e Tecnológico, Brazil.
We are indebted to B. R. Ruiz-Palacios, D. Newburg, A. Nieto, P. S. Cisalpino, D. M. M. Queiroz, and E. Calva for revision of the manuscript and for technical help. We thank the staffs of the Department of Infectious Diseases, National Institute of Medical Science and Nutrition Salvador Zubirán, Mexico, and the Center for Pediatric Research, Eastern Virginia Medical School, Norfolk.
REFERENCES
- 1.Akopyanz N, Bukanov N O, Westblom T U, Kresovich S, Berg D E. DNA among clinical isolates of Helicobacter pylori detected by PCR-based RAPD fingerprinting. Nucleic Acids Res. 1992;20:5137–5142. doi: 10.1093/nar/20.19.5137. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Berden J H M, Mutjens H L, Van Pute L B A. Reactive arthritis associated with Campylobacter jejuni enteritis. Br Med J. 1979;1:380–381. doi: 10.1136/bmj.1.6160.380-a. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Burucoa C, Frémaux C, Pei Z, Tummuru M, Blaser M J, Cenatiempo Y, Fauchère J L. Nucleotide sequence and characterization of peb4A encoding an antigenic protein in Campylobacter jejuni. Res Microbiol. 1995;146:467–476. doi: 10.1016/0923-2508(96)80292-0. [DOI] [PubMed] [Google Scholar]
- 4.Calva J J, Ruiz-Palacios G M, Lopez-Vidal A B, Ramos A, Bojalil R. Cohort study of intestinal infection with Campylobacter in Mexican children. Lancet. 1988;i:503–506. doi: 10.1016/s0140-6736(88)91297-4. [DOI] [PubMed] [Google Scholar]
- 5.Darling W M, Peel R N, Skirrow M B. Campylobacter cholecystitis. Lancet. 1979;i:1302. doi: 10.1016/s0140-6736(79)92269-4. [DOI] [PubMed] [Google Scholar]
- 6.Fauchère J L, Rosenau A, Veron M, Moyen E N, Richard S, Pfister A. Association with HeLa cells of Campylobacter jejuni and Campylobacter coli isolated from human feces. Infect Immun. 1986;54:283–297. doi: 10.1128/iai.54.2.283-287.1986. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Fleischmann R D, Adams M D, White O, Clayton R A, Kirkness E F, Kerlavage A R, Bult C J, Tomb J, Dougherty B A, Merrick J M, McKenney K, Sutton G G, Fitzhugh W, Fields C A, Gocayne J D, Scott J D, Shirley R, Liu I I, Glodek A, Kelley J M, Weidman J F, Phillips C A, Spriggs T, Hedblom E, Cotton M D, Utterback T, Hanna M C, Nguyen D T, Saudek D M, Brandon R C, Fine L D, Fritchman J L, Fuhrmann J L, Geoghagen N S, Gnehm C L, McDonald I A, Small K V, Fraser C M, Smith H O, Venter J C. Whole-genome random sequencing and assembly of Haemophilus influenzae Rd. Science. 1995;269:496–512. doi: 10.1126/science.7542800. [DOI] [PubMed] [Google Scholar]
- 8.Fry B N, Feng S, Chen Y Y, Newell D G, Coloe P J, Korolik V. The galE gene of Campylobacter jejuni is involved in lipopolysaccharide synthesis and virulence. Infect Immun. 2000;68:2594–2601. doi: 10.1128/iai.68.5.2594-2601.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Fujimoto S, Mishu A, Misawa N, Patton C M, Blaser M J. Restriction fragment length polymorphism analysis and random amplified polymorphic DNA analysis of Campylobacter jejuni strains isolated from patients with Guillain-Barré syndrome. J Infect Dis. 1997;176:1105–1108. doi: 10.1086/516522. [DOI] [PubMed] [Google Scholar]
- 10.Garvis S G, Puzon G J, Konkel M E. Molecular characterization of a Campylobacter jejuni 29-kilodalton periplasmic binding protein. Infect Immun. 1996;64:3537–3543. doi: 10.1128/iai.64.9.3537-3543.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Goodwin P H, Annis S L. Rapid identification of genetic variation and pathotype of Leptosphaeria maculans by random amplified polymorphic DNA assay. Appl Environ Microbiol. 1991;57:2482–2486. doi: 10.1128/aem.57.9.2482-2486.1991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Goossens H, Henocque G, Kremp L. Nosocomial outbreak of Campylobacter jejuni meningitis in newborn infants. Lancet. 1986;ii:146. doi: 10.1016/s0140-6736(86)91956-2. [DOI] [PubMed] [Google Scholar]
- 13.Guerry P, Logan S M, Thornton S, Trust T. Genomic organization and expression of Campylobacter flagellin genes. J Bacteriol. 1990;172:1853–1860. doi: 10.1128/jb.172.4.1853-1860.1990. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Ketley J M. Pathogenesis of enteric infection by Campylobacter. Microbiology. 1997;143:5–21. doi: 10.1099/00221287-143-1-5. [DOI] [PubMed] [Google Scholar]
- 15.Konkel M E, Garvis S G, Tipton S L, Anderson J R, Cieplak W., Jr Identification and molecular cloning of a gene encoding a fibronectin-binding protein (CadF) from Campylobacter jejuni. Mol Microbiol. 1997;24:953–963. doi: 10.1046/j.1365-2958.1997.4031771.x. [DOI] [PubMed] [Google Scholar]
- 16.Konkel M E, Kim B J, Rivera-Amill V, Garvis S G. Bacterial secreted proteins are required for the internalization of Campylobacter into cultured mammalian cells. Mol Microbiol. 1999;32:691–701. doi: 10.1046/j.1365-2958.1999.01376.x. [DOI] [PubMed] [Google Scholar]
- 17.Kuroki S, Haruta T, Yoshioka M, Kobayashi Y, Nukina M, Nakanishi H. Guillain-Barré syndrome associated with Campylobacter infection. Pediatr Infect Dis. 1991;10:149–151. doi: 10.1097/00006454-199102000-00014. [DOI] [PubMed] [Google Scholar]
- 18.Lindblom G, Cervantes L E, Sjögren E, Kaijser B, Ruiz-Palacios G M. Adherence, enterotoxigenicity, invasiveness and serogroups in Campylobacter jejuni and Campylobacter coli strains from adult humans with acute enterocolitis. APMIS. 1990;98:179–184. [PubMed] [Google Scholar]
- 19.Mahenthiralingam E, Simpson D A, Speert D P. Identification and characterization of a novel DNA marker associated with epidemic Burkholderia cepacia strains recovered from patients with cystic fibrosis. J Clin Microbiol. 1997;35:808–816. doi: 10.1128/jcm.35.4.808-816.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Mazurier S, van de Giessen A, Heuvelman K, Wernars K. RAPD analysis of Campylobacter isolates: DNA fingerprinting without the need to purify DNA. Lett Appl Microbiol. 1992;14:260–262. doi: 10.1111/j.1472-765x.1992.tb00700.x. [DOI] [PubMed] [Google Scholar]
- 21.Mead P S, Slutsker L, Dietz V, McCaig L F, Bresee J S, Shapiro C, Griffin P M, Tauxe R V. Food-related illness and death in the United States. Emerg Infect Dis. 1999;5:607–625. doi: 10.3201/eid0505.990502. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Melo M A, Pechère J. Identification of Campylobacter jejuni surface protein that binds to eukaryotic cells in vitro. Infect Immun. 1990;58:1749–1756. doi: 10.1128/iai.58.6.1749-1756.1990. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Misawa N, Allos B M, Blaser M J. Differentiation of Campylobacter jejuni serotype O19 strains from non-O19 strains by PCR. J Clin Microbiol. 1998;36:3567–3573. doi: 10.1128/jcm.36.12.3567-3573.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Mishu B, Ilyas A A, Koski C L, Vriesendorp F, Cook S D, Mithen F A, Blazer M J. Serologic evidence of previous Campylobacter jejuni infection in patients with the Guillain-Barré syndrome. Ann Intern Med. 1993;118:947–953. doi: 10.7326/0003-4819-118-12-199306150-00006. [DOI] [PubMed] [Google Scholar]
- 25.Mondon P, Thélu J, Lebeau B, Ambroise-Thomas P, Guillot R. Virulence of Aspergillus fumigatus strains investigated by random amplified polymorphic DNA analysis. J Med Microbiol. 1995;42:299–303. doi: 10.1099/00222615-42-4-299. [DOI] [PubMed] [Google Scholar]
- 26.Pei Z, Blaser M J. PEB1, the major cell-binding factor of Campylobacter jejuni, is a homolog of the binding component in gram-negative nutrient transport systems. J Biol Chem. 1993;268:18717–18725. [PubMed] [Google Scholar]
- 27.Pitcher D G, Saunders N A, Owen R J. Rapid extraction of bacterial genomic DNA with guanidinium thiocyanate. Lett Appl Microbiol. 1989;8:151–156. [Google Scholar]
- 28.Ruiz-Palacios G M, Torres J, Torres N I, Escamilla E, Ruiz-Palacios B R, Tamayo J. Cholera-like enterotoxin produced by Campylobacter jejuni. Lancet. 1983;i:250–253. doi: 10.1016/s0140-6736(83)90234-9. [DOI] [PubMed] [Google Scholar]
- 29.Ruiz-Palacios G M, Cervantes L E, Newburg D S, Lopez-Vidal Y, Calva J J. In vitro models for studying Campylobacter infections. In: Nachamkin I, Blaser M J, Tompkins L S, editors. Campylobacter jejuni. Current status and future trends. Washington, D.C.: American Society for Microbiology; 1992. pp. 176–183. [Google Scholar]
- 30.Russell R G, Blake D C., Jr Cell association and invasion of Caco-2 cells by Campylobacter jejuni. Infect Immun. 1994;62:3773–3779. doi: 10.1128/iai.62.9.3773-3779.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Schröder W, Moser I. Primary structure analysis and adhesion studies on the major outer membrane protein of Campylobacter jejuni. FEMS Microbiol Lett. 1997;150:141–147. doi: 10.1111/j.1574-6968.1997.tb10362.x. [DOI] [PubMed] [Google Scholar]
- 32.Skirrow M B, Jones D M. Campylobacter bacteremia in England and Wales, 1981-91. Epidemiol Infect. 1993;110:567–573. doi: 10.1017/s0950268800050986. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Tomb J F, White O, Kerlavage A R, Clayton R A, Sutton G G, Fleischmann R D, Ketchum K A, Klenk H P, Gill S, Dougherty B A, Nelson K, Quackenbush J, Zhou L, Kirkness E F, Peterson S, Loftus B, Richardson D, Dodson R, Khalak H G, Godek A, McKenney K, Fitzgerald L M, Lee N, Adams M D, Hickey E K, Berg D E, Gocayne J D, Utterback T R, Peterson J D, Kelley J M, Cotton M D, Weidman J M, Fuji C, Bowman C, Watthey L, Wallin E, Hayes W S, Borodovsky M, Karp P D, Smith H O, Fraser C M, Venter J C. The complete genome sequence of the gastric pathogen Helicobacter pylori. Science. 1997;388:539–547. doi: 10.1038/41483. [DOI] [PubMed] [Google Scholar]
- 34.Wassenaar T M, Bleumink-Pluym N M, Newell D G, Nuijten P J M, van de Zeijst B A M. Differential flagellin expression in a flaA flaB+ mutant of Campylobacter jejuni. Infect Immun. 1994;62:3901–3906. doi: 10.1128/iai.62.9.3901-3906.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Wassenaar T M. Toxin production by Campylobacter spp. Clin Microbiol Rev. 1997;10:466–476. doi: 10.1128/cmr.10.3.466. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Welsh J, McClelland M. Fingerprinting genomes using PCR with arbitrary primers. Nucleic Acids Res. 1990;18:7213–7218. doi: 10.1093/nar/18.24.7213. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Williams J G K, Kubelik A R, Livak K J, Rafalski J A, Tingey S T. DNA polymorphism amplified by arbitrary primers are useful genetic markers. Nucleic Acids Res. 1990;18:6531–6535. doi: 10.1093/nar/18.22.6531. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Williams J G K, Hanafey M K, Rafalski J A, Tingey S V. Genetic analysis using random amplified polymorphic DNA markers. Methods Enzymol. 1993;218:704–740. doi: 10.1016/0076-6879(93)18053-f. [DOI] [PubMed] [Google Scholar]
- 39.Woodburn M A, Yousten A A, Hilu K H. Random amplified polymorphic DNA fingerprinting of mosquito-pathogenic and nonpathogenic strains of Bacillus sphaericus. Int J Syst Bacteriol. 1995;45:212–217. doi: 10.1099/00207713-45-2-212. [DOI] [PubMed] [Google Scholar]