Abstract
Nonalcoholic steatohepatitis (NASH) is induced by steatosis and metabolic inflammation. While involvement of the innate immune response has been shown, the role of the adaptive immune response in NASH remains controversial. Likewise, the role of regulatory T cells (Treg) in NASH remains unclear although initial clinical trials aim to target these regulatory responses. High‐fat high‐carbohydrate (HF‐HC) diet feeding of NASH‐resistant BALB/c mice as well as the corresponding recombination activating 1 (Rag)‐deficient strain was used to induce NASH and to study the role of the adaptive immune response. HF‐HC diet feeding induced strong activation of intrahepatic T cells in BALB/c mice, suggesting an antigen‐driven effect. In contrast, the effects of the absence of the adaptive immune response was notable. NASH in BALB/c Rag1−/− mice was substantially worsened and accompanied by a sharp increase of M1‐like macrophage numbers. Furthermore, we found an increase in intrahepatic Treg numbers in NASH, but either adoptive Treg transfer or anti‐cluster of differentiation (CD)3 therapy unexpectedly increased steatosis and the alanine aminotransferase level without otherwise affecting NASH. Conclusion: Although intrahepatic T cells were activated and marginally clonally expanded in NASH, these effects were counterbalanced by increased Treg numbers. The ablation of adaptive immunity in murine NASH led to marked aggravation of NASH, suggesting that Tregs are not regulators of metabolic inflammation but rather enhance it.
Abbreviations
- Actb
actin beta
- ALT
alanine transaminase
- anti‐CD3 F(ab)2’
non‐Fc receptor binding anti‐CD3 monoclonal antibody
- AST
aspartate aminotransferase
- AT
adipose tissue
- Ccl2
chemokine (C‐C motif) ligand 2
- CD
cluster of differentiation
- cDNA
complementary DNA
- Gapdh
glyceraldehyde‐3‐phosphate dehydrogenase
- HCC
hepatocellular carcinoma
- HE
hematoxylin and eosin
- HFD
high‐fat diet
- HF‐HC
high‐fat high‐carbohydrate
- IFN
interferon
- IHL
intrahepatic lymphocyte
- IL
interleukin
- KC
Kupffer cell
- MoMF
monocyte‐derived macrophage
- NAFLD
nonalcoholic fatty liver disease
- NAS
nonalcoholic fatty liver disease activity score
- NASH
nonalcoholic steatohepatitis
- NCD
normal chow diet
- NK
natural killer
- NKT
natural killer T
- ORO
Oil Red O
- Rag
recombination activating 1
- Rag− /−
knockout of recombination activating 1
- RT‐PCR
real‐time polymerase chain reaction
- TCR
T‐cell receptor
- TNF
tumor necrosis factor
- Treg
regulatory T cell
One third of the Western population is affected by nonalcoholic fatty liver disease (NAFLD), and 10%‐20% of the affected population suffers from nonalcoholic steatohepatitis (NASH). The presence of NASH accelerates disease progression, leading to cirrhosis and hepatocellular carcinoma (HCC).( 1 , 2 , 3 ) In North America, NASH‐cirrhosis has evolved to be the second leading cause of liver transplantation,( 4 , 5 ) but there is currently no established therapy. Therefore, therapeutic modulation of intrahepatic immune responses might be a future therapeutic option. While there are strong associations between NASH and an activated innate immune response in mice( 6 , 7 ) and humans,( 8 ) little is known about the role of adaptive immunity in NASH.( 9 ) There is an association between increased T helper 17 immune responses and the severity of NASH( 10 , 11 ) and a reduction of hepatic regulatory T cells (Tregs) in mice by apoptosis.( 12 ) Moreover, recent evidence suggests driving roles for cluster of differentiation (CD)8+ T cells and natural killer T (NKT) cells( 13 ) and a relative depletion of CD4+ T cells.( 14 ) These experiments have also raised the point that the modulation of the adaptive immune response might be a double‐edged sword. While there was some control of NASH by T‐cell depletion, this approach led to an increase in HCC occurrence.( 14 ) The ablation of the T‐cell and B‐cell responses in C57BL/6 recombination activating 1 (Rag)‐knockout (Rag−/− ) mice completely prevented NASH development, suggesting a driving role for the adaptive immune response in the pathophysiology of NASH.( 13 ) This finding sharply contrasts with the effects of the adaptive immune response on adipose tissue (AT) inflammation as C57BL/6‐background Rag−/− mice are much more susceptible to AT inflammation and insulin resistance development than wild‐type C57BL/6 mice.( 15 ) The latter results suggest a protective role for the adaptive immune system in metabolic inflammation, and this phenotype can be rescued by adoptively transferring CD4+ T cells and Treg numbers.
Little is known about the role of intrahepatic Tregs in NASH. AT inflammation after a high‐fat diet (HFD) is characterized by a marked reduction in Treg numbers, and the restoration of Treg numbers by interleukin (IL)‐2 therapy or adoptive transfer can down‐regulate AT inflammation and restore insulin sensitivity.( 16 )
Some of the abovementioned discrepancies might be due to the use of methacholine‐ and choline‐deficient diets. However, while these diets induce steatosis and NASH, these induced conditions do not resemble human NASH as those mice are lean, insulin sensitive, and without metabolic syndrome.( 17 , 18 )
We therefore wanted to investigate the role of adaptive immunity in disease‐relevant models of HF‐high‐carbohydrate (HC) diets in mice that are more resistant to NASH development. Moreover, we wanted to investigate the intrahepatic Treg response and the impact of therapeutic interventions that increase intrahepatic Treg numbers.
Materials and Methods
Ethics Statement
Animal care and experiments were performed in accordance with institutional and national guidelines. All animal experiments were performed according to protocols approved by the animal welfare commission of the Hannover Medical School and local ethics animal review board (Lower Saxony State Office for Consumer Protection and Food Safety, Oldenburg, Germany).
Mice
Animals were purchased and maintained under specific pathogen‐free conditions at the central animal facility of the Hannover Medical School (Hannover, Germany). BALB/c Rag1−/− mice were bred in‐house. We fed 6‐8‐week‐old mice a normal chow diet (NCD) from ssniff (Altromin standard diet #1324 total pathogen free; Altromin) or an HF‐HC diet (ssniff EF R/M D12330 mod. */Surwit + 1% cholesterol; ssniff) with 45 g/L 55% fructose/45% sucrose (Sigma) in the drinking water for 16 weeks. Fifty micrograms of a non‐Fc receptor binding anti‐CD3 monoclonal antibody (anti‐CD3 F[ab])2’) (145‐2C11; Bio‐X‐Cell) or isotype control (Bio‐X‐Cell) was intraperitoneally injected into mice for 5 consecutive days during week 16 of diet feeding. For the glucose tolerance test, 16‐hour‐fasted mice received 1 mg glucose/g body weight. Blood glucose level was measured in blood collected from the mouse tail vein with OneTouchUltra strips (LifeScan).
Serum Analysis
Blood samples were collected by a retro‐orbital puncture. Aspartate aminotransferase (AST) and alanine transaminase (ALT) levels were determined by photometric enzymatic activity assays using an Olympus AU400 Chemistry Analyzer and serum as described.( 19 , 20 )
Protein Detection in Serum
Proteins were measured using the Olink MOUSE EXPLORATORY panel (Olink Proteomics AB, Uppsala, Sweden) according to the manufacturer's instructions as described.( 20 , 21 , 22 ) The Proximity Extension Assay technology used for the Olink protocol has been well described( 23 ) and enables 92 analytes to be analyzed simultaneously (further details can be found in the Supporting Material).
Histology
Murine liver tissue was fixed in formalin and embedded in paraffin or Tissue‐Tek optimal cutting temperature compound (Sakura) for cryosections (8 µm). Paraffin‐embedded sections (5 µm) were prepared for hematoxylin and eosin (HE) staining, Oil Red O (ORO) triglyceride staining, and picrosirius red collagen staining.( 24 ) After staining, the sections were examined in a blinded manner by a pathologist using the approved modified NAFLD activity scoring (NAS) system for NASH.( 25 ) The NAS total score represents the sum of scores for steatosis, lobular inflammation, and ballooning and ranges from 0 to 8. NAS scores of 0‐2 are largely considered nondiagnostic for NASH; scores of 3‐4 are evenly distributed among those considered nondiagnostic, borderline, or positive for NASH; and scores of 5‐8 are considered mostly confirmed diagnosis for NASH.
Real‐Time Polymerase Chain Reaction
Nucleic acid isolation, complementary DNA (cDNA) synthesis, preamplification, quantitative real‐time polymerase chain reaction (RT‐PCR), and bioinformatical analysis were performed as described.( 22 , 26 ) In brief, total RNA was isolated from frozen liver samples, quantified using a NanoDrop 1000 Spectrophotometer, and the RNA was reverse transcribed into cDNA. Preamplification of cDNA and quantitative RT‐PCR were performed in a 48.48 Dynamic Array integrated fluidic circuit using the preamplified samples. Ct normalization was performed by subtracting the mean values of the housekeeping genes glyceraldehyde‐3‐phosphate dehydrogenase (Gapdh) and actin beta (Actb) from the genes of interest. A heat map and principal component analysis of the −ΔCT values were plotted by Qlucore software (P < 0.05 and q < 0.2).
Statistics
An unpaired Student two‐tailed t test was performed using Prism 8 software (GraphPad), and heat map analyses were performed using Glucore Omics Explorer 3.5. Additional methods are described in the Supporting Material.
Results
HF‐HC Diet Induces a Mild to Severe NASH Phenotype
Sixteen weeks of constant ad libitum feeding of our HF‐HC diet led to significantly higher weight gain and NASH development. This was assessed by the NAS system in BALB/c mice fed the HF‐HC diet compared to animals fed an NCD (Fig. 1A). HE and ORO staining of paraffin‐embedded liver sections showed increased steatosis, mild inflammation, and characteristic hepatocyte ballooning in mice fed the HF‐HC diet (Fig. 1B). As assumed, the BALB/c animals were relatively resistant to inflammation; only the intrahepatic triglycerides increased significantly while the transaminases did not (Fig. 1C). Glucose tolerance was also not impaired by the HFD. (Fig. 1C). In agreement with our hypothesis, mice on the BALB/c background gained weight, with increased hepatic triglycerides with a mild steatosis.
FIG. 1.
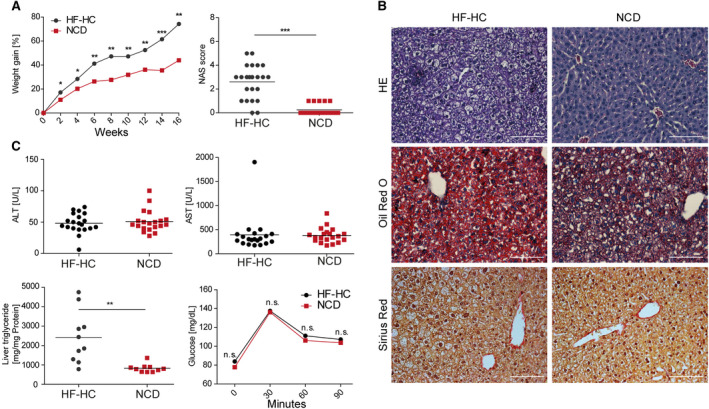
Histology, biochemical analysis, and glucose tolerance tests show the stages of NASH phenotype severity in HF‐HC diet‐fed BALB/c mice. (A) Longitudinal weight gain of HF‐HC‐fed (black dots) and NCD‐fed (red squares) BALB/c mice was followed over 16 weeks. Liver sections were analyzed at week 16 using the approved blinded NAS. Data show mean (left panel) and individual scores with mean (horizontal line) (right panel). (B) HE, ORO, and sirius staining were performed with liver sections harvested after 16 weeks of HF‐HC and NCD feeding. (C) BALB/c mice fed an HF‐HC diet (black dots) or NCD (red squares) were analyzed at week 16 regarding ALT and AST levels in their blood serum, triglyceride content in the liver, and glucose intolerance. Data show individual values with mean (horizontal line) (top panels, lower left panel) and mean (lower right panel); *P < 0.05, **P < 0.01, ***P < 0.001. Abbreviation: n.s., not significant.
Strong Activation but No Clonal Expansion of Intrahepatic T Cells
A loss of CD4+ T cells has been reported in mice fed a methionine/choline‐deficient diet or a pure HFD given a carcinogen,( 14 ) but other groups have reported an increase in intrahepatic CD4+ T cell numbers in mice fed a choline‐deficient HFD.( 13 ) While we could also see a relative decrease in the CD4+forkhead box P3 (Foxp3)− T‐cell proportion (Fig. 2A), there was no change in the absolute number of CD4+ T cells in mice fed the HF‐HC diet (Fig. 2B). This finding can be explained by relative (Fig. 2A; Supporting Fig. S1A) and absolute increases in intrahepatic Treg numbers (Fig. 2B). The increase in intrahepatic Tregs is in sharp contrast to the decrease observed in the visceral AT under HFD feeding.( 15 )
FIG. 2.
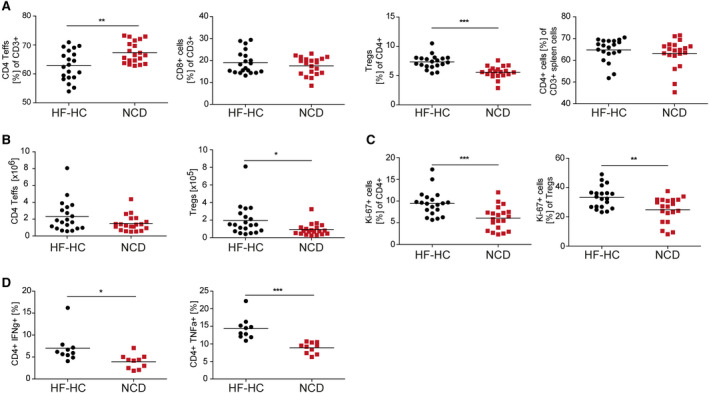
Intrahepatic CD4+ T cells show hyperactivity and proliferative characteristics, and Foxp3+ Treg‐cell numbers are significantly increased in HF‐HC diet‐treated mice. (A) BALB/c mice received an HF‐HC diet (black dots) or NCD (red squares) for 16 weeks. The frequencies of CD4+ and CD8+ Teffs and Tregs among IHLs and CD4+ splenocytes. (B) Absolute numbers of intrahepatic CD4+ Teffs and Tregs. (C) Percentage of proliferating Ki‐67+ cells among intrahepatic CD4+ Teffs and Tregs. (D) Intracellular staining for IHLs determined the content of IFN‐γ+ and TNF‐α+ cells among the CD4+ Teffs and Tregs isolated from BALB/c HF‐HC‐fed or NCD‐fed mice. Data show individual values with mean (horizontal line); *P < 0.05, **P < 0.01, ***P < 0.001. Abbreviations: FoxP3, forkhead box P3; IHL, intrahepatic lymphocyte; Teffs, effector T cells.
However, intrahepatic CD4+ T cells were strongly activated, as assessed by increased proliferation (Fig. 2C) and more interferon (IFN)‐γ‐producing and tumor necrosis factor (TNF)‐α‐producing T cells (Fig. 2D; Supporting Fig. S1B,C) that was not observed for CD8+ T cells (Supporting Fig. S1D). We then wanted to assess whether local intrahepatic T‐cell activation could be an antigen‐driven process, as suggested for AT inflammation.( 15 ) To this end, we performed an analysis of the T‐cell receptor (TCR) repertoire by 5‘‐rapid amplification of cDNA ends of the complementarity‐determining region 3 of the TCR beta chains, followed by high‐throughput sequencing. While the HF‐HC diet was expected to lead to the emergence of some dominant T‐cell clones, this was far less pronounced than hypothesized in the spleen and only marginally present in the liver T‐cell compartment (Fig. 3A). Taken together, these data reveal that even in the absence of steatosis, adaptive immunity was activated with an activated and expanded Treg compartment.
FIG. 3.
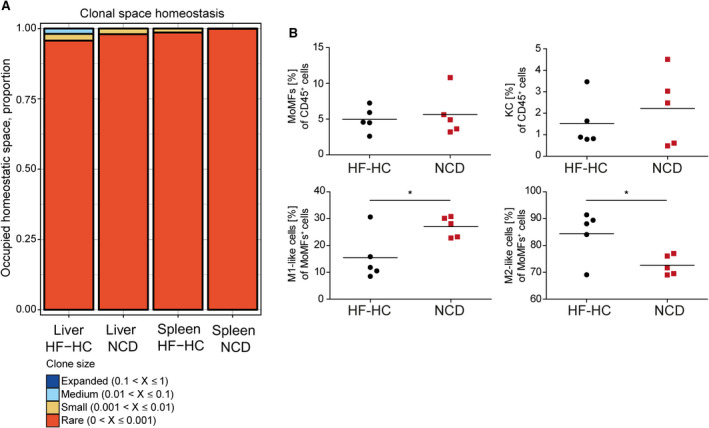
HF‐HC diet feeding induces marginal clonal TCR expansion and leads to an increase of M2‐like macrophages. (A) IHLs and intrasplenic T cells were isolated from BALB/c mice fed for 16 weeks with an HF‐HC diet or NCD, and the portion of the occupied homeostatic clonal space was determined. (B) Percentile distributions of CD11b+F4/80+ MoMFs, CD11b−F4/80+ KCs, Ly6Chigh MoMFs, and Ly6Clow MoMFs in HF‐HC‐fed (black dots) and NCD‐fed (red squares) BALB/c mice. Data show individual values with mean (horizontal line); *P < 0.05.
Macrophage Phenotype Correlates With NASH Resistance
Macrophages in the liver can be separated into monocyte‐derived macrophages (MoMFs) with CD11bhigh and F4/80high expression and Kupffer cells (KCs) with CD11bnegative/low and F4/80high expression.( 27 ) MoMFs can be further subgrouped into Ly6Chigh (proinflammatory M1‐like phenotype) or Ly6Clow (anti‐inflammatory M2‐like phenotype) cells.( 28 ) While BALB/c mice with NASH had no changes in NKT cell, natural killer (NK) cell, KC, granulocyte, or dendritic cell numbers, they showed a marked intrahepatic expansion of MoMFs with an anti‐inflammatory M2‐like phenotype (Fig. 3B; Supporting Fig. S2A). These data suggest that NASH resistance of the BALB/c strain derives from M2‐like MoMFs.
Molecular Characterization of Inflammation Induced by an HF‐HC Diet Reveals an Inflammatory Phenotype
Analysis of NASH severity by the NAS system is a crude estimation of disease activity that does not predict the subsequent disease course in patients with NASH. We therefore investigated several parameters of fibrosis, inflammation, inflammasomes, T‐cell homeostasis, endoplasmic reticulum stress, fatty acid metabolism, and reactive oxygen species pathways by quantitative PCR analysis of liver tissue to obtain a more nuanced molecular profile (see the gene list in Supporting Materials and Methods, Supporting Table S1). Compared with BALB/c mice fed the control diet, HF‐HC diet‐fed BALB/c mice showed an up‐regulation of the expression of genes belonging to inflammation and inflammasome pathways by Tnf‐α, toll‐like receptor 4 (Tlr4), chemokine (C‐C motif) ligand 2 (Ccl2), and NLR family, pyrin domain containing 3 (Nlpr3) expression up‐regulation (Fig. 4A). An up‐regulation of the expression of genes belonging to fatty acid metabolism by fibroblast growth factor 21 (Fgf21) and CD36 could also be detected (Fig. 4A). CCL2 and TNF‐α were also up‐regulated protein levels in sera (Fig. 4B), together with IL‐6, IL‐17, IL‐23, and other inflammatory molecules, while caspase‐3, for example, was down‐regulated. Again, in the absence of steatosis and elevated transaminases, these data reveal an ongoing inflammation that is driven by CCL2 and TNF‐α.
FIG. 4.
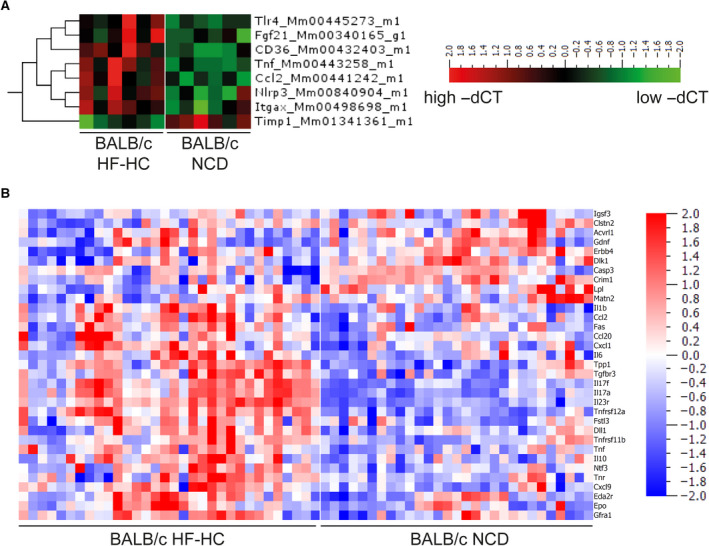
Distinct gene expression and serum protein profiles resulting in inflammatory pattern after HF‐HC feeding. (A) Heat map shows gene expression quantified by −ΔCT normalized to Actb and Gapdh expression from BALB/c mice fed for 16 weeks with an HF‐HC diet or NCD. (B) Heat map of serum protein expression of BALB/c mice fed for 16 weeks with an HF‐HC diet or NCD. The shown heat maps take into account multiplicity correction after calculating the P (<0.05) and q values (<0.05) for all 92 proteins. Abbreviations: Acvrl1, activin A receptor like type 1; Ccl20, C‐C motif chemokine ligand 20; Cxcl1, C‐X‐C motif chemokine ligand 1; Cxcl9, C‐X‐C motif chemokine ligand 9; Clstn2, calsyntenin 2; Casp3, caspase 3; Crim1, cysteine rich transmembrane BMP regulator 1; Dll1, delta like canonical notch ligand 1; Dlk1, delta like non‐canonical notch ligand 1; Eda2r, ectodysplasin A2 receptor; Erbb4, Erb‐B2 receptor tyrosine kinase 4; Epo, erythropoietin; Fas, fas cell surface death receptor; Fgf21, fibroblast growth factor 21; Fstl3, follistatin like 3; Gfra1, GDNF family receptor alpha 1; Gdnf, glial cell, derived neurotrophic factor; Igsf3, immunoglobulin superfamily member 3; Itgax, integrin subunit alpha X; Lpl, lipoprotein lipase; Matn2, matrilin 2; Ntf3, neurotrophin 3; Nlpr3, NLR family, pyrin domain containing 3; Tnr, tenascin R; Timp1, tissue inhibitor of metalloproteinases 1; Tnfrsf11b, TNF receptor superfamily member 11B; Tnfrsf12a, TNF receptor superfamily member 12A; Tlr4, toll‐like receptor 4; Tgfbr3, transforming growth factor beta receptor 3; Tpp1, tripeptidyl peptidase 1.
Absence of Adaptive Immunity in Rag−/− Mice Worsens Inflammation in NASH
To specifically analyze the role of adaptive immune response in the induction of NASH, BALB/c Rag1−/− mice lacking B, T, and NKT cells were fed an HF‐HC diet. Our data demonstrated attenuated weight gain in the Rag‐deficient animals over 16 weeks (Fig. 5A).
FIG. 5.
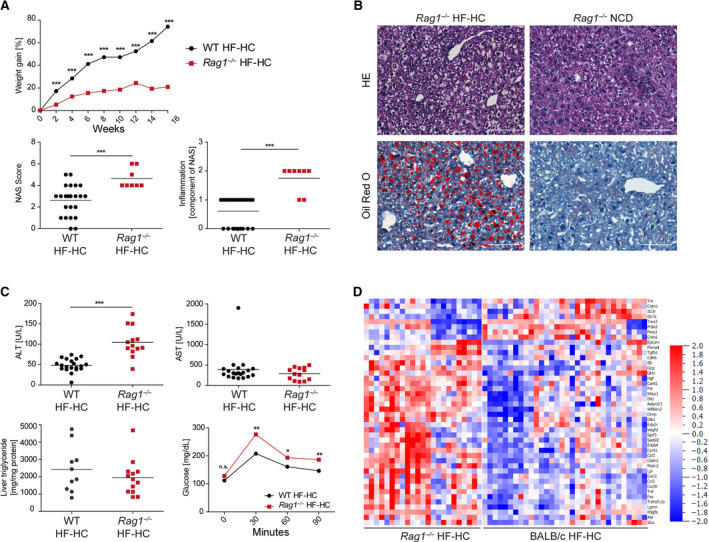
Histology, biochemical analyses, and glucose tolerance test show severe NASH induction in BALB/c Rag1−/− mice fed an HF‐HC diet. (A) Longitudinal weight gain of HF‐HC‐fed BALB/c (black dot) and BALB/c Rag1−/− (red squares) mice was followed over 16 weeks. Week‐16 liver sections were analyzed using the approved blinded NAS system. Inflammation scores (0‐3 points) are also shown separately. Data show mean (top panel) and individual values with mean (horizontal line) (lower panels). (B) HE and ORO staining of liver sections from HF‐HC‐fed and NCD‐fed BALB/c Rag1−/− mice were used to determine the NAS. (C) HF‐HC‐fed BALB/c wild‐type (black dots) and BALB/c Rag1−/− (red squares) mice were analyzed at week 16 regarding ALT and AST levels in the blood serum, triglyceride content in the liver, and glucose intolerance. Data show individual values with mean (horizontal line) (upper panels, lower left panel) and mean (lower right panel). (D) Heat map shows serum protein expression of BALB/c and BALB/c Rag1−/− mice fed for 16 weeks with an HF‐HC diet. The shown heat maps take into account multiplicity correction after calculating the P (<0.05) and q values (<0.05) for all 92 proteins; *P < 0.05, **P < 0.01, ***P < 0.001. Abbreviations: Adam23, ADAM metallopeptidase domain 23; Ahr, aryl hydrocarbon receptor; Cant1, calcium activated nucleotidase 1; Cdh6, cadherin 6; Clmp, CXADR like membrane protein; Clstn2, calsyntenin 2; Cntn1, contactin‐1; Crim1, cysteine rich transmembrane BMP regulator 1; Csf2, colony stimulating factor 2; Cxcl1, C‐X‐C motif chemokine ligand 1; Cxcl9, C‐X‐C motif chemokine ligand 9; Cyr61, cellular communication network factor 1; Dlk1, delta like non‐canonical notch ligand 1; Dll1, delta like canonical notch ligand 1; Eda2r, ectodysplasin A2 receptor; Epcam, epithelial cell adhesion molecule; Erbb4, Erb‐B2 receptor tyrosine kinase 4; Fas, fas cell surface death receptor; Fst, follistatin; Gcg, glucagon; Ghrl, ghrelin and obestatin prepropeptide; Hgf, hepatocyte growth factor; Igsf3, immunoglobulin superfamily member 3; Lgmn, legumain; Lpl, lipoprotein lipase; Matn2, matrilin 2; n.s., not significant; Parp1, poly(ADP‐ribose) polymerase 1; Pdgfb, platelet derived growth factor subunit B; Plxna4, plexin A4; Prdx5, peroxiredoxin 5; Sez6l2, seizure related 6 homolog like 2; Tgfb1, transforming growth factor beta 1; Tnfrsf11b, TNF receptor superfamily member 11B; Tnni3, troponin I3; Tnr, tenascin R; Vedfd, vascular endothelial growth factor D; Wfikkn2, WAP, follistatin/kazal, immunoglobulin, kunitz and netrin domain containing 2; Wisp1, WNT1‐inducible‐signaling pathway protein 1; WT, wild type.
Notably, the relatively NASH‐resistant BALB/c strain showed a marked increase in NASH severity as assessed by NAS that is mainly caused by more inflammation even if mice lack an adaptive immune system (Fig. 5A,B). Increased ALT levels in the BALB/c Rag1−/− animals were accompanied by pathologic glucose tolerance (Fig. 5C). Molecular analysis reflected the more severe inflammation measured by the NAS system (Fig. 5D). This result was accompanied by strong increases in proinflammatory M1‐like MoMF, NK cell, and granulocyte numbers in the BALB/c Rag1−/− animals and a decrease in anti‐inflammatory M2‐like MoMF numbers (Fig. 6A,B). The inflammatory genes Tnf‐α, Ccl2, and IL‐23, which are produced by inflammatory macrophages,( 29 ) were up‐regulated in the HF‐HC diet‐fed BALB/c Rag1−/− mice (Fig. 6C) as well as on the protein level (Figs. 5D and 6D). The worsening of NASH in the BALB/c Rag1−/− mice was so dramatic that proteins belonging to fibrosis induction were also up‐regulated. Cytokine analysis of blood serum highlighted significantly higher systemic levels of inflammatory cytokines, such as IL‐1β, TNF‐α, IL‐12, IL‐17, and IL‐23, in the BALB/c Rag1−/− mice than in the BALB/c wild‐type mice fed the HF‐HC diet (Fig. 6D). In summary, the worsening of the histologic NASH phenotype in the BALB/c Rag1−/− mice is reflected by an inflammatory molecular pattern at the genetic and protein levels.
FIG. 6.
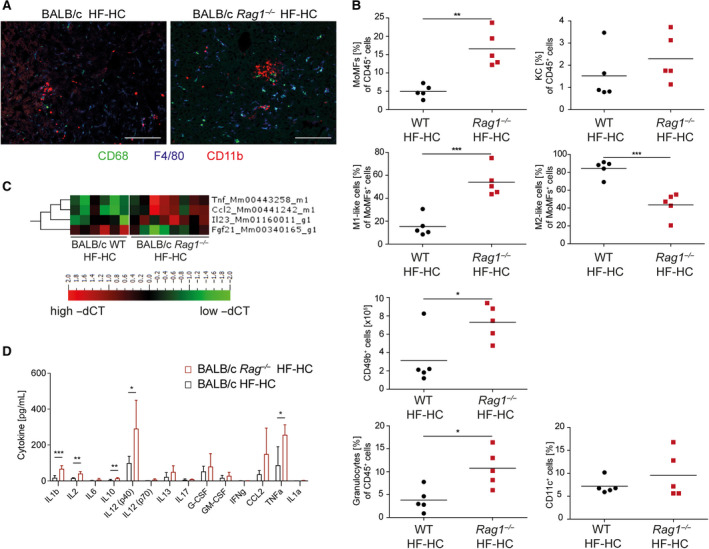
HF‐HC diet feeding in RAG‐knockout mice disturbs macrophage heterogeneity, as strongly shown by the induction of a distinct gene expression profile. (A) Cryopreserved liver sections from HF‐HC diet‐fed BALB/c and BALB/c Rag1−/− mice were immunohistochemically stained for CD86, F4/80, and CD11b. (B) Percentile distributions of CD11b+F4/80+ MoMFs, CD11b−F4/80+ KCs, Ly6Chigh MoMFs, Ly6Clow MoMFs, and absolute numbers of NK cells, CD11b+Ly6G+ granulocytes, and CD11c+ dendritic cells in HF‐HC‐fed BALB/c (black dots) and BALB/c Rag1−/− (red squares) mice. Data show individual values with mean (horizontal line). (C) Heat map shows the gene expression quantified by −ΔCT normalized to Actb and Gapdh expression. (D) Bar graph of the quantified cytokine content in the blood serum from HF‐HC‐fed BALB/c Rag1−/− (red, n = 5) and BALB/c WT (black, n = 5) mice determined by multiplex analyses. Data show mean + SD; *P < 0.05, **P < 0.01, ***P < 0.001. Abbreviations: Fgf21, fibroblast growth factor 21; G‐CSF, granulocyte colony‐stimulating factor; GM‐CSF, granulocyte‐macrophage colony‐stimulating factor; WT, wild type.
Adoptive Transfer of Tregs Increases Steatosis
We wanted to investigate which T‐cell population could be responsible for the aggravated NASH phenotype in BALB/c Rag1−/− animals fed an HF‐HC diet by adoptively transferring CD4+, CD8+, and Tregs. This strategy was important because the rescue of AT inflammation in Rag−/− mice is dependent on Tregs.( 15 , 16 ) The aggravated NASH phenotype could not be rescued by the adoptive transfer of CD4+ or CD8+ T cells. However, the transfer‐recipient mice developed colitis accompanied by weight loss (data not shown), which could have influenced the NASH phenotype. Although the adoptive transfer of Tregs into BALB/c Rag1−/− animals had no influence on the aggravated NASH phenotype, it significantly increased the hepatic triglyceride content (Fig. 7A). The effect on protein expression in the sera was limited by any T‐cell transfer (Fig. 7B), but transfer of CD4+ T cells clearly increased IL‐17 levels, which changed the most (P = 0.000012).
FIG. 7.
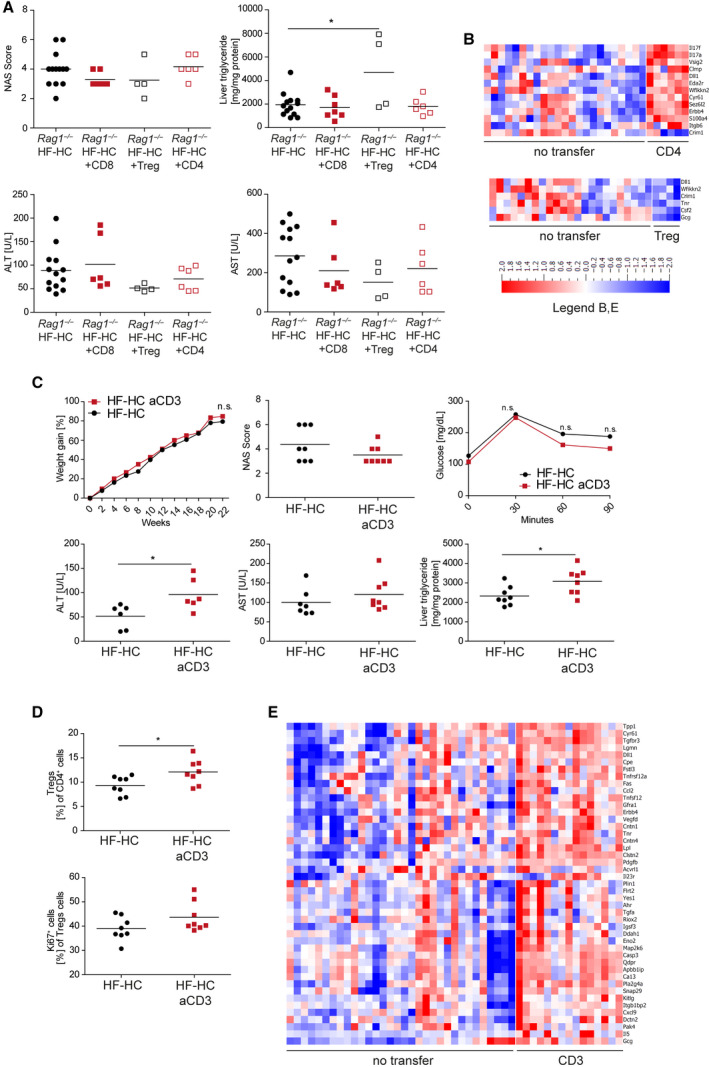
Adoptive transfer of Teffs is not sufficient to revert the Rag1−/− NASH phenotype, while Treg transfer or anti‐CD3 intervention worsens the inflammation. (A) HF‐HC‐fed BALB/c Rag1−/− mice (black dots), BALB/c Rag1−/− mice with CD8+ T‐cell transfer (red squares), BALB/c Rag1−/− mice with Treg transfer (black open squares), and BALB/c Rag1−/− mice with CD4+ T‐cell transfer (red open squares) were analyzed at week 16 regarding their NAS, hepatic triglyceride content, and ALT and AST levels in the blood serum. Data show individual values with mean (horizontal line); P < 0.05. (B) Heat maps show serum protein expression of BALB/c Rag1−/− mice fed for 16 weeks with an HF‐HC diet and either adoptive CD4+ T‐cell (upper panel) or Treg transfer (lower panel). (C) HF‐HC diet‐fed BALB/c mice were treated at week 16 with anti‐CD3 and followed for 6 weeks (HF‐HC full black dots, HF‐HC with anti‐CD3 full red squares). Longitudinal weight gain of anti‐CD3‐treated HF‐HC diet‐fed BALB/c mice was followed over 22 weeks. Treatment efficacy was analyzed by measuring the NAS, ALT, and AST levels and hepatic triglyceride content as well as glucose intolerance. Data show mean (weight gain, glucose) or individual values with mean (horizontal line); P < 0.05. (D) Frequencies of Tregs and their Ki‐67 expression were determined by flow cytometry analysis of CD4+ cells in the IHL population from HF‐HC diet‐fed BALB/c and HF‐HC diet‐fed anti‐CD3‐treated BALB/c mice after 22 weeks. Data show individual values with mean (horizontal line); P < 0.05. (E) Heat maps show serum protein expression of HF‐HC diet‐fed BALB/c mice fed for 22 weeks with an HF‐HC diet or treated with anti‐CD3 at week 16. Abbreviations: aCD3, anti‐CD3; Acvrl1, activin A receptor like type 1; Ahr, aryl hydrocarbon receptor; Apbb1ip, amyloid beta precursor protein binding family B member 1 interacting protein; Ca13, carbonic anhydrase 13; Casp3, caspase 3; Clmp, CXADR like membrane protein; Clstn2, calsyntenin 2; Cntn1, contactin 1; Cntn4, contactin 4; Cpe, carboxypeptidase E; Crim1, cysteine rich transmembrane BMP regulator 1; Csf2, colony stimulating factor 2; Cxcl9, C‐X‐C motif chemokine ligand 9; Cyr61, cellular communication network factor 1; Dctn2, dynactin subunit 2; Ddah1, dimethylarginine dimethylaminohydrolase 1; Dll1, delta like canonical notch ligand 1; Eda2r, ectodysplasin A2 receptor; Eno2, enolase 2; Erbb4, Erb‐B2 receptor tyrosine kinase 4; Fas, fas cell surface death receptor; Flrt2, fibronectin leucine rich transmembrane protein 2; Fstl3, follistatin like 3; Gcg, glucagon; Gfra1, GDNF family receptor alpha 1; Igsf3, immunoglobulin superfamily member 3; Itgb1bp2, integrin subunit beta 1 binding protein 2; Itgb6, integrin subunit beta 6; Kitlg, KIT ligand; Lgmn, legumain; Lpl, lipoprotein lipase; Map2k6, mitogen‐activated protein kinase kinase 6; n.s., not significant; Pdgfb, platelet derived growth factor subunit B; Pla2g4a, phospholipase A2 group 4A; Plin1, perilipin 1; Qdpr, quinoid dihydropteridine reductase; Riox2, ribosomal oxygenase 2; S100a4, S100 calcium binding protein A4; Sez6l2, seizure related 6 homolog like 2; Snap29, synaptosome associated protein 29; Tgfbr3, transforming growth factor beta receptor 3; Tgfa, transforming growth factor alpha; Tnfsf12, TNF superfamily member 12; Tnfrsf12a, TNF receptor superfamily member 12A; Tnr, tenascin R; Tpp1, tripeptidyl peptidase 1; Vedfd, vascular endothelial growth factor D; Vsig, v‐set and immunoglobulin domain‐containing protein; Wfikkn2, WAP, follistatin/kazal, immunoglobulin, kunitz and netrin domain containing 2; Yes1, src family tyrosine kinase.
In summary, surprisingly, the transfer of CD4+ T cells and Tregs ameliorated the NASH phenotype. In contrast, CD4 T‐cell transfer increased inflammation on the molecular level and Treg administration augmented hepatic triglyceride content.
Aggravation of NASH in Response to Therapy With Anti‐CD3 F(ab)2’ Fragments
Immunomodulatory therapy with low doses of anti‐CD3 F(ab)2’ fragments increased Treg numbers and has been used successfully to treat autoimmune diseases, such as type 1 diabetes in nonobese diabetic mice.( 30 ) More importantly, the increase in Treg numbers is sufficient to control AT and metabolic inflammation and increase insulin sensitivity in mice fed an HFD.( 15 ) HF‐HC diet‐fed BALB/c mice received 50 µg anti‐CD3 F(ab)2’ fragments on 5 consecutive days to modulate T cells. The animals were analyzed 6 weeks after treatment for body weight, glucose metabolism, liver enzymology, liver histology, and T cells. Therapy with the anti‐CD3 F(ab)2’ fragments increased intrahepatic Treg numbers and slightly improved glucose tolerance (Fig. 7C,D). Surprisingly, this treatment did not result in amelioration of NASH. Instead, compared with untreated mice, the anti‐CD3 F(ab)2’ fragment‐treated mice showed a more severe NASH phenotype with increased ALT levels and hepatic triglyceride content (Fig. 7A) and increased Fas, CCl2, IL‐5, IL‐23, and other inflammatory molecules (Fig. 7E). In summary, strengthening the Treg compartment by anti‐CD3 F(ab)2’ therapy worsened inflammation at the biochemical and molecular levels.
Discussion
The development of NASH is closely associated with obesity, metabolic syndrome, and insulin resistance.( 31 ) Animal models should therefore mimic these disease conditions to promote our knowledge of pathophysiology and new therapeutic interventions. The steatosis and inflammation observed with methacholine or choline deficiency lack these disease characteristics, and even combinations of toxic and high‐fat diets might fuel inflammation but do not resemble the NASH phenotype seen in patients. Therefore, the HF‐HC diet including fructose is currently one of the best ways to induce a NASH phenotype in otherwise healthy inbred mouse strains. Although approximately 30% of the Western population has NAFLD, only 10%‐20% of the affected population will continue to develop NASH, suggesting that genetic factors in addition to environmental factors might modulate susceptibility and progression of the disease.
As adaptive immunity seems to be involved in metabolic inflammation in AT and liver tissue, we studied the role of the adaptive immune response in an HF‐HC diet model using mice on the less NASH‐susceptible BALB/c background. We showed that the HF‐HC diet was able to induce NASH after 16 weeks in mice. We observed increased intrahepatic activation and proliferation, especially of CD4+ T cells, with more IFN‐γ‐producing and TNF‐α‐producing T cells showing an activation of the intrahepatic T‐cell response, suggesting antigen‐driven local activation. The observed relative decrease in the CD4+ T‐cell numbers, which was also observed by Ma et al.,( 14 ) cannot be seen in the absolute number of CD4+ T cells per liver. Wolf et al.( 13 ) also reported an increase in the CD4+ T‐cell number in their NASH model. Activation of the T‐cell response was accompanied by an increase in the intrahepatic Treg number, which had not been previously reported.( 9 ) This increase in the Treg number within the liver sharply contrasts with the strong decrease in Treg numbers in the visceral AT in animals fed an HF diet.( 15 , 16 ) Therefore, Treg numbers seem to react to ectopic fat accumulation and inflammation in a tissue‐specific pattern, with a strong decrease in AT and an increase in NASH liver tissue. In addition, an increase in fully functional intrahepatic Tregs has been described for various other inflammatory situations in patients and mice.( 32 , 33 , 34 , 35 ) Mechanistic studies showed that Tregs are more susceptible to apoptosis in response to oxidative stress in steatosis.( 12 ) In contrast, we observed more Treg proliferation and absolute increases in intrahepatic Treg numbers. Together with the increased anti‐inflammatory macrophages, this might explain the reduced susceptibility to NASH in BALB/c mice.
The NASH phenotype in the BALB/c Rag1−/− animals showed a strongly enhanced NASH phenotype with a strong accumulation of proinflammatory M1‐like MoMFs. This finding reveals the rather protective role of adaptive immune cells in limiting innate inflammation in wild‐type BALB/c animals. Furthermore, it has been described that CCL2+ neutrophils mediated immune evasion by promoting anti‐inflammatory macrophages and Tregs in HCC.( 36 ) This perfectly complements our data as we measured a strong enrichment of CCL2 at both the hepatic RNA and protein levels. These data could be one of the reasons for the difficulties associated with inducing NASH in wild‐type BALB/c animals.( 9 , 37 ) It is interesting to note the tissue‐specific effects of the adaptive immune response. While C57BL/6 Rag−/− animals show a marked increase in AT inflammation,( 15 ) the effects on the NASH phenotype in mice on the BALB/c background were modest. It is important to note that NASH could be induced in BALB/c Rag−/− animals. Therefore, we could not recapitulate the lack of steatosis, intrahepatic inflammation, and NASH in Rag−/− mice reported by Wolf et al.( 13 ) However, that group used a mixed choline‐deficient and HF diet that does not resemble the metabolic situation in patients with NASH. In a recent and extensive study by Malehmir et al.( 7 ), seven different NASH‐inducing diets were reported with the development of HCC and the role of platelets but not under immunodeficient conditions. Our results suggest that the adaptive immune response modifies NASH severity dependent on the genetic background but is not an essential component of NASH development. Instead, the strong activation of the innate immune response in BALB/c Rag−/− mice with concomitant impaired glucose tolerance suggests that the innate immune response might be more important than the adaptive immune response for the development of insulin resistance. Proinflammatory cytokines, such as TNF‐α and IL‐1, secreted by M1‐polarized macrophages can inhibit insulin signaling,( 38 ) thereby leading to the severe phenotype in the knockout animals.
It has been reported that Tregs can control AT inflammation,( 15 , 16 ) yet their role in NASH seems to be less clear. NASH is not caused by a numeric lack of Tregs, as has been suggested for AT inflammation. Instead, we observed a supraproportional increase in intrahepatic Treg accumulation in NASH mice, which could counteract the activated T cells. On the other hand, adoptively transferring Tregs into Rag−/− BALB/c mice was not able to correct the enhanced NASH or control activation of the adaptive immune response. Instead, the transfer of Tregs increased steatosis, which could be due to a steatosis‐promoting effect of transforming growth factor beta.( 39 ) This increase in steatosis is consistently also seen with an increase in intrahepatic Treg numbers after anti‐CD3 therapy. Therefore, the role of Tregs in controlling metabolic inflammation seems to differ between AT and liver tissue.
The same observation seems to be true for therapy with anti‐CD3 F(ab)2’ fragments. While Winer and coworkers( 15 ) demonstrated an increase in AT‐Treg numbers and amelioration of AT inflammation, we also saw an increase in intrahepatic Treg numbers, but this change was accompanied by an increase in steatosis and transaminase levels.
In summary, the adaptive immune response is activated during NASH and points toward local antigen‐driven T‐cell activation. This activation is partly counterbalanced by a supraproportional increase in Treg accumulation within the liver. The overall influence of the adaptive immune system is dependent on the genetic background, yet adaptive immunity is not essential for the development of NASH. Tregs and immunomodulatory therapies, such as anti‐CD3 F(ab)2’ fragments, can ameliorate AT inflammation( 15 ) but also increase steatosis and transaminase levels in NASH. Therefore, the effect of immunomodulatory therapies might differ depending on the tissue experiencing metabolic inflammation. This information will be important for the planning of future therapeutic trials to treat patients with NASH. However, lifestyle interventions and weight loss have beneficial effects on adipose and liver tissue inflammation.( 40 , 41 )
Supporting information
Fig S1
Fig S2
Supplementary Material
Supported by the German Research Foundation (grants BU 2722/2‐3 to L.E.B.‐M., HA 6880/2‐1, and HA 6880/2‐3 to M.H.‐W.).
Potential conflict of interest: Dr. Hupa‐Breier is on the speakers’ bureau for Gilead and Falk Pharma. The other authors have nothing to report.
References
Author names in bold designate shared co‐first authorship.
- 1. Younossi ZM, Koenig AB, Abdelatif D, Fazel Y, Henry L, Wymer M. Global epidemiology of nonalcoholic fatty liver disease‐meta‐analytic assessment of prevalence, incidence, and outcomes. Hepatology 2016;64:73‐84. [DOI] [PubMed] [Google Scholar]
- 2. Younossi Z, Anstee QM, Marietti M, Hardy T, Henry L, Eslam M, et al. Global burden of NAFLD and NASH: trends, predictions, risk factors and prevention. Nat Rev Gastroenterol Hepatol 2018;15:11‐20. [DOI] [PubMed] [Google Scholar]
- 3. Anstee QM, Targher G, Day CP. Progression of NAFLD to diabetes mellitus, cardiovascular disease or cirrhosis. Nat Rev Gastroenterol Hepatol 2013;10:330‐344. [DOI] [PubMed] [Google Scholar]
- 4. Wong RJ, Aguilar M, Cheung R, Perumpail RB, Harrison SA, Younossi ZM, et al. Nonalcoholic steatohepatitis is the second leading etiology of liver disease among adults awaiting liver transplantation in the United States. Gastroenterology 2015;148:547‐555. [DOI] [PubMed] [Google Scholar]
- 5. Golabi P, Bush H, Stepanova M, Locklear CT, Jacobson IM, Mishra A, et al. Liver transplantation (LT) for cryptogenic cirrhosis (CC) and nonalcoholic steatohepatitis (NASH) cirrhosis: data from the Scientific Registry of Transplant Recipients (SRTR): 1994 to 2016. Medicine (Baltimore) 2018;97:e11518. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6. Tosello‐Trampont AC, Landes SG, Nguyen V, Novobrantseva TI, Hahn YS. Kuppfer cells trigger nonalcoholic steatohepatitis development in diet‐induced mouse model through tumor necrosis factor‐alpha production. J Biol Chem 2012;287:40161‐40172. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7. Malehmir M, Pfister D, Gallage S, Szydlowska M, Inverso D, Kotsiliti E, et al. Platelet GPIbalpha is a mediator and potential interventional target for NASH and subsequent liver cancer. Nat Med 2019;25:641‐655. [DOI] [PubMed] [Google Scholar]
- 8. Gadd VL, Skoien R, Powell EE, Fagan KJ, Winterford C, Horsfall L, et al. The portal inflammatory infiltrate and ductular reaction in human nonalcoholic fatty liver disease. Hepatology 2014;59:1393‐1405. [DOI] [PubMed] [Google Scholar]
- 9. Sutti S, Albano E. Adaptive immunity: an emerging player in the progression of NAFLD. Nat Rev Gastroenterol Hepatol 2020;17:81‐92. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10. Rau M, Schilling A‐K, Meertens J, Hering I, Weiss J, Jurowich C, et al. Progression from nonalcoholic fatty liver to nonalcoholic steatohepatitis is marked by a higher frequency of Th17 cells in the liver and an increased Th17/resting regulatory T cell ratio in peripheral blood and in the liver. J Immunol 2016;196:97‐105. [DOI] [PubMed] [Google Scholar]
- 11. Harley ITW, Stankiewicz TE, Giles DA, Softic S, Flick LM, Cappelletti M, et al. IL‐17 signaling accelerates the progression of nonalcoholic fatty liver disease in mice. Hepatology 2014;59:1830‐1839. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12. Ma X, Hua J, Mohamood AR, Hamad AR, Ravi R, Li Z. A high‐fat diet and regulatory T cells influence susceptibility to endotoxin‐induced liver injury. Hepatology 2007;46:1519‐1529. [DOI] [PubMed] [Google Scholar]
- 13. Wolf M, Adili A, Piotrowitz K, Abdullah Z, Boege Y, Stemmer K, et al. Metabolic activation of intrahepatic CD8+ T cells and NKT cells causes nonalcoholic steatohepatitis and liver cancer via cross‐talk with hepatocytes. Cancer Cell 2014;26:549‐564. [DOI] [PubMed] [Google Scholar]
- 14. Ma C, Kesarwala AH, Eggert T, Medina‐Echeverz J, Kleiner DE, Jin P, et al. NAFLD causes selective CD4(+) T lymphocyte loss and promotes hepatocarcinogenesis. Nature 2016;531:253‐257. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15. Winer S, Chan Y, Paltser G, Truong D, Tsui H, Bahrami J, et al. Normalization of obesity‐associated insulin resistance through immunotherapy. Nat Med 2009;15:921‐929. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16. Feuerer M, Herrero L, Cipolletta D, Naaz A, Wong J, Nayer A, et al. Lean, but not obese, fat is enriched for a unique population of regulatory T cells that affect metabolic parameters. Nat Med 2009;15:930‐939. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17. Rinella ME, Elias MS, Smolak RR, Fu T, Borensztajn J, Green RM. Mechanisms of hepatic steatosis in mice fed a lipogenic methionine choline‐deficient diet. J Lipid Res 2008;49:1068‐1076. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18. Rinella ME, Green RM. The methionine‐choline deficient dietary model of steatohepatitis does not exhibit insulin resistance. J Hepatol 2004;40:47‐51. [DOI] [PubMed] [Google Scholar]
- 19. Hardtke‐Wolenski M, Fischer K, Noyan F, Schlue J, Falk CS, Stahlhut M, et al. Genetic predisposition and environmental danger signals initiate chronic autoimmune hepatitis driven by CD4+ T cells. Hepatology 2013;58:718‐728. [DOI] [PubMed] [Google Scholar]
- 20. Römermann D, Ansari N, Schultz‐Moreira AR, Michael A, Marhenke S, Hardtke‐Wolenski M, et al. Absence of Atg7 in the liver disturbed hepatic regeneration after liver injury. Liver Int 2020;40:1225‐1238. [DOI] [PubMed] [Google Scholar]
- 21. Buitrago‐Molina LE, Dywicki J, Noyan F, Trippler M, Pietrek J, Schlue J, et al. Splenectomy prior to experimental induction of autoimmune hepatitis promotes more severe hepatic inflammation, production of IL‐17 and apoptosis. Biomedicines 2021;9:58. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22. Buitrago‐Molina LE, Pietrek J, Noyan F, Schlue J, Manns MP, Wedemeyer H, et al. Treg‐specific IL‐2 therapy can reestablish intrahepatic immune regulation in autoimmune hepatitis. J Autoimmun 2021;117:102591. [DOI] [PubMed] [Google Scholar]
- 23. Assarsson E, Lundberg M, Holmquist G, Björkesten J, Thorsen SB, Ekman D, et al. Homogenous 96‐plex PEA immunoassay exhibiting high sensitivity, specificity, and excellent scalability. PLoS One 2014;9:e95192. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24. Schultz Moreira AR, Rüschenbaum S, Schefczyk S, Hendgen‐Cotta U, Rassaf T, Broering R, et al. 9‐PAHSA prevents mitochondrial dysfunction and increases the viability of steatotic hepatocytes. Int J Mol Sci 2020;21:8279. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25. Kleiner DE, Brunt EM, Van Natta M, Behling C, Contos MJ, Cummings OW, et al.; Nonalcoholic Steatohepatitis Clinical Research Network . Design and validation of a histological scoring system for nonalcoholic fatty liver disease. Hepatology 2005;41:1313‐1321. [DOI] [PubMed] [Google Scholar]
- 26. Dywicki J, Buitrago‐Molina LE, Pietrek J, Lieber M, Broering R, Khera T, et al. Autoimmune hepatitis induction can occur in the liver. Liver Int 2020;40:377‐381. [DOI] [PubMed] [Google Scholar]
- 27. Ju C, Tacke F. Hepatic macrophages in homeostasis and liver diseases: from pathogenesis to novel therapeutic strategies. Cell Mol Immunol 2016;13:316‐327. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28. Rahman K, Vengrenyuk Y, Ramsey SA, Vila NR, Girgis NM, Liu J, et al. Inflammatory Ly6Chi monocytes and their conversion to M2 macrophages drive atherosclerosis regression. J Clin Invest 2017;127:2904‐2915. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29. Duvallet E, Semerano L, Assier E, Falgarone G, Boissier MC. Interleukin‐23: a key cytokine in inflammatory diseases. Ann Med 2011;43:503‐511. [DOI] [PubMed] [Google Scholar]
- 30. Chatenoud L. Immune therapy for type 1 diabetes mellitus‐what is unique about anti‐CD3 antibodies? Nat Rev Endocrinol 2010;6:149‐157. [DOI] [PubMed] [Google Scholar]
- 31. Adams LA, Lindor KD. Nonalcoholic fatty liver disease. Ann Epidemiol 2007;17:863‐869. [DOI] [PubMed] [Google Scholar]
- 32. Peiseler M, Sebode M, Franke B, Wortmann F, Schwinge D, Quaas A, et al. FOXP3+ regulatory T cells in autoimmune hepatitis are fully functional and not reduced in frequency. J Hepatol 2012;57:125‐132. [DOI] [PubMed] [Google Scholar]
- 33. Taubert R, Hardtke‐Wolenski M, Noyan F, Wilms A, Baumann AK, Schlue J, et al. Intrahepatic regulatory T cells in autoimmune hepatitis are associated with treatment response and depleted with current therapies. J Hepatol 2014;61:1106‐1114. [DOI] [PubMed] [Google Scholar]
- 34. Taubert R, Pischke S, Schlue J, Wedemeyer H, Noyan F, Heim A, et al. Enrichment of regulatory T cells in acutely rejected human liver allografts. Am J Transplant 2012;12:3425‐3436. [DOI] [PubMed] [Google Scholar]
- 35. Hardtke‐Wolenski M, Taubert R, Noyan F, Sievers M, Dywicki J, Schlue J, et al. Autoimmune hepatitis in a murine autoimmune polyendocrine syndrome type 1 model is directed against multiple autoantigens. Hepatology 2015;61:1295‐1305. [DOI] [PubMed] [Google Scholar]
- 36. Zhou S‐L, Zhou Z‐J, Hu Z‐Q, Huang X‐W, Wang Z, Chen E‐B, et al. Tumor‐associated neutrophils recruit macrophages and T‐regulatory cells to promote progression of hepatocellular carcinoma and resistance to sorafenib. Gastroenterology 2016;150:1646‐1658.e17. [DOI] [PubMed] [Google Scholar]
- 37. Farrell GC, Mridha AR, Yeh MM, Arsov T, Van Rooyen DM, Brooling J, et al. Strain dependence of diet‐induced NASH and liver fibrosis in obese mice is linked to diabetes and inflammatory phenotype. Liver Int 2014;34:1084‐1093. [DOI] [PubMed] [Google Scholar]
- 38. Chung KJ, Nati M, Chavakis T, Chatzigeorgiou A. Innate immune cells in the adipose tissue. Rev Endocr Metab Disord 2018;19:283‐292. [DOI] [PubMed] [Google Scholar]
- 39. Wei Y, Tian Q, Zhao X, Wang X. Serum transforming growth factor beta 3 predicts future development of nonalcoholic fatty liver disease. Int J Clin Exp Med 2015;8:4545‐4550. [PMC free article] [PubMed] [Google Scholar]
- 40. Promrat K, Kleiner DE, Niemeier HM, Jackvony E, Kearns M, Wands JR, et al. Randomized controlled trial testing the effects of weight loss on nonalcoholic steatohepatitis. Hepatology 2010;51:121‐129. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41. Vilar‐Gomez E, Martinez‐Perez Y, Calzadilla‐Bertot L, Torres‐Gonzalez A, Gra‐Oramas B, Gonzalez‐Fabian L, et al. Weight loss through lifestyle modification significantly reduces features of nonalcoholic steatohepatitis. Gastroenterology 2015;149:367‐378.e5. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Fig S1
Fig S2
Supplementary Material


